Abstract
Organisms living in polar regions have evolved a series of antifreeze (glyco) proteins (AFGPs) to enable them to survive by modulating the structure of ice. These proteins have huge potential for use in cellular cryopreservation, ice-resistant surfaces, frozen food, and cryosurgery, but they are limited by their relatively low availability and questions regarding their mode of action. This has triggered the search for biomimetic materials capable of reproducing this function. The identification of new structures and sequences capable of inhibiting ice growth is crucial to aid our understanding of these proteins. Here, we show that plant c-type lectins, which have similar biological function to human c-type lectins (glycan recognition) but no sequence homology to AFPs, display calcium-dependent ice recrystallization inhibition (IRI) activity. This IRI activity can be switched on/off by changing the Ca2+ concentration. To show that more (nonantifreeze) proteins may exist with the potential to display IRI, a second motif was considered, amphipathicity. All known AFPs have defined hydrophobic/hydrophilic domains, rationalizing this choice. The cheap, and widely used, antimicrobial Nisin was found to have cation-dependent IRI activity, controlled by either acid or addition of histidine-binding ions such as zinc or nickel, which promote its amphipathic structure. These results demonstrate a new approach in the identification of antifreeze protein mimetic macromolecules and may help in the development of synthetic mimics of AFPs.
Introduction
Antifreeze proteins and glycoproteins (AF(G)Ps) are found in a wide variety of different organisms that habitually experience freezing temperatures. They were initially discovered in Arctic and Antarctic fish species but have since been reported in a wide range of plants and insects.1−5 AFPs provide protection through two main methods: thermal hysteresis (TH) and ice recrystallization inhibition (IRI).6 The former is the depression of the freezing point relative to that of the melting point,7 and the latter is the slowing of the rate of the ice crystal growth (Ostwald ripening) process.8 The underlying mechanisms for each action are still under investigation, and it has emerged that TH and IRI activities are not necessarily linked. In 2001, Enaide et al. reported that simplified AFGPs could retain potent IRI activity while displaying essentially zero TH activity.9 Furthermore, Ben and co-workers have developed a range of structures that possess only IRI activity, including carbohydrate-based surfactants, synthetic antifreeze proteins, and several small molecules.10−12 Davies et al. have also shown that the magnitude of TH does not always scale with IRI.13
IRI-active compounds have tremendous potential for applications in frozen foods, cryosurgery, and cryopreservation.14−16 For example, ice growth (recrystallization) during thawing has been shown to be a major contributor to cell death during cellular cryopreservation. Carpenter et al. showed that AFPs could enhance cryopreservation of red blood cells, but the benefit was limited by the onset of dynamic ice shaping (DIS) (associated with TH), which actually reduced cell recovery.17 There are various conflicting results on AF(G)P-supplemented cryopreservation that show both benefit and detriment, normally due to the DIS property.16,18,19 Despite these challenges, there is an urgent need for new cryopreservatives as alternatives to the current state of the art, which employs (toxic) organic solvents, and also for the preservation of cell types that currently cannot be frozen. Ben et al. have shown that IRI-specific glycopeptides and alkyl glycosides can enhance cellular cryopreservation.20 Gibson et al. have demonstrated that IRI-active synthetic polymers enhance nonvitreous cryopreservation of red blood cells.21 Despite these structurally diverse IRI mimetics, an understanding of the structural motifs essential for activity remains elusive. Another outstanding challenge is the development of smart AFP derivatives with switchable activity, enabling them to be activated both spatially and temporally.
Current understanding has revealed that amphiphilicity (segregated hydrophilic and hydrophobic domains) is needed for IRI activity, with most AFPs showing defined regions of each.22 Ben et al. have shown that hydrophobically modified saccharides have (rather weak) IRI activity associated with their amphiphilicity.10 Despite this progress, there are still few compounds that are potent IRI inhibitors, which, in turn, has limited their application and the biophysical studies required to understand their remarkable ability to slow ice growth.
AFPs display a wide variety of secondary and tertiary structures and amino acid sequences while all binding to the same ice substrate.23 AFPs have been grouped into several different subtypes based on structure, for example, type 1 AFPs consist of a long amphipathic α helix,24 whereas type 2 AFPs are cysteine-rich globular proteins containing several disulfide bonds.25 In an attempt to understand the evolutionary origins of AFPs, sequence alignment studies have been undertaken. This has revealed strong similarity to other families of proteins such as lectins and apolipoproteins.26 A defining characteristic of these protein families is that they discriminate between related classes of molecules; for example, lectins bind specific carbohydrates. Due to this similarity, it may be possible that there exists many protein structures that, subject to the correct evolutionary conditions, could have become AFPs and hence might display some latent activity. To this end, a c-type lectin from rattlesnake Crotalus atrox has been evaluated for specific ice crystal face binding, although not for IRI activity.27
The aim of this study was to investigate if other nonantifreeze proteins can still display ice recrystallization inhibition activity, based on their functional and structural similarities to known AFPs, rather than the traditional chemical biology approaches based on sequence similarities. The ability to modulate AFP function by application of external stimuli is also studied as a route to smart AFP mimetics.
Experimental Section
Materials
Poly(ethylene glycol), HEPES, sodium acetate, glacial acetic acid, sodium hydroxide pellets, calcium chloride, glyceraldehyde, Nisin A, zinc acetate, and nickel acetate were purchased from Sigma-Aldrich (UK). Lectins concanavalin A (Con-A), soybean agglutinin (SBA), and Ricinus communis agglutinin 120 (RCA120) were purchased from Vector Laboratories (USA). Nisin was dialyzed against PBS buffer for 24 h (5 buffer changes) to ensure that all salts were removed before use. HEPES buffer solution (10 mM) was prepared using solid powder and adjusted to pH 7.4 using sodium hydroxide pellets. Sodium acetate buffer (0.2M) was prepared by mixing sodium acetate and glacial acetic acid in deionized water and adjusted to pH 5 with sodium hydroxide pellets.
