Background: BpV(phen) is an insulin-mimetic small molecule.
Results: We demonstrate that bpV(phen) reduces the stability of p62 in a proteasome-dependent way to activate HDAC6 to inhibit autophagy and induce apoptosis and pyroptosis.
Conclusion: We propose that bpV(phen) inhibits autophagy through p62.
Significance: We provide insights into a novel function of bpV(phen) and p62 in autophagy.
Keywords: apoptosis, autophagy, caspase 1 (CASP1), histone deacetylase 6 (HDAC6), microtubule, p62 (sequestosome 1(SQSTM1)), tubulin, bpV(phen), lactate dehydrogenase, pyroptosis
Abstract
Autophagy is a cellular process that controls and executes the turnover of dysfunctional organelles and misfolded or abnormally aggregated proteins. Phosphatase and tensin homologue deleted on chromosome 10 (PTEN) activates the initiation of autophagy. Autophagosomes migrate along acetylated microtubules to fuse with lysosomes to execute the degradation of the engulfed substrates that usually bind with sequestosome 1 (SQSTM1, p62). Microtubule-associated protein 1 light chain 3 (LC3) traces the autophagy process by converting from the LC3-I to the LC3-II isoform and serves as a major marker of autophagy flux. Potassium bisperoxo(1,10-phenanthroline)oxovanadate (bpV(phen)) is an insulin mimic and a PTEN inhibitor and has the potential to treat different diseases. Here we show that bpV(phen) enhances the ubiquitination of p62, reduces the stability of p62, disrupts the interaction between p62 and histone deacetylase 6 (HDAC6), activates the deacetylase activity of HDAC6 on α-tubulin, and impairs stable acetylated microtubules. Microtubular destabilization leads to the blockade of autophagosome-lysosome fusion and accumulation of autophagosomes. Autophagy defects lead to oxidative stress and lysosomal rupture, which trigger different types of cell death, including apoptosis and pyroptosis. The consistent results from multiple systems, including mouse and different types of mammalian cells, are different from the predicted function of bpV(phen) as a PTEN inhibitor to activate autophagy flux. In addition, levels of p62 are reduced but not elevated when autophagosomal degradation is blocked, revealing a novel function of p62 in autophagy regulation. Therefore, it is necessary to pay attention to the roles of bpV(phen) in autophagy, apoptosis, and pyroptosis when it is developed as a drug.
Introduction
Mammalian cells primarily use the autophagy-lysosome pathway to degrade dysfunctional organelles, misfolded/aggregated proteins, and other macromolecules (1). The autophagy process is regulated by a class III PI3K complex functioning in vesicle nucleation; a serine-threonine kinase complex involved in autophagic induction; and a pair of novel ubiquitin-like protein-conjugating systems, the ATG12 and ATG8 systems, promoting the extension and completion of vesicles (2). Phosphatase and tensin homologue deleted on chromosome 10 (PTEN),3 a phosphotyrosine phosphatase, activates autophagy initiation by inhibiting PI3K (3). Microtubule-associated protein 1 light chain 3 (LC3) is the mammalian homologue of ATG8, one of the key autophagy markers (4). The LC3 precursor is truncated to form the cytosolic LC3-I and further conjugated with phosphatidylethanolamine to create the membrane-associated LC3-II with the assistance of ATG5 and ATG12 (5, 6). The LC3-II-associated isolation membranes target and then completely envelop individual organelles or macromolecules that bind to polyubiquitin-binding protein p62/SQSTM 1 (p62) to form autophagosomes (7, 8). Autophagosomes migrate along acetylated microtubules to fuse with lysosomes to form autolysosomes, in which substrates are degraded (9–11). Inhibition of autophagy initiation reduces the generation of autophagosomes, so the levels of LC3-II and p62 are reduced when lysosomal activity is suppressed. However, inhibition of the degradation of autophagosomes leads to accumulation of LC3-II and p62.
Potassium bisperoxo(1,10-phenanthroline)oxovanadate (K[VO(O2)2C12H8N2]·3H2O or bpV(phen)) is a peroxovanadium small molecule with insulin-mimetic properties (12). It has been demonstrated to activate insulin receptor kinase and to inhibit the dephosphorylation of autophosphorylated insulin receptors and cause significant decreases in circulating insulin and plasma glucose levels (13). It has also been characterized as a potent protein phosphotyrosine phosphatase inhibitor exhibiting anti-tumor activity (14). It inhibits PTEN to induce cardioprotection against ischemia-reperfusion injury (15).
Because of the potential of bpV(phen) to treat different diseases and its inhibition of PTEN which has been suggested to regulate autophagy through PI3K, we wanted to investigate whether bpV(phen) affects autophagy. Here we report that bpV(phen) promotes not only cell apoptosis but also pyroptosis, resulting in overall cell death. Surprisingly, it exhibited no impact on autophagy initiation but blocked autophagosomal degradation by reducing the stability of p62 to activate HDAC6 to impair the fusion of autophagosomes and lysosomes, supported by acetylated microtubules.
