Background: Kinetochore microtubule dynamics orchestrate proper chromosome movement in mitosis.
Results: CSPP1 is a novel kinetochore protein that regulates chromosome oscillation during cell division.
Conclusion: CSPP1 associates with CENP-H to control chromosome oscillation by regulating kinetochore microtubule dynamics.
Significance: Our findings provide insight into the molecular mechanism of chromosome movements.
Keywords: kinetochore, microtubule, microtubule-associated protein (MAP), mitosis, mitotic spindle, phosphorylation, kinase
Abstract
Mitotic chromosome segregation is orchestrated by the dynamic interaction of spindle microtubules with the kinetochores. During chromosome alignment, kinetochore-bound microtubules undergo dynamic cycles between growth and shrinkage, leading to an oscillatory movement of chromosomes along the spindle axis. Although kinetochore protein CENP-H serves as a molecular control of kinetochore-microtubule dynamics, the mechanistic link between CENP-H and kinetochore microtubules (kMT) has remained less characterized. Here, we show that CSPP1 is a kinetochore protein essential for accurate chromosome movements in mitosis. CSPP1 binds to CENP-H in vitro and in vivo. Suppression of CSPP1 perturbs proper mitotic progression and compromises the satisfaction of spindle assembly checkpoint. In addition, chromosome oscillation is greatly attenuated in CSPP1-depleted cells, similar to what was observed in the CENP-H-depleted cells. Importantly, CSPP1 depletion enhances velocity of kinetochore movement, and overexpression of CSPP1 decreases the speed, suggesting that CSPP1 promotes kMT stability during cell division. Specific perturbation of CENP-H/CSPP1 interaction using a membrane-permeable competing peptide resulted in a transient mitotic arrest and chromosome segregation defect. Based on these findings, we propose that CSPP1 cooperates with CENP-H on kinetochores to serve as a novel regulator of kMT dynamics for accurate chromosome segregation.
Introduction
Mitosis is an orchestration of dynamic interactions between chromosomes and spindle microtubules by which genomic materials are equally distributed into two daughter cells (1). Deregulation of the accurate chromosome segregation during mitosis results in aneuploidy of daughter cells, which further promotes chromosome instability and tumorigenesis (2, 3). Accurate chromosome alignment at the spindle equator is a precondition for equal segregation of sister chromatids. The fine regulation of kinetochore microtubule (kMT)4 dynamics and kinetochore-MT attachment is essential for full chromosome alignment (4, 5). Moreover, aligned chromosomes oscillate back and forth within a confined region around the metaphase plate to achieve a synchronized separation at the following anaphase onset (6). Several kinetochore proteins such as CENP-H, SKAP, CENP-E, Kif18A, and MCAK participate in the regulation of chromosome oscillation (7–12). However, the precise mechanisms underlying the oscillation are not well understood. It has been proposed that oscillations are driven by coordinated MT polymerization at the trailing kinetochores and MT depolymerization at the leading kinetochores (13). Again, how the coordination between the dynamic MTs at sister kinetochores is achieved remains elusive. Furthermore, tubulin turnover at plus ends of kMTs is reported to be an order of magnitude slower than that of non-kMTs (14), suggesting a regulation of kMT dynamics by kinetochores.
The centrosome and spindle pole-associated protein 1 (CSPP1) gene was first identified as a gene overexpressed in the malignant transformation of large B-cell lymphoma (15). Multiple CSPP1 transcript isoforms exist with the cell type-specific expression profile (15, 16). So far, two CSPP1 protein isoforms, CSPP-S and CSPP-L (In this work, we study the longer isoform, CSPP-L.), were biochemically characterized (15–18). Although both isoforms localize at the centrosome in interphase and at the mitotic spindle in mitosis, HeLa cells predominantly express the CSPP1-L isoform (16–18). In addition, CSPP1 isoforms exhibit cell cycle-dependent expression. For example, CSPP-S expression is highest in G1 phase. However, CSPP1 mRNA peaks in G2/M phase, suggesting that the level of CSPP1 protein in mitosis is precisely regulated. Either suppression or overexpression of CSPP1 causes mitotic defects, suggesting that a precisely regulated CSPP1 activity is essential for accurate mitotic progression (16). CSPP1 also promotes the completion of cytokinesis by recruiting guanine nucleotide exchange factor MyoGEF to the central spindle (17). Beyond mitosis and cytokinesis, CSPP1 is required for ciliogenesis (18). Consistent with its role in ciliogenesis, mutations in CSPP1 were detected in individuals with ciliopathy phenotypes, including Joubert syndrome and Meckel-Gruber syndrome (19–21).
In this study, we elucidated the molecular mechanism by which CSPP1 functions in mitosis. We found that CSPP1 is a novel kinetochore protein, and its kinetochore localization requires Aurora B kinase. Consistent with our finding that the kinetochore recruitment of CSPP1 depends on CENP-H, depletions of both CENP-H and CSPP1 enormously impair chromosome oscillation. Importantly, our study showed that CSPP1 suppresses kMT dynamics as depletion of CSPP1 increases the velocity of kinetochore movement, and overexpression of CSPP1 suppressed the kinetochore velocity. Thus, CSPP1 participates in the regulation of faithful mitotic progression by coordinating fine chromosome oscillation.
Materials and Methods
Plasmids
GFP-CSPP1 was a kind gift from Dr. Hans-Christian Aasheim. For 3×FLAG-CSPP1, full-length cDNA was amplified with a primer set of 5′-gccagatctgatgctgttcccgctccaggtggccg-3′ and 5′-gccctcgagttaaccatgtgcagtcgacaggccctg-3′, followed by cloning into p3×FLAG-myc-CMV-24 vector (Sigma). GFP-tagged CENP-H and CENP-B (in pEGFP-C2 vector), GST-CENP-H (in pGEX-6p-1 vector), GFP-Aurora B, GFP-Mad2, GFP-tagged α-tubulin, and mCherry-tagged H2B (mCherry-H2B fusion was cloned into pcDNA3.1-B vector) were also constructed and used in this study.
Expression and Purification of Recombinant Proteins
Purification of recombinant proteins was carried out as described previously (11). Briefly, the GST fusion proteins from bacteria in the soluble fraction were purified using glutathione-agarose chromatography, Histidine-tagged proteins were purified using nickel-nitrilotriacetic acid-agarose beads.
For introducing TAT-GFP fusion proteins to probe the functional relevance of the CENP-H/CSPP1 interaction, aliquots of synchronized HeLa cells (50% confluency) were released into G2 phase (8 h after thymidine wash-off) before the addition of TAT-GFP peptides (2.5 μm; TAT-GFP as control; TAT-GFP-CSPP1-C as disrupting peptide) at 37 °C for 1 h before image collection. After incubation, the cells were washed with PBS and then examined directly under a fluorescence microscopy as described previously (22).
Antibodies and siRNAs
To generate antibody against human CSPP1, mice were immunized with His6-tagged CSPP1-M (295–848 amino acids) purified from Escherichia coli Rosetta (DE3) strain by affinity purification and gel filtration. IgG was purified from diluted serum with protein A/G beads (Thermo Scientific) followed by acid elution with 0.1 m glycine, pH 2.5, and neutralization with Tris buffer. Purified IgG was concentrated with Amicon Ultra-4 device (Millipore) and stored in the presence of 30% glycerol. Other primary antibodies used in this study are as follows: ACA (gift from Don W. Cleveland, University of California at San Diego); anti-CENP-H antibody (ab77207, Abcam); anti-CENP-I rabbit antibody (A303-374A, Bethyl Laboratories Inc.); anti-CENP-F rabbit antibody (ab5, Abcam); anti-CENP-A-pS7 rabbit antibody (A13968, Invitrogen); and anti-Hec1 mouse antibody (9G3.23, GeneTex). Anti-FLAG antibody (F1804), anti-tubulin antibody (DM1A, T9026), anti-GFP antibody (G6539), and anti-Mad2 mouse antibody (M8694) were purchased from Sigma. Anti-Aurora B antibody (611082) and anti-BubR1 antibody (612503) were purchased from BD Biosciences.
CSPP1 siRNA (synthesized by Qiagen) targeting to 5′-GAAGATTTGCGCAGTGGAC-3′ was used, as described previously (15). Another CSPP1 siRNA purchased from Santa Cruz Biotechnology (sc-77463) was also used in our work. Hec1 siRNA (AAGUUCAAAAGCUGGAUGAUCUU), Aurora B siRNA (AACGCGGCACUUCACAAUUGA), and CENP-F siRNA (AAGAGAAGACCCCAAGUCAUC) were purchased or synthesized in Dharmacon. CENP-H siRNA (CAGAGAGGAUAAAGAUCAUACGACA) and CENP-I siRNA (AACAAACCAUUUCGUGTGAGA) were purchased or synthesized in Qiagen.
Cell Culture, Transfection, and Drug Treatments
HeLa cells, from American Type Culture Collection (ATCC), were cultured and maintained in advanced Dulbecco's modified Eagle's medium (DMEM, Gibco) with 10% fetal bovine serum (FBS, HyClone) and 2 mm glutamine (Gibco) at 37°C with 8% CO2. Cells were transfected with plasmids or siRNA (usually 100 nm), using Lipofectamine 2000 (Invitrogen). For mitotic synchronization, cells were blocked at G1/S phase in 2 mm thymidine (Sigma) for 16 h, followed by release in fresh medium for 7–9 h. In some specific experiments, cells were treated with the indicated inhibitors for another 1 or 2 h, wherein 100 ng/ml nocodazole was used to depolymerize MT; 10 ng/ml nocodazole was then applied to generate misaligned kinetochores. Hesperadins was used at 100 nm; MG132 was used at 20 μm; and okadaic acid (OA) was used at 100 nm.
Immunofluorescence Microscopy and Data Analysis
Cells grown on coverslips were fixed with cold methanol at −20 °C for 3 min or with 3.7% formaldehyde at 37 °C for 10 min. After formaldehyde fixation, cells were subsequently permeabilized with 0.1% Triton X-100 in PBS at room temperature for 10 min. Following a 30-min block in 1% BSA and incubation with primary antibodies at room temperature for 1 h, cells were incubated with secondary antibodies for another 1 h. After 1 min of staining with 4,6-diamidino-2-phenylindole (DAPI, Sigma), cells were mounted. Images were acquired using Olympus 60×/1.42 Plan APO N objective on an Olympus IX71 microscope (Applied Precision Inc.) or LSM 710 confocal microscope (Carl Zeiss). Deconvoluted images from each focal plane were projected into a single stack montage using SoftWorx software (Applied Precision). In some cases, images of a single focal plane are shown in the figures. For fluorescence intensity quantification, the kinetochore staining signals were measured with ImageJ software, according to the procedures described previously (8, 23).
