Background: NFS1-ISD11 complex is essential for the Fe-S cluster assembly process and the homozygous R68L ISD11 mutation causes the mitochondrial disorder, COXPD19.
Results: Putative helix-3 and the C terminus of ISD11 are critical for NFS1-ISD11 subcomplex formation and Fe-S cluster biogenesis.
Conclusion: ISD11 interaction is critical for NFS1 stability and its steady-state levels, in vivo.
Significance: Identified important ISD11 residues will provides valuable information to understand COXPD19 progression.
Keywords: electron transfer, iron-sulfur protein, mitochondria, mitochondrial disease, molecular chaperone, protein-protein interaction, reactive oxygen species (ROS), respiration
Abstract
Biogenesis of the iron-sulfur (Fe-S) cluster is an indispensable process in living cells. In mammalian mitochondria, the initial step of the Fe-S cluster assembly process is assisted by the NFS1-ISD11 complex, which delivers sulfur to scaffold protein ISCU during Fe-S cluster synthesis. Although ISD11 is an essential protein, its cellular role in Fe-S cluster biogenesis is still not defined. Our study maps the important ISD11 amino acid residues belonging to putative helix 1 (Phe-40), helix 3 (Leu-63, Arg-68, Gln-69, Ile-72, Tyr-76), and C-terminal segment (Leu-81, Glu-84) are critical for in vivo Fe-S cluster biogenesis. Importantly, mutation of these conserved ISD11 residues into alanine leads to its compromised interaction with NFS1, resulting in reduced stability and enhanced aggregation of NFS1 in the mitochondria. Due to altered interaction with ISD11 mutants, the levels of NFS1 and Isu1 were significantly depleted, which affects Fe-S cluster biosynthesis, leading to reduced electron transport chain complex (ETC) activity and mitochondrial respiration. In humans, a clinically relevant ISD11 mutation (R68L) has been associated in the development of a mitochondrial genetic disorder, COXPD19. Our findings highlight that the ISD11 R68A/R68L mutation display reduced affinity to form a stable subcomplex with NFS1, and thereby fails to prevent NFS1 aggregation resulting in impairment of the Fe-S cluster biogenesis. The prime affected machinery is the ETC complex, which showed compromised redox properties, causing diminished mitochondrial respiration. Furthermore, the R68L ISD11 mutant displayed accumulation of mitochondrial iron and reactive oxygen species, leading to mitochondrial dysfunction, which correlates with the phenotype observed in COXPD19 patients.
Introduction
Biological activity of proteins often requires the assistance of a non-protein chemical moiety called cofactors. Iron-sulfur (Fe-S) clusters are one of the most versatile and evolutionarily conserved cofactors that are present ubiquitously in all life forms ranging from ancient microorganisms to present day genera (1, 2). Typically, these low molecular mass moieties bind to conserved sequence motifs of proteins essentially through cysteine or histidine residues (3). More than 200 proteins require Fe-S clusters for their biological function, including members of the ETC complexes, enzymes, and several transcription factors to regulate gene expression. Moreover, Fe-S clusters can also act as sensors of iron and oxygen (4). Although the chemical structures of Fe-S clusters moieties appear simple, their biogenesis in a living cell is highly complex and coordinated process. Based on the number of iron and sulfur atoms, Fe-S clusters are assembled into two major types, [2Fe-2S] and [4Fe-4S] clusters. Besides, the presence of [3Fe-4S] clusters have also been reported in some Fe-S cluster proteins (5).
In higher eukaryotes, the majority of Fe-S cluster biogenesis takes place in mitochondria and consists of a series of complex events that can be categorized into three major steps. Initially, iron, sulfur, and electrons are transferred to a scaffold protein as raw materials for the biogenesis process. Second, assembling the Fe-S clusters in the scaffold protein. Finally, transfer of Fe-S clusters from the scaffold protein to a recipient apoprotein (1). In humans, the process of sulfur transfer is assisted by the cysteine desulfurase protein NFS1 (IscS in bacteria and Nfs1 in yeast), whereas the precise source of iron is still elusive. However, the iron-binding protein frataxin (CyaY in bacteria and Yfh1 in yeast) is speculated to function as the putative iron donor by interacting with the sulfur donor and scaffold protein (6). Ferredoxin reductase (Arh1 in yeast) transfers electrons from NADH to the scaffold protein via ferredoxin 2 (Fdx in bacteria and Yah1 in yeast). A conserved protein ISCU (IscU in bacteria and Isu1/2 in yeast) acts as a scaffold for the assembly of Fe-S cluster before its transfer to apoproteins. The transfer process is mediated by chaperone machinery comprising the mtHsp70/Mortalin (HscA in bacteria and Ssq1 in yeast) and the J-protein co-chaperone, HSCB (HscB in bacteria and Jac1 in yeast) along with monothiol glutaredoxin, AND GLRX5 (GrxD in bacteria and Grx5 in yeast) (6). Based on the types of Fe-S clusters to be incorporated in the final recipient proteins, additional targeting factors are involved in the transfer process in higher eukaryotes (6–8). Although the functional importance of individual proteins in the biogenesis pathway has been reported, the molecular chemistry of Fe-S cluster assembly and transfer are not firmly established.
The donation of the sulfur atoms is brought about by the NFS1 protein, which belongs to the aminotransferase subgroup of the pyridoxal 5-phosphate-dependent enzymes and is highly conserved from bacteria to humans (9–10). NFS1 acts as a cysteine desulfurase to obtain sulfur from cysteine and forms alanine in this process. A conserved cysteine residue of NFS1 serves as a nucleophile to attack the γ-sulfhydryl group (γ-SH) of free cysteine amino acid and obtains the γ-SH to form a persulfide intermediate at the active site cysteine (11–14). The crystal structures of bacterial cysteine desulfurase protein highlights that it exists as a dimer and each monomer consists of two subdomains. The larger domain binds to the pyridoxal phosphate, and the smaller domain harbors the active site cysteine that transiently holds the sulfur as a persulfide released from cysteine (13, 15). In bacteria, the crystal structure of the NFS1 ortholog IscS shows that it forms a tight complex with IscU, thereby accelerating sulfur transfer from IscS to IscU (16, 17). Biochemical and genetic analysis involving yeast Nfs1 showed that it stabilizes the scaffold protein Isu1 and modulates its level in different growth conditions (18). Moreover, both Nfs1 and Jac1 exhibit binding to mutually exclusive sites in Isu1 (19). However, the mechanism of sulfur transfer from Nfs1 to the scaffold protein is not yet fully established.
In yeast, it has been reported that the Nfs1 protein remains in a complex with an 11-kDa protein named Isd11 (ISD11 in humans), which has no obvious sequence homologs in bacteria. Isd11 is an essential protein in yeast and depletion of ISD11 results in impairment of Fe-S cluster biogenesis (20, 21). Additionally, it has been suggested that the Nfs1 protein is prone to aggregation and degradation in the absence of Isd11 (20). Although Isd11 is not required for desulfurase activity of Nfs1 in vitro, the Nfs1-Isd11 complex represents the functional sulfur donor, in vivo (21). In humans, the Isd11 ortholog ISD11 is believed to play a similar conserved role in Fe-S cluster biogenesis (22) and loss of ISD11 function results in a mitochondrial disorder termed as combined oxidative phosphorylation deficiency 19 (COXPD19)5 (23).
Recent studies on human ISD11 have shown that a homozygous mutation in the LYRM4 gene on chromosome 6p25, which codes for the ISD11 protein, converts a conserved arginine residue at position 68 into leucine (R68L) implicated in the mitochondrial genetic disorder COXPD19. This disorder is characterized by respiratory distress, hypotonia, gastroesophageal reflux, hepatomegaly, and severe lactic acidosis in the neonates. Studies on patient liver and muscle samples have demonstrated decreased activities of mitochondrial ETC complexes I–IV. Postmortem examination of COXPD19 patients had shown widened fiber size in the skeletal muscle, increased lipid content in muscle and liver along with the presence of abnormal mitochondria as detected through electron microscopic analysis. It is known that the R68L mutation in the ISD11 protein leads to development of COXPD19, although detailed molecular mechanisms associated with the cellular defects have not been elucidated.
Because of the crucial importance of the ISD11 protein in Fe-S cluster biogenesis, we have delineated the role of indispensable ISD11 residues for mitochondrial function by utilizing yeast and cell lines as model systems. In this report, we have mapped key residues on the ISD11 protein that are essential for NFS1 interaction to maintain its levels by forming a stable subcomplex that is critical for Fe-S cluster biogenesis. Additionally, our study uncovers the cellular defects associated with the R68L ISD11 mutation, thus revealing biochemical insights into COXPD19 progression. Our findings highlight that the R68L mutation results in impaired Fe-S cluster biogenesis, elevated mitochondrial iron, and oxidative stress, which contribute significantly toward the development of COXPD19.
Experimental Procedures
Cell Culture and Transfection
HeLa cells were cultured in modified Eagle's medium (MEM) (Invitrogen) containing 10% fetal bovine serum (Invitrogen) and 1% penicillin-streptomycin (Invitrogen) and grown at 37 °C in 5% CO2. The cells were transfected with pCI-Neo vector carrying a WT copy or R68L mutant copy of LYRM4 using Lipofectamine 2000 (Invitrogen) and harvested after 48 h for mitochondria isolation and different experiments.
Yeast Strains, Genetic Analysis, Plasmid Construction and Mutagenesis
For yeast genetic analysis, full-length human WT LYRM4 was amplified from a HeLa cell cDNA library (Stratagene). WT LYRM4 and LYRM4 mutants with a C-terminal FLAG tag were cloned in pRS414 yeast expression vector under the TEF promoter containing a Trp marker for selection. The Δisd11 strain (derived from YPH499) carrying a URA3-based plasmid pRS316 with the ISD11 ORF was transformed with pRS414-LYRM4 and the transformants were selected on tryptophan omission plates at 30 °C followed by spot test analysis using serial dilutions of the cells on medium containing 5-fluoroorotic acid (U. S. Biological) to eliminate the WT ISD11 containing plasmid pRS316.
