Background: During intracellular replication, Legionella pneumophila produces effector proteins that exploit host cell processes.
Results: The effector GobX is an E3 ubiquitin ligase that targets to the Golgi where it is post-translationally modified with a palmitoyl moiety.
Conclusion: Bacteria can hijack host-mediated S-palmitoylation for subcellular localization of their effectors.
Significance: Pharmacological inhibition of S-palmitoylation enzymes may interfere with effector function and thus microbial virulence.
Keywords: bacterial pathogenesis, click chemistry, Golgi, protein palmitoylation, ubiquitylation (ubiquitination), DHHC protein, Legionella pneumophila, translocated effector
Abstract
The facultative intracellular pathogen Legionella pneumophila, the causative agent of Legionnaires disease, infects and replicates within human alveolar macrophages. L. pneumophila delivers almost 300 effector proteins into the besieged host cell that alter signaling cascades and create conditions that favor intracellular bacterial survival. In order for the effectors to accomplish their intracellular mission, their activity needs to be specifically directed toward the correct host cell protein or target organelle. Here, we show that the L. pneumophila effector GobX possesses E3 ubiquitin ligase activity that is mediated by a central region homologous to mammalian U-box domains. Furthermore, we demonstrate that GobX exploits host cell S-palmitoylation to specifically localize to Golgi membranes. The hydrophobic palmitate moiety is covalently attached to a cysteine residue at position 175, which is part of an amphipathic α-helix within the C-terminal region of GobX. Site-directed mutagenesis of cysteine 175 or residues on the hydrophobic face of the amphipathic helix strongly attenuated palmitoylation and Golgi localization of GobX. Together, our study provides evidence that the L. pneumophila effector GobX exploits two post-translational modification pathways of host cells, ubiquitination and S-palmitoylation.
Introduction
The capability of pathogens to modulate host cell processes for their own advantage is fundamental to the success of any microbial infection. To accomplish this goal, bacteria often utilize specialized nano-machines, such as the type IV secretion system (T4SS),3 to deliver effector proteins into the host cell cytosol (1). Once translocated, the effectors unfold their biochemical activities toward a particular host factor or compartment and manipulate host cell processes such as vesicular trafficking, cell cycle regulation, transcription, translation, or innate immunity to create conditions favorable for bacterial survival (2, 3).
The Gram-negative bacterium Legionella pneumophila encodes almost 300 effectors that are translocated by the Dot/Icm T4SS into the cytosol of the host cell, which are freshwater amoeba in the environment, and alveolar macrophages during colonization of the human lung, resulting in Legionnaires pneumonia (2, 4). Most L. pneumophila effectors lack notable sequence homology to other proteins and have not been characterized in detail. Although in-frame deletions in individual effector-encoding genes are mostly tolerated by L. pneumophila (5), mutations that disable the Dot/Icm system render the bacterium avirulent (6), underscoring the critical importance of translocated effectors for L. pneumophila pathogenicity.
Although our mechanistic understanding of L. pneumophila effector function is mostly incomplete, it has become increasingly clear that the effectors often represent molecular mimics of eukaryotic proteins both with respect to their function and subcellular targeting mechanisms. Bioinformatics approaches contributed to the discovery of a variety of effectors with eukaryotic-like motifs or domains such as ankyrin or leucine-rich repeats, coiled-coils, guanine nucleotide exchange factors or GTPase-activating proteins, and ubiquitin-related domains such as F- and U-boxes (7, 8). Most of these domains are general protein-protein interaction modules that reveal little if any information about the exact host target of an effector. F- and U-box domains are found in eukaryotic E3 ubiquitin ligases, which catalyze the final step in an enzymatic cascade that results in the transfer of the small protein ubiquitin from E2 ubiquitin-conjugating enzymes to a particular target protein (9, 10). Polyubiquitination of target proteins alters their cellular fate, often resulting in their proteasomal degradation. Thus, it is not surprising that pathogens like L. pneumophila exploit this pathway by delivering their own E3 ligases through the Dot/Icm system into the infected host cell. E3 ligase activity has thus far been experimentally confirmed for only four L. pneumophila effectors, namely LegAU13/AnkB, LubX, LegU1, and SidC (11–14), although it is believed that additional L. pneumophila effectors with ubiquitin ligase activity exist.
Equally unclear as the effectors' biological activities are the molecular mechanisms that help them reach their correct subcellular location where they encounter their natural targets. The few cases that have been studied in detail suggest that here, too, molecular mimicry is a recurring theme. Several L. pneumophila effectors target to lipid bilayers by specifically binding to the (poly)phosphorylated forms of phosphatidylinositols (PtdIns), the main structural phospholipid present in the cytosolic leaflet of eukaryotic membranes. SidM and SidC, for example, interact with PtdIns(4)P, a phospholipid enriched within the Legionella-containing vacuole (LCV) membrane of infected cells but also within the Golgi, plasma membrane, and late endosomes (15–17). In contrast, LidA preferentially binds to PtdIns(3)P, a phospholipid commonly found on early endosomal membranes as well as on ER subdomains involved in autophagosome formation (15, 18). Other L. pneumophila effectors exploit protein-protein interaction for their subcellular targeting. For example, VipD, a phospholipase A1 that aids in the prevention of early endosomal fusion with the LCV, localizes to early endosomes by specifically binding to the active form of Rab5, a small GTPase enriched on endosomal membranes (19–21). The disturbance of this protein-protein interaction, by exchanging critical amino acid residues within the VipD-Rab5 interface, prevents VipD endosomal targeting and phospholipase A1 activity (21). Another group of L. pneumophila effectors exploits post-translational modifications, more precisely lipidation, to increase their hydrophobicity thus facilitating their association with host cell membranes. A common lipidation is prenylation, the covalent and irreversible conjugation of an isoprenoid moiety through a thioether bond to cysteine residues. Prenylation can be further classified into farnesylation and geranylgeranylation, each of which occurs on cysteine residues located within a consensus motif (CAAX for farnesylation; CXC or CC for geranylgeranylation; with C = cysteine, A = any aliphatic residue, X = Met, Ser, Gln, Ala, or Cys) at or near the C terminus of proteins. Bioinformatics analyses identified multiple L. pneumophila effectors with a CAAX motif at their C-terminal end, and several of them were subsequently confirmed to exploit host-mediated prenylation for membrane association and localization within eukaryotic cells (22, 23). The other post-translational lipidation involving cysteine residues is S-acylation, or S-palmitoylation, which is the covalent yet reversible addition of a 16-carbon palmitate moiety to an internal cysteine via a thioester linkage. Unlike CAAX-specific prenylation, there is no strict consensus sequence known for S-palmitoylation (24), and L. pneumophila effectors that exploit S-palmitoylation for their intracellular localization have yet to be discovered.
Here, we provide the first evidence for the existence of such an effector (Lpg2455), which we named GobX. We demonstrate that S-palmitoylation contributes to precise subcellular localization of GobX and reveal that an unexpectedly short region within GobX is sufficient for this lipidation and thus subcellular localization to occur.
Experimental Procedures
Strains, Media, and Reagents
L. pneumophila strains were grown and maintained as described (25). Thymidine was supplemented at 100 μg/ml. L. pneumophila strains Lp02 (thyA hsdR rpsL) and Lp03 (Lp02 dotA3 (T4SS−)) are thymidine-auxotroph derivatives of L. pneumophila strain Philadelphia-1 (6). An in-frame deletion of lpg2455 in L. pneumophila strain Lp02 was generated as described (27). Saccharomyces cerevisiae strain INVSc1 (MATa his3D1 leu2 trp1-289 ura3-52) was grown in YPD media containing 2% (w/v) glucose. HEK293T, COS-1, RAW264.7, and J774A.1 tissue culture cells (American Type Culture Collection, ATCC) were grown in DMEM supplemented with 10% FBS and incubated in 5% CO2 at 37 °C. U937 cells were grown in RPMI 1640 medium supplemented with 10% FBS, incubated in 5% CO2 at 37 °C, and differentiated in the same media containing 10 ng/ml 12-O-tetradecanoylphorbol-13-acetate for 48 h.
Antibodies were purchased from Abcam (giantin, EEA-1, and LAMP-1), Sigma (TGN46), Santa Cruz Biotechnology (TOM70 and GST), Enzo Life Sciences (calnexin), and Life Technologies, Inc. (streptavidin-HRP conjugate, GFP, and secondary fluorophores). Antibody directed against Lpg2455 was generated by immunizing rabbits with purified recombinant full-length protein (GenScript). Antibody directed against hemagglutinin (HA) was a kind gift of Ramanujan Hegde (MRC Laboratory of Molecular Biology, Cambridge, UK).
Construction of Expression Clones
All the plasmids and oligonucleotides used in this study are listed in supplemental Tables S1 and S2. The plasmid pDONR221TM-lpg2455 was a kind gift of Ralph Isberg (Tufts University). The GatewayTM-compatible plasmid pJB908D was generated by introducing the attR1-attR2 cassette from pDESTTM17 and PCR-amplified with the respective primers into pJB908 at KpnI and SacI restriction sites. lpg2455 (gobX) was cloned into pJB908D via the GatewayTM cloning technology. The plasmid pXDC61 was a kind gift of Howard Shuman (University of Chicago). pXDC61.1-HA was generated by digesting pXDC61 with NdeI and KpnI restriction enzymes, and a single hemagglutinin (HA) tag was inserted using the annealed oligonucleotide-cloning method as described by Addgene. gobX was cloned into pXDC61 and pXDC61.1-HA at KpnI and XbaI restriction sites. pXDC61.1-HA-GobX with the C175A mutant was generated using the QuikChangeTM site-directed mutagenesis procedure (Agilent Technologies). gobX was cloned into the yeast expression vector pYES2/NTA (Life Technologies, Inc.) at BamHI and XbaI sites, and the mutants were generated using the QuikChangeTM site-directed mutagenesis procedure.
Full-length gobX and truncations were PCR-amplified from their respective primers and cloned into the eukaryotic expression vector pcDNATM6.2/N-EmGFP-DEST via GatewayTM cloning technology (Life Technologies, Inc.). gobX mutants were generated using the QuikChangeTM site-directed mutagenesis procedure with the appropriate primers. gobX(171–190) homologs from L. pneumophila strains Philadelphia-1, Lens, and Lorraine, gobX(172–186), and gobX(175–184) were cloned into the vector pEGFP-C1 (Clontech) at BamHI and EcoRI restriction sites using the annealed oligonucleotide-cloning method. pmCherry-gobX was generated by subcloning gobX at BamHI and SalI restriction sites into the vector pmCherry-C1 (Clontech). The 23 HA-tagged mouse Asp-His-His-Cys (DHHC) palmitoyltransferases were generously provided by Masaki Fukata (National Institutes of Natural Sciences, Japan), and the respective DHHS variants were generated by the QuikChangeTM site-directed mutagenesis with the appropriate primers.
gobX was cloned into the destination vector pDESTTM17 by the GatewayTM cloning technology to generate the His6-tagged fusion. GST-tagged gobX was generated by subcloning gobX into the expression vector pGEX-6p-1 (GE Healthcare) at BamHI and EcoRI restriction sites.
Digitonin Fractionation of Legionella-infected Macrophage Cells
L. pneumophila strains Lp02 and Lp03 containing the plasmid pJB908D-Lpg2455 were grown to A600 = 1–2 in the presence of 0.5 mm isopropyl β-dithiogalactopyranoside (IPTG) and incubated with J774A.1 macrophages (1 × 108 cells) at an m.o.i. of 50 for 1 h at 37 °C. Digitonin fractionation was performed as described previously (28). Briefly, infected cells were resuspended in PBS containing 1% digitonin and incubated at room temperature for 20 min with constant mixing. The cell lysate was centrifuged at 15,000 rcf for 10 min. The digitonin-soluble fraction was mixed with SDS sample buffer and subjected to SDS-PAGE separation followed by immunoblot analysis using antibody directed against Lpg2455.