Ice Recrystallization Inhibition (Splat) Assay
Ice recrystallization inhibition was measured using a modified splay assay.28 A 10 μL sample of polymer dissolved in PBS buffer (pH 7.4) was dropped 1.40 m onto a chilled glass coverslip sitting on a piece of polished aluminum placed on dry ice. Upon hitting the chilled glass coverslip, a wafer with a diameter of approximately 10 mm and thickness of 10 μm was formed instantaneously. The glass coverslip was transferred onto the Linkam cryostage and held at −8 °C under N2 for 30 min. Photographs were obtained using an Olympus CX 41 microscope with a UIS-2 20×/0.45/∞/0-2/FN22 lens and crossed polarizers (Olympus Ltd., Southend-on-Sea, UK), equipped with a Canon DSLR 500D digital camera. Images were taken of the initial wafer (to ensure that a polycrystalline sample had been obtained) and after 30 min. Image processing was conducted using ImageJ, which is freely available.29 In brief, 10 of the largest ice crystals were measured, and the single longest length in any axis was recorded. This was repeated for at least three wafers, and the average (mean) value was calculated to find the largest grain dimension along any axis. The average of this value from three individual wafers was calculated to give the mean largest grain size (MLGS). This average value was then compared to that of a PBS buffer negative control, providing a way of quantifying the amount of IRI activity.
Analysis of Sequence Alignment
Sequences of proteins were analyzed using the online BLAST sequence alignment tool,30 whereas three-dimensional structures of proteins were imaged using PyMOL.31 A type 2 AFP from Sea raven Hemitripterus americanu (PDB code 2AFP, accession no. P05140) was searched against using the Protein Data Bank proteins database and blastp algorithm. The sequences of those proteins defined as having significant alignments by BLAST were used for further sequence alignment. Multiple sequence alignment was conducted using Jalview Desktop, with Muscle with defaults multiple sequence alignment web service for alignment.32
Use of Lectins and Nisin
The lectins used in this study were kept as a solid lyophilized powder until required for testing; then, they were prepared in the appropriate buffer at 5 mg mL–1 solutions and diluted as needed. Denaturation of the protein was achieved by incubation of the protein at 80 °C for 15 min.
Results and Discussion
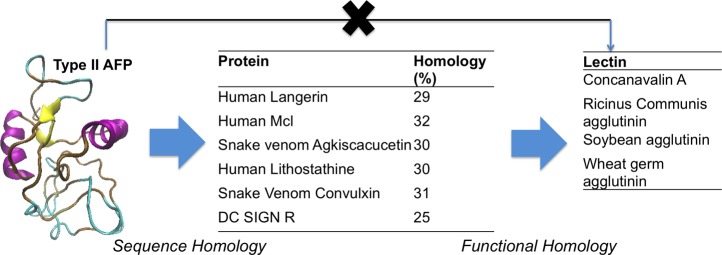
Inspired by the work of Graham et al. and others,25,33 which unravelled the genetic history of AFPs, sequence alignment of type 2 AFP from the Sea raven, H. americanus, against all known proteins within the Protein Data Bank (PBD) was conducted to identify those proteins with high sequence identity. This search agreed with previous reports, showing that, in addition to other type 2 AFPs, a range of c-type lectins also had high sequence similarity and that the overall sequence was identified as a member of the c-type lectin superfamily. The c-type lectins most closely related to type 2 AFP include human langerin (c-type transmembrane protein on Langerhans cells), human lithostathine, and a rat mannose binding protein (Figure 1). Interestingly, lithostathine has been found to play a role in the prevention of pancreatic stones by preventing calcite crystal growth, a somewhat similar role to an AFP, suggesting that common crystallization-inhibiting motifs have evolved in both cases. As reported by Rubinsky et al., a c-type lectin from the rattle snake, C. atrox, has Ca2+-dependent ice shaping activity, but no reports on IRI activity have been made.27
Figure 1.
Schematic of homology between type 2 antifreeze proteins and c-type lectins and the functional relationship to plant lectins.
While the lectins described above are most similar to type 2 AFP in sequence, they can be challenging to produce in large quantities, requiring (mammalian cell) recombinant expression. Furthermore, the use of human lectins (or rattlesnake venom) in biomedical applications would be limited by immunogenicity and associated problems. Therefore, we decided to move on from sequence similarity to functional similarities, specifically, the c-type lectins found in plants; some plant lectins have found use in medical applications, such as banana lectin in HIV inhibition.34 These have structural similarities (particularly in the carbohydrate recognition domain, CRD) to human c-type lectins, but they do not have a significant relationship to native AFPs. A range of plant lectins was selected based their commercial availability: concanavalin A (Con A, Mw 104 kDa), soybean agglutinin (SBA, Mw 120 kDa), and R. communis agglutinin 120 (RCA120, Mw 120 kDa).
To determine if the lectins demonstrate any antifreeze protein associated activity, a modified splat assay was employed to screen for ice recrystallization inhibition (IRI). Briefly, buffered 10 μL solutions of the analyte of interest were dropped onto a glass slide chilled to −80 °C to produce a wafer of small ice crystals. These were annealed at −8 °C, and the average ice crystal size was recorded after a fixed period of time and compared to a negative control to give the mean largest grain size (MLGS). The smallest size (as a percentage) from this assay is ∼20% (as an ice crystal cannot have zero size). HEPES buffer was employed in place of the normal PBS to enable the addition of divalent ions (Ca2+), which cannot be used in PBS (due to calcium phosphate precipitation). Assessment of the IRI activity of Con A at 5 mg mL–1 is shown in Figure 2A. As can be seen, Con A showed no activity, being statistically identical to our negative control, PEG (poly(ethylene glycol)).35 However, for Con A to bind its native glycans (α-mannose/glucose), it is essential for Ca2+ to be present (or similar divalent ions) in the carbohydrate recognition domain, inspiring us to repeat the experiment in the presence of CaCl2. Under these conditions, there was definite IRI activity, suggesting that Con A is also a c-type antifreeze protein, which can be turned on (or off) by addition of calcium ions. Calcium-dependent ice binding also has been reported for rainbow smelt and Atlantic herring fish species and Antarctic bacteria Marinomonas primoryensis.(36,37) Control experiments of CaCl2-doped buffer revealed that the IRI activity was not due to the metal ions (unlike, for example, ZrAc, which has some unique activity to inhibit ice growth) (see Supporting Information).38 Concentration dependence of the IRI activity was also measured (Figure 2B). While far weaker than native AFPs or PVA, this activity is stronger than that for synthetic analogues such as poly(ampholytes), which have sufficient IRI to act as cryopreservation enhancers.39−41
Figure 2.
