Background: The biosynthesis of deoxypentoses appended to bacterial secondary metabolites is poorly understood.
Results: Characterization of CalS8, a putative sugar dehydrogenase in calicheamicin biosynthesis, reveals unique base permissivity with a bias toward TDP-glucose.
Conclusion: CalS8 contains a modified intersubunit loop implicated as the substrate specificity factor in this prototype dehydrogenase.
Significance: This work presents a new blueprint for base specificity annotation among putative UGDHs.
Keywords: biosynthesis, carbohydrate, crystal structure, dehydrogenase, natural product, sugar nucleotide, UDP-glucose dehydrogenase (UGDH), enediyne, deoxysugar, calicheamicin
Abstract
Classical UDP-glucose 6-dehydrogenases (UGDHs; EC 1.1.1.22) catalyze the conversion of UDP-α-d-glucose (UDP-Glc) to the key metabolic precursor UDP-α-d-glucuronic acid (UDP-GlcA) and display specificity for UDP-Glc. The fundamental biochemical and structural study of the UGDH homolog CalS8 encoded by the calicheamicin biosynthetic gene is reported and represents one of the first studies of a UGDH homolog involved in secondary metabolism. The corresponding biochemical characterization of CalS8 reveals CalS8 as one of the first characterized base-permissive UGDH homologs with a >15-fold preference for TDP-Glc over UDP-Glc. The corresponding structure elucidations of apo-CalS8 and the CalS8·substrate·cofactor ternary complex (at 2.47 and 1.95 Å resolution, respectively) highlight a notably high degree of conservation between CalS8 and classical UGDHs where structural divergence within the intersubunit loop structure likely contributes to the CalS8 base permissivity. As such, this study begins to provide a putative blueprint for base specificity among sugar nucleotide-dependent dehydrogenases and, in conjunction with prior studies on the base specificity of the calicheamicin aminopentosyltransferase CalG4, provides growing support for the calicheamicin aminopentose pathway as a TDP-sugar-dependent process.
Introduction
UDP-glucose 6-dehydrogenases (UGDHs;3 EC 1.1.1.22) catalyze the conversion of UDP-α-d-glucose (UDP-Glc) to the key metabolic precursor UDP-α-d-glucuronic acid (UDP-GlcA) (1, 2). In higher eukaryotes and humans, UDP-GlcA serves as a biosynthetic precursor to important proteoglycans and glycosaminoglycans and is a substrate for UDP-glucuronosyltransferase-catalyzed glucuronidation, a reaction estimated to account for ∼35% of all drugs metabolized by phase II drug-metabolizing enzymes (3). In bacteria and fungi, UDP-GlcA is a key substrate in primary metabolism for the production of glycoconjugates that contribute to pathogenicity, virulence, and antibiotic resistance (4–7). Nucleotide diphosphate (NDP)-GlcA has also recently been implicated as a key precursor to novel functionalized pentoses appended to bioactive bacterial secondary metabolites including enediynes (calicheamicin, esperamicin, and maduropeptin) (8–10), indolocarbazoles (AT2433) (11), and orthosomycins (avilamycin and everninomicin) (12–14) (Fig. 1A). Biochemical and structural studies of representative bacterial UDGHs involved in primary metabolism revealed these enzymes to exert a specificity for UDP-Glc, to exist as dimers and, in some cases, to be regulated by bacterial tyrosine kinases (2, 15–22). In contrast, the only UGDH from secondary metabolism biochemically characterized thus far (CalS8) revealed turnover with both UDP-Glc and TDP-α-d-glucose (TDP-Glc) (9). To further probe the potential unique nucleotide specificity of CalS8, herein we report the CalS8 kinetic parameters and x-ray structure determination. This work highlights CalS8 to display the key structural/catalytic signatures of UGDH family members with a modified intersubunit loop structure caused by the insertion of three amino acid residues (Gly259-Ser260-Ser261) and deletion of a conserved UGDH Phe/Tyr residue as potential contributors to the determined >20-fold lower Km for TDP-Glc. As the first biochemical and structural characterization of a base-permissive UGDH family member with preference for thymidine, this study presents a basis for annotation of CalS8 as a TDP-α-d-glucose dehydrogenase (TGDH) and a potential structural blueprint for divergent base specificity among putative UGDHs in bacterial secondary metabolism.
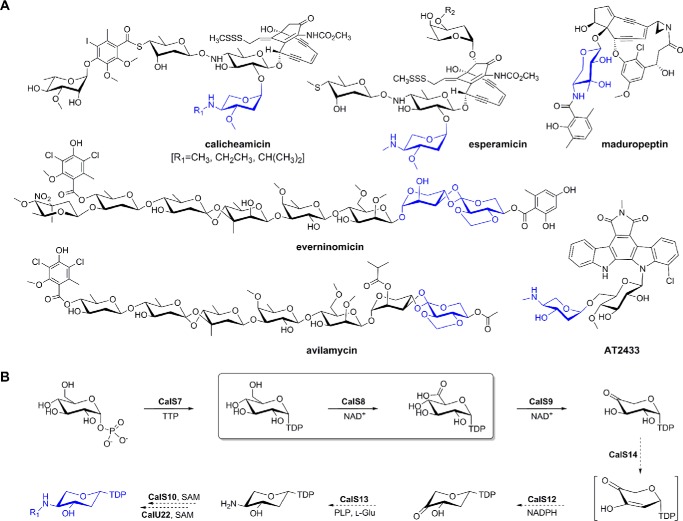
FIGURE 1.
Aminopentose-containing metabolites and corresponding biosynthetic pathways. A, pentose-containing bacterial secondary metabolites where corresponding pentoses are highlighted in blue. B, proposed biosynthetic pathway of aminodideoxypentose shared by calicheamicin, esperamicin, and AT2433 where steps for which biochemical characterization exists are denoted with solid arrows and putative steps (based upon sequence-guided annotation) are illustrated with dotted arrows. The CalS8-catalyzed reaction is highlighted within the box.
Experimental Procedures
Materials
Escherichia coli B834 (DE3) and BL21 (DE3)-Gold strain competent cells were purchased from Stratagene (La Jolla, CA). The pET-28b E. coli expression vector and thrombin were purchased from Novagen (Madison, WI). Primers were purchased from Integrated DNA Technologies (Coralville, IA). Pfu DNA polymerase was purchased from Stratagene (La Jolla, CA). Restriction enzymes and T4 DNA ligase were purchased from New England Biolabs (Ipswich, MA). Ni-NTA superflow column and gel filtration column HiLoad 16/600 were purchased from GE Healthcare. Amicon Ultra® centrifugal filters were purchased from EMD Millipore (Merck KGaA, Darmstadt, Germany), and l-selenomethionine was purchased from Medicilon, Inc. (Shanghai, China). Crystal screen kits were purchased from Hampton Research (Aliso Viejo, CA), Molecular Dimensions (Altamonte Springs, FL), Rigaku (Seattle, WA), and Microlytic (Burlington, MA). All other chemicals were reagent grade or better and purchased from Sigma. X-ray data were collected at beamline 21-ID-F (LS-CAT) of the Advanced Photon Source at Argonne National Laboratory (Chicago, IL) and at the 19-ID and 19-BM beamlines of the Structural Biology Center at the Advanced Photon Source (Chicago, IL). Analytical reverse-phase HPLC (Varian Pro-Star equipped with photodiode array detector, Model 330, Santa Clara, CA) was conducted using a Gemini NX C-18 (5 μm, 250 × 4.6 mm) column (from Phenomenex, Torrance, CA).
