Background: Posttranslational modifications of E2F1 alter its transcriptional activity.
Results: UCH37 binds E2F1 and deubiquitinates its UbK63-specific linkages to enhance the transcriptional activation of E2F1 target genes, particularly upon DNA damage.
Conclusion: Ubiquitination and deubiquitination of UbK63-specific linkages provide an additional layer of regulation for E2F1 transcriptional activity.
Significance: UCH37 is the first known deubiquitinating enzyme to directly regulate E2F1.
Keywords: apoptosis, deubiquitylation (deubiquitination), DNA damage, transcription regulation, ubiquitylation (ubiquitination), E2F1, UCH37/UCHL5
Abstract
E2F1 is tightly controlled by multiple mechanisms, but whether ubiquitination regulates its transcriptional activity remains unknown. Here we identify UCH37 as the first, to our knowledge, deubiquitinating enzyme for E2F1. UCH37 does not deubiquitinate UbK48 chains or affect E2F1 protein stability. Instead, UCH37, but not a catalytically dead mutant, decreases the Lys-63-linked ubiquitination of E2F1 and activates its transcriptional activity. UCH37 depletion reduces the gene expression of both proliferative and pro-apoptotic E2F1 target genes. UCH37 depletion also decreases both cell proliferation and apoptosis induction in functional assays. Interestingly, UCH37 expression is induced by E2F1, and its level rises in G1/S transition and S phase, suggesting a positive feedback loop between UCH37 and E2F1. UCH37 protein and mRNA levels are also induced after DNA damage. UCH37 localizes to the promoters of E2F1 pro-apoptotic target genes such as caspase 3, caspase 7, PARP1, and Apaf-1 and activates their expression after DNA damage. Moreover, the expression of E2F1 proliferative and pro-apoptotic genes is correlated with the levels of UCH37 in many primary tumors. These results uncover a novel mechanism for E2F1 transcriptional activation through removal of its Lys-63-linked ubiquitination by UCH37.
Introduction
Ubiquitination is a critical posttranslational modification that regulates a myriad of cell processes. Studying the removal of ubiquitin, known as deubiquitination, has shown to have an equally important role in regulation. In the human genome, there are 95 putative deubiquitinating enzymes (DUBs),5 classified into five subfamilies, that are responsible for cleaving the covalent linkage of ubiquitin from substrates (1). Ubiquitin C-terminal hydrolase 37 (UCH37, also known as UCHL5, ubiquitin C-terminal hydrolase isoenzyme L5) is a member of the UCH family of DUBs, with the members UCHL1, UCHL3, and BAP1, all of which have been associated with oncogenesis (2).
UCH37 can be recruited to the 19S proteasome by Rpn13, which activates UCH37-mediated ubiquitin recycling before substrates undergo proteasomal degradation (3–5). Compared with other family members, UCHL1 and UCHL3, UCH37 has a unique short C-terminal tail extension. The C-terminal tail of UCH37 acts to autoinhibit the catalytic activity of UCH37. However, this autoinhibition is released when UCH37 interacts with Rpn13 (5). Interestingly, nuclear UCH37 can associate with the Ino80 chromatin-remodeling complex (6), implying a role for UCH37 in the regulation of either DNA repair or transcription. For the former, UCH37 has been shown to protect nuclear factor related to κB-binding protein (a subunit of the Ino80 complex) from degradation and promote double-stranded break resection and repair by homologous recombination (7). Nevertheless, a role in transcriptional regulation for UCH37 has not been demonstrated.
E2F1 is an important transcription factor involved in the regulation of cell cycle progression, DNA repair, and apoptosis response (8). Regulation of a transcription factor that plays such paradoxical roles is crucial, and the differential regulation of E2F1 activities is, at least in part, dependent on its posttranslational modifications, including phosphorylation, acetylation, neddylation, and methylation (9). Under normal growth conditions, E2F1 is bound to a hypophosphorylated retinoblastoma protein (pRb), inhibiting the ability of E2F1 to be transcriptionally active. When the cell is ready to progress through the cell cycle, Cdk4/Cyclin D phosphorylates pRb, and pRb then releases E2F1 to transcriptionally activate genes needed for G1/S phase transition. Upon DNA damage, phosphorylation by DNA damage-dependent kinases such as ataxia telangiectasia mutated (10) and Chk2 (11) stabilize E2F1. Subsequently, E2F1 transcriptionally activates an apoptotic response in a p53-dependent pathway by activating p14ARF (12) and also in a p53-independent manner by activating p73 (13), caspases (14), and Apaf-1 (15). Although ubiquitination has been shown to regulate E2F1 protein stability, whether E2F1 transcriptional activity can be regulated by its ubiquitination remains unknown. Here we show that E2F1 activity can be regulated by Lys-63 linkage-specific ubiquitin chain formation. We also identify the first deubiquitinating enzyme for E2F1 and demonstrate its role in activating the transcriptional function of E2F1.
Experimental Procedures
Cell Culture, Transfection, and Treatment
HEK293T, HEK293, H1299, and U2OS cells and primary human foreskin fibroblasts (HFFs), passage 7 (ATCC), were maintained in DMEM supplemented with 10% FBS, penicillin (50 IU/ml), and streptomycin (50 μg/ml). All cells were grown in a humidified incubator at 37 °C with 5% CO2 and 95% air. A standard method using calcium chloride or polyethylenimine was used for transfection for HEK293T and HEK293 cells. Lipofectamine 2000 (Life Technologies) was used for transfection of H1299 and U2OS cells. After transfection, cells were incubated for 36–48 h and treated with 5 μm Adriamycin (EMD Millipore) or 1 μm b-AP15 (Calbiochem) for the indicated times as described for each experiment.
Plasmid Construction
To construct pRK5-HA-His-UbK63, His-UbK63 was amplified from pRK5-HA-UbK63 (purchased from Addgene, Plasmid 17606) using the primers 5′-GTCGACCCATCATCATCATCATCATATGCAGATCTTCGTCAGAACG-3′ and 5′-AGCGGCCGCTCAACCACCTC-3′. The product was digested with SalI/NotI and then subcloned to the SalI/NotI-digested pRK5-HA vector. The HA-UbK63R mutant was a gift from Dr. Ze'ev Ronai (16). pcDNA3.0-HA-UCH37, UCH37(C88A), and pcDNA3.0-HA-UbcH13 were purchased from Addgene (plasmids 19415, 19416, and 12461, respectively). The pGEX-6P1-UCH37 construct was a gift from Dr. Chittaranjan Das. The Lentiviral UCH37 shRNAs 2, 4, 5, and 6 were produced from pGIPZ human UCH37 shRNA (clone IDs V2LHS_134650, V2LHS_254565, V2LHS_255042, and V3LHS_385962, respectively; Open Biosystems). The GIPZ non-silencing lentiviral shRNA control plasmid was also purchased from Open Biosystems (catalog no. RH4346).
In Vivo Ubiquitination Assay
A denaturing immunoprecipitation was performed by transfecting HEK293T or H1299 cells with either vector or HA-UbK63, an ubiquitin construct that can only have Lys-63 linkages. Cells were harvested in SDS lysis buffer (60 mm Tris-HCl (pH 6.8), 1% SDS) and then boiled for 5 min. Lysates were diluted 1:10 in TNN buffer (50 mm Tris (pH 7.5), 0.25 m NaCl, 5 mm EDTA, and 0.5% Nonidet P-40), sonicated, and clarified by 10-min centrifugation at 14,000 rpm. Lysates were then immunoprecipitated with either an E2F1 antibody or control mouse IgG bound to protein G beads overnight. The next morning, the beads were washed four times with radioimmune precipitation assay lysis buffer and once with 0.5 m LiCl, followed by one last wash with radioimmune precipitation assay buffer. For nickel pulldown assays, HEK293T or stable UCH37 knockdown (U2OS) cells were transfected with His-UbK63 and appropriate plasmids, depending on the experimental design. After transfection, cells were treated with 5 μm Adriamycin for 5 h. We then followed the nickel pulldown protocol (17) with minor modifications. Cells were lysed in nickel lysis buffer (6 m guanidinium HCl, 100 mm Na2HPO4/Na2HPO4 (pH 8.0), 10 mm Tris-HCl (pH 8.0), and 5 mm imidazole). 20% of cells were lysed separately in SDS lysis buffer for input. The lysates were then sonicated and clarified by 10-min centrifugation at 14,000 rpm in a microfuge. Equivalent amounts of lysates were incubated overnight with Ni-NTA-agarose beads at 4 °C. Beads were washed consecutively with nickel lysis buffer and buffer 2 (8 m urea, 100 mm Na2HPO4/Na2HPO4 (pH 8.0), and 10 mm Tris-HCl (pH 8.0)). Finally, the beads were washed with buffer A (8 m urea, 100 mm Na2HPO4/Na2HPO4 (pH 8.0), and 10 mm Tris-HCl (pH 6.3)) with various amounts of Triton X-100 (one time with 0.2%, one wash with 0%, and then one wash with 0.1%). All working buffers except the elution buffer had their final pH values adjusted to 7.4 using HCl because pH is important for controlling the Ni-NTA pulldown background. The beads were then boiled in nickel elution buffer (5% SDS, 150 mm Tris-HCl (pH 6.7), 30% glycerol, 720 mm β-mercaptoethanol, 200 mm imidazole, and 0.3% bromphenol blue) and analyzed by SDS-PAGE, followed by Western blot analysis with the appropriate antibodies. Fractionation prior to the UbK63 assay was done following the protocol developed by Andegeko et al. (18).
In Vitro Deubiquitination Assay
HEK293T cells were transfected with HA-UbK63 and FLAG-E2F1, treated with 5 μm Adriamycin, and lysed in radioimmune precipitation assay (RIPA) buffer (50 mm Tris (pH 8.0), 150 mm NaCl, 0.5% sodium deoxycholate, 1% NP-40, and 0.1% SDS). Cell lysates were incubated with FLAG-agarose beads overnight at 4 °C to obtain purified UbK63-E2F1. Beads were washed four times with NETN wash buffer (150 mm NaCl, 5 mm EDTA, 50 mm Tris (pH 7.5), and 0.5% NP-40). Alongside this immunoprecipitation, HEK293T cells were transfected with HA-UCH37 or mutant HA-UCH37(C88A). These cells were harvested in TNN lysis buffer and immunoprecipitated with HA-agarose beads to obtain purified HA-UCH37 or HA-UCH37(C88A). Immobilized UbK63-E2F1 and HA-UCH37 were then combined in an in vitro DUB buffer (50 mm Tris-HCl (pH 7.5), 150 mm NaCl, 2 mm EDTA (pH 8.0), and 2 mm DTT) and incubated at 37 °C for 1 h. The beads were then boiled in Laemmli buffer and analyzed by SDS-PAGE, followed by Western blot analysis using HA antibody.
