Background: GRASP65 and GM130 play a key role in Golgi stacking.
Results: The crystal structure of the GRASP65-GM130 complex reveals an unexpected binding mode.
Conclusion: Both PDZ domains of GRASP65 interact with GM130. A novel mechanism for Golgi stacking is proposed.
Significance: A novel interaction mode permits us to propose a new molecular basis for Golgi structure.
Keywords: Golgi; protein assembly; protein complex; protein-protein interaction; vesicles; GM130, GRASP65, PDZ domain, Golgi stacking, vesicle tethering
Abstract
GM130 and GRASP65 are Golgi peripheral membrane proteins that play a key role in Golgi stacking and vesicle tethering. However, the molecular details of their interaction and their structural role as a functional unit remain unclear. Here, we present the crystal structure of the PDZ domains of GRASP65 in complex with the GM130 C-terminal peptide at 1.96-Å resolution. In contrast to previous findings proposing that GM130 interacts with GRASP65 at the PDZ2 domain only, our crystal structure of the complex indicates that GM130 binds to GRASP65 at two distinct sites concurrently and that both the PDZ1 and PDZ2 domains of GRASP65 participate in this molecular interaction. Mutagenesis experiments support these structural observations and demonstrate that they are required for GRASP65-GM130 association.
Introduction
The Golgi apparatus is a major organelle in most eukaryotic cells. It maintains a unique morphology that is critical for its role in highly dynamic trafficking (1). The closely aligned, flattened, disc-shaped cisternal stacks of this organelle are often laterally linked, forming a ribbon-like structure in mammalian cells (2, 3). Under electron microscopy, the proteinaceous intercisternal bridges are visible between the stacked Golgi cisternae and may hold the cisternae together (4). The proteins GRASP652 and GM130, which were identified from a Golgi matrix fraction, are proposed to be the main structural components of the intercisternal bridges (5, 6), although no direct evidence for this proposition been found to date.
A mitotic Golgi disassembly and reassembly cell-free assay previously revealed that GRASPs mediate the stacking of Golgi cisternae (6–8). Two isoforms of GRASPs are found in vertebrates, GRASP65 and GRASP55, which are named according to their relative molecular weights. These two isomers have alternative roles in Golgi stack formation. Knockdown of either isoform unlinks the ribbon structure and causes defective glycosylation, whereas knockdown of both isoforms disrupts the Golgi stacked structure altogether (9–14). GM130 is a coiled-coil, rod-like protein localized on the cis side of the Golgi cisternae. It was the first protein to be identified as an autoantigen of the Golgi matrix protein and is now used as a specific marker for the Golgi apparatus. Its C-terminal coiled-coil regions interact with t-SNARE protein-syntaxin 5 and are regulated by vesicle docking protein p115, which interacts with the N-terminal region of GM130 (15). Previous studies have revealed that GM130 is also involved in the maintenance of the Golgi structure and plays a major role in the disassembly and reassembly of the Golgi apparatus during mitosis (5, 16). GM130 thus plays a key role in vesicle tethering and fusion at the cis-cisternae to facilitate transit between transport vesicles and the stacked cisternae (10, 15, 17–20). Previous biochemical studies have demonstrated that the C terminus of GM130 interacts with the PDZ domain of GRASP65 (21–24). The GM130 and GRASP65 proteins are tightly bound together under physiological conditions (8) and most of their functions require this interaction (10, 25). GRASP65 is membrane anchored to the Golgi apparatus through myristoylation of its N terminus (glycine at position 2) and is localized to the cis-cisternae through an interaction with GM130 (24, 26). In addition, it was found that GRASP65 did not localize to the Golgi apparatus in GM130 knockdown cells (25).
The crystal structures of the conserved N-terminal domains of both GRASP65 and GRASP55 have been elucidated (27, 28) and indicate that they are composed of two circularly permuted PDZ-like domains, PDZ1 and PDZ2. Previous biochemical studies determined that GM130 only interacts with the PDZ2 domain of GRASP65 (24, 26, 29). For a long time, PDZ1 domains have been thought to interact with each other and play a substantial role in GRASP protein clustering to mediate cisternae stacking (27, 28, 30). Although GM130 and GRASP65 have been extensively studied, their mediation of Golgi stacking and vesicle tethering remains unclear, and structural information regarding their molecular recognition is currently lacking.
We present here the crystal structure of the GRASP65 PDZ domains in complex with the C-terminal tail of GM130. Our structural study reveals that the specific interactions between GRASP65 and GM130 involve both the PDZ1 and PDZ2 domains. Mutagenesis experiments support these structural observations and demonstrate that these interactions are requisite for GRASP65-GM130 association. Our crystal structure implies a new model for Golgi and molecular stacking.
Experimental Procedures
Construct Generation
The GRASP domain (residues 2–210) of human GRASP65 was amplified from the full-length gene (NM_031899) and cloned into the modified pRSFDuet-1 vector (Novagen) with an N-terminal His6 tag and a PreScission protease cleavage site. Human GM130 C-terminal peptide (residues 968–990) was amplified from the full-length gene (NM_004486), cloned into the pGEX-6P-1 (Novagen) vector and expressed as a GST fusion protein. The human GM130 coiled-coil region (resides 447–897) was amplified and cloned into the pRSFDuet-1 vector with an N-terminal His6 tag.
For expression in HeLa cells, GM130 (residues 890–990) was ligated into eukaryotic GFP fusion vectors, and the mitochondrial target sequence Tom20 (from NM_014765) was added according to the reference (31). Briefly, four rounds of loop-in PCR yielded the mitochondrial target sequence TOM20 (residues 1–47) upstream of EGFP in a modified pEGFP-C1 vector (Clontech Laboratories, Inc.). GM130 (residues 890–990) was inserted into the BglII and KpnI sites to yield TOM20-GFP-GM130Cterm, and mCherry was substituted for GFP in TOM20-GFP-GM130Cterm to yield TOM20-mCherry-GM130Cterm. Point mutations were introduced using the QuikChange protocol (Stratagene). All of the constructs were confirmed by DNA sequencing.
Protein Expression and Purification
The GRASP65-GM130 complex protein was obtained by co-expression of GRASP65 and the GM130 C-terminal peptide in BL21(DE3) Escherichia coli (Novagen) cells were induced with 0.2 mm isopropyl β-d-thiogalactopyranoside for 16 h at 16 °C. After lysis (PBS, pH 7.4, containing 1 mm β-mercaptoethanol and 1 mm PMSF, Sigma), sonication, and centrifugation, the clarified cell lysate was incubated with glutathione-Sepharose beads (GE Healthcare) and washed with PBS. The untagged GRASP65-GM130 complex protein was obtained after the beads were incubated with PreScission protease at 4 °C overnight and further separated by size exclusion chromatography on a Superdex-200 column (GE Healthcare) pre-equilibrated in a buffer with 20 mm Tris-HCl (pH 8.0), 150 mm NaCl, and 1 mm DTT. The peak fraction was concentrated to 20 mg/ml for crystallization. The GM130 coiled-coil region (amino acids 447 to 897) protein was obtained by following a procedure similar to that of the complex protein with the exception of the lysis buffer (50 mm Tris-HCl, pH 8.0, and 100 mm NaCl containing 1 mm β-mercaptoethanol and 1 mm PMSF) and affinity beads (Ni2+-nitrilotriacetic acid agarose).
