Background: Nicotinamide riboside (NR) and nicotinic acid riboside (NAR) can serve as precursors of NAD in human cells.
Results: Human cells generate and release NR and NAR.
Conclusion: NR and NAR are authentic intermediates of human NAD metabolism.
Significance: Different cell populations might support each other's NAD pools by providing ribosides as NAD precursors.
Keywords: molecular cell biology, NAD biosynthesis, nicotinamide, nicotinamide adenine dinucleotide (NAD), nucleoside/nucleotide metabolism, 5′-nucleotidase, nicotinic acid, nicotinic acid riboside
Abstract
NAD is essential for cellular metabolism and has a key role in various signaling pathways in human cells. To ensure proper control of vital reactions, NAD must be permanently resynthesized. Nicotinamide and nicotinic acid as well as nicotinamide riboside (NR) and nicotinic acid riboside (NAR) are the major precursors for NAD biosynthesis in humans. In this study, we explored whether the ribosides NR and NAR can be generated in human cells. We demonstrate that purified, recombinant human cytosolic 5′-nucleotidases (5′-NTs) CN-II and CN-III, but not CN-IA, can dephosphorylate the mononucleotides nicotinamide mononucleotide and nicotinic acid mononucleotide (NAMN) and thus catalyze NR and NAR formation in vitro. Similar to their counterpart from yeast, Sdt1, the human 5′-NTs require high (millimolar) concentrations of nicotinamide mononucleotide or NAMN for efficient catalysis. Overexpression of FLAG-tagged CN-II and CN-III in HEK293 and HepG2 cells resulted in the formation and release of NAR. However, NAR accumulation in the culture medium of these cells was only detectable under conditions that led to increased NAMN production from nicotinic acid. The amount of NAR released from cells engineered for increased NAMN production was sufficient to maintain viability of surrounding cells unable to use any other NAD precursor. Moreover, we found that untransfected HeLa cells produce and release sufficient amounts of NAR and NR under normal culture conditions. Collectively, our results indicate that cytosolic 5′-NTs participate in the conversion of NAD precursors and establish NR and NAR as integral constituents of human NAD metabolism. In addition, they point to the possibility that different cell types might facilitate each other's NAD supply by providing alternative precursors.
Introduction
Nicotinamide adenine dinucleotide (NAD) is an essential coenzyme of redox reactions in central metabolic pathways in which it functions as a carrier of electrons and hydrogen ions. Moreover, NAD is used as a substrate and is degraded by several families of regulatory proteins such as protein deacetylases (sirtuins), ADP-ribosyltransferases, poly(ADP-ribose) polymerases, and ADP-ribosyl cyclases, which govern vital processes including gene expression, progression of the cell cycle, insulin secretion, DNA repair, apoptosis, and aging (1–6). Proper control of these NAD-dependent metabolic and signaling processes depends on how efficiently cells can maintain their NAD contents. Dysregulation of NAD homeostasis can lead to a variety of pathologies such as neurodegenerative diseases, diabetes, metabolic syndrome, and cancer (2, 7–9).
Generally, human cells regulate their NAD supply through biosynthesis using various precursors delivered with the diet. Although NAD can be generated from tryptophan via the kynurenine pathway, the major precursors in humans are nicotinamide (Nam)4 and nicotinic acid (NA). Both nicotinamide riboside (NR) and nicotinic acid riboside (NAR) also serve as NAD precursors. However, their role in physiological NAD generation is less clear. The bases Nam and NA are converted to the corresponding mononucleotides nicotinamide mononucleotide (NMN) and nicotinic acid mononucleotide (NAMN) by the phosphoribosyltransferases Nam phosphoribosyltransferase (NamPRT) and NA phosphoribosyltransferase (NAPRT), respectively. NMN and NAMN are then adenylylated by NMN adenylyltransferase to form the corresponding dinucleotide NAD and nicotinic acid adenine dinucleotide, respectively. Nicotinic acid adenine dinucleotide is converted to NAD by NAD synthetase. NMN and NAMN can also be generated from the nucleosides NR and NAR via their phosphorylation by nicotinamide riboside kinases (Fig. 1A) (2, 8, 10, 11).
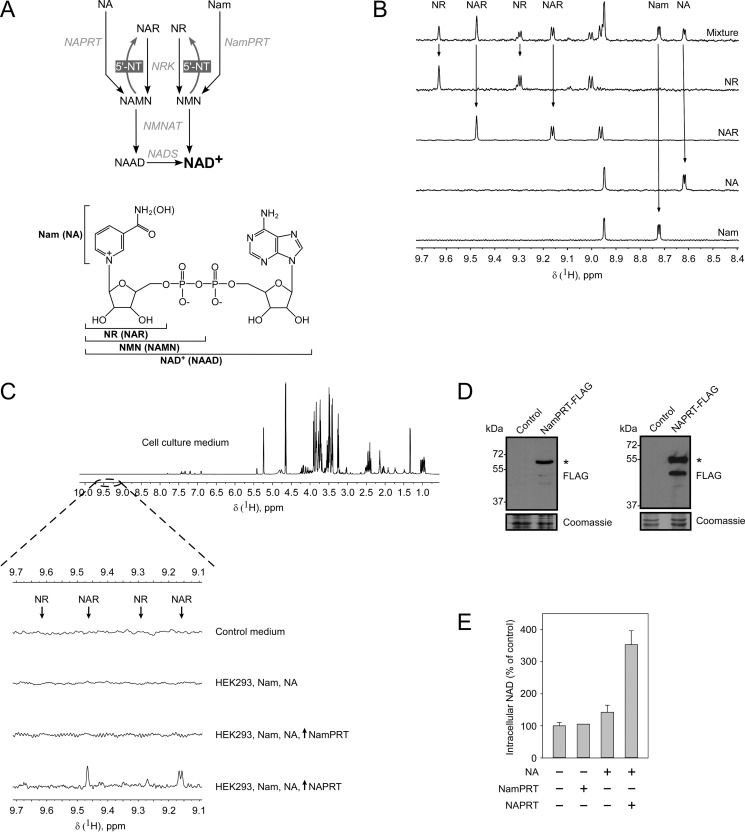
FIGURE 1.
