Abstract
Formaldehyde has been used for decades to probe macromolecular structure and function and to trap complexes, cells, and tissues for further analysis. Formaldehyde crosslinking is routinely employed for detection and quantification of protein-DNA interactions, interactions between chromatin proteins, and interactions between distal segments of the chromatin fiber. Despite widespread use and a rich biochemical literature, important aspects of formaldehyde behavior in cells have not been well described. Here, we highlight features of formaldehyde chemistry relevant to its use in analyses of chromatin complexes, focusing on how its properties may influence studies of chromatin structure and function.
Keywords: chromatin immunoprecipitation (ChIP), chromatin structure, nucleic acid chemistry, protein cross-linking, transcription factor, formaldehyde chemistry
Introduction
Prior to its use in the chromatin field, formaldehyde use had a long history in a number of fields, including vaccine production (1, 2) and histology (3). In this review, we focus on its use in chromatin immunoprecipitation approaches and protein-protein interaction studies applied to understand the location and abundance of transcription factor binding along DNA. A recent complementary perspective highlights gaps in knowledge with a particular focus on how formaldehyde crosslinking data have been used to interpret aspects of chromatin three-dimensional organization (4). Here, we briefly review prior work describing formaldehyde reactivity toward proteins, DNA, and their constituent monomers. This information provides a basis for understanding how formaldehyde functions in widely used assays in the chromatin field, and conversely, highlights less well understood aspects of formaldehyde behavior in cells. These issues are of significance for designing crosslinking-based studies as well as for properly interpreting the resulting data. The analysis of formaldehyde-fixed chromatin has provided fundamental insights into where and when regulatory factors associate with the DNA template in vivo, but it in general does not provide unambiguous information about chromatin binding kinetics. A major goal of ongoing work is to understand kinetic and thermodynamic aspects of chromatin complex assembly at single copy loci in vivo. Development of experimental strategies to achieve these goals will require a deeper and more comprehensive understanding of the effects mediated by formaldehyde in cells.
The following discussion provides a framework for understanding aspects of formaldehyde function when used to trap macromolecular complexes in cells, with the main features shown in Fig. 1. Beginning with basic chemical reactivity, this review will explore the ability of formaldehyde to crosslink with proteins and DNA to form protein-protein or protein-DNA complexes, common molecular quenchers, and the potential for crosslink reversal. Progress in capturing crosslinked complexes will also be discussed with an emphasis on the impact of a better understanding of formaldehyde chemistry in vivo.
FIGURE 1.
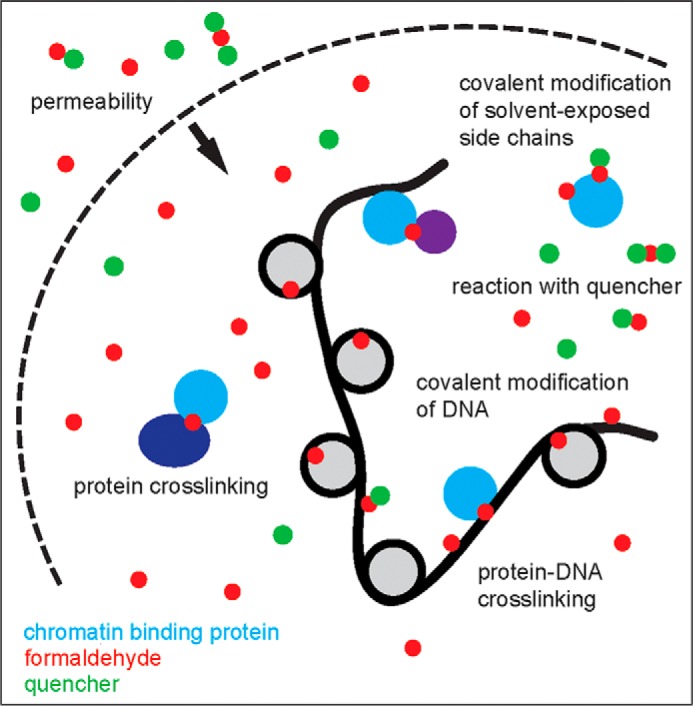
Graphic depicting the main aspects of formaldehyde reactivity in cells. The dashed arc represents cell or nuclear membranes, which are thought to be highly permeable to formaldehyde (red circles). The thick black curved line represents DNA, shown assembled as nucleosomes (light gray circles). A chromatin-interacting factor is schematized in cyan, with other partner proteins shown in dark blue and purple. Small molecules such as glycine and Tris that react with formaldehyde and can therefore quench reactivity with cellular constituents are shown as green circles. Formaldehyde can crosslink macromolecules together as well as modify exposed groups on macromolecules, forming a product species potentially stabilized by reactivity with a quencher. Quenchers are ordinarily added to the extracellular milieu and may exert their main effects outside the cell.
Basic Chemistry
Formaldehyde is the smallest aldehyde, an electrophilic molecule susceptible to chemical attack by a wide range of nucleophilic species of biological interest. The chemical complexity of formaldehyde-mediated reaction products was appreciated 70 years ago (5). Initially using amino acids, and subsequently proteins and other substrates, it was shown that formaldehyde reacts in vitro with a wide range of functional groups, forming a complex array of products (6, 7). It has been known since the 1940s that such products can include intramolecular and intermolecular crosslinked species and that the reaction conditions (e.g. pH, temperature) can strongly influence the nature, yield, and half-life of chemical modifications (8). The concentration of formaldehyde used, incubation times, and other conditions can vary substantially among different applications employing formaldehyde fixation, yielding very different chemical products (reviewed in Ref. 9).
Formaldehyde reacts with macromolecules in several steps (Fig. 2). In the first step, a nucleophilic group on an amino acid or DNA base (for example) forms a covalent bond with formaldehyde, resulting in a methylol adduct, which is then converted to a Schiff base. Methylols and Schiff bases can decompose rapidly (10–12) or may be stabilized in a second chemical step involving another functional group, often on another molecule, leading to formation of a methylene bridge (13). A methylene bridge might form between a solvent-exposed group on a macromolecule and a small molecule in solution such as glycine, which is frequently used as a formaldehyde quencher. Alternatively, and of most interest to biologists, is the formation of a covalent bond linking functional groups in two different macromolecules. The small size of formaldehyde dictates its linkage of groups that are ∼2 Å apart, making it well suited for capture of interactions between macromolecules that are in close proximity (14, 15). (Commercial preparations of formaldehyde may also contain formaldehyde aggregates (16) whose reactivities and distance-spanning capabilities are unclear.)
FIGURE 2.