Experimental Procedures
Antibodies, siRNAs, Plasmids, and Other Reagents
Antibodies against human LC3 (catalog no. NB 100-2331) were purchased from Novus Biologicals. The IgG control antibody from rabbit (catalog no. sc-2027), primary antibodies against PARP (catalog no. sc-7150), proliferating cell nuclear antigen, catalog no. sc-9707), α-tubulin (catalog no. sc-12462), β-actin (catalog no. sc-47778), HDAC6 (catalog no. sc-11420), acetylated α-tubulin (catalog no. sc-23950), random sequence control siRNA (catalog no. sc-44234), and siRNA specific to p62 (catalog no. sc-29679) were from Santa Cruz Biotechnology, Inc. Antibody against p62 (SQSTN1, catalog no. BWL-PW9860) was from Enzo Life Sciences International Inc. HRP-conjugated secondary antibodies against mouse (catalog no. 172-1011) and rabbit (catalog no. 172-1019) were from Bio-Rad. FITC goat anti-rabbit IgG (catalog nos. R6393 and A-21070) and Oligofectamine (catalog no. 12252-011) were from Invitrogen. RFP-LC3 was supplied by Dr. Mizushima (16). Antibody against caspase 1 (catalog no. PRS3459), MG-132, bafilomycin A1, bpV(phen), and the lactate dehydrogenase activity assay kit (catalog no. MAK066–1KT) were from Sigma-Aldrich. The protein G beads were from Amersham Biosciences. Antibody against phospho-AKT (Ser-473) (p-AKT) was from Cell Signaling Technology. The CellTiter 96® non-radioactive cell proliferation assay (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide) was from Promega. The Annexin V/FITC apoptosis detection kit was from BD Biosciences.
Cell Culture, Cell Transfection with siRNA, Coimmunoprecipitation Assay, Immunoblot Analysis, and Fluorescence Microscopy
HepG2 cells, HeLa cells, HeLa cells stably expressing ERFP-LC3, and mouse embryonic fibroblasts (MEFs) were established and cultured as described previously (17, 18). HeLa cells were transfected with random or p62-specific siRNA molecules packed with Oligofectamine in a similar way as we described previously (17, 18). Cell lysates were prepared from attached cells, and coimmunoprecipitation was performed as described previously (17, 18). Cell lysates with the same amounts of total proteins were subjected to immunoprecipitation with equal amounts of specific antibodies and control antibodies (IgG) from the same species. Lysates and/or immunoprecipitates were subjected to immunoblot analyses as reported previously (17, 18). Immunofluorescence staining was performed, and images were captured with a laser-scanning microscope in a similar way as described previously (17, 18).
Cell Proliferation Assay
The cell proliferation rate was assessed using the CellTiter 96® non-radioactive cell proliferation assay (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide, Promega) according to the protocols of the manufacturer. Briefly, 10,000 cells were seeded in a well on 96-well plates (Corning) containing 100 μl of culture medium plus different concentrations of bpV(phen) and grown at 37 °C for 24 h. Then 15 μl of dye solution was added to each well, plates were incubated continuously at 37 °C for 4 h, the reaction was terminated with 100 μl of stop solution, and optical densities at 570 nm were determined.
Detection of Apoptotic Cells
Apoptosis was evaluated using the Annexin V/FITC apoptosis detection kit. At first, cells were treated with different concentrations of bpV(phen) for 48 h and harvested by centrifugation twice at 1000 rpm (5 min for each spin). Cells were then washed twice (3 min for each wash) in binding buffer. 1 × 106 cells were resuspended in 1 ml of binding buffer containing 1.25 μl of Annexin V-FITC (BD Biosciences) and 10 μl of propidium iodide (PI), and incubated for 15 min at room temperature in the dark. Finally, cell cycle analysis was performed by flow cytometry (BD Calibar). Scatter plots were performed against the intensities of FITC fluorescence and PI fluorescence. The scatter plots were divided into four quadrants: the left lower quadrant (Annexin V-FITC (−) and PI (−)) representing viable cells, the left upper quadrant (Annexin V-FITC (−) and PI (+)) representing necrotic cells, the right lower quadrant (Annexin V-FITC (+) and PI (−)) representing early apoptotic cells, and the right upper quadrant (Annexin V-FITC (+) and PI (+)) representing late apoptotic cells.
Lactate Dehydrogenase Release Assay
To quantify the extent of pyroptosis after bpV(phen) exposure, release of lactate dehydrogenase in cell culture supernatants was determined using the lactate dehydrogenase activity assay kit from Sigma-Aldrich according to the instructions of the manufacturer. Briefly, HeLa cells were cultured in the presence of different concentration of bpV(phen) for 48 h. Then 50 μl of cell culture supernatant was collected, added to the kit reagent, and incubated for 10 min at room temperature. After addition of stop solution, fluorescence intensity was measured using a microplate reader at an excitation wavelength of 560 nm and an emission wavelength of 590 nm.
Animal Experimentation
Animal protocols were approved by the Institutional Animal Care and Use Committee, Institute of Biosciences and Technology, Texas A&M Health Science Center. All animals received humane care according to the criteria outlined in the Guide for the Care and Use of Laboratory Animals prepared by the National Academy of Sciences and published by the National Institutes of Health (Publication 86-23, revised 1985). Three pairs of 1-month-old, freely fed female wild-type C57BL/6 mice were subjected to subcutaneous injection of 2.5 μmol/30 g body weight in a total of 250 μl of saline (or saline only for the control group) as described previously (19).
