Background: Levoglucosan kinase converts the cellulosic biomass pyrolysis product levoglucosan to the common metabolite glucose 6-phosphate.
Results: Crystallographic structures demonstrate two metal binding sites and an alternate binding arrangement for levoglucosan.
Conclusion: The unusual mode of levoglucosan binding provides a rationale for the high Km of LGK for its substrate.
Significance: These results provide a greater understanding of the underlying biochemistry of anhydrosugar processing.
Keywords: biofuel, carbohydrate processing, crystal structure, hexokinase, structural biology, anhydrosugar, levoglucosan
Abstract
The most abundant carbohydrate product of cellulosic biomass pyrolysis is the anhydrosugar levoglucosan (1,6-anhydro-β-d-glucopyranose), which can be converted to glucose 6-phosphate by levoglucosan kinase (LGK). In addition to the canonical kinase phosphotransfer reaction, the conversion requires cleavage of the 1,6-anhydro ring to allow ATP-dependent phosphorylation of the sugar O6 atom. Using x-ray crystallography, we show that LGK binds two magnesium ions in the active site that are additionally coordinated with the nucleotide and water molecules to result in ideal octahedral coordination. To further verify the metal binding sites, we co-crystallized LGK in the presence of manganese instead of magnesium and solved the structure de novo using the anomalous signal from four manganese atoms in the dimeric structure. The first metal is required for catalysis, whereas our work suggests that the second is either required or significantly promotes the catalytic rate. Although the enzyme binds its sugar substrate in a similar orientation to the structurally related 1,6-anhydro-N-acetylmuramic acid kinase (AnmK), it forms markedly fewer bonding interactions with the substrate. In this orientation, the sugar is in an optimal position to couple phosphorylation with ring cleavage. We also observed a second alternate binding orientation for levoglucosan, and in these structures, ADP was found to bind with lower affinity. These combined observations provide an explanation for the high Km of LGK for levoglucosan. Greater knowledge of the factors that contribute to the catalytic efficiency of LGK can be used to improve applications of this enzyme for levoglucosan-derived biofuel production.
Introduction
The anhydrosugar levoglucosan (1,6-anhydro-β-d-glucopyranose) is a well recognized product of biomass burning that is often used as a molecular tracer of such events in soils, aerosols, snow pits, and even human urine (1–6). Although not as abundant as levoglucosan, other anhydrosugars such as mannosan, galactosan, levogalactosan, levomannosan, and cellobiosan have also been detected in aerosol and soil samples (1–4). It has been estimated that 4 billion metric tons of carbon are released globally by biomass burning every year (7). A characterization of wildfire smoke demonstrated that anhydrosugars were the second most abundant form of organic carbon in given samples, at a concentration of 24 mg/g of organic carbon (8), leading to an estimated annual production of 90 million metric tons of anhydrosugars. These anhydrosugars thus represent a significant yet undercharacterized portion of the global carbon cycle. Although the radical-mediated degradation of aerosol-associated levoglucosan has been observed (9, 10), it has also been reported that levoglucosan, and presumably other anhydrosugars, can return to the soil via rainwater (11). Once in the soil, these sugars are subjected to further degradation (12). Two distinct biological pathways have been identified during levoglucosan degradation. The ATP-dependent levoglucosan kinase (LGK)2 pathway (Fig. 1), which is present in some fungi, produces glucose 6-phosphate (G6P), a common metabolic intermediate (13). The modularity of this pathway has been demonstrated by its functional expression in bacteria such as Escherichia coli (14). A bacterial levoglucosan dehydrogenase pathway that converts levoglucosan to 3-keto-levoglucosan in an NAD+-dependent reaction has also been described (15). However, the detailed mechanisms of LGK and levoglucosan dehydrogenase, associated metabolic pathways, and the identity of levoglucosan membrane transporters remain unknown. An increased understanding of the biological pathways associated with the microbial degradation of these sugars can be realized by identifying and characterizing the enzymes and cofactors required for these specific reactions.
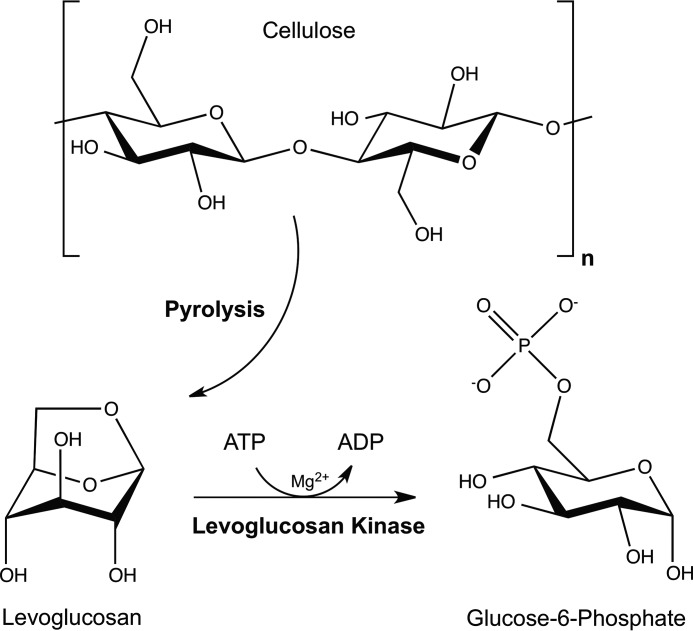
FIGURE 1.
Conversion of cellulose to levoglucosan through pyrolysis is followed by enzymatic cleavage and phosphorylation through the action of levoglucosan kinase to produce G6P.
Beyond revealing the fundamental science of anhydrosugar metabolism, the potential applications of these sugars are interesting from the perspective of bioengineering. Biomass is an attractive source of carbon and energy for the production of renewable biofuels and chemicals. The use of corn- and sugar cane-derived glucose for fermentative production of ethanol is an example of this biomass-to-biofuel pathway. The use of lignocellulosic biomass, such as switchgrass or crop residues, shows several advantages when compared with these feedstocks due to issues related to food/fuel conflicts, land usage and economic viability (16, 17). With the inherent recalcitrance of lignocellulosic biomass to efficient depolymerization into clean, fermentable sugars (18, 19), it is interesting to consider that thermochemical processing of such biomass by fast pyrolysis results in the recovery of a significant portion of biomass-associated sugars as anhydrosugars (20). Knowledge of the existing biological pathways related to these sugars must be expanded if they are to be used effectively as carbon and energy sources for the fermentative production of biorenewable fuels and chemicals.
Anhydrosugars are also produced naturally during depolymerization of the bacterial cell wall for the purpose of peptidoglycan recycling (21). The resulting 1,6-anhMurNAc sugar is cleaved and phosphorylated by AnmK, which is expressed by many Gram-negative bacteria, to produce MurNAc-6-phosphate (22). LGK and AnmK form a sub-family of anhydrosugar kinases in the hexokinase family (23). They share significant sequence homology and a similar fold and domain architecture, including a nucleotide binding domain and a sugar binding domain that is separated by a dynamic hinge region (23). We previously determined the crystal structures of AnmK from Pseudomonas aeruginosa bound to its sugar substrate and a product ADP (24). An aspartate residue (Asp-182) was identified as the general base that initiates the attack of a nucleophilic water molecule on the anomeric carbon (C1) of anhMurNAc, thereby promoting cleavage of the 1,6-anhydro bond and transfer of the γ-phosphate of ATP to the O6 oxygen of the sugar. Subsequent structural studies of AnmK in the open conformation bound to the ATP analog, AMPPCP, provided the basis for the conformational dynamics of the enzyme during its catalytic cycle, whereby it cycles between a catalytically competent closed state and an open state (25).
LGK was first isolated from fungal sources and was subsequently determined to require ATP and magnesium for phosphorylation of levoglucosan (13). From the perspective of metabolic engineering, levoglucosan is not within the native substrate range of common industrial biocatalysts. Recently, codon-optimized LGK from Lipomyces starkeyi was introduced into ethanologenic E. coli by chromosome integration. The engineered strain was found to be able to use levoglucosan as the only carbon source for ethanol production (14). However, LGK enzymes display a relatively low binding affinity for levoglucosan (Km = 69–105 mm (26, 27)), consistent with the residual levoglucosan left unutilized during fermentation (14). Although it was discovered almost 25 years ago and clearly has potential as a catalyst in advanced biofuels production, relatively little is known about LGK. Crystallographic structures of L. starkeyi LGK, the object of this study, provide detailed insights in to the mechanism of LGK, including binding of two divalent metal ions and an unusual binding mode of levoglucosan.
