Background: 20-Hydroxyecdysone (20E) and juvenile hormone (JH) coordinately control insect molting and metamorphosis.
Results: Induced by 20E and suppressed by JH, E93 acts through GAGA-containing motifs and up-regulates a subset of 20E response genes.
Conclusion: E93 transcriptionally modulates 20E signaling to promote Bombyx metamorphosis.
Significance: Our study helps in understanding the complicated molecular mechanisms of insect metamorphosis.
Keywords: apoptosis, autophagy, Bombyx mori, juvenile hormone (JH), transcription factor, 20-hydroxyecdysone, E93, fat body, metamorphosis, wing disc
Abstract
As revealed in a previous microarray study to identify genes regulated by 20-hydroxyecdysone (20E) and juvenile hormone (JH) in the silkworm, Bombyx mori, E93 expression in the fat body was markedly low prior to the wandering stage but abundant during larval-pupal metamorphosis. Induced by 20E and suppressed by JH, E93 expression follows this developmental profile in multiple silkworm alleles. The reduction of E93 expression by RNAi disrupted 20E signaling and the 20E-induced autophagy, caspase activity, and cell dissociation in the fat body. Reducing E93 expression also decreased the expression of the 20E-induced pupal-specific cuticle protein genes and prevented growth and differentiation of the wing discs. Importantly, the two HTH domains in E93 are critical for inducing the expression of a subset of 20E response genes, including EcR, USP, E74, Br-C, and Atg1. By contrast, the LLQHLL and PLDLSAK motifs in E93 inhibit its transcriptional activity. E93 binds to the EcR-USP complex via a physical association with USP through its LLQHLL motif; and this association is enhanced by 20E-induced EcR-USP interaction, which attenuates the transcriptional activity of E93. E93 acts through the two HTH domains to bind to GAGA-containing motifs present in the Atg1 promoter region for inducing gene expression. In conclusion, E93 transcriptionally modulates 20E signaling to promote Bombyx larval-pupal metamorphosis.
Introduction
Insect ecdysteroids include 20-hydroxyecdysone (20E)2, the most active form of molting hormone, and ecdysone, the immediate precursor of 20E (1). 20E binds to a heterodimer in the nuclear receptor complex, ecdysone receptor (EcR) and ultraspiracle protein (USP). With the assistance of a molecular chaperone complex, EcR-USP binds to 20E response elements (EcRE) present in the promoter regions of 20E primary response genes. In the absence of 20E, EcR-USP associates with transcriptional co-repressors. When 20E binds to EcR-USP, the transcriptional co-repressors are dissociated, and multiple transcriptional co-activators are recruited, resulting in the induction of gene expression through EcRE. 20E-EcR-USP triggers a transcriptional cascade, including transcription of the 20E primary response genes (i.e. the transcription factor genes Br-C, E74, E75, and E93) and subsequently the 20E secondary response genes (1, 2). E93 was first identified as a 20E primary response gene in the fruit fly, Drosophila melanogaster (3), and it encodes a member of the helix-turn-helix (HTH) transcription factor family (4). A recent study suggests that E93 expression is suppressed by juvenile hormone (JH) via the JH receptor Met and a JH primary-response gene Kr-h1 (5, 6). However, how E93 expression is coordinately regulated by 20E and JH remains unclear.
20E initiates and orchestrates larval-larval molting and larval-pupal-adult metamorphosis (7). During metamorphosis, 20E induces programmed cell death (PCD), including apoptosis (type I PCD) and autophagy (type II PCD), and remodeling of larval tissues as well as the generation of adult tissues from imaginal discs (8–10). In Drosophila, 20E is both necessary and sufficient to induce autophagy and apoptosis of larval tissues in a tissue- and stage-specific manner primarily via up-regulating most Atg genes and many apoptosis genes (8, 11, 12). Among the 20E primary-response genes, E93 is a key player in 20E signaling during metamorphosis. E93 predominately transduces 20E signaling to induce autophagy and caspase activity in the fat body during the larval-prepupal metamorphosis (13), in the midgut a few hours after pupariation (14, 15), and in the salivary gland immediately after pupation (16–18). Meanwhile, E93 is expressed widely in adult cells during the pupal stage and is required for many patterning processes during adulthood (19). Later studies suggest that E93 is the key determinant promoting metamorphosis, thus acting as the adult specifier in insects (6).
In general, Br-C, E74, E75, and E93 positively affect 20E signaling (8). As a potent HTH transcription factor, E93 binds several 20E response genes and PCD genes on polytene chromosomes in Drosophila (16). The expression of these genes is reduced in E93 mutants, whereas E93 overexpression up-regulates some of the genes (13, 16). However, little is known regarding the underlying molecular mechanisms involved in E93 regulation of 20E signaling and insect metamorphosis (8, 13, 16).
The silkworm, Bombyx mori, is a model insect for lepidopterans (20). During larval-pupal metamorphosis in Bombyx, the fat body undergoes significant remodeling (involving progressive autophagy, caspase activity, and cell dissociation) that is mainly controlled by the 20E-triggered transcriptional cascade (21–25). The wing discs grow and differentiate during this period, and the pupal-specific wing disc cuticle protein genes (WCPs) apparently contribute to wing disc differentiation. The expression of pupal-specific WCPs is induced by 20E signaling, and 20E induction is prevented by JH (26–31).
Our previous microarray studies to identify genes regulated by 20E and JH in Bombyx (21, 22) revealed that E93 is abundantly expressed in the fat body during larval-pupal metamorphosis. In the current study, we determined that E93 is regulated by 20E and JH at the transcriptional level, confining its expression to a specific period during larval-pupal metamorphosis. Moreover, E93 acts through GAGA-containing motifs and modulates 20E signaling to promote larval tissue remodeling and adult tissue formation during Bombyx larval-pupal metamorphosis. Interestingly, the expression level of E93 is induced by 20E-EcR-USP, but the transcriptional activity of E93 is attenuated by its physical association with USP.
Experimental Procedures
Silkworms and Genetics
The silkworms of the P50 strain (Chinese variant, Dazao, wild-type tetramolter allele, 4M) were provided by the Sericultural Research Institute, Chinese Academy of Agricultural Sciences, Zhenjiang, China, and used for most of the studies unless specified otherwise. The silkworms of the trimolter allele (3M; number 19–350 at the silkworm gene bank at Southwest University, Chongqing, China) and pentamolter allele (5M; number 15–010) were presented by Southwest University and also used in the in vivo JH injection experiments. All silkworm larvae were reared on fresh mulberry leaves in the laboratory at 25 °C under 14 h light/10 h dark cycles.
The SP2-GAL4 and UAS-EcRDN transgenic lines were produced via germline transformation using the p50 strain as donors (24). The transgenic silkworm SP2-GAL4>UAS-EcRDN was obtained by crossing SP2-GAL4 with UAS-EcRDN as described previously; SP2-GAL4 was used as a control here (24).
Collection of Fat Body Tissues, Wing Discs, and Hemolymph Samples
The peripheral fat body tissues from the 5th abdominal segment, wing discs, and hemolymph samples were collected at various developmental stages or after various treatments.
Conventional Molecular, Biochemical, and Cellular Methods
Full-length E93 cDNA was cloned using rapid amplification of cDNA ends (RACE) (25, 32). Details of quantitative real-time PCR (qPCR) and Western blotting have been previously described (21–25, 32). Caspase 3 activity was determined according to the manufacturer's instructions (Beyotime, China) with a Multiskan Flash Microplate Reader (Thermo Fisher Scientific) (23, 25).
Full-length E93 was cloned into the pcDNA 3.1(+) vector (Invitrogen) to create the expression constructs (25). Truncated E93 derivatives were generated from the wild-type E93 using site-directed mutagenesis (Invitrogen). The E93ΔHTH1, E93ΔHTH2, E93ΔLLQHLL, E93ΔPLDLSAK, and E93ΔHTH1ΔHTH2 mutants have deletions in amino acids 258–298, 439–503, 145–149, 218–222, and 258–298 plus 439–503, respectively.
Multiple fragments of the Atg1 promoter (−1820 to −1 bp, −1000 to −1 bp, −1000 to −400 bp, −600 to −1bp, −800 to −1820, and −1820 to −1421 bp) were cloned. The three putative GAGA sequences (−961 to −946 bp, −717 to −700 bp, and −50 to −33 bp) and two unrelated sequences (−260 to −244 bp and −777 to −761 bp) in the 1-kb Atg1 promoter region were individually detected using site-directed mutagenesis. The above DNA sequences and four copies of GAGA sequences were individually cloned into the pGL3 basic vector containing the hsp27 mini-promoter.
Radioimmunoassay for Measuring Ecdysteroids Titers
Total ecdysteroid titers of the hemolymph samples were determined by radioimmunoassay as described previously (33). Briefly, hemolymph samples collected from larvae were mixed with 30 volumes of methanol, vortexed vigorously, and centrifuged at 7000 × g for 5 min. A radioimmunoassay was performed to evaluate the supernatants using 20E (Sigma) as a standard. The rabbit antiserum used was raised against 20E conjugated with human serum albumin. [3H]Ecdysone (∼60 Ci/mmol) was obtained from New England Nuclear (Boston, MA). Cross-reactions of the antiserum between ecdysone and 20E occurred at a ratio of 1:2.5.
Fluorescence Microscopy and Transmission Electron Microscopy
TUNEL (Beyotime) labeling and LysoTracker Red (Invitrogen) staining were used to estimate caspase activity and autophagy, respectively, using an Olympus FluoView FV1000 confocal microscope (23–25). Transmission electron microscopy (TEM) analysis was performed to observe autophagic components as previously described (24). For TUNEL staining, LysoTracker Red staining, or TEM analysis, each kind of observations were performed under similar conditions.
