Background: Type IVa pilus (T4aP) assembly is primed by minor pilins.
Results: Non-core subunit PilE interacts with core minor pilins and is incorporated into pili; PilE is structurally similar to Neisseria PilX and PilV.
Conclusion: PilE connects the priming complex and the major pilin.
Significance: This function may be broadly conserved for non-core minor components in T4aP.
Keywords: bacterial adhesion, bacterial pathogenesis, Pseudomonas aeruginosa (P. aeruginosa), type IV pili, X-ray crystallography, Neisseria, PilC1, PilY1, bacterial two-hybrid, twitching motility, protein-protein interaction, minor pilins, pilus assembly, Neisseria meningitidis
Abstract
Many bacterial pathogens, including Pseudomonas aeruginosa, use type IVa pili (T4aP) for attachment and twitching motility. T4aP are composed primarily of major pilin subunits, which are repeatedly assembled and disassembled to mediate function. A group of pilin-like proteins, the minor pilins FimU and PilVWXE, prime pilus assembly and are incorporated into the pilus. We showed previously that minor pilin PilE depends on the putative priming subcomplex PilVWX and the non-pilin protein PilY1 for incorporation into pili, and that with FimU, PilE may couple the priming subcomplex to the major pilin PilA, allowing for efficient pilus assembly. Here we provide further support for this model, showing interaction of PilE with other minor pilins and the major pilin. A 1.25 Å crystal structure of PilEΔ1–28 shows a typical type IV pilin fold, demonstrating how it may be incorporated into the pilus. Despite limited sequence identity, PilE is structurally similar to Neisseria meningitidis minor pilins PilXNm and PilVNm, recently suggested via characterization of mCherry fusions to modulate pilus assembly from within the periplasm. A P. aeruginosa PilE-mCherry fusion failed to complement twitching motility or piliation of a pilE mutant. However, in a retraction-deficient strain where surface piliation depends solely on PilE, the fusion construct restored some surface piliation. PilE-mCherry was present in sheared surface fractions, suggesting that it was incorporated into pili. Together, these data provide evidence that PilE, the sole P. aeruginosa equivalent of PilXNm and PilVNm, likely connects a priming subcomplex to the major pilin, promoting efficient assembly of T4aP.
Introduction
Type IV pili (T4P)2 are long, thin, fibrous surface appendages found on Gram-negative and Gram-positive bacteria, as well as archaea (1, 2). They function in attachment, twitching motility, DNA uptake, electron transfer, and biofilm formation (1, 3). There are two main classes of T4P, type IVa and type IVb, distinguished by differences in their subunits and assembly machineries (1, 3, 4). The pili are composed of thousands of major pilin subunits, but minor (low abundance) pilin subunits are also present, potentially at the tip of the pilus due to their role in priming of pilus assembly (5–10).
Major pilin subunits are expressed as pre-pilins, which are processed by a bi-functional pre-pilin peptidase/N-methylase into assembly-competent mature subunits by removal of their type III signal sequence and methylation of the new N terminus (11–14). Although diverse in sequence, T4a major pilins share a conserved fold, consisting of an extended N-terminal α-helix connected to a four-stranded antiparallel β-sheet (6). The N-terminal α-helices, which form the hydrophobic inner core of an assembled pilus fiber, can be divided into two segments, α1-N and α1-C, with the highly conserved, hydrophobic α1-N segment retaining the monomers in the inner membrane prior to assembly. The globular C-terminal domains decorate the exterior of the pilus and typically contain a disulfide bond connecting the C terminus to the conserved β-sheet, forming a disulfide-bonded loop, also known as the D-region (3). Consistent with their incorporation into pili, the minor pilins are also processed by the pre-pilin peptidase and, based on the limited number of structures available, have architectures similar to major pilins (7, 9, 10, 15, 16).
The T4P system is evolutionarily related to the type II secretion (T2S) system, proposed to form a short pilus-like fiber in the periplasm that acts as a piston during the secretion of select exoproteins (17, 18). The T2S system has a set of four minor subunits, called the minor pseudopilins, which prime pseudopilus assembly (19). These four minor (pseudo)pilins are conserved between T2S and T4aP systems, suggesting that they are core components of (pseudo)pilus assembly (20). In support of this idea, Escherichia coli K-12 T4aP core minor pilins can prime heterologous assembly of Klebsiella oxytoca pseudopili and vice versa (21), and Pseudomonas aeruginosa T4aP assembly can be primed by either its minor pilins or its T2S minor pseudopilins (10).
Unlike E. coli, P. aeruginosa and Neisseria spp. T4aP systems include additional, non-core minor pilins (20). In P. aeruginosa, the core minor pilins are called FimU, PilV, PilW, and PilX, and are encoded in an operon with the large putative adhesin, PilY1 (22), and the non-core minor pilin, PilE (23). We showed (10) that PilVWX and PilY1 depend on one another for incorporation into pili, suggesting that they form a subcomplex and that this putative subcomplex was required for PilE to be recovered in the pilus fraction. We proposed that (among other functions) the minor pilins prime pilus assembly and are thus assembled into the pilus fiber (10).
Although P. aeruginosa has only one non-core minor pilin (PilE), Neisseria meningitidis has three, called ComP, PilXNm, and PilVNm. ComP is incorporated into T4aP of Neisseria and is required for competence (24, 25), binding directly to DNA via an electropositive region extending across the surface-exposed β-sheet region (16). The PilXNm protein (called PilLNg in Neisseria gonorrhoeae) is potentially orthologous to P. aeruginosa PilE, based on synteny (both genes are located at the 3′ ends of their respective minor pilin operons) and their limited sequence identity. However, PilVNm, another non-core subunit encoded elsewhere in the Neisseria genome, is also a potential PilE orthologue. It is more similar to P. aeruginosa PilE than is PilXNm (35% identity, versus 27% for PilXNm) and of similar length (122 residues for mature PilVNm, 134 for PilE, and 152 for PilXNm), raising the possibility that N. meningitidis encodes two PilE equivalents. PilXNm and PilVNm share 25% identity (Fig. 1), can be incorporated into pili, and modulate a number of pilus-associated phenotypes, although neither protein is essential for assembly (8, 15). Previous studies reported only subtle differences in piliation status of single mutants, but pilVNm pilXNm double mutants have no surface pili (9, 25–27). The x-ray crystal structure of the C-terminal domain of PilXNm revealed a canonical type IV pilin fold, with α1-C connected to a four-stranded antiparallel β-sheet ending in a disulfide-bonded loop (15). Its D-region contained an additional short helix and a hook-like loop implicated in opposing pilus retraction, potentially via inter-subunit interactions between antiparallel pilus fibers (15).
FIGURE 1.
Sequence alignment of PilE with PilXNm and PilVNm. Sequence alignment of P. aeruginosa PilE from strain PAO1 and N. meningitidis PilX and PilV from strain 8013 using MUSCLE (49) is shown. Numbering is according to the mature pilin, with the leader sequence shown in gray. The first 28 residues that were removed for structural studies are colored red. Pa, P. aeruginosa.
A recent study suggested that PilXNm and PilVNm do not have to be incorporated into pili to function (27). Imhaus and Duménil fused the fluorescent mCherry protein to the minor pilins' C termini, with the assumption that the fusion proteins would become too bulky to traverse the outer membrane secretin, trapping them in the periplasm. However, both fusions complemented their cognate mutants (27). They further showed that pilus-related functions are dependent on the number of pili expressed on the surface and suggested that PilXNm and PilVNm optimize the initiation of pilus assembly, thus influencing the number of surface pili produced and the resulting biological outputs (27).