Experimental Procedures
LGK Expression and Purification
The codon optimized lgk gene from L. starkeyi YZ-215 (GenBank accession number EU751287) was originally synthesized by GenScript (Piscataway, NJ) as described (14). The gene was then PCR-amplified from its pUC57 construct using Phusion DNA polymerase (New England Biolabs) and oligonucleotide primers 5′-GATATACATATGCCGATTGCGACGTCTACCG-3′ and 5′-GATATAGGATCCCGCCCAGTTGTTGGTGATAAC-3′ (Alpha DNA, Montreal, Quebec, Canada). The PCR amplicon and a modified version of a pET28 plasmid (Novagen) containing a reduced multicloning site (NdeI/SpeI/BamHI) and a C-terminal His6 tag were restricted with NdeI and BamHI and ligated using T4 DNA ligase (New England Biolabs). The ligation product, recombinant plasmid pLSlgk, was then used to transform E. coli strain BL21(DE3) GOLD cells (Stratagene) for further expression and purification.
Recombinant E. coli BL21(DE3) GOLD cells harboring pLSlgk were grown to an A600 of ∼ 0.5 at 37 °C, with shaking, in 500-ml volumes of LB medium supplemented with 35 μg/ml kanamycin. Expression of LGK was induced with 1 mm isopropyl-1-thio-β-d-galactopyranoside for 3 h at 30 °C, with shaking. Cells were pelleted by centrifugation and stored at −80 °C. Pellets were thawed in 20 ml of ice-cold lysis buffer (0.5 m NaCl, 20 mm Tris-HCl, pH 7.5, 0.1 mm PMSF, 2 mm imidazole) and lysed using a French pressure cell press (Ameco). The lysate was clarified by centrifugation and mixed with 2 ml of TALON metal affinity resin (Clontech) with gentle shaking for 30 min at room temperature. The TALON beads were centrifuged and re-suspended in binding buffer (500 mm NaCl, 20 mm Tris, pH 7.5, 0.5 mm TCEP) before being poured into a 20-ml gravity column. The column was washed with 20 ml of binding buffer supplemented with 5 mm imidazole (Sigma), followed by 20 ml of binding buffer supplemented with 10 mm imidazole. The LGK protein was eluted from the column with 10 ml of binding buffer supplemented with 250 mm imidazole. The eluate was dialyzed overnight against 20 mm Tris, pH 7.5, 100 mm NaCl, 1 mm EDTA, 5 mm DTT. The protein was further purified by gel filtration (Superdex 200) in 20 mm Tris, pH 7.5, 50 mm NaCl, 0.5 mm TCEP prior to concentration using an Amicon Ultra-15 concentrator with a 10,000 Da cut-off (Millipore). Chromatographic steps were performed using an ÄKTA FPLC (GE Healthcare).
LGK Crystallization, Data Collection, and Structure Determination
LGK crystals were grown at room temperature using the hanging drop vapor diffusion method by mixing equal volumes of reservoir buffer (22% PEG 4000, 100 mm sodium acetate, 100 mm Tris, pH 7.5) and LGK (7 mg/ml) in magnesium crystallization buffer (50 mm NaCl, 2 mm ADP, 4 mm MgCl2, 0.5 mm TCEP, 20 mm Tris, pH 7.5) for the structure bound to magnesium. For the structure bound to manganese, equal volumes of reservoir buffer (18% PEG 4000, 100 mm sodium acetate, 100 mm Tris, pH 7.2) and LGK (7 mg/ml) in a manganese crystallization buffer (50 mm NaCl, 2 mm ADP, 5 mm manganese(II) chloride tetrahydrate, 25 mm levoglucosan, 0.5 mm TCEP, 20 mm Tris, pH 7.5) were mixed. For the structure bound to magnesium, ADP, and levoglucosan, equal volumes of reservoir buffer (19% PEG 4000, 100 mm sodium acetate, 100 mm Tris, pH 6.8) and LGK (7 mg/ml) in crystallization buffer (50 mm NaCl, 2 mm ADP, 4 mm MgCl2, 0.5 mm TCEP, 1 mm aluminum nitrate, 10 mm sodium fluoride, 20 mm Tris, pH 7.5) were mixed, and a resultant crystal was then soaked directly in the crystallization drop with 100 mm levoglucosan for 10 min. The monomeric structure bound to levoglucosan in the productive conformation was determined by mixing equal amounts of LGK (9 mg/ml) in crystallization buffer (50 mm NaCl, 0.5 mm TCEP, 20 mm Tris, pH 7.5, 200 mm levoglucosan) and reservoir buffer (1.8 m ammonium sulfate, Hepes 7.0, and 100 mm sodium acetate). The monomeric structure bound to ADP and levoglucosan (alternate orientation only) was determined by mixing equal amounts of LGK (25 mg/ml) in crystallization buffer (100 mm NaCl, 0.5 mm TCEP, 20 mm Tris, pH 7.5, 2 mm ADP, 4 mm MgCl2, 1 mm aluminum nitrate, and 10 mm sodium fluoride) and reservoir buffer (1.3 m sodium malonate, 93 mm Bis-Tris propane, pH 7.0) and then soaking a resultant crystal in the drop with 200 mm levoglucosan. Crystals were cryoprotected by dragging them through a drop containing cryoprotectant solution (reservoir buffer supplemented with 9% sucrose (w/v), 2% glucose (w/v), 8% glycerol (v/v), and 8% ethylene glycol (v/v)) prior to being flash-cooled in liquid nitrogen. X-ray diffraction data for all of the structures, except the monomeric structure with levoglucosan in the alternate orientation, were collected at beamline 08ID-1 at the Canadian Light Source (Saskatoon, Saskatchewan, Canada), integrated using XDS (28), and scaled and merged using SCALA (29). For the LGK structure bound to manganese, a single-wavelength anomalous diffraction dataset was collected (λ = 1.8920; energy = 6.55306; f″ = 8.42; f′ = −7.46). The monomeric structure with levoglucosan bound in the alternate orientation was collected at the Stanford Synchrotron Radiation Lightsource beamline BL7-1, integrated using MOSFLM (30), and scaled and merged using SCALA.
Phase estimates for the structure bound to magnesium were initially obtained by molecular replacement using PHASER (31) and the structure of AnmK from P. aeruginosa as a search model (PDB identifier: 3QBW). The model was subsequently rebuilt using the PHENIX autobuild routine (32) for initial structural analysis. For the structure bound to manganese, HySS (33) was used to locate four manganese atoms (1.9 Å resolution cut-off used, anomalous signal = 0.0781; occupancies = 1.00, 0.96, 0.96, 0.94). Phasing was done using PHASER-EP (final figure of merit = 0.39) prior to density modification using the resolve routine in AUTOSOL, and the output density modified map file was used as an initial map file for subsequent automated model building using the PHENIX autobuild routine (32), which successfully built 862 of 894 protein residues (correlation coefficient = 0.84). This structure was then subsequently used as the starting model for the structure bound to magnesium and ADP to reduce model bias with the previously built model using the AnmK structure for molecular replacement. The remainder of the LGK structures were built using the experimentally determined LGK structure as a starting model, with rounds of iterative model building and refinement performed using Coot and PHENIX (34). The stereochemical quality of the final models was assessed using MolProbity (35). Refinement statistics are presented in Table 1. All structural figures were prepared using PyMOL (36).
TABLE 1.
