Background: The structure and flexibility of hydrophilic LHCII domains in an aqueous environment are only partially known.
Results: The structure and flexibility of the N-terminal and lumenal loop domains are analyzed by EPR distance measurements.
Conclusion: The N terminus covers only part of the stromal LHCII surface, and the lumenal loop contains a highly flexible section.
Significance: The structural information presented helps to understand regulatory LHCII functions in photosynthesis.
Keywords: electron paramagnetic resonance (EPR), light-harvesting complex (antenna complex), membrane protein, molecular modeling, protein structure, LHCII, N -terminus, lumenal loop
Abstract
The major light harvesting complex II (LHCII) of green plants plays a key role in the absorption of sunlight, the regulation of photosynthesis, and in preventing photodamage by excess light. The latter two functions are thought to involve the lumenal loop and the N-terminal domain. Their structure and mobility in an aqueous environment are only partially known. Electron paramagnetic resonance (EPR) has been used to measure the structure of these hydrophilic protein domains in detergent-solubilized LHCII. A new technique is introduced to prepare LHCII trimers in which only one monomer is spin-labeled. These heterogeneous trimers allow to measure intra-molecular distances within one LHCII monomer in the context of a trimer by using double electron-electron resonance (DEER). These data together with data from electron spin echo envelope modulation (ESEEM) allowed to model the N-terminal protein section, which has not been resolved in current crystal structures, and the lumenal loop domain. The N-terminal domain covers only a restricted area above the superhelix in LHCII, which is consistent with the “Velcro” hypothesis to explain thylakoid grana stacking (Standfuss, J., van Terwisscha Scheltinga, A. C., Lamborghini, M., and Kühlbrandt, W. (2005) EMBO J. 24, 919–928). The conformation of the lumenal loop domain is surprisingly different between LHCII monomers and trimers but not between complexes with and without neoxanthin bound.
Introduction
The major light harvesting complex II (LHCII)2 of green plants has its main function in collecting light energy for the photosynthetic reaction centers, extending the absorption capacity of their core pigments and thereby increasing the rate of photosynthesis. Besides this task the LHCII is involved in regulatory processes. First of all, under excess light conditions, light-harvesting complexes are able to switch from their light-harvesting to an energy-dissipating state, rapidly converting the excitation energy into heat and thus protecting the photosynthetic apparatus from photochemical damage. This process is known as non-photochemical quenching (NPQ) (1). Second, LHCII is able to balance the energy flow between PSII and PSI in a process called state transition (2). If PSII is excited more frequently than PSI, the plastoquinone pool in the thylakoid is in a reduced state. This activates a kinase that phosphorylates Thr-5 or Thr-6 near the N terminus of LHCP, which in turn causes LHCII to dissociate from the PSII holocomplex, to migrate from the appressed grana domains of the thylakoid to the non-stacked stromal lamellae where PSI resides, and to enlarge the absorption cross-section of PSI.
The crystal structure of LHCII is known at a resolution of 2.4 Å (0.24 nm) (3, 4). It exhibits the LHCII apoprotein (LHCP) non-covalently binding 14 chlorophyll (Chl) and 4 carotenoid molecules. For the light-harvesting function of LHCII these pigments need to be positioned relative to each other such that rapid and efficient transfer of excitation energy is ensured. This would require a rigid structure of the pigment-protein complex. The regulatory functions of LHCII, by contrast, involve different functional states adopted by the protein, necessitating a more flexible structure of at least parts of the complex (5). In fact, measurements by pulsed electron paramagnetic resonance (EPR) have shown that LHCII possesses a rigid core and flexible protein domains in its periphery (6).
One of these flexible domains is the N-terminal protein section. The first 11 or so amino acids have not been resolved in the crystal structures, which has been attributed to their non-uniform positioning even in the crystal. This protein domain contains the phosphorylation site involved in state transition (see above); therefore, its structural behavior is of interest for understanding the function of LHCII. In this article, the position of the N-terminal domain with regard to the rest of LHCII has been assessed by triangulation, measuring molecular distances by double electron-electron resonance (DEER).
The other flexible section in the LHCII apoprotein analyzed in this study is the lumenal loop domain exposed to the aqueous compartment of the thylakoid lumen in plant chloroplasts. Conformational changes of this hydrophilic protein domain have been proposed to be involved in the regulatory process of NPQ (see above) (7). The lumenal loop may be a pH sensor monitoring low pH values in the lumen that are an indicator of light-stress situations (8). A structural transition of the carotenoid neoxanthin (NX) has been observed in connection with the transition of the photosynthetic apparatus from a light-harvesting to an energy-dissipating state (9). To test whether structural changes of the NX moiety can in principle trigger structural changes in the lumenal loop, we compared the lumenal loop structure in the presence and absence of NX.
Intra-molecular distances have been measured in the present study by DEER (10) to assess the structure and flexibility of the N-terminal and stromal loop domains in LHCII. This required placing spin labels site specifically in the protein domains to be analyzed. In previous studies, EPR has similarly been used to measure intra-molecular distances in monomeric complexes of LHCII (6, 11, 12). Functional LHCII in vivo is predominantly organized in trimers; therefore, it was desirable to extend intra-molecular distance measurements to trimeric complexes. This is not trivial because a trimer carrying two spin labels per monomer yields so many DEER signals according to the numerous spin couplings between the 6 spins present that evaluating these signals becomes very difficult. Therefore, we are presenting a procedure here to construct recombinant LHCII trimers that carry pairs of spin labels only in one of the monomers in each trimer.
Experimental Procedures
LHCP Versions
All LHCP versions used in this study were derivatives of Lhcb1*2 (AB80) (13) having the native cysteine at position 79 replaced with serine. In LHCP versions containing mutations, at positions 3, 11, 34, 59, and 102 serine, at positions 124 and 143 isoleucine, and at 96 valine and 113 lysine were replaced with cysteine. LHCP versions for the production of heterogeneous trimers carried two affinity tags, a His6 tag and a Strep tag. In LHCP versions 3/34, 3/59, 7/34, 7/59, 11/34, and 11/59, a C-terminal Strep tag was followed by a 2-glycine spacer and a His6 tag, in the case of 96/143, 102/143, and 113/143 the His6 tag was on the C terminus, whereas the Strep tag was N-terminal.
The genes were inserted in the pDS12-RBSII vector and overexpressed overnight in Escherichia coli strain JM101. After purification LHCP was labeled with 20-fold excess of 3-(2-iodacetamido)-PROXYL (IA-Proxyl, Sigma, free radical) per labeling position as described (6).
LHCII Reconstitution and Trimerization
Reconstitution of monomers was performed according to Ref. 6. Samples lacking NX where reconstituted with a mixture of purified pigments containing Chl a:Chl b:lutein at a molar ratio of 8:6:2. For EPR samples, the standard reconstitution buffer was changed against a buffer containing 50 mm Tris-HCl, pH 7.8, 0.2% DM, and 12.5% sucrose in D2O using Amicon 30-kDa centrifugal filters. Buffer exchange was performed by reducing the volume of the monomer solution by half and refilling it with buffer; this procedure was repeated five times. The solution was then centrifuged in the filter units until a monomer concentration of 120 μm was reached. 80% Glycerol-D8 was added, so the final monomer samples contained 80 μm LHCII monomers in 50 mm Tris-HCl, 0.2% DM, 12.5% sucrose, 27% glycerol-D8, in D2O, pH 7.8.
For heterogeneous trimers, parallel reconstitutions of 5 mg of labeled LHCP of the desired version and 40 mg of C79Sh were performed. The reconstitution solutions were mixed in a 1:8 ratio if the reconstitution yield was similar between labeled and unlabeled proteins, otherwise the mixing ratio was adjusted to match the 1:8 ratio of labeled and unlabeled monomers. Trimerization was then performed according to the standard procedure (6). The trimer solution was applied to 5 × 500-μl streptactine-Macroprep® columns and was washed and eluted according to the manual of the Macroprep material (IBA GmbH, Goettingen). The heterogeneous trimers were then purified by sucrose gradient ultracentrifugation.
For EPR measurements of heterogeneous trimers, the buffer was exchanged as described above for monomers. The final heterogeneous trimer concentration was between 10 and 20 μm LHCII trimers in 50 mm Tris-HCl, 0.2% DM, 12.5% sucrose, 12.5% glycerol-D8, in D2O, pH 7.8.
Pigment compositions of all LHCII samples were checked by UV-visual spectroscopy in comparison to spectra of reconstituted wild type LHCII with its native pigment composition. The absorption spectra indicated that, apart from NX and violaxanthin (VX) (see below), all LHCII preparations had the same pigment composition ±0.5 Chl a or Chl b. Monomeric and trimeric LHCII complexes were separated by sucrose density gradient centrifugation as described before and verified (i) by their different sedimentation behavior in the density gradient (14), and (ii) by their CD spectra in the visible domain with trimeric LHCII exhibiting a characteristic negative peak at 474 nm (14, 15) (see Fig. 1 for an example). In neither respect were differences seen between heterogeneous and wild type trimers.
FIGURE 1.