Immunoprecipitation and Western Blot Analysis
Cells were harvested in TNN buffer 36–48 h after transfection, and immunoprecipitation was performed as described previously (19). For endogenous immunoprecipitation, cells were cross-linked using 1% formaldehyde and quenched with 125 mm glycine. Cells were then lysed with nuclear extraction buffer (10 mm Tris HCl (pH 8.0), 85 mm KCl, 5 mm EGTA (pH 8.0), and 0.5% Nonidet P-40). The cells were spun down and then resuspended in TNN buffer and immunoprecipitated as above. The specific signals were detected with appropriate antibodies. The antibodies specific to HA (Y11), E2F1 (C-20 or KH95), GST, and GAPDH were purchased from Santa Cruz Biotechnology. Histone 3, Ser(P)-15-p53, and Lys-63 linkage-specific polyubiquitin rabbit monoclonal antibody (D7A11) antibodies were purchased from Cell Signaling Technology. The HSP90 antibody (ADI-SPA-846-D) was purchased from Enzo Life Sciences. The FLAG antibody was purchased from Sigma. The His6 antibody was purchased from Clontech. The UCH37 antibody was purchased from Abcam (catalog no. ab38528).
Dual-Luciferase Reporter Assay
HEK293 or stable knockdown shUCH37 (U2OS) cells were transfected with p14ARF promoter-Luc (firefly luciferase) and pRL(Renilla luciferase)-TK (a constitutively active plasmid for controlling transfection efficiency containing only a portion of the herpes simplex virus TK promoter and lacking E2F1 binding sites) and other plasmids needed for each independent experiment. Firefly and Renilla luciferase activities were measured using the Promega Dual-Luciferase reporter assay system, and the firefly luciferase activities were normalized against the Renilla activities as described previously (20).
GST Pulldown Assay
The GST fusion proteins were induced by 1.0 mm isopropyl β-d-thiogalactopyranoside in Escherichia coli strain DH5α and purified. The GST portion on GST-UCH37 was excised by PreScission protease (Pharmacia). Approximately 5 μg of GST-E2F1 full-length was immobilized on glutathione-Sepharose beads, incubated with 0.5–1 μg of UCH37, and rotated at 4 °C for 4 h with NETN wash buffer. Beads were washed five times with NETN wash buffer, subjected to SDS-PAGE, and analyzed by Western blotting with anti-UCH37 antibody.
RNA Extraction and Real-time RT-PCR
RNA was extracted using TRIzol reagent (Invitrogen). Quantitative PCR was performed in triplicate on an MX3005P thermal cycler using the SYBR Green dye method to track the progress of the reactions, with ROX dye added as reference. GAPDH was run in parallel with test genes. The PCR condition were as follows: 95 °C denaturation for 30 s and 55 °C annealing for 1 min and 72 °C for 2 min. The results were analyzed with MxPro 4.0 QPCR software (Stratagene). The primers used for quantitative PCR were as follows: Apaf-1 (Apaf1–5, 5′-AATGGACACCTTCTTGGACG-3′; Apaf1–3, 5′-GCACTTCATCCTCATGA GCC-3′), caspase 3 (Casp3-5, 5′-TCGGTC TGGTACAGATGTCG-3′; Casp3-3, 5′-CATACAAGAAGTCGGCCTCC-3′), p73 (p73-5, 5′-TTTAACAGGATTGGGGTGTC-3′; p73-3, 5′-CGTGAACTCCTCCTTGATGG-3′), E2F1 (E2F1-5′, 5′-GAGGGCATCCAGCTCATT G-3′; E2F1-3′, 5′-GGTCCCCAAAGTC ACAGTC-3′), CycE (CycE5, 5′-CTCCAG GAAGAGGAAGGCAA-3′; CycE3, 5′-TCGATTTTGGCCATTTCTTCA-3′), Cdc6 (Cdc6-F, 5′-GCCAAGAAGGAGCACAAGAT-3′; Cdc6-R, 5′-GGGTGGGGTGTAA GAGAAGAA-3′); TK3 (5′-ATGTGT GCAGAAGCTGCTGC-3′), TK5 (5′-ATG AGCTGCATTAACCTGCCCACT-3′); TopBP1 (TopBP1F, 5′-TGGTTGCTAG AGTGTTTCA-3′; TopBP1R, 5′-TTTCAGTATAAGATGAGTA-3′), and UCH37 (UCH37-5, 5′-TGGTTCAGGACTCCCGACTT-3′; UCH37-3, 5′-CTGGTGGGTACAGTT CAGTAAC-3′).
ChIP Assay
HEK293T cells were grown in 15-cm-diameter plates, cross-linked with 1% formaldehyde, washed, and scraped with PBS, and then nuclei were extracted with nuclear extraction buffer supplemented with protease inhibitors. Lysates were spun down, and pellets were resuspended in radioimmune precipitation assay lysis buffer. Lysates were then sonicated six times for 20 s, with 1-min intervals on ice. HA-agarose beads were precleared with salmon sperm DNA for 3 h before lysates were added and rotated overnight at 4 °C. Beads were rotated for 5 min at 4 °C for the following consecutive washes: ice-cold low-salt buffer (0.1% SDS, 1% Triton X-100, 2 mm EDTA, 20 mm Tris-HCl (pH 8.0), and 150 mm NaCl), high-salt buffer (0.1% SDS, 1% Triton X-100, 2 mm EDTA, 20 mm Tris-HCl (pH 8.0), and 500 mm NaCl), LiCl buffer (0.25 m LiCl, 1% Nonidet P-40, 1% deoxycholic acid, 1 mm EDTA, and 10 mm Tris-HCl (pH 8.0)), and twice with TE buffer (10 mm Tris-HCl (pH 8.0) and 1 mm EDTA). Chromatin was eluted in fresh elution buffer (0.1 m NaHCO3). Cross-links were then reversed by incubating samples under high-salt conditions for 4 h at 65 °C, followed by digestion of RNA by RNase A and protein by proteinase K. DNA was purified via phenol/chloroform extraction and ethanol precipitation. The ChIP primer sequences that flank the E2F-binding sites within the promoters of Caspase 3, Caspase 7, and Thymidine Kinase 1 have been described previously (21). The ChIP primers for Cyclin E1 were cCycE-F (5′-TTGGCCCCGCCCTGTCCGCC-3′) and cCycE-R (5′-GGCGGCCGCGCCGGCTGCTC-3′); for PARP1 were c-PARP1-F (5′-GCGGCACTGCACTCCAGCG-3′) and c-PARP1-R (5′-TGATGCCTGGCCGCGGGAA-3′); and for APAF1 were c-Apaf1-F (5′-GGAGACCCTAGGACGACAAG-3′) and c-Apaf1-R (5′-CAGTGAAGCAACGAGGATGC-3′).
Cell Proliferation Assay
Cell proliferation was measured in shUCH37 stable knockdown cells. 10,000 cells were plated in triplicate and counted every other day for a week with a Beckman Coulter counter.
Recombinant Adenovirus Infection
The recombinant adenovirus expressing E2F1 (AdE2F1) and an empty vector (AdCMV) have been described previously (29). The viruses were purified by CsCl banding. Virus titers were measured with an Adeno-X rapid titer kit from Clontech. HFF cells were starved in 0.1% FBS for 48 h. Cells were either infected with AdCMV or AdE2F1 for 18 h and then harvested in SDS lysis buffer, and proteins were detected via immunoblotting.
Apoptosis Assays
Apoptosis was assayed by several independent assays: Annexin-V allophycocyanin staining followed by flow cytometry and a Caspase 3/7 activity assay performed according to the instructions of the manufacturer (Promega Caspase-GloTM 3/7 assay).
Statistical Analyses
Two-tailed Student's t test was performed to evaluate the differences between experimental groups. p Values of less than 0.05 were considered statistically significant. Gene expression data in the TCGA RNA sequencing database were extracted through the cBioPortal server, and Pearson correlation coefficients were calculated to evaluate correlations. The information of breast cancer subtypes classified according to PAM50 RNA sequencing in the TCGA breast cancer cohort was obtained from the UCSC Cancer Browser.
Results
UCH37, a Deubiquitinating Enzyme, Interacts with E2F1, and That Interaction Is Enhanced upon DNA Damage
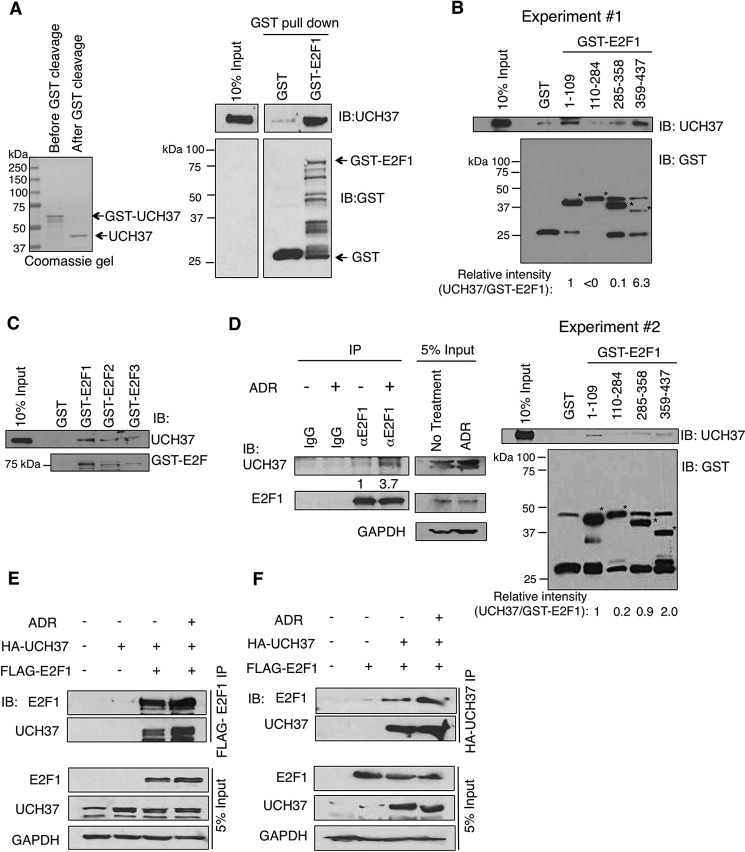
An immunoprecipitation/MS assay of FLAG-E2F1 in HEK293T cells revealed UCH37, a deubiquitinating enzyme, as a putative E2F1-interacting protein. Therefore, we characterized UCH37 for E2F1 regulation in the subsequent studies. We first investigated the interaction between E2F1 and UCH37 through an in vitro approach. Purified recombinant UCH37 directly interacted with purified GST-E2F1 protein in a GST pulldown assay (Fig. 1A). We also mapped the domains of E2F1 that directly interact with UCH37. Incubation of recombinantly purified UCH37 with GST-E2F1 domain fragments immobilized on glutathione-Sepharose beads revealed that UCH37 binds the N terminus (amino acids 1–109) and C terminus (amino acids 359–437) of E2F1 (Fig. 1B). This suggests that UCH37 requires multiple domains of E2F1 for optimal binding. Because of the high homology between the E2F family members, we decided to determine whether UCH37 could bind to other family members. Using the same in vitro approach as described above, we determined that UCH37 could also bind to E2F2 and E2F3 (Fig. 1C).
