Background: Reversible assembly is a major mode of V-ATPase regulation.
Results: Amino acid starvation reversibly increases V-ATPase assembly.
Conclusion: Amino acids are a novel regulator of V-ATPase assembly.
Significance: Learning how amino acids are sensed and influence cellular physiology is important to understanding the basic processes of cellular homeostasis.
Keywords: amino acid, lysosomal acidification, mechanistic target of rapamycin (mTOR), proton transport, vacuolar ATPase, nutrient sensing, regulated assembly
Abstract
The vacuolar H+-ATPase (V-ATPase) is an ATP-dependent proton pump composed of a peripheral ATPase domain (V1) and a membrane-integral proton-translocating domain (V0) and is involved in many normal and disease processes. An important mechanism of regulating V-ATPase activity is reversible assembly of the V1 and V0 domains. Increased assembly in mammalian cells occurs under various conditions and has been shown to involve PI3K. The V-ATPase is necessary for amino acid-induced activation of mechanistic target of rapamycin complex 1 (mTORC1), which is important in controlling cell growth in response to nutrient availability and growth signals. The V-ATPase undergoes amino acid-dependent interactions with the Ragulator complex, which is involved in recruitment of mTORC1 to the lysosomal membrane during amino acid sensing. We hypothesized that changes in the V-ATPase/Ragulator interaction might involve amino acid-dependent changes in V-ATPase assembly. To test this, we measured V-ATPase assembly by cell fractionation in HEK293T cells treated with and without amino acids. V-ATPase assembly increases upon amino acid starvation, and this effect is reversed upon readdition of amino acids. Lysosomes from amino acid-starved cells possess greater V-ATPase-dependent proton transport, indicating that assembled pumps are catalytically active. Amino acid-dependent changes in both V-ATPase assembly and activity are independent of PI3K and mTORC1 activity, indicating the involvement of signaling pathways distinct from those implicated previously in controlling assembly. By contrast, lysosomal neutralization blocks the amino acid-dependent change in assembly and reactivation of mTORC1 after amino acid starvation. These results identify an important new stimulus for controlling V-ATPase assembly.
Introduction
The vacuolar proton-translocating adenosine triphosphatase (V-ATPase)2 is an ATP-dependent proton pump that controls the pH of intracellular organelles such as lysosomes and endosomes and transports protons across the plasma membrane (1–3). It is composed of two multisubunit domains: the peripheral V1 domain, which is responsible for ATP hydrolysis, and the membrane-integral V0 domain, which carries out proton translocation. The V-ATPase is critical for many pH-dependent processes, including membrane trafficking, protein turnover, bone resorption, and urinary acidification (1, 2).
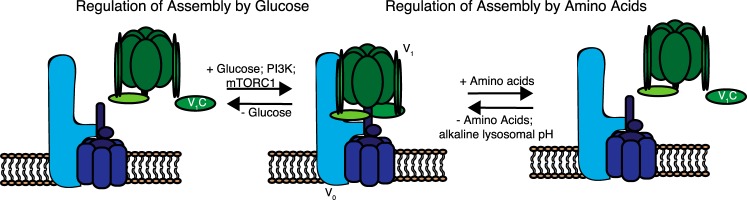
V-ATPase activity is carefully regulated, particularly by the process of reversible assembly, whereby the equilibrium between the V-ATPase holoenzyme and the separate V1 and V0 domains is controlled (1, 4, 5). When not in complex, the V1 and V0 domains are unable to perform their functions, and V-ATPase-dependent acidification is prevented (6, 7). Control of V-ATPase assembly in mammalian cells is not well understood. There is emerging evidence that glucose influences assembly (8–10), as has been described previously in yeast (4). PI3K activity may also play a role downstream of various stimuli, including dendritic cell maturation (11), elevation of glucose concentration (8–10), and influenza infection (9).
Recently, the V-ATPase was identified as a critical component of the amino acid-sensing machinery that communicates amino acid availability to mTORC1 (12). The V-ATPase has been shown to directly associate with the Ragulator complex on the lysosomal membrane (12). The Ragulator serves as a platform for the RagGTPases, which, when activated by the presence of amino acids, recruit mTORC1 to lysosomes, allowing it to be activated by lysosomally localized Ras homolog enriched in brain (Rheb) (13, 14). V-ATPase inhibition blocks the amino acid signal by an unknown mechanism.
Intriguingly, the association of the V-ATPase with the Ragulator is amino acid-dependent (12). Interaction between the cytosolic V1 domain and the Ragulator is weakened upon amino acid addition, whereas the V0/Ragulator association is unaffected by amino acids (12). We hypothesized that this differential association with the Ragulator may be due to amino acid-dependent changes in V-ATPase assembly. Increased V-ATPase assembly would lower lysosomal pH values, increasing protein turnover and, therefore, increasing amino acid levels. An amino acid-responsive V-ATPase could couple lysosomal protein turnover to amino acid availability. Here we demonstrate, for the first time, that V-ATPase assembly is responsive to amino acid availability, increasing during amino acid starvation. Further, we sought to describe the signaling that controls this change and identify the critical amino acids driving it.
Experimental Procedures
Materials and Equipment
Minimal essential medium, DMEM/F-12, FBS, penicillin/streptomycin, and Prolong Gold anti-fade mounting medium were obtained from Invitrogen. DMEM/F-12 cell culture medium lacking amino acids was from US Biologicals. Amino acid powders, FITC-Dextran, anti-vinculin antibody, and chloroquine were purchased from Sigma. LY294002 was from Selleck Chemicals, Rapamycin was from Alfa Aesar, and concanamycin A was from Bioviotica. The DAMP (3-(2,4-dinitroanilino) 3-amino-N-methyldipropylamine), staining kit was purchased from Oxford Biomedical Research. Antibodies to Akt, Ser(P)-473-Akt, p70 S6 kinase, and Thr(P)-389-p70 S6 kinase were from Cell Signaling Technology. Antibody for V-ATPase subunit A was from Abnova, and antibody for subunit d1 was from Abcam. Antibody against HSP90α was from Enzo Life Sciences. Peroxidase-conjugated anti-mouse and anti-rabbit antibodies were from Abcam, and FITC-conjugated anti-goat antibody was obtained from Sigma. Precast 4–15% mini-Protean gels from Bio-Rad were used for SDS-PAGE. Primary antibody concentrations used were as follows: Akt, Ser(P)-473-Akt, p70 S6 kinase, Thr(P)-389-p70 S6 kinase, subunit A, and subunit d1 (1:1000); vinculin (1:100,000); and HSP90α (1:5000). Chemiluminescence substrate for horseradish peroxidase was from GE, and luminescence was detected with Kodak Biomax film.
Cell homogenization was performed with a ball bearing homogenizer from Isobiotec. Cytosolic fractions were concentrated in Amicon Ultra 10K centrifugal filters from Merck Millipore. FITC quenching assays were performed using a PerkinElmer Life Sciences luminescence spectrophotometer, model LS50B, with Flwinlab software. Confocal microscopy was performed on a Leica SPE system.