Injection of dsRNA and Methoprene in Vivo
The detailed method for injecting dsRNA and methoprene into the silkworm was previously described (21). dsRNA was generated for GFP and E93 using the T7 RiboMAXTM Express RNAi System (Promega). E93 RNAi was administered at the initiation of the wandering stage (IW). Fat body tissues, wing discs, and hemolymph samples were collected for bioassays at the indicated time after the injection of 30 μg of dsRNA (21–25, 32).
Methoprene (Dr. Ehrenstorfer GmbH, Germany) was dissolved in DMSO to make a stock solution (5 mg/ml), then diluted with DMSO for the working solution (1 mg/ml). A 5-μl aliquot of the working solution was injected into the hemolymph of a larva at the initiation of the wandering stage through an abdominal leg in the last pair. Forty-eight hours after methoprene injection (or DMSO as a control), samples of fat body tissue were collected for qPCR analysis (21–25).
Culture of Fat Body Tissues and Bm-12 Cells
Fat body tissues collected from L5-2 larvae were cultured in Grace's medium (Invitrogen) at 27 °C. Bm-12 cells (34) were maintained in TNM-FH (Sigma) medium supplemented with 10% heat-inactivated fetal bovine serum (HyClone) at 27 °C. After preincubation (1 h for fat body tissue and 1 day for Bm-12 cells), the medium was removed and replaced with fresh medium with 2 μm 20E and/or 2 μm methoprene. After 2 h of incubation, mRNA was isolated for qPCR analysis. To determine whether E93 is a 20E primary response gene, 2 μm 20E and/or 10 μg/ml of CHX (Enzo Life Science) was applied (24, 25, 32).
RNAi and Transient Transfection Assay in Bm-12 Cells
RNAi in Bm-12 cells was performed using the Effectene transfection reagent (Qiagen) for 48 h at a final concentration of 2 μg/ml of dsRNA as described previously, followed by qPCR analysis (24, 25).
The transient transfection assay in Bm-12 cells was performed for 48 h using Effectene according to the manufacturer's instructions (Qiagen). The final DNA concentration was 2 μg/ml, and the DNA:Effectene ratio was 1:25. The vector used to overexpress EGFP, E93, and the E93 mutants in Bm-12 cells was pEGFP-N1 (Clontech) under the control of the BmNPV ie1 promoter (24, 25, 32). Co-transfection was followed by qPCR analysis.
Immunoprecipitation
The transient transfection assay in Bm-N cells was performed the same as in Bm-12 cells. After co-transfection of the pEGFP-N1 vectors overexpressing HA-EcR, FLAG-USP, and V5-E93 (or V5-E93ΔLLQHLL) in Bm-N cells, cells were treated with 2 μm 20E for 12 h, followed by immunoprecipitation. In other experiments, HA-EcR (or FLAG-USP) and V5-E93 (or V5-E93ΔLLQHLL) were co-transfected in Bm-N cells without 20E treatment. Bm-N cells were harvested and lysed in ice-cold Nonidet P-40 lysis buffer (Beyotime). Lysates were incubated with HA (SAB1411733, Sigma), FLAG (F3165, Sigma), V5 (R960–25, Invitrogen), or IgG (Beyotime) (as a negative control) for 4 h, followed by incubation with protein A/G (Pierce) overnight at 4 °C. After extensive washing with cold Nonidet P-40 buffer, the samples were treated with RIPA lysis buffer (Beyotime) about 15 min on the ice. Then immunoprecipitates were separated by SDS-PAGE and analyzed by Western blots (35).
Dual Luciferase Assay in HEK293 Cells
HEK293T cells were maintained in Dulbecco's modified Eagle's medium (HyClone) supplemented with 10% heat-inactivated fetal bovine serum, and transfection was performed using Lipofectamine 2000 reagent (Invitrogen). To identify the parts of the Atg1 promoter region responsive to E93, the 1.82-kb region of the Atg1 promoter upstream of the transcription start site was cloned into the SacI and EcoRI sites of the pGL3 basic vector containing the hsp70 minimal promoter (Promega). The deletions and mutations of the promoter regions were also constructed in pGL3. The pRL vector (Promega) carrying Renilla luciferase driven by the Actin3 promoter was used for normalization. Meanwhile, Bombyx E93 (or its mutant constructs) was cloned into the pcDNA 3.1(+) vector (Invitrogen) to create the expression constructs. After transfection of the E93 (or its mutants) expression construct, a reporter pGL3 vector, and the reference pRL vector into HEK293T cells for 48 h, the cells were collected. The relative luciferase activity was calculated by normalizing the reporter firefly luciferase level to the reference Renilla luciferase level. Dual luciferase assays were conducted using the Dual Luciferase Assay System (Promega) and a Modulus Luminometer (Turner BioSystems) (24, 25, 32, 35).
Chromatin Immunoprecipitation (ChIP) Assay
Bm-N cells were grown in 10-cm dishes (70% confluent) and transfected with FLAG-E93 expression plasmid for 48 h. Then the cells were fixed and subjected to ChIP using the Agarose ChIP Kit (Pierce) and the FLAG antibody (F3165, Sigma). Mock immunoprecipitations with preimmune serum were used for negative controls. The precipitated DNA and input were analyzed by qPCR to detect the binding between FLAG-E93 and fragments of the Atg1 promoter (32, 35). For ChIP-qPCR, the primers are listed in Table 1.
TABLE 1.
Primers for ChIP-qPCR
S, sense; A, antisense.
| Gene name | Primer sequence |
|---|---|
| Atg1 promoter | S: CCGATTGAGTTCAGGAGTTG |
| −1785 to −1733b | A: GAGTTATGCGCTGTACGCTA |
| Atg1 promoter | S: TTACCTACGGCATGGTAAGC |
| −1677 to −1613b | A: AAATCAACTGCCCAACATTT |
| Atg1 promoter | S: AGGAGGTAGAACGGACTGCT |
| −793 to −703b | A: TTAAGGTTCCGCTGGAATAG |
| Atg1 promoter | S: AGGATCCGAGATGACGTGT |
| −419 to −372b | A: ATCCTGCAGAAAGAATGCAG |
| Atg1 promoter | S: GCAGGATCGGAAATGACTAC |
| −359 to −285b | A: TCCCGAATTCGTATTTCTGT |
| Atg1 promoter | S: CGTCCTGCGACAGATTATTA |
| −121 to −49b | A: AGCCGGTTCGTTTGAAGATA |
Statistics
The experimental data were analyzed using Student's t test and analysis of variance: for the t test: * (decrease) or # (increase), p < 0.05; ** (decrease) or ## (increase), p < 0.01. For analysis of variance: bars labeled with different lowercase letters are significantly different (p < 0.05). Throughout the study, values are represented as the mean ± S.D. of at least three independent experiments.
Results
E93 Expression during Larval-Pupal Metamorphosis
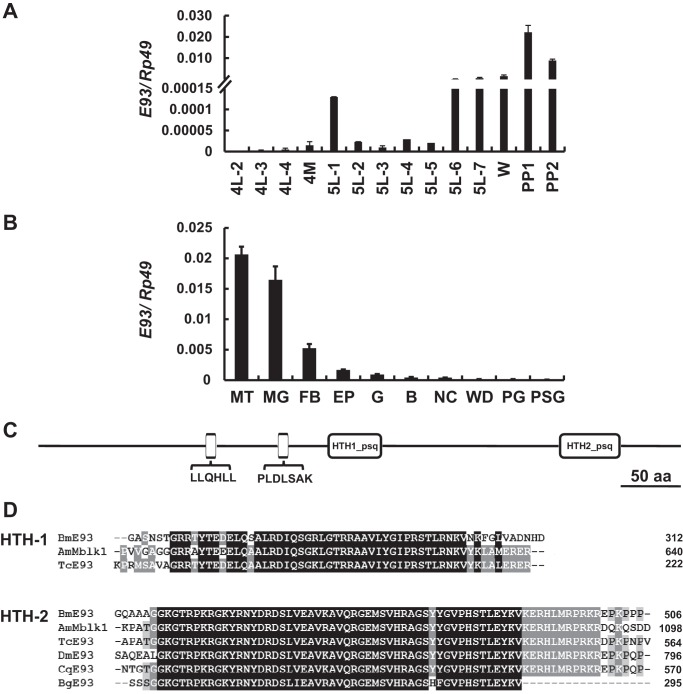
In a previous study, we performed a microarray to identify genes potentially regulated by 20E and JH in the Bombyx fat body (21, 22). Notably, BGIBMGA010815 (SilkDB code number) showed markedly higher expression during larval-pupal metamorphosis in comparison to its levels during the 4th larval molting stage and the 5th larval feeding stage, suggesting that BGIBMGA010815 expression might be induced by 20E and suppressed by JH. To confirm the developmental profile of BGIBMGA010815, we further performed qPCR analysis using total RNA isolated from fat body tissues dissected on day 2 of the 4th larval instar (4L-2) to day 2 of the prepupal stage (PP2). BGIBMGA010815 mRNA levels were undetectable from 4L-2 to the end of the feeding stage during the 5th larval instar, except for a small increase on 5L-1. However, BGIBMGA010815 mRNA levels began to increase on 5L-6 and peaked on PP1 (Fig. 1A). A spatial distribution analysis during the wandering stage revealed that BGIBMGA010815 mRNA levels were high in the Malpighian tubules, midgut, and fat body compared with other tissues (Fig. 1B).
FIGURE 1.
The Bombyx E93 gene. A, the developmental changes in E93 mRNA levels in the fat body were determined by qPCR from day 2 of the fourth instar (4L-2) to day 2 of the prepupal stage (PP2). 4M, the 4th larval molting stage; W, the wandering stage. B, spatial distribution analysis of E93 expression at the wandering stage. MT, malpighian tubules; MG, midgut; FB, fatbody; EP, epidermis; G, gonad; B, brain; NC, nerve cord; WD, wing disc; PG, prothoracic gland; PSG, post silk gland. C, significant domains of Bobmyx E93: LLQHLL, PLSLSAK, HTH-1, and HTH-2. D, alignment of the predicted amino acid sequences of the HTH-1 and HTH-2 domains of E93 proteins from B. mori (Bm), Apis mellifera (Am; E93 was early cognized as Mblk1), Tribolium castaneum (TC), D. melanogaster (Dm), Culex quinquefasciatus (Cq), and Blattella germanica (Bg). Black and light gray shading indicate identical and similar amino acid residues, respectively.