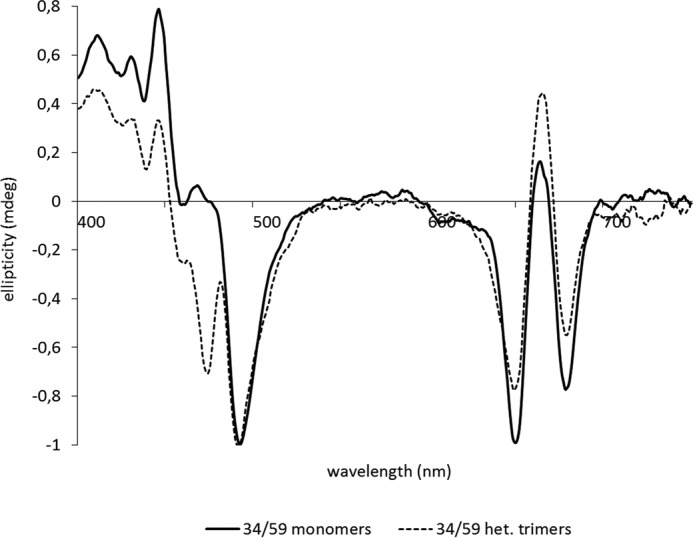
CD spectra of monomeric and trimeric LHCII used for biochemical characterization of LHCII labeled in positions 34/59 as an example for all other LHCII versions. CD spectra are normalized to 492 nm.
DEER Distance Measurements
Distance measurements were performed using 40–50 μl of protein solution in 3-mm (outer diameter) quartz tubes at a temperature of 50 K at Q-band (∼34.5 GHz) frequencies using the four-pulse DEER sequence (16). Samples were shock-frozen in liquid nitrogen. Measurements were performed on a home built Q-band spectrometer at 150 W nominal output power using a home built TE102 rectangular resonator suitable for oversized 3-mm sample tubes (17). All pulse lengths were set to 12 ns, the pump pulse was applied at the maximum of the nitroxide spectrum with a frequency offset of 100 MHz for observer pulses. Samples were prepared with deuterated buffer and 27% deuterated glycerol (v/v) as a cryoprotectant. Deuterium modulations were suppressed by an 8-step nuclear modulation averaging cycle with 16-ns increments (17). DEER traces were analyzed with DeerAnalysis (18). For modeling of N-terminal residues 3–13 in trimers, the new data were augmented by distance distributions for trimers consisting of singly labeled protomers at labeling positions 3, 4, 7, 9, 10, 11, and 12 and with electron spin echo envelope modulation (ESEEM) water accessibility parameters π(D2O) for the same residues from previous work (6).
Simulation and Modeling
Simulated distance distributions were based on the LHCII crystal structure with Protein Data Bank identifier 2BHW (4) and were obtained using the home written open-source software package MMM. Water molecules were disregarded, whereas all cofactors were included in computations for the monomer. All computations with monomers were performed on chain A of the crystal structure. We checked that computations for chains B and C lead to very similar results. Likewise, trimer simulations consider only two spin labels attached to chain A, whereas the presence of chains B and C may influence the distance distributions due to additional label-protein interactions. In trimers, computations for site 143 were performed after removing CHL612 in the same protomer of the trimer, as this improved agreement of all simulated distance distributions including this residue with experimental data. Spin label rotamers and their populations were computed using a rotamer library for IA-Proxyl that was generated from a molecular dynamics trajectory computed at 298 K (19). The interaction between spin-labeled atoms and protein atoms was modeled with a Lennard-Jones potential as implemented in the MMM software.
Ensemble models of N-terminal residues 3–13 were computed by adapting a Monte Carlo approach for loop modeling. The coordinates of residues 14–232 were first transformed to a frame where the C3 symmetry axis of the trimer is the z axis. This axis is also assumed to be the lipid bilayer normal. The z coordinate of the center of the bilayer and the optimal bilayer thickness were determined by minimizing free energy for inserting residues into the bilayer, based on propensities from Refs. 20 and 21. A more detailed description of the algorithm has been given in Ref. 22. Pairs of backbone dihedrals φi,ψi were randomly selected so that their distribution conformed to residue-specific Ramachandran plots (23). Backbone N, Cα, and carbonyl C coordinates were computed from the backbone dihedrals using the Sugeta-Miyazawa algorithm (24) and assuming standard peptide bond lengths and angles. The initial rotation matrix was computed according to Ref. 25 from the backbone coordinates of residue 14 in structure 2BHW. At each residue for which restraints were defined, mean spin label coordinates were predicted by transforming the mean N-O midpoint coordinate in a rotamer library of an unrestricted IA-Proxyl side chain (19) from the peptide standard frame to the local residue frame. For each distance restraint with mean distance 〈ri〉 and standard deviation σr,i of the peak in the distance distribution we computed a probability density pi = exp{−[(r − 〈ri〉)/σr,i]2}, which is related to probability of fulfilling the restraint via the error function. Here r is the distance in the model. A threshold pi ≥ 0.8 corresponds to accepting 50% of all conformations that are in agreement with a restraint, providing an intuitive visualization at the same probability level as used for thermal ellipsoids in crystal structures. The corresponding probability density for all distance restraints is then given by p = Πi, pi with an acceptance threshold p ≥ 0.8n, where n is the number of restraints. Water accessibility restraints were defined as lower and upper bounds for the predicted z coordinate zα of the Cα atom. A linear fit of experimental ESEEM water accessibility parameters π(D2O) for residues 10, 11, 12, 14, 22, 29, 34, and 59 to the water population p(zα) in a lipid bilayer (26) was performed (correlation coefficient 0.517). Using the error estimate of this linear fit we computed bounds for zα for residues 3, 4, 7, 9, 10, 11, and 12. Given the low correlation coefficient we set the bounds at 1.5 times the standard deviation. With this choice, no upper bounds could be established, i.e. within experimental uncertainty the residues might be fully water exposed. For all residues the restrictions are imposed by the lower bounds. Models were rejected if any water accessibility violated its respective lower bound. For accepted models, carbonyl O atoms were added in the peptide plane and a test for self-clashes or clashes with atoms in residues 14–232 in all three chains of the trimer was performed. A clash was defined as an approach of two heavy atoms of non-consecutive residues closer than 2.5 Å (0.25 nm). Models with clashes were rejected. Modeling runs were stopped after 100 models had been generated.
Biased homology models of the lumenal loop domain were generated with Modeler 9.9 (27) using MMM as an interface and structure 2BHW with 100% sequence identity as a template. The additional distance restraint between residues 113 and 143 (2.23 nm) was implemented as described in Ref. 22. A secondary structure was explicitly restrained for the short α-helices 102–105 and 112–114 as well as the short β-strands 110–111 and 119–121. This choice corresponds to modeling the minimal movement of the domain that leads to fulfillment of the distance restraint while maintaining structural integrity of the peptide chain.
Results
Two flexible domains of the LHCII apoprotein, the N-proximal section and the lumenal loop, have been characterized with respect to their position and mobility in detergent-solubilized LHCII. Distances have been measured by DEER between several points in both flexible domains and reference points in the rigid part of LHCII (Fig. 2). In the case of the loop domain, these distances were then compared with those read from the LHCII crystal structure (4), and the widths of the peaks yielded information about the local mobilities. For the latter, the mobilities of the spin labels at their individual attachment sites had to be taken into account. This was done by modeling their conformational distribution by an approach that considers the interaction of precomputed spin label rotamers with the protein based on the LHCII crystal structure (6, 19). Any peak width significantly exceeding that predicted by the molecular models was assigned to local mobility of the protein domains. The N-proximal 9–11 amino acids in LHCII have not been resolved in the crystal structures. Therefore, their position with respect to the rigid core of LHCII was mapped by triangulation, i.e. by measuring distances to two positions in the structure whose localization is known.
FIGURE 2.
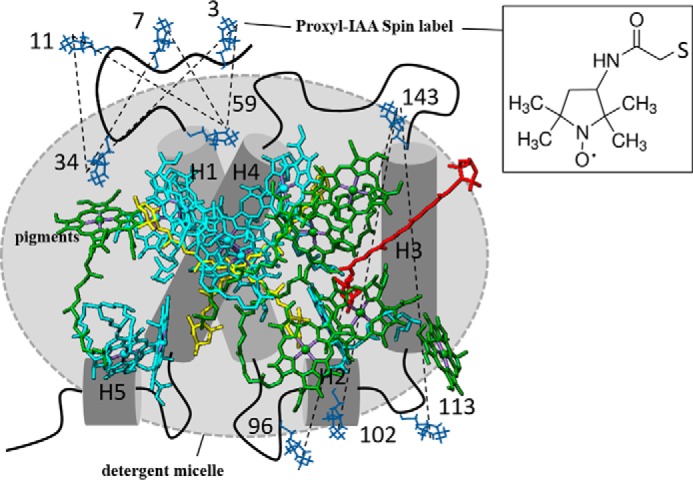
Model of an LHCII monomer showing the PROXYL labeling sitesand the measured intramolecular distances defined by these labels. Thenumbers of the labeled amino acids are given. The dotted lines show the intramolecular distances measured by DEER. H1-H5, α helices 1–5. PROXYL labels, blue. Chl a, cyan. Chl b, green. Lutein, yellow. NX, red. The inset shows the chemical structure of the PROXYL label.
The measurement of intra-molecular distances in LHCII required the preparation of doubly spin-labeled derivatives of the apoprotein LHCP. Cysteines were introduced at the positions to be measured, replacing Ser in most cases, but slightly less conservative exchanges against Val and Ile were necessary at some positions. Because the only endogenous Cys in position 79 in these LHCP derivatives had been replaced by Ser, the newly introduced Cys residues were highly specifically labeled by the sulfhydryl-reactive spin label iodoacetamido-PROXYL (6, 12). Labeling was 80% efficient in each position so that an estimated 60% of the proteins in each sample were doubly labeled. This was checked by comparison of the double integral of continuous wave EPR spectra of labeled LHCP at known protein concentrations and of known quantities of free PROXYL-IAA (data not shown). The proteins carrying single labels were not expected to give rise to DEER signals but lowered the modulation depth in the DEER measurements. Due to partial reduction of the N-O group during reconstitution the effective ratio of doubly labeled to singly labeled protein was significantly lower than 80% in some cases, which did not influence the quality of the measurements, except for requiring longer measurement times to reach sufficient signal-to-noise ratio. All labeled LHCII monomers and trimers were characterized by absorption, fluorescence (28), and CD spectroscopy (14) to make sure that neither mutation nor labeling had significantly affected the structure.