Cell Culture
HEK293T cells were maintained in minimal essential medium supplemented with 10% FBS and 1% penicillin/streptomycin. MCF10CA1a cells were maintained in DMEM/F-12 medium supplemented with 10% FBS and 1% penicillin/streptomycin. Cells were grown in a humidified incubator at 37 °C with 5% CO2.
Amino Acid Starvation and Stimulation
HEK293T (3.0 × 106) cells were seeded into 10-cm poly-d-lysine-coated plates. The following day, the monolayer was rinsed twice with warm PBS, and medium was replaced with amino acid-free medium. The amino acid concentration was immediately restored to that normally found in the medium for baseline (+/+) samples by addition of a 10× solution prepared in PBS. The final amino acid concentrations were as follows: 0.05 mm l-alanine, 0.699 mm l-arginine hydrochloride, 0.05 mm l-asparagine, 0.05 mm l-aspartic acid, 0.2 mm l-cysteine, 0.05 mm l-glutamic acid monosodium salt, 2.5 mm l-glutamine, 0.25 mm glycine, 0.15 mm l-histidine hydrochloride-H2O, 0.416 mm l-isoleucine, 0.451 mm l-leucine, 0.499 mm l-lysine hydrochloride, 0.116 mm l-methionine, 0.215 mm l-phenylalanine, 0.15 mm l-proline, 0.25 mm l-serine, 0.449 mm l-threonine, 0.0442 mm l-tryptophan, 0.214 mm l-tyrosine, and 0.452 mm l-valine. Solutions lacking single amino acids were prepared similarly without the indicated amino acid. After 50 min at 37 °C, amino acids or an equivalent volume of PBS were added to the amino acid readdition (−/+) and starved (−/−) samples, respectively. Incubation at 37 °C was continued for 15 min. Where applicable, inhibitors were used at the following concentrations: 5 μm concanamycin A, 50 μm LY294002, 2 nm rapamycin, and 100 μm chloroquine.
Cellular Fractionation and Western Blotting
To prepare subcellular membrane and cytosol fractions, cells were placed on ice, and the monolayer was rinsed twice with ice-cold PBS. Cells were scraped into 650 μl of homogenization buffer (250 mm sucrose, 1 mm EDTA, 10 mm HEPES, 1 mm PMSF, 2 μg/ml aprotinin, 5 μg/ml leupeptin, and 1 μg/ml pepstatin) and lysed by repeated passage through a ball bearing homogenizer. The homogenate was cleared of intact cells and nuclei by centrifugation at 500 × g for 10 min. The supernatant was subjected to ultracentrifugation at 100,000 × g for 30 min to pellet the membrane fraction. The supernatant containing the cytosolic fraction was concentrated using Amicon Ultra 10K centrifugal filters according to the instructions of the manufacturer. Resulting cytosolic samples were supplemented with 1% SDS to yield the final cytosolic fractions. Membrane pellets were rinsed once with homogenization buffer and resuspended in homogenization buffer containing 1% SDS to yield the final membrane fractions.
To prepare whole cell lysates, cells were kept on ice or at 4 °C for the duration of their preparation. The monolayer was rinsed twice with ice-cold PBS. Cells were collected by scraping into 150 μl of lysis buffer (150 mm NaCl, 1% Triton X-100, 50 mm Tris-HCl (pH 8.0), 1 mm PMSF, 2 μg/ml aprotinin, 5 μg/ml leupeptin, 1 μg/ml pepstatin, 1 mm NaF, 1 mm NaN3, and 1 mm β-glycerophosphate). Cell suspensions were agitated continuously for 30 min to allow complete lysis. Crude lysates were cleared of unbroken cells and nuclei by centrifugation at 16,100 × g for 20 min.
Protein concentrations of lysates and fractions were determined by the Lowry method (15). Samples were diluted in SDS-containing buffer, and proteins were separated by SDS-PAGE on 4–15% gradient gels before transfer to a nitrocellulose membrane. Subunit A of the V1 domain, subunit d1 of the V0 domain, vinculin, and HSP90α were detected by Western blotting using specific antibodies as noted above. Band intensities were quantified using ImageJ software. Subunit A intensities were normalized for membrane (subunit d1) and cytosol (HSP90α or vinculin), and the ratio of membrane:cytosolic subunit A was calculated. Ratios were normalized for each individual experiment, with the baseline (+/+) sample set as 1.
FITC-Dextran Loading of Lysosomes and Concanamycin A-dependent Proton Pumping
HEK293T cells (4.0 × 106) were plated in 10-cm poly-d-lysine-coated plates. The following day, the medium was replaced with medium containing 2.2 mg/ml FITC-Dextran, and cells were incubated overnight to allow uptake of the dye by endocytosis. The following day, FITC-Dextran-containing medium was replaced with unlabeled medium, and cells were incubated to allow all dye to progress to the lysosomal compartment (16). Cells were treated with amino acids and inhibitors as described above. After treatment, cells were placed on ice and rinsed with ice-cold PBS. Cells were collected by scraping into 500 μl of fractionation buffer (125 mm KCl, 1 mm EDTA, 50 mm sucrose, 20 mm HEPES, 1 mm PMSF, 2 μg/ml aprotinin, 5 μg/ml leupeptin, and 1 μg/ml pepstatin) and collected by centrifugation at 1200 × g for 5 min. Cells were resuspended in 750 μl of fractionation buffer and lysed by spraying through a 27-gauge needle 10 times. Lysates were cleared of nuclei and intact cells by centrifugation at 2000 × g for 10 min. The resulting supernatant was then centrifuged for 15 min at 16,100 × g to sediment the FITC-Dextran-containing lysosomes and other light organelles (12, 17). The resulting pellets were resuspended in 100 μl of fractionation buffer, and protein concentration was determined by the Lowry method (15).
To measure concanamycin A-dependent proton pumping, 20 μg of protein was added to 500 μl of fractionation buffer prewarmed to 37 °C. Sample fluorescence was excited at 490 nm, and emission fluorescence at 520 nm was measured continuously in a luminescence spectrophotometer and recorded with Flwinlab software. After the initial fluorescence stabilized, 1 mm ATP and 2 mm magnesium were added to the sample to initiate ATPase activity and proton pumping into the lysosomes, causing quenching of FITC in the lysosomal lumen. The V-ATPase dependence of quenching was verified by performing the assay in the presence of 1 μm concanamycin A, which completely inhibited fluorescence quenching. GraphPad Prism software was used to fit the data by linear regression.
DAMP Staining to Detect Acidic Intracellular Compartments
HEK293T cells (0.3 × 106) were plated into 6-well plates containing poly-d-lysine-coated glass coverslips and allowed to attach overnight. The following day, cells were treated with medium containing 100 μm chloroquine (or an equivalent volume of PBS) for 65 min. DAMP reagent was added after 15 min, and incubation at 37 °C continued. Cells were rinsed with PBS and fixed for 15 min with 4% paraformaldehyde. Free aldehydes were quenched by a 5-min incubation of samples in 50 mm ammonium chloride. Fixed cells were permeabilized with PBS containing 0.1% Triton X-100 for 10 min. DAMP was detected with a polyclonal goat-anti-DNP (dinitrophenol) antibody. Secondary anti-goat antibodies were FITC-conjugated to allow detection of a DAMP signal. Coverslips were mounted with Prolong Gold anti-fade with DAPI. Samples were imaged by confocal microscopy.