Live Cell Microscopy and Data Analysis
Live cell imaging was performed as described previously (24, 25). Cells were cultured in CO2-independent medium plus 10% FBS and 2 mm glutamine at 37 °C. In cell cycle progression imaging, HeLa cells expressing mCherry-H2B and GFP-α-tubulin were transfected with the indicated siRNA(s) and synchronized. Images were acquired from nuclear envelope breakdown (NEBD) with 3- or 5-min intervals under the Olympus DeltaVision microscope. In the tracking of oscillatory kinetochores around the metaphase plate, siRNA-transfected cells expressing GFP-CENP-B, as kinetochore marker, were imaged every 3 or 10 s under the Olympus DeltaVision microscope or LSM confocal microscope (Carl Zeiss). Sister kinetochore positions of each kinetochore pair were calculated and presented as relative to the mean position of the sister kinetochore center within the imaging duration. The amplitude of oscillations were assessed by calculating the standard deviations of relative sister kinetochore positions and sister kinetochore separations. The velocity of sister kinetochore movement along the spindle axis was also calculated, as an indicator for the rate of kMT plus end turnover.
Immunoprecipitation and GST Pulldown
For immunoprecipitations, HeLa or HEK293T cells, transfected with indicated plasmids, were collected and lysed in lysis buffer (50 mm Tris-HCl, pH 7.4, 150 mm NaCl, 1 mm EDTA, and 0.1% Triton X-100), in the presence of protease inhibitor mixture (Sigma). After clarification by centrifugation, cell lysates were incubated with FLAG M2 beads or GFP-Trap microbeads at 4 °C rotating for 4 h. In immunoprecipitation of endogenous CSPP1, HeLa cell lysates were incubated with IgG or CSPP1 antibody at 4 °C rotating for 4 h, followed by extended incubation with protein A/G microbeads for another 1 h.
As to pulldown assays, transiently transfected HEK293T cells were collected and lysed in the lysis buffer. Cleared cell lysates or bacterially purified proteins were incubated with purified GST or GST-fusion proteins bound on glutathione-Sepharose 4B beads at 4 °C rotating for 2 h. After three washes in PBS containing 0.2% Triton X-100, beads were boiled and applied to SDS-PAGE, followed by subsequent Western blotting analysis.
Purification of Recombinant TAT-GFP Proteins and Interrogation of CENP-H-CSPP1 Interaction in Vivo
To directly assess the functional effect of the CENP-H/CSPP1 interaction in mitosis, a membrane-permeable peptide containing CSPP1(934–1221) was constructed. This was achieved by introducing an 11-amino acid peptide derived from the TAT protein transduction domain into a fusion protein containing amino acids involving binding interface between CSPP1 and CENP-H, as described previously (22, 26). Trial experiments were employed to determine the optimal concentration to perturb CSPP1/CENP-H interaction in HeLa cells, which has identified an optimal concentration of TAT-CSPP1-C peptide at 2.5 μm (22).
For introducing TAT-GFP fusion proteins to probe for the functional relevance of CENP-H/CSPP1 interaction in live cell division, HeLa cells were cultured to 50–60% confluency before thymidine synchronization. Just before introduction, the cells were washed with serum-free media and incubated with TAT-GFP fusion peptides at various concentrations for 30 min. After incubation, the mcherry-H2B-expressing cells were washed with PBS and then examined directly under fluorescence microscopy as detailed previously (22). Mitotic delay was quantified by the time intervals between nuclear envelope breakdown to metaphase alignment and to anaphase onset (24).
Results
CSPP1 Is a Novel Kinetochore Component
CSPP1 localizes at centrosomes and the mitotic spindle and plays a key function in accurate mitosis (15–17). To address the molecular mechanism of CSPP1 function in mitosis, we first examined the precise localization of CSPP1 in mitotic HeLa cells. To this end, aliquots of HeLa cells were transiently transfected to express GFP-CSPP1. Exogenously expressed GFP-CSPP1 localizes at the centrosomes in prophase cells. In prometaphase cells, besides mitotic spindle and spindle poles, we noticed that GFP-CSPP1 also decorated the kinetochores (Fig. 1A). However, during metaphase, the kinetochore localization of GFP-CSPP1 decreased sharply. Consistent with its essential role in cytokinesis (17), the GFP-CSPP1 signal is apparent at the central spindle in anaphase cells (Fig. 1A). Although the kinetochore localization of CSPP1 decreases once chromosome alignment is achieved, the kinetochore localization of GFP-CSPP1 is still visible in metaphase cells with moderate expression levels, despite the background from strong decoration of spindle MTs (Fig. 1B). In contrast, kinetochore localization of GFP-CSPP1 is readily apparent under a confocal microscope in cells expressing a low level of GFP-CSPP1 (Fig. 1C).
FIGURE 1.
CSPP1 is a novel kinetochore protein. A, localization of exogenous GFP-CSPP1 during mitosis. HeLa cells transfected with GFP-tagged CSPP1 were fixed with formaldehyde and staining for kinetochores (anti-centromere auto-antibody (ACA), red) and DNA (blue). Scale bars, 5 μm. B, representative mitotic cell overexpressing GFP-CSPP1. HeLa cell transiently transfected with GFP-CSPP1 was fixed and stained for CENP-F (red) and DNA (blue). The boxed areas are shown magnified. Scale bar, 5 μm. C, representative confocal image of a mitotic cell with a low expression of GFP-CSPP1. HeLa cell expressing a low level of GFP-CSPP1 was fixed with formaldehyde and immunostained for kinetochores (ACA, red) and DNA (blue). Image was acquired under an LSM 710 confocal microscope. The magnified areas are shown. D, characterization of CSPP1 antibody by Western blot. HeLa cell lysate was separated by SDS-PAGE and probed with CSPP1 antibody. Both chemiluminescence images of long exposure (exp.) and short exposure are shown. E, representative immunofluorescence images of HeLa cells at different mitotic stages. Untreated HeLa cells were fixed with cold methanol and immunostained with CSPP1 antibody (green), ACA antiserum (red, and DNA (blue). Images are shown in slices. Scale bars, 5 μm. F, kinetochore localization of CSPP1 is independent of MTs. HeLa cell was treated with MT poison nocodazole for 2 h and then was fixed. The cells were stained for CSPP1 (green), ACA (red), and DNA (blue). Scale bar, 5 μm. G, CSPP1 localizes at kinetochores in taxol-treated cells. Mitotic HeLa cells treated with 100 nm taxol for 2 h were fixed and stained for CSPP1, ACA, MTs, and DNA. Note that centrosome localization (arrowhead) of CSPP1 is evident in taxol-treated cells but not in nocodazole-treated cells (F). Scale bar, 5 μm.
To verify the kinetochore localization of endogenous CSPP1, a CSPP1 antibody was generated in-house. The antibody specifically recognized a protein of 150 kDa, corresponding to the size of CSPP1, in HeLa cell lysates (Fig. 1D). For unknown reasons, we failed to detect the CSPP-S of 100 kDa. Immunostaining of mitotic HeLa cells with the CSPP1 antibody displayed centrosome localization in prophase cells, spindle pole and kinetochore localizations in prometaphase cells, and central spindle localization in anaphase cells (Fig. 1E), similar to the observations for GFP-CSPP1. To address whether the kinetochore localization of CSPP1 depends on spindle MTs, we employed the MT depolymerization drug nocodazole and the MT stabilizer taxol. As shown (Fig. 1F), in MT-depolymerized mitotic cells the kinetochore localization of CSPP1 is evident, suggesting that kinetochore targeting of CSPP1 does not requires spindle MTs. The kinetochore residence of CSPP1 is also obvious in the presence of kMT attachment (Fig. 1G), showing that MT occupancy does not affect its kinetochore localization. Thus, we conclude that CSPP1 is a bona fide component of kinetochore.
Aurora B Kinase Activity Determines the Residence of CSPP1 on Kinetochores
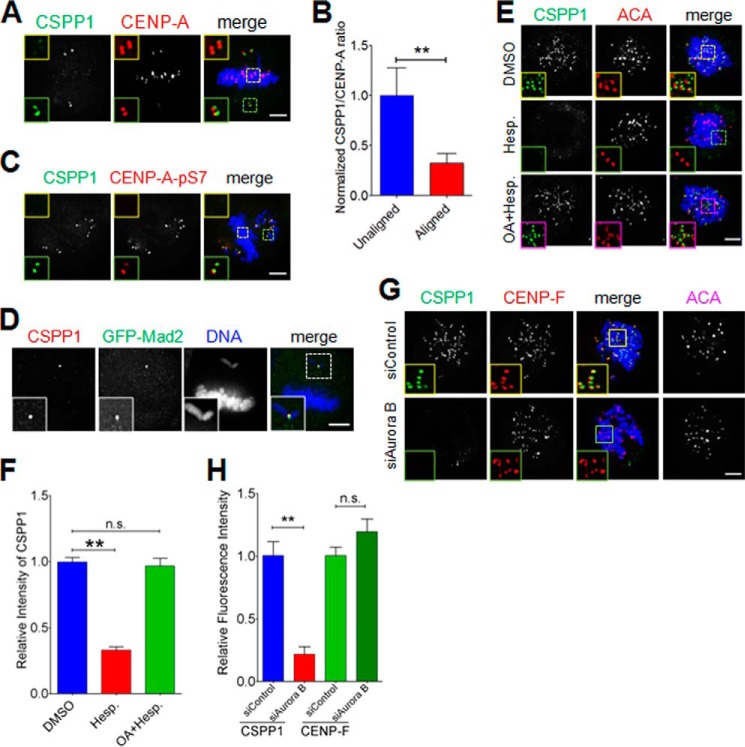
In our investigation, we noticed that CSPP1 preferentially localized on the unaligned kinetochores in low concentration (10 ng/ml) nocodazole-treated cells, which bear both aligned kinetochores and misaligned kinetochores (Fig. 2A). Quantification analysis of the CSPP1/CENP-A intensity ratio indicated a significant decrease of CSPP1 protein levels by 67% on aligned kinetochores compared with unaligned kinetochores (Fig. 2B). The major difference between unaligned and aligned kinetochores is the phosphorylation of a series of outer kinetochore proteins by Aurora B in a tension-regulated manner (27). We therefore asked whether the preferential association of CSPP1 with unaligned kinetochore correlates with the higher Aurora B activity. Indeed, CSPP1 and Ser(P)-7-CENP-A (a marker of Aurora B activity (28)) showed perfect superimposition at unaligned kinetochores (Fig. 2C). Also, CSPP1 readily colocalized with Mad2, a commonly used marker for unattached kinetochores where Aurora B activity is high (Fig. 2D). To further verify the dependence of CSPP1 kinetochore localization on Aurora B activity, we examined the CSPP1 localization in cells treated with hesperadin, a known Aurora B-specific inhibitor (29). In control cells, CSPP1 readily localized at the kinetochores, although in the cells treated with hesperadins, kinetochore localization of CSPP1 was almost invisible. Inhibition of protein phosphatases with OA prior to the addition of hesperadins precluded the loss of CSPP1 from the kinetochore upon Aurora B inhibition (Fig. 2, E and F), further supporting our notion that kinetochore localization of CSPP1 depends on the Aurora B kinase signaling. Consistently, kinetochore residence of CSPP1 was apparently affected in Aurora B-depleted cells, although CENP-F localization was not altered (Fig. 2, G and H), suggesting that CSPP1 function is under the control of the Aurora B signaling cascade. Thus, we reasoned that the kinetochore localization of CSPP1 requires Aurora B kinase activity.