Intra-molecular distances had to be measured in monomeric LHCII and also in trimeric complexes, the native form of LHCII whose crystal structure has been elucidated. The measurement in trimers is not a trivial task. If each apoprotein in an LHCII trimer carries two spin labels, then six labels are expected to give rise to DEER signals which, in most cases, will make the analysis of these signals impossible. Therefore it was necessary to construct LHCII trimers in which only one subunit was spin-labeled, termed heterogeneous trimers (Fig. 3). To this end, labeled monomers were mixed with an 8-fold excess of non-labeled ones. All apoproteins carried His6 tags, which are necessary for trimerizing LHCII via immobilized metal affinity chromatography (29). Those LHCII monomers carrying spin labels were additionally equipped with another affinity tag, a Strep tag (30). This additional tag was used to isolate LHCII trimers containing only one spin-labeled monomer (see below). Most doubly spin-labeled LHCP derivatives carried a C-terminal His6 tag and an N-terminal Strep tag. Only in those derivatives that had their spin labels near their N termini were both affinity tags attached to their C termini, so as not to interfere with the folding of the N-terminal domain. All affinity-tagged LHCII monomers and trimers were checked by absorption, fluorescence, and CD spectroscopy to determine that the tags did not interfere with protein folding toward the native LHCII structure. After trimer formation, the majority of the complexes (an estimated 85.7% after the first purification step, Fig. 4) did not contain any spin label at all and were removed by Strep tag affinity chromatography. The trimers carrying two (11.2%, after the second purification step) or three (1.4%) labeled monomers are expected to exert only a minor influence on the obtained distance distributions.
FIGURE 3.
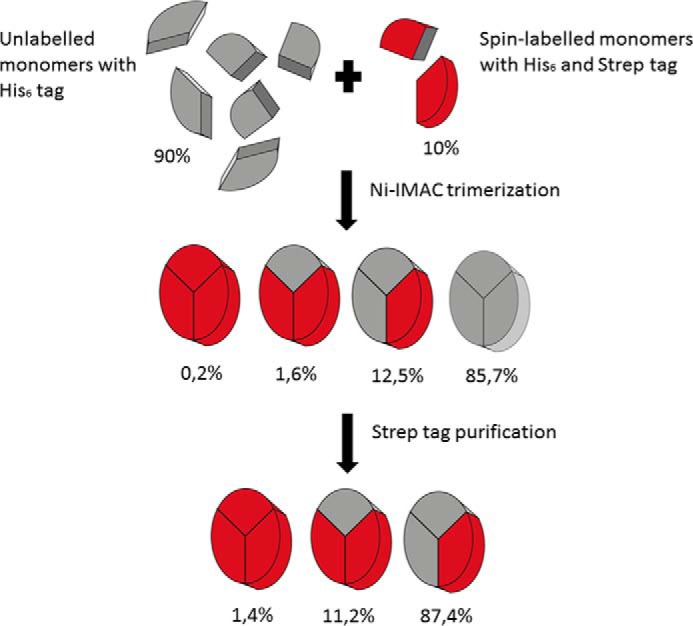
Production of heterogeneous trimers. Spin-labeled recombinant LHCII monomers with two affinity tags, one His6 tag and one Strep tag, were mixed with unlabeled recombinant LHCII monomers carrying only a His6 tag at a ratio of 1:8. After trimerization on a nickel column utilizing the His6 tag, trimeric complexes carrying no spin label and no Strep tag were removed from the solution by purification using the Strep tag II system from IBA. The resulting solution consisted mostly of trimeric complexes in which only one carried a label.
FIGURE 4.
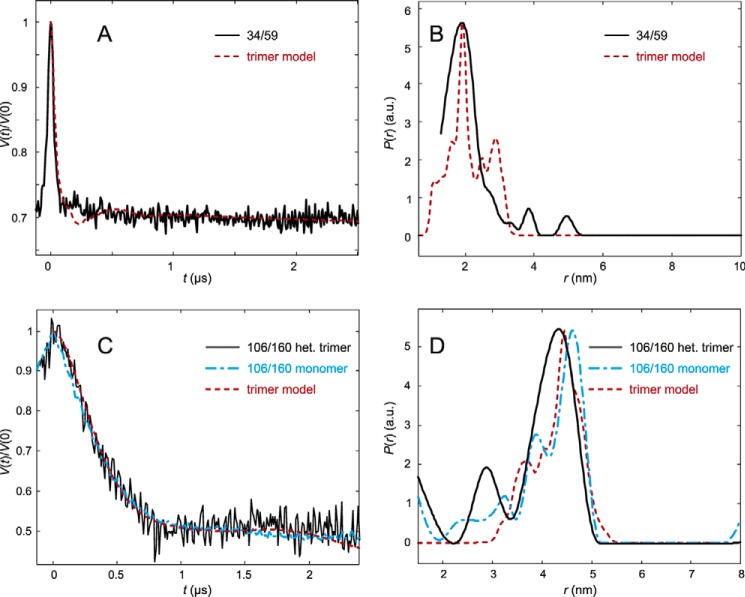
DEER analysis for LHCP spin labeled at residues 34 and 59 (A and B) and 106 and 160 (C and D) in trimeric LHCII in β-d-octylglucoside micelles. A, LHCII version 34/59. Primary experimental (DEER) data (black) and rotamer library prediction (red) based on the crystal structure (PDB code 2BHW). B, LHCII version 34/59. Experimental (black) distribution of interspin distances. C, LHCII version 106/160. Primary experimental (DEER) data (black) and rotamer library prediction (red) based on the crystal structure (PDB code 2BHW). Data from a control experiment on monomeric LHCP is shown as a blue line. D, LHCII version 106/160. Experimental (black, trimer; blue, monomer control) distributions of interspin distances are shown.
DEER Measurements and Data Analysis
Disordered domains can be structurally characterized by a sufficient number of site to site distance distributions. The DEER experiment provides access to such distance distributions in the required distance range between about 1.5 and 6 nm (10). To perform this experiment, amino acid residues at two sites in the system were mutated to cysteines and labeled by a thiol-reactive spin label. The two labels can be separately excited by two microwave frequencies resulting in a signal trace that is modulated as a function of time by the dipole-dipole interaction between the two unpaired electrons (primary DEER data). Using the inverse cube dependence of the dipole-dipole interaction on distance, the primary data can be converted to a label to label distance distribution. This conversion is an ill-posed mathematical problem that can be stabilized by the requirement that the distribution must be non-negative at all distances and by Tikhonov regularization. Although the detailed shape of the distribution varies between different preparations and different measurements as a consequence of ill-posedness, key parameters such as the mean distance and width of the distribution are stable even for broad distributions (31). For structural modeling, the label to label distribution needs to be related to a distance distribution between backbone sites, usually between the Cα atoms of the labeled residues. This can be done fast and with sufficient accuracy for disordered domains (32) by precomputing a library of spin label rotamers and attaching them in silico to a structure or structural model, considering the label-protein interaction with pairwise Lennard-Jones potentials between label and protein atoms (19). Structural models generated in this way have an uncertainty of the order of 0.8 nm (33).
Structural Restraints from ESEEM Water Accessiblity Measurements
For symmetry reasons, distance measurements in LHCII trimers consisting of three single spin-labeled monomers (6) provide information only about their positions relative to each other in the membrane plane but not with regard to the C3 symmetry axis (referred to as z axis hereafter). Water accessibility measurements by ESEEM spectroscopy (34) have revealed that residues in the N-terminal domain in LHCII trimers are all positioned near the membrane surface, in an almost coplanar arrangement (6). These findings have been used as an additional restraint to localize the N-proximal amino acids.
By analyzing the data from labeling sites in the N-terminal domain that are resolved in the crystal structure, we established a linear correlation between the water population pwater(zCα) at the z coordinate of the Cα atom zCα and the water accessibility parameter π(D2O), using a model for water distribution in the lipid bilayer that resulted from neutron scattering data (26). We found pwater(zCα) = 4.01 π(D2O) − 0.163 with a correlation coefficient of 0.517. The relatively poor correlation probably results from the difference between the spin label position and Cα coordinate and from application of a lipid bilayer model to measurements on detergent-solubilized protein. By fitting a bilayer model to LHCII as described in Ref. 22 and scaling the membrane width in the water distribution model to the fitted bilayer for LHCII, we could obtain estimates for zCα from π(D2O). We used the predicted error of the linear fit to establish lower (zmin) and upper (zmax) bounds on zCα (Table 1). If the upper bound is not specified, the residue could be fully water exposed within uncertainty of the model. For comparison, residues in the structurally well resolved section of the N-terminal domain have the following zCα coordinates: 14, 2.56 nm; 22, 1.99 nm; 29, 1.86 nm; 34, 2.77 nm; and 59, 2.22 nm. We note that all residues in the structurally unresolved section have rather high water accessibility suggesting localization on the surface of the membrane or, in our case, the detergent micelle.
TABLE 1.
