Background: Proteinase-activated receptor 2 (PAR2) correlated with cancer metastasis.
Results: Down-regulation of miR-125b, which targets Gab2, mediates PAR2-induced cancer cell migration.
Conclusion: PAR2 promotes cancer cell migration through RNA methylation-mediated repression of miR-125b.
Significance: This study suggests a novel epigenetic mechanism by which miRNA expression is altered to regulate cancer cell migration.
Keywords: cell biology, cell signaling, microRNA (miRNA), migration, tumor metastasis, Gab2, proteinase-activated receptor 2, RNA methylation, mi-125b
Abstract
Proteinase activated-receptor 2 (PAR2) participates in cancer metastasis promoted by serine proteinases. The current study aimed to test the molecular mechanism by which PAR2 promotes cancer cell migration. In different cancer cells, activation of PAR2 by activating peptide (PAR2-AP) dramatically increased cell migration, whereas knock down of PAR2 inhibited cellular motility. The PAR2 activation also repressed miR-125b expression while miR-125b mimic successfully blocked PAR2-induced cell migration. Moreover, Grb associated-binding protein 2 (Gab2) was identified as a novel target gene of miR-125b and it mediated PAR2-induced cell migration. The correlation of PAR2 with miR-125b and Gab2 was further supported by the findings obtained from human colorectal carcinoma specimens. Remarkably, knock down of NOP2/Sun domain family, member 2 (NSun2), a RNA methyltransferase, blocked the reduction in miR-125b induced by PAR2. Furthermore, PAR2 activation increased the level of N6-methyladenosine (m6A)-containing pre-miR-125b in NSun2-dependent manner. Taken together, our results demonstrated that miR-125b mediates PAR2-induced cancer cell migration by targeting Gab2 and that NSun2-dependent RNA methylation contributes to the down-regulation of miR-125b by PAR2 signaling. These findings suggest a novel epigenetic mechanism by which microenvironment regulates cancer cell migration by altering miRNA expression.
Introduction
Metastasis is the main cause of mortality in cancer patients. Besides the genetic background of cancer cells, alteration in microenvironment has emerged as a vital factor to regulate the progression of cancer. Proteases are important component in microenvironment. Serine proteases, such as trypsin, mast cell-derived tryptase, matriptase, tissue factors, and KLKs, are strongly associated with cancer metastasis (1–3). Because of the complexity of microenvironment, the molecular mechanisms by which proteinases promote metastasis have not been fully understood. Notably, all these proteinases can selectively activate protease-activated receptor 2 (PAR2)2 through proteolysis of the receptor (4).
PAR2 belongs to the PAR family of G-protein-coupled receptors (GPCRs). PAR2 is highly expressed in various cancers (5–7), and blocking PAR2 activity with monoclonal antibodies effectively suppresses tumor growth in vivo (8). It has been reported that PAR2 activation not only promotes cell proliferation (7, 9–12) but also contributes to hypoxia-induced angiogenesis as well (13). Interestingly, PAR2 and its activating proteinases are also expressed at the leading edge of cancer invasion (10, 14), and its expression levels are tightly correlated with lymphatic metastasis in breast cancer and colorectal carcinoma (15, 16). All these observations suggest that activation of PAR2 promotes cell motility and facilitates tumor cell invasion. However, the mechanism by which PAR2 regulates cancer metastasis has not been fully understood.
MicroRNAs (miRNAs) are small ∼22 nucleotide (nt) RNA molecules that modulate gene expression by binding target mRNA usually in the 3′-untranslated region, leading to translational repression and/or degradation of mRNA (17). Increasing evidence indicate that given miRNAs including miR-125b are actively involved in the progress of tumorigenesis including metastasis and invasion (18–20). Expression of the miR-125b is suppressed in different cancers, and decreased level of miR-125b is associated with increased metastasis and the poor prognosis in patients with hepatocellular, breast cancers, and small cell carcinoma of the cervix (21–23). In addition, miR-125b is shown to modulate cell migration by targeting specific genes such as MTA1 (metastasis-associated gene 1) (24), Erbb3 (erythroblastic leukemia oncogene homolog 3) (25), and Lin28B(Lin-28 homolog B) (21).
In the present study, we demonstrated that miR-125b mediates cell migration induced by PAR2 activation and identified Gab2 (Grb2-associated-binding protein 2) as a novel target gene of miR-125b. Moreover, it was found that PAR2 decreased the level of miR-125b through NSun2 (NOP2/Sun RNA methyltransferase family, member 2)-mediated pre-miR-125b2 methylation. Our findings not only reveal a novel epigenetic mechanism underlying promotion of cancer metastasis by PAR2 but also provide potential target for development of new therapeutic approaches in human cancer.
Experimental Procedures
Cell Culture and Treatment
Human colonic epithelial cell lines HT-29, SW620, HCT-116, RKO, and human lung adenocarcinoma A549 cells were purchased from ATCC (Manassas, VA). These cells were grown in Dulbecco's modified Eagle's medium/F12 (Hyclone) supplemented with 10% fetal bovine serum (Gibco). Stable transfectant cell lines with PAR2 knockdown were developed and cultured as described previously (16).
To selectively activate PAR2, cells were treated with PAR2 activating peptide (SLIGRL-NH2, 100 μm) (Shanghai Apeptide Co. Ltd., China). Same amount of reverse peptide (LRGILS-NH2) was used as control. MiR-125b mimic, miR-125b inhibitor, and negative control oligos were obtained from GeneCopoeia (GuangZhou Ribobio Co. Ltd., China). siRNA targeting Gab2(5′-GAG ACA GCG AAG AGA ACU ATT-3′); siRNA targeting BMPR1B (5′-GGG CAA ACU UCC UUG AUA ATT-3′) and NSun2 (5′-GCC UCA UCA UAA GAU CUU ATT-3′) were purchased from Genepharma (Suzhou, China). For 5′-azacytidine (5-aza-CR) treatment, A549 cells were seeded at 2 × 105 per well in 6-well plates and incubated with 5 μmol/liter 5-aza-CR (Sigma-Aldrich) for 72 h before the treatment with PAR2-AP. The medium containing 5-aza-CR was replaced every 24 h.
RNA Isolation and Real-time PCR
Total RNA was isolated from cell lines, human, or animal samples by using Trizol reagent (Invitrogen). After treatment with DNase I, RNA was reverse transcribed into cDNA with Thermo scientific maxima first strand cDNA synthesis kit for mRNA or pri-miR-125b detection, or with TakaraTM microRNA transcription kit for mature and pre- microRNA detection. Real-time quantitative PCR was carried out on Bio-Rad S1000 PCR instrument, and each sample was analyzed in triplicate. PCR data were normalized to GAPDH or U6 snRNA expression for mRNA and miRNA, respectively.