FRET-based Translocation Assay
Translocation of Lpg2455in infected macrophages using fluorescence resonance energy transfer (FRET)-based detection was performed using the method described previously (29) with modifications. Briefly, T4SS-competent or -deficient L. pneumophila strains carrying β-lactamase fusions were grown to post-exponential phase in the presence of 0.5 mm IPTG. Monolayers of RAW264.7 macrophages were seeded in black clear-bottom 96-well plates at 1 × 105 cells/well and challenged with the appropriate strain of L. pneumophila at an m.o.i. of 50. CCF4-AM substrates (Life Technologies, Inc.) were mixed with medium in the wells 1 h post-infection. The cells were further incubated for 2 h at 25 °C and visually inspected under a Zeiss Observer.Z1 fluorescence microscope with a Coumarin/Pacific Blue long pass filter (Chroma Technology).
Intracellular Growth Curve
The intracellular growth assay was performed as described previously (30). Briefly, U937 macrophages were challenged by the indicated L. pneumophila strains for 2 h at an m.o.i. of 0.05. The macrophages were permeabilized with 0.02% digitonin at 2, 24, 48, and 72 h post-infection, and the supernatant was collected. The wells were washed once with PBS, and the solution was combined with the initial lysate. Dilutions of the combined cell lysate were plated on CYET plates, and the mean colony-forming units (CFU) were calculated and converted to fold growth to generate the growth curve.
Separation of Macrophage Organelles
To detect the intracellular localization of GobX, RAW264.7 macrophages were seeded at a density of 1 × 107 cells per dish in 10 10-cm dishes. L. pneumophila strain Lp02 containing HA-tagged GobX or GobXC175A was grown overnight and induced with 0.5 mm IPTG for 4 h. The macrophages were infected with post-exponential phase L. pneumophila at an m.o.i. of 50 for 4 h. The macrophages were washed three times with buffer HM (0.25 m sucrose, 10 mm Tris, pH 7.4) to remove extracellular bacteria, and cells were harvested using a cell scraper and resuspended in a total volume of 3 ml of HM. The cells were lysed by douncing five times in a ball bearing dounce homogenizer. The concentration of sucrose in the lysate was adjusted to 1.6 m with the addition of 3 m sucrose in 10 mm Tris, pH 7.4.
Separation of Golgi by sucrose density gradient ultracentrifugation was performed as described previously (31) with modifications. The lysate was overlaid with 45, 35, and 25% sucrose and underlaid with 65% sucrose and centrifuged at 100,000 rcf for 4 h at 4 °C (Fig. 3C). The Golgi fraction was extracted at the 45/35% interface. The Golgi fraction and total lysate were separated by SDS-PAGE and analyzed by immunoblot.
FIGURE 3.
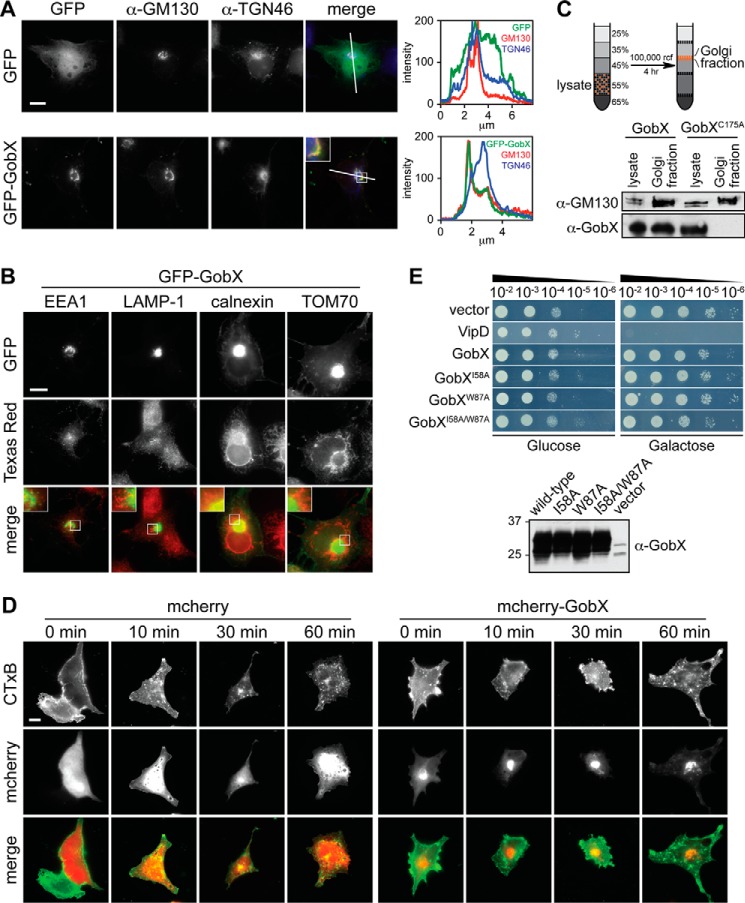
GobX localizes to the Golgi region. A, intracellular localization of GobX. Transiently transfected COS-1 cells producing either GFP (control) or GFP-GobX were labeled with antibody directed against the medial Golgi marker GM130 or the trans-Golgi network marker TGN46 and imaged by fluorescence microscopy. Scale bar, 2 μm. The pixel intensity of the green (GFP), red (GM130), and blue (TGN46) fluorescent signals along the line indicated in the image is shown at the right. Enlarged view of the boxed region is shown in the inset. B, GobX does not co-localize with endosomes, lysosomes, ER, or mitochondria. COS-1 cells producing GFP-GobX were labeled with antibody directed against different organelle markers and imaged by fluorescence microscopy. EEA-1, early endosome; LAMP-1, lysosome; calnexin, ER; TOM70, mitochondria. Scale bar, 2 μm. The boxed regions are magnified in insets for each panel. C, GobX was enriched in the Golgi fraction in infected macrophages. RAW264.7 macrophages challenged with Lp02 producing HA-GobX or HA-GobXC175A was fractionated by sucrose discontinuous gradient centrifugation. The total cell lysate and the fraction enriched in Golgi membranes (45/35% interface) were probed by immunoblot using anti-GM130 and anti-GobX antibody. D, GobX does not interfere with retrograde trafficking. Transiently transfected COS-1 cells producing either mCherry or mCherry-GobX were incubated with CTxB for 10 min, and CTxB traffic was monitored after 0, 10, 30, and 60 min within the cell. Scale bar, 2 μm. E, GobX does not interfere with yeast growth. Serial dilutions (10−2–10−6) of S. cerevisiae INVSc1 cells containing plasmids encoding GobX or GobX variants with attenuated E3 ligase activity were plated on repressing (glucose) or inducing (galactose) synthetic medium plates and analyzed for growth after 48 h. VipD served as a growth defect control (20). Production of GobX and GobX variants in INVSc1 cells was analyzed in liquid culture of synthetic media containing 2% (w/v) galactose and detected on an immunoblot using anti-GobX antibody. All immunofluorescence images, immunoblots, and yeast growth assays are representative of at least two independent experiments with similar outcomes.
Yeast Growth Assay
The yeast expression vector pYES2 contains a GAL1 promoter for inducible protein expression. GobX or GobX mutants were cloned into pYES2 and transformed to S. cerevisiae strain INVSc1 using the lithium acetate/PEG transformation method as described in the manufacturer's protocol (Life Technologies, Inc.). Transformants were grown in synthetic media containing 2% (w/v) glucose lacking uracil to saturation at 30 °C. The cultures were normalized to an A600 = 1, and 10-fold serial dilutions were spotted on glucose-containing (repressing) or galactose-containing (inducing) synthetic media plates lacking uracil, and the growth was monitored after 48 h.
Immunofluorescence Microscopy
Mammalian expression clones were transiently transfected into semi-confluent COS-1 cells in 24-well plates. All transfections were performed using Lipofectamine® 2000 following the manufacturer's protocol (Life Technologies, Inc.). After overnight transfection, cells were fixed with 3.8% formaldehyde, permeabilized with ice-cold methanol, blocked with 5% goat serum, and immunostained with the indicated antibodies as described previously (32). Coverslips were mounted in ProLong® Gold antifade reagent (Life Technologies, Inc.) and imaged with a Zeiss Axio Observer.Z1 inverted light microscope.
For cholera toxin trafficking, COS-1 cells were transiently transfected overnight with the plasmid encoding mCherry or mCherry-gobX. Cells were incubated on ice with 1 mg/ml cholera toxin subunit B (CTxB with Alexa Fluor® 488 conjugate) for 10 min, washed with PBS, and incubated in regular DMEM in 5% CO2 at 37 °C, and the traffic of CTxB was chased after 10, 30, and 60 min using fluorescence microscopy. For 2-bromopalmitic acid (2-BP) treatment, COS-1 cells were transiently transfected overnight with GFP-gobX. 2-Bromohexadecanoic acid (100 mm stock in DMSO, Sigma) or DMSO vehicle was diluted with DMEM and added to the transfected cells to a final concentration of 40 μm. Cells were incubated for 5 h in 5% CO2 at 37 °C, immunostained as described above, and visualized using fluorescence microscopy. The localization of DHHC palmitoyltransferases was detected by transiently transfecting plasmids encoding respective DHHC palmitoyltransferases with or without GFP, GFP-gobX, or GFP-gobX(171–190) into COS-1 cells. DHHC palmitoyltransferases were immunostained using anti-HA antibody and visualized using fluorescence microscopy.
Subcellular Fractionation
1 × 107 HEK293T cells were transiently transfected with GFP-gobX or variants. Cells were scraped, washed, and resuspended in PBS with protease inhibitors, and lysed by 20 strokes in a Dounce homogenizer. Cell debris was pelleted by centrifugation at 15,000 rcf for 10 min at 4 °C. The post-nuclear supernatant was collected and centrifuged at 55,000 rpm for 45 min at 4 °C, and the final supernatant was collected as the cytosolic fraction. The pellet was resuspended with an equal volume of 2% IGEPAL in PBS and collected as the membrane fraction. SDS sample buffer was added to each fraction, and the original post-nuclear supernatant was resolved on the SDS-PAGE and analyzed by immunoblot.
17-ODYA Labeling and Click Chemistry
0.5–1 × 106 HEK293T cells were transiently transfected with GFP-gobX or variants or co-transfected with DHHC palmitoyltransferases or respective DHHS variants. Prior to 17-ODYA labeling, cells were incubated with 10 mg/ml fatty acid-free BSA in DMEM in 5% CO2 at 37 °C for 30 min. Cells were then metabolically labeled with 50 μm 17-ODYA (25 mm stock in DMSO, Santa Cruz Biotechnology) for 30 min or 2 h as indicated. Cells were harvested, washed, and lysed with lysis buffer (10 mm Tris-HCl, pH 7.5, 150 mm NaCl, 0.5 mm EDTA, 0.5% IGEPAL) on ice for 30 min. Cell debris was pelleted by centrifugation at 15,000 rcf for 10 min at 4 °C, and the supernatant was diluted 5-fold with dilution buffer (10 mm Tris-HCl, pH 7.5, 150 mm NaCl, 0.5 mm EDTA). GFP-GobX was immunoprecipitated using GFP-Trap® magnetic beads following the manufacturer's protocol (Chromotek). The beads were washed two times with dilution buffer and resuspended in PBS. Cu(I)-assisted click chemistry was performed by adding 100 μm biotin-azide (10 mm stock in DMSO), 1 mm tris(2-carboxyethyl)phosphine hydrochloride (freshly prepared 50 mm solution in deionized water), 100 μm tris[(1-benzyl-1H-1,2,3-triazol-4-yl)methyl]amine, 10 mm stock in DMSO), and 1 mm CuSO4·H2O (50 mm stock in deionized water). The reaction was incubated at 30 °C with shaking for 1 h and stopped with SDS sample buffer. The sample was incubated at 70 °C for 5 min, resolved on the SDS-PAGE, and analyzed by immunoblot with streptavidin-HRP conjugate and anti-GFP antibody.