Here, we investigated the role of the P. aeruginosa minor pilin PilE in pilus assembly. We show that PilE interacts with the major pilin subunit and other core minor pilins, supporting its proposed role as a connector of a minor pilin subcomplex to the major pilin-containing fiber during initiation of pilus assembly. Our 1.25 Å high-resolution structure of PilEΔ1–28 revealed a high level of structural similarity to PilXNm despite their limited sequence identity, and structural modeling suggested that PilVNm is more similar to PilE than to PilXNm. A PilE-mCherry fusion failed to complement a pilE single mutant for piliation and motility, but expression of PilE-mCherry in a retraction-deficient mutant (engineered so that pilus assembly depends solely on PilE) revealed that the fusion was functional, and incorporated into surface pili. Based on these data, we suggest that Neisseria expresses two PilE orthologues with partly redundant function, whereas P. aeruginosa has only a single (and thus essential) non-core subunit.
Experimental Procedures
Bacterial Strains and Plasmids
Bacterial strains and plasmids used in this study are listed in Table 1. Bacterial strains were stored at −80 °C in LB supplemented with 15% glycerol. E. coli strains were grown at 37 °C, unless otherwise stated, on LB agar containing ampicillin (100 μg/ml), kanamycin (50 μg/ml), gentamicin (15 μg/ml), or chloramphenicol (30 μg/ml), as appropriate. P. aeruginosa strains were grown at 37 °C on LB agar containing gentamicin (30 μg/ml).
TABLE 1.
Strains and plasmids used in this study
| Strains and plasmids | Description | Reference |
|---|---|---|
| E. coli strains | ||
| DH5α | F′ Phi80dlacZ ΔM15 Δ (lacZYA-argF)U169 deoR recA1 endA1 hsdR17(rK-mK+) phoA supE44 λ-thi-1 | Invitrogen |
| TOP10 | F-mcrA Δ (mrr-hsdRMS-mcrBC) ϕ80lacZΔM15 ΔlacX74 recA1 araD139 Δ(ara-leu) 7697 galU galK rpsL endA1 nupG | Invitrogen |
| Origami B (DE3) | F− ompT hsdSB(rB− mB−) gal dcm lacY1 ahpC (DE3) gor522::Tn10 trxB (KanR, TetR) | Novagen |
| BTH101 | F−, cya-99, araD139, galE15, galK16, rpsL1 (StrR), hsdR2, mcrA1, mcrB1 | Euromedex |
| P. aeruginosa strains | ||
| mPAO1 | Wild-type laboratory strain | 50 |
| mPAO1 pilA::Tn | ISphoA/hah transposon insertion at position 163 of pilA | 50 |
| mPAO1 pilT::Tn | ISphoA/hah transposon insertion at position 885 of pilT | 50 |
| mPAO1 pilE::Tn | ISphoA/hah transposon insertion at position 183 of pilE | 50 |
| mPAO1 pilT::Tn ΔMP ΔMPP::FRT | ISphoA/hah transposon insertion at position 885 of pilT and deletion of fimTUpilVWXY1Y2E and deletion of xcpUVWX replaced with an FRT scar | 10 |
| mPAO1 pilE::Tn pilT::FRT ΔfimU ΔMPP::FRT | ISphoA/hah transposon insertion at position 183 of pilE with FRT insertion in the NruI site of pilT and deletion of fimU and deletion of xcpUVWX replaced with an FRT scar | 10 |
| Plasmids | ||
| pBADGr | pMLBAD with aacC1 gene (gentamicin resistance) disrupting dhfrII (trimethoprim resistance), arabinose inducible | 33 |
| pBADGrpilE | PAO1 pilE cloned in pBADGr, GmR | 7 |
| pBADGrpilE mCherry | PAO1 pilE cloned in pBADGr encoding a C-terminal mCherry fluorescent tag | This study |
| pUT18C | pUC19-derived vector with T18 fragment (residues 225–399) of CyaA under control of lac promoter, MCS at 3′ end of T18 ORF, AmpR | 51 |
| pUT18CpilE | mature PAO1 pilE cloned in pUT18C, AmpR | This study |
| pKT25 | pSU40-derived vector with T25 fragment (residues 1–224) of CyaA under control of a lac promoter; MCS at 3′ end of T25 ORF, KanR | 51 |
| pKT25pilA | Mature PAO1 pilA cloned in pKT25, KanR | 10 |
| pKT25fimU | Mature PAO1 fimU cloned in pKT25, KanR | This study |
| pKT25pilV | Mature PAO1 pilV cloned in pKT25, KanR | 10 |
| pKT25pilW | Mature PAO1 pilW cloned in pKT25, KanR | 10 |
| pKT25pilX | Mature PAO1 pilX cloned in pKT25, KanR | 10 |
| pT18zip | pT18 plasmid with 35-residue leucine zipper cloned into KpnI site, AmpR | 28 |
| pT25zip | pT25 plasmid with 35-residue leucine zipper cloned into KpnI site, CmR | 28 |
| pET151pilE Δ1–28 | PilE (Δ1–28 of mature protein) expression vector with N-terminal His6 and V5 epitope tag | This study |
Bacterial Adenylate Cyclase Two-hybrid Assay
A bacterial adenylate cyclase two-hybrid assay (28) was used to test for protein-protein interactions between PilE and other pilins. The DNA sequence encoding full-length, mature P. aeruginosa PAO1 PilE was PCR-amplified using the forward and reverse primers: 5′-GCATCTAGACTTCACGTTGCTGGAAATGGTGGTGGT-3′ and 5′-CATGGTACCTCAGCGCCAGCAGTCGTTGAC-3′, respectively, followed by restriction digest with XbaI and KpnI for directional cloning into pUT18C for a T18 N-terminally tagged protein. Similarly, full-length mature FimU was N-terminally tagged with T25 by PCR amplifying fimU with the forward and reverse primers: 5′-GCATCTAGACTTCACCCTGATCGAGTTGCTGAT-3′ and 5′-CATGAATTCTCAATAGCATGACTGGGGCGCCT-3′, respectively, followed by digestion with XbaI and EcoRI for cloning into pKT25. Plasmids were confirmed by sequencing. pUT18C-pilE was co-transformed with pKT25-fimU or pKT25-pilA/V/W/X (10) into E. coli BTH101 for interaction experiments. Briefly, a single colony from each transformation was grown in LB supplemented with kanamycin (50 μg/ml) and ampicillin (100 μg/ml) overnight at 30 °C, followed by subculturing into fresh medium with antibiotics and 1 mm isopropyl-1-thio-β-d-galactopyranoside for induction. Cells were grown to A600 = 0.6 and spot plated in triplicate on LB agar + X-gal and MacConkey agar + maltose and incubated at 30 °C for 24 h. A T18- and T25-tagged leucine zipper expressed from pT18-zip and pT25-zip (28) was used a positive control. The experiment was performed in triplicate, and representative images were taken.
Protein Expression and Purification
PAO1 pilE encoding N-terminally truncated mature PilEΔ1–28 was PCR-amplified using forward primer 5′-CACCATCCGCTCCAACCGC-3′ and reverse primer 5′-TCAGCGCCAGCAGTCGTT-3′. This fragment was ligated into pET151/D-TOPO (Invitrogen) and transformed into TOP10 cells for propagation. The correct construction of pET151 pilEΔ1–28 encoding N-terminal His6 V5 epitope-tagged PilEΔ1–28 with a tobacco etch virus protease cleavage site to remove the tags was verified by DNA sequencing. The construct was transformed into E. coli Origami B (DE3) cells, and protein was expressed and purified as described previously (10). Briefly, selenomethionine (SeMet)-labeled PilEΔ1–28 was expressed from E. coli Origami cells in SeMet high-yield M9 minimal medium (Shanghai Medicilon) following the manufacturer's instructions. Cells were harvested by centrifugation at 3,200 × g, and the pellet was resuspended in lysis buffer (20 mm Tris, pH 8, 500 mm NaCl, and 0.1% lauryldimethylamine oxide) with 1× benzamidine. Cells were lysed by three passages through a French press, and after centrifugation to remove cell debris, the clarified lysate was applied on an ÄKTA FPLC system to a 5-ml Ni HiTrap Chelating HP column (GE Healthcare, Mississauga, Ontario Canada) pre-charged with 100 mm NiCl2. The column was washed in a stepwise manner with 15, 30, and 45 mm imidazole followed by elution of bound proteins with 300 mm imidazole. The elution fraction was dialyzed into 20 mm Tris, pH 8, 100 mm NaCl, treated with tobacco etch virus protease at a final concentration of 0.04 mg/ml for 3 h at room temperature, and applied to a second nickel affinity chromatography column as above. Untagged PilEΔ1–28 was collected in the flow-through fraction, buffer-exchanged into 20 mm Tris, pH 8, 50 mm NaCl, and concentrated to 4 mg/ml.
