Background: Eisosomes are protein-based structures of unknown function at the plasma membrane of yeast cells.
Results: Eisosomes are shown to organize lipid domains that promote signal transduction during environmental stress.
Conclusion: Environmental stress induces clustering of signal transduction lipids and proteins.
Significance: A new mechanism to relay extracellular cues for intracellular responses is provided.
Keywords: cell biology, lipid signaling, plasma membrane, Rho (Rho GTPase), Schizosaccharomyces pombe, stress response, TORC2, eisosome
Abstract
Eisosomes are multiprotein structures that generate linear invaginations at the plasma membrane of yeast cells. The core component of eisosomes, the BAR domain protein Pil1, generates these invaginations through direct binding to lipids including phosphoinositides. Eisosomes promote hydrolysis of phosphatidylinositol 4,5 bisphosphate (PI(4,5)P2) by functioning with synaptojanin, but the cellular processes regulated by this pathway have been unknown. Here, we found that PI(4,5)P2 regulation by eisosomes inhibits the cell integrity pathway, a conserved MAPK signal transduction cascade. This pathway is activated by multiple environmental conditions including osmotic stress in the fission yeast Schizosaccharomyces pombe. Activation of the MAPK Pmk1 was impaired by mutations in the phosphatidylinositol (PI) 5-kinase Its3, but this defect was suppressed by removal of eisosomes. Using fluorescent biosensors, we found that osmotic stress induced the formation of PI(4,5)P2 clusters that were spatially organized by eisosomes in both fission yeast and budding yeast cells. These cortical clusters contained the PI 5-kinase Its3 and did not assemble in the its3-1 mutant. The GTPase Rho2, an upstream activator of Pmk1, also co-localized with PI(4,5)P2 clusters under osmotic stress, providing a molecular link between these novel clusters and MAPK activation. Our findings have revealed that eisosomes regulate activation of MAPK signal transduction through the organization of cortical lipid-based microdomains.
Introduction
Multiprotein structures at the plasma membrane have the potential to generate cortical microdomains through association with membrane-based lipids. The dynamics of such lipoprotein microdomains provide spatiotemporal control over a range of cellular processes. The yeast plasma membrane is highly compartmentalized by protein-lipid microdomains including prominent structures called eisosomes (1–3). In both budding and fission yeasts, protein-based eisosomes sit on the cytosolic face of the plasma membrane to generate linear invaginations, which have been observed at the ultrastructural level by both freeze-fracture and deep-etch electron microscopy (4, 5). Recent studies have also uncovered eisosome structures at the plasma membrane of microalgae and lichens (6). The core component of yeast eisosomes is a BAR domain protein called Pil1, which directly binds to lipids and provides structural curvature to these plasma membrane invaginations (7–9).
Recent studies have begun to elucidate the cellular function of Pil1 and eisosomes. Genetic screens in fission yeast demonstrated that eisosome proteins function with Syj1, a synaptojanin protein that hydrolyzes PI(4,5)P2,2 and its putative ligand Tax4 (8). Furthermore, budding yeast eisosomes were shown to recruit synaptojanin to the plasma membrane, and eisosome mutants exhibited elevated levels of PI(4,5)P2 (10). These studies have provided a framework to understand how eisosomes, which bind tightly to lipids at the plasma membrane, contribute to PI(4,5)P2 hydrolysis with specific synaptojanin isoforms. The connection between eisosomes and PI(4,5)P2 has opened a key question, What cellular processes are regulated by this phosphoinositide regulatory mechanism?
PI(4,5)P2 contributes to a wide variety of cellular activities despite its low abundance in the plasma membrane. The local abundance of PI(4,5)P2 is set by the balanced activities of PI 5-kinase, which phosphorylates PI 4-phosphate to generate PI(4,5)P2, versus counteracting synaptojanin-related lipid phosphatases, which hydrolyze PI(4,5)P2 to generate PI 4-phosphate (11). Mutations in the yeast PI 5-kinase revealed a role for PI(4,5)P2 in organization of actin cables for cell polarity and also for cortical actin patches that mediate endocytosis (12, 13). Consistent with these early findings, work in diverse cell types has shown that the formation of endocytic vesicles requires dynamic formation and hydrolysis of PI(4,5)P2 (14–16). Beyond this structural role, PI(4,5)P2 has the ability to control signal transduction. In budding yeast, PI(4,5)P2 promotes signal transduction through the cell wall integrity pathway, a classical MAP kinase cascade that transmits stress signals from the cell surface for adaptation responses (17). In this pathway PI(4,5)P2 acts upstream of Rho1 GTPase and its effector protein kinase C ortholog PKC1, which then feeds into the MAP kinase kinase kinase Bck1 (17). The spatiotemporal connection between PI(4,5)P2 generation and Rho1 activation is not fully known but is proposed to involve recruitment of the activating Rho GEF Rom2 to the plasma membrane upon heat stress (17). Activating signals for this pathway result in dual phosphorylation and activation of the MAP kinase Slt2/Mpk1.
The same MAPK pathway is conserved in the fission yeast Schizosaccharomyces pombe, where it is called the cell integrity pathway. In the fission yeast cell integrity pathway, the MAPK Pmk1 becomes activated in response to many environmental conditions. In contrast to budding yeast, osmotic stress is the most potent inducer of the cell integrity pathway in fission yeast (18). The phosphorylation and activation of Pmk1 during osmotic stress is transient, peaking around 15 min, and returning to basal levels after 60 min. Pmk1 dynamics are driven primarily by inactivating MAPK phosphatases, whose transcriptional expression is mediated by the stress-activated protein kinase pathway (19). Importantly, MAPK activation under osmotic stress is completely dependent on the activity of both Rho2 and Pck2 (20). Rgf1, a Rho GEF, is also required for Pmk1 activation upon osmotic stress (21), and its membrane localization is altered in cells expressing the hypomorphic allele of PI 5-kinase its3-1 (22). This raises the possibility that PI(4,5)P2 might regulate the fission yeast cell integrity activity, but the connection has remained unclear.
In budding yeast, activation of cell integrity MAPK signaling can suppress mutations in the conserved TORC2 complex (23–27), which controls the cellular stress response through AKT and its orthologs (28–31). This suppression has been observed in budding yeast through environmental conditions that trigger cell integrity MAPK activation or through genetic manipulation to increase cellular PI(4,5)P2 levels, thereby activating Rho-PKC-MAPK signaling. Eisosome mutants similarly suppressed mutations in the catalytic subunit of fission yeast TORC2 (8), raising the possibility for a connection between eisosomes and cell integrity signaling. Here, we explored this possibility and found that eisosomes inhibit cell integrity signaling upon osmotic stress through PI(4,5)P2. Osmotic stress induced the formation of stable PI(4,5)P2 cortical clusters that were organized by eisosomes in both fission yeast and budding yeast. These clusters appeared to concentrate Rho2 for cell integrity signaling, providing a potential mechanism linking PI(4,5)P2 and cell integrity signaling upon osmotic stress.
Experimental Procedures
Yeast Strains and Methods
Standard S. pombe media and methods were used (32), and strains are listed in supplemental Table S1. Gene tagging and deletion were performed using PCR and homologous recombination, and integrations were verified by colony PCR (33). All yeast strains were generated by tetrad dissection when applicable. GFP-2xPH(PLCδ) was cloned from pRS214-GFP-2xPH(PLCδ) (a gift from Scott Emr). GFP-2xPH(PLCδ) was cloned into pJK148-PAct1-TAdh1 using XmaI and NotI to produce pJK148-PAct1-GFP-2xPH(PLCδ)-TAdh1. The resulting plasmid was linearized using NruI and transformed into yeast strain JM837 (leu1-32 h−). mCherry-2xPH(PLCδ) was cloned from pRS415-PGPD-GFP-2xPH(PLCδ) (a gift from Scott Emr). mCherry-2xPH(PLCδ) was cloned into pREP42 using XhoI and BamHI to produce pREP42-mCherry-2xPH(PLCδ). This plasmid (pJM1125) was transformed into yeast strain JM3309 (rho2Δ::kanMX6 GFP-Rho2::leu1+ ade6-M21X ura4-D18 leu1-32 h+) and grown on EMM-URI+Thi plates. Expression was induced by growth in the absence of thiamine for 20 h at 25 °C.
Microscopy
For Figs. 5, A--C, 6, A and D, 7A, 8, 9, and 12, cells were imaged in liquid medium under a coverslip with a 0.5-μm step size using a Deltavision Imaging System (Applied Precision/GE Healthcare) comprised of a customized Olympus IX-71 inverted wide-field microscope, a Photometrics CoolSNAP HQ2 camera, and Insight solid state illumination unit. Images were captured as Z-series and processed by iterative deconvolution in SoftWoRx (Applied Precision/GE Healthcare) and then analyzed in ImageJ (National Institutes of Health). Maximum intensity projections were generated using 5–6 focal planes from the top to the middle of the Z-series. For Figs. 10 and 11 single focal planes of the cell middle were taken. For stresses tested in Fig. 5A, cells were grown to log phase at 25 °C and washed with minimal media containing indicated stress and resuspended in fresh stress media and shaken at 25 °C for the indicated times.
FIGURE 5.
