Background: NFE2L3 is involved in carcinogenesis, stress response, differentiation, and inflammatory processes.
Results: NFE2L3 is polyubiquitinated via the E3 ubiquitin ligase FBW7, which regulates its turnover. This process requires prior phosphorylation by GSK3.
Conclusion: NFE2L3 is tightly regulated by FBW7 and GSK3 through polyubiquitination.
Significance: Our data highlight the regulation of NFE2L3 by FBW7 and GSK3 and its potential role in cellular stress response.
Keywords: E3 ubiquitin ligase, oxidative stress, protein degradation, ubiquitin, ubiquitin ligase, FBW7, GSK3, NFE2L3, ubiquitination
Abstract
The NFE2L3 transcription factor has been implicated in various cellular processes, including carcinogenesis, stress response, differentiation, and inflammation. Previously it has been shown that NFE2L3 has a rapid turnover and is stabilized by proteasomal inhibitors. The mechanisms regulating the degradation of this protein have not been investigated. Here we report ubiquitination of NFE2L3 and demonstrate that F-box/WD repeat-containing protein 7 (FBW7 or FBWX7), a component of Skp1, Cullin 1, F-box containing complex (SCF)-type E3 ligase, is the E3 ligase mediating the degradation of NFE2L3. We showed that FBW7 interacts with NFE2L3 and that dimerization of FBW7 is required for the degradation of the transcription factor. We also demonstrate that the kinase glycogen synthase kinase 3 (GSK3) mediates the FBW7-dependent ubiquitination of NFE2L3. We show phosphorylation of NFE2L3 by GSK3 and its significance in the regulation of NFE2L3 by the tumor suppressor FBW7. FBW7 abrogated NFE2L3-mediated repression of the NAD(P)H:quinone oxidoreductase 1 (NQO1) gene antioxidant response element (ARE). Our findings reveal FBW7 and GSK3 as novel regulators of the NFE2L3 transcription factor and a potential mechanism by which FBW7 might regulate detoxification and the cellular response to stress.
Introduction
The cap 'n' collar (CNC)3 proteins are a subgroup of basic leucine zipper (bZIP) transcription factors that play a key role in several cellular processes through the regulation of mammalian gene expression (1, 2). Vertebrate CNC members include NF-E2 (nuclear factor-Erythroid derived 2) (3, 4), NRF1/NFE2L1 (nuclear factor erythroid 2-related factor-1) (5), NRF2/NFE2L2 (6), and NRF3/NFE2L3 (nuclear factor, erythroid 2-like 3) (7, 8) as well as the more distantly related BACH1 and BACH2 proteins (9). This family of proteins is characterized by a short (43 amino acids) and highly conserved CNC domain important for unique DNA binding (10) as well as a basic leucine zipper domain required for heterodimerization and DNA binding (11, 12). CNC proteins have been shown to play important roles in the oxidative stress response, carcinogenesis, and development (2).
As other members of the CNC family, the NFE2L3 transcription factor binds to MAREs (Maf recognition elements) or AREs (antioxidant response elements) (7, 8) by heterodimerizing with members of the small MAF (musculoaponeurotic fibrosarcoma) family, MAFF, MAFK, and MAFG (7, 8, 14, 15). NFE2L3 comprises a transactivation domain in the central region of the protein and has been described as both a positive (7, 8) and negative (14, 16) transcriptional regulator.
Nfe2l3−/− mice develop increased number of T-cell lymphoblastic lymphoma after treatment with the carcinogen benzo[a]pyrene (17). These mice also exhibit increased weight loss when treated with the antioxidant butylated hydroxytoluene (BHT) (18). Of note, a series of gene expression array studies have linked NFE2L3 to various malignancies including metastatic breast cancer (19), lymphoma (20, 21), colorectal (22), and testicular cancer (23).
Biochemical studies have shown that NFE2L3 is a stringently regulated and post-translationally modified protein that exists in three distinct forms in the cell, which are denoted as the A, B, and C forms (14). Transfection experiments showed that all three forms arise from the same cDNA. Each form is localized in specific cellular compartments: the A form is associated with the endoplasmic reticulum where it is N-glycosylated, and the B form is mainly cytoplasmic, whereas the shorter, likely proteolytically cleaved C form is predominantly nuclear (21, 22).
Ubiquitination is a fundamental regulatory post-translational modification that not only maintains protein homeostasis within the cell but also controls several pathways and signaling cascades (15). Ubiquitin molecules get covalently linked to lysine residues of target proteins. There are three key steps involved in the ubiquitination process. Ubiquitin activating enzyme (E1) activates ubiquitin. The activated ubiquitin is then transferred to the ubiquitin conjugating enzyme (E2). Ubiquitin ligases (E3) act as adaptors linking the target protein to E2, where ubiquitin is transferred onto the protein either directly or through an E3-ubiquitin intermediate stage (16, 17). Ubiquitin itself possesses seven lysine residues, and any of these lysines could be further ubiquitinated, forming a chain, known as polyubiquitination, which often serves as a signal for proteasome-dependent degradation (18). However recently, linkage-specific ubiquitination (e.g. lysine-48/48 and lysine-63/63) has been shown to have specific functions and regulatory roles. Lys-48-linked ubiquitination is primarily associated with 26S proteasome-dependent degradation (19, 20) and Lys-63-linked ubiquitination has been implicated in DNA damage repair, stress response, inflammatory pathways, intracellular trafficking, endocytosis, and lysosomal degradation of membrane proteins (21–26).
FBW7 (F-box and WD repeat domain-containing 7), also known as FBXW7, CDC4, AGO, and SEL10, is a well established tumor suppressor that has been shown to regulate several oncoproteins, such as, cyclin E, c-MYC, cJUN, Notch, and mTOR through ubiquitin-mediated degradation (27). FBW7 is a component of the SCF (Skp1, Cullin 1, F-box containing complex) complex E3 ubiquitin ligase (28, 29). FBW7 comprises an F-box domain that interacts directly with SKP1 to recruit ubiquitin-conjugating enzymes and WD40 repeats that physically bind its substrates (30, 31). FBW7 is one of the most mutated ubiquitin ligases in cancer, and loss of function has been associated with tumorigenesis (32) and chromosomal instability (33).
Glycogen synthase kinase 3 (GSK3) is encoded by two genes known as GSK3α (GSK3A or GSK3α) and GSK3β (GSK3B or GSK3β), which differ in size at 51 and 47 kDa, respectively, due to a proline-rich N-terminal extension present in GSK3A. This kinase was first identified in the 1980s for its role in negatively regulating the activity of glycogen synthase in response to insulin signaling (34, 35). Since this discovery, more than 30 years of research has revealed that GSK3 plays critical roles in a plethora of cellular events including cell metabolism, polarity, apoptosis, development, and transcriptional regulation (36). Due to its various critical cellular functions, it is not surprising that this kinase has been associated with many pathologies ranging from cancer to neurodegenerative disorders. Over the years a pattern has emerged revealing that many proteins targeted by FBW7 require prior phosphorylation by GSK3, implicating a role for GSK3 in the regulation of proteolysis (29, 37, 38). GSK3 dramatically increases the affinity between FBW7 ubiquitin ligase and its substrates by first phosphorylating the FBW7 binding site(s), often referred to as the phosphodegron. GSK3 and FBW7 can thus work in concert to mediate ubiquitination of many important protein targets thereby regulating pinnacle cellular events such as oncogenesis, apoptosis, DNA repair, and embryogenesis (29).
In this article we present data showing a role of the FBW7 tumor suppressor and GSK3 in the regulation of the NFE2L3 transcription factor. This is relevant for a better understanding of the regulatory mechanisms linking NFE2L3 to cellular stress and cancer.