Primers for mature miR-125b and PAR2 were obtained from GeneCopoeia. Other primers were as follows: Gab2 sense, 5′-CGC TGC TA5′-GAC AAC AGC CGA CTT CAC C-3′ and antisense, 5′-GCC CAC AAT CAT TTT CCC T-3′; BMPR1B sense, 5′-TGA TGG ACC TAT ACA CCA CAG G-3′ and antisense, 5′-ATA GTC CTT TGG ACC AGC AGA G-3′; NSun2 sense, 5′-CGC TGC TAC CTG CTC GTC-3′ and antisense, 5′-TTT CTC ATA GTG CCG TCT CCA-3′; pri-miR-125b1 sense, 5′-CCA TAC CAC CTG TTT GTT GCA TCT-3′ and antisense, 5′-CTG AGA GGA GCG CAA CAA TGT-3′; pri-miR-125b2 sense, 5′-GAA GAA TTC TAC CGC ATC AAA CCA-3′ and antisense, 5′-CTG CAG ACA ATC AAT AAG GTC CAA-3′; pre-miR-125b1 sense, 5′-TGG GAG CTG CGA TGC T-3′; pre-miR-125b2 sense, 5′-GCT CTT GGG ACC TAG GCG GA-3′. The antisense primer for miRNAs was bought from Takara (Dalian, China).
For overexpression study, full-length of Gab2 and BMPR1B sequence were purchased from Vigene (Rockville, MD, USA) and subcloned into pENTER expression vector, respectively.
Western Blot
Western blot was conducted as described earlier (16). Briefly, proteins were extracted from cultured cells with RIPA buffer (0.01% EDTA, 0.1% Triton X-100, and 10% proteinase inhibitor mixture). Protein concentrations were quantified using a protein assay kit (Bio-Rad). Lysates were separated on 10% SDS-PAGE gel and transferred to polyvinylidene difluoride membranes. The membranes were probed overnight at 4 °C with primary antibody against human Gab2 (Cell Signaling Technology, 1:1000), β-catenin (Cell Signaling Technology, 1:1000), or NSun2 (Abcam, 1:2000), followed by incubation with peroxidase-conjugated secondary antibody (Cell Signaling Technology) for 1 h. The signal was visualized with ECL (Millipore).
Migration Assay
After starving in serum-free media for 12 h, 5 × 104 cells were seeded on the top chamber of transwell (8.0-μm pore; Corning, MA) and incubated with 100 μm PAR2-AP, while 600 μl media containing 10% FBS was added to the bottom chamber. Ten hours later, the cells remaining in the upper chamber were removed with cotton swabs, and the cells on the undersurface of transwells were fixed and stained with crystal violet solution. The number of migratory cells was calculated by counting five different fields under a phase-contrast microscope of each transwell filter.
Gab2 3′-UTR Luciferase Reporter
3′-UTR sequence of Gab2 containing the putative miR-125b target sites was amplified from A549 genomic DNA using Gab2 sense, 5′-CGC TAG CCG TTC TTC CTC CCA TCC AC-3′ and antisense, 5′-CTC TAG ACC ACT GCT GAG CCT CCG TC-3′ and cloned immediately into the downstream of the luciferase reading frame in the plasmid PIS0-Report-vector. To generate the 3′-UTR fragments mutating the miR-125b recognition elements by KOD-plus-mutagenesis kit (TOYOBO Co., Ltd. Japan), the following primer pairs were used: 5′-TGA CAA CTG CGT CCC TGA ACC CTT CCC CTG GGA GGT G-3′ and antisense, 5′- AGT GCT TTG AAG GCT GAG AC-3′. The sequence of PCR product was confirmed by DNA sequencing.
Transient Transfection and Luciferase Assay
The cells in 24-well plates were transfected with or without miR-125b mimic (100 nm) together with wild type or mutant Gab2 3′UTR reporter (200 ng) and pRL-TK (2 ng). The transfection agent Lipofectamine 2000 (Invitrogen) was incubated with DNA or RNA in serum-free medium for 20 min before being added to cells and then incubating for an additional 2 h. After treatment with PAR2-AP, cell lysate was collected for luciferase activity assay, which was performed with a Dual-Luciferase assay kit (Promega). All data were normalized by pRL-TK.
Measurement of m6A RNA
To detect M6A RNA, cell extract was prepared with EZ-Magna RIPTM Kit (Millipore) according to the manufacturer's instructions. Briefly, after treatment with PAR2-AP for a half hour, A549 cells were collected and lysed in RIP lysis buffer. To pull down m6A RNA, whole-cell extract was immunoprecipitated with anti-m6A antibody (Synaptic Systems) conjugated with magnetic beads. After incubation with proteinase K, RNA was recovered followed by treatment with DNase I to avoid DNA contamination. The RNA samples were then subjected to RT and real-time quantitative PCR to detect pri-miR-125, pre-miR-125b and miR-125b. Normal mouse IgG was used as negative control. Sample without immunoprecipitation was used as input and positive control.
Human Subjects Tissue Specimen
All the colorectal cancer tissues and the paired normal tissues (≥5 cm from cancer tissue) were obtained from Cancer hospital, Chinese Academy of Medical Sciences, Beijing, China. All specimens were classified according to the tumor node metastasis (TNM) classification system. No patients received chemotherapy or radiotherapy before resection. Tissues were submerged in RNA laterTM (Ambion) for 24 h and then stored at −70 °C until use. The use of human tissues was approved by the Institutional Review Board of the Chinese Academy of Medical Sciences Cancer Institute.
Statistical Analysis
Statistical analyses were performed on data collected from at least three independent experiments. Data were presented as means ± S.D. and analysis using GraphPad Prism 5 software. Comparison of >2 groups was made using ANVOA with Tukey test. Comparison of 2 groups was made using Student's t test for unpaired data. The differences were considered statistically significant when p value less than 0.05.
Results
MiR-125b Mediates PAR2-induced Cell Migration
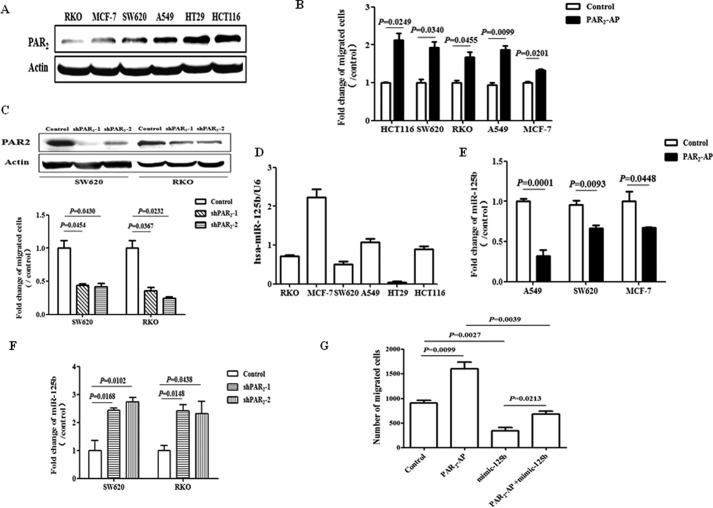
In the first set of experiments, we found that PAR2 protein was expressed in all the human cancer cell lines we tested (Fig. 1A). To determine whether PAR2 activation induce cell migration, activating peptide (PAR2-AP) was used to selectively activate PAR2 on cell surface. As shown, activation of PAR2 by PAR2-AP for 10 h significantly stimulated cell migration as measured by the transwell assay in different cancer cell lines (Fig. 1B). And there was no any effect on cell proliferation as examined by MTT assay (data not shown). In addition, knockdown of PAR2 expression with shRNA remarkably attenuated cell migration compared with that observed in scramble RNA control group in different cancer cells (Fig. 1C). These results strongly suggested that PAR2 activation promotes migration in different cell lines.