Gene Cloning and Expression and Protein Purification
For biochemical studies, the wild-type calS8 gene (National Center for Biotechnology Information (NCBI) accession, gi: 21637116; accession: AAM70332.1) from the genomic DNA of calicheamicin producer, Micromonospora echinospora LL6600, was amplified by PCR using designed primers containing suitable NdeI (forward) and EcoRI (reverse) restriction sites (5′-AATTCCATATGTTCCTTCCCGACCC-3′ forward and 5′-AATTCGAATTCTCACCTTCCAATGCCGCGGTAGG-3′ reverse primers). The PCR products were digested and ligated into pET28a to provide a production construct, confirmed via sequencing, for the N-terminal-His6-CalS8 fusion protein (referred to simply as CalS8 in the current study). Production of CalS8 in E. coli BL21(DE3) was accomplished by inoculation of LB containing 35 μg ml−1 of kanamycin and subsequent growth at 37 °C at 200 rpm to an A600 of ∼0.6, at which time the temperature was lowered to 25 °C and expression was induced with 0.5 mm isopropyl-β-d-thiogalactoside. Cells were allowed to continue to grow at 25 °C for ∼18 h at 220 rpm. The cells were harvested by centrifugation (6,000 × g at 4 °C for 20 min), resuspended in buffer A (20 mm NaH2PO4, 300 mm NaCl, 10 mm imidazole, pH 7.8), and purified as described above via Ni-NTA affinity chromatography using an ÄKTA Purifier 10 (GE Healthcare). Buffer exchange of pooled fractions containing the purified CalS8 was accomplished using a PD-10 column (GE Healthcare) eluted with 25 mm Tris buffer, pH 8, to provide recovered protein at a final concentration of 20 mg ml−1. Final purified CalS8 was drop-frozen in liquid nitrogen and stored at −80 °C. Protein purity was confirmed by SDS-PAGE, and protein concentration was determined using the Bradford protein assay kit (from Bio-Rad) using BSA as a standard.
For structural studies, calS8 gene was amplified by PCR from M. echinospora genomic DNA with KOD Hot Start DNA polymerase using the 5′-TACTTCCAATCCAATGCCATGCCGTTCCTTCCCGACCC-3′ forward and 5′-TTATCCACTTCCAATGTTACCTTCCAATGCCGCGGTAGG-3′ reverse primers. The amplification buffer was supplemented with betaine to a final 2.5 m concentration. PCR products were purified, treated with T4 polymerase (23), and cloned into the pMCSG68 vector according to ligation-independent procedures (24, 25), and the corresponding final clone was determined via sequencing as a CalS8 P294S mutant. This N-terminal tobacco etch virus protease-cleavable StrepII/His6-CalS8 dual-tagged production vector was transformed into E. coli BL21(DE3)-Gold strain. The clone was subsequently validated in small scale production, Ni-NTA affinity purification, and tobacco etch virus protease cleavage propensity of the affinity tag. For large-scale protein production, bacterial culture was grown at 37 °C and 190 rpm in 1 liter of enriched M9 medium (26). When the A600 reached ∼1, the culture was air-cooled to 4 °C over a 60-min period and supplied with 90 mg liter−1 of l-selenomethionine (Se-Met) and 25 mg liter−1 of each of the methionine biosynthetic inhibitory amino acids (l-valine, l-isoleucine, l-leucine, l-lysine, l-threonine, l-phenylalanine). Gene expression was induced by the addition of isopropyl-β-d-thiogalactoside to a final concentration of 0.5 mm, and the cultures were subsequently grown overnight at 18 °C. The cells were harvested and resuspended in lysis buffer (500 mm NaCl, 5% (v/v) glycerol, 50 mm HEPES, pH 8.0, 20 mm imidazole, and 10 mm β-mercaptoethanol). Cells were disrupted by lysozyme treatment (1 mg ml−1) and sonication, and the insoluble cellular material was removed by centrifugation. The corresponding N-His6-CalS8 was purified via Ni-NTA affinity chromatography using the ÄKTAxpress system (GE Health Systems) following a protocol with linear imidazole (10–250 mm) elution gradient. This was followed by the cleavage of the His6 tag using recombinant His6-tagged tobacco etch virus protease at 4 °C for 24–48 h, with an additional Ni-NTA purification to remove the protease, uncut protein, and affinity tag. The pure protein was concentrated using Amicon Ultra®-15 concentrators (Millipore, Bedford, MA) in 20 mm HEPES, pH 8.0, buffer, 250 mm NaCl, and 2 mm DTT. Protein concentrations were determined from the absorbance at 280 nm using a molar absorption coefficient (ϵ280 = 33,960 M−1 cm−1) calculated by using the method developed by Gill and von Hippel (27). The concentration of protein sample used for crystallization was 42.3 mg ml−1. A second batch was purified for co-crystallization with various ligands. This batch was grown the same except that inhibitory amino acids and selenomethionine were not added. The concentration of native protein samples used for crystallization was 43.0 mg ml−1. Individual aliquots of purified protein were stored at −80 °C until needed.
Protein Crystallization
Enzyme from the above-mentioned production for crystallography was screened against crystallization conditions with the help of the Mosquito liquid dispenser (TTP Labtech, Cambridge, MA) using the sitting-drop vapor-diffusion technique in 96-well CrystalQuick plates (Greiner Bio-One, Monroe, NC). For each condition, 0.4 μl of protein and 0.4 μl of crystallization formulation were mixed; the mixture was equilibrated against 140 μl of the reservoir in the well. Protein from the second batch was used to prepare protein-ligand complexes by mixing protein with the following combinations at 4 °C for several hours before setting up crystallizations: 4.51 mm UDP-Glc and 4.51 mm NAD+; 4.51 mm UDP-Glc and 4.51 mm NADH; 4.51 mm TDP-Glc and 4.51 mm NAD+; or 4.51 mm UDP-GlcA acid and 4.51 mm NAD+. Several commercially available crystallization screens were used including: MCSG-1-3 (Microlytic Inc.) at 24 and 4 °C for the apo-protein and MCSG-1-4 at 24 °C for the protein-ligand complexes. Crystals were obtained under several conditions. The best crystals for the apo-form appeared at 24 °C under the condition 15% PEG 3350 and 0.1 m magnesium formate, which corresponds to condition G10 from the MCSG-1 screen. The crystals grew within 2 weeks. The best crystals for the protein-ligand complexes appeared with 4.51 mm TDP-Glc and 4.51 mm NAD+ at 24 °C under the condition 16% PEG 4000, 0.1 m Tris, pH 8.5, and 0.2 m magnesium chloride, which corresponds to condition F4 from the MCSG-4 screen. The crystals grew within 2 weeks. The apo and substrate-bound forms of P294S crystals were cryoprotected by a brief transfer to the crystallization solution plus 20% glycerol and flash-frozen in liquid nitrogen.