Lower bounds for the z coordinate of Cα atoms of N-terminal residues derived from the ESEEM water accessibility parameters π(D2O) reported in Ref. 6
The z axis is perpendicular to the membrane and has its origin in the center of the lipid bilayer.
| Site | 3 | 4 | 7 | 9 | 10 | 11 | 12 |
|---|---|---|---|---|---|---|---|
| zmin/nm | 1.87 | 1.75 | 1.79 | 1.75 | 1.89 | 1.73 | 2.06 |
N Terminus
Labeling positions to assess the localization of the N-terminal region in LHCII trimers were 3, 7, and 11 in relationship to positions 34 and 59. The latter positions clearly belong to the rigid core of LHCII according to our earlier measurements of distances between monomers in a trimer with each monomer carrying a single spin label in the position to be characterized for its mobility (6). To confirm this by intramolecular distance measurements, we constructed heterotrimers with one monomer carrying spin labels at positions 34 and 59. A second control with spin labels at positions 106 and 160 was analyzed to confirm the similarity between detergent-solubilized heterotrimeric LHCII and the crystal structure. Fig. 4 shows that in both cases the primary DEER data and the distance distributions agree quite well with predictions from molecular modeling. For the distance distribution between residues 34 and 59, the measured maximum coincides almost precisely with that taken from the rotamer model, whereas the measured maximum of the 106–160 distance distribution is at a slightly (0.2 nm) shorter distance than from the rotamer model. These data indicate that the conformations seen here of the N-terminal domain between residues 59 and 34 and of the LHCII core between residues 106 and 160 in detergent micelles do not differ significantly from observed in the crystal.
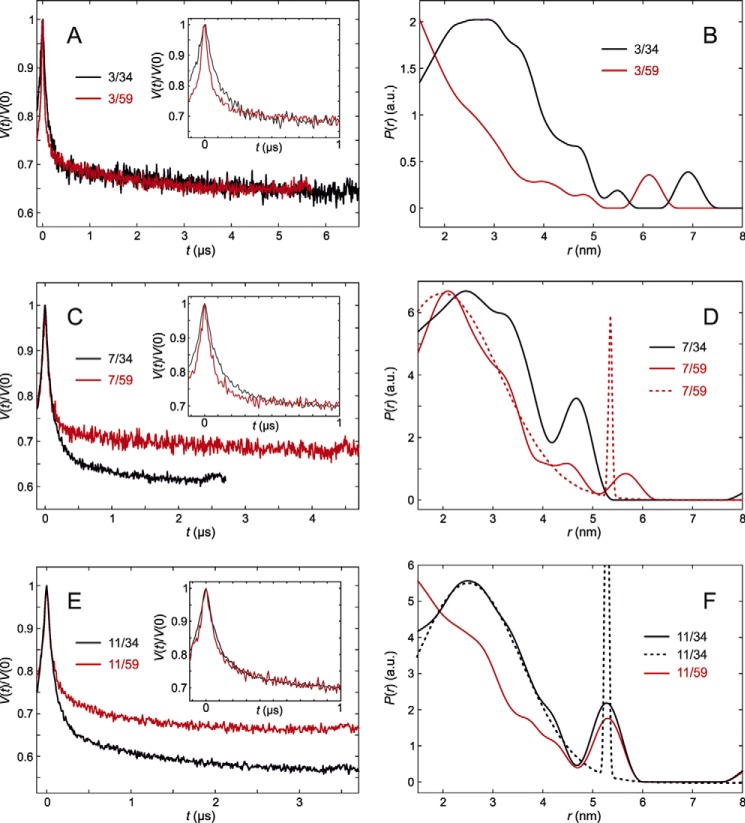
The measurement between site 3 next to the N terminus and site 34 showed a broad distance distribution (Fig. 5, A and B). Part of the distribution between sites 3 and 59 extends to shorter distances than can be measured by the DEER technique. Nevertheless, both the primary data and the distance distributions allow for safely concluding that residue 3 is closer to residue 59 than to residue 34.
FIGURE 5.
DEER analysis for trimeric LHCII versions with spin labels near the N terminus. Insets in A, C, and E show modulation depth-scaled zooms into the first 1 μs to highlight changes. A, primary experimental (DEER) data for trimeric LHCII versions 3/34 (black) and 3/59 (red) in n-octyl-β-d-glucoside (OG) micelles. B, inter-spin distance distributions between labels at residues 3/34 (black) and 3/59 (red). C, primary experimental (DEER) data for LHCII versions 7/34 (black) and 7/59 (red). D, distance distributions for LHCII versions 7/34 (black) and 7/59 (red). The dashed red line is the fit by a bimodal Gaussian distribution used for deriving the distance restraint for 7/59. The width of the long-distance aggregate peak is an unstable parameter at the restricted length of the time domain data. E, primary experimental (DEER) data for LHCII versions 11/34 (black) and 11/59 (red). F, distance distributions for LHCII versions 11/34 (black) and 11/59 (red). The dashed black line is the fit by a bimodal Gaussian distribution used for deriving the distance restraint for 11/34.
For position 7 the distributions regarding both anchor positions are rather similar, extending from less than 1.5 to about 4 nm (Fig. 5, C and D). This means that in detergent solution position 7 has, on average, a similar distance from positions 34 and 59. Together with earlier water accessibility data (6) that locate this residue near the surface of the detergent micelle, this allows for positioning residue 7 roughly on top of the superhelix formed by helices H1 and H4. Finally we checked site 11 (Fig. 5, E and F), which is again found to be significantly more remote from anchor site 34 than from anchor site 59.
To derive distance restraints for modeling the conformational distribution of residues 3–13 of the N-terminal domain, background corrected DEER data were analyzed in terms of bimodal Gaussian fits (Table 2). Note that the standard deviation σr,2 of the minor long-distance peak, corresponds to aggregates or unfolded protein, and thus its width and amplitude are not stable parameters in these fits, whereas the position and width of the major short-distance peak as well as its relative fraction f1 and the integral intensity of the long-distance peak are stable.
TABLE 2.
Distance restraints obtained by bimodal Gaussian fits
Mean distances 〈ri〉 and standard deviations σ4,i for the two components are given as well as the fraction f1 of the major component that was assigned to the distance distribution in well folded LHCII.
| Site pair | 〈r1〉/nm | σr,1/nm | 〈r2〉/nm | σr,2/nm | f1 |
|---|---|---|---|---|---|
| 3/34 | 2.66 | 1.60 | 5.56 | 0.05 | 0.97 |
| 3/59 | 1.50 | 1.66 | 5.90 | 0.93 | 0.91 |
| 7/34 | 2.67 | 1.62 | 6.60 | 0.05 | 0.96 |
| 7/59 | 1.92 | 1.61 | 5.36 | 0.05 | 0.96 |
| 11/34 | 2.45 | 1.56 | 5.23 | 0.05 | 0.92 |
| 11/59 | 1.50 | 1.73 | 4.73 | 1.13 | 0.82 |
Lumenal Loop
As described for the experiments addressing the N-terminal section, heterogeneous trimers were used to study the position and flexibility of the lumenal loop in a single monomer within a trimeric assembly. Distances were measured from three different positions (96, 102, and 113) in the loop region to an anchor position (143) on the stromal side of helix 2 in the rigid core of LHCII. These measurements were performed on monomeric and trimeric LHCII with and without bound NX.
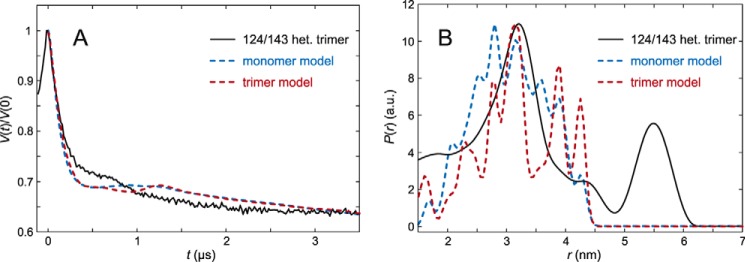
The rigidity of reference site 143 was tested against site 124 at the lumenal end of helix 2 (Fig. 6). The distance distribution obtained from the primary DEER data showed a peak with moderate width around 3.3 nm that is in reasonable agreement with the peak position predicted by the rotamer library approach from the crystal structure as a slightly narrower peak. This indicates that the two sites are fixed at a very similar distance in micelles as in the crystal structure. The width of the main peak of less than 1 nm qualifies residue 143 as an anchor for measuring the flexibility of the three labeled positions in the lumenal loop. The peak near 5.6 nm can again be assigned to the presence of protein aggregates. Note that residues 124 and 143 are located close to the interface between two monomers in the trimer, which leads to the differences in prediction for the monomeric and trimeric protein. In fact, the prediction for the trimer disregards the interaction of spin labels at site 143 with chlorophyll CHL612 in the same monomer, as otherwise the agreement with all measured distance distributions between site 143 and other sites deteriorates. We assume that the label can pack against CHL612 with minor influence on the LHCII structure, whereas the coarse-grained rotamer library prediction fails to find the tight fit of the label into a small cavity. Such cases have been observed before (35, 36).
FIGURE 6.
DEER analysis for trimeric LHCII version 124/143 in n-octyl-β-d)-glucoside (OG) micelles. A, primary experimental (DEER) data (black) and predictions by rotamer library simulations based on the crystal structure (PDB code 2BHW) assuming only a monomer (blue) or all three protomers in the trimer (red). B, corresponding distance distribution.