Crystallographic data processing and refinement statistics for levoglucosan kinase crystallographic structures
| PDB ID (ligand) |
|||||
|---|---|---|---|---|---|
| 4ZFV (ADP/Mg) | 4YH5 (ADP/Mn) | 4ZLU (ADP/Mg/LG) | 5BVC (ADP/Mg/LG (alternate)) | 5BSB (LG) | |
| Data collectiona | |||||
| Space group | P41212 | P41212 | P41212 | P41212 | P41212 |
| Unit cell (Å) | a = b = 114.40, c = 232.50 | a = b = 114.44, c = 232.48 | a = b = 114.86, c = 234.77 | a = b = 70.25, c = 264.19 | a = b = 70.32, c = 263.33 |
| α = β = γ = 90.00 | α = β = γ = 90.00 | α = β = γ = 90.00 | α = β = γ = 90.00 | α = β = γ = 90.00 | |
| Wavelength (Å) | 0.9795 | 1.8920 | 0.9795 | 1.1271 | 0.9795 |
| Resolution range (Å) | 47.20–1.50 (1.58–1.50) | 47.21–1.90 (2.00–1.90) | 47.57–1.80 (1.90–1.80) | 43.27–2.00 (2.11–2.00) | 48.86–1.85 (1.95–1.85) |
| Total observations | 1,365,581 | 2,394,174 | 567,803 | 148,841 | 325,310 |
| Total unique observations | 244,767 | 121,746 | 145,247 | 44,939 | 57,761 |
| I/σI | 10.3 (2.1) | 21.6 (4.2) | 9.1 (2.2) | 6.2 (2.0) | 9.8 (1.9) |
| Completeness (%) | 99.9 (100.0) | 99.9 (99.1) | 99.9 (99.7) | 98.5 (99.9) | 100.0 (100.0) |
| Rmerge | 0.099 (0.787) | 0.104 (0.569) | 0.101 (0.719) | 0.100 (0.592) | 0.096 (0.861) |
| Rpim | 0.045 (0.363) | 0.024 (0.175) | 0.059 (0.397) | 0.062 (0.368) | 0.043 (0.395) |
| Redundancy | 5.6 (5.5) | 19.7 (11.2) | 3.9 (4.2) | 3.3 (3.4) | 5.6 (5.7) |
| Anomalous completeness (%) | 99.8 (98.5) | ||||
| Anomalous multiplicity | 10.2 (5.6) | ||||
| Refinement statistics | |||||
| Resolution (Å) | 47.20–1.50 | 47.21–1.90 | 47.10–1.80 | 43.3–2.00 | 48.86–1.85 |
| Reflections (total) | 244,650 | 121,618 | 145,033 | 44,796 | 57,602 |
| Reflections (test) | 12,339 | 6081 | 7286 | 2271 | 2897 |
| Total atoms refined | 8309 | 7824 | 7762 | 3610 | 3680 |
| Solvent | 1440 | 1032 | 1001 | 292 | 359 |
| Rwork (Rfree) | 0.16 (0.17) | 0.16 (0.17) | 0.19 (0.22) | 0.20 (0.23) | 0.19 (0.22) |
| RMSDsb (bond lengths (Å)/angles (°)) | 0.007/1.095 | 0.007/1.069 | 0.008/1.145 | 0.008/1.117 | 0.006/1.035 |
| Ramachandran plot (favored/allowed (%)) | 98.1/1.7 | 98.3/1.5 | 98.3/1.5 | 97.7/2.1 | 97.2/2.6 |
| Mean B values (Å2) | |||||
| Overall protein chain (A/B) | 15.7/17.0 | 20.1/21.6 | 20.4/27.2 | 28.0 | 29.6 |
| ADP | 13.0/15.5 | 18.3/20.6 | 21.5/74.9 | 50.6 | |
| Mg or Mn | 12.6,12.4/13.6,13.7 | 27.3,28.3/29.2,29.5 | 23.8, 20.8 | 42.0 | |
| LG | 30.3/25.5 | 20.8 | 26.4 | ||
| Active site Tris | 26.9 | ||||
a Values in parentheses refer to the high-resolution shell.
b RMSDs, root mean square deviations.
Enzyme Kinetics
LGK activity assays were performed using the G6P dehydrogenase coupling system (37). For the reaction velocity as a function of ATP concentration, all assays were carried out in at least triplicate in a buffered solution of 4 or 8 mm MgCl2, 50 mm TAPS, pH 9.0, 1 mm NADP+, 1 unit of G6P dehydrogenase from Saccharomyces cerevisiae (Sigma), and 2.4 μg of LGK. The reaction was initiated by the addition of levoglucosan (Carbosynth) at a concentration of 100 mm, bringing the final volume to 1 ml in a quartz reaction vessel. The change in A340 over a 2-min time period at 30 °C was measured using a Cary 300 spectrophotometer equipped with a Peltier temperature controller. The graph was plotted using Prism 6 software (GraphPad Software, La Jolla, CA). LGK reaction velocity as a function of levoglucosan concentration and buffer type was assayed using 100-μl microplates in the presence of HEPES and Tris at pH 7.6 at a concentration of 55 mm. All assays were performed with 99 mm NaCl, 1.5 mm NAD+, 2 mm ATP, 20 mm MgCl2, and 0.8 units of G6P from Leuconostoc mesenteroides (Sigma). Levoglucosan was added to each assay well in a 1:2 serial dilution at final concentrations ranging from 550 to 17.2 mm. Assay components were sealed and incubated on a 30 °C pebble bath for 5 min prior to the addition of 10 μl of purified LGK for an assay enzyme concentration of 0.1 μm. A340 was monitored over a 6-min time period by a Synergy H1 spectrophotometer (BioTek, Winooski, VT) using a kinetic read method every 21 s at a controlled temperature of 30 °C. Prism 6 software was used to calculate the Michaelis-Menten parameters using a non-linear fit of the datasets.
1H NMR Studies
All spectra were recorded on a 700-MHz Bruker AVANCE III instrument equipped with a TCI cryoprobe at 20 °C. 38 μl of enzyme solution was added to a total reaction volume of 600 μl of solution containing all other reagents (20 mm levoglucosan, 4.5–7 units of lactic dehydrogenase, 3–5 units of pyruvate kinase from rabbit muscle (Sigma), 15 mm phosphoenolpyruvic acid, 1 mm ATP, 10 mm magnesium, 60 mm NaCl, and 100 mm Bis-Tris, pD 6.9). The sample was mixed, the time of mixing was recorded, and the sample was placed on the magnet. Spectra were recorded at predefined intervals with a single scan recorded for each spectrum to obtain the exact time stamp and to avoid partial saturation of resonances due to insufficient relaxation delay. Spectra were processed using the MNova 9.1 suite (Mestrelab Research), and intensities were determined from spectral deconvolution. Buffer resonance at 1.88 ppm was also quantified to ensure consistency of measurements. Intensities were fitted to a single exponential growth/decay function, and plots were generated using Origin 7 software.
Results
Overall Structures, Metal Binding, and Active Site Coordination
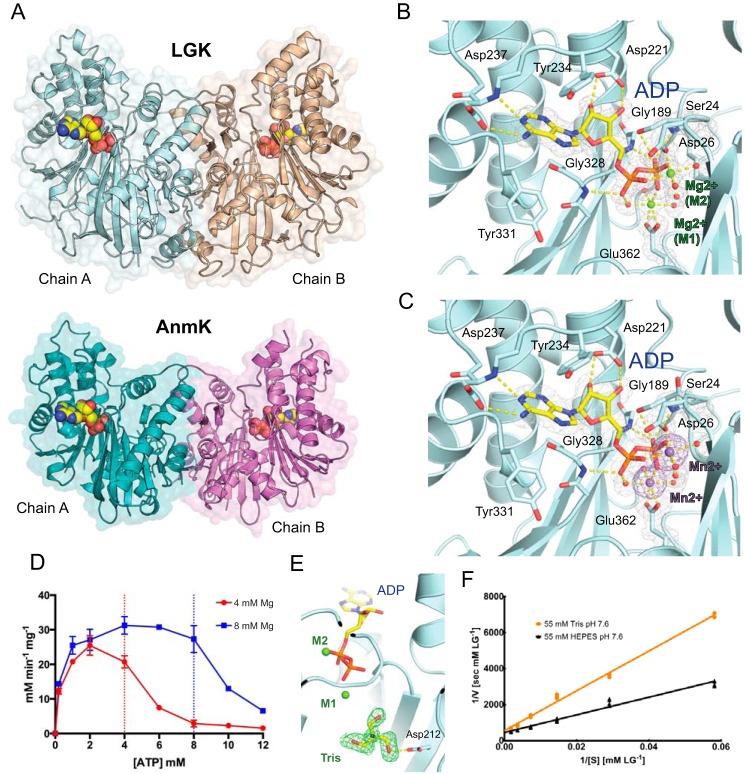
L. starkeyi LGK was targeted for crystallographic studies to gain a better understanding of its structure, mechanism, and substrate interactions. LGK was first crystallized in the presence of ADP and magnesium, and the structure was solved to 1.5 Å in the tetragonal space group P41212 as a non-crystallographic dimer (Fig. 2A; Table 1). The structure was initially solved using the closed AnmK structure (3QBW) as the molecular replacement search model. Each monomer of LGK contains a deep hinge between the two domains where the substrates bind. In many hexokinases, including AnmK, these two domains have often been shown to rotate apart from one another (25), whereas in this structure, the domains appear to be closed. Although longer in amino acid sequence, the overall tertiary structure of LGK is similar to the structure of AnmK in the closed conformation (root mean square deviation of C-alphas = 1.8 Å), including a conserved dimeric interface. LGK is somewhat larger than AnmK, with extra helical and loop arrangements at the bottom of the hinge region (Fig. 2A).