Human cells convert nicotinic acid to nicotinic acid riboside, which is subsequently released into the culture medium. A, schematic overview of NAD biosynthetic pathways in humans. Nam is salvaged by a NamPRT to form NMN, which is adenylylated by NMN adenylyltransferase (NMNAT) to form NAD+. NA is salvaged by NAPRT to form NAMN. NAMN is then adenylylated by NMN adenylyltransferase to form nicotinic acid adenine dinucleotide (NAAD), which is converted to NAD+ by NAD synthetase (NADS). NR and NAR are salvaged by nicotinamide riboside kinase (NRK) to form NMN or NAMN. NMN and NMN are dephosphorylated to NR and NAR by cytosolic 5′-NTs. The lower panel indicates the structures of NAD+ and its metabolites. B, 700-MHz 1H NMR spectra of major NAD precursors: Nam, NA, NR, NAR, and a mixture of them. Arrows indicate peaks that were chosen for the identification and quantification of corresponding metabolites in the mixture. C, HEK293 cells were transiently transfected with vectors encoding FLAG-tagged NamPRT or NAPRT in the presence of Nam and NA in the culture medium. 3 days after transfection, culture media from control and transfected cells were analyzed by NMR spectroscopy. A 700-MHz 1H NMR spectrum of cell culture medium is shown in the uppermost panel. The region of the NMR spectra that contains peaks corresponding to NR or NAR (indicated by arrows) is highlighted by a dashed line. Neither NR nor NAR were detected in the conditioned medium from cells expressing NamPRT or from untransfected cells. The expression of NAPRT leads to release of nucleoside NAR from cells to the culture medium. D, the expression of FLAG-tagged NamPRT and NAPRT (indicated by asterisks) in HEK293 cells was confirmed by immunoblotting using antibody to FLAG peptide. Coomassie Blue staining served as a loading control. E, relative NAD levels in HEK293 cells transiently expressing NamPRT or NAPRT. The NAD level in untransfected cells (control) was taken as 100%. Data are presented as mean ± S.D. (error bars) (n = 3). The expression of NAPRT, but not NamPRT, significantly increases cellular NAD level.
Despite the considerable progress in the understanding of the mechanisms of NAD biosynthesis during the past decade, several fundamental questions still remain unanswered. So far, little is known about the molecular mechanisms underlying the interconversions of the key NAD intermediates and the relationships between their intra- and extracellular pools. Recent studies have established that all known NAD metabolites can serve as extracellular precursors of intracellular NAD (12). However, most likely, extracellular nucleotides need to be degraded to their corresponding ribosides (NR or NAR), which then enter cells as NAD precursors.
Over the last few years, NR has been in the focus of many studies, which demonstrated that dietary supplementation of this riboside can efficiently enhance NAD levels in animal tissues and attenuate the development of various pathologies. For example, in a mouse model of Alzheimer disease, NR treatment significantly increased the NAD level in the cerebral cortex and improved cognitive function (13). Moreover, NR protected from noise-induced hearing loss and spiral ganglia neurite degeneration in mice (14). The nucleoside also prevented weight gain in mice challenged with a high fat diet (15). Similarly, dietary NR supplementation effectively delayed the progression of early and late stage mitochondrial myopathy, caused increased mitochondrial biogenesis, and improved insulin sensitivity (16). The beneficial action of NR on mitochondrial biology was further highlighted in a mouse model of mitochondrial disease characterized by impaired cytochrome c oxidase biogenesis. Supplementation with NR led to marked improvement of the respiratory chain defect and exercise intolerance (17). These findings suggest that NR might serve as a potent agent for the treatment of neurodegenerative diseases and metabolic disorders associated with mitochondrial dysfunction.
It has recently been shown that, in yeast, NR and NAR are authentic intracellular intermediates. That is, these ribosides are produced within the cells and can serve as additional sources of NAD precursors. NR and NAR are generated from the mononucleotides NMN and NAMN, respectively, through their dephosphorylation by the cytosolic 5′-nucleotidases (5′-NTs) Isn1 and Sdt1 (18) or the phosphatase Pho8 (19). Moreover, NR is released from yeast cells into the growth medium (18–21).
In this study, we tested whether NR or NAR can be generated in human cells and thereby represent an integral part of NAD metabolism. Our findings indicate that previously identified human cytoplasmic 5′-nucleotidases are capable of dephosphorylating NAMN and (to a lesser extent) NMN, thereby generating a pool of ribosides in human cells. Thus, NAR can be generated from NA via NAMN formation (by NAPRT). NAMN, in turn, is then dephosphorylated to NAR by 5′-NTs (Fig. 1A). Strikingly, we observed that the generation and release of NAR from a fraction of cells in the culture are sufficient to maintain viability of neighboring cells that are unable to utilize NAD precursors other than ribosides. These results indicate that NAR and NR represent an integral part of the human NAD metabolome. Moreover, they suggest that different cell types might support each other's NAD pools by providing ribosides as NAD precursors.
Experimental Procedures
Chemicals and Reagents
Unless otherwise specified, all chemicals and reagents were of analytical grade and were purchased from Sigma and Amresco. Nicotinamide riboside was obtained from Biosynth. Nicotinic acid riboside was obtained from Toronto Research Chemicals. Cell culture reagents were from Gibco, HyClone, Greiner Bio-One, and Orange Scientific. The following antibodies were used: mouse anti-FLAG M2 (Sigma), mouse anti-SOD2 D10 (Santa Cruz Biotechnology), fluorescence-conjugated secondary antibodies (Invitrogen), and HRP-conjugated rabbit anti-mouse antibodies (Sigma). Enhanced chemiluminescence (ECL) reagents were from GE Healthcare. DNA-modifying and restriction enzymes were purchased from Thermo Scientific and New England Biolabs.
Cell Culture
Cells were cultivated in Ham's F-12 medium for HeLa S3 and Dulbecco's modified Eagle's medium (DMEM) for HEK293 and HepG2 cells. Media were supplemented with 10% (v/v) FBS (Gibco), 2 mm l-glutamine, and 20 μg/ml gentamicin. 2 μm FK866 and/or 100 μm NA were added to the cell culture medium as indicated. Transient transfection of eukaryotic cells was performed using Effectene reagent (Qiagen) or the calcium phosphate precipitation method. Co-culture experiments were performed using ThinCert cell culture inserts produced from polyethylene terephthalate capillary pore membrane (pore size, 0.4 μm; Greiner Bio-One). Cell viability was determined using 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay.
