Background: A soluble form of IL-6 receptor mediates pathogenic IL-6 trans-signaling.
Results: ADAM10 and ADAM17 release IL-6 receptor from both human and murine monocytes/macrophages, whereas in the blood IL-6 receptor is also present on microvesicles.
Conclusion: Shedding of endogenous IL-6 receptor is similar in humans and mice.
Significance: Microvesicle release represents a novel mode of soluble IL-6 receptor generation with potential clinical implications.
Keywords: ADAM, interleukin 6 (IL-6), interleukin 6 receptor (IL6R), protease, shedding, ADAM10, ADAM17, exosomes, microvesicles
Abstract
Generation of the soluble interleukin-6 receptor (sIL-6R) is a prerequisite for pathogenic IL-6 trans-signaling, which constitutes a distinct signaling pathway of the pleiotropic cytokine interleukin-6 (IL-6). Although in vitro experiments using ectopically overexpressed IL-6R and candidate proteases revealed major roles for the metalloproteinases ADAM10 and ADAM17 in IL-6R shedding, the identity of the protease(s) cleaving IL-6R in more physiological settings, or even in vivo, remains unknown. By taking advantage of specific pharmacological inhibitors and primary cells from ADAM-deficient mice we established that endogenous IL-6R of both human and murine origin is shed by ADAM17 in an induced manner, whereas constitutive release of endogenous IL-6R is largely mediated by ADAM10. Although circulating IL-6R levels are altered in various diseases, the origin of blood-borne IL-6R is still poorly understood. It has been shown previously that ADAM17 hypomorphic mice exhibit unaltered levels of serum sIL-6R. Here, by quantification of serum sIL-6R in protease-deficient mice as well as human patients we also excluded ADAM10, ADAM8, neutrophil elastase, cathepsin G, and proteinase 3 from contributing to circulating sIL-6R. Furthermore, we ruled out alternative splicing of the IL-6R mRNA as a potential source of circulating sIL-6R in the mouse. Instead, we found full-length IL-6R on circulating microvesicles, establishing microvesicle release as a novel mechanism for sIL-6R generation.
Introduction
The pleiotropic cytokine IL-6 can exert its signal via two extracellular routes. The first route, termed “classic” IL-6 signaling, relies on IL-6 binding the membrane IL-6R receptor (IL-6R), which in turn leads to dimerization of the second receptor subunit glycoprotein 130 (gp130) and initiation of intracellular signaling cascades (1). In contrast to gp130, which is present on almost every cell of the body, IL-6R expression is much more restricted with mainly hepatocytes and certain leukocyte subtypes bearing IL-6R. Signaling via membrane-bound IL-6R is rather anti-inflammatory and has been shown to be critically involved in plasma cell differentiation, the acute phase response, and fever induction (2). Alternatively, IL-6 can engage a soluble form of the IL-6R in solution with the same affinity as the membrane-bound receptor. The resulting binary cytokine complex exhibits agonistic properties and is able to activate cells that only express gp130 but lack membrane-bound IL-6R. This alternative IL-6 signaling pathway has been termed “IL-6 trans-signaling” and greatly expands the spectrum of IL-6 responsive cells (3). It has become evident in recent years that most of the pro-inflammatory and deleterious actions of IL-6, which includes generation of autoimmune Th17 cells, inhibition of T cell apoptosis, or proliferation of malignant epithelial cells, can in fact be attributed to IL-6 trans-signaling (4). In humans, a soluble form of the IL-6R is generated by proteolytic release of the ectodomain as well as transcription from an alternatively spliced mRNA lacking the transmembrane region (5, 6). Ectodomain shedding of transmembrane proteins represents a swift and highly regulable response of the cell toward extracellular stimuli and is therefore ideally suited to counteract inflammatory insults (7). As IL-6R shedding can initiate pathogenic IL-6 trans-signaling, it is of high clinical relevance to identify the protease responsible for sIL-6R release. Although previous studies on IL-6R cleavage almost exclusively relied on ectopically overexpressed receptors or proteases, the closely related proteases ADAM103 and ADAM17 were identified as IL-6R sheddases in vitro (8, 9). However, the identity of the enzyme that processes IL-6R in vivo, whether under physiological or pathological conditions, remains elusive. In this study, we used cells naturally expressing high levels of IL-6R, as opposed to cells ectopically expressing IL-6R after transfection with an IL-6R cDNA, to elucidate the roles of ADAM10 and ADAM17 in IL-6R cleavage. In contrast to a previous report showing that upon stimulation ectopically expressed murine IL-6R is shed by ADAM10, we report that endogenous IL-6R in human and murine cells is cleaved by ADAM17 in an induced fashion. On the other hand, constitutive release of endogenous IL-6R, regardless of the species, is clearly mediated by ADAM10.
A soluble form of the IL-6R is present in human serum at relatively high levels and several reports have connected serum sIL-6R generation to alternative mRNA splicing (10, 11). However, whereas serum sIL-6R levels attributed to alternative splicing vary considerably between studies, the bulk of serum sIL-6R (>65%) originates from processes other than differential mRNA splicing (12, 13). A recently identified SNP in the human IL-6R gene located within the ADAM17 cleavage site, which leads to increased sIL-6R protein amounts in the circulation, strongly suggests involvement of a metalloprotease of the ADAM family in serum sIL-6R generation (14–16). Surprisingly, hypomorphic ADAM17 mutant mice showing only residual proteolytic activity (5%) exhibit unaltered sIL-6R serum levels (17). This indicates that either an alternative protease, e.g. ADAM10, or even an entirely different mechanism is responsible for serum sIL-6R generation. Here, we show that conditional ablation of ADAM10 in myeloid cells, which have been reported to generate the bulk (63%) of blood-borne sIL-6R (18), also resulted in unaltered sIL-6R serum levels. We further excluded neutrophil elastase, cathepsin G, and proteinase 3, which are highly abundant in myeloid cells, from contributing to sIL-6R generation in mouse as well as human blood. Moreover, by employing an elaborate PCR strategy we ruled out alternative splicing as a mechanism generating serum sIL-6R in the mouse. Instead, we were able to locate membrane-associated IL-6R on circulating microvesicles, challenging the prevailing dogma that only metalloproteinases and, in humans, alternative splicing account for the release of sIL-6R into the circulation.
Experimental Procedures
Cell Lines and Reagents
THP-1, U937, RAW264.7, and J774.A1 cells were obtained from the American Type Culture Collection (ATCC; LGC Standards, Wesel, Germany) and cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% FCS (PAN Biotech, Aidenbach, Germany), 2 mm l-glutamine, 100 units/ml of penicillin, and 100 μg/ml of streptomycin. Cells were stimulated with PMA or LPS from Escherichia coli O111:B4 (both Sigma) as depicted in the figure legends. Generation of neutralizing, bivalent ADAM17 antibody D1(A12) and recombinant murine ADAM10 prodomain were described previously (19, 20). The hydroxamate-based ADAM inhibitors GI254023X and GW280264X were synthesized by Iris Biotech (Marktredwitz, Germany). Marimastat (BB-2516) was obtained from Sigma.
Mice, Generation of Murine Bone Marrow-derived Macrophages (BMDMs), and Human Monocyte-derived Macrophages
Hypomorphic ADAM17 mice (ADAM17ex/ex) and floxed ADAM10 mice (ADAM10flox/flox) have been previously described (17, 21). LysM_Cre×ADAM10flox/flox mice expressed Cre recombinase under the control of the endogenous lysozyme 2 promotor. Littermates, which are homozygous for floxed ADAM10 but lack Cre recombinase, served as controls. Generation of ADAM8 and DPPI knock-out mice have been described elsewhere (22, 23). As the DPPI knock-out mice used in this study were on an ApoE-deficient background, we used ApoE knock-out mice as controls. To generate BMDMs bone marrow was collected from femur and tibia of 8–12-week-old mice and BMDMs were differentiated in DMEM containing 10% FCS, 1 mm sodium pyruvate, 2 mm l-glutamine, 10 mm HEPES, 100 units/ml of penicillin, 100 μg/ml of streptomycin, and 40 ng/ml of M-CSF (Immunotools, Friesoythe, Germany) for 7–10 days. BMDMs were detached using Accutase (PAA Laboratories), seeded on 6-well plates at a density of 0.5 × 106 cells/ml, and stimulated the next day as indicated. Primary human macrophages were generated as previously described (24). Briefly, peripheral blood mononuclear cells were isolated from human blood by density gradient centrifugation and monocytes were subsequently purified with CD14 MicroBeads according to the manufacturer's instructions (Miltenyi Biotec, Bergisch-Gladbach, Germany). CD14+ monocytes were differentiated into macrophages by culture in supplemented RPMI 1640 medium (10% FCS, 100 units/ml of penicillin, 100 μg/ml of streptomycin, 2 mm l-glutamine) containing human M-CSF (100 ng/ml, Immunotools, Friesoythe, Germany) for at least 10 days. All individuals underwent a written, informed-consent process approved by the ethics commission of the Medical Faculty of Kiel University (A 102/14). Nine patients suffering from Papillon Lefèvre syndrome and 9 healthy gender and age (±2 years; in 1 pair 3 years and 6 months) matched control individuals were recruited at the Dept. of Periodontology, Goethe-University, Frankfurt, Germany. Twenty ml of blood were sampled from arm veins and sera was obtained by centrifugation. The serum was stored at −80 °C and after all patients had been sampled sent to Kiel University. All patients underwent a written, informed-consent process approved by the ethics commission of the Medical Faculty of the Goethe-University Frankfurt (31/05).