We then cloned full-length cDNA of BGIBMGA010815 using RACE. BGIBMGA010815 encodes a protein with 549 amino acid residues that forms the typical structure of a HTH transcription factor containing two HTH domains (Fig. 1C) (4). Its amino acid sequence only shows 12–14% similarity to five E93 proteins identified from other insect species. However, the HTH domains are highly conserved (Fig. 1D). Moreover, each of these proteins contains a nuclear receptor interaction motif (LXXLL or LLXXL; LLQHLL in Bombyx) (36) and co-repressor C-terminal-binding protein interaction motif (PXDL(S/T)X(K/R); PLDLSAK in Bombyx) (37) (Fig. 1C). Based on sequence similarity and the conservation of important functional motifs, we identified BGIBMGA010815 as Bombyx E93 (GenBankTM accession number KJ913673).
E93 Expression in the Fat Body Is Induced by 20E and Suppressed by JH
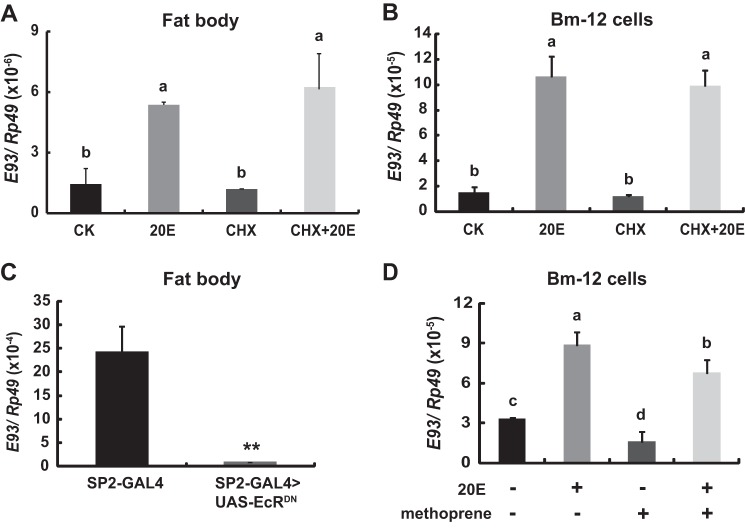
We then verified the hypothesis that E93 expression is induced by 20E and suppressed by JH. The addition of 20E to the cultured fat body tissues isolated from 5L-2 larvae resulted in a greater than 3-fold increase in E93 mRNA levels within 2 h. Cycloheximide (CHX), an inhibitor of protein synthesis, did not block the 20E induction of E93 expression (Fig. 2A). Similar results were obtained in Bombyx Bm-12 cells (Fig. 2B), confirming that E93 is a 20E primary response gene. To compensate for the gain-of-function studies, a dominant-negative mutant of EcR (EcRDN) was overexpressed to inhibit 20E signaling specifically in the larval fat body using the binary Bombyx GAL4/UAS system (24). The E93 mRNA levels in SP2-GAL4>UAS-EcRDN at the wandering stage were reduced by 95% compared with those in control SP2-GAL4 larvae (Fig. 2C), demonstrating that 20E induces E93 expression through EcR-USP during larval-pupal metamorphosis. The addition of methoprene (a JH analog) to the Bm-12 cell culture medium for 2 h suppressed both the basal and 20E-induced expression of E93 (Fig. 2D), suggesting that E93 expression may be suppressed by JH.
FIGURE 2.
E93 expression is induced by 20E and suppressed by JH. A and B, E93 is a 20E primary response gene. Fat body tissues isolated from L5-2 larvae (B) and Bm-12 cells (C) were cultured in Grace's medium (CK) for 2 h, whereas the experimental fat body tissues or Bm-12 cells were cultured in Grace's medium containing 2 μm 20E (20E), 10 μg/ml of CHX, or 2 μm 20E and 10 μg/ml of CHX (20E + CHX). C, E93 expression in the SP2-GAL4>UAS-EcRDN larval fat body at the wandering stage is decreased in comparison with SP2-GAL4 (control). D, E93 expression is induced by 20E and suppressed by the JH analog methoprene. Bm-12 cells were treated by 2 μm 20E or/and 2 μm methoprene for 2 h.
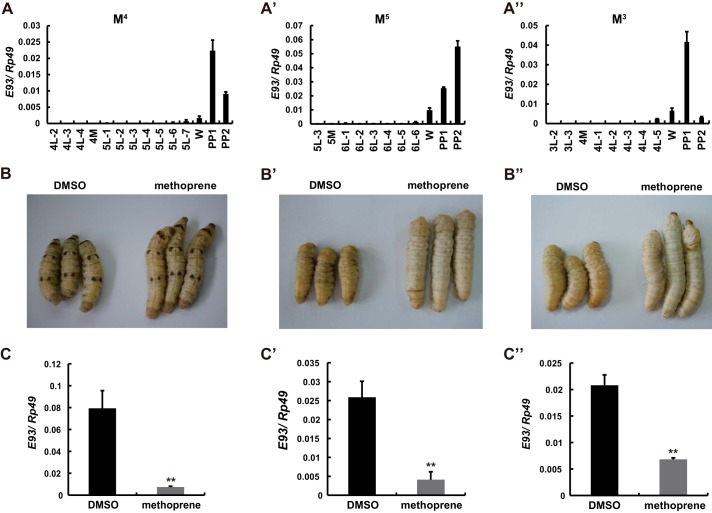
We also examined whether the unique developmental profile of E93 expression in the tetramolter allele is similar to the expression patterns of other silkworm alleles. Interestingly, in both the pentamolter and trimolter alleles, E93 mRNA levels in the fat body were low prior to the wandering stage, began to increase during the wandering stage, and reached maximum levels during the prepupal stage (Fig. 3, A–A″). We further investigated whether JH is able to suppress 20E-induced E93 expression in the fat body during larval-pupal metamorphosis in all three silkworm alleles. Notably, 48 h after injection of methoprene into the larvae of all three alleles at IW, methoprene not only delayed pupation (Fig. 3, B–B″) but also suppressed E93 expression in the fat body (Fig. 3, C–C′). These data indicate that, when the JH level is low, E93 expression in the prepupal fat body is induced by 20E.
FIGURE 3.
Methoprene application delays pupation and suppresses E93 expression in the fat body. A–A″, developmental changes in E93 mRNA levels in the fat body of the wild-type tetramolter allele (4M) (A), the pentamolter allele (5M) (A′), and the trimolter allele (3M). B–C″, pupation is delayed (B–B″) and E93 expression in the fat body is suppressed (C–C″) 48 h after injection of the JH analog methoprene (5 μg/larva) into the larvae in comparison with DMSO (control).
Reducing E93 Expression via RNAi Causes Lethality and Prevents Fat Body Remodeling
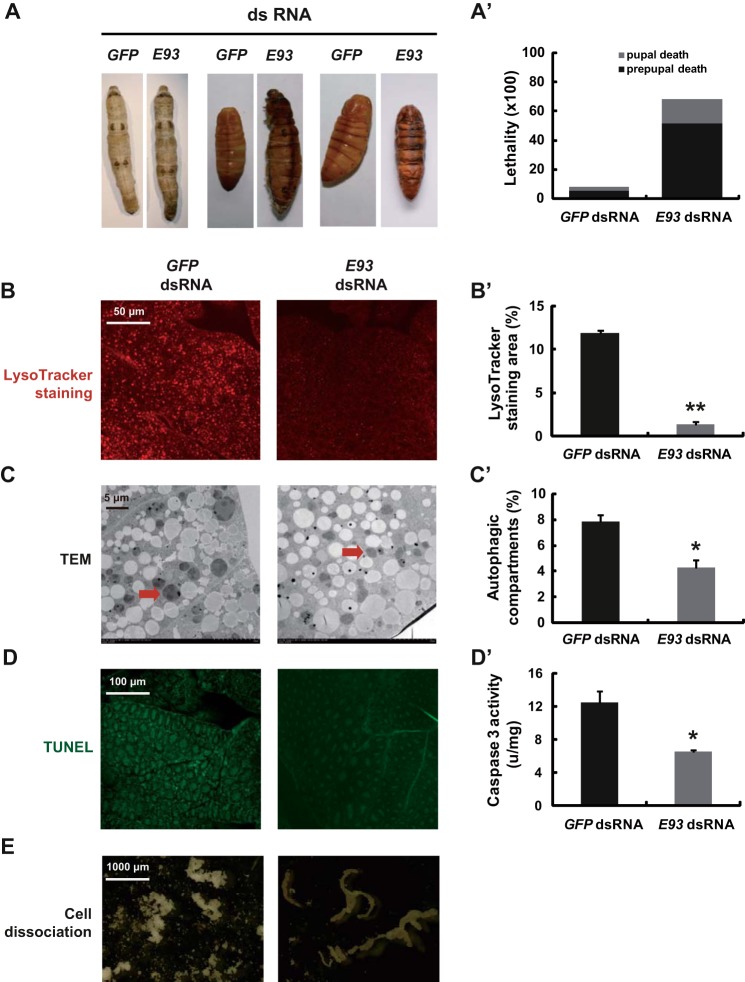
To determine the function of E93 during larval-pupal metamorphosis, E93 expression was suppressed using RNAi (E93 RNAi) at IW. Twenty-four hours after injection with E93 dsRNA, E93 expression in the fat body decreased by 85% compared with the control levels (GFP RNAi) (not shown). Moreover, E93 RNAi caused lethal phenotypes, with ∼50 and 20% lethality during the prepupal and pupal stages, respectively. Some E93 RNAi larvae died during the wandering stage, others failed to form normal pupae and died as larval-pupal intermediates, and others arrested during the pupal stage (Fig. 4, A and A′).
