Background: The enterotoxigenic E. coli (ETEC) Type IVb pilus systems each possess a single minor pilin.
Results: We show that these minor pilins are required for pilus assembly and report the x-ray crystal structure of CofB from the CFA/III pilus.
Conclusion: ETEC minor pilins initiate pilus assembly.
Significance: The CofB structure has implications for understanding assembly in more complex Type IV pilus systems.
Keywords: bacterial pathogenesis, membrane protein, Type II secretion (T2S) system, type IV pili, X-ray crystallography, ETEC, enterotoxigenic E. coli, minor pilin, pili, pilus assembly
Abstract
Type IV pili are extracellular polymers of the major pilin subunit. These subunits are held together in the pilus filament by hydrophobic interactions among their N-terminal α-helices, which also anchor the pilin subunits in the inner membrane prior to pilus assembly. Type IV pilus assembly involves a conserved group of proteins that span the envelope of Gram-negative bacteria. Among these is a set of minor pilins, so named because they share their hydrophobic N-terminal polymerization/membrane anchor segment with the major pilins but are much less abundant. Minor pilins influence pilus assembly and retraction, but their precise functions are not well defined. The Type IV pilus systems of enterotoxigenic Escherichia coli and Vibrio cholerae are among the simplest of Type IV pilus systems and possess only a single minor pilin. Here we show that the enterotoxigenic E. coli minor pilins CofB and LngB are required for assembly of their respective Type IV pili, CFA/III and Longus. Low levels of the minor pilins are optimal for pilus assembly, and CofB can be detected in the pilus fraction. We solved the 2.0 Å crystal structure of N-terminally truncated CofB, revealing a pilin-like protein with an extended C-terminal region composed of two discrete domains connected by flexible linkers. The C-terminal region is required for CofB to initiate pilus assembly. We propose a model for CofB-initiated pilus assembly with implications for understanding filament growth in more complex Type IV pilus systems as well as the related Type II secretion system.
Introduction
Bacterial Type IV pili (T4P)3 are long thin filaments of the major pilin subunit. This small protein has an extended 53-amino acid α-helix, α1, of which the C-terminal half, α1C, is embedded in the globular C-terminal domain of the pilin, and the N-terminal half, α1N, forms a hydrophobic stalk that both anchors the subunit in the inner membrane prior to pilus assembly and holds the subunits together in the assembled pilus filament (1–3). Within the hydrophobic α1N, there is an invariant acidic residue, Glu5. The globular C-terminal domain has a central 4- or 5-stranded antiparallel β-sheet and a pair of disulfide-bonded cysteines. Vibrio cholerae and enterotoxigenic Escherichia coli (ETEC) produce Type IV pilins of the IVb (T4b) class, which are distinguished from those of the Type IVa (T4a) class by having longer signal peptides (25–30 amino acids), longer mature proteins (>200 amino acids), and a variable hydrophobic amino acid at their mature N-terminal position (4). T4b pilins also have longer D-regions, which lie between the disulfide-bonded cysteines in the C-terminal region of the globular domain. In contrast, the T4a pilins of Neisseria gonorrhoeae, Neisseria meningitidis, and Pseudomonas aeruginosa have a 6–8-amino acid signal peptide, a ∼150-amino acid mature pilin, and an N-terminal phenylalanine. The structures of the T4a and T4b pilins differ primarily in the αβ-loop that connects α1 with the central β-sheet, in the D-region, and in the connectivity of the β-sheet itself. The T4b pilus machinery is much simpler, requiring less than a dozen proteins all encoded on the same gene cluster, whereas the T4a assembly machinery utilizes 40 or more proteins encoded on genes distributed throughout the genome (5). Despite these differences, all Type IV pilins share the canonical ladle-shaped pilin structure and helical arrangement within the pilus filament in which the N-terminal α-helices associate to form a hydrophobic core, and subunits are related by an axial rise of 8–10 Å and an azimuthal rotation of ∼100° (6, 7). Importantly, the conserved Glu5, which is critical for efficient pilus assembly (7–11), is positioned in this hydrophobic core to neutralize the positively charged N-terminal amino group of the neighboring pilin subunit (6, 7).
T4P assembly occurs in the inner membrane where the pilin subunits are anchored via their hydrophobic N-terminal α-helix, α1N. Subunits are thought to add to the growing pilus at its base, with the filament growing through the periplasm and across the outer membrane via the secretin channel. Pilus assembly requires a core assembly machinery composed of the major pilin subunit, a prepilin peptidase that removes the signal peptide and adds a methyl group to the N-terminal amine of residue 1 (12–14), a cytoplasmic assembly ATPase that powers the addition of each subunit to the growing pilus (15–17), an inner membrane core protein (sometimes called the platform protein) of unknown function (18–20), and an outer membrane secretin channel (21–25). This core assembly machinery is also conserved in the bacterial Type II secretion (T2S) system, which polymerizes “pseudopilin” subunits into a periplasmic “pseudopilus” that extrudes protein substrates across the outer membrane without itself forming an extracellular filament (26, 27). The genes encoding the T2S machinery, like those of the T4b pilus systems, are encoded on a single operon (28).
Most Type IVa pili, as well as the bundle-forming pili of the Type IVb class, also utilize a second “retraction” ATPase that catalyzes filament disassembly. Retraction is necessary for key T4P functions, such as twitching motility, DNA uptake, phage transduction, and bacterial dissemination (29–35). No such retraction ATPase has been identified for V. cholerae and ETEC Type IVb pili, and these pili have not been shown to mediate twitching motility or DNA uptake. The T2S systems also lack a retraction ATPase. T2S is thought to occur via a piston-like movement of the pseudopilus (36–38), a mechanism that is apparently independent of a retraction ATPase. The T4P and T2S systems are functionally related as several T4P systems have secretory functions. The V. cholerae toxin coregulated pilus (TCP) apparatus secretes a protein, TcpF, which is required for colonization of the infant mouse (39, 40), and CFA/III secretes CofJ (41).
In addition to the major pilin, which is the structural unit for the Type IV pilus filament, all Type IV pilus systems possess one or more minor pilins, which share the N-terminal α-helix with the major pilins but are expressed in much lower levels. The V. cholerae TCP and ETEC CFA/III and Longus pilus systems of the T4b class each possess a single minor pilin encoded on the pilus operon immediately following the major pilin gene. In contrast, the bundle-forming pilus T4b system has three minor pilins that are encoded at the 3′ end of the bfp operon. This arrangement is also seen for the T2S systems, which have four minor pilins (26), and for the Type IV pilus systems of some Gram-positive Clostridia species (3). The more complex T4a pilus systems also have multiple minor pilins that are typically encoded on their own gene clusters. Most minor (pseudo)pilins are similar in size to their respective major (pseudo)pilins and possess a position 5 glutamate. However, within each T4a pilus system, one of the minor pilins has a hydrophobic residue instead of glutamate at position 5, such as P. aeruginosa PilX and N. meningitidis PilK. This is also observed in the T2S system, but the minor pseudopilins lacking Glu5 are typically much larger than their respective minor and major pseudopilins and are classified as GspK family members (42). These include ETEC GspK, Klebsiella oxytoca PulK, and P. aeruginosa XcpX. The sole minor pilins of the V. cholerae and ETEC T4b pilus systems, TcpB and CofB, respectively, have Glu5 but, like the T2S GspK proteins, are substantially larger than their respective major pilins.
Minor (pseudo)pilins are involved in pilus assembly and functions, but their precise mechanisms have been challenging to identify due to their multiplicity and functional redundancy and, in the case of the T4a pili, the presence of retraction ATPases. The minor pilins of N. gonorrhoeae and P. aeruginosa are required for wild type levels of T4P assembly, but assembly can proceed at reduced levels in minor pilin mutants that also lack the retraction ATPase (43–46). In addition to their role in pilus assembly, minor pilins act in adhering to and signaling of host cells (47–49), autoaggregation (50), and swarming motility (51).
Several minor (pseudo)pilin structures have been solved, all lacking their N-terminal α1N segment (52–58). Most of these resemble the structures of their respective major (pseudo)pilins, suggesting that they can incorporate into the pilus, but some possess additional features. The N. meningitidis T4a minor pilin PilX is similar in structure to the N. gonorrhoeae major pilin, PilE, but with a 2-turn α-helix instead of a β-hairpin in the D-region (54). This feature is expected to be exposed on the pilus surface, and immunogold transmission EM demonstrated a low level of PilX incorporation into N. meningitidis Type T4a pili. PilX is involved in pilus-mediated autoaggregation and adhesion to host cells, and these functions require the 2-turn α-helix (50, 54). The P. aeruginosa minor pilins PilV, PilW, and PilX along with a non-pilin protein, PilY, are proposed to form a priming complex that is connected via minor pilins PilE and FimU to the major pilin subunits in the pilus shaft (44, 46). PilE and FimU have pilin-like structures, but FimU has a second β-sheet within the αβ-loop (46).
A ternary crystal structure of the ETEC T2S minor pseudopilins GspI, GspJ, and GspK provides critical insight into the role of these proteins in pseudopilus assembly (55). Whereas GspI is similar in structure to the major pseudopilin GspG, GspJ is larger, with two β-sheets in the globular domain, similar to P. aeruginosa FimU. GspK is the largest of the three, having a discrete pilin domain with an N-terminal α-helix and a β-sheet, but with a large α-helical domain inserted between strands β1 and β2. These GspI-GspJ-GspK minor pilins are staggered with respect to one another within the ternary complex (55), similar to the helical arrangement of the major pilins in the N. gonorrhoeae Type IVa pilus (6). This GspI-GspJ-GspK complex is predicted to cap the pseudopilus, with GspK located at the tip. Indeed, the globular domain of GspK is likely too large to fit anywhere but at the tip of the pilus and may serve as a steric block to prevent the pseudopilus from growing across the outer membrane secretin. Consistent with this tip localization, the minor pseudopilins of the K. oxytoca Pul T2S system are required for efficient pseudopilus assembly (59). The Type IVa and T2S minor (pseudo)pilins are interchangeable to some degree, because E. coli K12 minor pseudopilins initiate K. oxcytoca pseudopilus assembly but not secretion (60), and P. aeruginosa minor pseudopilins restore low levels of T4a pilus assembly in a minor pilin deletion strain when the retraction ATPase is absent (46).
The complexity of the T2S and T4a pilus systems, with multiple minor (pseudo)pilins and, in the case of the T4a pili, a retraction ATPase, make it challenging to decipher the molecular mechanism by which the minor pilins influence pilus assembly and functions. The V. cholerae and ETEC T4b pilus systems represent comparatively simple systems with only a single minor pilin and no retraction ATPase. We report here a high resolution x-ray crystal structure of N-terminally truncated CofB, the sole minor pilin from the ETEC CFA/III pilus system, and show that CofB mediates CFA/III pilus assembly.
Experimental Procedures
Bacterial Strains
Bacterial strains, plasmids, and primers are listed in Table 1. E. coli strains were grown with antibiotics appropriate for plasmid selection at the following concentrations: ampicillin (Ap), 100 μg/ml; chloramphenicol (Cm), 20 μg/ml; streptomycin, 100 μg/ml; kanamycin (Km), 45 μg/ml.
TABLE 1.