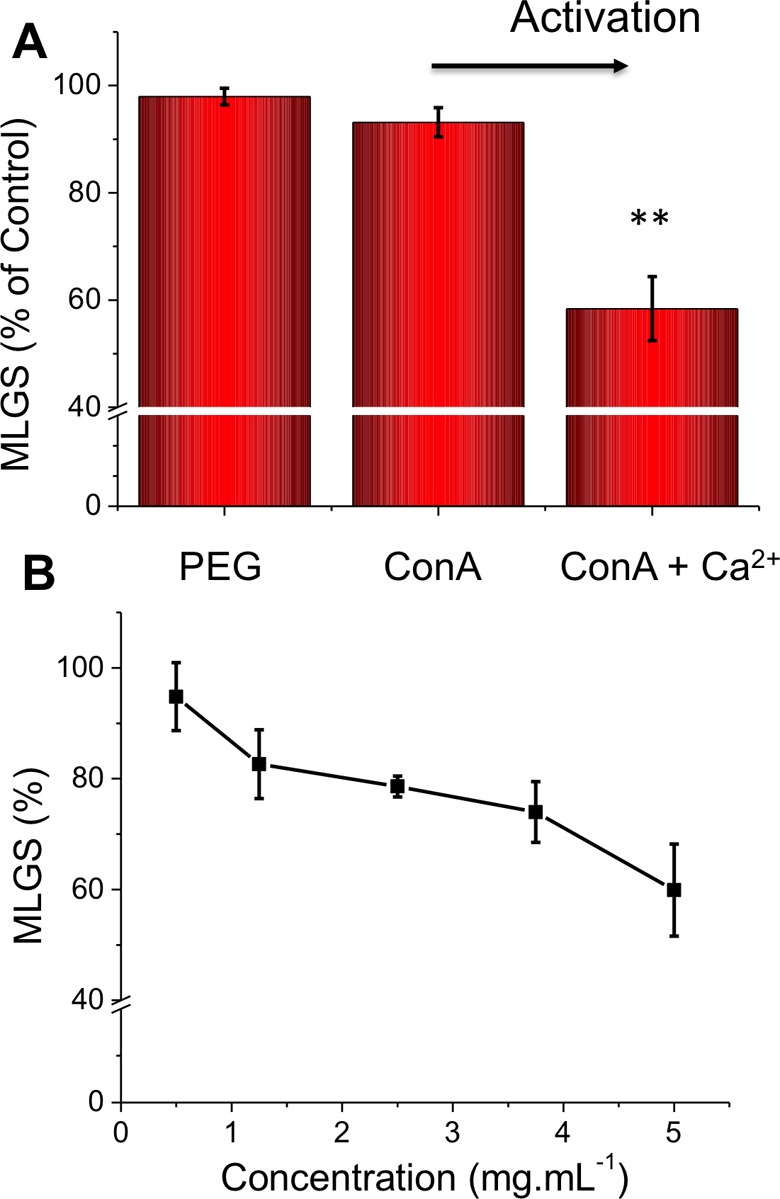
IRI activity of Con A in HEPEs buffer. (A) Effect of (5 mg mL–1) CaCl2 addition. [Con A] = 5 mg mL–1, [CaCl2] = 5 mg mL–1, [PEG] = 5 mg mL–1. (B) Concentration dependence of Con A with 5 mg mL–1 CaCl2. Error bars represent ± SD from a minimum of three repeats. MLGS = mean largest grain size relative to HEPES buffer or HEPES and calcium chloride buffer control. **, p < 0.01 relative to PEG control.
Con A exists as a tetramer around pH 7, but at lower pH, it dissociates into its monomeric or dimeric units, providing an accessible route to study the IRI activity in more detail. In pH 5 acetate buffer, Con A is a dimer and showed statistically significant, but only slightly enhanced, activity relative to that of the native tetramer measured at pH 7.4 (see Supporting Information). This implies that the overall tertiary/quaternary structure of the protein might not be essential for activity and that the carbohydrate recognition domain is the crucial component; it should be stressed this is only speculation at this point. To evaluate if this is a general phenomena or unique to Con A, lectins SBA and RCA120 were also evaluated by the splat assay, with and without Ca2+, and the results are shown in Figure 3.
Figure 3.