Crystallization and Structure Determination
SeMet PilEΔ1–28 crystals were grown using the hanging drop/vapor diffusion method in a 1:1 ratio of protein (4 mg/ml SeMet PilEΔ1–28 in 20 mm Tris, pH 8, 50 mm NaCl) and precipitant (0.2 m ammonium tartrate dibasic, 20% (w/v) PEG 3350) over 1.5 m ammonium sulfate at 20 °C. Crystals were flash-frozen in a nitrogen cold stream with no further cryo-protection. Diffraction data were collected at the X25 beamline of National Synchrotron Light Source (NSLS) in Brookhaven, NY with a wavelength of 0.979 Å.
Single anomalous diffraction data were processed using the HKL2000 program suite (29). The HySS submodule was used to locate the single SeMet site followed by phasing, density modification, automated model building, and refinement in the Phenix suite of programs (30, 31). Iterative rounds of manual model building and refinement were performed in Coot (32) until Rwork and Rfree values converged and could no longer be improved. Further details of data collection and model refinement statistics are listed in Table 2.
TABLE 2.
PilE data collection and refinement statistics
ASU, asymmetric unit.
| SeMet-PilE | ||
|---|---|---|
| Data collection | ||
| Beamline | NSLS X25 | |
| Wavelength | 0.979 | |
| Space group | C2 | |
| Unit cell parameters | ||
| a, b, c (Å) | 76.16, 35.56, 43.54 | |
| α, β, γ (°) | 90.0, 97.32, 90.0 | |
| No. of molecules in ASU | 1 | |
| Resolution range (Å)a | 50.0–1.25 (1.27–1.25) | |
| Unique reflections | 30,749 | |
| Data redundancya | 6.4 (4.5) | |
| Completeness (%)a | 95.4 (89.6) | |
| I/σ(I)a | 20.7 (7.6) | |
| Rmerge (%)a | 8.4 (21.0) | |
| Wilson B | 8.28 | |
| Model and refinement | ||
| Resolution range (Å) | 43.21–1.25 | |
| Rwork (%) | 15.90 | |
| Rfree (%) | 17.53 | |
| No. of reflections | 30,645 | |
| No. of amino acid residues/atoms | 831 | |
| No. of waters | 248 | |
| RMSD bond lengths (Å) | 0.005 | |
| RMSD bond angles (°) | 0.965 | |
| Average B (Å2) | 11.51 | |
| Ramachandran statistics (%) | ||
| Favored | 99.12 | |
| Allowed | 0.88 | |
| PDB code | 4NOA | |
a Values in parentheses represent highest resolution shell.
Construction of Fluorescently Tagged PilE
PAO1 pilE with the gene encoding mCherry fused on the 3′ end was synthesized by GenScript (Piscataway, NJ) with flanking EcoRI and HindIII restriction sites. The insert was subcloned into the EcoRI and HindIII sites of the P. aeruginosa-compatible and arabinose-inducible vector pBADGr (33) to generate pBADGr-pilE-mCherry.
Twitching Motility Assay
Twitching motility stab assays were performed as described previously (7). Briefly, strains of interest were stab-inoculated in duplicate to the plastic-agar interface of an LB 1% agar plate, which was incubated at 37 °C for 24 h. The agar was carefully removed, and adherent bacteria were stained with 1% crystal violet. The experiment was performed in triplicate.
Sheared Surface Protein Preparation
Proteins were sheared from the surface of P. aeruginosa cells as described previously (10). Briefly, bacterial strains were streaked in a cross-hatched manner on a 150-mm diameter LB agar (1.5%) plate containing gentamicin (30 μg/ml) and grown overnight at 37 °C. Cells were scraped using a glass coverslip, resuspended in 1× PBS, and vortexed for 30 s to shear off surface proteins. Bacterial cells were pelleted by centrifugation at 16,100 × g for 5 min followed by a second spin of the supernatant for 20 min. Sheared surface proteins in the clarified supernatant were precipitated on ice for 1 h using 0.4 m NaCl and 2.4% (w/v) PEG 8000 followed by centrifugation at 16,100 × g for 30 min. The pellets containing surface proteins (pilin and flagellin) were resuspended in 150 μl of 1× SDS loading buffer and boiled for 10 min. Samples were separated on 15% SDS-PAGE gels and stained with Coomassie Brilliant Blue for visualization. Densitometry was performed using ImageJ (34), where pilin levels were standardized against the flagellin band.
For Western blot analysis of PilE in surface fractions, the sheared surface protein samples were separated by SDS-PAGE and transferred to nitrocellulose as described (10), followed by detection with 1:1000 dilution of rabbit polyclonal PilE antibody (7) and 1:3000 dilution of goat anti-rabbit IgG-alkaline phosphatase-conjugated secondary antibody, developed using nitro-blue tetrazolium/5-bromo-4-chloro-3-indolylphosphate (NBT/BCIP).
Intracellular PilE Protein Levels
Bacterial cell pellets, recovered after removal of surface proteins by mechanical shearing as described above, were resuspended in 1× PBS to an A600 of 0.6. Two ml of bacterial suspension were centrifuged at 16,100 × g for 2 min, and the pellets were resuspended in 200 μl of 1× SDS loading buffer and boiled for 10 min. The lysates were separated by SDS-PAGE and transferred to nitrocellulose for immunoblot analysis with anti-PilE polyclonal antibody as described above.
Fluorescence Microscopy
Overnight cultures of the strains of interest were stab-inoculated into individual chambers of 1.0 borosilicate chambered cover glass slides (Lab-Tek) containing 1% LB agar. Slides were incubated in the dark for 1 h at 37 °C. Cells were then visualized using an EVOS FL Auto microscope (Life Technologies) with a Plan Apochromat 60× oil immersion objective, using either transmitted (white) light or a Texas Red LED light cube (emission 585/29, excitation 624/40). Images were acquired using the EVOS FL Auto Cell Imaging System software (Life Technologies) and exported as TIFF files. TIFF images were processed in ImageJ (34) by cropping representative regions of interest and then adjusting their brightness to improve visualization of mCherry.
Results
PilE Interacts with Major and Minor Pilins
We showed previously (10) that incorporation of the P. aeruginosa minor pilin PilE into T4aP depended on the presence of the putative PilVWXY1 priming subcomplex and that pilus assembly required (at a minimum) PilVWXY1 plus either FimU or PilE as putative connectors of the priming subcomplex to the major subunit, PilA. We tested potential interactions of full-length, mature PilE with PilA and the other minor pilins using a bacterial adenylate cyclase two-hybrid assay (28). PilE was N-terminally tagged with the T18 fragment of the Bordetella pertussis adenylate cyclase, whereas PilA, FimU, PilV, PilW, and PilX were N-terminally tagged with the T25 fragment, and interactions were identified on LB + X-gal and MacConkey + maltose plates (Fig. 2). PilE and FimU interactions were positive on both media, consistent with previous results (10). Similar to the other putative connector, FimU, PilE interacted with the major pilin PilA and the minor pilins PilV and PilX, but not with PilW, supporting its proposed role in stably linking the minor pilin PilVWXY1 priming subcomplex to the major subunit PilA.
FIGURE 2.