For purification of WT and mutant ISD11 proteins using a bacterial expression system, we inserted ORFs (without signal sequence predicted using PSORT program) of WT and mutant LYRM4 constructs (amino acids 32–91) with a C-terminal His6 tag between the EcoRI-XhoI restriction sites of the pGEX-KG vector. In the case of NFS1 (amino acids 56–457), the ORF lacking the N-terminal signal sequence was cloned between the BamHI-SalI restriction sites of the pRSFDuet-1 vector, carrying an N-terminal His6 tag sequence in a vector backbone. The point mutations were created through QuikChange site-directed mutagenesis, using high-fidelity Pfu Turbo DNA polymerase (Stratagene). All the clones were verified by DNA sequencing reactions.
Protein Expression and Purification
For purification of GST-tagged ISD11 proteins with C-terminal His6 tag sequences, pGEX-KG-LYRM4 constructs were expressed in the BL21(DE3) strain and allowed to grow at 25 °C to an optical density (A600) of 0.6, followed by induction using 0.5 mm isopropyl 1-thio-β-d-galactopyranoside for 6 to 10 h. Cells were harvested by centrifugation and lysed in buffer A (50 mm Tris-Cl, pH 7.5, 100 mm NaCl, 1 mm dithiothreitol, 0.5 mm EDTA, 1 mm PMSF, 5 μm leupeptin, 5 μm pepstatin, and 10% glycerol) along with 0.2 mg/ml of lysozyme at 4 °C for 1 h. The samples were gently treated with 0.2% deoxycholate, followed by DNase I (10 μg/ml) treatment for 15 min at 4 °C. The cell lysates were separated into supernatant and pellet by centrifugation at 22,000 × g for 30 min at 4 °C. The supernatant was incubated with Ni-NTA-Sepharose (GE Healthcare) for 2 h at 4 °C. Unbound proteins and nonspecific contaminants were removed by multiple washes of buffer A alone, followed by sequential single washes of buffer B (buffer A along with 0.05% Tween 20), buffer C (buffer A along with 1 mm ATP, 10 mm MgCl2), buffer D (buffer A along with 1 m NaCl), and buffer E (buffer A along with 40 mm imidazole). Finally, the bound proteins were eluted with buffer containing 50 mm Tris-Cl, pH 7.5, 100 mm NaCl, 250 mm imidazole, and 10% glycerol. For GST pulldown analysis, the proteins purified by Ni-NTA chromatography were subjected to binding with GST-Sepharose beads (GE Healthcare) followed by washing in a conventional manner. The bound proteins were stored in buffer containing 50 mm Tris-Cl, pH 7.5, and 100 mm NaCl at 4 °C.
For purification of human NFS1 protein, the pRSFDuet-1-NFS1 construct was transformed into a chaperone competent BL21 strain harboring a pGro7 plasmid, which expresses the groES-groEL chaperone upon l-arabinose induction. The transformed competent cells were allowed to grow at 24 °C to an A600 of 0.6. The bacterial culture was subjected to groES-groEL chaperone induction using l-arabinose (1 mg/ml) for 30 min, followed by NFS1 induction using 0.5 mm isopropyl 1-thio-β-d-galactopyranoside for 8 h. Cells were harvested and subjected to NFS1 purification using Ni-NTA chromatography following the aforementioned protocol.
GST Pulldown Analysis
Purified WT and mutant GST-ISD11 (3 μm) proteins were incubated with a 10-μl bed volume of glutathione-Sepharose beads in 200 μl of GST buffer A (50 mm Tris-Cl, pH 7.5, 100 mm NaCl, 150 mm imidazole, 50 μm pyridoxal phosphate, 0.1% Triton X-100, 10% glycerol) for an interaction study with NFS1. Unbound proteins were removed by washing the beads three times with GST buffer A. The samples were then blocked with 0.2% BSA for 30 min at 10 °C, followed by washing two times with GST buffer A to remove excess unbound BSA. The beads were resuspended in 200 μl of GST buffer A and incubated with increasing concentrations of NFS1 for 25 min at 10 °C. The GST beads were washed three times with GST buffer A and resolved on SDS-PAGE followed by Coomassie dye staining.
For GST pulldown analysis using mitochondrial lysate, GST-NFS1 protein was purified using glutathione-Sepharose beads following conventional methods. The purified GST-NFS1 slurry was stored in buffer containing 50 mm Tris-Cl, pH 7.5, and 100 mm NaCl at 4 °C. Mitochondria isolated from HeLa cells expressing FLAG-tagged WT or mutant ISD11 (amount corresponding to 1 mg of mitochondrial protein) were lysed for 3 min on ice in 1 ml of buffer B (50 mm Tris-Cl, pH 7.5, 150 mm NaCl, 0.2% Tween 20, 10 μm leupeptin, 10 μm pepstatin, 2 mm phenylmethylsulfonyl fluoride, 50 μm pyridoxal phosphate, and 1 mm ascorbic acid). Membrane debris was removed by centrifugation for 10 min at 18,000 × g and the supernatant was incubated with 2 μm GST-NFS1 slurry for 2 h on a rotary shaker at 4 °C. Beads were collected by centrifugation for 5 min at 800 × g and washed three times with 1 ml of buffer B. Bound proteins were subjected to SDS-PAGE and identified by immunostaining.
Measurement of Mitochondrial and Cellular ROS
The extent of superoxide generation in the WT and mutant mitochondria was measured by FACS analysis, utilizing MitoSOX Red dye (Molecular Probes) according to a previously described protocol in both the yeast model system and HeLa cell lines (24, 25). The respiratory inhibitor rotenone (1 mm) was used as a positive control, which produces higher superoxide levels by inhibiting complex I.
Production of mitochondrial superoxide was further validated by fluorescence microscopy following previously published methods (24, 26). Briefly, HeLa cells were treated with 1 μm MitoSOX Red for 20 min and washed with 1× PBS, followed by mounting in ProLong Gold Antifade reagent with DAPI (Invitrogen). A similar strategy was used for H2DCFDA staining to measure the overall cellular ROS. HeLa cells were treated with 3 μm H2DCFDA for 15 min and mounted in ProLong antifade reagent (Invitrogen). Fluorescence images were acquired using a ×63 objective lens in a Zeiss Axio Observer Z1 Apotome 2.0 (Jena, Germany) microscope. For the microscopic analysis in yeast, 0.1 A600 of WT and mutant strain from mid-log phase (grown at 30 °C and then shifted to 37 °C) were incubated with 5 μm MitoSOX Red for 15 min or 5 μm H2DCFDA for 20 min, washed, and fluorescence images were captured using ×100 objective of Delta Vision Fluorescence Microscope (DV Elite, GE Healthcare).
Enzymatic Activity Analysis and Mitochondrial ATP Level Quantification
Activity of complex I, complex II, and complex IV of Electron Transport Chain and aconitase were measured according to a protocol described previously (27, 28). Mitochondrial ATP was quantified using the Mitochondrial ToxGloTM assay kit (Promega) as per the manufacturer's instructions. Briefly, 2 μg of protein corresponding to HeLa cells mitochondria and 4 μg of protein corresponding to yeast cells mitochondria were resuspended in 80 μl of SE buffer (250 mm sucrose, 1 mm EDTA, 10 mm MOPS-KOH, pH 7.2). Subsequently, 100 μl of ATP detection reagent was added to each reaction, mixed, and the luminescence was measured using a microplate counter (model 2450, PerkinElmer Life Sciences).
Aggregation Analysis of NFS1
Aggregation assay using purified NFS1 was performed by monitoring absorbance of NFS1 at 320 nm at 37 °C. Briefly, 1 μm purified NFS1 protein was incubated alone or with GST-ISD11 (2 μm) in 500 μl of buffer F (50 mm Tris-Cl, pH 7.5, 50 mm NaCl, 250 mm imidazole, 5% glycerol) at 37 °C and absorbance at 320 nm was measured with time. NFS1 with 2 μm GST protein alone was used as a control.
Aggregation of NFS1 in mitochondrial lysate was performed following previously described protocols (20, 21). Briefly, the mitochondria isolated from WT or mutant ISD11 expressing HeLa cells or yeast strains were subjected to heat shock for 20 min at 37 °C. Mitochondria was subsequently lysed with 0.5% Triton X-100 in buffer containing 50 mm Tris-Cl, pH 7.5, 100 mm NaCl, 10 μm leupeptin, 10 μm pepstatin, 1 mm PMSF. The aggregate fraction was pelleted by centrifugation for 10 min at 30,000 × g. Total and the supernatant fractions were precipitated with trichloroacetic acid (TCA), and the samples were analyzed by immunodecoration.
Miscellaneous
Analysis of mitochondrial mass and membrane potential was performed following previously reported techniques (24, 25). Overall cellular and mitochondrial iron levels were estimated using iron-specific colorimetric assay and atomic absorption spectroscopy according to earlier described procedures (24, 29, 30). In the case of flow cytometric analysis, 10,000 events were analyzed, and the relative fluorescence intensity values obtained from three independent (n = 3) experiments were plotted. For statistical analysis, one-way (non-parametric) analysis of variance was performed, followed by Tukey post-test (compare all pairs of columns) using GraphPad Prism. p values have been represented comparing columns with WT. Columns with no p value denotes nonsignificant as compared with WT. A p value of <0.05 were defined as significant, and asterisks were used to denote significance as follows: *, p < 0.05; **, p < 0.01; ***, p < 0.001.