Crystallization and Data Collection
The purified complex proteins were crystallized using a sitting drop vapor diffusion method with a 1:1 mixture of sample and reservoir solution. Crystals appeared in the reservoir buffer containing 0.1 m Tris, pH 8.5, 0.2 m Li2SO4, and 30% PEG4K after 1 week at 18 °C and were frozen in a cryoprotectant consisting of the reservoir solution supplemented with 20% PEG400. Data were collected on the BL17U1 station at the Shanghai Synchrotron Radiation Facility (SSRF) and processed using the HKL2000 and XDS software programs (32, 33).
Structure Determination and Refinement
The crystal structures were determined by the molecular replacement program PHASER (34) using the two PDZ domain structures of GRASP65 as an initial search model. Model building and iterative refinement were then performed with the COOT and PHENIX refinement programs (35, 36). The orientations of the amino acid side chains and bound water molecules were modeled on the basis of 2Fobs − Fcalc and Fobs − Fcalc difference Fourier maps. Detailed data collection and refinement statistics are listed in Table 1. The model figures were generated with PyMol and Chimera (37). The interactions were analyzed with PyMol and LigPlus (38).
TABLE 1.
Data collection and refinement statistics
| GRASP65-GM130 complex | |
|---|---|
| Data collection | |
| Space group | P61 |
| Cell dimensions | |
| a, b, c (Å) | 129.7 129.7 37.1 |
| α, β, γ (°) | 90.0 90.0 120.0 |
| Resolution (Å) | 64.86−1.96 (2.03−1.96)a |
| Rmerge | 0.1033 (0.4814) |
| I/σI | 13.78 (4.39) |
| Completeness (%) | 99.56 (100) |
| Redundancy | 13.1 (13.4) |
| Refinement | |
| Resolution (Å) | 64.86−1.96 |
| No. reflections | 340004 |
| Rwork/Rfree | 0.15/0.19 |
| No. atoms | 1964 |
| Protein | 1758 |
| Ligand/ion | 5 |
| Water | 201 |
| B-factors | 47.90 |
| Protein | 47.50 |
| Ligand/ion | 61.10 |
| Water | 50.90 |
| Root mean square deviations | |
| Bond lengths (Å) | 0.0180 |
| Bond angles (°) | 1.770 |
| Ramachandran plot | |
| Favored regions (%) | 97 |
| Allowed refions (%) | 3 |
| Outliers (%) | 0 |
a Values in parentheses are statistics for highest resolution shell.
Isothermal Titration Calorimetry (ITC)
The measurements were taken using an ITC-200 microcalorimeter (MicroCal) at 25 °C. The sample cell (300 μl in volume) was filled with GM130 peptide or mutants at a concentration of 100 μm in a PBS, pH 7.4, buffer containing 5 mm β-mercaptoethanol. The injection syringe (40 μl) was filled with GRASP65 (residues 2–210) or mutants at a concentration of 10 μm. The experimental parameters were: 30 injections, 1.3 μl, 1-s per injection, a 150-s interval, and 1000 rpm stirring speed. The data were analyzed and fitted using the MicroCal Origin software suite.
Native and SDS-PAGE
The affinity-purified recombinant GM130 coiled-coil domain (amino acids 447 to 897) was analyzed by 6% native and SDS-PAGE. The sample buffers consisted of the native loading buffer (with neither SDS nor a reducing agent) and the SDS loading buffer, respectively. The resulting gels were stained with Coomassie Blue.
Cell Culture, Transfection, and Image Capture
HeLa cells were grown in RPMI 1640 medium containing 10% fetal bovine serum and maintained at 37 °C in a 5% CO2 incubator. Transient transfection was performed with TurboFect (Thermo Scientific) according to the manufacturer's specifications. At 24 h after transfection, the cells were washed twice with PBS and fixed with Immunol Staining Fix Solution (Beyotime, China) at room temperature for 15 min. The cells were analyzed using an Olympus FluoViewTM FV1000 confocal laser scanning microscope (Melville, NY) coupled to an inverted microscope with a ×60 differential interference contrast oil immersion objective lens. The cell specimens were excited by laser light (λ = 543 nm for mCherry). Image processing was performed using FV10-ASW 2.0 Viewer and ImageJ (rsbweb.nih.gov/ij).
Results
Overall Structure of GRASP65 in Complex with the GM130 Peptide
The structure of the PDZ domains of GRASP65 (residues 2–210) in complex with the GM130 C-terminal peptide (residues 968–990) was determined and refined at 1.96-Å resolution (Rwork = 15% and Rfree = 19%). Of the 209 amino acids corresponding to the first and second PDZ domains, only residues 12–208 were clearly visible in the electron density map, whereas 20 of the 23 residues in the GM130 C-terminal peptide could be clearly assigned in the structure.
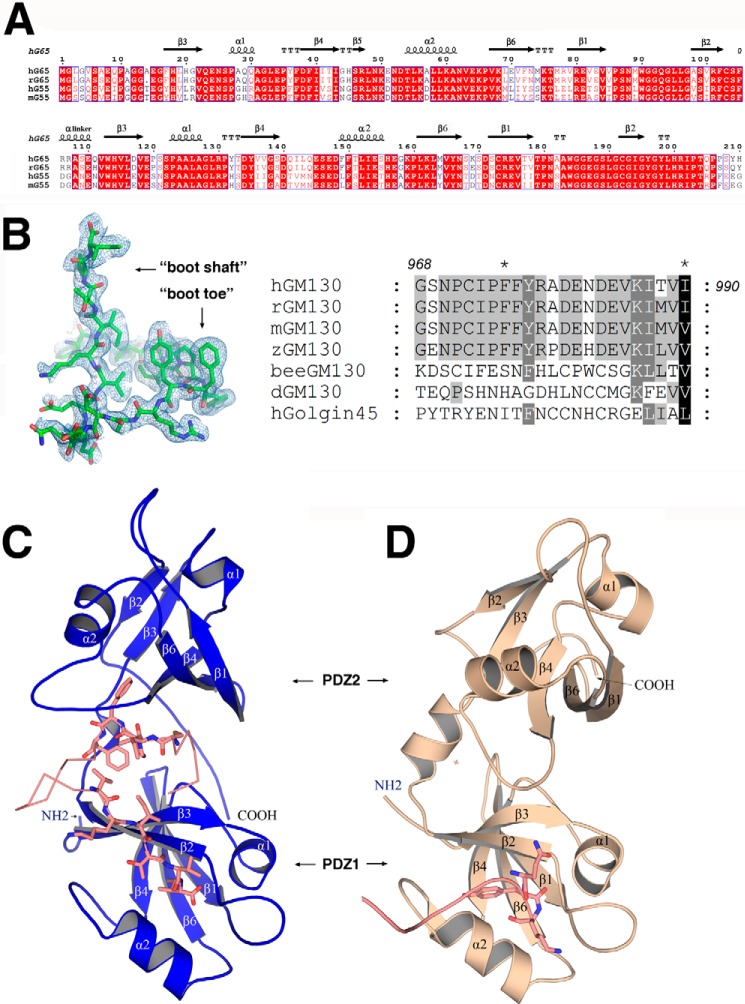
The two PDZ-like domains (PDZ1 and PDZ2) are connected by a flexible linker containing a short α-helix. PDZ1 and PDZ2 can be well aligned, with each PDZ-fold consisting of a six-stranded anti-parallel barrel capped on each end by two α-helices. The topology of the PDZ domains of the GRASPs differs from the topology typically observed in eukaryotes, presenting a circularly permuted architecture (NH2-β3α1β4β5α2β6β1β2-COOH) compared with typical eukaryotic PDZ domains (NH2-β1β2β3α1β4β5α2β6-COOH), as previously reported (27) (Fig. 1, A and C). The bound GM130 peptide exhibits a boot-like shape in the complex, and its amino acid sequence is highly conserved among vertebrates (Fig. 1B). The “boot shaft” binds to the PDZ1 conventional peptide-binding groove formed between the α2-helix and the β2-strand, whereas the “boot toe” is inserted into the cleft between PDZ1 and PDZ2.