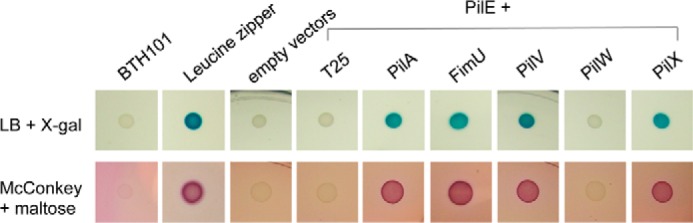
Interactions of PilE with minor pilins and PilA. Protein-protein interactions were tested using a bacterial adenylate cyclase two-hybrid system (BACTH). Mature PilE was N-terminally tagged with T18, whereas PilA, FimU, PilV, PilW, and PilX were N-terminally tagged with T25. Interactions were tested in E. coli cya mutant strain BTH101 on LB agar + X-gal and MacConkey + maltose indicator plates, which result in blue or red colonies if there is an interaction. A leucine zipper was used as a positive control.
The High-resolution Crystal Structure of PilE Reveals Characteristic Pilin Architecture
To gain further insight into the function of PilE, we solved a 1.25 Å high-resolution x-ray crystal structure of SeMet-labeled PilEΔ1–28. The structure lacks the first 28 N-terminal hydrophobic residues of the mature minor pilin, which were removed to improve its solubility. We also obtained native PilEΔ1–28 crystals and collected data; the protein crystallized in the same space group, C2, as the SeMet form, but the data were of lower resolution. Crystallographic data collection and refinement statistics are detailed in Table 2.
PilEΔ1–28 has a typical type IV pilin fold, characterized by an N-terminal α-helix connected to a four-stranded antiparallel β-sheet, terminating with a disulfide-bonded loop (Fig. 3A). The N-terminal residues from Asn-32 to Ser-52 form the α1-C helix, which likely extends in the full-length protein from the N-terminal hydrophobic α1-N-helix of a full-length pilin. The α1-C helix is packed against a four-stranded antiparallel β-sheet, to which it is connected by a 26-residue loop containing a 310 helix. Between the β2 and β3 strands of the β-sheet, residues Ile-97 to Lys-108 form a long loop with a 310 helix. Cys-106 and Cys-132 form a disulfide bond encompassing 25 residues that make up the D-region of PilE. In type IV pilins, the D-region is hypervariable in sequence, and in major pilins, the Cys residues typically staple the C terminus to the last β-strand (20). In contrast, the C terminus of PilE is linked by a disulfide bond to the β2-β3 loop, and the D-region encompasses β3, β4, and a short two-turn helix. Mutation of Cys-132 to Ala resulted in protein instability and loss of pilus assembly and twitching motility (data not shown), suggesting that the disulfide bond is a critical stabilizing feature of PilE. Similar results have been reported for Cys point mutants of the major pilin, PilA (33).
FIGURE 3.
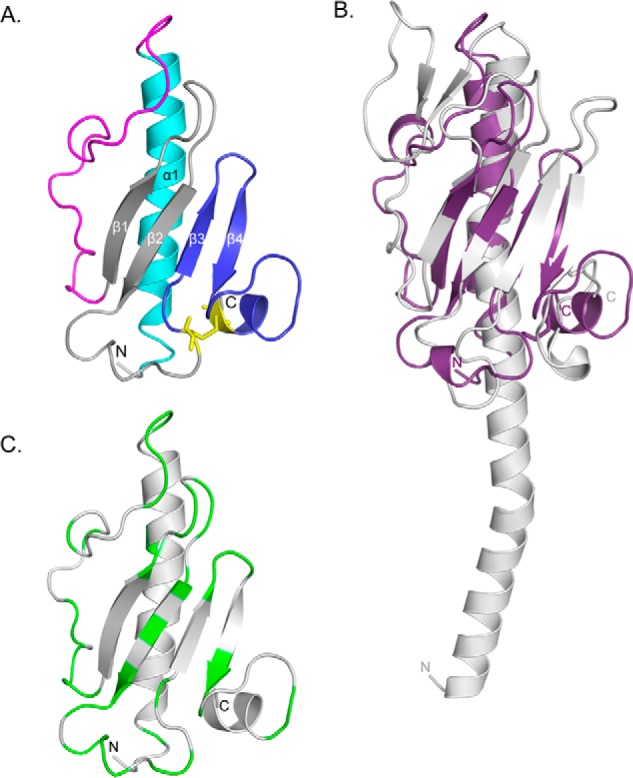
X-ray crystal structure of PilEΔ1–28. A, the x-ray crystal structure of selenomethionine-labeled PilEΔ1–28 was solved to 1.25 Å (PDB code: 4NOA). The N-terminal α-helix is colored cyan, αβ-loop is in magenta, β-sheet is in gray, and D-region is in blue. Cys residues are represented as sticks and colored yellow. B, structural alignment between PilEΔ1–28 (purple) and PilAPAK (gray, PDB code: 1OQW). 96 residues aligned with an RMSD of 3.8 Å. The arrow indicates the hook-like protrusion in both structures. C, mapping of residues differing between PilEPAO1 and PilEPA14. Non-conservative residues are colored green. Structural illustrations and alignments were generated with PyMOL (version 1.3, Schrödinger, LLC).
Of the P. aeruginosa minor pilins, PilE is the most similar to PilA, with 38% sequence similarity between full-length proteins. Despite the sequence differences and the differences in the connectivity of the disulfide bond in the D-region, 96 of 108 residues of PilEΔ1–28 could be aligned with the structure of PilAPAK (Protein Data Bank (PDB) 1OQW), with a root mean square deviation (RMSD) of 3.8 Å (Fig. 3B). The α1-helix of major pilins has a characteristic shallow S-shaped curve created by residues Pro-22 and Pro- or Gly-42 (35–37). Mature PilE has an Asp residue at position 42, and its truncated α1-C helix was not curved in our structure (Fig. 3, A and B). However, there is a Pro-22 residue in the PilE sequence that likely creates a kink in the α1-N helix region of PilE. The αβ-loops of major pilins are involved in inter-subunit interactions between pilin subunits in the pilus (38). In P. aeruginosa PilAPAK, this region forms a minor β-sheet, whereas in PilEΔ1–28, this region has a 310 helix and is less extended, possibly to accommodate interactions with the other minor pilins. Although β3 and β4 of PilEΔ1–28 are part of the D-region, the length and orientation of the four β-strands of central β-sheet are generally conserved between PilAPAK and PilEΔ1–28, as is the packing of the β-sheet against α1-C.
Each P. aeruginosa strain carries one of two different sets of T4aP minor pilin genes (exemplified by those of common laboratory strains PAO1 and PA14), which are encoded with specific major pilin genes in a “pilin island” that bears signatures of horizontal gene transfer (39). The sequence similarities between the minor pilin orthologues encoded by the two sets of genes range from ∼60 to 75%, with higher similarity in the N termini and lower in the C termini of each pair (40). With the exception of pilXPA14, cross-complementation of PAO1 minor pilin mutants with PA14 minor pilin genes restored surface piliation and twitching motility to various degrees, suggesting that most subunits can make functional interactions with heterologous partners (39). PilEPAO1 and PilEPA14 share 61% amino acid sequence similarity (39), and we found that most of the divergent residues map to loops or solvent-exposed surfaces (Fig. 3C). These results suggest that overall conservation of PilE architecture is important for its function.
Comparison of PilE and PilXNm Structures
The top hit from a structural comparison of PilEΔ1–28 with others in the PDB using DaliLite (41) was the PilXNm minor pilin from N. meningitidis (15). PilXNm is encoded with the Neisseria PilHIJK equivalents of the P. aeruginosa core minor pilins FimU-PilVWX and has been implicated in controlling efficient pilus biogenesis, as well as in attachment and aggregation of surface-exposed pili (8, 15, 27). PilE and PilXNm share 25% overall sequence identity, concentrated in the α1-N region (18 of 28 residues, 64%) that was deleted for structural studies (Fig. 1). The structure of N-terminally truncated PilXNm is composed of the typical N-terminal α1-C helix connected to a four-stranded antiparallel β-sheet (15) (Fig. 4A). Although there is only 14% sequence identity between the C-terminal domains of PilE and PilXNm, the critical pilin structural elements are maintained, and the Cα molecules align over 104 residues with an RMSD of 4.3 Å (Fig. 4B).