Experimental Procedures
Cell Culture, Treatments, and Transfections
MCF7, MDA-MB-231, and HEK293T were purchased from ATCC and maintained in high glucose Dulbecco's modified Eagle's medium (Invitrogen) supplemented with 10% fetal bovine serum, 100 units/ml penicillin, and 100 μg/ml streptomycin at 37 °C with 10% CO2. HCT116 was a kind gift from Dr. Moulay Alaoui-Jamali (McGill University, Montreal). These cells were cultured in RPMI 1640 media (Invitrogen) supplemented with 10% fetal bovine serum, 100 units/ml penicillin, and 100 μg/ml streptomycin at 37 °C with 10% CO2. Transient transfections in MCF7 and HCT116 cells were performed with Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions using 2 μg of expression vector in a 100-mm dish seeded with 2 million cells 16 h before transfection. Cells were maintained in media without FBS or penicillin/streptomycin during transfection. The medium was changed to normal growth medium 6 h post transfection. In HEK293T cells, transfection was performed using calcium phosphate-based method as described previously (39). All treatments with lithium chloride (Sigma) were done for 16 h at the indicated concentrations in regular growth medium. Treatment with 10 μm MG-132 (Millipore) was carried out in regular growth medium for 6 h.
siRNA-mediated Knockdown
For inhibition of GSK3 expression, GSK3 siRNA (Cell Signaling) was transfected in MCF7 and MDA-MB-231 cells using INTERFERin transfection reagent (Polyplus transfection) following the manufacturer's protocol. Results were validated using combinations of GSK3α and -β siRNAs (Qiagen).
Immunoblotting and Antibodies
Cells were lysed with whole cell lysis buffer (10 mm Tris-HCl, pH 8.0, 420 mm NaCl, 250 mm sucrose, 2 mm MgCl2, 1% Triton X-100) for 20 min on ice. Protein extracts were separated on CriterionTM XT (Bio-Rad) 4–12% Bis-Tris gradient gels according to manufacturer's instructions. Proteins were then transferred to the PVDF membrane (Immobilon®-P, Millipore Corp.) in a wet transfer system in the absence of methanol. Membrane was blocked for 2–6 h at room temperature in 5% milk in 1× TBST (50 mm Tris-HCl, pH 7.6, 200 mm NaCl, 0.05% Tween 20) and incubated with primary antibody overnight at 4 °C. After 3 washes with 2× TBST, membrane was incubated for 1 h with secondary antibody and washed at least 5 times with 2× TBST. The proteins were detected by ImmobilonTM Western Chemiluminescent HRP substrate from Millipore according to instructions. Quantification of band intensity was carried out using Image LabTM software (Bio-Rad). We used an antiserum specific for a peptide located in the C-terminal portion of NFE2L3 described previously (8). We also utilized an additional antiserum raised against another peptide (KENSLQQNDDDENKIAEKPDWEAE) of NFE2L3. The peptide was coupled to keyhole limpet hemocyanin and used to immunize female New Zealand White rabbits (Pocono Farms). The serum was purified using peptide coupled to Affi-Gel 10 as previously described (8). HA antibody was purchased from ABCAM (12CA5), and B-catenin antibody was purchased from Santa Cruz Biotechnology (sc-7963). GSK3 A/B antibody was purchased from Cell Signaling (9331L).
Plasmids
NFE2L3 plasmid was created using pcDNA3.1+ Hygro as the vector backbone. Expression plasmids coding for pcDNA 3.1 HA-tagged FBW1a, FBW2, FBW4, and FBW7 were kind gifts from Dr. Jefferson Chan's laboratory (University of California, Irvine, CA). pFLAG-FBW7α and pFLAG-FBW7ΔD constructs were kind gifts from Dr. Bruce Clurman's laboratory (University of Washington). pRetroSuper FBW7 shRNA-1 and -2 plasmids were purchased from Addgene (Addgene plasmids 15660 and 15661). HA-tagged ubiquitin plasmids as well as GSK3B shRNA constructs were kind gifts from Dr. Marc Servant (University of Montreal). GSK3 wild type, constitutively active, and kinase dead pcDNA3 plasmids were purchased from Addgene (Addgene plasmids 14753, 14754, and 14755). All NFE2L3 mutant constructs were created by site-directed mutagenesis (Agilent) as per the manufacturer's instructions. Lys-48 and Lys-63-Ub constructs were kind gifts from Dr. Rongtuan Lin (McGill University). Luciferase constructs with NQO1 AREs were kind gifts from Dr. Jawed Alam (Ochsner Medical Center).
Co-immunoprecipitation
HEK293T cells were transfected with NFE2L3 and other expression vectors as indicated using the calcium phosphate transfection method and then lysed 48 h after transfection with immunoprecipitation buffer (10 mm Tris-HCl, pH 8.0, 150 mm NaCl, 250 mm sucrose, 2 mm MgCl2, 1% Triton X-100). The lysates were incubated at 4 °C overnight with either antiserum specific for NFE2L3 or preimmune serum in the presence of protein A-agarose beads (EMD Millipore). The following day supernatant was removed, and beads were washed extensively with immunoprecipitation buffer. The proteins were eluted in XT sample buffer by heating at 95 °C for 10 min and analyzed as outlined in the immunoblotting section.
Mass Spectrometry
Mass spectrometry was carried out at the Institute for Research in Immunology and Cancer, University of Montreal. Briefly, affinity-purified protein complexes were resuspended in 50 μl of 50 mm ammonium bicarbonate. Tris(2-carboxyethyl)phosphine was added to the protein samples at a final concentration of 5 mm. Samples were incubated at 37 °C at 650 rpm for 30 min. 30 μl of chloroacetamide at 55 mm was added, and the samples were incubated at 37 °C at 650 rpm for 30 min. Tryptic digestion (1 μg each; Promega) was performed overnight at 37 °C, and samples were dried down in a SpeedVac concentrator and resolubilized in 50 μl of aqueous 5% acetonitrile (formic acid, 0.2%). Samples (20 μl each) were injected onto a C18 precolumn (0.3 mm inner diameter × 5 mm), and peptides were separated on a C18 analytical column (150-μm inner diameter × 100 mm) using an Eksigent nanoLC-2D system. A 56-min gradient from 10% to 60% acetonitrile (0.2% formic acid) was used to elute peptides at a flow rate of 600 nl/min.
The LC system was coupled to an LTQ-Orbitrap Velos mass spectrometer (Thermo Fisher). MS analyses were performed using data-dependent acquisition in which each full MS spectrum was followed by six collision-induced dissociation MS/MS spectra in the linear ion trap for the most abundant multiply charged ions. The conventional MS spectra (survey scan) were acquired in the Orbitrap at a resolution of 60,000 for m/z 400 after the accumulation of 106 ions in the linear ion trap. Mass calibration used a lock mass from ambient air (protonated (Si(CH3)2O)6; m/z 445.120029) and provided mass accuracy within 15 ppm for precursor ion mass measurements. The dynamic exclusion of previously acquired precursor ions was enabled (repeat count 1, repeat duration of 15 s, exclusion duration of 15 s). MS/MS spectra were acquired in collision-induced dissociation mode using an isolation window of 2 Da, and precursor ions were sequentially isolated and accumulated to a target value of 10,000 with a maximum injection time of 100 ms.
Luciferase Activity Assay
Firefly and Renilla luciferase activity was measured sequentially from a single cell lysate on a Lumat LB 9507 Luminometer (Berthold Technologies) by using the dual luciferase assay system (Promega) according to the manufacturer's protocol. The results were expressed as a ratio of firefly luciferase (Fluc) activity to Renilla luciferase (Rluc) activity. Data were collected from at least three triplicate experiments, and standard deviation was calculated.
In Vitro Kinase Assay
In vitro kinase assays were performed using 0.5 μg of GST-NFE2L3 and 0.5 μg of GST-GSK3B in the presence of 5 μCi of γ-32P-labeled ATP and kinase buffer (20 mm Tris, pH 7.5, 10 mm MgCl2, 2 mm DDT, 20 μm cold ATP, 5 mm β-glycerol phosphate, 0.1 m sodium orthovanadate) at 30 °C for 30 min. The reactions were terminated by the addition of 8 μl of 4× SDS sample buffer, denatured, resolved by SDS-PAGE, and imaged by autoradiography.
Statistical Analysis
The t test was used to compare mean values in groups of samples. All reported p values were calculated for groups with unequal variance using the Excel software program (Microsoft). p < 0.05 was considered significant.