Results
Identification of Critical Residues of Human ISD11 (LYRM4) Required for Cell Growth and Survival
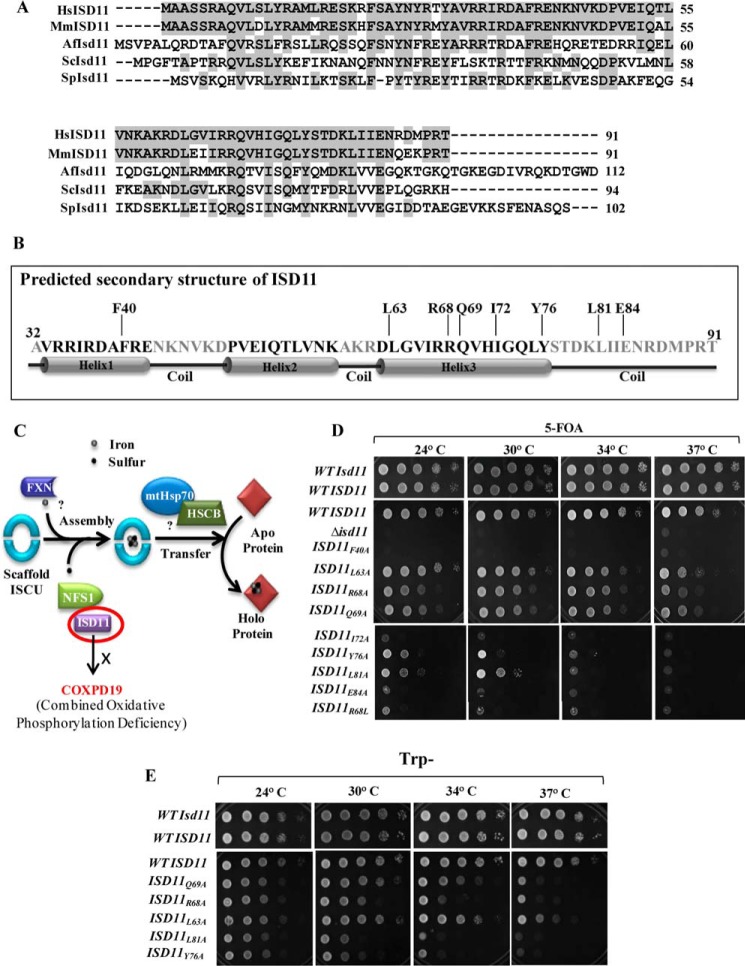
Human mitochondrial matrix protein ISD11 (Isd11 in yeast) assists the function of the sulfur donor protein NFS1 (Nfs1 in yeast) (22, 23, 31). Although there is no bacterial ortholog of the ISD11 protein, it is well conserved in eukaryotes, ranging from yeast to humans as highlighted in multiple sequence alignment (Fig. 1A). Previous reports highlight that yeast Isd11 binds to a hydrophobic cysteine desulfurase protein, Nfs1, and stabilizes the complex, thereby preventing its aggregation (20, 21). To uncover the essential function of human ISD11 in assisting NFS1 activity; we tested the ability of the wild type (WT) LYRM4 gene in restoring the cellular viability of a Δisd11 yeast strain. To determine a centromeric plasmid, pRS414 carrying a WT copy of LYRM4 under TEF promoter was transformed into the Δisd11 strain harboring yeast WT ISD11 in pRS316 vector with URA3 as a selectable marker. The transformed strains were selected on tryptophan omitted plates and tested further for their viability by drop dilution analysis on a media containing 5-fluoroorotic acid (5-FOA). 5-FOA acts as a counter selection for the cells that have lost the pRS316 plasmid carrying the yeast WT ISD11. We observed that WT LYRM4 could complement inviability of THE Δisd11 strain at all the tested temperatures (Fig. 1D, top panel).
FIGURE 1.
Multiple sequence alignment of ISD11 orthologs and effect of mutations on cell viability. A, multiple sequence alignment of ISD11 protein orthologs from different species: Homo sapiens (Hs), Mus musculus (Mm), Aspergillus fumigatus (Af), S. cerevisiae (Sc), and Schizosaccharomyces pombe (Sp) was performed using ClustalW multiple sequence alignment program. Identical residues are highlighted in gray. B, secondary structure of matured form of ISD11 (amino acids 32–91) was predicted using Robetta Full-chain Protein Structure Prediction Server, depicting three helices and position of critical amino acid residues along the length of the protein. C, a schematic representation of Fe-S cluster biogenesis indicating the essential steps. Sulfur donation is assisted by NFS1-ISD11 protein complex, whereas frataxin acts as the putative iron donor. ATP-dependent mtHsp70 chaperone GRP75 and co-chaperone HSCB with the help of other transfer factors aids in the incorporation of Fe-S clusters into recipient apoproteins. R68L point mutations in ISD11 lead to development of COXPD19. D, Δisd11 yeast strain carrying a yeast WT ISD11 in pRS316 plasmid was transformed with WT LYRM4 or LYRM4 mutants (F40A, Q63A, R68A, Q69A, I72A, Y76A, L81A, E84A, and R68L) in pRS414 vector under TEF promoter and subjected to drop test analysis on 5-FOA medium followed by incubation at the indicated temperatures for 72 h. E, Δisd11 yeast strain harboring a copy of WT LYRM4 or ts LYRM4 mutants (Q69A, R68A, L63A, L81A, and Y76A) in a centromeric plasmid pRS414 TEF were subjected to spot test analysis on Trp− plates, followed by incubation for 72 h at the indicated temperatures. Δisd11 yeast strain carrying a WT ISD11 was taken as positive control.
Based on secondary structure prediction it is evident that the ISD11 protein is a small α-helical protein and consists of 3 putative helix forming regions (Fig. 1B). To reveal the functional significance of amino acid residues of the human ISD11 protein that are critical for the NFS1 activity, an alanine substitution mutagenesis was performed to retain the structural integrity by causing minimal perturbations to the protein. 8 mutations at different positions with alanine substitutions were generated, these mutants showed significant growth phenotypes, including amino acid residues Phe-40, Leu-63, Arg-68, Gln-69, Ile-72, Tyr-76, Leu-81, and Glu-84. Of these, the Arg-68 residue was identified to be critical for ISD11 function because an R68L conversion leads to development of combined oxidative phosphorylation deficiency (COXPD19) in humans (Fig. 1C) (23). Interestingly, none of the 8 mutants of ISD11 could completely rescue the lethal phenotype of the Δisd11 strain on 5-FOA media. The mutant ISD11F40A, ISD11I72A, and ISD11E84A strains were found to be lethal at all temperatures. Whereas, ISD11Y76A and ISD11L81A showed severe temperature-sensitive (ts) phenotype, as they grew slowly at a lower temperature but were inviable at non-permissive temperature conditions. The other three mutants namely, ISD11L63A, ISD11R68A, and ISD11Q69A supported the growth at lower temperatures but exhibit ts phenotypes at 37 οC (Fig. 1D, middle and lower panel). Importantly, the COXPD19-associated ISD11R68L mutant failed to rescue the inviability of Δisd11 on 5-FOA containing media at all temperatures, suggesting the critical ability in vivo of the Arg-68 residue for ISD11 function (Fig. 1D, lower panel). The ts phenotype of ISD11L63A, ISD11R68A, ISD11Q69A, ISD11Y76A, and ISD11L81A mutants was further verified by spot analysis on tryptophan omitted minimal media by incubating at different temperatures (Fig. 1E). Strikingly, most of the mutants isolated were clustered around part of helix 3 and the C-terminal loop region except the Phe-40 residue, which belongs to helix 1, thus underlining the importance of these regions for the function of ISD11 (Fig. 1B).
To test whether the phenotype of lethal or ts mutants is not due to direct consequence of their stability in vivo, we determined the steady-state expression levels after FLAG tagging ISD11 in the presence of wild type before 5-FOA treatment. All lethal as well as ts mutants showed equivalent levels indicating their stable expression in normal growth conditions (Fig. 2A). In addition, the mitochondria from ts mutants were isolated by growing them at permissive temperature conditions. The steady-state levels were analyzed after inducing the phenotype in isolated mitochondria and subjected for immunodecoration using antibodies against different components of the core Fe-S assembly machinery. As indicated in Fig. 2B, the levels of ISD11 (Fig. 2E) and Jac1 (Fig. 2F) in the organelle relatively remained unaffected in all ts mutants as compared with cells expressing WT ISD11. However, a reduction in the levels of Nfs1 (Fig. 2C) and the scaffold protein, Isu1 (Fig. 2D), was observed for the ts mutants highlighting the critical ability of these residues of the ISD11 protein in a Fe-S assembly process. Mitochondrial matrix protein Mge1 was taken as loading control (Fig. 2G).
FIGURE 2.
Estimation of expression levels of proteins involved in Fe-S cluster biogenesis pathway. A, Δisd11 yeast strain carrying yeast WT ISD11 in pRS316 plasmid was transformed with human WT LYRM4 or LYRM4 mutants (F40A, Q63A, R68A, Q69A, I72A, Y76A, L81A, and E84A) in pRS414 TEF vector and subjected to immunodetection of FLAG-tagged ISD11. B, WT and ts mutants (L63A, R68A, Q69A, Y76A, and L81A) grown at permissive temperature were subjected to heat shock at 37 °C, followed by mitochondria isolation. The mitochondrial lysates were separated on SDS-PAGE and subjected to Western blot analysis to compare protein levels of Nfs1, ISD11, Isu1, and Jac1. Mitochondrial matrix protein Mge1 was taken as loading control. C–G, relative protein levels of Nfs1 (C), Isu1 (D), ISD11 (E), Jac1 (F), and Mge1 (G) from WT and ts mutants were quantified by densitometry. Data represented as mean ± S.E. The values were plotted based on three independent experiments (n = 3). p value of <0.05 was defined as significant, and asterisks are used to denote significance, where: *, p < 0.05; **, p < 0.01; ***, p < 0.001. A.U., arbitrary unit.
Critical ISD11 Residues Required for the Formation of Subcomplex with NFS1
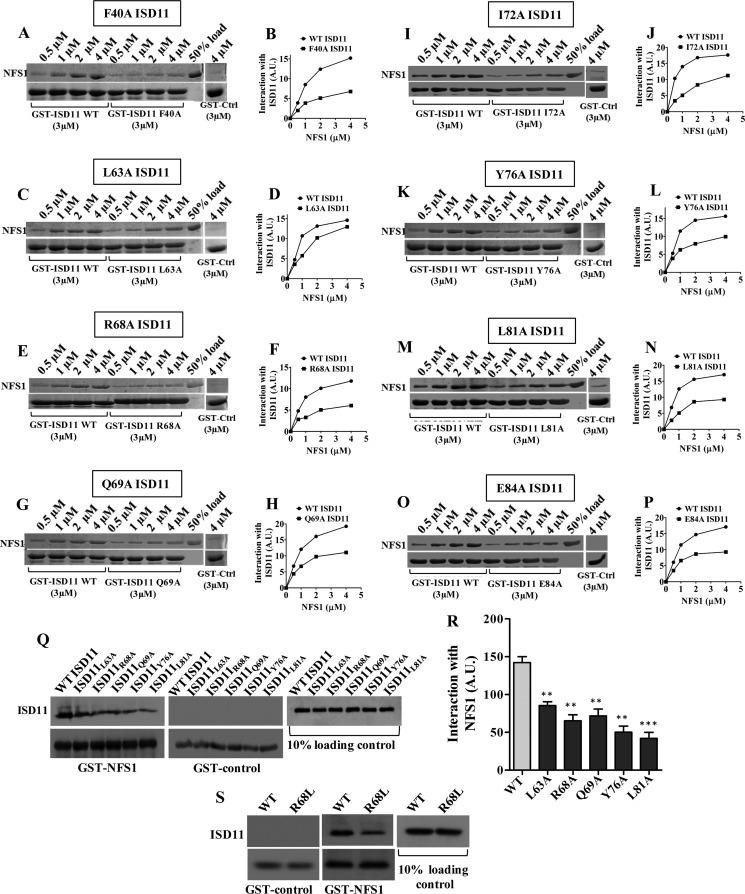
Studies in Saccharomyces cerevisiae suggest that Nfs1 and Isd11 are involved in early steps of Fe-S cluster biogenesis (32). However, the precise function of Isd11 is still not defined (20, 21, 33). To functionally characterize human ISD11, we utilized ts mutants and analyzed their interaction with human NFS1. To analyze the in vitro interaction, we first recombinantly purified human NFS1 to homogeneity from Escherichia coli (BL21 cells) expressing the groES-groEL chaperone system. Similarly, human ISD11 was recombinantly expressed and purified from E. coli (BL21 cells) using a double tagged system (N-terminal GST tag and C-terminal His6 tag).