FIGURE 2.
Aurora B kinase activity regulates kinetochore localization of CSPP1. A, CSPP1 preferentially resides on misaligned kinetochores. To induce misaligned chromosomes, single thymidine treatment released HeLa cells were treated with 10 ng/ml nocodazole for 1 h. After methanol fixation, cells were stained for CSPP1 (green), CENP-A (red), and DNA (blue). Scale bar, 5 μm. B, quantitative analysis of CSPP1 kinetochore signal intensity (normalized to CENP-A intensity) as in A. Error bars indicate mean ± S.E. from analysis of more than 40 kinetochores in five cells. Student's t test was used to calculate the p value for comparison of aligned kinetochores and unaligned kinetochores. **, p < 0.001. C, CSPP1 enriched misaligned kinetochores show high Aurora B activity. HeLa cells as described in A were stained for CSPP1 (green), CENP-A-Ser(P)-7 (red), and DNA (blue). Scale bar, 5 μm. D, preferential enrichment of CSPP1 on unaligned kinetochore that is positive for Mad2. HeLa cells expressing GFP-Mad2 were treated as described in A and stained for CSPP1 (red) and DNA (blue). Scale bar, 5 μm. E, Aurora B activity is required for proper localization of CSPP1 on kinetochores. Representative immunofluorescence images of prometaphase cells treated with DMSO (control) or hesperadins for 2 h or first with OA (1 h) followed by OA + hesperadins (Hesp.) (2 h) were fixed by methanol and stained for CSPP1 (green), ACA (red), and DNA (blue). Images are shown in 3-μm projection of z stacks. Scale bar, 5 μm. F, quantitative analysis of CSPP1 kinetochore signal intensity (normalized to ACA intensity) as in E. Error bars indicate mean ± S.E. from analysis of more than 50 kinetochores in five cells. Student's t test was used to calculate p value for comparison of aligned kinetochores and unaligned kinetochores. **, p < 0.0001; n.s., not significant. G, representative immunofluorescence images of prometaphase cells transfected with control or Aurora B siRNA. Before fixation, cells were treated with nocodazole. Then cells were stained for CSPP1 (green), CENP-F (red), DNA (blue), and ACA (shown as grayscale images). The boxed areas are shown magnified. Scale bar, 5 μm. H, quantitative analysis of CSPP1 or CENP-F kinetochore signal intensity (normalized to ACA intensity) as in G. Error bars represent mean ± S.E. from analysis of more than 50 kinetochores in five cells. Student's t test was used to calculate p value for comparison of aligned kinetochores and unaligned kinetochores. **, p < 0.0001; n.s., not significant.
CSPP1 Resides on Kinetochores by Binding to CENP-H Complex
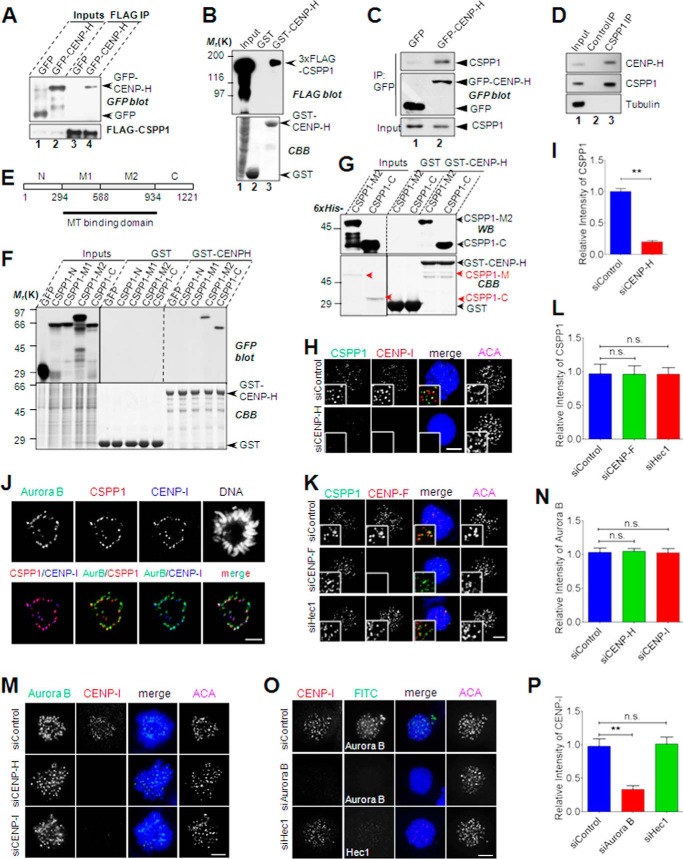
To probe the precise function of CSPP1 on kinetochores, we searched for its binding partners. To this end, we conducted a pulldown assay followed by mass spectrometry identification. Among the candidates, a well known kinetochore component, CENP-H, was paid special attention as the rest do not localize to the kinetochore as does CSPP1. Subsequently, we confirmed that CSPP1 interacts with CENP-H in vivo and in vitro by immunoprecipitation and pulldown assays (Fig. 3, A and B) where FLAG-tagged CSPP1 readily binds to CENP-H. Not only exogenous CSPP1 but also endogenous CSPP1 could be immunoprecipitated by CENP-H and vice versa (Fig. 3, C and D), thus confirming their interaction. The MT binding domain of CSPP1 lies in the middle region of the protein. To identify the domain that mediates its association with CENP-H, we constructed four CSPP1 deletion mutants according to previously reported information (15, 16) and predicted secondary structure of the protein (Fig. 3E). Pulldown assay indicated that the C-terminal half of CSPP1 contributes to its association with CENP-H (Fig. 3F). However, their interaction is direct, because bacterially expressed and purified CSPP1 and CENP-H also bind to each other in vitro (Fig. 3G).
FIGURE 3.
CSPP1 interacts with CENP-H complex. A, CSPP1 interacts with CENP-H in vivo. HEK293T cells cotransfected with 3×FLAG-CSPP1 and GFP or GFP-CENP-H were collected and lysed. The lysates were incubated with anti-FLAG M2 beads. After extensive washes, immunoprecipitations (IP) were probed with anti-FLAG and anti-GFP blots, respectively. Arrows indicate GFP or GFP-CENP-H protein. B, CSPP1 interacts with CENP-H in vitro. GST- and GST-CENP-H-bound glutathione 4B beads were used as affinity matrices to absorb 3×FLAG-CSPP1 expressing cell lysate. Pulldowns were analyzed by SDA-PAGE and probed with anti-FLAG antibody. GST and GST-CENP-H proteins were visualized by Coomassie Blue (CBB) staining. C, CENP-H forms a complex with endogenous CSPP1. Mitotic cells expressing GFP or GFP-CENP-H were lysed and subjected to immunoprecipitation with anti-GFP microbeads. CSPP1 was visualized by anti-CSPP1 antibody. For immunoprecipitation of endogenous CSPP1, mitotic cell lysates were incubated with IgG or CSPP1 antibody followed by extended incubation with protein A/G microbeads. D, endogenous CSPP1 and CENP-H forms a cognate complex. Aliquots of HeLa cells were synchronized by nocodazole treatment followed by incubation of anti-CSPP1 antibody with clarified cell lysate. CSPP1 immunoprecipitates were then fractionated by SDS-PAGE followed by Western blotting analyses of CSPP1, CENP-H, and tubulin. CENP-H was pulled down by an anti-CSPP1 antibody (lane 3) but not control IgG (lane 2). E, schematic showing CSPP1 domains and design for recombinant proteins for mapping CENP-H interactions. F, CSPP1 binds to CENP-H through its C-terminal half. GST- and GST-CENP-H-bound glutathione 4B beads were used as affinity matrices to pull down GFP-tagged CSPP1 deletion mutants as indicated in D. Pulldowns were analyzed by SDA-PAGE and probed with anti-GFP antibody. GST and GST-CENP-H proteins were visualized by Coomassie Blue (CBB) staining. G, direct interaction between CSPP1 and CENP-H. Bacterially expressed and purified CSPP1 deletion mutants were incubated with GST- or GST-CENP-H-bound glutathione 4B beads. After extensive washing, samples were boiled and applied to SDS-PAGE followed by Western blot analyses or Coomassie Blue staining. H, CENP-H complex is required for the kinetochore localization of CSPP1. Representative immunofluorescence images of prometaphase cells transfected with control or CENP-H siRNA. After 2 h of nocodazole treatment, cells were fixed by cold methanol and stained for CSPP1 (green), CENP-I (red), DNA (blue), and ACA (shown as grayscale images). Scale bar, 5 μm. I, quantitative analysis of CSPP1 kinetochore signal intensity (normalized to ACA intensity) as in G. Error bars indicate mean ± S.E. from analysis of more than 50 kinetochores in five cells. Student's t test was used to calculate p value for comparison. **, p < 0.0001. J, spatial localization of CSPP1, CENP-H complex (shown as CENP-I), and Aurora B. HeLa cell expressing GFP-Aurora B was fixed by methanol and subjected to immunostaining for CSPP1 (red), CENP-I (pseudocolor blue), and DAPI. Images were shown for each channel and two-channel merges. Scale bar, 5 μm. K, CENP-F and Hec1 do not contribute to kinetochore localization of CSPP1. HeLa cells transfected with the indicated siRNA were processed as described in H and immunostained for CSPP1 (green), CENP-F (red), DNA (blue), and ACA (shown as grayscale images). Scale bar, 5 μm. L, quantitative analysis of CSPP1 kinetochore signal intensity (normalized to ACA intensity) as in K. Error bars indicate mean ± S.E. from analysis of more than 50 kinetochores in five cells. Student's t test was used to calculate p value for comparison. n.s., not significant. M, depletion of CENP-H complex does not affect kinetochore localization of Aurora B kinase. HeLa cells transfected with indicated siRNA were processed as in H and immunostained for Aurora B kinase (green), CENP-I (red), DNA (blue), and ACA (shown as grayscale images). Scale bar, 5 μm. N, quantitative analysis of Aurora B kinetochore signal intensity (normalized to ACA intensity) as in M. Error bars indicate mean ± S.E. from analysis of more than 50 kinetochores in five cells. Student's t test was used to calculate p value for comparison. n.s., not significant. O, Aurora B kinase also controls kinetochore localization of CENP-H complex. HeLa cells transfected with indicated siRNA were processed as in H followed by immunostaining for CENP-I (red), Aurora B or Hec1 (green, Hec1 knockdown as a negative control), DNA (blue), and ACA. Scale bar, 5 μm. P, quantitative analysis of CENP-I kinetochore signal intensity (normalized to ACA intensity). Error bars indicate mean ± S.E. from analysis of more than 50 kinetochores in five cells. Student's t test was used to calculate p value for comparison. **, p < 0.0001; n.s., not significant.