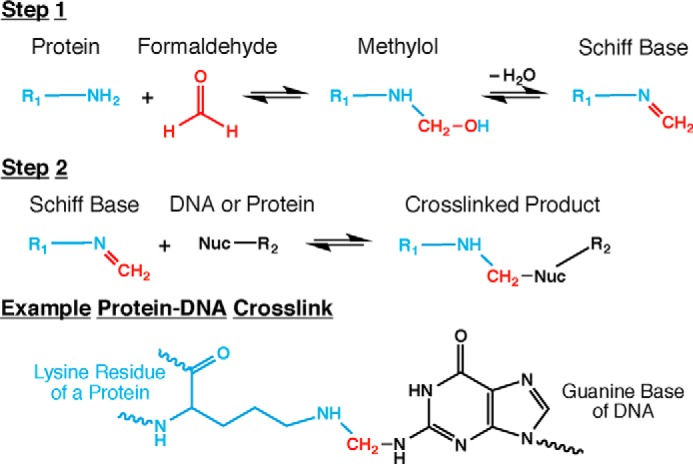
Chemical reactions occurring during formaldehyde crosslinking of biomolecules. Formaldehyde crosslinking of biomolecules occurs in two steps. First, formaldehyde reacts with a relatively strong nucleophile, most commonly a lysine ϵ-amino group from a protein. This reaction forms a methylol intermediate that can lose water to yield a Schiff base (an imine). Second, the Schiff base reacts with another nucleophile, possibly an amino group of a DNA base, to generate a crosslinked product. This second nucleophile might also be from another protein, the same protein as the first nucleophile, a quencher molecule, or another endogenous small molecule, and therefore a protein-DNA crosslink is only one of many possible products. All of the reactions in this two-step process are reversible, which is a key feature of formaldehyde crosslinking for chromatin capture. A specific example of a protein-DNA crosslink is shown. The atoms are color coded to match those of Fig. 1: cyan, protein; red, formaldehyde; and black, DNA.
Formaldehyde Reactivity with Proteins
As studies of formaldehyde reactivity became more sophisticated, it was found that conditions that more closely resemble those used for crosslinking components in cells yield a subset of the products identified in the earlier studies (17). Using model peptides, formaldehyde was found to react with N-terminal amino groups and side chains of cysteine, histidine, lysine, tryptophan, and arginine (10). Reaction products were in some cases influenced by the peptide sequence, yielding intramolecular crosslinks as well as linkages of the N terminus and histidine, asparagine, glutamine, tryptophan, tyrosine, and arginine residues to glycine molecules added to the reaction (10). Despite the long incubation time (48 h), adducts were not detected between glycine and peptide cysteine or lysine residues. Subsequent work employing model substrates along with formaldehyde concentrations and reaction times more in line with those used with cells identified a smaller subset of formaldehyde reaction products involving lysine, tryptophan, and cysteine side chains as well as the peptide N terminus (17). Such studies have often been motivated by interest in developing techniques for analysis of native protein complex subunit composition. As discussed in more detail below, the rapid reactivity of formaldehyde with cellular constituents suggests that cells are highly permeable to formaldehyde, and the requirement for crosslinked groups to be closely apposed makes formaldehyde a good candidate for capturing macromolecular complexes in vivo containing specific but unstably bound subunits, which can then be analyzed by mass spectrometry (18).
In discussing the complexity of crosslinked complexes formed by incubation of cells with formaldehyde, it is important to distinguish between two types of complexity. The first is the chemical complexity arising from the multiplicity of macromolecular functional groups that can potentially react with formaldehyde, and the second is the complexity associated with the types and numbers of macromolecules crosslinked to each other. Although formaldehyde can potentially generate a great variety of chemically distinct products in vitro, the biologically relevant chemical complexity is in all likelihood simpler under incubation conditions more typically used for analyses of macromolecular complexes in vivo. This is due to several factors, including a lowered effective formaldehyde concentration in cells when compared with most model experiments in vitro, limiting the ability of formaldehyde to locate and interact with a functional group. Although there is much greater macromolecular diversity in cells than in typical in vitro experiments, native macromolecules likely provide a smaller range of chemically reactive groups than model substrates used in vitro. As discussed below, N-terminal amino groups may be less available and side chains are less accessible to formaldehyde crosslinking due to protein tertiary structure in native proteins. These factors would decrease the proportion of potentially chemically reactive groups and allow for a smaller, less diverse set of chemical products in vivo. For instance, reactivity with native proteins is limited to those nucleophilic groups that are accessible to formaldehyde, and indeed, studies exploring differential formaldehyde reactivity have been used to provide insight into enzyme structure and catalytic function (19). Solvent-accessible lysine residues have been found to provide the most reactive functional groups in native proteins, and moreover, modification of native proteins by formaldehyde does not appear to perturb tertiary structure very much (20). This is consistent with early work in the chromatin field that established that lysine residues are the predominant sites of formation of methylene bridges in histone complexes; such studies led to the suggestion as well that formaldehyde crosslinking does not in general perturb protein structure (21). The apparent preference of formaldehyde for accessible lysine residues may explain in part why formaldehyde has emerged as the crosslinker of choice for trapping protein-DNA complexes, as lysine residues are common mediators of interactions with DNA (22). The differential reactivity of accessible groups on protein surfaces has also been explored to understand how formaldehyde fixation impacts epitope recognition by antibodies (23). Of note, the potential for formaldehyde to affect antibody recognition could possibly impact a wide range of experiments that require quantification of recovered fixed material by immunoprecipitation. Conditions can often be worked out such that formaldehyde treatment does not adversely impact antibody recognition (24, 25), but to our knowledge, this has not been examined in great detail in the chromatin field. Importantly, the apparent predominance of a subset of reactive sites on macromolecules under typical experimental conditions does not suggest that overall crosslinking complexity in cells is necessarily simple. Although in vivo crosslinking is probably predominated by a subset of the chemical products observed in vitro, there is potential for macromolecules to become crosslinked together in multiple ways and in multiple combinations, forming larger daisy-chained structures that complicate in vivo crosslinking results. Indeed, there is some evidence that formaldehyde treatment of cells can result in higher order chromatin or nuclear structures whose formation may yield misleading interpretations of chromatin association data by trapping factors within dense crosslinked networks (4, 26).