For hydroxylamine treatment, GFP-gobX produced in HEK293T cells was labeled with 17-ODYA, immunoprecipitated, and performed with the click chemistry. The beads were then incubated with 1 m hydroxylamine at room temperature for 1 h, followed by SDS-PAGE and immunoblot analyses. For acyl-protein thioesterase 1 (APT1) inhibitor treatment, 1 μm palmostatin B (Calbiochem) was added to DMEM containing fatty acid-free BSA 30 min prior to the 17-ODYA labeling. The same amount of inhibitor was also added during the 17-ODYA labeling and the 1-h chase period in regular DMEM.
In Vitro Ubiquitination Assay
The bacterial expression clones were introduced into Escherichia coli strain BL21(DE3), and the His6- or GST-tagged gobX was overexpressed in LB media in the presence of 0.2 mm IPTG at 20 °C for 16 h. His6-tagged GobX was purified with TALON® metal affinity resin (Clontech), and GST-tagged GobX was purified with glutathione-Sepharose® 4B (GE Healthcare) following the manufacturer's protocol. The purified proteins were dialyzed against PBS with 1 mm 2-mercaptoethanol.
The in vitro ubiquitination assay was performed using the ubiquitinylation kit (Enzo Life Sciences). A 10-μl reaction mixture (1× ubiquitinylation buffer, 5 mm Mg-ATP, 1 mm DTT, 2.5 μm biotinylated ubiquitin, and 100 nm E1) was combined with ∼1–2.5 μm of the respective E2s and 1 μm of the purified GobX. The reaction mixture was incubated at 37 °C for 1 h, quenched by adding an equal volume of SDS sample buffer, and incubated at 94 °C for 10 min. The proteins were resolved on the SDS-PAGE, and the ubiquitinated products were detected with streptavidin-HRP conjugate on an immunoblot.
Results
Lpg2455 Is a Translocated Substrate of the Dot/Icm T4SS
The protein Lpg2455 from L. pneumophila strain Philadelphia-1 was proposed to be a translocated substrate of the Dot/Icm T4SS based on the presence of an E-block translocation motif at its C terminus (33). To experimentally validate its T4SS-dependent delivery into host cells during infection, we used two different methods. In the first approach, digitonin was used to detergent-extract proteins from J774A.1 macrophages challenged with L. pneumophila producing plasmid-encoded Lpg2455. Immunoblot analysis confirmed the presence of Lpg2455 in lysate from macrophages challenged with the virulent L. pneumophila strain Lp02 but not with the T4-defective strain Lp03 (Fig. 1A). In the second approach, we used a fluorescence resonance energy transfer (FRET)-based translocation assay in which a cell membrane-penetrating CCF4-AM compound was used that changes color from green to blue when cleaved by β-lactamase. As shown in Fig. 1B, when RAW264.7 macrophages were challenged with Lp02 containing β-lactamase-tagged Lpg2455, ∼50% of the cells emitted blue color, meaning that β-lactamase-Lpg2455, but not β-lactamase, was translocated into the host cytosol. The translocation was T4SS-dependent because macrophages infected with Lp03 producing the fusion protein remained green. Together, these results confirmed that Lpg2455 is indeed a Dot/Icm-translocated effector from L. pneumophila.
FIGURE 1.
Lpg2455 is a Dot/Icm translocated effector. A, Lpg2455 is translocated into host cells during infection. J774A.1 macrophages were challenged with either L. pneumophila strain Lp02 or the T4-defective strain Lp03 at an m.o.i. of 50 for 1 h. Digitonin-extracted macrophage lysate was analyzed by immunoblot using antibody directed against Lpg2455. The band representing Lpg2455 is indicated with an arrow. The anti-actin immunoblot served as a loading control. Equal production of the plasmid-encoded Lpg2455 by the bacteria used for the macrophage challenge was confirmed by analyzing Lp02 and Lp03 (Lp) lysate. B, Lpg2455 is translocated into host cells during infection. RAW264.7 macrophages were challenged with either Lp02 or Lp03 producing β-lactamase (β-Lac)-fused Lpg2455 at an m.o.i. of 50 for 1 h. Translocation of the effector protein is monitored by the cleavage of CCF4-AM, which changes color from green (520 nm) to blue (447 nm). Macrophages challenged with Lp02 or Lp03 producing β-lactamase served as a negative control. C, production of Lpg2455 is growth phase-dependent. L. pneumophila strain Lp02 or Lp02Δlpg2455 was cultured in liquid media to the indicated cell density (OD600). Equal amounts of bacterial lysate were loaded in each lane and analyzed by immunoblot using antibody directed against Lpg2455 or LidA. D, Lpg2455 is dispensable for intracellular replication. U937 macrophages were challenged with L. pneumophila strain Lp02, the T4-defective strain Lp03, or Lp02Δlpg2455 at an m.o.i. of 0.05. Intracellular growth of L. pneumophila was monitored after 2, 24, 48, and 72 h. The average fold growth was taken from two independent experiments. Images shown are representative of at least two independent experiments with similar outcomes.
The expression profile of most effector-encoding genes from L. pneumophila studied to date was synchronized with the growth phase of the bacterium. Monitoring the production of Lpg2455 during L. pneumophila growth in rich liquid media can serve as an indicator for the effector's most probable time of Dot/Icm-mediated translocation during host cell infection. Lpg2455 was almost undetectable within lysate from exponentially replicating bacteria grown to low density (A600 = 1–2), which is equivalent to the intracellular phase during which L. pneumophila replicates within the LCV (Fig. 1C). In contrast, the production of Lpg2455 was strongly enhanced during late exponential and stationary growth in liquid media, which correlates to the transmissive (virulent) form of L. pneumophila that prepares for host cell lysis and infection of neighboring cells. Lysate from the L. pneumophila mutant Lp02Δlpg2455 did not contain detectable amounts of Lpg2455, indicating that the signal detected in the Lp02-derived lysate was specific. Notably, proliferation of the strain Lp02Δlpg2455 within U937 macrophages was not significantly impaired compared with the parental strain Lp02 (Fig. 1D), indicating that Lpg2455 is dispensable for intracellular survival and replication of L. pneumophila within the mammalian host.
Lpg2455 Is a U-box E3 Ubiquitin Ligase
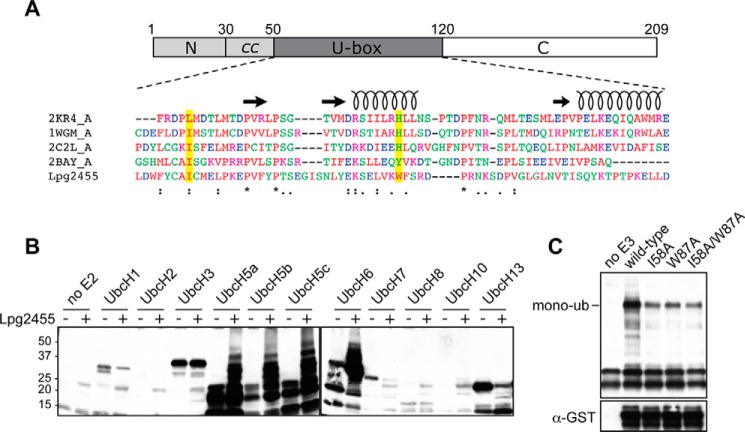
Lpg2455 possesses no primary sequence homology to other proteins, yet based on remote secondary structure homology, the central region (residue 51–120) was predicted by HHpred (34) to be similar to U-box E3 ubiquitin ligases, enzymes that facilitate the covalent attachment of ubiquitin to target proteins within eukaryotes. The most notable homologs include S. cerevisiae Prp19, Homo sapiens ubiquitin conjugation factor E4, and Mus musculus CHIP (Fig. 2A). Given that E3 ligases often exhibit auto-ubiquitination activity in the absence of their substrates, we tested whether this attribute was preserved within the U-box domain of Lpg2455. Upon supplementing Lpg2455 with a panel of human E2 ubiquitin-conjugating enzymes, we detected robust auto-ubiquitination activity in reactions containing Lpg2455 and human E2 UbcH5a, -5b, -5c, or UbcH6 but not in control reactions lacking either an E2 enzyme or Lpg2455, as determined by streptavidin-HRP detection of biotinylated ubiquitin species within the immunoblot (Fig. 2B).
FIGURE 2.
Lpg2455 is a U-box E3 ligase. A, central region of Lpg2455 shows similarity to U-box E3 ligase domains. Top, schematic representation of Lpg2455 with numbers indicating amino acid positions. Bottom, sequence of Lpg2455 U-box domain was aligned with U-boxes from the indicated E3 ligases using ClustalW. Secondary structural elements of U-box domains are illustrated as spirals (α-helices) and arrows (β-strands). The two conserved hydrophobic residues, Ile-58 and Trp-87, are highlighted in yellow. 2KR4, E3 ubiquitin ligase E4B (E-value = 0.21); 1WGM, human ubiquitin conjugation factor E4A (E-value = 0.37); 2C2L, CHIP U-box E3 ubiquitin ligase (E-value = 0.048); 2BAY, Prp19 U-box E3 ubiquitin ligase (E-value = 0.003) (E-value calculated from HHpred). B, Lpg2455 exhibits auto-ubiquitination activity in vitro. Purified His6-tagged Lpg2455 was incubated with biotinylated ubiquitin in the presence of each of the indicated E2 enzymes. Proteins were separated by SDS-PAGE, and ubiquitinated products were detected by immunoblot using streptavidin-conjugated HRP. C, mutations in the E2 binding interface attenuate auto-ubiquitination of Lpg2455. GST-tagged Lpg2455 and Lpg2455 mutants I58A, W87A, or I58A/W87A were incubated in the ubiquitination reaction with the E2 enzyme UbcH5a. Proteins were separated by SDS-PAGE and analyzed by immunoblot. Ubiquitinated products were detected in the streptavidin-conjugated HRP immunoblot with the mono-ubiquitinated GST-Lpg2455 indicated. The total amount of GST-Lpg2455 in each ubiquitination reaction was detected with anti-GST antibody. The immunoblots are representatives of at least two independent experiments with similar outcomes.
Despite the limited primary sequence homology of Lpg2455 to other U-box E3 ubiquitin ligases, its conserved secondary structure indicated that Ile-58 and Trp-87 were similar to conserved hydrophobic/aromatic residues involved in E2 binding by other U-box E3 ligases (Fig. 2A). Upon substitution of Ile-58 or Trp-87 with alanine, the resulting variants Lpg2455I58A, Lpg2455W87A, or Lpg2455I58A/W87A were strongly attenuated in auto-ubiquitination activity as compared with wild-type Lpg2455, consistent with Ile-58 and Trp-87 being required for E2 binding (Fig. 2C). These studies confirm that the central domain of Lpg2455 functions as an E3 ligase.
Lpg2455 Localizes to the Golgi Region
We tagged Lpg2455 with green fluorescence protein (GFP) and analyzed its subcellular localization within transiently transfected COS-1 cells (Fig. 3A). Unlike GFP, which showed a cytosolic distribution, GFP-tagged Lpg2455 was highly enriched at the juxtanuclear locale, with low detectable signal in the cytosol. Immunostaining of cells with antibodies specific for a variety of different organelle markers revealed extensive overlap of GFP-Lpg2455 with giantin and GM130, the cis/medial Golgi markers, but not with TGN46 (trans-Golgi network), EEA1 (endosomes), LAMP-1 (lysosomes), calnexin (endoplasmic reticulum; ER), or TOM70 (mitochondria) (Fig. 3, A and B).