To verify the kinetochore localization of CSPP1 is dependent on CENP-H, we examined the kinetochore localization of CSPP1 in CENP-H-depleted cells. CENP-H suppression, as indicated by kinetochore loss of CENP-I, resulted in a significant decrease of the CSPP1 signal on kinetochores (Fig. 3, H and I), showing that CENP-H is required for CSPP1 localization. Based on the colocalization (Fig. 3J), we conclude that both Aurora B kinase and CENP-H are required to target CSPP1 onto kinetochores, where CENP-H might be the direct receptor. Previous studies showed that CENP-H interacted with and recruited the NDC80 complex (7, 30). To test whether the abrogation of CSPP1 localization by CENP-H depletion is a direct result of decreased kinetochore recruitment of the NDC80 complex, we tested the kinetochore localization of CSPP1 in the absence of Hec1, a key component of the NDC80 complex. As shown in Fig. 3, K and L, kinetochore residence of CSPP1 was unaffected in Hec1-depleted cells. Depletion of CENP-F, another outer kinetochore protein, does not have an effect on CSPP1 localization either (Fig. 3, K and L). We have shown that Aurora B kinase is required for kinetochore localization of CSPP1 (Fig. 2). To rule out the possibility that loss of kinetochore CSPP1 in CENP-H-depleted cells is due to decreased Aurora B kinase localization, we checked whether kinetochore recruitment of Aurora B kinase was affected by CENP-H depletion. As shown, kinetochore localization of Aurora B kinase in CENP-H- or CENP-I-depleted cells is indistinguishable from that in control cells (Fig. 3, M and N). Furthermore, it was reported previously that kinetochore organization is not obviously affected by CENP-H depletion (8). Taken together, we concluded that kinetochore recruitment of CSPP1 is directly mediated by CENP-H.
Intriguingly, kinetochore targeting of the CENP-H complex, as indicated by CENP-I, is greatly affected in Aurora B-depleted cells but not in Hec1-depleted cells (Fig. 3, O and P). This seems a paradox because the CENP-H complex constitutively resides on kinetochores during mitosis, whereas Aurora B kinase is absent from kinetochores during chromosome alignment and is no longer on kinetochores in anaphase. Thus, Aurora B kinase seems to be required for only initial targeting of the CENP-H complex to the kinetochores but not for later maintenance.
CSPP1 Regulates Mitotic Progression
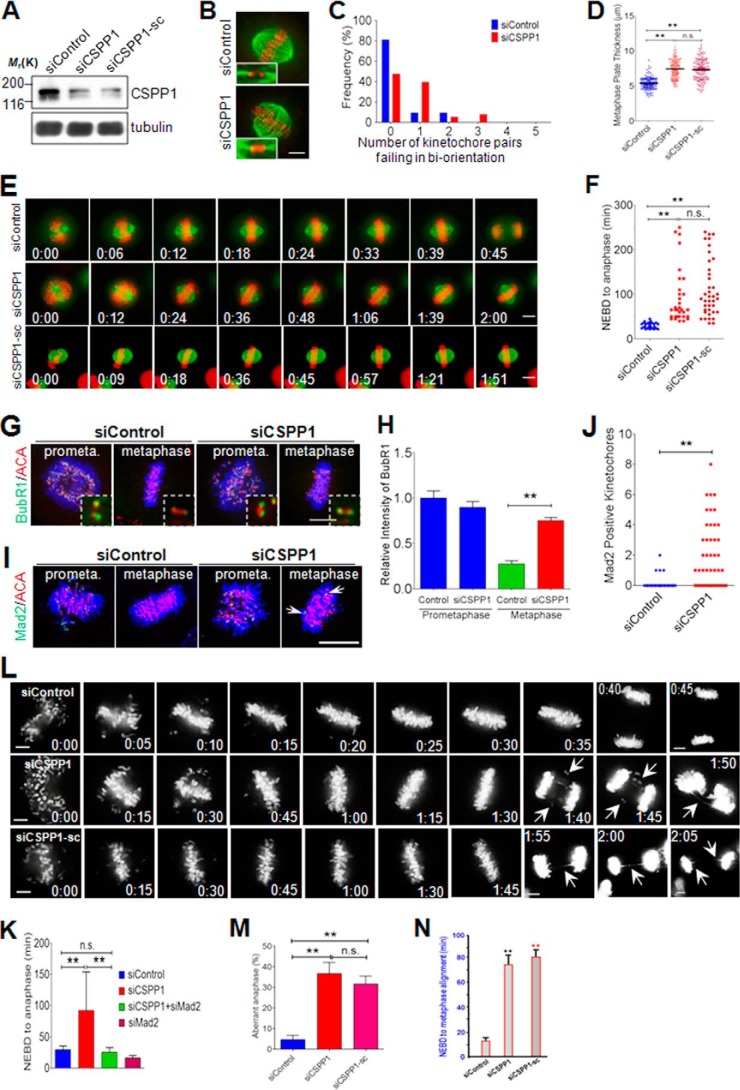
Previous studies show that suppression of CSPP1 via siRNA-mediated knockdown resulted in metaphase arrest (17), suggesting that CSPP1 is essential for metaphase events and perhaps metaphase-anaphase transition. To probe the precise function of CSPP1 during mitosis, aliquots of HeLa cells were transiently transfected with siRNA from Qiagen. As shown in Fig. 4A, the specificity and efficiency of CSPP1 siRNA were confirmed by Western blotting analyses. Compared with control depleted cells, although spindle morphology is normal, broader chromosome plates at the equator were observed, suggesting inefficient chromosome congression (Fig. 4B). To check whether the defect of metaphase plate compaction is due to aberrant kinetochore-MT attachment, we quantified the number of misaligned kinetochores. Similar to control cells, kinetochore/MT interactions in CSPP1-depleted cells are not significantly affected, as most CSPP1-depleted cells harbor only 1–3 non-bi-oriented kinetochore pairs (Fig. 4, B and C). To ascertain the phenotype observed, we quantified the metaphase plate thickness in CSPP1-depleted cells, where a different siRNA (purchased from Santa Cruz Biotechnology, siCSPP1-sc) was included (Fig. 4A). In control cells, the average thickness of the metaphase plate is 5.39 ± 0.07 μm, and in CSPP1-depleted cells, the thickness is up to 7.46 ± 0.1 and 7.32 ± 0.12 μm for each of the two siRNAs, respectively (Fig. 4D).
FIGURE 4.