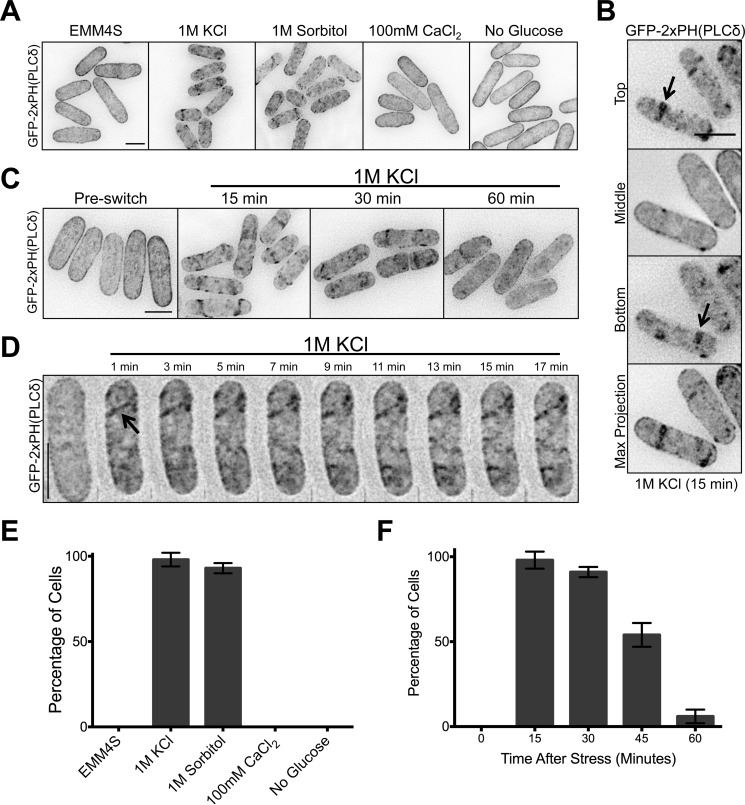
PI(4,5)P2 forms clusters on the plasma membrane upon osmotic stress. A, localization of the PI(4,5)P2 probe GFP-2xPH(PLCδ) after 15 min in the indicated environmental stresses. Images are inverted maximum projections for 0.5-μm-spaced Z-planes in the top half of the cell. B, single focal planes and maximum projection images of cells after 15 min in 1 m KCl. Arrows point to cortical clusters. C, localization of the PI(4,5)P2 probe GFP-2xPH(PLCδ) after exposure to 1 m KCl for the indicated times. Images are inverted maximum projections for 0.5-μm-spaced Z-planes in the top half of cells. D, time-lapse microscopy of GFP-2xPH(PLCδ) in a single cell during osmotic stress in a microfluidic device. Images are inverted maximum projections for 0.2 μm-spaced Z-planes at the top of the cell. The arrow highlights a newly formed cortical cluster. E, quantification of cells containing PI(4,5)P2 clusters after exposure to indicated environmental stress. Plots represent the averages of three biological replicate experiments, and error bars are S.D. between replicates. F, quantification of cells containing PI(4,5)P2 clusters at the indicated time points after exposure to 1 m KCl. Values and error bars were derived as in panel E from three biological replicate experiments. All scale bars are 5 μm.
FIGURE 6.
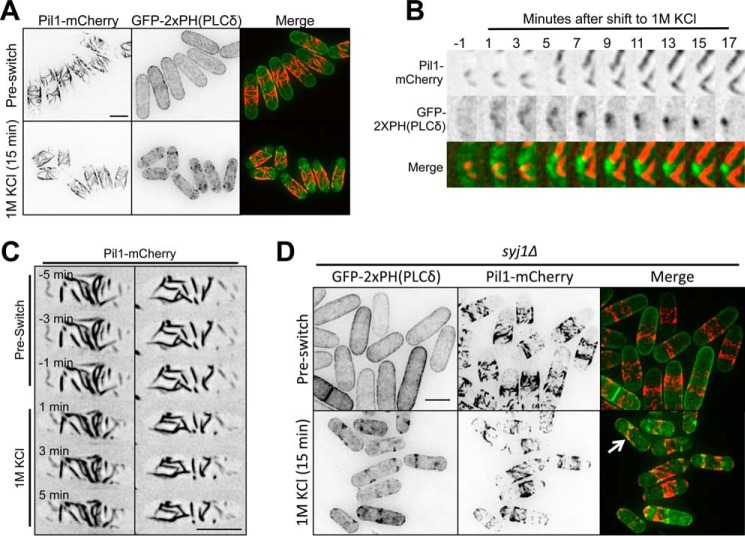
Localization of PI(4,5)P2 clusters relative to eisosomes upon osmotic stress. A, localization of GFP-2xPH(PLCδ) and Pil1-mCherry upon osmotic stress. Images are inverted maximum projections for 0.5-μm-spaced Z-planes of the top of the cell. B, magnified view of time-lapse microscopy for GFP-2xPH(PLCδ) and Pil1-mCherry during osmotic stress. Images are inverted maximum projections for 0.5-μm-spaced Z-planes of the top of the cell. C, time-lapse microscopy of Pil1-mCherry during osmotic stress in a microfluidic device. Each column represents time points for a single cell. Images are inverted maximum projections for 0.2-μm-spaced Z-planes of the top of the cell. D, localization of GFP-2xPH(PLCδ) and Pil1-mCherry upon osmotic stress in syj1Δ cells. Images are inverted maximum projections for 0.5-μm-spaced Z-planes of the top of the cell. Thr arrow points to a cell with some colocalization. All scale bars are 5 μm.
FIGURE 7.
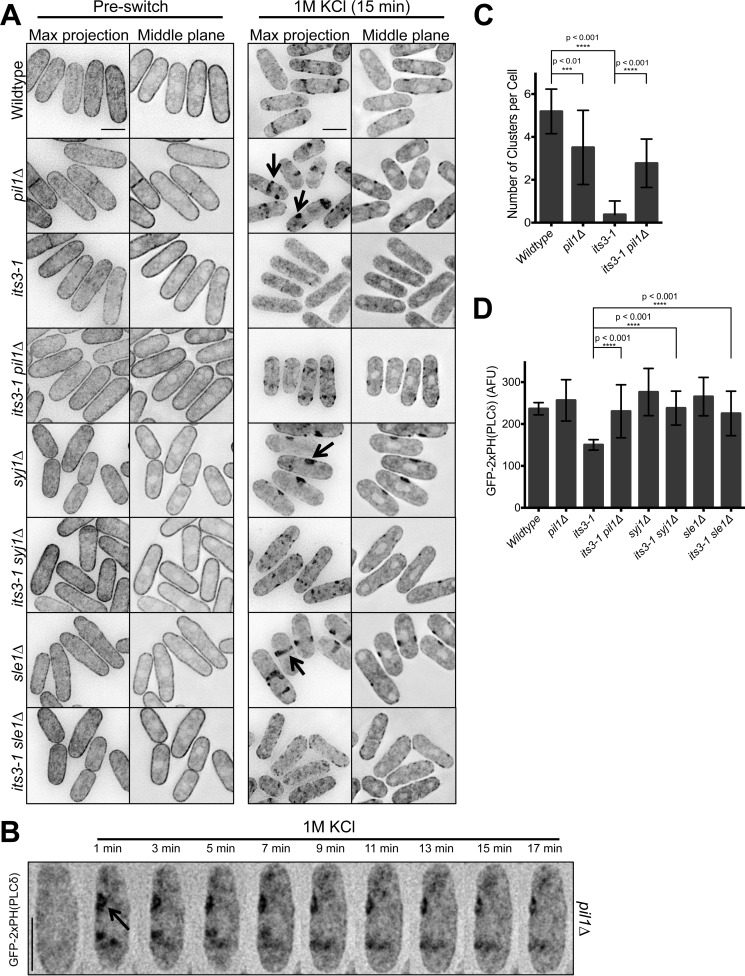
Eisosomes regulate the abundance and localization of PI(4,5)P2 clusters. A, localization of GFP-2xPH(PLCδ) in the indicated mutants upon osmotic stress. Images are inverted maximum projections for 0.5-μm-spaced Z-planes at the top of the cell or a single focal plane of the cell middle. The arrows highlight large cortical clusters in the middle of pil1Δ, sle1Δ, and syj1Δ cells. B, time-lapse microscopy of GFP-2xPH(PLCδ) in a pil1Δ cell during osmotic stress in a microfluidic device. Images are inverted maximum projections for 0.2-μm-spaced Z-planes in the top cortical membrane. The arrow highlights a newly formed cortical cluster. C, quantification of the number of PI(4,5)P2 patches in the indicated strains after 15 min of exposure to 1 m KCl. n = 100 for each time point. p values indicate statistical significance by unpaired Student's t test. D, quantification of relative PI(4,5)P2 concentration at the cortex of the indicated strains after 15 min of exposure to 1 m KCl. Values were measured as fluorescence intensity (arbitrary fluorescent units (AFU)/pixel) of GFP-2xPH(PLCδ) at the cell cortex. n = 100 cells for each strain; p values indicate statistical significance by unpaired Student's t test.
FIGURE 8.
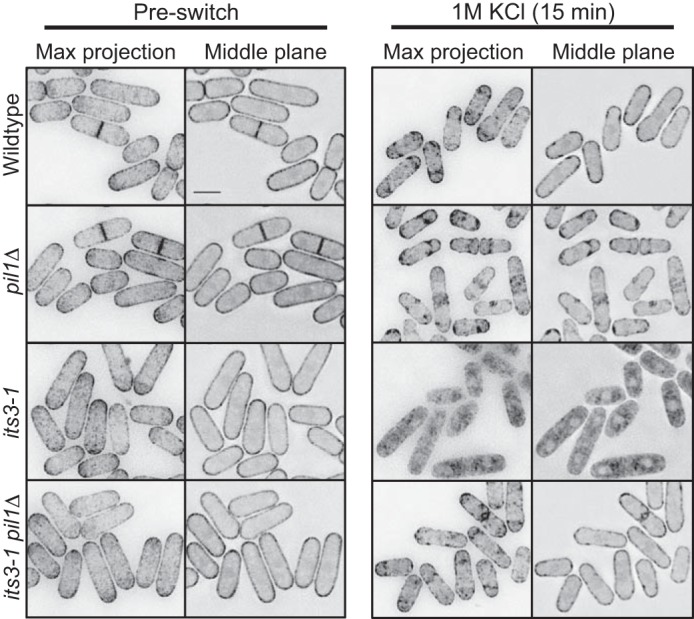
A different PI(4,5)P2 biosensor confirms regulation of PI(4,5)P2 clusters by eisosomes. Shown is localization of PH(Num1)-GFP in the indicated mutants upon osmotic stress. Images are inverted maximum projections for 0.5-μm-spaced Z-planes of the top of the cell or a single focal plane of the cell middle. The scale bar is 5 μm.
FIGURE 9.