FIGURE 1.
Sequence alignment and crystal structures of GRASP65. A, sequence alignment of GRASP domains from human (hG65, NP_114105; hG55, Q9H8Y8.3), rat (rG65, AAI61850.1), and mouse (mG55, NP_081628.3). The strands are numbered according to their GRASP55 PDZ domain structure (27). The figure was generated with ESPript (47). B, electron density map of the bound GM130 C-terminal peptide (left) and a sequence alignment of GM130 from human (AAF65550.1), rat (NP_072118.2), mouse (NP_598613), zebra fish fish (XP_005168139.1), honey bee(XP_623918.3), and Drosophila (CAB77666.1) as well as human GOLGIN45 (NP_003657.1) C-terminal peptides (right). C, schematic representation of the overall structure of GRASP65 (colored blue) bound with the GM130 C-terminal peptide (colored salmon). D, schematic representation of the unbound structure of GRASP65 (Protein Data Bank code 4KFV, colored wheat) illustrating the neighboring molecular C terminus (colored salmon) interacting with the PDZ1 classic peptide binding groove.
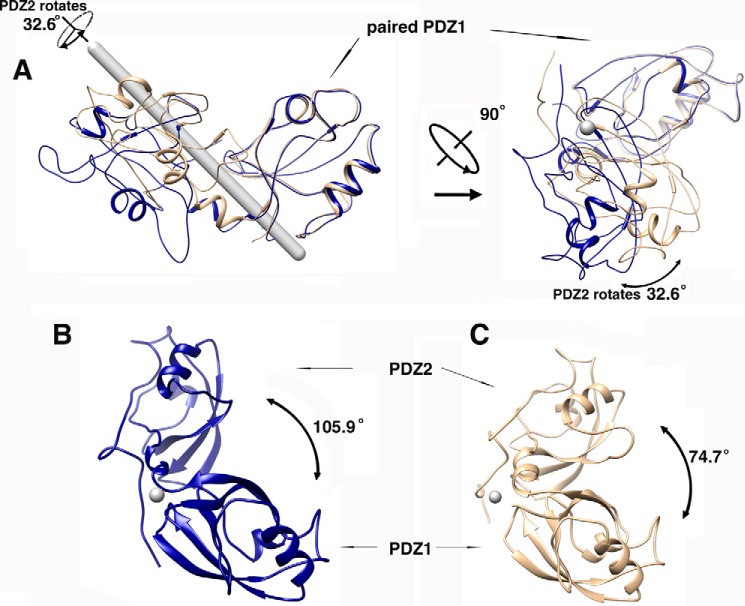
Upon GM130 C-terminal peptide binding, GRASP65 conformation changes dramatically. Although PDZ1 and PDZ2 within the complex can still be correctly aligned with their corresponding individual domains in the unbound GRASP65 structure, the relative orientations of PDZ1 and PDZ2 undergo a rigid-body conformational change upon GM130 binding (Fig. 1, C and D). In its bound conformation, GRASP65 presents a much wider cleft between PDZ1 and PDZ2 to accommodate the GM130 C-terminal peptide. A superimposition of the two GRASP structures using PDZ1 as a template illustrates a 32.6° rotation of the PDZ2 domain upon GM130 peptide binding, allowing the opening of the cleft (Fig. 2).
FIGURE 2.
Conformational changes of GRASP65 upon GM130 C-terminal peptide binding. The software program Chimera was used for superimposing the structures, measuring the angles of rotation of the domains and generating the figures. The gray rod denotes the rotation axis parallel to the paper plane, and the gray spots denote the rotation axis perpendicular to the paper plane. A, the superimposed structures of GRASP65 with and without the GM130 C-terminal peptide using the PDZ1 domain to pinpoint a rotation of 32.6 degrees at the PDZ2 along its axis. The two representations are rotated by 90 degrees. The unbound GRASP65 structure is colored wheat; the bound GRASP65 structure is colored blue. B, measurement of the angle between the PDZ1 and PDZ2 domains of GRASP65 upon GM130 binding. C, measurement of the angle between the PDZ1 and PDZ2 domains of GRASP65 without GM130 binding.
In the unbound conformation (free form of the GRASP65 PDZ domains), the peptide-binding groove of the first PDZ domain (PDZ1) is occupied by the C-terminal residues (SQYK) from a neighboring molecule. This nonspecific interaction is outcompeted by GM130 binding when the C-terminal residues of GM130 (boot shaft) interact with GRASP at the same PDZ1 peptide-binding groove. The displaced C-terminal residues of GRASP65 then become disordered in the bound conformation.
Molecular Basis of GM130 C-terminal Peptide Recognition
Previous biochemical and mutational studies concluded that GM130 interacts with the PDZ2 domain of GRASP65 (24, 27). Our complex structure, however, clearly indicates that the GM130 C-terminal peptide interacts with both the PDZ1 and PDZ2 domains of GRASP65. In the bound structure, the GM130 C-terminal residues (KITVI) interact with the GRASP65 PDZ1 binding groove through a canonical PDZ binding motif recognition mode. These five GM130 C-terminal residues form a 6th anti-parallel β-strand that extends the β-sheet of the GRASP65 PDZ1 domain, whereas the carboxyl-terminal residue (Ile990) is coordinated by a network of hydrogen bonds to main chain amide groups in the “carboxylate binding loop” of PDZ1 (following the nomenclature of Lee and Zheng (39)). This interaction map of the GRASP65-GM130 complex confirms that the circularly permuted PDZ domain-containing protein GRASP65 (PDZ1-PDZ2) contains the classic PDZ binding motif. This motif possesses the well conserved PDZ peptide-binding groove motif φ-G-φ (here Leu96-Gly97-Ala98), where φ represents a hydrophobic amino acid that recognizes the C-terminal peptide motif -φ-φ-φ-φ-COOH (39). Two carboxylate oxygen atoms from Ile990 of the GM130 C terminus form 4 hydrogen bonds with the amide nitrogen atoms of Leu95, Leu96, Gly97, and Ala98 of the carboxylate-binding loop. Gly97 adopts the identical left-handed α-helical conformation observed in other PDZ structures to allow the amide groups in the loop region to serve as H-bonding donors. The side chain of Ile990 is stabilized by Leu55 and Lys56 from the second α helix (α2) via hydrophobic interactions. The detailed interactions between GM130 and PDZ1 at the binding groove are shown in Fig. 3A.