Statistical Analysis
Graphed data are means of the indicated number of independent experiments. Paired Student's t test was used to calculate significance. p < 0.05 was considered significant.
Results
Amino Acids Alter V-ATPase Assembly
To test whether amino acid availability alters V-ATPase assembly, we subjected HEK293T cells to complete amino acid starvation for 65 min (−/−) or starvation for 50 min followed by stimulation for 15 min (−/+). Baseline assembly was assessed in cells maintained in amino acids for the duration of the experiment (+/+). After treatment, cells were lysed, and the resulting homogenate was separated into membrane and cytosolic fractions by sedimentation. The fractions were subjected to SDS-PAGE followed by Western blotting to assess the distribution of V1 (specifically, subunit A of the V1 domain) in the fractions. Amino acid starvation leads to an increase in membrane-associated subunit A, indicating increased V-ATPase assembly, whereas readdition of amino acids returns the assembly to near baseline levels (Fig. 1A). Western blots were quantified using ImageJ software, and assembly was calculated as the ratio of membrane to cytosolic subunit A, normalized to baseline (+/+) assembly levels for each experiment. Amino acid starvation increases V-ATPase assembly ∼2-fold, and readdition of amino acids restores near baseline assembly (Fig. 1B). We confirmed that this effect is not specific to HEK293T cells by subjecting the breast cancer cell line MCF10CA1a to the same amino acid starvation and stimulation and observed a comparable increase in assembly during amino acid starvation and a return to near baseline after amino acid readdition (data not shown).
FIGURE 1.
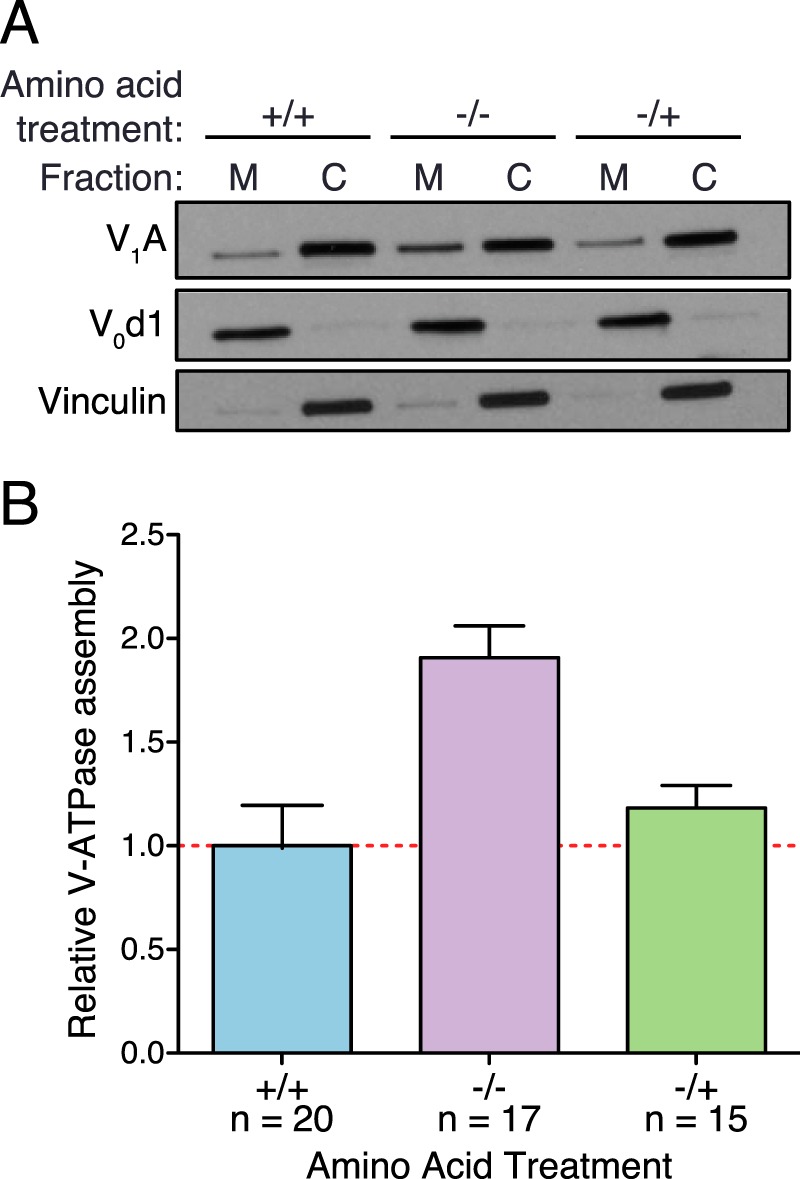
Amino acids alter V-ATPase assembly. A, HEK293T cells were maintained in amino acids for 65 min (+/+), starved of amino acids for 65 min (−/−), or starved of amino acids for 50 min followed by readdition of amino acids for 15 min (−/+). Cell homogenates were prepared, separated into membrane (M) and cytosolic (C) fractions by sedimentation, and subjected to SDS-PAGE and Western blotting using antibodies against subunit A as a measure of V1, subunit d1 as a loading control for the membrane fraction, and vinculin as a loading control for the cytosolic fraction, as described under “Experimental Procedures.” The amount of subunit A present in the membrane fraction indicates the amount of assembled V-ATPase. A representative Western blot is shown. B, Western blots performed as in A were quantified by ImageJ analysis to assess relative V-ATPase assembly. Subunit A band intensities were normalized to membrane and cytosolic loading controls, and the ratio of membrane to cytosolic subunit A was calculated. To combine results from independent experiments, values were normalized to baseline (+/+) assembly levels (defined as 1.0 for each trial). The average ratio for (+/+) conditions was 0.37 (± 0.05). p < 0.05, +/+ versus −/−. The number of independent trials is shown as n, and the error bars represent mean ± S.E.