FIGURE 1.
PAR2 activation promotes cell migration through the down-regulation of miR-125b. A, expression of PAR2 protein was measured with Western blot in different cell lines. B, cell migration was evaluated with transwell assay. Cells were treated with the activating peptide of PAR2 (PAR2-AP, 100 μm) for 10 h and then migrated cells were stained and quantified. C, upper panel, Western blot shows the protein level of PAR2 in SW620 and RKO cells stably transfected with or without shPAR2. ShPAR2–1 and shPAR2–2 represent different clones from the same parental cells. Lower panel, cell migration was tested with transwell assay in these cells. D, expression level of miR-125b was measured with real time PCR in different cell lines. E, after the treatment with PAR2-AP (100 μm) for 6 h, cells were collected and tested for miR-125b expression by real-time PCR. F, level of miR-125b was measured in SW620 and RKO cells stably transfected with or without shRNA-PAR2 (shPAR2). G, cells were transfected with miR-125b mimic (50 μm). For the migration assay, thirty-six hours later, cells were seeded into transwell and treated with PAR2-AP for another 10 h. The data showed the means ± S.D. from three independent experiments.
To test whether miR-125b involves in PAR2-related migration, we measured the level of miR-125b and found that it negatively correlated with PAR2 expression in all cell lines we used (Fig. 1D). In addition, the expression of miR-125b was significantly repressed after the stimulation with PAR2-AP (Fig. 1E). On the contrary, inhibition of PAR2 expression dramatically elevated miR-125b expression compared with scramble RNA control group (Fig. 1F). Importantly, over expression of miR-125b blocked cell migration induced by PAR2 activation in A549 cells (Fig. 1G). Similar results were observed in SW620 cells (data not shown). Taken together, these findings indicate that down-regulation of miR-125b mediates PAR2-induced cell migration.
Prediction of miR-125b Target Genes
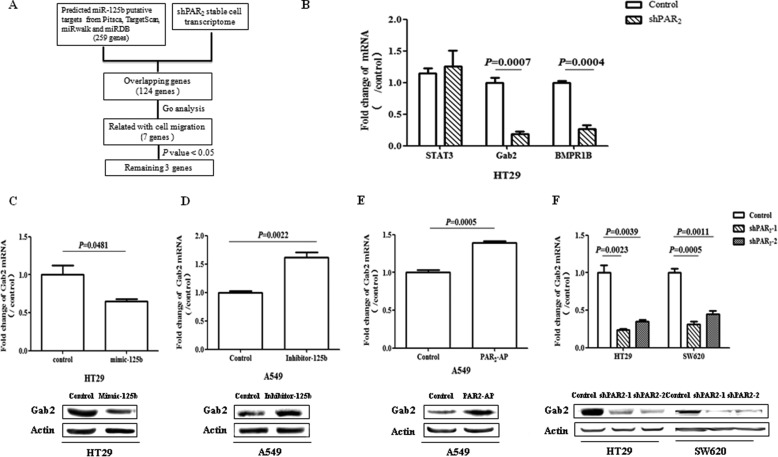
To inquire into the mechanism by which miR-125b mediates PAR2-induced cell migration, we identified the downstream targets of miR-125b. First, the target mRNAs of miR-125b were predicted with different database including Pitsca, TargetScan, miRwalk, and miRDB. There were 259 potential target transcripts by at least two databases. And 124 of them were overlapped with PAR2-related transcriptome, which was conducted with Human Gene Expression Microarray V3.0 in HT-29 cells. Among them, 7 genes were annotated involved in cell migration according to the DAVID Annotation Bioinformatics Database. Furthermore, 3 of them showed significant difference between stably knockdown of PAR2 group and scramble control group (p < 0.05) (Fig. 2A). At last, validating with real-time PCR, Gab2, and BMPR1B were significantly down-regulated with the knock down of PAR2 in HT29 cells (Fig. 2B).
FIGURE 2.
miR-125b target genes associated with PAR2 signaling. A, schematic illustration of experimental setup. B, mRNA expression of potential target genes were measured with real time-PCR in HT29 cells with or without shPAR2. C, after the transfection with miR-125b mimic (mimic-125b, 50 μm), the level of Gab2 was measured by real-time PCR and Western in HT29 cells. D, after the transfection with miR-125b inhibitor (Inhibitor-125b, 100 μm), the mRNA and protein levels of Gab2 were measured in A549 cells. E, Gab2 mRNA and protein expression was measured in A549 cells after the treatment with PAR2-AP for 6 h. F, levels of Gab2 mRNA and protein were measured in HT29 and SW620 with or without shPAR2. The data showed the means ± S.D. from three independent experiments.
Since BMPR1B has been reported as the target gene of miR-125b (Ref. 49), we verified the regulation of Gab2 by miR-125b. It was found that forced expression of miR-125b achieved by the transfection with the mimic of miR-125b inhibited Gab2 expression at both mRNA and protein levels (Fig. 2C). In contrast, the inhibitor of miR-125b elevated the expression of Gab2 (Fig. 2D). Furthermore, activation of PAR2 increased Gab2 expression (Fig. 2E) while disruption of PAR2 signaling repressed Gab2 (Fig. 2F) at both mRNA and protein levels. All these results strongly suggest that Gab2 is a possible target gene of miR-125b.
Gab2 Was Identified as a Novel Target Gene of miR-125b
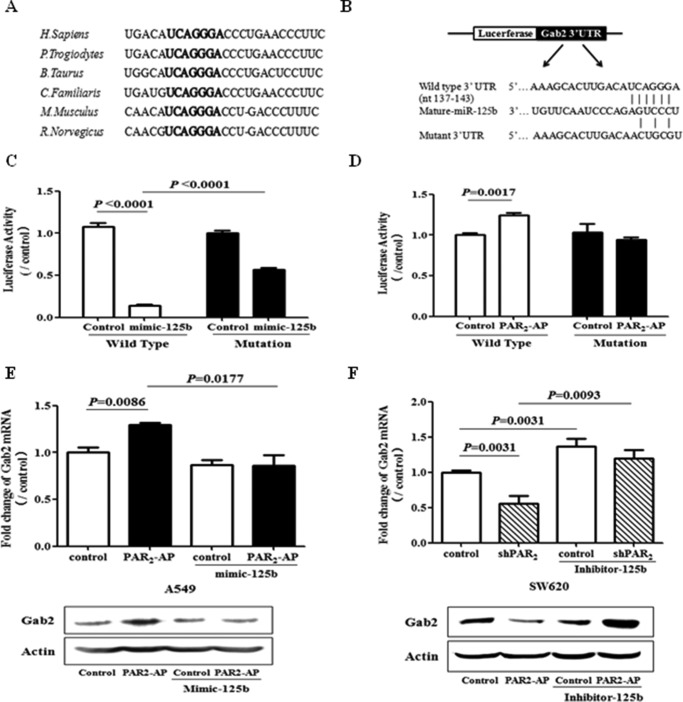
To confirm Gab2 is a direct target gene of miR125b, the possible binding site of miR-125 in the Gab2 mRNA was analyzed. TargetScan analysis indicated that Gab2 contains miR-125b binding site within its 3′-UTR, and the sequence of the binding site is highly conserved across different species (Fig. 3A). To further validate that Gab2 is regulated by miR-125b through interaction of the complementary 3′-UTR region, luciferase reporters containing wild-type or mutant 3′-UTR of Gab2 were constructed (Fig. 3B). MiR-125b mimic significantly reduced the activity of the luciferase reporter, which was partially abolished by the mutation of miR-125b binding site in 3′-UTR of Gab2 (Fig. 3C). Furthermore, PAR2 activation up-regulated the activity of wild type Gab2 3′-UTR-derived luciferase reporter but not the mutated one (Fig. 3D). Consistent with luciferase activity, PAR2 activation also elevated Gab2 mRNA and protein levels, which were reduced by overexpression of miR-125b (Fig. 3E). Contrarily, inhibition of PAR2 repressed the levels of Gab2 mRNA and protein, which was recovered by the inhibitor of miR-125b (Fig. 3F). All these results suggest that miR125b partially regulates Gab2 expression through the binding site within 3′-UTR of Gab2 mRNA.