Cloning and Generation of Expression Vectors
For expression of FLAG-tagged proteins in human cells, their corresponding open reading frames were inserted into pFLAG-CMV-5a (Sigma). For expression of His6-tagged proteins in Escherichia coli, their corresponding open reading frames were inserted into pET-24a (Novagen). All cloned DNA sequences were verified by DNA sequence analysis.
NAD Measurement
HEK293 cells were cultivated in 96-well plates. Intracellular NAD was measured using an NAD+/NADH Cell-Based Assay kit (Cayman Chemical) according to the manufacturer's protocol.
NMR Analysis
Culture media from transfected and untransfected HEK293 or HepG2 cells were collected and stored at −80 °C. To precipitate proteins, the samples were incubated on ice with 2 volumes of acetonitrile (Biosolve) for 30 min and then centrifuged at 15,000 × g for 30 min at 4 °C. Supernatants were lyophilized and resuspended in D2O-based buffer containing 50 mm NaPi (pH 6.5) and 1 mm sucrose as a chemical shift reference (δ(1H), 5.42 ppm) and internal standard for quantification. 100 μm standard solutions of Nam, NA, NR, and NAR were prepared using the same buffer. Samples were stored at −80 °C until NMR analysis. All NMR experiments were performed using a Varian DirectDrive NMR System 700-MHz spectrometer equipped with a 5-mm z-gradient salt-tolerant hydrogen/carbon/nitrogen probe at 25 °C. The PRESAT pulse sequence from a standard sequence library (Varian, ChemPack 4.1) was used for acquisition of 1H spectra. The following acquisition parameters were used: relaxation delay, 2.0 s; acquisition time, 3.9 s; and number of scans, 13,800. The NMR data were processed using the Varian VNMRJ software, version 4.2 and Mestrelab Mestrenova 8.1. The concentrations of metabolites were determined by integration of the corresponding non-overlapping proton signals with the following chemical shifts (δ(1H)): 8.72 ppm for Nam, 8.61 ppm for NA, 9.62 or 9.29 ppm for NR, and 9.47 or 9.16 ppm for NAR.
Protein Determination, SDS-PAGE, and Western Blotting
Protein concentration was determined using Quick Start Bradford 1× Dye Reagent (Bio-Rad) or the BCA Protein Assay kit (Thermo Scientific). Cell lysates were prepared in SDS sample buffer (50 mm Tris/HCl (pH 6.8), 2% (w/v) SDS, 10% (v/v) glycerol, 100 mm β-mercaptoethanol, and 0.01% (w/v) bromphenol blue). Gel electrophoresis and immunoblotting were carried out according to standard procedures. ECL was used for immunodetection. Equal protein loading was confirmed by SOD2 immunodetection or Coomassie Blue staining.
Immunocytochemistry
Cells were fixed with 4% (v/v) formaldehyde in PBS and permeabilized using 0.5% (v/v) Triton X-100 in PBS. Nuclei were stained with DAPI. Images were taken using a Leica DMI6000B epifluorescence microscope (Leica Microsystems) equipped with ×10, ×40, and ×100 objectives.
Affinity Purification of His6-tagged Proteins
Chemically competent E. coli cells (Rosetta, DE3) were transformed with vectors encoding His6-tagged protein CN-IA, CN-II, CN-III, or Sdt1. The expression of recombinant proteins was induced by 0.1 mm isopropyl β-d-thiogalactopyranoside (for CN-II-His, CN-III-His, and Sdt1-His) or by 0.5 mm isopropyl β-d-thiogalactopyranoside (for CN-IA-His). Cells were agitated for 3 h at room temperature (for CN-II-His, CN-III-His, and Sdt1-His) or at 37 °C (for CN-IA-His) and then harvested by centrifugation. The pellets obtained from cells expressing CN-II-His, CN-III-His, or Sdt1-His protein were resuspended in buffer containing 50 mm Tris-HCl (pH 8.0), 300 mm NaCl, and 10 mm imidazole. The pellet obtained from cells expressing CN-IA-His protein was resuspended in buffer containing 50 mm Tris-HCl (pH 8.0), 300 mm NaCl, 10 mm imidazole, 10% glycerol, and 10 mm β-mercaptoethanol. Cells were lysed by adding lysozyme to a final concentration of 1 mg/ml for 20 min at 4 °C. The obtained cell lysates were sonicated on ice and centrifuged at 15,000 × g for 30 min at 4 °C. CN-IA-His, CN-II-His, CN-III-His, and Sdt1-His proteins were then affinity-purified from supernatants using nickel-nitrilotriacetic acid-agarose (Qiagen) according to the manufacturer's protocol. Purified proteins were dialyzed against buffer containing 50 mm Tris-HCl (pH 7.5), 150 mm NaCl, and 1 mm DTT. Then glycerol (to a final concentration of 10%) and BSA (to a final concentration of 250 ng/μl) were added to the samples. Samples were aliquoted and stored at −80 °C.
5′-Nucleotidase Activity Assay
Enzymatic activity of purified recombinant proteins was determined by measurement of inorganic phosphate (Pi) released during 5′-nucleotidase reaction. Pi quantification was performed using an ATPase Assay kit (Innova Biosciences) according to the manufacturer's protocol. Briefly, CN-IA-His was incubated with substrate in buffer containing 50 mm HEPES (pH 7.0), 5 mm MgCl2, 100 mm KCl, 1 mm ADP, and 100 ng/μl BSA. CN-II-His was incubated with substrate in buffer containing 50 mm MES (pH 6.5), 5 mm MgCl2, 1 mm DTT, 1 mm ATP, and 100 ng/μl BSA. CN-III-His was incubated with substrate in buffer containing 50 mm Tris (pH 7.5), 5 mm MgCl2, 1 mm DTT, and 100 ng/μl BSA. Sdt1-His was incubated with substrate in buffer containing 50 mm HEPES (pH 7.0), 5 mm MgCl2, 1 mm DTT, and 100 ng/μl BSA. Enzymatic reactions were initiated by the addition of enzyme and run in a final volume of 40 μl for 10 min at room temperature. Then malachite green reagent was added to the reaction mixture, and absorbance was measured at 650 nm using a Nanodrop 2000. Activity was expressed as micromoles of formed Pi/minute/milligram of protein (μmol/min/mg).