Data Collection and Structure Refinement
X-ray diffraction data of Se-Met (apo) and native (complexed with TDP-Glc and NAD+) enzymes were collected at the 19-BM and 19-ID beamlines of the Structural Biology Center at the Advanced Photon Source at the Argonne National Laboratory. The data for apo-enzyme were collected at beamline 19-BM, whereas for substrate-bound enzyme, 19-ID was used. In both cases, the datasets were collected at 100 K at the wavelength corresponding to the selenium K-absorption edge. All data were processed and scaled using HKL3000 (28). Intensities were converted to structure factor amplitudes in the Ctruncate program (29, 30) from the CCP4 package (31). As the selenium anomalous signal was too weak for experimental phasing, the structure of the apo-enzyme form was solved by molecular replacement with UGDH from Porphyromonas gingivalis as the template (Protein Data Bank (PDB): 3GG2) using MR_Rosetta (32, 33). The initial models were further built in COOT (34) and refined in BUSTER (35) with NCS and translation/libration/screw (TLS) restraints (6 TLS groups per protein chain). The structure of substrate-bound enzyme was solved by molecular replacement with the apo-form of P294S as a search model. The structure was manually rebuilt and refined in BUSTER with TLS parameters (10 groups for chain A and 4 groups for chain B). The final models have been validated with the MolProbity server (36). The refinement statistics are shown in Table 1. The atomic coordinates and structure factors have been deposited in the Protein Data Bank under accession codes 4XRR (apo) and 4XR9 (substrate/NAD+-bound). Analysis and validation of structures were performed with the aid of MolProbity (36) and COOT (34) validation tools. Figures were prepared using PyMOL (62). Analysis of the corresponding structures revealed the presence of a P294S mutation introduced as a PCR artifact during initial cloning.
TABLE 1.
Summary of CalS8 (P294S) crystal parameters, data collection, and refinement statistics
Values in parentheses are for the highest resolution shell.
| apo-CalS8 | CalS8·TMP·NAD+ complex | |
|---|---|---|
| Crystal parameters | ||
| Space group | P21 | P21 |
| Unit-cell parameters a, b, c , β (Å, °) | 70.39, 88.20, 70.94 103.4 | 70.68, 87.04, 70.66 103.4 |
| Data collection statistics | ||
| Wavelength (Å) | 0.9793 | 0.9792 |
| Resolution range (Å) | 40.0–2.55 | 30.0–1.95 |
| Unique reflections | 27,596 (1,381) | 60,172 (2,978) |
| Completeness (%) | 99.9 (100) | 99.9 (100) |
| Rmergea | 0.16 (0.93) | 0.07 (0.72) |
| Multiplicity | 3.8 (3.8) | 3.9 (3.9) |
| Mean I/σ(I) | 10.6 (1.74) | 22.10 (2.22) |
| CC1/2(CC*) | 0.619 (0.875) | 0.746 (0.924) |
| Refinement and model statistics | ||
| Rcrystb/Rfreec | 0.184/0.225 | 0.171/0.205 |
| Resolution (Å) | 37.16–2.55 | 28.19–1.95 |
| Reflections work/test set | 26,452/1,128 | 57,108/3,044 |
| RMSD bond lengths (Å) | 0.010 | 0.009 |
| RMSD bond angles (°) | 0.810 | 0.74 |
| B factor - protein/GOL/TMP/NAD/Na/solvent (Å2) | 49.7/52.3/-/-/-/34.3 | 40.7/52.2/42.7/61.9/47.7/41.0 |
| No. of protein atoms | 6,640 | 6,726 |
| No. of waters | 85 | 293 |
| No. of auxiliary molecules in the asymmetric unit | 1 GOL | 1 GOL, 1 TMP, 1 NAD, 1 Na+ |
| Ramachandran plot (%) | ||
| Favorable region | 97.9 | 97.9 |
| Outliers | 0 | 0 |
| Clashscored | 2.25 | 1.40 |
| PDB | 4XRR | 4XR9 |
a Rmerge = Σh Σi | Ii(h) − 〈I(h)〉|/ΣhΣi Ii(h), where Ii(h) is the intensity of an individual measurement of the reflection and 〈I(h)〉 is the mean intensity of the reflection.
b Rcryst = Σh ‖Fobs| − |Fcalc‖/Σh |Fobs|, where Fobs and Fcalc are the observed and calculated structure-factor amplitudes, respectively.
c Rfree was calculated as Rcryst using 5.0% of randomly selected unique reflections that were omitted from the structure refinement.
d As defined by MolProbity.
Enzymatic Activity and Kinetic Characterization
CalS8 activity assays were performed in a total volume of 100 μl in 50 mm Tris-HCl buffer, pH 8, containing 2 mm (T/U)DP-Glc, 5 mm MgCl2, and 20 mm NAD+ and 4 μm CalS8 or P294S at 30 °C. The progress of the reaction was monitored by the change in absorbance at 340 nm (ϵ = 6,220 m−1) on a BMG Labtech FLUOstar Omega plate reader (Cary, NC), which accompanies the reduction of NAD+ to NADH. Product formation was monitored via analytical reverse-phase HPLC (gradient of 1% B to 50% B over 30 min, 50% B for 5 min, 50% B to 1% over 1 min, 1% B for 7 min (A = 50 mm phosphate buffer, pH 6, with 5 mm tetrabutylammonium bisulfate; B = acetonitrile); 1 ml min−1; A254). Prior to HPLC analysis, the reaction mixture was mixed with an equal volume of methanol and then centrifuged at 14,000 × g for 15 min to remove the precipitated protein. Subsequent confirmation of the product was achieved by NMR. Kinetic parameters for TDP-Glc and UDP-Glc were measured under standard assay conditions by holding NAD+ constant (20 mm) and varying (T/U)DP-Glc (from 0.02 to 10 mm). All reactions were performed in triplicate, and concentration of product formed was calculated using NADH standard. The initial velocity data were fitted to the Michaelis-Menten equation by nonlinear regression using GraphPad Prism4 software.
Results and Discussion
Overall Structure
Crystal structures of apo (P294S·Apo) and ternary ligand-bound complex (P294S·TDP-Glc·NAD+) CalS8 were determined at 2.47 and 1.95 Å resolution, respectively (PDB 4XRR and 4XR9, respectively). Both belong to the P21 space group (Table 1) and contain two subunits in the asymmetric unit with a large dimer subunit interaction surface area of ∼2,700 Å2 (Fig. 2). Although crystals of the ternary P294S·TDP-Glc·NAD+ complex were obtained by co-crystallization in the presence of TDP-Glc and NAD+, electron density for only the nucleotide monophosphate (TMP) portion of the sugar nucleotide (Fig. 3C) and NAD+ cofactor (Fig. 3A) was observed in one of the two subunits (ligands have occupancy of 0.9). The absence of glucose 1-phosphate moiety could be a result of disorder in the crystal or partial hydrolysis during crystallization. The remaining TMP moiety occupies essentially the same position as that of UDP-Glc in other representative ligand-bound UGDH structures such the hUGDH·UDP-Glc-NADH complex (PDB 2Q3E) (where hUGDH is human UGDH) (37). It is intriguing that the root mean square deviation (RMSD) between two monomers within the dimeric structures is ∼2.5 Å, likely indicative of allosteric interaction between two monomers. However, because the RMSD between the apo and ternary complex structures is 0.37 Å, the remaining discussion refers to the high-resolution structure of ternary ligand-bound complex (P294S·TDP-Glc·NAD+) of CalS8.
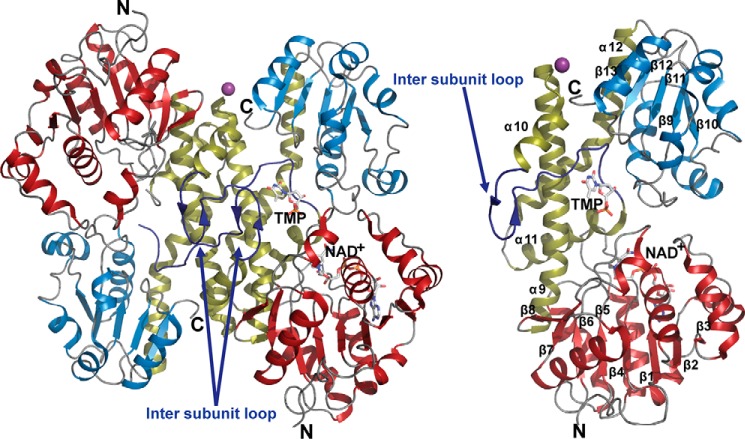
FIGURE 2.