The in vitro interaction was performed by a GST pulldown analysis using His-tagged WT NFS1 and GST-bound WT ISD11 or mutant proteins. The interaction was analyzed by incubating a fixed concentration of immobilized GST-bound WT or mutant ISD11 proteins with increasing concentrations of NFS1 at 10 οC for 25 min. A lower temperature was used to prevent self-aggregation of the NFS1 protein. Immobilized GST alone was taken as a negative control. As indicated in Fig. 3, the GST pulldown analysis showed a robust interaction of NFS1 with WT ISD11. However, the lethal mutant proteins, ISD11F40A (Fig. 3, A and B), ISD11I72A (Fig. 3, I and J), and ISD11E84A (Fig. 3, O and P) showed dramatic reduction in their interaction with NFS1. At the same time, the ts mutant proteins, ISD11R68A (Fig. 3, E and F), ISD11Q69A (Fig. 3, G and H), ISD11Y76A (Fig. 3, K and L), and ISD11L81A (Fig. 3, M and N) showed a moderately reduced interaction with NFS1. Strikingly, on the other hand, the ISD11L63A protein, which exhibits the ts phenotype only at 37 οC, showed a trivial decrease in the interaction as compared with WT ISD11 (Fig. 3, C and D). To further investigate these findings, the subcomplex formation was further assessed in the mitochondrial lysates for ts mutants (Fig. 3Q). To determine this, the GST-bound NFS1 was incubated with the mitochondrial lysate prepared after inducing the phenotype and subjected to pulldown analysis. The GST-bound protein beads were washed and separated on SDS-PAGE followed by immunodetection using the respective antibodies. As indicated in the Fig. 3R, wild type ISD11 showed a robust interaction, whereas the ISD11L63A mutant showed a minor decrease and other ts mutants showed reduced subcomplex formation with NFS1, which is consistent with in vitro GST pulldown analysis using purified proteins. In summary, these findings provide conclusive evidence highlighting a linear correlation with the severity of the growth phenotype with their relative affinity to form a stable subcomplex with NFS1, which is essential in vivo for the Fe-S cluster biogenesis. Additionally, we have also analyzed NFS1-ISD11 subcomplex formation in the COXPD19-associated R68L ISD11 mutant. Disease variant R68L ISD11 was overexpressed in the mammalian system (HeLa cells), and the subcomplex formation ability was examined using the mitochondrial lysate. Notably, our results demonstrate a reduction in the subcomplex formation between R68L ISD11 with NFS1 as compared with WT ISD11, which supports yeast mitochondrial pulldown analysis (Fig. 3S).
FIGURE 3.
Interaction analysis of NFS1 with ISD11 mutants. A-P, glutathione-Sepharose bound WT GST-ISD11 (3 μm) or mutant GST-ISD11 (3 μm) was incubated with increasing concentrations of purified NFS1, as indicated. The unbound proteins were washed and analyzed by SDS-PAGE followed by Coomassie staining. GST alone (3 μm) was used as a negative control, and 50% input of NFS1 was taken as a loading control. The band intensities of bound NFS1 were analyzed by densitometry using ImageJ software. Interaction of NFS1 with WT ISD11 and F40A ISD11 (A and B), L63A ISD11 (C and D), R68A ISD11 (E and F), Q69A ISD11 (G and H), I72A ISD11 (I and J), Y76A ISD11 (K and L), L81A ISD11 (M and N), and E84A ISD11 (O and P) are represented. Q and R, interaction analysis of NFS1 with ts mutants in mitochondrial lysates. Mitochondria isolated from WT and ts mutant strains (Q69A, R68A, L63A, L81A, and Y76A) were subjected to lysis and the supernatant fractions were incubated with GST-NFS1 (2 μm). Unbound proteins were washed, and FLAG-tagged ISD11 was detected by Western blotting using the anti-FLAG antibodies. GST alone was taken as negative control and ISD11 as loading control (Q). The band intensities shown in Q were analyzed by densitometry using ImageJ software (R). S, mitochondria isolated from WT or R68L ISD11 expressing HeLa cells were lysed, and the supernatant fractions were incubated with GST-NFS1 (2 μm). Unbound proteins were removed by washing, followed by SDS-PAGE and immunostaining using an anti-FLAG antibody to detect the FLAG-tagged ISD11. GST alone was taken as negative control and ISD11 as loading control. Data are represented as mean ± S.E. The values were obtained from three independent experiments (n = 3). p value of <0.05 was defined as significant, and asterisks are used to denote significance, where: *, p < 0.05; **, p < 0.01; ***, p < 0.001. A.U., arbitrary unit.
ISD11 Interaction Is Essential for the Prevention of NFS1 Aggregation
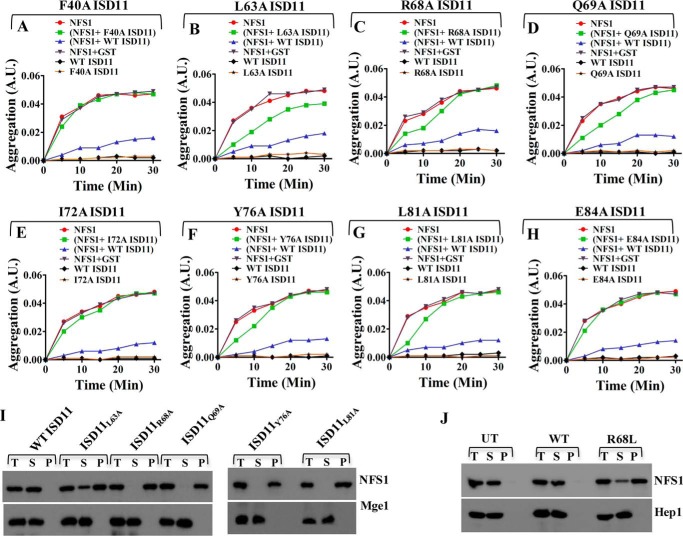
Previous reports have shown that yeast Isd11 forms a stable subcomplex with the sulfur donor Nfs1 and prevents it from self-aggregation, in vivo (20, 21). To firmly establish a role of ISD11 in the prevention of NFS1 aggregation, we attempted in vitro aggregation analysis of NFS1 in the presence and absence of ISD11 by a light scattering method. We observed that, incubating NFS1 with the GST-tagged WT ISD11 protein maintained NFS1 in the soluble state, whereas NFS1 alone formed aggregates at 37 °C, in vitro. At the same time, the negative control GST-ISD11 did not show aggregation under the same condition. Furthermore, the GST tag alone did not influence in the prevention of aggregation of NFS1 suggesting that subcomplex formation is essential for the stability of NFS1 (Fig. 4). Interestingly, none of the ISD11 mutant proteins could protect the NFS1 from aggregation robustly like the WT, at the same temperature. Lethal mutants, ISD11F40A (Fig. 4A), ISD11I72A (Fig. 4E), and ISD11E84A (Fig. 4H) exhibit a minimal ability to prevent NFS1 aggregation. Notably, ISD11L63A, a comparatively weaker mutant in terms of its ability to form a subcomplex with NFS1 showed a partial suppression of aggregation (Fig. 4B). Whereas, other ts mutants ISD11R68A (Fig. 4C), ISD11Q69A (Fig. 4D), ISD11Y76A (Fig. 4F), and ISD11L81A (Fig. 4G) were partially prevented by NFS1 aggregation only at the initial incubation time but failed to protect for a longer period. The mutant ISD11 proteins alone as negative controls, in all the aforementioned experiments, did not show any aggregation under the same experimental conditions.
FIGURE 4.
Effect of ISD11 mutations in preventing aggregation of NFS1. A-H, aggregation of NFS1 was monitoring by measuring absorbance of NFS1 (1 μm) at different time intervals time at 320 nm at 37 °C. Red lines indicate aggregation of NFS1 (1 μm) alone. Aggregation of WT GST-ISD11 (2 μm) alone is indicated in black lines. Aggregation of NFS1 (1 μm) in presence of WT GST-ISD11 (2 μm) showed in purple lines. Gray lines indicate aggregation of NFS1 in the presence of 1 μm GST as a control. Green lines depict aggregation of NFS1 (1 μm) in presence of 2 μm of F40A GST-ISD11 (A) or L63A GST-ISD11 (B) or R68A GST-ISD11 (C) or Q69A GST-ISD11 (D) or I72A GST-ISD11 (E) or Y76A GST-ISD11 (F) or L81A GST-ISD11 (G) or E84A GST-ISD11 (H). Yellow lines are used to indicate aggregation of mutants alone as controls. I, in vivo aggregation analysis of NFS1 in WT and ts mutants (Q69A, R68A, L63A, L81A, and Y76A). Mitochondria isolated from WT and ts mutants were subjected to heat shock at 37 °C, followed by lysis and separation of pellet and supernatant fractions. Total (T), supernatant (S), and pellet (P) fractions were immunoprobed using NFS1-specific antibody. Mitochondrial matrix protein Mge1 was taken as soluble protein control. J, to analyze aggregation of NFS1 in HeLa cells, a similar experiment was performed in mitochondria isolated from UT (untransfected) HeLa cells and HeLa cells expressing WT and R68L ISD11. Total (T), supernatant (S), and pellet (P) fractions were probed for NFS1 through Western blotting. Mitochondrial matrix protein Hep1 was taken as soluble protein control. A.U., absorbance unit.