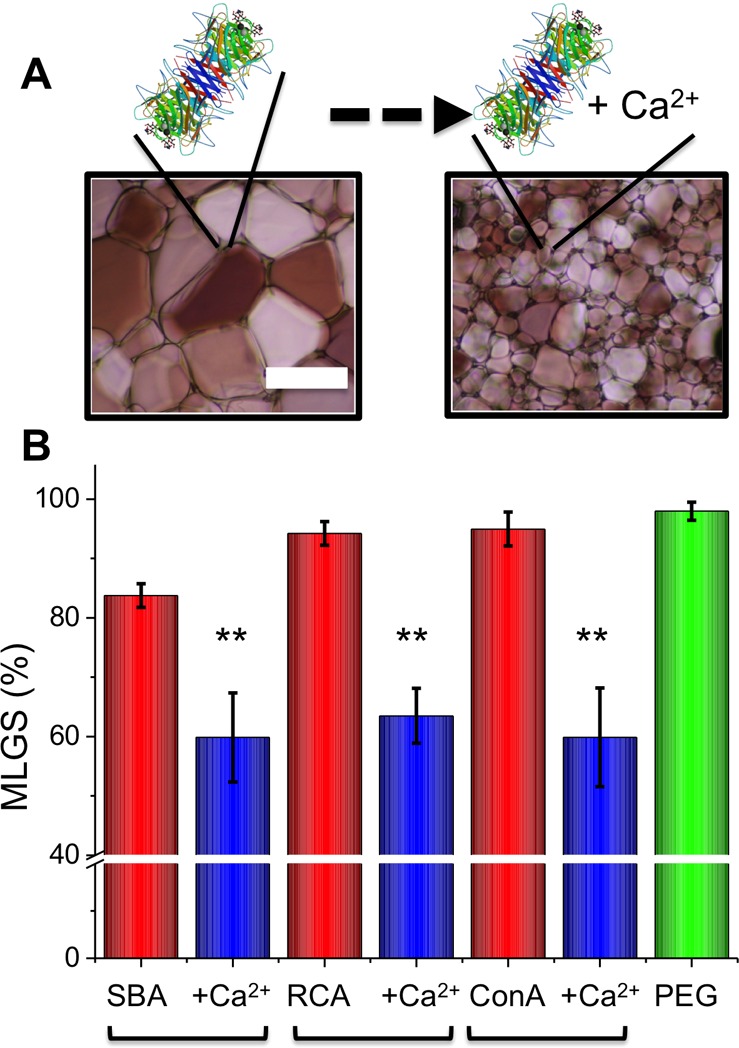
IRI activity of c-type lectins with and without 5 mg mL–1 CaCl2. Error bars represent ± SD from a minimum of three repeats. MLGS = mean largest grain size relative to HEPES buffer or HEPES and calcium chloride buffer control. **, p < 0.01 relative to PEG control.
Both SBA and RCA inhibited ice crystal growth at just 5 mg mL–1, with both requiring the addition of Ca2+ to switch on the activity, to a similar extent as that with Con A. To rule out this being a nonspecific effect, Con A and SBA were both denatured (confirmed by circular dichroism; Supporting Information) by boiling and being retested. This reduced their activity in both cases, confirming that the activity results from a specific structural feature of the proteins, not a broad macromolecular effect (Figure 4A). It should be noted that heating at 80 °C is not expected to completely denature the protein (and there will be some refolding); hence, the MLGS value does not return to 100%. The effect of adding in competing carbohydrate ligands, which will bind the CRD (in the presence of Ca2+), was measured, and the results are shown in Figure 4B. There was a very weak enhancement in activity upon addition of the sugars, which, in the case of SBA/GalNAc, was statistically significant but was small. This would appear to rule out the CRD directly binding to the ice (as the sugars would be competing ligands). Alternatively, it might be that the sugar has a similar hydrogen-bonding network as the water and therefore it is tolerated. A detailed site-directed mutagenesis study would be required to undercover this, which is beyond the scope of the present study. No ice shaping was observed (which would imply specific ice face interaction), but this effect is normally seen only at high AFP concentrations (>20 mg mL–1); it was not possible to obtain homogeneous protein solutions at these concentrations. Therefore, direct ice interaction cannot be ruled out, but there is building evidence that many (macro)molecules can inhibit ice growth without ice binding by disruption of the quasi-liquid layer/eutectic phase interface.35,40,42
Figure 4.
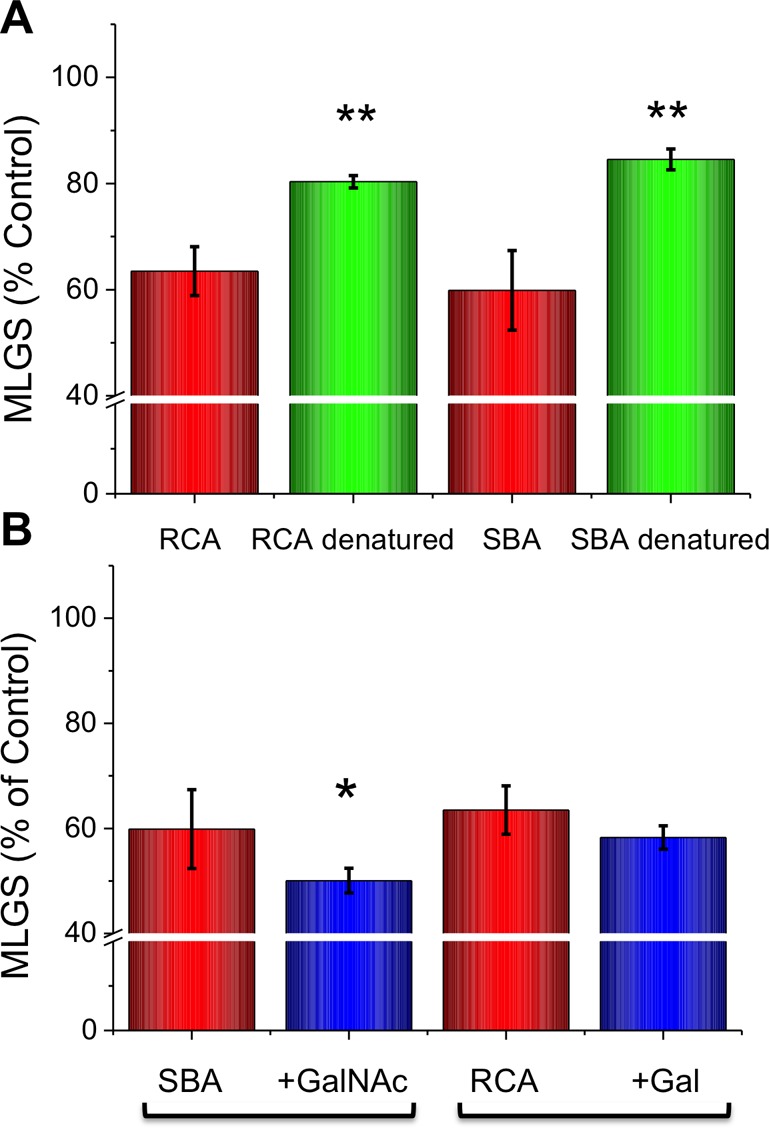
IRI inhibition/activation studies of SBA and RCA. (A) Effect of denaturation. (B) IRI effect of the addition of competitive carbohydrates (10 mM). GalNac = N-acetylgalactosamine. Gal = galactose. Error bars represent ± SD from a minimum of three repeats. MLGS = mean largest grain size relative to HEPES buffer or HEPES and calcium chloride buffer control. **, p < 0.01 relative to respective denatured sample; *, p < 0.05 relative to respective sugar-free lectin solution.