Bacterial strains, plasmids, and primers
| Reagent | Description or nucleotide sequence | Source/Reference |
|---|---|---|
| Strains | ||
| ETEC 31-10 | Wild type ETEC LT, CFA/III | Ref. 92 |
| ETEC 31-10P | CFA/III-negative plasmidless derivative of strain 31-10; LT | Ref. 81 |
| ETEC E9034A | Wild type ETEC LT, ST, CFA/III, CS3 | Ref. 93 |
| ETEC E9034A ΔlngA | ΔlngA | This study |
| ETEC E9034A ΔlngB | ΔlngB | This study |
| E. coli MC4100 | [araD139]B/r Δ(argF-lac)169 λ− e14- flhD5301 Δ(fruK-yeiR)725(fruA25) relA1 rpsL150(StrR) rbsR22 Δ(fimB-fimE)632(::IS1) deoC1 | Coli Genetic Stock Center |
| E. coli MC4100 pcof | pcof | This study |
| E. coli MC4100 pcofΔcofA | pcof ΔcofA | This study |
| E. coli MC4100 pcofΔcofB | pcof ΔcofB | This study |
| E. coli DH5α | fhuA2 lac(del)U169 phoA glnV44 Φ80′ lacZ(del)M15 gyrA96 recA1 relA1 endA1 thi-1 hsdR17 | Laboratory collection |
| E. coli DH5α pcof | pcof | Ref. 41 |
| E. coli SW105 | DH10B[λcl857(cro-bioA)<>araC-PBADflpe] galK | NCI, National Institutes of Health |
| E. coli Origami DE3 | Δ(ara-leu)7697 ΔlacX74 ΔphoA PvuII phoR araD139 ahpC galE galK rpsL F′[lac+ lacI q pro] (DE3) gor522::Tn10 trxB (KanR, StrR, TetR)4 | Novagen |
| Plasmids | ||
| pACYC184 | p15A, CmR and TcR | ATCC |
| pcof | pACYC184 with cof operon cloned in TcR gene, CmR | Ref. 41 |
| pcofΔcofA | pcof plasmid with cofA gene disrupted | Ref. 41 |
| pcofΔcofB | pcof plasmid with cofB gene disrupted | This study |
| pJMA10.1 | pBAD22 derivative; araC replaced with PrhaB NcoI removed; bla, ApR | R. K. Taylor, J. Marles (Geisel School of Medicine) and this study |
| pcofA | pJMA10.1 containing the cofA gene | This study |
| pcofB | pJMA10.1 containing the cofB gene | This study |
| pcofB259 | pJMA10.1 containing the cofB gene fragment encoding residues up to 259 | This study |
| plngA | pJMA10.1 containing the lngA gene | This study |
| plngB | pJMA10.1 containing the lngB gene | This study |
| pKD46 | repA 101(ts), contains RED recombinase genes γ, exo, β, ApR | M.S. Donnenberg (University of Maryland) |
| pKD4 | Ori6Kγ, contains kan cassette, ApR | M.S. Donnenberg (University of Maryland) |
| pKD3 | Ori6Kγ, contains cat cassette, ApR | M.S. Donnenberg (University of Maryland) |
| pCP20 | Contains FLP recombinase gene, CmR and ApR, temperature sensitive replication. | M.S. Donnenberg (University of Maryland) |
| pet15b | T7 promoter, N-terminal His tag, isopropyl-β-d-1-thiogalactopyranoside-inducible, ApR | Novagen |
| pet15b-cofB | pet15b containing the cofB gene fragment encoding residues 25–518 | This study |
| Primers | ||
| CofB-RED-For | GCAAGTATTGCATAAGTACGGGAACTCTGTGTATGTGTAGGCTGGAGCTGCTTC | This study |
| CofB-RED-Rev | CATATGAATATCCTCCTTAGACAATCCTGCACATTAAGTTCAATACGTC | This study |
| CofB-RED-H1P1 | GTATCTAATGCTCTGATTTCAGAAATCGCCGGTGTAGGCTGGAGCTGCTTC | This study |
| CofB-RED-H2P2 | AACCTGAATGGCTGTACCAGATAAATATCCTCATATGAATATCCTCCTTAG | This study |
| CofA-F-KpnI | CGGGGTACCATGCTTTCGGTTTATAACAGAAC | This study |
| CofA-R-HindIII | CCCAAGCTTTTAACGGCTCGCCAAAG | This study |
| CofB-F-KpnI | CCGGGGTACCATGAATATGAGGGGTTTCACG | This study |
| CofB-R-XbaI | ATATATTCTAGATTAGGTTTGTGGTTCTGTACTGCACC | This study |
| CofB259-R-BamHI | CCGCGGATCCTTATTTTCCAAGATTAGGATCGATAC | This study |
| LngA-RED-For | ATGCTATCCGTGTATAACCGG | This study |
| LngA-RED-Rev | TTAACGGCTACCTAAAGTAATTG | This study |
| LngA-RED-H1P1 | ATCGGTACGATTGCAGCCGGTGTCGTGATTCTGGCTCAGCGTGTGTAGGCTGGA GCTGCTTC | This study |
| LngA-RED-H2P2 | AACTTGCCCAAGAATAGAACGGCACTGCTCTTGGCTTAATCCATGGGAATTAGCCATGGTCC | This study |
| LngB-RED-For | ATGAAAATGAGAGGCTTCACAC | This study |
| LngB-RED-Rev | TTAGGTTTGTGGTTCTGTACTG | This study |
| LngB-RED-H1P1 | AACAACAAAGAAAAAGAAATTACAAATCCACTTTATGATCAGGTGTAGGCTGGAGCTGCTTC | This study |
| LngB-RED-H2P2 | GACAATATCACTGTCAGTGCTGTTTCCAAAAGAAGCATATGTATGGGAATTAGCCATGGTCC | This study |
| LngA-F-KpnI | GGGGGGTACCATGCTATCCGTGTATAACCGG | This study |
| LngA-R-HindIII | CCCCAAGCTTTTAACGGCTACCTAAAGTAATTG | This study |
| LngB-F-KpnI | GGGGGGTACCATGAAAATGAGAGGCTTCACAC | This study |
| LngB-R-HindIII | CCCCAAGCTTTTAGGTTTGTGGTTCTGTACTG | This study |
| Ec-cofB-fpcr | GGAATTCCATATGTATAAAGAGAAAGAAGCAGATGAAGCCAGA | This study |
| Ec-cofB-rpcr | CGCGGATCCTTAGGTTTGTGGTTCTGTACTGCACCATG | This study |
| Ec-cofb-l40m-i41m-fpcr | CAAATTGTATCTAATGCTATGATGTCAGAAATCGCCGGCATT | This study |
| Ec-cofb-l40m-i41m-rpcr | AATGCCGGCGATTTCTGACATCATAGCATTAGATACAATTTG | This study |
Expression of pcof in E. coli MC4100
The vector pcof contains the entire ETEC cof operon inserted into the cloning vector pACYC184 (ATCC) between restriction enzyme sites EcoNI and EagI-HF (41). pcof was transformed into electrocompetent E. coli MC4100 cells, and cells containing this plasmid were selected with Cm. To induce CFA/III pilus expression, cells were grown overnight at 37 °C on CFA (1% casamino acids, 0.15% yeast extract, pH 7.4) agar-Cm plates (61).
Construction of pcofA, pcofB, pcofB259, plngA, and plngB
Vectors expressing the major pilin CofA, the minor pilin CofB, and the truncated minor pilin CofB259 were derived from a plasmid with a pBAD22 backbone in which the arabinose-inducible araC promoter had been replaced with the rhamnose-inducible promoter PrhaB (pJMA10.1). pJMA10.1 contains an ApR marker. Genes cofA, cofB, and cofB(1–259) were PCR-amplified from ETEC 31-10 genomic DNA with the Q5 DNA polymerase (New England Biolabs) using primers CofA-F-KpnI/-R-HindIII, CofB-F-KpnI/-R-XbaI, and CofB-F-KpnI/CofB259-R-BamHI, respectively. PCR products were purified, digested with KpnI and XbaI or BamHI, and ligated into pJMA10.1 at the corresponding restriction sites using T4 DNA ligase (New England Biolabs). All constructs were verified by DNA sequencing (GENEWIZ). Plasmids were transformed into MC4100-pcof and -pcof deletion strains, and cells were grown on LB-Cm/Ap plates. Genes for lngA and lngB were PCR-amplified from ETEC E9034A genomic DNA using primers LngA-F-KpnI/-R-HindIII and LngB-F-KpnI/-R-HindIII, respectively. PCR products were purified, digested with KpnI and HindIII, and ligated into pJMA10.1 at the KpnI/HindIII restriction sites using T4 DNA ligase. All constructs were verified by DNA sequencing. Plasmids were transformed into ETEC E9034A-ΔlngA and -ΔlngB deletion strains, and cells were grown on LB-Ap plates.
Deletion of Minor Pilin Genes
The cofB gene was disrupted in pcof using the λ-RED recombinase system (62) used previously to generate the pcofΔcofA construct (41). The central portion of the cofB gene (residues 45–462) was targeted for deletion using the CofB-RED-For/-Rev primers to PCR-amplify the kan cassette in plasmid pKD4 using Q5 DNA polymerase. The amplified kan cassette is flanked by a short stretch of nucleotides that correspond to the sequence flanking the targeted deletion site. PCR products were purified using the QIAquick PCR purification kit (Qiagen). E. coli DH5α-pcof cells were transformed with pKD46 (ApR) encoding the λ-RED recombinase. Electrocompetent DH5α-pcof-pKD46 cells were prepared and transformed with the kan cassette PCR product. Cells with successful integration of the kan cassette were selected by plating on LB-Cm-Km and incubated at 30 °C and further screened for sensitivity to ampicillin corresponding to the loss of pKD46. ApS, KmR, and CmR colonies were screened by PCR to confirm insertion of the kan cassette, and pcof-cofB::Km was purified by plasmid miniprep (Qiagen). This plasmid was transformed into competent E. coli SW105 cells expressing the FLP recombinase, which recognizes and cleaves the FRT sequence flanking the kan cassette. Expression of the FLP recombinase was induced with arabinose, and colonies were screened on agar plates with and without Km for loss of the kan cassette. KmS clones were screened for deletion of the cofB gene fragment by PCR using primers CofB-F-KpnI/-R-XbaI for the 5′ and 3′ ends of cofB and confirmed by DNA sequencing. A positive pcofΔcofB plasmid was amplified in E. coli DH5α, purified, and transformed into electrocompetent E. coli MC4100 cells.
lngA and lngB genes were disrupted in ETEC E9034A using the λ-RED recombinase system (62, 63). The CmR-encoding cat cassette was PCR-amplified from pKD3 with primer sets LngA-RED-For/-Rev and LngB-RED-For/-Rev to replace the lngA gene fragment (encoding residues 57–155) and the lngB gene fragment (encoding residues 80–381), respectively. The cassettes were transformed into electrocompetent ETEC E9034A-pKD46 cells expressing λ-RED recombinase, and clones were screened for resistance to Cm and sensitivity to Ap. Positive clones were made electrocompetent and transformed with pCP20, which encodes the genes for the FLP recombinase. After excision of the cat cassette by FLP recombinase, positive clones were screened for sensitivity to Cm and PCR-amplified with LngA-For-KpnI/-Rev-HindIII or LngB-For-KpnI/-Rev-HindIII to confirm gene disruption. The gene deletions were further confirmed by DNA sequencing.
Assessing CofB Expression and CFA/III Assembly in ETEC and MC4100-pcof Strains
ETEC and E. coli MC4100 pcof cells were grown overnight under CFA/III-inducing conditions on CFA agar plates. Cells were overlaid with 5 ml of phosphate-buffered saline (PBS; 10 mm Na2HPO4, 2 mm KH2PO4, 137 mm NaCl, 2.7 mm KCl) with gentle agitation on a rocking unit for 15 min. Cells were gently washed off the plates, and A600 was measured. Cell suspensions were normalized to an A600 of 0.1; this mixture constitutes the whole cell culture (WCC) fraction used to assess total protein levels. Cells were removed by centrifugation at 3000 × g for 10 min, and the supernatant was further filtered through a 0.22-μm syringe drive filter. This filtered supernatant (Sup) fraction was used to assess CofA in the shed CFA/III pili. Samples were mixed with Laemmli sample buffer (60 mm Tris, pH 6.8, 5% 2-mercaptoethanol, 2% SDS, 10% glycerol, 0.02% bromphenol blue) and boiled for 10 min prior to being loaded onto 15% SDS-polyacrylamide gels. Proteins were transferred onto polyvinylidene difluoride (PVDF) membrane (Bio-Rad) for immunoblotting and detected by rabbit polyclonal antisera raised against N-terminally truncated CofA (64) or CofB (N-terminal peptide 61–74 and C-terminal peptide 403–419; Pacific Immunology). Goat anti-rabbit secondary antibodies conjugated to horseradish peroxidase (Jackson ImmunoResearch) were used to detect primary antibody. Immunoblots were visualized by enhanced chemiluminescence (ECL) with the SuperSignal West Pico chemiluminescent substrate (Thermo Scientific) for CofA or the SuperSignal West Femto chemiluminescent substrate (Thermo Scientific) for CofB. A Fujifilm LAS4000 imager (FujiFilm) was used to capture images of the immunoblots.