Homodimer (left) and monomer (right) structure of CalS8 (PDB 4XR9). The N terminus NAD+-binding and C terminus substrate-binding domains are colored brick-red and blue, respectively. The dimerization domain and intersubunit loops are colored gold and blue, respectively. The secondary structural elements are labeled within the monomer structure.
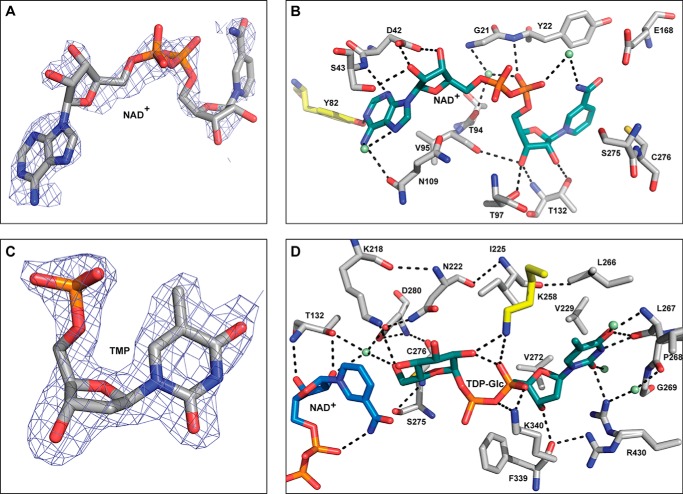
FIGURE 3.
CalS8 (PDB 4XR9) active site interactions. A, electron density map of NAD+ within CalS8 structure (PDB 4XR9; NAD+ occupancy is 0.9). B, residues involved in cofactor binding where the putative regulatory tyrosine is highlighted in yellow and water molecules are represented as green spheres. C, 2mFo-DFc electron density map contoured at 1σ of TMP within CalS8 structure (PDB 4XR9; TMP occupancy is 0.9). D, residues involved in binding the sugar-α-1-phosphate portion of the substrate highlighting key residues involved in catalysis. Water molecules are represented as green spheres, and lysine from the adjacent subunit is colored yellow.
The overall fold of CalS8 is similar to typical nucleotide sugar dehydrogenases (2, 38–39), bearing a bilobal structure consisting of a large N-terminal NAD+-binding domain and a small C-terminal nucleotide sugar-binding domain, each adopting an α/β Rossmann fold (40) (Fig. 2, right panel). The dimer assembly is created through a head-to-tail arrangement of the individual subunits in a back-to-back orientation. Each subunit of CalS8 contains one active site at the cleft formed by the interface of the two domains. The dimer interface is adjacent to the active site, mainly formed by hydrophobic and hydrogen-bonding interactions involving a four-helical bundle (α9–α12) connecting the N- and C-terminal domains (Fig. 2, right panel). The N-terminal NAD+-binding domain has a mixed β-sheet formed by eight β-strands (strand order β3, β2, β1, β4, β5, β6, β7, β8) where β7 and β8 are antiparallel. This large β-sheet is flanked by α-helices α1–α9 on both sides. The C-terminal sugar nucleotide-binding domain is formed by a five-stranded parallel β-sheet (strand order β13, β12, β11, β9, β10) flanked by helices α12–α18 on both sides. The N- and C-terminal domains are connected by a linker domain consisting of a long loop that extends to the adjacent subunit (referred to as the intersubunit loop henceforth) (Fig. 2) flanked on both sides by helices α10 (residues Asp245-Asn254) and α11 (residues Gly274-Arg291) along with two long interdomain helices, α9 (residues Ser210-Gly243) and α12 (residues Leu295-Leu323), respectively (Fig. 2). When compared with UGDH structures, the most distinguishing feature of CalS8 is the modified intersubunit loop containing the β-hairpin composed of two antiparallel β-strands (Ser251-Met252 and Pro257-Lys258) (Fig. 2) formed by the insertion of three extra amino acid residues (Gly259-Ser260-Ser261) when compared with other UGDHs.
Active Site
The NAD+ cofactor is situated on the top of the β-sheet of the N-terminal domain containing a classical glycine-rich Rossmann fold (residues Gly19-Ala20-Gly21-Tyr22-Val23-Gly24) commonly implicated in NAD+ binding (Fig. 3B). NAD+ also interacts with helix α11, a region containing the characteristic GGXCXXKD sequence signature of nucleotide sugar dehydrogenases (residues Gly273-Gly274-Ser275-Cys276-Leu277-Thr278-Lys279-Asp280) known to participate in catalysis (Fig. 3B and residues colored yellow in Fig. 4) (41). The endocyclic N1 from the adenine ring of NAD+ makes a hydrogen-bonding interaction with the Tyr82 side-chain phenolic hydroxyl of putative tyrosine residue (colored yellow in Fig. 3B), a residue conserved among UGDHs that has been demonstrated to be modified via bacterial regulatory tyrosine kinases (2, 15–22). Consistent with the observed preference for NAD+ (9), CalS8 also contains a conserved Asp42 (Fig. 3B), known to be the determinant for specificity for NAD+ versus NADP+ (37).
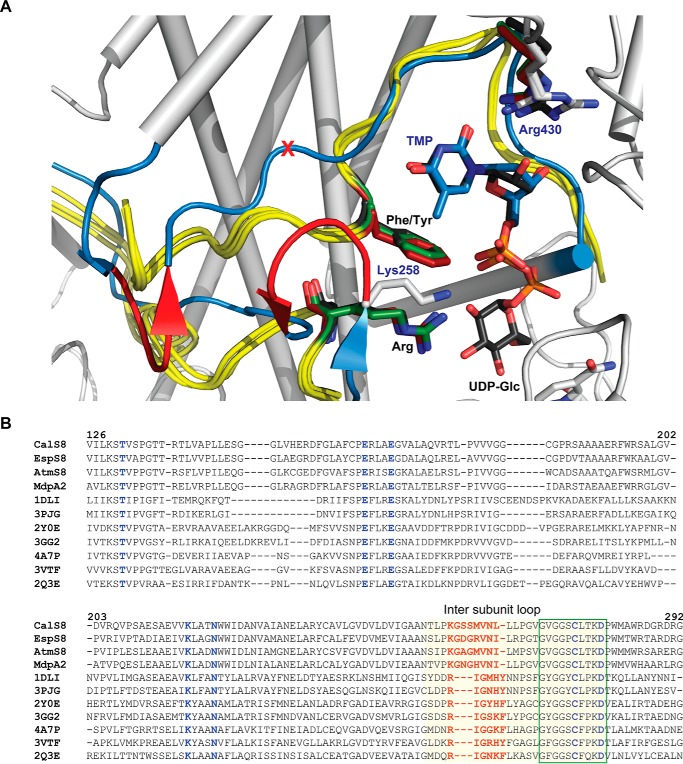
FIGURE 4.