FIGURE 2.
A, crystal structures of LGK (top) and AnmK (bottom) bound to ADP. B, the nucleotide binding site demonstrates octahedral coordination of two magnesium atoms. Residues involved in binding are shown in stick format with carbons in cyan, nitrogens in blue, and oxygens in red. For the ADP, the carbon and phosphorous atoms are shown in yellow and orange, respectively. Waters and magnesiums are shown as red and green spheres, respectively. The gray mesh represents the 2mFo − DFc electron density map contoured around the ADP and magnesiums at 1σ. Molecular bonds are shown as yellow dashed lines. C, the nucleotide binding site in the presence of ADP and manganese. Anomalous difference Fourier map contoured around the manganese atoms is shown as a purple mesh at 3σ, while the manganese atoms are shown as purple spheres. D, LGK reaction velocity as a function of varying ATP concentrations and two constant magnesium concentrations. The vertical dotted lines delineate the concentration at which ATP and magnesium are equal. Error bars indicate means ± S.E. E, a molecule of Tris bound in the LGK active site. The mFo − DFc omit map is shown as a green mesh contoured at 2.5σ. F, Lineweaver-Burk plots of LGK reaction.
LGK binds the reaction product ADP through multiple hydrogen bonds (Fig. 2B), including bonds between the adenyl moiety and Asp-237, between the nucleotide ribose and Asp-221, and between the ADP phosphates and protein residues Ser-24, Gly-189, and Gly-328. Similarly to the vast majority of other kinases, magnesium is required for LGK catalysis (13). A striking observation found in the electron density map for the LGK structures is the apparent binding of two magnesium ions in the active site. The magnesium ions are observed in ideal octahedral coordination with Glu-362 and Asp-26, several water molecules, and the ADP (Fig. 2B). The first of the bound metals, designated M1, forms an electrostatic interaction with the β-phosphate, and its positioning suggests that it plays a direct role in phosphoryl transfer. The second of these metals, designated M2, likely plays a role in coordinating the position of the α- and β-phosphates because it binds to both of these phosphates, whereas a role in modulation of electrostatic charges is also plausible.
The binding of two divalent metal ions by LGK was further verified by co-crystallizing the enzyme with manganese instead of magnesium and solving the structure de novo using the anomalous signal of the four manganese atoms present in the dimeric structure (Fig. 2C). The initial molecular replacement solution for the LGK structure bound to ADP and magnesium was subsequently rebuilt using the experimentally determined LGK structure as a starting model to reduce phase error and model bias from the AnmK structure. Although magnesium has previously been shown to be required for the LGK reaction, we sought to determine whether both magnesiums are required for catalysis. Considering a dissociation constant (KD) of 50 μm for the MgATP complex (38), we analyzed the LGK reaction rate holding the magnesium concentration at two constant values (4 or 8 mm) and varying the ATP concentrations. Using this reasoning, a second magnesium atom would be available for catalysis when ATP concentrations are less than the magnesium concentrations, whereas free magnesium would be depleted when ATP concentrations exceeded the magnesium concentration. The resulting data are shown in Fig. 2D, with a dramatic drop-off in activity when the ATP concentration exceeds the magnesium concentration at both 4 mm and 8 mm magnesium. Given this result and our structural observations, we favor a model whereby either both magnesiums are required for catalysis or the second magnesium greatly enhances the rate of activity. An alternate explanation for the observed rates of activity could be that free ATP is acting as an inhibitor of the reaction when free magnesium is depleted, although our structural work shows that the nucleotide binds much more strongly when magnesium is also bound (see below). It should also be considered that although both metals play a role in catalysis, the use of two magnesiums may also limit the rate of ADP product release following catalysis, and further kinetic studies will be required to more thoroughly investigate the kinetic consequences of two magnesium binding sites.
Interestingly, a molecule of Tris, which was present in the crystallization buffer, was also observed bound in the active site in one of the LGK monomers bound to magnesium and ADP (Fig. 2E). Because the Tris molecule forms a hydrogen bond with the catalytic aspartate (Asp-212), it occupies roughly the same position where it is expected that levoglucosan would be coordinated. Interestingly, analysis of the Lineweaver-Burk plots of the reaction in the presence of either Tris or HEPES as the buffer demonstrated that Tris is a competitive inhibitor of the reaction (Fig. 2F). We determined a Km of 119 ± 12 mm for levoglucosan in HEPES buffer and an apparent Km of 255 ± 20 mm in Tris-HCl buffer, resulting in a KI for Tris of 48 ± 27 mm. Tris has been shown to inhibit the catalytic activity of other enzymes (39–41), and although its KI appears relatively high, the comparably high Km of LGK for levoglucosan suggests that the inhibition is significant.
Mechanism of Sugar Cleavage
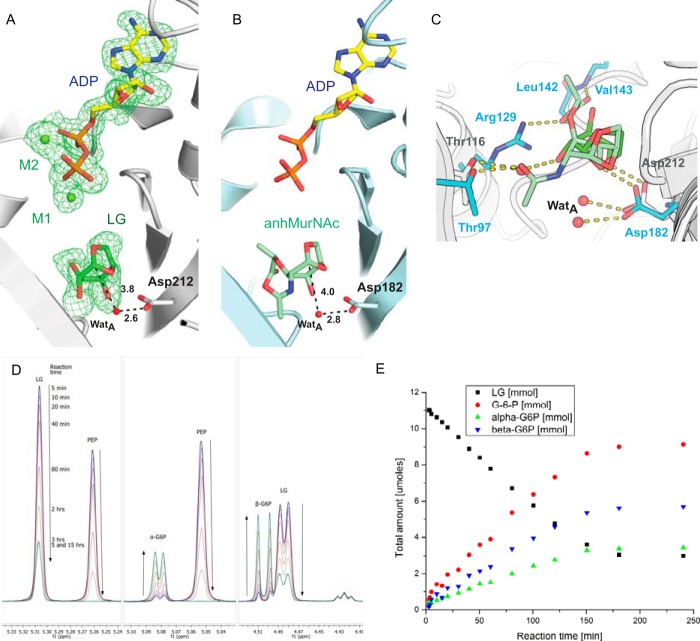
To gain insights into the mechanism of 1,6-anhydro bond cleavage and to evaluate sugar binding interactions including a possible rationale for the high intrinsic Km of LGK for levoglucosan, we co-crystallized LGK with ADP and magnesium and then soaked the grown crystals with 100 mm levoglucosan. Despite the fact that levoglucosan is smaller than anhMurNAc because it has hydroxyls in place of the bulkier lactyl and acetamido moieties of anhMurNAc, the positioning of levoglucosan in the A-chain monomer is very similar to the anhMurNAc in the binding site of AnmK (Fig. 3, A–C). LGK exhibits a very high Km for its substrate when compared with AnmK (0.2 mm) and, accordingly, forms fewer bonding interactions (24). Levoglucosan forms only a single hydrogen bond with LGK (Asp-212), although the substrate also appears to be stabilized by hydrogen bonds with water molecules in the active site. Specifically, a water molecule appears to mediate a hydrogen-bonding network between Thr-116 and O1 of LG (Fig. 3C). In AnmK, a similar bonding arrangement is observed between the analogous threonine residue and the acetamido group of anhMurNAc, suggesting that the threonine residue plays a conserved role in substrate binding. Although aluminum nitrate and sodium fluoride were also present in the crystallization buffer, the expected aluminum fluoride could not be unambiguously modeled in the resulting data. However, the ADP and two magnesium ions in this monomer (Fig. 3A) are observed with similar bonding arrangements to those shown in Fig. 2.
FIGURE 3.