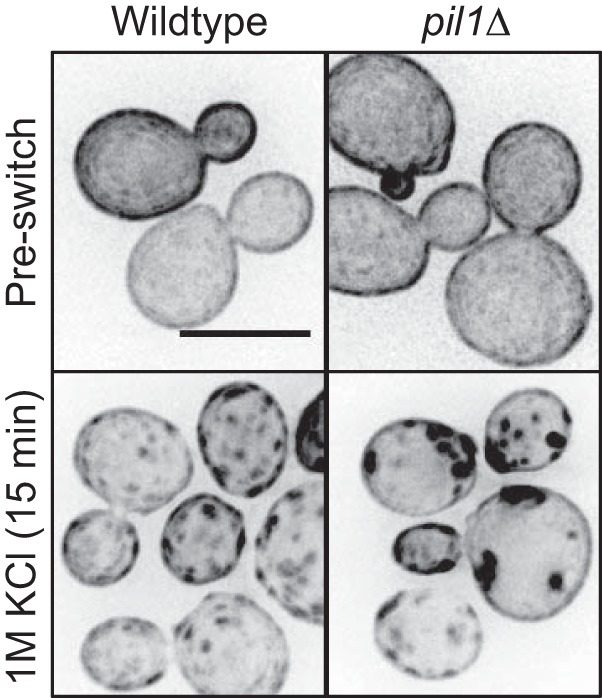
Eisosomes regulate PI(4,5)P2 clusters in budding yeast. Shown is localization of GFP-2xPH(PLCδ) in budding yeast upon hyperosmotic stress. Note that clusters are larger and darker in pil1Δ cells. Images are inverted maximum projections for 0.5 μm spaced Z-planes at the top of the cell. All scale bars are 5 μm.
FIGURE 12.
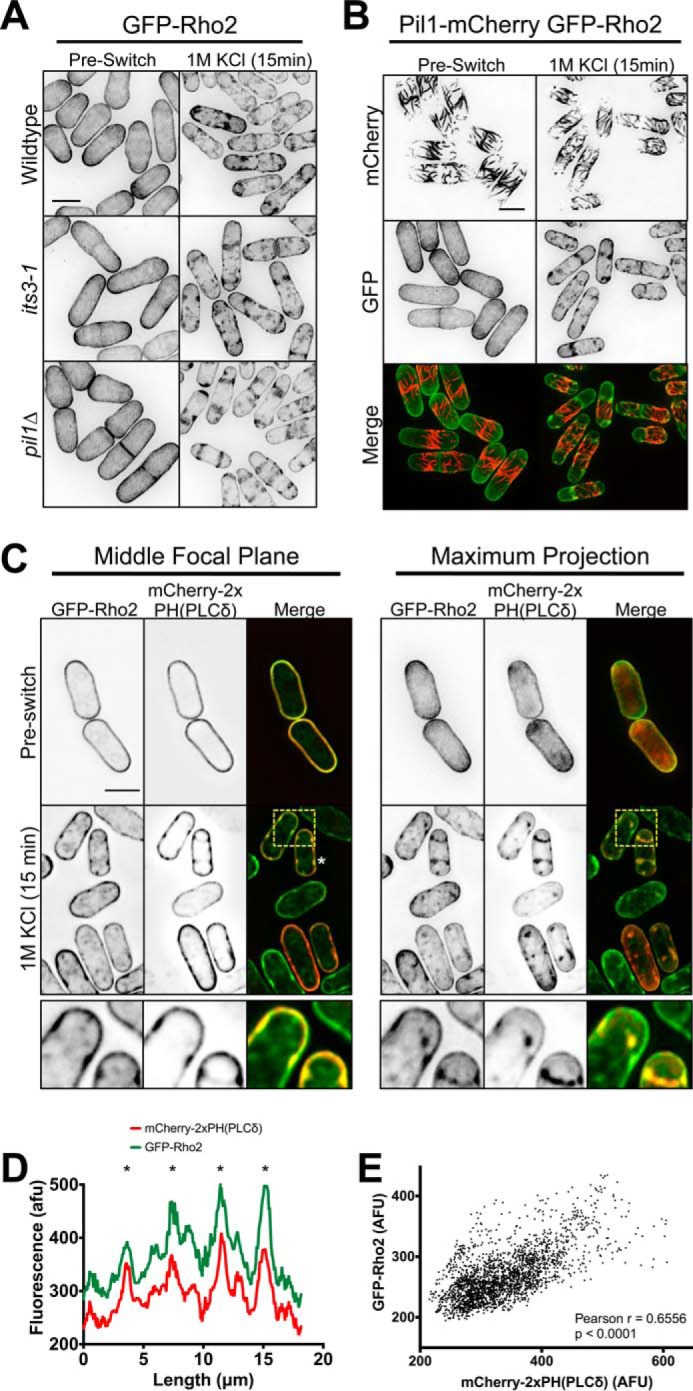
Formation of Rho2 clusters at the plasma membrane during osmotic stress. A, GFP-Rho2 clusters are not PI(4,5)P2-dependent. Images are inverted maximum projections for 0.5-μm-spaced Z-planes for GFP-Rho2 in the indicated strains before and during osmotic stress. B, localization of GFP-Rho2 and Pil1-mCherry upon osmotic stress. Images are inverted maximum projections for 0.5-μm-spaced Z-planes in the top half of the cell. C, co-localization of GFP-Rho2 and mCherry-PH(PLCδ) after 15 min of osmotic stress. Images are inverted maximum projections for 0.5-μm-spaced Z-planes of the top of the cell or a single focal plane of the cell middle. The lower row shows a zoomed image of the yellow-boxed region. D, line scan of GFP-Rho2 and mCherry-2xPH(PLCδ) signal from cell marked with a white asterisk in panel C. A line was drawn around the cell perimeter, and the fluorescence intensity was measured in each channel. Overlapping peaks that represent colocalized clusters are marked with asterisks. afu, arbitrary fluorescent units. E, Scatterplot of GFP-Rho2 and mCherry-PH(PLCδ) signal for each pixel at the cortex of 10 different cells after 15 min of osmotic stress. In each cell a line was drawn around the cell perimeter, and the fluorescence was measured in both channels for each pixel. All scale bars are 5 μm.
FIGURE 10.
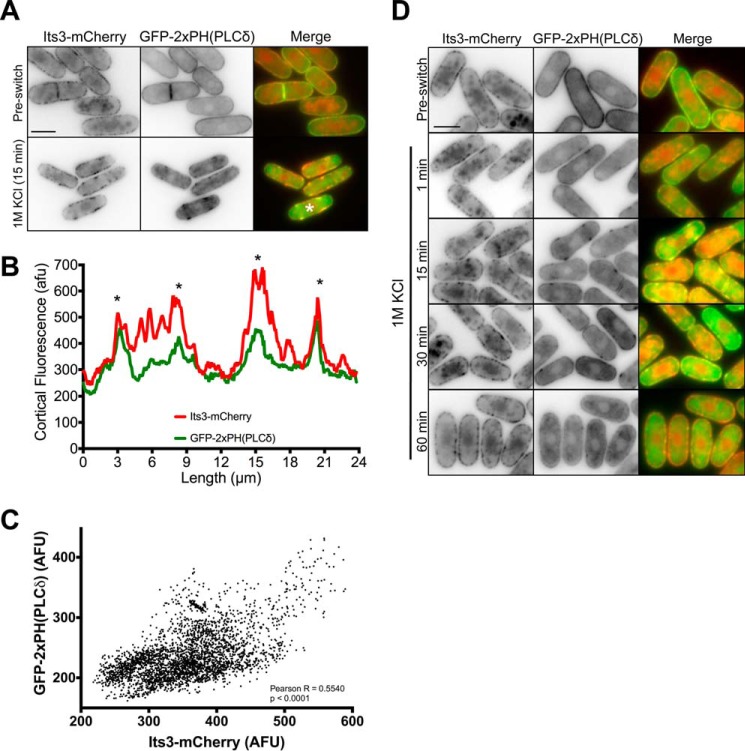
PI(4,5)P2 clusters contain the PI5K, Its3. A, co-localization of Its3-mCherry and GFP-2xPH(PLCδ) before and after 15 min of osmotic stress. Images are inverted single focal planes. B, line scan of Its3-mCherry and GFP-2xPH(PLCδ) signal from cell marked with a white asterisk in panel A. A line was drawn around the cell perimeter, and the fluorescence intensity was measured. Overlapping peaks that represent colocalized clusters are marked with asterisks. afu, arbitrary fluorescent units. C, Scatterplot of Its3-mCherry and GFP-PH(PLCδ) signal for each pixel at the cortex of 10 individual cells after 15 min of osmotic stress. In each cell a line was drawn around the cell perimeter, and the fluorescence was measured in both channels for each pixel. D, timing of Its3-mCherry and GFP-2xPH(PLCδ) clusters during osmotic stress. Note co-localized clusters within 1 min. Images are inverted single focal planes. All scale bars are 5 μm.
FIGURE 11.
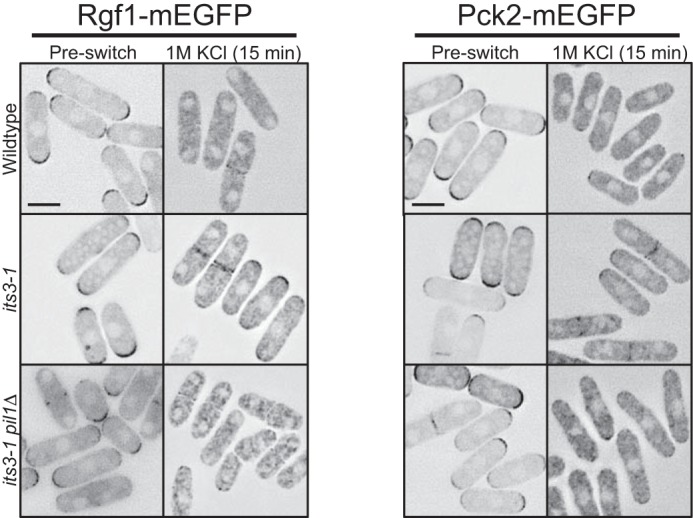
PI(4,5)P2-dependent dispersal of Rgf1 and Pck2 to the cytoplasm upon osmotic stress. Localization of Rgf1 and Pck2 after 15 min of osmotic stress. Both proteins are partially retained at periphery in its3-1 mutant cells but localize to the cytoplasm in both wild type and its3-1 pil1Δ double mutant cells upon osmotic stress. Images are inverted single focal planes. All scale bars are 5 μm.