Comparison of CalS8 and representative known UGDH structures highlighting the residues implicated in base specificity. A, overlay highlighting the intersubunit loop of representative UGDH structures with CalS8·TMP·NAD+ ternary complex (colored white, PDB 4XR9). The overlaid UGDH structures include human (PDB 2Q3E), P. gingivalis (PDB 3GG2), and B. cepacia (PDB 2Y0E). The intersubunit loop of UGDH and CalS8 are colored yellow and blue respectively; black- and blue-colored labels correspond to residues of UGDH and CalS8 residues, respectively. The putative base specificity-modulating residues of TGDH are colored red (red part of the loop corresponds to amino acid residues Gly259-Ser260-Ser261, and a red X corresponds to the position of deletion of a residue in CalS8 (which in UGDHs is a Phe or Tyr)). B, sequence alignment of base binding sites of putative TGDHs (CalS8 from the calicheamicin producer M. echinospora, PDB 4XR9; EspS8 from the esperamicin producer Actinomadura verrucosospora; AtmS8 from the AT2433 producer Actinomadura melliaura; and MdpA2 from the maduropeptin producer Actinomadura madurae) and UGDHs with known structure (S. pyogenes (PDB 1DLI), K. pneumoniae (PDB 3PJG), B. cepacia (PDB 2Y0E), P. gingivalis (PDB 3GG2), S. elodea (PDB 4A7P), P. islandicum (PDB 3VTF), and human (PDB 2Q3E)). The UGDHs are represented via PDB IDs with conserved UGDH/TGDH signature motif (green box) and intersubunit (light yellow-shaded), and putative base specificity-modulating and catalytic residues are highlighted (red and blue, respectively).
To model sugar nucleotide binding, the α-1-sugar-phosphate of UDP-Glc from the crystal structure of hUGDH (PDB 2Q3E) (37) was appended to the determined CalS8-bound TMP to afford a properly oriented sugar nucleotide ligand-bound model. This docked model and the structure bound to TMP suggest that the nucleotide base of TDP-Glc is surrounded by the intersubunit loop and anchored to the enzyme through direct and indirect water-mediated hydrogen-bonding interactions primarily originating from residues in the C-terminal domain, whereas N-terminal domain residues contribute to corresponding substrate sugar and phosphate interactions. Key interactions include: substrate thymidine O2 and O4 hydrogen-bonding with the main-chain and side-chain amide NHs of Leu267 and Arg430, respectively (Fig. 3D); thymidine endocyclic N3 hydrogen-bonding with the main-chain carbonyl of Leu267; hydrophobic interactions between the substrate thymidine C5 methyl and the hydrophobic surface created by the side chains of Ile225, Val229, Leu266, and Val272; substrate nucleotide ribose C3′-OH hydrogen-bonding with the main-chain amide and carbonyl of Phe339, (Fig. 3D); and substrate nucleotide pyrophosphate hydrogen-bonding to the side-chain amine of Lys340 from the same subunit and Lys258 from the adjacent subunit (Fig. 3D).
The active site cleft, formed by the interface of NAD+- and TDP-Glc-binding domains, contains a number of residues conserved among NDP-glucose-dehydrogenase family members (Cys276, Asp280, Lys218, Asn222, Thr132, Glu164, and Glu168; Fig. 3, B and D). Within this set, Cys276 is the catalytic residue that forms covalent substrate-enzyme thiohemiacetal/ester intermediates in the oxidative half-reactions en route to NDP-GlcA (2, 37, 42–44). Within this context, Lys218 has been proposed to act as general base and in oxyanion stabilization, whereas Asp280 has been implicated in Cys276 deprotonation and oxyanion stabilization via coordination with an active site water molecule. In addition, Glu164/Glu168 are proposed to function as Brønsted bases in promoting the thioester hydrolysis, whereas Asn222 and Thr132 are believed to also help to stabilize the substrate C6″-oxygen negative charge over the course of the reaction (2, 37, 42–44). In the CalS8 docked model, the Cys276 side-chain thiol is 1.9 Å from the C6″-OH of the sugar (Fig. 3D), consistent with the requisite covalent bond in catalysis. A water molecule also implicated in catalysis (37, 44) is observed in the CalS8 complex 3.2 Å from the substrate C6″-OH, which hydrogen-bonds to side-chain carboxylate of Asp280 and side-chain hydroxyl of Thr132 (distance ∼2.3 and 2.8 Å, respectively) (Fig. 3D). This core water molecule and the side-chain hydroxyl of Thr132 also hydrogen-bond with the C2′-OH of ribose of NAD+ (2.9 and 3.2 Å, respectively), facilitating the association of the reactive groups of substrate and cofactor in the active site (Fig. 3D).
Enzyme Activity and Base Specificity
Preliminary in vitro end point assays for CalS8 revealed the enzyme to accept both UDP-Glc and TDP-Glc, and importantly, provided the first evidence in support of glucose rather than ribose as a progenitor of functionalized pentoses in bacterial secondary metabolism (9). When compared with most UGDHs that display specificity toward UDP-Glc (18–22, 44–48), a broader selectivity for the nucleotide of CalS8 is uncommon and is consistent with the demonstrated base specificity of the calicheamicin TDP-aminodideoxypentosyl transferase CalG4 (58–60). To further probe the base specificity of CalS8, the kinetic parameters of CalS8 with TDP-Glc and UDP-Glc as substrates were determined (Table 2). This analysis revealed the kcat of CalS8 in the presence of UDP-Glc as only slightly better (1.2-fold) to that with TDP-Glc. In contrast, a marked difference in Km was observed with the Km of TDP-Glc (>20-fold improved over that of UDP-Glc, translating to a >15-fold enhancement of specificity constant (kcat/Km) for TDP-Glc). Thus, although CalS8 is capable of accepting both TDP-Glc and UDP-Glc, it is formally considered a TGDH based upon this kinetic analysis. Importantly, similar analysis of the corresponding P294S mutant with TDP-Glc revealed the mutant to be a catalytically competent model of the wild-type enzyme (Table 2).
TABLE 2.
Determination of kinetic parameters for CalS8 and CalS8 mutant P294S mutant using standard conditions with saturating (20 mm) NAD+ and varied (from 0.02 to 10 mm) UDP/TDP-Glc concentrations at 30 °C
| Enzyme/Substrate | kcat | Km | kcat/Km × 10−3 |
|---|---|---|---|
| min−1 | μm | ||
| CalS8/UDP-Glc | 1.6 ± 0.1 | 930.0 ± 128.0 | 1.7 |
| CalS8/TDP-Glc | 1.3 ± 0.1 | 44.9 ± 7.4 | 29.0 |
| P294S/TDP-Glc | 1.0 ± 0.1 | 31.9 ± 3.3 | 31.4 |
To date, the PDB contains UGDH structures from eight different sources including: human (PDB 2Q3E, 2QG4, 3ITK, 3KHU, 3PRJ, 3PTZ, 3TDK, 3TF5, 4EDF, 4QEJ, 4RJT) (37, 42, 49–51); thermophilic archaea, Pyrobaculum islandicum (3VTF) (52); metazoan, Caenorhabditis elegans (PDB 2O3J); and pathogenic bacteria such as Sphingomonas elodea (PDB 4A7P) (48), Burkholderia cepacia (PDB 2Y0C, 2Y0D, 2Y0E) (53), Streptococcus pyogenes (1DLI, 1DLJ) (44), Klebsiella pneumoniae (3PHL, 3PID, 3PJG, 3PLN, 3PLR) (41), and P. gingivalis (PDB 3GG2). All adopt the typical UGDH fold containing conserved residues necessary for cofactor binding and for catalysis (Figs. 2 and 3). From structural and sequence alignments (Fig. 4), there are two distinguishing features in CalS8 that may contribute to base specificity. The first is the insertion of three amino acid residues (Gly259-Ser260-Ser261, colored red within the intersubunit loop in Fig. 4A, and red-colored residues in Fig. 4B) to form β-hairpin containing antiparallel β-strands (Ser251-Met252 and Pro257-Lys258) in the intersubunit loop (Fig. 4A and highlighted in yellow in Fig. 4B). The second is the deletion of the conserved UGDH aromatic residue (F/Y, red cross in Fig. 4A and highlighted in red in Fig. 4B) from the signature motif ((F/Y)X5GG(F/Y)CXXKD, residues colored blue in Fig. 4B). Specifically, the insertion of three residues resulting in the reorientation of the intersubunit loop and deletion of the conserved UGDH Phe/Tyr enables creation of extra space to accommodate the thymidine C5 methyl. In particular, the deletion of the UGDH aromatic residue (which is Phe265, Tyr249, Tyr242, Phe333, and Tyr253 of human, S. pyogenes, K. pneumoniae, P. gingivalis, and P. islandicum UGDH, respectively) in CalS8 is anticipated to relieve the potential for steric infringement upon the thymidine C5 methyl. In addition, a conservative CalS8 Arg to Lys258 substitution (Fig. 4 and yellow-colored residue in Fig. 3D) was observed in the context of the key side-chain residues that contribute to substrate nucleotide pyrophosphate hydrogen-bonding from the adjacent subunit β-hairpin in the intersubunit loop (boxed in Fig. 1B). Although this conservative side-chain substitution is less likely to impact upon base specificity, it is noteworthy that arginine is conserved at this position among UGDHs. However, it is interesting to note the UGDH arginine residue that hydrogen-bonds to C2′-OH of ribose is also conserved in CalS8 (Arg430) and may explain the promiscuity of CalS8 toward UDP-Glc as a substrate. Importantly, of the other putative TDGHs encoded by secondary metabolite gene clusters (Fig. 4B, At2433 AtmS8, esperamicin EspS8, and maduropeptin MdpA2), AtmS8 was also recently demonstrated to display a preference for TDP-Glc in vitro (63).