Formaldehyde Reactivity with DNA
Formaldehyde reacts with amino and imino groups of DNA bases, and extensive studies have been performed to document the kinetic and thermodynamic aspects of such reactions (11, 12, 27–29). Although formaldehyde reactivity with proteins does not appear to perturb protein tertiary structure, formaldehyde reactivity with DNA is notably different as covalent modification of DNA bases requires disruption of base pairing in duplex DNA, and in fact, formaldehyde was used in pioneering studies to probe DNA melting (30–34). Modified bases are thus precluded from base pairing and promote further DNA denaturation (30). This likely occurs to some extent in stretches of naked DNA in cells treated with formaldehyde, although under typical conditions employed for in vivo studies, the recovered DNA is by and large suitable for enzymatic manipulation (35). Formaldehyde modification of naked DNA in vitro may be more extensive (36). Conformational changes in DNA that promote formaldehyde reactivity have been referred to as DNA “breathing” or base flipping. Measurement of the rates of such spontaneous conformational changes is an active area of investigation (37), and it is unclear what specific DNA conformational changes are required to allow reaction of DNA bases with formaldehyde (i.e. full extrahelical extrusion of a DNA base may not be required). The rates of formaldehyde reactivity with naked DNA in vitro were found to be orders of magnitude below diffusion-limited rates, although these studies make it clear that reaction conditions can have large effects on reactivity. Indeed, it was recognized early on that it would be difficult to extrapolate rates of reaction obtained in relatively simple in vitro systems to other more complex systems, let alone in vivo (11).
Capture of Protein-DNA Complexes
The early use of formaldehyde as a probe of macromolecular structure led to the discovery that formaldehyde can crosslink histones to DNA (38). Retrieval of the crosslinked complexes and analysis of the associated DNA then gave birth to the ChIP assay (14, 39, 40), which has become ubiquitous in the chromatin field in a multitude of variations (41–49). Although ChIP assays performed without crosslinking have proven valuable for analyses of stable chromatin complexes (50), crosslinking has made it possible to identify interactions that would not otherwise withstand the isolation procedure. Given the central utility of crosslinking and its critical role in establishing many of the principles underlying the current understanding of chromatin structure and function, a clear picture of formaldehyde chemistry is critical to ensure that any biases resulting from formaldehyde crosslinking are taken into account.
The ability of formaldehyde to crosslink amino acids to DNA bases has been examined systematically in vitro. In comparing the products of reactions containing lysine, cysteine, histidine, or tryptophan with each of the four DNA bases, the highest yield of crosslinked product was obtained with lysine and deoxyguanosine (51), consistent with lysine being the most reactive among residues in native proteins as described above. Similar results were obtained using short peptides and trinucleotides (51). In the context of protein-DNA interactions, the first chemical step could involve reaction with an amino acid side chain in a protein, the protein N terminus, or an amino or imino group on a DNA base; importantly, however, the ϵ-amino group on the lysine side chain is a better nucleophile than are the amino/imino groups on DNA bases whose lone pair electrons are delocalized in the aromatic ring. For this reason, it seems reasonable to speculate that in most crosslinked protein-DNA complexes, a Schiff base is formed on a lysine residue first, followed by nucleophilic attack by the DNA base held in proximity to the side chain, resulting in a methylene bridge.
Interestingly, and in line with this idea, formaldehyde reactivity with DNA was stimulated substantially by adding amino acids or histones to an in vitro reaction, resulting in stable products that in some cases contained both DNA and the protein or amino acid (52). The ∼20–30-fold stimulation in the reaction rates observed in these early experiments by the addition of glycine or lysine (for example) was striking; furthermore, formaldehyde crosslinking of proximal functional groups on specific, stable macromolecular complexes presumably can occur even faster because of the constrained physical proximity of the reacting species (53). In addition to the ubiquity of lysine side chains in DNA-binding proteins (for interaction with the phosphate backbone), the DNA bases provide a high density of amino and imino groups along the length of the nucleic acid. These two features may contribute to the relatively higher yield of protein-DNA crosslinks when compared with protein-protein crosslinks as measured by conjugation of chromatin regulatory complexes that interact indirectly with DNA (54). It has been observed that for some transcriptional co-regulators, protein-protein crosslinks are not efficiently detected between factors that interact with chromatin indirectly when using chromosome conformation capture (3C) methods; this could be due to either inefficiencies in formaldehyde crosslinking between proteins (due to non-optimal reactive side chain availability) or the rarity of these protein-protein interactions (4).
Several different crosslinking agents have been used in ChIP (55), but all of the features described above, including cell permeability, short spacer length, rapid reactivity, as well as reversibility (discussed below), have led to formaldehyde becoming the crosslinking agent of choice for ChIP. This utility has been borne out by many genome-wide studies that have shown how profiles of crosslinked complexes capture transcription factor binding to physiologically significant DNA sites (for example, see Refs. 56–58). Because DNA site-specific transcription factors can also bind to nonspecific sites (59–62), crosslinking of non-specifically bound proteins to DNA would be expected to occur and may account in part for binding events detected in genome-wide studies that cannot be readily explained physiologically. Non-DNA-binding proteins are not crosslinked to chromatin (14, 63), and non-specifically bound factors are presumably bound to a multitude of disparate sites at low levels consistent with their relative occupancies (64, 65). Analyses of chromatin binding by a series of mutants in the methyl CpG-binding protein 2 gene led to the conclusion that there is a threshold interaction lifetime of about 5 s required for crosslinking (26). However, it has been possible to perform ChIP on transcription factors whose interactions with chromatin are known from imaging studies to be highly transient (time scale of a few seconds) (61, 66–69). Importantly, in vivo ChIP signals have been found to correlate with DNA binding specificities and affinities measured in vitro (70, 71), supporting the use of formaldehyde for measuring chromatin binding interactions in cells with quantitative rigor and over a broad thermodynamic range. A better understanding of formaldehyde's effects in cells could potentially be obtained by biochemical studies of protein-DNA complex crosslinking in vitro. However, it is noteworthy that there are examples in which in vitro and in vivo binding behaviors differ when assessed using formaldehyde in one system or another (14, 55, 70).
As mentioned above, formaldehyde crosslinking can be used to isolate physiologically relevant complexes for analysis by mass spectrometry (9, 18). Formaldehyde crosslinking has been exploited to identify constituents of some cellular complexes, including the proteasome, for example, (72, 73), and to identify protein-protein interactions in tissues (74). Unbiased efforts to identify proteins associated with specific DNA sequences in vivo have employed formaldehyde to stabilize such complexes during their isolation. It is technically challenging to obtain sufficient material for identification of the DNA-associated proteins by mass spectrometry, but this strategy has nonetheless been successfully employed for repeated DNA sequences such as telomeres (75–77) and in model in vitro systems (36), as well as for single copy and multicopy regions in yeast (75, 76, 78–80). The relatively mild formaldehyde reaction conditions used to maximize crosslinking of physiologically relevant associations and avoid spurious ones may yield a relatively low proportion of formaldehyde crosslinked material and thereby contribute to the technical challenges of this kind of approach (36).