FIGURE 4.
E93 RNAi causes lethality and prevents fat body remodeling. dsRNA (30 μg/larva) was injected into staged larvae at the initiation of the wandering stage. GFP dsRNA (30 μg per larva) was used as a control. A and A′, some E93 RNAi treated silkworms died during the wandering stage (A; left panel), some failed to form normal pupae and died as larval-pupal intermediates (A; middle panel), and some arrested at the pupal stage (A; right panel). The chart (A′) shows the quantification of the lethality in A. B and B′, LysoTracker Red staining (red, ×40) in the fat body 24 h after dsRNA treatment. The chart (B′) shows the quantification of LysoTracker Red staining in B. C and C′, TEM analysis (7,500×) 24 h after dsRNA treatment. The red arrow denotes an autolysosome. The chart (C′) shows the quantification of autophagosomes and autolysosomes in C. D and D′, TUNEL labeling (green, ×20) (D) and caspase 3 activity (D′) in the fat body 24 h after dsRNA treatment. E, a comparison of fat body cell dissociation 24 h after pupation.
Importantly, E93 RNAi inhibited autophagy, caspase activity, and cell dissociation in the fat body during larval-pupal metamorphosis. Twenty-four hours after injection with E93 dsRNA, LysoTracker Red staining was dramatically decreased (Fig. 4, B and B′), and both the number and size of autophagosomes were reduced according to TEM analysis (Fig. 4, C and C′). A TUNEL assay revealed a significant reduction in labeling (Fig. 4D), and caspase 3 activity was decreased to 50% of the GFP RNAi control level (Fig. 4D′). In addition, the fat body cell dissociation that occurred 24 h after pupation in the GFP RNAi control pupae was completely inhibited in the E93 RNAi pupae (Fig. 4E).
Overall, E93 RNAi caused larval and pupal lethality and prevented fat body remodeling. These phenotypes are similar to those observed in Bombyx larvae injected with dsRNA of key genes in the 20E signal transduction pathway, including βftz-F1 (38), EcR and USP (21), and Met1 and Met2 (25). The effects of E93 RNAi suggest that E93 plays an important role in modulating 20E signaling, which promotes larval-pupal metamorphosis in Bombyx.
E93 RNAi Decreases Ecdysteroid Titers and Disrupts 20E Signaling in the Fat Body
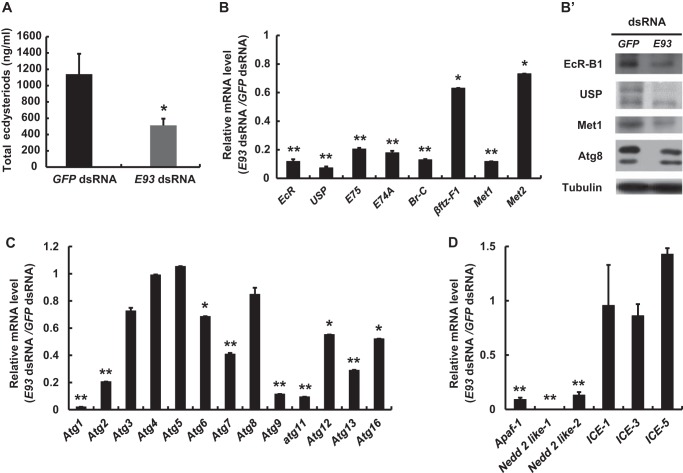
Several 20E response genes, including E75 (39), βftz-F1 (40), and Br-C (41, 42), are essential for maintaining the titer of ecdysteroids in Drosophila. According to a radioimmunoassay performed 24 h after dsRNA treatment, ecdysteroid titers decreased by 50% in E93 RNAi larvae compared with those in GFP RNAi control larvae (Fig. 5A), suggesting that E93 may regulate the production of ecdysteroids in Bombyx.
FIGURE 5.
E93 RNAi decreases ecdysteroid titer and disrupts 20E signaling in the fat body. dsRNA (30 μg/larva) was injected into staged larvae at the initiation of the wandering stage. GFP dsRNA (30 μg/larva) was used as a control. All the experiments were performed 24 h after dsRNA treatment. A, a comparison of ecdysteroid titer. B–B′, qPCR analysis of 20E-response genes (B) and Western blot analysis of the protein levels of some 20E-response genes (B′) in the fat body. C and D, qPCR analysis of 20E-induced Atg genes (C) and apoptosis genes (D) in the fat body.
The expression of several key genes in the 20E-triggered transcriptional cascade was determined in the fat body at 24 h after E93 dsRNA injection. Among the eight genes tested, the mRNA levels of EcR, USP, E75, E74, Br-C, and Met1 decreased by 80–90% compared with those in the control larvae, whereas βftz-F1 and Met2 exhibited a 20–40% decrease (Fig. 5B). Moreover, Western blots for EcR-B1, USP, and Met1 revealed decreased protein levels in the E93 RNAi larvae (Fig. 5B′), suggesting that E93 RNAi disrupts the 20E-triggered transcriptional cascade in the fat body during larval-pupal metamorphosis.
The mRNA levels of 20E-induced Atg and apoptosis genes in the fat body were measured 24 h after E93 dsRNA injection. The mRNA levels of most of the Atg genes tested, including Atg1, Atg2, Atg6, Atg7, Atg9, Atg11, Atg12, Atg13, and Atg16, were decreased in the E93 RNAi larvae compared with those in the control larvae, whereas the mRNA levels of Atg3, Atg4, Atg5, and Atg8 did not change (Fig. 5C). According to Western blot analysis, Atg8 protein levels were also not affected by E93 RNAi (Fig. 5B′). Among the apoptosis genes tested, Apaf-1, Nedd 2 like-1, and Nedd 2 like-2 mRNA levels decreased in E93 RNAi larvae, whereas the mRNA levels of ICE1, ICE3, and ICE5 did not decrease (Fig. 5D). These data demonstrate that E93 regulates certain genes in 20E signaling and 20E-induced autophagy and apoptosis pathways involved in fat body remodeling during larval-pupal metamorphosis.
E93 Regulates Wing Disc Growth and Differentiation and 20E-induced Pupal-specific WCPs
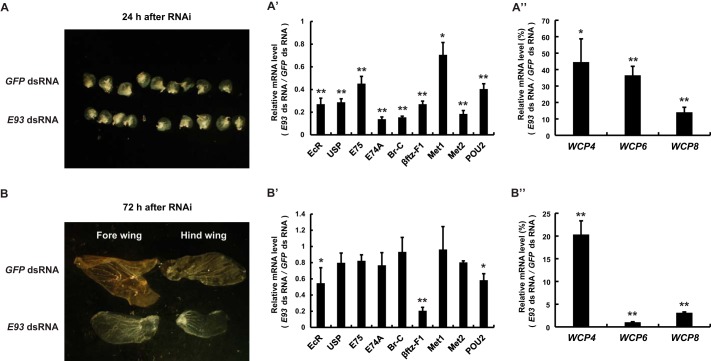
We also examined whether E93 RNAi affects wing disc growth and differentiation during larval-pupal metamorphosis. No changes were observed in the wing discs at 24 h after E93 dsRNA injection (Fig. 6A), but their development was significantly retarded by 72 h after E93 dsRNA injection (Fig. 6B). At the later time point, the size of the wing discs was much smaller in the E93 RNAi silkworms than in the GFP RNAi control silkworms. The wing discs in the control silkworms already differentiated into pupal-specific structures, but those in the E93 RNAi silkworms remained in the larval form.
FIGURE 6.
E93 RNAi retards wing disc growth and differentiation and down-regulates 20E-induced pupal-specific WCPs. dsRNA (30 μg/larva) was injected into staged larvae at the initiation of the wandering stage. GFP dsRNA (30 μg/larva) was used as a control. A–A″, comparisons of morphology of the fore wing disc (A), qPCR analyses of 20E-response genes (A′) and pupal-specific WCPs (A″) in the wing discs 24 h after dsRNA treatment. B–B″, comparisons of wing disc morphology (B), qPCR analyses of 20E-response genes (B′) and pupal-specific WCPs (B″) in the wing discs 72 h after dsRNA treatment.
We then investigated whether the 20E-triggered transcriptional cascade and the 20E-induced pupal-specific WCPs were affected by E93 RNAi in the wing discs. At 24 h after E93 dsRNA injection, all nine tested genes in the 20E-triggered transcriptional cascade showed decreased mRNA levels in E93 RNAi larvae compared with control larvae (Fig. 6A′). Similarly, WCP4, WCP6, and WCP8 showed moderately decreased mRNA levels in E93 RNAi larvae compared with control larvae (Fig. 6A″). Interestingly, most genes in the 20E-triggered transcriptional cascade except βftz-F1 returned to the control levels at 72 h after E93 dsRNA injection (Fig. 6B′), but the mRNA levels of WCPs further decreased (Fig. 6B″), mirroring the phenotypic changes observed in the wing discs. These data showed that E93 RNAi disrupts the 20E-triggered transcriptional cascade and down-regulates the 20E-induced pupal-specific WCPs, resulting in failures in the growth and differentiation of pupal-specific structures in the wing discs during larval-pupal metamorphosis.