CSPP1 is essential for accurate mitotic progression. A, Western blot shows the efficiency of CSPP1 siRNAs (from Qiagen and Santa Cruz Biotechnology). HeLa cells transfected with control or CSPP1 siRNA were lysed 48 h post-transfection and subjected to Western blots using indicated antibodies. Comparable knockdown efficiency of two independent siRNAs from Qiagen and Santa Cruz Biotechnology, respectively, was shown. B, representative immunofluorescence images (maximal projection) of HeLa cells transfected with control or CSPP1 siRNA for 48 h. To enrich metaphase cells, 8 h after release from thymidine block, cells were treated with MG132 for another 2 h. Afterward, cells were fixed and immunostained for MT (green) and ACA (red). Scale bar, 5 μm. C, CSPP1 depletion does not broadly affect kinetochore-MT attachment. Quantification of the unattached kinetochores in metaphase cells is as treated in B. Histogram shows the percentage of cells with fully aligned kinetochores and cells bearing unattached kinetochore pairs. n = 32 (siControl); n = 38 (siCSPP1). D, CSPP1 knockdown impairs further compaction of metaphase plate during chromosome alignment. Quantitative analysis of metaphase plate thickness of HeLa cells as described in B. **, p < 0.0001 (two-tailed t test). About 150 cells for each group were analyzed. Error bars represent S.E. Note the similar phenotype in the two groups of cells treated with two different sources of siRNA. E, CSPP1 depletion leads to metaphase arrest. Representative real time images show typical mitotic progression in HeLa cells expressing mCherry-H2B and GFP-tubulin. 48 h post-transfection with the indicated siRNAs, images were acquired at the indicated time points. Scale bar, 5 μm. F, quantification of time lapsed from NEBD to anaphase onset for HeLa cells as described in E. **, p < 0.0001 (two-tailed t test); n.s., not significant. n = 32 cells. Note the similar phenotype in the two groups of cells treated with two different sources of siRNA. G, CSPP1 knockdown activated mitotic checkpoint. Representative immunofluorescence images of Hela cells treated with control or CSPP1 siRNA. To enrich prometaphase cells, cells were fixed 8 h post-release from thymidine block; to enrich metaphase cells, after 8 h release from thymidine block, cells were treated with MG132 for an extra 2 h. Cells were fixed and stained for BubR1 (green), ACA (red), and DAPI (blue). Insets are enlarged views of representative kinetochore pairs. Scale bar, 5 μm. H, quantification of BubR1 intensity at the kinetochore (normalized against ACA signals) in cells treated as in G. **, p < 0.0001 (two-tailed t test). n = 100 kinetochores from five cells. Error bars represent S.E. I, representative immunofluorescence images of Mad2 staining in HeLa cells as described in G. In contrast to control metaphase cells, CSPP1-depleted metaphase cells bear Mad2-positive kinetochores (arrows). Scale bar, 5 μm. J, quantitative analysis of Mad2-positive kinetochores in cells as described in I. **, p < 0.0001 (two-tailed t test). n > 50 cells. Error bars represent S.E. K, Mad2 depletion releases mitotic arrest induced by knockdown of CSPP1. Quantification of time for mitotic progression from NEBD to anaphase onset. HeLa cells were treated as described in E. n (siControl) = 32; n (siCSPP1) = 32; n (siCSPP1 + siMad2) = 11; n (siMad2) = 19. **, p < 0.0001 (two-tailed t test); n.s., not significant. Error bars represent S.E. L, live cell images show defect of chromosome segregation for CSPP1-depleted cells. HeLa cells expressing mCherry-H2B were transfected with two different CSPP1 siRNAs for 48 h, separately. 8 h after released from thymidine block, the cells were subjected to live cell imaging. Arrows indicate chromosome bridges or lagging chromosomes. Scale bar, 5 μm. M, statistical analysis of aberrant chromosome segregation among anaphase cells treated with CSPP1 siRNAs, as described in L. Cells that were arrested throughout the whole imaging time without entering into anaphase were excluded from statistics. **, p < 0.0001 (two-tailed t test); n.s., not significant. Three independent imaging experiments are shown. Error bars represent S.E. N, statistical analysis of prometaphase-metaphase delay in CSPP1-depleted cells treated with CSPP1. The prometaphase-metaphase delay was judged by the intervals between NEBD and metaphase alignment in experiment shown in L. Cells that were arrested throughout the whole imaging time without entering into anaphase were excluded from statistics. **, p < 0.0001 (two-tailed t test); n.s., not significant. Three independent imaging experiments are shown. Error bars represent S.E.
To investigate the role of CSPP1 in mitotic progression, we next applied live cell imaging for cells after knocking down CSPP1. Consistent with a previous study, knockdown of CSPP1 led to mitotic arrest (Fig. 4, E and F) (17). To answer whether the arrest is dependent on the spindle assembly checkpoint (SAC), we evaluated the kinetochore localization of BubR1 and Mad2, two key components of the SAC, in control or CSPP1-depleted cells. Both BubR1 and Mad2 localize to the kinetochores in prometaphase cells transfected with control or CSPP1 siRNA (Fig. 4, G and I). In line with the SAC satisfaction, BubR1 staining on kinetochores diminished sharply in control metaphase cells. In striking contrast, the BubR1 signal was still evident on kinetochores in metaphase cells after depletion of CSPP1 (Fig. 4, G and H), suggesting a persistent activation of SAC. Consistently, Mad2-positive kinetochores could readily be seen in metaphase cells transfected with CSPP1 siRNA, which is in sharp contrast to control cells (Fig. 4, I and J). This is consistent with the finding that CSPP1-depleted cells often bear 1–3 unattached kinetochore pairs (Fig. 4C). Together, these data suggested that knocking down CSPP1 disrupted anaphase entry by precluding the satisfaction of SAC. Indeed, mitotic arrest induced by knockdown of CSPP1 could be reversed by codepletion of Mad2 (Fig. 4K).
Although most of the CSPP1-depleted cells could eventually enter anaphase after a period of arrest, a large portion of these anaphase cells often bears lagging chromosome and/or chromosome bridges (Fig. 4, L and M), further demonstrating an essential function of CSPP1 in proper mitotic progression as depletion of CSPP1 resulted in a significant delay between prometaphase and metaphase alignment (Fig. 4, L and N). The extended delay of prometaphase-metaphase transition in CSPP1-depleted cells indicated that CSPP1 functions at accurate kinetochore-microtubule attachment.
CSPP1 Regulates Chromosome Oscillation
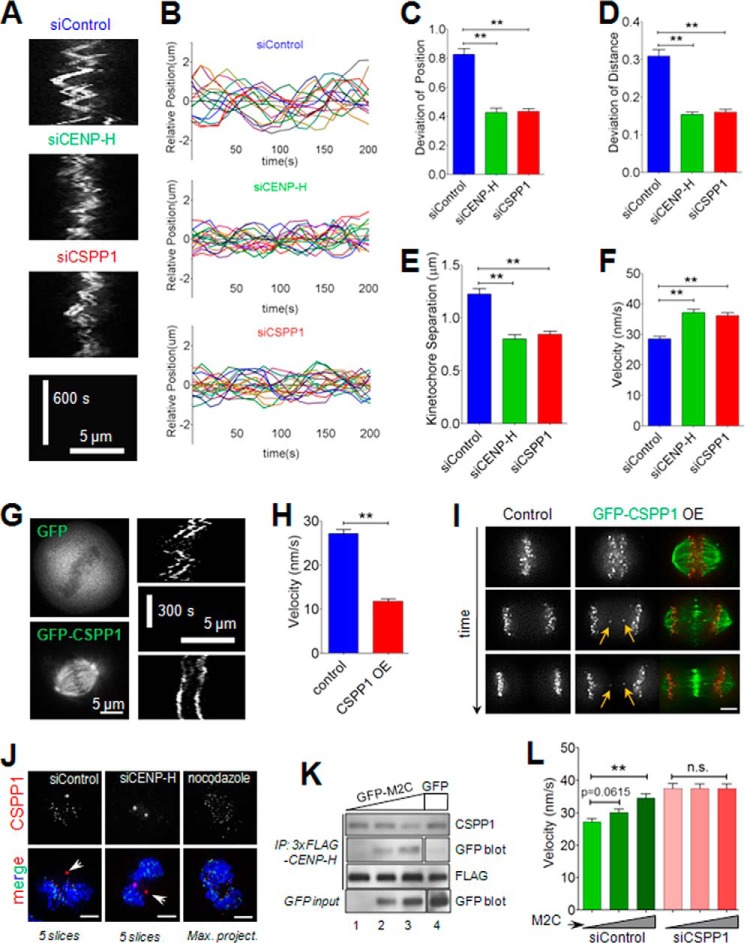
In a recent study, CENP-H complex was shown to control kMT dynamics and oscillatory movement of kinetochores along the spindle axis (8). This prompted us to investigate whether CSPP1 plays a similar role. To verify the potential role of CSPP1 in chromosome oscillations in complex with CENP-H, we tracked the oscillatory movements of sister kinetochores in live cells expressing GFP-CENP-B. In control cells, kinetochores oscillate back and forth regularly. In contrast, kinetochore oscillations in CENP-H- or CSPP1-depleted cells are greatly impaired (Fig. 5A), suggesting that CSPP1 regulates proper oscillation as does CENP-H. To analyze chromosome oscillation accurately, we tracked and plotted trajectories of more than 15 moving kinetochores, which were randomly selected from each category. To facilitate comparison, all the trajectories were precisely plotted around y = 0 axis. Oscillations in CENP-H- or CSPP1-depleted cells are obviously attenuated in contrast to that in control cells (Fig. 5B). For numerical comparison, we measured three parameters of oscillatory movement for each kinetochore pair as follows: (a) deviation from average position, which reflects the amplitude of oscillation for either sister kinetochore (31); (b) deviation from average sister kinetochore distance, which is indicative of the changes in sister kinetochore separation, commonly named as “breathing” (9); (c) average of sister kinetochore distance, which is usually used to indicate kinetochore tension. Statistically, deviation from average position in control cells is 0.83 ± 0.04, which is consistent with previous results (32), although the value is significantly decreased in CENP-H-depleted cells (0.43 ± 0.03) or CSPP1-depleted cells (Fig. 5C; 0.43 ± 0.02). Not only for the oscillatory movement, sister kinetochore breathing (0.31 ± 0.02 for deviation of kinetochore distances in control cells) is also dampened in the absence of CENP-H (0.15 ± 0.01) or CSPP1 (0.16 ± 0.01) (Fig. 5D). Consistently, the average distance between sister kinetochores is also decreased (Fig. 5E; 1.22 ± 0.05 μm in control cells, 0.80 ± 0.04 μm in CENP-H-depleted cells, and 0.84 ± 0.03 μm in CSPP1-depleted cells), demonstrating a lack of inter-kinetochore tension in the absence of CSPP1. Taken together, we demonstrated that CSPP1 plays a pivotal role in regulating kinetochore oscillations.
FIGURE 5.