Results
NFE2L3 Is Regulated by the Ubiquitin-Proteasome Pathway
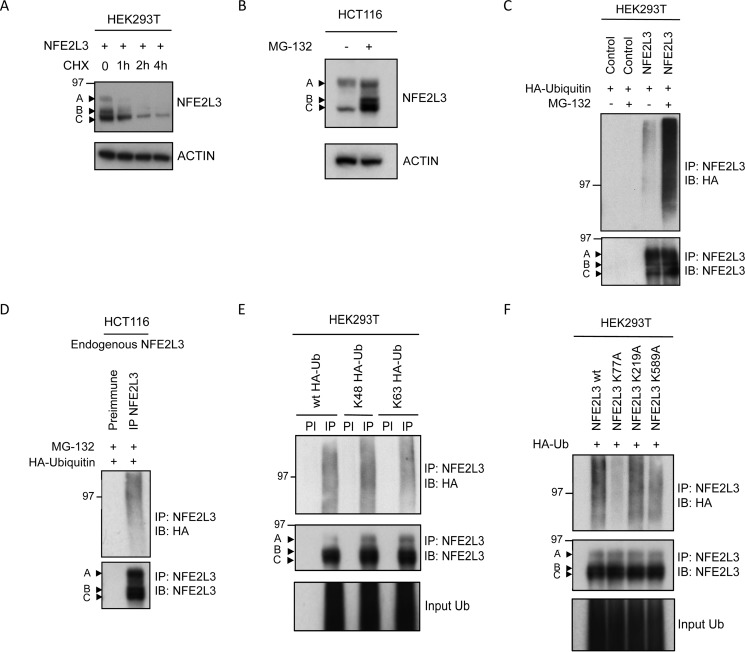
Cycloheximide chase assay in HEK293T cells overexpressing NFE2L3 revealed a very short half-life (∼20–40 min) (Fig. 1A). Inhibition of the proteasome pathway with MG-132 stabilized all three forms of endogenous NFE2L3 in HCT116 cells; however, the B and C forms were significantly more stabilized than the A form (Fig. 1B). These results further support our previously published data showing stabilization of endogenous NFE2L3 with MG-132, epoxomicin, or β-lactacystin in JAR choriocarcinoma cells (14). As the ubiquitin-proteasome pathway mediates the degradation of many short-lived transcription factors, we investigated ubiquitination of NFE2L3. To assess this, we performed ubiquitination assays to detect the polyubiquitinated forms of NFE2L3, often detected as a smear. Plasmids encoding NFE2L3 and HA-tagged ubiquitin were contransfected into HEK293T cells. Immunoprecipitation (IP) was performed with NFE2L3 antiserum (IP-NFE2L3) or preimmune serum. Immunoblot analysis using an antibody specific for ubiquitin (HA) revealed polyubiquitination of NFE2L3, which was further enhanced with MG-132 treatment (Fig. 1C). There was no ubiquitination observed in the control samples, transfected with expression vector without insert (Fig. 1C).
FIGURE 1.
NFE2L3 is ubiquitinated. A, NFE2L3-expressing plasmid was transfected in HEK293T cells and treated with cycloheximide (CHX; 50 μg/ml) and collected at specified time points. Indicated by arrows are the A, B, and C forms of NFE2L3. B, HCT116 cells were treated with MG-132 (10 μm) or DMSO (control) for 6 h and immunoblotted using antibody specific for NFE2L3. C, HA-tagged ubiquitin-expressing plasmid along with control vector or construct expressing NFE2L3 was transfected in HEK293T cells and treated with MG-132 (10 μm) or DMSO (Control) for 6 h. These precipitates were immunoblotted (IB) with antibody specific to HA to detect ubiquitin as well as NFE2L3 antibody. D, HCT116 cells were transfected with plasmid encoding HA-ubiquitin, and endogenous NFE2L3 was immunoprecipitated. These samples were immunoblotted with antibodies specific to HA as well as NFE2L3. E, NFE2L3-expressing plasmid was cotransfected with either HA-tagged Lys-48 or Lys-63 plasmids. NFE2L3 was immunoprecipitated, and these samples were immunoblotted with antibody specific to NFE2L3 as well as HA antibody to detect NFE2L3 Lys-48 or Lys-63 ubiquitination. PI, preimmune. F, mutant NFE2L3 constructs containing alanines at potential Lys-77, Lys-219, and Lys-589 ubiquitination sites were transfected in HEK293T cells along with HA-ubiquitin plasmid. NFE2L3 was immunoprecipitated from lysates and immunoblotted with antibodies specific for NFE2L3 as well as HA.
HCT116 colon cancer cells express high levels of NFE2L3. To analyze ubiquitination of endogenous NFE2L3, we performed IP analyses and carried out immunoblot analysis against ubiquitin (HA). We observed ubiquitination in the IP-NFE2L3 sample but not in the preimmune sample, confirming ubiquitination of endogenous NFE2L3 in HCT116 cells (Fig. 1D).
Although Lys-48 linked ubiquitination remains the most documented form of polyubiquitination, it is now clear that polyubiquitination could also occur through alternative lysine residues present on ubiquitin. To further investigate the type of ubiquitin linkage that is involved in NFE2L3 polyubiquitination, we performed ubiquitination assays in HEK293T cells cotransfected with NFE2L3 and wild type ubiquitin or ubiquitin bearing a single lysine at either position 48 or 63 (Lys-48 or Lys-63). We observed polyubiquitination with both Lys-48 and Lys-63 ubiquitin linkages in IP-NFE2L3 samples and no ubiquitination in corresponding preimmune samples (Fig. 1E).
To determine the ubiquitin sites in NFE2L3 protein, we used a mass spectrometry (MS)-based approach (40). Briefly, trypsin digestion of ubiquitinated proteins cleaves off all but the two C-terminal glycine residues of ubiquitin. These C-terminal glycine (GG) residues remain linked to the ϵ-amino group of the modified lysine residue after digestion of the substrate protein. The GG residues prevent cleavage by trypsin at that site and can be distinguished by MS. MS analysis of the NFE2L3 protein by others (41) and us revealed three potential ubiquitination sites at Lys-77, Lys-219, and Lys-589. We then mutated the potential lysine sites in NFE2L3 to alanines and performed ubiquitin assays comparing ubiquitination levels of mutant NFE2L3 (K77A, K219A, or K589A) to wild type. We found that K77A mutant showed remarkably less ubiquitination compared with the wild type control, suggesting that Lys-77 functions as a ubiquitination site in NFE2L3 (Fig. 1F).
NFE2L3 Turnover Is Regulated by FBW7 Tumor Suppressor
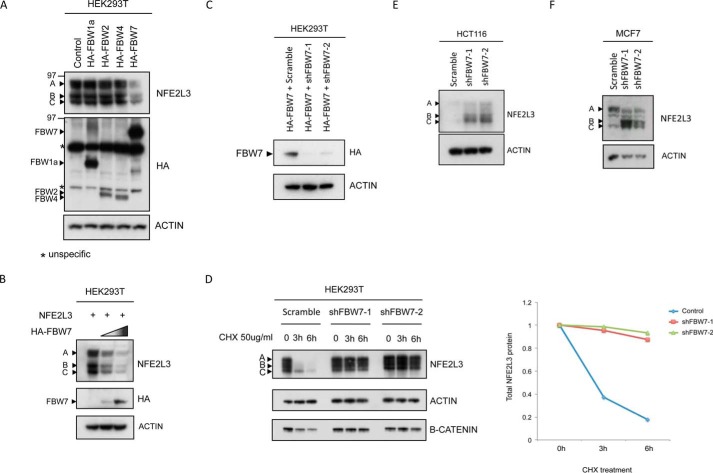
To analyze the role of FBW proteins in NFE2L3 degradation, NFE2L3 was cotransfected with plasmids encoding FBW1a, -2, -4, and -7, respectively, in HEK293T cells. Intriguingly, FBW7 overexpression led to a decrease in NFE2L3 levels, whereas the other FBW proteins had no effect on NFE2L3 (Fig. 2A). Furthermore, NFE2L3 protein levels decreased in a dose-dependent manner when cotransfected with increasing amounts of FBW7 plasmid (Fig. 2B). To further elucidate the role of FBW7 in NFE2L3 regulation, we used two independent shRNA constructs against FBW7, both of which effectively depleted FBW7 (Fig. 2C). HEK293T cells were cotransfected with NFE2L3 and either control shRNA plasmid or two distinct shRNAs against FBW7, and a cycloheximide chase assay was performed. We found that depletion of FBW7 significantly stabilized NFE2L3 (Fig. 2D). Endogenous NFE2L3 was also stabilized upon FBW7 knockdown in two independent cell lines (Fig. 2, E and F).
FIGURE 2.