Next, we tested whether the analysis of GFP-Lpg2455 in transiently transfected cells properly reflected the localization of endogenous Lpg2455 translocated by L. pneumophila into infected cells. Several attempts to visually detect translocated Lpg2455 using immunofluorescence microscopy were unsuccessful, possibly due to low abundance of the effector protein (data not shown). We thus took a biochemical approach to confirm Golgi localization of Lpg2455 in infected macrophages. Lysate from macrophages challenged with L. pneumophila strain Lp02 producing HA-tagged Lpg2455 was subjected to sucrose gradient fractionation to separate organelles according to their density. The fraction containing Golgi membranes was collected, separated by SDS-PAGE, and probed by immunoblot for the presence of HA-Lpg2455. As shown in Fig. 3C, HA-Lpg2455 was highly enriched in the Golgi-containing sucrose fraction. Most importantly, the Lpg2455 mutant C175A, which is attenuated in Golgi targeting (described below), was not present in the Golgi fraction, demonstrating that Lpg2455 specifically targets to Golgi membranes upon translocation by the L. pneumophila T4SS. Based on its intracellular localization and the U-box E3 ligase activity, we will from here on refer to Lpg2455 as GobX (Golgi-localized U-box E3 ligase).
Despite the abundance of GFP-GobX within the Golgi region, we observed no obvious cytotoxicity or Golgi fragmentation in transfected COS-1 cells (Fig. 3A), indicating that GobX did not negatively impact early secretory vesicle transport or Golgi integrity. Likewise, using fluorescently labeled CTxB as a reporter, we found no inhibitory effect of GFP-GobX on either endocytosis or retrograde transport of this cargo through the Golgi compartment to the ER (Fig. 3D). Furthermore, production of either GobX or the E3 ligase-attenuated mutants in S. cerevisiae did not cause any obvious growth defect, whereas the previously reported VipD inhibited growth of yeast cells (Fig. 3E) (20). Together, these data are a strong indication that GobX does not significantly impact Golgi function.
20-Amino Acid Segment and the Cysteine at Position 175 in GobX Are Required for Golgi Localization
Given the robust accumulation of GobX within the Golgi region (Fig. 3A), we decided to determine which part of GobX was responsible for this behavior. We divided GobX into three regions (Fig. 4A) as follows: an N-terminal region (residues 1–50) that contained a short segment predicted to form coiled coils (residues 30–50); the central region (residues 51–120) that possesses U-box E3 ligase activity; and a C-terminal region (residues 121–209) with no noticeable homology to other proteins.
FIGURE 4.
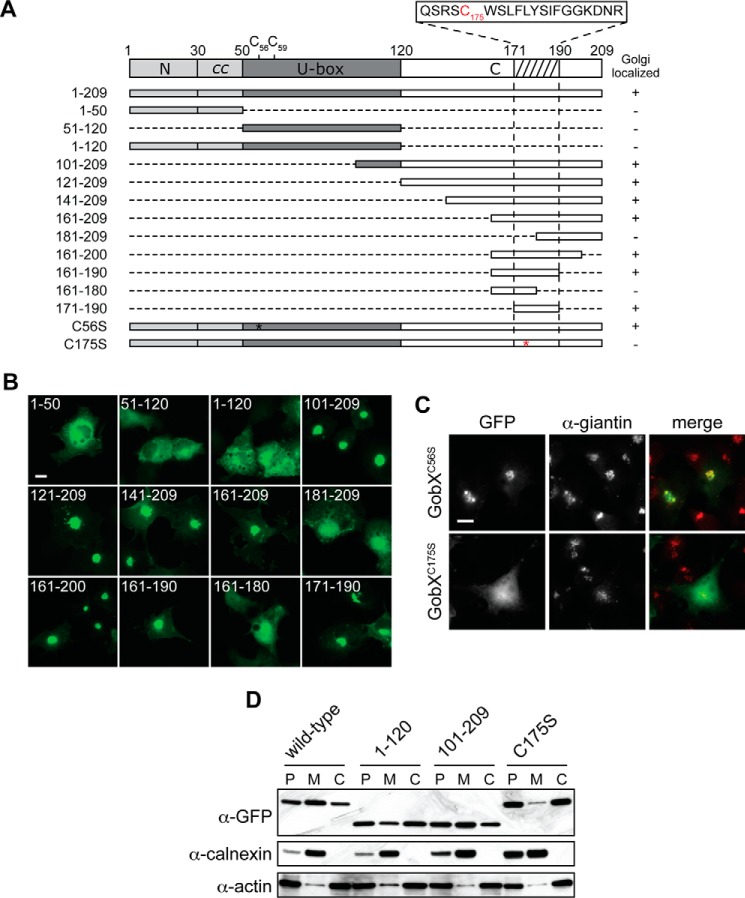
Cys-175 is required for the Golgi localization of GobX. A, schematic illustration of the GobX variants. Dashed lines indicate regions deleted in the truncated GobX variants. The asterisk denotes the position of cysteine to serine substitution in GobX. The minimal Golgi-localizing fragment (residue 171–190) is indicated by the hatched box, and the amino acid sequence of this region is shown with Cys-175 highlighted in red. The other two cysteine residues located within the U-box domain (Cys-56 and Cys-59) are also shown. Asterisk, position of C175S substitution in GobX. GobX variants that localize to the Golgi region (as shown in B and C are marked with +, and variants that showed cytosolic distribution are marked −. B, 20-amino acid region is sufficient to target GobX to the Golgi region. The GobX variants described in A were produced as GFP fusions in transiently transfected COS-1 cells and analyzed for their subcellular distribution. Scale bar, 2 μm. C, subcellular localization of GobX is affected by C175S substitution. The GFP-GobX mutants C56S and C175S were produced in transiently transfected COS-1 cells. Cells were chemically fixed and labeled with anti-giantin antibody. Scale bar, 2 μm. D, HEK293T cells producing wild-type GFP-GobX or the indicated GobX variants were homogenized. The post-nuclear (P) supernatant was subsequently fractionated into the membrane (M) and cytosolic (C) fraction. The presence of GobX in each fraction was detected by immunoblot using anti-GFP antibody. Calnexin and actin served as markers for the membrane and cytosolic fraction, respectively. All immunofluorescence images and immunoblots are representatives of at least two independent experiments with similar outcomes.
We generated genetic fusions between GFP and four GobX variants as follows: the N-terminal region (GobX(1–50)), the U-box domain (GobX(51–120)), the C-terminal region (GobX(101–209)), as well as GobX(1–120) comprising both the N-terminal and U-box domain (Fig. 4A). Upon production in transiently transfected COS-1 cells, only GobX(101–209) localized to the Golgi region in a manner similar to full-length GobX, whereas the remaining variants (GobX(1–50), GobX(51–120), and GobX(1–120)) were dispersed throughout the cytosol (Fig. 4B). We created further truncations in GobX(101–209) either from the N-terminal end (proteins starting at residues 121, 141, 161, or 181) or from the C-terminal end (proteins ending at residues 200, 190, or 180). Among all constructs tested, only GobX(181–209) and GobX(161–180) failed to target to the Golgi region (Fig. 4B). Finally, we generated a GFP-tagged GobX fragment composed of residues 171–190 (GobX(171–190)) and found that its localization was indistinguishable to that of full-length GobX (Fig. 4B). Therefore, region 171–190 was named the minimal Golgi localization region.
To decipher the molecular mechanism underlying Golgi targeting of GobX, we closely examined the amino acid composition of the minimal Golgi localization region (Fig. 4A). The presence of an isolated cysteine residue at position 175 (Cys-175) seemed unusual. Although GobX contained two additional cysteine residues (Cys-56 and Cys-59), they were proposed to be part of a zinc finger motif typically found in U-box domains. To evaluate whether Cys-175 was involved in Golgi targeting, we substituted it with a serine residue and examined the effect on the subcellular distribution of GobX (Fig. 4C). Upon production in transiently transfected COS-1 cells, GFP-GobXC175S showed a clear redistribution to the cytosol with only residual enrichment at the perinuclear locale. The Golgi targeting defect of GobXC175S was further studied in a subcellular fractionation assay where membrane compartments were separated from the cytosol by ultracentrifugation (Fig. 4D). Using immunoblot analysis, we found that GobX and GobX(101–209), the two variants that efficiently localized to the Golgi region (Figs. 3A and 4B), were enriched in the membrane fraction of transiently transfected HEK293T cell lysate, whereas GobXC175S partitioned preferentially with the cytosolic fraction, as did GobX(1–120), a fragment that lacks the region containing Cys-175. Most importantly, unlike HA-GobX, which was enriched in the Golgi membrane-containing sucrose fraction, the cysteine mutant HA-GobXC175A was not, indicating that it failed to localize to the Golgi region upon translocation by L. pneumophila into infected macrophages (Fig. 3C). These results indicated that Cys-175 is required for Golgi membrane association of GobX both in transiently transfected and infected cells.
Cys-175 of GobX Is the Target of S-Palmitoylation within Eukaryotic Cells
Within eukaryotic cells, cysteine residues are often the target of post-translational lipidations that increase the hydrophobicity of the recipient protein, thus facilitating its association with membranes. Of the two possible post-translational lipidations on cysteine residues, we excluded prenylation because Cys-175 of GobX was not part of a typical CAAX or CX(C/C)C motif. Thus, we experimentally examined the role of S-palmitoylation in subcellular targeting of GobX. 2-BP is an irreversible pharmacological inhibitor of palmitoyl acyltransferases (PATs) (35), a family of enzymes known to catalyze palmitoylation (36, 37). Upon incubation of transiently transfected COS-1 cells with 2-BP, the cytosolic pool of GFP-GobX was noticeably increased (Fig. 5A), whereas the enrichment of GFP-GobX at the Golgi region was unaffected in vehicle-treated cells. Thus, Golgi localization of GobX seemed to be dependent on the activity of PATs.
FIGURE 5.
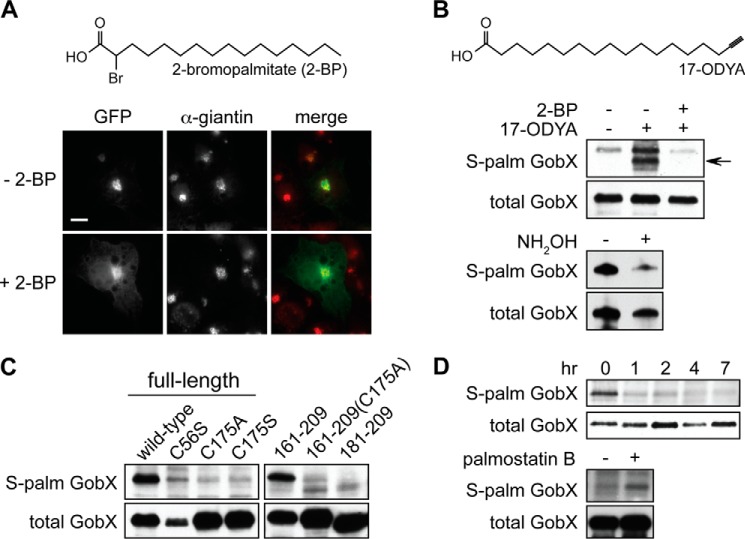
GobX is S-palmitoylated at Cys-175. A, palmitoylation inhibitor 2-BP interferes with GobX Golgi localization. Transiently transfected COS-1 cells producing GFP-GobX were treated with 40 μm 2-bromopalmitate (+2-BP) or the DMSO vehicle (−2-BP) for 5 h followed by immunofluorescence microscopy. The chemical structure of 2-BP is shown on top. B, top, HEK293T cells producing GFP-GobX were metabolically labeled for 2 h with 17-ODYA in the presence or absence of 2-BP. GFP-GobX was immunoprecipitated from the cell lysate and labeled via click chemistry with biotin-azide. Palmitoylated GobX (arrow) was detected by streptavidin-HRP in an immunoblot, whereas the total amount of GobX was detected in an anti-GFP immunoblot. The chemical structure of 17-ODYA is shown on top. Bottom, palmitoylated GFP-GobX was treated with or without 1 m hydroxylamine followed by immunoblot analyses. C, HEK293T cells producing the indicated GFP-GobX variants were metabolically labeled with 17-ODYA for 2 h. S-Palmitoylated and total GobX were detected as described in B. D, top, HEK293T cells producing GFP-GobX were pulse-labeled with 17-ODYA for 30 min and chased in regular DMEM for up to 7 h. S-Palmitoylated and total GobX was detected as described in B. Bottom, HEK293T cells producing GFP-GobX were pulse-labeled with 17-ODYA and chased in regular DMEM for 1 h in the presence or absence of 1 μm APT1 inhibitor palmostatin B. S-Palmitoylated and total GobX was detected as described in B. All immunofluorescence images and immunoblots are representatives of at least two independent experiments with similar outcomes.