Assessing LngB Expression and Longus Assembly in ETEC E9034A
ETEC E9034A cells were grown overnight on LB agar plates at 37 °C. Single colonies were inoculated in 2 ml of Terrific Broth (1.2% tryptone, 2.4% yeast extract, 1.7 mm KH2PO4, 7.2 mm K2HPO4, pH 7.2) and grown for 4 h in an upright position on a shaking incubator at 250 rpm and 37 °C. Cells were inoculated 1:100 (v/v) in 2 ml of Terrific Broth and grown for a further 2 h. The WCC fraction was used to assess total protein levels. Cells were removed by centrifugation at 3000 × g for 10 min, and the Sup was used to assess LngA within the shed Longus pili. Samples were mixed with Laemmli sample buffer and boiled for 10 min prior to being loaded onto 15% SDS-polyacrylamide gels. Proteins were transferred onto PVDF membrane (Bio-Rad) for immunoblotting. Due to their close sequence similarity, LngA and LngB could be detected using the anti-CofA and -CofB (peptide 61–74) antibodies, respectively. Blots were developed as described for CofA and CofB.
Cloning, Expression, and Purification of N-terminally Truncated CofB
The gene fragment encoding CofB residues 25–518 was PCR-amplified from ETEC 31-10 genomic DNA using primers Ec-cofB-fpcr and Ec-cofB-rpcr and cloned into expression vector pET-15b (Novagen) at the NdeI and BamHI sites to provide an N-terminal His6 tag for purification by nickel-Sepharose column chromatography (GE Healthcare). CofB was expressed in E. coli Origami(DE3) cells (Novagen). Cells were grown to an A600 of 0.1–0.2 in LB-Ap broth at 37 °C. CofB expression was induced with 0.2 mm isopropyl-β-d-1-thiogalactopyranoside, and cells were grown for a further 20 h at 14 °C. Cells were pelleted by centrifugation for 30 min at 5000 × g at 4 °C and subjected to two cycles of flash-freezing in liquid nitrogen followed by thawing in a water bath to partially lyse the cells. Cells were resuspended in lysis buffer containing 50 mm NaH2PO4/Na2HPO4 (pH 7.4), 100 mm NaCl, and EDTA-free protease inhibitor mixture (Roche Applied Science). The suspension was incubated at room temperature with lysozyme for 1 h, gently stirring, and then cells were lysed by sonication. Cell debris was removed by centrifugation for 60 min at 40,000 × g at 4 °C. The supernatant was filtered using a 0.4-μm filter and then loaded onto a HisTrap column (GE Healthcare) pre-equilibrated with buffer A (50 mm NaH2PO4/Na2HPO4, pH 7.4, 30 mm imidazole, pH 7.4, 500 mm NaCl). The column was washed with buffer A, and CofB was eluted with buffer B (20 mm Tris-HCl, pH 7.4, 100 mm NaCl, 250 mm imidazole). Fractions containing CofB were pooled and dialyzed against buffer C (20 mm Tris-HCl, pH 7.4, 100 mm NaCl, 1 mm EDTA, 1 mm EGTA). The His6 tag was removed by thrombin cleavage, and CofB was further purified on a Sephacryl S-100 HR size exclusion column (GE Healthcare) pre-equilibrated with buffer C. Fractions were shown by SDS-PAGE to be greater than 95% pure, and peak fractions were combined and concentrated to 15 mg/ml using an Amicon stirred cell concentrator with a 10,000-Da molecular mass cut-off filter (Millipore). Protein was flash-frozen in liquid nitrogen and stored at −80 °C.
For selenomethionine (SeMet)-substituted CofB (residues 25–518), Leu40 and Ile41 were changed to Met by site-directed mutagenesis on pET-15b-pcofB using primers Ec-cofb-l40m-i41m-fpcr and Ec-cofb-l40m-i41m-rpcr. SeMet-CofB was expressed in E. coli Origami(DE3) cells (Novagen) by inhibition of the methionine biosynthesis pathway (65). Briefly, cells were grown at 37 °C in M9 minimal medium supplemented with ampicillin, glucose, MgSO4, CaCl2, thiamine, and all naturally occurring amino acids except for Gly, Ala, Pro, Asn, Cys, and Met. When the A600 reached 0.1–0.2, amino acids Thr, Leu, Phe, Leu, Ile, Val, and SeMet were added. After incubating the cell culture for a further 15 min, the temperature was reduced to 14 °C, and isopropyl-β-d-1-thiogalactopyranoside was added to 0.2 mm to induce expression of SeMet-CofB, which was purified using the same procedure as for native CofB.
Crystallization of CofB
Initial CofB crystallization conditions were obtained from the high throughput screening laboratory at the Hauptman-Woodward Medical Research Institute (66). Both native and SeMet CofB crystals were grown by hanging drop vapor diffusion at 20 °C. Native CofB crystals were grown in the presence of 100 mm MES, pH 5.6, and 1.6 m ammonium sulfate. SeMet CofB crystals were obtained in 100 mm HEPES, pH 7.5, 700 mm NaH2PO4, 800 mm KH2PO4. All crystals were cryocooled in mother liquor containing 30% glycerol and stored in liquid nitrogen for x-ray diffraction data collection at the Stanford Synchrotron Radiation Lightsource (SSRL).
Collection and Processing of CofB X-ray Diffraction Data
A diffraction data set for a native CofB crystal was collected on Beamline 14-1 at 100 K. The crystal belongs to the rhombohedral system, as determined by Web-Ice (67). The raw data set was processed using XDS (94). The space group was determined by POINTLESS in the CCP4 suite (68), and the data set was scaled to 2 Å resolution by AIMLESS (68). SeMet CofB crystals were tested on SSRL Beamline 7-1, and two-wavelength multiple anomalous diffraction (MAD) data were collected after deciding on wavelengths, inflection point, and high energy remote, based on the x-ray fluorescence scan output. The MAD data set was processed by iMOSFLM (69) and scaled by SCALA (70) in the CCP4 suite (68).
Structure Determination of CofB
The SeMet-CofB structure was solved by MAD phasing using SOLVE/RESOLVE (71, 72). SOLVE located seven selenium atoms and determined the initial phases of the structure. Phases were improved by density modification procedures by RESOLVE. This 3 Å SeMet-CofB structure was used as a model to solve the 2 Å native CofB structure by molecular replacement. The initial model was built by ARP/wARP (73), examined in COOT (74), and further refined using REFMAC5 (75). COOT was used to locate water oxygens, glycerol, and sulfate ions from the difference map as well as the composite annealed omit map, calculated by CNS (76, 77). Final TLS and restrained refinement of the structure with water oxygens and ligands brought Rcryst and Rfree values to 20.5 and 22.9%, respectively. The final structure validation was performed using COOT (74) and MolProbity (78).
Results
The ETEC Minor Pilin CofB Is Required for CFA/III Assembly
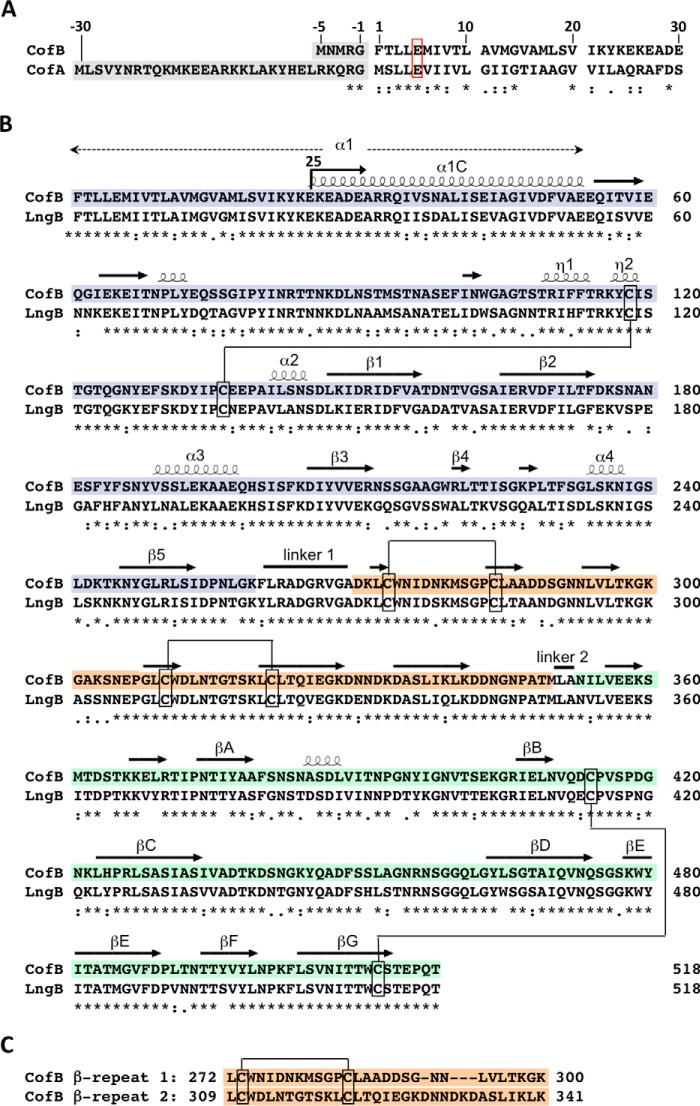
The ETEC minor pilin CofB is encoded on the cof operon immediately downstream of the gene encoding the major pilin, CofA, the structural subunit for the CFA/III Type IVb pilus (64, 79, 80). CofB is a 518-amino acid protein with a predicted 5-amino acid signal peptide (Fig. 1, A and B). The CofB signal peptide is much shorter than the 30-amino acid signal peptide of CofA, which is probably processed by the prepilin peptidase CofP encoded on the cof operon. The N-terminal 26-amino acid region of the mature CofB protein shares sequence similarity with CofA (61%), including the conserved Glu5 (Fig. 1A). This region corresponds to α1N, the inner membrane anchor and polymerization domain of the major pilins. Beyond the N terminus, CofB has no sequence homology with CofA. CofB is more than twice the length of the 208-amino acid CofA. CofB is, however, identical in length and highly similar in sequence to LngB, the minor pilin for the ETEC Longus pilus, with 78% sequence identity between the two proteins, including 8 cysteines (Fig. 1B). Both proteins have a ∼30-residue tandem repeat (Fig. 1C).
FIGURE 1.
CofB amino acid sequence and alignment with CofA and LngB. A, alignment of the signal peptides (shaded gray) and N-terminal 30 residues of the minor pilin CofB and the major pilin CofA from the ETEC CFA/III pilus system. B, sequence alignment of CofB (GenBankTM accession number BAB62898) with the minor pilin LngB (ABU50041) from the ETEC Longus pilus system. Residues 25–518 were expressed recombinantly for crystallization, as indicated by the arrow at residue 25. The secondary structure elements are indicated above the CofB sequence, based on its crystal structure, shown in Fig. 4. The pilin domain is shown with a blue background, the β-repeat domain is orange, and the β-sandwich domain is green. The disulfide bond connectivity is indicated. C, alignment of the CofB tandem repeats 1 and 2, which form the β-repeat domain.