CSPP1 cooperates with CENP-H complex to regulate chromosome oscillation. A, CSPP1 is required for proper chromosome oscillation. Representative kymograph images showing chromosome movement in cells transfected with control siRNA, CENP-H siRNA, or CSPP1 siRNA. HeLa cells expressing GFP-CENP-B were treated with the indicated siRNA for 48 h. 8 h after released from thymidine block, cells were subjected to real time imaging. Movements of selected kinetochores are presented in kymograph images. B, kinetochore movements are weakened in CENP-H- or CSPP1-depleted cells. Trajectories of moving kinetochores in cells as described in A were plotted around y = 0 axis according to time (x axis). n (siControl) = 15; n (siCENP-H) = 18; n (siCSPP1) = 19. C, statistical analysis of Deviation of Position derived from one of each kinetochore pairs as in A. D, statistical analysis of deviation of inter-kinetochore distance derived from the kinetochore pairs as in A. E, statistical analysis of inter-kinetochore distance of the kinetochore pairs in A. C–E, n (siControl) = 18 kinetochore pairs from eight cells; n (siCENP-H) = 16 kinetochore pairs from seven cells; n (siCSPP1) = 24 kinetochore pairs from 11 cells; mean ± S.E.; **, p < 0.0001 (two-tailed t test). F, velocity of kinetochore pair movement is increased in the absence of CSPP1. HeLa cells were treated and imaged as described in A. The speed of kinetochore pair movement along the spindle axis was measured and calculated. **, p < 0.0001, two-tailed t test; n = 105 kinetochores from seven cells; mean ± S.E. G, CSPP1 overexpression weakens chromosome movement. Kymograph images of selected kinetochore pairs from GFP- or GFP-CSPP1-overexpressing cells are shown. HeLa cells were transfected with GFP or GFP-CSPP1 plasmids together with mCherry-CENP-B(1–137) plasmid. After 8 h release from thymidine block, transfected cells were subjected to real time imaging. Scale bar, 5 μm. H, overexpression of CSPP1 down-regulates velocity of kinetochore movement. Quantitative analyses of kinetochore velocity of cells overexpressing CSPP1 are shown. OE, overexpression. **, p < 0.0001, two-tailed t test; n = 105 kinetochores; mean ± S.E. I, overexpression of CSPP1 perturbed proper mitotic progression. HeLa cells as described in G were imaged through anaphase. Arrows indicate kinetochores on lagging chromosomes or chromosome bridges. Scale bar, 5 μm. J, CENP-H depletion does not affect centrosome localization of CSPP1, which is sensitive to MT depolymerizer nocodazole. Mitotic HeLa cells transfected with control or CENP-H siRNA were fixed and stained for CSPP1 (red), ACA (green), and DNA (blue). A positive control treated with 10 ng/ml nocodazole for 2 h before fixation was also included. Scale bars, 5 μm. K, CENP-H interacting domain of CSPP1 competes with endogenous CSPP1 to bind to CENP-H. HeLa cells transfected with 5 μg of 3×FLAG-CENP-H plasmid together with 0 μg (lane 1), 5 μg (lane 2), or 10 μg (lane 3) GFP-CSPP1-M2C(588–1221) plasmid were synchronized to prometaphase followed by immunoprecipitation with anti-FLAG microbeads. Note that binding of endogenous CSPP1 to CENP-H decreases as GFP-CSPP1-M2C increases. GFP (lane 4), as control, was also included. L, intervention of CSPP1/CENP-H interaction affect kinetochore movement. Mitotic HeLa cells expressing GFP-CSPP1-M2C and mCherry-CENP-B(1–137) were transfected with control or CSPP1 siRNA followed by live cell imaging as described in A. Velocity of kinetochore movement in cells expressing different amounts of GFP-CSPP1-M2C was quantitatively analyzed. To better interpret the result, we selected cells with mean fluorescent intensity of GFP-CSPP1-M2C of 0 (non-transfected), 350–450 (medium expression), or 750–850 (high expression). **, p < 0.0001; n.s., not significant, two-tailed t test; n >60 kinetochores; mean ± S.E.
Aberrant oscillation is usually a result of perturbed kMT dynamics (8, 10, 31, 33). In this scenario, we measured the velocities of kinetochore movement in control or CSPP1-depleted cells. In normal cells, the average speed is 28.55 ± 0.82 nm/s, and in CENP-H-depleted cells, the velocity is up to 37.16 ± 1.11 nm/s, which is consistent with previous results (8). Similarly, the value is increased up to 36.21 ± 0.97 nm/s in the absence of CSPP1 (Fig. 5F), demonstrating a potential role of CSPP1 in restricting kMT dynamics. If this is the case, overexpression of CSPP1 would down-regulate kMT dynamics. Indeed, CSPP1 overexpression also dampens chromosome oscillation (Fig. 5G). Consistently, velocity of kinetochore movement in CSPP1-overexpressing cells decreases to 11.77 ± 0.58 nm/s (Fig. 5H). CSPP1 overexpression also leads to chromosome mis-segregation (Fig. 5I). These results demonstrate that CSPP1 functions to inhibit kMT dynamics.
We then asked whether the role of CSPP1 in regulating kMT dynamics is based on its interaction with CENP-H on kinetochores. We have shown that chromosome oscillation in CENP-H-depleted cells was affected to a similar extent as that in CSPP1-depleted cells. As shown in Fig. 5J, CENP-H depletion only leads to loss of CSPP1 from kinetochores without affecting centrosomal CSPP1, but it seems that loss of CSPP1 from centrosomes in CSPP1-depleted cells does not worsen the situation, which highlights the role of kinetochore CSPP1 in chromosome oscillation. To address this question in a more direct way, aliquots of HeLa cells were transiently transfected to express the CENP-H-interacting domain of CSPP1 (CSPP1-M2C, amino acids 588–1221) to compete with endogenous CSPP1 in order to bind to CENP-H. As shown (Fig. 5K), binding of endogenous CSPP1 to CENP-H decreases as the expression level of CSPP1-M2C increases, suggesting that CSPP1-M2C is a useful tool to interrogate the CSPP1/CENP-H interaction. Live cell imaging revealed that the higher the expression level of CSPP1-M2C, the faster the kinetochores move, demonstrating that disruption of CSPP1/CENP-H interaction leads to aberrant kMT dynamics. In contrast, when endogenous CSPP1 is knocked down, expression of CSPP1-M2C no longer affects kinetochore movement (Fig. 5L), suggesting that CSPP1-M2C alone does not affect kinetochore movement. Taken together, we conclude that CSPP1 interaction with CENP-H on kinetochores is basic to its function in regulating chromosome oscillation.
CSPP1/CENP-H Interaction Is Essential for Accurate Chromosome Oscillation and Segregation
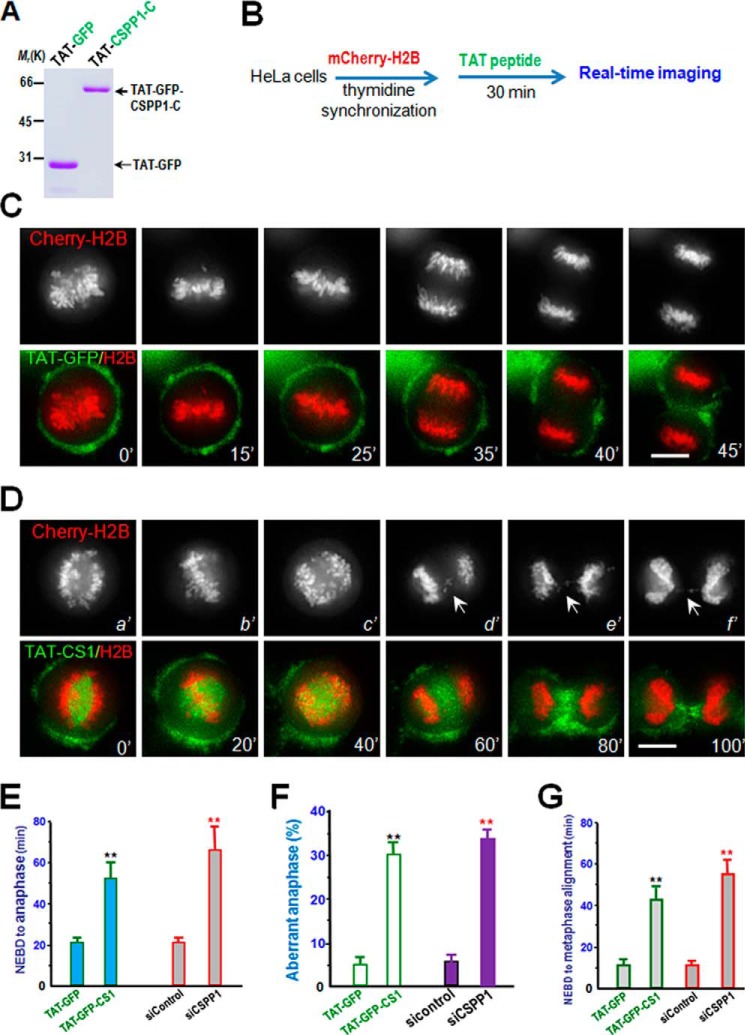
To directly evaluate the functional role of the CSPP1 interaction with CENP-H in chromosome oscillation and segregation in mitosis, a membrane-permeable peptide containing CSPP1 (934–1221 amino acids; annotated as TAT-GFP-CSPP1-C) was constructed. This was achieved by introducing an 11-amino acid peptide derived from the TAT protein transduction domain into a fusion protein containing amino acids involving binding interface between CSPP1 and CENP-H, as described previously (22, 26).
The recombinant protein was histidine-tagged and purified to homogeneity by use of nickel-affinity beads (Fig. 6A, TAT-GFP peptide). As predicted, the recombinant GFP-TAT peptide disrupted the CSPP1-CENP-H association in vivo. Neither GFP nor TAT-GFP interfered with the interaction of CSPP1 with Hec1, which is involved in the regulation of microtubule dynamics. The results demonstrate the effect of the TAT-GFP peptide in competing with full-length CSPP1 for association with CENP-H.
FIGURE 6.
Perturbation of CENP-H/CSPP1 interaction attenuates accurate chromosome segregation. A, Coomassie Blue staining of SDS-polyacrylamide gel was used to assess the quality of purified recombinant TAT-GFP-His6 protein and TAT-GFP-CSPP1(934–1221) (CSPP1-C) proteins. Bacteria expressed TAT-GFP-CSPP1-C and TAT-GFP-H6 (control) were purified with nickel-nitrilotriacetic acid affinity chromatography and desalted into DMEM. Protein concentration was determined by Bradford assays. B, schematic diagram of experimentation employing TAT peptide to perturb CENP-H/CSPP1 interaction. Aliquots of HeLa cells stably expressing mCherry-H2B were synchronized with thymidine and released into early prophase as detailed under “Materials and Methods.” TAT-GFP-CSPP1-C and TAT-GFP were added into synchronized HeLa cells followed by real time imaging. Aliquots of monastrol and MG132 (20 μm) were added with TAT peptide to enrich prometaphase cells during TAT treatment. C and D, HeLa cells expressing mCherry-H2B were synchronized with thymidine and released for 8 h to reach G2/M. Cells were cultured in DMEM with 2.5 μm TAT-GFP-CSPP1-C (D) or TAT-GFP (C) at 37 °C for 30 min before image collection. Live cell observation and imaging were performed every 5 min. Note that chromosomes in TAT-GFP-CSPP1-C-treated cells failed to oscillate (panel c′) before entry into premature anaphase with sister chromatid inter-connected (arrows; panels d′–f′). Scale bar, 10 μm. E, quantitative analyses of the mitotic progression as a function of CSPP1-CENP-H association. The timing of cell division from NEBD to anaphase onset was delayed in cells treated with TAT-GFP-CSPP1-C (2.5 μm), which is comparable with what was seen in CSPP1-suppressed cells. **, p < 0.001. Error bars represent S.E. F, quantitative analyses of aberrant chromosome segregation among anaphase cells treated with TAT-GFP-CSPP1-C (2.5 μm) as described in C and D. Cells that were arrested throughout the whole imaging time without entering into anaphase were excluded from statistics. **, p < 0.0001. Error bars represent S.E. G, quantitative analyses of the prometaphase-metaphase progression as a function of CSPP1-CENP-H association. The timing of cell division from NEBD to metaphase alignment was delayed in cells treated with TAT-GFP-CSPP1-C (2.5 μm), which is comparable with what was seen in CSPP1-depleted cells. **, p < 0.001. Error bars represent S.E.