To biochemically verify S-palmitoylation of GobX, we combined metabolic 17-ODYA (or Alk-16) labeling with copper(I)-assisted azide-alkyne cycloaddition (click chemistry) as a reporter assay to detect this lipidation (38–40). GFP-GobX was produced in the presence or absence of 17-ODYA in HEK293T cells depleted of natural fatty acids, immunoprecipitated, and subjected to click chemistry resulting in the cyclo-addition of the biotin tag, which can be detected via immunoblot with streptavidin-conjugated HRP. As seen in Fig. 5B, the labeling of GFP-GobX with the palmitate analog was observed only in the presence of 17-ODYA, but not in cells lacking 17-ODYA or in cells containing both 17-ODYA and the PAT inhibitor 2-BP. Treatment of palmitoylated GobX with hydroxylamine, a reagent that chemically reverses the thioester linkage between a palmitate moiety and the cysteine residue it is linked to, diminished the 17-ODYA label on GobX (Fig. 5B). Substitution of Cys-175 with either alanine (C175A) or serine (C175S) also abrogated metabolic labeling of GobX with 17-ODYA, whereas a GobX variant with Cys-56 replaced by serine (C56S) was still palmitoylated, although the signal was reduced due to a lower transfection efficiency of the encoding plasmid (Fig. 5C). Similarly, although GobX(161–209) was efficiently palmitoylated, the GobX(161–209) mutant C175A was not and neither was GobX(181–209), which lacked the region containing Cys-175. Taken together, our biochemical analysis confirmed that Cys-175 within the minimal Golgi localization region of GobX is S-palmitoylated within mammalian cells.
In eukaryotic cells, S-palmitoylation is a reversible post-translational modification, with the forward reaction catalyzed by PATs, and the reverse reaction mediated by palmitoyl protein thioesterases (41, 42). To determine the reversibility of GobX palmitoylation, we performed a pulse-chase experiment. GFP-GobX produced in transfected HEK293T cells was metabolically labeled with 17-ODYA for 30 min followed by a 7-h “chase” period with unlabeled fatty acids as part of the regular medium. As shown in Fig. 5D, 17-ODYA was efficiently incorporated into GFP-GobX during the pulse period but disappeared as soon as 1 h after the removal of 17-ODYA. To further examine the de-palmitoylation of GobX by palmitoyl thioesterases, we used the APT1 inhibitor, palmostatin B, which efficiently inhibited palmitate removal from Ras proteins in vivo (43). As seen in Fig. 5D, in the presence of the APT1 inhibitor, removal of 17-ODYA from GFP-GobX was noticeably delayed 1 h into the chase period as compared with untreated cells. Together, these results indicate that palmitoylated GobX is a substrate of palmitoyl thioesterases and that its S-palmitoylation is transient and rapidly reversed in vivo.
GobX Palmitoylation Is Catalyzed by Specific DHHC Palmitoyl Transferases
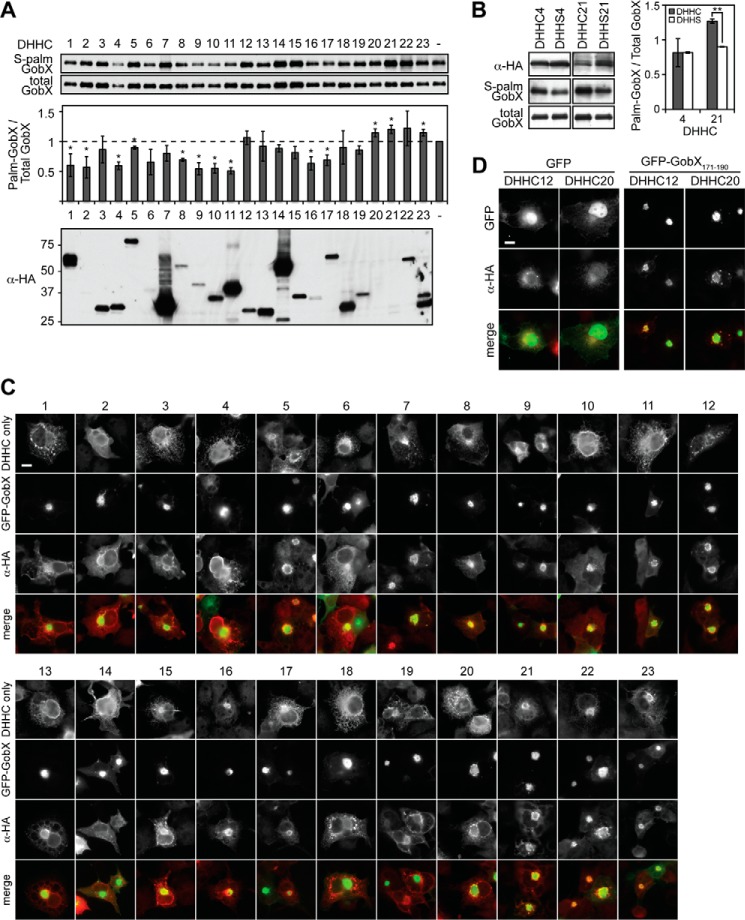
Eukaryotic S-palmitoylation is catalyzed by a conserved family of proteins that have a DHHC motif-containing domain with palmitoyltransferase activity. Human cells encode at least 23 DHHC motif-containing PATs. These enzymes are multipass membrane proteins that reside within a variety of cellular compartments, including the ER, Golgi, and plasma membrane where they catalyze protein palmitoylation (24). The factors that determine substrate specificity of DHHC PATs are largely unknown, which makes it challenging to predict palmitoylated target proteins and their cognate PAT(s) (24, 44). To investigate which DHHC proteins contribute to GobX palmitoylation, we monitored the efficiency of 17-ODYA labeling of GobX in doubly transfected HEK293T cells simultaneously producing each individual member of the mouse DHHC PAT family, which is highly homologous to the human DHHC PATs (Fig. 6A). Only three of the DHHC enzymes (DHHC20, -21, and -23) caused a noticeable stimulatory effect on GFP-GobX palmitoylation, whereas 10 of the DHHC enzymes, when produced together with GobX, decreased its level of palmitoylation most likely by competing with other DHHC proteins for the donor molecule palmitoyl-CoA. This increase or decrease did not appear to be correlated with the expression level of the DHHC enzymes (Fig. 6A, bottom), indicating that GobX palmitoylation within cells is mediated by specific rather than the most abundant DHHC proteins.
FIGURE 6.
GobX palmitoylation requires the activity of host DHHC palmitoyltransferases. A, GobX palmitoylation is altered by multiple DHHC proteins. HEK293T cells were co-transfected with plasmids encoding GFP-GobX and each of the 23 mouse DHHC palmitoyltransferases. Cells were metabolically labeled with 17-ODYA for 30 min. GFP-GobX was immunoprecipitated, click-labeled with biotin-azide, and subjected to immunoblot analyses as described in Fig. 5B. The signal of S-palmitoylated and total GobX in cells singly transfected with GFP-GobX (−) were set as 1 in both immunoblots, and the signal of the DHHC co-transfected cells were normalized relative to it. The ratio of S-palmitoylated GobX over total GobX is plotted in the graph, and the average was taken from two independent experiments. (Statistical analysis was performed using a two-tailed Student t test. *, p < 0.1 relative to GFP-GobX single transfected cells.) Production of the 23 mouse DHHC palmitoyltransferases in the HEK293T cell lysate was determined by immunoblot using anti-HA antibody. B, catalytically inactive PATs do not enhance GobX palmitoylation. HEK293T cells were co-transfected with plasmids encoding GFP-GobX and the indicated DHHC palmitoyltransferases in their active (DHHC) or inactive (DHHS) form. 17-ODYA labeling and detection was performed as in A. The average was taken from two independent experiments. (Statistical analysis was performed using a two-tailed Student t test. **, p < 0.01.) The presence of DHHC or DHHS palmitoyltransferases in HEK293T cell lysate was determined by immunoblot using anti-HA antibody. C, GobX recruits DHHC proteins to the Golgi region. The subcellular localization of the 23 DHHC proteins in the absence (top panel) or presence (bottom panels) of GobX was visualized in transiently transfected COS-1 cells. Scale bar, 2 μm. D, GobX(171–190) recruits DHHC palmitoyltransferases to the Golgi. The subcellular localization of DHHC 12 and 20 was visualized in COS-1 cells co-transfected with GFP or GFP-GobX(171–190). Scale bar, 2 μm. All immunofluorescence images are representatives of at least two independent experiments with similar outcomes.
The DHHC motif within PATs is essential for their catalytic activity (37, 45). We constructed catalytically inactive variants by cysteine to serine substitution within the DHHC motif, and we compared 17-ODYA labeling of GFP-GobX in transiently transfected HEK293T cells co-producing either the DHHC or the DHHS form of PATs (Fig. 6B). As described above, simultaneous production of active DHHC4, for which GobX is not a substrate (Fig. 6A), or inactive DHHS4 did not alter GobX palmitoylation. In contrast, the presence of inactive DHHS21 did not recapitulate the enhancement of GobX palmitoylation caused by its active counterpart, further supporting the idea that GobX palmitoylation is specifically catalyzed by a selective group of DHHC PATs.
In order for DHHC proteins to catalyze palmitoylation, they need to reside, at least temporarily, within the same membrane compartment as their target proteins. When we examined the localization of all 23 DHHC proteins within transiently transfected COS-1 cells, we found that many DHHC proteins, including DHHC20, -21, and -23 (which stimulated GobX palmitoylation), localized to the ER and/or Golgi region, consistent with earlier reports (Fig. 6C) (46). Interestingly, in the presence of either GFP-GobX or GFP-GobX(171–190), but not GFP alone, did we observe a dramatic relocation of many of the DHHC proteins, such as DHHC20, from their subcellular compartment to the Golgi region where GFP-GobX or GFP-GobX(171–190) is located (Fig. 6, C and D). The relocation was also observed in DHHC proteins that had no stimulatory effect on GobX palmitoylation (Fig. 6, A and C). Moreover, the fluorescent signal of the relocated DHHC proteins was primarily detected at the periphery of the Golgi region and overlapped only partially with GFP-GobX, which was concentrated on membranes in the center of the Golgi region, suggesting that the DHHC proteins and GobX inhabit adjacent yet not identical membrane domains. These results indicate that the subcellular localization of DHHC proteins is of a dynamic nature and can be directly influenced by the presence of palmitoylated target proteins or indirectly by the re-distribution of other proteins, most likely other DHHC proteins, meaning that they might exist or traffic in a larger DHHC protein complex or in membrane subdomains.