FBW7 regulates NFE2L3 turnover. A, constructs encoding HA-tagged FBW1a, -2, -4, and -7 were transfected along with NFE2L3 plasmid in HEK293T cells. Lysates were immunoblotted with HA antibody to detect the different FBWs as well as NFE2L3 antibody. B, NFE2L3 expressing plasmid was transfected with increasing amounts of HA-tagged FBW7 plasmid in HEK293T cells and immunoblotted with antibodies specific for HA or NFE2L3. C, plasmid encoding HA-tagged FBW7 was transfected along with two FBW7 shRNAs in HEK293T cells. Exogenous FBW7 levels were visualized by immunoblotting with HA antibody. D, NFE2L3 expressing construct was cotransfected with control shRNA, FBW7 shRNA 1, and FBW7 shRNA 2 in HEK293T cells. Cycloheximide (CHX, 50 μg/ml) chase assay was performed collecting at the indicated time points. B-catenin levels are shown as a positive control. Quantification of the A, B, and C forms of NFE2L3 was done individually, and the cumulative intensities are represented at each time point on the graph on the right. HCT116 (E) and MCF7 (F) cells were transfected with control shRNA or two shRNAs targeting FBW7, and endogenous NFE2L3 was visualized by immunoblotting using antibody specific for NFE2L3.
FBW7 Physically Interacts with NFE2L3 and Facilitates Its Ubiquitination
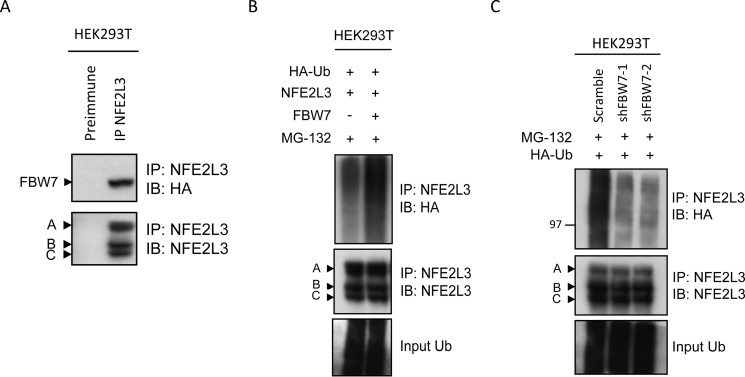
Because we showed that FBW7 destabilizes NFE2L3, we were interested in whether FBW7 could interact with the transcription factor. Plasmids encoding HA-FBW7 and NFE2L3 were cotransfected in HEK293T cells, and the cells were treated with MG-132 to inhibit NFE2L3 degradation. Lysates were immunoprecipitated with NFE2L3 antiserum (IP-NFE2L3) or preimmune serum and analyzed by immunoblot. Precipitates probed with HA antibody revealed a band corresponding to HA-FBW7 only in the IP-NFE2L3 sample (Fig. 3A). This suggests that these proteins are able to interact.
FIGURE 3.
FBW7 physically interacts with NFE2L3 and facilitates ubiquitination. A, HA-FBW7 and NFE2L3 expression constructs were cotransfected in HEK293T cells, and NFE2L3 was immunoprecipitated from these lysates with NFE2L3 antiserum. Precipitates were immunoblotted (IB) with antibodies specific for NFE2L3 as well as HA to visualize FBW7. B, NFE2L3, HA-Ubiquitin and HA-FBW7 constructs were cotransfected in HEK293T cells and treated with MG-132 (10 μm) for 6 h before collection. These lysates were immunoprecipitated with NFE2L3 antiserum and immunoblotted with NFE2L3 or HA antibodies to detect NFE2L3 ubiquitination. C, NFE2L3 plasmid and two different shRNAs targeting FBW7 together with HA-ubiquitin expressing plasmid were cotransfected in HEK293T cells and treated with MG-132 (10 μm) for 6 h before collection. NFE2L3 was immunoprecipitated from lysates and immunoblotted with NFE2L3 or HA antibodies.
We were then interested in determining whether FBW7 could promote NFE2L3 ubiquitination. Indeed, expression of FBW7 led to an increase in NFE2L3 polyubiquitination (Fig. 3B). Correspondingly, knockdown of FBW7 with two independent shRNAs led to a decrease in NFE2L3 ubiquitination (Fig. 3C). Immunoblot showing whole cell levels of input ubiquitin (HA) are shown as control. Together, these results demonstrate a link between FBW7 and NFE2L3 polyubiquitination.
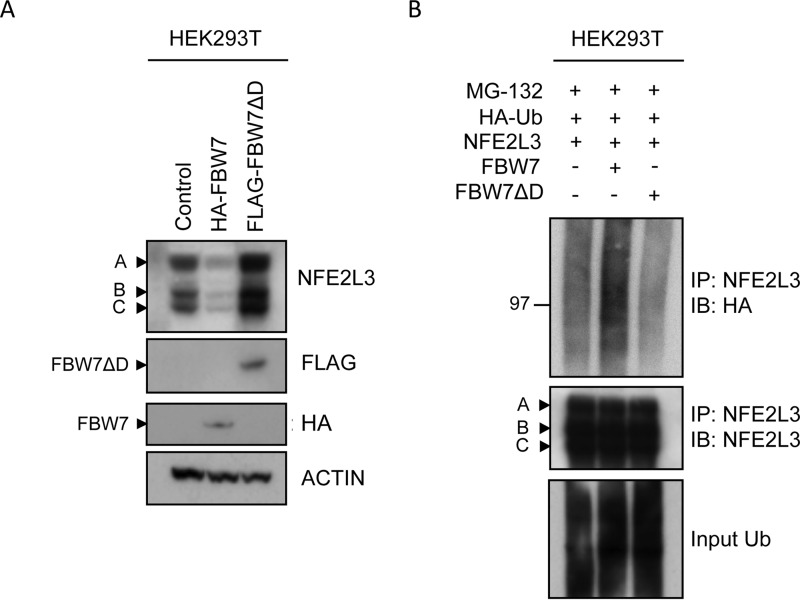
FBW7 Dimerization Is Required for NFE2L3 Degradation
Recently, FBW7 dimerization has been shown to be important for binding to various substrates (29). We, therefore, wanted to determine whether dimerization of FBW7 is required for its regulation of NFE2L3. We found that although wild type FBW7 destabilized NFE2L3, a mutated FBW7 construct lacking its dimerization domain was unable to promote NFE2L3 degradation but, on the contrary, substantially stabilized NFE2L3 by likely competing with the endogenous FBW7 (Fig. 4A). Furthermore, expression of wild type FBW7 enhanced NFE2L3 polyubiquitination as expected, but this effect was not observed with the dimerization mutant construct (Fig. 4B). Rather, polyubiquitination of NFE2L3 was decreased in the presence of the FBW7 dimerization mutant. Thus, FBW7 dimerization is a critical mechanistic aspect necessary for efficient degradation of NFE2L3 by FBW7.
FIGURE 4.
FBW7 dimerization is required for NFE2L3 degradation. A, NFE2L3 expressing construct was cotransfected with either HA-FBW7 or FLAG-FBW7ΔD constructs in HEK293T cells. These samples were analyzed by immunoblotting with antibodies specific for NFE2L3, HA, and FLAG. B, NFE2L3-expressing plasmid was cotransfected with either control vector, HA-FBW7, or FLAG-FBW7ΔD and HA-ubiquitin constructs in HEK293T cells and treated with MG-132 (10 μm) for 6 h before collection. NFE2L3 was immunoprecipitated from these lysates, and these samples were immunoblotted with NFE2L3 and HA antibodies to detect NFE2L3 ubiquitination.
GSK3 Binds and Phosphorylates NFE2L3
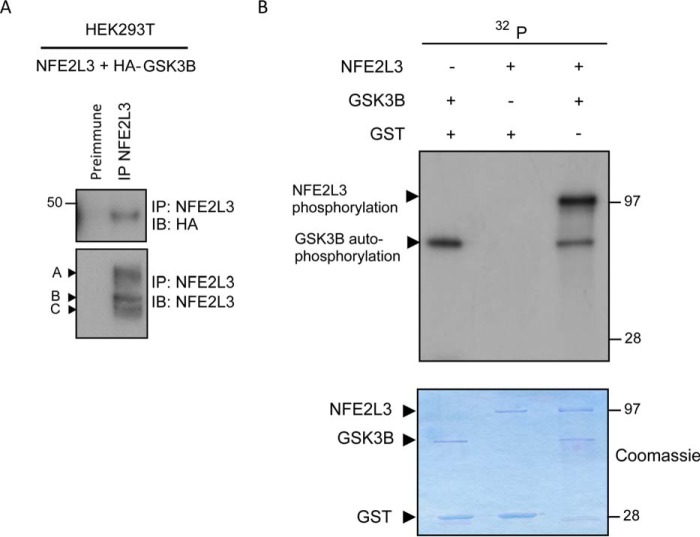
Many FBW7 substrates require binding and phosphorylation by GSK3 to facilitate FBW7 recognition. Thus, we were interested in determining whether GSK3 was linked to FBW7-dependent degradation of NFE2L3 via a similar mechanism. To assess GSK3 binding, NFE2L3 and HA-tagged GSK3B constructs were cotransfected in HEK293T cells. Cells were treated with MG-132, and NFE2L3 was immunoprecipitated with NFE2L3 antiserum (IP-NFE2L3) or preimmune serum as a control (Fig. 5A). Probing with HA revealed a band corresponding to GSK3B that was only present in the IP-NFE2L3 sample. This showed that GSK3B is able to interact with NFE2L3.