E93 Directly Induces Expression of a Subset of 20E Response Genes
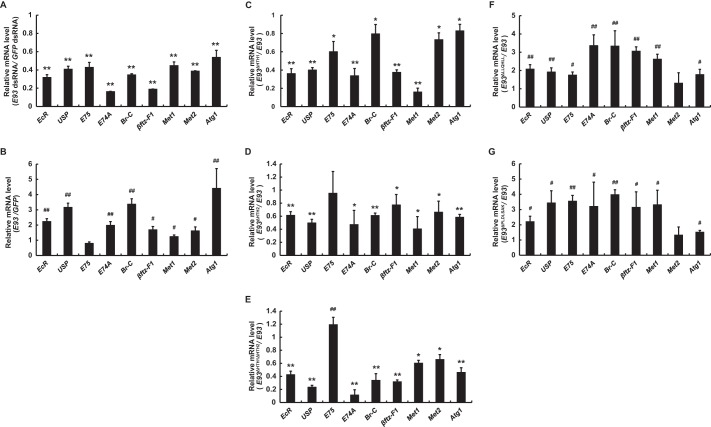
We further investigated whether E93 could directly regulate the basal expression levels of 20E response genes in Bm-12 cells. At 48 h after dsRNA treatment, E93 expression in E93 RNAi cells decreased by ∼70% of the control level in the GFP RNAi cells (not shown). Interestingly, the expression of all nine tested genes in the 20E-triggered transcriptional cascade decreased by 50–85% of their control levels at 48 h after E93 dsRNA treatment (Fig. 7A). Notably, Atg1, an essential Atg gene and a 20E primary-response gene (24), was also down-regulated by E93 dsRNA treatment (Fig. 7A). We also tested the effect of E93 overexpression on the mRNA levels of 20E response genes. Among the genes tested, Atg1 showed the most sensitivity to E93 overexpression and was up-regulated ∼4-fold by E93 overexpression (Fig. 7B). Br-C, USP, EcR, E74a, βftz-F1, Met2, and Met1 also showed increased mRNA levels in E93-overexpressed cells. However, E75 expression was not affected by E93 overexpression (Fig. 7B). These data in Bm-12 cells indicated that E93 is directly involved in the expression of a subset of 20E response genes.
FIGURE 7.
E93 directly induces expression of a subset of 20E-response genes. A, RNAi in Bm-12 cells was conducted for 48 h at a final concentration of 2 μg/ml of E93 or GPF dsRNA (control), followed by qPCR analysis of the 20E-response genes. B, transient transfection assay in Bm-12 cells was carried out for 48 h at a final concentration of 2 μg/ml of E93 or GPF pEGFP-N1 expression vectors (control), followed by qPCR analysis of the 20E-response genes. The transient transfection assays below were similarly performed. C–E, comparisons of the effects of E93ΔHTH1 (C), E93ΔHTH2 (D), and E93ΔHTH1ΔHTH2 (E) overexpression on 20E-response gene expression with E93 overexpression. F and G, comparisons of the effects of E93ΔLLQHLL (F) and E93ΔPLDLSAK (G) overexpression on 20E-response gene expression with E93 overexpression.
We then examined the possible roles of the two HTH domains in gene regulation by E93. Three mutant E93 constructs with deleted HTH1 and/or HTH2 domains, namely E93ΔHTH1, E93ΔHTH2, and E93ΔHTH1ΔHTH2, were individually transfected into Bm-12 cells. Compared with that of wild-type E93, the ability of E93ΔHTH1, E93ΔHTH2, and E93ΔHTH1ΔHTH2 to induce 20E response genes was reduced by 20–80, 30–60, and 50–90%, respectively (Fig. 7, C–E).
We also examined whether the LLQHLL and PLDLSAK motifs in Bombyx E93 are required for its transcriptional activity. Two mutant E93 constructs with deleted LLQHLL or PLDLSAK motifs (E93ΔLLQHLL and E93ΔPLDLSAK) were transfected into Bm-12 cells. Compared with that of wild-type E93, the ability of E93ΔLLQHLL and E93ΔPLDLSAK to induce the expression of a subset of 20E response genes was increased 1.2–4-fold (Fig. 7, F and G).
In conclusion, as a HTH transcription factor, E93 can directly induce the expression of a subset of 20E response genes. The two HTH domains of E93 are critical for its transcriptional activity, whereas the LLQHLL and PLDLSAK motifs of E93 play inhibitory roles.
E93 Binds to EcR-USP via a Physical Interaction with USP through Its LLQHLL Motif
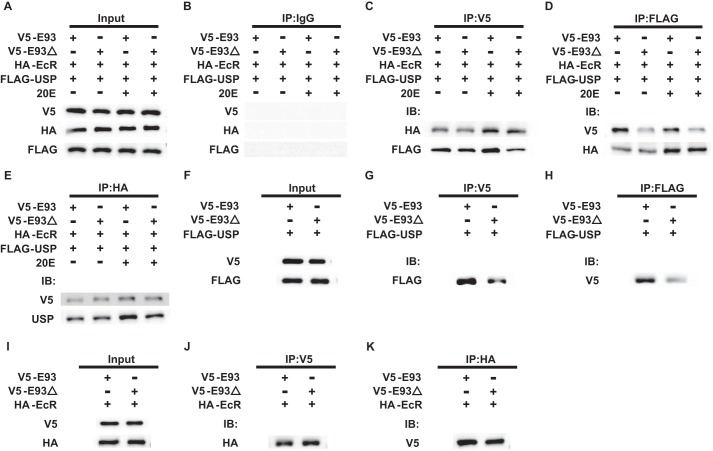
Because E93 contains the nuclear receptor interaction motif LLQHLL, we have investigated whether E93 directly binds to EcR and/or USP via LLQHLL and whether 20E affects this binding. HA-EcR, FLAG-USP, and V5-E93 (or V5-E93ΔLLQHLL) were co-transfected into Bm-N cells and the transfected cells were treated with 20E (or DMSO as a control). Co-immunoprecipitation experiments revealed that IgG (a negative control) binds to neither HA-EcR, FLAG-USP, V5-E93, nor V5-E93ΔLLQHLL; however, HA-EcR, FLAG-USP, and V5-E93 (or V5-E93ΔLLQHLL) bound to each other but the binding among these three proteins varied depending on the partners and the presence of 20E (Fig. 8, A–E). When V5-E93 was used as an input, its association with HA-EcR and FLAG-USP was enhanced by 20E. Interestingly, deletion of the LLQHLL motif from E93 reduced its interaction with both EcR and USP. Moreover, the 20E-enhanced interaction between EcR-USP with E93 was abolished when E93ΔLLQHLL was used in place of V5-E93. Compared with the association between E93 and USP, the association between E93 and EcR was not much affected by the deletion of LLQHLL motif of E93 (Fig. 8C). When FLAG-USP was used as an input, its association with HA-EcR was enhanced by 20E but not affected by the deletion of LLQHLL motif of E93. By contrast, the association between USP and E93 was not affected by 20E, whereas the association was much weaker when E93ΔLLQHLL was used instead of E93 (Fig. 8D). When HA-EcR was used as an input, its association with FLAG-USP or V5-E93 was enhanced by 20E but not affected by the LLQHLL motif of E93 (Fig. 8E). The co-immunoprecipitation experiments suggest that E93 binds to EcR-USP via a physical association with USP through its LLQHLL motif and that this association is enhanced by 20E-induced EcR-USP interaction.
FIGURE 8.
E93 binds to EcR-USP via a physical interaction with USP through its LLQHLL motif. A–E, Bm-N cells were co-transfected with the pEGFP-N1 constructs of HA-EcR, FLAG-USP, and V5-E93 (or V5-E93ΔLLQHLL) under the control of the BmNPV ie1 promoter for 48 h and treated with 2 μm 20E (or DMSO) for 12 h. Cell lysates were subjected to immunoprecipitation (IP) using IgG as a control (B), a V5 antibody for V5-E93 and V5-E93ΔLLQHLL (C), a FLAG antibody for FLAG-USP (D), and a HA antibody for HA-EcR (E), respectively; input panels represent 10% of the initial material (A). IB, immunoblot. F–H, Bm-N cells were co-transfected FLAG-USP and V5-E93 (or V5-E93ΔLLQHLL) for 48 h. Cell lysates were subjected to immunoprecipitation with a V5 antibody for V5-E93 (G) and a FLAG antibody for FLAG-USP (H), respectively; input panels represent 10% of the initial material (F). I–K, Bm-N cells were co-transfected HA-EcR and V5-E93 (or V5-E93ΔLLQHLL) for 48 h. Cell lysates were subjected to immunoprecipitation with a V5 antibody for V5-E93 (J) and a HA antibody for HA-EcR (K), respectively; input panels represent 10% of the initial material (I). Each experiment was repeated five times and a representative image from all the repeats is shown.
To verify the hypothesis that the LLQHLL motif of E93 directly binds to USP but not EcR, HA-EcR (or FLAG-USP) and V5-E93 (or V5-E93ΔLLQHLL) were co-transfected into Bm-N cells. Co-immunoprecipitation experiments confirmed that the physical association between V5-E93 and FLAG-USP (Fig. 8, F–H), but not between V5–93 and HA-EcR (Fig. 8, I–K), was dependent on the LLQHLL motif of E93. In addition, the composite co-immunoprecipitation experiments also showed that EcR and USP interact with E93 even in the absence of LLQHLL motif, albeit with lower affinities suggesting that other motifs of E93 also may play a role in these interactions.
EcR-USP Attenuates the Transcriptional Activity of E93 in the 1-kb Atg1 Promoter Region
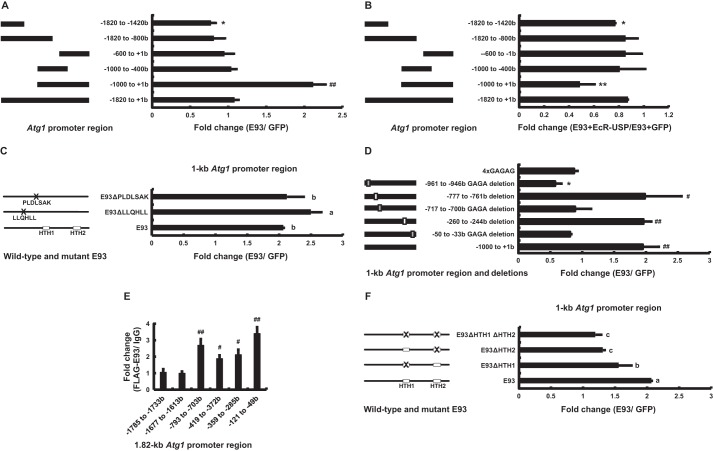
Using a dual luciferase assay system and HEK293 cells, we identified regions of the Atg1 promoter (24) that are essential for E93-induced Atg1 expression. The 1.8-kb Atg1 promoter region (−1820 to +1 bp upstream of the transcriptional start site) and a series of deletion constructs (−1820 to −1420 bp; −1820 to −800 bp; −600 to +1 bp; −1000 to −400 bp; −1000 to +1 bp) were cloned into the pGL3 vector. Upon E93 overexpression, only the −1000 to +1 bp region supported an ∼210% increase in luciferase activity (Fig. 9A). This −1000 to +1 bp region is referred to as the 1-kb Atg1 promoter region and was used in all further studies.