Amino Acids Alter Lysosomal V-ATPase-dependent Proton Transport
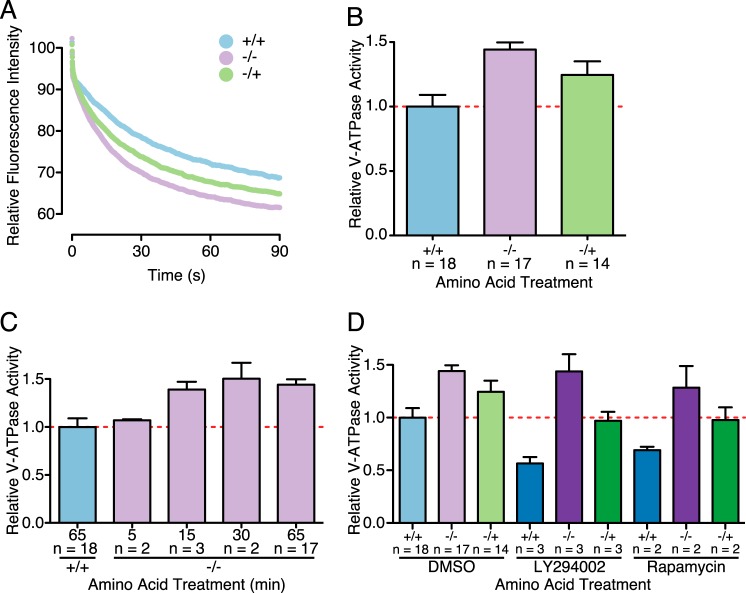
To independently confirm the effect of amino acids on V-ATPase assembly and to determine whether the assembled complexes were active and present in lysosomes, we utilized the pH-dependent fluorescence probe FITC-Dextran to measure the rate of proton pumping in lysosomes. FITC-Dextran added to the medium of cells is taken up by fluid-phase endocytosis and is trafficked through the endocytic pathway to lysosomes, where it accumulates (16, 17). Following the loading period with a chase of at least 30 min ensures that FITC-Dextran is localized only to lysosomes (16). Because FITC fluorescence is quenched at low pH, the fluorescence signal permits the measurement of lysosomal acidification. V-ATPase-dependent acidification is defined as the concanamycin-sensitive portion of lysosomal acidification observed following the addition of magnesium-ATP to cell lysates containing FITC-loaded lysosomes (11). Lysosomes derived from cells starved of amino acids display increased concanamycin-sensitive, ATP-dependent fluorescence quenching, indicating increased proton transport (Fig. 2A). This increase was partially reversed upon amino acid readdition. The initial rate of fluorescence quenching was determined by linear regression analysis of each condition. Rates of fluorescence quenching from independent experiments were normalized to the value observed for lysosomes isolated from cells maintained in amino acids throughout (+/+). As shown in Fig. 2B, amino acid starvation led to an increase in the rate of V-ATPase-dependent fluorescence quenching, which was partially reversed upon amino acid readdition for 15 min. Therefore, V-ATPase complexes assembled in response to amino acid depletion are at least partially localized to lysosomes and measurably catalytically active.
FIGURE 2.
Amino acids alter V-ATPase-dependent proton transport, as measured by ATP-dependent fluorescence quenching of FITC-loaded lysosomes. A, HEK293T cells were allowed to take up FITC-Dextran by endocytosis, and the dye was chased to the lysosomal compartment as described under “Experimental Procedures.” Cells were lysed mechanically, and a fraction containing FITC-Dextran-loaded lysosomes was isolated by sedimentation. Fluorescence was measured over time to assess pH-dependent quenching following addition of 1 mm magnesium-ATP. The representative traces shown are for lysosomes isolated from HEK293T cells maintained in amino acids (+/+), starved of amino acids (−/−), or starved of amino acids for 50 min followed by readdition of amino acids for 15 (−/+). ATP-dependent fluorescence quenching was not observed for samples preincubated in the presence of 5 μm concanamycin A. B, the initial rate of magnesium-ATP-dependent fluorescence quenching was determined by a linear regression analysis for each condition. Rates of fluorescence quenching from independent experiments were normalized to the value observed for lysosomes isolated from cells maintained in amino acids throughout (+/+). p < 0.05, +/+ versus −/− and −/+. C, the time course of amino acid-dependent increase in V-ATPase-dependent proton transport after amino acid starvation. HEK293T cells were loaded with FITC-Dextran and starved of amino acids for 5, 15, 30, or 65 min before cell lysis and isolation of the lysosome-containing fraction as described above. p < 0.05, +/+ versus −/− (15 min) and −/− (65 min). D, HEK293T cells were subjected to amino acid starvation and readdition in the presence of 50 μm LY294002 (LY) to inhibit PI3K, 2 nm rapamycin to inhibit mTORC1, or in the absence of inhibitors, as indicated, followed by measurement of ATP-dependent fluorescence quenching as described above. p < 0.05; +/+ versus −/− and −/+; +/+ LY versus −/− LY and −/+ LY; −/− LY versus −/+ LY. In B—D, the number of independent trials is shown as n, and the error bars represent mean ± S.E.
We next sought to characterize the time course of the increase in V-ATPase assembly following amino acid starvation. Cells were loaded with FITC-Dextran and starved of amino acids for 5, 15, 30, or 65 min before cell homogenization and sedimentation to obtain the fraction containing FITC-Dextran-loaded lysosomes. V-ATPase-dependent proton transport, as measured by the rate of concanamycin-sensitive fluorescence quenching following addition of magnesium-ATP, increased following amino acid depletion, being measurable at 5 min and maximal at 30 min (Fig. 2C).
Concanamycin A Treatment Blocks the Amino Acid-dependent Change in Assembly
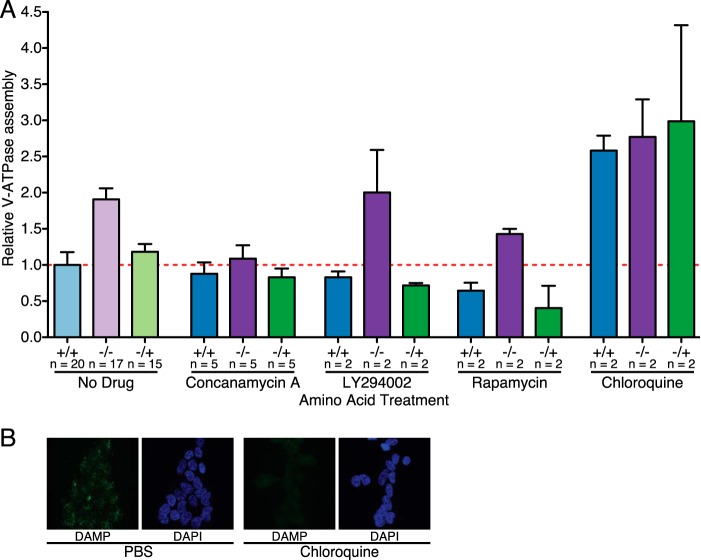
V-ATPase assembly has been shown previously to change in response to glucose levels, and this change, at least in yeast, is dependent on V-ATPase activity (18). We therefore sought to assess whether addition of concanamycin A, a V-ATPase-specific inhibitor, would alter the change in V-ATPase assembly in response to amino acid starvation and readdition. Cells were treated as described above but with the addition of 5 μm concanamycin A or an equivalent volume of DMSO to the culture medium for the duration of the experiment. This concentration was selected because it is sufficient to alter mTORC1 signaling, as described previously (Ref. 12 and data not shown). As shown in Fig. 3A, although concanamycin slightly decreased assembly, the amino acid-dependent change in V-ATPase assembly is largely blocked in the presence of concanamycin. Therefore, like the glucose-dependent change in assembly, the amino acid-dependent assembly change is prevented by treatment with a V-ATPase inhibitor.
FIGURE 3.