Estimation of Kinetic Parameters
CN-II-His was incubated with different concentrations of IMP, NMN, or NAMN. The enzyme concentration was 1 ng/μl when assayed with IMP and NAMN and 5 ng/μl when assayed with NMN. CN-III-His was incubated with different concentrations of CMP, NMN, or NAMN. The enzyme concentration was 0.5 ng/μl when assayed with CMP, 5 ng/μl when assayed with NMN, and 2.5 ng/μl when assayed with NAMN. Sdt1-His was incubated with different concentrations of CMP, NMN, or NAMN. The enzyme concentration was 0.125 ng/μl when assayed with CMP, 0.25 ng/μl when assayed with NMN, and 1 ng/μl when assayed with NAMN. Data were graphed using SigmaPlot 12.0, and Km and Vmax were calculated using nonlinear regression (ligand binding; one-site saturation equation).
HPLC Analysis
CN-II-His, CN-III-His, or Sdt1-His was incubated with 5 mm NMN or NAMN in their corresponding reaction buffers for 50 min at room temperature (see “5′-Nucleotidase Activity Assay”). Reactions were stopped by heat inactivation (3 min at 95 °C), filtered, and subjected to HPLC analysis. The nucleotides NMN and NAMN and their respective ribosides NR and NAR were separated by reversed phase HPLC on a CC 250/3 Nucleosil 100-3 C18 HD column (Machery and Nagel) and eluted using an acetonitrile gradient (buffer A, 10 mm KPi (pH 5.0), 2 mm tetrabutylammonium bromide, and 3% acetonitrile; buffer B, 10 mm KPi (pH 7.5), 2 mm tetrabutylammonium bromide, and 30% acetonitrile). Nucleotides and ribosides were detected at 259 nm and quantified by peak integration based on analysis of standard nucleotides.
Results
Human Cells Convert Nicotinic Acid to Nicotinic Acid Riboside, Which Is Subsequently Released into the Culture Medium
To study the release of NR and NAR from human cells, we first optimized a 1H NMR-based experimental approach to detect NAD precursors in culture medium (see “Experimental Procedures” and Fig. 1, B and C). Peaks in the NMR spectra for both the bases Nam and NA as well as their corresponding nucleosides NR and NAR were clearly separated in the range of 8.5–9.7 ppm (Fig. 1B) and were also outside the range of peaks originating from other compounds in the culture medium (Fig. 1C, uppermost spectrum). Thus, for each of these compounds, specific peaks were identified that could be used for detection and quantification. The detection limit (when the signal to noise ratio is ≥3) was ∼150–200 nm for all tested metabolites.
We then explored whether human cells growing in the presence of Nam and NA as NAD precursors can produce and release NR or NAR into the culture medium. Using NMR, we were not able to detect these ribosides in culture medium obtained after 3 days of cultivation of HEK293 cells (Fig. 1C). In yeast, NR and NAR are generated from the corresponding mononucleotides NMN and NAMN (18, 19). Therefore, we hypothesized that the absence of NR and NAR release from human cells growing under normal conditions could be due to low intracellular levels of NMN and NAMN. To test this hypothesis, HEK293 cells were transiently transfected with vectors encoding the FLAG-tagged phosphoribosyltransferases NamPRT and NAPRT (Fig. 1D), which convert Nam and NA to NMN and NAMN, respectively (Fig. 1A). After 3 days of incubation, the culture medium still did not contain any measurable NR or NAR following expression of NamPRT (Fig. 1C). However, expression of NAPRT led to readily detectable accumulation of NAR in the culture medium (Fig. 1C, lowermost spectrum). We also measured the intracellular NAD contents of the transfected cells (Fig. 1E). The results paralleled the observations of the nucleoside analyses. Namely, the NAD level in cells transfected with the NAPRT construct was significantly elevated, whereas overexpression of NamPRT did not change the cellular NAD contents. The absence of an effect on the cellular NAD level by NamPRT overexpression can be ascribed to the inhibition of NamPRT by physiological NAD concentrations (22). Taken together, these results indicate that human cells have the potential to convert NA to NAR, which can be released into the medium.
Cytosolic 5′-Nucleotidases CN-IA, CN-II, and CN-III Generate Nicotinic Acid Riboside in Human Cells
The observation of NAR production and release prompted us to address the question as to how this riboside is generated in human cells. Both NR and NAR are authentic intracellular metabolites in yeast and can be generated from NMN and NAMN, respectively, by dephosphorylation (18, 19). Therefore, we wondered whether the known human cytosolic 5′-nucleotidases CN-IA, CN-II, and CN-III (23, 24) can dephosphorylate NMN and NAMN and thus generate NR and NAR. We also examined the 5′-nucleotidase activity of the yeast enzyme Sdt1, which is known to produce NR and NAR from the corresponding mononucleotides in yeast cells (18). We transfected HEK293 and HeLa S3 cells with vectors encoding CN-IA, CN-II, CN-III, and Sdt1 proteins fused to a C-terminal FLAG epitope. Transient expression of recombinant proteins was confirmed by immunoblotting (Fig. 2A). As shown in Fig. 2B, all overexpressed enzymes localized to the cytoplasm as expected. Next, we analyzed the culture medium of the transfected cells for the presence of NR or NAR (Fig. 2C). However, none of the samples contained any detectable riboside (not shown), similar to untransfected cells (Fig. 1C).