Crosslinking Kinetics, Stability, and Reversal
Formaldehyde crosslinking in chromatin studies typically employs relatively low formaldehyde concentrations (1% or less). The facile detection of protein-DNA complexes following incubation times of 30 min or less suggests that macromolecular crosslinking occurs relatively rapidly, as suggested by the earliest ChIP experiments (14, 40, 81). Relatively rapid formaldehyde reactivity in cells is also consistent with the ability to distinguish ChIP signals over short time intervals (seconds to minutes) (63, 82, 83). Formaldehyde crosslinks are quite stable in vivo when compared with the durations of most crosslinking experiments, with crosslink half-lives of ∼10–20 h depending on the cell type and conditions (15). In ChIP experiments, crosslinks are most often reversed by heat (21). The reversibility of formaldehyde crosslinking has been explored in some detail in an effort to recover proteins from fixed tissue and cell samples (84, 85). The temperature and salt concentration dependence of the formaldehyde crosslink reversal rate has been established, revealing a crosslink half-life consistent with the estimate of crosslink half-life in cells (tens of hours at 37 °C) (86). That study also quantitatively showed the extent to which heat can increase the crosslink reversal rate. More such measurements on other aspects of crosslinking chemistry will be useful in developing a quantitative understanding of how the abundance of a particular crosslinked species obtained under some set of conditions (i.e. the effect of pH, quencher choice and concentration, and formaldehyde concentration) relates to dynamic aspects of complex assembly/disassembly and stability (83).
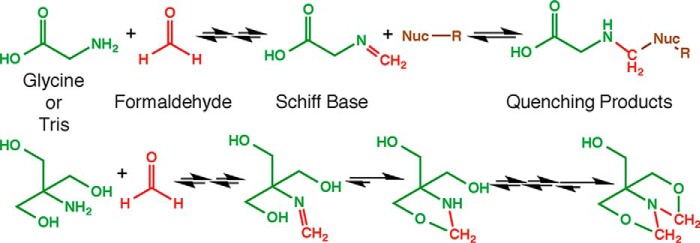
To limit formaldehyde reactivity to a particular time interval, unreacted formaldehyde is quenched with an excess of a small reactive molecule added to the reaction (Fig. 3). Quenching is important but not well understood (9). Glycine has been typically used as a sink for unreacted formaldehyde in ChIP (44, 55) as well as in approaches to map higher order chromatin structure (87–90). The efficacy of glycine is improved by reduced pH, but detailed studies of the quenching reaction have not been reported (9). In principle, formaldehyde crosslinking could be quenched by reaction of the quencher with formaldehyde molecules in solution or reaction with formaldehyde conjugates on other molecules in the cell, if the quencher is readily cell-permeable. As discussed above, formaldehyde-mediated glycine conjugates have been detected or inferred in vitro, although there was no evidence for such conjugates seen in proteins analyzed from formaldehyde-treated cells (9, 25, 83). On the other hand, evidence suggests that glycine-DNA conjugates are formed in an in vitro reaction (36). Despite the fact that glycine has been used routinely to quench crosslinking, Tris is a more efficient quencher (9), which can be explained chemically by the ability of Tris to form a cyclic product upon reaction with formaldehyde (36) (Fig. 3). However, at higher concentrations of Tris, which would likely be used for quenching, Tris can also facilitate crosslink reversal (91), thereby potentially impacting the yield of crosslinked material.
FIGURE 3.
Formaldehyde quenching reactions with glycine and Tris, the two most common quenchers. The chemical reactions are analogous to those shown in Fig. 2 with the amino group of glycine or Tris acting as the primary nucleophile. The Schiff base formed from glycine may or may not react with a second nucleophile, but regardless, the crosslinking between macromolecules has been quenched. The Tris molecule has readily available second nucleophiles (hydroxyl groups) that create stable intramolecular five-membered rings. It is also possible for Tris to react with two formaldehyde molecules, leading to the final product shown. The propensity for Tris to form these stable intramolecular products likely allows it to scavenge formaldehyde from other molecules and thereby facilitate crosslink reversal. The atoms are color-coded: green, quencher; red, formaldehyde; brown, miscellaneous nucleophile.
Complex Effects in the Cellular Milieu
Given the pivotal role of this reagent in understanding chromatin biology and the continuing evolution of technologies exploiting its properties, it is critical that a deeper understanding is achieved of formaldehyde crosslinking as it occurs in cells. Although formaldehyde can mediate myriad chemical reactions in vitro, the conditions used for crosslinking in cells suggest that, with respect to chromatin, the chemical complexity of macromolecule-containing reaction products is more limited, with reactions occurring mainly with solvent-exposed lysine residues and endo- and exocyclic amino groups on bases. Formaldehyde has a number of other properties that make it well suited for trapping macromolecular complexes in cells, including cell permeability and the temperature-dependent stability of methylene bridge-containing adducts. Macromolecules that do not interact are in general not crosslinked together efficiently, and methylol/Schiff base intermediates are reversibly formed and appear to be inefficiently trapped by reaction with quenchers in cells. This explains why proteins and DNA isolated from formaldehyde-treated cells appear unmodified in general (14, 83, 92). Within minutes of formaldehyde incubation, there is very little detectable free DNA (<10%) (36, 86), and crosslinking appears to occur uniformly along DNA as well (14).
ChIP has been developed by empirical determination of seemingly optimal crosslinking conditions, with low recoveries occurring for either too little or too much crosslinking (93). If the formaldehyde concentration is too low or the incubation time is too short, not enough crosslinked material will be produced. On the other hand, a formaldehyde concentration that is too high or an incubation time that is too long also reduces recovery, presumably reflecting the formation of complexes that are insoluble or the masking of epitopes recognized by the antibody used for immunoprecipitation. We have observed little effect of formaldehyde incubation time on chromatin protein yield over a broad range of formaldehyde concentrations and incubation times (92), but elevated formaldehyde concentrations can impact yield even after moderate incubation times, suggesting the formation of such complexes. Given the dense concentration of macromolecules in the nucleus, it is plausible that formaldehyde may cause the formation of higher order networks of crosslinked chromatin (4), as illustrated in Fig. 4. This is an area worthy of additional investigation, particularly because it may explain nonspecific DNA crosslinking that occurs in ChIP or ChIP-related methods (94). Conversely, other spurious enrichment phenomena, such as localization of unrelated proteins, can occur with ChIP at highly expressed genes; these, too, warrant deeper investigation (95, 96) to ensure that apparent ChIP signals in fact represent true association with the loci of interest.
FIGURE 4.