Figs. 5D, 6, B and C, and 7B were imaged in a CellASIC ONIX Microfluidics Plate using the microscope setup described above at 25 °C. Maximum intensity projections were generated using three 0.2-μm focal planes at the top cortex of cell. Before osmotic shock, log phase cells in minimal media were loaded into a microfluidic flow cell (CellASIC) at 8 p.s.i. for 5 s. CellASIC ONIX Y04C plates were used. Before loading cells into the imaging chamber of the flow cell, the chamber was primed with minimal media lacking stress using the ONIX microfluidic perfusion platform (CellASIC). To ensure that cells were at a steady state, cells were loaded into the imaging chamber and grown for 2 h with fresh medium flowing at 1 p.s.i. before imaging. During osmotic shock, the medium in the flow cell was exchanged using the ONIX system at 1 p.s.i. Changes in cell shape and morphology were visible 1 min after media change, indicating that a 1-min lag time occurs between media chamber change. Control experiments of changing from unstressed media to unstressed media were performed to ensure that changes in cell shape and morphology were due to media change and not due to changes in media chambers. 1-min time points of cells were taken using the Deltavision Imaging System (Applied Precision/GE Healthcare) described above with UltimateFocus.
For Figs. 10B and 12D, a 2-pixel wide line was drawn around the cortex of cells using ImageJ (National Institutes of Health), and fluorescence intensity values were graphed for each channel. Peaks of fluorescence intensity correlated with visible clusters, and asterisks in plots indicate overlapping clusters. For scatterplots in Figs. 10C and 12E, a similar 2-pixel wide line was drawn around the cortex of 10 different cells. For each pixel, the fluorescence intensity in the mCherry and GFP channel was measured and plotted, with GFP on the y axis and mCherry on the x axis. Values for Pearson correlation coefficient and p value in these plots were measured in Prism (Graphpad Software, La Jolla, CA). mCherry-2xPH(PLCδ) was expressed from a plasmid, leading to cell-to-cell variation in expression levels. Therefore, measurements in Fig. 12E were derived from cells that were selected based on similar mCherry expression levels.
For Fig. 7D, the indicated GFP-2xPH(PLCδ) cells were stressed for 15 min in 1 m KCl and then imaged as serial 0.5-μm Z-stacks with the same exposure time and illumination power. In each case, the focal plane representing the top cortex of the cell was identified, and a 30 × 132-pixel box was drawn. Background-subtracted GFP signal (arbitrary fluorescent units) per pixel was measured in this box. This measurement was performed for 100 cells per strain using ImageJ.
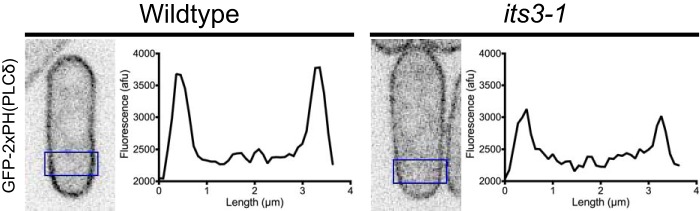
For Fig. 4, cells were imaged in liquid medium under a coverslip by spinning disk confocal microscopy on a Nikon Eclipse Ti equipped with a Yokogawa spinning disk, a Nikon 100 × 1.4NA Plan Apo VC objective, and a Hamamatsu ImagEM C9100–13 EM-CCD camera. This system was controlled by MetaMorph 7 and assembled by Quorum Technologies. Single focal plane images of the cell middle were taken. GFP-2xPH(PLCδ) and its3-1 GFP-2xPH(PLCδ) cells were imaged under identical conditions. GFP intensity through cells (18 pixel × 42 pixel box) was measured, and the background signal from image was subtracted. Quantification of 25 cells was done in ImageJ (National Institutes of Health). A ratio of cortical to cytoplasmic signal was done by averaging the two cortical peaks and dividing by the average of 10 data points in the cell middle.
FIGURE 4.
PI(4,5)P2 localizes to the plasma membrane in fission yeast. Localization of the PI(4,5)P2 probe GFP-2xPH(PLCδ) in wild type and the PI5K mutant, its3-1. Both strains were grown at the permissive temperature of 25 °C. Images are inverted single focal planes of the cell middle. Fluorescence intensity plots were generated from regions boxed in blue; note reduced cortical enrichment in its3-1 mutant cells.
To quantify the number of PI(4,5)P2 clusters in the middle of wild type versus pil1Δ cells after osmotic stress, the length of each cell was divided into three equal zones. The central zone was considered the cell middle. We counted the number of cells containing patches in this middle zone and also the number of patches in this middle zone. This procedure was applied to 100 cells per strain.
Detection of Total and Activated Pmk1
Cells from logarithmic-phase cultures (A600 = 0.5) of either control or mutants expressing a genomic Pmk1-HA-His6-tagged fusion and growing in YE4S medium were collected during unperturbed growth or after treatment with 1 m KCl (Sigma). Plasmid pREP41X-its3+ was employed in Its3 overexpression experiments, and the transformants were grown in EMM2 minimal medium for 24 h with or without 5 g/ml thiamine (Sigma). Preparation of cell extracts and purification of HA-tagged Pmk1 with Ni2+-nitrilotriacetic acid-agarose beads (Qiagen) was performed as previously described (18). Dually phosphorylated and total Pmk1 were detected with rabbit polyclonal anti-phospho-p44/42 (Cell Signaling) and mouse monoclonal anti-HA antibody (12CA5, Roche Molecular Biochemicals), respectively. Membrane blots were revealed with an anti-mouse-HRP-conjugated secondary antibody (Sigma) and the ECL system (Amersham Biosciences-Pharmacia). All Pmk1 activation experiments are biological triplicates. Relative units for Pmk1 activation were calculated by determining the signal ratio of the anti-phospho-P44/42 blot (activated Pmk1) with respect to the anti-HA blot (total Pmk1) at each time point using ImageJ. In each experiment mean relative units ± S.D. are shown. p values <0.05 were determined by unpaired Student's t test.
Gad8 Ser(P)-546 Phosphorylation
Cells were grown to exponential phase in YE4S at 25 °C. A fraction was collected before switching cells to 37 °C for 4 h. Western blots were probed with anti-Cdc2 (Santa Cruz sc-53) as a loading control. Anti-Ser(P)-562 was generated in rabbits against the phosphopeptide QRFANWpSYQRPT (pS is Ser(P); 21st Century Biochemicals, Inc.).
Results
Eisosome Mutants Suppress TORC2 Mutations through Cell Integrity Signaling
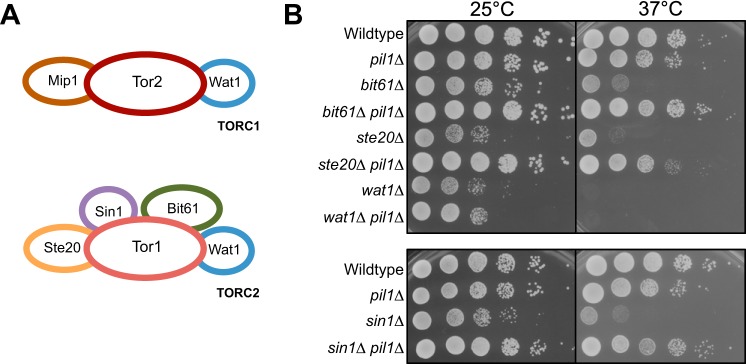
We first examined the genetic connection between eisosomes and TORC2, a conserved multiprotein complex that functions in stress signaling. Fission yeast cells contain two Tor protein complexes, TORC1 and TORC2 (34) (Fig. 1A). The catalytic subunit of TORC2 is the protein kinase Tor1, and the growth defects of tor1 mutants are suppressed by mutations in the Pil1-eisosome pathway (8). To test if this genetic interaction extends to other components of TORC2, we generated double mutants between pil1Δ and other components of TORC2 (bit61Δ, ste20Δ, wat1Δ, and sin1Δ). We found that pil1Δ strongly suppressed the temperature-sensitive growth defects of bit61Δ, ste20Δ, and sin1Δ but not wat1Δ (Fig. 1B). Bit61, Ste20, and Sin1 are specific to TORC2 complex, whereas Wat1 is shared between TORC1 and TORC2 complexes. These data indicate that pil1Δ is a general suppressor of TORC2-specific mutants.
FIGURE 1.
pil1Δ is a general suppressor of TORC2-specific mutants. A, schematic of TORC1 and TORC2. B, 10-fold serial dilutions of the indicated strains grown at different temperatures.
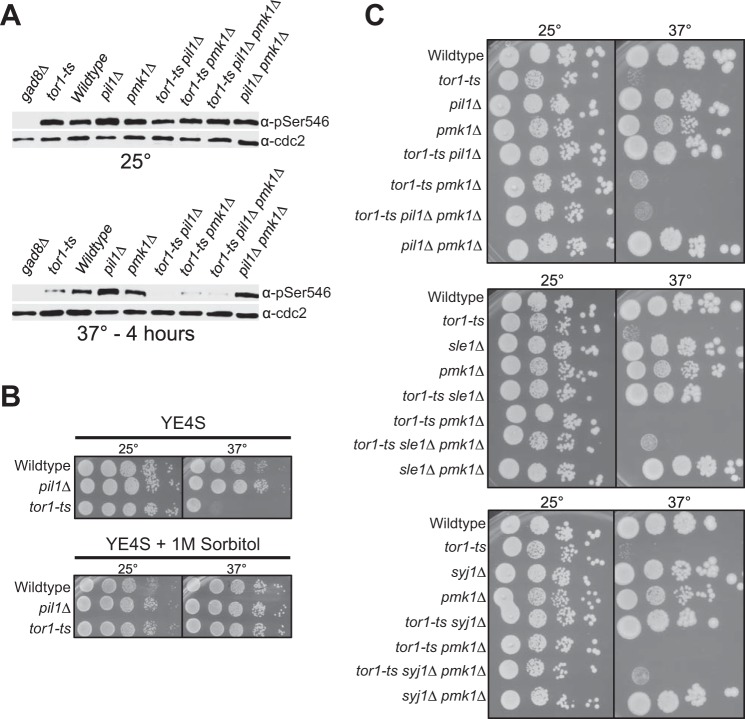
The most well characterized target of TORC2 is the Akt ortholog Gad8 (30, 35). The TORC2 complex activates Gad8 by phosphorylating Gad8-Ser-546. We tested the possibility that pil1Δ suppressed TORC2 mutants by inducing TORC2-independent phosphorylation of Gad8-Ser-546. Using a phospho-specific antibody, we found that pil1Δ did not restore Gad8-Ser-546 phosphorylation to TORC2 mutants (Fig. 2A). Thus, eisosome removal does not suppress TORC2 mutants through alternative Akt phosphorylation mechanisms.