Control of amino acid-induced changes in V-ATPase assembly. A, quantification of V-ATPase assembly in HEK293T cells maintained in amino acids (+/+), starved of amino acids (−/−), or starved of amino acids for 50 min followed by readdition of amino acids for 15-min (−/+) was performed as described for Fig. 1 except, where indicated, with the addition of 5 μm concanamycin A to inhibit the V-ATPase, 50 μm LY294002 to inhibit PI3K, 2 nm rapamycin to inhibit mTORC1, or 100 μm chloroquine to neutralize the lysosomal pH. Ratios for each set of conditions are normalized to cells maintained in amino acids (+/+) in the absence of inhibitors. p < 0.05, +/+ versus −/− and −/+. The number of independent trials is shown as n, and the error bars represent mean ± S.E. B, 100 μm chloroquine neutralizes lysosomes in HEK293T cells, as assessed by DAMP staining. HEK293T cells were grown on glass coverslips and treated with 100 μm chloroquine or medium containing an equivalent volume of PBS for 65 min. DAMP was added to the culture medium 50 min before cells were fixed to allow accumulation in acidic compartments. DAMP was detected by anti-DNP antibodies (green fluorescence), and immunocytochemistry was performed. Nuclei were stained with DAPI (blue fluorescence). Staining was assessed by confocal fluorescence microscopy.
Amino Acid Sensing Is Independent of PI3K and mTOR Signaling
PI3K has been shown to be involved in increased V-ATPase assembly in mammalian systems in response to various stimuli, including maturation of dendritic cells (11), elevated glucose concentrations (8–10), and influenza infection (9). We therefore wished to determine whether inhibition of PI3K by treatment of cells with the inhibitor LY294002 would alter V-ATPase assembly in response to amino acid availability. We first established the minimal dose of LY294002 necessary to inhibit PI3K signaling by measuring the inhibition of phosphorylation of Akt, a major downstream target of PI3K (19). Treatment with 50 μm LY294002 completely blocked Akt phosphorylation (data not shown). We then proceeded to subject cells to amino acid starvation and readdition in the presence of 50 μm LY294002. Although PI3K inhibition slightly lowers baseline assembly, it does not prevent increased assembly in response to amino acid starvation or the reduction in assembly after amino acid readdition (Fig. 3A). The effect of PI3K inhibition on V-ATPase assembly was confirmed by V-ATPase-dependent proton transport as measured by ATP-dependent, concanamycin-sensitive fluorescence quenching (Fig. 2D). Cells were subjected to amino acid starvation and stimulation in the presence of 50 μm LY294002, followed by measurement of proton transport as described above. V-ATPase-dependent proton pumping in FITC-Dextran-loaded lysosomes was reduced by treatment with LY294002, but the change in proton transport upon amino acid starvation and readdition was undiminished (Fig. 2D). Therefore, unlike the change in assembly of the mammalian V-ATPase in response to other stimuli, the amino acid-dependent change in assembly does not appear to involve PI3K.
mTORC1 activity has also been linked to regulation of V-ATPase assembly (11). We first established the minimal effective dose of rapamycin, an inhibitor of mTORC1, to fully inhibit S6 kinase phosphorylation by mTORC1 as 2 nm (data not shown). We then tested whether the addition of 2 nm rapamycin would alter V-ATPase assembly in response to amino acid availability. As with PI3K inhibition, rapamycin treatment decreased baseline assembly but did not prevent the increased assembly observed upon amino acid depletion or the subsequent decreased assembly upon amino acid readdition (Fig. 3A). To confirm this result, the effect of rapamycin on V-ATPase-dependent proton transport in FITC-Dextran-loaded lysosomes was measured. As shown in Fig. 2D, amino acid-dependent changes in V-ATPase-dependent proton transport were unchanged by inhibition of mTORC1. Therefore, unlike the increased assembly observed during dendritic cell maturation, the amino acid-dependent change in assembly was not dependent upon mTORC1.
Amino Acid Sensing Requires an Acidic Lysosomal Compartment
The V-ATPase is responsible for maintaining an acidic lysosomal pH and is also known to sense and respond to changes in luminal pH (20, 21). Further, chloroquine treatment blocks disassembly of the yeast V-ATPase in response to glucose depletion (20). We therefore tested whether addition of 100 μm chloroquine, which neutralizes acidic intracellular compartments as detected by DAMP staining (Fig. 3B), would alter the V-ATPase response to changes in amino acid availability. Consistent with our previous results in yeast (20), treatment with 100 μm chloroquine significantly increased V-ATPase assembly under all conditions but completely blocked changes in assembly in response to amino acid availability (Fig. 3A). Therefore, the amino acid-induced change in assembly requires an acidic lysosomal pH.
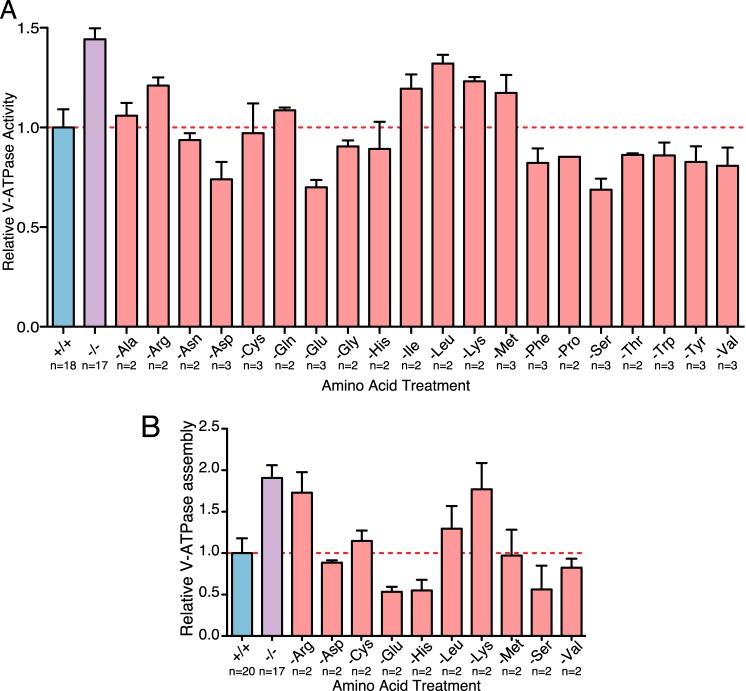
Starvation of Specific Amino Acids Modulates V-ATPase Activity and Assembly
We have described an increase in V-ATPase assembly and activity during starvation of all 20 amino acids. To test whether these changes were driven by a few key amino acids or were only observable following starvation of the complete complement of amino acids, we tested the effect of starvation of each individual amino acid on V-ATPase-dependent acidification of lysosomes. Withdrawal of several amino acids, including arginine, isoleucine, leucine, lysine, and methionine, caused an increase in activity, whereas starvation for several others, including aspartate, glutamate, and serine, actually decreased activity (Fig. 4A). For several of the amino acids showing changes in activity, we also investigated the effects of withdrawal on V-ATPase assembly. As shown in Fig. 4B, for most amino acids, the observed changes in assembly generally parallel the changes in activity. Therefore, arginine, leucine, and lysine starvation increases both activity and assembly, whereas glutamate and serine starvation decreases both activity and assembly. For other amino acids, including aspartate and methionine, there may be changes in activity that are not accounted for by changes in assembly, although additional work will be required to establish whether these differences are significant.
FIGURE 4.