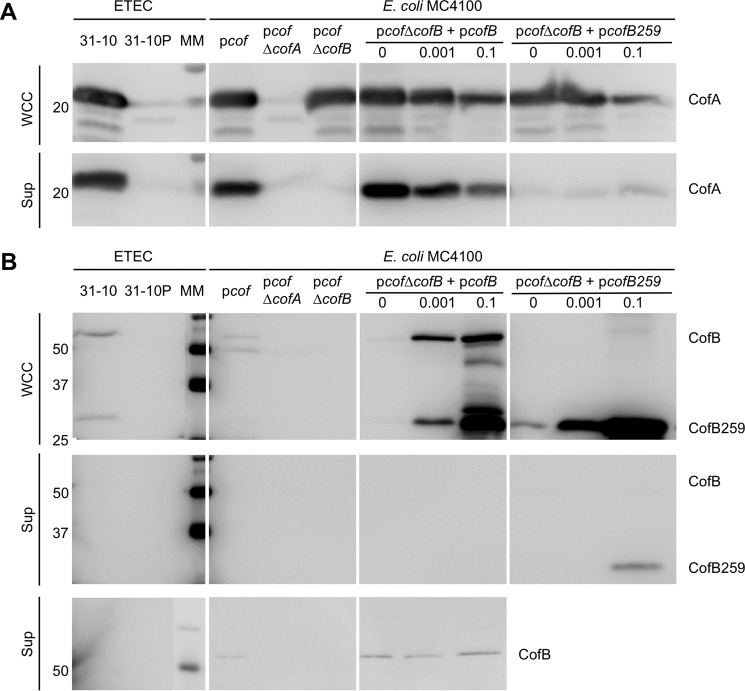
The role of CofB in CFA/III assembly was examined using a heterologous E. coli CFA/III expression system, which allows manipulation of genes within the cof operon. In ETEC strain 31–10, the cof operon is located on a 55-kb virulence plasmid (81) that is unstable and not amenable to genetic manipulation (41). The cof operon was cloned into plasmid pACYC184, producing pcof, which was transformed into several E. coli expression strains (DH5α, HB101, and MC4100 (41)). These strains express CFA/III and secrete the soluble protein CofJ, also encoded on the cof operon, in a pilus-dependent manner. To test the role of CofB in CFA/III pilus assembly, the cofB gene in pcof was deleted using the λ-RED recombinase method (62), and pcofΔcofB was transformed into E. coli MC4100. Cells were grown on CFA agar plates (61) and then washed off of the plates with PBS, and their numbers were normalized. This mixture, referred to as the WCC fraction, was analyzed by SDS-PAGE and immunoblotting using an anti-CofA antibody (64) to determine total CofA levels relative to the “wild type” MC4100-pcof strain. Additional controls included this strain lacking the major pilin, CofA (MC4100-pcofΔcofA), wild type ETEC 31-10 (82), and ETEC 31-10P, which lacks the 55-kb virulence plasmid (81). To determine CFA/III pilus assembly levels, cells were removed from the WCC by centrifugation and filtration, and the amount of CofA in the Sup fraction was analyzed by SDS-PAGE and immunoblotting. Total CofA levels in the WCC fraction of MC4100-pcofΔcofB were comparable with those of the positive controls: ETEC strain 31-10 and MC4100-pcof (Fig. 2A). As expected, no CofA was present in negative control strains ETEC 31-10P and MC4100-pcofΔcofA. However, whereas CofA levels in the Sup fraction, representing CFA/III pili, were comparable for ETEC 31-10 and MC4100-pcof, no CofA was present in this fraction for MC4100-pcofΔcofB, suggesting that CofB is required for pilus assembly.
FIGURE 2.
Immunoblots of ETEC CFA/III proteins in whole cell culture and supernatant fractions. The indicated strains were grown on CFA plates, harvested and resuspended, and aliquots of this WCC fraction and the Sup fraction, in which cells were removed by centrifugation and filtration, were analyzed by SDS-PAGE and immunoblotting. The WCC fraction represents the total protein, and the Sup fraction contains CFA/III pili that have been shed from the cells. Blots were probed with anti-CofA (A) and anti-CofB (peptide 61–74) (two top panels) and anti-CofB (peptide 403–419) (bottom panel) (B). Expression of CofB from pcofB and CofB259 from pcofB259 was induced with 0, 0.001, or 0.1% rhamnose (w/v) as indicated. Molecular masses of markers (MM) are indicated on the left along with protein bands. Each panel represents a single blot, allowing comparison of protein levels across the panel.
To confirm that the loss of CFA/III in MC4100-pcof-ΔcofB is due to disruption of the cofB gene and not a downstream effect on the cof operon, cofB was cloned into expression vector pJMA10.1. pcofB was transformed into MC4100-pcofΔcofB. Pilus assembly was rescued to approximately wild type levels when no rhamnose was added, presumably due to low level CofB expression from a leaky promoter (Fig. 2A). However, induction with rhamnose, even at very low levels (0.001%), resulted in reduced levels of CofA in both the WCC and Sup fractions, indicating that pilus assembly is optimal with very low levels of CofB expression. This observation cannot be explained by competition between CofA and CofB for the signal peptidase because we saw no accumulation of unprocessed CofA under these conditions. Instead, the reduced CofA levels observed upon overexpression of CofB suggest a feedback mechanism that limits the total amount of pilin in the inner membrane.
To verify CofB expression and test its cellular localization, WCC and Sup fractions were blotted with antibody against an N-terminal CofB peptide (residues 61–74). CofB was not detected in ETEC 31-10 or MC4100-pcof WCC using the SuperSignal West Pico chemiluminescent substrate (Thermo Scientific) used for detection of the major pilin, CofA, but a faint band at ∼57 kDa, corresponding to CofB, was observed for ETEC 31-10 and MC4100-pcof WCC when the more sensitive Femto substrate was used, consistent with CofB being expressed at very low levels in the wild type strains (Fig. 2B, top). As expected, the CofB band was absent in MC4100-pcofΔcofB WCC, but CofB was detected in uninduced MC4100-pcofΔcofB+pcofB WCC, confirming that it is produced under these conditions and at a level that is sufficient for pilus assembly (Fig. 2A). No CofB was detected in the Sup fraction using the N-terminal antibody against CofB peptide 61–74 (Fig. 2B, middle), but an antibody against a C-terminal peptide (residues 403–419) detected CofB in the Sup fraction of MC4100-pcof and MC4100-pcof-ΔcofB+pcofB samples using the Femto substrate kit with a long (60-s) exposure time (Fig. 2B, bottom), suggesting that CofB is incorporated into the pili but at very low levels.
Significant proteolysis of CofB was observed in the WCC when the minor pilin was expressed at high levels (Fig. 2B, top). The stable proteolytic fragments are N-terminal because they are detected by antibodies specific for the N-terminal peptide. The most abundant fragment has a mass of ∼29 kDa, which corresponds to a CofB fragment spanning residues 1 to ∼260. This fragment is also present in ETEC 31-10 WCC.
The ETEC Minor Pilin LngB Is Required for Pilus Assembly
To examine the role of the ETEC minor pilins in a native T4b pilus system, we turned to ETEC strain E9034A, which produces the Longus T4b pilus (83, 84). We deleted the minor pilin gene lngB from the E9034A chromosome using the λ-RED recombinase system (62). As with the MC4100-pcofΔcofB strain, pilus assembly was abrogated in ETEC E9034A-ΔlngB, comparable with that of the ΔlngA mutant, and assembly was rescued when LngB was expressed ectopically (Fig. 3A). LngB expression from plngB in ETEC E9034A-ΔlngB does not appear to be as high as CofB expression from pcofB in MC4100-pcofΔcofB and requires a higher level of induction (0.001–0.1% rhamnose) to restore wild type Longus levels. Consistent with lower LngB levels, no inhibitory effect on LngA expression/stability or Longus assembly was observed when LngB was overexpressed, in contrast to the CFA/III system.
FIGURE 3.
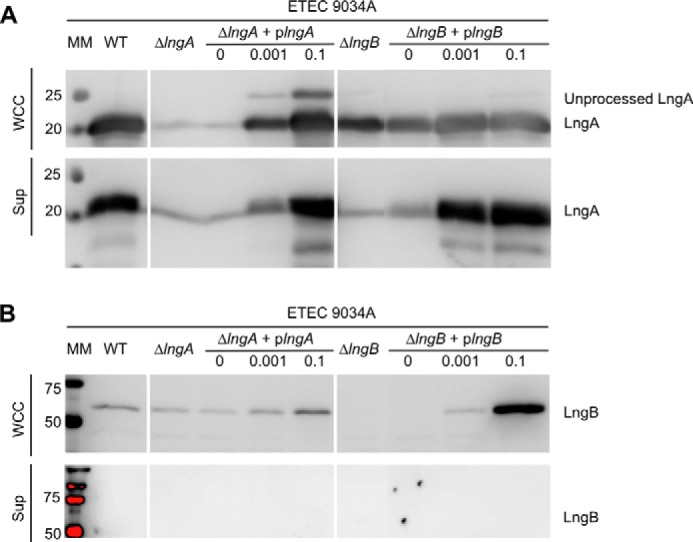
The ETEC minor pilin LngB is required for Longus pilus assembly. The indicated strains were grown in Terrific Broth. WCC and culture Sup were analyzed by SDS-PAGE and immunoblotting. The WCC fraction represents the total protein, and the Sup fraction contains Longus pili that have been sheared off or have naturally shed from the cells. Blots were probed with anti-CofA (A) and anti-CofB antibodies (peptide 61–74) (B), which cross-react with LngA and LngB, respectively. Expression of LngB from plngB was induced with 0, 0.001, or 0.1% rhamnose (w/v), as indicated. Molecular masses of markers (MM) are indicated on the left along with protein bands. Each panel represents a single blot, allowing comparison of protein levels across the panel.
We were unable to detect LngB in the pilus (Sup) fraction even when LngB was overexpressed, and the ultrasensitive Femto chemiluminescent substrate was used to develop the blot (Fig. 3B). Given that our results with the CFA/III pilus system showed that the pilus assembly was most efficient when only low levels of CofB were produced, we tested whether increasing the level of the major pilin, LngA, might disrupt pilus assembly by altering the major/minor pilin stoichiometry. However, we found that overexpressing LngA in ETEC E9034A-ΔlngA had no effect on pilus assembly, probably because the excess LngA was not processed by the prepilin peptidase (Fig. 3A). These results parallel those shown for the MC4100 heterologous CFA/III expression system and confirm that the ETEC minor pilins are necessary for T4b pilus assembly.
Crystal Structure of ETEC CofB
To understand how CofB might initiate pilus assembly, we solved its x-ray crystal structure. We expressed a recombinant form of CofB (CofB(25–518)) lacking its hydrophobic N-terminal 24 residues corresponding to the protruding half of α1, α1N, of the major pilins (4, 6, 85, 86). Both native and SeMet-CofB proteins were expressed and purified and crystals were grown. A 3 Å resolution x-ray crystal structure was solved for SeMet-CofB by MAD methods, and this structure was used as a model for molecular replacement to solve a 2 Å structure of the native N-terminally truncated CofB. Data collection and refinement statistics are shown in Table 2. Crystallization of CofB has also been reported elsewhere (87). In that study, CofB (residues 29–518) was expressed using a different plasmid (pTT240), purified using an additional anion exchange step, and crystallized under different conditions (sodium formate, sodium acetate), but the space group and unit cell dimensions and resolution are essentially the same as those reported here.
TABLE 2.