FIGURE 2.
Suppression of TORC2 mutants by pil1Δ requires the cell integrity pathway. A, Western blot showing TORC2-dependent phosphorylation of Gad8. Note that suppression of phenotype by pil1Δ is not due to Gad8 phosphorylation. B, 10-fold serial dilutions of the pil1Δ and tor1-ts cells at different temperatures with or without sorbitol, which activates the cell integrity pathway. C, 10-fold serial dilutions of the indicated strains at 25 °C and 37 °C.
In budding yeast activation of the cell integrity pathway suppresses TORC2 mutations (23–27). The cell integrity pathway senses and relays cell stress through a classical MAP kinase cascade, which in fission yeast culminates with phospho-activation of the MAPK Pmk1. We found that growth on sorbitol suppressed the growth defects of a temperature-sensitive tor1-L2045D mutant (hereafter tor1-ts) (Fig. 2B). This suggests that cell integrity pathway activation also suppresses TORC2 mutants in fission yeast and raises the possibility that eisosome mutants might activate this pathway to suppress defective TORC2 activity. Consistent with this model, growth of the double mutant tor1-ts pil1Δ at the restrictive temperature was abolished by pmk1Δ (Fig. 2C). Although Pmk1 was essential for suppression of tor1-ts by pil1Δ, the pmk1Δ mutation did not alter phosphorylation of the Tor1 target Gad8-S546 (Fig. 2A). We extended this connection between eisosomes and cell integrity signaling by examining Sle1 and Syj1, which function in a genetic pathway with Pil1 (8). sle1Δ and syj1Δ also suppressed the tor1-ts mutant, and as with pil1Δ this suppression required Pmk1 (Fig. 2C). We conclude that TORC2 mutants are suppressed either by activation of the cell integrity pathway or by removal of eisosomes. Our data suggest that eisosomes might act as upstream inhibitors of cell integrity activation.
PI(4,5)P2 Is Important for Cell Integrity Pathway Activation
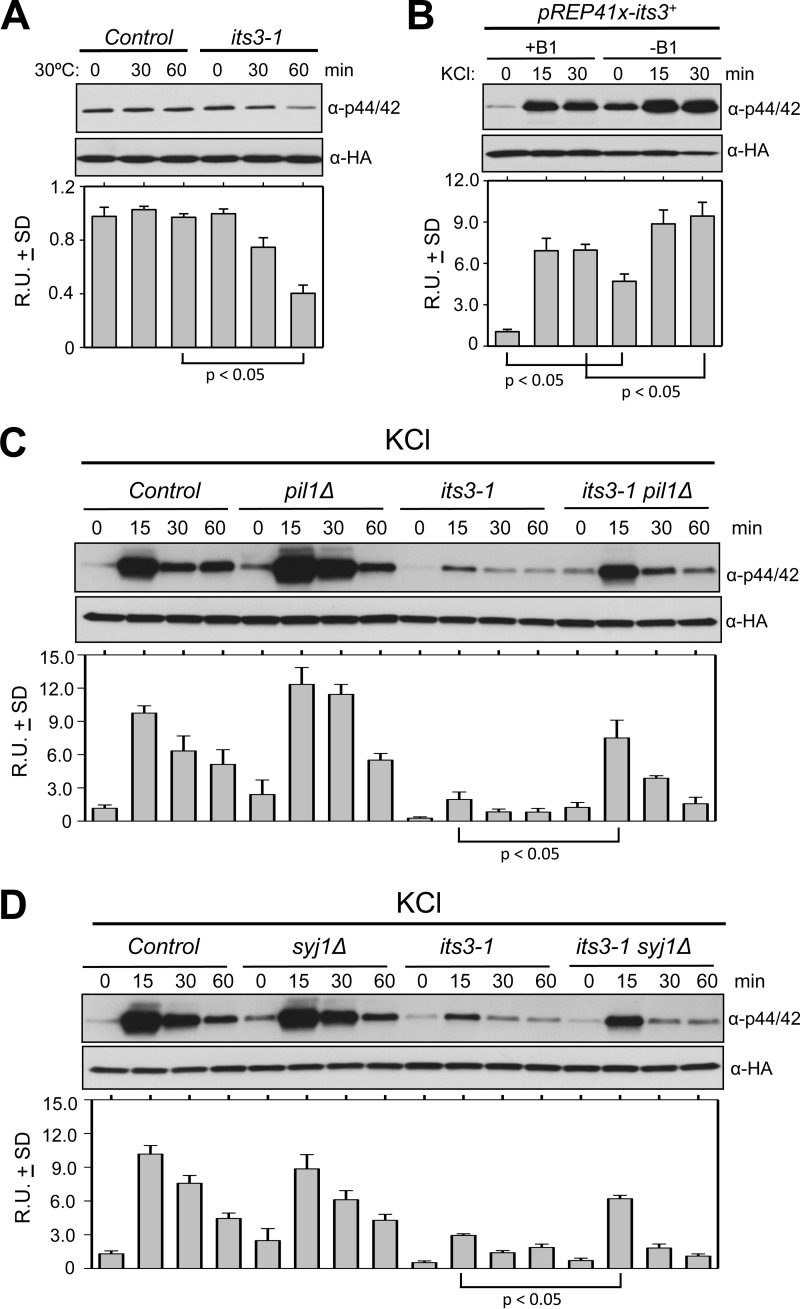
Eisosomes function with the lipid phosphatase synaptojanin to down-regulate PI(4,5)P2 levels in both budding yeast and fission yeast (8, 10). We hypothesized that eisosomes control the cell integrity pathway through their role in PI(4,5)P2 hydrolysis. As a first test of this hypothesis, we altered PI(4,5)P2 levels and measured activation of MAPK Pmk1. Mutations in the essential PI 5-kinase Its3 decrease cellular PI(4,5)P2 levels (36). Upon shift to the semi-permissive temperature, the temperature-sensitive its3-1 mutant exhibited significant reduction in Pmk1 basal phosphorylation when compared with wild type cells, which were unaffected by the temperature shift (Fig. 3A). Moreover, controlled overexpression of Its3 generated a significant increase in both basal Pmk1 activity and activation upon osmotic stress (Fig. 3B). Thus, Pmk1 activity correlates with cellular PI(4,5)P2 levels, consistent with previously demonstrated role for PI(4,5)P2 in activating this pathway in budding yeast (22, 23, 37).
FIGURE 3.
PI(4,5)P2 regulation by eisosomes inhibits Pmk1 activation. A, control or its3-1 mutant cells were incubated at 30 °C and assayed for Pmk1 activation at the indicated time points. B, cells carrying the pREP41x-its3+ plasmid were grown under conditions to repress (+B1) or induce (−B1) overexpression of Its3 and then tested for Pmk1 activation at the indicated time points after the addition of 1 m KCl. C and D, the indicated strains were tested for Pmk1 activation at the indicated time points after the addition of 1 m KCl. All strains carry a HA6H-tagged chromosomal version of pmk1+. For all panels cells were harvested, and Pmk1-HA6H was affinity-purified. Each sample was then immunoblotted for total Pmk1 with anti-HA and for activated Pmk1 with anti-phospho-p42/44 antibodies. R.U. indicates relative units of activated versus total Pmk1. All experiments were performed in triplicate, and graphs show mean relative units ± S.D. p values were determined by unpaired Student's t test.
Eisosomes function with the PI(4,5)P2 phosphatase Syj1 to counter Its3 function, suggesting that they might negatively regulate Pmk1 activation. Indeed, removal of eisosomes by pil1Δ partially restored Pmk1 activation in the its3-1 mutant upon osmotic stress, whereas pil1Δ alone promoted a slight increase in MAPK activity (Fig. 3C). Similarly, syj1Δ partially restored Pmk1 activation during osmostress in this sensitized its3-1 background (Fig. 3D). Thus, Pil1 and Syj1 regulate Pmk1 activation when PI(4,5)P2 levels drop below a certain threshold, which is revealed in the its3-1 mutant. These combined data indicate that PI(4,5)P2 promotes activation of the cell integrity pathway upon osmotic stress, and eisosomes and Syj1 inhibit this PI(4,5)P2-dependent activation.
Eisosomes Organize PI(4,5)P2 Clusters after Osmotic Stress
Our genetic and biochemical data led us to examine the cellular distribution of PI(4,5)P2 upon osmotic stress. We used the biosensor GFP-2xPH(PLCδ), which has been used in diverse cell types due to its high specificity for PI(4,5)P2 versus other phospholipids (38–41). In fission yeast cells, GFP-2xPH(PLCδ) localized to the plasma membrane uniformly (Fig. 4). We tested this sensor in its3-1 cells at the permissive temperature of 25 °C, when cellular PI(4,5)P2 levels are reduced (36). In its3-1 cells, GFP-2xPH(PLCδ) still localized to the plasma membrane but at reduced levels (Fig. 4). The ratio of cortical-to-cytoplasmic fluorescence was reduced in its3-1 cells (1.23 ± 0.12 for its3-1 cells; 1.82 ± 0.11 for wild type cells; n = 25 cells each, mean ± S.D.), indicating that this sensor reports on PI(4,5)P2 levels in fission yeast cells similar to other systems.
We found a striking redistribution of GFP-2xPH(PLCδ) to clusters on the plasma membrane upon hyperosmotic shock (Fig. 5, A, B, and E). This redistribution was observed upon multiple osmotic stresses but did not occur in response to ionic stress (CaCl2) or in the absence of glucose (Fig. 5, A and E). We visualized the kinetics of PI(4,5)P2 cluster assembly and disassembly by testing time points during osmotic stress. Upon osmotic stress, PI(4,5)P2 clusters peaked in number at 15 min and were mostly gone after 60 min (Fig. 5, C and F). Notably, this timing follows the kinetics of Pmk1 activation and deactivation during osmotic stress (Fig. 3) (18). We next observed the formation of PI(4,5)P2 clusters in single cells. A burst of PI(4,5)P2 appeared at the cortex within 1 min after osmotic stress followed by organization of PI(4,5)P2 into immobile clusters (Fig. 5D). We conclude that PI(4,5)P2 is redistributed to cortical clusters on the plasma membrane upon osmotic stress.