FIGURE 9.
The transcriptional activity of E93 in the Atg1 promoter region is both dependent and independent of 20E-EcR-USP. A, HEK293 cells were co-transfected with the E93 (GFP as a control) expression construct, the pGL3 basic plasmids containing the indicated promoter regions of Atg1, the hsp70 basal promoter regulating expression of firefly luciferase (Fluc) and a reference reporter plasmid carrying Renilla luciferase (Rluc). After 48 h of transfection, the dual luciferase assays were performed. Luciferase activity fold-change is defined as the relative luciferase activity induced by E93/GFP. B, HEK293 cells were co-transfected with the E93, EcR, and USP (E93 and GFP as a control) expression constructs, Fluc with the indicated promoter regions of Atg1, and Rluc. Luciferase activity fold-change is defined as the relative luciferase activity induced by E93 + EcR-USP/E93 + GFP. The others are the same as in A. C, HEK293 cells were co-transfected with the wild-type E93 or E93 mutant expression constructs (E93ΔLLQHLL and E93ΔPLDLSAK), Fluc with the 1-kb Atg1 promoter, and Rluc. The others are the same as in A. D, the mutated 1-kb Atg1 promoter regions, but not different Atg1 promoter regions, were used here, whereas the others are the same as in A. E, a ChIP assay of FLAG-E93 binding to the 1.82-kb region of ATG1 promoter. Bm-N cells were transfected with FLAG-E93 expression plasmid for 48 h, and cells were immunoprecipitated with IgG or antibodies against FLAG. Results of qPCR analyses are presented as FLAG-E93/IgG. F, HEK293 cells were co-transfected with the wild-type E93 or E93 mutant expression constructs (E93ΔHTH1, E93ΔHTH2, and E93ΔHTH1ΔHTH2), Fluc with the 1-kb Atg1 promoter, and Rluc. The others are the same as in A.
Importantly, EcR-USP attenuated the transcriptional activity of E93 mainly in the 1-kb Atg1 promoter region (Fig. 9B). Moreover, the transcriptional activity of E93ΔPLDLSAK in the 1-kb Atg1 promoter region was higher than E93 (Fig. 9C). Altogether, 20E-EcR-USP binds to E93 to attenuate its transcriptional activity, providing a negative feedback regulation loop.
E93 Acts through the Two HTH Domains to Bind to GAGA-containing Motifs Present in the Atg1 Promoter to Induce Gene Expression
A previous study (43) has shown that Pipsqueak, another HTH transcription factor in Drosophila, binds to GAGA sequences through its HTH domains, but it remains unknown whether Pipsqueak acts through GAGA-containing motifs in promoter regions to induce gene expression. Three GAGA sequences are present in the 1-kb Atg1 promoter region, and the deletion of any one of the GAGA sequences abolished E93-induced luciferase activity. By contrast, individual deletions of two unrelated sequences (−260 to −244 bp; −777 to −761 bp) in the 1-kb Atg1 promoter region had no effect on E93-induced luciferase activity. Notably, E93 was not able to activate luciferase activity driven by four copies of GAGA sequences (Fig. 9D). These experimental data reveal that the GAGA sequences alone in the 1-kb Atg1 promoter region are necessary but insufficient for E93-induced gene expression.
We then performed ChIP in Bm-N cells to examine whether E93 binds to the GAGA-containing nucleotides in the Atg1 promoter region. The binding of FLAG-E93 to DNA was detected using the FLAG antibody and cross-linked chromatin isolated from Bm-N cells, which were transfected with the FLAG-E93 expression plasmid. As measured by qPCR, the FLAG antibody increased precipitations of all four DNA fragments in the 1-kb Atg1 promoter region (−793 to −703 bp; −419 to −372 bp; −359 to −285 bp; −121 to −49 bp), but not those in the −1820 to −800 bp region (−1785 to −1733 bp; −1677 to −1613 bp) (Fig. 9E). Taken both the dual luciferase assays and ChIP-qPCR data together, we conclude that E93 acts through the GAGA-containing nucleotides in the 1-kb Atg1 promoter region to induce gene expression.
Furthermore, we confirmed the roles of the two HTH domains, as well as the LLQHLL and PLDLSAK motifs, in E93 in the induction of luciferase activity driven by the 1-kb Atg1 promoter region. In comparison with E93 overexpression, luciferase activity was significantly lower during overexpression of E93ΔHTH1 and E93ΔHTH2, and was nearly abolished during overexpression of E93ΔHTH1ΔHTH2 (Fig. 9F). These data suggest that E93 acts through the two HTH domains to bind to GAGA-containing motifs present in the Atg1 promoter to induce gene expression.
In summary, E93 is induced by 20E and suppressed by JH at the transcriptional level, and E93 transcriptionally modulates the 20E-triggered transcriptional cascade to promote larval tissue remodeling and adult tissue formation during Bombyx larval-pupal metamorphosis. Notably, the E93 action to modulate 20E signaling is both dependent and independent on 20E-EcR-USP (Fig. 10).
FIGURE 10.
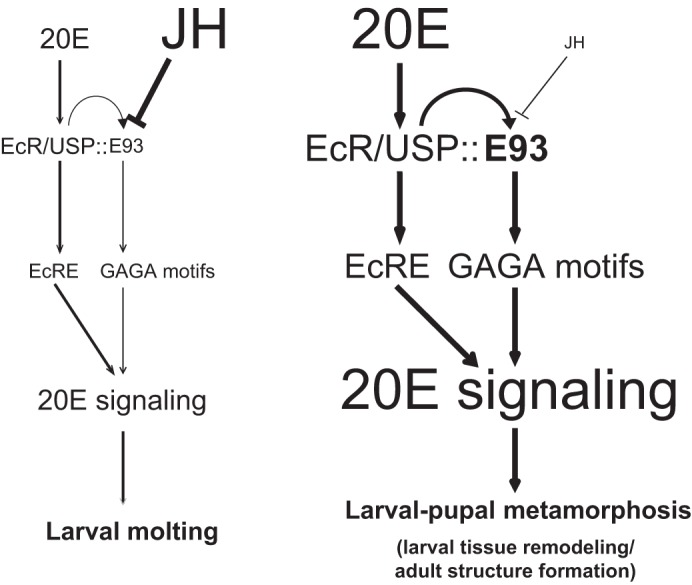
A model: E93 transcriptionally modulates 20E signaling to promote Bombyx larval-pupal metamorphosis. Induced by 20E and suppressed by JH, E93 is abundantly expressed during the larval-pupal metamorphosis. At this stage, E93 acts through GAGA-containing motifs to induce expression of a subset of 20E-response genes, positively affects 20E signaling, and promotes larval tissue remodeling and adult tissue formation. The expression level of E93 is induced by 20E-EcR-USP, but the transcriptional activity of E93 is attenuated by its physical association with USP. Notably, the E93 action to modulate 20E signaling is both dependent and independent on 20E-EcR-USP. Text and arrow sizes convey magnitude of signal transduction.
Discussion
The microarray (21, 22) and qPCR (Fig. 1A) analyses revealed a unique developmental profile of E93 expression in the Bombyx fat body, with markedly low expression prior to the wandering stage and abundant expression during larval-pupal metamorphosis. The unique expression pattern of E93 in the fat body was not limited to the tetramolter allele but was also observed in the pentamolter and trimolter alleles (Fig. 3, A–A″). Similarly, E93 mRNA levels in Drosophila were underdetectable from the embryonic stage to the late third instar, whereas E93 is expressed from the wandering stage to the adult stage with a peak at the prepupal stage (3, 5, 6). Detailed studies revealed that the developmental expression pattern of E93 is tissue-specific, peaking in the fat body during larval-pupal metamorphosis (13), in the midgut a few hours after pupariation (14, 15), and in the salivary gland immediately after pupation (16–18). Previous studies in Drosophila indicate that E93 is a 20E primary-response gene and its expression is suppressed by JH (3, 5, 6).
Similarly, induced by 20E and suppressed by JH, E93 is abundantly expressed in the fat body during Bombyx larval-pupal metamorphosis. Along with EcR-USP, E93 is also involved in the primary response to 20E (Fig. 2, A–C). A gene structure analysis of Bombyx E93 revealed six exons. The first and third introns are large, composed of 18.4 and 16.6 kb, respectively (not shown). A potential EcRE (AGTTCAATGGCCT) is present in the 0.3-kb promoter region of E93 (not shown), supporting that E93 is a 20E primary response gene.
Moreover, JH suppresses both the basal and 20E-induced expression levels of E93 in both cultured Bombyx cells (Fig. 2D) and fat body tissues isolated from silkworms of multiple alleles (Fig. 3). E93 expression appears to be suppressed by JH via Met and Kr-h1 in several other insects (5, 6). Because one major function of JH is to suppress 20E action to prevent premature metamorphosis during larval molting (44), the detailed molecular mechanism of JH suppression of 20E-induced E93 expression merits further investigation.
Our experimental data in Bombyx suggest that a high level of JH during larval molting inhibits 20E-induced E93 expression and that a low level of JH during larval-pupal metamorphosis fails to inhibit 20E-induced E93 expression. In conclusion, E93, which is coordinately controlled by both 20E and JH, is abundantly expressed in the fat body during larval-pupal metamorphosis and promotes 20E-induced larval-pupal metamorphosis (Fig. 10).