Results
Treatment with bpV(phen) Induces Cell Apoptosis and Pyroptosis
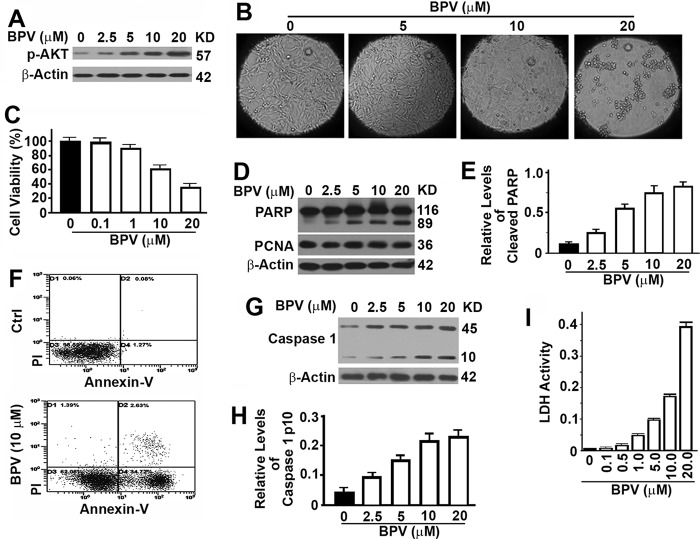
To test the function of bpV(phen), we treated MEF cells with increasing concentrations of bpV(phen) for 48 h. As a PTEN inhibitor, bpV(phen) enhanced the levels of phosphorylated AKT (Fig. 1A). A dose-dependent reduction in cell population and cell viability was detected (Fig. 1, B and C). Examination of the treated cells by immunoblot revealed an even level of proliferating cell nuclear antigen and increasing levels of 89-kD PARP fragments (Fig. 1, D and E). Proliferating cell nuclear antigen is a cellular marker for proliferation. PARP is cleaved into an 89-kD fragment by the apoptosis-specific caspase 3, and levels of such fragments serve as markers of apoptosis. These data suggest that bpV(phen) exposure helps the cell maintain a constant rate of cell proliferation but an increasing rate of apoptosis so that the cell population is reduced. Cell apoptosis was further confirmed by Annexin-V flow cytometry (Fig. 1F). Pyroptosis is another type of cell death that is different from apoptosis and is characterized by the activation of caspase 1 and release of cellular contents. Treatment with bpV(phen) led to dose-dependent increases in cellular levels of active caspase 1 with a molecular weight of 10 kDa and levels of lactate dehydrogenase released in the medium (Fig. 1, G–I). Therefore, bpV(phen) induces both apoptosis and pyroptosis.
FIGURE 1.
Treatment with bpV(phen) induces cell apoptosis and pyroptosis. A, representative immunoblots showing the impact of bpV(phen) (BPV) on the levels of phosphorylated AKT in HeLa cells treated with different concentration of bpV(phen) dissolved in dimethyl sulfoxide for 48 h. KD, kilodalton. B, representative morphology of cultured MEF cells after treatment with different concentrations of bpV(phen) for 48 h. C, plots of viabilities (percent) of HeLa cells incubated with increasing concentrations of bpV(phen) for 48 h by 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide assay. Data here and throughout are the mean ± S.D. of at least three repeats. D, immunoblot analysis of the levels of PARP and proliferating cell nuclear antigen (PCNA) in MEF cells treated with different concentrations of bpV(phen) for 48 h. In immunoblot analyses shown here and later, cell lysates with the same amounts of total proteins were loaded, and the level of β-Actin served as another loading control. E, plots of relative intensities of the cleaved 89-kD PARP fragment to β-Actin as shown in D. The plots here and shown later are the mean ± S.D. of at least three repeats. F, representative dot plots of apoptotic cells in HeLa cells treated with increasing concentrations of bpV(phen) by flow cytometry. Ctrl, control; PI, propidium iodide. G, immunoblot analysis of the levels of caspase 1 in MEF cells treated with different concentrations of bpV(phen) for 48 h. H, plots of relative intensities of the cleaved 10-kD active caspase 1 to β-Actin as shown in G. I, plots of lactate dehydrogenase (LDH) activity released in medium from cultured HeLa cells.
Treatment with BpV(phen) Results in Blockade of the Degradation of Autophagosomes
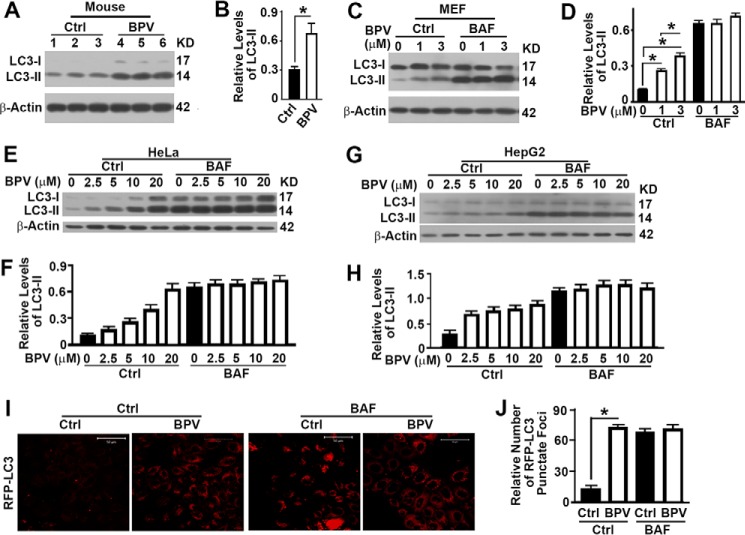
Because of the close relationship between autophagy and the cell death pathways of apoptosis and pyroptosis (20, 21), we examined the impact of bpV(phen) on autophagy. We injected freely fed wild-type mice with bpV(phen) and collected liver tissues for autophagy analysis. We found that bpV(phen) induced a significant increase in the levels of LC3-II (Fig. 2, A and B), suggesting either an activation of autophagy initiation or an inhibition of autophagosomal degradation. To determine the exact mechanism, we treated MEF cells with bpV(phen) in the absence or presence of bafilomycin A1 (BAF) to block the degradation of autophagosomes and observed similar increases of LC3-II in the absence of BAF but no increase of LC3-II in the presence of BAF (Fig. 2, C and D), indicating a blockade of the degradation of autophagosomes. The same trends were observed when either HeLa cells (Fig. 2, E and F) or HepG2 cells (Fig. 2, G and H) were exposed to bpV(phen). When HeLa cells stably expressing RFP-LC3 were exposed to bpV(phen), we detected a significant increase in the number of RFP-LC3 punctate foci in the absence of BAF but no increase in the presence of BAF (Fig. 2, I and J). All results indicate that bpV(phen) induces a blockade of the degradation of autophagosomes.
FIGURE 2.