The positioning of PI(4,5)P2 clusters led us to examine their spatial relationship with eisosomes. During interphase, eisosome filaments are restricted to the cell middle due to their exclusion from growing cell tips. At this stage, PI(4,5)P2 localizes throughout the plasma membrane, and we did not observe any clear enrichment at eisosomes (Fig. 6A). During osmotic stress, PI(4,5)P2 clusters did not colocalize with eisosomes but, rather, formed at the boundary of the “eisosome domain” in the cell middle (Fig. 6A). We considered the possibility that osmotic stress induces a structural change in the cell cortex, allowing eisosomes to release phospholipids for cluster assembly. In some cases, the initial stages of PI(4,5)P2 cluster assembly occurred at the edge of individual eisosome filaments (Fig. 6B), but we also observed cluster assembly at sites distinct from eisosomes. Moreover, we did not observe any reproducible changes in eisosomes upon osmotic stress (Fig. 6C). Intriguingly, we found a small degree of colocalization between Pil1 and PI(4,5)P2 clusters in osmotically stressed syj1Δ mutant cells, which typically display shorter and disorganized eisosomes (Fig. 6D). This association might reflect dysfunctional eisosomes in the syj1Δ mutant or, alternatively, a role for Syj1 in hydrolyzing PI(4,5)P2 in the vicinity of eisosomes. These and other possibilities remain presently unresolved. From these collective experiments, we conclude that PI(4,5)P2 clusters are positioned at the edge of eisosomes, but these structures do not overlap in wild type cells.
Because PI(4,5)P2 clusters formed at the edge of eisosomes, we next tested if eisosomes contribute to spatial and temporal regulation of these clusters. In wild type cells, PI(4,5)P2 clusters were excluded from the cell middle (Fig. 7A). By contrast, PI(4,5)P2 clusters were observed in the middle of pil1Δ cells. To quantify this effect, we segmented interphase cells into three equally sized domains along their long axis. In the domain that represents the cell middle, where eisosomes localize, we observed PI(4,5)P2 clusters for 89% of pil1Δ cells but for only 7% of wild type cells (n > 100 cells per strain). This indicates that eisosomes in the cell middle restrict the spatial distribution of these clusters. Compared with wild type cells, pil1Δ mutants had fewer PI(4,5)P2 clusters per cell (3.51 ± 1.73 per pil1Δ cell; 5.19 ± 1.04 per wild type cell; p < 0.01), but they were larger (1.31 ± 0.89 μm2 in pil1Δ; 0.92 ± 0.59 μm2 in wild type) (Fig. 7C). These large clusters formed in pil1Δ cells within 1 min after osmotic stress and appeared largely immobile (Fig. 7B). Beyond pil1Δ, similar defects were observed in sle1Δ and syj1Δ cells (Fig. 7A), indicating a general role for this eisosome pathway in PI(4,5)P2 cluster organization. As expected, clusters were largely absent upon osmotic stress in the its3-1 mutant, which impairs generation of PI(4,5)P2 (Fig. 7A). However, they were restored in its3-1 pil1Δ double mutants (Fig. 7, A and C) as well as in its3-1 syj1Δ and its3-1 sle1Δ double mutants. We also measured cortical GFP-2xPH(PLCδ) fluorescence in these strains and found that pil1Δ, syj1Δ, and sle1Δ all suppressed the defect of its3-1 mutant upon osmotic stress (Fig. 7D). Thus, removal of eisosomes restores both PI(4,5)P2 clusters and Pmk1 activation to its3-1 mutant cells.
To ensure that these results were not specific to the GFP-2xPH(PLCδ) construct, we also examined a distinct fluorescent marker for PI(4,5)P2. The PH domain of budding yeast Num1 protein has binding specificity for PI(4,5)P2 (42) such that PH(Num1)-GFP biosensor has previously been used to localize PI(4,5)P2 in fission yeast (43). Using PH(Num1)-GFP, we again observed the formation of cortical PI(4,5)P2 clusters upon osmotic stress (Fig. 8). Similar to the GFP-2xPH(PLCδ) construct, these clusters were spatially organized by eisosomes. Moreover, these clusters were absent in the its3-1 mutant, but this defect was suppressed in the its3-1 pil1Δ mutant (Fig. 8). We conclude that PI(4,5)P2 clusters appear as a physiological response to osmotic stress and are not caused by the GFP-2xPH(PLCδ) construct.
We also tested if the formation and organization of PI(4,5)P2 clusters is conserved in budding yeast. Using the GFP-2xPH(PLCδ) biosensor, we observed the assembly of cortical PI(4,5)P2 clusters upon osmotic stress in budding yeast cells (Fig. 9), consistent with a recent report (44). We further found that clusters were larger in budding yeast pil1Δ cells (Fig. 9), indicating that organization of PI(4,5)P2 clusters is a conserved feature of eisosomes.
PI(4,5)P2 Clusters Contain the PI5K, Its3
The lack of colocalization between eisosomes and PI(4,5)P2 clusters suggested that other proteins might contribute to their assembly. Because cluster formation depends on the PI 5-kinase Its3, we tagged the endogenous protein with a functional C-terminal mCherry epitope. Its3-mCherry localized in small puncta throughout the plasma membrane in unstressed cells (Fig. 10A), similar to previous results using plasmid-based Its3 overexpression (36). Upon osmotic stress, Its3-mCherry localized to more concentrated clusters that colocalized with PI(4,5)P2 clusters (Figs. 10, A and B). We traced line scans around the periphery of 10 cells and observed significant correlation between the fluorescence intensities of Its3-mCherry and the PI(4,5)P2 biomarker (Fig. 10C; Pearson's correlation coefficient of 0.55; p < 0.001) Thus, these clusters contain the PI 5-kinase that generates PI(4,5)P2. We next tested if Its3-mCherry is present at the early stages of PI(4,5)P2 cluster assembly. Within 1 min of osmotic stress, we observed colocalization of Its3-mCherry and PI(4,5)P2 clusters (Fig. 10D). Similar to PI(4,5)P2 redistribution, Its3 clusters peak at 15 min and are gone by 60 min (Figs. 5C and 10D). These data indicate that Its3 localizes to PI(4,5)P2 clusters as well as being necessary for their assembly (Fig. 7A).
PI(4,5)P2 Controls Redistribution of Cell Integrity Components
Finally, we sought to identify molecular connections between PI(4,5)P2 clusters and the cell integrity signaling pathway. Rho GTPases represent molecular “switches” upstream of MAP kinases in the cell integrity pathway. In fission yeast, Rho2 is the specific activator of the pathway by acting through the PKC protein Pck2 in response to osmotic stress (20, 45). The Rho-GEF Rgf1 is also required for Pmk1 activation upon osmotic stress and acts upstream of Pck2 (21). We found that both Rgf1 and Pck2 localize to growing cell tips in unstressed cells but then re-localize to the cytoplasm after osmotic stress (Fig. 11). This removal from the cortex is impaired in the its3-1 mutant and restored in its3-1 pil1Δ double mutants (Fig. 11). Thus, these critical upstream regulators re-localize during osmotic stress in a PI(4,5)P2-dependent manner, but their removal from the cortex indicates that Rgf1 and Pck2 do not associate with PI(4,5)P2 clusters.
We next visualized Rho2. GFP-Rho2 localized in a diffuse pattern along the cell periphery in unstressed cells and then redistributed to cortical clusters upon osmotic shock (Fig. 12A). Formation of these Rho2 clusters was not impaired in its3-1 or pil1Δ mutants (Fig. 12A). Some clusters partially overlapped with eisosomes, but they were primarily positioned at the boundary of the eisosome domain in the cell middle (Fig. 12B). Because this Rho2 positioning pattern was strongly reminiscent of PI(4,5)P2 clusters, we next tested for colocalization of GFP-Rho2 and mCherry-2xPH(PLCδ), a red version of the PI(4,5)P2 biosensor. Both Rho2 and PI(4,5)P2 localized homogenously around the cell periphery in unstressed cells but assembled into colocalized clusters upon osmotic stress (Fig. 12C). Line scans along the cell periphery of single cells revealed clear correlation in the peaks of Rho2 and PI(4,5)P2 signal in these clusters (Fig. 12D). By plotting signal intensity for each pixel along the periphery of 10 cells, we calculated a Pearson's correlation coefficient of 0.656 (p < 0.0001) for the two signals (Fig. 12E), confirming the colocalization of Rho2 and PI(4,5)P2 upon osmotic stress. Importantly, although Rho2 clusters are not PI(4,5)P2-dependent, the strong colocalization of Rho2 and PI(4,5)P2 raises the possibility that both the kinetics and magnitude of downstream signaling through the cell integrity pathway during osmotic stress are regulated by their association.
Discussion
Our work has uncovered eisosomes as a novel and unanticipated player in the connection between PI(4,5)P2 and the MAPK cell integrity signaling pathway. We found that overexpression of the PI 5-kinase increased MAPK activation, whereas mutations in the PI 5-kinase inhibited MAPK activation. These and other data indicate that PI(4,5)P2 participates in activation of the MAPK cell integrity pathway in both budding yeast and fission yeast (22, 23, 37). The recent discovery that eisosomes act with synaptojanin-related lipid phosphatases to decrease PI(4,5)P2 levels in both budding yeast and fission yeast thus raised the possibility that eisosomes inhibit cell integrity signaling through lipids. Our data indicate that either removal of eisosomes or deletion of syj1+ can restore MAPK activation in the its3-1 mutant. This demonstrates that eisosomes act as inhibitors of Pmk1 activation. We also note that Slt2, the budding yeast ortholog of Pmk1, was recently shown to phosphorylate Pil1 (46), raising the possibility for feedback in this eisosome-MAPK regulatory system.