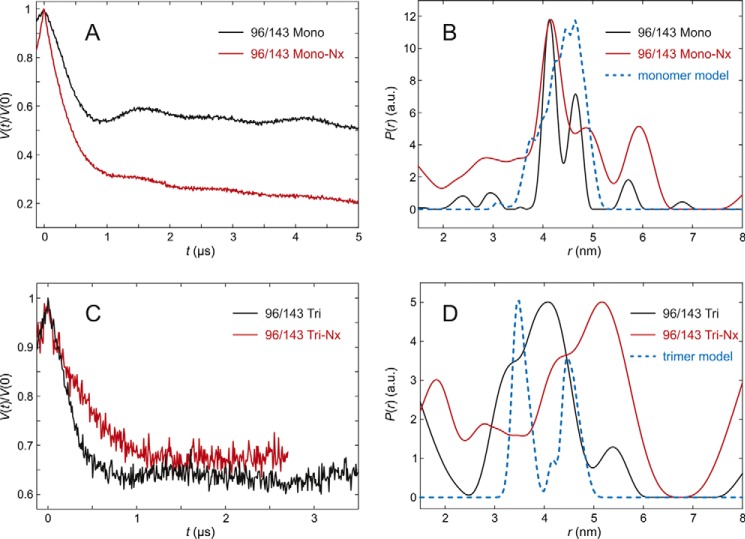
The flexibility of the first section of the lumenal loop was checked with LHCP doubly spin labeled at positions 96 and 143 (Fig. 7, A–D). In fully pigmented LHCII monomers, both the width of the distance distribution and the peak position are matched quite nicely by the rotamer library simulation (MMM). The small deviation can be explained by a slight difference in predicted and experimental distribution of spin label side group conformers, without invoking a difference in backbone conformation between LHCII crystals and LHCII solubilized in micelles. In monomers lacking NX, the width of the distribution is broader but the peak position remains the same. The modulation depth is too large for a two-spin system, which together with the broad component of the distance distribution suggests a significant fraction of aggregates. For the trimeric assembly, the distance distribution of the fully pigmented complex is nearly superimposable with the simulation but slightly broader, whereas the complex lacking NX had a higher population of longer distances indicated by the more prominent peak at 5.5 nm, most likely because of a higher tendency to form aggregates.
FIGURE 7.
DEER analysis for LHCII version 96/143 in n-octyl-β-d)-glucoside (OG) micelles. A, primary experimental (DEER) data in monomeric LHCII, fully pigmented (Mono, black) and lacking NX and VX (Mono-NX, red). B, distance distributions in monomeric LHCII in the presence (black) and absence (red) of NX and VX and prediction by the rotamer library (blue) based on chain A only in the crystal structure (PDB code 2BHW). C, primary experimental (DEER) data in trimeric LHCII, fully pigmented (Tri), and lacking NX and VX (Tri-NX). D, distance distribution in trimeric LHCII in the presence (black) and absence (red) of NX and VX and prediction by the rotamer library based on the labels attached to chain A with chains B and C contributing to label-protein interactions (blue).
The larger contamination of aggregates may have been caused by a lower reconstitution yield of the heterogenous trimer sample without NX of this LHCP version. This resulted in a less efficient purification of this particular sample, because as much volume as possible had to be collected from the sucrose density gradient used for isolating trimers to achieve sufficient yields. In both monomers and trimers of LHCII labeled at positions 96 and 143 and lacking NX, the main distance peak positions are both at 4.1 nm, suggesting that the most frequent monomer conformation of the loop domain does not differ from the trimer at this position.
The midsection of the lumenal loop was studied by LHCP doubly spin labeled at positions 102 and 143, with the label in the loop being placed within the short amphiphilic helix between residues 100 and 106. In monomeric LHCII, only a slight but significant shift of the peak position to shorter distances was observed between the fully pigmented complex and the one lacking NX (Fig. 8, A–D). The experimental distances are shorter by only 0.2 and 0.5 nm than the ones predicted by the rotamer library from the crystal structure. This may indicate a shift of the backbone at this position toward the core of the protein in detergent-solubilized LHCII compared with the crystal structure. However, using the “uniform rotamer distribution” feature of MMM we have found that the peak shift could also be explained by different side group conformations around the labeling position. The trimer sample exhibits broader distance distribution with some contributions at longer distances that are closer to the rotamer library prediction, but also a larger fraction of shorter distances. The apparent bimodality of the distribution is not significant, as it cannot be discerned by a kink in the primary data. The distribution in the sample lacking NX is also broad with a single maximum at 3.6 nm close to the maximum for the monomer sample. Comparing the monomer and trimer samples with NX bound, the difference is a broadening of the distribution that suggests larger flexibility in the trimer, whereas the position of the main peak is at the same position. This conclusion should be regarded with some caution as modulation depth and signal-to-noise ratio are lower than expected. However, the same conclusion can be drawn by comparison between monomers and trimers lacking NX for which data quality is higher for this LHCII version.
FIGURE 8.
DEER analysis for LHCII version 102/143 in n-octyl-β-d)-glucoside (OG) micelles. A, primary experimental (DEER) data in monomeric LHCII, fully pigmented (Mono, black) and lacking NX and VX (Mono-NX, red). B, distance distributions in monomeric LHCII in the presence (black) and absence (red) of NX and VX and prediction by the rotamer library (blue) based on chain A only in the crystal structure (PDB code 2BHW). C, primary DEER data in trimeric LHCII, fully pigmented (Tri) and lacking NX and VX (Tri-NX). D, distance distribution in trimeric LHCII in the presence (black) and absence (red) of NX and VX and prediction by the rotamer library based on the labels attached to chain A with chains B and C contributing to label-protein interactions (blue).
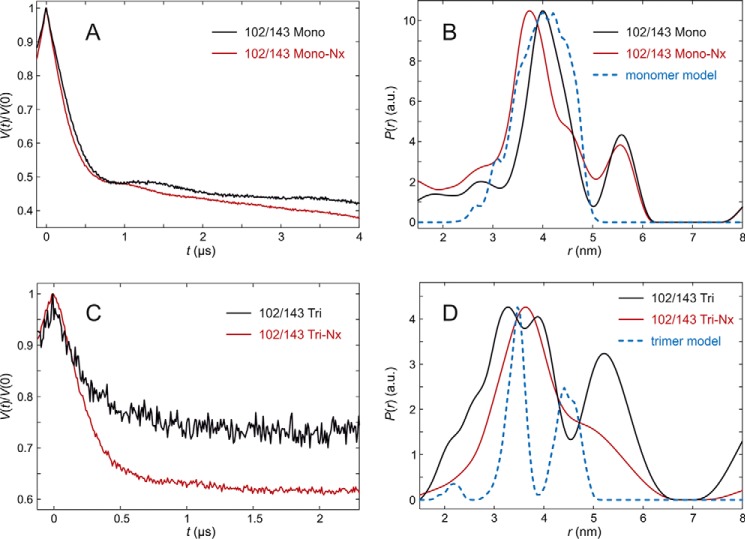
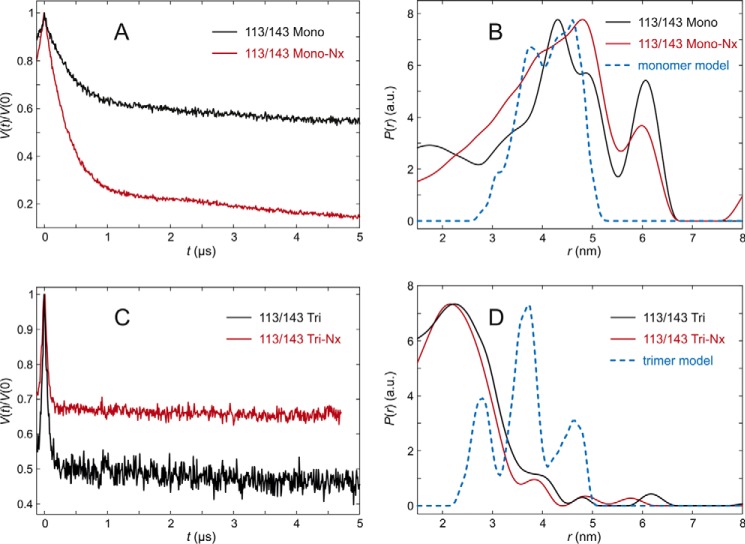
The C-proximal section of the lumenal loop domain, analyzed by LHCP labeled at positions 113 and 143, showed quite unexpected differences between the measured and modeled distance distributions of trimeric complexes (Fig. 9, A–D). The distance distribution measured with fully pigmented monomers was broad and peaked at about 4.3 nm, whereas modeling predicted a distribution that was also centered near 4.3 nm, but significantly narrower. In monomers lacking NX, the experimental distance distribution peaked at 4.8 nm and appeared even broader than that of the fully pigmented complex. Again the modulation depth is larger than expected, indicating some extent of aggregation that would explain the broadening. In the trimeric assembly state, the complexes plus and minus NX exhibited very similar distance distributions centered at about 2.2 nm. Surprisingly, the main peak maxima in the monomer and trimer measurements differed by about 2.1 nm. This deviation is far too large to be explained by altered label conformations and must have its origin in different conformations of the peptide. Furthermore, the peak observed in the trimeric assemblies is incompatible with predictions from the crystal structure for any set of rotamers computed by either excluding or including interaction with chlorophyll CHL612 in the same protomer. We can safely conclude that position 113 is closer to position 143 in solubilized LHCII than in the crystal structure. For this LHCII version the fraction of aggregated protein is insignificantly small. Therefore, a distance restraint for modeling was derived by a monomodal Gaussian fit of the data obtained in the presence of NX. We found a mean distance of 2.23 nm and a standard deviation of 1.4 nm.
FIGURE 9.