GobX Homologs from Various L. pneumophila Strains Possess Golgi Targeting Ability
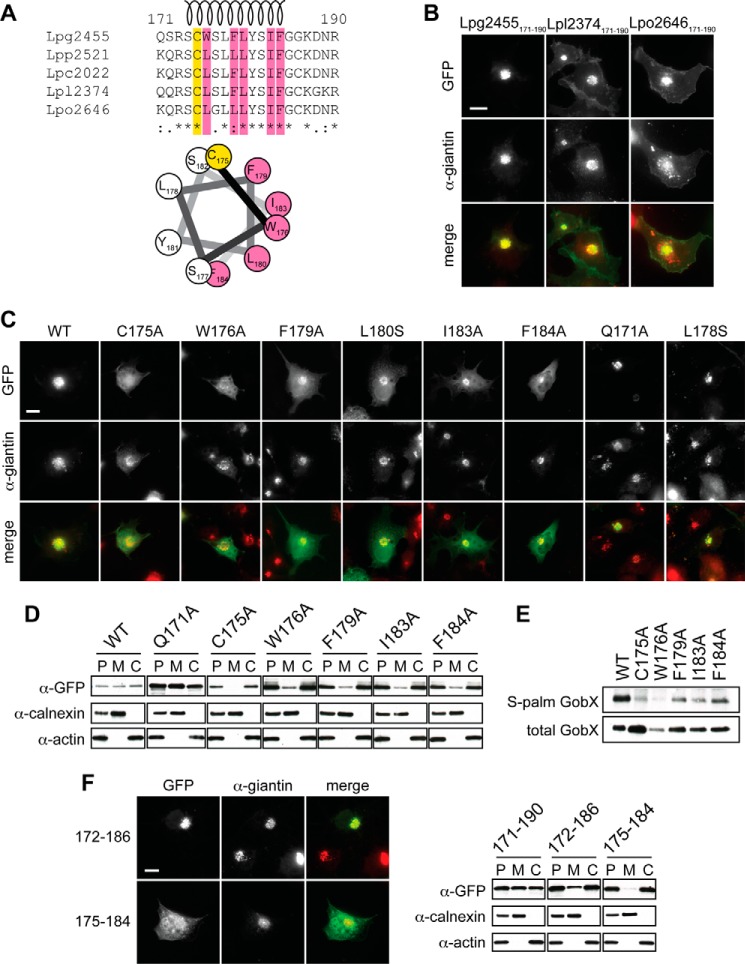
Homologs of gobX are present in several sequenced Legionella isolates, including L. pneumophila strain Paris (lpp2521), Corby (lpc2022), Lens (lpl2374), and Lorraine (lpo2646). The alignment of these homologs with GobX from L. pneumophila strain Philadelphia-1 revealed 93–95% sequence identity within the N-terminal region and the U-box domain (residues 1–170) but only 59–69% sequence identity within the C-terminal region (residues 171–209) (data not shown). Moreover, besides Cys-175 only 11 of the 20 residues within the minimal Golgi localization region (GobX(171–190)) were identical among all five homologs (Fig. 7A). We thus wondered whether region 171–190 of the GobX homologs possessed Golgi targeting capability despite the sequence variation. Upon production of GFP-tagged GobX(171–190) from strains Lens (Lpl2374) and Lorraine (Lpo2646) in transiently transfected COS-1 cells, both fusion proteins were found to be enriched at the Golgi region to the same extent as GobX(171–190) from strain Philadelphia-1 (Fig. 7B). Thus, the sequence variation within the minimal Golgi localization region did not affect subcellular targeting of the GobX homologs.
FIGURE 7.
Amphipathic helix in GobX is required for Golgi localization and palmitoylation. A, alignment of GobX(171–190) from five different L. pneumophila strains. Philadelphia-1, Lpg2455; Paris, Lpp2521; Corby, Lpc2022; Lens, Lpl2374; and Lorraine, Lpo2646. Conserved amino acid residues are marked with an asterisk at the bottom. The S-palmitoylated cysteine 175 is highlighted in yellow, and the residues critical for Golgi membrane localization of Lpg2455 (see below) are highlighted in pink. The predicted helical region is shown as a spiral. A helical wheel projection of residues Cys-175 to Phe-184 is shown at the bottom (26). B, Golgi targeting of GobX(171–190) from L. pneumophila isolates. The subcellular localization of GobX(171–190) from L. pneumophila strains Philadelphia-1 (Lpg2455), Lens (Lpl2374), and Lorraine (Lpo2646) was examined in transiently transfected COS-1 cells. The Golgi region was immunostained with anti-giantin antibody. Scale bar, 2 μm. C, identification of residues critical for Golgi targeting of GobX. The subcellular localization of the indicated GFP-GobX mutants was examined in transiently transfected COS-1 cells. The Golgi region was labeled with anti-giantin antibody. Scale bar, 2 μm. D, substitution of single residues in GobX interfered with membrane association. HEK293T cells producing the indicated GobX mutants were lysed and separated into the membrane and cytosolic fraction. The presence of GobX was detected as described in Fig. 4D. Calnexin and actin served as markers for the membrane and cytosolic fraction, respectively. E, substitution of single residues in GobX interfered with palmitoylation. GFP-GobX mutants were produced in transiently transfected HEK293T cells. S-Palmitoylation of GobX was determined by metabolic labeling with 17-ODYA and detected as described in Fig. 5B. F, further truncation of GobX(171–190) abrogates efficient Golgi targeting. Left, subcellular localization of GFP-tagged GobX(172–186) and GobX(175–184) was examined in transiently transfected COS-1 cells. Scale bar, 2 μm. Right, HEK293T cells producing GFP-tagged GobX(171–190), GobX(172–186), and GobX(175–184) were homogenized and separated into the membrane and cytosolic fraction, and the presence of GobX was detected as described in Fig. 4D. All immunofluorescence images and immunoblots are representatives of at least two independent experiments with similar outcomes.
Amphipathic Helix in GobX(171–190) Links Membrane Insertion to Palmitoylation
Given the unexpected tolerance of the minimal Golgi localization region toward amino acid substitutions, we hypothesized that the Golgi-targeting cues within GobX(171–190) may be structure-based rather than sequence-based. Upon closer examination, we noticed that region 174–184 is predicted to form an α-helix (Fig. 7A), with Cys-175 situated at its beginning. To define additional residues that contribute to palmitoylation and membrane association of GobX, we interrogated region 171–190 by alanine scanning. With the exception of Gly-185 and Gly-186, each residue of region 171–190 was replaced with alanine, and subcellular localization of the resulting GFP-GobX mutant was determined by fluorescence microscopy within transiently transfected COS-1 cells (Fig. 7C). Whereas alanine substitution of residues outside the helix region (e.g. Gln-171) had no obvious effect on Golgi localization of GFP-GobX, replacement of Trp-176, Phe-179, Ile-183, or Phe-184 with alanine resulted in a dispersed cytosolic distribution similar to the palmitoylation-deficient mutant GobXC175A (Fig. 7C). The effect of alanine substitutions to disrupt membrane association of GobX was further analyzed by cell fractionation assay (Fig. 7D). Alanine substitutions of Trp-176, Phe-179, Ile-183, or Phe-184 caused GobX to be highly enriched in the cytosolic fraction similar to GobXC175A, as opposed to wild-type GobX, or GobXQ171A, which was distributed equally between the membrane and cytosolic fraction. Thus, Trp-176, Phe-179, Ile-183, and Phe-184 are important for stable Golgi membrane association of GobX. Interestingly, all of these residues are hydrophobic in nature and predicted to be located on one side of the proposed α-helix in GobX(171–190), thus giving the helix an amphipathic character (Fig. 7A). Supporting this model was the fact that the serine residues at positions 174, 177, and 182 were positioned at the lipid-cytosol interface so that they could be available for interaction with polar lipid headgroups. The only residue on the hydrophobic face of the amphipathic helix that did not cause an obvious localization defect of GFP-GobX when replaced with an alanine was Leu-180. A possible explanation was that alanine is very similar in character to leucine. Thus, to validate that Leu-180 also contributed to the amphipathic character of the helix, we substituted it with a polar serine residue. Indeed, GFP-GobXL180S showed an enriched cytosolic distribution similar to alanine substitutions of the other hydrophobic residues in the amphipathic helix (Fig. 7C), whereas serine substitution of Leu-178, a residue predicted to face the cytosol, did not alter subcellular localization of GobX. These results further strengthened the hypothesis that an amphipathic helix adjacent to Cys-175 plays an important role in Golgi localization of GobX.
Alanine substitution of Trp-176, Phe-179, Ile-183, and Phe-184 attenuated Golgi membrane localization of GobX. We thus wondered whether these variants were still palmitoylated at Cys-175 but eventually dissociated from the Golgi membrane due to the reduced affinity of the amphipathic helix to lipid bilayers. Alternatively, the less hydrophobic amphipathic helix of the GobX variants could have prevented efficient palmitoylation of GobX by Golgi-resident DHHC proteins. To distinguish these two possibilities, we examined the palmitoylation efficiency of these GFP-GobX variants using 17-ODYA labeling and found a strong reduction in the level of palmitoylation for each of them (Fig. 7E). Therefore, without proper membrane association of the amphipathic helix, Cys-175 was not efficiently palmitoylated by Golgi-localized DHHC proteins thus preventing GobX from stably localizing to the Golgi membrane.
Given that the residues flanking the amphipathic helix seemed dispensable for Golgi targeting when replaced with alanine, we attempted to further truncate the minimal Golgi localization region by removing amino acid residues from both the N- and C-terminal end of GobX(171–190). Although visual analysis revealed that GFP-tagged GobX(172–186) displayed Golgi localization similar to GobX(171–190) (Fig. 7F), the subcellular fractionation assay indicated that this variant had a reduced membrane association capability compared with GobX(171–190) (Fig. 7F, right). The even shorter variant GobX(175–184) showed a cytosolic distribution in either assay suggesting that the residues flanking the amphipathic helix, though tolerant to alanine substitution, were still required for proper localization of GobX(171–190), probably by contributing to helix stability and subsequent palmitoylation of GobX.
Discussion
In this study, we provided evidence that the effector protein GobX from L. pneumophila utilizes host cell machineries for the catalysis of two different post-translational modifications, ubiquitination and S-palmitoylation. To our knowledge, GobX is only the second known example of a bacterial E3 ubiquitin ligase that exploits this type of lipidation, the other one being SspH2 from Salmonella (47).
Although S-palmitoylation has been detected in several hundred human proteins (48, 49), the only bacterial proteins besides GobX known to receive this reversible lipidation are the type III-translocated effectors SspH2, SseI, and SifA from Salmonella (47, 50) and a family of cysteine proteases from Pseudomonas syringae (51, 52). There are, however, several notable differences between these effectors and GobX. First, GobX was highly enriched at the Golgi region (Fig. 3A), whereas the Salmonella and Pseudomonas proteins were targeted preferentially to the plasma membrane in eukaryotic cells. Second, GobX exploited a different set of DHHC proteins than SspH2 and SseI (Fig. 6A) (47), which may explain their different subcellular localizations. Third, GobX was de-palmitoylated in less than an hour by the endogenous thioesterases in HEK293T cells (Fig. 5D), whereas the palmitoylation of SspH2 remained stable for several hours (47). Finally, the palmitoylation site in the Salmonella and Pseudomonas effectors was located within the very N-terminal region of the proteins, whereas palmitoylation of GobX occurred on Cys-175 within the C-terminal domain. Together, these differences highlight the level of flexibility in S-palmitoylation within eukaryotic cells and that it can be exploited by various bacterial pathogens to direct their effectors to different subcellular locations. Notably, while our study was under review, Frankel and co-workers (53) published data suggesting that the L. pneumophila effector LpdA may also exploit host-mediated S-palmitoylation, further strengthening the role of this post-translational modification in membrane targeting of microbial effectors.