CofB data collection and refinement statistics
1 Values in parenthesis represent the highest resolution shell.
2 Rp.i.m. = Σhkl{1/[N(hkl) − 1]}½ Σi |Ii(hkl) − I(hkl)|/Σhkl ΣiIi(hkl).
3 Rcryst = Σhkl |Fobs − Fcalc|/ΣhklFobs.
4 Rfree is the cross-validation R factor for 5% of the reflections against which the model was not refined.
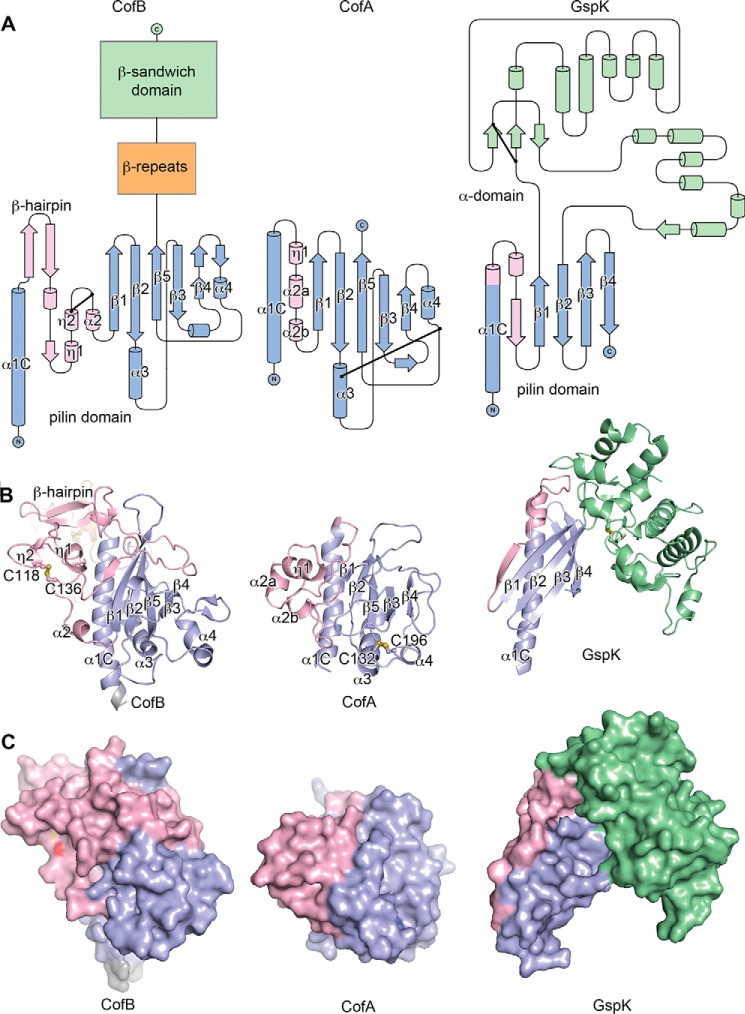
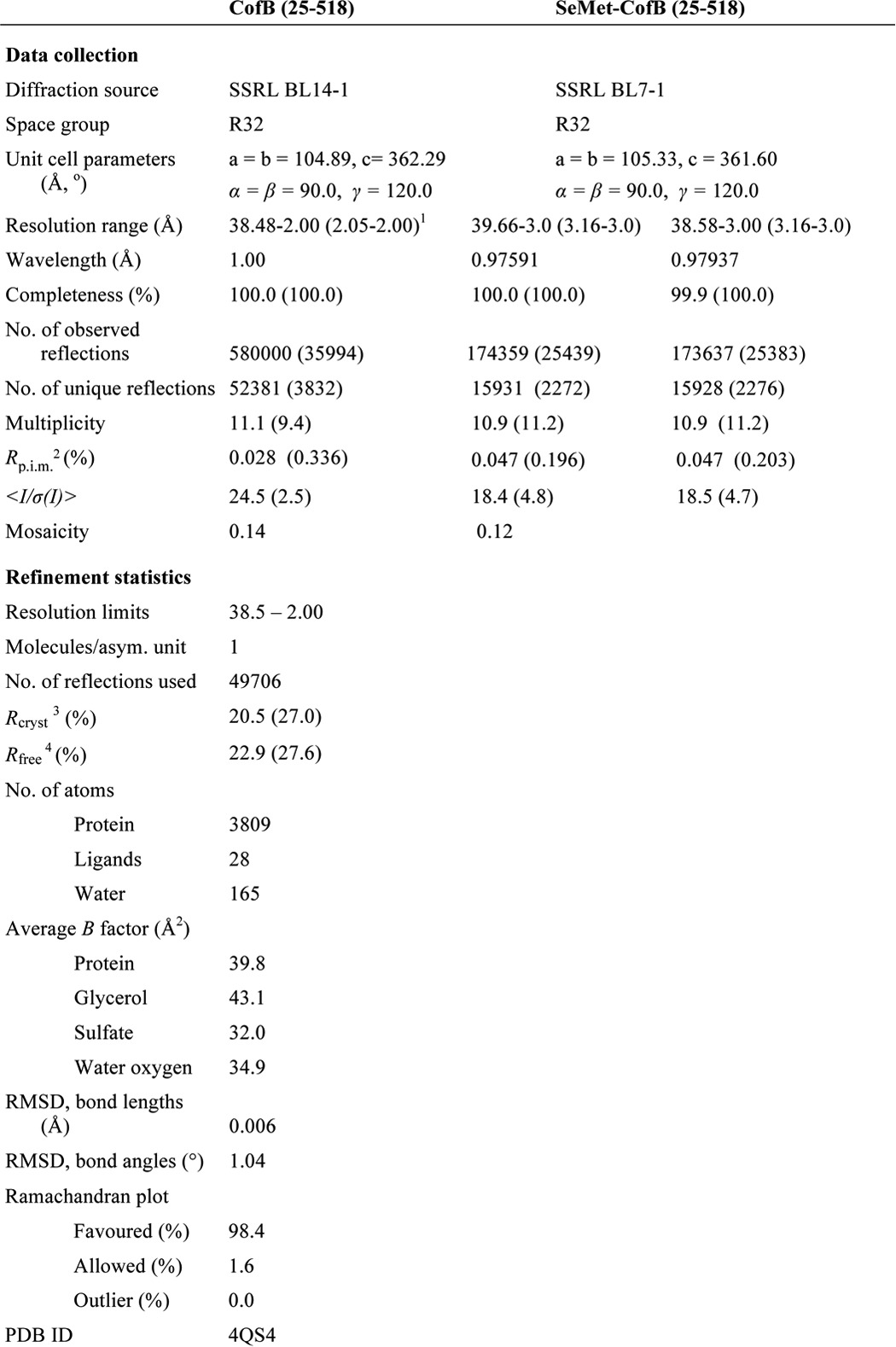
CofB has an unusual structure for a minor pilin; the N-terminal half of the protein, residues 25–259, is a pilin domain, and the C-terminal half, residues 260–518, is extended with two discrete domains: a central β-repeat domain (so-named because it has two small β-sheets, each corresponding to one of the tandem repeat sequences (Fig. 1C)) and a C-terminal elongated β-sandwich domain (Fig. 4). Linker segments connect the pilin domain to the first β-repeat and the second β-repeat to the β-sandwich domain. The C-terminal region extends from the “inside” face of the pilin domain that in the major pilins faces into the core of the assembled pilus. The pilin domain has the canonical T4b pilin fold seen in the major pilins, CofA (64) and TcpA (88, 89), with an N-terminal α-helical backbone, α1C, and a 5-stranded anti-parallel β-sheet, in which strand β5 forms the central β-strand. This β-sheet lies against α1C and two shorter α-helices, α3 and α4, all of which are located on the inside face of the pilin domain (64). α3 is situated between strands β2 and β3 and α4 is between β4 and β5.
FIGURE 4.
X-ray crystal structure of ETEC CofB. A, schematic representation of the CofB structure, residues 25–518. The pilin domain is colored blue with the its αβ-loop in pink, the central β-repeat domain in orange, the C-terminal β-sandwich domain in green (proximal β-sheet is light green, distal β-sheet is dark green), and linkers in gray. The eight cysteines participating in disulfide bonds are shown in a ball-and-stick representation with sulfurs colored yellow (Cys273–Cys284 in the first β-sheet and Cys310–Cys321 in the second sheet of the β-repeat, Cys414–Cys512 in the β-sandwich domain, and Cys118–Cys136 in the pilin domain are not visible). B, topology diagram of CofB, colored as in A. Disulfide bonds are indicated by a line between two dots. C, surface representation of CofB, shown in the same orientation and color scheme as in A. Cysteine sulfurs are colored yellow, nitrogens are blue, and oxygens are red.
The CofB pilin domain, which terminates at β5, is connected to the β-repeat domain via linker 1, a 10-amino acid segment that extends from the inside face of the pilin domain adjacent to the C-terminal end of α1C (Fig. 4, A and B). The β-repeat domain has two three-stranded anti-parallel β-sheets whose planes lie at ∼90° to each other and are connected by a tight lysine-rich 9-residue loop. Each β-sheet has a disulfide bond connecting the end of its first strand and the beginning of the second strand (Cys273–Cys284, Cys310–Cys321). A 13-residue loop connects the terminal β-strand of the second β-repeat with the first β-strand of a β-hairpin in the C-terminal β-sandwich domain, but the linker between these domains, linker 2, is defined by only 2 amino acids, Leu351 and Ala352, because all other residues in this loop are tightly associated with their respective domains. The short β-hairpin forms the proximal end of the β-sandwich domain, which has a twisted 6-stranded anti-parallel β-sandwich. The strands in the β-sandwich are labeled sequentially in Fig. 4, A and B, with βA-βG-βC forming the inner β-sheet, which is proximal to the β-repeats, and strand βF/B-βE-βD forming the outer or distal β-sheet. Large intersheet loops between strands βA and βB and strands βC and βD are splayed on either side at the distal end of the β-sandwich. Another loop between strands βB and βC has a disulfide bond to the end of βG near the C terminus of the protein (Cys414–Cys512). The pilin domain, β-repeat, β-sandwich domains, and linkers together measure 130 Å in length, whereas the pilin domain itself is 60 Å along its long axis parallel to α1C. Apart from the linkers, no interactions connect the three domains (Fig. 4C), implying conformational flexibility.
Comparison of CofB with CofA and GspK
The pilin domain of CofB shares the same core structure as the major pilin, CofA, with the α1C backbone, antiparallel β-sheet with the central β5 strand, and helices α3 and α4 (root mean square deviation for CofA and CofB(25–259) backbone atoms, 4.4 Å) (Fig. 5, A and B). However, in CofB, the αβ-loop that connects α1C to the β-sheet is longer (93 residues) and bulkier than in CofA. From α1C of CofB, a β-hairpin extends away from the top of the pilin domain, followed by a meandering loop that crosses the top front of this domain and then winds back to form three sequential single-turn helices, η1, η2, and α2. A disulfide bond between Cys118 and Cys136 stabilizes this helical cluster. The αβ-loop of CofA has only 50 residues and lacks the β-hairpin and meandering loop that crosses over the CofB pilin domain, but it has three short helices in approximately the same orientation as those of CofB. No disulfide bond stabilizes the CofA αβ-loop. Instead, CofA has a disulfide bond closer to its C terminus between α3 and the α4-β5 loop (Cys132–Cys196), which is more typical of the major pilins. Apart from the αβ-loop, the overall size and volume of the two pilin domains is comparable.
FIGURE 5.
Comparison of the CofB structure with the major pilin CofA and the minor pseudopilin GspK. A, topology diagrams of CofB (pilin domain), CofA, and GspK. Pilin domains are shown in blue with the αβ-loop in pink. The β-repeat and β-sandwich domains of CofB are shown as boxes. The GspK α-domain is shown in green. B, schematic representations of CofB, CofA (Protein Data Bank entry 3S0T) and GspK (3CI0) structures. CofB is rotated 90° relative to its orientation in Fig. 4, such that the C-terminal domains point away from the reader and are not visible. Disulfide bonds are shown in ball-and-stick representations with sulfurs colored yellow. C, surface representations of CofB, CofA, and GspK, colored as in A. Cysteine sulfurs are colored yellow, nitrogens are blue, and oxygens are red.