DEER analysis for LHCII version 113/143 in n-octyl-β-d)-glucoside (OG) micelles. A, primary experimental (DEER) data in monomeric LHCII, fully pigmented (Mono, black), and lacking NX and VX (Mono-NX, red). B, distance distributions in monomeric LHCII in the presence (black) and absence (red) of NX and VX and prediction by the rotamer library (blue) based on chain A only in the crystal structure (PDB code 2BHW). C, primary experimental (DEER) data in trimeric LHCII, fully pigmented (Tri), and lacking NX and VX (Tri-NX). D, distance distribution in trimeric LHCII in the presence (black) and absence (red) of NX and VX and prediction by the rotamer library based on the labels attached to chain A with chains B and C contributing to label-protein interactions (blue).
Discussion
DEER distance measurements have been used to analyze the positioning and motional freedom of two presumably flexible domains of LHCII, the N-terminal domain and the lumenal loop. In the following, these data sets will be discussed separately.
N Terminus
The crystal structures of LHCII give no information about the structure and localization of the N-terminal 9 (4) or 13 amino (3) acids. Most of this information comes from earlier EPR studies (6, 11, 12, 34). The present study extends our knowledge about this functionally important section of LHCII because we were able, for the first time, to measure intra-molecular distances in LHCP in the context of trimeric LHCII.
The present data confirms previous findings of a considerable flexibility of the N-proximal protein section, whereas the core of the protein is rigid (6). Controls (Fig. 4) proved that in the rigid part of LHCII in aqueous solution the intramolecular distances determined by DEER coincide well with the distances read from the crystal structure. This indicates that DEER is a reliable tool to determine the positioning with regard to the rest of the complex of N-terminal residues that are not resolved in the crystal structure. In the distance distributions between spin labels in this domain and labels in the rigid part of LHCII, the peak widths are significantly larger than the possible contribution of the local mobility of the label itself. On the other hand, virtually all distance distributions measured exhibit a clear maximum, indicating a preferred localization of that part of the N-terminal domain. Consequently, the orientation of the N terminus with regard to the rest of LHCII is far from random in its aqueous environment. Although not completely impossible, a selection of conformers during the shock freezing process is unlikely because it would only comprise those in structural equilibria that are faster than a few milliseconds, whereas our data on LHCII apoprotein folding show that the (re-)arrangement of entire domains of the protein takes seconds (12).
In previous EPR studies, the N-terminal protein structure in LHCII has been studied by measuring inter-molecular distances between LHCII subunits in trimers where each monomer carried a spin label near its N terminus (11). Some of these measurements were interpreted in terms of bimodal distance distributions, suggesting different preferred N-terminal protein structures between trimeric and monomeric LHCII. More recent measurements using additional labeling sites also indicated bimodal distance distributions between N termini in LHCII trimers, although they lacked baseline separation and, thus, the significance of this observation was questionable. In the present study, the intramolecular distance distributions within LHCII monomers in a trimer exhibited broad distributions but did not suggest two distinct families of conformations of the N terminus. In retrospect, the bimodality with baseline separation seen in our first set of experiments (11) can be traced back to an insufficient length of the primary data traces that lead to uncertainties in background correction. Newer data for the same LHCII version, obtained with a Q-band (34 GHz) spectrometer at much higher sensitivity (17), are all consistent with broad monomodal distributions. We are left with the conclusion that there is no definite proof for the existence of two preferred structures of the N-terminal domain in LHCII. We note, however, that water accessibility for some of the first 12 residues in the N-terminal loop is significantly larger in monomers than trimers (6), which suggests different conformational preferences of this loop in monomers and trimers.
The present results were used to model the position of the structurally unknown part of the N-terminal domain. Several positions in this domain were triangulated by measuring distances to two fixed positions in the rigid part of the complex, placing restraints on the possible localizations of the N terminus. Further restraints came from previous distance measurements mentioned above between single spin labels on each monomer in a trimer (6). To assess the “elevation” of the N-terminal section above the surface of the detergent micelle (which should be similar to the elevation above a membrane surface) we used ESEEM measurements of the D2O accessibility of N-proximal spin labels.
The ESEEM measurements showed that the accessibility parameter π(D2O) (34) of all residues lie between the value of a completely buried reference and complete water accessibility. Therefore all positions are located on the micellar surface, constraining the number of possible conformations of the N-terminal region significantly (6).
An ensemble model for residues 3–13 was created based on coordinates for residues 14–232 from the crystal structure of the LHCII (Protein Data Bank code 2BHW). The structure was first transformed to a frame where the C3-symmetry axis coincides with the z axis. A coarse-grained model for the lipid bilayer was modeled through minimizing the free energy of solvation of accessible amino acid residues using literature data (20). The middle of the lipid bilayer defines the origin of the z axis, so that all coordinates are relative to the middle. For calculation of the N-terminal model, residues 10–13 were removed from the 2BHW structure, as the rotamer library predictions from that structure were incompatible with the experimental data for residues in the 3–11 range and hardly compatible with the width of the distance distribution observed for residue 12, we had no experimental data for residue 13 in solubilized LHCII. Note also that residues 10 and 11 exhibit at best weak electron density in the map used to create the structure and that this density differs between the three monomers in the trimer.
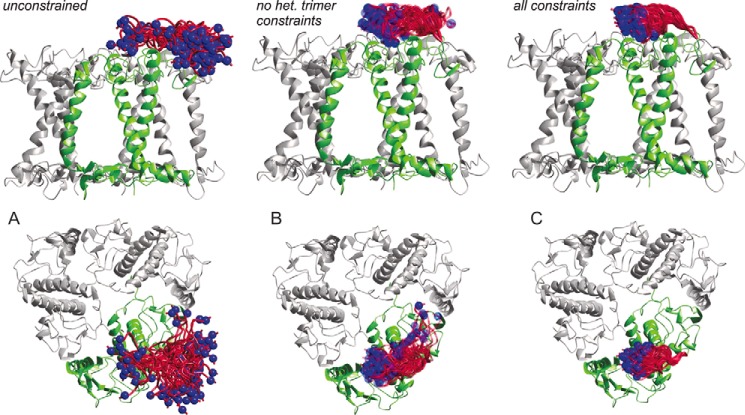
In Fig. 10, the N termini are modeled with increasing numbers of restraints from the data sources mentioned above. Fig. 10A shows the crystal structure of an LHCII trimer (2BHW) viewed from the membrane normal (top) and from above the membrane plane (bottom) with amino acids 3–13 modeled without any restraints except hindrance through steric clashes of amino acid side chains. The N-terminal section covers much of the aqueous space around position 14 and even dips into the lipid bilayer (or detergent micelle). Fig. 10B shows the ensemble of models with the structural restraints coming from measurements of distances between labels in distinct monomers within the trimer and from D2O accessibility. The residence of the N-terminal region is already largely confined to an area above its own monomer, whereas this set of restraints still allows for a second family of conformations where the N terminus reaches toward the second transmembrane helix of the adjacent monomer. Fig. 10C includes the restraints resulting from the present measurements of distances within the same monomer inside a trimer. These restraints exclude the second minor family of conformations and locate the N-terminal protein section in a narrow area above the superhelix, in an ellipsoid space with ∼2 nm in height and width and 4 nm in length.
FIGURE 10.
Model of 25 possible conformations of amino acid residues 3–13 of a LHCII monomer in a trimeric assembly viewed from the membrane plane (top) and from above the membrane normal (bottom). Blue spheres show a space-filling model of the Cα atom of residue 3. A, model without any restraints. B, restraints based on data published in Ref. 6. C, restraints based on data from Ref. 6 and heterogeneous trimer measurements.
On the one hand, some flexibility of the N-terminal domain is probably a prerequisite for the kinase involved in state transition, Stt7 (Chlamydomonas) or STN7 (Arabidopsis) (37, 38) to gain access to its target site at amino acid positions 5 and/or 6. For the minor light-harvesting complex CP29, a high mobility of its N terminus has also been detected by EPR and been interpreted to facilitate its phosphorylation (39). On the other hand, for the contribution of LHCII to the interaction between stacked membranes in the thylakoid grana, a limited mobility of this protein domain may be favorable. Grana stacking is thought to be stabilized by electrostatic interaction between positive charges in the N-proximal 15 amino acids and negative charges in the other stroma-exposed protein domains. Standfuss et al. (4) have put forward a Velcro model in which the stromal surface of LHCII exposes alternating patches of positive and negative charges. An electrostatic interaction of an N-terminal domain with negative charges of the same trimer would be counterproductive in this model because then these charges would be shielded from their interaction with the opposing surface of the next membrane in the stack. Our results obtained on detergent-solubilized LHCII safely exclude that in this environment the N terminus extends outwards from the C3 symmetry axis of the trimer, as modeled by Standfuss et al. (4), they suggest that it turns slightly inwards toward the negatively charged patch formed by residues Glu-150, Asp-153, Asp-162, Asp-168, Asp-169, and Glu-171. We note that the conformation modeled by Standfuss et al. (4) may be stabilized by the very grana stacking that it is hypothesized to support and may thus be less stable both in detergent-solubilized LHCII and, at least partially, in the crystal structure. The family of conformations that we detect would also be expected to be stable in monomeric LHCII, as the N-terminal domain contacts only residues within the same protomer. Our observations, together with the Velcro model, are also consistent with the notion that the conformation of the N-terminal loop can switch easily, as may be expected for regulation of photosynthesis by phosphorylation of residue Thr-5 or Thr-6. Further conformation of this model of a conformational switch of the N-terminal loop underlying state transition would require similar measurements as reported here, but in an environment that resembles stacked grana.