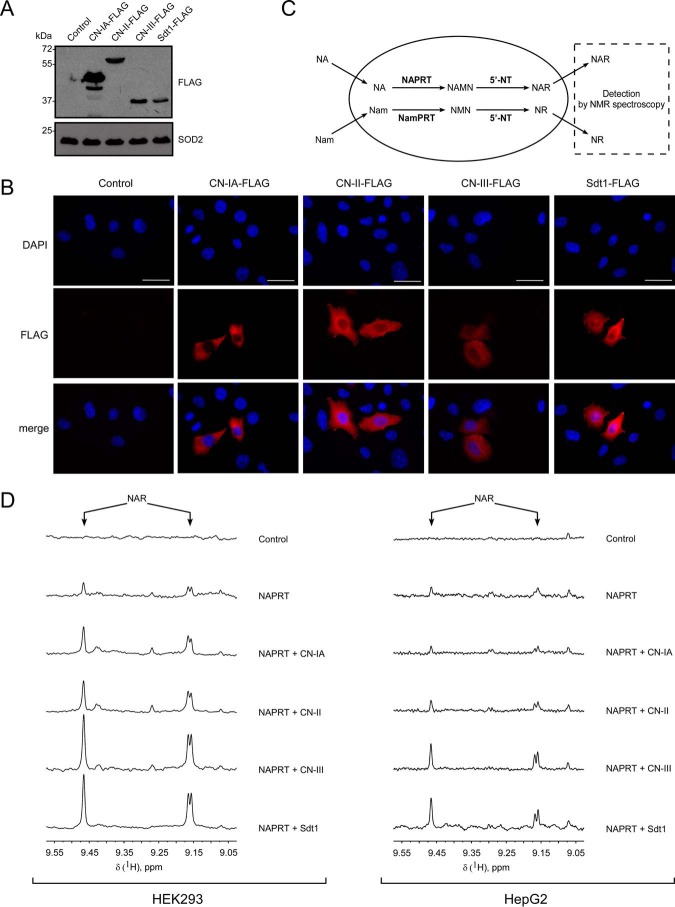
FIGURE 2.
Cytosolic 5′-nucleotidases CN-IA, CN-II, and CN-III generate nicotinic acid riboside in human cells. A and B, HEK293 (A) and HeLa S3 (B) cells were transiently transfected with vectors encoding FLAG-tagged 5′-NTs CN-IA, CN-II, CN-III, and Sdt1. The expression of FLAG-tagged 5′-NT was confirmed by immunoblotting using antibody to FLAG peptide. SOD2 served as a loading control (A). Subcellular distribution of FLAG-tagged proteins is shown in B. Cell nuclei were stained with DAPI. Scale bars, 10 μm. C, schematic representation of the experimental approach. 5′-NTs were expressed or co-expressed with NamPRT or NAPRT in human cells in the presence of Nam and NA. NR and NAR release from transfected cells to culture medium was analyzed by NMR spectroscopy. D, HEK293 (left panel) and HepG2 (right panel) cells were co-transfected with plasmid encoding NAPRT and vectors encoding the indicated 5′-NT or empty vector. 3 (for HEK293 cells) and 7 days (for HepG2 cells) after transfection, culture media from control and transfected cells were analyzed by NMR spectroscopy. 1H NMR spectra show NAR release from cells transiently expressing NAPRT. Co-expression with 5′-NT significantly increased the extracellular level of NAR.
Given that NAR accumulation in the medium was detected following NAPRT overexpression (Fig. 1C), we decided to perform co-transfections of NAPRT with each of the 5′-nucleotidases. As shown in Fig. 2D, in both HEK293 and HepG2 cells, the presence of CN-II, CN-III, or Sdt1 substantially increased the accumulation of NAR in the medium, compared with overexpression of NAPRT alone, when the cells were grown in the presence of Nam and NA. For CN-IA, this effect was weaker in HEK293 cells and undetectable in HepG2 cells. We also performed co-expressions of NamPRT with the 5′-nucleotidases in HEK293 cells. However, the obtained NMR spectra did not indicate the presence of NR (or NAR) in the culture medium (not shown). The capability of the 5′-nucleotidases to increase cellular NAR production and release suggests that at least NAMN is cleaved by these enzymes.
CN-II and CN-III Can Generate NR and NAR in Vitro by Dephosphorylation of NMN and NAMN
It has been reported that the three human 5′-nucleotidases have relatively broad substrate specificity (23, 24). CN-IA is characterized by a high affinity to AMP, its preferred substrate, and is known to be allosterically activated by ADP (25). CN-II preferentially hydrolyzes 6-hydroxypurine nucleotides such as IMP and GMP and is allosterically activated by ATP (26, 27). Pyrimidine 5′-nucleotidase CN-III most efficiently catalyzes the dephosphorylation of the mononucleotides CMP and UMP to the corresponding nucleosides (28). To test the ability of these enzymes to cleave NMN or NAMN, we constructed prokaryotic expression vectors encoding N-terminally His6-tagged CN-IA, CN-II, CN-III, and Sdt1 proteins. Following overexpression in E. coli, the proteins were purified using a nickel-nitrilotriacetic acid matrix (Fig. 3A). The specific NMN/NAMN 5′-nucleotidase activities of purified proteins were analyzed using a colorimetric method to detect Pi released during the reaction. The formation of NR and NAR in these reactions was verified by HPLC (Fig. 3B).
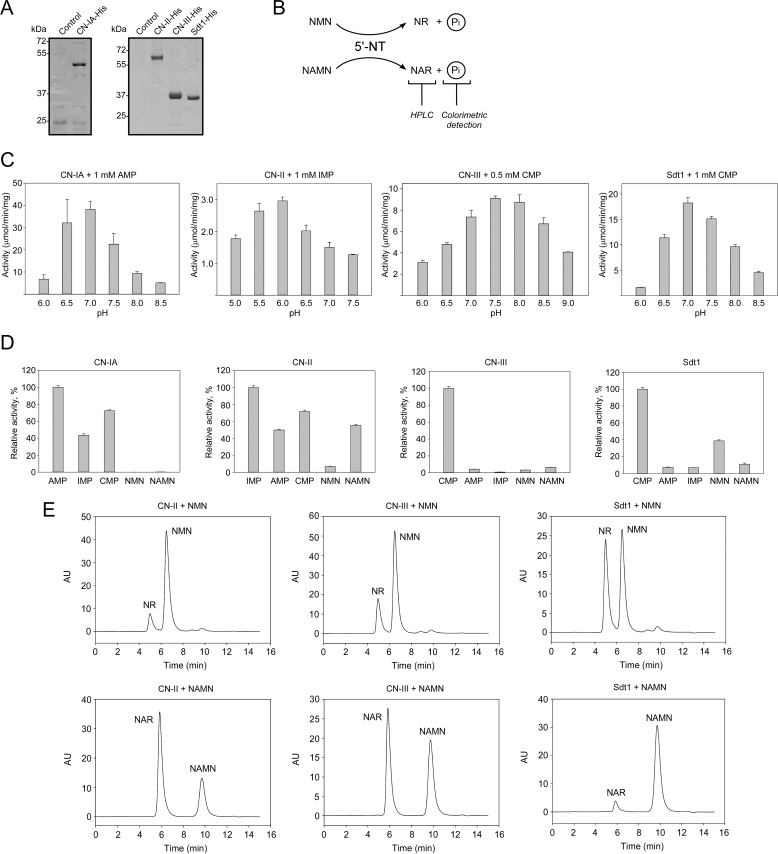
FIGURE 3.