Genetic results based on the suppression of TORC2 mutations further support our model for the regulation of MAPK signaling by eisosomes. Previous data in budding yeast (24, 26) as well as our results in this study have shown that activation of the cell integrity MAPK pathway through environmental stress suppresses the growth defects of TORC2 mutants. Furthermore, we have shown a similar suppression of multiple TORC2 mutants through mutations in the fission yeast eisosome-synaptojanin pathway. Based on this similarity, we hypothesized that the cell integrity pathway is activated in pil1Δ torc2 mutants, leading to survival. Consistent with this possibility, the cell integrity pathway is required for survival of pil1Δ tor1-ts mutants at the restrictive temperature. These genetic results combined with our biochemical data for Pmk1 activation strongly suggest that eisosomes act as upstream inhibitors of the cell integrity pathway.
By monitoring the spatial dynamics of PI(4,5)P2 during osmotic stress, we have identified a series of PI(4,5)P2 clusters at the plasma membrane. Their assembly and disassembly kinetics mirror the activation and deactivation of Pmk1 during osmotic stress. Furthermore, their absence in the its3-1 mutant and restoration in the pil1Δ its3-1 mutant correlates strongly with Pmk1 activation levels in these same mutants. We propose that these PI(4,5)P2 clusters play a critical role in the activation of MAPK signaling for the cell integrity pathway. Interestingly, these clusters are conserved in budding yeast (44), and we found that eisosomes are required for the organization of PI(4,5)P2 clusters in both budding yeast and fission yeast. This means that eisosomes play a conserved role in organizing PI(4,5)P2 clusters, and this function likely extends to other fungal species. It is interesting to note that similar PI(4,5)P2 clusters have been observed in vastly different systems such as neurons where they promote exocytosis by concentrating SNARE proteins (47, 48). Thus, PI(4,5)P2 clusters appear in a wide range of cell types and organisms and may represent a general mechanism for spatiotemporal control of cell biological processes.
Our identification of PI(4,5)P2 clusters raises two major questions. First, how are these clusters formed? We found that the PI 5-kinase Its3 colocalizes with clusters and, therefore, is likely to promote their assembly through local PI(4,5)P2 synthesis. It has been proposed that local tethering of PI 5-kinases can generate PI(4,5)P2 clusters as signaling platforms in mammalian systems as well (49). Our data also indicate that clusters form at the edges of eisosomes, suggesting that eisosomes act as boundary elements to prevent cluster formation in the cell middle. Beyond positioning, we speculate that eisosomes may play a role in cluster assembly and/or turnover. The lipids encased within a cellular eisosome are unknown, but the core eisosome protein Pil1 binds preferentially to negatively charged lipids including PI 4-phosphate and PI(4,5)P2 in vitro (4, 7–9). Thus, eisosomes may inhibit PI(4,5)P2 cluster formation by sequestering this phospholipid or its precursor PI 4-phosphate. The precise localization and synthesis of lipid isoforms during this process will require additional work, which may include the generation of new tools to visualize lipid species in cells.
A second key question is how PI(4,5)P2 clusters promote Pmk1 activation through the cell integrity pathway. We hypothesized that these clusters recruit and/or concentrate regulatory proteins to activate the pathway. Osmotic stress induced a PI(4,5)P2-dependent relocalization of the Rho GEF Rgf1 and the Rho effector Pck2, but these factors were not recruited to cortical clusters. By contrast, osmotic stress led to the formation of Rho2 cortical clusters that colocalized with PI(4,5)P2 clusters. Thus, the recruitment of Rho2, a key upstream molecular switch for the cell integrity pathway, to PI(4,5)P2 clusters may activate this pathway. We note that Rho2 still forms clusters in the its3-1 mutant, suggesting that Rho2 clusters do not require PI(4,5)P2. Rather, Rho2 and PI(4,5)P2 clusters may assemble through independent mechanisms and then merge to activate MAPK signaling. The requirement for independent but overlapping clusters to trigger pathway activation under osmotic stress has the potential to influence both kinetics and robustness of signaling as evidenced by the altered Pmk1 activation observed in either its3-1 cells or in response to Its3 overexpression. These independent clusters may also represent a form of “coincidence detection,” a well known mechanism to suppress spontaneous noise in neuronal signaling networks (50, 51). Such possibilities require future work to define the number of separate clusters and their mechanisms of assembly/association.
Our findings in the spatial regulation of fission yeast MAPK activation differ from the current model in budding yeast and may point to divergent environmental activations signals for this conserved pathway. The most surprising distinction is that the fission yeast Rho GEF Rgf1 is removed from the cell cortex in a PI(4,5)P2-dependent manner when osmotic stress triggers cell integrity activation. This contrasts the behavior of the budding yeast Rho GEF Rom2, which is recruited to PI(4,5)P2-dependent cortical clusters upon heat shock to activate Rho1 and MAPK signaling (17). Importantly, the budding yeast pathway is strongly activated by multiple stimuli including heat stress and cell wall damage but not by osmotic stress (17). In contrast, the fission yeast pathway is strongly activated by osmotic stress, whereas heat and cell wall stress lead to a less pronounced activation with slower kinetics (18, 20). We have recently shown that delayed Pmk1 activation under cell wall stress or glucose deprivation, but not during osmotic stress, is linked to enhanced Pck2 translation mediated by the TORC2 complex (52). Thus, different activation mechanisms may have evolved to use this conserved response pathway for distinct environmental conditions in different species.
Author Contributions
R. K. and J. B. M. conceived the study and wrote the paper. R. K. and M. M. performed the experiments. R. K., M. M., J. C., and J. B. M. analyzed the experiments, reviewed the results, and edited and approved the final version of the manuscript.
Supplementary Material
Acknowledgments
We thank members of the Moseley laboratory and the Biochemistry Department for discussions. We thank C. Barlowe, S. Emr, M. Kaksonen, T. Kuno, S. Martin, S. Oliferenko, P. Perez, C. Stefan, and M. Yanagida for strains and reagents.
This work was supported, in whole or in part, by National Institutes of Health Grant GM099774 (to J. B. M.). This work was also supported by The Norris Cotton Cancer Center, The Pew Scholars Program in the Biomedical Sciences, a Robert D. Watkins Fellowship from the American Society for Microbiology (to R. K.), Ministerio de Economía y Competitividad (Spain) Grant BFU2014–52828-P (to J. C.), and European Regional Development Fund (ERDF) co-funding from the European Union. The authors declare that they have no conflicts of interest with the contents of this article.

This article contains supplemental Table S1.
- PI(4,5)P2
- phosphatidylinositol (PI) 4,5 bisphosphate
- GEF
- guanine nucleotide exchange factor
- MAP
- mitogen-activated protein.
References
- 1.Ziółkowska N. E., Christiano R., and Walther T. C. (2012) Organized living: formation mechanisms and functions of plasma membrane domains in yeast. Trends Cell Biol. 22, 151–158 [DOI] [PubMed] [Google Scholar]
- 2.Olivera-Couto A., and Aguilar P. S. (2012) Eisosomes and plasma membrane organization. Mol. Genet. Genomics 287, 607–620 [DOI] [PubMed] [Google Scholar]
- 3.Douglas L. M., and Konopka J. B. (2014) Fungal membrane organization: the eisosome concept. Annu. Rev. Microbiol. 68, 377–393 [DOI] [PubMed] [Google Scholar]
- 4.Karotki L., Huiskonen J. T., Stefan C. J., Ziółkowska N. E., Roth R., Surma M. A., Krogan N. J., Emr S. D., Heuser J., Grünewald K., and Walther T. C. (2011) Eisosome proteins assemble into a membrane scaffold. J. Cell Biol. 195, 889–902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Strádalová V., Stahlschmidt W., Grossmann G., Blazíková M., Rachel R., Tanner W., and Malinsky J. (2009) Furrow-like invaginations of the yeast plasma membrane correspond to membrane compartment of Can1. J. Cell Sci. 122, 2887–2894 [DOI] [PubMed] [Google Scholar]
- 6.Lee J. H., Heuser J. E., Roth R., and Goodenough U. (2015) Eisosome ultrastructure and evolution in fungi, microalgae, and lichens. Eukaryot. Cell. EC.00106–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Olivera-Couto A., Graña M., Harispe L., and Aguilar P. S. (2011) The eisosome core is composed of BAR domain proteins. Mol. Biol. Cell 22, 2360–2372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kabeche R., Roguev A., Krogan N. J., and Moseley J. B. (2014) A Pil1-Sle1-Syj1-Tax4 functional pathway links eisosomes with PI(4,5)P2 regulation. J. Cell Sci. 127, 1318–1326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ziółkowska N. E., Karotki L., Rehman M., Huiskonen J. T., and Walther T. C. (2011) Eisosome-driven plasma membrane organization is mediated by BAR domains. Nat. Struct. Mol. Biol. 18, 854–856 [DOI] [PubMed] [Google Scholar]
- 10.Fröhlich F., Christiano R., Olson D. K., Alcazar-Roman A., DeCamilli P., and Walther T. C. (2014) A role for eisosomes in maintenance of plasma membrane phosphoinositide levels. Mol. Biol. Cell 25, 2797–2806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Strahl T., and Thorner J. (2007) Synthesis and function of membrane phosphoinositides in budding yeast, Saccharomyces cerevisiae. Biochim. Biophys. Acta 1771, 353–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Homma K., Terui S., Minemura M., Qadota H., Anraku Y., Kanaho Y., and Ohya Y. (1998) Phosphatidylinositol-4-phosphate 5-kinase localized on the plasma membrane is essential for yeast cell morphogenesis. J. Biol. Chem. 273, 15779–15786 [DOI] [PubMed] [Google Scholar]