Lumenal Loop
The second set of experiments was aimed at the lumenal loop domain with regard to its positioning and flexibility in dependence on the oligomerization state of LHCII and its pigment composition. Recombinant LHCII passes two major purification steps upon the reconstitution and trimerization procedure, one by nickel-immobilized metal affinity chromatography and one by sucrose gradient purification. Experiments have shown that after this procedure, the binding site of VX is mostly unoccupied due to the instable binding of this xanthophyll. NX, which is located near the lumenal loop, is reversibly bound (15). In highly diluted LHCII solutions the binding site is unoccupied, but it is under the experimental conditions of this study. Whether there is a physiological reason for the reversibility of the NX binding is not clear, but because NX is a precursor for abscisic acid it might provide a substrate pool for ABA synthesis as part of a heat and drought stress response (15). Besides that, a conformational change of the NX molecule is discussed to be a step in the switch of the LHCII to a dissipating state in NPQ, which might have consequences for the conformation of the lumenal loop as well. The recombinant LHCII versions lacking NX exhibited a fluorescence quantum yield reduced by 15% (15) indicating that they are closer to the energy-dissipating structure of LHCII than the fully pigmented complex.
To investigate flexibility of the lumenal loop and the influence of bound NX on its conformation, spin-labeled monomers and heterogeneous trimers were reconstituted with and without NX. Because VX dissociates very easily without any noticeable consequences for the overall LHCII structure, it was not included in the reconstitution mixtures. The complexes obtained without NX were functional with regard to their endogenous energy transfer from Chl b to Chl a and as stable as the fully pigmented ones. The absorption spectra of complexes minus NX were compared with those of fully pigmented complexes and indicated, apart from NX and VX, the same pigment compositions in both cases ±0.5 Chl a or Chl b (see “Experimental Procedures”). On this basis, measurements of the structural impact of NX via DEER were possible.
The first section of the lumenal loop was analyzed by measuring distances from position 96 to 143 in the rigid part of the structure (6). Both in monomers and heterogenous trimers, the distance distributions plus and minus NX were almost superimposable, showing that this section of the lumenal loop is not influenced by NX with regard to their position or flexibility. The localization of position 96 with respect to 143 in solubilized LHCII agrees within experimental and spin label prediction uncertainty with the crystal structure.
In the second section, represented by the distance distributions of LHCP spin labeled at positions 102 and 143, the presence of NX results in somewhat stronger broadening of the distance distribution in the heterogeneous trimer than observed in the absence of NX. For this section our data suggest slightly enhanced backbone flexibility in trimeric assemblies compared with the monomeric state. The mean localization of position 102 with respect to 143 agrees with the crystal structure within experimental and prediction uncertainty.
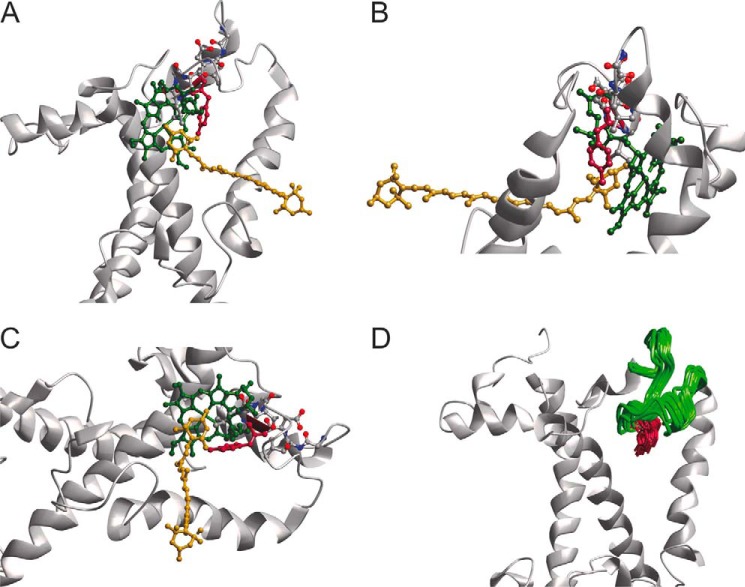
The last section, monitored by the spin pair 113/143, exhibited surprising differences between the monomer and trimer preparations. In monomers, the distance to reference position 143 on the stromal side is somewhat longer than predicted from the crystal structure, regardless of whether or not NX is bound. The difference is larger than experimental and prediction uncertainty and suggests a population of at least some conformations where position 113 is more remote from position 143 than in the crystal structure. Looking at the C-proximal loop section in a trimeric assembly, the distance between the loop and the anchor is only 2.2 nm, almost 2 nm shorter than predicted, with a larger difference of about 2.1 nm to the monomer. If one takes into account the mobility of the spin labels (11), position 113 is shifted at least 0.4 nm toward the micelle interior in comparison to the crystal structure, and most likely by more. This movement would cause steric clashes between Tyr-112 and the phytol chain of Chl a 606 in the conformation present in the crystal structure, so the pigment would have to alter its conformation as well. The same would be true for NX, as it is bound to Tyr-112 with an H-bond. Simulation shows, however, that only small movements would be required both for the phytol of Chl a 606 and for NX (Fig. 11). Comparing the distance distributions of position 113/143 of LHCII trimers with and without NX one can see almost no difference. The conformational change of the loop domain upon LHCII trimer formation as seen in our measurements is therefore independent of bound NX.
FIGURE 11.
Modeled lumenal loop domain with the restraint given by the trimer measurement of LHCII version 113/143. A–C, location of residue 113 if moved according to the measurement of 113/143 Tri (see Fig. 7) viewed from different angles. D, 20 models of residue 113 according to measurement of 113/143 Tri as only restraint with cofactors removed from the model. Obtained with Modeler (41) from the structure 2BHW (4).
Another possibility is that the label itself, in its function as a hydrophobic side chain, promotes alteration of the loop domain. We cannot safely exclude that this applies to our measurements. Nevertheless, if this is the case, it still demonstrates the high sensibility of the loop structure to its environment.
The three sections analyzed showed different mobilities. The section on the N-proximal side of the amphiphilic helix seems to be rigid, whereas a conformational change of the helix itself may depend on bound NX in a trimeric assembly. The section downstream of the helix is more mobile than the rest of the loop, which could be seen in increased peak widths in the distance distributions of LHCP versions 113/143. Here, two different conformations of the loop in monomers and trimers were detected, independently of the presence or absence of NX (see above).
A twist in NX associated to Tyr-112 via a hydrogen bond is thought to be involved in the induction of the energy dissipating state in NPQ (40), and exchanges of amino acid residue 111 resulted in an increased energy quenching capability. We detect a large structural flexibility of this section of the lumenal loop, which is in principle consistent with the notion of a local conformational change exerted by the NX twist. However, whereas we see a large difference in distance distributions between monomeric and trimeric LHCII, we see no appreciable differences in the complexes plus and minus NX. Either the binding of NX has no large effect on the lumenal loop structure, or we do not see this effect because of the high mobility of the loop domain.
Conclusion
The DEER measurements performed on the N-terminal and the lumenal loop domains of LHCII confirmed their mobilities seen in former studies (6, 34). The newly introduced technique of heterogeneously labeled trimers helped to reveal the conformation of these domains in the trimeric assembly state of LHCII. In detergent micelles lacking the grana stacks of the thylakoid environment, the N terminus adopts a conformation quite similar to that in the monomer. The lumenal loop domain revealed a rigid behavior on the N-proximal side of the amphiphilic helix, whereas the C-proximal section around the NX binding site adopted a conformation different from the one in the crystal structure. Both domains have in common that they show a high sensitivity to environmental changes, a feature that is likely to be related to the regulatory function of LHCII in photosynthesis.
Author Contributions
H. P. and G. J. conceived and coordinated the study. N. F. did the biochemical experiments. C. D. developed the method described in Fig. 3 and did some of the sample production and biochemical characterization of the LHCII samples concerning the N terminus. Y. P. and T. v. H. performed the EPR measurements and analysis of EPR spectra. G. J. analyzed the EPR spectra and modeled the N terminus and the lumenal loop. N. F., G. J., and H. P. wrote the manuscript. All authors reviewed the results and approved of the final version of the manuscript.
This work was supported by Schweizer Nationalfonds Grant 200020_14441 (to G. J.) and Deutsche Forschungsgemeinschaft Grant PA 324/9–1 (to H. P.). The authors declare that they have no conflicts of interest with the contents of this article.
- LHCII
- light harvesting complex II
- NPQ
- non-photochemical quenching
- Chl
- chlorophyll
- DEER
- double electron-electron resonance
- NX
- neoxanthin
- VX
- violaxanthin
- ESEEM
- electron spin echo envelope modulation
- LHCP
- light harvesting chlorophyll-binding protein
- DM
- n-dodecyl β-d-maltoside.