FIGURE 1.
E2F1 interacts with UCH37 in vitro and in vivo. A, recombinant UCH37 interacted in vitro with GST-E2F1 in a GST pulldown assay. IB, immunoblot. B, recombinant UCH37 was incubated with various GST-E2F1 fragments: N terminus (amino acids 1–109), DNA binding domain (amino acids 110–284), marked box domain (amino acids 285–358), and Rb binding domain (amino acids 359–437). A GST pulldown assay was performed. Shown are results from two independent experiments. The asterisks indicate the corresponding GST-E2F1 fragments. The signals of bound UCH37 and GST-E2F1 fragments were quantified using ImageJ software. The UCH37 signals bound to GST-E2F1 fragments were first subtracted by that bound to the control GST beads and then normalized by the abundance of each corresponding GST fusion protein. The relative intensity of UCH37/GST-E2F1 is shown at the bottom of each panel. C, UCH37 is able to directly interact with different E2F members. Recombinant UCH37 was incubated with GST-E2F1, GST-E2F2, or GST-E2F3 for a GST pulldown assay. D, HEK293T cells were left untreated or treated with 5 μm Adriamycin (ADR) for 5 h and then subjected to immunoprecipitation (IP) using an E2F1-specific antibody (KH95). The relative intensity of immunoprecipitated UCH37 was quantified by densitometry. E, HEK293T cells were transfected with empty vector, HA-UCH37 alone, or FLAG-E2F1 and then left untreated or treated with 5 μm Adriamycin for 5 h. Cell lysates were subjected to immunoprecipitation using anti-FLAG-agarose. F, HEK293T cells were transfected with empty vector, FLAG-E2F1 alone, or HA-UCH37 and then left untreated or treated with 5 μm Adriamycin for 5 h. Cell lysates were subjected to immunoprecipitation using anti-HA-agarose.
Although UCH37 can bind to multiple E2F family members, we decided to focus our study exclusively on E2F1. To determine whether E2F1 can interact with UCH37 in vivo, HEK293T cells in the absence or presence of the DNA-damaging agent Adriamcyin were subjected to endogenous immunoprecipitation assays. Cells were cross-linked and then harvested in nuclear extraction buffer and incubated with a control mouse IgG or mouse anti-E2F1 (KH95) antibody. The results demonstrated that E2F1 and UCH37 interacted and that the E2F1/UCH37 complex increased upon Adriamycin treatment in vivo (Fig. 1D). There was also a modest increase in UCH37 protein level inside the cells after Adriamycin treatment. The interaction was also seen by a reciprocal immunoprecipitation. HEK293T cells were transfected with FLAG-E2F1 and HA-UCH37 and left untreated or treated with Adriamycin. Then the lysates were immunoprecipitated with FLAG-agarose beads. As with the endogenous immunoprecipitation, E2F1 interacted with UCH37 with a slight enhancement after DNA damage, although not as robustly (Fig. 1E). A reciprocal immunoprecipitation with HA-agarose beads resulted in the same conclusion (Fig. 1F). These results demonstrate that UCH37 can interact directly with E2F1 and that their complex accumulates upon DNA damage.
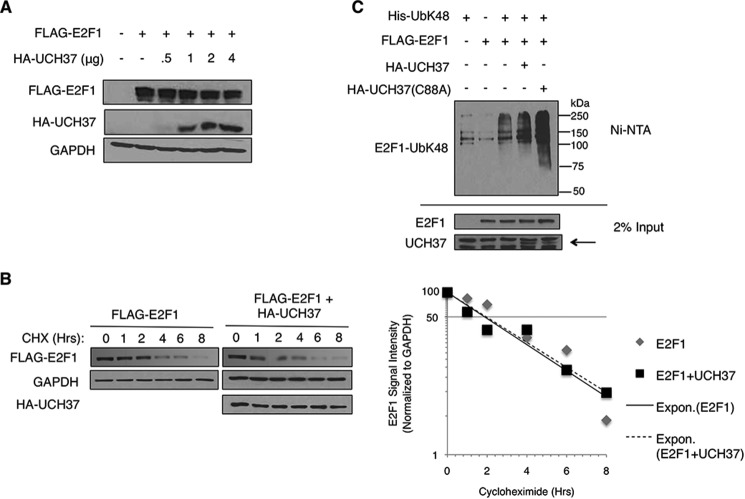
UCH37 Does Not Stabilize E2F1 Protein
Ubiquitin has seven lysines capable of forming covalent polyubiquitin chains. Certain lysine linkages, such as lysine 48-specific linkages (UbK48) and lysine 11-specific linkages (UbK11) are linked to targeting the substrate to proteasomal degradation. Previous studies have demonstrated that UCH37 can stabilize its substrate (7, 22, 23). Because UCH37 is associated with the proteasome, we wanted to study whether UCH37 has an effect on E2F1 protein stability via proteasomally mediated regulation. We first examined the E2F1 protein level with increasing levels of UCH37. HEK293T cells were transfected with FLAG-E2F1 and increasing amounts of UCH37. Then the cells were lysed with SDS lysis buffer, and proteins were observed through immunoblotting (Fig. 2A). We saw that increasing amounts of UCH37 did not stabilize E2F1. Analysis of HEK293T lysates with ectopically expressed FLAG-E2F1 and HA-UCH37 in the presence of cycloheximide at different time points showed that the half-life of E2F1 in the presence of UCH37 did not change significantly (Fig. 2B). Finally, we performed a nickel pulldown assay to determine whether UCH37 could decrease the UbK48-specific linkages on E2F1 (Fig. 2C). HEK293T cells were transfected with FLAG-E2F1 and His-UbK48 (all lysines were changed to arginine except Lys-48) and HA-UCH37 or HA-UCH37(C88A) (a catalytic mutant). Cells were harvested under denaturing conditions, followed by Ni-NTA pulldown. This result demonstrated that UCH37 was unable to deubiquitinate UbK48-specific linkages on E2F1. The UbK48 linkages on E2F1 increased in the sample transfected with the catalytic mutant UCH37(C88A). This could be either due to a higher expression of transfected His-UbK48 in that sample or, alternatively, due to a possible dominant negative effect of mutant UCH37 on the processing of ubiquitin chains in the proteasome. Together, these results suggest that UCH37 does not play a traditional role and does not regulate E2F1 protein stability through the deubiquitination of E2F1-UbK48.
FIGURE 2.
UCH37 does not stabilize E2F1 protein. A, HEK293T cells were transfected with FLAG-E2F1 and increasing amount of HA-UCH37. UCH37 and E2F1 protein levels were detected through immunoblotting. B, HEK293T cells were transfected with FLAG-E2F1 with vector control of FLAG-E2F1 with HA-UCH37. The cells were treated with cycloheximide (CHX, 20 μg/ml) for various times. HA-UCH37 and FLAG-E2F1 were detected by Western blot analysis. E2F1 signal intensities were measured by densitometry. Expon., exponential trendline. C, HEK293T cells were transfected with various combinations of FLAG-E2F1, His-UbK48, and HA-UCH37 or HA-UCH37(C88A) and then harvested under denaturing conditions. Lysates were subjected to a Ni-NTA pulldown assay, followed by immunoblotting for E2F1. The arrow indicates the bands specific to UCH37.