Starvation of individual amino acids alters V-ATPase-dependent proton transport and assembly. A, HEK293T cells were allowed to take up FITC-Dextran by endocytosis, and the dye was chased to the lysosomal compartment as described under “Experimental Procedures.” Loaded cells were maintained in amino acids (+/+), starved of all amino acids (−/−), or starved of the indicated amino acid for 65 min. Cells were then lysed mechanically, and a fraction containing FITC-Dextran-loaded lysosomes was isolated by sedimentation. Fluorescence was measured over time to assess pH-dependent quenching following addition of 1 mm magnesium-ATP. ATP-dependent fluorescence quenching was not observed for samples preincubated in the presence of 5 μm concanamycin A. The initial rate of magnesium ATP-dependent fluorescence quenching was determined by linear regression analysis for each condition. Rates of fluorescence quenching from independent experiments were normalized to the value observed for lysosomes isolated from cells maintained in amino acids throughout the experiment (+/+). p < 0.05, +/+ versus −/−, −Glu, −Pro, −Ser, and −Thr. The number of independent trials is shown as n, and the error bars represent mean ± S.E. B, quantification of V-ATPase assembly in HEK293T cells maintained in amino acids (+/+), starved of amino acids (−/−), or starved of the indicated amino acids for 65 min was performed as described for Fig. 1. p < 0.05, +/+ versus −/−. The number of independent trials is shown as n, and the error bars represent mean ± S.E.
V-ATPase Assembly and mTORC1 Activity
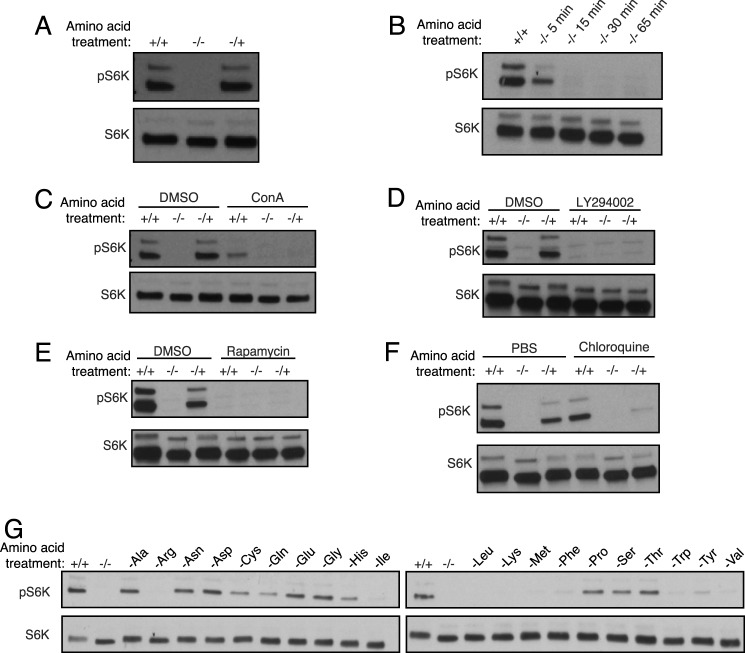
The V-ATPase has been shown previously to be important in the amino acid-dependent regulation of mTORC1 activity (12). We therefore wished to compare amino acid-dependent changes in V-ATPase assembly and amino acid-dependent activation of mTORC1. We first confirmed that amino acid depletion reduced and amino acid readdition stimulated mTORC1 activity by measuring the phosphorylation of p70 S6 kinase, a major downstream target of mTORC1 (22), by Western blot analysis using a phospho-p70 S6 kinase antibody. As shown in Fig. 5A, S6 kinase phosphorylation was abolished after amino acid depletion and restored following amino acid readdition. We next measured the time course of amino acid-dependent changes in mTORC1 activity. As shown in Fig. 5B, loss of S6 kinase phosphorylation was detectable at 5 min and nearly complete after 15 min of amino acid depletion. Comparison with the time course of amino acid-dependent changes in V-ATPase-dependent proton transport (Fig. 2C) suggests that if increased activity and assembly of the V-ATPase are necessary for inactivation of mTORC1, then only a modest fractional change in assembly is required.
FIGURE 5.
Activation of mTORC1 by amino acids. A, HEK293T cells were maintained in amino acids for 65 min (+/+), starved of amino acids for 65 min (−/−), or starved of amino acids for 50 min followed by a readdition of amino acids for 15-min (−/+). Cell homogenates were prepared and subjected to SDS-PAGE, and Western blotting was performed using antibodies against S6 kinase or Thr(P)-389-S6 kinase as indicated. The level of phospho-S6 kinase is a measure of mTORC1 activity. B, HEK293T cells were maintained in amino acids (+/+) or deprived of amino acids for the indicated times, followed by measurement of phospho-S6 kinase levels as described in A. C, the experiment described in A was performed in the absence (DMSO) or presence of 5 μm concanamycin A (ConA), a specific V-ATPase inhibitor, as indicated. D, the experiment described in A was performed in the absence (DMSO) or presence of 50 μm LY294002, a PI3K inhibitor, as indicated. E, the experiment described in A was performed in the absence (DMSO) or presence of 2 nm rapamycin, an inhibitor of mTORC1, as indicated. F, the experiment described in A was performed in the absence (PBS) or presence of 100 μm chloroquine, a weak base that neutralizes lysosomes, as indicated. G, HEK293T cells were maintained in amino acids for 65 min (+/+), starved of amino acids for 65 min (−/−), or starved of individual amino acids for 65 min, and homogenates were prepared and assessed as described in A.
As reported previously, concanamycin A significantly inhibits amino acid-dependent changes in S6 kinase phosphorylation (12) (Fig. 5C). Similarly, basal and amino acid-dependent changes in mTORC1 activity are blocked by PI3K inhibition and rapamycin (Fig. 5, D and E). Surprisingly, neutralization of the lysosomal compartment with chloroquine, which does not inhibit basal mTORC1 activity, inhibits mTORC1 reactivation by amino acid stimulation after starvation (Fig. 5F). Because the V-ATPase is highly assembled in the presence of chloroquine (Fig. 3A), these results suggest that lysosomal acidification may, in some cases, override any signal from V-ATPase assembly in controlling mTORC1 activity.
Full activation of mTORC1 requires all amino acids, and starvation of individual amino acids variably inhibits mTORC1 (Fig. 5G and Ref. 23).
Discussion
Nutrient sensing is fundamental to cellular physiology. The V-ATPase has emerged as an important component of cellular metabolic signaling and sensing. It has been shown previously to be regulated by glucose levels in yeast and mammalian cells (4, 8–10). It is also known that V-ATPase function enables amino acid sensing and subsequent activation of mTORC1 during amino acid sufficiency (12). The V-ATPase directly interacts with the Ragulator complex on the lysosomal surface in an amino acid-dependent manner. We therefore hypothesized that amino acids may regulate the reversible assembly of the V-ATPase, an important mode of control of the activity of the enzyme. Because cells deprived of amino acids may increase protein degradation in lysosomes to restore amino acid levels (24), we hypothesized that amino acid withdrawal would increase pump assembly, therefore lowering lysosomal pH values and increasing lysosomal protein degradation. Coupling mTORC1 activation to this change in V-ATPase assembly could allow the coordination of cellular growth, as controlled by mTORC1, with the presence of sufficient nutrients to sustain appropriate expansion.