Conclusions
In summary, the reported determination of fundamental kinetic parameters reveals CalS8 as one of the first characterized examples of a base-permissive TGDH and thereby extends understanding of progenitors to glycosylated bacterial secondary metabolites bearing functionalized pentoses (55–57). The corresponding structure elucidation of the apo and P294S·TDP-Glc·NAD+ ternary complexes highlights a remarkably high degree of conservation between classical UGDHs and this model TGDH where a modified intersubunit loop structure caused by insertion of three residues and deletion of a UGDH conserved aromatic residue likely contributes to the CalS8 base permissivity. As such, this study begins to provide a putative blueprint for base specificity among sugar nucleotide-dependent dehydrogenases. In addition, the demonstrated CalS8 preference for TDP-Glc is consistent with the previously reported specificity of the culminating calicheamicin aminopentosyltransferase CalG4 for TDP in the context of CalG4-catalyzed “reverse” reactions (58–60). Thus, the cumulative characterization of CalS8 and CalG4 provides growing support for the calicheamicin aminopentose pathway as a TDP-sugar-dependent process and contrasts the recently reported characterization of CalS9 as a UDP-GlcA decarboxylase (61).
Author Contributions
S. S. performed biochemical studies, analysis of structure and manuscript preparation; K. M. solved the crystal structure, analyzed the structure and contributed to manuscript preparation; L. B., M. E., G. B., R. M. Y., and A. J. contributed to data acquisition, analysis and/or manuscript proofing; C. A. B. contributed to database management, logistics and manuscript proofing; M. K. K. contributed materials; and J. S. T. and G. N. P. contributed to experimental design, data interpretation and manuscript preparation.
Acknowledgments
Use of the Structural Biology Center beamlines was supported by the U. S. Department of Energy, Office of Biological and Environmental Research, under Contract DE-AC02-06CH11357. The Life Sciences Collaborative Access Team (LS-CAT) has been supported by the Michigan Economic Development Corporation and the Michigan Technology Tri-Corridor.
This work was supported by National Institutes of Health Grants U01GM098248 (to G. N. P.), R01 CA84374 (to J. S. T.), and GM094585 (to A. J.) and the National Center for Advancing Translational Sciences (UL1TR000117). The authors declare the following competing financial interest: J. S. T. is a cofounder of Centrose (Madison, WI).
The atomic coordinates and structure factors (codes 4XRR and 4XR9) have been deposited in the Protein Data Bank (http://wwpdb.org/).
- UGDH
- UDP-glucose 6-dehydrogenase
- UDP-Glc
- UDP-α-d-glucose
- UDP-GlcA
- UDP-α-d-glucuronic acid
- TDP-Glc
- TDP-α-d-glucose
- TGDH
- TDP-α-d-glucose dehydrogenase
- NDP
- nucleotide diphosphate
- Ni-NTA
- nickel-nitrilotriacetic acid.
References
- 1.Strominger J. L., Kalckar H. M., Axelrod J., and Maxwell E. S. (1954) Enzymatic oxidation of uridine diphosphate glucose to uridine diphosphate glucuronic acid. J. Am. Chem. Soc. 76, 6411–6412 [Google Scholar]
- 2.Egger S., Chaikuad A., Kavanagh K. L., Oppermann U., and Nidetzky B. (2010) UDP-glucose dehydrogenase: structure and function of a potential drug target. Biochem. Soc. Trans. 38, 1378–1385 [DOI] [PubMed] [Google Scholar]
- 3.Guillemette C. (2003) Pharmacogenomics of human UDP-glucuronosyltransferase enzymes. Pharmacogenomics J. 3, 136–158 [DOI] [PubMed] [Google Scholar]
- 4.Raetz C. R., Reynolds C. M., Trent M. S., and Bishop R. E. (2007) Lipid A modification systems in Gram-negative bacteria. Annu. Rev. Biochem. 76, 295–329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bar-Peled M., Griffith C. L., and Doering T. L. (2001) Functional cloning and characterization of a UDP-glucuronic acid decarboxylase: the pathogenic fungus Cryptococcus neoformans elucidates UDP-xylose synthesis. Proc. Natl. Acad Sci. U.S.A. 98, 12003–12008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Broach B., Gu X., and Bar-Peled M. (2012) Biosynthesis of UDP-glucuronic acid and UDP-galacturonic acid in Bacillus cereus subsp. cytotoxis NVH 391–98. FEBS J. 279, 100–112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Alvarez D., Merino S., Tomás J. M., Benedí V. J., and Albertí S. (2000) Capsular polysaccharide is a major complement resistance factor in lipopolysaccharide O side chain-deficient Klebsiella pneumoniae clinical isolates. Infect. Immun. 68, 953–955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ahlert J., Shepard E., Lomovskaya N., Zazopoulos E., Staffa A., Bachmann B. O., Huang K., Fonstein L., Czisny A., Whitwam R. E., Farnet C. M., and Thorson J. S. (2002) The calicheamicin gene cluster and its iterative type I enediyne PKS. Science 297, 1173–1176 [DOI] [PubMed] [Google Scholar]
- 9.Bililign T., Shepard E. M., Ahlert J., and Thorson J. S. (2002) On the origin of deoxypentoses: evidence to support a glucose progenitor in the biosynthesis of calicheamicin. Chembiochem. 3, 1143–1146 [DOI] [PubMed] [Google Scholar]
- 10.Van Lanen S. G., Oh T. J., Liu W., Wendt-Pienkowski E., and Shen B. (2007) Characterization of the maduropeptin biosynthetic gene cluster from Actinomadura madurae ATCC 39144 supporting a unifying paradigm for enediyne biosynthesis. J. Am. Chem. Soc. 129, 13082–13094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gao Q., Zhang C., Blanchard S., and Thorson J. S. (2006) Deciphering indolocarbazole and enediyne aminodideoxypentose biosynthesis through comparative genomics: Insights from the AT2433 biosynthetic locus. Chem. Biol. 13, 733–743 [DOI] [PubMed] [Google Scholar]
- 12.Weitnauer G., Mühlenweg A., Trefzer A., Hoffmeister D., Süssmuth R. D., Jung G., Welzel K., Vente A., Girreser U., and Bechthold A. (2001) Biosynthesis of the orthosomycin antibiotic avilamycin A: Deductions from the molecular analysis of the avi biosynthetic gene cluster of Streptomyces viridochromogenes Tü57 and production of new antibiotics. Chem. Biol. 8, 569–581 [DOI] [PubMed] [Google Scholar]
- 13.Staffa A., Zazopoulos E., Mercure S., Nowacki P. P., and Farnet C. M. (July 31, 2003) Genetic locus for everninomicin biosynthesis. U. S. Patent 20,030,143,666