The finding of a minimal Golgi-localizing region GobX(171–190) was intriguing. It is currently unclear how the 20 amino acid stretch can be specifically delivered or targeted to the Golgi, and whether this is accomplished by chaperone-mediated delivery, preference for certain membrane lipids, or binding to specific Golgi-resident proteins. Nonetheless, palmitoylation at Cys-175 was required for the Golgi localization. According to the “two-signal hypothesis,” the membrane affinity of a singly palmitoylated (or prenylated) polypeptide is not sufficient to maintain stable membrane association (54). Thus, additional membrane-anchoring forces are required. Palmitoylated cysteines can be surrounded by membrane-targeting determinants like polybasic or hydrophobic residues, prenylation or myristoylation sites, or transmembrane domains, all of which are believed to further increase membrane affinity (54). Here, we provide evidence that a palmitoylated cysteine can also be paired with an amphipathic helix. However, this amphipathic helix in GobX did not entirely fit the criteria of a typical membrane-targeting determinant. It played a role not only as an enhancer to the membrane affinity of the palmitoylated form of GobX but rather functioned as a key element for GobX palmitoylation to occur. Substitution of individual hydrophobic residues on the nonpolar face of the helix disrupted GobX palmitoylation and, consequently, its subcellular localization (Fig. 7). We propose that theamphipathic helix has to be partially immersed within the lipid bilayer to bring Cys-175 into close proximity with the DHHC motif in PAT proteins that catalyze the lipidation reaction (Fig. 8). Although a similar arrangement, a palmitoylation site adjacent to an amphipathic helix, has been described for other membrane-associated proteins such as Irgm1 (Immunity-related GTPases) or PI4KIIa (phosphatidylinositol 4-kinase IIa) (55, 56), there is no evidence that the amphipathic helix in these proteins is required for palmitoylation. It will be interesting to see whether other bacterial or eukaryotic proteins exist that utilize a “two-signal” strategy similar to GobX to exploit S-palmitoylation.
FIGURE 8.
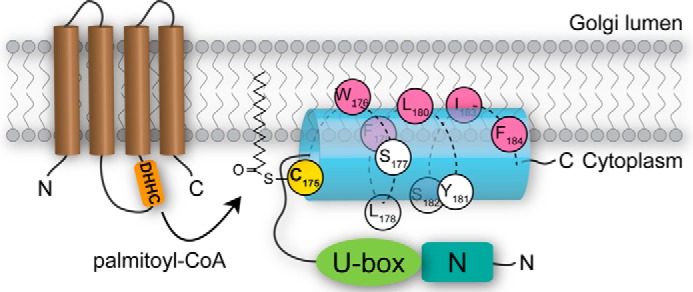
Model for membrane association of GobX. Upon arrival at the Golgi membrane, the non-polar face of the amphipathic helix (blue) of GobX(175–184) containing hydrophobic residues (pink) embeds into the lipid bilayer, whereas the polar residues (white) are facing the cytosol. Membrane association brings Cys-175 (yellow) into close proximity with the catalytic site of DHHC palmitoyltransferases (brown) for palmitoylation to occur.
Within transfected cells, we discovered a remarkable relocation of several of the 23 DHHC acyltransferases to the Golgi region in the presence of GobX (Fig. 6C). Even more surprising, the minimal Golgi localization region (amino acids 171–190) was sufficient to trigger this relocation process (Fig. 6D). The molecular mechanism underlying DHHC protein capture at the Golgi by GobX is unclear. However, because recruitment was not limited to DHHC proteins that enhanced GobX palmitoylation, this behavior could indicate that DHHC proteins are organized within membrane subdomains or larger protein complexes that traffic together between cellular compartments.
An important question was whether the localization of GobX within transiently transfected mammalian cells (Fig. 3A) adequately represented the effector's localization pattern during the course of an infection. More precisely, would GobX upon translocation by intracellular L. pneumophila associate with the LCV membrane rather than the Golgi membrane, as is the case for several other L. pneumophila effectors such as SidM or SidD (30, 32, 57). This is particularly relevant given that the LCV membrane displays several features reminiscent of the Golgi, most notably, the presence of PtdIns(4)P and the small GTPase Rab1 (58–60). Although we were unable to unambiguously visualize translocated GobX within infected cells by immunofluorescence microscopy, we were able to use sucrose gradient ultracentrifugation to separate Golgi membranes of macrophages infected with L. pneumophila. GobX, but not the palmitoylation-deficient cysteine mutant (C175A), was clearly enriched in the Golgi fraction (Fig. 3C), indicating that GobX after translocation by the Dot/Icm T4SS targets to Golgi membranes and that palmitoylation of Cys-175 is required for this localization. Although we cannot exclude the possibility that a fraction of GobX may be found on LCVs, this is an unlikely scenario given that PATs were absent from the LCV membrane proteome (61, 62).
In addition to S-palmitoylation, GobX also exploited host cell ubiquitination by functioning as an E3 ubiquitin ligase (Fig. 2). The central region of GobX (residues 51–120) showed functional similarity to mammalian U-box E3 ligases, and substitution of Ile-58 and Trp-87 within the predicted E2-binding site severely reduced auto-ubiquitination activity of GobX (Fig. 2C). How ubiquitin ligase activity of GobX at the Golgi contributes to L. pneumophila infection is currently unclear. We hypothesize that GobX ubiquitinates an unknown Golgi protein to change its fate, either by targeting it to a different subcellular location or by promoting its proteasomal degradation. Given that production of GobX did not seem to alter Golgi integrity or function (Fig. 3A), we excluded the possibility that GobX interfered with secretory traffic, which is considered essential in eukaryotic cells. Likewise, there was no noticeable reduction in the transport of CTxB from the plasma membrane to the ER (Fig. 3D), indicating that retrograde cargo flow was also normal in the presence of GobX. Moreover, producing GobX in yeast cells did not impact yeast proliferation (Fig. 3E). The most likely explanation for the absence of cell division or viability defect was that GobX targeted only one of several functionally redundant host proteins and that the remaining host factors were sufficient to prevent the emergence of a phenotype under the conditions tested here. This would be in line with the fact that most E3 ligases are highly selective enzymes that bind to and ubiquitinate a particular target protein. During infection, L. pneumophila might translocate additional unknown effector(s) that function in synergy with GobX and that target the redundant host factor(s), thus taking control of the host pathway. Future studies will need to determine the identity of the host target of GobX and how its ubiquitination activity benefits intracellular L. pneumophila.
Author Contributions
Y. L., A. G. D., E. C., and M. P. M. conceived and designed the experiments. Y. L., A. G. D., and E. C. performed the experiments. Y. L., A. G. D., E. C., and M. P. M. analyzed the data. B. K. and T. R. E. contributed reagents/materials/in silico data. Y. L. and M. P. M. wrote the paper.
Supplementary Material
Acknowledgments
We are grateful to members of the Machner laboratory for critical reading and insightful discussion. We thank Dr. Masaki Fukata (National Institutes of Natural Sciences, Japan) for providing the DHHC PAT plasmid library.
This work was supported, in whole or in part, by National Institutes of Health Grant 1ZIAHD008893-04 from the Intramural Research Program. The authors declare that they have no conflicts of interest with the contents of this article.

This article contains supplemental Tables S1 and S2.
- T4SS
- type IV secretion system
- LCV
- Legionella-containing vacuole
- CTxB
- cholera toxin subunit B
- PAT
- palmitoyl acyltransferase
- m.o.i.
- multiplicity of infection
- α
- anti
- 2-BP
- 2-bromopalmitic acid
- PtdIns
- phosphatidylinositol
- rcf
- relative centrifugal force
- IPTG
- isopropyl β-dithiogalactopyranoside
- 17-ODYA
- 17-octadecynoic acid
- ER
- endoplasmic reticulum.
References
- 1. Voth D. E., Broederdorf L. J., Graham J. G. (2012) Bacterial Type IV secretion systems: versatile virulence machines. Future Microbiol. 7, 241–257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Hubber A., Roy C. R. (2010) Modulation of host cell function by Legionella pneumophila type IV effectors. Annu. Rev. Cell Dev. Biol. 26, 261–283 [DOI] [PubMed] [Google Scholar]
- 3. Ge J., Shao F. (2011) Manipulation of host vesicular trafficking and innate immune defence by Legionella Dot/Icm effectors. Cell. Microbiol. 13, 1870–1880 [DOI] [PubMed] [Google Scholar]
- 4. Ensminger A. W., Isberg R. R. (2009) Legionella pneumophila Dot/Icm translocated substrates: a sum of parts. Curr. Opin. Microbiol. 12, 67–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. O'Connor T. J., Adepoju Y., Boyd D., Isberg R. R. (2011) Minimization of the Legionella pneumophila genome reveals chromosomal regions involved in host range expansion. Proc. Natl. Acad. Sci. U.S.A. 108, 14733–14740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Berger K. H., Isberg R. R. (1993) Two distinct defects in intracellular growth complemented by a single genetic locus in Legionella pneumophila. Mol. Microbiol. 7, 7–19 [DOI] [PubMed] [Google Scholar]
- 7. Cazalet C., Rusniok C., Brüggemann H., Zidane N., Magnier A., Ma L., Tichit M., Jarraud S., Bouchier C., Vandenesch F., Kunst F., Etienne J., Glaser P., Buchrieser C. (2004) Evidence in the Legionella pneumophila genome for exploitation of host cell functions and high genome plasticity. Nat. Genet. 36, 1165–1173 [DOI] [PubMed] [Google Scholar]
- 8. de Felipe K. S., Pampou S., Jovanovic O. S., Pericone C. D., Ye S. F., Kalachikov S., Shuman H. A. (2005) Evidence for acquisition of Legionella type IV secretion substrates via interdomain horizontal gene transfer. J. Bacteriol. 187, 7716–7726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hatakeyama S., Yada M., Matsumoto M., Ishida N., Nakayama K.-I. (2001) U box proteins as a new family of ubiquitin-protein ligases. J. Biol. Chem. 276, 33111–33120 [DOI] [PubMed] [Google Scholar]
- 10. Zheng N., Schulman B. A., Song L., Miller J. J., Jeffrey P. D., Wang P., Chu C., Koepp D. M., Elledge S. J., Pagano M., Conaway R. C., Conaway J. W., Harper J. W., Pavletich N. P. (2002) Structure of the Cul1-Rbx1-Skp1-F boxSkp2 SCF ubiquitin ligase complex. Nature 416, 703–709 [DOI] [PubMed] [Google Scholar]
- 11. Kubori T., Hyakutake A., Nagai H. (2008) Legionella translocates an E3 ubiquitin ligase that has multiple U-boxes with distinct functions. Mol. Microbiol. 67, 1307–1319 [DOI] [PubMed] [Google Scholar]
- 12. Ensminger A. W., Isberg R. R. (2010) E3 Ubiquitin ligase activity and targeting of BAT3 by multiple Legionella pneumophila translocated substrates. Infect. Immun. 78, 3905–3919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kubori T., Nagai H. (2011) Bacterial effector-involved temporal and spatial regulation by hijack of the host ubiquitin pathway. Front. Microbiol. 2, 145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hsu F., Luo X., Qiu J., Teng Y.-B., Jin J., Smolka M. B., Luo Z.-Q., Mao Y. (2014) The Legionella effector SidC defines a unique family of ubiquitin ligases important for bacterial phagosomal remodeling. Proc. Natl. Acad. Sci. U.S.A. 111, 10538–10543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Brombacher E., Urwyler S., Ragaz C., Weber S. S., Kami K., Overduin M., Hilbi H. (2009) Rab1 guanine nucleotide exchange factor SidM is a major phosphatidylinositol 4-phosphate-binding effector protein of Legionella pneumophila. J. Biol. Chem. 284, 4846–4856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ragaz C., Pietsch H., Urwyler S., Tiaden A., Weber S. S., Hilbi H. (2008) The Legionella pneumophila phosphatidylinositol-4 phosphate-binding type IV substrate SidC recruits endoplasmic reticulum vesicles to a replication-permissive vacuole. Cell. Microbiol. 10, 2416–2433 [DOI] [PubMed] [Google Scholar]