To directly address our hypothesis, we measured V-ATPase assembly by cell fractionation and Western blotting to determine the cellular distribution of the V1 domain during amino acid sufficiency and starvation. We observed that V-ATPase assembly increases upon amino acid starvation, an effect that is reversed upon amino acid readdition, indicating that this change is indeed due to alteration in the equilibrium of assembled and disassembled pumps. It is important to note that, during cell lysis, to facilitate fractionation of membrane and cytosolic cellular components, intact pumps may be disrupted, leading to an increased amount of V1 in the cytosolic fractions. Therefore, this method may lead to an underestimation of the absolute amount of assembled pumps in vivo. Nevertheless, we observe a clear difference in the relative amount of assembled pumps between the amino acid sufficient and starved states. We also observed that V-ATPase-dependent proton pumping in lysosomes is increased after amino acid starvation and decreased upon subsequent readdition, indicating that the newly assembled pumps are catalytically active and that at least part of the observed change in V-ATPase assembly occurs on lysosomes, the site of mTORC1 recruitment and activation. The increase in relative activity, as measured by the rate of initial quenching, is somewhat smaller than the change in relative assembly, which could indicate that not all of the newly assembled pumps are active. However, we do not know whether there is a linear relationship between changes in V-ATPase assembly and activity, as measured by the methods employed, and, therefore, cannot exclude the possibility that all newly assembled pumps are indeed catalytically active.
The biochemical signaling pathways that control the reversible assembly of the V-ATPase are not well understood. The system is best studied in yeast in the context of glucose starvation, which causes pump disassembly (4, 18). Inhibiting V-ATPase activity blocks glucose starvation-induced disassembly in yeast (18). We tested the effect of V-ATPase inhibition on the amino acid starvation-induced increase in assembly and found that concanamycin greatly diminishes this increased assembly. Interestingly, unlike the situation in yeast, inhibition of V-ATPase activity in HEK293T cells does not shift the equilibrium to an assembled state of the pump. V-ATPase inhibition also blocks amino acid sensing by mTORC1 (12). It could be that V-ATPase assembly must respond dynamically to amino acids and that, by preventing this dynamic change in assembly by V-ATPase inhibition, the amino acid signal is not transmitted. Alternately, binding of concanamycin to the pump may hinder the interactions necessary for amino acid sensing.
It is also possible that neutralization of the lysosome by concanamycin blocks amino acid sensing, although previous work has suggested that the proton gradient generated by the V-ATPase across the lysosomal membrane is dispensable for amino acid sensing (12). Here we show that, although mTORC1 signaling is maintained in the presence of chloroquine, which neutralizes the lysosomal pH, it is not reactivated by amino acid readdition after a period of starvation. It could be that neutralization of the lysosome prevents only the amino acid-dependent decrease in V-ATPase assembly, which may be required for amino acids to reactivate mTORC1 following amino acid depletion. Indeed, when the lysosome is neutralized by chloroquine treatment, V-ATPase assembly is elevated independent of amino acids. This result is consistent with glucose-dependent assembly in yeast, where vacuolar neutralization by chloroquine blocks glucose starvation-induced disassembly (20). Increased lysosomal pH may interfere with the binding of amino acids to their lysosomal sensors, perhaps mimicking the starved state, or it could be that the influence of luminal pH on assembly overrides any amino acid-regulated signal and becomes the primary determinant of V-ATPase assembly.
PI3K has been shown to promote V-ATPase assembly in a number of systems, including glucose-dependent increases in renal cells (8), increases during dendritic cell maturation (11), and increased assembly following influenza infection (9). Although we see a small decrease in pump assembly and activity upon inhibiting PI3K, inhibition has no effect on pump assembly in response to amino acids, indicating that amino acids control V-ATPase assembly by a pathway that is independent of PI3K. We also tested the involvement of mTORC1, which is a critical downstream effector of PI3K. Although mTORC1 inhibition decreases baseline assembly, the -fold increase in assembly observed upon amino acid starvation is unaffected. Therefore, unlike assembly changes in mammalian cells characterized previously, amino acid-dependent changes in V-ATPase assembly are controlled independent of both PI3K and mTORC1 (Fig. 6).
FIGURE 6.
Model of the regulated assembly of the V-ATPase in response to amino acids or changes in glucose concentrations. As demonstrated previously, changes in V-ATPase assembly in response to changes in glucose concentration, during dendritic cell maturation, or during influenza infection are dependent upon PI3K and, in the case of dendritic cell maturation, mTORC1 activity. By contrast, changes in assembly in response to changes in amino acid levels are independent of PI3K and mTORC1 activity but do depend upon the luminal pH and the catalytic activity of the V-ATPase. V1 subunits are depicted in green and V0 subunits in blue.
To determine whether starvation for particular amino acids could account for the observed increase in V-ATPase activity and assembly upon complete amino acid starvation, the effect of withdrawal of individual amino acids was determined. We found that starvation for some amino acids (arginine, isoleucine, leucine, lysine, and methionine) increased V-ATPase activity, although withdrawal of no single amino acid fully recapitulated the effect of total amino acid starvation. These changes in V-ATPase activity were largely paralleled by changes in assembly, although starvation for methionine appears to increase activity without increasing assembly. Interestingly, starvation for a number of amino acids, including aspartate, glutamate, and serine, led to a decrease in V-ATPase activity. Again, these changes in activity were generally paralleled by changes in assembly, although starvation for aspartate caused a decrease in activity without a change in assembly. These results suggest that there may be additional modes of regulation of V-ATPase activity in response to changes in individual amino acids. Moreover, the increase in V-ATPase assembly and activity observed during total amino acid starvation is not merely the sum of the independent effects of each amino acid, highlighting the complexity of the connection between the levels of individual amino acids, V-ATPase activity, and V-ATPase assembly, which will be important to explore in future studies.
V-ATPase activity is thought to be critical for mTORC1 activation, and, indeed, treatment of cells with V-ATPase inhibitors or knockdown of the V-ATPase by shRNA inhibits amino acid-dependent mTORC1 recruitment to lysosomes and subsequent activation (12). However, our data demonstrate that, when the V-ATPase is highly active, as during total amino acid starvation, mTORC1 is completely inhibited. Moreover, withdrawal of certain amino acids, including arginine, leucine, and lysine, although leading to complete loss of mTORC1 activity, nevertheless cause an increase in V-ATPase activity. This suggests that the link between V-ATPase activity and mTORC1 activation may be more complex than currently appreciated.
We hypothesize that there are two possible ways in which amino acids modulate V-ATPase assembly and activity. First, they may interact directly with the pump, causing structural rearrangements that lead to the observed changes in assembly and activity. Second, they may activate intermediate signaling pathways that affect regulators of the V-ATPase, leading to the observed changes. Distinguishing between these two possibilities will be the subject of future work.
Elucidating how amino acids affect the V-ATPase is an important step in understanding the basic processes that contribute to cellular homeostasis and the balance of cellular growth that is disrupted in disease states like cancer and diabetes. Future work will strive to further elucidate the signaling pathways controlling the amino acid starvation-induced increase in V-ATPase assembly and their link to activation of mTORC1.