To further investigate these findings in vivo, mitochondria from ts mutants were purified and incubated at 37 οC for 20 min and subjected to fractionation analysis. Samples were clarified by ultracentrifugation into insoluble fractions from the supernatant and analyzed on SDS-PAGE followed by immunodetection using protein-specific antibodies. Mitochondrial matrix protein Mge1 was used as a soluble control. As indicated in Fig. 4I, NFS1 was separated completely in the supernatant fraction like Mge1 due to stable interaction with ISD11 in the WT mitochondria. In contrast, in all mutant mitochondria, the majority of NFS1 remained in the pellet fraction, except for the ISD11L63A where a small amount of NFS1 was associated into the supernatant, thus supporting in vitro aggregation experiments. Similarly, we tested for the ability of the R68L mutant in maintaining the stability of NFS1 in human mitochondria. Intriguingly, the majority of the NFS1 was found associated with the pellet fraction, which is consistent with observations made in yeast mitochondria. In the case of untransfected (UT) and WT ISD11 expressing HeLa cell mitochondria, NFS1 remained in the soluble fraction, suggesting that R68L ISD11 fails to prevent NFS1 aggregation. Mitochondrial matrix Hep1 protein was used as a marker for the soluble protein (Fig. 4J). In conclusion, these results further confirm the role of key residues that are important for interaction of ISD11 with NFS1 to prevent its aggregation.
Defective Interaction between ISD11 and NFS1 Leads to Compromised Fe-S Cluster Biogenesis and Reduced Mitochondrial Respiration
Although ISD11 is not critical for the cysteine desulfurase activity of NFS1, however, it is essential for stabilizing NFS1 in eukaryotic systems. To delineate how reduced subcomplex formation between ISD11 and NFS1 influences the Fe-S cluster biogenesis in vivo, we measured ETC complex II and aconitase activity. Both yeast Isd11 and human ISD11 showed comparable complex II and aconitase activity. On the other hand, ISD11L63A mutant exhibits 25–30% reduction in complex II and aconitase activity consistent with its mild growth phenotype. The ts mutants ISD11R68A and ISD11Q69A showed about 55–60% reduction in the complex II and aconitase activity. Interestingly, ISD11Y76A and ISD11L81A have shown very drastic defects in the growth as well as subcomplex formation with NFS1, and exhibit maximum reduction of 70–75% complex II and aconitase activity (Fig. 5, A and B). The levels of aconitase and Rieske Fe-S proteins were further assessed by Western blotting using mitochondrial lysate. A significant reduction in Fe-S cluster containing proteins was observed for all ts mutants, especially ISD11Y76A and ISD11L81A showed a greater decrease in aconitase levels (Fig. 5C). Furthermore, to investigate the influence of reduced interaction of R68L ISD11 with NFS1 in the Fe-S cluster biogenesis, we measured activity of the Fe-S cluster containing enzymes in human mitochondria expressing the R68L ISD11 mutant. The levels of aconitase and Rieske Fe-S proteins were declined in the R68L ISD11 mitochondrial lysate as compared with WT ISD11 (Fig. 5E). Moreover, we observed reduction in ETC complex I activity performed in the presence and absence of the rotenone inhibitor (Fig. 5F). Activity of ETC complex II and aconitase in R68L mutant mitochondria were also decreased as compared with WT HeLa cells (Fig. 5, G and H).
FIGURE 5.
Measurement of Fe-S cluster containing enzymes activity and respiration efficiency in ts mutants and in R68L HeLa cells. A represents activity of ETC complex II in WT and ts mutants (Q69A, R68A, L63A, L81A, and Y76A). B, representation of enzymatic activity of aconitase in mitochondria isolated from WT and ts mutant strains. C, mitochondrial levels of Fe-S cluster proteins aconitase (Aco1) and Rieske Fe-S protein in WT ISD11 and ts mutants were analyzed by Western blotting using indicated specific antibodies. Mitochondrial matrix protein Mge1 was taken as loading control. D, the level of mitochondrial ATP was measured using the mitochondrial ToxGlo assay kit in WT and ts mutant mitochondria. E, the expression level of mitochondrial aconitase (Aco2) and Rieske Fe-S protein in WT and R68L HeLa cells was analyzed by SDS-PAGE and immunodecoration with antibodies against the indicated proteins. Mitochondrial protein Tim23 was taken as loading control. F, activity of ETC complex I was measured in isolated mitochondria obtained from untransfected (UT) or WT and R68L expressing HeLa cells. The reactions were performed in the presence or absence of complex I inhibitor rotenone to check the specificity. G, representation of ETC complex II activity in the presence of an inhibitor of complex I, III, and IV in HeLa cell mitochondria. H, measurement of activity of TCA cycle enzyme, aconitase in the mitochondrial lysate from HeLa cells. I, mitochondrial ATP was measured in HeLa cells using the Mitochondrial ToxGlo assay kit. Data are represented as mean ± S.E. The values were obtained from three independent experiments (n = 3). p value of <0.05 was defined as significant, and asterisks are used to denote significance, where: *, p < 0.05; **, p < 0.01; ***, p < 0.001. A.U., arbitrary unit.
To determine the overall mitochondrial respiration status in ISD11 ts mutants, the total ATP content was measured in the purified mitochondria. As anticipated, both yeast Isd11 and human WT ISD11 controls showed equivalent amounts of mitochondrial ATP content. On the other hand, ISD11L63A, ISD11R68A, and ISD11Q69A mutants exhibit around 40% reduction in the total ATP levels. At the same time, ISD11Y76A and ISD11L81A mutants showed greater than a 50% decrease in the ATP levels thereby highlighting the significance of subcomplex formation between NFS1 and ISD11 in Fe-S cluster synthesis and for the constitutive mitochondrial functions (Fig. 5D). In concurrence with observations made in the yeast system, HeLa cells expressing the R68L ISD11 mutant demonstrated reduced mitochondrial ATP production as compared with WT controls, suggesting diminished respiration efficiency (Fig. 5I).
Compromised Fe-S Cluster Biogenesis in COXPD19 Disease Variant, R68L ISD11 leads to Iron Accumulation, Oxidative Stress, and Mitochondrial Dysfunction
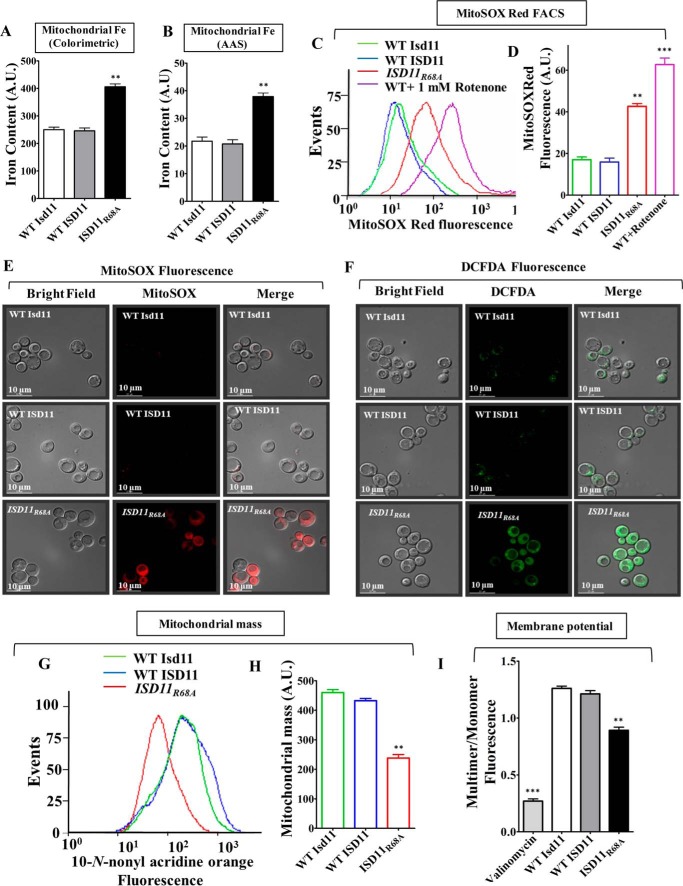
To delineate the disease progression mechanism of COXPD19, the ISD11R68A yeast strain was utilized for in vivo analysis, which closely mimics the genotype of COXPD19 patients. Based on our biochemical analysis it is evident that ISD11R68A showed defective Fe-S cluster biogenesis and reduced mitochondrial respiration due to compromised interaction with NFS1. To establish a direct correlation between observed patient phenotypes and cellular etiology, iron homeostasis was tested in the ISD11R68A mutant. The accumulation of mitochondrial iron levels was analyzed by iron-specific colorimetric assay and atomic absorption spectroscopy (AAS) (24, 29, 30). Mitochondrial lysates from yeast cells expressing ISD11R68A showed a substantially higher iron content as compared WT controls (Fig. 6, A and B). Enhanced iron load as a result of improper Fe-S cluster biogenesis generates mitochondrial oxidative stress (34, 35). To test the hypothesis, we measured superoxide levels using MitoSOX Red dye. This dye is a fluorogenic dye specifically targeted to mitochondria of living cells, where it reacts with superoxide radicalsand produces red fluorescence (36). Yeast strains expressing ISD11R68A showed a significant increment in the MitoSOX Red fluorescence measured by flow cytometric analysis as compared with WT controls suggesting an enhancement in the levels of mitochondrial superoxide (Fig. 6, C and D). The elevation of mitochondrial superoxide level in mutant cells was further verified by fluorescence microscopy. As indicated in Fig. 6E, ISD11R68A expressing yeast cells display robust increments in the MitoSOX Red fluorescence as compared with WT yeast strains, thus supporting the flow cytometric data. Besides, the ISD11R68A mutant also exhibit elevation in total cellular peroxide levels as measured by H2DCFDA dye, which gives green fluorescence upon reacting with the peroxides. Fig. 6F represents the microscopic imaging analysis wherein ISD11R68A yeast cells showed robust fluorescence compared with WT controls as an indication of enhanced ROS.
FIGURE 6.