CofB is unusual for a minor pilin with its large size and extended multidomain structure. Most minor pilins are similar in size and structure to their corresponding major pilins. CofB shares characteristics with the ETEC minor pseudopilin GspK, which, like CofB, is approximately twice the size of its respective major pseudopilin GspG and has a canonical pilin domain plus a second non-pilin domain (Fig. 5) (55). However, in GspK, the non-pilin domain is an α-helical region, the “α-domain,” inserted between strands β1 and β2 of the pilin domain β-sheet. The α-domain is covalently attached to the pilin domain at both its N and C terminus but also has extensive noncovalent interactions, making it an integral part of the pilin domain (Fig. 5C). This rigid single domain structure contrasts with the extended flexible three-domain arrangement of CofB.
Docking of CofB into CFA/III Pilus Filament Model
The similarity of the pilin domain to CofA implies that CofB might incorporate into the growing pilus filament. However, insertion of CofB would be blocked by its C-terminal region, which is located on the inner face of the pilin domain, as shown in the CFA/III model in Fig. 6A (64). CofB does, however, fit well onto the tip of the filament (Fig. 6B), where there is room for the C-terminal region. Such a tip location is observed for GspK in the ternary crystal structure of the ETEC minor pseudopilins GspI, GspJ, and GspK (55) and is consistent with the role of CofB as an initiator of pilus assembly.
FIGURE 6.
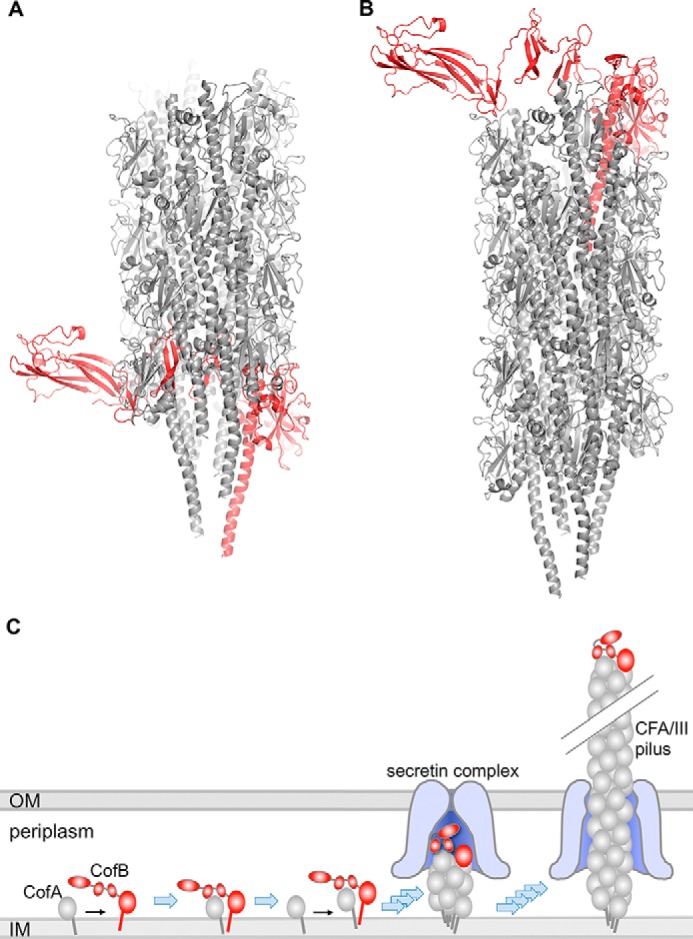
Models of CofB incorporation into CFA/III pilus. A, CofB (red) was superimposed via its pilin domain upon the terminal CofA major pilin subunit in the CFA/III pilus model (gray), derived by fitting the CofA crystal structure (64) onto the V. cholerae TCP helical reconstruction (7), with the N-terminal 28 residues of CofA modeled from the full-length P. aeruginosa PilA crystal structure (4). The β-repeat and β-sandwich domains would clash with adjacent CofA subunits in the pilus. B, CofB superimposed onto the first CofA subunit of the CFA/III pilus, which results in no steric clashes. C, model for CofB-mediated initiation of CFA/III assembly. CofB may recruit the first minor pilin in the pilus and/or may signal opening of the outer membrane (OM) secretin channel. CofB folding would allow it to pass through the secretin channel, to be displayed on the tip of the pilus. OM, outer membrane; IM, inner membrane.
The C-terminal Region Is Required for Initiation of Pilus Assembly
Because CofB has such discrete and well defined domains, we tested the requirement for the extended C-terminal region in initiating pilus assembly. We generated a CofB variant truncated at residue 259, comprising the pilin domain only. This site was chosen because residues 260 and beyond do not interact with the pilin domain in our CofB crystal structure. Furthermore, a stable proteolytic fragment corresponding to the pilin domain is produced when CofB is overexpressed (Fig. 2B, top). CofB259 is stably expressed but, unlike full-length CofB, is unable to rescue pilus assembly in the MC4100-pcofΔcofB strain regardless of its expression level (Fig. 2A). These results demonstrate the requirement for the CofB C-terminal region for initiating pilus assembly.
Discussion
The ETEC minor pilins CofB and LngB, from the CFA/III and Longus T4b pilus systems, respectively, are highly similar in sequence and size. Both proteins have short leader sequences that are more similar to those of the T4a pilins and the minor pseudopilins than to their respective major pilins, suggesting that they are not processed by the prepilin peptidases encoded on their pilus operons. Both minor pilins have 8 cysteines and two tandem ∼30-amino acid sequence repeats. We show here that both CofB and LngB are required for pilus assembly and that very low expression levels are sufficient.
The CofB structure is like no other major or minor pilin structure published to date. Although it contains a canonical pilin domain, the extended flexible nature of the C-terminal region suggests an ability to adapt. The CofB pilin domain shares the overall fold of CofA, including the non-sequential arrangement of the β-strands within the central β-sheet of the globular domain. The additional bulk from the β-hairpin in the αβ-loop of CofB may affect its ability to pack into a growing pilus in place of CofA, even in the absence of the C-terminal region.
The structure of CofB, its role in initiating pilus assembly, and its similarity to the GspK minor pseudopilin together imply that it is located at the tip of the pilus. Our immunoblots show that CofB is present in the CFA/III fraction at very low levels, consistent with it being a tip-associated pilin rather than one that is distributed throughout the filament. CFA/III pili are several μm in length. Based on the transmission EM image reconstruction of the closely related V. cholerae TCP, which has an axial rise per subunit of 8.4 Å (7), a 5-μm-long pilus is composed of ∼6000 CofA subunits. We have thus far been unable to demonstrate CofB localization at the pilus tip using immunogold transmission EM with either of our anti-CofB antibodies. This may be because these antibodies, raised against peptides, are not capable of binding to the folded protein in the context of the pilus filament.
We propose that the CofB minor pilin is the first pilin subunit in a new pilus filament and that it recruits the first CofA major pilin via interactions between its C-terminal region and the CofA globular domain (Fig. 6C). This model is supported by our results showing that CFA/III pili are not made for the CofB259 construct that lacks the C-terminal region. The placement of the C-terminal region as shown in the CofB crystal structure, together with the bulky αβ-loop, would prohibit its insertion into a growing pilus filament (Fig. 6A), but CofB fits nicely at the tip of our CFA/III pilus model (Fig. 6B). When modeled as a rigid body, the C-terminal region of CofB spans the tip of the pilus and protrudes on the other side of the filament, where it could potentially clash with the secretin channel and associated periplasmic pilus assembly proteins (the secretin complex) as the pilus grows across the outer membrane. However, the linkers connecting the C-terminal domains may provide sufficient conformational flexibility to allow these domains to tuck into the end of the pilus instead of protruding from it, thus allowing the tip to pass through the secretin channel. This apparent flexibility distinguishes CofB from the bulky, rigid GspK minor pseudopilin, which may simply act as a mechanical blocker to prevent passage of the pseudopilus through the secretin, relegating this filament to the periplasm.
The CFA/III pilus functions like a T2S system in extruding CofJ through the secretin channel. Cryo-EM data provide evidence that T2S substrates can home to the vestibule of the secretin channel, ready to be pushed though the channel by the pseudopilus (90). Thus, the minor pilins located at the pseudopilins tip, as well as CofB and LngB at the tip of the ETEC T4P, may also be involved in signaling the secretin channel to open.
The ETEC and V. cholerae T4b pili are relatively simple systems compared with the T4a pili and even compared with the T2S systems. Our results demonstrate that a single minor pilin is capable of initiating pilus assembly for the ETEC T4P systems, with its pilin domain serving a structural role to anchor the C-terminal region at the tip of the pilus, where it functions to recruit major pilins and/or signal the secretin channel to open. Such tasks may require multiple minor pilins in the more complex T2S and T4a pilus systems. In the case of the T2S system, the pseudopili must rapidly assemble and disassemble, producing a piston-like motion to extrude substrate across the outer membrane. In the case of T4a pili (and EPEC T4b pili), pilus assembly must counter pilus disassembly facilitated by the retraction ATPase. Such complex pilus dynamics may require more sophisticated control that cannot be accomplished by a single minor pilin. The T2S and T4a pilus systems may have evolved from the primitive ETEC and V. cholerae T4b pilus systems to specialize in protein secretion or perform more complex pilus functions like twitching motility and host cell signaling.
Our CofB structure represents the first structure of a minor pilin from a T4b pilus system and of any minor pilin that can initiate pilus assembly single-handedly. This structure will inform experiments to further explore its mechanism for initiating pilus assembly in ETEC and the related V. cholerae TCP system and will provide insights into minor pilin functions for the more complex T2S and pilus systems. The ETEC and V. cholerae minor pilins may also provide new targets for antimicrobial agents designed to inhibit T4P assembly.
Author Contributions
S. K. cloned, expressed, purified, and crystallized CofB and solved its crystal structure. D. N. conceived and performed experiments shown in Fig. 2 and contributed to those of Fig. 3, which were performed by T. H. and G. Y. L. C. coordinated the study and analyzed data along with S. K. and D. N. S. K., D. N., and L. C. wrote the paper with editorial contributions from T. H. and G. Y. All authors approved the final version of the paper.
Acknowledgments
We thank Ronald Taylor for insightful discussions and the staff at SSRL Beamlines 7-1 and 14-1 for assistance with remote data collection. Molecular structure figures were prepared using the PyMOL Molecular Graphics System, version 1.7.4 (Schrödinger, LLC). Topology diagrams were prepared using TopDraw (91).
This work was supported by Canadian Institutes of Health Research Grant MOP-125959 (to L. C.). The authors declare that they have no conflicts of interest with the contents of this article.
The atomic coordinates and structure factors (code 4QS4) have been deposited in the Protein Data Bank (http://wwpdb.org/).
- T4P
- Type IV pilus/pili
- ETEC
- enterotoxigenic E. coli
- T4a
- Type IVa
- T4b
- Type IVb
- T2S
- Type II secretion
- TCP
- toxin coregulated pilus
- Ap
- ampicillin
- Cm
- chloramphenicol
- Km
- kanamycin
- WCC
- whole cell culture
- Sup
- supernatant
- SeMet
- selenomethionine
- MAD
- multiple anomalous diffraction.