FIGURE 4.
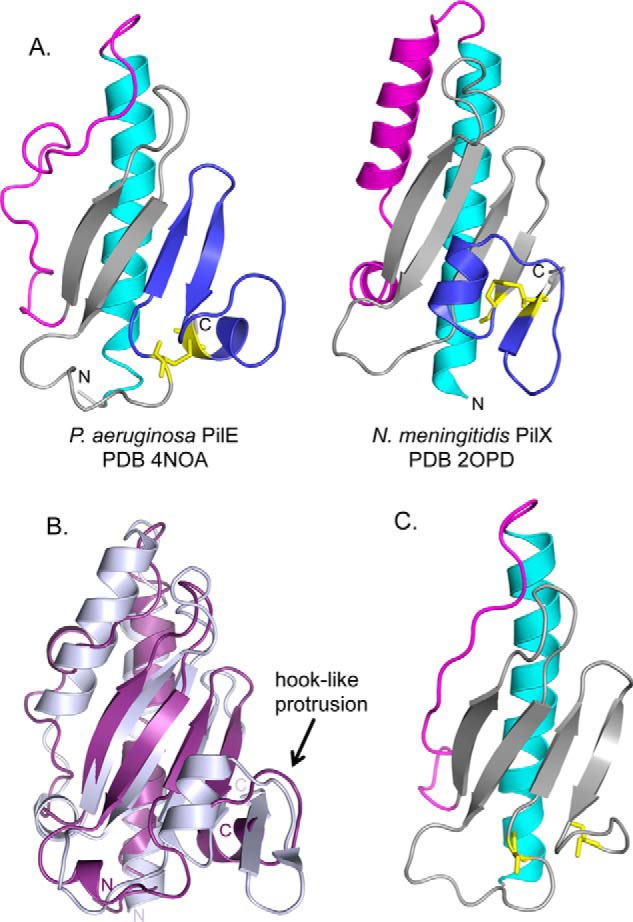
Comparison of PilEΔ1–28 with N. meningitidis PilXNm. A, side-by-side comparison of PilEΔ1–28 and PilXNm,Δ1–28 with the N-terminal α-helices colored in cyan, αβ-loops in magenta, β-sheets in gray, and D-regions in blue with the cysteines represented as sticks in yellow. B, structural alignment of PilEΔ1–28 (purple) and PilXNm,Δ1–28 (light blue). 104 residues are aligned with an RMSD of 4.3 Å. C, Phyre2-generated model of PilVNm based on PilE. Structural illustrations and alignments were generated with PyMOL (version 1.3, Schrödinger, LLC.).
Like PilE, the N-terminal α1-C helix in the PilXNm structure lacks a kink at position 42, although it has a Gly at this position (15), which in other pilin structures allows for a second bend in the S-shaped α1 helix (36–38). Of note, the D-regions of both PilE and PilXNm have hook-like protrusions (Fig. 4B). This feature was previously suggested to be important for protein-protein interactions between PilXNm subunits on neighboring but antiparallel pilus fibers, opposing pilus retraction and thereby promoting aggregation (15). Complementation of a pilE mutant with a construct encoding PilE with an in-frame deletion of the corresponding region (residues 120–127) restored wild-type pilus assembly and twitching motility (data not shown), suggesting that this region of PilE is not crucial for function in P. aeruginosa.
PilVNm Is Predicted to Be Structurally Similar to PilE
Although there is no structure yet available for PilVNm, it has higher sequence identity to PilE (35% overall identity) than the obvious structural homologue PilXNm (27% overall identity) (Figs. 1 and 4A). We used the Phyre2 structural prediction algorithm (42) to search for the best homology model for PilVNm, using only its C-terminal region (residues 29–122, mature PilVNm numbering). The top hit for PilVNm was our PilE structure, with a confidence level of 99.9% over an alignment of 90 residues (Fig. 4C). The next three hits were N. meningitidis minor pilin ComP (PDB 2MK3), P. aeruginosa major pilin PilA (PDB 1OQW), and N. gonorrhoeae major pilin PilE (PDB 2PIL), with decreasing levels of confidence. Of note, the PilXNm structure was not among the hits for the PilVNm C-terminal region, suggesting that the level of sequence identity between them was too low to generate even a low-confidence model. Repeating the search with the full-length mature PilVNm sequence returned similar results (data not shown).
Although PilVNm has Cys residues in the same position as PilXNm according to the alignment, no disulfide bond was present in the Phyre2-generated models of PilVNm. These structures do not model the last four residues of PilVNm after the last Cys, suggesting sufficient differences within this region to preclude high-confidence predictions. Nevertheless, PilVNm likely has a fold similar to PilE including a disulfide bond in its D-region, consistent with reports that it is incorporated into pili (26, 43).
PilE Incorporation into Pili Is Necessary for Function
Imhaus and Duménil (27) recently reported that PilXNm and PilVNm are required for efficient pilus biogenesis in N. meningitidis. They suggested that the functional pool of PilXNm and PilVNm was located in the periplasm rather than on the cell surface (as might be expected for integral components of assembled pili), because mCherry fusions considered too bulky to pass through the PilQ secretin were capable of complementing their cognate mutants. However, the proteins were unable to complement function unless processed by the pre-pilin peptidase (27), an essential prerequisite for pilus incorporation (11). This finding was consistent with other studies showing that minor pilins are present in sheared surface fractions, suggesting incorporation into pili (7, 9).
To reproduce this experiment in P. aeruginosa, mCherry was fused to the C terminus of PilE and its ability to complement a pilE mutant was tested. Analysis of cells complemented with the fusion protein by fluorescence microscopy revealed circumferential staining, confirming its expected periplasmic localization (Fig. 5A). However, complementation of a pilE mutant with PilE mCherry resulted in no recoverable surface pili or twitching motility, similar to the negative control (Fig. 5, B and C). The levels of PilE mCherry expressed from the pBADGr vector were intermediate between those of unmodified PilE expressed from the same plasmid and those expressed from the chromosomal locus (Fig. 5D), both of which restore similar levels of twitching motility. Therefore, the amount of fusion protein expression is unlikely to underlie the absence of twitching motility or pili in the strain expressing the PilE mCherry fusion. Consistent with our model (in which PilE stabilizes interactions between the major and a minor pilin subcomplex required for efficient pilus biogenesis), these data suggest that P. aeruginosa PilE cannot restore pilus biogenesis from a periplasmic location. Alternatively, attachment of a bulky fluorescent protein could impair the ability of PilE to function efficiently in the initiation of pilus assembly, leading to a lack of surface pili under circumstances where retraction is active, due to unbalanced extension/retraction dynamics.
FIGURE 5.
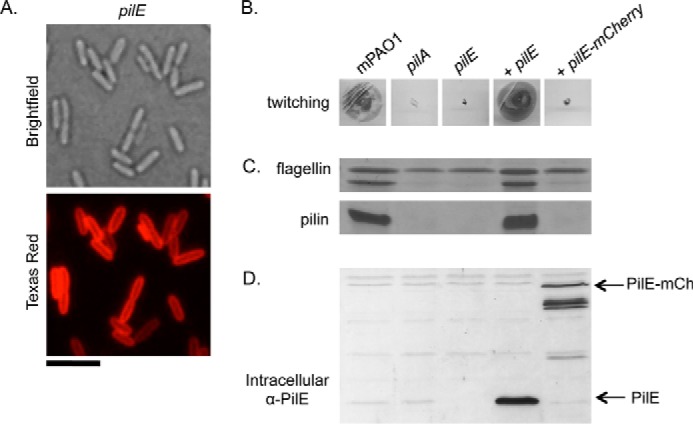
Complementation of pilE with PilE mCherry. mCherry was fused to the C-terminal end of PilE, and the level of complementation of a pilE mutant was assessed. A, fluorescence microscopy analysis of PilE mCherry localization. Scale bar represents 5 μm. B, twitching motility was tested by stab-inoculating to the bottom of an LB 1% agar plate and staining with 1% crystal violet after a 24-h incubation at 37 °C. C, pili were sheared from the surface of cells of interest and separated on a 15% SDS-PAGE gel. The flagellin band is used as a loading control. D, intracellular levels of PilE were probed by Western blot analysis with a α-PilE peptide antibody (1:1000 dilution). Arrows indicate the bands of interest.