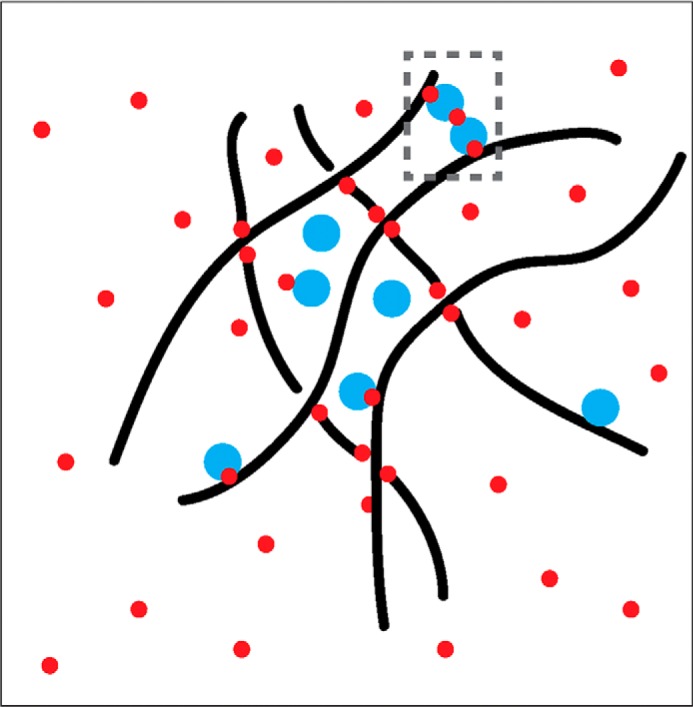
Potential effects of formaldehyde in mediating formation of higher order chromatin structures. The black wavy lines denote chromatin fibers, which may become a crosslinked meshwork in the presence of formaldehyde (red circles). The formation of these potentially confounding structures may or may not be mediated by physiologically relevant higher order interactions captured by crosslinking (dashed gray rectangle). Such a meshwork may define localized neighborhoods in the nucleus that trap proteins (cyan) that may or may not interact specifically with nearby DNA sequences in an unperturbed cell.
ChIP assays have been pivotal in establishing our current view of chromatin structure and function. As answers to deeper questions about chromatin binding dynamics and higher order structure are pursued, we feel it is imperative that we understand more fully how the procedures employed to obtain snapshots of chromatin state may perturb the very properties being measured, particularly as variations in experimental design can yield different interpretations (4, 97). In this regard, an understanding of the behavior and properties of formaldehyde in the cell is important for determining the best methods for measuring dynamic interactions using crosslinking. A better understanding of formaldehyde crosslinking may in turn lead to better quantitative models (98).
Footnotes
This work was supported by National Institutes of Health Grants R01 GM55763 (to D. T. A.) and P50 HG004952 (to L. M. S. and B. L. F.) and by NCI Cancer Training Grant T32 CA009109-38 (to E. A. H.). This is the second article in the Thematic Minireview series “Protein Interactions, Structures, and Networks.” The authors declare that they have no conflicts of interest with the contents of this article.
References
- 1.Eckels K. H., and Putnak R. (2003) Formalin-inactivated whole virus and recombinant subunit flavivirus vaccines. Adv. Virus Res. 61, 395–418 [DOI] [PubMed] [Google Scholar]
- 2.Nencioni L., Volpini G., Peppoloni S., Bugnoli M., De Magistris T., Marsili I., and Rappuoli R. (1991) Properties of pertussis toxin mutant PT-9K/129G after formaldehyde treatment. Infect. Immun. 59, 625–630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Werner M., Chott A., Fabiano A., and Battifora H. (2000) Effect of formalin tissue fixation and processing on immunohistochemistry. Am. J. Surg. Pathol. 24, 1016–1019 [DOI] [PubMed] [Google Scholar]
- 4.Gavrilov A., Razin S. V., and Cavalli G. (2015) In vivo formaldehyde cross-linking: it is time for black box analysis. Brief. Funct. Genomics 14, 163–165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fraenkel-Conrat H., Cooper M., and Olcott H. S. (1945) The reaction of formaldehyde with proteins. J. Am. Chem. Soc. 67, 950–954 [Google Scholar]
- 6.Fraenkel-Conrat H., and Olcott H. S. (1948) The reaction of formaldehyde with proteins. V. Cross-linking between amino and primary amide or guanidyl groups. J. Am. Chem. Soc. 70, 2673–2684 [DOI] [PubMed] [Google Scholar]
- 7.Fraenkel-Conrat H., and Olcott H. S. (1948) Reaction of formaldehyde with proteins. VI. Cross-linking of amino groups with phenol, imidazole, or indole groups. J. Biol. Chem. 174, 827–843 [PubMed] [Google Scholar]
- 8.French D., and Edsall J. T. (1945) The reactions of formaldehyde with amino acids and proteins. Adv. Protein Chem. 2, 277–335 [Google Scholar]
- 9.Sutherland B. W., Toews J., and Kast J. (2008) Utility of formaldehyde cross-linking and mass spectrometry in the study of protein-protein interactions. J. Mass Spectrom. 43, 699–715 [DOI] [PubMed] [Google Scholar]
- 10.Metz B., Kersten G. F. A., Hoogerhout P., Brugghe H. F., Timmermans H. A. M., de Jong A., Meiring H., ten Hove J., Hennink W. E., Crommelin D. J. A., and Jiskoot W. (2004) Identification of formaldehyde-induced modifications in proteins: reactions with model peptides. J. Biol. Chem. 279, 6235–6243 [DOI] [PubMed] [Google Scholar]
- 11.McGhee J. D., and von Hippel P. H. (1975) Formaldehyde as a probe of DNA Structure. I. Reaction with exocyclic amino groups of DNA Bases. Biochemistry 14, 1281–1296 [DOI] [PubMed] [Google Scholar]
- 12.McGhee J. D., and von Hippel P. H. (1975) Formaldehyde as a probe of DNA structure. II. Reaction with endocyclic imino groups of DNA bases. Biochemistry 14, 1297–1303 [DOI] [PubMed] [Google Scholar]
- 13.Feldman M. Y. (1973) Reactions of nucleic acids and nucleoproteins with formaldehyde. Prog. Nucleic Acid Res. Mol. Biol. 13, 1–49 [DOI] [PubMed] [Google Scholar]
- 14.Solomon M. J., and Varshavsky A. (1985) Formaldehyde-mediated DNA-protein crosslinking: a probe for in vivo chromatin structures. Proc. Natl. Acad. Sci. U.S.A. 82, 6470–6474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Quievryn G., and Zhitkovich A. (2000) Loss of DNA-protein crosslinks from formaldehyde-exposed cells occurs through spontaneous hydrolysis and an active repair process linked to proteosome function. Carcinogenesis 21, 1573–1580 [PubMed] [Google Scholar]
- 16.Hahnenstein I., Albert M., Hasse H., Kreiter C. G., and Maurer G. (1995) NMR spectroscopic and densimetric study of reaction kinetics of formaldehyde polymer formation in water, deuterium oxide, and methanol. Ind. Eng. Chem. Res. 34, 440–450 [Google Scholar]
- 17.Toews J., Rogalski J. C., Clark T. J., and Kast J. (2008) Mass spectrometric identification of formaldehyde-induced peptide modifications under in vivo protein cross-linking conditions. Anal. Chim. Acta 618, 168–183 [DOI] [PubMed] [Google Scholar]
- 18.Klockenbusch C., O'Hara J. E., and Kast J. (2012) Advancing formaldehyde cross-linking toward quantitative proteomic applications. Anal. Bioanal. Chem. 404, 1057–1067 [DOI] [PubMed] [Google Scholar]
- 19.Means G. E., and Feeney R. E. (1995) Reductive alkylation of proteins. Anal. Biochem. 224, 1–16 [DOI] [PubMed] [Google Scholar]
- 20.Toews J., Rogalski J. C., and Kast J. (2010) Accessibility governs the relative reactivity of basic residues in formaldehyde-induced protein modifications. Anal. Chim. Acta 676, 60–67 [DOI] [PubMed] [Google Scholar]