In this study, we determined that E93 promotes fat body remodeling (including autophagy, caspase activity, and cell dissociation) during Bombyx larval-pupal metamorphosis (Fig. 4, B–E). Similarly, E93 is the key determinant required for 20E-induced PCD in multiple larval tissues during Drosophila metamorphosis (13–18). We also showed that E93 determines the growth and differentiation of the adult wing discs during larval-pupal metamorphosis in Bombyx (Fig. 6, A and B), supporting the role of E93 as the adult specifier in insects (6). The importance of this role for E93 is reflected in the fact that silkworms subjected to E93 RNAi (Fig. 4, B and B′) and E93 null mutants of Drosophila die during metamorphosis (16). Overall, in Bombyx, the phenotypic changes caused by E93 RNAi are similar to those caused by RNAi of several key genes involved in the 20E signal transduction pathway (21, 23–25, 38). Consistently, E93 RNAi disrupted 20E signaling (Figs. 5, B and B′, and 6, A′ and B′) and decreased the expression of several 20E-induced Atg and apoptosis genes in the fat body (Fig. 5, C and D) as well as pupal-specific WCPs in the wing disc (Fig. 6, A″ and B″). In addition, E93 induces a large number of 20E response genes, including Atg genes and apoptosis genes, in Drosophila (13, 16).
Thus, Bombyx E93 appears to be a key component in the modulation of 20E signaling. We further determined that E93 modulates 20E signaling through two mechanisms in Bombyx. First, E93 modulates 20E signaling by maintaining ecdysteroid titers (Fig. 5A). Several 20E response genes in Drosophila, including E75 (39), βftz-F1 (40), and Br-C (41, 42), are essential for maintaining the ecdysteroid titer. Using the binary GAL4/UAS system to specifically reduce E93 expression or overexpress E93 in the prothoracic gland, one should be able to clarify whether E93 is also required for maintaining the ecdysteroid titer in Drosophila. Second, E93 is required for the basal and 20E-induced expression of 20E response genes (Figs. 5, B and B′, 6, A′ and B′, and 7A), and E93 directly induces the expression of a subset of 20E response genes in Bombyx (Fig. 7B). Likewise, E93 induces the expression of multiple 20E response genes in Drosophila (13, 16). Altogether, E93 likely modulates 20E signaling through these two mechanisms in all insects.
E93 binds to the regulatory regions of many 20E response genes on polytene chromosomes in Drosophila (16). All E93 orthologs share significant similarity within, but not outside, the two HTH domains (Fig. 1D). Because E93 directly induces the expression of a subset of 20E response genes without 20E-EcR-USP (Fig. 7B), we further investigated the detailed molecular mechanism by which the HTH transcription factor E93 regulates gene expression in Bombyx. Importantly, the two HTH domains of E93 were shown to be indispensable for its transcriptional activity. The deletion of one or both HTH domains attenuated the ability of E93 to induce gene expression (Fig. 7, C–E) and luciferase activity of the 1-kb Atg1 promoter region (Fig. 9F). As revealed by dual luciferase assays in HEK293 cells and ChIP-qPCR in Bm-N cells, E93 acts through the two HTH domains to bind to GAGA-containing motifs present in the 1-kb Atg1 promoter region to induce gene expression, although GAGA sequences alone are necessary but insufficient for E93-induced Atg1 expression (Fig. 9, D and E). These results indicate that E93 is a bona fide transcriptional factor in inducing Atg1 expression, and the transcriptional activity of E93 could be somehow independent on 20E-EcR-USP.
Importantly, 20E-EcR-USP not only induces E93 expression at the transcriptional level (Fig. 2), but also modulates the transcriptional activity of E93 via protein-protein interaction and protein-DNA interaction. First, E93 binds to EcR-USP via a physical association with USP through its LLQHLL motif and that this association is enhanced by 20E-induced EcR-USP interaction, presumably by affecting the conformation of E93 (Fig. 8). Second, EcR-USP attenuated the transcriptional activity of E93 in the 1-kb Atg1 promoter region (Fig. 9B). Third, deletion of the LLQHLL motif increased the ability of E93 to induce gene expression (Fig. 6F) and luciferase activity (Fig. 9C), indicating that the LLQHLL motif exerts an inhibitory effect on the transcriptional activity of E93. Finally, the 1.8-kb Atg1 promoter region contains both a EcRE (24) and four copies of GAGA sequences (Fig. 9D), indicating that 20E-EcR-USP and E93 co-occupy the Atg1 promoter. Taken together, we conclude that the expression level of E93 was induced by 20E-EcR-USP, but the transcriptional activity of E93 was attenuated by its physical association with USP (Fig. 10). The inhibitory effect of 20E-EcR-USP on E93 transcriptional activity could be protective for E93-exaggerated cell death and cell differentiation during larval-pupal metamorphosis. In addition, deletion of the PLDLSAK motif also increased the ability of E93 to induce gene expression (Fig. 7G), in agreement with a possible physical interaction between the PXDL(S/T)X(K/R) motif and the transcriptional co-repressor CtBP (37).
The GAGA factor and Pipsqueak in Drosophila, which were initially identified as two HTH transcriptional factors that bind to GAGA-containing motifs, were later also found to act through a chromatin remodeling mechanism (45, 46). Because each E93 contains a LXXLL motif, it will be interesting to determine whether E93 might act as a chromatin remodeling factor to modulate the transcriptional activity of EcR-USP and other nuclear receptors. In summary, E93 acts through the GAGA-containing motifs in promoter regions to directly induce the expression of a subset of 20E response genes, and thus, E93 positively affects 20E signaling during Bombyx larval-pupal metamorphosis (Fig. 10).
Author Contributions
S. L. conceived and coordinated the study and wrote the manuscript. X. L. performed and analyzed most of the experiments, wrote the first draft. D. F., E. G., K. L., L. M., L. T., Y. C., and G. Z. performed some experiments or analyzed some data, presented important reagents. S. P. and G. Z. helped design the study and improved the manuscript. All authors reviewed the results and approved the final version of the manuscript.
Acknowledgments
We thank researchers in Sericultural Research Institute, Chinese Academy of Agricultural Sciences and Southwest University for providing the silkworms. English was polished by Nature Publishing Group.
This study was supported by National Science Foundation of China Grant 31330072, 973 program Grant 2012CB114605, and National Program for the Development of New Transgenic Species of China Grant 2014ZX08010-016B (to S. L.), National Science Foundation of China Grants 31472042 (to L. T.) and 31201747 (to L. M.), 863 Program Grant 2013AA102507 (to F. D.), and State Key Laboratory of Silkworm Genome Biology Grant 20120003 (to S. L.). The authors declare that they have no conflicts of interest with the contents of this article.
- 20E
- 20-hydroxyecdysone
- EcRE
- 20E response elements
- JH
- juvenile hormone
- HTH
- helix-turn-helix
- PCD
- programmed cell death
- WCP
- wing disc cuticle protein genes
- IW
- the initiation of wandering
- EcRDN
- dominant-negative mutant of EcR
- RACE
- rapid-amplification of cDNA ends
- qPCR
- quantitative real-time PCR
- TEM
- transmission electron microscopy
- CHX
- cycloheximide
- USP
- ultraspiracle protein
- DMSO
- dimethyl sulfoxide.