FIGURE 3.
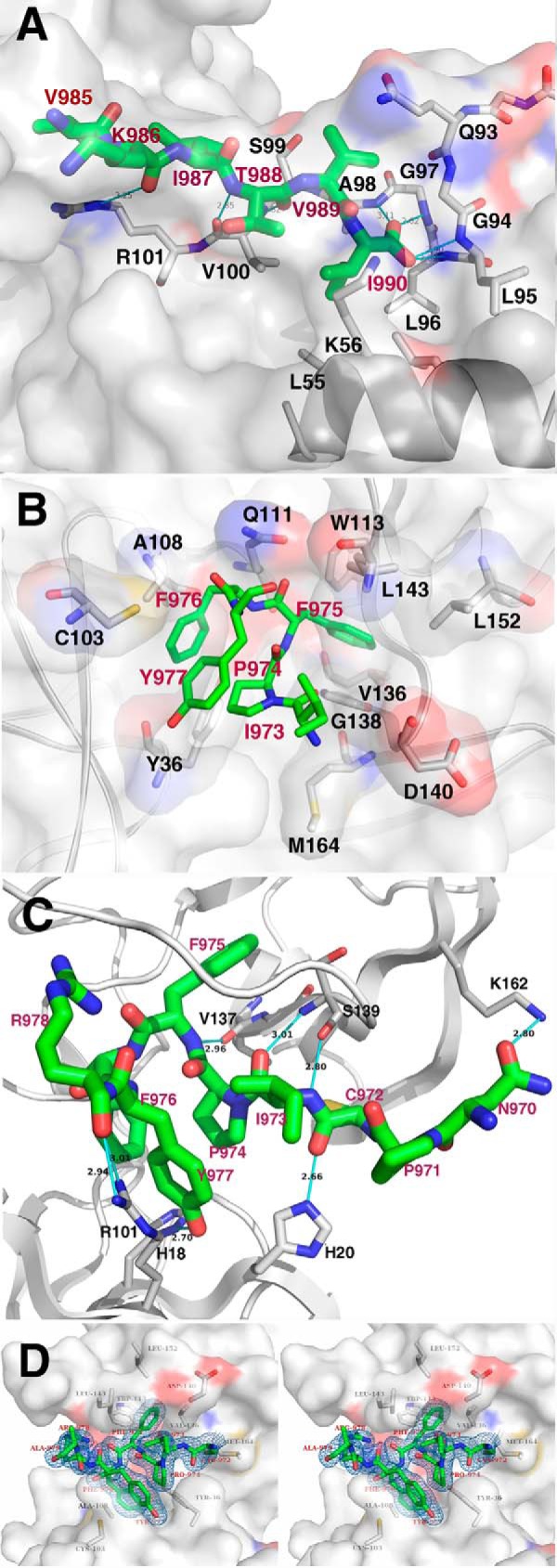
Detailed interactions between GRASP65 and the GM130 C-terminal peptide. A, detailed interactions between GRASP65 and the GM130 C-terminal residues at the conventional peptide-binding groove of the PDZ1 domain. The residues involved in the PDZ1-GM130 peptide interaction are shown with a stick model, in which the GRASP65 and GM130 peptide backbones are colored white and green, respectively, and the nitrogen and oxygen atoms are colored blue and red, respectively. The surface of the GRASP65 PDZ1 domain is also illustrated. B, detailed hydrophobic interactions between GRASP65 and the GM130 C-terminal peptide at the cleft between the PDZ1 and PDZ2 domains. The residues involved in the PDZ1-GM130 peptide hydrophobic interaction are shown with a stick model. The GRASP65 and GM130 peptide backbones are colored white and green, respectively, and the nitrogen and oxygen atoms are colored blue and red, respectively. The GRASP65 surface is also illustrated. C, detailed hydrogen bond interactions between GRASP65 and the GM130 C-terminal peptide at the cleft between the PDZ1 and PDZ2 domains. The schematic represents the backbone structure of GRASP65. The residues involved in the hydrogen bond interaction are shown as a stick model. The GRASP65 and GM130 peptide backbones are colored white and green, respectively, and the nitrogen and oxygen atoms are colored blue and red, respectively. D, stereo view of the GM130 C-terminal peptide inserting into the cleft between the PDZ1 and PDZ2 domains. The residues of the observable portion of the GM130 C-terminal peptide as well as the electron density of the omit map are illustrated (1.5 σ).
In addition to the interaction with PDZ1 at the conventional peptide-binding groove, the GM130 C-terminal peptide occupies a previously unknown, second binding site on GRASP65. The hydrophobic region of the peptide, consisting of residues Ile973, Pro974, Phe975, Phe976, and Tyr977, inserts into a hydrophobic cleft between PDZ1 and PDZ2, interacting with both PDZ domains. This hydrophobic cleft comprises residues Tyr36 and Cys103 from PDZ1, residues Gln111, Trp113, Asp140, Val136, Gly138, Leu143, Leu152, and Met164 from PDZ2, and residue Ala108 from the linker connecting the two PDZ domains. Strikingly, the hydrophobic side chain of Phe975 from the GM130 C-terminal peptide is deeply buried within a hydrophobic cavity at the center of PDZ2 (Fig. 3B), whereas the remainder of the peptide is stabilized by a combination of hydrophobic interactions and hydrogen bonds. The detailed hydrogen bonding interactions are shown in Fig. 3C. Notably, residue Leu152 lies at the bottom of this hydrophobic pocket and directly interacts with the side chain of Phe975 (Fig. 3B), which supplements a previous study that demonstrated that a GRASP65 L152A/I153A mutant could no longer bind GM130 (26).
Both Binding Sites Are Required for the Interaction of GM130 and GRASP65
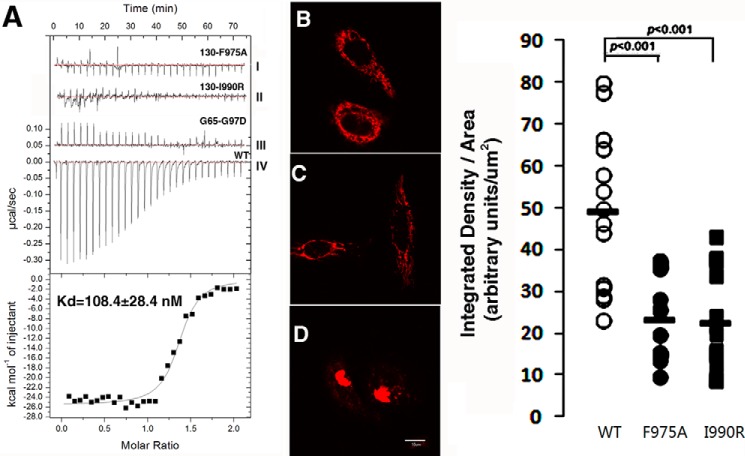
We used ITC and an in vivo organelle tethering assay to assess the relevance of the binding sites observed in our crystal structure. The ITC experiments exhibited a dissociation constant (Kd) of 108 ± 28 nm between wild type GRASP65 (2–210) and the GM130 peptide, whereas a single point mutation at the binding site cleft (F975A) or the C terminus (I990R) of the GM130 peptide dramatically decreased the binding affinity of GM130 for GRASP65. A similar observation occurred when the GM130 peptide was kept unchanged, whereas the highly conserved Gly97 of the carboxylate binding loop on PDZ1 was mutated to aspartic acid (Fig. 4A). These results indicate that the interaction between GRASP65 and GM130 requires not only the conventional PDZ domain recognition mode but also an additional cleft hydrophobic contact, as indicated by the crystal structure of the bound GRASP65.