- 21.Jackson V. (1978) Studies on histone organization in the nucleosome using formaldehyde as a reversible cross-linking agent. Cell 15, 945–954 [DOI] [PubMed] [Google Scholar]
- 22.Rohs R., Jin X., West S. M., Joshi R., Honig B., and Mann R. S. (2010) Origins of specificity in protein-DNA recognition. Ann. Rev. Biochem. 79, 233–269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vani K., Bogen S. A., and Sompuram S. R. (2006) A high throughput combinatorial library technique for identifying formalin-sensitive epitopes. J. Immunol. Methods 317, 80–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Klockenbusch C., and Kast J. (2010) Optimization of formaldehyde cross-linking for protein interaction analysis on non-tagged integrin β1. J. Biomed. Biotechnol. 2010, 927585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vasilescu J., Guo X., and Kast J. (2004) Identification of protein-protein interactions using in vivo cross-linking and mass spectrometry. Proteomics 4, 3845–3854 [DOI] [PubMed] [Google Scholar]
- 26.Schmiedeberg L., Skene P., Deaton A., and Bird A. (2009) A temporal threshold for formaldehyde crosslinking and fixation. PLoS ONE 4, e4636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fraenkel-Conrat H. (1954) Reaction of nucleic acid with formaldehyde. Biochim. Biophys. Acta 15, 307–309 [DOI] [PubMed] [Google Scholar]
- 28.Haselkorn R., and Doty P. (1961) The reaction of formaldehyde with polynucleotides. J. Biol. Chem. 236, 2738–2745 [PubMed] [Google Scholar]
- 29.Chang Y.-T., and Loew G. H. (1994) Reaction mechanisms of formaldehyde with endocyclic imino groups of nucleic acid bases. J. Am. Chem. Soc. 116, 3548–3555 [Google Scholar]
- 30.Utiyama H., and Doty P. (1971) Kinetic studies of denaturation and reaction with formaldehyde on polydeoxyribonucleotides. Biochemistry 10, 1254–1264 [DOI] [PubMed] [Google Scholar]
- 31.Von Hippel P. H., and Wong K.-Y. (1971) Dynamic aspects of native DNA structure: Kinetics of the formaldehyde reaction with calf thymus DNA. J. Mol. Biol. 61, 587–613 [DOI] [PubMed] [Google Scholar]
- 32.Shikama K., and Miura K.-I. (1976) Equilibrium studies on the formaldehyde reaction with native DNA. Eur. J. Biochem. 63, 39–46 [DOI] [PubMed] [Google Scholar]
- 33.McGhee J. D., and von Hippel P. H. (1977) Formaldehyde as a probe of DNA structure. 3. Equilibrium denaturation of DNA and synthetic polynucleotides. Biochemistry 16, 3267–3276 [DOI] [PubMed] [Google Scholar]
- 34.McGhee J. D., and von Hippel P. H. (1977) Formaldehyde as a probe of DNA structure. 4. Mechanism of the initial reaction of formaldehyde with DNA. Biochemistry 16, 3276–3293 [DOI] [PubMed] [Google Scholar]
- 35.Gavrilov A., and Razin S. V. (2009) Formaldehyde fixation of cells does not greatly reduce the ability to amplify cellular DNA. Anal. Biochem. 390, 94–96 [DOI] [PubMed] [Google Scholar]
- 36.Wu C.-H., Chen S., Shortreed M. R., Kreitinger G. M., Yuan Y., Frey B. L., Zhang Y., Mirza S., Cirillo L. A., Olivier M., and Smith L. M. (2011) Sequence-specific capture of protein-DNA complexes for mass spectrometric protein identification. PLoS ONE 6, e26217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yin Y., Yang L., Zheng G., Gu C., Yi C., He C., Gao Y. Q., and Zhao X. S. (2014) Dynamics of spontaneous flipping of a mismatched base in DNA duplex. Proc. Natl. Acad. Sci. U.S.A. 111, 8043–8048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Brutlag D., Schlehuber C., and Bonner J. (1969) Properties of formaldehyde-treated nucleohistone. Biochemistry 8, 3214–3218 [DOI] [PubMed] [Google Scholar]
- 39.Gilmour D. S., and Lis J. T. (1985) In vivo interactions of RNA polymerase II with genes of Drosophila melanogaster. Mol. Cell. Biol. 5, 2009–2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Solomon M. J., Larsen P. L., and Varshavsky A. (1988) Mapping protein-DNA interactions in vivo with formaldehyde: evidence that histone H4 is retained on a highly transcribed gene. Cell 53, 937–947 [DOI] [PubMed] [Google Scholar]
- 41.Collas P. (2010) The current state of chromatin immunoprecipitation. Mol. Biotechnol. 45, 87–100 [DOI] [PubMed] [Google Scholar]
- 42.Aparicio O. M., Geisberg J. V., Sekinger E. A., Yang A. S., Moqtaderi Z., and Struhl K. (2005) Chromatin immunoprecipitation for determining the association of proteins with specific genomic sequences in vivo. Curr. Protoc. Mol. Biol. Chapter 21, Unit 21.3, 10.1002/0471142727.mb2103s69 [DOI] [PubMed] [Google Scholar]
- 43.O'Neill L. P., and Turner B. M. (1996) Immunoprecipitation of chromatin. Methods Enzymol. 274, 189–197 [DOI] [PubMed] [Google Scholar]
- 44.Kuo M. H., and Allis C. D. (1999) In vivo cross-linking and immunoprecipitation for studying dynamic protein:DNA associations in a chromatin environment. Methods 19, 425–433 [DOI] [PubMed] [Google Scholar]
- 45.Park P. J. (2009) ChIP-seq: advantages and challenges of a maturing technology. Nat. Rev. Genetics 10, 669–680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kim T. H., and Ren B. (2006) Genome-wide analysis of protein-DNA Interactions. Annu. Rev. Genomics Hum. Genet. 7, 81–102 [DOI] [PubMed] [Google Scholar]