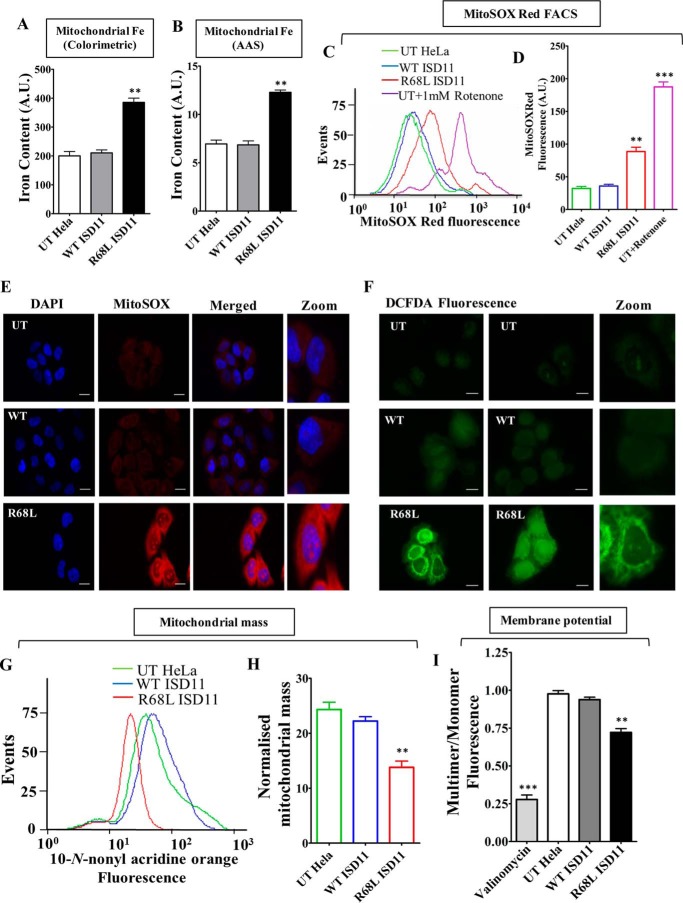
Analysis of mitochondrial iron, ROS levels and mitochondrial function in COXPD19 related ISD11R68A mutant. A and B, representation of iron levels in yeast mitochondria expressing WT Isd11 or WT ISD11 or ISD11R68A, measured by iron-specific colorimetric assay (A) and AAS (B). C and D, mitochondrial superoxide levels in WT and ISD11R68A yeast strains were estimated using MitoSOX Red dye by flow cytometry and represented in the overlaying histograms (C) and mean fluorescence intensity obtained by flow cytometry was quantitated (D). WT Isd11 cells treated with 1 mm rotenone was utilized as positive control. E, yeast strains were treated with MitoSOX Red dye (red fluorescence) and subjected to microscopic analysis using DeltaVision Fluorescence Microscope (×100 objective). Left column, bright field; middle column, MitoSOX red fluorescence; and right column, merged. F, overall cellular peroxide levels were measured by staining using H2DCFDA dye (green fluorescence), followed by fluorescence microscopy analysis using Delta Vision Fluorescence Microscope (×100 objective). Left, bright field; middle, H2DCFDA fluorescence, and right columns, merged. G and H, mitochondrial mass in yeast strains expressing WT Isd11 or WT ISD11 or ISD11R68A was assessed using NAO dye. The fluorescence was measured through flow cytometry and represented as histogram (G) or mean fluorescence intensity (MFI) obtained in flow cytomety was quantitated and represented as bar diagram (H). I, JC-1 staining was used for the estimation of mitochondrial membrane potential in yeasts strains. An emission scan from the 500 to 620 nm wavelength was performed in JC-1-stained yeast cells and the multimer (590 nm) to monomer (530 nm) ratio was quantified and represented. Mitochondria treated with valinomycin were used as a positive control. Data represented as mean ± S.E. The values were obtained from three independent experiments (n = 3). p value of <0.05 was defined as significant, and asterisks are used to denote significance, where: *, p < 0.05; **, p < 0.01; ***, p < 0.001. Scale bar, 10 μm. A.U., arbitrary unit.
To test whether the reduced respiration rate in ISD11R68A is due to a consequence of altered mitochondrial health, we measured the total mass and levels of functional mitochondria. The overall mitochondrial mass in WT and mutant cells was measured using a fluorescent dye, 10-N-nonyl acridine orange (NAO). NAO is a cell permeable dye that binds to cardiolipin of the mitochondrial membrane and upon excitation at 495 nm, fluoresces at an emission maximum of 519 nm, thus providing an estimate of the total mitochondrial mass present inside the cell (37). Notably, our results showed a significant decrease in mitochondrial mass in ISD11R68A mutant as an indicated shift in the peak toward loss of fluorescence as compared with human WT ISD11 and yeast WT Isd11 controls (Fig. 6G, compare peaks represented in red with green and blue). Upon quantification of the mean fluorescence intensity, the ISD11R68A mutant showed a 1.7-fold reduced mitochondrial mass than WT cells (Fig. 6H). The total functional mitochondria were estimated using membrane polarity sensitive cationic dye, JC-1. In the presence of an intact mitochondrial membrane potential JC-1 diffuses inside mitochondrial matrix and forms J-aggregates that give a red florescence with an emission maximum of 590 nm. However, upon loss of the mitochondrial membrane potential, JC-1 remains outside the organelle as monomer and fluoresces in the green region with an emission maximum of 530 nm. The ratio of red fluorescence (multimer) to green (monomer) gives an estimation of the inner mitochondrial membrane potential (38, 39). Upon quantification of red (590 nm) to green (530 nm) fluorescence ratio, about 25–30% depolarized mitochondria was accumulated in ISD11R68A as compared with WT controls (Fig. 6I). Together, these results suggest that Arg-68 of ISD11 plays an important role in the maintenance of normal mitochondrial function and cellular respiration.
To directly validate the observations obtained in the yeast model system, the disease variant R68L ISD11 mutant was overexpressed in HeLa cells and subjected to functional analysis. The R68L mutant mitochondria showed iron accumulation as estimated by colorimetric based method (Fig. 7A) and AAS analysis (Fig. 7B). As a result, we observed enhanced superoxide levels in mutant mitochondria as compared with untransfected and WT ISD11 as determined by MitoSOX Red dye using flow cytometric analysis (Fig. 7, C and D). The increment in the mitochondrial superoxide level in the R68L mutant was further validated by fluorescence microscopy using MitoSOX Red dye staining. Supporting our flow cytometric data and previous yeast-based observations, we monitored elevated MitoSOX Red fluorescence in the R68L mutant as compared with controls (Fig. 7E). Moreover, the R68L mutant also showed elevated cellular peroxide levels when analyzed by fluorescence microscopy using peroxide-specific H2DCFDA dye (Fig. 7F). To uncover whether increased mitochondrial iron and ROS levels impose adverse effects on mitochondrial functionality, we tested for the maintenance of mitochondrial mass and inner membrane potential of the R68L variant. The disease variant exhibits a decline in total mitochondrial mass (Fig. 7, G and H) and membrane potential (Fig. 7I). As a consequence, reduced mitochondrial ATP production was observed in R68L mutant mitochondria, which suggests diminished respiration efficiency. In summary, our results conclusively demonstrate in yeast and mammalian cell lines the importance Arg-68 of ISD11 for subcomplex formation with NFS1 and its possible pathological implications in the maintenance of normal mitochondrial function and cellular respiration in COXPD19 patients.
FIGURE 7.
Estimation of mitochondrial iron content, ROS levels, and evaluation of mitochondrial functionality in R68L ISD11 mutant. A and B, mitochondrial iron level was estimated in untranslated (UT), WT, and R68L HeLa cells using iron specific colorimetric assay (A) and AAS (B). C, flow cytometry analysis to measure the mitochondrial superoxide level in HeLa cells using MitoSOX Red staining. D, representation of mean fluorescence intensity of MitoSOX Red fluorescence in UT, WT, and R68L HeLa cells. The UT cells treated with 1 mm rotenone were used as a positive control. E, untransfected HeLa cells (UT) or cells expressing WT or R68L mutant proteins were stained with MitoSOX Red dye. The fluorescence was analyzed using Zeiss Apotome fluorescence microscope using ×63 objective lens (scale bar: 10 μm). F, to measure the overall cellular ROS level, fluorescence images of HeLa cells stained with H2DCFDA dye (green fluorescence) were obtained from Zeiss Apotome fluorescence microscope using ×63 objective lens (scale bar: 10 μm). G and H, mitochondrial mass was estimated by staining HeLa cells with NAO dye. The total MFI values obtained from flow cytometric analyses of NAO staining are presented as histogram (G) and mean fluorescence values obtained in the flow cytometry were quantitated (H). I, mitochondria isolated from HeLa cells was stained with the JC-1 dye to measure the mitochondrial membrane potential. The emission spectrum was scanned from wavelength 500 to 620 nm, and the ratio between JC-1 multimer (590 nm) and JC-1 monomer (530 nm) was represented. Data are represented as mean ± S.E. The values were obtained from three independent experiments (n = 3). p value of <0.05 was defined as significant, and asterisks are used to denote significance, where: *, p < 0.05; **, p < 0.01; ***, p < 0.001. A.U., arbitrary unit.
Discussion
In the present study, we have delineated the importance of the ISD11 protein as a part of the sulfur-transfer protein complex, NFS1-ISD11 in Fe-S cluster biogenesis pathway. Our results indicate that the ISD11 protein interacts with the cysteine desulfurase protein NFS1 and prevents its self-aggregation, thus assisting a functional Fe-S cluster biogenesis process in the mitochondrial compartment. Importantly, this is a first report that maps the crucial residues of ISD11 protein that involves in interaction with NFS1. Furthermore, this report delivers mechanistic insights on the possible explanation of COXPD19 development and progression as a result of R68L mutation in ISD11 protein.