References
- 1.Pan X., Liu Z., Li M., and Chang W. (2013) Architecture and function of plant light-harvesting complexes II. Curr. Opin. Struct. Biol. 23, 515–525 [DOI] [PubMed] [Google Scholar]
- 2.Minagawa J. (2011) State transitions: the molecular remodeling of photosynthetic supercomplexes that controls energy flow in the chloroplast. Biochim. Biophys. Acta 1807, 897–905 [DOI] [PubMed] [Google Scholar]
- 3.Liu Z., Yan H., Wang K., Kuang T., Zhang J., Gui L., An X., and Chang W. (2004) Crystal structure of spinach major light-harvesting complex at 2.72-Å resolution. Nature 428, 287–292 [DOI] [PubMed] [Google Scholar]
- 4.Standfuss J., Terwisscha van Scheltinga A. C., Lamborghini M., and Kühlbrandt W. (2005) Mechanisms of photoprotection and nonphotochemical quenching in pea light-harvesting complex at 2.5-Å resolution. EMBO J. 24, 919–928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ilioaia C., Johnson M. P., Liao P.-N., Pascal A. A., van Grondelle R., Walla P. J., Ruban A. V., and Robert B. (2011) Photoprotection in plants involves a change in lutein 1 binding domain in the major light-harvesting complex of photosystem II. J. Biol. Chem. 286, 27247–27254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dockter C., Müller A. H., Dietz C., Volkov A., Polyhach Y., Jeschke G., and Paulsen H. (2012) Rigid core and flexible terminus: structure of solubilized light-harvesting chlorophyll a/b complex (LHCII) measured by EPR. J. Biol. Chem. 287, 2915–2925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sunku K., de Groot H. J., and Pandit A. (2013) Insights into the photoprotective switch of the major light-harvesting complex II (LHCII): a preserved core of arginine-glutamate interlocked helices complemented by adjustable loops. J. Biol. Chem. 288, 19796–19804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Belgio E., Duffy C. D., and Ruban A. V (2013) Switching light harvesting complex II into photoprotective state involves the lumen-facing apoprotein loop. Phys. Chem. Chem. Phys. 15, 12253–12261 [DOI] [PubMed] [Google Scholar]
- 9.Zubik M., Luchowski R., Grudzinski W., Gospodarek M., Gryczynski I., Gryczynski Z., Dobrucki J. W., and Gruszecki W. I. (2011) Light-induced isomerization of the LHCII-bound xanthophyll neoxanthin: possible implications for photoprotection in plants. Biochim. Biophys. Acta 1807, 1237–1243 [DOI] [PubMed] [Google Scholar]
- 10.Jeschke G. (2012) DEER distance measurements on proteins. Annu. Rev. Phys. Chem. 63, 419–446 [DOI] [PubMed] [Google Scholar]
- 11.Jeschke G., Bender A., Schweikardt T., Panek G., Decker H., and Paulsen H. (2005) Localization of the N-terminal domain in light-harvesting chlorophyll a/b protein by EPR measurements. J. Biol. Chem. 280, 18623–18630 [DOI] [PubMed] [Google Scholar]
- 12.Dockter C., Volkov A., Bauer C., Polyhach Y., Joly-Lopez Z., Jeschke G., and Paulsen H. (2009) Refolding of the integral membrane protein light-harvesting complex II monitored by pulse EPR. Proc. Natl. Acad. Sci. U.S.A. 106, 18485–18490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cashmore A. R. (1984) Structure and expression of a pea nuclear gene encoding a chlorophyll a/b-binding polypeptide. Proc. Natl. Acad. Sci. U.S.A. 81, 2960–2964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hobe S., Prytulla S., Kühlbrandt W., and Paulsen H. (1994) Trimerization and crystallization of reconstituted light-harvesting chlorophyll a/b complex. EMBO J. 13, 3423–3429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hobe S., Trostmann I., Raunser S., and Paulsen H. (2006) Assembly of the major light-harvesting chlorophyll-a/b complex: thermodynamics and kinetics of neoxanthin binding. J. Biol. Chem. 281, 25156–25166 [DOI] [PubMed] [Google Scholar]
- 16.Pannier M., Veit S., Godt A., Jeschke G., and Spiess H. W. (2000) Dead-time free measurement of dipole–dipole interactions between electron spins. J. Magn. Reson. 142, 331–340 [DOI] [PubMed] [Google Scholar]
- 17.Polyhach Y., Bordignon E., Tschaggelar R., Gandra S., Godt A., and Jeschke G. (2012) High sensitivity and versatility of the DEER experiment on nitroxide radical pairs at Q-band frequencies. Phys. Chem. Chem. Phys. 14, 10762–10773 [DOI] [PubMed] [Google Scholar]
- 18.Jeschke G., Chechik V., Ionita P., Godt A., Zimmermann H., Banham J., Timmel C. R., Hilger D., and Jung H. (2006) DeerAnalysis2006: a comprehensive software package for analyzing pulsed ELDOR data. Appl. Magn. Reson. 30, 473–498 [Google Scholar]
- 19.Polyhach Y., Bordignon E., and Jeschke G. (2011) Rotamer libraries of spin labelled cysteines for protein studies. Phys. Chem. Chem. Phys. 13, 2356–2366 [DOI] [PubMed] [Google Scholar]
- 20.Adamian L., Nanda V., DeGrado W. F., and Liang J. (2005) Empirical lipid propensities of amino acid residues in multispan α helical membrane proteins. Proteins 59, 496–509 [DOI] [PubMed] [Google Scholar]
- 21.Alonso-García N., García-Rubio I., Manso J. A., Buey R. M., Urien H., Sonnenberg A., Jeschke G., and de Pereda J. M. (2015) Combination of x-ray crystallography, SAXS and DEER to obtain the structure of the FnIII-3,4 domains of integrin α6β4. Acta Crystallogr. D Biol. Crystallogr. 71, 969–985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Raba M., Dunkel S., Hilger D., Lipiszko K., Polyhach Y., Jeschke G., Bracher S., Klare J. P., Quick M., Jung H., and Steinhoff H.-J. (2014) Extracellular loop 4 of the proline transporter PutP controls the periplasmic entrance to ligand binding sites. Structure 22, 769–780 [DOI] [PubMed] [Google Scholar]
- 23.Hovmöller S., Zhou T., and Ohlson T. (2002) Conformations of amino acids in proteins. Acta Crystallogr. D Biol. Crystallogr. 58, 768–776 [DOI] [PubMed] [Google Scholar]
- 24.Sugeta H., and Miyazawa T. (1967) General method for calculating helical parameters of polymer chains from bond lengths, bond angles, and internal-rotation angles. Biopolymers 5, 673–679 [Google Scholar]
- 25.Shimanouchi T., and Mizushima S. (1955) On the helical configuration of a polymer chain. J. Chem. Phys. 23, 707 [Google Scholar]
- 26.Kucerka N., Nagle J. F., Sachs J. N., Feller S. E., Pencer J., Jackson A., and Katsaras J. (2008) Lipid bilayer structure determined by the simultaneous analysis of neutron and x-ray scattering data. Biophys. J. 95, 2356–2367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sali A., and Blundell T. L. (1993) Comparative protein modelling by satisfaction of spatial restraints. J. Mol. Biol. 234, 779–815 [DOI] [PubMed] [Google Scholar]
- 28.Hobe S., Niemeier H., Bender A., and Paulsen H. (2000) Carotenoid binding sites in LHCIIb: relative affinities towards major xanthophylls of higher plants. Eur. J. Biochem. 267, 616–624 [DOI] [PubMed] [Google Scholar]
- 29.Rühle W., and Paulsen H. (2004) Preparation of native and recombinant light-harvesting chlorophyll-a/b complex. Methods Mol. Biol. 274, 93–103 [DOI] [PubMed] [Google Scholar]
- 30.Schmidt T. G., and Skerra A. (2007) The Strep-tag system for one-step purification and high-affinity detection or capturing of proteins. Nat. Protoc. 2, 1528–1535 [DOI] [PubMed] [Google Scholar]
- 31.Jeschke G., Panek G., Godt A., Bender A., and Paulsen H. (2004) Data analysis procedures for pulse ELDOR measurements of broad distance distributions. Appl. Magn. Reson. 26, 223–244 [Google Scholar]
- 32.Jeschke G. (2013) Conformational dynamics and distribution of nitroxide spin labels. Prog. Nucl. Magn. Reson. Spectrosc. 72, 42–60 [DOI] [PubMed] [Google Scholar]
- 33.Bleicken S., Jeschke G., Stegmueller C., Salvador-Gallego R., García-Sáez A. J., and Bordignon E. (2014) Structural model of active Bax at the membrane. Mol. Cell. 56, 496–505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Volkov A., Dockter C., Bund T., Paulsen H., and Jeschke G. (2009) Pulsed EPR determination of water accessibility to spin-labeled amino acid residues in LHCIIb. Biophys. J. 96, 1124–1141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lillington J. E., Lovett J. E., Johnson S., Roversi P., Timmel C. R., and Lea S. M. (2011) Shigella flexneri Spa15 crystal structure verified in solution by double electron electron resonance. J. Mol. Biol. 405, 427–435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hagelueken G., Ward R., Naismith J. H., and Schiemann O. (2012) MtsslWizard: in silico spin-labeling and generation of distance distributions in PyMOL. Appl. Magn. Reson. 42, 377–391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Depège N., Bellafiore S., and Rochaix J.-D. (2003) Role of chloroplast protein kinase Stt7 in LHCII phosphorylation and state transition in Chlamydomonas. Science 299, 1572–1575 [DOI] [PubMed] [Google Scholar]
- 38.Bellafiore S., Barneche F., Peltier G., and Rochaix J.-D. (2005) State transitions and light adaptation require chloroplast thylakoid protein kinase STN7. Nature 433, 892–895 [DOI] [PubMed] [Google Scholar]
- 39.Shabestari M. H., Wolfs C. J., Spruijt R. B., van Amerongen H., and Huber M. (2014) Exploring the structure of the 100 amino-acid residue long N-terminus of the plant antenna protein CP29. Biophys. J. 106, 1349–1358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ruban A. V., Berera R., Ilioaia C., van Stokkum I. H., Kennis J. T., Pascal A. A., van Amerongen H., Robert B., Horton P., and van Grondelle R. (2007) Identification of a mechanism of photoprotective energy dissipation in higher plants. Nature 450, 575–578 [DOI] [PubMed] [Google Scholar]
- 41.Eswar N., Webb B., Marti-Renom M. A., Madhusudhan M. S., Eramian D., Shen M.-Y., Pieper U., and Sali A. (2006) Comparative protein structure modeling using Modeller. Curr. Protoc. Bioinformat. 5, 5.6.1–5.6.30 [DOI] [PMC free article] [PubMed] [Google Scholar]