The reasons for the observed activity remain unclear (as with many features of AF(G)Ps), but given the evolutionary origins of type 2 AFPs from lectins, the activity is perhaps not surprising. Lectins evolved to recognize the difference between sugars and water, a challenging task because both are essentially hydrated −OH groups. An antifreeze protein could be considered to discriminate between water and ice, which is a similar problem. The current understanding of antifreeze proteins suggests that amphipathic character is required, i.e., spatially defined hydrophilic and hydrophobic faces. Our initial analysis of the lectins’ crystal structures did not show an obvious amphiphilic structure, but crystallography provides only a static, not dynamic, description of protein structure and function and hence will require more investigation.
Given the amphipathic structure of many AFPs, it would seem that other amphipathic proteins would make good lead molecules. Host-defense, cationic antimicrobial peptides are well-known amphipathic proteins, and several are commercially available.43 To test our hypothesis that these peptides might retain latent antifreeze activity, Nisin A was selected as a model antimicrobial peptide. Nisin is readily available, of low molecular weight (34 amino acids, 3354 g mol–1), and widely used in the dairy industry. It has also been recently reported that the Ixodes scapularis tick antifreeze glycoprotein has antimicrobial activity, providing additional support to the hypothesis of repurposing antimicrobial peptides.44 In order to function as an antimicrobial, Nisin A requires a sufficiently low pH to protonate histidines 27 and 31, which then induces the correct amphipathic conformation to enable cell membrane disruption/pore formation.45−47Figure 5 shows a space-filling model illustrating the hydrophilic/hydrophobic domains of Nisin A compared to the type 1 AFP from winter flounder. This illustrates the spatiality segregated hydrophilic and hydrophobic domains, which we propose is a key component in order to obtain IRI activity. This peptide provides a convenient tool to test the amphipathic hypothesis. Therefore, the IRI activity of Nisin A was tested as a function of pH and concentration (Figure 6). It should be noted that in Figure 6A activity at each pH is reported relative to a blank of that pH solution.
Figure 5.
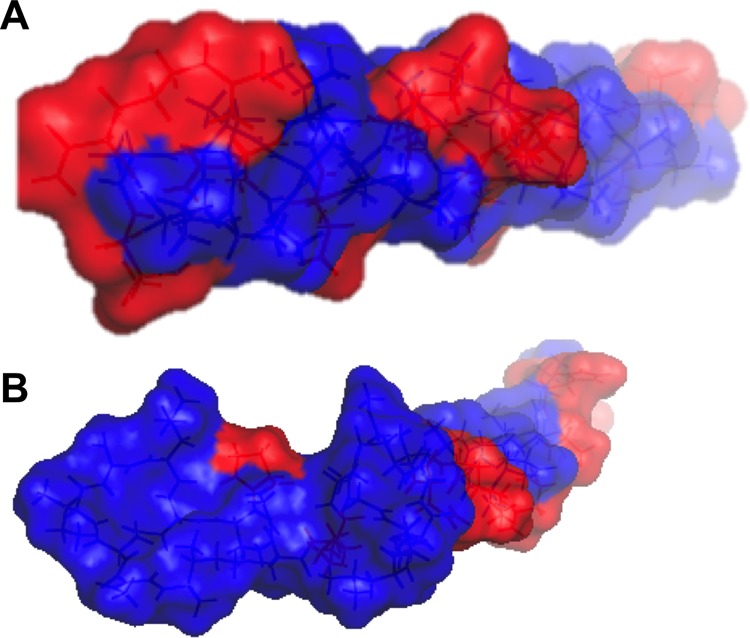
Space filling models of peptides, with hydrophilic (red) and hydrophobic (blue) domains indicated. (A) Winter flounder type I AFP; (B) Nisin A.
Figure 6.
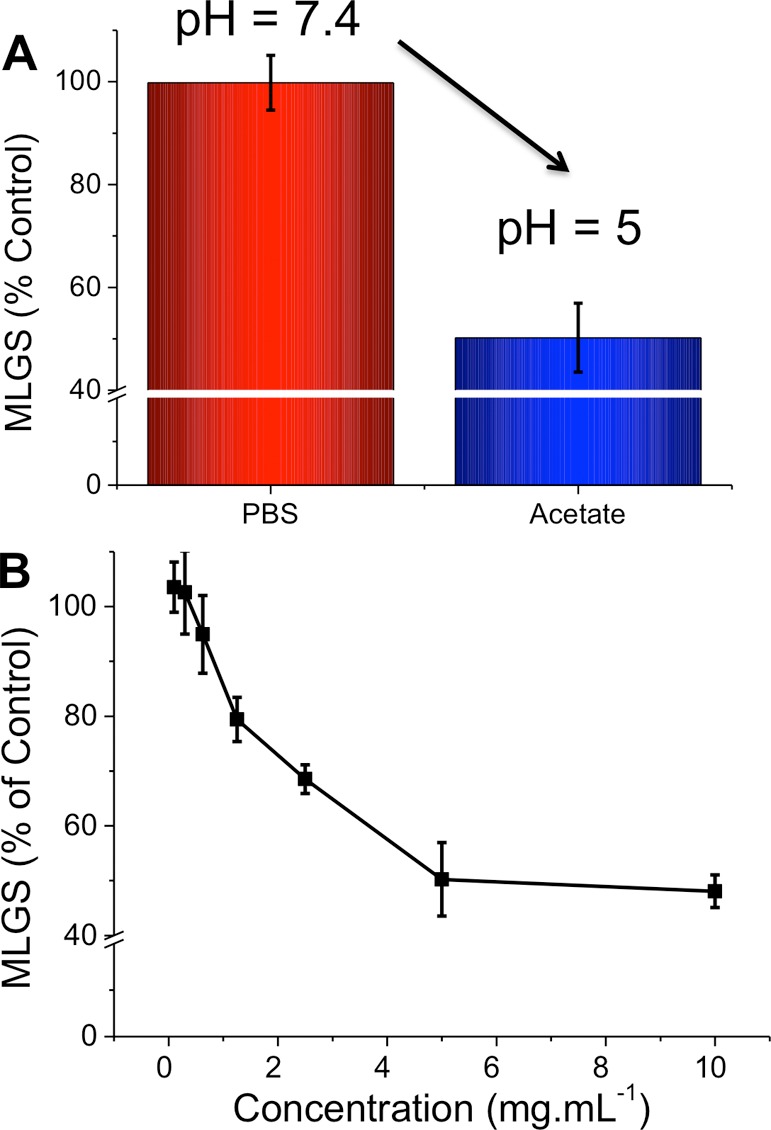
IRI activity of Nisin A. (A) pH dependence. (B) Concentration dependence at pH 5. Error bars represent ± SD from a minimum of three repeats. MLGS = mean largest grain size relative to PBS buffer or pH 5 acetate buffer controls. **, p < 0.01 relative to control.