References
- 1.Berry J. L., and Pelicic V. (2015) Exceptionally widespread nanomachines composed of type IV pilins: the prokaryotic Swiss Army knives. FEMS Microbiol. Rev. 39, 134–154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Giltner C. L., Nguyen Y., and Burrows L. L. (2012) Type IV pilin proteins: versatile molecular modules. Microbiol. Mol. Biol. Rev. 76, 740–772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Melville S., and Craig L. (2013) Type IV pili in Gram-positive bacteria. Microbiol. Mol. Biol. Rev. 77, 323–341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Craig L., Pique M. E., and Tainer J. A. (2004) Type IV pilus structure and bacterial pathogenicity. Nat. Rev. Microbiol. 2, 363–378 [DOI] [PubMed] [Google Scholar]
- 5.Pelicic V. (2008) Type IV pili: e pluribus unum? Mol. Microbiol. 68, 827–837 [DOI] [PubMed] [Google Scholar]
- 6.Craig L., Volkmann N., Arvai A. S., Pique M. E., Yeager M., Egelman E. H., and Tainer J. A. (2006) Type IV pilus structure by cryo-electron microscopy and crystallography: implications for pilus assembly and functions. Mol. Cell 23, 651–662 [DOI] [PubMed] [Google Scholar]
- 7.Li J., Egelman E. H., and Craig L. (2012) Structure of the Vibrio cholerae Type IVb pilus and stability comparison with the Neisseria gonorrhoeae Type IVa pilus. J. Mol. Biol. 418, 47–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Aas F. E., Winther-Larsen H. C., Wolfgang M., Frye S., Løvold C., Roos N., van Putten J. P., and Koomey M. (2007) Substitutions in the N-terminal α helical spine of Neisseria gonorrhoeae pilin affect Type IV pilus assembly, dynamics and associated functions. Mol. Microbiol. 63, 69–85 [DOI] [PubMed] [Google Scholar]
- 9.Horiuchi T., and Komano T. (1998) Mutational analysis of plasmid R64 thin pilus prepilin: the entire prepilin sequence is required for processing by type IV prepilin peptidase. J. Bacteriol. 180, 4613–4620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pasloske B. L., and Paranchych W. (1988) The expression of mutant pilins in Pseudomonas aeruginosa: fifth position glutamate affects pilin methylation. Mol. Microbiol. 2, 489–495 [DOI] [PubMed] [Google Scholar]
- 11.Strom M. S., and Lory S. (1991) Amino acid substitutions in pilin of Pseudomonas aeruginosa: effect on leader peptide cleavage, amino-terminal methylation, and pilus assembly. J. Biol. Chem. 266, 1656–1664 [PubMed] [Google Scholar]
- 12.Kaufman M. R., Seyer J. M., and Taylor R. K. (1991) Processing of TCP pilin by TcpJ typifies a common step intrinsic to a newly recognized pathway of extracellular protein secretion by Gram-negative bacteria. Genes Dev. 5, 1834–1846 [DOI] [PubMed] [Google Scholar]
- 13.Strom M. S., and Lory S. (1992) Kinetics and sequence specificity of processing of prepilin by PilD, the type IV leader peptidase of Pseudomonas aeruginosa. J. Bacteriol. 174, 7345–7351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang H. Z., Lory S., and Donnenberg M. S. (1994) A plasmid-encoded prepilin peptidase gene from enteropathogenic Escherichia coli. J. Bacteriol. 176, 6885–6891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Anantha R. P., Stone K. D., and Donnenberg M. S. (1998) Role of BfpF, a member of the PilT family of putative nucleotide-binding proteins, in type IV pilus biogenesis and in interactions between enteropathogenic Escherichia coli and host cells. Infect. Immun. 66, 122–131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nunn D. N., and Lory S. (1991) Product of the Pseudomonas aeruginosa gene pilD is a prepilin leader peptidase. Proc. Natl. Acad. Sci. U.S.A. 88, 3281–3285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tripathi S. A., and Taylor R. K. (2007) Membrane association and multimerization of TcpT, the cognate ATPase ortholog of the Vibrio cholerae toxin-coregulated-pilus biogenesis apparatus. J. Bacteriol. 189, 4401–4409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Blank T. E., and Donnenberg M. S. (2001) Novel topology of BfpE, a cytoplasmic membrane protein required for type IV fimbrial biogenesis in enteropathogenic Escherichia coli. J. Bacteriol. 183, 4435–4450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tønjum T., Freitag N. E., Namork E., and Koomey M. (1995) Identification and characterization of pilG, a highly conserved pilus-assembly gene in pathogenic Neisseria. Mol. Microbiol. 16, 451–464 [DOI] [PubMed] [Google Scholar]
- 20.Takhar H. K., Kemp K., Kim M., Howell P. L., and Burrows L. L. (2013) The platform protein is essential for type IV pilus biogenesis. J. Biol. Chem. 288, 9721–9728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Collins R. F., Frye S. A., Kitmitto A., Ford R. C., Tønjum T., and Derrick J. P. (2004) Structure of the Neisseria meningitidis outer membrane PilQ secretin complex at 12 Å resolution. J. Biol. Chem. 279, 39750–39756 [DOI] [PubMed] [Google Scholar]
- 22.Schmidt S. A., Bieber D., Ramer S. W., Hwang J., Wu C. Y., and Schoolnik G. (2001) Structure-function analysis of BfpB, a secretin-like protein encoded by the bundle-forming-pilus operon of enteropathogenic Escherichia coli. J. Bacteriol. 183, 4848–4859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gold V. A., Salzer R., Averhoff B., and Kuhlbrandt W. (2015) Structure of a type IV pilus machinery in the open and closed state. eLife 10.7554/eLife.07380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jain S., Mościcka K. B., Bos M. P., Pachulec E., Stuart M. C., Keegstra W., Boekema E. J., and van der Does C. (2011) Structural characterization of outer membrane components of the type IV pili system in pathogenic Neisseria. PLoS One 6, e16624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lieberman J. A., Frost N. A., Hoppert M., Fernandes P. J., Vogt S. L., Raivio T. L., Blanpied T. A., and Donnenberg M. S. (2012) Outer membrane targeting, ultrastructure, and single molecule localization of the enteropathogenic Escherichia coli type IV pilus secretin BfpB. J. Bacteriol. 194, 1646–1658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Korotkov K. V., Sandkvist M., and Hol W. G. (2012) The type II secretion system: biogenesis, molecular architecture and mechanism. Nat. Rev. Microbiol. 10, 336–351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McLaughlin L. S., Haft R. J., and Forest K. T. (2012) Structural insights into the Type II secretion nanomachine. Curr. Opin. Struct. Biol. 22, 208–216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sandkvist M. (2001) Type II secretion and pathogenesis. Infect. Immun. 69, 3523–3535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chiang P., Sampaleanu L. M., Ayers M., Pahuta M., Howell P. L., and Burrows L. L. (2008) Functional role of conserved residues in the characteristic secretion NTPase motifs of the Pseudomonas aeruginosa type IV pilus motor proteins PilB, PilT and PilU. Microbiology 154, 114–126 [DOI] [PubMed] [Google Scholar]
- 30.Misic A. M., Satyshur K. A., and Forest K. T. (2010) P. aeruginosa PilT structures with and without nucleotide reveal a dynamic type IV pilus retraction motor. J. Mol. Biol. 400, 1011–1021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Park H. S., Wolfgang M., van Putten J. P., Dorward D., Hayes S. F., and Koomey M. (2001) Structural alterations in a type IV pilus subunit protein result in concurrent defects in multicellular behaviour and adherence to host tissue. Mol. Microbiol. 42, 293–307 [DOI] [PubMed] [Google Scholar]
- 32.Aroeti B., Friedman G., Zlotkin-Rivkin E., and Donnenberg M. S. (2012) Retraction of enteropathogenic E. coli type IV pili promotes efficient host cell colonization, effector translocation and tight junction disruption. Gut Microbes 3, 267–271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Burrows L. L. (2005) Weapons of mass retraction. Mol. Microbiol. 57, 878–888 [DOI] [PubMed] [Google Scholar]
- 34.Bieber D., Ramer S. W., Wu C. Y., Murray W. J., Tobe T., Fernandez R., and Schoolnik G. K. (1998) Type IV pili, transient bacterial aggregates, and virulence of enteropathogenic Escherichia coli. Science 280, 2114–2118 [DOI] [PubMed] [Google Scholar]
- 35.Morand P. C., Bille E., Morelle S., Eugène E., Beretti J. L., Wolfgang M., Meyer T. F., Koomey M., and Nassif X. (2004) Type IV pilus retraction in pathogenic Neisseria is regulated by the PilC proteins. EMBO J. 23, 2009–2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Douzi B., Filloux A., and Voulhoux R. (2012) On the path to uncover the bacterial type II secretion system. Philos. Trans. R. Soc. Lond. B Biol. Sci. 367, 1059–1072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Durand E., Bernadac A., Ball G., Lazdunski A., Sturgis J. N., and Filloux A. (2003) Type II protein secretion in Pseudomonas aeruginosa: the pseudopilus is a multifibrillar and adhesive structure. J. Bacteriol. 185, 2749–2758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sauvonnet N., Vignon G., Pugsley A. P., and Gounon P. (2000) Pilus formation and protein secretion by the same machinery in Escherichia coli. EMBO J. 19, 2221–2228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kirn T. J., and Taylor R. K. (2005) TcpF is a soluble colonization factor and protective antigen secreted by El Tor and classical O1 and O139 Vibrio cholerae serogroups. Infect. Immun. 73, 4461–4470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Megli C. J., Yuen A. S., Kolappan S., Richardson M. R., Dharmasena M. N., Krebs S. J., Taylor R. K., and Craig L. (2011) Crystal structure of the Vibrio cholerae colonization factor TcpF and identification of a functional immunogenic site. J. Mol. Biol. 409, 146–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yuen A. S., Kolappan S., Ng D., and Craig L. (2013) Structure and secretion of CofJ, a putative colonization factor of enterotoxigenic E. coli. Mol. Microbiol. 90, 898–918 [DOI] [PubMed] [Google Scholar]
- 42.Bleves S., Voulhoux R., Michel G., Lazdunski A., Tommassen J., and Filloux A. (1998) The secretion apparatus of Pseudomonas aeruginosa: identification of a fifth pseudopilin, XcpX (GspK family). Mol. Microbiol. 27, 31–40 [DOI] [PubMed] [Google Scholar]
- 43.Carbonnelle E., Helaine S., Nassif X., and Pelicic V. (2006) A systematic genetic analysis in Neisseria meningitidis defines the Pil proteins required for assembly, functionality, stabilization and export of type IV pili. Mol. Microbiol. 61, 1510–1522 [DOI] [PubMed] [Google Scholar]
- 44.Giltner C. L., Habash M., and Burrows L. L. (2010) Pseudomonas aeruginosa minor pilins are incorporated into type IV pili. J. Mol. Biol. 398, 444–461 [DOI] [PubMed] [Google Scholar]
- 45.Winther-Larsen H. C., Wolfgang M., Dunham S., van Putten J. P., Dorward D., Løvold C., Aas F. E., and Koomey M. (2005) A conserved set of pilin-like molecules controls type IV pilus dynamics and organelle-associated functions in Neisseria gonorrhoeae. Mol. Microbiol. 56, 903–917 [DOI] [PubMed] [Google Scholar]