FACS, Western Blotting, and ELISA
105 THP-1 cells/96-well were stimulated with PMA ± inhibitors as indicated in the figure legends. Cells were subsequently washed with FACS buffer (1% BSA, PBS) and Fc receptors were blocked with anti-CD16/CD32 (eBioscience, San Diego, CA) according to the manufacturer. Human IL-6R was detected with mouse mAb 4–11 (1:400) and the stained cells were visualized with allophycocyanin-coupled rat anti-mouse IgG1 (eBioscience) by flow cytometry (FACS Canto I, BD Bioscience, San Jose, CA). EDTA mouse blood was blocked with anti-mouse CD16/32 antibody (BioLegend, San Diego, CA) and the different immune cell subsets were detected with anti-CD45, anti-CD11b, anti-Ly6G, anti-Ly6C, anti-CD115, anti-B220, and anti-CD3 (all BioLegend). Murine IL-6R surface expression was assessed using anti-mouse IL-6R mAb (D7715A7, BioLegend). Red blood cells were subsequently lysed and the remaining leukocytes were fixed with RBC lysis/fixation solution (BioLegend), followed by flow cytometry. Heparinized whole human blood was diluted (1:10) in RPMI medium and stimulated with LPS ± inhibitors as indicated in the figure legends. After Fc receptors were blocked (anti-CD16/CD32, eBioscience), human IL-6R expression on circulating monocytes was gauged by flow cytometry using anti-human IL-6R mAb (clone UV4, BioLegend) and anti-human CD14 mAb (clone M5E2, BioLegend). Red blood cells were removed and leukocytes were fixed with RBC lysis/fixation solution (BioLegend) after antibody staining. Western blot analysis was performed as described in Ref. 25 using the following antibodies: human IL-6R C terminus (sc-661; Santa Cruz Biotechnology, Heidelberg, Germany), human IL-6R N terminus mAb 4–11 (26), anti-IL-11R (sc-993, Santa Cruz), Hsp70 (number 4872; Cell Signaling, Danvers, MA), and Tsg101 (sc-7964; Santa Cruz Biotechnology). Rabbit TrueBlot secondary antibody reagent (Rockland, Limerick, PA) was used to specifically detect the primary antibody against human IL-6R C terminus by avoiding cross-reaction with serum antibodies. Generation of a rabbit antiserum against murine ADAM10 was described elsewhere (17). Microvesicle-free serum was generated by ultracentrifugation (2 h, 100,000 × g) and subsequently used for ELISA analysis. Levels of murine and human sIL-6R were measured using DuoSet ELISA kits from R&D Systems.
Microvesicle Isolation from Serum
1 ml of human serum was diluted with 1 volume of PBS and serum antibodies were depleted by incubating diluted serum twice with a 1:1 mixture of Protein A- and G-Sepharose at 4 °C overnight. Samples were cleared from dead cells and debris by low-speed centrifugation (30 min, 2,000 × g) and afterward spun for 45 min at 12,000 × g to precipitate larger vesicles. The supernatant was then loaded onto a 2-ml 40% sucrose/PBS cushion and subjected to ultracentrifugation at 100,000 × g for 2 h at 4 °C (Optima LE-80K, Type 70 Ti Rotor; Beckman Coulter, Krefeld, Germany). Microvesicle-containing sucrose was diluted with 5 volumes of PBS and again centrifuged for 2 h at 100,000 × g. For Western blotting the microvesicle pellet was lysed in 100 μl of RIPA buffer containing protease inhibitors, whereas microvesicles were suspended in PBS for analysis by dynamic light scattering (DLS). DLS measurements were performed on a Laser Spectroscatter 201 (RiNA) at an angle of 90 and 22 °C. Ten μl of microvesicle suspension were applied to a quartz cuvette and size distribution was depicted as the median occurrence out of 100 measurements.
RT-PCR
RNA was extracted from cells and tissues using the GeneJET RNA purification kit (Fermentas, Thermo Scientific). cDNA was synthesized from 1 μg of RNA with RevertAidTM Moloney MuLV reverse transcriptase (Fermentas, Thermo Scientific) using oligo(dT)18 primer. Murine IL-6R mRNA transcripts were amplified via PCR using the following primers: mu IL-6R forward 1, 5′-AAG GAG GAG CTT GAC CTT GG-3′ (Exon 6); mu IL-6R reverse 1, 5′-GTG GAG GAG AGG TCG TCT TG-3′ (Exon 10); mu IL-6R forward 5UTR, 5′-GTG CGA GCT GAG TGT GGA G-3′; mu IL-6R forward Exon1, 5′-GGC TGC ACG CTG TTG GTC-3′; and mu IL-6R forward Exon4, 5′-ACA GTG TGG GAA GCA AGT CC-3′. PCR fragments were separated by agarose gel electrophoresis. Primer sequences for detection of human alternatively spliced IL-6R were described previously (27).
Statistical Analysis
Data are presented as the mean ± S.D. from at least two independent experiments. Statistical significance was calculated using Student's t test and depicted in the figures when n ≥ 3. Values of p < 0.05 were considered significant.
Results
Endogenous Soluble IL-6 Receptor Is Released from Human Monocytic Cells in an ADAM-dependent Manner
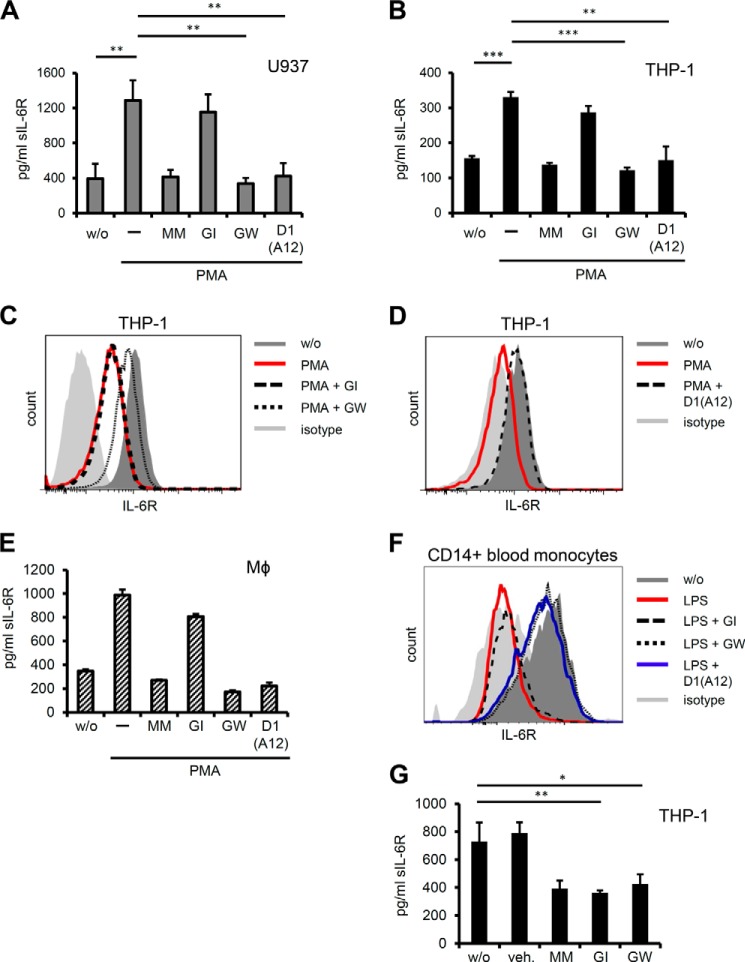
As most, if not all, studies on sIL-6R release so far were carried out in somewhat artificial cellular systems, where either IL-6R or a candidate protease, often genetically engineered, were ectopically introduced into cells, we sought to address IL-6R shedding in a more physiological setting using cells that endogenously express IL-6R. THP-1 as well as U937 cells are human cancer cell lines of monocytic origin that naturally harbor substantial amounts of membrane-bound IL-6R, as evidenced by flow cytometry (Fig. 1C, data not shown). The phorbol ester PMA strongly activates protein kinases PKC and ERK and has widely been used as an activator of ADAM17 (28). ELISA measurements revealed that sIL-6R protein accumulated in the supernatant of THP-1 and U937 cells upon PMA stimulation, which could be blocked when metalloproteases were globally inhibited by Marimastat (Fig. 1, A and B). To narrow down the metalloproteases involved in endogenous IL-6R shedding we took advantage of the specific inhibitor pair GI254023X/GW280264X, which distinguishes between ADAM17- and ADAM10-mediated proteolysis. Whereas GW280264X blocks both ADAM17 and ADAM10, GI254023X inhibits ADAM10 only (for conciseness inhibitors will be abbreviated to GW and GI hereafter). In both monocytic cell lines PMA-induced shedding was strongly reduced after administration of GW, whereas GI had almost no effect, implicating ADAM17 in stimulated IL-6R proteolysis (Fig. 1, A and B). Small molecule inhibitors tend to have off-target effects and indeed both GI and GW have been reported to additionally inhibit certain matrix metalloproteases (29). Therefore, we sought to additionally validate our results and took advantage of a recently developed ADAM17 inhibitory antibody, which due to its ”cross-domain“ architecture is highly specific for human ADAM17 (19). IL-6R shedding after PMA stimulation was significantly reduced in the presence of the antagonistic ADAM17 antibody D1(A12) confirming that cleavage of the endogenous human receptor is in fact relying on ADAM17 (Fig. 1, A and B). In accordance with this, FACS analysis of PMA-treated THP-1 cells showed drastically reduced levels of cell surface IL-6R compared with untreated cells. Consistent with ADAM17-mediated ectodomain shedding, the PMA-induced decrease in IL-6R surface expression was inhibited by addition of GW, but not GI (Fig. 1C). Importantly, incubation of THP-1 cells with D1(A12) antibody blocked PMA-induced loss of surface IL-6R to even pre-treatment levels (Fig. 1D). We sought to put our results into a more physiological context and treated in vitro differentiated primary human macrophages with PMA in order to activate ADAM17. Upon stimulation human macrophages released increased amounts of soluble IL-6R, which could be inhibited by GW and D1(A12), but not GI (Fig. 1E). Although PMA is frequently used to trigger ADAM17 activity, we sought to include a more physiological and well defined stimulus of ADAM17-mediated proteolysis. LPS is commonly used to mimic bacterial infection and has been shown to strongly trigger ADAM17-mediated TNFα release (29). We hypothesized that LPS might also lead to proteolytic release of endogenous IL-6R from LPS-responsive cells and thus treated human whole blood with LPS followed by assessment of IL-6R surface expression. Flow cytometry revealed that levels of surface IL-6R on CD14+ monocytes were strongly diminished after LPS stimulation, but could be restored by concomitant treatment with GW as well as D1(A12), but not GI (Fig. 1F). These data closely reflect the results obtained with THP-1 and U937 cells and implicates ADAM17 in the proteolytic cleavage of endogenous IL-6R from primary human macrophages and circulating monocytes.