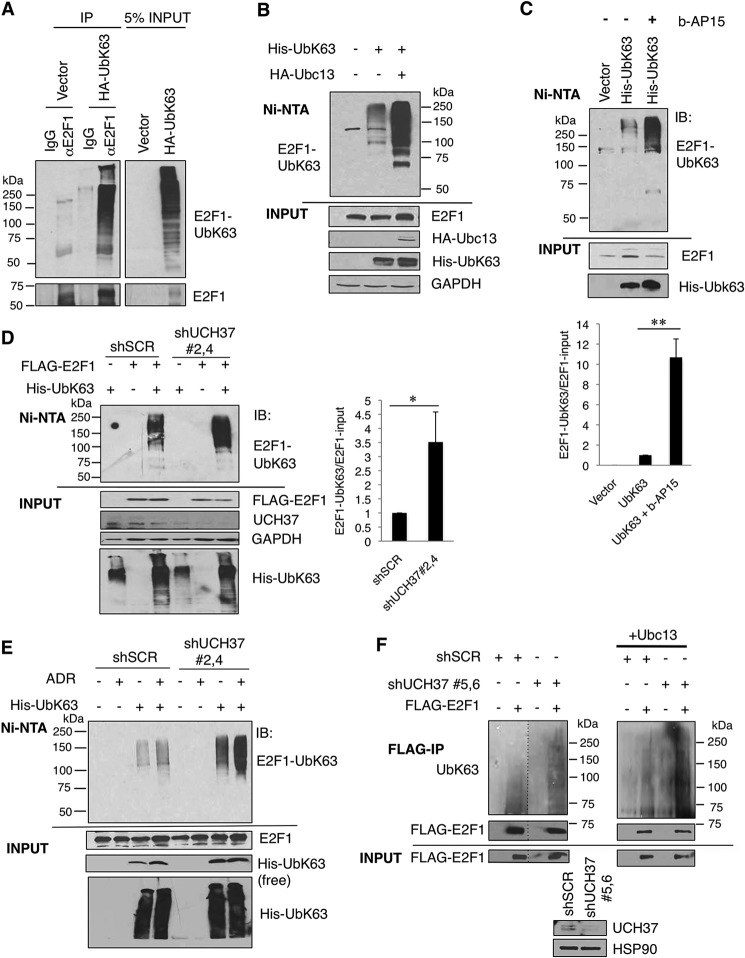
E2F1 Is Modified via UbK63-specific Ubiquitin Linkages, and Inhibition or Knockdown of UCH37 Results in an Increase in UbK63-E2F1
We next sought to discover whether UCH37 could deubiquitinate any other ubiquitin linkage. Ubiquitination linkages through the lysine 63 residue (UbK63) are known to play a role in cell signaling, protein localization, and DNA damage response. E2F1 has been shown previously to be ubiquitinated via UbK48-specific (24) and UbK11-specific (19) linkages for proteasomal degradation. However, ubiquitin Lys-63-specific linkages on E2F1 have yet to be examined. We transfected HEK293T cells with either empty vector or HA-UbK63, in which all lysines except Lys-63 were mutated to arginines. SDS and heat-denatured cell lysates were immunoprecipitated with either IgG or an E2F1-specific antibody. We observed an UbK63 signal only when immunoprecipitating with an E2F1 antibody, demonstrating that endogenous E2F1 can be ubiquitinated through UbK63-specific linkages (Fig. 3A). These semiendogenous results were verified by subjecting His-UbK63-transfected HEK293T cells to denaturing guanidinium HCl lysis, followed by Ni-NTA pulldown. To further support the specificity of this assay, cotransfection of Ubc13 (the E2-conjugating enzyme known to promote UbK63 ubiquitination (25)) increased Lys-63-linked ubiquitination of E2F1 (Fig. 3B). To interrogate a physiological role for UCH37 in E2F1 deubiquitination, we first used a DUB inhibitor, b-AP15, that inhibits two deubiquitinating enzymes interacting on the proteasomal lid, USP14, and UCH37 (26). We observed that, when HEK293T cells were treated with b-AP15, UbK63-E2F1 increased (Fig. 3C), supporting a role for UCH37 or USP14 in actively removing Lys-63 linkage-specific ubiquitin chains from E2F1 under the steady state. We then knocked down UCH37 using shRNA constructs and saw an increase in Lys-63 ubiquitination on transfected E2F1 compared with the scrambled shRNA control (Fig. 3D). We saw a similar result when performing a semiendogenous UbK63 assay to detect Lys-63 ubiquitination on endogenous E2F1 in both the absence and presence of Adriamycin treatment (Fig. 3E). Next we used endogenous ubiquitin to further prove that E2F1 can be ubiquitinated through UbK63-specific linkages (Fig. 3F). In HEK293T cells, we transfected FLAG-E2F1 and pooled shUCH37 or shSCR. We then immunoprecipitated FLAG-E2F1 and probed for ubiquitin using a UbK63-specific antibody. This result again demonstrated Lys-63 linkage-specific ubiquitination of E2F1. Consistent with the data in Fig. 3, D and E, UbK63-E2F1 increased when UCH37 was knocked down.
FIGURE 3.
E2F1 is ubiquitinated through UbK63-specific linkages, and reduced UCH37 shows increased UbK63-E2F1 linkages. A, HEK293T cells were transfected with either empty vector or HA-UbK63. The lysates were denatured by boiling in SDS lysis buffer as described under “Experimental Procedures” and then immunoprecipitated (IP) with either control mouse IgG control or E2F1-specific antibody (KH95). B, HEK293T cells were transfected with His-UbK63 and the E2 for UbK63 conjugation, Ubc13. The lysates were then subjected to a Ni-NTA pulldown assay. C, HEK293T cells were transfected with His-UbK63 and then treated with b-AP15 (1 μm for 5 h), which specifically inhibits UCH37 and USP14. The cells were harvested and subjected to a Ni-NTA pulldown assay. The intensity of UbK63-E2F1 in each sample was normalized by its corresponding E2F1 signal. The graph in the bottom panel represents the mean ± S.D. from three independent experiments. **, p < 0.001 (two-tailed Student's t test). IB, immunoblot. D, UCH37 was transiently knocked down in HEK293T cells by combination of two shUCH37 hairpins (shUCH37#2 and shUCH37#4) and cotransfected with His-UbK63 and FLAG-E2F1. The intensities of UbK63-E2F1 and E2F1 in each sample were quantified through ImageJ software, and the signal of UbK63-E2F1 was then normalized by the corresponding E2F1 signal. The data represent the mean ± S.D. from three independent experiments (biological replicates). *, p < 0.02 (two-tailed Student's t test). E, HEK293T cells were transfected with His-UbK63 alone with or without 5 μm ADR treatment for a semiendogenous UbK63 assay. Cells were harvested for the Ni-NTA pulldown assay. F, UCH37 was transiently knocked down in HEK293T cells by combination of two shUCH37 hairpins (shUCH37#5 and shUCH37#6) and cotransfected with FLAG-E2F1. Cell lysates were denatured by boiling in SDS lysis buffer as in A and then subjected to anti-FLAG immunoprecipitation. The ubiquitinated E2F1 was detected through immunoblotting with a UbK63-specific antibody. The dotted lines in F indicate the junctions of the images assembled from the same exposure films which was excised for brevity. The result was reproduced in the presence of Ubc13 (right panel). The HSP90 immunoblot served as a loading control. We quantified the intensities of UbK63-E2F1 and E2F1 and normalized the signal of UbK63-E2F1 by the corresponding E2F1 signal. The UbK63-E2F1/E2F1 in shUCH37 was 3.3- + 0.9-fold (mean ± S.D.) higher than that in the shSCR control (n = 3 biological replicates, p < 0.02, two-tailed Student's t test).
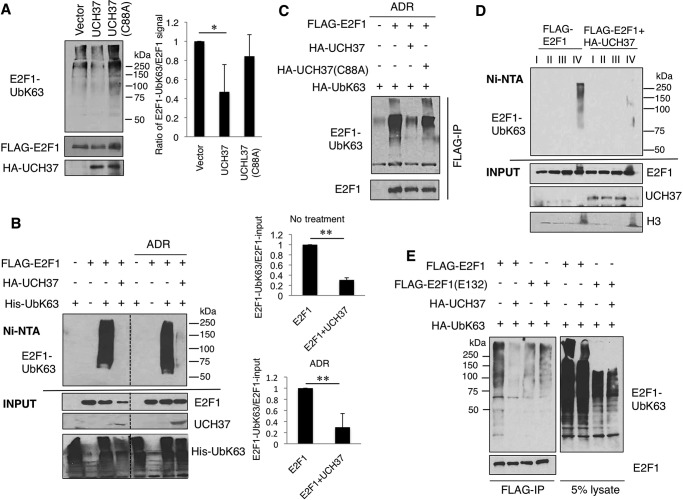
E2F1 Deubiquitination Depends on the Catalytic Activity of UCH37 and Localization to Chromatin
Next we investigated the catalytic role for UCH37 in deubiquitination of UbK63-E2F1. Using an in vitro approach as described under “Experimental Procedures,” we observed that UbK63-E2F1 was reduced significantly when purified UCH37 was added compared with the control. However, when a catalytic mutant, UCH37(C88A), was added instead, there was no reduction in UbK63-E2F1 (Fig. 4A). This result indicates that the decrease in UbK63 on E2F1 is due to the catalytic activity of UCH37. Next, overexpression of UCH37 in HEK293T cells led to a dramatic decrease in Lys-63-linked ubiquitin chain formation of E2F1 both in untreated cells and in cells treated with Adriamycin (Fig. 4B). The DUB activity of UCH37, but not UCH37(C88A), for UbK63-E2F1 was also reproduced in H1299 cells (Fig. 4C). Lys-63 linkage-specific ubiquitin chains often provide additional anchorage for protein-protein interactions. Therefore, UbK63-E2F1 might be tightly bound to chromatin. We performed a detergent extraction assay to separate loosely bound and tightly bound chromatin fractions (18). As seen in Fig. 4D, unlike the bulk of E2F1, Lys-63-ubiquitinated E2F1 was mainly located tightly on chromatin (fraction IV), and overexpression of UCH37 led to its deubiquitination on this fraction. Further supporting deubiquitination of the DNA-bound E2F1 by UCH37 is the observation that a DNA binding-defective mutant, E2F1(E132) (27), was not deubiquitinated by UCH37 (Fig. 4E). Taken together, these results provide evidence for UCH37 as a deubiquitinating enzyme for UbK63-E2F1. To our knowledge, this is the first DUB identified for E2F1.
FIGURE 4.
E2F1 deubiquitination depends on the catalytic activity of UCH37, and the deubiquitination is localized to chromatin. A, FLAG-E2F1 with HA-UbK63 and HA-UCH37 or the catalytic mutant HA-UCH37(C88A) were transfected into HEK293T cells and purified through separate immunoprecipitations. An in vitro deubiquitination assay was performed by combining purified HA-UCH37 or HA-UCH37(C88A) with FLAG-E2F1-UbK63. The signal intensity from three independent experiments was measured with ImageJ software. The intensity of UbK63-E2F1 in each sample was normalized by its corresponding FLAG-E2F1 signal. *, p < 0.01 (two-tailed Student's t test). B, HEK293T cells were transfected with various combinations of FLAG-E2F1, His-UbK63, and UCH37 with or without 5 μm ADR and then harvested under denaturing conditions. Lysates were subjected to Ni-NTA pulldown assay, followed by immunoblotting for FLAG-E2F1 using an HRP-conjugated anti-FLAG antibody. The dotted lines in B indicate the junctions of the images assembled from the same exposure films which was excised for brevity. This experiment was replicated three times. Signal intensity was quantified and normalized as above, and is shown in the right panels. No treatment, **, p = 9.005e-06; ADR, **, p = 0.0078; two-tailed Student's t test). C, H1299 cells were transfected with HA-UbK63, FLAG-E2F1, HA-UCH37, or HA-UCH37(C88A) and then treated with 5 μm ADR for 5 h. Lysates were immunoprecipitated (IP) with FLAG-agarose beads under denaturing conditions and then detected by immunoblotting for HA-UbK63. D, HEK293T cells were transfected with FLAG-E2F1 and His-UbK63 without or with UCH37. Cell lysates were extracted in an increasing percentage of Nonidet P-40 in different lysis buffers. Fraction III represents loosely bound chromatin, and fraction IV represents tightly bound chromatin. Each fraction was then subjected to Ni-NTA pulldown assay. The result was replicated in another independent experiment. E, HEK293T cells were transfected with HA-UbK63, FLAG-E2F1, or FLAG-E2F1(E132) (a DNA binding mutant) with and without HA-UCH37. Lysates were immunoprecipitated with FLAG-agarose under denaturing conditions and then detected by immunoblotting for HA-UbK63.