FIGURE 4.
ITC experiments and GM130-induced mitochondrial clustering analysis of the GRASP65-GM130 C-terminal peptide interaction. A, the top graph illustrates the raw data of GRASP65 with GM130 C-terminal peptides and their mutants (GM130(F975A), GM130(I990R), and GRASP65(G97D)), and the y axis indicates the heat released per second during GM130 and GRASP65 binding. The bottom graph illustrates the integrated heat for each injection of GRASP65 together with a fit, whereas the y axis represents the heat released per mole for each injection. The dissociation constants were obtained from curves obtained by the titration of GM130 with GRASP65 (WT). 24 h after transfection, HeLa cells expressing the TOM20-Cherry-GM130 F975A mutant (B), the TOM20-Cherry-GM130 I990R mutant (C), and TOM20-Cherry-GM130 WT (D) were analyzed by confocal microscopy. The bar scale is 10 μm. The pixel density and surface area of GM130 signal were quantified using NIH ImageJ version 1.48 software. The results are plotted for the wild type GM130-expressed cells (n = 13), the GM130 F975A mutant expressed cells (n = 14), and the GM130 I990R mutant expressed cells (n = 15).
An in vivo organelle tethering assay was used in a previous study to demonstrate that mitochondria expressing GM130 can recruit endogenous GRASP65, thus artificially clustering the mitochondria together (31). Using a similar approach, we expressed the GM130 peptide with the Tom20 mitochondria location signal fragment and cherry fluorescent protein in HeLa cells. The HeLa cells were transfected with the wild type GM130 peptide (Fig. 4D), the F975A GM130 mutant (Fig. 4B), or the I990R GM130 mutant (Fig. 4C). Single point mutations at both locations prevented the mitochondria from recruiting endogenous GRASP65 and blocked the clustering of mitochondria (Fig. 4, B and C). These results are consistent with our ITC experiments and validate our structural model as well as biophysical observations in physiologically relevant contexts.
Analysis of the Oligomeric State of GM130 by Native and SDS-PAGE
The coiled-coil region (amino acids 447 to 897) of GM130 was expressed and purified. It migrated as a monomer (Fig. 5B, lane 3) on SDS-PAGE, and several dimers were also observed when using the native loading buffer (Fig. 5B, lane 2). The GM130 oligomer was not stable enough to tolerate the SDS in the gel and separated into dimers and monomers. The multiple bands observed surrounding the dimer positions on the SDS-PAGE gel may be due to heterogeneity in the SDS-bound proteins. To check its oligomeric state under native conditions, the same GM130 construct was analyzed using native PAGE. A major band was evidenced at a molecular mass of 300–400 kDa, indicating that GM130 forms a hexamer under native conditions. A trace amount of the monomer, which is larger than the calculated molecular mass (52 kDa) due to its rod-like asymmetric shape, is also revealed in the figure (Fig. 5A, lane 1).
FIGURE 5.
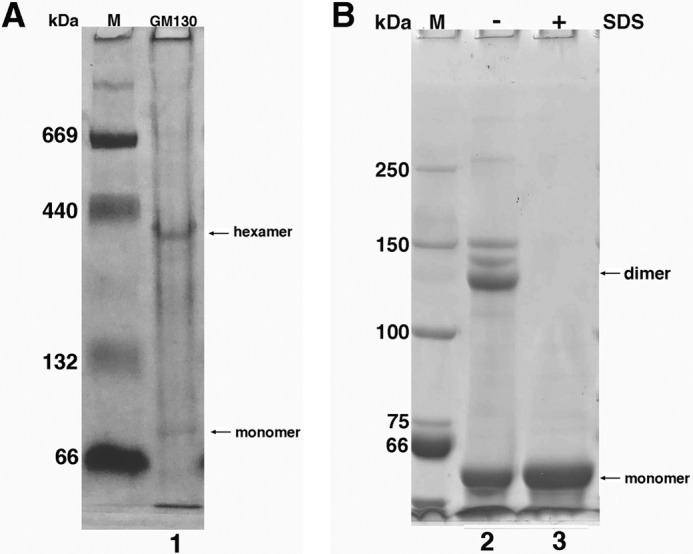
Oligomerization of the GM130 coiled-coil domain. The coiled-coil region (amino acids 447 to 897) of GM130 was analyzed by 6% Native and SDS-PAGE. A, the GM130 coiled-coil domain analyzed by 6% native PAGE evidences the formation of hexamers. A trace amount of the monomeric form is also evidenced (lane 1). B, the GM130 coiled-coil domain analyzed by 6% SDS-PAGE evidences a monomeric and dimeric form using native loading buffer (lane 2) and a solely monomeric form using SDS loading buffer (lane 3).
Discussion
GRASP65 and GM130 are thought to be structural candidates for the intercisternal bridge between the Golgi cisternae and are thought to mediate Golgi stacking and vesicle tethering and maintain a unique, highly dynamic stacked, ribbon-like structure in mammalian cells. The molecular details of this organization remain unknown.
Our three-dimensional structure of the complex between GRASP65 and the GM130 C-terminal peptide reveals functionally relevant details about their molecular interactions. The GRASP conserved region has two tandemly arranged PDZ domains (PDZ1 and PDZ2). Our structure indicates that the GM130 C-terminal peptide unexpectedly binds to the conventional PDZ binding groove on PDZ1. In addition, PDZ1 and PDZ2 generate an unforeseen interdomain pocket after GM130 recognition. This pocket defines the second interaction site. The boot shaft of the bound GM130 peptide binds to the PDZ1 conventional peptide-binding groove formed between the α2-helix and the β2-strand, whereas the boot toe is inserted into the newly formed cleft between PDZ1 and PDZ2. A similar binding mode in the polarity protein Scribble has been recently reported (40). A single mutation (G97D) in PDZ1 of GRASP65 abolished the GRASP65-GM130 interaction, whereas a previous biochemical study reported that ΔPDZ1-GRASP65 can remain bound to GM130 (24). These conflicting findings are yet to be explained.
In contrast with previously reported structures of GRASP65 and GRASP55 (27, 28), our GRASP65 construct undergoes substantial conformational changes. Although the individual PDZ domains are well aligned with previously reported structures, their relative position is rotated by 32.6° after GM130 binding. The binding site at the interdomain pocket formed by this conformational change is dominated by hydrophobic residues. A single mutation (F975A, mainly interacts with the PDZ2 domain of GRASP65) in GM130 abolishes the interaction of GRASP65 with it. This observation could explain why previous biochemical studies concluded that GM130 only interacts with the PDZ2 domain of GRASP65 (24, 26, 29). Structural studies cannot only validate biochemical results by showing the direct three-dimensional organization of biomolecules but can also modify the erroneous conclusions of biochemical studies, providing new insights into the molecular characteristics of the physiological role of a protein. The three-dimensional structure provided in this study exemplifies this concept.