FIGURE 1.
A, U937 cells were treated for 1 h with 100 nm of the ADAM17 activator PMA. Inhibitors Marimastat (MM, 3 μm), GI (3 μm), GW (3 μm), and neutralizing α-ADAM17 antibody (D1(A12), 100 nm) were added 30 min prior to stimulation. Conditioned supernatants were collected and sIL-6R levels assessed by ELISA. B, THP-1 cells were stimulated as in A and sIL-6R in cell supernatants was quantified by ELISA. C and D, THP-1 cells were treated as in B and IL-6R surface expression was analyzed by flow cytometry. Representative histograms are shown (n = 6). E, human M-CSF-differentiated macrophages were treated with 100 nm PMA for 2 h ± protease inhibitors (concentrations as in A). sIL-6R levels in conditioned supernatants were determined by ELISA. Error bars represent S.D., n = 2 independent experiments. F, whole human blood was treated with LPS (1 μg/ml for 2 h) and IL-6R surface levels on circulating CD14+ monocytes were determined by flow cytometry. Inhibitors GI (3 μm), GW (3 μm), and D1(A12) α-ADAM17 antibody (200 nm) were added 30 min prior to stimulation. One representative histogram (n = 3) is depicted. G, THP-1 cells were incubated overnight without stimulus ± protease inhibitors (concentrations as A) and sIL-6R was measured by ELISA. Error bars represent S.D. of 3 independent experiments, except stated otherwise. *, p < 0.05; **, p < 0.01; ***, p < 0.001. veh. = dimethyl sulfoxide.
To investigate the constitutive release of endogenous IL-6R we incubated THP-1 and U937 cells overnight with or without GI and GW and measured accumulating sIL-6R protein in the cell supernatant by ELISA. Both cell lines showed robust IL-6R shedding in the absence of stimulation, which could be blocked by GI as well as GW (Fig. 1G and data not shown). These results indicate that ADAM10 is critically involved in the constitutive release of endogenous IL-6R from human cells, which confirms previous studies using ectopically expressed human IL-6R (8, 9).
ADAM17 Is the Major Sheddase of PMA- and LPS-induced IL-6R Shedding in Murine Monocytic Cells Lines and BMDMs
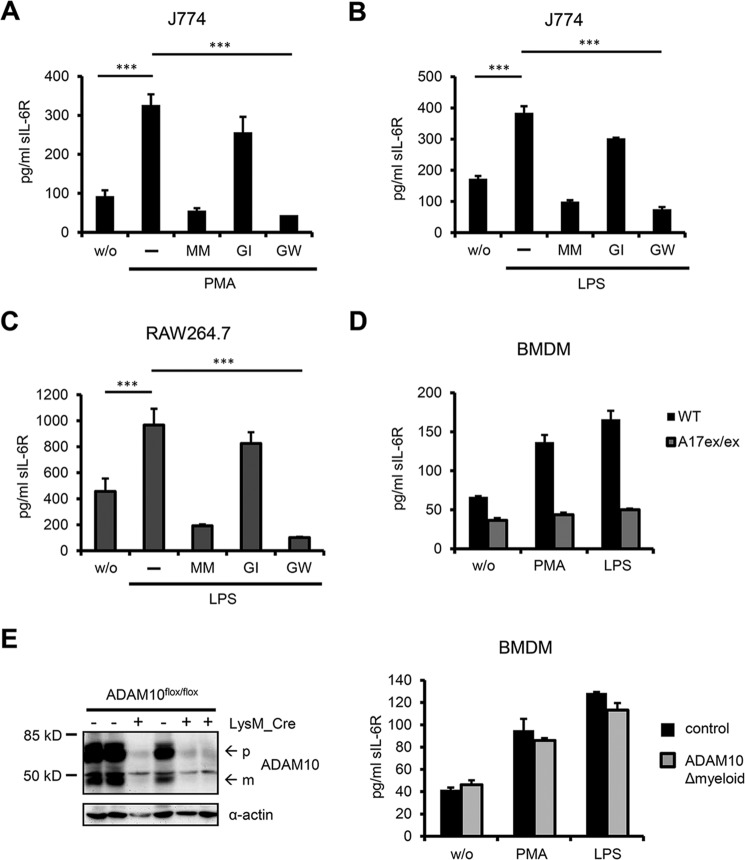
To initially characterize endogenous IL-6R shedding in the mouse we used the monocytic cell lines J774 and RAW264.7 and treated both cell lines with a short PMA pulse. Similar to human cells PMA potently induced IL-6R shedding in J774 and RAW264.7 cells, which could be inhibited by the pan-metalloprotease inhibitor Marimastat as well as the combined ADAM10/ADAM17 inhibitor GW, but not the ADAM10-specific inhibitor GI alone (Fig. 2A, data not shown). These results implicate ADAM17 as the main sheddase responsible for PMA-induced IL-6R cleavage in the mouse, which is in apparent contrast to a previous study reporting ADAM10 as the major IL-6R sheddase activated upon stimulation (9). This discrepancy can be explained by the fact that in the aforementioned report only ectopically expressed murine IL-6R was used to assess inducible IL-6R shedding, whereas in our study murine cells inherently bearing both protease and receptor were employed. We decided to employ a more physiological stimulus also in the murine system and thus treated J774 and RAW264.7 cells for 2 h with LPS, which led to a >100% rise of sIL-6R protein in the supernatant (Fig. 2, B and C). Pharmacological inhibition using GI/GW revealed ADAM17 as principal sheddase of LPS-induced IL-6R cleavage. Next, we sought to substantiate our findings on endogenous IL-6R shedding in the mouse by using primary cells derived from protease-deficient mice. ADAM10 and ADAM17 knock-out mice are embryonic lethal (although few ADAM17 knock-out mice survive until birth, but die shortly after) (30, 31). We therefore used hypomorphic ADAM17ex/ex mice, which exhibit less than 5% residual enzyme activity in all investigated tissues, and generated BMDMs for further analysis of murine IL-6R shedding (17). Levels of sIL-6R in the supernatant of wild type BMDMs increased upon treatment with PMA and LPS, which is in line with our results from J774 and RAW264.7 cells. In contrast, in BMDMs derived from ADAM17ex/ex mice no increase in sIL-6R release was observed after PMA as well as LPS stimulation (Fig. 2D). This clearly shows that ADAM17 is responsible for induced IL-6R shedding in murine BMDMs, which represent highly physiological cells naturally expressing both protease and substrate. Next, we generated BMDMs from conditional ADAM10 knock-out mice lacking ADAM10 in myeloid cells (ADAM10Δmyeloid mice). BMDMs from these animals were effectively devoid of ADAM10 but still released sIL-6R upon stimulation with PMA or LPS (Fig. 2E). This finding lends further support to the notion that ADAM10 is not involved in stimulated shedding of endogenous IL-6R in the mouse.
FIGURE 2.
A, murine J774 cells were treated for 1 h with 100 nm PMA. Inhibitors Marimastat (MM, 3 μm), GI (3 μm), and GW (3 μm) were added 30 min prior to stimulation and sIL-6R was measured by ELISA. B, J774 cells were stimulated for 2 h with 1 μg/ml of LPS with or without inhibitors (3 μm). sIL-6R levels in conditioned supernatant were determined by ELISA. C, murine RAW 264.7 cells were treated as in B and conditioned supernatants were assessed for sIL-6R levels by ELISA. D, BMDMs from ADAM17ex/ex and wild type mice were treated with 100 nm PMA or 1 μg/ml of LPS for 2 h. sIL-6R was quantified by ELISA. Data from one ADAM17ex/ex mouse and corresponding wild type littermate are shown representatively (n = 3 mice). Error bars represent S.D., n = 2 independent experiments. E, left: ADAM10 expression in BMDMs from ADAM10flox/flox mice ± LysM_Cre was visualized by Western blotting. ADAM10flox/flox mice transgenic for LysM_Cre will be designated as ADAM10Δmyeloid mice hereafter, whereas ADAM10flox/flox littermates will serve as corresponding control mice. Pro- and mature form of ADAM10 are designated with p and m, respectively. 3 mice per genotype are shown. α-Actin expression serves as loading control. Right, BMDMs from ADAM10Δmyeloid and control mice (n = 2 mice per group) were treated as in D and sIL-6R was measured by ELISA. Results from one corresponding mouse pair are shown. Error bars are S.D., n = 2 independent experiments. A–C, error bars represent S.D. of 3 independent experiments. ***, p < 0.001.
Constitutive Release of Endogenous Murine IL-6R Is Mediated by ADAM10 but Can Be Compensated for by ADAM17
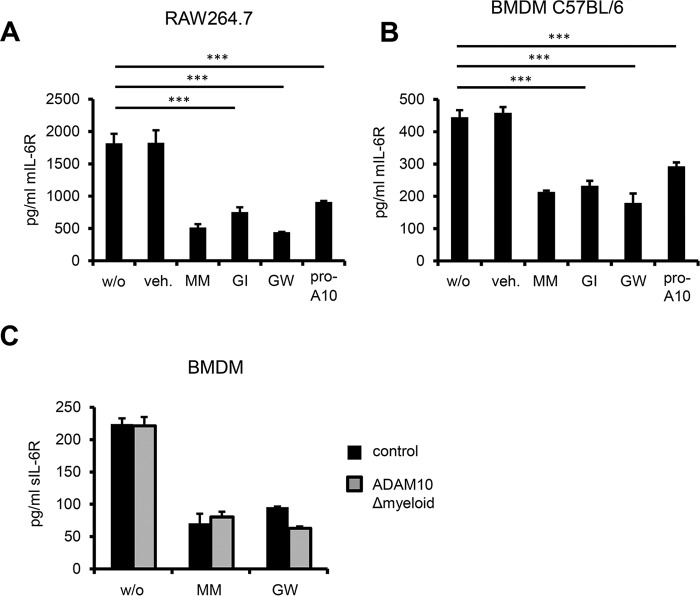
As we were able to show critical involvement of ADAM10 in constitutive release of endogenous human IL-6R, we likewise sought to assess the role of ADAM10 in constitutive IL-6R shedding in the murine system. RAW264.7 and J774 cells showed a strong reduction in constitutive IL-6R release when treated with the pan-metalloprotease inhibitor Marimastat, the combined ADAM10/17 inhibitor GW and, most importantly, the ADAM10-specific inhibitor GI (Fig. 3A, data not shown). This strongly suggests that ADAM10 is also responsible for the constitutive release of endogenous IL-6R in the mouse. We also assessed constitutive IL-6R shedding in the presence of recombinant murine ADAM10 prodomain, which has been reported to effectively block ADAM10 activity in the mouse (20). Supporting our results with pharmacological inhibitors we also noticed a substantial reduction of constantly released IL-6R when murine ADAM10 was blocked by the inhibitory prodomain (Fig. 3, A and B). Furthermore, we treated freshly isolated and differentiated C57BL/6 BMDMs with the same inhibitor panel and yielded similar results, confirming the prominent role of ADAM10 in constitutive IL-6R release also in primary murine cells (Fig. 3B). To our surprise, when we genetically ablated ADAM10 in BMDMs (ADAM10Δmyeloid), we did not observe a reduction in constitutively produced sIL-6R, which possibly indicates the presence of a compensatory protease making up for the loss of ADAM10 (Fig. 3C). To address this question we treated ADAM10-deficient BMDMs with the combined ADAM10/17 inhibitor GW to additionally block ADAM17 activity and assessed constitutive IL-6R shedding. As we observed a strong decrease in constitutive IL-6R shedding in ADAM10-deficient BMDMs treated with GW, the protease taking over constitutive IL-6R shedding from ADAM10 appears to be ADAM17 (Fig. 3C).