FIGURE 3.
Gab2 is a direct downstream target of miR-125b. A, potential binding site of miR-125b in Gab2 3′-UTR in different species. B, diagram showed the putative miR-125b-targeting seed sequence on Gab2–3′-UTR and illustrated the structure of wild type and mutant luciferase reporter of Gab2 3′-UTR. C, A549 cells were transfected with or without miR-125b mimic combined with wild type or mutant luciferase reporter of Gab2–3′-UTR. Twenty-four hours later, the cells were collected for luciferase assay. D, after the transfection with wild type or mutant luciferase reporter of Gab2–3′-UTR, A549 cells were stimulated with PAR2-AP for 6 h and then collected for luciferase assay. E, A549 cells were transfected with miR-125b mimic (mimic-125b) or control RNA. One day after transfection, the cells were treated with PAR2-AP for 6 h, and the cell lysates were collected for Gab2 mRNA and protein expression assay. F, SW620 cells with or without stable transfection of shPAR2 were transfected with inhibitor of miR-125b (Inhibitor-125b) or control RNA. Twenty-four hours later, the cell lysate was collected for Gab2 mRNA and protein expression assay. The data showed the means ± S.D. from three independent experiments.
Gab2 Mediates PAR2-induced Cell Migration
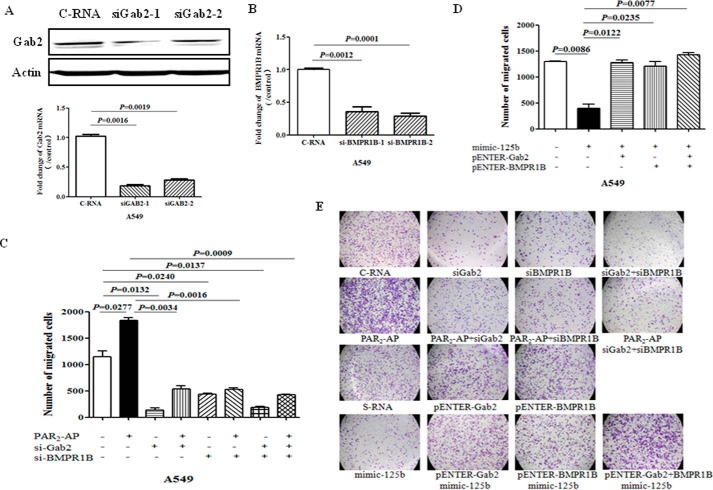
To test whether Gab2 mediates PAR2-induced cell migration, specific siRNAs against Gab2 were applied to reduce the expression of Gab2 at both mRNA and protein levels (Fig. 4A). Knockdown of Gab2 significantly blocked PAR2-induced cell migration (Fig. 4, C and E). Since BMPR1B, a known target gene of miR-125b, had been changed after PAR2 activation, we checked the effect of BMPR1B on PAR2-induced cell migration. Knockdown of BMPR1B with specific siRNA (Fig. 4B) also blocked cell migration stimulated by PAR2 activation (Fig. 4, C and E). In addition, overexpression of Gab2 or/and BMPR1B totally reversed the repression of cell migration induced by miR-125b (Fig. 4, D and E). These findings indicate that repression of miR-125b mediates cell migration induced by PAR2 through targeting Gab2 and BMPR1B expression in vitro.
FIGURE 4.
Gab2 and BMPR1B mediate PAR2-induced cell migration. A549 cells were transfected with siRNA against Gab2 or BMPR1B. Forty hours after transfection, the cells were collected to measure the expression of (A) Gab2 or (B) BMPR1B at protein and/or mRNA levels. C, for cell migration assay, forty hours after transfection, the cells were seeded on transwell and treated with PAR2-AP. After 10-h treatment, migrated cells were quantified. D, after transfection with mimic miR-125b with or without Gab2 or/and BMPR1B overexpression plasmid, the ability of cell migration was measured with transwell assay. E, representative pictures for the migration assay shown for 10 h. The data showed the means ± S.D. from three independent experiments.
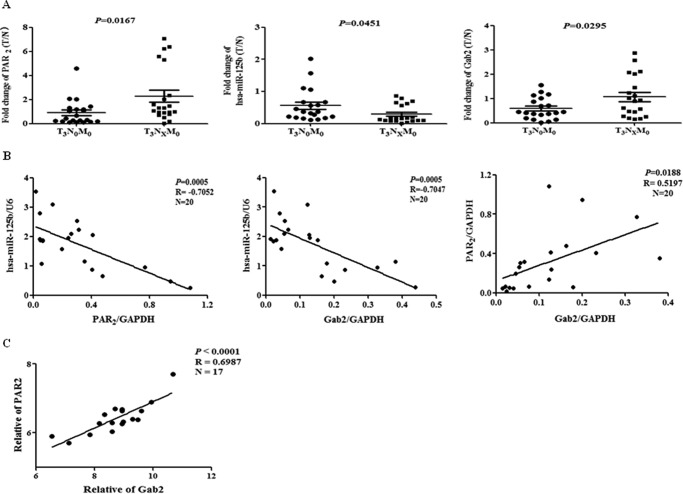
Correlation of PAR2, miR-125b, and Gab2 in CRC Specimens
To confirm the findings in vivo, we tested the expression of PAR2, miR-125b and Gab2 in 40 paired human CRC tissue specimens measured by real-time PCR. As shown in Fig. 5A, both PAR2 and Gab2 were up-regulated while miR-125b was down-regulated in lymphatic metastasis group compared with those of non-metastasis group. Importantly, in lymphatic metastasis group, the level of PAR2 was negatively correlated with that of miR-125b and positively correlated with that of Gab2 (Fig. 5B). Furthermore, miR-125b expression was negatively correlated with Gab2 levels (Fig. 5B). In addition, the level of PAR2 was also positively correlated with Gab2 in gene expression profiling of lung adenocarcinoma (NCBI GEO accession number GSE31546) uploaded by Hamon Center for Therapeutic Oncology Research of UT Southwestern Medical Center, which provided 17 primary lung cancer specimens expression profiling (Fig. 5C). These results strongly suggest that PAR2 mediates metastasis through miR-125b/Gab2 in human CRC.