Previous studies have shown that deletion of Isd11 in yeast or silencing of ISD11 in mammalian cells leads to impairment of Fe-S cluster biogenesis and reduction in Fe-S cluster containing enzyme activity (20, 22). Isd11/ISD11 is an essential protein but its actual role in Fe-S cluster biogenesis by interacting with the sulfur donor Nfs1 is not clear. Importantly, the amino acid residues are critical for Isd11/ISD11 function and are not well characterized. Our genetic analysis maps 8 prominent residues throughout the protein critical for ISD11 function. Intriguingly, one residue (Phe-40) belongs to the predicted N-terminal helix 1 region, whereas the remaining 7 were from helix 3 and the C-terminal region (Leu-63, Arg-68, Gln-69, Ile-72, Tyr-76, Leu-81, and Glu-84), thus highlighting important segments of ISD11 for its in vivo function. Based on the severity of growth phenotype and associated biochemical defects, the ISD11 mutants demonstrate a linear correlation of ISD11 function with its ability to form a stable subcomplex with NFS1, thus firmly establishing the indispensable nature of their interaction for a functional Fe-S cluster biogenesis. Interestingly, these observations indicate that ISD11 stabilizes NFS1, predominantly through hydrophobic interaction, considering the nature of the critical amino acid residues. Most of the essential residues mapped in ISD11, which are required for a stable interaction with NFS1 are highly conserved across phylogeny, thereby indicating the critical nature of these interacting residues across genera.
Based on our results, it is reasonable to believe that ISD11 stabilizes NFS1 through physical interaction, probably shielding the exposed hydrophobic surface. So, the mutual co-dependence of these two proteins might be essential for the initial rate of scaffolding reaction on Isu1/ISCU during Fe-S cluster biogenesis. In line with this argument, in yeast it has been reported that, an inadequate amount of Isd11 results in the steady-state reduction of mitochondrial levels of Nfs1 (20, 21). A similar result was obtained for the ts ISD11 mutants as well as in the COXPD19 disease variant (R68A), where Nfs1 levels were reduced as compared with the WT. A plausible reason for the decrease in the Nfs1 levels could be attributed to its relatively lower stability due to the compromised ISD11 interaction, causing aggregation and degradation by proteases in the mitochondrial compartment. Such regulatory mechanisms have been demonstrated in yeast Isu1 mutants, which are defective in the Nfs1 interaction and undergo degradation by Pim1 protease, therefore, modulating the protein levels (18). In addition to reduced Nfs1 expression, diminished levels of Isu1 was also observed in mutant strains, especially with a stronger ts phenotype and R68A disease variant, implying the paramount nature of these residues for maintaining Isu1 levels as well. However, the steady-state level of the J-protein Jac1, which is involved in the cluster transfer process, was unaltered in mutants, suggesting that impairment of ISD11 function affects the early steps of Fe-S cluster assembly not the late transfer process.
Sulfur donation is an indispensable and rate-limiting early event during the Fe-S assembly process (6). Because ISD11 assists the function of sulfur transfer protein NFS1 by preventing its aggregation, it is reasonable to predict that loss of ISD11 function will influence the rate of Fe-S cluster assembly reaction by impeding the sulfur transfer process. Consistent with this argument, compromised function of ISD11 mutants demonstrates decreased activity of the Fe-S cluster containing enzymes and reduction in their expression levels. An inefficient Fe-S cluster assembly process overall deregulates the maturation of the Fe-S cluster containing proteins, including crucial ETC multienzyme complexes. As a consequence, defective ETC enzymes lead to reduce pumping of protons across the mitochondrial inner membrane, thus decreasing the effective proton motive force required for synthesis of ATP inside mitochondria. Reinforcing the idea, the ISD11 ts mutants and COXPD19-associated disease variant (R68A/R68L) showed reduced mitochondrial membrane potential and diminished respiratory activity with lower ATP levels in the mitochondria. Based on these findings, it is reasonable to believe that reduced respiratory activity associated with the R68L ISD11 mutation in COXPD19 patients may be responsible for the manifestation of disease symptoms, especially the most common symptom of respiratory distress (23).
The inerrant mitochondrial Fe-S cluster biogenesis process is an effective mode of maintenance of cellular iron homeostasis and its toxicity (40). Defects in this process lead to the elevated mitochondrial iron load. For instance, in case of the neurodegenerative disorder Friedreich's ataxia or exercise intolerance disorder ISCU myopathy where the rate of Fe-S clusters biogenesis process is impaired, there is an increased iron level in the mitochondria (24, 41, 42). In agreement with this view, our results highlight that the reduction in Fe-S cluster synthesis in the R68A/R68L mutant leads to accumulation of iron in yeast and human mitochondria. Notably, we also observed a significant enhancement of ROS levels in the R68A/R68L mutant mitochondria in both model systems. This is probably due to the loss of ETC complex activity, causing leakage of free electrons during electron transport and producing superoxide radicals (43–45). Additionally, enrichment of the reduced form of iron in the mitochondria leads to production of superoxide radicals and highly toxic hydroxyl radicals through Fenton's reaction (46, 47). These superoxide and hydroxyl radicals are detrimental to the cellular components as they inflict further damage to the pre-existing Fe-S clusters present in the ETC and other enzyme complexes. This initiates a vicious cycle of ROS production causing oxidative stress, which has been implicated in several pathological conditions (48, 49). Moreover, acute oxidative stress also damages the mitochondrial inner membrane potential leading to membrane depolarization, thereby contributing toward reduced respiration and compromised mitochondrial quality control. Based on our experimental findings from both the yeast and mammalian systems, we believe that free radical-mediated oxidative damage plays an additive effect toward the manifestation of complex disease symptoms in COXPD19 patients.
In conclusion, this report indicates that ISD11 plays a significant role to stabilize NFS1. However, E. coli does not have an ortholog of Isd11 for IscS function, probably attributing to its better intrinsic stability. On the other hand, the reduced intrinsic stability associated with eukaryotic Nfs1/NFS1 may demand an additional interacting partner such as Isd11/ISD11 to attain maximum stability through physical association. This ensures that the steady-state level is maintained in intricate physiological conditions. It will be interesting to have the structure of ISD11 bound to NFS1, as it will be helpful in co-relating the functional aspects to structural details of the protein. This might allow further elucidation of the macromolecular defects associated with the progression of COXPD19 disease as a result of R68L mutation.
Author Contributions
P. P. S., P. D. S., and P. K. S. K. designed the study and analyzed the data. P. P. S. performed the experiments and prepared all the figures. S. S. and D. S. performed and analyzed the experiment shown in Fig. 7, E and F. P. P. S., P. D. S., S. S., and D. S wrote the manuscript. All authors reviewed the results and approved the final version of manuscript.
Acknowledgments
We thank Dr. Nicholas Pfanner for providing the Δisd11 yeast strain and antibody against aconitase and Rieske Fe-S prteins. We thank Dr. Roland Lill and Dr. Wing-Hang Tong for anti-yeast Nfs1 and anti-human NFS1 antibodies, respectively. We also thank the Flow Cytometry Facility of the Indian Institute of Science, Bangalore, for FACS experiments and the Solid State and Structural Chemistry Unit (SSCU) of the Indian Institute of Science, Bangalore, for AAS analysis.
This work was supported in part by Council of Scientific and Industrial Research Grant CSIR 37(1534)/12/EMR-II, India (to P. D. S.), and Swarnajayanthi Fellowship from Department of Science and Technology (DST), Ministry of Science and Technology, India (to P. D. S.). The authors declare that they have no conflicts of interest with the contents of this article.
- COXPD19
- combined oxidative phosphorylation deficiency 19
- H2DCFDA
- 2′,7′-dichlorodihydrofluorescein diacetate
- 5-FOA
- 5-fluoroorotic acid
- AAS
- atomic absorption spectroscopy
- NAO
- 10-N-nonyl acridine orange
- Ni-NTA
- nickel-nitrilotriacetic acid
- ROS
- reactive oxygen species.