FIGURE 3.
A, RAW264.7 cells were cultivated overnight without stimulus ± inhibitors (MM, GI, and GW at 3 μm, recombinant pro-ADAM10 domain 10 μm) and sIL-6R was measured by ELISA. B, C57BL/6 BMDMs cells were treated as RAW264.7 cells in A and sIL-6R was quantified by ELISA. C, BMDMs from ADAM10Δmyeloid mice (n = 2 mice per group) were cultivated overnight without stimulus ± inhibitors (MM, GW 3 μm) and sIL-6R was measured by ELISA. Error bars represent S.D., n = 2 independent experiments. A and B, error bars are S.D., n = 3 independent experiments. ***, p < 0.001. veh. = dimethyl sulfoxide.
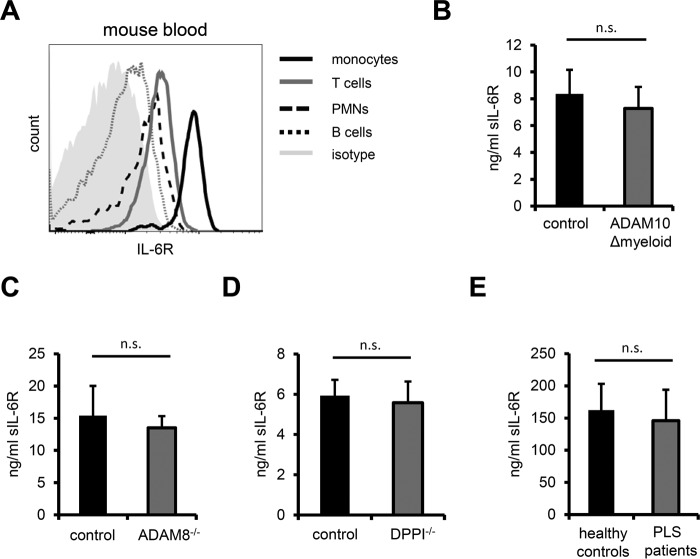
Serum sIL-6R Is Not Released by ADAM10, ADAM8, Neutrophil Elastase, Cathepsin G, or Proteinase 3
A soluble form of the IL-6R is present in the blood of humans as well as mice and genome-wide association studies have reported a correlation between serum sIL-6R levels and the outcome of autoimmune diseases like asthma, rheumatoid arthritis, and type I diabetes (15, 32). In that respect, it has been proposed that sIL-6R in conjunction with soluble gp130 (sgp130) in the blood operates as a buffer against IL-6 preventing global inflammation by overshooting IL-6 (33). Serum sIL-6R levels in hypomorphic ADAM17ex/ex mice are not different from their wild type counterparts (9). This indicates that either a different protease or an entirely different mode of generation contributes to circulating sIL-6R. Over 60% of serum sIL-6R in the mouse stem from myeloid cells, whereas the remainder can be attributed to hepatocytes and T cells (18). Given the role of ADAM10 as constitutive sheddase of endogenous IL-6R in monocytes and macrophages we reasoned that ADAM10 might be a good candidate for the protease generating circulating sIL-6R. Initially, we established that mouse monocytes bear high levels of surface IL-6R, largely exceeding the levels observed for neutrophils, T- and B-cells (Fig. 4A). These results are in line with observations showing similarly high IL-6R expression on human monocytes (Fig. 1, C and D, and Refs. 34 and 35). To support our hypothesis of ADAM10 being the main sheddase that generates circulating IL-6R we collected serum from a cohort of ADAM10Δmyeloid mice and determined sIL-6R serum levels. However, sIL-6R levels in ADAM10Δmyeloid mice were not significantly different from control animals (Fig. 4B). ADAM8 and ADAM10 exhibit overlapping substrate spectra, as exemplified by cleavage of CD23 (36), while ADAM8 expression in mouse skin is strongly up-regulated in the absence of ADAM10 (37). We speculated that ADAM8 might likewise compensate for the loss of ADAM10 in myeloid cells, but observed similar sIL-6R serum levels in ADAM8 knock-out mice as compared to wild type animals (Fig. 4C). The serine proteases neutrophil elastase, cathepsin G, and proteinase 3 are highly abundant in neutrophils and a subpopulation of monocytes (38). Furthermore, it has been reported that in human inflammatory lesions IL-6R is proteolytically cleaved by cathepsin G (39). All three proteases must be activated by N-terminal removal of two amino acids, which is accomplished by dipeptidylpeptidase I (DPPI, or alternatively cathepsin C) (38). To analyze whether neutrophil elastase, cathepsin G, or proteinase 3 are involved in the generation of circulating sIL-6R generation we analyzed sIL-6R levels in the serum of DPPI knock-out mice, which lack neutrophil elastase, cathepsin G, and proteinase 3 activity (23). Again, serum IL-6R levels were comparable in unchallenged DPPI knock-out and control mice (Fig. 4D). Humans deficient in DPPI suffer from Papillon-Lefèvre syndrome (PLS), which represents a rare autosomal recessive genetic disorder characterized by severe periodontitis as well as palmoplantar keratoderma (40). In keeping with the serum levels of DPPI knock-out mice, serum sIL-6R levels from PLS patients did not differ from healthy age-matched individuals (Fig. 4E). In summary, we show that deficiencies in ADAM10, ADAM8, and DPPI, which entails the loss of activity of neutrophil elastase, cathepsin G, and proteinase 3, did not result in a reduction of serum sIL-6R levels.
FIGURE 4.
A, steady state surface expression of murine IL-6R on major circulating immune cell subsets was analyzed by flow cytometry. Note that IL-6R expression on inflammatory monocytes (CD11b+, Ly6Chigh) is considerably higher than on neutrophils (CD11b+, Ly6G+), T and B cells (CD3+ and B220+, respectively). Leukocytes were pre-gated on CD45 prior to gating on different leukocyte subsets. One representative histogram is shown (n = 3 C57BL/6 mice). B, serum from ADAM10Δmyeloid mice (n = 17) was prepared from whole blood and murine sIL-6R was quantified by ELISA. ADAM10flox/flox littermates (n = 21) served as controls. C and D, serum sIL-6R levels in ADAM8−/− (n = 5, control n = 6) and DPPI−/− (n = 4, control n = 4) mice were measured as in B. As DPPI−/− animals were on an ApoE−/− background, ApoE−/− mice served as control. E, circulating sIL-6R in PLS patients and age-matched healthy individuals was quantified by ELISA (n = 9). Error bars represent S.D. n.s. = not significant.
Absence of Alternative IL-6R Splicing in Mouse Tissues Relevant for Circulating sIL-6R Generation
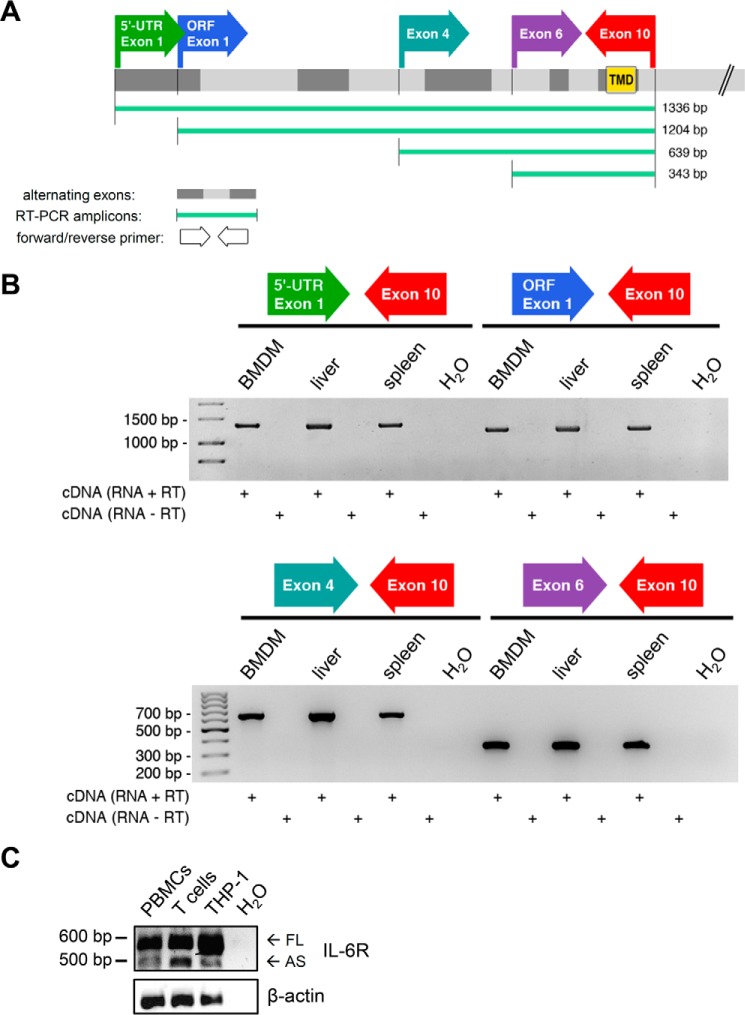
Alternative splicing of the human IL-6R mRNA can also lead to the generation of sIL-6R (6). Mechanistically, skipping the exon coding for the transmembrane region results in a soluble isoform that harbors a newly formed, unique C terminus (11). Antibodies targeting this novel C terminus have enabled the detection of alternatively spliced sIL-6R (AS-sIL-6R) in human serum. In mice, sIL-6R generation based on alternative splicing has not been described yet. We devised a PCR strategy that should inform us if alternative splicing is also employed as a mechanism to generate sIL-6R in the mouse. Assuming a similar splicing mechanism as in humans we designed PCR primers flanking exon 9, which encodes the transmembrane region of murine IL-6R. Moreover, we designed additional 5′-primers that would also detect splicing events occurring further upstream of the exon coding for the transmembrane region (Fig. 5A). RNA used for cDNA synthesis was isolated from organs or cells, which exhibit high IL-6R expression and have been shown to contribute to circulating sIL-6R levels (18). However, in contrast to human sIL-6R (Refs. 6, 27, and 41 and Fig. 5C), we could not detect an alternatively spliced murine sIL-6R mRNA in BMDMs, liver, and spleen of C57BL/6 mice (Fig. 5B). This strongly suggests that alternative splicing does not contribute to serum sIL-6R generation in the mouse.