- 17. Hammond G. R., Machner M. P., Balla T. (2014) A novel probe for phosphatidylinositol 4-phosphate reveals multiple pools beyond the Golgi. J. Cell Biol. 205, 113–126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Neunuebel M. R., Mohammadi S., Jarnik M., Machner M. P. (2012) Legionella pneumophila LidA affects nucleotide binding and activity of the host GTPase Rab1. J. Bacteriol. 194, 1389–1400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ku B., Lee K.-H., Park W. S., Yang C.-S., Ge J., Lee S.-G., Cha S.-S., Shao F., Heo W. D., Jung J. U., Oh B.-H. (2012) VipD of Legionella pneumophila targets activated Rab5 and Rab22 to interfere with endosomal trafficking in macrophages. PLoS Pathog. 8, e1003082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Gaspar A. H., Machner M. P. (2014) VipD is a Rab5-activated phospholipase A1 that protects Legionella pneumophila from endosomal fusion. Proc. Natl. Acad. Sci. U.S.A. 111, 4560–4565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lucas M., Gaspar A. H., Pallara C., Rojas A. L., Fernández-Recio J., Machner M. P., Hierro A. (2014) Structural basis for the recruitment and activation of the Legionella phospholipase VipD by the host GTPase Rab5. Proc. Natl. Acad. Sci. U.S.A. 111, E3514–E3523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ivanov S. S., Charron G., Hang H. C., Roy C. R. (2010) Lipidation by the host prenyltransferase machinery facilitates membrane localization of Legionella pneumophila effector proteins. J. Biol. Chem. 285, 34686–34698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Price C. T., Jones S. C., Amundson K. E., Kwaik Y. A. (2010) Host-mediated post-translational prenylation of novel Dot/Icm-translocated effectors of Legionella pneumophila. Front. Microbiol. 1, 131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Greaves J., Chamberlain L. H. (2011) DHHC palmitoyltransferases: substrate interactions and (patho)physiology. Trends Biochem. Sci. 36, 245–253 [DOI] [PubMed] [Google Scholar]
- 25. Feeley J. C., Gibson R. J., Gorman G. W., Langford N. C., Rasheed J. K., Mackel D. C., Baine W. B. (1979) Charcoal-yeast extract agar: primary isolation medium for Legionella pneumophila. J. Clin. Microbiol. 10, 437–441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Larkin M. A., Blackshields G., Brown N. P., Chenna R., McGettigan P. A., McWilliam H., Valentin F., Wallace I. M., Wilm A., Lopez R., Thompson J. D., Gibson T. J., Higgins D. G. (2007) Clustal W and Clustal X version 2.0. Bioinformatics 23, 2947–2948 [DOI] [PubMed] [Google Scholar]
- 27. Luo Z.-Q., Isberg R. R. (2004) Multiple substrates of the Legionella pneumophila Dot/Icm system identified by interbacterial protein transfer. Proc. Natl. Acad. Sci. U.S.A. 101, 841–846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Neunuebel M. R., Chen Y., Gaspar A. H., Backlund P. S. Jr., Yergey A., Machner M. P. (2011) De-AMPylation of the small GTPase Rab1 by the pathogen Legionella pneumophila. Science 333, 453–456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. de Felipe K. S., Glover R. T., Charpentier X., Anderson O. R., Reyes M., Pericone C. D., Shuman H. A. (2008) Legionella eukaryotic-like type IV substrates interfere with organelle trafficking. PLoS Pathog. 4, e1000117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Conover G. M., Derré I., Vogel J. P., Isberg R. R. (2003) The Legionella pneumophila LidA protein: a translocated substrate of the Dot/Icm system associated with maintenance of bacterial integrity. Mol. Microbiol. 48, 305–321 [DOI] [PubMed] [Google Scholar]
- 31. Graham J. M. (2001) Isolation of Golgi membranes from tissues and cells by differential and density gradient centrifugation. Curr. Protoc. Cell Biol. Chapter 3 Unit 3.9 [DOI] [PubMed] [Google Scholar]
- 32. Machner M. P., Isberg R. R. (2006) Targeting of host Rab GTPase function by the intravacuolar pathogen Legionella pneumophila. Dev. Cell 11, 47–56 [DOI] [PubMed] [Google Scholar]
- 33. Huang L., Boyd D., Amyot W. M., Hempstead A. D., Luo Z.-Q., O'Connor T. J., Chen C., Machner M., Montminy T., Isberg R. R. (2011) The E Block motif is associated with Legionella pneumophila translocated substrates. Cell. Microbiol. 13, 227–245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Söding J., Biegert A., Lupas A. N. (2005) The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res. 33, W244–W248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Davda D., El Azzouny M. A., Tom C. T., Hernandez J. L., Majmudar J. D., Kennedy R. T., Martin B. R. (2013) Profiling targets of the irreversible palmitoylation inhibitor 2-bromopalmitate. ACS Chem. Biol. 8, 1912–1917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Webb Y., Hermida-Matsumoto L., Resh M. D. (2000) Inhibition of protein palmitoylation, raft localization, and T cell signaling by 2-bromopalmitate and polyunsaturated fatty acids. J. Biol. Chem. 275, 261–270 [DOI] [PubMed] [Google Scholar]
- 37. Fukata M., Fukata Y., Adesnik H., Nicoll R. A., Bredt D. S. (2004) Identification of PSD-95 palmitoylating enzymes. Neuron 44, 987–996 [DOI] [PubMed] [Google Scholar]
- 38. Yount J. S., Zhang M. M., Hang H. C. (2013) Emerging roles for protein S-palmitoylation in immunity from chemical proteomics. Curr. Opin. Chem. Biol. 17, 27–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Charron G., Wilson J., Hang H. C. (2009) Chemical tools for understanding protein lipidation in eukaryotes. Curr. Opin. Chem. Biol. 13, 382–391 [DOI] [PubMed] [Google Scholar]
- 40. Charron G., Zhang M. M., Yount J. S., Wilson J., Raghavan A. S., Shamir E., Hang H. C. (2009) Robust fluorescent detection of protein fatty-acylation with chemical reporters. J. Am. Chem. Soc. 131, 4967–4975 [DOI] [PubMed] [Google Scholar]
- 41. Hernandez J. L., Majmudar J. D., Martin B. R. (2013) Profiling and inhibiting reversible palmitoylation. Curr. Opin. Chem. Biol. 17, 20–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Conibear E., Davis N. G. (2010) Palmitoylation and depalmitoylation dynamics at a glance. J. Cell Sci. 123, 4007–4010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Dekker F. J., Rocks O., Vartak N., Menninger S., Hedberg C., Balamurugan R., Wetzel S., Renner S., Gerauer M., Schölermann B., Rusch M., Kramer J. W., Rauh D., Coates G. W., Brunsveld L., et al. (2010) Small-molecule inhibition of APT1 affects Ras localization and signaling. Nat. Chem. Biol. 6, 449–456 [DOI] [PubMed] [Google Scholar]
- 44. Ohno Y., Kashio A., Ogata R., Ishitomi A., Yamazaki Y., Kihara A. (2012) Analysis of substrate specificity of human DHHC protein acyltransferases using a yeast expression system. Mol. Biol. Cell 23, 4543–4551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Roth A. F., Feng Y., Chen L., Davis N. G. (2002) The yeast DHHC cysteine-rich domain protein Akr1p is a palmitoyltransferase. J. Cell Biol. 159, 23–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Ohno Y., Kihara A., Sano T., Igarashi Y. (2006) Intracellular localization and tissue-specific distribution of human and yeast DHHC cysteine-rich domain-containing proteins. Biochim. Biophys. Acta 1761, 474–483 [DOI] [PubMed] [Google Scholar]
- 47. Hicks S. W., Charron G., Hang H. C., Galán J. E. (2011) Subcellular targeting of Salmonella virulence proteins by host-mediated S-palmitoylation. Cell Host Microbe 10, 9–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Martin B. R., Cravatt B. F. (2009) Large-scale profiling of protein palmitoylation in mammalian cells. Nat. Methods 6, 135–138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Martin B. R., Wang C., Adibekian A., Tully S. E., Cravatt B. F. (2012) Global profiling of dynamic protein palmitoylation. Nat. Methods 9, 84–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Reinicke A. T., Hutchinson J. L., Magee A. I., Mastroeni P., Trowsdale J., Kelly A. P. (2005) A Salmonella typhimurium effector protein SifA is modified by host cell prenylation and S-acylation machinery. J. Biol. Chem. 280, 14620–14627 [DOI] [PubMed] [Google Scholar]
- 51. Dowen R. H., Engel J. L., Shao F., Ecker J. R., Dixon J. E. (2009) A family of bacterial cysteine protease type III effectors utilizes acylation-dependent and -independent strategies to localize to plasma membranes. J. Biol. Chem. 284, 15867–15879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Nimchuk Z., Marois E., Kjemtrup S., Leister R. T., Katagiri F., Dangl J. L. (2000) Eukaryotic fatty acylation drives plasma membrane targeting and enhances function of several type III effector proteins from Pseudomonas syringae. Cell 101, 353–363 [DOI] [PubMed] [Google Scholar]
- 53. Schroeder G. N., Aurass P., Oates C. V., Tate E. W., Hartland E. L., Flieger A., Frankel G. (2015) The Legionella pneumophila effector LpdA is a palmitoylated phospholipase D virulence factor. Infect. Immun. 10.1128/IAI.00785-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Resh M. D. (2006) Trafficking and signaling by fatty-acylated and prenylated proteins. Nat. Chem. Biol. 2, 584–590 [DOI] [PubMed] [Google Scholar]
- 55. Henry S. C., Schmidt E. A., Fessler M. B., Taylor G. A. (2014) Palmitoylation of the immunity related GTPase, Irgm1: impact on membrane localization and ability to promote mitochondrial fission. PLoS ONE 9, e95021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Zhou Q., Li J., Yu H., Zhai Y., Gao Z., Liu Y., Pang X., Zhang L., Schulten K., Sun F., Chen C. (2014) Molecular insights into the membrane-associated phosphatidylinositol 4-kinase IIα. Nat. Commun. 5, 3552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Murata T., Delprato A., Ingmundson A., Toomre D. K., Lambright D. G., Roy C. R. (2006) The Legionella pneumophila effector protein DrrA is a Rab1 guanine nucleotide-exchange factor. Nat. Cell Biol. 8, 971–977 [DOI] [PubMed] [Google Scholar]
- 58. Derré I., Isberg R. R. (2004) Legionella pneumophila replication vacuole formation involves rapid recruitment of proteins of the early secretory system. Infect. Immun. 72, 3048–3053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Kagan J. C., Stein M.-P., Pypaert M., Roy C. R. (2004) Legionella subvert the functions of Rab1 and Sec22b to create a replicative organelle. J. Exp. Med. 199, 1201–1211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Weber S. S., Ragaz C., Reus K., Nyfeler Y., Hilbi H. (2006) Legionella pneumophila exploits PI(4)P to anchor secreted effector proteins to the replicative vacuole. PLoS Pathog. 2, e46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Shevchuk O., Batzilla C., Hägele S., Kusch H., Engelmann S., Hecker M., Haas A., Heuner K., Glöckner G., Steinert M. (2009) Proteomic analysis of Legionella-containing phagosomes isolated from Dictyostelium. Int. J. Med. Microbiol. 299, 489–508 [DOI] [PubMed] [Google Scholar]
- 62. Urwyler S., Nyfeler Y., Ragaz C., Lee H., Mueller L. N., Aebersold R., Hilbi H. (2009) Proteome analysis of Legionella vacuoles purified by magnetic immunoseparation reveals secretory and endosomal GTPases. Traffic 10, 76–87 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.