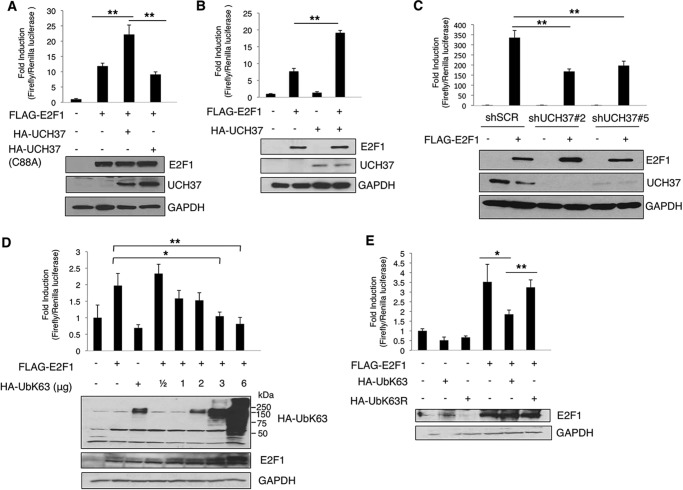
UCH37 Enhances E2F1 Transcriptional Activity
To investigate the effect of UCH37 on E2F1 transcriptional activity, we performed E2F1 activity reporter assays. Expression of UCH37, but not its catalytic mutant UCH37(C88A), in HEK293 cells promoted E2F1 transcriptional activity (Fig. 5A). The enhancing effect of E2F1 activity by UCH37 was also replicated in another cell line, H1299, a lung cancer cell line (Fig. 5B). Conversely, stable knockdown of UCH37 by two shRNAs in U2OS cells led to a decrease in E2F1 activity (Fig. 5C). We then examined the effect of Lys-63-linked ubiquitination on E2F1 transcriptional activity with a p14ARF promoter luciferase activity assay in H1299 cells. When E2F1 was cotransfected with increasing amounts of HA-UbK63, its transcriptional activity was reduced progressively (Fig. 5D). The repression was specific to Lys-63 linkage because HA-UbK63R, in which only Lys-63 was mutated to arginine, did not exert the same effect on E2F1 transcriptional activity (Fig. 5E). Together, these data suggest that Lys-63-linked ubiquitination of E2F1 can inhibit its transcriptional activity, whereas the deubiquitination activity of UCH37 activates E2F1 transcriptional activity.
FIGURE 5.
UCH37 increases E2F1 transcriptional activation, whereas overexpressing UbK63 decreases the transcriptional activation of E2F1. A–E, E2F1 transcriptional activity was analyzed by p14ARF promoter Dual-Luciferase assay. A, HEK293 cells were transfected with empty vector, FLAG-E2F1, and HA-UCH37 or HA-UCH37(C88A) along with promoter reporter constructs. The normalized activities of E2F1 are shown as -fold induction relative to that of the empty vector control. HA-UCH37 and FLAG-E2F1 protein levels were detected through immunoblotting. Results shown are the mean ± S.D. from three biological replicate experiments. **, p = 0.005 (for UCH37) and 0.002 (for UCH37(C88A)) (two-tailed Student's t test). B, H1299 cells were transfected with empty vector, FLAG-E2F1, and HA-UCH37 with promoter reporter constructs. HA-UCH37 and FLAG-E2F1 protein levels were detected through immunoblotting. **, p = 5.92163E-05, two-tailed Student's t test from three biological replicates. C, U2OS cells with a stable knockdown of UCH37 (shUCH37#2 or shUCH37#5) or a scrambled shRNA control (shSCR) were transfected with FLAG-E2F1 or empty vector with promoter reporter constructs. E2F1 activity was measured as in A. The data are the mean ± S.D. from three biological replicates. **, p = 0.0088 (shUCH37#2 versus shSCR) and 0.0046 (shUCH37#5 versus shSCR) (two-tailed Student's t test). D and E, HEK293 cells were transfected (D) with FLAG-E2F1 and increasing amounts of UbK63 (*, p = 0.0144; **, p = 0.0085) or (E) with HA-UbK63 or HA-UbK63R (*, p = 0.036; **, p = 0.0056). The data represent mean ± S.D. from three biological replicates. The p values are based on a two-tailed Student's t test.
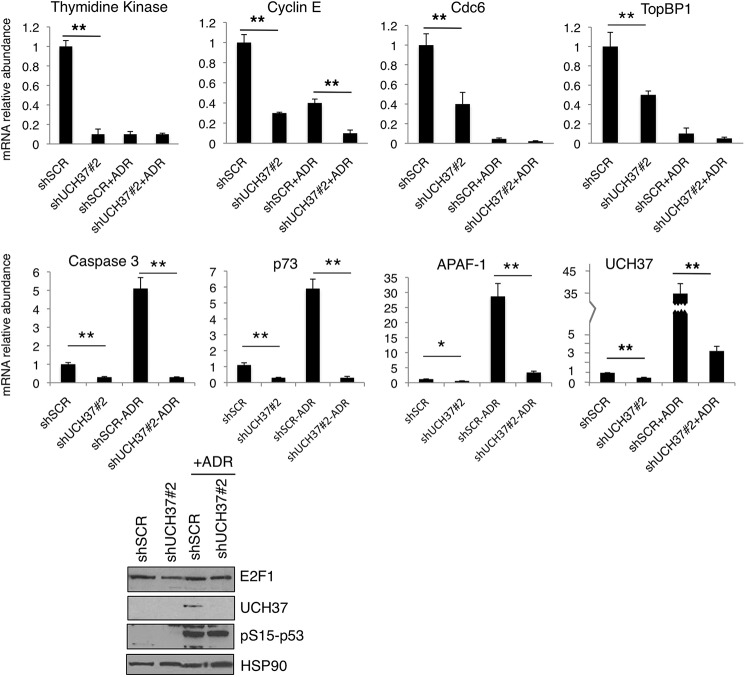
UCH37 Regulates the Expression of E2F1 Target Genes
We examined the expression of E2F1 target genes in UCH37-depleted U2OS cells. UCH37 depletion led to a decrease in the expression of both E2F1 cell proliferation and pro-apoptotic genes. Under normal growth conditions, UCH37 depletion led to a decrease in E2F1 target gene expression, including proliferative targets (thymidine kinase, cyclin E, Cdc6, and TopBP1) and pro-apoptotic targets (caspase 3, p73, and Apaf-1) (Fig. 6). After Adriamycin treatment, in the scrambled shRNA control cells, the expression of proliferative genes decreased, and that of E2F1 pro-apoptotic targets was induced as expected. However, the induction of pro-apoptotic genes such as caspase 3, p73, and Apaf-1 after Adriamycin treatment was inhibited greatly by UCH37 depletion. We probed the immunoblot of these U2OS cell lysates for phospho-p53 (Ser(P)-15) to determine whether DNA damage was sensed by ataxia telangiectasia mutated/ATR sensor kinases. The induction of Ser(P)-15-p53 by Adriamycin treatment was indeed indistinguishable between control and UCH37-depleted cells (Fig. 6, bottom panel). However, the shUCH37 cells were unable to induce E2F1-specific apoptotic genes. Together with the reporter assay, these results demonstrate a role for UCH37 in the up-regulation of the proliferative and pro-apoptotic target genes of E2F1. Interestingly, UCH37 mRNA was also induced by Adriamycin treatment, similar to E2F1 pro-apoptotic target genes (Fig. 6). This raised a possibility that UCH37 could be an E2F1 target and their reciprocal regulation might form a positive feedback loop.
FIGURE 6.
UCH37 regulates E2F1-specific target genes required for cell proliferation and apoptosis induction. U2OS knockdown cells (shUCH37#2 and shSCR) were treated with Adriamcyin, and then mRNA was harvested and E2F1 target genes were measured by quantitative PCR for the indicated genes. All samples were normalized to GAPDH expression, and the mean ± S.D. is expressed relative to the expression of genes in control cells. All statistics were done using two-tailed Student's t test from triplicate experiments. *, p < 0.05; **, p < 0.01. Aliquots of cell lysates were analyzed by immunoblotting (bottom panel). HSP90 immunoblot served as a loading control.
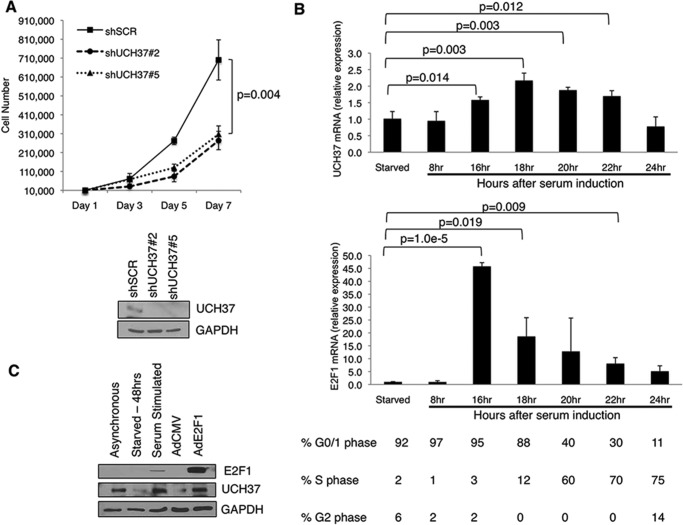
UCH37 Is an E2F1 Target, and Knockdown Results in Growth Suppression
To further investigate the regulation between UCH37 and E2F1, we next investigated the role of UCH37 in cell proliferation. Using the stable knockdown of UCH37 by two shRNAs in U2OS, cells were plated and counted every other day for a week. Both cell lines with depletion of UCH37 grew significantly slower compared with the scrambled control (Fig. 7A). This result is consistent with a similar study in A549 cells (28). To determine whether UCH37 is cell cycle-regulated, we used primary HFF cells to synchronize the cells and observe UCH37 mRNA expression through the cell cycle. HFF cells were serum-starved for 48 h in medium containing 0.1% FBS, and then entry of the cell cycle was induced by addition of 20% FBS. We found that UCH37 mRNA expression was induced at 16 h and stayed elevated until 22 h after serum addition (Fig. 7B). The induction corresponds to G1/S transition and S phase, when E2F1 mRNA rises. Finally, we wanted to investigate whether UCH37 is an E2F1 target. We starved HFF cells in 0.1% FBS for 48 h and infected cells with AdCMV or AdE2F1, a recombinant adenovirus expressing E2F1 (29). We then checked the UCH37 protein level. Indeed, UCH37 was induced by serum and by AdE2F1 (Fig. 7C). Previous studies of promoter analysis using ChIP-chip (30) and ChIP sequencing (31) show that E2F1 is bound to the promoter of UCH37. Taken together, these data confirm UCH37 as an E2F1 target.