References
- 1.Craig E. A., and Marszalek J. (2002) A specialized mitochondrial molecular chaperone system: a role in formation of Fe/S centers. Cell Mol. Life Sci. 59, 1658–1665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Craig E. A., Voisine C., and Schilke B. (1999) Mitochondrial iron metabolism in the yeast Saccharomyces cerevisiae. Biol. Chem. 380, 1167–1173 [DOI] [PubMed] [Google Scholar]
- 3.Meyer J. (2008) Iron-sulfur protein folds, iron-sulfur chemistry, and evolution. J. Biol. Inorg. Chem. 13, 157–170 [DOI] [PubMed] [Google Scholar]
- 4.Lill R., Dutkiewicz R., Elsässer H. P., Hausmann A., Netz D. J., Pierik A. J., Stehling O., Urzica E., and Mühlenhoff U. (2006) Mechanisms of iron-sulfur protein maturation in mitochondria, cytosol and nucleus of eukaryotes. Biochim. Biophys. Acta 1763, 652–667 [DOI] [PubMed] [Google Scholar]
- 5.Beinert H., Holm R. H., and Münck E. (1997) Iron-sulfur clusters: nature's modular, multipurpose structures. Science 277, 653–659 [DOI] [PubMed] [Google Scholar]
- 6.Lill R. (2009) Function and biogenesis of iron-sulphur proteins. Nature 460, 831–838 [DOI] [PubMed] [Google Scholar]
- 7.Maio N., and Rouault T. A. (2015) Iron-sulfur cluster biogenesis in mammalian cells: new insights into the molecular mechanisms of cluster delivery. Biochim. Biophys. Acta 1853, 1493–1512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stehling O., Wilbrecht C., and Lill R. (2014) Mitochondrial iron-sulfur protein biogenesis and human disease. Biochimie 100, 61–77 [DOI] [PubMed] [Google Scholar]
- 9.Land T., and Rouault T. A. (1998) Targeting of a human iron-sulfur cluster assembly enzyme, nifs, to different subcellular compartments is regulated through alternative AUG utilization. Mol. Cell 2, 807–815 [DOI] [PubMed] [Google Scholar]
- 10.Biederbick A., Stehling O., Rösser R., Niggemeyer B., Nakai Y., Elsässer H. P., and Lill R. (2006) Role of human mitochondrial Nfs1 in cytosolic iron-sulfur protein biogenesis and iron regulation. Mol. Cell. Biol. 26, 5675–5687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mühlenhoff U., Balk J., Richhardt N., Kaiser J. T., Sipos K., Kispal G., and Lill R. (2004) Functional characterization of the eukaryotic cysteine desulfurase Nfs1p from Saccharomyces cerevisiae. J. Biol. Chem. 279, 36906–36915 [DOI] [PubMed] [Google Scholar]
- 12.Zheng L., White R. H., Cash V. L., and Dean D. R. (1994) Mechanism for the desulfurization of l-cysteine catalyzed by the nifS gene product. Biochemistry 33, 4714–4720 [DOI] [PubMed] [Google Scholar]
- 13.Cupp-Vickery J. R., Urbina H., and Vickery L. E. (2003) Crystal structure of IscS, a cysteine desulfurase from Escherichia coli. J. Mol. Biol. 330, 1049–1059 [DOI] [PubMed] [Google Scholar]
- 14.Zheng L., and Dean D. R. (1994) Catalytic formation of a nitrogenase iron-sulfur cluster. J. Biol. Chem. 269, 18723–18726 [PubMed] [Google Scholar]
- 15.Kaiser J. T., Clausen T., Bourenkow G. P., Bartunik H. D., Steinbacher S., and Huber R. (2000) Crystal structure of a NifS-like protein from Thermotoga maritima: implications for iron sulphur cluster assembly. J. Mol. Biol. 297, 451–464 [DOI] [PubMed] [Google Scholar]
- 16.Marinoni E. N., de Oliveira J. S., Nicolet Y., Raulfs E. C., Amara P., Dean D. R., and Fontecilla-Camps J. C. (2012) (IscS-IscU)2 complex structures provide insights into Fe2S2 biogenesis and transfer. Angew. Chem. Int. Ed. Engl. 51, 5439–5442 [DOI] [PubMed] [Google Scholar]
- 17.Kato S., Mihara H., Kurihara T., Takahashi Y., Tokumoto U., Yoshimura T., and Esaki N. (2002) Cys-328 of IscS and Cys-63 of IscU are the sites of disulfide bridge formation in a covalently bound IscS/IscU complex: implications for the mechanism of iron-sulfur cluster assembly. Proc. Natl. Acad. Sci. U.S.A. 99, 5948–5952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Song J. Y., Marszalek J., and Craig E. A. (2012) Cysteine desulfurase Nfs1 and Pim1 protease control levels of Isu, the Fe-S cluster biogenesis scaffold. Proc. Natl. Acad. Sci. U.S.A. 109, 10370–10375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Majewska J., Ciesielski S. J., Schilke B., Kominek J., Blenska A., Delewski W., Song J. Y., Marszalek J., Craig E. A., and Dutkiewicz R. (2013) Binding of the chaperone Jac1 protein and cysteine desulfurase Nfs1 to the iron-sulfur cluster scaffold Isu protein is mutually exclusive. J. Biol. Chem. 288, 29134–29142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Adam A. C., Bornhövd C., Prokisch H., Neupert W., and Hell K. (2006) The Nfs1 interacting protein Isd11 has an essential role in Fe/S cluster biogenesis in mitochondria. EMBO J. 25, 174–183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wiedemann N., Urzica E., Guiard B., Müller H., Lohaus C., Meyer H. E., Ryan M. T., Meisinger C., Mühlenhoff U., Lill R., and Pfanner N. (2006) Essential role of Isd11 in mitochondrial iron-sulfur cluster synthesis on Isu scaffold proteins. EMBO J. 25, 184–195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shi Y., Ghosh M. C., Tong W. H., and Rouault T. A. (2009) Human ISD11 is essential for both iron-sulfur cluster assembly and maintenance of normal cellular iron homeostasis. Hum. Mol. Genet. 18, 3014–3025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lim S. C., Friemel M., Marum J. E., Tucker E. J., Bruno D. L., Riley L. G., Christodoulou J., Kirk E. P., Boneh A., DeGennaro C. M., Springer M., Mootha V. K., Rouault T. A., Leimkühler S., Thorburn D. R., and Compton A. G. (2013) Mutations in LYRM4, encoding iron-sulfur cluster biogenesis factor ISD11, cause deficiency of multiple respiratory chain complexes. Hum. Mol. Genet. 22, 4460–4473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Saha P. P., Kumar S. K., Srivastava S., Sinha D., Pareek G., and D'Silva P. (2014) The presence of multiple cellular defects associated with a novel G50E iron-sulfur cluster scaffold protein (ISCU) mutation leads to development of mitochondrial myopathy. J. Biol. Chem. 289, 10359–10377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Goswami A. V., Samaddar M., Sinha D., Purushotham J., and D'Silva P. (2012) Enhanced J-protein interaction and compromised protein stability of mtHsp70 variants lead to mitochondrial dysfunction in Parkinson's disease. Hum. Mol. Genet. 21, 3317–3332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Srivastava S., Sinha D., Saha P. P., Marthala H., and D'Silva P. (2014) Magmas functions as a ROS regulator and provides cytoprotection against oxidative stress-mediated damages. Cell Death Dis. 5, e1394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Spinazzi M., Casarin A., Pertegato V., Salviati L., and Angelini C. (2012) Assessment of mitochondrial respiratory chain enzymatic activities on tissues and cultured cells. Nat. Protoc. 7, 1235–1246 [DOI] [PubMed] [Google Scholar]
- 28.Pierik A. J., Netz D. J., and Lill R. (2009) Analysis of iron-sulfur protein maturation in eukaryotes. Nat. Protoc. 4, 753–766 [DOI] [PubMed] [Google Scholar]
- 29.Garland S. A., Hoff K., Vickery L. E., and Culotta V. C. (1999) Saccharomyces cerevisiae ISU1 and ISU2: members of a well-conserved gene family for iron-sulfur cluster assembly. J. Mol. Biol. 294, 897–907 [DOI] [PubMed] [Google Scholar]
- 30.Schilke B., Voisine C., Beinert H., and Craig E. (1999) Evidence for a conserved system for iron metabolism in the mitochondria of Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. U.S.A. 96, 10206–10211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shan Y., Napoli E., and Cortopassi G. (2007) Mitochondrial frataxin interacts with ISD11 of the NFS1/ISCU complex and multiple mitochondrial chaperones. Hum. Mol. Genet. 16, 929–941 [DOI] [PubMed] [Google Scholar]
- 32.Lill R., and Mühlenhoff U. (2008) Maturation of iron-sulfur proteins in eukaryotes: mechanisms, connected processes, and diseases. Annu. Rev. Biochem. 77, 669–700 [DOI] [PubMed] [Google Scholar]
- 33.Richards T. A., and van der Giezen M. (2006) Evolution of the Isd11-IscS complex reveals a single α-proteobacterial endosymbiosis for all eukaryotes. Mol. Biol. Evol. 23, 1341–1344 [DOI] [PubMed] [Google Scholar]
- 34.Gutteridge J. M., Maidt L., and Poyer L. (1990) Superoxide dismutase and Fenton chemistry: reaction of ferric-EDTA complex and ferric-bipyridyl complex with hydrogen peroxide without the apparent formation of iron(II). Biochem. J. 269, 169–174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dixon S. J., and Stockwell B. R. (2014) The role of iron and reactive oxygen species in cell death. Nat. Chem. Biol. 10, 9–17 [DOI] [PubMed] [Google Scholar]
- 36.Mukhopadhyay P., Rajesh M., Yoshihiro K., Haskó G., and Pacher P. (2007) Simple quantitative detection of mitochondrial superoxide production in live cells. Biochem. Biophys. Res. Commun. 358, 203–208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gallet P. F., Maftah A., Petit J. M., Denis-Gay M., and Julien R. (1995) Direct cardiolipin assay in yeast using the red fluorescence emission of 10-N-nonyl acridine orange. Eur. J. Biochem. 228, 113–119 [DOI] [PubMed] [Google Scholar]
- 38.Nowikovsky K., Reipert S., Devenish R. J., and Schweyen R. J. (2007) Mdm38 protein depletion causes loss of mitochondrial K+/H+ exchange activity, osmotic swelling and mitophagy. Cell Death Differ. 14, 1647–1656 [DOI] [PubMed] [Google Scholar]
- 39.Johnson L. V., Walsh M. L., Bockus B. J., and Chen L. B. (1981) Monitoring of relative mitochondrial membrane potential in living cells by fluorescence microscopy. J. Cell Biol. 88, 526–535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Richardson D. R., Lane D. J., Becker E. M., Huang M. L., Whitnall M., Suryo Rahmanto Y., Sheftel A. D., and Ponka P. (2010) Mitochondrial iron trafficking and the integration of iron metabolism between the mitochondrion and cytosol. Proc. Natl. Acad. Sci. U.S.A. 107, 10775–10782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Babcock M., de Silva D., Oaks R., Davis-Kaplan S., Jiralerspong S., Montermini L., Pandolfo M., and Kaplan J. (1997) Regulation of mitochondrial iron accumulation by Yfh1p, a putative homolog of frataxin. Science 276, 1709–1712 [DOI] [PubMed] [Google Scholar]
- 42.Cavadini P., Gellera C., Patel P. I., and Isaya G. (2000) Human frataxin maintains mitochondrial iron homeostasis in Saccharomyces cerevisiae. Hum. Mol. Genet. 9, 2523–2530 [DOI] [PubMed] [Google Scholar]
- 43.Batandier C., Fontaine E., Kériel C., and Leverve X. M. (2002) Determination of mitochondrial reactive oxygen species: methodological aspects. J. Cell Mol. Med. 6, 175–187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kudin A. P., Bimpong-Buta N. Y., Vielhaber S., Elger C. E., and Kunz W. S. (2004) Characterization of superoxide-producing sites in isolated brain mitochondria. J. Biol. Chem. 279, 4127–4135 [DOI] [PubMed] [Google Scholar]
- 45.Liu Y., Fiskum G., and Schubert D. (2002) Generation of reactive oxygen species by the mitochondrial electron transport chain. J. Neurochem. 80, 780–787 [DOI] [PubMed] [Google Scholar]
- 46.Levi S., and Rovida E. (2009) The role of iron in mitochondrial function. Biochim. Biophys. Acta 1790, 629–636 [DOI] [PubMed] [Google Scholar]
- 47.Toyokuni S. (2002) Iron and carcinogenesis: from Fenton reaction to target genes. Redox. Rep. 7, 189–197 [DOI] [PubMed] [Google Scholar]
- 48.Kirkinezos I. G., and Moraes C. T. (2001) Reactive oxygen species and mitochondrial diseases. Semin. Cell Dev. Biol. 12, 449–457 [DOI] [PubMed] [Google Scholar]
- 49.de Moura M. B., dos Santos L. S., and Van Houten B. (2010) Mitochondrial dysfunction in neurodegenerative diseases and cancer. Environ. Mol. Mutagen 51, 391–405 [DOI] [PubMed] [Google Scholar]