The results shown reveal that Nisin A has IRI activity comparable to that of several other synthetic macromolecules described in the literature.39,40 On the basis of its known pH-dependent antimicrobial activity, we surmise that protonation of the histidine residues (pKa value ∼6) in acetate buffer (pH 5) was the controlling factor. This switchable activity is unique and may provide a tool for targeted activation of AFP properties in applications such as cryosurgery.48 However, pH itself may not always be a useful (or desirable) trigger. Histidines are well-known to bind metal ions such as nickel or zinc (forming the basis of His-tag purifications, for example),49 and we therefore reasoned that addition of these ions might also be able to promote the formation of the correct conformation for IRI activity. Figure 7 shows the IRI activity of Nisin A with addition of these metal ions compared to that of control solutions containing the metal ions alone.
Figure 7.
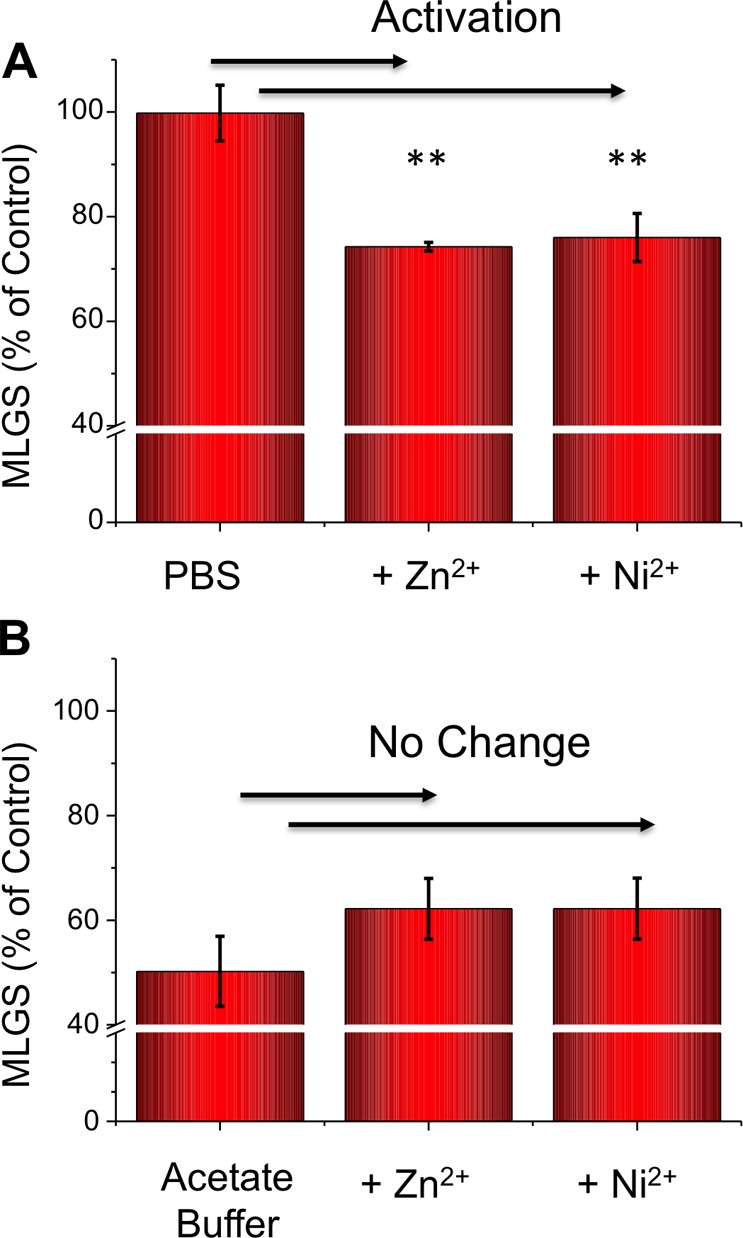
Metal ion activation of Nisin A IRI activity upon addition of nickel and zinc acetate at 5 mg mL–1. (A) PBS buffer, pH 7.4. (B) Acetate buffer, pH 5. Error bars represent ± SD from a minimum of three repeats. MLGS = mean largest grain size relative to PBS buffer, pH 5, acetate buffer controls, or buffer with nickel acetate added only. **, p < 0.01 relative to Nisin in PBS buffer only.
For both Zn and Ni additives in PBS (pH 7.4, no protonation of his-residues), there was a switching on of activity compared to the buffer alone. This demonstrates that metal ions can be used in place of H+ to induce the conformational changes in Nisin A that are required for activity, although the magnitude of the activity is reduced in this case. A control experiment conducted at pH 5 (acetate buffer), where histidines were already protonated, showed no effect by the metal ions, ruling out other interactions. These results show that the easily available, relatively small, antimicrobial peptide Nisin A has some latent IRI activity that can be activated by the use of specific metal ions. Considering the significant progress that has been made in the rational design of antimicrobial peptide mimics, it is anticipated that a similar approach will yield new IRI active compounds, without the need to mimic the structure of an AFP.