A, LGK active site in the presence of ADP, magnesium, and levoglucosan. Black dashed lines indicate distances in angstroms between moieties that are important for catalysis. The green mesh represents an mFo − DFc omit map calculated after the removal of ADP, magnesium, and levoglucosan shown at 2.5σ. WatA indicates water molecule. B, model of AnmK bound to ADP and anhMurNAc. The structure was modeled by superposing the ADP-bound crystallographic structure onto the anhMurNAc-bound crystallographic structure. C, superposition of the LGK and AnmK sugar binding sites. Protein carbon atoms of residues involved in binding of the sugars are shown in gray and cyan for LGK and AnmK, respectively. Carbon atoms for the sugar are shown in light green (anhMurNAc) or green (levoglucosan). D, 1H NMR analysis of the LGK reaction. Peaks corresponding to the anomeric protons of the substrate (levoglucosan, 5.31 ppm) and products (α and β anomers of G6P) are labeled. Peaks due to the presence of phosphoenolpyruvic acid (PEP) are also labeled. e, rates of levoglucosan consumption and G6P formation measured using 1H NMR are shown over time.
The similar arrangement of levoglucosan in the active site A-chain when compared with the closed AnmK structure suggests that the sugar cleavage mechanism is conserved between LGK and AnmK. For AnmK, Asp-182 acts as a base to enhance the nucleophilicity of a water molecule (WatA), which then attacks the anomeric carbon of the sugar resulting in cleavage of the 1,6-anhydro bond (24). WatA also appears ideally positioned to attack the anomeric carbon in the LGK structure (Fig. 3, A and C). Consistent with this observation, mutation of the predicted catalytic base in LGK, Asp-212, to an asparagine results in complete abrogation of LGK activity as evidenced by an NADP+-coupled assay. As another means to follow the progress of the LGK reaction, 1H NMR experiments were performed using a coupled system whereby ATP is regenerated from the LGK reaction ADP product using pyruvate kinase as the phosphotransfer enzyme and phosphoenolpyruvic acid as the phosphoryl donor. Importantly, this method allows for the direct observation of levoglucosan consumption and G6P production during the reaction. The reaction showed fast initial build-up of G6P (within 2 min) followed by a slower linear phase until the phosphoenolpyruvic acid was depleted (Fig. 3, D and E). Both α and β anomers of G6P appear simultaneously in the reaction mixture and at a ratio corresponding to the equilibrium ratio of anomers for G6P. The turnover rate of the LGK reaction calculated using this technique is 0.14/s at 20 °C, which is faster than the reported anomerization rates of G6P of 0.04/s at 20 °C (42). This suggests that the initial product of the LGK reaction is G6P in its linear form, because we would expect an accumulation of the α or β anomer following catalysis if either anomer were formed during the reaction. These results are in contrast to those obtained using AnmK as the enzyme, which demonstrated an accumulation of the α anomer of anhMurNAc following catalysis using similar techniques (24). It is apparent that levoglucosan is less stable during the catalytic reaction, because it is held in the active site through fewer bonding interactions when compared with anhMurNAc bound to AnmK. We propose that in addition to cleavage of the 1,6-anhydro ring, the pyranose ring of the levoglucosan is opened when the anomeric carbon of levoglucosan migrates from its position in a 1C4 to a 4C1 conformation. It is plausible that increased conformational strain during this reaction results in the linear form of G6P as the initial product, which then equilibrates to a mixture of both anomeric forms (Fig. 3E).
LGK Binds Levoglucosan in Two Distinct Orientations
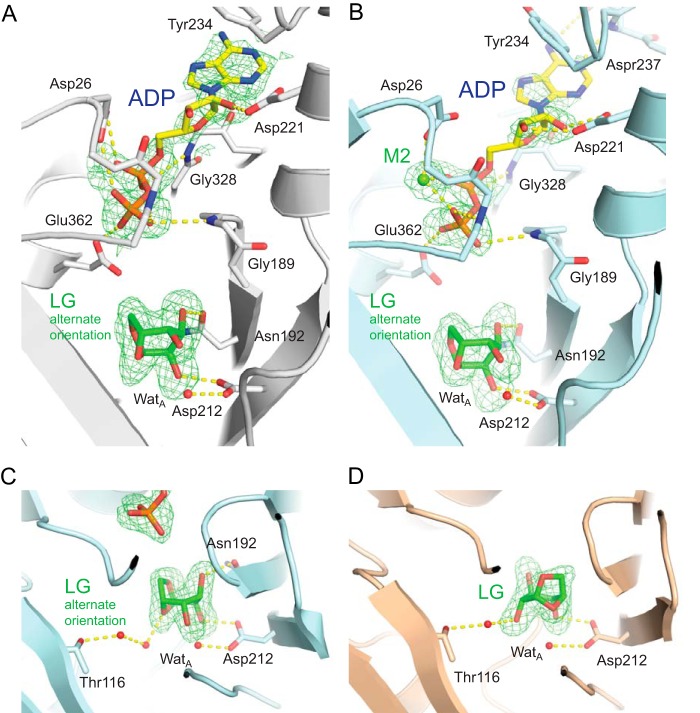
In contrast to the A-chain monomeric structure, the levoglucosan sugar substrate was also found to bind to LGK in an alternate orientation in the B-chain monomeric subunit, whereby it is rotated by ∼135° with regard to the plane of the pyranose sugar (Figs. 3A and 4A). Both conformations form hydrogen bonds with Asp-212, whereas the alternate conformation in the B-chain is supported by an additional hydrogen bond between Asn-192 and the O3 oxygen of levoglucosan. Interestingly, the magnesium ions are not bound in this structure and the B-factors (Table 1) and corresponding electron density for ADP are much poorer than in the A-chain structure. In the lower affinity ADP binding site, when compared with the other structures, several bonds either are abrogated or show longer bonding distances. Amino acids Asp-26 and Glu-362 are no longer bound to magnesium ions, and these residues instead form bonds directly with the phosphates of ADP (Fig. 4A). The two domains of the B-chain in this structure also appear slightly more open than the A-chain, with a 3° rotation of the two domains calculated using the program DynDom (43), suggesting a possible reciprocal effect between ligand binding and protein domain movements.
FIGURE 4.
A–C, LGK active site structures with levoglucosan bound in an alternate orientation. D, additional crystal form with levoglucosan bound in a catalytically conducive orientation. mFo − DFc omit maps are shown at 2.5 (A and B) or 3.0σ (C and D).
Our initial efforts to determine the structure of LGK with bound levoglucosan thus demonstrated two binding modes for the sugar. Because this was an intriguing result, we sought to determine additional crystal forms of LGK with levoglucosan bound to determine whether these results could be duplicated in another crystal form. After extensive crystallization screening, we found another crystallization condition for LGK in the presence of ADP and magnesium that uses either ammonium sulfate or sodium malonate as the precipitant, instead of PEG 4000 that was used for the original structures. We pursued this crystal form with the intention to determine additional structures of LGK bound to levoglucosan in either of the previous observed orientations. LGK in this form crystallizes in the same P41212 space group as the originally determined structures, although with different unit cell dimensions (Table 1), and with its dimeric interface lying on a crystal symmetry axis. The structure in this case was determined to 2.0 Å as a monomer in the asymmetric unit, with both monomers of the physical dimer essentially being determined as identical structures due to the crystal symmetry. Our initial attempts to solve the structure with ADP, aluminum fluoride, and magnesium present during co-crystallization followed by soaking with 200 mm levoglucosan resulted in a structure similar to that observed in the B-chain of the dimeric structure, with levoglucosan bound in the alternate orientation, ADP showing poor electron density, and no visible bound aluminum fluoride (Fig. 4B). Only the M2 magnesium appears bound in this structure, although with higher B-factors than in the other Mg-containing structures (Table 1).
Although levoglucosan in the alternate orientation makes two direct hydrogen bonds to LGK (Asp-212 and Asn-192), a similar water-bonding arrangement with Thr-116 to that observed in the catalytically conducive orientation appears to be maintained. However, in this orientation, two water molecules are required to bridge the gap to the levoglucosan ring oxygen, which may only act as a hydrogen bond acceptor (Fig. 4C). We were eventually also able to solve another structure of LGK, to 1.85 Å, with levoglucosan bound in the catalytically conducive conformation by co-crystallizing the enzyme with 200 mm levoglucosan in the monomeric crystal form without the nucleotide or magnesium present (Fig. 4D). These additional structures support the initial observations of two distinct binding modes of levoglucosan to LGK.