To examine the latter possibility, we tested whether pBADGr-pilE-mCherry could complement piliation in a previously characterized P. aeruginosa ΔfimU pilE ΔMPP pilT mutant (10). This strain is retraction-deficient due to inactivation of the pilT gene encoding the retraction ATPase, and non-piliated because it lacks the putative connector proteins FimU and PilE as well as the T2S minor pseudopilins (MPP), which can prime pilus assembly in the absence of the T4aP minor pilins (10). In this strain, either pilE or fimU can restore surface piliation (10). When expressed in this mutant, PilE-mCherry restored ∼20% of surface piliation when compared with wild type PilE, where levels were commensurate with the pilT control (Fig. 6A). The decrease in surface piliation is likely not due to differences in the levels of PilE (Fig. 6C). These data suggest that PilE-mCherry is partially functional in terms of initiation of pilus assembly, but do not reveal whether it is incorporated into pili. To address that question, we probed sheared surface protein fractions using a PilE-specific antibody and readily detected a band with a mass corresponding to PilE-mCherry (Fig. 6B). The presence of PilE-mCherry in sheared surface fractions was not the result of cell lysis, as another inner membrane protein with a single transmembrane segment and mainly periplasmic C-terminal domain, PilO, was undetectable in surface fractions (Fig. 6, B and C). Attempts to image PilE-mCherry in assembled pili by immunogold labeling were unsuccessful, probably because it is present in very small amounts. In total, the results suggest that PilE-mCherry is incorporated into pili during pilus biogenesis and that the presence of the fusion tag may reduce the frequency of its incorporation (and thus efficiency of assembly initiation) to the point where surface piliation and motility are lost when retraction is active.
FIGURE 6.
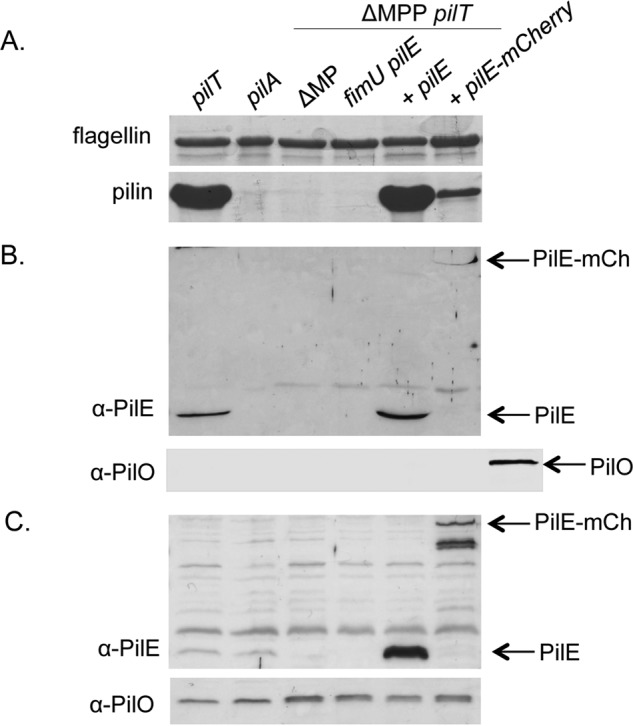
Pilus assembly by PilE mCherry in a retraction-deficient background. The ability of PilE mCherry to support pilus assembly in a retraction-deficient strain was tested in the ΔfimU pilE ΔMPP pilT mutant that lacks surface piliation in the absence of fimU and pilE. A, pilus assembly was probed by shearing proteins from the surface of the cells and analyzing the surface fractions by SDS-PAGE. The flagellin band is used as a loading control. B, incorporation of PilE mCherry into pili was examined by Western blot analysis of the surface fractions above and probing for PilE using an α-PilE peptide antibody (1:1000). Arrows indicate the bands of interest. The samples were probed with an antibody to inner membrane protein PilO (1:5000) as a control for cell lysis; the last lane is PAO1 lysate as a positive control for PilO. C, intracellular levels of PilE and PilO were probed by Western blot analysis with α-PilE peptide antibody (1:1000 dilution) and α-PilO antibody (1:5000), respectively.
Discussion
Our earlier results (10) suggested that the minor pilins prime T4aP assembly by forming a priming complex, analogous to that formed by minor pseudopilins (19), to initiate fiber polymerization. In our model, PilVWXY1 first form a subcomplex, which is then bound by PilE and coupled to the major subunit PilA through its interactions with both PilE and FimU, which interact with one another (10). Here we show that PilE interacts with the major pilin PilA and minor pilins FimU, PilV, and PilX (Fig. 2), supporting its role as a connector.
PilE has a typical type IV pilin structure (Fig. 3), likely facilitating its interactions with the major pilin and incorporation into the pilus. However, despite its structural and 38% sequence similarity to PilA, PilE does not form pili on its own, even when overexpressed (data not shown). The assembly process tolerates a wide range of PilE expression levels, while still supporting similar amounts of twitching motility (Fig. 5), likely due to the dependence of PilE on PilVWXY1 for incorporation into pili (10). Simply increasing intracellular levels of PilE would not necessarily affect the amount of PilE in the pilus if stoichiometric interactions with PilVWXY1 are necessary for its inclusion in pili. In addition, the architecture of the minor pilins may preclude their polymerization into a fiber. Major pilins have a Gly or Pro residue at position 42, creating a kink in their α1-C-helices that allows for inter-subunit interactions and flexibility of the fiber (35, 37, 44, 45). Putative connectors PilE (Fig. 3) and FimU (10) lack kinks in their α-1C helices. The absence of such curvature may have implications for the packing of these minor pilins with the priming subcomplex, or possibly their recognition by the assembly machinery, differentiating them from the major pilin proteins.
Imhaus and Duménil (27) created mCherry fusions to prevent the incorporation of PilXNm or PilVNm into surface-exposed pili, with the assumption that the fusions would be too large to fit through the secretin pore. However, it is difficult to differentiate that phenotype from one in which pilus assembly is impaired due to changes in interactions among major and minor pilin subunits because of the presence of the fusion protein. In P. aeruginosa, complementation of a pilE mutant with pilE-mCherry failed to restore surface piliation (Fig. 5C); however, in a retraction-deficient background, we recovered a small amount of pili in which PilE-mCherry could be detected (Fig. 6, A and B). This result is consistent with a model of pilus assembly initiation, in which only one PilE subunit per pilus is required (10). Pilus assembly is likely less efficient with PilE-mCherry versus wild type PilE, potentially due to suboptimal interactions with the fusion protein, inefficient priming, and thus no recoverable surface pili if retraction is active.
Neisseria non-core minor pilins PilXNm (PilLNg),PilVNm, and ComP all co-purify with pili, suggesting that they are part of the fiber (8, 9, 15, 16, 26, 43). PilXNm and PilVNm variants that cannot be processed by the pre-pilin peptidase were defective for complementation, suggesting that they need to be incorporated into pili for function (27). PilXNm is structurally similar to PilE (Fig. 4). Both are proposed to be involved in efficient initiation of pilus assembly (10, 27). In the absence of PilXNm or its N. gonorrhoeae homologue, PilLNg, piliation is reduced (9, 25), whereas without PilE, P. aeruginosa cells are non-piliated (7, 46). In trying to understand this difference between model species, we noticed that PilE and the N. meningitidis non-core minor pilin PilVNm were potentially orthologous. A Phyre2 analysis of the C-terminal domain of PilVNm yielded a high-confidence structural model on the PilE template, but returned no match with PilXNm, although DaliLite (41) analysis suggested that PilXNm is the top structural match for PilE (Fig. 4A). Based on these data, we propose that both PilVNm and PilXNm are PilE orthologues, possibly explaining why single P. aeruginosa pilE mutants lack surface pili, but both pilVNm and pilXNm must be deleted in Neisseria before piliation is lost (27).