Previous biochemical studies determined that the clustering of GRASP proteins is mediated by PDZ1 domains in vivo or in vitro. These interactions have long been thought to play a major role in Golgi stacking (27, 28, 30). The analysis of GRASP65 and GRASP55 apo structures implies that GRASP clustering is mediated by an interaction between the end of the GRASP domain and the conventional peptide-binding groove on PDZ1. In our complex structure, the conventional peptide-binding groove on PDZ1 is occupied by GM130 and cannot interact with the end of the GRASP domain. This new finding implies that GM130 may participate in GRASP clustering and play a direct role in Golgi stacking. This proposition is supported by the following observations: GRASP65 and GM130 always co-purify as a macromolecular complex in stacking Golgi (8) and GM130 knockdown prevents the lateral linking of Golgi stacks (10). Direct evidence for this proposition has not yet been obtained.
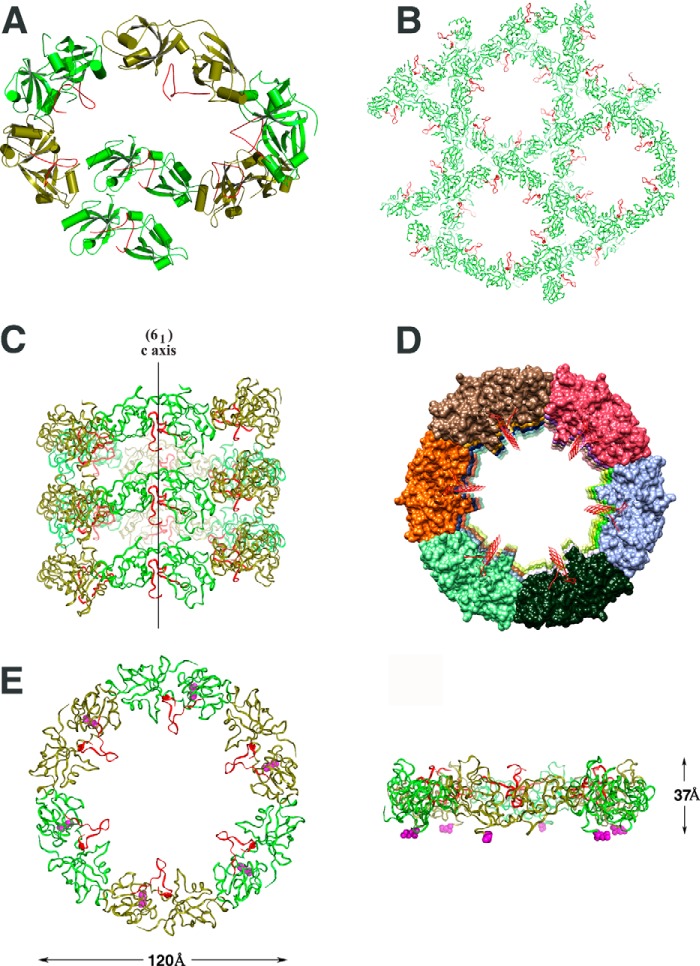
Does the conformational change of GRASP65 after GM130 binding have physiological consequences? An analysis of the protein packing in the crystal structure of the bound form revealed the mode of action of GRASP65 clustering after GM130 binding. The complex structure illustrates that GRASP65 can be packed into a superhelical structure upon GM130 binding (Figs. 6, A–D), and the helical axis of this structure coincides with the crystallographic 61 screw axis. Each helical turn contains six GRASP65 monomers with a pitch equal to the length of the crystallographic c axis (37 Å). Essentially, the conformational change of GRASP65 upon GM130 binding initiates the formation of this superhelical structure.
FIGURE 6.
The GRASP65 superhelix in the crystal of the complex and the GRASP65 hexameric ring model. The GM130 C-terminal peptide is colored red; GRASP65 is colored green. A, GRASP65 generates a superhelix arranged from top to tail in a circular manner along the crystallographic 61 screw axis. The schematic illustrates one full turn plus one additional molecule of the superhelix, which contains 7 GRASP65 complex molecules in total. B, crystal packing of the bound form of GRASP65 is illustrated above the 61 screw axis. The superhelical arrangement extends along the entire length of the crystal, resulting in long, solvent-filled central channels. C, side view of one superhelix in the crystal of the bound GRASP65. The crystallographic 61 screw axis is illustrated. D, top view of one superhelix. The aligned GM130 C-terminal peptides point to the ring center. For clarity, GRASP65 is represented in various colors. E, ribbon representation of the GRASP65 ring model containing 6 GRASP65 bound molecules. Both the front and side views of the GRASP65 ring model are illustrated. The N termini of GRASP65 are represented as magenta spheres located at the same side of the ring.
Because our crystallized construct was not myristoylated (GRASP65 is N-myristoylated at the highly conserved glycine at position 2, which anchors the GRASP65 N terminus to the Golgi cisternae membrane), the superhelical N terminus of the crystallized GRASP65 was free to drive the formation of a superhelix. In this superhelical structure, all of the N termini were uniformly oriented. If the N terminus of all of the GRASP65 molecules in each helical turn were constrained by a membrane-anchoring phenomenon and tethered to a flat membrane (as observed in intact cells), the six GRASP65 monomers in one helical turn of the superhelix would form a hexameric ring. This GRASP65 hexameric ring would maintain the 6-fold symmetry in the crystal structure and have an overall diameter of ∼120 Å and a height of ∼37 Å (Fig. 6E).
Based on the above information, Golgi stacking may be mediated by GRASP65 in two different modes. The first mode involves a homo-oligomeric form of GRASP65, whereas the second mode involves a hetero-oligomeric form of GRASP65 bound to GM130. The homo-oligomer localizes to the cisternal rims without binding GM130 and contributes to Golgi cisternae (cis-interaction) and ribbon formation (trans-interaction). The hetero-oligomer is localized between the cisternae and provides the main force for cisternae stacking. The binding of GM130 triggers conformational changes in GRASP65, which facilitates the formation of a hexameric ring-like hetero-oligomeric structure on the flat membrane of the cisterna.
GM130 is evidenced in the Golgi matrix fraction and has a molecular mass of ∼1,000 kDa after its extraction from the Golgi membrane with detergent and high salt (5). Our native gel results illustrate that GM130 tends to form hexamers. It has been proposed that synthetic peptides with designed coiled-coil motifs can form stable six-helix bundles (41). In a recent report, the authors used Blue Native SDS-PAGE to show that GM130 most likely forms a homo-tetramer rather than the previously proposed homo-dimer. They observed a band on their native gel containing a fragment (amino acids 447 to 897) that was smaller than and similar to the band we observed in the present study. GM130 can thus form a homo-oligomer, although the exact oligomer number needs to be investigated further.