FIGURE 5.
GSK3 binds and phosphorylates NFE2L3. A, NFE2L3 and HA-tagged GSK3B constructs were cotransfected in HEK293T cells and treated with MG-132 (10 μm) for 6 h before collection. NFE2L3 was immunoprecipitated, and these samples were analyzed by immunoblot (IB) using antibody specific for NFE2L3 as well as HA antibody to detect GSK3B. B, GST-NFE2L3 was incubated with either GST-GSK3B (third lane) or GST alone as a control (second lane). GST-GSK3B was also incubated with GST alone as another control (first lane). Reactions were performed by combining respective proteins in kinase buffer with the addition of [γ-32P]ATP. Samples were resolved by SDS-PAGE, Coomassie-stained, and visualized by autoradiography. The Coomassie stain is shown as a loading control; arrows indicate respective proteins.
We were next interested in determining whether GSK3 could specifically phosphorylate NFE2L3. This was assessed by an in vitro kinase assay utilizing recombinant GST-tagged NFE2L3 and GSK3B. In the sample containing only GSK3B and GST as a control (Fig. 5B, first lane), we detected phosphorylation of GSK3B, which likely corresponds to GSK3 autophosphorylation shown previously (42) (Fig. 5B, first lane). When GSK3B and NFE2L3 were combined (Fig. 5B, third lane), we found significant NFE2L3 phosphorylation. However, no phosphorylation was observed in the sample where NFE2L3 was combined with GST (Fig. 5B, second lane) suggesting that the observed phosphorylation (Fig. 5B, third lane) is specifically facilitated by GSK3B. Together, our findings show that GSK3 is capable of binding and phosphorylating NFE2L3.
GSK3 Is Involved in NFE2L3 Degradation
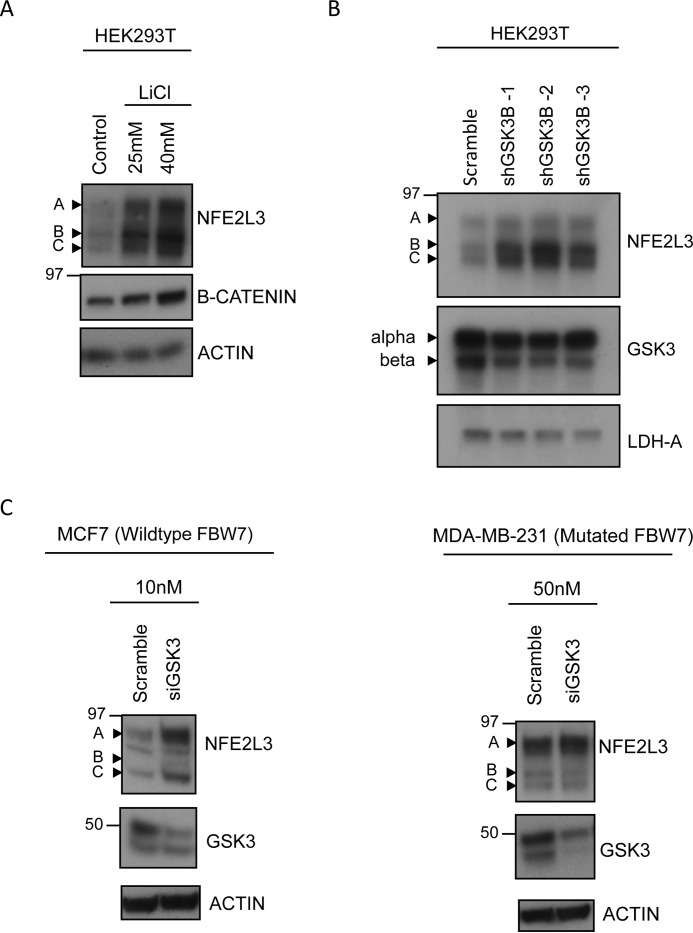
Based on the latter results, we wanted to further determine whether GSK3 plays a role in NFE2L3 degradation. Thus, we employed lithium chloride, which is a well characterized, specific inhibitor of GSK3 (43). Treatment with lithium chloride stabilized NFE2L3 expression in HEK293T cells in a dose-dependent manner (Fig. 6A). Moreover, this treatment also stabilized B-catenin levels in these cells, a well documented target of GSK3 (44).
FIGURE 6.
GSK3 is involved in NFE2L3 degradation. A, NFE2L3 expressing construct was transfected in HEK293T cells and treated with either control (NaCl) or lithium chloride at the indicated concentrations for 16 h. Lysates were immunoblotted using antibodies specific for NFE2L3 or B-catenin. B, plasmid coding for NFE2L3 was cotransfected with either control shRNA or one of three independent GSK3B shRNAs in HEK293T cells. Lysates were immunoblotted with NFE2L3 and GSK3 antibodies. C, MCF7 and MDA-MB-231 cells were transfected with the indicated amounts of control or GSK3 siRNA and collected 48 h later. Samples were immunoblotted with antibodies specific for NFE2L3 and GSK3.
For further validation, we sought to determine whether GSK3 knockdown would also stabilize NFE2L3. We found that three independent shRNA constructs targeting GSK3B were capable of stabilizing exogenous NFE2L3 in HEK293T cells (Fig. 6B). Furthermore, siRNA-mediated GSK3 knockdown in MCF7 cells also stabilized endogenous levels of NFE2L3 in this cell line, further supporting a role for GSK3 in controlling NFE2L3 turnover (Fig. 6C). This effect was absent in FBW7-mutant MDA-MB-231 cells.
FBW7-mediated Degradation of NFE2L3 Is GSK3-dependent
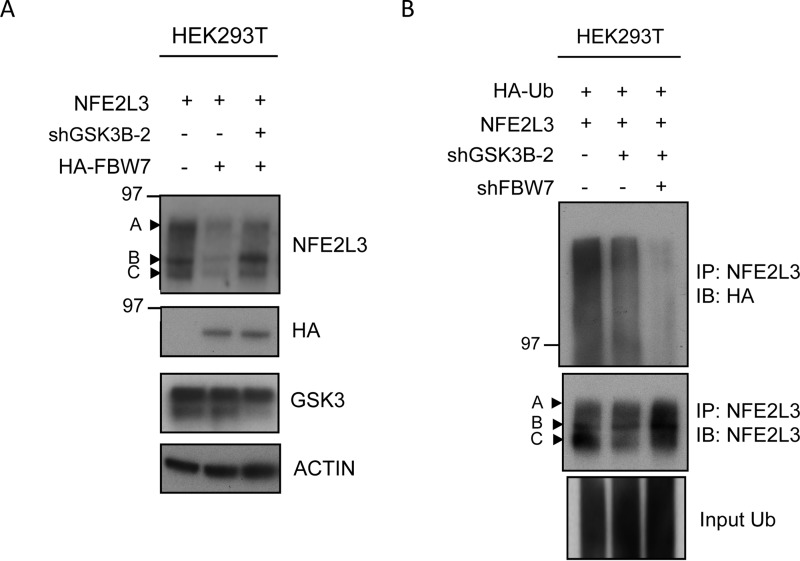
Finally, we wanted to investigate whether FBW7-mediated polyubiquitination of NFE2L3 was dependent on upstream GSK3 signaling. For this purpose NFE2L3 was expressed with either HA-FBW7 alone or in combination with GSK3B shRNA. As expected, we noticed a decrease in the levels of NFE2L3 upon co-transfection with HA-FBW7 (Fig. 7A). However, NFE2L3 levels were rescued when GSK3B was knocked down, suggesting that the effect of FBW7 on the stability of this transcription factor is dependent on the presence of GSK3.