- 13.Desrivières S., Cooke F. T., Parker P. J., and Hall M. N. (1998) MSS4, a phosphatidylinositol-4-phosphate 5-kinase required for organization of the actin cytoskeleton in Saccharomyces cerevisiae. J. Biol. Chem. 273, 15787–15793 [DOI] [PubMed] [Google Scholar]
- 14.Posor Y., Eichhorn-Grünig M., and Haucke V. (2015) Phosphoinositides in endocytosis. Biochim. Biophys. Acta 1851, 794–804 [DOI] [PubMed] [Google Scholar]
- 15.Weinberg J., and Drubin D. G. (2012) Clathrin-mediated endocytosis in budding yeast. Trends Cell Biol. 22, 1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Goode B. L., Eskin J. A., and Wendland B. (2015) Actin and endocytosis in budding yeast. Genetics 199, 315–358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Levin D. E. (2011) Regulation of cell wall biogenesis in Saccharomyces cerevisiae: the cell wall integrity signaling pathway. Genetics 189, 1145–1175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Madrid M., Soto T., Khong H. K., Franco A., Vicente J., Pérez P., Gacto M., and Cansado J. (2006) Stress-induced response, localization, and regulation of the Pmk1 cell integrity pathway in Schizosaccharomyces pombe. J. Biol. Chem. 281, 2033–2043 [DOI] [PubMed] [Google Scholar]
- 19.Madrid M., Núñez A., Soto T., Vicente-Soler J., Gacto M., and Cansado J. (2007) Stress-activated protein kinase-mediated down-regulation of the cell integrity pathway mitogen-activated protein kinase Pmk1p by protein phosphatases. Mol. Biol. Cell 18, 4405–4419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Barba G., Soto T., Madrid M., Núñez A., Vicente J., Gacto M., Cansado J., and Yeast Physiology Group (2008) Activation of the cell integrity pathway is channelled through diverse signalling elements in fission yeast. Cell. Signal. 20, 748–757 [DOI] [PubMed] [Google Scholar]
- 21.Garcia P., Tajadura V., and Sanchez Y. (2009) The Rho1p exchange factor Rgf1p signals upstream from the Pmk1 mitogen-activated protein kinase pathway in fission yeast. Mol. Biol. Cell 20, 721–731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Deng L., Sugiura R., Ohta K., Tada K., Suzuki M., Hirata M., Nakamura S., Shuntoh H., and Kuno T. (2005) Phosphatidylinositol-4-phosphate 5-kinase regulates fission yeast cell integrity through a phospholipase C-mediated protein kinase C-independent pathway. J. Biol. Chem. 280, 27561–27568 [DOI] [PubMed] [Google Scholar]
- 23.Morales-Johansson H., Jenoe P., Cooke F. T., and Hall M. N. (2004) Negative regulation of phosphatidylinositol 4,5-bisphosphate levels by the INP51-associated proteins TAX4 and IRS4. J. Biol. Chem. 279, 39604–39610 [DOI] [PubMed] [Google Scholar]
- 24.Schmidt A., Bickle M., Beck T., and Hall M. N. (1997) The yeast phosphatidylinositol kinase homolog TOR2 activates RHO1 and RHO2 via the exchange factor ROM2. Cell 88, 531–542 [DOI] [PubMed] [Google Scholar]
- 25.Schmelzle T., Helliwell S. B., and Hall M. N. (2002) Yeast protein kinases and the RHO1 exchange factor TUS1 are novel components of the cell integrity pathway in yeast. Mol. Cell. Biol. 22, 1329–1339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bickle M., Delley P. A., Schmidt A., and Hall M. N. (1998) Cell wall integrity modulates RHO1 activity via the exchange factor ROM2. EMBO J. 17, 2235–2245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Helliwell S. B., Schmidt A., Ohya Y., and Hall M. N. (1998) The Rho1 effector Pkc1, but not Bni1, mediates signalling from Tor2 to the actin cytoskeleton. Curr. Biol. 8, 1211–1214 [DOI] [PubMed] [Google Scholar]
- 28.Sarbassov D. D., Guertin D. A., Ali S. M., and Sabatini D. M. (2005) Phosphorylation and regulation of Akt/PKB by the rictor-mTOR complex. Science 307, 1098–1101 [DOI] [PubMed] [Google Scholar]
- 29.Hresko R. C., and Mueckler M. (2005) mTOR.RICTOR is the Ser-473 kinase for Akt/protein kinase B in 3T3-L1 adipocytes. J. Biol. Chem. 280, 40406–40416 [DOI] [PubMed] [Google Scholar]
- 30.Matsuo T., Kubo Y., Watanabe Y., and Yamamoto M. (2003) Schizosaccharomyces pombe AGC family kinase Gad8p forms a conserved signaling module with TOR and PDK1-like kinases. EMBO J. 22, 3073–3083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kamada Y., Fujioka Y., Suzuki N. N., Inagaki F., Wullschleger S., Loewith R., Hall M. N., and Ohsumi Y. (2005) Tor2 directly phosphorylates the AGC kinase Ypk2 to regulate actin polarization. Mol. Cell. Biol. 25, 7239–7248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Moreno S., Klar A., and Nurse P. (1991) Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol. 194, 795–823 [DOI] [PubMed] [Google Scholar]
- 33.Bähler J., Wu J. Q., Longtine M. S., Shah N. G., McKenzie A. 3rd, Steever A. B., Wach A., Philippsen P., and Pringle J. R. (1998) Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast 14, 943–951 [DOI] [PubMed] [Google Scholar]
- 34.Cybulski N., and Hall M. N. (2009) TOR complex 2: a signaling pathway of its own. Trends Biochem. Sci. 34, 620–627 [DOI] [PubMed] [Google Scholar]
- 35.Ikeda K., Morigasaki S., Tatebe H., Tamanoi F., and Shiozaki K. (2008) Fission yeast TOR complex 2 activates the AGC-family Gad8 kinase essential for stress resistance and cell cycle control. Cell Cycle 7, 358–364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang Y., Sugiura R., Lu Y., Asami M., Maeda T., Itoh T., Takenawa T., Shuntoh H., and Kuno T. (2000) Phosphatidylinositol 4-phosphate 5-kinase Its3 and calcineurin Ppb1 coordinately regulate cytokinesis in fission yeast. J. Biol. Chem. 275, 35600–35606 [DOI] [PubMed] [Google Scholar]
- 37.Helliwell S. B., Howald I., Barbet N., and Hall M. N. (1998) TOR2 is part of two related signaling pathways coordinating cell growth in Saccharomyces cerevisiae. Genetics 148, 99–112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kavran J. M., Klein D. E., Lee A., Falasca M., Isakoff S. J., Skolnik E. Y., and Lemmon M. A. (1998) Specificity and promiscuity in phosphoinositide binding by pleckstrin homology domains. J. Biol. Chem. 273, 30497–30508 [DOI] [PubMed] [Google Scholar]
- 39.Hammond G. R., Fischer M. J., Anderson K. E., Holdich J., Koteci A., Balla T., and Irvine R. F. (2012) PI4P and PI(4,5)P2 are essential but independent lipid determinants of membrane identity. Science 337, 727–730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yan Y., Denef N., Tang C., and Schüpbach T. (2011) Drosophila PI4KIIIalpha is required in follicle cells for oocyte polarization and Hippo signaling. Development 138, 1697–1703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Green R. A., Kao H. L., Audhya A., Arur S., Mayers J. R., Fridolfsson H. N., Schulman M., Schloissnig S., Niessen S., Laband K., Wang S., Starr D. A., Hyman A. A., Schedl T., Desai A., Piano F., Gunsalus K. C., and Oegema K. (2011) A high-resolution C. elegans essential gene network based on phenotypic profiling of a complex tissue. Cell 145, 470–482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yu J. W., Mendrola J. M., Audhya A., Singh S., Keleti D., DeWald D. B., Murray D., Emr S. D., and Lemmon M. A. (2004) Genome-wide analysis of membrane targeting by S. cerevisiae pleckstrin homology domains. Mol. Cell 13, 677–688 [DOI] [PubMed] [Google Scholar]
- 43.Zhang D., Vjestica A., and Oliferenko S. (2012) Plasma membrane tethering of the cortical ER necessitates its finely reticulated architecture. Curr. Biol. 22, 2048–2052 [DOI] [PubMed] [Google Scholar]
- 44.Guiney E. L., Goldman A. R., Elias J. E., and Cyert M. S. (2015) Calcineurin regulates the yeast synaptojanin Inp53/Sjl3 during membrane stress. Mol. Biol. Cell 26, 769–785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ma Y., Kuno T., Kita A., Asayama Y., and Sugiura R. (2006) Rho2 is a target of the farnesyltransferase Cpp1 and acts upstream of Pmk1 mitogen-activated protein kinase signaling in fission yeast. Mol. Biol. Cell 17, 5028–5037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mascaraque V., Hernáez M. L., Jiménez-Sánchez M., Hansen R., Gil C., Martín H., Cid V. J., and Molina M. (2013) Phosphoproteomic analysis of protein kinase C signaling in Saccharomyces cerevisiae reveals Slt2 mitogen-activated protein kinase (MAPK)-dependent phosphorylation of eisosome core components. Mol. Cell. Proteomics 12, 557–574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Honigmann A., van den Bogaart G., Iraheta E., Risselada H. J., Milovanovic D., Mueller V., Müllar S., Diederichsen U., Fasshauer D., Grubmüller H., Hell S. W., Eggeling C., Kühnel K., and Jahn R. (2013) Phosphatidylinositol 4,5-bisphosphate clusters act as molecular beacons for vesicle recruitment. Nat. Struct. Mol. Biol. 20, 679–686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.van den Bogaart G., Meyenberg K., Risselada H. J., Amin H., Willig K. I., Hubrich B. E., Dier M., Hell S. W., Grubmüller H., Diederichsen U., and Jahn R. (2011) Membrane protein sequestering by ionic protein-lipid interactions. Nature 479, 552–555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kwiatkowska K. (2010) One lipid, multiple functions: how various pools of PI(4,5)P2 are created in the plasma membrane. Cell. Mol. Life Sci. 67, 3927–3946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bi G., and Poo M. (2001) Synaptic modification by correlated activity: Hebb's postulate revisited. Annu. Rev. Neurosci. 24, 139–166 [DOI] [PubMed] [Google Scholar]
- 51.Spitzer N. C. (2004) Coincidence detection enhances appropriate wiring of the nervous system. Proc. Natl. Acad. Sci. U.S.A. 101, 5311–5312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Madrid M., Jiménez R., Sánchez-Mir L., Soto T., Franco A., Vicente-Soler J., Gacto M., Pérez P., and Cansado J. (2015) Multiple layers of regulation influence cell integrity control by the PKC ortholog Pck2 in fission yeast. J. Cell Sci. 128, 266–280 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.