- 47.Rhee H. S., and Pugh B. F. (2012) ChIP-exo method for identifying genomic location of DNA-binding proteins with near-single-nucleotide accuracy. Curr. Protoc. Mol. Biol. Chapter 21, Unit 21.24, 10.1002/0471142727.mb2124s100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schmidt D., Wilson M. D., Spyrou C., Brown G. D., Hadfield J., and Odom D. T. (2009) ChIP-seq: using high-throughput sequencing to discover protein-DNA interactions. Methods 48, 240–248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Komashko V. M., Acevedo L. G., Squazzo S. L., Iyengar S. S., Rabinovich A., O'Geen H., Green R., and Farnham P. J. (2008) Using ChIP-chip technology to reveal common principles of transcriptional repression in normal and cancer cells. Genome Res. 18, 521–532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kasinathan S., Orsi G. A., Zentner G. E., Ahmad K., and Henikoff S. (2014) High-resolution mapping of transcription factor binding sites on native chromatin. Nat. Methods 11, 203–209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lu K., Ye W., Zhou L., Collins L. B., Chen X., Gold A., Ball L. M., and Swenberg J. A. (2010) Structural characterization of formaldehyde-induced cross-links between amino acids and deoxynucleosides and their oligomers. J. Am. Chem. Soc. 132, 3388–3399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Siomin Y. A., Simonov V. V., and Poverenny A. M. (1973) The reaction of formaldehyde with deoxynucleotides and DNA in the presence of amino acids and lysine-rich histone. Biochim. Biophys. Acta 331, 27–32 [DOI] [PubMed] [Google Scholar]
- 53.Jencks W. P. (1997) From chemistry to biochemistry to catalysis to movement. Annu. Rev. Biochem. 66, 1–18 [DOI] [PubMed] [Google Scholar]
- 54.Zeng P.-Y., Vakoc C. R., Chen Z.-C., Blobel G. A., and Berger S. L. (2006) In vivo dual cross-linking for identification of indirect DNA-associated proteins by chromatin immunoprecipitation. BioTechniques 41, 694–698 [DOI] [PubMed] [Google Scholar]
- 55.Nowak D. E., Tian B., and Brasier A. R. (2005) Two-step cross-linking method for identification of NF-κB gene network by chromatin immunoprecipitation. BioTechniques 39, 715–725 [DOI] [PubMed] [Google Scholar]
- 56.Harbison C. T., Gordon D. B., Lee T. I., Rinaldi N. J., Macisaac K. D., Danford T. W., Hannett N. M., Tagne J.-B., Reynolds D. B., Yoo J., Jennings E. G., Zeitlinger J., Pokholok D. K., Kellis M., Rolfe P. A., Takusagawa K. T., Lander E. S., Gifford D. K., Fraenkel E., and Young R. A. (2004) Transcriptional regulatory code of a eukaryotic genome. Nature 431, 99–104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Venters B. J., Wachi S., Mavrich T. N., Andersen B. E., Jena P., Sinnamon A. J., Jain P., Rolleri N. S., Jiang C., Hemeryck-Walsh C., and Pugh B. F. (2011) A comprehensive genomic binding map of gene and chromatin regulatory proteins in Saccharomyces. Mol. Cell 41, 480–492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ren B., Robert F., Wyrick J. J., Aparicio O., Jennings E. G., Simon I., Zeitlinger J., Schreiber J., Hannett N., Kanin E., Volkert T. L., Wilson C. J., Bell S. P., and Young R. A. (2000) Genome-wide location and function of DNA binding proteins. Science 290, 2306–2309 [DOI] [PubMed] [Google Scholar]
- 59.Hammar P., Leroy P., Mahmutovic A., Marklund E. G., Berg O. G., and Elf J. (2012) The lac repressor displays facilitated diffusion in living cells. Science 336, 1595–1598 [DOI] [PubMed] [Google Scholar]
- 60.Elf J., Li G. W., and Xie X. S. (2007) Probing transcription factor dynamics at the single-molecule level in a living cell. Science 316, 1191–1194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hager G. L., McNally J. G., and Misteli T. (2009) Transcription dynamics. Mol. Cell 35, 741–753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mirny L., Slutsky M., Wunderlich Z., Tafvizi A., Leith J., and Kosmrlj A. (2009) How a protein searches for its site on DNA: the mechanism of facilitated diffusion. J. Phys. A: Math. Theor. 42, 434013 [Google Scholar]
- 63.Hall D. B., and Struhl K. (2002) The VP16 activation domain interacts with multiple transcriptional components as determined by protein-protein crosslinking in vivo. J. Biol. Chem. 277, 46043–46050 [DOI] [PubMed] [Google Scholar]
- 64.Buck M. J., and Lieb J. D. (2004) ChIP-chip: Considerations for the design, analysis, and application of genome-wide chromatin immunoprecipitation experiments. Genomics 83, 349–360 [DOI] [PubMed] [Google Scholar]
- 65.Struhl K. (2007) Interpreting chromatin immunoprecipitation experiments. in Evaluating Techniques in Biochemical Research (Zuk D., ed.) pp. 29–33, Cell Press, Cambridge, MA [Google Scholar]
- 66.Mazza D., Mueller F., Stasevich T. J., and McNally J. G. (2013) Convergence of chromatin binding estimates in live cells. Nat. Methods 10, 691–692 [DOI] [PubMed] [Google Scholar]
- 67.Chen J., Zhang Z., Li L., Chen B.-C., Revyakin A., Hajj B., Legant W., Dahan M., Lionnet T., Betzig E., Tjian R., and Liu Z. (2014) Single-molecule dynamics of enhanceosome assembly in embryonic stem cells. Cell 156, 1274–1285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Stasevich T. J., Hayashi-Takanaka Y., Sato Y., Maehara K., Ohkawa Y., Sakata-Sogawa K., Tokunaga M., Nagase T., Nozaki N., McNally J. G., and Kimura H. (2014) Regulation of RNA polymerase II activation by histone acetylation in single living cells. Nature 516, 272–275 [DOI] [PubMed] [Google Scholar]
- 69.Voss T. C., and Hager G. L. (2008) Visualizing chromatin dynamics in intact cells. Biochim. Biophys. Acta 1783, 2044–2051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Toth J., and Biggin M. D. (2000) The specificity of protein–DNA crosslinking by formaldehyde: in vitro and in Drosophila embryos. Nucleic Acids Res. 28, e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kaplan T., Li X.-Y., Sabo P. J., Thomas S., Stamatoyannopoulos J. A., Biggin M. D., and Eisen M. B. (2011) Quantitative models of the mechanisms that control genome-wide patterns of transcription factor binding during early Drosophila development. PLoS Genet. 7, e1001290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Guerrero C., Tagwerker C., Kaiser P., and Huang L. (2006) An integrated mass spectrometry-based proteomic approach: quantitative analysis of tandem affinity-purified in vivo cross-linked protein complexes (qtax) to decipher the 26s proteasome-interacting network. Mol. Cell. Proteomics 5, 366–378 [DOI] [PubMed] [Google Scholar]
- 73.Tagwerker C., Flick K., Cui M., Guerrero C., Dou Y., Auer B., Baldi P., Huang L., and Kaiser P. (2006) A tandem affinity tag for two-step purification under fully denaturing conditions: application in ubiquitin profiling and protein complex identification combined with in vivo cross-Linking. Mol. Cell. Proteomics 5, 737–748 [DOI] [PubMed] [Google Scholar]