FIGURE 7.
FBW7-mediated degradation of NFE2L3 is GSK3-dependent. A, NFE2L3 expression plasmid was cotransfected with either HA-FBW7 alone or in combination with shRNA targeting GSK3B in HEK293T cells. Lysates were analyzed by immunoblot with antibodies specific for NFE2L3 and HA antibody to detect FBW7 as well as GSK3. B, plasmid coding for NFE2L3 was cotransfected with HA-ubiquitin construct and GSK3B shRNA or in combination with FBW7 shRNA in HEK293T cells and treated with MG-132 (10 μm) for 6 h before collection. NFE2L3 was immunoprecipitated, and these samples were immunoblot-analyzed with antibodies specific for NFE2L3, HA, and GSK3.
To determine whether GSK3 acts in concert with FBW7 to regulate NFE2L3 polyubiquitination, ubiquitination assays were performed in HEK293T cells with either GSK3B shRNA alone or in combination with FBW7 shRNA. Immunoprecipitation of NFE2L3 and subsequent probing for HA-ubiquitin showed a decrease in NFE2L3 polyubiquitination with GSK3B shRNA and a further cumulative decrease in the sample where both FBW7 and GSK3B shRNAs were combined (Fig. 7B). These data support the hypothesis that FBW7-driven degradation of NFE2L3 requires the upstream actions of GSK3 to efficiently facilitate degradation of NFE2L3.
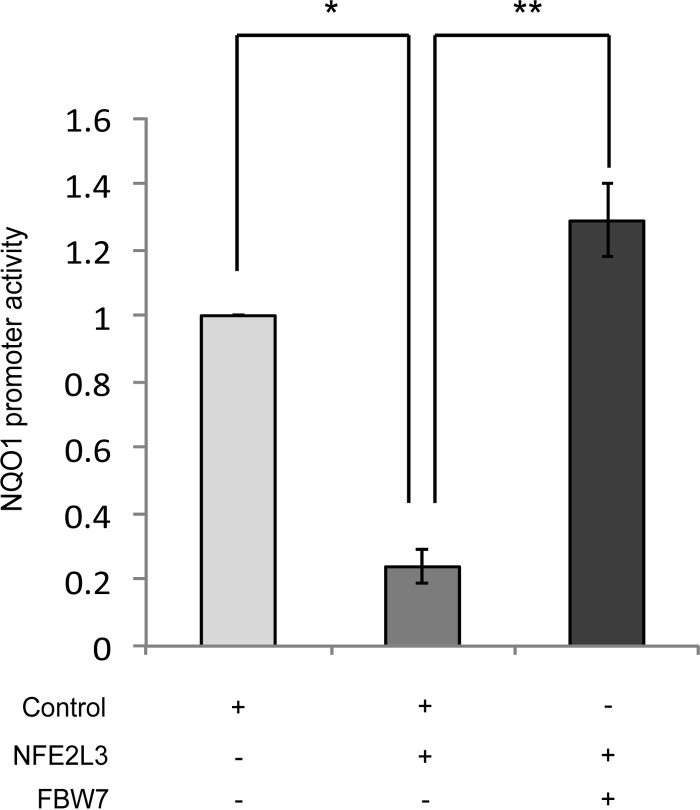
FBW7 Abolishes NFE2L3-mediated ARE Gene Repression
To determine the physiological relevance of FBW7-mediated regulation of NFE2L3, we investigated its effect on NFE2L3-mediated repression of NQO1 promoter activity, a gene coding for a detoxification enzyme. NFE2L3 and a luciferase construct comprising the AREs of the NQO1 promoter were cotransfected with either control vector or vector expressing FBW7 in MCF7 cells. NQO1 ARE-driven luciferase activity was significantly repressed by NFE2L3 (Fig. 8). On the contrary, in the presence of FBW7, NFE2L3 was unable to repress the luciferase reporter activity. This result suggests that FBW7 controls the transcriptional activity of NFE2L3 in the cellular response to stress.
FIGURE 8.
FBW7 abolishes NFE2L3-mediated ARE gene repression. Luciferase reporter assay showing activity of NQO1 gene ARE sequence when HEK293T cells were cotransfected with either NFE2L3 expressing construct and empty vector control or with a FBW7 expression construct. Promoter activity was analyzed by Dual-Luciferase assay. Data represent luciferase activities normalized to Renilla luciferase. * indicates an error < 0.05, and ** indicates an error < 0.01.
Discussion
Our study demonstrates NFE2L3 as a novel substrate of the tumor suppressor FBW7 that has been shown to regulate several other proteins, such as cyclin E, c-MYC, Notch, c-Jun, mTOR, and HIF-1A (29). It also illustrates GSK3-dependent ubiquitination and subsequent degradation of NFE2L3. NFE2L3 is the latest addition to the CNC family of transcription factors that is involved in a variety of cellular processes, including cancer, stress response/detoxification, and inflammation (45); however, its regulation at the cellular level has not yet been investigated in detail. Proteasome inhibitors stabilized NFE2L3, leading us to hypothesize that it is processed by the ubiquitin proteasome system (46). Here, we showed that (i) NFE2L3 is ubiquitinated, (ii) NFE2L3 turnover is facilitated by the FBW7 E3 ubiquitin ligase, (iii) FBW7-mediated ubiquitination process is dependent on upstream GSK3 signaling, and (iv) FBW7-mediated degradation controls the transcriptional activity of NFE2L3.
NFE2L3 has a rapid turnover and is stabilized by blocking proteasomal degradation. This effect was consistent across several cell lines, such as MCF7 breast cancer cells, JAR choriocarcinoma cells, HCT116 colon cancer cells, and human embryonic kidney cells (14). We then confirmed polyubiquitination of NFE2L3. Covalent attachment of ubiquitin molecules to proteins via Lys-48-linked ubiquitin chains and their subsequent degradation by the 26S proteasome is the classic polyubiquitination observed in several proteins. However, different cellular functions have been associated with particular types of ubiquitin linkages. NFE2L3 was polyubiquitinated by both Lys-48- and Lys-63-linked ubiquitin. Interestingly, NF-E2, a member of the CNC family of transcription factors, has also been shown to be ubiquitinated via Lys-48 and Lys-63 linkages. The Lys-48 linkage promotes degradation, whereas Lys-63-linked ubiquitination restricts its localization to the cytoplasm and decreases its transactivation capacity (47). Lys-63-linked ubiquitination of NFE2L3 does not change its localization (data not shown), and its significance is yet to be elucidated. Mass spectrometry analysis revealed three potential ubiquitination sites for NFE2L3 (Lys-77, Lys-219, and Lys-589). Mutation of Lys-77 significantly decreased NFE2L3 ubiquitination, suggesting that this residue is targeted by ubiquitin. Because Lys-77 is present only in the full form and not in the shorter C form of NFE2L3, we assume that there might be additional ubiquitination sites in the protein. Indeed, we observed a slight decrease in ubiquitination when Lys-589 was mutated; however, other sites might be present that are simultaneously ubiquitinated.
We found that the tight regulation of NFE2L3 and its rapid turnover is mediated by FBW7. Exogenous expression of F-box proteins other than FBW7 did not affect NFE2L3 levels. Furthermore, depletion of FBW7 not only stabilized NFE2L3 but also decreased ubiquitination. Similarly, exogenous FBW7 expression had the opposite effect, decreasing NFE2L3 levels in the cell in a dose-dependent manner. In MCF7 as well as HCT116 cells, we noticed stabilization of endogenous NFE2L3 upon FBW7 knockdown. We did not observe stabilization of the A form of NFE2L3 in these cell lines possibly because it may be degraded through a mechanism independent of the proteasome. It is to be noted that the A form is localized in the endoplasmic reticulum and is N-glycosylated and hence might be processed through a different mechanism with or without the involvement of a deglycosylation step. In agreement with this hypothesis, our present and previous data showed that the endogenous and exogenous levels of the A form increased substantially less upon MG-132 treatment compared with the B and C forms (14).