Discussion
Enzyme-driven phosphoryl transfer is a fundamental biochemical reaction that is carried out through a surprisingly variable array of mechanisms, the intricacies of which are often only made apparent when examined at the structural level. A detailed understanding of these reactions is crucial to understanding the cellular processes they regulate or control. The crystallographic structures of LGK described here provide insight into the unique reaction mechanism carried out by anhydrosugar kinases that involves both cleavage and phosphorylation of their sugar substrate. A somewhat unexpected observation for LGK was the binding of two magnesium ions in the enzyme active site. The binding of two magnesiums has been observed in other kinases and is common in protein kinases (i.e. CDK2 kinase (44, 45)). However, to our knowledge, this is the first observation of two magnesium binding sites in the hexokinase family. The M2 site appears to assist in stabilizing the positions and geometrical alignment of the nucleotide phosphate groups, because it forms bonding interactions with both the α- and β-phosphates of the ADP, although a role in charge stabilization or modulation during the catalytic reaction is also plausible. It is probable that the presence of the M1 site is also required for maintenance of charges that develop during the reaction by enhancing the electrophilicity of the departing γ-phosphate or by acting as a transient bridge between the β-phosphate of ADP and the phosphoryl group of the product during the reaction as suggested for other kinases (46). The binding of magnesium in other hexokinases such as Sulfolobus tokodaii hexokinase is also roughly equivalent in structural rearrangement to the M1 site, with magnesium binding to a β-phosphate oxygen of ADP, suggesting a shared functional evolution (47). For other kinases that use two magnesiums for catalysis such as the protein kinase CDK2, it has been shown that the resulting product ADP is slowly released from the active site following catalysis (45). The need by LGK to utilize two magnesium ions for catalysis may also come at a cost of lowering the rate of product release, although further more extensive kinetic studies will be required to evaluate the rate. In CDK2, it was shown that magnesium binds less tightly to the M1 site, which is consistent with the observation that in LGK, only the M2 magnesium is bound in the monomeric structure bound to ADP and levoglucosan (45). Although the comparative binding affinities of the two magnesiums remain unclear, it is likely that higher concentrations of magnesium would increase the relative occupancy of both magnesium sites in LGK and thereby limit ADP product release.
In our previous work, the position of magnesium could not unambiguously be resolved in the closed AnmK structure bound to ADP (24). However, a single magnesium appeared to be bound in one of the open AnmK structures bound to AMPPCP (25). It is plausible that AnmK also uses two magnesiums during catalysis, because the nucleotide binding site structures are highly conserved between the two enzymes and the one magnesium observed in AnmK was coordinated by the same glutamate residue (Glu-326 and Glu-362 in AnmK and LGK, respectively) that is key to the M1 binding. The inability to view both magnesiums in the closed AnmK structures may be due to several factors including the lower resolution of the AnmK structure (2.23 Å) or the relatively high B-factors for ADP bound to AnmK (41.3 and 56.9 for either monomer). The protein and water interactions that govern magnesium binding may also be influenced by the pH of the crystallization conditions, which was 5.6 for the closed AnmK structure and 7.5 for the LGK structure bound to two magnesiums, the latter of which represents a form of the structure at physiological pH. Taken together our observations suggest that ADP is bound to the AnmK structure in a lower affinity state, similar to the LGK structures in which magnesium ions were not observed.
Additional unusual observations for the LGK structures are the two binding orientations for the levoglucosan substrate, which were both observed in two different crystal forms, and the relatively few direct hydrogen bonds the enzyme makes with the substrate. Although the related enzyme AnmK binds its sugar substrate with a total of five hydrogen bonds (24), LGK mediates only one or two hydrogen bonds (catalytically conducive or the alternate arrangement, respectively) with its sugar substrate. These observations are also supported by the markedly differing Km values of the respective sugar substrates of AnmK and LGK enzymes. However, this does not explain why LGK has not evolved residues in the active site that would improve its affinity for levoglucosan by imposing more bonding interactions with the substrate. Although we cannot rule out that the unusually high Km for levoglucosan is not rate-limiting to the catalytic efficiency of the enzyme, we propose that the comparatively high Km for LGK is intrinsically linked to the ability of the enzyme to alternate the levoglucosan binding orientations. This structural requirement may prohibit selective mutations of residues in the active site that would support additional direct bonds between LGK and the sugar, which might enhance binding affinity, but would limit its ability to bind the sugar in two orientations. This property may also make the enzyme more prone to competitive inhibition, as evidenced by the binding of Tris in the active site and our kinetic analyses (Fig. 2, E and F). Other enzymes that have been shown to bind their substrate in multiple orientations do so with varying affinity and functionally are quite diverse, making it difficult to compare the mechanism of substrate binding for these enzymes (48–50).
The high Km of LGK for levoglucosan may also at least partially be due to the abundance of levoglucosan in the natural environment, particularly in areas that have been subjected to biomass burning events. In this context, the possibility of weak selection pressure to improve catalytic efficiency could also be influenced by the use of levoglucosan as only a secondary metabolite that is not absolutely required for growth or survival. LGK may also be somewhat promiscuous toward or have significant catalytic activity for other anhydrosugars. Although AnmK has been shown not to be able to use levoglucosan as a substrate, recombinant growth studies using E. coli have shown that LGK has weak activity for anhGlcN (1,6-anhydro-β-d-glucosamine) and anhGlcNAc 1,6-anhydro-N-acetyl-β-d-glucosamine (27). However, it does not appear that these sugars can be used as a carbohydrate source by L. starkeyi, suggesting that they are not transported into the cell or that metabolic pathways do not exist in Lipomyces to further utilize the resulting products (27).
In LGK, the making and breaking of water-bonding arrangements and dipole interactions not only play an intrinsic role in coordinating the octahedral networks required for magnesium coordination, but they also play a key role in mediating interactions with levoglucosan. This is apparent from the water-bonding network between levoglucosan and Thr-116 that appears to be maintained in both levoglucosan orientations. To this extent, a possible rationale for the alternate orientation of levoglucosan may be to abrogate water-bonding networks in the active site that are also required for the octahedral coordination with magnesium, displacing the metals and also promoting the release of ADP. With the knowledge that ADP is a product inhibitor of LGK (KI = 0.2 mm (51)) and G6P is not (13, 51), it is likely that G6P is quickly released following catalysis. This suggests the possibility that the active site can be reoccupied by another molecule of levoglucosan prior to ADP and magnesium release. A more detailed analysis of water orientations in the enzyme structure, including visualization of hydrogen atoms and their associated molecular interactions, using methods such as neutron crystallography would allow for a more thorough investigation into these mechanistic details (52).
Summary and Conclusions
In addition to a detailed examination of the structure and mechanism of anhydrosugar kinases that elaborates on the function of these enzymes in natural biological processes, these studies provide a blueprint for future efforts to engineer LGK enzymes as catalysts in practical applications, such as biofuel production. Mutations that alter the substrate or product affinity of the enzyme could lead to a greater catalytic efficiency for the bioconversion of levoglucosan. However, possible rational approaches may be hampered by the dual binding orientations of levoglucosan, and natural selection has not yet provided a better solution to the high intrinsic Km of LGK for its sugar substrate, suggesting that such efforts to improve affinity for the sugar may be very challenging. Combining structural data with advanced protein engineering and computational design may however yet hold promise for improving LGK to produce levoglucosan-derived biofuel.
Author Contributions
J. P. B., T. A. W., B. L. M., C. J. U., and R. M. designed the research; J. P. B., J. R. K., and R. M. performed the research; J. P. B., L. R. J., and R. M. wrote the manuscript. All authors contributed to analysis of the results and editing of the manuscript. The final version of the manuscript was approved by all of the authors.