Treatment with bpV(phen) results in blockade of the degradation of autophagosomes. A, immunoblot analysis of the impact of bpV(phen) (BPV) on the levels of the autophagy marker LC3-II in the liver tissues of wild-type mice. Ctrl, control. KD, kilodalton. B, plots of the relative intensities of LC3-II to β-Actin as shown in A. *, p < 0.05. C–H, immunoblot analyses (C, E, and G) and plots (D, F, and H) of the impact of bpV(phen) on the levels of the autophagy marker LC3-II in wild-type MEF (C and D), HeLa (E and F), or HepG2 cells (G and H) treated with different concentrations of bpV(phen) in the absence (Ctrl) or presence of BAF. I, fluorescence images of HeLa cells stably expressing RFP-LC3 treated with 20 μm of bpV(phen) for 48 h. J, quantification of GFP-LC3-labeled autophagosomes as shown in I. The data were the mean number of RFP-LC3 punctate foci ± S.D. for 10 randomly selected images in a field of 512 × 512 pixels.
Treatment with bpV(phen) Reduces the Stability of p62 in a Proteasome-dependent Way
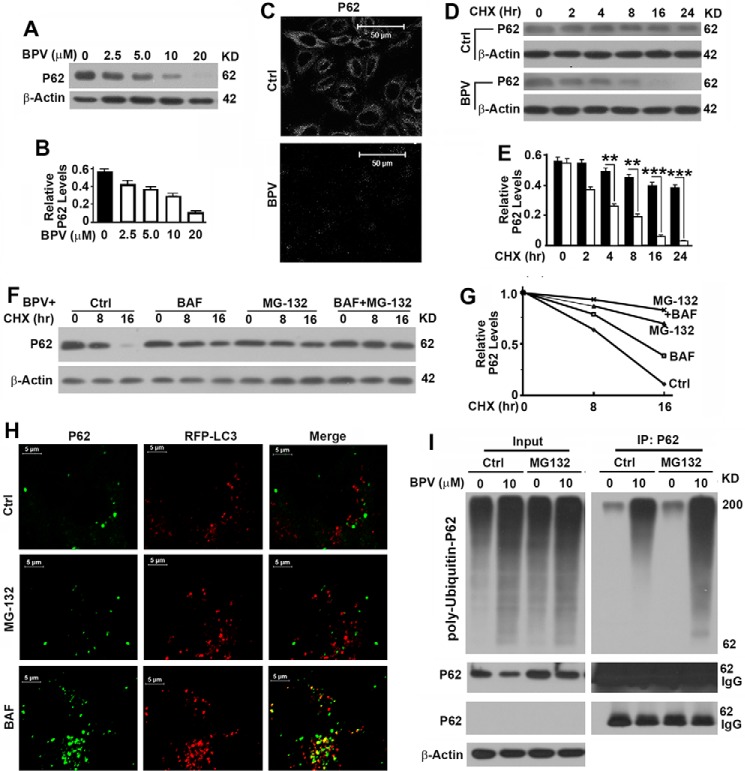
p62, another important marker of autophagy flux, has been widely considered to act as a receptor to bring ubiquitinated proteins into the autophagosome for degradation (7). It is expected to be elevated when the degradation of autophagosomes is inhibited. Surprisingly, we found a dose-dependent decrease in the levels of p62 on the basis of both immunoblot analysis (Fig. 3, A and B) and immunofluorescence staining (Fig. 3C) of HeLa cells. Further analysis revealed that it was the stability of p62 that was decreased significantly in the presence of bpV(phen) (Fig. 3, D and E). p62 has been reported to bind with polyubiquitinated protein aggregates and help them be packed into autophagosomes and then degraded with the bound aggregates in lysosomes (7, 22). Inhibition of lysosomal activity with BAF was predicted to increase the levels of p62. However, the decrease in levels of p62 in the presence of bpV(phen) was restored only partially by inhibition of the lysosomal activity with BAF but more dramatically by inhibition of the proteasomal activity of the 26S complex with MG-132 (Fig. 3, F and G). P62 was not associated with RFP-LC3 punctate foci in the presence of MG-132 but colocalized with punctate foci in the presence of BAF (Fig. 3H). Therefore, bpV(phen) exposure does not reduce the stability of p62 through autophagy. Exposure to bpV(phen) led to a nonspecific enhancement in the levels of total ubiquitinated proteins and a specific enhancement of p62 ubiquitination. Blocking proteasomal activity with MG-132 caused an accumulation of poly-ubiquitinated p62 (Fig. 3I). Compared with the control, MG-132 treatment led to accumulation of more ubiquitinated p62 with a low molecular weight, suggesting that ubiquitinated p62 with a high molecular weight cannot be degraded through proteasomes (Fig. 3I). Therefore, bpV(phen) treatment promoted the proteasomal degradation of poly-ubiquitinated p62.
FIGURE 3.
Treatment with bpV(phen) reduces the stability of p62 in a proteasome-dependent way. A, immunoblot analysis of the impact of bpV(phen) (BPV) on the levels of p62 in HeLa cells. KD, kilodalton. B, plots of relative intensities of p62 to β-Actin as shown in A. C, immunostaining analysis of p62 levels in HeLa cells left untreated or treated with 20 μm of bpV(phen) for 48 h with anti-p62 antibody. Ctrl, control. D, immunoblot analysis of the impact of bpV(phen) on the stability of p62. HeLa cells were grown to confluence, and protein translation was inhibited with cycloheximide (CHX) in the absence (Ctrl) or presence of 20 μm bpV(phen). Cells were collected at different time points, and the same amounts of total proteins were loaded. β-Actin served as another loading control. E, plots of relative intensities of p62 to β-Actin as shown in D. F, immunoblot analysis of the impact of MG-132 and bafilomycin A1 on the bpV(phen)-reduced stability of p62. HeLa cells were grown to confluence and treated with 20 μm of bpV(phen) and cycloheximide in the absence or presence of MG-132 and/or BAF. Cells were collected at different time points and analyzed as in D. **, p < 0.01; ***, p < 0.001. G, plots of relative intensities of p62 as shown in F. The initial intensities of p62 at time 0 under different conditions were set to be 1. H, colocalization analysis of p62 with RFP-LC3 punctate foci in untreated (Ctrl) and MG-132- or BAF-treated HeLa cells stably expressing RFP-LC3 in the presence of bpV(phen). p62 was visualized by staining with anti-p62 antibody. I, immunoblot analysis of the impact of bpV(phen) on the levels of poly-ubiquitinated p62. 293T cells overexpressing p62 and His-ubiquitin were cultured in the absence or presence of 10 μm bpV(phen) for 30 h and left untreated or treated with MG-132 for 6 h before cells were harvested. Cell lysates were subjected to immunoprecipitation (IP) with anti-p62 antibody and immunoblotted with anti-p62 and ubiquitin antibodies. Two panels of p62 immunoblot results were generated by long and short exposure. IgG and p62 overlapped each other.