CN-II and CN-III generate NR and NAR in vitro by dephosphorylation of NMN and NAMN, respectively. A, His-tagged proteins CN-IA, CN-II, CN-III, and Sdt1 were purified after overexpression in E. coli and analyzed by SDS-PAGE. B, schematic representation of the experimental approach. NMN/NAMN 5′-nucleotidase activities of purified proteins were measured using a colorimetric method to detect Pi released during the reaction. The formation of NR and NAR in these reactions was detected by HPLC. C, pH optima for the indicated 5′-nucleotidases were estimated using corresponding preferred substrates. The 5′-nucleotidase activity is presented as μmol of released Pi/min/mg of purified protein. Data are presented as mean ± S.D. (error bars) (n = 3). D, relative 5′-nucleotidase activities of the indicated 5′-NTs with various mononucleotides that were used as substrate at 5 mm. The specific activities (see Table 1) with corresponding preferred substrates were taken as 100%. Data are presented as mean ± S.D. (error bars) (n = 3). E, the generation of NR and NAR from mononucleotides NMN and NAMN by CN-II, CN-III, and Sdt1 proteins was confirmed by HPLC. AU, absorbance units.
To establish optimal reaction conditions of the individual 5′-nucleotidases, we first estimated the pH optima with their preferred substrates (Fig. 3C). Next, the efficiency to dephosphorylate the mononucleotides AMP, IMP, CMP, NMN, and NAMN (at 5 mm) was analyzed (Fig. 3D and Table 1). As expected, each of the human 5′-nucleotidases exhibited the highest activity toward their preferred substrate (AMP for CN-IA, IMP for CN-II, and CMP for CN-III). Besides AMP, CN-IA also efficiently dephosphorylated IMP and CMP but not NMN or NAMN. The rates of NMN or NAMN dephosphorylation by CN-IA were less than 1% compared with the rate obtained with AMP. CN-II showed the least substrate selectivity and cleaved all tested substrates. It exhibited a relative activity toward NAMN of about 50% compared with IMP, the preferred substrate, whereas the rate of NMN dephosphorylation was about 7% compared with that for IMP. CN-III appeared to be quite selective for CMP as substrate. Still, it dephosphorylated NMN, NAMN, and AMP with similar efficiency at about 3–6% of the rate obtained with CMP (Fig. 3D and Table 1). This substrate specificity resembled that of Sdt1. However, unlike all the human enzymes tested, Sdt1 preferred NMN over NAMN as substrate (Fig. 3D and Table 1). The activities toward NMN and NAMN detected by the colorimetric assay were confirmed using HPLC (Fig. 3E).
TABLE 1.
Substrate specificities of human cytosolic 5′-nucleotidases
5′-Nucleotidase activities of the indicated 5′-NT with various mononucleotides that were used as substrate at 5 mm are given. Enzyme activities were determined using the phosphate release assay (see “Experimental Procedures”). Data are presented as mean ± S.D. (n = 3).
| 5′-NT | Substrate | Specific activity | Relative activity |
|---|---|---|---|
| μmol/min/mg | % | ||
| CN-IA | AMP | 148.57 ± 3.74 | 100.0 ± 2.5 |
| IMP | 64.65 ± 2.67 | 43.5 ± 1.8 | |
| CMP | 107.74 ± 1.60 | 72.5 ± 1.1 | |
| NMN | 0.16 ± 0.01 | 0.1 ± 0.01 | |
| NAMN | 0.38 ± 0.03 | 0.3 ± 0.02 | |
| CN-II | IMP | 3.25 ± 0.08 | 100.0 ± 2.5 |
| AMP | 1.62 ± 0.02 | 50.0 ± 0.6 | |
| CMP | 2.34 ± 0.05 | 72.0 ± 1.5 | |
| NMN | 0.23 ± 0.01 | 7.0 ± 0.3 | |
| NAMN | 1.80 ± 0.04 | 55.5 ± 1.2 | |
| CN-III | CMP | 9.11 ± 0.23 | 100.0 ± 2.5 |
| AMP | 0.38 ± 0.02 | 4.2 ± 0.2 | |
| IMP | 0.07 ± 0.01 | 0.8 ± 0.1 | |
| NMN | 0.29 ± 0.01 | 3.2 ± 0.1 | |
| NAMN | 0.59 ± 0.02 | 6.5 ± 0.2 | |