Acknowledgments
We thank Shaun Labiuk, Pavel Afonine, and the Canadian Light Source as well as the staff at the SLAC National Accelerator Laboratory for assistance in data collection and processing. We thank the Los Alamos National Laboratory Principal Associate Directorate for Science, Technology and Engineering for institutional investment in key equipment that enabled NMR measurements reported here. The Canadian Light Source is supported by Natural Sciences and Engineering Research Council of Canada (NSERC), the National Research Council, Canadian Institutes of Health Research, and the University of Saskatchewan. Use of the Stanford Synchrotron Radiation Lightsource, SLAC National Accelerator Laboratory, is supported by the U. S. Department of Energy (DOE), Office of Science, Office of Basic Energy Sciences under Contract DE-AC02-76SF00515. The Stanford Synchrotron Radiation Lightsource Structural Molecular Biology Program is supported by the DOE Office of Biological and Environmental Research, and by the National Institutes of Health, NIGMS (including Grant P41GM103393). We also thank Brian Broom-Peltz for technical assistance and Dr. Virginia A. Unkefer for help with editing the manuscript.
This work was supported in part by the Natural Sciences and Engineering Research Council of Canada (NSERC) (Grant 311775-2010) (to B. L. M.), a Manitoba Research Chair (to B. L. M.), and a Manitoba Health Research Council Postdoctoral Fellowship (to J. P. B.). This work was also partially funded through the Protein Crystallography Station from the Department of Energy Office of Biological and Environmental Research (to J. P. B. and C. J. U.) and National Science Foundation (NSF) CAREER Award 1254238 (to J. R. K. and T. A. W.). The authors declare that they have no conflicts of interest with the contents of this article.
- LGK
- levoglucosan kinase
- LG
- levoglucosan
- G6P
- glucose 6-phosphate
- TAPS
- 3-{[2-hydroxy-1,1-bis(hydroxymethyl)ethyl]amino}-1-propanesulfonic acid
- TCEP
- tris(2-carboxyethyl)phosphine
- Bis-Tris
- 2-(bis(2-hydroxyethyl)amino)-2-(hydroxymethyl)propane-1,3-diol
- Bis-Tris propane
- 1,3-bis[tris(hydroxymethyl)methylamino]propane
- AMPPCP
- adenosine 5′-(β,γ-methylene)triphosphate
- anhMurNAc
- 1,6-anhydro-N-acetylmuramic acid
- AnmK
- 1,6-anhydro-N-acetylmuramic acid kinase.
References
- 1.Vancampenhout K., De Vos B., Wouters K., Swennen R., Buurman P., and Deckers J. (2012) Organic matter of subsoil horizons under broadleaved forest: highly processed or labile and plant-derived? Soil Biol. Biochem. 50, 40–46 [Google Scholar]
- 2.Chuang M.-T., Chou C. C. K., Sopajaree K., Lin N.-H., Wang J.-L., Sheu G.-R., Chang Y.-J., and Lee C.-T. (2013) Characterization of aerosol chemical properties from near-source biomass burning in the northern Indochina during 7-SEAS/Dongsha experiment. Atmos. Environ. 78, 72–81 [Google Scholar]
- 3.Hopmans E. C., dos Santos R. A. L., Mets A., Damste J. S. S., and Schouten S. (2013) A novel method for the rapid analysis of levoglucosan in soils and sediments. Org. Geochem. 58, 86–88 [Google Scholar]
- 4.Tsai Y. I., Sopajaree K., Chotruksa A., Wu H.-C., and Kuo S.-C. (2013) Source indicators of biomass burning associated with inorganic salts and carboxylates in dry season ambient aerosol in Chiang Mai Basin, Thailand. Atmos. Environ. 78, 93–104 [Google Scholar]
- 5.Wallner P., Kundi M., Moshammer H., Scharf S., Schmutzer M., Weiss S., Hohenblum P., and Hutter H. P. (2013) Urinary levoglucosan levels in Austrian communities differing in agrarian quota. Int. J. Hyg. Environ. Health 216, 280–283 [DOI] [PubMed] [Google Scholar]
- 6.Kehrwald N., Zangrando R., Gabrielli P., Jaffrezo J.-L., Boutron C., Barbante C., and Gambaro A. (2012) Levoglucosan as a specific marker of fire events in Greenland snow. Tellus B 64, 18196 [Google Scholar]
- 7.Levine J. S. (1990) Global biomass burning: atmospheric, climatic, and biospheric implications. EOS Trans. Am. Geophys. Union 71, 1075–1077 [Google Scholar]
- 8.Vicente A., Alves C. l., Calvo A. I., Fernandes A. P., Nunes T., Monteiro C., Almeida S. M., and Pio C. (2013) Emission factors and detailed chemical composition of smoke particles from the 2010 wildfire season. Atmos. Environ. 71, 295–303 [Google Scholar]
- 9.Bai J., Sun X., Zhang C., Xu Y., and Qi C. (2013) The OH-initiated atmospheric reaction mechanism and kinetics for levoglucosan emitted in biomass burning. Chemosphere 93, 2004–2010 [DOI] [PubMed] [Google Scholar]
- 10.Lai C., Liu Y., Ma J., Ma Q., and He H. (2014) Degradation kinetics of levoglucosan initiated by hydroxyl radical under different environmental conditions. Atmos. Environ. 91, 32–39 [Google Scholar]
- 11.Mullaugh K. M., Byrd J. N., Avery G. B. Jr., Mead R. N., Willey J. D., and Kieber R. J. (2014) Characterization of carbohydrates in rainwater from the southeastern North Carolina. Chemosphere 107, 51–57 [DOI] [PubMed] [Google Scholar]
- 12.Knicker H., Hilscher A., de la Rosa J. M., Gonzalez-Perez J. A., and Gonzalez-Vila F. J. (2013) Modification of biomarkers in pyrogenic organic matter during the initial phase of charcoal biodegradation in soils. Geoderma 197, 43–50 [Google Scholar]
- 13.Kitamura Y., and Yasui T. (1991) Purification and some properties of levoglucosan (1,6-anhydro-β-d-glucopyranose) kinase from the yeast Sporobolomyces salmonicolor. Agr. Biol. Chem. 55, 523–529 [Google Scholar]
- 14.Layton D. S., Ajjarapu A., Choi D. W., and Jarboe L. R. (2011) Engineering ethanologenic Escherichia coli for levoglucosan utilization. Bioresour. Technol. 102, 8318–8322 [DOI] [PubMed] [Google Scholar]
- 15.Nakahara K., Kitamura Y., Yamagishi Y., Shoun H., and Yasui T. (1994) Levoglucosan dehydrogenase involved in the assimilation of levoglucosan in Arthrobacter Sp I-552. Biosci. Biotechnol. Biochem. 58, 2193–2196 [DOI] [PubMed] [Google Scholar]
- 16.Tilman D., Socolow R., Foley J. A., Hill J., Larson E., Lynd L., Pacala S., Reilly J., Searchinger T., Somerville C., and Williams R. (2009) Energy. Beneficial biofuels–the food, energy, and environment trilemma. Science 325, 270–271 [DOI] [PubMed] [Google Scholar]
- 17.Perlack R. D., and Stokes B. J. (2011) US billion-ton update: biomass supply for a bioenergy and bioproducts industry. SciTech Connect 10.2172/1023318 [DOI] [Google Scholar]
- 18.Chundawat S. P., Beckham G. T., Himmel M. E., and Dale B. E. (2011) Deconstruction of lignocellulosic biomass to fuels and chemicals. Annu. Rev. Chem. Biomol. Eng. 2, 121–145 [DOI] [PubMed] [Google Scholar]
- 19.Phitsuwan P., Sakka K., and Ratanakhanokchai K. (2013) Improvement of lignocellulosic biomass in planta: a review of feedstocks, biomass recalcitrance, and strategic manipulation of ideal plants designed for ethanol production and processability. Biomass Bioenerg. 58, 390–405 [Google Scholar]
- 20.Jarboe L. R., Wen Z., Choi D., and Brown R. C. (2011) Hybrid thermochemical processing: fermentation of pyrolysis-derived bio-oil. Appl. Microbiol. Biotechnol. 91, 1519–1523 [DOI] [PubMed] [Google Scholar]
- 21.Park J. T., and Uehara T. (2008) How bacteria consume their own exoskeletons (turnover and recycling of cell wall peptidoglycan). Microbiol Mol. Biol. Rev. 72, 211–227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Uehara T., Suefuji K., Valbuena N., Meehan B., Donegan M., and Park J. T. (2005) Recycling of the anhydro-N-acetylmuramic acid derived from cell wall murein involves a two-step conversion to N-acetylglucosamine-phosphate. J. Bacteriol. 187, 3643–3649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hurley J. H. (1996) The sugar kinase/heat shock protein 70/actin superfamily: implications of conserved structure for mechanism. Annu. Rev. Biophys. Biomol. Struct. 25, 137–162 [DOI] [PubMed] [Google Scholar]