Suppression of p62 Causes Deacetylation of α-Tubulin and Inhibition of the Degradation of Autophagosomes
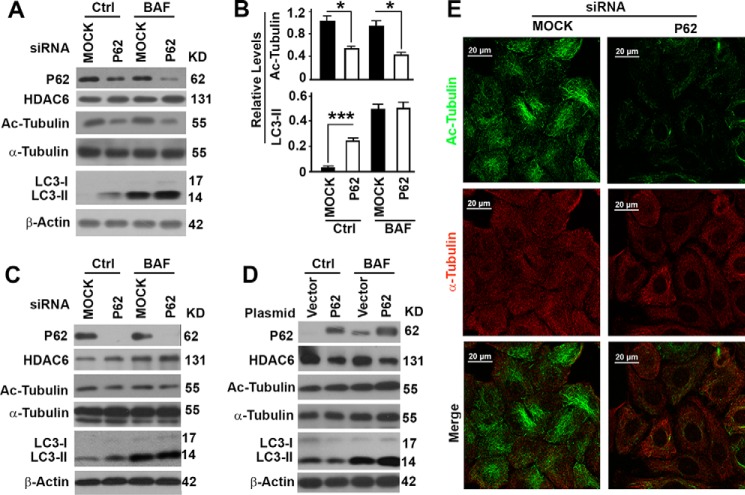
In addition to serving as a substrate receptor, p62 has recently been reported to interact with HDAC6 to suppress its deacetylase activity on α-tubulin (23). We detected no change in the levels of total α-tubulin but significant decreases in the levels of acetylated α-tubulin when p62 levels were suppressed in HeLa cells with two different p62-specific siRNA molecules (Fig. 4, A–C). Overexpression of p62 exhibited the opposite impact on the levels of acetylated tubulin (Fig. 4D). The levels of acetylated microtubules were reduced dramatically, whereas the general microtubules remained constant (Fig. 4E). Acetylated microtubules are stable and required for the trafficking of autophagosomes to fuse with lysosomes (11). The reduction in levels of acetylated microtubules led to an increase of LC3-II in the absence of BAF but even levels of LC3-II in the presence of BAF (Fig. 4, A–C). Overexpression of p62 accelerated the degradation of autophagosomes and led to a reduction in LC3-II levels in the absence of BAF (Fig. 4D). Therefore, degradation of p62 leads to a release of HDAC6 activity that causes deacetylation of α-tubulin, destabilization of microtubules, and blockade of the degradation of autophagosomes.
FIGURE 4.
Suppression of p62 causes deacetylation of α-tubulin and inhibition of the degradation of autophagosomes. A, representative immunoblot results showing the impact of p62 suppression with a p62-specific siRNA on the levels of p62, HDAC6, total and acetylated α-tubulin (Ac-Tubulin), and LC3-II in the absence or presence of bafilomycin A1. Ctrl, control. KD, kilodalton. B, plots of relative intensities of acetylated α-tubulin or LC3-II as shown in A. *, p < 0.05; ***, p < 0.001. C, representative immunoblot results showing the impact of p62 suppression with another p62-specific siRNA. D, representative immunoblot results showing the impact of p62 overexpression. E, immunofluorescence analyses of the impact of p62 suppression on the acetylated microtubules and general microtubules visualized with antibody against acetylated α-tubulin or α-tubulin, respectively.
Treatment with bpV(phen) Leads to Reduction of p62 and Release of HDAC6 to Deacetylate the Acetylated α-Tubulin
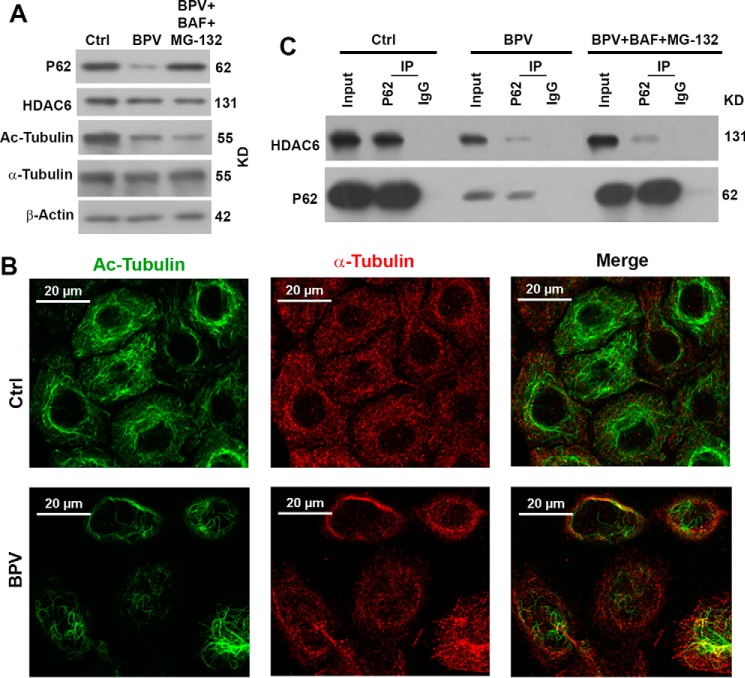
To decipher how bpV(phen) impacts autophagy through p62, we tested the impact of bpV(phen) on the interaction of p62 with HDAC6. When HeLa cells were exposed to bpV(phen), the levels of HDAC6 were reduced slightly, but the levels of acetylated α-tubulin were reduced dramatically, although the total levels of α-tubulin were unchanged (Fig. 5A). The levels of acetylated microtubules in HeLa cells were also impaired in the presence of bpV(phen) (Fig. 5B). We detected a much weaker interaction between p62 and HDAC6 in the presence of bpV(phen) than in the absence of bpV(phen) (Fig. 5C). We reasoned that such a weak interaction might have been caused by reduced input levels of HDAC6 and p62. We treated the cells with BAF and MG-132 and restored the levels of both proteins to be close to the levels of untreated cells, but the weak interaction was not at all improved (Fig. 5C). Therefore, bpV(phen) exposure disrupted the interaction between p62 and HDAC6 and led to the release of HDAC6 to deacetylate α-tubulin, destabilized microtubules, and impaired the degradation of autophagosomes.