The T2S system lacks both PilE and PilY1 equivalents. We showed previously that PilY1 is required for PilVWX incorporation into pili and that all four members of this putative subcomplex must be present for PilE to be incorporated into pili, suggesting that it recognizes a novel subcomplex interface (10). PilE may act as a quality control point to ensure the incorporation of the important non-pilin protein PilY1 into each pilus fiber. Like P. aeruginosa, other species such as Xylella fastidiosa, Ralstonia solanacearum, and Chromobacterium violaceum carry T4aP minor pilin operons that encode both PilY1-like proteins and PilE equivalents (9), suggesting a functional link.
Consistent with having two putative PilE equivalents, Neisseria spp. encode two PilY1-like proteins, PilC1 and PilC2 (47, 48). However, they are encoded separately from the minor pilins PilHIJK(X/L) (9), and their expression and regulation are not well understood. In N. gonorrhoeae, incorporation of PilLNg into pili depended on the core minor pilins PilHIJK as well as PilC1/2 (9). Similarly, PilVNg was dependent on PilC1/2 for pilus incorporation, although dependence on other minor pilins was not tested (43). Like PilY1, which does not require PilE for pilus incorporation, PilC is still present in surface pilus fractions in the absence of PilXNm, PilLNg, or PilVNm,Ng (8, 9, 26, 43). Based on the insights provided by characterization of PilE, we suggest that PilXNm and PilVNm may interact with one or more of the core minor pilins, plus PilC1 and/or PilC2, to initiate pilus assembly, and that the function of non-core PilE-like minor pilins may be conserved across T4aP-producing species that express PilY1-like proteins.
Author Contributions
Y. N. and L. L. B. designed the study, and Y. N., M. S. J., and L. L. B. wrote the paper. Y. N. and S. D. B. purified and crystallized PilE protein, and S. S. M., Y. N., and M. S. J. determined its X-ray structure. Y. N. and H. H. designed and constructed mutants and fusion constructs, and analyzed PilE function. Y. N. and R. N. C. B. performed microscopy experiments. All authors analyzed the results and approved the final version of the manuscript.
This work was supported by Canadian Institutes of Health Research (CIHR) Operating Grants MOP 86639 (to L. L. B.) and MOP 89903 (to M. S. J.). This work was also supported by a Graduate Studentship from Cystic Fibrosis Canada and a Banting Scholarship from the CIHR (to Y. N.). The authors declare that they have no conflicts of interest with the contents of this article.
The atomic coordinates and structure factors (code 4NOA) have been deposited in the Protein Data Bank (http://wwpdb.org/).
- T4P
- type IV pili
- T4aP
- type IVa pilus
- T2S
- type II secretion
- RMSD
- root mean square deviation
- X-gal
- 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside
- SeMet
- selenomethionine
- Nm
- Neisseria meningitidis.
References
- 1.Pelicic V. (2008) Type IV pili: e pluribus unum? Mol. Microbiol. 68, 827–837 [DOI] [PubMed] [Google Scholar]
- 2.Albers S. V., and Pohlschröder M. (2009) Diversity of archaeal type IV pilin-like structures. Extremophiles 13, 403–410 [DOI] [PubMed] [Google Scholar]
- 3.Craig L., Pique M. E., and Tainer J. A. (2004) Type IV pilus structure and bacterial pathogenicity. Nat. Rev. Microbiol. 2, 363–378 [DOI] [PubMed] [Google Scholar]
- 4.Ayers M., Howell P. L., and Burrows L. L. (2010) Architecture of the type II secretion and type IV pilus machineries. Future Microbiol. 5, 1203–1218 [DOI] [PubMed] [Google Scholar]
- 5.Mattick J. S. (2002) Type IV pili and twitching motility. Annu. Rev. Microbiol. 56, 289–314 [DOI] [PubMed] [Google Scholar]
- 6.Craig L., and Li J. (2008) Type IV pili: paradoxes in form and function. Curr. Opin. Struct. Biol. 18, 267–277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Giltner C. L., Habash M., and Burrows L. L. (2010) Pseudomonas aeruginosa minor pilins are incorporated into type IV pili. J. Mol. Biol. 398, 444–461 [DOI] [PubMed] [Google Scholar]
- 8.Hélaine S., Carbonnelle E., Prouvensier L., Beretti J. L., Nassif X., and Pelicic V. (2005) PilX, a pilus-associated protein essential for bacterial aggregation, is a key to pilus-facilitated attachment of Neisseria meningitidis to human cells. Mol. Microbiol. 55, 65–77 [DOI] [PubMed] [Google Scholar]
- 9.Winther-Larsen H. C., Wolfgang M., Dunham S., van Putten J. P., Dorward D., Løvold C., Aas F. E., and Koomey M. (2005) A conserved set of pilin-like molecules controls type IV pilus dynamics and organelle-associated functions in Neisseria gonorrhoeae. Mol. Microbiol. 56, 903–917 [DOI] [PubMed] [Google Scholar]
- 10.Nguyen Y., Sugiman-Marangos S., Harvey H., Bell S. D., Charlton C. L., Junop M. S., and Burrows L. L. (2015) Pseudomonas aeruginosa minor pilins prime type IVa pilus assembly and promote surface display of the PilY1 adhesin. J. Biol. Chem. 290, 601–611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Strom M. S., and Lory S. (1991) Amino acid substitutions in pilin of Pseudomonas aeruginosa: effect on leader peptide cleavage, amino-terminal methylation, and pilus assembly. J. Biol. Chem. 266, 1656–1664 [PubMed] [Google Scholar]
- 12.Strom M. S., Nunn D. N., and Lory S. (1993) A single bifunctional enzyme, PilD, catalyzes cleavage and N-methylation of proteins belonging to the type IV pilin family. Proc. Natl. Acad. Sci. U.S.A. 90, 2404–2408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sastry P. A., Finlay B. B., Pasloske B. L., Paranchych W., Pearlstone J. R., and Smillie L. B. (1985) Comparative studies of the amino acid and nucleotide sequences of pilin derived from Pseudomonas aeruginosa PAK and PAO. J. Bacteriol. 164, 571–577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Szabó Z., Stahl A. O., Albers S. V., Kissinger J. C., Driessen A. J., and Pohlschröder M. (2007) Identification of diverse archaeal proteins with class III signal peptides cleaved by distinct archaeal prepilin peptidases. J. Bacteriol. 189, 772–778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Helaine S., Dyer D. H., Nassif X., Pelicic V., and Forest K. T. (2007) 3D structure/function analysis of PilX reveals how minor pilins can modulate the virulence properties of type IV pili. Proc. Natl. Acad. Sci. U.S.A. 104, 15888–15893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cehovin A., Simpson P. J., McDowell M. A., Brown D. R., Noschese R., Pallett M., Brady J., Baldwin G. S., Lea S. M., Matthews S. J., and Pelicic V. (2013) Specific DNA recognition mediated by a type IV pilin. Proc. Natl. Acad. Sci. U.S.A. 110, 3065–3070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hobbs M., and Mattick J. S. (1993) Common components in the assembly of type 4 fimbriae, DNA transfer systems, filamentous phage and protein-secretion apparatus: a general system for the formation of surface-associated protein complexes. Mol. Microbiol. 10, 233–243 [DOI] [PubMed] [Google Scholar]
- 18.Durand E., Bernadac A., Ball G., Lazdunski A., Sturgis J. N., and Filloux A. (2003) Type II protein secretion in Pseudomonas aeruginosa: the pseudopilus is a multifibrillar and adhesive structure. J. Bacteriol. 185, 2749–2758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cisneros D. A., Bond P. J., Pugsley A. P., Campos M., and Francetic O. (2012) Minor pseudopilin self-assembly primes type II secretion pseudopilus elongation. EMBO J. 31, 1041–1053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Giltner C. L., Nguyen Y., and Burrows L. L. (2012) Type IV pilin proteins: versatile molecular modules. Microbiol. Mol. Biol. Rev. 76, 740–772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cisneros D. A., Pehau-Arnaudet G., and Francetic O. (2012) Heterologous assembly of type IV pili by a type II secretion system reveals the role of minor pilins in assembly initiation. Mol. Microbiol. 86, 805–818 [DOI] [PubMed] [Google Scholar]