To determine whether the CSPP1/CENP-H interaction is required for mitotic progression, cells expressing mCherry-H2B were synchronized with thymidine and were then exposed to the TAT-GFP peptide or TAT-GFP 30 min before NEBD (illustrated in Fig. 6B); real time imaging of the cells began 5 min before NEBD (Fig. 6, C and D). HeLa cells with TAT-GFP peptide did not alter cell cycle progression into mitosis relative to cells treated with the TAT-GFP control (Fig. 6C). However, in the presence of 2.5 μm TAT-GFP-CSPP1-C, HeLa cells took an average of 53.3 ± 8.3 min (n = 9 cells) to transit from NEBD to the anaphase onset of sister chromatid separation (Fig. 6, D and E). This delay is comparable with what was observed in CSPP1-suppressed cells (Fig. 6E), confirming that perturbation of the CSPP1/CENP-H interaction and suppression of CSPP1 exhibited a similar delay in chromosome segregation. Careful examination of real time chromosome dynamics in TAT-GFP-CSPP1-C-treated cells revealed that perturbation of the CSPP1/CENP-H interaction abolished the chromosome oscillation (Fig. 6D, panel c′) and resulted in premature anaphase bridge formation (Fig. 6D, panels d′–f′). The inhibitory action of TAT peptide is relatively specific, as a low concentration of TAT-GFP peptide did not interfere with mitotic progression. Our statistical analyses show that perturbation of the CSPP1/CENP-H interaction resulted in premature anaphase bridge formation similar to what was observed in CSPP1-suppresed cells (Fig. 6F). Further analyses of the time interval between NEBD and metaphase alignment demonstrate an extended delay of prometaphase-metaphase transition in TAT-GFP-CSPP1-C-treated cells, which indicates the function of the CSPP1/CENP-H interaction in accurate kinetochore-microtubule attachment. Thus, we concluded that the CSPP1/CENP-H interaction is required for accurate chromosome oscillation and segregation in mitosis.
Discussion
Proper chromosome oscillation and kMT dynamics is required for accurate chromosome segregation. Although great progress had been achieved in the field, the molecular mechanism of the coordination of chromosome oscillation and kMT dynamics remains poorly understood.
In this study, we provide evidence that centrosome and spindle pole protein CSPP1 is a novel kinetochore component. By binding to the CENP-H complex, CSPP1 resides on kinetochores and regulates chromosome oscillation. CENP-H complex and CSPP1 possibly act in the same pathway, which is based on three observations as follows. 1) CENP-H directly recruit CSPP1 on kinetochores. 2) CENP-H depletion and CSPP1 depletion result in dampened oscillation to a similar extent (8). 3) kMT dynamics or velocity of kinetochore movement is up-regulated in both CENP-H- and CSPP1-depleted cells. Despite the finding of CENP-H on kMT dynamics, no work, to our knowledge, has reported that CENP-H had any MT association activity. In this setting, CSPP1 would be the direct regulator in controlling kMT dynamics downstream of CENP-H.
Although the direct interaction between CENP-H and CSPP1 was confirmed, we found that CENP-H binding alone is necessary but not sufficient for CSPP1 localization because CSPP1 deletion mutants bearing CENP-H binding activity failed to localize to the kinetochores. Thus, it would be of great interest to identify other kinetochore proteins that are required for accurate recruitment of CSPP1 to the kinetochore.
Compared with astral MTs, kMT dynamics is much slower, indicating the regulation of kMT dynamics by kinetochore (14). However, kinetochore components that contribute to this regulation are largely unknown. During chromosome movement, MCAK was reported to elevate kMT dynamics (9, 10). In contrast, we and others showed that CENP-H functions in an opposite way, as its depletion significantly enhances kMT dynamics. Here, we demonstrated that CSPP1 also possesses an inhibitory effect on kMT dynamics. Thus, suppression of kMT dynamics by kinetochore is a function of, at least in part, the CENP-H·CSPP1 complex.
Kinetochore movement relies on MT polymerization at trailing kinetochore and depolymerization at leading kinetochore. But how this coordination between sister kinetochores is achieved remains largely unknown. CENP-H complex was reported to asymmetrically localize to sister kinetochores (8). Specifically, more CENP-H complex was found on trailing kinetochores, on which kMTs were growing. We propose that more CENP-H/CSPP1 on trailing kinetochores inhibits MT depolymerization, although less CENP-H/CSPP1 on leading kinetochores allows kMTs to depolymerize, achieving a coordinated chromosome movement. Many additional studies will be required to further address this hypothesis, such as whether CSPP1 localizes asymmetrically on sister kinetochores as does CENP-H. If this is the case, Aurora B kinase, which senses kinetochore tension and corrects aberrant kinetochore-MT attachment (27, 29, 34, 35), might acts upstream to CENP-H/CSPP1. It was demonstrated that Aurora B phosphorylation depends on the tension-dependent separation of the kinase from its substrates (27, 35, 36), and mechanical deformations happen within inter-kinetochore and intra-kinetochore during chromosome movement (37). Thus, in this hypothesis, different tensions within moving sister kinetochores may trigger the asymmetric distribution of CENP-H/CSPP1. Our earlier study (38) demonstrated that CENP-E is also preferentially associated with (or accessible at) the stretched, leading kinetochore known to provide the primary power for chromosome movement in congressing chromosomes. This evidence strongly supports a model in which CENP-E functions in congression to tether kinetochores to the disassembling microtubule plus ends. It would be of great interest in the future to visualize how the activities of two mitotic kinesins CENP-E and MCAK coordinate chromosome dynamics at the leading and trailing kinetochores, respectively. The recent development of an intra-molecular FRET sensor may facilitate a better understanding on how the conformational changes of MCAK and perhaps CENP-E play in the aforementioned process (39).
CSPP1 was originally identified as a gene involved in the malignant transformation of B-cell lymphoma (15). Recently, CSPP1 was identified as a molecular marker for the classification of distinct subgroups of basal-like breast carcinoma (40). Considering the role of aneuploidy in the promotion of tumorigenesis, our finding that CSPP1 functions in ensuring accurate mitosis may provide a good explanation for the relevance of CSPP1 with tumorigenesis. Numerical and structural aberrations of centrosomes contribute to genomic instability and tumor formation (41, 42). As CSPP1 is a centrosome protein in interphase, the function of CSPP1 in the centrosome may also account for the correlation of CSPP1 expression with tumor transformation. Recently, mutations of CSPP1 were identified in patients with Joubert syndrome, a disease with aberrant cilia function (19–21)., Further work should definitely be done to investigate the role of CSPP1 in centrosomes and ciliogenesis.
In conclusion, we demonstrated that the kinetochore localization of CSPP1 depends on CENP-H and requires Aurora B kinase activity. CSPP1 is required for the normal chromosome oscillation. Our findings provide novel insights into a better understanding of the function and molecular mechanism of CSPP1 in accurate mitotic progression and suggest a possible molecular explanation of the relevance of CSPP1 functional perturbation in tumor formation.
Author Contributions
S. H., Z. W., and X. Y. conceived the project. L. Z., Z. W., W. W., C. W., S. H., and Z. S. designed and performed most biochemical experiments. L. Z., Z. W., W. W., X. L., and L. L. designed and performed cell biological characterization. M. Y., X. W., and H. Z. performed in vitro reconstitution experiments and data analyses. M. G.-B., L. B., X. D., X .L., and X. Y. wrote the manuscript. All authors have read and approved the manuscript.
Acknowledgments
We are grateful to Dr. Hans-Christian Aasheim for kindly providing CSPP1 cDNA. We thank members of our groups for insightful discussions during the course of this study.
This work was supported by Chinese 973 Project Grants 2012CB917204, 2012CB945002, and 2002CB713700, Anhui Province Key Project Grant 08040102005, International Collaboration Grant 2009DFA31010, Chinese Natural Science Foundation Grants 90508002, 31301120, 91129714, 31301121, 31071184, 81270466, 90913016, 31501095, and MOE20113402130010, Fundamental Research Funds for Central Universities Grant WK2060190018, Anhui Provincial Natural Science Foundation Grant 1508085SMC213, China Postdoctoral Science Foundation Grant 2014M560517, and National Institutes of Health Grants DK56292, CA164133, and G12RR03034. The authors declare that they have no conflicts of interest with the contents of this article.
- kMT
- kinetochore microtubule
- MT
- microtubule
- NEBD
- nuclear envelope breakdown
- OA
- okadaic acid
- SAC
- spindle assembly checkpoint
- MCAK
- mitotic chromosome-associated kinesin
- ACA
- anti-centromere auto-antibody.