| Sdt1 | CMP | 20.62 ± 0.43 | 100.0 ± 2.1 |
| AMP | 1.51 ± 0.09 | 7.3 ± 0.4 | |
| IMP | 1.43 ± 0.03 | 6.9 ± 0.2 | |
| NMN | 7.99 ± 0.25 | 38.8 ± 1.2 | |
| NAMN | 2.27 ± 0.29 | 11.0 ± 1.4 |
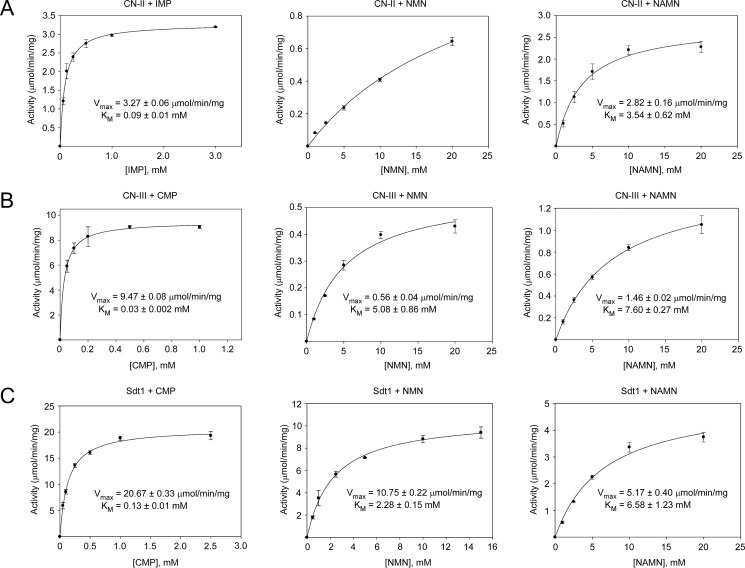
Next, we conducted a detailed kinetic analyses of the three enzymes found capable to cleave NMN and NAMN, namely CN-II, CN-III, and Sdt1. We determined Km and Vmax values for each of the enzymes with NMN, NAMN, and their respective preferred substrates (Fig. 4). Generally, the affinities of these 5′-nucleotidases were considerably higher for their preferred substrates compared with those for NMN or NAMN. In fact, in the range of substrate concentrations used (up to 20 mm), CN-II activity with NMN as substrate kept increasing almost linearly, thereby precluding a proper calculation of kinetic constants (Fig. 4A, middle panel). Thus, our results indicate that CN-II and CN-III require millimolar concentrations of NMN or NAMN for efficient catalysis in vitro. The low affinities of CN-II and CN-III toward NMN and NAMN suggest that these nucleotides, at their physiological concentrations, are unlikely to be substantially dephosphorylated in cells. Only under conditions where NMN or NAMN concentrations rise would the 5′-nucleotidases be activated. In line with this assumption, overexpression of FLAG-tagged CN-II and CN-III in human cells resulted in NAR formation only under conditions that led to increased NAMN production from nicotinic acid (Fig. 2D). Interestingly, Sdt1 exhibited Km and Vmax values for NMN and NAMN very similar to those obtained for CN-II and CN-III. That is, even though Sdt1 has been identified as being a major NR and NAR producer in yeast (18), its affinity toward NMN and NAMN is, in fact, relatively low (Fig. 4C).
FIGURE 4.
CN-II and CN-III require millimolar concentrations of NMN or NAMN for efficient catalysis in vitro. Michaelis-Menten kinetics of CN-II (A), CN-III (B), and Sdt1 (C) for the indicated substrates are shown. The Km and Vmax values were determined by nonlinear regression using SigmaPlot. Data are presented as mean ± S.D. (error bars) (n = 3).
Nicotinic Acid Riboside Released from HepG2 Cells Is Taken Up and Used as NAD Precursor by Neighboring Cells
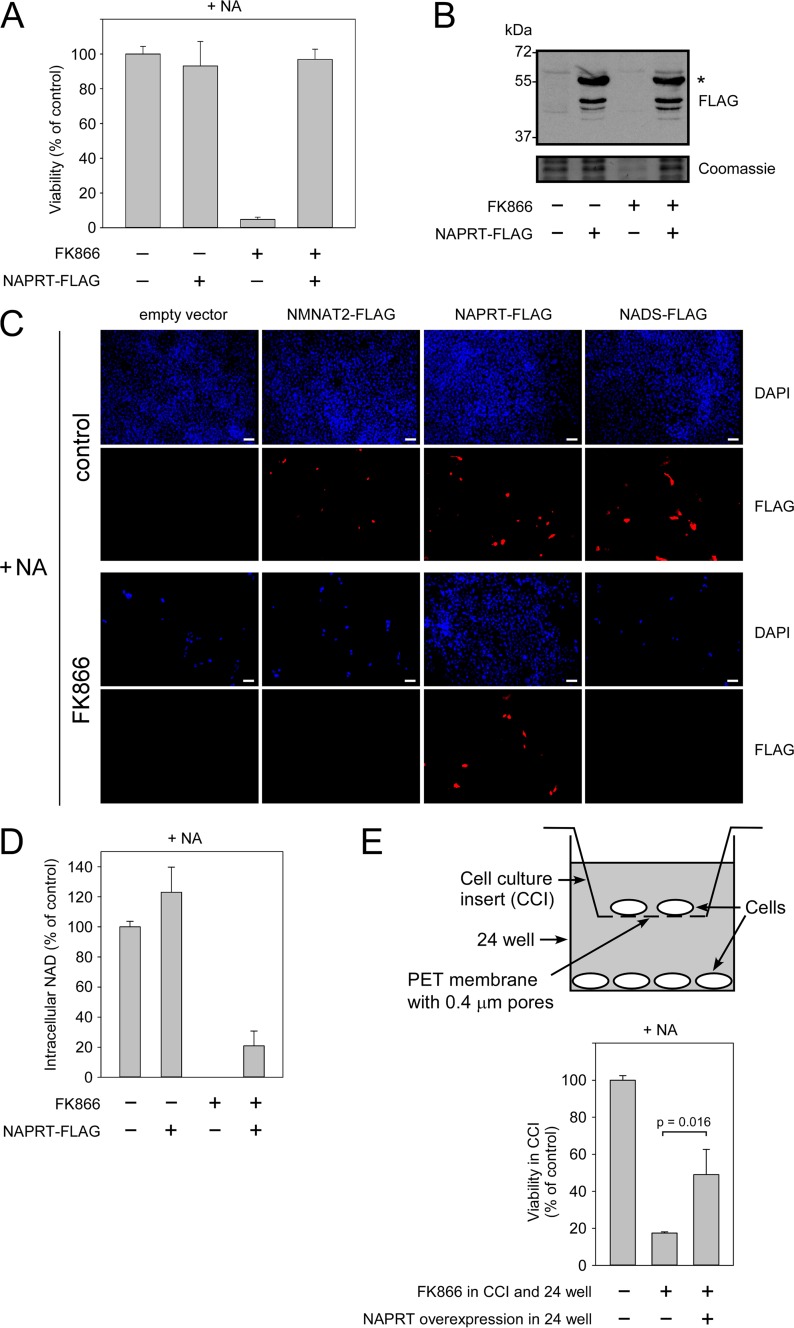
Given that human cells can produce and release at least the riboside NAR, we next wished to explore whether this capability might be of physiological relevance. To address this question, we made use of the HepG2 cell line as an experimental model because these cells lack NAPRT activity. Consequently, HepG2 cells are unable to survive in the presence of NA as the sole NAD precursor (12, 22). Nam is present in normal culture medium, but its conversion to NMN can be prevented by adding FK866, a potent inhibitor of NamPRT (29). Thus, unlike other human cell lines, HepG2 cells die in the presence of FK866 even when the medium is supplemented with NA (Fig. 5A). Surprisingly, transient overexpression of FLAG-tagged NAPRT (Fig. 5B) led to full recovery of cell survival (Fig. 5A) even though not all cells were transfected. Overexpression of other enzymes involved in NAD biosynthesis from NA had no effect on cell survival as visualized in Fig. 5C: treatment of control-transfected HepG2 cells with FK866 led to massive cell death even in the presence of NA. Transfection with vectors encoding NMN adenylyltransferase 2 or NAD synthetase activities (besides NAPRT, these enzyme activities constitute the pathway of NAD generation from NA; Fig. 1A) had no effect on cell viability. However, overexpression of NAPRT rescued