FIGURE 5.
Correlation of PAR2, miR-125b and GAB2 in human specimens. A, expression of PAR2, Gab2 and miR-125b in human CRC samples was tested with real time PCR. The data are shown as the fold change of tumor sample (T) to paired normal tissue (N). B, Pearson's correlation analysis was conducted for PAR2/miR-125b, Gab2/miR-125b and PAR2/Gab2. Black line represents linear regression line. C, Pearson's correlation analysis was conducted for PAR2/Gab2 in human lung cancer specimens microarray, which provided 17 primary lung cancer specimens gene expression profiling (NCBI GEO accession number GSE31546).
PAR2 Suppresses miR-125b through NSun2-mediated Repression of miR-125b Processing
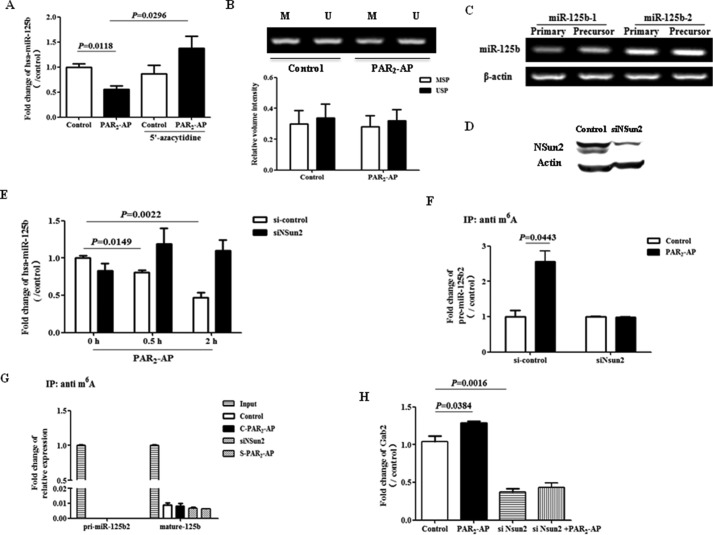
Since miR-125b mediate PAR2-induced cell migration both in vitro and in vivo, it is important to understand how PAR2 signaling regulates the level of miR-125b. Recent studies showed that the methylation state of CpG-rich regions is correlated with the expression pattern of miR-125b in breast cancer (22). To investigate whether hypermethylation of miR-125b promoter contribute to PAR2-induced repression of miR-125b, 5-azacytidine, a demethylation agent, was used. The data showed that pretreatment with 5-azacytidine completely reverse the level of miR-125b reduced by PAR2-AP (Fig. 6A). However, there was no obvious change in methylation status of pri-miR-125b1 promoter as measured by MSP after PAR2 activation (Fig. 6B). Moreover, there was no significant change of primary miR-125b after PAR2 activation (data not shown).
FIGURE 6.
PAR2 regulates miR-125b expression through NSun2-dependent mechanism. A, after the pretreatment with 5′-azacytidine for 72 h, A549 cells were stimulated with PAR2-AP for 6 h. The level of miR-125b was measured by real time PCR. B, A549 cells were treated with PAR2-AP for 6 h and then collected for MSP PCR to detect the methylation of miR-125b1 promoter. Upper panel, representative pictures of MSP PCR; lower panel, quantification of three independent MSP PCR. M, methylated form. U, unmethylated form. C, levels of pri- and pre-miR-125b1, pri- and pre-miR-125b2 were measured with PCR. D, Western blot shows the expression of Nsun2 after transfection with control RNA or siRNA against Nsun2 (siNSun2) in A549 cells. E, 40 h after transfection with si-Nsun2, cells were stimulated with PAR2-AP for 0.5 or 2 h and then collected to measure miR-125b expression. F and G, after transfection with siNSun2, A549 cells were treated with PAR2-AP for 0.5 h. RNA was isolated and pulled down with an anti-m6A antibody. Pull down material was collected to measure the levels of (F) pre-miR-125b2 and (G) pri-miR-125b2 and miR-125b with real-time PCR. H, after transfection with siNSun2, A549 cells were treated with PAR2-AP for 6 h and then collected to detect Gab2 mRNA by real time PCR. The data showed the means ± S.D. from three independent experiments.
Actually, mature miR-125b comes from two primary forms, pri-miR-125b1 and pri-miR-125b2. Most recently, NSun2, a RNA methyltransferase, is shown to interfere in the mature processing of miR-125b from pri- and pre-miR-125b2. Interestingly, we found that pri- and pre-miR-125b2 was the predominant form of miR-125 precursor in A549 cells but not pri- and pre-miR-125b1 (Fig. 6C). Furthermore, knockdown of NSun2 with specific siRNA (Fig. 6D) blocked the repression of miR-125b induced by PAR2 activation (Fig. 6E).
To test whether PAR2 activation induces methylation of miR-125b and its precursors, methylated RNA was pull down with anti-m6A antibody and miRNA and its precursors were measured with qPCR. The data showed that PAR2 activation induced m6A methylation of pre-miR-125b2 (Fig. 6F) but not pri-miR-125b2 (Fig. 6G). Moreover, knockdown of NSun2 completely blocked both PAR2-induced m6A methylation of pre-miR-125b2 (Fig. 6F) and PAR2-induced Gab2 mRNA expression (Fig. 6H). These findings strongly suggest that PAR2 reduces miR-125b expression through an NSun2-dependent and m6A methylation-involved mechanism.
Discussion
Both PAR2 and its-activating proteinases, including tissue factor, trypsin, KLKs, are aberrantly overexpressed in a variety of malignancies, and shown to be involved in many aspects of cancer biology including cell proliferation, angiogenesis, and metastasis (26, 27). In addition, PAR2 contributes to the tumorigenesis through the induction of Cox-2 (28), MMP 2/9 (29), and interleukin-8 (30). Inhibition of PAR2 not only effectively suppresses tumor growth (31) but also prevents tumor formation in mouse model (8). Therefore, PAR2 is an attractive and potential therapeutic target for tumor. Here, we demonstrated that miR-125b mediates PAR2-promoted cell migration by targeting Gab2 in cancer cells. Moreover, PAR2 suppressed the processing of miR-125b in Nsun2-dependent and miRNA methylation-mediated manner (Fig. 7). Thus, the interaction between PAR2 signaling and miRNAs provides novel intrinsic molecular mechanism by which PAR2 participates in tumorigenesis.
FIGURE 7.