FIGURE 7.
UCH37 is an E2F1 transcriptional target, and its expression is induced at G1/S transition and S phase of the cell cycle. A, U2OS knockdown cells (shSCR, shUCH37#2, and shUCH37#5) were plated and counted for 7 days. Shown are means ± S.D. from three biological replicates (p = 0.004, two-tailed Student's t test). B, primary HFFs were brought to quiescence by serum starvation (0.25% FBS) for 48 h and then stimulated by 20% FBS for the indicated periods of time. mRNA was collected, and UCH37 and E2F1 were measured by quantitative PCR. All samples were normalized to GAPDH expression. The numbers below each time point indicate the percentage of cells in each phase of the cell cycle, as assayed by propidium iodide staining/DNA histogram analysis. C, HFF cells were starved for 48 h and then infected with either AdCMV or AdE2F1 for 18 h. Cells lysates were analyzed by immunoblotting.
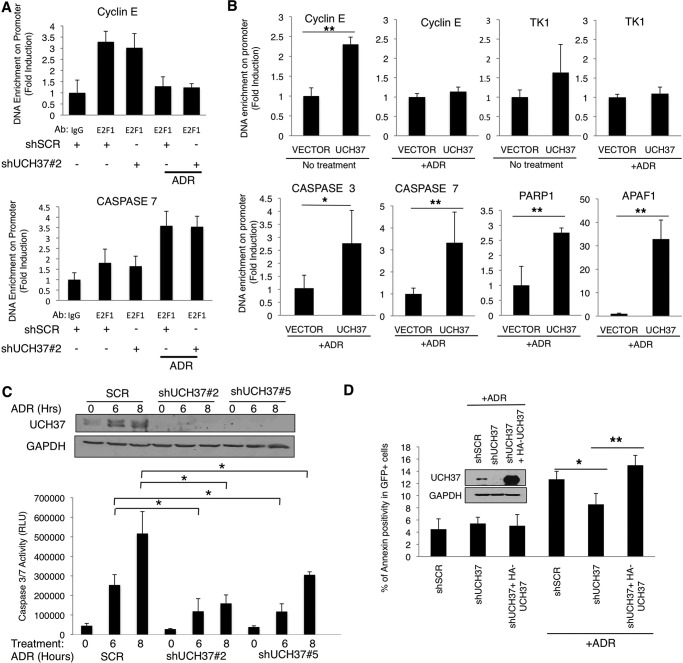
UCH37 Binds E2F1 Target Gene Promoters and Enhances Apoptotic Response after DNA Damage
Taking into consideration that UCH37 alters E2F1 target genes, we wanted to determine whether UCH37 is changing the binding of E2F1 to target gene promoters. Using U2OS stable knockdown cells (shSCR and shUCH37#2), we performed ChIP with and without Adriamycin treatment. We used mouse IgG and an E2F1-specific antibody for the ChIP assay to determine whether depletion of UCH37 affects E2F1 binding to a proliferative promoter (cyclin E) or a pro-apoptotic promoter (caspase 7). Under normal growth conditions, E2F1 binds equally to the cyclin E promoter in both the scrambled control and shUCH37 cells (Fig. 8A). Similarly, after Adriamycin treatment, E2F1 binds equally to the caspase 7 promoter in both the scrambled control and UCH37 knockdown cells. On the basis of this experiment, we conclude that UCH37 does not affect the binding of E2F1 to the promoter. Together with the result shown in Fig. 4E, these data show that UCH37, although not affecting E2F1 DNA-binding activity, can deubiquitinate the promoter-bound E2F1 and activate its transcriptional activity. To obtain further evidence for deubiquitination of E2F1 by UCH37 on chromatin, we investigated whether UCH37 bound to E2F1 target gene promoters. Limited by lack of a specific antibody for an UCH37 ChIP assay, we expressed HA-UCH37 in HEK293T cells and carried out ChIP assays using well characterized anti-HA-agarose beads. First, we examined proliferative genes such as cyclin E and thymidine kinase. Indeed, HA-UCH37 was enriched on both of these promoters under normal growth conditions. However, after DNA damage, HA-UCH37 was no longer enriched (Fig. 8B). On the basis of our quantitative PCR results, shown in Fig. 6, UCH37 also regulates pro-apoptotic genes after DNA damage. Therefore, we examined whether HA-UCH37 was enriched on these pro-apoptotic gene promoters after DNA damage. Indeed, HA-UCH37 was significantly enriched on the E2F1 pro-apoptotic-specific targets such as caspase 3, caspase 7, Apaf-1, and PARP1 after Adriamycin treatment (Fig. 8B). A physiological role for UCH37 in the apoptotic response to Adriamycin was supported further by measuring Adriamycin-induced apoptosis in UCH37-depleted U2OS cells (Fig. 8C). U2OS cells with a stable knockdown of UCH37 were either left untreated or treated with Adriamycin for 6 or 8 h. The scrambled control had an increase in caspase 3/7 activity when treated with Adriamycin, whereas shUCH37 cells have a diminished response to treatment. An independent apoptosis analysis was performed using Annexin-V-allophycocyanin staining followed by flow cytometry. H1299 cells were transiently transfected with shSCR, shUCH37, or shUCH37 with HA-UCH37. Because these shRNA constructs (pGIPZ) all contain GFP, we could identify the transfected cells in the flow cytometry analysis by gating GFP positivity and measure apoptosis in these cells. Consistent with the result obtained from stable U2OS cell lines, UCH37 depletion in H1299 cells also decreased apoptosis after Adriamycin treatment, and the effect was reversed by overexpressing HA-UCH37 (Fig. 8D). Taken together, we conclude that UCH37 has a role in promoting the E2F1-specific apoptotic response to DNA damage.
FIGURE 8.
UCH37 binds E2F1 target gene promoters and enhances apoptotic response after DNA damage. A, U2OS knockdown cells (shUCH37#2 or shSCR control) were treated with 5 μm ADR for 5 h, harvested, and subjected to chromatin immunoprecipitation with either IgG or E2F1 antibody (Ab). B, HEK293T cells were transfected with empty vector or HA-UCH37. Chromatin immunoprecipitation was performed through HA immunoprecipitation and subjected to quantitative PCR for the indicated E2F1-responsive promoters. *, p < 0.05; **, p < 0.01 (two-tailed Student's t test, n = 3). C, U2OS knockdown cells (shUCH37#2, shUCH37#5, or shSCR control) were treated with ADR for the indicated periods and then harvested following the Promega caspase 3/7 activity protocol. Shown are mean ± S.D. from three biological replicates. *, p < 0.05 (two-tailed Student's t test). Depletion of UCH37 in U2OS cells was confirmed by immunoblotting (top panel). RLU, relative luminescence units. D, H1299 cells were transiently transfected with shSCR, shUCH37, or shUCH37 rescued with HA-UCH37. Cells were then treated with ADR (5 μm) for 24 h and then stained with Annexin-V-allophycocyanin, followed by flow cytometry. GFP positivity was used to gate cells that expressed shRNAs because pGIPZ shRNA constructs contain GFP. The data represent the mean ± from three independent experiments. *, p = 0.024; **, p = 0.01 (two-tailed t test).
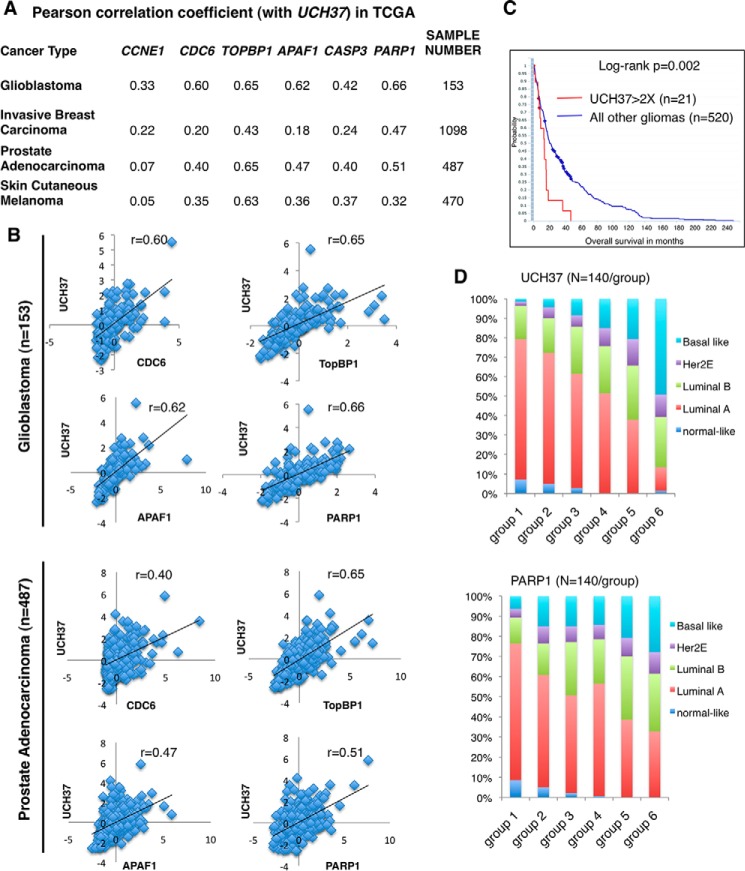
In our cell line systems, we found a role for UCH37 in the expression of E2F1 proliferative and pro-apoptotic target genes. We wondered whether the expression of UCH37 would be correlated with some of these genes in primary tumors. Using the TCGA database, we performed a Pearson correlation analysis between the expression of UCH37 and cell proliferation genes such as CCNE1 (Cyclin E), CDC6, and TopBP1. We also analyzed UCH37 and pro-apoptotic genes, such as APAF1, CASP3, and PARP1, in a variety of carcinoma tissues (Fig. 9A). Despite the heterogeneity of tumor tissues, UCH37 is positively correlated with both cell proliferation genes and pro-apoptotic genes in many carcinoma databases, with the best correlation seen in glioblastoma (Fig. 9B). We sought to investigate its clinical relevance in gliomas using the NCI/National Institutes of Health REMBRANDT database. Indeed, glioma patients with elevated UCH37 expression in tumors had shorter survival times (Fig. 9C). The correlation between UCH37 and PARP1 exists across most tumor types. When we ranked TCGA breast cancer by the expression of UCH37 or PARP1 and calculated the frequency of each breast cancer subtype in each group, the distribution profiles according to UCH37 and PARP1 levels were similar; the lowest expression group of either UCH37 or PARP1 is mainly the luminal A subtype. As expression increases, the frequencies of luminal A and normal-like subtypes decrease, but the frequency of the basal-like subtype increases (Fig. 9D). Although these correlative data could result from many factors, they do suggest the clinical significance of our study.