FIGURE 5.
A, murine IL-6R consists of 10 exons, with exon 9 encoding the membrane-spanning region. The schematic shows the position of the primers used for RT-PCR as well as the expected amplicon lengths. A 3′-primer located in the last exon was combined with four different 5′-primers to additionally cover potentially larger splice variants. Exon 1 of murine IL-6R consists of both the 5′-UTR and the ORF start. TMB, transmembrane domain. B, cDNA originating from C57BL/6 BMDMs, liver, and spleen was subjected to RT-PCR employing the primer combinations depicted in A. Analyzed tissues and cells have been reported to contribute to serum sIL-6R generation. With all PCR primer combinations only one distinct band was detected, which corresponds to the size of murine IL-6R still harboring the transmembrane region. RT-PCR including only RNA (RNA − RT) or H2O as template served as negative controls, showing the absence of genomic DNA contamination. C, alternatively spliced human IL-6R was detected by RT-PCR using a validated, previously published primer pair that flanks the transmembrane region. The larger and more abundant fragment (592 bp) represents full-length human IL-6R, whereas the 498-bp fragment lacks 94 nucleotides including the transmembrane coding region of human IL-6R (27). cDNA was generated from human peripheral blood mononuclear cells, T cells, and THP-1 cells. FL, full-length; AS, alternatively spliced.
A Full-length IL-6R Isoform Is Associated with Circulating Microvesicles in Humans
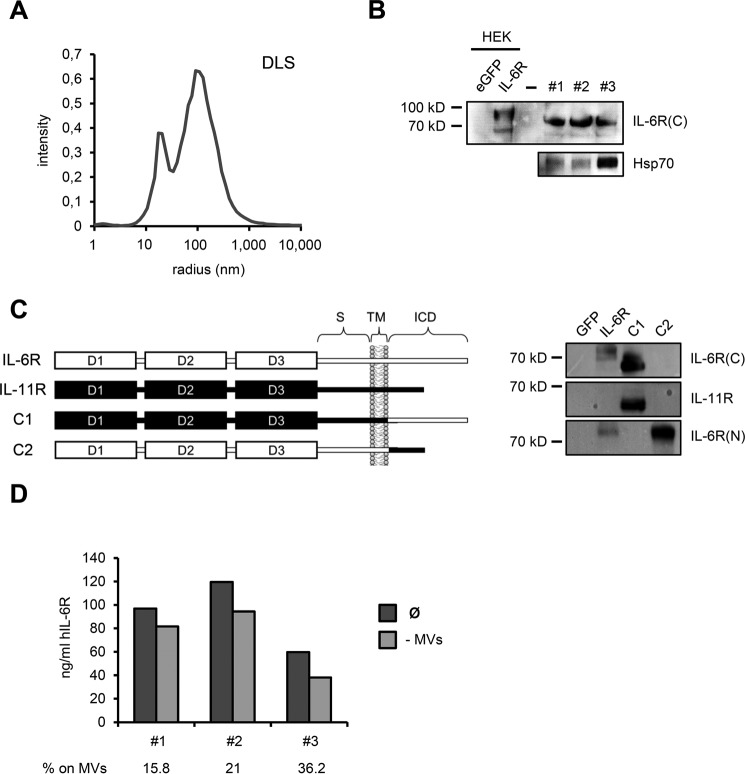
Apart from ectodomain shedding and alternative splicing, soluble cytokine receptors can also be generated by the release of extracellular microvesicles that carry the full-length receptor protein (42). It has in fact been shown that circulating TNFRI in humans is associated with exosomes, which represent a subgroup of extracellular microvesicles originating from endosomal compartments (43). As mice deficient for several candidate proteases show unaltered serum sIL-6R levels and at the same time alternative splicing does not appear to contribute to circulating sIL-6R levels in the mouse (Ref. 9 and this study), we reasoned that the bulk of serum sIL-6R might be produced by a different mechanism, such as microvesicle release. To that end, we isolated microvesicles from the serum of three healthy individuals by serial ultracentrifugation and visualized membrane-associated IL-6R by Western blotting. As microvesicle preparations from serum tend to be crude and frequently include protein contaminations such as protein aggregates and antibodies potentially interfering with subsequent applications, we applied an optimized isolation protocol (see “Experimental Procedures”). Quality confirmation of the microvesicle suspension by DLS revealed the presence of vesicles with an average hydrodynamic size of 100 nm, which is consistent with the reported diameter of exosomes (Fig. 6A). Detection of the exosomal marker protein Hsp70 by Western blotting also suggested that the microvesicle preparation contained exosomes (Fig. 6B). Importantly, Western blot analysis using an antibody against the C-terminal, intracellular portion of human IL-6R revealed the presence of full-length IL-6R protein on circulating microvesicles (Fig. 6B). Full-length IL-6R on microvesicles exhibited a higher electrophoretic mobility than its cellular counterpart in HEK293 cells transiently transfected with a human IL-6R cDNA, which is, however, consistent with a previous report showing less glycosylated TNFRI isoforms on exosomes (44). Moreover, we confirmed the specificity of the employed antibody toward the human IL-6R C terminus using a panel of IL-6R/IL-11R chimeras (Fig. 6C). Western blot analysis with three independent antibodies against the human IL-6R N terminus additionally verified the presence of IL-6R protein on circulating microvesicles (data not shown). To evaluate the proportion of circulating sIL-6R that is associated with microvesicles we subjected serum of the same three healthy individuals to ultracentrifugation to pellet microvesicles and then measured the amount of residual sIL-6R in the microvesicle-free supernatant by ELISA. Comparison of sIL-6R levels prior and post-ultracentrifugation revealed that a clearly detectable fraction sIL-6R is present on circulating microvesicles, which ranged from 15.8 to 36.2% between the three individuals (Fig. 6D). In summary, we established microvesicle release as a novel mechanism of sIL-6R generation and showed that a considerable fraction of serum sIL-6R in humans localizes to circulating microvesicles.
FIGURE 6.
A, microvesicle suspension was analyzed by DLS measurements and revealed two predominant fractions with hydrodynamic sizes of 19 and 100 nm. Whereas an average size of 100 nm is consistent with exosomes, the smaller fraction at 19 nm most likely corresponds to vesicle fragments resulting from the microvesicle enrichment procedure. B, Western blot analysis showing full-length IL-6R on circulating microvesicles of three healthy individuals, with the employed antibody specifically recognizing the intracellular C terminus of human IL-6R. cDNAs encoding GFP and human IL-6R were transiently transfected into HEK293 cells and served as negative and positive controls, respectively. Note that microvesicle-associated IL-6R appeared slightly smaller than cellular IL-6R in HEK293 lysates. Circulating microvesicles also contained substantial amounts of exosomal marker protein Hsp70. C, left: schematic showing composition of human IL-6R/IL-11R chimeras used to confirm antibody specificity. Portions of human IL-6R are depicted in white, whereas human IL-11R parts are highlighted in black. S, stalk region; TM, transmembrane region; ICD, intracellular domain. Construction of IL6R/IL-11R chimeras was described previously (54). Right: HEK cells were transiently transfected with IL-6R/IL-11R chimeras, along with GFP as negative control. IL-6R-transfected HEK cells served as positive control. The antibody used in this study to detect microvesicle-associated IL-6R only detects the human IL-6R C terminus, but not the IL-11R intracellular portion. IL-6R(C), antibody recognizing intracellular IL-6R C terminus; IL-6R(N), antibody against extracellular IL-6R N terminus. D, serum of the same individuals as in B was subjected to ultracentrifugation and sIL-6R levels prior and after ultracentrifugation were quantified by ELISA. Serum after ultracentrifugation was regarded as microvesicle-free (−MVs). Proportion of circulating sIL-6R associated with microvesicles is depicted below the diagram in %.
Discussion
Understanding the molecular mechanisms of sIL-6R generation is of paramount importance, as the blockade of IL-6 trans-signaling represents a valid therapeutic strategy for many IL-6-driven diseases (45). A soluble form of the human IL-6R can be generated by proteolytic cleavage and alternative splicing, with in inflammatory settings ectodomain shedding apparently being the prevailing mechanism that leads to sIL-6R release (11). In vitro experiments have revealed a crucial role for ADAM10 as constitutive and ADAM17 as inducible IL-6R sheddase (8, 9). However, in these studies cell lines have been used that do not inherently express IL-6R, necessitating ectopic expression. Whether results obtained with cell lines artificially overexpressing IL-6R readily translate into more physiological settings is not known. We therefore sought to investigate IL-6R shedding in cells of monocytic origin, as these cells exhibit high levels of endogenous IL-6R. This allowed us to conduct shedding experiments in a more physiological context, where protease and substrate are expressed in the same cell. Using pharmacological inhibitors as well as a specific ADAM17-neutralizing antibody we showed for the first time that endogenous human IL-6R is proteolytically processed by ADAM17 after stimulation with PMA as well as LPS. However, this is in line with previous studies using ectopically overexpressed human IL-6R (8, 9). We additionally corroborated our results from human monocytic cell lines with primary human macrophages and circulating CD14+ monocytes, further adding to the physiological relevance of our data.