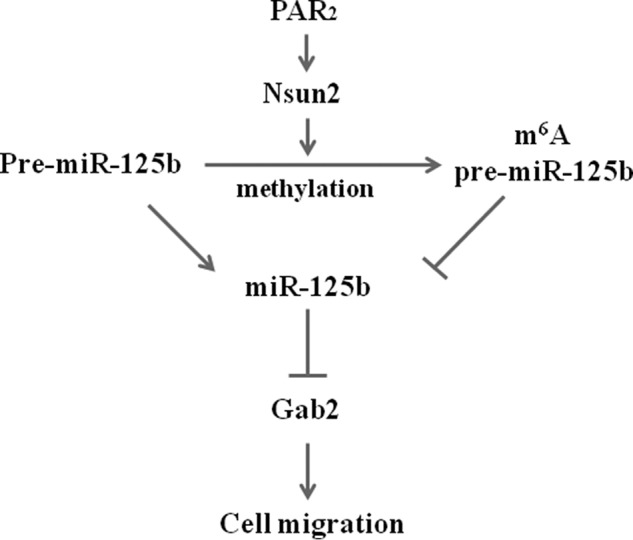
Diagram of signaling pathways responsible for PAR2-regulated miR-125b expression and function. PAR2 (Proteinase activated receptor 2) activation promotes Nsun2 (NOP2/Sun domain family, member 2)-dependent m6A methylation of pre-miR-125b2, which interferes with the processing of miR-125b from pre-miR-125b and results in the down-regulation of miR-125b. Gab2 is a downstream target of miR-125b and mediates the cell migration induced by PAR2. According to the ending of lines, “⊥” represents inhibitory effect while “↓” represents stimulatory effect.
In the last decade, miRNAs have emerged as critical regulators in various cancers, including colorectal carcinoma (33, 34). The role of miR-125b in tumorigenesis is controversial since it can act as oncogene or tumor suppressor, depending on types of different tumors. Deregulations of miR-125b are observed commonly in breast (22, 32), ovarian (36), and liver cancers (21), while its overexpression is implicated in pancreatic (37), and prostate (38) cancers. With regard to the expression of miR-125b in CRC, the evidence is conflicting. A study from Japan shows that high levels of miR-125b expression are associated with reduced survival rates in 89 colorectal tumors specimens (39). However, due to the absence of paired adjacent non-tumor tissue for each CRC sample, it is unclear whether the level of miR-125b in tumor is lower than that in non-tumor tissue. Compared with paired non-tumor tissue, our results clearly showed that miR-125b expression was down-regulated in tumor site, and it was also associated with lymphatic metastasis in CRC specimens (Fig. 5A).
Although deregulation of miR-125b plays a role in tumor development, the regulation of miR-125b expression is not well understood. Promoter methylation is a common mechanism for silencing miRNAs in cancer (40). Hypermethylation of miR-125b-1 promoter is involved in miR-125b down-regulation in invasive breast cancer (22). Although the inhibition of methylation by 5-aza recovered the level of miR-125b (Fig. 6A), the expression of pri-miR-125b-1 and pri-miR125b-2 were not altered by PAR2 activation (data not shown). Thus, our evidence does not support the transcriptional suppression of miR-125b by PAR2.
Except for DNA methylation, RNA methylation is also implicated in the control of miR-125b processing (41). Methylation is a prevalent modification of almost all species of RNA. Methylation of RNA regulates the stability of RNAs and the biogenesis/processing of RNAs (42). Recently, the processing of miRNA-125b was found to be regulated by Nsun2-mediated RNA methylation in mammalian cells (41). NSun2 (NOP2/Sun domain family, member 2 is a nucleolar RNA methyltransferase implicating in cell proliferation, stem cell differentiation, and human cancers (42). NSun2 is highly expressed in cancer (43) and validated as a transcriptional target of the Lef1/β-catenin complex in hair follicles (44) and c-myc in skin (43). NSun2 methylates the 3′-untranslated region (UTR) of p16 mRNA and induces the stabilization of p16 mRNA (45). Methylation by Nsun2 decreases the levels miR-125b and disrupts its function. NSun2 is also shown to methylate precursor of miR-125b, interferes with its processing and reduces the level of mature miR-125b (41). In our study, Nsun2 silencing not only inhibited m6A-containing pre-miR-125b-2 (Fig. 6F) but also rescued the down-regulation of miR-125b induced by PAR2 (Fig. 6E). These findings indicate that PAR2 signaling interferes miR-125b processing through miRNA methylation in Nsun2-dependent manner.
Gab2 was identified as a novel target of miR-125b in the current study. Gab2, as a scaffolding protein, integrates and amplifies signals in a variety of sources including growth factors, cytokine, and antigen receptors as well as cell adhesion molecules (46). Recently, Gab2 is emerging as an oncoprotein, and its protein is frequently overexpressed in various cancers (46). Here we showed that Gab2 expression was associated with lymph node metastasis in CRC, and the findings are consistent with other recent reports (46, 47). Besides gene amplification, E2F and estrogen signaling have also been reported to transcriptionally up-regulate Gab2 expression (35). In the present study, we provided a novel epigenetic mechanism by which Gab2 was up-regulated by PAR2 signaling. Our previous study demonstrated that PAR2 signaling also activates the transcriptional activity of E2F in colon cancer cells (16). Therefore, our results strongly suggest that PAR2 up-regulates Gab2 expression at both transcriptional and post-transcriptional levels in vivo. Moreover, as a scaffolding protein, Gab2 mediates the crosstalk between various signaling pathways (46). This also provides the basis for the synergistic action of PAR2 signaling. For example, Gab2 acts as a downstream of EGFR (35, 47), which is transactivated by PAR2 (7). However, further study is needed to test whether Gab2 mediates transactivation of EGFR by PAR2 activation. Therefore, considering the simultaneous exposure to plenty of stimuli in the real in vivo condition, Gab2 is considered as a key molecule for the crosstalk and integration of PAR2 signaling and other pathways.
In conclusion, our results indicate that activation of PAR2 promotes cell migration through the down-regulation of miR-125b via m6A-related mechanisms. Our study provides a novel epigenetic mechanism by which microenvironment regulates the level of miRNAs and promotes cancer cell migration.
Author Contributions
H. W. and L. Y. designed the study and wrote the paper. Y. M. and W. H. performed array data. W. L. and L. C. performed human data analysis. Z. Z., Y. T., and X. Z. collected human samples. W. W. designed the study on m6A methylation. All authors analyzed the results and approved the final version of the manuscript.
This work was supported by funding from the 973 National Key Fundamental Research Program of China (2012CB967003) and the National Nature Science Foundation of China (81172034 and 91129717). The authors declare that they have no conflicts of interest with the contents of this article.
- PAR
- protease-activated receptor
- GPCR
- G-protein-coupled receptor
- miRNA
- microRNA
- Gab
- Grb2-associated-binding protein
- m6A
- N6-methyladenosine.