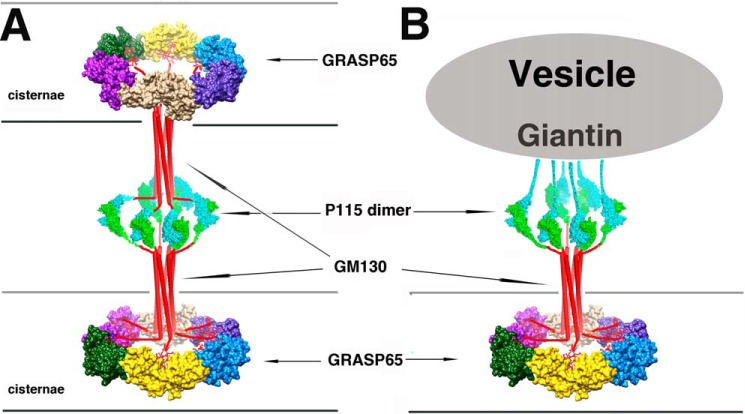
Assuming the presence of hexamers, we propose that each GM130 C terminus in the hexamer binds to one GRASP65 molecule in the GRASP65 ring on the flat membrane of cisternae, whereas each of its N termini binds to one p115 molecule of the dimer. In cisternae stacking, both p115 molecules in the dimer bind to GM130 and GM130 interacts with the GRASP65 ring on the opposing cisternae membranes. The p115 dimer acts as a cisternae connector and mediates Golgi stacking in this model (Fig. 7A). In the vesicle tethering model, one p115 molecule in the dimer binds to GM130, whereas the other p115 molecule binds to vesicle-bound Giantin (Fig. 7B). The partner of p115 can be switched under control of Rab1 GTPase, the conserved oligomeric Golgi complex (42, 43) and the phosphorylation of GM130, p115, and Giantin (21, 44, 45). Although the GRASP65 ring-like configuration in our model is a deduced structure that might represent an artifact caused by packing forces in the crystal, it is well supported by previous biochemical and electron microscopy study results (4, 8, 41, 46). Further studies and direct in cellulo evidence are required to validate the GRASP65 ring-like configuration in our model.
FIGURE 7.
Molecular models of GRASP65/GM130/P115-mediated cis-cisternae membrane stacking and vesicle tethering. A, “double wheel” model of GRASP65/GM130/P115-mediated cis-cisternae membrane stacking. The P115 dimer and GM130 hexamer bridge the two rings formed by the six GRASP65 molecules at opposing cisternae membranes. B, “single wheel” model of GRASP65/GM130/P115/Giantin-mediated vesicle tethering. The structure at one side of the cisternae membrane is represented by a double wheel model. At the other side, P115 binds to the Giantin N terminus on the vesicle instead of GM130 and bridges the vesicles and the cisterna membrane.
Author Contributions
F. H., X. S., and X. H. completed the protein purification, crystallization, and biochemical experiments; N. S. carried out structure determination and refinement; B. L. made the constructs; F. H., X. M., and N. S. analyzed the data, designed the study, and wrote the paper. All of the authors have discussed the results and commented on the manuscript.
Acknowledgments
The x-ray data were collected at the BL17U1 beamline of the Shanghai Synchrotron Radiation Facility. We are grateful to Prof. Jiahuai Han for the plasmids; Youxing Jiang, Nam Nguyen, and Mingdong Huang for their thoughtful discussions and reviews of the manuscript; and Jinping Xue for helping with confocal microscopy.
This work was supported by the National Natural Science Foundation of China Grant 31370738, the One Hundred Person Project of the Chinese Academy of Sciences, the National Science Foundation of Fujian Province (2013J01150) and Campus France (PHC CAI YUANPEI 2013, Project 26203WD). The authors declare that they have no conflict of interest with the contents of this article.
The atomic coordinates and structure factors (code 4REY) have been deposited in the Protein Data Bank (http://wwpdb.org/).
- GRASP65
- Golgi reassembly stacking protein of 65 kDa
- GM130
- Golgi matrix protein of 130 kDa
- ITC
- isothermal titration calorimetry.
References
- 1.Farquhar M. G., and Palade G. E. (1981) The Golgi apparatus (complex)-(1954–1981)-from artifact to center stage. J. Cell Biol. 91, 77s-103s [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rambourg A., and Clermont Y. (1990) Three-dimensional electron microscopy: structure of the Golgi apparatus. Eur. J. Cell Biol. 51, 189–200 [PubMed] [Google Scholar]
- 3.Klumperman J. (2011) Architecture of the mammalian Golgi. Cold Spring Harbor Perspect. Biol. 3, 1–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cluett E. B., and Brown W. J. (1992) Adhesion of Golgi cisternae by proteinaceous interactions: intercisternal bridges as putative adhesive structures. J. Cell Sci. 103, 773–784 [DOI] [PubMed] [Google Scholar]
- 5.Nakamura N., Rabouille C., Watson R., Nilsson T., Hui N., Slusarewicz P., Kreis T. E., and Warren G. (1995) Characterization of a cis-Golgi matrix protein, GM130. J. Cell Biol. 131, 1715–1726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shorter J., Watson R., Giannakou M. E., Clarke M., Warren G., and Barr F. A. (1999) GRASP55, a second mammalian GRASP protein involved in the stacking of Golgi cisternae in a cell-free system. EMBO J. 18, 4949–4960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rabouille C., Misteli T., Watson R., and Warren G. (1995) Reassembly of Golgi stacks from mitotic Golgi fragments in a cell-free system. J. Cell Biol. 129, 605–618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Barr F. A., Puype M., Vandekerckhove J., and Warren G. (1997) GRASP65, a protein involved in the stacking of Golgi cisternae. Cell 91, 253–262 [DOI] [PubMed] [Google Scholar]
- 9.Feinstein T. N., and Linstedt A. D. (2008) GRASP55 regulates Golgi ribbon formation. Mol. Biol. Cell 19, 2696–2707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Puthenveedu M. A., Bachert C., Puri S., Lanni F., and Linstedt A. D. (2006) GM130 and GRASP65-dependent lateral cisternal fusion allows uniform Golgi-enzyme distribution. Nat. Cell Biol. 8, 238–248 [DOI] [PubMed] [Google Scholar]
- 11.Xiang Y., and Wang Y. (2010) GRASP55 and GRASP65 play complementary and essential roles in Golgi cisternal stacking. J. Cell Biol. 188, 237–251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xiang Y., Zhang X., Nix D. B., Katoh T., Aoki K., Tiemeyer M., and Wang Y. (2013) Regulation of protein glycosylation and sorting by the Golgi matrix proteins GRASP55/65. Nat. Commun. 4, 1659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jarvela T., and Linstedt A. D. (2014) Isoform-specific tethering links the Golgi ribbon to maintain compartmentalization. Mol. Biol. Cell 25, 133–144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Duran J. M., Kinseth M., Bossard C., Rose D. W., Polishchuk R., Wu C. C., Yates J., Zimmerman T., and Malhotra V. (2008) The role of GRASP55 in Golgi fragmentation and entry of cells into mitosis. Mol. Biol. Cell 19, 2579–2587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Diao A., Frost L., Morohashi Y., and Lowe M. (2008) Coordination of golgin tethering and SNARE assembly: GM130 binds syntaxin 5 in a p115-regulated manner. J. Biol. Chem. 283, 6957–6967 [DOI] [PubMed] [Google Scholar]
- 16.Nakamura N. (2010) Emerging new roles of GM130, a cis-Golgi matrix protein, in higher order cell functions. J. Pharmacol. Sci. 112, 255–264 [DOI] [PubMed] [Google Scholar]