Conclusions
Here, we demonstrate that proteins and peptides with little structural homology to antifreeze proteins can have surprisingly potent ice recrystallization inhibition activity, based on two motifs. C-type plant lectins are shown to be calcium-dependent ice recrystallization inhibitors, with their activity modulated by the addition of calcium, providing some switching capability. The activity was shown to be present in three different plant lectins, which all had similar magnitude. As antifreeze proteins are known to be amphipathic, a second class of peptides was also tested, antimicrobial peptides. The 34 amino acid peptide Nisin A was found to have surprisingly strong IRI activity that was activated by a change in pH and protonation of its histidine residues. To enable antifreeze activity at physiological pH, nickel or zinc ions were employed as the stimulus, which can bind the histidine residues and promote the same conformational change. These results show that new antifreeze protein mimetics could be discovered in non-cold-acclimatized species and that these may provide a route to switchable antifreeze materials whose function can be tuned by externally applied stimuli.
Acknowledgments
Equipment used was supported by the Innovative Uses for Advanced Materials in the Modern World (AM2), with support from Advantage West Midlands (AWM) and part funded by the European Regional Development Fund (ERDF). This work was supported by a Research Project Grant from the Royal Society. D.E.M. acknowledges the EPSRC for a Ph.D. studentship grant through the MOAC Doctoral Training Centre, grant no. EP/F500378/1. The Biotechnology and Biological Sciences Research Council (grant no. BB/F011199/1) supported the CD spectrometer. M.I.G. thanks the European Research Council for a Starter grant (CRYOMAT 638661).
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.biomac.5b01118.
Sequence alignment data, pH dependence of IRI activity of Con A, circular dichroism spectra for Con A and SBA, and circular dichroism spectra of Nisin A (PDF).
Author Contributions
M.I.G. designed and led the project, and D.E.M. undertook all experimental work. D.E.M. and M.I.G. wrote the paper.
The authors declare no competing financial interest.
Supplementary Material
References
- Ahlgren J. A.; Cheng C. C.; Schrag J. D.; Devries A. L. Comp. Biochem. Physiol. B 2006, 144, 290–300. [DOI] [PubMed] [Google Scholar]
- Cheng C. C.; DeVries A. L. In Life Under Extreme Conditions: Biochemical Adaptation; di Prisco G, Ed; Springer: Berlin, 1991; pp 1–14. [Google Scholar]
- Duman J. G.; Bennett V.; Sformo T.; Hochstrasser R.; Barnes B. M. J. Insect Physiol. 2004, 50, 259–266 10.1016/j.jinsphys.2003.12.003. [DOI] [PubMed] [Google Scholar]
- Middleton A. J.; Marshall C. B.; Faucher F.; Bar-Dolev M.; Braslavsky I.; Campbell R. L.; Walker V. K.; Davies P. L. J. Mol. Biol. 2012, 416, 713–724 10.1016/j.jmb.2012.01.032. [DOI] [PubMed] [Google Scholar]
- Smallwood M.; Worrall D.; Byass L.; Elias L.; Ashford D.; Doucet C. J.; Holt C.; Telford J.; Lillford P.; Bowles D. J. Biochem. J. 1999, 340, 385–391 10.1042/bj3400385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett J. Int. J. Biochem. Cell Biol. 2001, 33, 105–117 10.1016/S1357-2725(00)00083-2. [DOI] [PubMed] [Google Scholar]
- Celik Y.; Graham L. A.; Mok Y. F.; Bar M.; Davies P. L.; Braslavsky I. Proc. Natl. Acad. Sci. U. S. A. 2010, 107, 5423–5428 10.1073/pnas.0909456107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson M. I. Polym. Chem. 2010, 1, 1141–1152 10.1039/c0py00089b. [DOI] [Google Scholar]
- Eniade A.; Murphy A. V.; Landreau G.; Ben R. N. Bioconjugate Chem. 2001, 12, 817–823 10.1021/bc0155059. [DOI] [PubMed] [Google Scholar]
- Balcerzak A. K.; Febbraro M.; Ben R. N. RSC Adv. 2013, 3, 3232–3236 10.1039/c3ra23220d. [DOI] [Google Scholar]
- Balcerzak A. K.; Capicciotti C. J.; Briard J. G.; Ben R. N. RSC Adv. 2014, 4, 42682–42696 10.1039/C4RA06893A. [DOI] [Google Scholar]
- Balcerzak A. K.; Ferreira S. S.; Trant J. F.; Ben R. N. Bioorg. Med. Chem. Lett. 2012, 22, 1719–1721 10.1016/j.bmcl.2011.12.097. [DOI] [PubMed] [Google Scholar]
- Yu S. O.; Brown A.; Middleton A. J.; Tomczak M. M.; Walker V. K.; Davies P. L. Cryobiology 2010, 61, 327–334 10.1016/j.cryobiol.2010.10.158. [DOI] [PubMed] [Google Scholar]
- Griffith M.; Ewart K. V. Biotechnol. Adv. 1995, 13, 375–402 10.1016/0734-9750(95)02001-J. [DOI] [PubMed] [Google Scholar]
- Koushafar H.; Pham L.; Lee C.; Rubinsky B. J. Surg. Oncol. 1997, 66, 114–121. [DOI] [PubMed] [Google Scholar]
- Chao H.; Davies P. L.; Carpenter J. F. J. Exp. Biol. 1996, 199, 2071–2076. [DOI] [PubMed] [Google Scholar]
- Carpenter J. F.; Hansen T. N. Proc. Natl. Acad. Sci. U. S. A. 1992, 89, 8953–8957 10.1073/pnas.89.19.8953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beirao J.; Zilli L.; Vilella S.; Cabrita E.; Schiavone R.; Herraez M. P. Biol. Reprod. 2012, 86, 59. 10.1095/biolreprod.111.093401. [DOI] [PubMed] [Google Scholar]