- 14.Gaisser S., Trefzer A., Stockert S., Kirschning A., and Bechthold A. (1997) Cloning of an avilamycin biosynthetic gene cluster from Streptomyces viridochromogenes Tü57. J. Bacteriol. 179, 6271–6278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lacour S., Doublet P., Obadia B., Cozzone A. J., and Grangeasse C. (2006) A novel role for protein-tyrosine kinase Etk from Escherichia coli K-12 related to polymyxin resistance. Res. Microbiol. 157, 637–641 [DOI] [PubMed] [Google Scholar]
- 16.Petranovic D., Grangeasse C., Macek B., Abdillatef M., Gueguen-Chaignon V., Nessler S., Deutscher J., and Mijakovic I. (2009) Activation of Bacillus subtilis Ugd by the BY-kinase PtkA proceeds via phosphorylation of its residue tyrosine 70. J. Mol. Microbiol. Biotechnol. 17, 83–89 [DOI] [PubMed] [Google Scholar]
- 17.Schiller J. G., Bowser A. M., and Feingold D. S. (1973) Partial purification and properties of UDPG dehydrogenase from Escherichia coli. Biochim. Biophys. Acta 293, 1–10 [DOI] [PubMed] [Google Scholar]
- 18.Turner W., and Botha F. C. (2002) Purification and kinetic properties of UDP-glucose dehydrogenase from sugarcane. Arch. Biochem. Biophys. 407, 209–216 [DOI] [PubMed] [Google Scholar]
- 19.Granja A. T., Popescu A., Marques A. R., Sá-Correia I., and Fialho A. M. (2007) Biochemical characterization and phylogenetic analysis of UDP-glucose dehydrogenase from the gellan gum producer Sphingomonas elodea ATCC 31461. Appl. Microbiol. Biotechnol. 76, 1319–1327 [DOI] [PubMed] [Google Scholar]
- 20.Jiang S. S., Lin T. Y., Wang W. B., Liu M. C., Hsueh P. R., and Liaw S. J. (2010) Characterization of UDP-glucose dehydrogenase and UDP-glucose pyrophosphorylase mutants of Proteus mirabilis: defectiveness in polymyxin B resistance, swarming, and virulence. Antimicrob. Agents Chemother. 54, 2000–2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mainprize I. L., Bean J. D., Bouwman C., Kimber M. S., and Whitfield C. (2013) The UDP-glucose dehydrogenase of Escherichia coli K-12 displays substrate inhibition by NAD that is relieved by nucleotide triphosphates. J. Biol. Chem. 288, 23064–23074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Grangeasse C, Obadia B, Mijakovic I, Deutscher J, Cozzone AJ, Doublet P. (2003) Autophosphorylation of the Escherichia coli protein kinase Wzc regulates tyrosine phosphorylation of Ugd, a UDP-glucose dehydrogenase. J. Biol. Chem. 278, 39323–39329 [DOI] [PubMed] [Google Scholar]
- 23.Kim Y., Babnigg G., Jedrzejczak R., Eschenfeldt W. H., Li H., Maltseva N., Hatzos-Skintges C., Gu M., Makowska-Grzyska M., Wu R., An H., Chhor G., and Joachimiak A. (2011) High-throughput protein purification and quality assessment for crystallization. Methods 55, 12–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Aslanidis C., and de Jong P. J. (1990) Ligation-independent cloning of PCR products (LIC-PCR). Nucleic Acids Res. 18, 6069–6074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Eschenfeldt W. H., Lucy S., Millard C. S., Joachimiak A., and Mark I. D. (2009) A family of LIC vectors for high-throughput cloning and purification of proteins. Methods Mol. Biol. 498, 105–115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Donnelly M. I., Zhou M., Millard C. S., Clancy S., Stols L., Eschenfeldt W. H., Collart F. R., and Joachimiak A. (2006) An expression vector tailored for large-scale, high-throughput purification of recombinant proteins. Protein Expr. Purif. 47, 446–454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gill S. C., and von Hippel P. H. (1989) Calculation of protein extinction coefficients from amino acid sequence data. Anal. Biochem. 182, 319–326 [DOI] [PubMed] [Google Scholar]
- 28.Minor W., Cymborowski M., Otwinowski Z., and Chruszcz M. (2006) HKL-3000: the integration of data reduction and structure solution: from diffraction images to an initial model in minutes. Acta Crystallogr. D. Biol. Crystallogr. 62, 859–866 [DOI] [PubMed] [Google Scholar]
- 29.French S., and Wilson K. (1978) Treatment of negative intensity observations. Acta Crystallogr. A. 34, 517–525 [Google Scholar]
- 30.Padilla J. E., and Yeates T. O. (2003) A statistic for local intensity differences: robustness to anisotropy and pseudo-centering and utility for detecting twinning. Acta Crystallogr. D. Biol. Crystallogr. 59, 1124–1130 [DOI] [PubMed] [Google Scholar]
- 31.Winn M. D., Ballard C. C., Cowtan K. D., Dodson E. J., Emsley P., Evans P. R., Keegan R. M., Krissinel E. B., Leslie A. G., McCoy A., McNicholas S. J., Murshudov G. N., Pannu N. S., Potterton E. A., Powell H. R., Read R. J., Vagin A., and Wilson K. S. (2011) Overview of the CCP4 suite and current developments. Acta Crystallogr. D. Biol. Crystallogr. 67, 235–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.DiMaio F. (2013) Advances in Rosetta structure prediction for difficult molecular-replacement problems. Acta Crystallogr. D. Biol. Crystallogr. 69, 2202–2208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Terwilliger T. C., Dimaio F., Read R. J., Baker D., Bunkóczi G., Adams P. D., Grosse-Kunstleve R. W., Afonine P. V., and Echols N. (2012) Phenix.mr_rosetta: Molecular replacement and model rebuilding with Phenix and Rosetta. J. Struct. Funct. Genomics 13, 81–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Emsley P., and Cowtan K. (2004) Coot: Model-building tools for molecular graphics. Acta Crystallogr. D. Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 35.Bricogne G., Blanc E., Brandl M., Flensburg C., Keller P., Paciorek W., Roversi P., Sharff A., Smart O.S., Vonrhein C. T. Womack (2011) BUSTER, Version 2.10.0, Global Phasing Ltd., Cambridge, UK [Google Scholar]
- 36.Chen V. B., Arendall W. B. 3rd, Headd J. J., Keedy D. A., Immormino R. M., Kapral G. J., Murray L. W., Richardson J. S., and Richardson D. C. (2010) MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D. Biol. Crystallogr. 66, 12–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Egger S., Chaikuad A., Kavanagh K. L., Oppermann U., and Nidetzky B. (2011) Structure and mechanism of human UDP-glucose 6-dehydrogenase. J. Biol. Chem. 286, 23877–23887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Perozich J., Leksana A., and Hempel J. (1995) UDP-glucose dehydrogenase: structural characteristics. Adv. Exp. Med. Biol. 372, 79–84 [PubMed] [Google Scholar]