FIGURE 5.
Treatment with bpV(phen) leads to a reduction of p62 and release of HDAC6 to deacetylate the acetylated α-tubulin. A, immunoblot analysis of the impact of bpV(phen) (BPV) on the levels of p62, HDAC6, and total and acetylated α-tubulin (Ac-Tubulin) HeLa cells left untreated or treated with 20 μm of bpV(phen) for 48 h in the absence or presence of both MG-132 and bafilomycin A1. Ctrl, control. KD, kilodalton. B, immunofluorescence analyses of the impact of bpV(phen) on the acetylated microtubules, visualized with antibodies against acetylated α-tubulin (green) and total α-tubulin (red). C, immunoblot analyses of the interaction between HDAC6 and p62. HeLa cells were left untreated or treated with 20 μm of bpV(phen) for 48 h in the absence or presence of both MG-132 and bafilomycin A1. Equal amounts of cell lysates were subjected to immunoprecipitation (IP) with equal amounts of antibodies against p62 or control IgG and immunoblotted with the respective antibodies.
Discussion
p62 protein contains multiple domains involving different functions, such as the LC3-interacting region and ubiquitin-associated domain (24). It is generally considered a substrate receptor that connects the polyubiquitinated protein aggregates and dysfunctional mitochondria to LC3-II and facilitates their packaging into autophagosomes (7, 8, 25). Specific phosphorylation of serine 403 in the ubiquitin-associated domain leads to enhancement of its association with the polyubiquitinated cargo for autophagosomal engulfment (24). The sequence 163–225 of p62 has been identified as the HDAC6-interactive domain (23). Serine 207 in the HDAC6-interactive domain of p62 is highly phosphorylated in the presence of a proteasomal inhibitor, and inhibition of autophagy with bafilomycin A1 has been reported to show no accumulation of phosphorylated p62 at serine 207 (24). Treatment with bpV(phen) seems to enhance the phosphorylation of p62 at serine 207 and disrupt the interaction with HDAC6. Further analysis of the impact of bpV(phen) on the p62-HDAC6 interaction will clarify the mechanism in detail. Interestingly, proteasomal inhibition did not cause any alternation in the phosphorylation of the tyrosine residue in p62 protein (24). Therefore, the impact of bpV(phen) on autophagy seems to be not through its PTEN inhibition because PTEN inhibition may activate autophagy initiation.
p62 has been reported to bind with ubiquitinated proteins and be engulfed and degraded in the autophagosome. It is unknown whether p62 is required to be ubiquitinated for lysosomal degradation (26). Cullin-3 (Cul-3) is an E3 ubiquitin ligase that catalyzes the ubiquitination of p62 and promotes the degradation of p62 through proteasomes (27). Exposure to bpV(phen) may activate E3 ubiquitin ligases such as Cul-3 to promote the ubiquitination of p62 or promote the modification of p62 into a better substrate of ubiquitin ligase. The report that proteasomal inhibition with MG-132 did not cause any alteration in the ubiquitination of p62 protein (24) suggests that the ubiquitination of p62 is a specific effect of bpV(phen) exposure.
Overactive autophagy leads to the depletion of cellular contents and, eventually, cell death. In this sense, autophagy is classified as type II programmed cell death (28). Instead, autophagy has a pro-survival function when cells are under metabolic stress (29, 30). Its dysfunction causes failure of the cells to sustain metabolic homeostasis and survive (31). If the autophagic process is blocked before autophagosomal formation, then the fragmented mitochondria will release cytochrome c and other small molecules to induce conventional apoptosis, which is usually associated with diverse forms of aggregation and perinuclear clustering of the dysfunctional mitochondria. The dead cell debris generated from apoptosis and autophagy is still contained in an intact plasma membrane and taken up by phagocytosis (32, 33). If the process is blocked before autolysosome formation or if autophagosomes are not degraded efficiently, then the accumulated mitochondria may become damaged by their own production of superoxide, start to leak electrons, lose their membrane potentials, and even induce robust oxidative stress (1, 34) or lysosomal rupture (35). Both oxidative stress and lysosomal rupture, in turn, activate the NLRP3 inflammasome, which results in direct activation of caspase 1 (36). Activation of caspase 1 subsequently induces the secretion of potent pro-inflammatory cytokines and, eventually, an inflammatory form of cell death referred to as pyroptosis of the cell and other cells in the environment (21, 37–41). Although bpV(phen) has been suggested as a drug for different types of diseases, it is necessary to consider the potential side effects caused by its impact on autophagy, apoptosis, and pyroptosis.