FIGURE 9.
UCH37 is positively correlated with E2F1 target genes in a variety of carcinomas. A and B, Pearson correlation coefficients between the expression of UCH37 and that of E2F1 target genes, including cell cycle progression genes (Cyclin E, CDC6, and TOPBP1) and pro-apoptotic genes (APAF1, CASP3, and PARP1) in the TCGA database (data extracted from cBioPortal, TCGA Provisional). The correlations in 153 glioblastoma patients (33) and 487 prostate cancer patients from the TCGA database are also shown as scatter plots in B. C, high UCH37 expression correlates with poor survival of glioma patients. Data shown are Kaplan-Meier survival curves for 541 glioma patients retrieved from the NCI/National Institutes of Health REMBRANDT database via the G-DOC Plus server at Georgetown University. Patients were sorted into two groups based on the -fold difference of UCH37 expression compared with mean values. D, 840 breast cancer samples in the TCGA database with gene expression information for UCH37 and PARP1 and breast cancer molecular subtypes (on the basis of PAM50 RNA sequencing, UCSC Cancer Browser) were analyzed. Patients were ranked according to the levels of expression of UCH37 (top panel) or PARP1 (bottom panel) in their breast tumors. We then equally divided the patients into either six groups according to expression level (group 1 < group 2 < group 3 < group 4 < group 5 < group 6), with 140 samples/group. The percentages of breast cancer subtypes within each group were calculated.
Discussion
In this report, we identify a deubiquitinating enzyme, UCH37, that binds to and deubiquitinates UbK63-E2F1 (Figs. 1, 3, and 4), which, in turn, transcriptionally activates E2F1 target genes (Figs. 5 and 6). These data unveil the first deubiquitinating enzyme that regulates the transcriptional activity of E2F1.
Although UCH37 regulates the protein turnover of its other substrates, such as nuclear factor related to κB-binding protein (7), we did not observe a change in E2F1 protein level by overexpression or depletion of UCH37 (Fig. 2). Because Lys-63-linked ubiquitination does not target proteins for degradation, a lack of effect on E2F1 levels by UCH37 is consistent with its DUB activity toward UbK63 of E2F1. Our study also shows, for the first time, a DUB activity toward UbK63 chains for UCH37. It is quite likely that there are other DUBs for UbK48 or UbK11 of E2F1 to regulate its turnover.
The data presented here support the idea that the ubiquitinated form of E2F1 (UbK63-E2F1) plays a repressive role in the transcriptional activity of E2F1. Although the exact mechanism remains unclear, it is thought that polyubiquitination of a substrate can form a docking site for multiprotein complexes. In our example, it is possible that UbK63-E2F1 can facilitate the binding of corepressor complexes; for example, KAP1 and HDAC1 (32). When UCH37 binds and deubiquitinates E2F1, corepressors are then displaced and allow for coactivators to bind and activate the transcriptional activity of E2F1.
UCH37 has been shown to be at the site of transcriptional regulation with the chromatin-remodeling complex Ino80 (6). Our study is the first to show the role of UCH37 in the regulation of a transcription factor and its transcriptional activity. Whether E2F1 and Ino80 cooperate and lead to the activation of these proliferative and pro-apoptotic genes would be worthy to investigate. Interestingly, UCH37 is present on the proliferative gene promoters under normal conditions, but, under DNA damage conditions, it selectively binds to the pro-apoptotic promoters. The mechanism(s) underlying the selectivity remain(s) unclear. Perhaps it is the difference in cofactors bound to UCH37 that allows this selectivity to occur. Future investigations to determine the exact mechanism for selective promoter occupancy of UCH37 and to identify other cofactors involved are warranted.
Although this study focuses on E2F1, UCH37 likely has a broader effect on transcriptional regulation. As suggested by the domain-mapping experiment, UCH37 binds to multiple domains of E2F1, including its C-terminal pRb-binding domain, which is conserved in other E2F proteins, UCH37 can also bind to E2F2 and E2F3 in vitro (Fig. 1C). It would be interesting to investigate in the future whether binding to E2F2 and E2F3 by UCH37 leads to their deubiquitination and changes in activities.
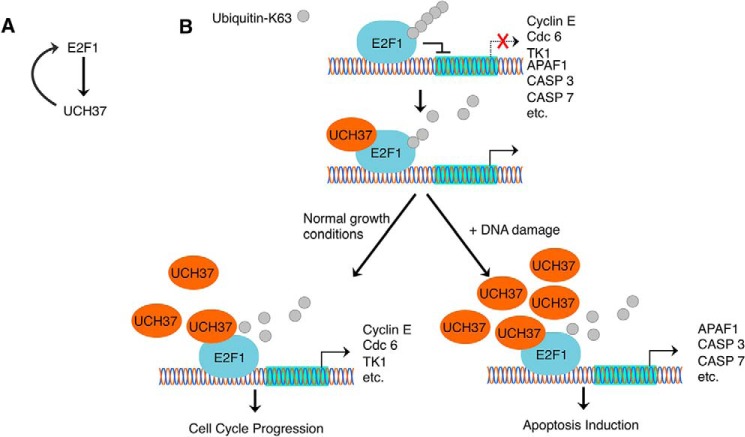
Our study also suggests a positive feedback loop in the regulation of E2F1 and UCH37 (Fig. 10A). We identified UCH37 as an E2F1 target. E2F1 transcriptionally activates UCH37, which, in turn, deubiquitinates E2F1, allowing E2F1 to transcriptionally activate gene targets. During DNA damage, UCH37 protein and mRNA levels are induced. This up-regulation of UCH37 can promote an increase in the E2F1 transcriptional activation of pro-apoptotic genes, creating a stronger induction of apoptosis after DNA damage.
FIGURE 10.
Reciprocal regulation of UCH37 and E2F1. A, E2F1 and UCH37 create a positive feedback mechanism. E2F1 transcriptionally activates UCH37, and UCH37 is able to deubiquitinate E2F1 to allow E2F1 to further transcriptionally activate genes. B, proposed model depicting how UCH37 regulates E2F1. Lys-63-linked ubiquitin chain formation plays a repressive role in E2F1 transcriptional activation. UCH37 can deubiquitinate E2F1 and activates E2F1 activity on target gene promoters. Lys-63-linked ubiquitination and deubiquitination of E2F1 occur constitutively, but the intracellular levels of UCH37 can tip the balance toward deubiquitination. For example, upon DNA damage, more UCH37 is available to interact with and deubiquitinate E2F1. Deubiquitination of UbK63-E2F1, along with other modifications of E2F1, activates E2F1 pro-apoptotic function during DNA damage.
In summary, we identified the first deubiquitinating enzyme for E2F1 and a new mechanism to regulate its activity. As depicted in Fig. 10B, E2F1 is ubiquitinated through UbK63-specific linkage, which represses its transcriptional activity. The repression can be relieved by UCH37 through removal of Lys-63-ubiquitin chains from E2F1. Our data are consistent with the notion that both Lys-63 ubiquitination and deubiquitination occur constitutively under normal growing conditions to activate the cell proliferation genes of E2F1. Upon DNA damage, UCH37 and E2F1 levels increase, and more UCH37 can bind and deubiquitinate E2F1 and activate its pro-apoptotic genes.
Author Contributions
C. S. M. and W. C. L. designed the research, analyzed the data, and wrote the paper with comments from all authors. C. S. M. performed most experiments and generated all data except Fig. 9. V. B. performed the immunoprecipitation/MS experiment and identified the interaction between UCH37 and E2F1. J. D. G. set up the Ni-NTA pulldown in vivo ubiquitination assay. G. L. constructed the pRK5-HA-His-UbK63 plasmid. W. C. L. performed data mining (Fig. 9).
Acknowledgments
We thank Joan Conaway, Chittaranjan Das, Dong-Er Zhang, and Ze'ev Ronai for plasmids and members of the W. C. L. and Fang-Tsyr (Fannie) Lin laboratories for discussions. We also thank the Cytometry and Cell Sorting Core at Baylor College of Medicine (with funding from the National Institutes of Health (AI036211, CA125123, and RR024574) and Joel M. Sederstrom for assistance.
This work was supported by National Institutes of Health Grants RO1CA100857 and RO1CA138641 and Department of Defense Grants W81XWH-14-1-0339 and W81XWH-14-1-0306. The authors declare that they have no conflicts of interest with the contents of this article.
- DUB
- deubiquitinating enzyme
- UCH
- ubiquitin carboxyl-terminal hydrolase
- HFF
- human foreskin fibroblast
- Ni-NTA
- nickel-nitrilotriacetic acid
- UbK63
- ubiquitination linkage through lysine 63
- ADR
- Adriamycin
- TCGA
- The Cancer Genome Atlas.