Author Contributions
M. F. and L. S. conceived the study, designed the experiments, and wrote the paper. L. S. performed and analyzed all experiments and prepared the figures. Both authors reviewed the results and approved the final version of the manuscript.
Acknowledgments
We thank Kristina Cotter and Christina McGuire for many helpful discussions.
This work was supported by National Institutes of Health Grants GM34478 (to M. F.) and CA189321 (to L. S.) and a Sackler Families Collaborative Cancer Biology Award from Tufts University (to L. S. and M. F.). The authors declare that they have no conflicts of interest with the contents of this article.
- V-ATPase
- vacuolar proton-translocating adenosine triphosphatase
- LY
- LY294002
- DMSO
- dimethyl sulfoxide
- DAMP
- 3-(2,4-dinitroanilino) 3-amino-N-methyldipropylamine
- DNP
- dinitrophenol.
References
- 1.Forgac M. (2007) Vacuolar ATPases: rotary proton pumps in physiology and pathophysiology. Nat. Rev. Mol. Cell Biol. 8, 917–929 [DOI] [PubMed] [Google Scholar]
- 2.Breton S., and Brown D. (2013) Regulation of luminal acidification by the V-ATPase. Physiology (Bethesda) 28, 318–329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kane P. M. (2012) Targeting reversible disassembly as a mechanism of controlling V-ATPase activity. Curr. Protein Pept. Sci. 13, 117–123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kane P. M. (1995) Disassembly and reassembly of the yeast vacuolar H+-ATPase in vivo. J. Biol. Chem. 270, 17025–17032 [PubMed] [Google Scholar]
- 5.Sumner J. P., Dow J. A., Earley F. G., Klein U., Jäger D., and Wieczorek H. (1995) Regulation of plasma membrane V-ATPase activity by dissociation of peripheral subunits. J. Biol. Chem. 270, 5649–5653 [DOI] [PubMed] [Google Scholar]
- 6.Zhang J., Myers M., and Forgac M. (1992) Characterization of the V0 domain of the coated vesicle (H+)-ATPase. J. Biol. Chem. 267, 9773–9778 [PubMed] [Google Scholar]
- 7.Puopolo K., and Forgac M. (1990) Functional reassembly of the coated vesicle proton pump. J. Biol. Chem. 265, 14836–14841 [PubMed] [Google Scholar]
- 8.Sautin Y. Y., Lu M., Gaugler A., Zhang L., and Gluck S. L. (2005) Phosphatidylinositol 3-kinase-mediated effects of glucose on vacuolar H+-ATPase assembly, translocation, and acidification of intracellular compartments in renal epithelial cells. Mol. Cell Biol. 25, 575–589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Marjuki H., Gornitzky A., Marathe B. M., Ilyushina N. A., Aldridge J. R., Desai G., Webby R. J., and Webster R. G. (2011) Influenza A virus-induced early activation of ERK and PI3K mediates V-ATPase-dependent intracellular pH change required for fusion. Cell Microbiol. 13, 587–601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kohio H. P., and Adamson A. L. (2013) Glycolytic control of vacuolar-type ATPase activity: a mechanism to regulate influenza viral infection. Virology 444, 301–309 [DOI] [PubMed] [Google Scholar]
- 11.Liberman R., Bond S., Shainheit M. G., Stadecker M. J., and Forgac M. (2014) Regulated assembly of the V-ATPase is increased during cluster disruption-induced maturation of dendritic cells through a PI-3 kinase/mTOR-dependent pathway. J. Biol. Chem. 289, 1355–1363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zoncu R., Bar-Peled L., Efeyan A., Wang S., Sancak Y., and Sabatini D. M. (2011) mTORC1 senses lysosomal amino acids through an inside-out mechanism that requires the vacuolar H(+)-ATPase. Science 334, 678–683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bar-Peled L., Schweitzer L. D., Zoncu R., and Sabatini D. M. (2012) Ragulator is a GEF for the rag GTPases that signal amino acid levels to mTORC1. Cell 150, 1196–1208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sancak Y., Bar-Peled L., Zoncu R., Markhard A. L., Nada S., and Sabatini D. M. (2010) Ragulator-Rag complex targets mTORC1 to the lysosomal surface and is necessary for its activation by amino acids. Cell 141, 290–303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lowry O. H., Rosebrough N. J., Farr A. L., and Randall R. J. (1951) Protein measurement with the Folin phenol reagent. J. Biol. Chem. 193, 265–275 [PubMed] [Google Scholar]
- 16.Galloway C. J., Dean G. E., Marsh M., Rudnick G., and Mellman I. (1983) Acidification of macrophage and fibroblast endocytic vesicles in vitro. Proc. Natl. Acad. Sci. U.S.A. 80, 3334–3338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Steinberg B. E., Huynh K. K., Brodovitch A., Jabs S., Stauber T., Jentsch T. J., and Grinstein S. (2010) A cation counterflux supports lysosomal acidification. J. Cell Biol. 189, 1171–1186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Parra K. J., and Kane P. M. (1998) Reversible association between the V1 and V0 domains of yeast vacuolar H+-ATPase is an unconventional glucose-induced effect. Mol. Cell Biol. 18, 7064–7074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Franke T. F., Kaplan D. R., Cantley L. C., and Toker A. (1997) Direct regulation of the Akt proto-oncogene product by phosphatidylinositol-3,4-bisphosphate. Science 275, 665–668 [DOI] [PubMed] [Google Scholar]
- 20.Shao E., and Forgac M. (2004) Involvement of the nonhomologous region of subunit A of the yeast V-ATPase in coupling and in vivo dissociation. J. Biol. Chem. 279, 48663–48670 [DOI] [PubMed] [Google Scholar]
- 21.Hurtado-Lorenzo A., Skinner M., El Annan J., Futai M., Sun-Wada G.-H., Bourgoin S., Casanova J., Wildeman A., Bechoua S., Ausiello D. A., Brown D., and Marshansky V. (2006) V-ATPase interacts with ARNO and Arf6 in early endosomes and regulates the protein degradative pathway. Nat. Cell Biol. 8, 124–136 [DOI] [PubMed] [Google Scholar]
- 22.Chung J., Kuo C. J., Crabtree G. R., and Blenis J. (1992) Rapamycin-FKBP specifically blocks growth-dependent activation of and signaling by the 70 kd S6 protein kinases. Cell 69, 1227–1236 [DOI] [PubMed] [Google Scholar]
- 23.Hara K., Yonezawa K., Weng Q.-P., Kozlowski M. T., Belham C., and Avruch J. (1998) Amino acid sufficiency and mTOR regulate p70 S6 kinase and eIF-4E BP1 through a common effector mechanism. J. Biol. Chem. 273, 14484–14494 [DOI] [PubMed] [Google Scholar]
- 24.Blommaart P. J., Zonneveld D., Meijer A. J., and Lamers W. H. (1993) Effects of intracellular amino acid concentrations, cyclic AMP, and dexamethasone on lysosomal proteolysis in primary cultures of perinatal rat hepatocytes. J. Biol. Chem. 268, 1610–1617 [PubMed] [Google Scholar]