Author Contributions
Q. C., J. L., and L. L. conceived and coordinated the study and wrote the paper. Q. C., T. X., C. H., and Y. Z. designed, performed, and analyzed the experiments shown in Figs. 1 and 2. Q. C. and W. L. designed, performed, and analyzed the experiments shown in Fig. 3. Q. C., and F. Y. designed, performed, and analyzed the experiments shown in Figs. 4 and 5. J. Z., K. S., G. X., and H. H. provided technical assistance and contributed to the preparation of the figures. All authors reviewed the results and approved the final version of the manuscript.
This work was supported, in whole or in part, by NCI/National Institutes of Health Grant CA142862 (to L. L.). This work was also supported by Department of Defense New Investigator Award W81XWH (to L. L.). The authors declare that they have no conflicts of interest with the contents of this article.
- PTEN
- phosphatase and tensin homologue deleted on chromosome 10
- bpV(phen)
- potassium bisperoxo(1,10-phenanthroline)oxovanadate
- MEF
- mouse embryonic fibroblast
- PI
- propidium iodide
- PARP
- poly(ADP-ribose) polymerase
- BAF
- bafilomycin 1A.
References
- 1.Mizushima N., Noda T., Yoshimori T., Tanaka Y., Ishii T., George M. D., Klionsky D. J., Ohsumi M., and Ohsumi Y. (1998) A protein conjugation system essential for autophagy. Nature 395, 395–398 [DOI] [PubMed] [Google Scholar]
- 2.Thompson C. B. (1995) Apoptosis in the pathogenesis and treatment of disease. Science 267, 1456–1462 [DOI] [PubMed] [Google Scholar]
- 3.Arico S., Petiot A., Bauvy C., Dubbelhuis P. F., Meijer A. J., Codogno P., and Ogier-Denis E. (2001) The tumor suppressor PTEN positively regulates macroautophagy by inhibiting the phosphatidylinositol 3-kinase/protein kinase B pathway. J. Biol. Chem. 276, 35243–35246 [DOI] [PubMed] [Google Scholar]
- 4.Kabeya Y., Mizushima N., Ueno T., Yamamoto A., Kirisako T., Noda T., Kominami E., Ohsumi Y., and Yoshimori T. (2000) LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J. 19, 5720–5728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tanida I., Ueno T., and Kominami E. (2004) LC3 conjugation system in mammalian autophagy. Int. J. Biochem. Cell Biol. 36, 2503–2518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hanada T., Noda N. N., Satomi Y., Ichimura Y., Fujioka Y., Takao T., Inagaki F., and Ohsumi Y. (2007) The Atg12-Atg5 conjugate has a novel E3-like activity for protein lipidation in autophagy. J. Biol. Chem. 282, 37298–37302 [DOI] [PubMed] [Google Scholar]
- 7.Bjørkøy G., Lamark T., Brech A., Outzen H., Perander M., Overvatn A., Stenmark H., and Johansen T. (2005) p62/SQSTM1 forms protein aggregates degraded by autophagy and has a protective effect on huntingtin-induced cell death. J. Cell Biol. 171, 603–614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bjørkøy G., Lamark T., and Johansen T. (2006) p62/SQSTM1: a missing link between protein aggregates and the autophagy machinery. Autophagy 2, 138–139 [DOI] [PubMed] [Google Scholar]
- 9.Mizushima N., Levine B., Cuervo A. M., and Klionsky D. J. (2008) Autophagy fights disease through cellular self-digestion. Nature 451, 1069–1075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Levine B., and Kroemer G. (2008) Autophagy in the pathogenesis of disease. Cell 132, 27–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xie R., Nguyen S., McKeehan W. L., and Liu L. (2010) Acetylated microtubules are required for fusion of autophagosomes with lysosomes. BMC Cell Biol. 11, 89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Posner B. I., Faure R., Burgess J. W., Bevan A. P., Lachance D., Zhang-Sun G., Fantus I. G., Ng J. B., Hall D. A., and Lum B. S. (1994) Peroxovanadium compounds: a new class of potent phosphotyrosine phosphatase inhibitors which are insulin mimetics. J. Biol. Chem. 269, 4596–4604 [PubMed] [Google Scholar]
- 13.Yale J. F., Lachance D., Bevan A. P., Vigeant C., Shaver A., and Posner B. I. (1995) Hypoglycemic effects of peroxovanadium compounds in Sprague-Dawley and diabetic BB rats. Diabetes 44, 1274–1279 [DOI] [PubMed] [Google Scholar]
- 14.Caron D., Savard P. E., Doillon C. J., Olivier M., Shink E., Lussier J. G., and Faure R. L. (2008) Protein tyrosine phosphatase inhibition induces anti-tumor activity: evidence of Cdk2/p27 kip1 and Cdk2/SHP-1 complex formation in human ovarian cancer cells. Can. Let. 262, 265–275 [DOI] [PubMed] [Google Scholar]
- 15.Zu L., Shen Z., Wesley J., and Cai Z. P. (2011) PTEN inhibitors cause a negative inotropic and chronotropic effect in mice. Eur. J. Pharmacol. 650, 298–302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mizushima N., Yamamoto A., Matsui M., Yoshimori T., and Ohsumi Y. (2004) In vivo analysis of autophagy in response to nutrient starvation using transgenic mice expressing a fluorescent autophagosome marker. Mol. Biol. Cell 15, 1101–1111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zou J., Yue F., Jiang X., Li W., Yi J., and Liu L. (2013) Mitochondrion-associated protein LRPPRC suppresses the initiation of basal levels of autophagy via enhancing Bcl-2 stability. Biochem. J. 454, 447–457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zou J., Yue F., Li W., Song K., Jiang X., Yi J., and Liu L. (2014) Autophagy inhibitor LRPPRC suppresses mitophagy through interaction with mitophagy initiator Parkin. PloS ONE 9, e94903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pouliot P., Camateros P., Radzioch D., Lambrecht B. N., and Olivier M. (2009) Protein tyrosine phosphatases regulate asthma development in a murine asthma model. J. Immunol. 182, 1334–1340 [DOI] [PubMed] [Google Scholar]