References
- 1.Yao X., and Fang G. (2009) Visualization and orchestration of the dynamic molecular society in cells. Cell Res. 19, 152–155 [DOI] [PubMed] [Google Scholar]
- 2.Bakhoum S. F., and Compton D. A. (2012) Kinetochores and disease: keeping microtubule dynamics in check! Curr. Opin. Cell Biol. 24, 64–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Holland A. J., and Cleveland D. W. (2009) Boveri revisited: chromosomal instability, aneuploidy and tumorigenesis. Nat. Rev. Mol. Cell Biol. 10, 478–487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cheeseman I. M., and Desai A. (2008) Molecular architecture of the kinetochore-microtubule interface. Nat. Rev. Mol. Cell Biol. 9, 33–46 [DOI] [PubMed] [Google Scholar]
- 5.Foley E. A., and Kapoor T. M. (2013) Microtubule attachment and spindle assembly checkpoint signalling at the kinetochore. Nat. Rev. Mol. Cell Biol. 14, 25–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Walczak C. E., Cai S., and Khodjakov A. (2010) Mechanisms of chromosome behavior during mitosis. Nat. Rev. Mol. Cell Biol. 11, 91–102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cheeseman I. M., Hori T., Fukagawa T., and Desai A. (2008) KNL1 and the CENP-H/I/K complex coordinately direct kinetochore assembly in vertebrates. Mol. Biol. Cell 19, 587–594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Amaro A. C., Samora C. P., Holtackers R., Wang E., Kingston I. J., Alonso M., Lampson M., McAinsh A. D., and Meraldi P. (2010) Molecular control of kinetochore-microtubule dynamics and chromosome oscillations. Nat. Cell Biol. 12, 319–329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jaqaman K., King E. M., Amaro A. C., Winter J. R., Dorn J. F., Elliott H. L., McHedlishvili N., McClelland S. E., Porter I. M., Posch M., Toso A., Danuser G., McAinsh A. D., Meraldi P., and Swedlow J. R. (2010) Kinetochore alignment within the metaphase plate is regulated by centromere stiffness and microtubule depolymerases. J. Cell Biol. 188, 665–679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wordeman L., Wagenbach M., and von Dassow G. (2007) MCAK facilitates chromosome movement by promoting kinetochore microtubule turnover. J. Cell Biol. 179, 869–879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huang Y., Wang W., Yao P., Wang X., Liu X., Zhuang X., Yan F., Zhou J., Du J., Ward T., Zou H., Zhang J., Fang G., Ding X., Dou Z., and Yao X. (2012) CENP-E kinesin interacts with SKAP protein to orchestrate accurate chromosome segregation in mitosis. J. Biol. Chem. 287, 1500–1509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang X., Zhuang X., Cao D., Chu Y., Yao P., Liu W., Liu L., Adams G., Fang G., Dou Z., Ding X., Huang Y., Wang D., and Yao X. (2012) Mitotic regulator SKAP forms a link between kinetochore core complex KMN and dynamic spindle microtubules. J. Biol. Chem. 287, 39380–39390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Skibbens R. V., Skeen V. P., and Salmon E. D. (1993) Directional instability of kinetochore motility during chromosome congression and segregation in mitotic newt lung cells: a push-pull mechanism. J. Cell Biol. 122, 859–875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhai Y., Kronebusch P. J., and Borisy G. G. (1995) Kinetochore microtubule dynamics and the metaphase-anaphase transition. J. Cell Biol. 131, 721–734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Patzke S., Hauge H., Sioud M., Finne E. F., Sivertsen E. A., Delabie J., Stokke T., and Aasheim H. C. (2005) Identification of a novel centrosome/microtubule-associated coiled-coil protein involved in cell-cycle progression and spindle organization. Oncogene 24, 1159–1173 [DOI] [PubMed] [Google Scholar]
- 16.Patzke S., Stokke T., and Aasheim H. C. (2006) CSPP and CSPP-L associate with centrosomes and microtubules and differently affect microtubule organization. J. Cell. Physiol. 209, 199–210 [DOI] [PubMed] [Google Scholar]
- 17.Asiedu M., Wu D., Matsumura F., and Wei Q. (2009) Centrosome/spindle pole-associated protein regulates cytokinesis via promoting the recruitment of MyoGEF to the central spindle. Mol. Biol. Cell 20, 1428–1440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Patzke S., Redick S., Warsame A., Murga-Zamalloa C. A., Khanna H., Doxsey S., and Stokke T. (2010) CSPP is a ciliary protein interacting with Nephrocystin 8 and required for cilia formation. Mol. Biol. Cell 21, 2555–2567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Akizu N., Silhavy J. L., Rosti R. O., Scott E., Fenstermaker A. G., Schroth J., Zaki M. S., Sanchez H., Gupta N., Kabra M., Kara M., Ben-Omran T., Rosti B., Guemez-Gamboa A., Spencer E., et al. (2014) Mutations in CSPP1 lead to classical Joubert syndrome. Am. J. Hum. Genet. 94, 80–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shaheen R., Shamseldin H. E., Loucks C. M., Seidahmed M. Z., Ansari S., Ibrahim Khalil M., Al-Yacoub N., Davis E. E., Mola N. A., Szymanska K., Herridge W., Chudley A. E., Chodirker B. N., Schwartzentruber J., Majewski J., et al. (2014) Mutations in CSPP1, encoding a core centrosomal protein, cause a range of ciliopathy phenotypes in humans. Am. J. Hum. Genet. 94, 73–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tuz K., Bachmann-Gagescu R., O'Day D. R., Hua K., Isabella C. R., Phelps I. G., Stolarski A. E., O'Roak B. J., Dempsey J. C., Lourenco C., Alswaid A., Bönnemann C. G., Medne L., Nampoothiri S., Stark Z., et al. (2014) Mutations in CSPP1 cause primary cilia abnormalities and Joubert syndrome with or without Jeune asphyxiating thoracic dystrophy. Am. J. Hum. Genet. 94, 62–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ward T., Wang M., Liu X., Wang Z., Xia P., Chu Y., Wang X., Liu L., Jiang K., Yu H., Yan M., Wang J., Hill D. L., Huang Y., Zhu T., and Yao X. (2013) Regulation of a dynamic interaction between two microtubule-binding proteins, EB1 and TIP150, by the mitotic p300/CBP-associated factor (PCAF) orchestrates kinetochore microtubule plasticity and chromosome stability during mitosis. J. Biol. Chem. 288, 15771–15785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yuan K., Li N., Jiang K., Zhu T., Huo Y., Wang C., Lu J., Shaw A., Thomas K., Zhang J., Mann D., Liao J., Jin C., and Yao X. (2009) PinX1 is a novel microtubule-binding protein essential for accurate chromosome segregation. J. Biol. Chem. 284, 23072–23082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xia P., Wang Z., Liu X., Wu B., Wang J., Ward T., Zhang L., Ding X., Gibbons G., Shi Y., and Yao X. (2012) EB1 acetylation by P300/CBP-associated factor (PCAF) ensures accurate kinetochore-microtubule interactions in mitosis. Proc. Natl. Acad. Sci. U.S.A. 109, 16564–16569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xia P., Zhou J., Song X., Wu B., Liu X., Li D., Zhang S., Wang Z., Yu H., Ward T., Zhang J., Li Y., Wang X., Chen Y., Guo Z., and Yao X. (2014) Aurora A orchestrates entosis by regulating a dynamic MCAK-TIP150 interaction. J. Mol. Cell Biol. 6, 240–254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cao D., Su Z., Wang W., Wu H., Liu X., Akram S., Qin B., Zhou J., Zhuang X., Adams G., Jin C., Wang X., Liu L., Hill D. L., Wang D., Ding X., and Yao X. (2015) Signaling scaffold protein IQGAP1 interacts with microtubule plus-end tracking protein SKAP and links dynamic microtubule plus-end to steer cell migration. J. Biol. Chem. 290, 23766–23780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu D., Vader G., Vromans M. J., Lampson M. A., and Lens S. M. (2009) Sensing chromosome bi-orientation by spatial separation of aurora B kinase from kinetochore substrates. Science 323, 1350–1353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zeitlin S. G., Shelby R. D., and Sullivan K. F. (2001) CENP-A is phosphorylated by Aurora B kinase and plays an unexpected role in completion of cytokinesis. J. Cell Biol. 155, 1147–1157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hauf S., Cole R. W., LaTerra S., Zimmer C., Schnapp G., Walter R., Heckel A., van Meel J., Rieder C. L., and Peters J. M. (2003) The small molecule Hesperadins reveals a role for Aurora B in correcting kinetochore-microtubule attachment and in maintaining the spindle assembly checkpoint. J. Cell Biol. 161, 281–294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mikami Y., Hori T., Kimura H., and Fukagawa T. (2005) The functional region of CENP-H interacts with the Nuf2 complex that localizes to centromere during mitosis. Mol. Cell. Biol. 25, 1958–1970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stumpff J., von Dassow G., Wagenbach M., Asbury C., and Wordeman L. (2008) The kinesin-8 motor Kif18A suppresses kinetochore movements to control mitotic chromosome alignment. Dev. Cell 14, 252–262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Magidson V., O'Connell C. B., Lončarek J., Paul R., Mogilner A., and Khodjakov A. (2011) The spatial arrangement of chromosomes during prometaphase facilitates spindle assembly. Cell 146, 555–567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Manning A. L., Bakhoum S. F., Maffini S., Correia-Melo C., Maiato H., and Compton D. A. (2010) CLASP1, astrin and Kif2b form a molecular switch that regulates kinetochore-microtubule dynamics to promote mitotic progression and fidelity. EMBO J. 29, 3531–3543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fuller B. G., Lampson M. A., Foley E. A., Rosasco-Nitcher S., Le K. V., Tobelmann P., Brautigan D. L., Stukenberg P. T., and Kapoor T. M. (2008) Midzone activation of aurora B in anaphase produces an intracellular phosphorylation gradient. Nature 453, 1132–1136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lampson M. A., and Cheeseman I. M. (2011) Sensing centromere tension: Aurora B and the regulation of kinetochore function. Trends Cell Biol. 21, 133–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Welburn J. P., Vleugel M., Liu D., Yates J. R. 3rd, Lampson M. A., Fukagawa T., and Cheeseman I. M. (2010) Aurora B phosphorylates spatially distinct targets to differentially regulate the kinetochore-microtubule interface. Mol. Cell 38, 383–392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dumont S., Salmon E. D., and Mitchison T. J. (2012) Deformations within moving kinetochores reveal different sites of active and passive force generation. Science 337, 355–358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yao X., Anderson K. L., and Cleveland D. W. (1997) The microtubule-dependent motor centromere-associated protein E (CENP-E) is an integral component of kinetochore corona fibers that link centromeres to spindle microtubules. J. Cell Biol. 139, 435–447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ems-McClung S. C., Hainline S. G., Devare J., Zong H., Cai S., Carnes S. K., Shaw S. L., and Walczak C. E. (2013) Aurora B inhibits MCAK activity through a phosphoconformational switch that reduces microtubule association. Curr. Biol. 23, 2491–2499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sternemalm J., Russnes H. G., Zhao X., Risberg B., Nord S., Caldas C., Børresen-Dale A. L., Stokke T., and Patzke S. (2014) Nuclear CSPP1 expression defined subtypes of basal-like breast cancer. Br. J. Cancer 111, 326–338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nigg E. A. (2002) Centrosome aberrations: cause or consequence of cancer progression? Nat. Rev. Cancer 2, 815–825 [DOI] [PubMed] [Google Scholar]
- 42.Nigg E. A., and Raff J. W. (2009) Centrioles, centrosomes, and cilia in health and disease. Cell 139, 663–678 [DOI] [PubMed] [Google Scholar]