GSK3 has been implicated as the upstream kinase regulating the turnover of most substrates targeted by FBW7 (29). Herein, we found that GSK3 binds to and phosphorylates NFE2L3. GSK3 inhibition with lithium chloride stabilized NFE2L3. We also observed that GSK3 knockdown in MCF7 cells, which retain functional FBW7, stabilized endogenous NFE2L3 levels. In contrast, GSK3 knockdown in the MDA-MB-231 cells harboring mutant non-functional FBW7 did not lead to stabilization of NFE2L3. These data showed that the effect of GSK3 on NFE2L3 degradation is dependent on an intact FBW7. As further validation of this codependence, we showed that FBW7-mediated destabilization of NFE2L3 was rescued upon GSK3 knockdown. In addition, knockdown of GSK3 decreased NFE2L3 polyubiquitination, whereas combined knockdown of GSK3 and FBW7 led to an even more significant decrease. Together, these findings clearly indicate the interdependence of GSK3 and FBW7 in NFE2L3 polyubiquitination and consequent degradation.
FBW7 binds mostly at conserved phosphodegron motifs having the general consensus (S/T)PXX(S/T/D/E) (29). Based on our co-immunoprecipitation assays, FBW7 binds NFE2L3; however, the exact number of potential low affinity phosphodegrons is difficult to determine as there are a large number of possible weak signals that FBW7 could bind to in NFE2L3. Interestingly, a recent study has shown that although FBW7 binds some substrates as a monomer, dimerization of FBW7 enables binding to substrates with weaker phosphodegrons (48). By dimerization, FBW7 can bind a protein with two or more weak phosphodegron sequences thereby stabilizing the interaction with its substrate and mediating ubiquitination (49). This has been suggested for some substrates of FBW7 such as MCL-1 and cyclin E (29). In our analysis we found that an FBW7 construct lacking the dimerization domain does not promote NFE2L3 degradation and fails to enhance its ubiquitination. Thus, it is highly likely that FBW7 binds to NFE2L3 as a dimer. Finally, we showed that FBW7 could mediate ARE-driven gene expression through modulation of NFE2L3 levels. AREs act as key DNA recognition sites that are present in the regulatory regions of genes coding for detoxification enzymes and stress response proteins (13, 50–52). Transfection of a construct coding for FBW7 alone did not change NQO1 promoter activity (data not shown), indicating that the observed effect is exclusively NFE2L3-dependent. We, therefore, hypothesize that degradation of NFE2L3 by FBW7 or stabilization of NFE2L3 in FBW7-mutated cell lines might play a role in the cellular response to stress.
Author Contributions
Under the supervision of V. B., M. B. K. performed the majority of the experiments presented in this article and wrote the manuscript. I. D.-F. carried out the GSK3 related experiments and assisted in the preparation of the manuscript.
Acknowledgments
We thank Marina Bury, Anna Derjuga, Jadwiga Gasiorek, and Sean Seltzer for critical reading of the manuscript and/or fruitful discussions. We thank Marc Servant for providing ubiquitin/GSK3 shRNA constructs, Jefferson Y. Chan for FBW7 constructs, and Bruce Clurman for the FBW7 dimerization mutant construct. We thank Rongtuan Lin for guidance and the Lys-48 and Lys-63 plasmids. We also thank Jawed Alam for the luciferase constructs.
This research was funded by Canadian Institutes of Health Research Grant MOP-97932 (to V. B.). The authors declare that they have no conflicts of interest with the contents of this article.
- CNC
- cap 'n' collar
- NF-E2
- nuclear factor-erythroid derived 2
- NRF
- nuclear factor erythroid 2-related factor
- FBW7
- F-box/WD repeat-containing protein 7
- GSK3
- glycogen synthase kinase 3
- NQO1
- NAD(P)H:quinone oxidoreductase 1
- ARE
- antioxidant response element
- Bis-Tris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol
- IP
- immunoprecipitation
- Ub
- ubiquitin.
References
- 1.Blank V. (2008) Small Maf proteins in mammalian gene control: mere dimerization partners or dynamic transcriptional regulators? J. Mol. Biol. 376, 913–925 [DOI] [PubMed] [Google Scholar]
- 2.Sykiotis G. P., and Bohmann D. (2010) Stress-activated cap'n'collar transcription factors in aging and human disease. Sci. Signal. 3, re3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Andrews N. C., Kotkow K. J., Ney P. A., Erdjument-Bromage H., Tempst P., and Orkin S. H. (1993) The ubiquitous subunit of erythroid transcription factor NF-E2 is a small basic-leucine zipper protein related to the v-maf oncogene. Proc. Natl. Acad. Sci. U.S.A. 90, 11488–11492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Andrews N. C. (1994) Erythroid transcription factor NF-E2 coordinates hemoglobin synthesis. Pediatr. Res. 36, 419–423 [DOI] [PubMed] [Google Scholar]
- 5.Chan J. Y., Han X. L., and Kan Y. W. (1993) Cloning of Nrf1, an NF-E2-related transcription factor, by genetic selection in yeast. Proc. Natl. Acad. Sci. U.S.A. 90, 11371–11375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Moi P., Chan K., Asunis I., Cao A., and Kan Y. W. (1994) Isolation of NF-E2-related factor 2 (Nrf2), a NF-E2-like basic leucine zipper transcriptional activator that binds to the tandem NF-E2/AP1 repeat of the β-globin locus control region. Proc. Natl. Acad. Sci. U.S.A. 91, 9926–9930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kobayashi A., Ito E., Toki T., Kogame K., Takahashi S., Igarashi K., Hayashi N., and Yamamoto M. (1999) Molecular cloning and functional characterization of a new Cap'n' collar family transcription factor Nrf3. J. Biol. Chem. 274, 6443–6452 [DOI] [PubMed] [Google Scholar]
- 8.Chénais B., Derjuga A., Massrieh W., Red-Horse K., Bellingard V., Fisher S. J., and Blank V. (2005) Functional and placental expression analysis of the human NRF3 transcription factor. Mol. Endocrinol. 19, 125–137 [DOI] [PubMed] [Google Scholar]
- 9.Oyake T., Itoh K., Motohashi H., Hayashi N., Hoshino H., Nishizawa M., Yamamoto M., and Igarashi K. (1996) Bach proteins belong to a novel family of BTB-basic leucine zipper transcription factors that interact with MafK and regulate transcription through the NF-E2 site. Mol. Cell. Biol. 16, 6083–6095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kerppola T. K., and Curran T. (1994) Maf and Nrl can bind to AP-1 sites and form heterodimers with Fos and Jun. Oncogene 9, 675–684 [PubMed] [Google Scholar]
- 11.Landschulz W. H., Johnson P. F., and McKnight S. L. (1988) The leucine zipper: a hypothetical structure common to a new class of DNA binding proteins. Science 240, 1759–1764 [DOI] [PubMed] [Google Scholar]
- 12.Itoh K., Igarashi K., Hayashi N., Nishizawa M., and Yamamoto M. (1995) Cloning and characterization of a novel erythroid cell-derived CNC family transcription factor heterodimerizing with the small Maf family proteins. Mol. Cell. Biol. 15, 4184–4193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jeyapaul J., and Jaiswal A. K. (2000) Nrf2 and c-Jun regulation of antioxidant response element (ARE)-mediated expression and induction of γ-glutamylcysteine synthetase heavy subunit gene. Biochem. Pharmacol. 59, 1433–1439 [DOI] [PubMed] [Google Scholar]
- 14.Nouhi Z., Chevillard G., Derjuga A., and Blank V. (2007) Endoplasmic reticulum association and N-linked glycosylation of the human Nrf3 transcription factor. FEBS Lett. 581, 5401–5406 [DOI] [PubMed] [Google Scholar]
- 15.Hershko A., and Ciechanover A. (1998) The ubiquitin system. Annu. Rev. Biochem. 67, 425–479 [DOI] [PubMed] [Google Scholar]
- 16.Dye B. T., and Schulman B. A. (2007) Structural mechanisms underlying posttranslational modification by ubiquitin-like proteins. Annu. Rev. Biophys. Biomol. Struct. 36, 131–150 [DOI] [PubMed] [Google Scholar]
- 17.Pickart C. M. (2001) Mechanisms underlying ubiquitination. Annu. Rev. Biochem. 70, 503–533 [DOI] [PubMed] [Google Scholar]