References
- 1.Nijman S. M., Luna-Vargas M. P., Velds A., Brummelkamp T. R., Dirac A. M., Sixma T. K., and Bernards R. (2005) A genomic and functional inventory of deubiquitinating enzymes. Cell 123, 773–786 [DOI] [PubMed] [Google Scholar]
- 2.Fang Y., Fu D., and Shen X. Z. (2010) The potential role of ubiquitin c-terminal hydrolases in oncogenesis. Biochim. Biophys. Acta 1806, 1–6 [DOI] [PubMed] [Google Scholar]
- 3.Hamazaki J., Iemura S., Natsume T., Yashiroda H., Tanaka K., and Murata S. (2006) A novel proteasome interacting protein recruits the deubiquitinating enzyme UCH37 to 26S proteasomes. EMBO J. 25, 4524–4536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Qiu X. B., Ouyang S. Y., Li C. J., Miao S., Wang L., and Goldberg A. L. (2006) hRpn13/ADRM1/GP110 is a novel proteasome subunit that binds the deubiquitinating enzyme, UCH37. EMBO J. 25, 5742–5753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yao T., Song L., Xu W., DeMartino G. N., Florens L., Swanson S. K., Washburn M. P., Conaway R. C., Conaway J. W., and Cohen R. E. (2006) Proteasome recruitment and activation of the Uch37 deubiquitinating enzyme by Adrm1. Nat. Cell Biol. 8, 994–1002 [DOI] [PubMed] [Google Scholar]
- 6.Yao T., Song L., Jin J., Cai Y., Takahashi H., Swanson S. K., Washburn M. P., Florens L., Conaway R. C., Cohen R. E., and Conaway J. W. (2008) Distinct modes of regulation of the Uch37 deubiquitinating enzyme in the proteasome and in the Ino80 chromatin-remodeling complex. Mol. Cell 31, 909–917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nishi R., Wijnhoven P., le Sage C., Tjeertes J., Galanty Y., Forment J. V., Clague M. J., Urbé S., and Jackson S. P. (2014) Systematic characterization of deubiquitylating enzymes for roles in maintaining genome integrity. Nat. Cell Biol. 16, 1016–1026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.DeGregori J., and Johnson D. G. (2006) Distinct and overlapping roles for E2F family members in transcription, proliferation and apoptosis. Curr. Mol. Med. 6, 739–748 [DOI] [PubMed] [Google Scholar]
- 9.Poppy Roworth A., Ghari F., and La Thangue N. B. (2015) To live or let die: complexity within the E2F1 pathway. Mol. Cell. Oncol. 2, e970480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lin W. C., Lin F. T., and Nevins J. R. (2001) Selective induction of E2F1 in response to DNA damage, mediated by ATM-dependent phosphorylation. Genes Dev. 15, 1833–1844 [PMC free article] [PubMed] [Google Scholar]
- 11.Stevens C., Smith L., and La Thangue N. B. (2003) Chk2 activates E2F-1 in response to DNA damage. Nat. Cell Biol. 5, 401–409 [DOI] [PubMed] [Google Scholar]
- 12.Bates S., Phillips A. C., Clark P. A., Stott F., Peters G., Ludwig R. L., and Vousden K. H. (1998) p14ARF links the tumour suppressors RB and p53. Nature 395, 124–125 [DOI] [PubMed] [Google Scholar]
- 13.Irwin M., Marin M. C., Phillips A. C., Seelan R. S., Smith D. I., Liu W., Flores E. R., Tsai K. Y., Jacks T., Vousden K. H., and Kaelin W. G. (2000) Role for the p53 homologue p73 in E2F-1-induced apoptosis. Nature 407, 645–648 [DOI] [PubMed] [Google Scholar]
- 14.Nahle Z., Polakoff J., Davuluri R. V., McCurrach M. E., Jacobson M. D., Narita M., Zhang M. Q., Lazebnik Y., Bar-Sagi D., and Lowe S. W. (2002) Direct coupling of the cell cycle and cell death machinery by E2F. Nat. Cell Biol. 4, 859–864 [DOI] [PubMed] [Google Scholar]
- 15.Moroni M. C., Hickman E. S., Lazzerini Denchi E., Caprara G., Colli E., Cecconi F., Müller H., and Helin K. (2001) Apaf-1 is a transcriptional target for E2F and p53. Nat. Cell Biol. 3, 552–558 [DOI] [PubMed] [Google Scholar]
- 16.Laine A., Topisirovic I., Zhai D., Reed J. C., Borden K. L., and Ronai Z. (2006) Regulation of p53 localization and activity by Ubc13. Mol. Cell. Biol. 26, 8901–8913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rodriguez M. S., Desterro J. M., Lain S., Midgley C. A., Lane D. P., and Hay R. T. (1999) SUMO-1 modification activates the transcriptional response of p53. EMBO J. 18, 6455–6461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Andegeko Y., Moyal L., Mittelman L., Tsarfaty I., Shiloh Y., and Rotman G. (2001) Nuclear retention of ATM at sites of DNA double strand breaks. J. Biol. Chem. 276, 38224–38230 [DOI] [PubMed] [Google Scholar]
- 19.Budhavarapu V. N., White E. D., Mahanic C. S., Chen L., Lin F. T., and Lin W. C. (2012) Regulation of E2F1 by APC/C Cdh1 via K11 linkage-specific ubiquitin chain formation. Cell Cycle 11, 2030–2038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chowdhury P., Lin G. E., Liu K., Song Y., Lin F. T., and Lin W. C. (2014) Targeting TopBP1 at a convergent point of multiple oncogenic pathways for cancer therapy. Nat. Commun. 5, 5476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Paik J. C., Wang B., Liu K., Lue J. K., and Lin W. C. (2010) Regulation of E2F1-induced apoptosis by the nucleolar protein RRP1B. J. Biol. Chem. 285, 6348–6363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fang Y., Fu D., Tang W., Cai Y., Ma D., Wang H., Xue R., Liu T., Huang X., Dong L., Wu H., and Shen X. (2013) Ubiquitin C-terminal hydrolase 37, a novel predictor for hepatocellular carcinoma recurrence, promotes cell migration and invasion via interacting and deubiquitinating PRP19. Biochim. Biophys. Acta 1833, 559–572 [DOI] [PubMed] [Google Scholar]
- 23.Wicks S. J., Haros K., Maillard M., Song L., Cohen R. E., Dijke P. T., and Chantry A. (2005) The deubiquitinating enzyme UCH37 interacts with Smads and regulates TGF-β signalling. Oncogene 24, 8080–8084 [DOI] [PubMed] [Google Scholar]
- 24.Peart M. J., Poyurovsky M. V., Kass E. M., Urist M., Verschuren E. W., Summers M. K., Jackson P. K., and Prives C. (2010) APC/C(Cdc20) targets E2F1 for degradation in prometaphase. Cell Cycle 9, 3956–3964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hofmann R. M., and Pickart C. M. (1999) Noncanonical MMS2-encoded ubiquitin-conjugating enzyme functions in assembly of novel polyubiquitin chains for DNA repair. Cell 96, 645–653 [DOI] [PubMed] [Google Scholar]
- 26.D'Arcy P., Brnjic S., Olofsson M. H., Fryknäs M., Lindsten K., De Cesare M., Perego P., Sadeghi B., Hassan M., Larsson R., and Linder S. (2011) Inhibition of proteasome deubiquitinating activity as a new cancer therapy. Nat. Med. 17, 1636–1640 [DOI] [PubMed] [Google Scholar]
- 27.Helin K., Wu C. L., Fattaey A. R., Lees J. A., Dynlacht B. D., Ngwu C., and Harlow E. (1993) Heterodimerization of the transcription factors E2F-1 and DP-1 leads to cooperative trans-activation. Genes Dev. 7, 1850–1861 [DOI] [PubMed] [Google Scholar]
- 28.Chen Z., Niu X., Li Z., Yu Y., Ye X., Lu S., and Chen Z. (2011) Effect of ubiquitin carboxy-terminal hydrolase 37 on apoptotic in A549 cells. Cell Biochem. Funct. 29, 142–148 [DOI] [PubMed] [Google Scholar]
- 29.Liu K., Lin F. T., Ruppert J. M., and Lin W. C. (2003) Regulation of E2F1 by BRCT domain-containing protein TopBP1. Mol. Cell. Biol. 23, 3287–3304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xu X., Bieda M., Jin V. X., Rabinovich A., Oberley M. J., Green R., and Farnham P. J. (2007) A comprehensive ChIP-chip analysis of E2F1, E2F4, and E2F6 in normal and tumor cells reveals interchangeable roles of E2F family members. Genome Res. 17, 1550–1561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cao A. R., Rabinovich R., Xu M., Xu X., Jin V. X., and Farnham P. J. (2011) Genome-wide analysis of transcription factor E2F1 mutant proteins reveals that N- and C-terminal protein interaction domains do not participate in targeting E2F1 to the human genome. J. Biol. Chem. 286, 11985–11996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang C., Rauscher F. J. 3rd, Cress W. D., and Chen J. (2007) Regulation of E2F1 function by the nuclear corepressor KAP1. J. Biol. Chem. 282, 29902–29909 [DOI] [PubMed] [Google Scholar]
- 33.Brennan C. W., Verhaak R. G., McKenna A., Campos B., Noushmehr H., Salama S. R., Zheng S., Chakravarty D., Sanborn J. Z., Berman S. H., Beroukhim R., Bernard B., Wu C. J., Genovese G., Shmulevich I., Barnholtz-Sloan J., Zou L., Vegesna R., Shukla S. A., Ciriello G., Yung W. K., Zhang W., Sougnez C., Mikkelsen T., Aldape K., Bigner D. D., Van Meir E. G., Prados M., Sloan A., Black K. L., Eschbacher J., Finocchiaro G., Friedman W., Andrews D. W., Guha A., Iacocca M., O'Neill B. P., Foltz G., Myers J., Weisenberger D. J., Penny R., Kucherlapati R., Perou C. M., Hayes D. N., Gibbs R., Marra M., Mills G. B., Lander E., Spellman P., Wilson R., Sander C., Weinstein J., Meyerson M., Gabriel S., Laird P. W., Haussler D., Getz G., and Chin L. (2013) The somatic genomic landscape of glioblastoma. Cell 155, 462–477 [DOI] [PMC free article] [PubMed] [Google Scholar]