- 24.Bacik J.-P., Whitworth G. E., Stubbs K. A., Yadav A. K., Martin D. R., Bailey-Elkin B. A., Vocadlo D. J., and Mark B. L. (2011) Molecular basis of 1,6-anhydro bond cleavage and phosphoryl transfer by Pseudomonas aeruginosa 1,6-anhydro-N-acetylmuramic acid kinase. J. Biol. Chem. 286, 12283–12291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bacik J. P., Tavassoli M., Patel T. R., McKenna S. A., Vocadlo D. J., Khajehpour M., and Mark B. L. (2014) Conformational itinerary of Pseudomonas aeruginosa 1,6-anhydro-N-acetylmuramic acid kinase during its catalytic cycle. J. Biol. Chem. 289, 4504–4514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ning J. Y., Yu Z. S., Xie H. J., Zhang H. X., Zhuang G. Q., Bai Z. H., Yang S. M., and Jiang Y. M. (2008) Purification and characterization of levoglucosan kinase from Lipomyces starkeyi YZ-215. World J. Microb. Biotechnol. 24, 15–22 [Google Scholar]
- 27.Dai J. H., Yu Z. S., He Y. Z., Zhang L., Bai Z. H., Dong Z. Y., Du Y. G., and Zhang H. X. (2009) Cloning of a novel levoglucosan kinase gene from Lipomyces starkeyi and its expression in Escherichia coli. World J. Microb. Biotechnol. 25, 1589–1595 [Google Scholar]
- 28.Kabsch W. (2010) Integration, scaling, space-group assignment and post-refinement. Acta Crystallogr. D 66, 133–144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Evans P. (2006) Scaling and assessment of data quality. Acta Crystallogr. D. 62, 72–82 [DOI] [PubMed] [Google Scholar]
- 30.Leslie A. G. (2006) The integration of macromolecular diffraction data. Acta Crystallogr D. 62, 48–57 [DOI] [PubMed] [Google Scholar]
- 31.McCoy A. J., Grosse-Kunstleve R. W., Storoni L. C., and Read R. J. (2005) Likelihood-enhanced fast translation functions. Acta Crystallogr. D. 61, 458–464 [DOI] [PubMed] [Google Scholar]
- 32.Adams P. D., Afonine P. V., Bunkóczi G., Chen V. B., Davis I. W., Echols N., Headd J. J., Hung L.-W., Kapral G. J., Grosse-Kunstleve R. W., McCoy A. J., Moriarty N. W., Oeffner R., Read R. J., Richardson D. C., Richardson J. S., Terwilliger T. C., and Zwart P. H. (2010) PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D. 66, 213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zwart P. H., Afonine P. V., Grosse-Kunstleve R. W., Hung L. W., Ioerger T. R., McCoy A. J., McKee E., Moriarty N. W., Read R. J., Sacchettini J. C., Sauter N. K., Storoni L. C., Terwilliger T. C., and Adams P. D. (2008) Automated structure solution with the PHENIX suite. Methods Mol. Biol. 426, 419–435 [DOI] [PubMed] [Google Scholar]
- 34.Emsley P., and Cowtan K. (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr. D. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 35.Chen V. B., Arendall W. B. 3rd, Headd J. J., Keedy D. A., Immormino R. M., Kapral G. J., Murray L. W., Richardson J. S., and Richardson D. C. (2010) MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr D 66, 12–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.DeLano W. L. (2010) The PyMOL Molecular Graphics System, version 1.3r1, Schrödinger, LLC, New York [Google Scholar]
- 37.Hirai M., Ohtani E., Tanaka A., and Fukui S. (1977) Glucose-phosphorylating enzymes of Candida yeasts and their regulation in vivo. Biochim. Biophys. Acta 480, 357–366 [DOI] [PubMed] [Google Scholar]
- 38.Gupta R. K., Gupta P., Yushok W. D., and Rose Z. B. (1983) Measurement of the dissociation constant of MgATP at physiological nucleotide levels by a combination of 31P NMR and optical absorbance spectroscopy. Biochem. Biophys. Res. Commun. 117, 210–216 [DOI] [PubMed] [Google Scholar]
- 39.Gordon-Weeks R., Koren'kov V. D., Steele S. H., and Leigh R. A. (1997) Tris is a competitive inhibitor of K+ activation of the vacuolar H+-pumping pyrophosphatase. Plant Physiol 114, 901–905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Desmarais W. T., Bienvenue D. L., Bzymek K. P., Holz R. C., Petsko G. A., and Ringe D. (2002) The 1.20 Å resolution crystal structure of the aminopeptidase from Aeromonas proteolytica complexed with tris: a tale of buffer inhibition. Structure 10, 1063–1072 [DOI] [PubMed] [Google Scholar]
- 41.Ghalanbor Z., Ghaemi N., Marashi S. A., Amanlou M., Habibi-Rezaei M., Khajeh K., and Ranjbar B. (2008) Binding of Tris to Bacillus licheniformis α-amylase can affect its starch hydrolysis activity. Protein Pept. Lett. 15, 212–214 [DOI] [PubMed] [Google Scholar]
- 42.Salas M., Vinuela E., and Sols A. (1965) Spontaneous and enzymatically catalyzed anomerization of glucose 6-phosphate and anomeric specificity of related enzymes. J. Biol. Chem. 240, 561–568 [PubMed] [Google Scholar]
- 43.Hayward S., and Lee R. A. (2002) Improvements in the analysis of domain motions in proteins from conformational change: DynDom version 1.50. J. Mol. Graph Model 21, 181–183 [DOI] [PubMed] [Google Scholar]
- 44.Bao Z. Q., Jacobsen D. M., and Young M. A. (2011) Briefly bound to activate: transient binding of a second catalytic magnesium activates the structure and dynamics of CDK2 kinase for catalysis. Structure 19, 675–690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jacobsen D. M., Bao Z. Q., O'Brien P., Brooks C. L. 3rd, and Young M. A. (2012) Price to be paid for two-metal catalysis: magnesium ions that accelerate chemistry unavoidably limit product release from a protein kinase. J. Am. Chem. Soc. 134, 15357–15370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bernstein B. E., Michels P. A., and Hol W. G. (1997) Synergistic effects of substrate-induced conformational changes in phosphoglycerate kinase activation. Nature 385, 275–278 [DOI] [PubMed] [Google Scholar]
- 47.Nishimasu H., Fushinobu S., Shoun H., and Wakagi T. (2007) Crystal structures of an ATP-dependent hexokinase with broad substrate specificity from the hyperthermophilic archaeon Sulfolobus tokodaii. J. Biol. Chem. 282, 9923–9931 [DOI] [PubMed] [Google Scholar]
- 48.Kupfer R., Liu S. Y., Allentoff A. J., and Thompson J. A. (2001) Comparisons of hydroperoxide isomerase and monooxygenase activities of cytochrome P450 for conversions of allylic hydroperoxides and alcohols to epoxyalcohols and diols: probing substrate reorientation in the active site. Biochemistry 40, 11490–11501 [DOI] [PubMed] [Google Scholar]
- 49.Regni C., Naught L., Tipton P. A., and Beamer L. J. (2004) Structural basis of diverse substrate recognition by the enzyme PMM/PGM from P. aeruginosa. Structure 12, 55–63 [DOI] [PubMed] [Google Scholar]
- 50.Sachett L. G., and Verli H. (2011) Dynamics of different arachidonic acid orientations bound to prostaglandin endoperoxide synthases. Eur. J. Med. Chem. 46, 5212–5217 [DOI] [PubMed] [Google Scholar]
- 51.Zhuang X., and Zhang H. (2002) Identification, characterization of levoglucosan kinase, and cloning and expression of levoglucosan kinase cDNA from Aspergillus niger CBX-209 in Escherichia coli. Protein Expr. Purif. 26, 71–81 [DOI] [PubMed] [Google Scholar]
- 52.Chen J. C., Hanson B. L., Fisher S. Z., Langan P., and Kovalevsky A. Y. (2012) Direct observation of hydrogen atom dynamics and interactions by ultrahigh resolution neutron protein crystallography. Proc. Natl. Acad. Sci. U.S.A. 109, 15301–15306 [DOI] [PMC free article] [PubMed] [Google Scholar]