- 18.Komander D., and Rape M. (2012) The ubiquitin code. Annu. Rev. Biochem. 81, 203–229 [DOI] [PubMed] [Google Scholar]
- 19.Finley D., Sadis S., Monia B. P., Boucher P., Ecker D. J., Crooke S. T., and Chau V. (1994) Inhibition of proteolysis and cell cycle progression in a multiubiquitination-deficient yeast mutant. Mol. Cell. Biol. 14, 5501–5509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chau V., Tobias J. W., Bachmair A., Marriott D., Ecker D. J., Gonda D. K., and Varshavsky A. (1989) A multiubiquitin chain is confined to specific lysine in a targeted short-lived protein. Science 243, 1576–1583 [DOI] [PubMed] [Google Scholar]
- 21.Spence J., Sadis S., Haas A. L., and Finley D. (1995) A ubiquitin mutant with specific defects in DNA repair and multiubiquitination. Mol. Cell. Biol. 15, 1265–1273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Arnason T., and Ellison M. J. (1994) Stress resistance in Saccharomyces cerevisiae is strongly correlated with assembly of a novel type of multiubiquitin chain. Mol. Cell. Biol. 14, 7876–7883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sun L., Deng L., Ea C. K., Xia Z. P., and Chen Z. J. (2004) The TRAF6 ubiquitin ligase and TAK1 kinase mediate IKK activation by BCL10 and MALT1 in T lymphocytes. Mol. Cell 14, 289–301 [DOI] [PubMed] [Google Scholar]
- 24.Ikeda F., and Dikic I. (2008) Atypical ubiquitin chains: new molecular signals. “Protein Modifications: Beyond the Usual Suspects” review series. EMBO Rep. 9, 536–542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xu P., Duong D. M., Seyfried N. T., Cheng D., Xie Y., Robert J., Rush J., Hochstrasser M., Finley D., and Peng J. (2009) Quantitative proteomics reveals the function of unconventional ubiquitin chains in proteasomal degradation. Cell 137, 133–145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nathan J. A., Kim H. T., Ting L., Gygi S. P., and Goldberg A. L. (2013) Why do cellular proteins linked to Lys-63-polyubiquitin chains not associate with proteasomes? EMBO J. 32, 552–565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nakayama K. I., and Nakayama K. (2006) Ubiquitin ligases: cell-cycle control and cancer. Nat. Rev. Cancer 6, 369–381 [DOI] [PubMed] [Google Scholar]
- 28.Nakayama K. I., and Nakayama K. (2005) Regulation of the cell cycle by SCF-type ubiquitin ligases. Semin Cell Dev. Biol. 16, 323–333 [DOI] [PubMed] [Google Scholar]
- 29.Welcker M., and Clurman B. E. (2008) FBW7 ubiquitin ligase: a tumour suppressor at the crossroads of cell division, growth, and differentiation. Nat. Rev. Cancer 8, 83–93 [DOI] [PubMed] [Google Scholar]
- 30.Hao B., Oehlmann S., Sowa M. E., Harper J. W., and Pavletich N. P. (2007) Structure of a Fbw7-Skp1-cyclin E complex: multisite-phosphorylated substrate recognition by SCF ubiquitin ligases. Mol. Cell 26, 131–143 [DOI] [PubMed] [Google Scholar]
- 31.Orlicky S., Tang X., Willems A., Tyers M., and Sicheri F. (2003) Structural basis for phospho-dependent substrate selection and orientation by the SCFCdc4 ubiquitin ligase. Cell 112, 243–256 [DOI] [PubMed] [Google Scholar]
- 32.Mao J. H., Perez-Losada J., Wu D., Delrosario R., Tsunematsu R., Nakayama K. I., Brown K., Bryson S., and Balmain A. (2004) Fbxw7/Cdc4 is a p53-dependent, haploinsufficient tumour suppressor gene. Nature 432, 775–779 [DOI] [PubMed] [Google Scholar]
- 33.Rajagopalan H., and Lengauer C. (2004) hCDC4 and genetic instability in cancer. Cell Cycle 3, 693–694 [DOI] [PubMed] [Google Scholar]
- 34.Embi N., Rylatt D. B., and Cohen P. (1980) Glycogen synthase kinase-3 from rabbit skeletal muscle: separation from cyclic-AMP-dependent protein kinase and phosphorylase kinase. Eur. J. Biochem. 107, 519–527 [PubMed] [Google Scholar]
- 35.Woodgett J. R., and Cohen P. (1984) Multisite phosphorylation of glycogen synthase: molecular basis for the substrate specificity of glycogen synthase kinase-3 and casein kinase-II (glycogen synthase kinase-5). Biochim. Biophys. Acta 788, 339–347 [DOI] [PubMed] [Google Scholar]
- 36.Beurel E., Grieco S. F., and Jope R. S. (2015) Glycogen synthase kinase-3 (GSK3): regulation, actions, and diseases. Pharmacol. Ther. 148, 114–131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Welcker M., Orian A., Jin J., Grim J. E., Grim J. A., Harper J. W., Eisenman R. N., and Clurman B. E. (2004) The Fbw7 tumor suppressor regulates glycogen synthase kinase 3 phosphorylation-dependent c-Myc protein degradation. Proc. Natl. Acad. Sci. U.S.A. 101, 9085–9090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ye X., Nalepa G., Welcker M., Kessler B. M., Spooner E., Qin J., Elledge S. J., Clurman B. E., and Harper J. W. (2004) Recognition of phosphodegron motifs in human cyclin E by the SCF(Fbw7) ubiquitin ligase. J. Biol. Chem. 279, 50110–50119 [DOI] [PubMed] [Google Scholar]
- 39.Kingston R. E., Chen C. A., and Okayama H. (2003) Calcium phosphate transfection. Curr. Protoc. Cell Biol. Chapter 20, Unit 20.3 [DOI] [PubMed] [Google Scholar]
- 40.Udeshi N. D., Mani D. R., Eisenhaure T., Mertins P., Jaffe J. D., Clauser K. R., Hacohen N., and Carr S. A. (2012) Methods for quantification of in vivo changes in protein ubiquitination following proteasome and deubiquitinase inhibition. Mol. Cell. Proteomics 11, 148–159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kim W., Bennett E. J., Huttlin E. L., Guo A., Li J., Possemato A., Sowa M. E., Rad R., Rush J., Comb M. J., Harper J. W., and Gygi S. P. (2011) Systematic and quantitative assessment of the ubiquitin-modified proteome. Mol. Cell 44, 325–340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cole A., Frame S., and Cohen P. (2004) Further evidence that the tyrosine phosphorylation of glycogen synthase kinase-3 (GSK3) in mammalian cells is an autophosphorylation event. Biochem. J. 377, 249–255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Klein P. S., and Melton D. A. (1996) A molecular mechanism for the effect of lithium on development. Proc. Natl. Acad. Sci. U.S.A. 93, 8455–8459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Doble B. W., and Woodgett J. R. (2003) GSK-3: tricks of the trade for a multi-tasking kinase. J. Cell Sci. 116, 1175–1186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chevillard G., and Blank V. (2011) NFE2L3 (NRF3): the Cinderella of the cap'n'collar transcription factors. Cell. Mol. Life Sci. 68, 3337–3348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hershko A., and Ciechanover A. (1992) The ubiquitin system for protein degradation. Annu. Rev. Biochem. 61, 761–807 [DOI] [PubMed] [Google Scholar]
- 47.Lee T. L., Shyu Y. C., Hsu T. Y., and Shen C. K. (2008) Itch regulates p45/NF-E2 in vivo by Lys-63-linked ubiquitination. Biochem. Biophys. Res. Commun. 375, 326–330 [DOI] [PubMed] [Google Scholar]
- 48.Welcker M., Larimore E. A., Swanger J., Bengoechea-Alonso M. T., Grim J. E., Ericsson J., Zheng N., and Clurman B. E. (2013) Fbw7 dimerization determines the specificity and robustness of substrate degradation. Genes Dev. 27, 2531–2536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Welcker M., and Clurman B. E. (2007) Fbw7/hCDC4 dimerization regulates its substrate interactions. Cell Div. 2, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Biswas C., Shah N., Muthu M., La P., Fernando A. P., Sengupta S., Yang G., and Dennery P. A. (2014) Nuclear heme oxygenase-1 (HO-1) modulates subcellular distribution and activation of Nrf2, impacting metabolic and anti-oxidant defenses. J. Biol. Chem. 289, 26882–26894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Biswas M., and Chan J. Y. (2010) Role of Nrf1 in antioxidant response element-mediated gene expression and beyond. Toxicol. Appl. Pharmacol. 244, 16–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jaiswal A. K. (2004) Nrf2 signaling in coordinated activation of antioxidant gene expression. Free Radic. Biol. Med. 36, 1199–1207 [DOI] [PubMed] [Google Scholar]