- 20.Bortoluci K. R., and Medzhitov R. (2010) Control of infection by pyroptosis and autophagy: role of TLR and NLR. Cell Mol. Life Sci. 67, 1643–1651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ryter S. W., Mizumura K., and Choi A. M. (2014) The impact of autophagy on cell death modalities. Int. J. Cell Biol. 2014, 502676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Komatsu M., Waguri S., Koike M., Sou Y. S., Ueno T., Hara T., Mizushima N., Iwata J., Ezaki J., Murata S., Hamazaki J., Nishito Y., Iemura S., Natsume T., Yanagawa T., Uwayama J., Warabi E., Yoshida H., Ishii T., Kobayashi A., Yamamoto M., Yue Z., Uchiyama Y., Kominami E., and Tanaka K. (2007) Homeostatic levels of p62 control cytoplasmic inclusion body formation in autophagy-deficient mice. Cell 131, 1149–1163 [DOI] [PubMed] [Google Scholar]
- 23.Yan J., Seibenhener M. L., Calderilla-Barbosa L., Diaz-Meco M. T., Moscat J., Jiang J., Wooten M. W., and Wooten M. C. (2013) SQSTM1/p62 interacts with HDAC6 and regulates deacetylase activity. PloS ONE 8, e76016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Matsumoto G., Wada K., Okuno M., Kurosawa M., and Nukina N. (2011) Serine 403 phosphorylation of p62/SQSTM1 regulates selective autophagic clearance of ubiquitinated proteins. Mol. Cell 44, 279–289 [DOI] [PubMed] [Google Scholar]
- 25.Geisler S., Holmström K. M., Skujat D., Fiesel F. C., Rothfuss O. C., Kahle P. J., and Springer W. (2010) PINK1/Parkin-mediated mitophagy is dependent on VDAC1 and p62/SQSTM1. Nat. Cell Biol. 12, 119–131 [DOI] [PubMed] [Google Scholar]
- 26.Pankiv S., Clausen T. H., Lamark T., Brech A., Bruun J. A., Outzen H., Øvervatn A., Bjørkøy G., and Johansen T. (2007) p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J. Biol. Chem. 282, 24131–24145 [DOI] [PubMed] [Google Scholar]
- 27.Lee J., Kim H. R., Quinley C., Kim J., Gonzalez-Navajas J., Xavier R., and Raz E. (2012) Autophagy suppresses interleukin-1β (IL-1β) signaling by activation of p62 degradation via lysosomal and proteasomal pathways. J. Biol. Chem. 287, 4033–4040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Klionsky D. J. (2007) Autophagy: from phenomenology to molecular understanding in less than a decade. Nat. Rev. Mol. Cell Biol. 8, 931–937 [DOI] [PubMed] [Google Scholar]
- 29.Gozuacik D., and Kimchi A. (2007) Autophagy and cell death. Curr. Top. Dev. Biol. 78, 217–245 [DOI] [PubMed] [Google Scholar]
- 30.Levine B., and Yuan J. (2005) Autophagy in cell death: an innocent convict? J. Clin. Invest. 115, 2679–2688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mathew R., Kongara S., Beaudoin B., Karp C. M., Bray K., Degenhardt K., Chen G., Jin S., and White E. (2007) Autophagy suppresses tumor progression by limiting chromosomal instability. Genes Dev. 21, 1367–1381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cryns V., and Yuan J. (1998) Proteases to die for. Genes Dev. 12, 1551–1570 [DOI] [PubMed] [Google Scholar]
- 33.Desagher S., and Martinou J. C. (2000) Mitochondria as the central control point of apoptosis. Trends Cell Biol. 10, 369–377 [DOI] [PubMed] [Google Scholar]
- 34.Liu L., McKeehan W. L., Wang F., and Xie R. (2012) MAP1S enhances autophagy to suppress tumorigenesis. Autophagy 8, 278–280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hornung V., Bauernfeind F., Halle A., Samstad E. O., Kono H., Rock K. L., Fitzgerald K. A., and Latz E. (2008) Silica crystals and aluminum salts activate the NALP3 inflammasome through phagosomal destabilization. Nat. Immunol. 9, 847–856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lamkanfi M., and Dixit V. M. (2014) Mechanisms and functions of inflammasomes. Cell 157, 1013–1022 [DOI] [PubMed] [Google Scholar]
- 37.Strowig T., Henao-Mejia J., Elinav E., and Flavell R. (2012) Inflammasomes in health and disease. Nature 481, 278–286 [DOI] [PubMed] [Google Scholar]
- 38.Lamkanfi M., and Dixit V. M. (2012) Inflammasomes and their roles in health and disease. Annu. Rev. Cell Dev. Biol. 28, 137–161 [DOI] [PubMed] [Google Scholar]
- 39.Zhou R., Yazdi A. S., Menu P., and Tschopp J. (2011) A role for mitochondria in NLRP3 inflammasome activation. Nature 469, 221–225 [DOI] [PubMed] [Google Scholar]
- 40.Gurung P., Lukens J. R., and Kanneganti T. D. (2015) Mitochondria: diversity in the regulation of the NLRP3 inflammasome. Trends Mol. Med. 21, 193–201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yu J., Nagasu H., Murakami T., Hoang H., Broderick L., Hoffman H. M., and Horng T. (2014) Inflammasome activation leads to Caspase-1-dependent mitochondrial damage and block of mitophagy. Proc. Nat. Acad. Sci. U.S.A. 111, 15514–15519 [DOI] [PMC free article] [PubMed] [Google Scholar]