In contrast to a recent report showing that in the mouse induced shedding of (ectopically expressed) IL-6R is solely mediated by ADAM10 (9), we provided clear evidence that ADAM17 mediates PMA- and LPS-induced cleavage of endogenous murine IL-6R. In addition to murine cell lines we employed bone marrow-derived macrophages from ADAM17- and ADAM10-deficient mice to demonstrate that induced shedding of endogenous IL-6R shedding in the mouse depends on ADAM17 rather than ADAM10. The observed differences in murine IL-6R shedding between endogenous and ectopically overexpressed receptor emphasize the general need to routinely include cells that naturally express both protease and analyzed substrate when carrying out shedding experiments.
Pharmacological inhibition as well as blockade with recombinant murine ADAM10 prodomain revealed that constitutive IL-6R shedding in murine monocytic cell lines and wild type BMDMs is reliant on ADAM10. Surprisingly, constitutive IL-6R release in ADAM10-deficient BMDMs was not impaired suggesting compensatory shedding by an alternative IL-6R sheddase. Indeed, when we additionally blocked ADAM17 activity in ADAM10-deficient BMDMs with GW (ADAM10/17 inhibitor) we observed a strong reduction in constitutive IL-6R shedding. This indicates that, whereas ADAM10 appears to be the principal sheddase constitutively processing IL-6R, in the absence of ADAM10 constitutive IL-6R release can be made up for by ADAM17. Our results are in line with a previous report describing a similar compensatory relationship between ADAM10 and ADAM17 for several ADAM substrates (46). Taken together, we showed in cell lines as well as primary cells expressing high levels of endogenous IL-6R that stimulated IL-6R shedding, regardless of the species, is mediated by ADAM17, whereas ADAM10 controls constitutive IL-6R release.
Soluble IL-6R is present in human and murine blood in high amounts, with serum sIL-6R levels ranging from 25 to 50 ng/ml in healthy human individuals (11). The functional role of circulating sIL-6R is not fully understood, but it is likely that it serves a different purpose than the sIL-6R isoform that is generated in local tissues and initiates IL-6 trans-signaling. It has been proposed that blood-borne sIL-6R in conjunction with a soluble form of the second IL-6 receptor subunit gp130 (sgp130) acts as a buffer to prevent potentially overshooting IL-6 from acting systemically (33). Whereas in humans a substantial fraction of serum sIL-6R can be attributed to alternative splicing of the IL-6R mRNA, the bulk of circulating sIL-6R appears to be generated by mechanisms distinct from differential mRNA splicing (13). A SNP within the ADAM17 cleavage site leads to increased serum sIL-6R levels in SNP carriers suggesting a decisive role of ectodomain shedding in serum sIL-6R generation (14, 15). Surprisingly, hypomorphic ADAM17ex/ex mice, which almost completely lack ADAM17 protein, exhibit unaltered serum sIL-6R levels pointing to the involvement of a different protease in serum sIL-6R generation. Because we confirmed ADAM10 as a constitutive sheddase of endogenous IL-6R and steady-state levels of circulating IL-6R are likely to originate from a constitutive proteolytic process, we first sought to determine serum IL-6R levels in ADAM10-deficient mice. ADAM10 knock-out mice are embryonic lethal thus precluding the analysis of circulating sIL-6R (31). Conditional knock-out mice lacking IL-6R only in myeloid cells show greatly reduced sIL-6R serum levels (>60%), thereby establishing myeloid cells as the main cellular source of serum sIL-6R (18). However, conditional ADAM10 knock-out animals devoid of the protease in myeloid cells (ADAM10Δmyeloid mice) exhibited equal levels of circulating sIL-6R essentially ruling out ADAM10 as underlying sheddase. Moreover, while increased ADAM8 expression has been reported in ADAM10-deficient mouse skin (37), comparable serum sIL-6R levels in ADAM8 knock-out mice also excluded ADAM8 as compensatory IL-6R sheddase. DPPI activates neutrophil elastase, cathepsin G as well as proteinase 3 by excising dipeptides from the N terminus (38). Notably, cathepsin G has previously been implicated in human IL-6R shedding in an inflammatory setting (39). Although all three serine proteases are highly abundant in myeloid cells and therefore represent ideal candidate sheddases mediating the release of sIL-6R into the bloodstream, we were not able to detect significant differences between DPPI knock-out mice and control animals in terms of circulating sIL-6R levels. PLS is a rare autosomal recessive genetic disorder caused by a deficiency in human DPPI (40). Consistent with our results in DPPI knock-out mice, we observed similar levels of circulating sIL-6R when we compared a small cohort of PLS patients with healthy age-matched controls. Taken together, we showed unaltered sIL-6R serum levels in several mouse lines deficient for IL-6R candidate sheddases as well as humans carrying a DPPI null allele. This suggests that either an as yet unknown IL-6R sheddase or a mechanism that is distinct from limited proteolysis accounts for the bulk of serum sIL-6R. Whereas alternative IL-6R mRNA splicing contributes to appreciable levels of circulating sIL-6R in humans, a systematic analysis focusing on alternative IL-6R splicing in the mouse is still lacking. As in humans alternative splicing of the IL-6R mRNA leads to a 94-bp deletion containing the transmembrane region, we devised a PCR strategy that would detect similar skipping of the murine transmembrane domain. Using cDNA from tissues that express high levels of endogenous IL-6R and have been shown to contribute to circulating sIL-6R we were, however, not able to detect an alternatively spliced isoform of murine IL-6R. In support of our data, no splice variant of murine IL-6R appears in the Ensembl genome database, whereas eight different mRNA transcripts have been found for human IL-6R (Ensembl release 77).
Apart from ectodomain shedding and alternative splicing soluble cytokine receptors can also be generated by the release of extracellular microvesicles, such as exosomes (42). Exosomes are relatively small in size (50–180 nm) and originate from endosomal compartments by inward budding (47). In contrast, the two other major classes of extracellular microvesicles, shedding microvesicles and apoptotic blebs, are shed directly from the plasma membrane and are considerably larger (up to 1 μm in diameter) than exosomes (48). It has been shown that full-length isoforms of transmembrane proteins such as TNFRI, Amphiregulin, and ICAM-1 are located on exosomes, with exosomal TNFRI even accounting for the majority of circulating human TNFRI (43, 49–51). As mice deficient for several candidate proteases, including the two main IL-6R sheddases ADAM17 (17) and ADAM10 (this study), exhibited unchanged serum levels of sIL-6R and alternative splicing did not appear to contribute to circulating sIL-6R levels in the mouse, we hypothesized that serum sIL-6R might also be associated with circulating microvesicles. As microvesicle enrichment from serum requires appreciable amounts of starting material, we decided to use human rather than mouse serum. We detected full-length IL-6R on circulating human microvesicles from three healthy donors by Western blot analysis using an antibody specific for the IL-6R C terminus. Comparison of sIL-6R levels of microvesicle-free serum (post-ultracentrifugation) with sIL-6R levels prior to ultracentrifugation allowed us to estimate the proportion of circulating sIL-6R that is localized to microvesicles, which ranged from 15.8 to 36.2%. However, a larger cohort of individuals is required to more accurately assess the fraction of circulating sIL-6R associated with microvesicles. In view of the proposed function of circulating sIL-6R as a buffer toward overshooting IL-6 it will also be crucial to establish if microvesicle-associated IL-6R can bind IL-6 (and sgp130) equally well compared with the shed or alternatively spliced sIL-6R isoforms. In this respect, IL-6R complex formation might be impaired due to possible sterical restraints imposed on exosomal IL-6R, which in turn might entail altered functional properties of microvesicle-associated sIL-6R compared with the other two sIL-6R variants. Although in humans microvesicle-associated IL-6R (up to 36% according to our data) and alternative IL-6R mRNA splicing together contribute to a substantial fraction of circulating sIL-6R, a large portion of sIL-6R in human serum is still unaccounted for, warranting further clarification. Several potential scenarios are conceivable, with an as yet unidentified sheddase releasing sIL-6R into the circulation probably being the most obvious one. However, as some transmembrane proteins, such as L1, CD44, and EGFR, can additionally be shed from microvesicles (52, 53), it is possible that the proportion of microvesicle-associated IL-6R, as established in this study, is in fact an underestimate. To be more specific, we can envision a scenario where IL-6R is initially released into the bloodstream on microvesicles and only subsequently undergoes ectodomain shedding, which would result in a circulating sIL-6R isoform that is basically indistinguishable from directly shed sIL-6R. Experiments addressing this hypothesis are currently being pursued in our laboratory. In summary, by linking a full-length IL-6R isoform to circulating microvesicles we identified a third major mechanism, apart from ectodomain shedding and alternative splicing, of sIL-6R generation. This novel mode of sIL-6R release might have potential clinical relevance, as the increase in serum sIL-6R levels observed in various disease conditions could in theory simply be due to an accelerated release of IL-6R containing microvesicles.
Author Contributions
B. R. conceived and coordinated the study and wrote the manuscript. N. S. (Figs. 1, 4, and 6), D. M. (Figs. 2 and 3), A. M. (Fig. 1), J. v. d. H. (Fig. 5), J. S. (Fig. 2), K. K. (Fig. 6), M. M. (Fig. 6), and J. W. (Fig. 5) designed, performed, and analyzed the experiments. G. M., C. G., J. W. B., S. G., B. S., and P. E. provided reagents and contributed to study design, manuscript writing, and revision. A. C. and S. R. J. co-coordinated the study and provided critical comments for the manuscript. All authors reviewed the results and approved the final version of the manuscript.