- Lee J.; Kim S. K.; Youm H. W.; Kim H. J.; Lee J. R.; Suh C. S.; Kim S. H. PLoS One 2015, 10, e0126252. 10.1371/journal.pone.0126252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capicciotti C. J.; Poisson J. S.; Boddy C. N.; Ben R. N. Cryobiology 2015, 70, 79–89 10.1016/j.cryobiol.2015.01.002. [DOI] [PubMed] [Google Scholar]
- Deller R. C.; Vatish M.; Mitchell D. A.; Gibson M. I. Nat. Commun. 2014, 5, 3244–3249 10.1038/ncomms4244. [DOI] [PubMed] [Google Scholar]
- Davies P. L.; Hew C. L. FASEB J. 1990, 4, 2460–2468. [DOI] [PubMed] [Google Scholar]
- Davies P. L.; Sykes B. D. Curr. Opin. Struct. Biol. 1997, 7, 828–834 10.1016/S0959-440X(97)80154-6. [DOI] [PubMed] [Google Scholar]
- Harding M. M.; Ward L. G.; Haymet A. D. J. Eur. J. Biochem. 1999, 264, 653–665 10.1046/j.1432-1327.1999.00617.x. [DOI] [PubMed] [Google Scholar]
- Gronwald W.; Loewen M. C.; Lix B.; Daugulis A. J.; Sonnichsen F. D.; Davies P. L.; Sykes B. D. Biochemistry 1998, 37, 4712–4721 10.1021/bi972788c. [DOI] [PubMed] [Google Scholar]
- Ewart K.; Lin Q.; Hew C. Cell. Mol. Life Sci. 1999, 55, 271–283 10.1007/s000180050289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubinsky B.; Coger R.; Ewart K. V.; Fletcher G. L. Nature 1992, 360, 113–114 10.1038/360113b0. [DOI] [PubMed] [Google Scholar]
- Knight C. A.; Hallett J.; DeVries A. Cryobiology 1988, 25, 55–60 10.1016/0011-2240(88)90020-X. [DOI] [PubMed] [Google Scholar]
- Schindelin J.; Arganda-Carreras I.; Frise E.; Kaynig V.; Longair M.; Pietzsch T.; Preibisch S.; Rueden C.; Saalfeld S.; Schmid B.; Tinevez J.-Y.; White D. J.; Hartenstein V.; Eliceiri K.; Tomancak P.; Cardona A. Nat. Methods 2012, 9, 676–682 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul S. F.; Gish W.; Miller W.; Myers E. W.; Lipman D. J. J. Mol. Biol. 1990, 215, 403–410 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- The PyMOL Molecular Graphics System, version 1.3r1; Schrödinger, LLC: New York, 2010.
- Edgar R. C. Nucleic Acids Res. 2004, 32, 1792–1797 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham L. A.; Lougheed S. C.; Ewart K. V.; Davies P. L. PLoS One 2008, 3, e2616. 10.1371/journal.pone.0002616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson M. D.; Winter H. C.; Goldstein I. J.; Markovitz D. M. J. Biol. Chem. 2010, 285, 8646–8655 10.1074/jbc.M109.034926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Congdon T.; Notman R.; Gibson M. I. Biomacromolecules 2013, 14, 1578–1586 10.1021/bm400217j. [DOI] [PubMed] [Google Scholar]
- Gilbert J. A.; Davies P. L.; Laybourn-Parry J. FEMS Microbiol. Lett. 2005, 245, 67–72 10.1016/j.femsle.2005.02.022. [DOI] [PubMed] [Google Scholar]
- Ewart K. V.; Yang D. S. C.; Ananthanarayanan V. S.; Fletcher G. L.; Hew C. L. J. Biol. Chem. 1996, 271, 16627–16632 10.1074/jbc.271.28.16627. [DOI] [PubMed] [Google Scholar]
- Deville S.; Viazzi C.; Leloup J.; Lasalle A.; Guizard C.; Maire E.; Adrien J.; Gremillard L. PLoS One 2011, 6, e26474. 10.1371/journal.pone.0026474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capicciotti C. J.; Leclere M.; Perras F. A.; Bryce D. L.; Paulin H.; Harden J.; Liu Y.; Ben R. N. Chem. Sci. 2012, 3, 1408–1416 10.1039/c2sc00885h. [DOI] [Google Scholar]
- Mitchell D. E.; Lilliman M.; Spain S. G.; Gibson M. I. Biomater. Sci. 2014, 2, 1787–1795 10.1039/C4BM00153B. [DOI] [PubMed] [Google Scholar]
- Mitchell D. E.; Cameron N. R.; Gibson M. I. Chem. Commun. 2015, 51, 12977–12980 10.1039/C5CC04647E. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tam R. Y.; Rowley C. N.; Petrov I.; Zhang T.; Afagh N. A.; Woo T. K.; Ben R. N. J. Am. Chem. Soc. 2009, 131, 15745–15753 10.1021/ja904169a. [DOI] [PubMed] [Google Scholar]
- Brogden K. A. Nat. Rev. Microbiol. 2005, 3, 238–250 10.1038/nrmicro1098. [DOI] [PubMed] [Google Scholar]
- Heisig M.; Abraham N. M.; Liu L.; Neelakanta G.; Mattessich S.; Sultana H.; Shang Z.; Ansari J. M.; Killiam C.; Walker W.; Cooley L.; Flavell R. A.; Agaisse H.; Fikrig E. Cell Rep. 2014, 9, 417–424 10.1016/j.celrep.2014.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Periago P. M.; Moezelaar R. Int. J. Food Microbiol. 2001, 68, 141–148 10.1016/S0168-1605(01)00461-5. [DOI] [PubMed] [Google Scholar]
- Rollema H. S.; Kuipers O. P.; Both P.; de Vos W. M.; Siezen R. J. Appl. Environ. Microbiol. 1995, 61, 2873–2878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao F. H.; Abee T.; Konings W. N. Appl. Environ. Microbiol. 1991, 57, 2164–2170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koushafar H.; Pham L.; Lee C.; Rubinsky B. J. Surg. Oncol. 1997, 66, 114–121. [DOI] [PubMed] [Google Scholar]
- Bornhorst J. A.; Falke J. J. Methods Enzymol. 2000, 326, 245–254 10.1016/S0076-6879(00)26058-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.