- 22.Siryaporn A., Kuchma S. L., O'Toole G. A., and Gitai Z. (2014) Surface attachment induces Pseudomonas aeruginosa virulence. Proc. Natl. Acad. Sci. U.S.A. 111, 16860–16865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Belete B., Lu H., and Wozniak D. J. (2008) Pseudomonas aeruginosa AlgR regulates type IV pilus biosynthesis by activating transcription of the fimU-pilVWXY1Y2E operon. J. Bacteriol. 190, 2023–2030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wolfgang M., van Putten J. P., Hayes S. F., and Koomey M. (1999) The comP locus of Neisseria gonorrhoeae encodes a type IV prepilin that is dispensable for pilus biogenesis but essential for natural transformation. Mol. Microbiol. 31, 1345–1357 [DOI] [PubMed] [Google Scholar]
- 25.Brown D. R., Helaine S., Carbonnelle E., and Pelicic V. (2010) Systematic functional analysis reveals that a set of seven genes is involved in fine-tuning of the multiple functions mediated by type IV pili in Neisseria meningitidis. Infect. Immun. 78, 3053–3063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Takahashi H., Yanagisawa T., Kim K. S., Yokoyama S., and Ohnishi M. (2012) Meningococcal PilV potentiates Neisseria meningitidis type IV pilus-mediated internalization into human endothelial and epithelial cells. Infect. Immun. 80, 4154–4166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Imhaus A. F., and Duménil G. (2014) The number of Neisseria meningitidis type IV pili determines host cell interaction. EMBO J. 33, 1767–1783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Karimova G., Pidoux J., Ullmann A., and Ladant D. (1998) A bacterial two-hybrid system based on a reconstituted signal transduction pathway. Proc. Natl. Acad. Sci. U.S.A. 95, 5752–5756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Otwinowski Z., and Minor W. (1997) Processing of x-ray diffraction data collected in the oscillation mode. Methods Enzymol. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 30.Adams P. D., Grosse-Kunstleve R. W., Hung L. W., Ioerger T. R., McCoy A. J., Moriarty N. W., Read R. J., Sacchettini J. C., Sauter N. K., and Terwilliger T. C. (2002) PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr. D Biol. Crystallogr. 58, 1948–1954 [DOI] [PubMed] [Google Scholar]
- 31.McCoy A. J., Grosse-Kunstleve R. W., Adams P. D., Winn M. D., Storoni L. C., and Read R. J. (2007) Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Emsley P., and Cowtan K. (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 33.Harvey H., Habash M., Aidoo F., and Burrows L. L. (2009) Single-residue changes in the C-terminal disulfide-bonded loop of the Pseudomonas aeruginosa type IV pilin influence pilus assembly and twitching motility. J. Bacteriol. 191, 6513–6524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Abràmoff M. D., Magalhães P. J., and Ram S. J. (2004) Image processing with ImageJ. Biophotonics Int. 11, 36–42 [Google Scholar]
- 35.Craig L., Taylor R. K., Pique M. E., Adair B. D., Arvai A. S., Singh M., Lloyd S. J., Shin D. S., Getzoff E. D., Yeager M., Forest K. T., and Tainer J. A. (2003) Type IV pilin structure and assembly: x-ray and EM analyses of Vibrio cholerae toxin-coregulated pilus and Pseudomonas aeruginosa PAK pilin. Mol. Cell 11, 1139–1150 [DOI] [PubMed] [Google Scholar]
- 36.Hartung S., Arvai A. S., Wood T., Kolappan S., Shin D. S., Craig L., and Tainer J. A. (2011) Ultrahigh resolution and full-length pilin structures with insights for filament assembly, pathogenic functions, and vaccine potential. J. Biol. Chem. 286, 44254–44265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Parge H. E., Forest K. T., Hickey M. J., Christensen D. A., Getzoff E. D., and Tainer J. A. (1995) Structure of the fibre-forming protein pilin at 2.6 Å resolution. Nature 378, 32–38 [DOI] [PubMed] [Google Scholar]
- 38.Craig L., Volkmann N., Arvai A. S., Pique M. E., Yeager M., Egelman E. H., and Tainer J. A. (2006) Type IV pilus structure by cryo-electron microscopy and crystallography: implications for pilus assembly and functions. Mol. Cell 23, 651–662 [DOI] [PubMed] [Google Scholar]
- 39.Giltner C. L., Rana N., Lunardo M. N., Hussain A. Q., and Burrows L. L. (2011) Evolutionary and functional diversity of the Pseudomonas type IVa pilin island. Environ. Microbiol. 13, 250–264 [DOI] [PubMed] [Google Scholar]
- 40.Asikyan M. L., Kus J. V., and Burrows L. L. (2008) Novel proteins that modulate type IV pilus retraction dynamics in Pseudomonas aeruginosa. J. Bacteriol. 190, 7022–7034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Holm L., and Rosenström P. (2010) Dali server: conservation mapping in 3D. Nucleic Acids Res. 38, W545–W549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kelley L. A., and Sternberg M. J. (2009) Protein structure prediction on the Web: a case study using the Phyre server. Nat. Protoc. 4, 363–371 [DOI] [PubMed] [Google Scholar]
- 43.Winther-Larsen H. C., Hegge F. T., Wolfgang M., Hayes S. F., van Putten J. P., and Koomey M. (2001) Neisseria gonorrhoeae PilV, a type IV pilus-associated protein essential to human epithelial cell adherence. Proc. Natl. Acad. Sci. U.S.A. 98, 15276–15281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li J., Lim M. S., Li S., Brock M., Pique M. E., Woods V. L. Jr., and Craig L. (2008) Vibrio cholerae toxin-coregulated pilus structure analyzed by hydrogen/deuterium exchange mass spectrometry. Structure 16, 137–148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Burrows L. L. (2008) A nice return on the “stalk” exchange. Structure 16, 19–20 [DOI] [PubMed] [Google Scholar]
- 46.Russell M. A., and Darzins A. (1994) The pilE gene product of Pseudomonas aeruginosa, required for pilus biogenesis, shares amino acid sequence identity with the N-termini of type 4 prepilin proteins. Mol. Microbiol. 13, 973–985 [DOI] [PubMed] [Google Scholar]
- 47.Jonsson A. B., Pfeifer J., and Normark S. (1992) Neisseria gonorrhoeae PilC expression provides a selective mechanism for structural diversity of pili. Proc. Natl. Acad. Sci. U.S.A. 89, 3204–3208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Morand P. C., Tattevin P., Eugene E., Beretti J. L., and Nassif X. (2001) The adhesive property of the type IV pilus-associated component PilC1 of pathogenic Neisseria is supported by the conformational structure of the N-terminal part of the molecule. Mol. Microbiol. 40, 846–856 [DOI] [PubMed] [Google Scholar]
- 49.Edgar R. C. (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jacobs M. A., Alwood A., Thaipisuttikul I., Spencer D., Haugen E., Ernst S., Will O., Kaul R., Raymond C., Levy R., Chun-Rong L., Guenthner D., Bovee D., Olson M. V., and Manoil C. (2003) Comprehensive transposon mutant library of Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. U.S.A. 100, 14339–14344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Karimova G., Ullmann A., and Ladant D. (2001) Protein-protein interaction between Bacillus stearothermophilus tyrosyl-tRNA synthetase subdomains revealed by a bacterial two-hybrid system. J. Mol. Microbiol. Biotechnol. 3, 73–82 [PubMed] [Google Scholar]