Acknowledgments
We thank Stefanie Schnell and Annett Lickert for excellent technical assistance. The murine ADAM10-prodomain was a kind gift of Marcia L. Moss (BioZyme Inc.). ADAM10flox/flox; LysM_Cre mice were kindly provided by Marjo Donners (Department of Pathology, Maastricht University, Netherlands).
This work was supported in part by Deutsche Forschungsgemeinschaft, Bonn, Germany, Project Grant RA 2404/1-1 (to B. R.), SFB 877, project A1 (to A. C. and S. R.-J.), and project A10 (to C. G.), the Cluster of Excellence ”Inflammation at Interfaces“ (to S. R.-J., B. R., and A. C.), and Cancer Research UK (to G. M.). The authors declare that they have no conflicts of interest with the contents of this article.
- ADAM
- a disintegrin and metalloproteinase
- apoE
- apolipoprotein E
- BMDM
- bone marrow-derived macrophage
- DPPI
- dipeptidylpeptidase I
- PMA
- phorbol 12-myristate 13-acetate
- TNFRI
- tumor necrosis factor receptor 1
- TNFα
- tumor necrosis factor α
- DLS
- dynamic light scattering
- PLS
- Papillon-Lefèvre syndrome
- gp130
- glycoprotein 130.
References
- 1.Heinrich P. C., Behrmann I., Haan S., Hermanns H. M., Müller-Newen G., and Schaper F. (2003) Principles of interleukin (IL)-6-type cytokine signalling and its regulation. Biochem. J. 374, 1–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chalaris A., Garbers C., Rabe B., Rose-John S., and Scheller J. (2011) The soluble interleukin 6 receptor: generation and role in inflammation and cancer. Eur J. Cell Biol. 90, 484–494 [DOI] [PubMed] [Google Scholar]
- 3.Rose-John S., and Heinrich P. C. (1994) Soluble receptors for cytokines and growth factors: generation and biological function. Biochem. J. 300, 281–290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Scheller J., Garbers C., and Rose-John S. (2014) Interleukin-6: from basic biology to selective blockade of pro-inflammatory activities. Semin. Immunol. 26, 2–12 [DOI] [PubMed] [Google Scholar]
- 5.Müllberg J., Schooltink H., Stoyan T., Günther M., Graeve L., Buse G., Mackiewicz A., Heinrich P. C., and Rose-John S. (1993) The soluble interleukin-6 receptor is generated by shedding. Eur. J. Immunol. 23, 473–480 [DOI] [PubMed] [Google Scholar]
- 6.Lust J. A., Donovan K. A., Kline M. P., Greipp P. R., Kyle R. A., and Maihle N. J. (1992) Isolation of an mRNA encoding a soluble form of the human interleukin-6 receptor. Cytokine 4, 96–100 [DOI] [PubMed] [Google Scholar]
- 7.Khokha R., Murthy A., and Weiss A. (2013) Metalloproteinases and their natural inhibitors in inflammation and immunity. Nat. Rev. Immunol. 13, 649–665 [DOI] [PubMed] [Google Scholar]
- 8.Matthews V., Schuster B., Schütze S., Bussmeyer I., Ludwig A., Hundhausen C., Sadowski T., Saftig P., Hartmann D., Kallen K. J., and Rose-John S. (2003) Cellular cholesterol depletion triggers shedding of the human interleukin-6 receptor by ADAM10 and ADAM17 (TACE). J. Biol. Chem. 278, 38829–38839 [DOI] [PubMed] [Google Scholar]
- 9.Garbers C., Jänner N., Chalaris A., Moss M. L., Floss D. M., Meyer D., Koch-Nolte F., Rose-John S., and Scheller J. (2011) Species specificity of ADAM10 and ADAM17 proteins in interleukin-6 (IL-6) trans-signaling and novel role of ADAM10 in inducible IL-6 receptor shedding. J. Biol. Chem. 286, 14804–14811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Honda M., Yamamoto S., Cheng M., Yasukawa K., Suzuki H., Saito T., Osugi Y., Tokunaga T., and Kishimoto T. (1992) Human soluble IL-6 receptor: its detection and enhanced release by HIV infection. J. Immunol. 148, 2175–2180 [PubMed] [Google Scholar]
- 11.Jones S. A., Horiuchi S., Topley N., Yamamoto N., and Fuller G. M. (2001) The soluble interleukin 6 receptor: mechanisms of production and implications in disease. FASEB J. 15, 43–58 [DOI] [PubMed] [Google Scholar]
- 12.Dimitrov S., Lange T., Benedict C., Nowell M. A., Jones S. A., Scheller J., Rose-John S., and Born J. (2006) Sleep enhances IL-6 trans-signaling in humans. FASEB J. 20, 2174–2176 [DOI] [PubMed] [Google Scholar]
- 13.Müller-Newen G., Köhne C., Keul R., Hemmann U., Müller-Esterl W., Wijdenes J., Brakenhoff J. P., Hart M. H., and Heinrich P. C. (1996) Purification and characterization of the soluble interleukin-6 receptor from human plasma and identification of an isoform generated through alternative splicing. Eur. J. Biochem. 236, 837–842 [DOI] [PubMed] [Google Scholar]
- 14.Reich D., Patterson N., Ramesh V., De Jager P. L., McDonald G. J., Tandon A., Choy E., Hu D., Tamraz B., Pawlikowska L., Wassel-Fyr C., Huntsman S., Waliszewska A., Rossin E., Li R., Garcia M., Reiner A., Ferrell R., Cummings S., Kwok P.-Y. Y., Harris T., Zmuda J. M., Ziv E., and Health, Aging and Body Composition (Health ABC) Study. (2007) Admixture mapping of an allele affecting interleukin 6 soluble receptor and interleukin 6 levels. Am. J. Hum. Genet. 80, 716–726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ferreira R. C., Freitag D. F., Cutler A. J., Howson J. M., Rainbow D. B., Smyth D. J., Kaptoge S., Clarke P., Boreham C., Coulson R. M., Pekalski M. L., Chen W.-M., Onengut-Gumuscu S., Rich S. S., Butterworth A. S., Malarstig A., Danesh J., and Todd J. A. (2013) Functional IL6R 358Ala allele impairs classical IL-6 receptor signaling and influences risk of diverse inflammatory diseases. PLoS Genet. 9, e1003444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Garbers C., Monhasery N., Aparicio-Siegmund S., Lokau J., Baran P., Nowell M. A., Jones S. A., Rose-John S., and Scheller J. (2014) The interleukin-6 receptor Asp358Ala single nucleotide polymorphism rs2228145 confers increased proteolytic conversion rates by ADAM proteases. Biochim. Biophys. Acta 1842, 1485–1494 [DOI] [PubMed] [Google Scholar]
- 17.Chalaris A., Adam N., Sina C., Rosenstiel P., Lehmann-Koch J., Schirmacher P., Hartmann D., Cichy J., Gavrilova O., Schreiber S., Jostock T., Matthews V., Häsler R., Becker C., Neurath M. F., Reiss K., Saftig P., Scheller J., and Rose-John S. (2010) Critical role of the disintegrin metalloprotease ADAM17 for intestinal inflammation and regeneration in mice. J. Exp. Med. 207, 1617–1624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McFarland-Mancini M. M., Funk H. M., Paluch A. M., Zhou M., Giridhar P. V., Mercer C. A., Kozma S. C., and Drew A. F. (2010) Differences in wound healing in mice with deficiency of IL-6 versus IL-6 receptor. J. Immunol. 184, 7219–7228 [DOI] [PubMed] [Google Scholar]
- 19.Tape C. J., Willems S. H., Dombernowsky S. L., Stanley P. L., Fogarasi M., Ouwehand W., McCafferty J., and Murphy G. (2011) Cross-domain inhibition of TACE ectodomain. Proc. Natl. Acad. Sci. U.S.A. 108, 5578–5583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Moss M. L., Bomar M., Liu Q., Sage H., Dempsey P., Lenhart P. M., Gillispie P. A., Stoeck A., Wildeboer D., Bartsch J. W., Palmisano R., and Zhou P. (2007) The ADAM10 prodomain is a specific inhibitor of ADAM10 proteolytic activity and inhibits cellular shedding events. J. Biol. Chem. 282, 35712–35721 [DOI] [PubMed] [Google Scholar]
- 21.Jorissen E., Prox J., Bernreuther C., Weber S., Schwanbeck R., Serneels L., Snellinx A., Craessaerts K., Thathiah A., Tesseur I., Bartsch U., Weskamp G., Blobel C. P., Glatzel M., De Strooper B., and Saftig P. (2010) The disintegrin/metalloproteinase ADAM10 is essential for the establishment of the brain cortex. J. Neurosci. 30, 4833–4844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kelly K., Hutchinson G., Nebenius-Oosthuizen D., Smith A. J., Bartsch J. W., Horiuchi K., Rittger A., Manova K., Docherty A. J., and Blobel C. P. (2005) Metalloprotease-disintegrin ADAM8: expression analysis and targeted deletion in mice. Dev. Dyn. 232, 221–231 [DOI] [PubMed] [Google Scholar]
- 23.Adkison A. M., Raptis S. Z., Kelley D. G., and Pham C. T. (2002) Dipeptidyl peptidase I activates neutrophil-derived serine proteases and regulates the development of acute experimental arthritis. J. Clin. Invest. 109, 363–371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Möller-Hackbarth K., Dewitz C., Schweigert O., Trad A., Garbers C., Rose-John S., and Scheller J. (2013) A disintegrin and metalloprotease (ADAM) 10 and ADAM17 are major sheddases of T cell immunoglobulin and mucin domain 3 (Tim-3). J. Biol. Chem. 288, 34529–34544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rabe B., Chalaris A., May U., Waetzig G. H., Seegert D., Williams A. S., Jones S. A., Rose-John S., and Scheller J. (2008) Transgenic blockade of interleukin 6 transsignaling abrogates inflammation. Blood 111, 1021–1028 [DOI] [PubMed] [Google Scholar]
- 26.Chalaris A., Rabe B., Paliga K., Lange H., Laskay T., Fielding C. A., Jones S. A., Rose-John S., and Scheller J. (2007) Apoptosis is a natural stimulus of IL6R shedding and contributes to the proinflammatory trans-signaling function of neutrophils. Blood 110, 1748–1755 [DOI] [PubMed] [Google Scholar]