- 46.Nguyen Y., Sugiman-Marangos S., Harvey H., Bell S. D., Charlton C. L., Junop M. S., and Burrows L. L. (2015) Pseudomonas aeruginosa minor pilins prime type IVa pilus assembly and promote surface display of the PilY1 adhesin. J. Biol. Chem. 290, 601–611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Imhaus A. F., and Duménil G. (2014) The number of Neisseria meningitidis type IV pili determines host cell interaction. EMBO J. 33, 1767–1783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bernard S. C., Simpson N., Join-Lambert O., Federici C., Laran-Chich M. P., Maïssa N., Bouzinba-Ségard H., Morand P. C., Chretien F., Taouji S., Chevet E., Janel S., Lafont F., Coureuil M., Segura A., Niedergang F., Marullo S., Couraud P. O., Nassif X., and Bourdoulous S. (2014) Pathogenic Neisseria meningitidis utilizes CD147 for vascular colonization. Nat. Med. 20, 725–731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Brissac T., Mikaty G., Duménil G., Coureuil M., and Nassif X. (2012) The meningococcal minor pilin PilX is responsible for type IV pilus conformational changes associated with signaling to endothelial cells. Infect. Immun. 80, 3297–3306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hélaine S., Carbonnelle E., Prouvensier L., Beretti J. L., Nassif X., and Pelicic V. (2005) PilX, a pilus-associated protein essential for bacterial aggregation, is a key to pilus-facilitated attachment of Neisseria meningitidis to human cells. Mol. Microbiol. 55, 65–77 [DOI] [PubMed] [Google Scholar]
- 51.Kuchma S. L., Griffin E. F., and O'Toole G. A. (2012) Minor pilins of the type IV pilus system participate in the negative regulation of swarming motility. J. Bacteriol. 194, 5388–5403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cehovin A., Simpson P. J., McDowell M. A., Brown D. R., Noschese R., Pallett M., Brady J., Baldwin G. S., Lea S. M., Matthews S. J., and Pelicic V. (2013) Specific DNA recognition mediated by a type IV pilin. Proc. Natl. Acad. Sci. U.S.A. 110, 3065–3070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Franz L. P., Douzi B., Durand E., Dyer D. H., Voulhoux R., and Forest K. T. (2011) Structure of the minor pseudopilin XcpW from the Pseudomonas aeruginosa type II secretion system. Acta Crystallogr. D Biol. Crystallogr. 67, 124–130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Helaine S., Dyer D. H., Nassif X., Pelicic V., and Forest K. T. (2007) 3D structure/function analysis of PilX reveals how minor pilins can modulate the virulence properties of type IV pili. Proc. Natl. Acad. Sci. U.S.A. 104, 15888–15893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Korotkov K. V., and Hol W. G. (2008) Structure of the GspK-GspI-GspJ complex from the enterotoxigenic Escherichia coli type 2 secretion system. Nat. Struct. Mol. Biol. 15, 462–468 [DOI] [PubMed] [Google Scholar]
- 56.Yanez M. E., Korotkov K. V., Abendroth J., and Hol W. G. (2008) The crystal structure of a binary complex of two pseudopilins: EpsI and EpsJ from the type 2 secretion system of Vibrio vulnificus. J. Mol. Biol. 375, 471–486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nguyen Y., Jackson S. G., Aidoo F., Junop M., and Burrows L. L. (2010) Structural characterization of novel Pseudomonas aeruginosa type IV pilins. J. Mol. Biol. 395, 491–503 [DOI] [PubMed] [Google Scholar]
- 58.Piepenbrink K. H., Maldarelli G. A., de la Peña C. F., Mulvey G. L., Snyder G. A., De Masi L., von Rosenvinge E. C., Günther S., Armstrong G. D., Donnenberg M. S., and Sundberg E. J. (2014) Structure of Clostridium difficile PilJ exhibits unprecedented divergence from known type IV pilins. J. Biol. Chem. 289, 4334–4345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cisneros D. A., Bond P. J., Pugsley A. P., Campos M., and Francetic O. (2012) Minor pseudopilin self-assembly primes type II secretion pseudopilus elongation. EMBO J. 31, 1041–1053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Cisneros D. A., Pehau-Arnaudet G., and Francetic O. (2012) Heterologous assembly of type IV pili by a type II secretion system reveals the role of minor pilins in assembly initiation. Mol. Microbiol. 86, 805–818 [DOI] [PubMed] [Google Scholar]
- 61.Evans D. G., Evans D. J. Jr., and Tjoa W. (1977) Hemagglutination of human group A erythrocytes by enterotoxigenic Escherichia coli isolated from adults with diarrhea: correlation with colonization factor. Infect. Immun. 18, 330–337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Datsenko K. A., and Wanner B. L. (2000) One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U.S.A. 97, 6640–6645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mazariego-Espinosa K., Cruz A., Ledesma M. A., Ochoa S. A., and Xicohtencatl-Cortes J. (2010) Longus, a type IV pilus of enterotoxigenic Escherichia coli, is involved in adherence to intestinal epithelial cells. J. Bacteriol. 192, 2791–2800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kolappan S., Roos J., Yuen A. S., Pierce O. M., and Craig L. (2012) Structure of CFA/III and longus Type IV pili from enterotoxigenic Escherichia coli. J. Bacteriol. 194, 2725–2735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Doublié S. (1997) Preparation of selenomethionyl proteins for phase determination. Methods Enzymol. 276, 523–530 [PubMed] [Google Scholar]
- 66.Luft J. R., Collins R. J., Fehrman N. A., Lauricella A. M., Veatch C. K., and DeTitta G. T. (2003) A deliberate approach to screening for initial crystallization conditions of biological macromolecules. J. Struct. Biol. 142, 170–179 [DOI] [PubMed] [Google Scholar]
- 67.Gonzáles A., Moorhead P., McPhillips S. E., Song J., Sharp K., Taylor J. R., Adams P. D., Sauter N. K., and Soltis S. M. (2008) Web-Ice: integrated data collection and analysis for macromolecular crystallography. J. Appl. Crystallogr. 41, 176–184 [Google Scholar]
- 68.Collaborative Computational Project, Number 4 (1994) The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 50, 760–763 [DOI] [PubMed] [Google Scholar]
- 69.Battye T. G., Kontogiannis L., Johnson O., Powell H. R., and Leslie A. G. (2011) iMOSFLM: a new graphical interface for diffraction-image processing with MOSFLM. Acta Crystallogr. D Biol. Crystallogr. 67, 271–281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Evans P. (2006) Scaling and assessment of data quality. Acta Crystallogr. D Biol. Crystallogr. 62, 72–82 [DOI] [PubMed] [Google Scholar]
- 71.Terwilliger T. C. (2000) Maximum-likelihood density modification. Acta Crystallogr. D Biol. Crystallogr. 56, 965–972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Terwilliger T. C., and Berendzen J. (1999) Automated MAD and MIR structure solution. Acta Crystallogr. D Biol. Crystallogr. 55, 849–861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Langer G., Cohen S. X., Lamzin V. S., and Perrakis A. (2008) Automated macromolecular model building for x-ray crystallography using ARP/wARP version 7. Nat. Protoc. 3, 1171–1179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Emsley P., and Cowtan K. (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 75.Murshudov G. N., Vagin A. A., and Dodson E. J. (1997) Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 53, 240–255 [DOI] [PubMed] [Google Scholar]
- 76.Brunger A. T. (2007) Version 1.2 of the Crystallography and NMR system. Nat. Protoc. 2, 2728–2733 [DOI] [PubMed] [Google Scholar]
- 77.Brünger A. T., Adams P. D., Clore G. M., DeLano W. L., Gros P., Grosse-Kunstleve R. W., Jiang J. S., Kuszewski J., Nilges M., Pannu N. S., Read R. J., Rice L. M., Simonson T., and Warren G. L. (1998) Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 54, 905–921 [DOI] [PubMed] [Google Scholar]
- 78.Chen V. B., Arendall W. B. 3rd, Headd J. J., Keedy D. A., Immormino R. M., Kapral G. J., Murray L. W., Richardson J. S., and Richardson D. C. (2010) MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 66, 12–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Honda T., Arita M., and Miwatani T. (1984) Characterization of new hydrophobic pili of human enterotoxigenic Escherichia coli: a possible new colonization factor. Infect. Immun. 43, 959–965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Taniguchi T., Fujino Y., Yamamoto K., Miwatani T., and Honda T. (1995) Sequencing of the gene encoding the major pilin of pilus colonization factor antigen III (CFA/III) of human enterotoxigenic Escherichia coli and evidence that CFA/III is related to type IV pili. Infect. Immun. 63, 724–728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Taniguchi T., Akeda Y., Haba A., Yasuda Y., Yamamoto K., Honda T., and Tochikubo K. (2001) Gene cluster for assembly of pilus colonization factor antigen III of enterotoxigenic Escherichia coli. Infect. Immun. 69, 5864–5873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Honda T., Wetprasit N., Arita M., and Miwatani T. (1989) Production and characterization of monoclonal antibodies to a pilus colonization factor (colonization factor antigen III) of human enterotoxigenic Escherichia coli. Infect. Immun. 57, 3452–3457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Girón J. A., Levine M. M., and Kaper J. B. (1994) Longus: a long pilus ultrastructure produced by human enterotoxigenic Escherichia coli. Mol. Microbiol. 12, 71–82 [DOI] [PubMed] [Google Scholar]
- 84.Gomez-Duarte O. G., Chattopadhyay S., Weissman S. J., Giron J. A., Kaper J. B., and Sokurenko E. V. (2007) Genetic diversity of the gene cluster encoding longus, a type IV pilus of enterotoxigenic Escherichia coli. J. Bacteriol. 189, 9145–9149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Hartung S., Arvai A. S., Wood T., Kolappan S., Shin D. S., Craig L., and Tainer J. A. (2011) Ultra-high resolution and full-length pilin structures with insights for filament assembly, pathogenic functions, and vaccine potential. J. Biol. Chem. 286, 44254–44265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Parge H. E., Forest K. T., Hickey M. J., Christensen D. A., Getzoff E. D., and Tainer J. A. (1995) Structure of the fibre-forming protein pilin at 2.6 Å resolution. Nature 378, 32–38 [DOI] [PubMed] [Google Scholar]
- 87.Kawahara K., Oki H., Fukakusa S., Maruno T., Kobayashi Y., Motooka D., Taniguchi T., Honda T., Iida T., Nakamura S., and Ohkubo T. (2015) Cloning, expression, purification, crystallization and X-ray crystallographic analysis of CofB, the minor pilin subunit of CFA/III from human enterotoxigenic Escherichia coli. Acta Crystallogr. F Struct. Biol. Commun. 71, 663–667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Craig L., Taylor R. K., Pique M. E., Adair B. D., Arvai A. S., Singh M., Lloyd S. J., Shin D. S., Getzoff E. D., Yeager M., Forest K. T., and Tainer J. A. (2003) Type IV pilin structure and assembly: x-ray and EM analyses of Vibrio cholerae toxin-coregulated pilus and Pseudomonas aeruginosa PAK pilin. Mol. Cell 11, 1139–1150 [DOI] [PubMed] [Google Scholar]
- 89.Lim M. S., Ng D., Zong Z., Arvai A. S., Taylor R. K., Tainer J. A., and Craig L. (2010) Vibrio cholerae El Tor TcpA crystal structure and mechanism for pilus-mediated microcolony formation. Mol. Microbiol. 77, 755–770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Reichow S. L., Korotkov K. V., Gonen M., Sun J., Delarosa J. R., Hol W. G., and Gonen T. (2011) The binding of cholera toxin to the periplasmic vestibule of the type II secretion channel. Channels 5, 215–218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Bond C. S. (2003) TopDraw: a sketchpad for protein structure topology cartoons. Bioinformatics 19, 311–312 [DOI] [PubMed] [Google Scholar]
- 92.Honda T., Lertpocasombat K., Hata A., Miwatani T., and Finkelstein R. A. (1989) Purification and characterization of a protease produced by Vibrio cholerae non-O1 and comparison with a protease of V. cholerae O1. Infect. Immun. 57, 2799–2803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Levine M. M., Ristaino P., Marley G., Smyth C., Knutton S., Boedeker E., Black R., Young C., Clements M. L., and Cheney C. (1984) Coli surface antigens 1 and 3 of colonization factor antigen II-positive enterotoxigenic Escherichia coli: morphology, purification, and immune responses in humans. Infect. Immun. 44, 409–420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Kabsch H. (1993) Automatic processing of rotation diffraction data from crystals of initially unknown symmetry and cell constants. J. Appl. Crystallogr. 26, 795–800 [Google Scholar]