References
- 1.Yamanaka N., Rewitz K. F., and O'Connor M. B. (2013) Ecdysone control of developmental transitions: lessons from Drosophila research. Annu. Rev. Entomol. 58, 497–516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Riddiford L. M., Cherbas P., and Truman J. W. (2000) Ecdysone receptors and their biological actions. Vitam. Horm. 60, 1–73 [DOI] [PubMed] [Google Scholar]
- 3.Baehrecke E. H., and Thummel C. S. (1995) The Drosophila E93 gene from the 93F early puff displays stage- and tissue-specific regulation by 20-hydroxyecdysone. Dev. Biol. 171, 85–97 [DOI] [PubMed] [Google Scholar]
- 4.Siegmund T., and Lehmann M. (2002) The Drosophila Pipsqueak protein defines a new family of helix-turn-helix DNA-binding proteins. Dev. Genes Evol. 212, 152–157 [DOI] [PubMed] [Google Scholar]
- 5.Belles X., and Santos C. G. (2014) The MEKRE93 (Methoprene tolerant-Krüppel homolog 1-E93) pathway in the regulation of insect metamorphosis, and the homology of the pupal stage. Insect Biochem. Mol. Biol. 52, 60–68 [DOI] [PubMed] [Google Scholar]
- 6.Ureña E., Manjón C., Franch-Marro X., and Martín D. (2014) Transcription factor E93 specifies adult metamorphosis in hemimetabolous and holometabolous insects. Proc. Natl. Acad. Sci. U.S.A. 111, 7024–7029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zitnan D., Kim Y. J., Zitnanová I., Roller L., and Adams M. E. (2007) Complex steroid-peptide-receptor cascade controls insect ecdysis. Gen. Comp. Endocrinol. 153, 88–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yin V. P., and Thummel C. S. (2005) Mechanisms of steroid-triggered programmed cell death in Drosophila. Semin. Cell Dev. Biol. 16, 237–243 [DOI] [PubMed] [Google Scholar]
- 9.Hay B. A., and Guo M. (2006) Caspase-dependent cell death in Drosophila. Annu. Rev. Cell Dev. Biol. 22, 623–650 [DOI] [PubMed] [Google Scholar]
- 10.Ryoo H. D., and Baehrecke E. H. (2010) Distinct death mechanisms in Drosophila development. Curr. Opin. Cell Biol. 22, 889–895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee C. Y., Clough E. A., Yellon P., Teslovich T. M., Stephan D. A., and Baehrecke E. H. (2003) Genome-wide analyses of steroid- and radiation-triggered programmed cell death in Drosophila. Curr. Biol. 13, 350–357 [DOI] [PubMed] [Google Scholar]
- 12.Liu H., Jia Q., Tettamanti G., and Li S. (2013) Balancing crosstalk between 20-hydroxyecdysone-induced autophagy and caspase activity in the fat body during Drosophila larval-prepupal transition. Insect Biochem. Mol. Biol. 43, 1068–1078 [DOI] [PubMed] [Google Scholar]
- 13.Liu H., Wang J., and Li S. (2014) E93 predominantly transduces 20-hydroxyecdysone signaling to induce autophagy and caspase activity in Drosophila fat body. Insect Biochem. Mol. Biol. 45, 30–39 [DOI] [PubMed] [Google Scholar]
- 14.Lee C. Y., and Baehrecke E. H. (2001) Steroid regulation of autophagic programmed cell death during development. Development 128, 1443–1455 [DOI] [PubMed] [Google Scholar]
- 15.Lee C. Y., Cooksey B. A., and Baehrecke E. H. (2002) Steroid regulation of midgut cell death during Drosophila development. Dev. Biol. 250, 101–111 [DOI] [PubMed] [Google Scholar]
- 16.Lee C. Y., Wendel D. P., Reid P., Lam G., Thummel C. S., and Baehrecke E. H. (2000) E93 directs steroid-triggered programmed cell death in Drosophila. Mol. Cell 6, 433–443 [DOI] [PubMed] [Google Scholar]
- 17.Lee C. Y., Simon C. R., Woodard C. T., and Baehrecke E. H. (2002) Genetic mechanism for the stage- and tissue-specific regulation of steroid triggered programmed cell death in Drosophila. Dev. Biol. 252, 138–148 [DOI] [PubMed] [Google Scholar]
- 18.Berry D. L., and Baehrecke E. H. (2007) Growth arrest and autophagy are required for salivary gland cell degradation in Drosophila. Cell 131, 1137–1148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mou X., Duncan D. M., Baehrecke E. H., and Duncan I. (2012) Control of target gene specificity during metamorphosis by the steroid response gene E93. Proc. Natl. Acad. Sci. U.S.A. 109, 2949–2954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xia Q., Li S., and Feng Q. (2014) Advances in silkworm studies accelerated by the genome sequencing of Bombyx mori. Annu. Rev. Entomol. 59, 513–536 [DOI] [PubMed] [Google Scholar]
- 21.Tian L., Guo E., Diao Y., Zhou S., Peng Q., Cao Y., Ling E., and Li S. (2010) Genome-wide regulation of innate immunity by juvenile hormone and 20-hydroxyecdysone in the Bombyx fat body. BMC Genomics 11, 549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tian L., Guo E., Wang S., Liu S., Jiang R. J., Cao Y., Ling E., and Li S. (2010) Developmental regulation of glycolysis by 20-hydroxyecdysone and juvenile hormone in fat body tissues of the silkworm, Bombyx mori. J. Mol. Cell Biol. 2, 255–263 [DOI] [PubMed] [Google Scholar]
- 23.Tian L., Liu S., Liu H., and Li S. (2012) 20-hydroxyecdysone upregulates apoptotic genes and induces apoptosis in the Bombyx fat body. Arch. Insect Biochem. Physiol. 79, 207–219 [DOI] [PubMed] [Google Scholar]
- 24.Tian L., Ma L., Guo E., Deng X., Ma S., Xia Q., Cao Y., and Li S. (2013) 20-Hydroxyecdysone upregulates Atg genes to induce autophagy in the Bombyx fat body. Autophagy 9, 1172–1187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guo E., He Q., Liu S., Tian L., Sheng Z., Peng Q., Guan J., Shi M., Li K., Gilbert L. I., Wang J., Cao Y., and Li S. (2012) MET is required for the maximal action of 20-hydroxyecdysone during Bombyx metamorphosis. PLoS ONE 7, e53256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Noji T., Ote M., Takeda M., Mita K., Shimada T., and Kawasaki H. (2003) Isolation and comparison of different ecdysone-responsive cuticle protein genes in wing discs of Bombyx mori. Insect Biochem. Mol. Biol. 33, 671–679 [DOI] [PubMed] [Google Scholar]
- 27.Wang H. B., Iwanaga M., and Kawasaki H. (2009) Activation of BMWCP10 promoter and regulation by BR-C Z2 in wing disc of Bombyx mori. Insect Biochem. Mol. Biol. 39, 615–623 [DOI] [PubMed] [Google Scholar]
- 28.Wang H. B., Nita M., Iwanaga M., and Kawasaki H. (2009) betaFTZ-F1 and Broad-Complex positively regulate the transcription of the wing cuticle protein gene, BMWCP5, in wing discs of Bombyx mori. Insect Biochem. Mol. Biol. 39, 624–633 [DOI] [PubMed] [Google Scholar]
- 29.Deng H., Zheng S., Yang X., Liu L., and Feng Q. (2011) Transcription factors BmPOUM2 and BmβFTZ-F1 are involved in regulation of the expression of the wing cuticle protein gene BmWCP4 in the silkworm, Bombyx mori. Insect Mol. Biol. 20, 45–60 [DOI] [PubMed] [Google Scholar]
- 30.Deng H., Zhang J., Li Y., Zheng S., Liu L., Huang L., Xu W. H., Palli S. R., and Feng Q. (2012) Homeodomain POU and Abd-A proteins regulate the transcription of pupal genes during metamorphosis of the silkworm, Bombyx mori. Proc. Natl. Acad. Sci. U.S.A. 109, 12598–12603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ali M. S., Iwanaga M., and Kawasaki H. (2013) Ecdysone-responsive transcriptional regulation determines the temporal expression of cuticular protein genes in wing discs of Bombyx mori. Gene 512, 337–347 [DOI] [PubMed] [Google Scholar]
- 32.Hossain M. S., Liu Y., Zhou S., Li K., Tian L., and Li S. (2013) 20-Hydroxyecdysone-induced transcriptional activity of FoxO upregulates brummer and acid lipase-1 and promotes lipolysis in Bombyx fat body. Insect Biochem. Mol. Biol. 43, 829–838 [DOI] [PubMed] [Google Scholar]
- 33.Li K., Guo E., Hossain M. S., Li Q., Cao Y., Tian L., Deng X., and Li S. (2015) Bombyx E75 isoforms display stage- and tissue-specific response to 20-hydroxyecdysone. Sci. Rep. 5, 12114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Khurad A. M., Zhang M. J., Deshmukh C. G., Bahekar R. S., Tiple A. D., and Zhang C. X. (2009) A new continuous cell line from larval ovaries of silkworm, Bombyx mori. In Vitro Cell Dev. Biol. Anim. 45, 414–419 [DOI] [PubMed] [Google Scholar]
- 35.He Q., Wen D., Jia Q., Cui C., Wang J., Palli S. R., and Li S., (2014) Heat shock protein 83 (Hsp83) facilitates methoprene-tolerant (Met) nuclear import to modulate juvenile hormone signaling. J. Biol. Chem. 289, 27874–27885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Heery D. M., Kalkhoven E., Hoare S., and Parker M. G. (1997) A signature motif in transcriptional coactivators mediates binding to nuclear receptors. Nature 387, 733–736 [DOI] [PubMed] [Google Scholar]
- 37.Vo N., Fjeld C., and Goodman R. H. (2001) Acetylation of nuclear hormone receptor-interacting protein RIP140 regulates binding of the transcriptional corepressor CtBP. Mol. Cell Biol. 21, 6181–6188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cheng D., Xia Q., Duan J., Wei L., Huang C., Li Z., Wang G., and Xiang Z. (2008) Nuclear receptors in Bombyx mori: insights into genomic structure and developmental expression. Insect Biochem. Mol. Biol. 38, 1130–1137 [DOI] [PubMed] [Google Scholar]
- 39.Bialecki M., Shilton A., Fichtenberg C., Segraves W. A., and Thummel C. S. (2002) Loss of the ecdysteroid-inducible E75A orphan nuclear receptor uncouples molting from metamorphosis in Drosophila. Dev. Cell 3, 209–220 [DOI] [PubMed] [Google Scholar]
- 40.Parvy J. P., Blais C., Bernard F., Warren J. T., Petryk A., Gilbert L. I., O'Connor M. B., and Dauphin-Villemant C. (2005) A role for βFTZ-F1 in regulating ecdysteroid titers during post-embryonic development in Drosophila melanogaster. Dev. Biol. 282, 84–94 [DOI] [PubMed] [Google Scholar]
- 41.Xiang Y., Liu Z., and Huang X. (2010) br regulates the expression of the ecdysone biosynthesis gene npc1. Dev. Biol. 344, 800–808 [DOI] [PubMed] [Google Scholar]
- 42.Moeller M. E., Danielsen E. T., Herder R., O'Connor M. B., and Rewitz K. F. (2013) Dynamic feedback circuits function as a switch for shaping a maturation-inducing steroid pulse in Drosophila. Development 140, 4730–4739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lehmann M., Siegmund T., Lintermann K. G., and Korge G. (1998) The pipsqueak protein of Drosophila melanogaster binds to GAGA sequences through a novel DNA-binding domain. J. Biol. Chem. 273, 28504–28509 [DOI] [PubMed] [Google Scholar]
- 44.Jindra M., Palli S. R., and Riddiford L. M. (2013) The juvenile hormone signaling pathway in insect development. Annu. Rev. Entomol. 58, 181–204 [DOI] [PubMed] [Google Scholar]
- 45.Benyajati C., Mueller L., Xu N., Pappano M., Gao J., Mosammaparast M., Conklin D., Granok H., Craig C., and Elgin S. (1997) Multiple isoforms of GAGA factor, a critical component of chromatin structure. Nucleic Acids Res. 25, 3345–3353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schwendemann A., and Lehmann M. (2002) Pipsqueak and GAGA factor act in concert as partners at homeotic and many other loci. Proc. Natl. Acad. Sci. U.S.A. 99, 12883–12888 [DOI] [PMC free article] [PubMed] [Google Scholar]