References
- 1.Soreide K., Janssen E. A., Körner H., and Baak J. P. (2006) Trypsin in colorectal cancer: molecular biological mechanisms of proliferation, invasion, and metastasis. J. Pathol. 209, 147–156 [DOI] [PubMed] [Google Scholar]
- 2.Melillo R. M., Guarino V., Avilla E., Galdiero M. R., Liotti F., Prevete N., Rossi F. W., Basolo F., Ugolini C., de Paulis A., Santoro M., and Marone G. (2010) Mast cells have a protumorigenic role in human thyroid cancer. Oncogene 29, 6203–6215 [DOI] [PubMed] [Google Scholar]
- 3.Dorn J., Harbeck N., Kates R., Gkazepis A., Scorilas A., Soosaipillai A., Diamandis E., Kiechle M., Schmalfeldt B., and Schmitt M. (2011) Impact of expression differences of kallikrein-related peptidases and of uPA and PAI-1 between primary tumor and omentum metastasis in advanced ovarian cancer. Ann. Oncol. 22, 877–883 [DOI] [PubMed] [Google Scholar]
- 4.Gieseler F., Ungefroren H., Settmacher U., Hollenberg M. D., and Kaufmann R. (2013) Proteinase-activated receptors (PARs) - focus on receptor-receptor-interactions and their physiological and pathophysiological impact. Cell Commun. Signal 11, 86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Black P. C., Mize G. J., Karlin P., Greenberg D. L., Hawley S. J., True L. D., Vessella R. L., and Takayama T. K. (2007) Overexpression of protease-activated receptors-1,-2, and-4 (PAR-1, -2, and -4) in prostate cancer. Prostate 67, 743–756 [DOI] [PubMed] [Google Scholar]
- 6.Massi D., Naldini A., Ardinghi C., Carraro F., Franchi A., Paglierani M., Tarantini F., Ketabchi S., Cirino G., Hollenberg M. D., Geppetti P., and Santucci M. (2005) Expression of protease-activated receptors 1 and 2 in melanocytic nevi and malignant melanoma. Hum. Pathol. 36, 676–685 [DOI] [PubMed] [Google Scholar]
- 7.Caruso R., Pallone F., Fina D., Gioia V., Peluso I., Caprioli F., Stolfi C., Perfetti A., Spagnoli L. G., Palmieri G., Macdonald T. T., and Monteleone. G. (2006) Protease-activated receptor-2 activation in gastric cancer cells promotes epidermal growth factor receptor trans-activation and proliferation. Am. J. Pathol. 169, 268–278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Versteeg H. H., Schaffner F., Kerver M., Ellies L. G., Andrade-Gordon P., Mueller B. M., and Ruf W. (2008) Protease-activated receptor (PAR) 2, but not PAR1, signaling promotes the development of mammary adenocarcinoma in polyoma middle T mice. Cancer Res. 68, 7219–7227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Darmoul D., Gratio V., Devaud H., and Laburthe M. (2004) Protease-activated receptor 2 in colon cancer: trypsin-induced MAPK phosphorylation and cell proliferation are mediated by epidermal growth factor receptor transactivation. J. Biol. Chem. 279, 20927–20934 [DOI] [PubMed] [Google Scholar]
- 10.Nishibori M. , Mori S., and Takahashi H. K. (2005) Physiology and pathophysiology of proteinase-activated receptors (PARs): PAR-2-mediated proliferation of colon cancer cell. J. Pharmacol. Sci. 97, 25–30 [DOI] [PubMed] [Google Scholar]
- 11.Yada K., Shibata K., Matsumoto T., Ohta M., Yokoyama S., and Kitano S. (2005) Protease-activated receptor-2 regulates cell proliferation and enhances cyclooxygenase-2 mRNA expression in human pancreatic cancer cells. J. Surg. Oncol. 89, 79–85 [DOI] [PubMed] [Google Scholar]
- 12.Sánchez-Hernández P. E., Ramirez-Dueñas M. G., Albarran-Somoza B., García-Iglesias T., del Toro-Arreola A., Franco-Topete R., Daneri-Navarro A. (2008) Protease-activated receptor-2 (PAR-2) in cervical cancer proliferation. Gynecol. Oncol. 108, 19–26 [DOI] [PubMed] [Google Scholar]
- 13.Uusitalo-Jarvinen H., Kurokawa T., Mueller B. M., Andrade-Gordon P., Friedlander M., and Ruf W. (2007) Role of protease activated receptor 1 and 2 signaling in hypoxia-induced angiogenesis. Arterioscler. Thromb. Vasc. Biol. 27, 1456–1462 [DOI] [PubMed] [Google Scholar]
- 14.Ohta T., Shimizu K., Yi S., Takamura H., Amaya K., Kitagawa H., Kayahara M., Ninomiya I., Fushida S., Fujimura T., Nishimura G., and Miwa K. (2003) Protease-activated receptor-2 expression and the role of trypsin in cell proliferation in human pancreatic cancers. Int. J. Oncol. 23, 61–66 [PubMed] [Google Scholar]
- 15.Su S., Li Y., Luo Y., Sheng Y., Su Y., Padia R. N., Pan Z. K., Dong Z., and Huang S. (2009) Proteinase-activated receptor 2 expression in breast cancer and its role in breast cancer cell migration. Oncogene 28, 3047–3057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ma Y., Bao-Han W., Lv X., Su Y., Zhao X., Yin Y., Zhang X., Zhou Z., MacNaughton W. K., and Wang H. (2013) MicroRNA-34a mediates the autocrine signaling of PAR2-activating proteinase and its role in colonic cancer cell proliferation. PLoS ONE 8, e72383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ha M., and Kim V. N. (2014) Regulation of microRNA biogenesis. Nat. Rev. Mol. Cell Biol. 15, 509–524 [DOI] [PubMed] [Google Scholar]
- 18.Yang R., Dick M., Marme F., Schneeweiss A., Langheinz A., Hemminki K., Sutter C., Bugert P., Wappenschmidt B., Varon R., Schott S., Weber B. H., Niederacher D., Arnold N., Meindl A., Bartram C. R., Schmutzler R. K., Müller H., Arndt V., Brenner H., Sohn C., and Burwinkel B. (2011) Genetic variants within miR-126 and miR-335 are not associated with breast cancer risk. Breast Cancer Res. Treat. 127, 549–554 [DOI] [PubMed] [Google Scholar]
- 19.Yang S., Li Y., Gao J., Zhang T., Li S., Luo A., Chen H., Ding F., Wang X., and Liu Z. (2013) MicroRNA-34 suppresses breast cancer invasion and metastasis by directly targeting Fra-1. Oncogene 32, 4294–4303 [DOI] [PubMed] [Google Scholar]
- 20.Zhou W., Fong M. Y., Min Y., Somlo G., Liu L., Palomares M. R., Yu Y., Chow A., O'Connor S. T., Chin A. R., Yen Y., Wang Y., Marcusson E. G., Chu P., Wu J., Wu X., Li A. X., Li Z., Gao H., Ren X., Boldin M. P., Lin P. C., and Wang S. E. (2014) Cancer-secreted miR-105 destroys vascular endothelial barriers to promote metastasis. Cancer Cell 25, 501–515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liang L., Wong C. M., Ying Q., Fan D. N., Huang S., Ding J., Yao J., Yan M., Li J., Yao M., Ng I. O., and He X. (2010) MicroRNA-125b suppressesed human liver cancer cell proliferation and metastasis by directly targeting oncogene LIN28B2. Hepatology 52, 1731–1740 [DOI] [PubMed] [Google Scholar]