- 74.Schmitt-Ulms G., Hansen K., Liu J., Cowdrey C., Yang J., DeArmond S. J., Cohen F. E., Prusiner S. B., and Baldwin M. A. (2004) Time-controlled transcardiac perfusion cross-linking for the study of protein interactions in complex tissues. Nat. Biotechnol. 22, 724–731 [DOI] [PubMed] [Google Scholar]
- 75.Déjardin J., and Kingston R. E. (2009) Purification of proteins associated with specific genomic loci. Cell 136, 175–186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kennedy-Darling J., Guillen-Ahlers H., Shortreed M. R., Scalf M., Frey B. L., Kendziorski C., Olivier M., Gasch A. P., and Smith L. M. (2014) Discovery of chromatin-associated proteins via sequence-specific capture and mass spectrometric protein identification in Saccharomyces cerevisiae. J. Proteome Res. 13, 3810–3825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Antão J. M., Mason J. M., Déjardin J., and Kingston R. E. (2012) Protein landscape at Drosophila melanogaster telomere-associated sequence repeats. Mol. Cell. Biol. 32, 2170–2182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Byrum S. D., Raman A., Taverna S. D., and Tackett A. J. (2012) ChAP-MS: a method for identification of proteins and histone posttranslational modifications at a single genomic locus. Cell Rep. 2, 198–205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Byrum S. D., Taverna S. D., and Tackett A. J. (2013) Purification of a specific native genomic locus for proteomic analysis. Nucleic Acids Res. 41, e195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Byrum S. D., Taverna S. D., and Tackett A. J. (2015) Purification of specific chromatin loci for proteomic analysis. Methods Mol. Biol. 1228, 83–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Dedon P. C., Soults J. A., Allis C. D., and Gorovsky M. A. (1991) Formaldehyde cross-linking and immunoprecipitation demonstrate developmental changes in H1 association with transcriptionally active genes. Mol. Cell. Biol. 11, 1729–1733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Katan-Khaykovich Y., and Struhl K. (2002) Dynamics of global histone acetylation and deacetylation in vivo: rapid restoration of normal histone acetylation status upon removal of activators and repressors. Genes Dev. 16, 743–752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Poorey K., Viswanathan R., Carver M. N., Karpova T. S., Cirimotich S. M., McNally J. G., Bekiranov S., and Auble D. T. (2013) Measuring chromatin interaction dynamics on the second time scale at single-copy genes. Science 342, 369–372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Shi S.-R., Taylor C. R., Fowler C. B., and Mason J. T. (2013) Complete solubilization of formalin-fixed, paraffin-embedded tissue may improve proteomic studies. Proteomics Clin. Appl. 7, 264–272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Wiśniewski J. R., Duś K., and Mann M. (2013) Proteomic workflow for analysis of archival formalin-fixed and paraffin-embedded clinical samples to a depth of 10,000 proteins. Proteomics Clin. Appl. 7, 225–233 [DOI] [PubMed] [Google Scholar]
- 86.Kennedy-Darling J., and Smith L. M. (2014) Measuring the formaldehyde protein-DNA cross-link reversal rate. Anal. Chem. 86, 5678–5681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.de Wit E., and de Laat W. (2012) A decade of 3C technologies: insights into nuclear organization. Genes Dev. 26, 11–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Splinter E., Grosveld F., and de Laat W. (2004) 3C technology: analyzing the spatial organization of genomic loci in vivo. Methods Enzymol. 375, 493–507 [DOI] [PubMed] [Google Scholar]
- 89.Hagège H., Klous P., Braem C., Splinter E., Dekker J., Cathala G., de Laat W., and Forné T. (2007) Quantitative analysis of chromosome conformation capture assays (3C-qPCR). Nat. Protoc. 2, 1722–1733 [DOI] [PubMed] [Google Scholar]
- 90.Miele A., and Dekker J. (2009) Mapping cis- and trans-chromatin interaction networks using chromosome conformation capture (3C). Methods Mol. Biol. 464, 105–121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Kawashima Y., Kodera Y., Singh A., Matsumoto M., and Matsumoto H. (2014) Efficient extraction of proteins from formalin-fixed paraffin-embedded tissues requires higher concentration of tris(hydroxymethyl)aminomethane. Clin. Proteomics 11, 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Viswanathan R., Hoffman E. A., Shetty S. J., Bekiranov S., and Auble D. T. (2014) Analysis of chromatin binding dynamics using the crosslinking kinetics (CLK) method. Methods 70, 97–107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Orlando V. (2000) Mapping chromosomal proteins in vivo by formaldehyde-crosslinked-chromatin immunoprecipitation. Trends Biochem. Sci. 25, 99–104 [DOI] [PubMed] [Google Scholar]
- 94.Fan X., Lamarre-Vincent N., Wang Q., and Struhl K. (2008) Extensive chromatin fragmentation improves enrichment of protein binding sites in chromatin immunoprecipitation experiments. Nucleic Acids Res. 36, e125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Fan X., and Struhl K. (2009) Where does Mediator bind in vivo? PLoS ONE 4, e5029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Teytelman L., Thurtle D. M., Rine J., and van Oudenaarden A. (2013) Highly expressed loci are vulnerable to misleading ChIP localization of multiple unrelated proteins. Proc. Natl. Acad. Sci. U.S.A. 110, 18602–18607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Nacheva G. A., Guschin D. Y., Preobrazhenskaya O. V., Karpov V. L., Ebralidse K. K., and Mirzabekov A. D. (1989) Change in the pattern of histone binding to DNA upon transcriptional activation. Cell 58, 27–36 [DOI] [PubMed] [Google Scholar]
- 98.Voit E. O., Martens H. A., and Omholt S. W. (2015) 150 years of the mass action law. PLoS Comput. Biol. 11, e1004012. [DOI] [PMC free article] [PubMed] [Google Scholar]