- 17.Marra P., Salvatore L., Mironov A. Jr., Di Campli A., Di Tullio G., Trucco A., Beznoussenko G., Mironov A., and De Matteis M. A. (2007) The biogenesis of the Golgi ribbon: the roles of membrane input from the ER and of GM130. Mol. Biol. Cell 18, 1595–1608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Karakas E., and Furukawa H. (2014) Crystal structure of a heterotetrameric NMDA receptor ion channel. Science 344, 992–997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shorter J., Beard M. B., Seemann J., Dirac-Svejstrup A. B., and Warren G. (2002) Sequential tethering of Golgins and catalysis of SNAREpin assembly by the vesicle-tethering protein p115. J. Cell Biol. 157, 45–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shorter J., and Warren G. (1999) A role for the vesicle tethering protein, p115, in the post-mitotic stacking of reassembling Golgi cisternae in a cell-free system. J. Cell Biol. 146, 57–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nakamura N., Lowe M., Levine T. P., Rabouille C., and Warren G. (1997) The vesicle docking protein p115 binds GM130, a cis-Golgi matrix protein, in a mitotically regulated manner. Cell 89, 445–455 [DOI] [PubMed] [Google Scholar]
- 22.Linstedt A. D., Jesch S. A., Mehta A., Lee T. H., Garcia-Mata R., Nelson D. S., and Sztul E. (2000) Binding relationships of membrane tethering components: the Giantin N terminus and the GM130 N terminus compete for binding to the p115 C terminus. J. Biol. Chem. 275, 10196–10201 [DOI] [PubMed] [Google Scholar]
- 23.Puthenveedu M. A., and Linstedt A. D. (2001) Evidence that Golgi structure depends on a p115 activity that is independent of the vesicle tether components Giantin and GM130. J. Cell Biol. 155, 227–238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Barr F. A., Nakamura N., and Warren G. (1998) Mapping the interaction between GRASP65 and GM130, components of a protein complex involved in the stacking of Golgi cisternae. EMBO J. 17, 3258–3268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kodani A., and Sütterlin C. (2008) The Golgi protein GM130 regulates centrosome morphology and function. Mol. Biol. Cell 19, 745–753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bachert C., and Linstedt A. D. (2010) Dual anchoring of the GRASP membrane tether promotes trans pairing. J. Biol. Chem. 285, 16294–16301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Truschel S. T., Sengupta D., Foote A., Heroux A., Macbeth M. R., and Linstedt A. D. (2011) Structure of the membrane-tethering GRASP domain reveals a unique PDZ ligand interaction that mediates Golgi biogenesis. J. Biol. Chem. 286, 20125–20129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Feng Y., Yu W., Li X., Lin S., Zhou Y., Hu J., and Liu X. (2013) Structural insight into Golgi membrane stacking by GRASP65 and GRASP55 proteins. J. Biol. Chem. 288, 28418–28427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Truschel S. T., Zhang M., Bachert C., Macbeth M. R., and Linstedt A. D. (2012) Allosteric regulation of GRASP protein-dependent Golgi membrane tethering by mitotic phosphorylation. J. Biol. Chem. 287, 19870–19875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tang D., Yuan H., Vielemeyer O., Perez F., and Wang Y. (2012) Sequential phosphorylation of GRASP65 during mitotic Golgi disassembly. Biol. Open 1, 1204–1214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sengupta D., Truschel S., Bachert C., and Linstedt A. D. (2009) Organelle tethering by a homotypic PDZ interaction underlies formation of the Golgi membrane network. J. Cell Biol. 186, 41–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Otwinowski Z., and Minor W. (1997) Processing of x-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 33.Kabsch W. (2010) Xds. Acta Crystallogr. D Biol. Crystallogr. 66, 125–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McCoy A. J., Grosse-Kunstleve R. W., Adams P. D., Winn M. D., Storoni L. C., and Read R. J. (2007) Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Adams P. D., Afonine P. V., Bunkóczi G., Chen V. B., Davis I. W., Echols N., Headd J. J., Hung L. W., Kapral G. J., Grosse-Kunstleve R. W., McCoy A. J., Moriarty N. W., Oeffner R., Read R. J., Richardson D. C., Richardson J. S., Terwilliger T. C., and Zwart P. H. (2010) PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Emsley P., Lohkamp B., Scott W. G., and Cowtan K. (2010) Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pettersen E. F., Goddard T. D., Huang C. C., Couch G. S., Greenblatt D. M., Meng E. C., and Ferrin T. E. (2004) UCSF Chimera: a visualization system for exploratory research and analysis. J. Comp. Chem. 25, 1605–1612 [DOI] [PubMed] [Google Scholar]
- 38.Wallace A. C., Laskowski R. A., and Thornton J. M. (1995) LIGPLOT: a program to generate schematic diagrams of protein-ligand interactions. Protein Eng. 8, 127–134 [DOI] [PubMed] [Google Scholar]
- 39.Lee H. J., and Zheng J. J. (2010) PDZ domains and their binding partners: structure, specificity, and modification. Cell Commun. Signal. 8, 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ren J., Feng L., Bai Y., Pei H., Yuan Z., and Feng W. (2015) Interdomain interface-mediated target recognition by the Scribble PDZ34 supramodule. Biochem. J. 468, 133–144 [DOI] [PubMed] [Google Scholar]
- 41.Zaccai N. R., Chi B., Thomson A. R., Boyle A. L., Bartlett G. J., Bruning M., Linden N., Sessions R. B., Booth P. J., Brady R. L., and Woolfson D. N. (2011) A de novo peptide hexamer with a mutable channel. Nat. Chem. Biol. 7, 935–941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.An Y., Chen C. Y., Moyer B., Rotkiewicz P., Elsliger M. A., Godzik A., Wilson I. A., and Balch W. E. (2009) Structural and functional analysis of the globular head domain of p115 provides insight into membrane tethering. J. Mol. Biol. 391, 26–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Striegl H., Roske Y., Kümmel D., and Heinemann U. (2009) Unusual armadillo fold in the human general vesicular transport factor p115. PloS One 4, e4656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dirac-Svejstrup A. B., Shorter J., Waters M. G., and Warren G. (2000) Phosphorylation of the vesicle-tethering protein p115 by a casein kinase II-like enzyme is required for Golgi reassembly from isolated mitotic fragments. J. Cell Biol. 150, 475–488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Seemann J., Jokitalo E. J., and Warren G. (2000) The role of the tethering proteins p115 and GM130 in transport through the Golgi apparatus in vivo. Mol. Biol. Cell 11, 635–645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Orci L., Perrelet A., and Rothman J. E. (1998) Vesicles on strings: morphological evidence for processive transport within the Golgi stack. Proc. Natl. Acad. Sci. U.S.A. 95, 2279–2283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Robert X., and Gouet P. (2014) Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 42, W320–324 [DOI] [PMC free article] [PubMed] [Google Scholar]