- 27.Oh J. W., Revel M., and Chebath J. (1996) A soluble interleukin 6 receptor isolated from conditioned medium of human breast cancer cells is encoded by a differentially spliced mRNA. Cytokine 8, 401–409 [DOI] [PubMed] [Google Scholar]
- 28.Horiuchi K., Le Gall S., Schulte M., Yamaguchi T., Reiss K., Murphy G., Toyama Y., Hartmann D., Saftig P., and Blobel C. P. (2007) Substrate selectivity of epidermal growth factor-receptor ligand sheddases and their regulation by phorbol esters and calcium influx. Mol. Biol. Cell 18, 176–188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ludwig A., Hundhausen C., Lambert M. H., Broadway N., Andrews R. C., Bickett D. M., Leesnitzer M. A., and Becherer J. D. (2005) Metalloproteinase inhibitors for the disintegrin-like metalloproteinases ADAM10 and ADAM17 that differentially block constitutive and phorbol ester-inducible shedding of cell surface molecules. Comb. Chem. High Throughput Screen 8, 161–171 [DOI] [PubMed] [Google Scholar]
- 30.Peschon J. J., Slack J. L., Reddy P., Stocking K. L., Sunnarborg S. W., Lee D. C., Russell W. E., Castner B. J., Johnson R. S., Fitzner J. N., Boyce R. W., Nelson N., Kozlosky C. J., Wolfson M. F., Rauch C. T., Cerretti D. P., Paxton R. J., March C. J., and Black R. A. (1998) An essential role for ectodomain shedding in mammalian development. Science 282, 1281–1284 [DOI] [PubMed] [Google Scholar]
- 31.Hartmann D., de Strooper B., Serneels L., Craessaerts K., Herreman A., Annaert W., Umans L., Lübke T., Lena Illert A., von Figura K., and Saftig P. (2002) The disintegrin/metalloprotease ADAM 10 is essential for Notch signalling but not for α-secretase activity in fibroblasts. Hum. Mol. Genet. 11, 2615–2624 [DOI] [PubMed] [Google Scholar]
- 32.Ferreira M. A., Matheson M. C., Duffy D. L., Marks G. B., Hui J., Le Souëf P., Danoy P., Baltic S., Nyholt D. R., Jenkins M., Hayden C., Willemsen G., Ang W., Kuokkanen M., Beilby J., Cheah F., de Geus E. J., Ramasamy A., Vedantam S., Salomaa V., Madden P. A., Heath A. C., Hopper J. L., Visscher P. M., Musk B., Leeder S. R., Jarvelin M.-R., Pennell C., Boomsma D. I., Hirschhorn J. N., Walters H., Martin N. G., James A., Jones G., Abramson M. J., Robertson C. F., Dharmage S. C., Brown M. A., Montgomery G. W., Thompson P. J., and Australian Asthma Genetics Consortium. (2011) Identification of IL6R and chromosome 11q13.5 as risk loci for asthma. Lancet 378, 1006–1014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Scheller J., and Rose-John S. (2012) The interleukin 6 pathway and atherosclerosis. Lancet 380, 338. [DOI] [PubMed] [Google Scholar]
- 34.Oberg H. H., Wesch D., Grüssel S., Rose-John S., and Kabelitz D. (2006) Differential expression of CD126 and CD130 mediates different STAT-3 phosphorylation in CD4+CD25− and CD25 high regulatory T cells. Int. Immunol. 18, 555–563 [DOI] [PubMed] [Google Scholar]
- 35.Wognum A. W., van Gils F. C., and Wagemaker G. (1993) Flow cytometric detection of receptors for interleukin-6 on bone marrow and peripheral blood cells of humans and rhesus monkeys. Blood 81, 2036–2043 [PubMed] [Google Scholar]
- 36.Schlomann U., Koller G., Conrad C., Ferdous T., Golfi P., Garcia A. M., Höfling S., Parsons M., Costa P., Soper R., Bossard M., Hagemann T., Roshani R., Sewald N., Ketchem R. R., Moss M. L., Rasmussen F. H., Miller M. A., Lauffenburger D. A., Tuveson D. A., Nimsky C., and Bartsch J. W. (2015) ADAM8 as a drug target in pancreatic cancer. Nat. Commun. 6, 6175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Weber S., Niessen M. T., Prox J., Lüllmann-Rauch R., Schmitz A., Schwanbeck R., Blobel C. P., Jorissen E., de Strooper B., Niessen C. M., and Saftig P. (2011) The disintegrin/metalloproteinase Adam10 is essential for epidermal integrity and Notch-mediated signaling. Development 138, 495–505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Korkmaz B., Moreau T., and Gauthier F. (2008) Neutrophil elastase, proteinase 3 and cathepsin G: physicochemical properties, activity and physiopathological functions. Biochimie 90, 227–242 [DOI] [PubMed] [Google Scholar]
- 39.Bank U., Reinhold D., Schneemilch C., Kunz D., Synowitz H. J., and Ansorge S. (1999) Selective proteolytic cleavage of IL-2 receptor and IL-6 receptor ligand binding chains by neutrophil-derived serine proteases at foci of inflammation. J. Interferon Cytokine Res. 19, 1277–1287 [DOI] [PubMed] [Google Scholar]
- 40.Toomes C., James J., Wood A. J., Wu C. L., McCormick D., Lench N., Hewitt C., Moynihan L., Roberts E., Woods C. G., Markham A., Wong M., Widmer R., Ghaffar K. A., Pemberton M., Hussein I. R., Temtamy S. A., Davies R., Read A. P., Sloan P., Dixon M. J., and Thakker N. S. (1999) Loss-of-function mutations in the cathepsin C gene result in periodontal disease and palmoplantar keratosis. Nat. Genet. 23, 421–424 [DOI] [PubMed] [Google Scholar]
- 41.Horiuchi S., Koyanagi Y., Zhou Y., Miyamoto H., Tanaka Y., Waki M., Matsumoto A., Yamamoto M., and Yamamoto N. (1994) Soluble interleukin-6 receptors released from T cell or granulocyte/macrophage cell lines and human peripheral blood mononuclear cells are generated through an alternative splicing mechanism. Eur. J. Immunol. 24, 1945–1948 [DOI] [PubMed] [Google Scholar]
- 42.Levine S. J. (2008) Molecular mechanisms of soluble cytokine receptor generation. J. Biol. Chem. 283, 14177–14181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hawari F. I., Rouhani F. N., Cui X., Yu Z.-X., Buckley C., Kaler M., and Levine S. J. (2004) Release of full-length 55-kDa TNF receptor 1 in exosome-like vesicles: a mechanism for generation of soluble cytokine receptors. Proc. Natl. Acad. Sci. U.S.A. 101, 1297–1302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang J., Hawari F. I., Shamburek R. D., Adamik B., Kaler M., Islam A., Liao D.-W., Rouhani F. N., Ingham M., and Levine S. J. (2008) Circulating TNFR1 exosome-like vesicles partition with the LDL fraction of human plasma. Biochem. Biophys. Res. Commun. 366, 579–584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Calabrese L. H., and Rose-John S. (2014) IL-6 biology: implications for clinical targeting in rheumatic disease. Nat. Rev. Rheumatol. 10, 720–727 [DOI] [PubMed] [Google Scholar]
- 46.Le Gall S. M., Bobé P., Reiss K., Horiuchi K., Niu X.-D., Lundell D., Gibb D. R., Conrad D., Saftig P., and Blobel C. P. (2009) ADAMs 10 and 17 represent differentially regulated components of a general shedding machinery for membrane proteins such as transforming growth factor α, L-selectin, and tumor necrosis factor α. Mol. Biol. Cell 20, 1785–1794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Keller S., Sanderson M. P., Stoeck A., and Altevogt P. (2006) Exosomes: from biogenesis and secretion to biological function. Immunol. Lett. 107, 102–108 [DOI] [PubMed] [Google Scholar]
- 48.Cocucci E., Racchetti G., and Meldolesi J. (2009) Shedding microvesicles: artefacts no more. Trends Cell Biol. 19, 43–51 [DOI] [PubMed] [Google Scholar]
- 49.Higginbotham J. N., Demory Beckler M., Gephart J. D., Franklin J. L., Bogatcheva G., Kremers G.-J., Piston D. W., Ayers G. D., McConnell R. E., Tyska M. J., and Coffey R. J. (2011) Amphiregulin exosomes increase cancer cell invasion. Curr. Biol. 21, 779–786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Effenberger T., von der Heyde J., Bartsch K., Garbers C., Schulze-Osthoff K., Chalaris A., Murphy G., Rose-John S., and Rabe B. (2014) Senescence-associated release of transmembrane proteins involves proteolytic processing by ADAM17 and microvesicle shedding. FASEB J. 28, 4847–4856 [DOI] [PubMed] [Google Scholar]
- 51.Segura E., Nicco C., Lombard B., Véron P., Raposo G., Batteux F., Amigorena S., and Théry C. (2005) ICAM-1 on exosomes from mature dendritic cells is critical for efficient naive T-cell priming. Blood 106, 216–223 [DOI] [PubMed] [Google Scholar]
- 52.Stoeck A., Keller S., Riedle S., Sanderson M. P., Runz S., Le Naour F., Gutwein P., Ludwig A., Rubinstein E., and Altevogt P. (2006) A role for exosomes in the constitutive and stimulus-induced ectodomain cleavage of L1 and CD44. Biochem. J. 393, 609–618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sanderson M. P., Keller S., Alonso A., Riedle S., Dempsey P. J., and Altevogt P. (2008) Generation of novel, secreted epidermal growth factor receptor (EGFR/ErbB1) isoforms via metalloprotease-dependent ectodomain shedding and exosome secretion. J. Cell. Biochem. 103, 1783–1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Düsterhöft S., Höbel K., Oldefest M., Lokau J., Waetzig G. H., Chalaris A., Garbers C., Scheller J., Rose-John S., Lorenzen I., and Grötzinger J. (2014) A disintegrin and metalloprotease 17 dynamic interaction sequence, the sweet tooth for the human interleukin 6 receptor. J. Biol. Chem. 289, 16336–16348 [DOI] [PMC free article] [PubMed] [Google Scholar]