- 39.Singh S., Phillips G. N. Jr., Thorson J. S. (2012) The structural biology of enzymes involved in natural product glycosylation. Nat. Prod. Rep. 29, 1201–1237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rao S. T., and Rossmann M. G. (1973) Comparison of super-secondary structures in proteins. J. Mol. Biol. 76, 241–256 [DOI] [PubMed] [Google Scholar]
- 41.Chen Y. Y., Ko T. P., Lin C. H., Chen W. H., and Wang A. H. (2011) Conformational change upon product binding to Klebsiella pneumoniae UDP-glucose dehydrogenase: A possible inhibition mechanism for the key enzyme in polymyxin resistance. J. Struct. Biol. 175, 300–310 [DOI] [PubMed] [Google Scholar]
- 42.Egger S., Chaikuad A., Klimacek M., Kavanagh K. L., Oppermann U., and Nidetzky B. (2012) Structural and kinetic evidence that catalytic reaction of human UDP-glucose 6-dehydrogenase involves covalent thiohemiacetal and thioester enzyme intermediates. J. Biol. Chem. 287, 2119–2129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sommer B. J., Barycki J. J., and Simpson M. A. (2004) Characterization of human UDP-glucose dehydrogenase: Cys-276 is required for the second of two successive oxidations. J. Biol. Chem. 279, 23590–23596 [DOI] [PubMed] [Google Scholar]
- 44.Campbell R. E., Mosimann S. C., van De Rijn I., Tanner M. E., and Strynadka N. C. (2000) The first structure of UDP-glucose dehydrogenase reveals the catalytic residues necessary for the two-fold oxidation. Biochemistry 39, 7012–7023 [PubMed] [Google Scholar]
- 45.Hempel J., Perozich J., Romovacek H., Hinich A., Kuo I., and Feingold D. S. (1994) UDP-glucose dehydrogenase from bovine liver: Primary structure and relationship to other dehydrogenases. Protein Sci. 3, 1074–1080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Easley K. E., Sommer B. J., Boanca G., Barycki J. J., and Simpson M. A. (2007) Characterization of human UDP-glucose dehydrogenase reveals critical catalytic roles for lysine 220 and aspartate 280. Biochemistry 46, 369–378 [DOI] [PubMed] [Google Scholar]
- 47.Fontaine T., Lamarre C., Simenel C., Lambou K., Coddeville B., Delepierre M., and Latgé J. P. (2009) Characterization of glucuronic acid containing glycolipid in Aspergillus fumigatus mycelium. Carbohydr. Res. 344, 1960–1967 [DOI] [PubMed] [Google Scholar]
- 48.Rocha J., Granja A. T., Sá-Correia I., Fialho A., and Frazão C. (2010) Cloning, expression, purification, crystallization and preliminary crystallographic studies of UgdG, an UDP-glucose dehydrogenase from Sphingomonas elodea ATCC 31461. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 66, 69–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hung R. J., Chien H. S., Lin R. Z., Lin C. T., Vatsyayan J., Peng H. L., and Chang H. Y. (2007) Comparative analysis of two UDP-glucose dehydrogenases in Pseudomonas aeruginosa PAO1. J. Biol. Chem. 282, 17738–17748 [DOI] [PubMed] [Google Scholar]
- 50.Rajakannan V., Lee H. S., Chong S. H., Ryu H. B., Bae J. Y., Whang E. Y., Huh J. W., Cho S. W., Kang L. W., Choe H., and Robinson R. C. (2011) Structural basis of cooperativity in human UDP-glucose dehydrogenase. PLoS One 6, e25226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kadirvelraj R., Custer G. S., Keul N. D., Sennett N. C., Sidlo A. M., Walsh R. M. Jr., and Wood Z. A. (2014) Hysteresis in human UDP-glucose dehydrogenase is due to a restrained hexameric structure that favors feedback inhibition. Biochemistry 53, 8043–8051 [DOI] [PubMed] [Google Scholar]
- 52.Kadirvelraj R., Sennett N. C., Polizzi S. J., Weitzel S., and Wood Z. A. (2011) Role of packing defects in the evolution of allostery and induced fit in human UDP-glucose dehydrogenase. Biochemistry 50, 5780–5789 [DOI] [PubMed] [Google Scholar]
- 53.Sakuraba H., Kawai T., Yoneda K., and Ohshima T. (2012) Structure of a UDP-glucose dehydrogenase from the hyperthermophilic archaeon Pyrobaculum islandicum. Acta Crystallogr. Sect. F. Struct. Biol. Cryst. Commun. 68, 1003–1007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rocha J., Popescu A. O., Borges P., Mil-Homens D., Moreira L. M., Sá-Correia I., Fialho A. M., and Frazão C. (2011) Structure of Burkholderia cepacia UDP-glucose dehydrogenase (UGD) BceC and role of Tyr10 in final hydrolysis of UGD thioester intermediate. J. Bacteriol. 193, 3978–3987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Elshahawi S. I., Shaaban K. A., Kharel M. K., and Thorson J. S. (2015) A comprehensive review of glycosylated bacterial natural products. Chem. Soc. Rev. 10.1039/c4cs00426d [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Thibodeaux C. J., Melançon C. E. 3rd, and Liu H. W. (2008) Natural-product sugar biosynthesis and enzymatic glycodiversification. Angew. Chem. Int. Ed. Engl. 47, 9814–9859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kren V., and Rezanka T. (2008) Sweet antibiotics: the role of glycosidic residues in antibiotic and antitumor activity and their randomization. FEMS Microbiol. Rev. 32, 858–889 [DOI] [PubMed] [Google Scholar]
- 58.Zhang C., Griffith B. R., Fu Q., Albermann C., Fu X., Lee I. K., Li L., Thorson J. S. (2006) Exploiting the reversibility of natural product glycosyltransferase-catalyzed reactions. Science 313, 1291–1294 [DOI] [PubMed] [Google Scholar]
- 59.Zhang C., Bitto E., Goff R. D., Singh S., Bingman C. A., Griffith B. R., Albermann C., Phillips G. N. Jr., Thorson J. S. (2008) Biochemical and structural insights of the early glycosylation steps in calicheamicin biosynthesis. Chem. Biol. 15, 842–853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chang A, Singh S, Helmich KE, Goff RD, Bingman CA, Thorson JS, Phillips G. N. Jr. (2011) Complete set of glycosyltransferase structures in the calicheamicin biosynthetic pathway reveals the origin of regiospecificity. Proc. Natl. Acad. Sci. 108, 17649–17654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Simkhada D., Oh T. J., Pageni B. B., Lee H. C., Liou K., Sohng J. K. (2009) Characterization of CalS9 in the biosynthesis of UDP-xylose and the production of xylosyl-attached hybrid compound. Appl. Microbiol. Biotechnol. 83, 885–895 [DOI] [PubMed] [Google Scholar]
- 62.DeLano W. L. (2010) The PyMOL Molecular Graphics System, version 1.7.4.4, Schrödinger, LLC, New York [Google Scholar]
- 63.Peltier-Pain P., Singh S., Thorson J. S. (2015) Characterization of early enzymes involved in TDP-aminodideoxypentose biosynthesis en route to AT2433. Chembiochem 10.1002/cbic.201500365 [DOI] [PMC free article] [PubMed] [Google Scholar]