- 22.Zhang Y., Yan L. X., Wu Q. N., Du Z. M., Chen J., Liao D. Z., Huang M. Y., Hou J. H., Wu Q. L., Zeng M. S., Huang W. L., Zeng Y. X., and Shao J. Y. (2011) miR-125b is methylated and functions as a tumor suppressor by regulating the ETS1 proto-oncogene in human invasive breast cancer. Cancer Res. 71, 3552–3562 [DOI] [PubMed] [Google Scholar]
- 23.Huang L., Lin J. X., Yu Y. H., Zhang M. Y., Wang H. Y., and Zheng M. (2012) Downregulation of six microRNAs is associated with advanced stage, lymph node metastasis and poor prognosis in small cell carcinoma of the cervix. PLoS ONE 7, e33762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li Y., Chao Y., Fang Y., Wang J., Wang M., Zhang H., Ying M., Zhu X., and Wang H. (2013) MTA1 promotes the invasion and migration of non-small cell lung cancer cells by downregulating miR-125b. J Exp Clin Cancer Res. 32, 33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Scott G. K., Goga A., Bhaumik D., Berger C. E., Sullivan C. S., and Benz C. C. (2007) Coordinate suppression of ERBB2 and ERBB3 by enforced expression of micro-RNA miR-125a or miR-125b. J. Biol. Chem. 282, 1479–1486 [DOI] [PubMed] [Google Scholar]
- 26.Schaffner F., Yokota N., and Ruf W. (2012) Tissue factor proangiogenic signaling in cancer progression. Thromb Res. 129 Suppl. 1, S127-S131 [DOI] [PubMed] [Google Scholar]
- 27.Hu L., Xia L., Zhou H., Wu B., Mu Y., Wu Y., and Yan J. (2013) TF/FVIIa/PAR2 promotes cell proliferation and migration via PKCalpha and ERK-dependent c-Jun/AP-1 pathway in colon cancer cell line SW620. Tumour Biol. 34, 2573–2581 [DOI] [PubMed] [Google Scholar]
- 28.Wang H., Wen S., Bunnett N. W., Leduc R., Hollenberg M. D., and MacNaughton W. K. (2008) Proteinase-activated receptor-2 induces cyclooxygenase-2 expression through beta-catenin and cyclic AMP-response element-binding protein. J. Biol. Chem. 283, 809–815 [DOI] [PubMed] [Google Scholar]
- 29.Wilson S. R., Gallagher S., Warpeha K., and Hawthorne S. J. (2004) Amplification of MMP-2 and MMP-9 production by prostate cancer cell lines via activation of protease-activated receptors. Prostate 60, 168–174 [DOI] [PubMed] [Google Scholar]
- 30.Wang H., Moreau F., Hirota C. L., and MacNaughton W. K. (2010) Proteinase-activated receptors induce interleukin-8 expression by intestinal epithelial cells through ERK/RSK90 activation and histone acetylation. FASEB J. 24, 1971–1980 [DOI] [PubMed] [Google Scholar]
- 31.Versteeg H. H., Schaffner F., Kerver M., Petersen H. H., Ahamed J., Felding-Habermann B., Takada Y., Mueller B. M., and Ruf W. (2008) Inhibition of tissue factor signaling suppresses tumor growth. Blood 111, 190–199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Saetrom P., Biesinger J., Li S. M., Smith D., Thomas L. F., Majzoub K.., Rivas G. E., Alluin J., Rossi J. J., Krontiris T. G., Weitzel J., Daly M. B., Benson A. B., Kirkwood J. M., O'Dwyer P. J., Sutphen R., Stewart J. A., Johnson D., and Larson G. P. (2009) A risk variant in an miR-125b binding site in BMPR1B is associated with breast cancer pathogenesis. Cancer Res. 69, 7459–7465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lu J., Getz G., Miska E. A., Alvarez-Saavedra E., Lamb J., Peck D., Sweet-Cordero A., Ebert B. L., Mak R. H., Ferrando A. A., Downing J. R., Jacks T., Horvitz H. R., and Golub T. R. (2005) MicroRNA expression profiles classify human cancers. Nature 435, 834–838 [DOI] [PubMed] [Google Scholar]
- 34.Lee Y. S., and Dutta A. (2009) MicroRNAs in cancer. Annu.Rev. Pathol. 4, 199–227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gu H., and Neel B. G. (2003) The “Gab” in signal transduction. Trends Cell Biol. 13, 122–130 [DOI] [PubMed] [Google Scholar]
- 36.Guan Y., Yao H., Zheng Z., Qiu G., and Sun K. (2011) MiR-125b targets BCL3 and suppresses ovarian cancer proliferation. Int. J. Cancer 128, 2274–2283 [DOI] [PubMed] [Google Scholar]
- 37.Bera A., VenkataSubbaRao K., Manoharan M. S., Hill P., and Freeman J. W. (2014) A miRNA signature of chemoresistant mesenchymal phenotype identifies novel molecular targets associated with advanced pancreatic cancer. PLoS ONE 9:e106343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Amir S., Ma A. H., Shi X. B., Xue L., Kung H. J., and Devere White R. W. (2013) Oncomir miR-125b suppresses p14(ARF) to modulate p53-dependent and p53-independent apoptosis in prostate cancer. PLoS ONE 8, e61064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nishida N., Yokobori T., Mimori K., Sudo T., Tanaka F., Shibata K., Ishii H., Doki Y., Kuwano H., and Mori M. (2011) MicroRNA miR-125b is a prognostic marker in human colorectal cancer. Int J Oncol 38, 1437–1443 [DOI] [PubMed] [Google Scholar]
- 40.Tsai K. W., Kao H. W., Chen H. C., Chen S. J., and Lin W. C. (2009) Epigenetic control of the expression of a primate-specific microRNA cluster in human cancer cells. Epigenetics 4, 587–592 [DOI] [PubMed] [Google Scholar]
- 41.Yuan S., Tang H., Xing J., Fan X., Cai X., Li Q., Han P., Luo Y., Zhang Z., Jiang B., Dou Y., Gorospe M., and Wang W. (2014) Methylation by NSun2 represses the levels and function of microRNA 125b. Mol. Cell. Biol. 34, 3630–3641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ji L., and Chen X. (2012) Regulation of small RNA stability: methylation and beyond. Cell Res 22, 624–636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Frye M., and Watt F. M. (2006) The RNA methyltransferase Misu (NSun2) mediates Myc-induced proliferation and is upregulated in tumors. Curr. Biol. 16, 971–981 [DOI] [PubMed] [Google Scholar]
- 44.Blanco S., Kurowski A., Nichols J., Watt F. M., Benitah S. A., and Frye M. (2011) The RNA-methyltransferase Misu (NSun2) poises epidermal stem cells to differentiate. PLoS Genet. 7:e1002403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang X., Liu Z., Yi J., Tang H., Xing J., Yu M., Tong T., Shang Y., Gorospe M., and Wang W. (2012) The tRNA methyltransferase NSun2 stabilizes p16INK(4) mRNA by methylating the 3′-untranslated region of p16. Nat. Commun. 3, 712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wöhrle F. U., Daly R. J., and Brummer T. (2009) Function, regulation and pathological roles of the Gab/DOS docking proteins. Cell Commun. Signal 7, 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Matsumura T., Sugimachi K., Takahashi Y., Uchi R., Sawada G., and Ueda M. (2014) Clinical Significance of GAB2, a Scaffolding/Docking Protein Acting Downstream of EGFR in Human Colorectal Cancer. Ann. Surg. Oncol. 21, 743–749 [DOI] [PubMed] [Google Scholar]