both transfected and untransfected cells present in the same culture dish. Interestingly, overexpression of NAPRT in the presence of NA led only to a moderate increase of the NAD concentration (Fig. 5D). Addition of FK866 decreased NAD to undetectable levels, whereas NAPRT overexpression in the presence of NA maintained ∼20% of the overall NAD content (Fig. 5D). Note that this number includes both transfected and untransfected cells. The survival of untransfected cells in the presence of FK866 and NA suggested that the NAPRT-expressing cells somehow contributed to the NAD supply of the untransfected cells. To exclude that this effect was brought about by direct cell-cell contacts, we used a co-culture system (Fig. 5E, upper panel) in which the cell culture insert contained only untransfected cells. Overexpression of NAPRT in cells in the well plate significantly increased survival of the separated untransfected cells after treatment with FK866 and NA (Fig. 5E, lower panel), indicating that the effect did not require direct cell-cell contacts.
FIGURE 5.
HepG2 cells expressing NAPRT release NAD precursors, which thereby support neighboring cells that are unable to metabolize Nam or NA. HepG2 cells, which lack NAPRT activity, were cultivated in the presence of NA. Nam utilization was inhibited by FK866 addition as indicated. A, cells die in the presence of FK866. Overexpression of NAPRT restores the ability to use NA as an NAD precursor and recovers cell survival. Cell viability was measured by MTT assay 7 days after the transfection of cells with vector encoding FLAG-tagged NAPRT. Viability of untreated untransfected cells (control) was taken as 100%. Data are presented as mean ± S.D. (error bars) (n = 3). B, the expression of FLAG-tagged NAPRT (indicated by an asterisk) was confirmed by FLAG immunoblotting analysis. Coomassie Blue staining served as a loading control. C, cells transiently transfected with vectors encoding the indicated FLAG-tagged proteins or with empty vector. Cell nuclei were stained with DAPI. Scale bars, 100 μm. D, relative NAD levels in cells transiently expressing FLAG-tagged NAPRT. The NAD level in untreated untransfected cells (control) was taken as 100%. Data are presented as mean ± S.D. (error bars) (n = 3). The expression of NAPRT-FLAG partially recovered the NAD level in FK866-treated cells. E, two populations of cells were separated by growing in a co-culture system (upper panel). Overexpression of NAPRT-FLAG in cells growing in a 24-well plate considerably increased the viability of untransfected cells growing in cell culture inserts (CCI) in the presence of FK866 and NA (lower panel). Cell viability was measured by MTT assay 7 days after the transfection. Viability of untreated untransfected cells (control) growing in cell culture inserts was taken as 100%. Data are presented as mean ± S.D. (error bars) (n = 3). The p value was calculated using Student's t test. PET, polyethylene terephthalate; NMNAT2, NMN adenylyltransferase 2.
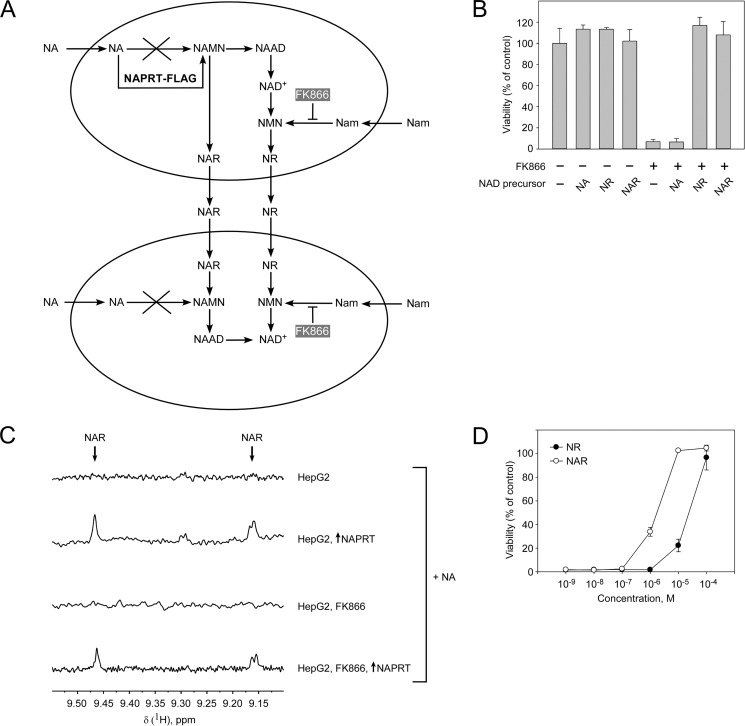
These results suggested that NAPRT-overexpressing cells must have produced and released NAR or NR to provide usable NAD precursors to untransfected cells because untransfected cells were unable to metabolize either Nam or NA (Fig. 6A). Consequently, HepG2 cells should be able to survive having only NR or NAR available as NAD precursors. This supposition was supported by experiments analyzing cell survival in the presence of FK866 and supplementation with NAR or NR (100 μm). Both ribosides maintained cell viability at control levels (Fig. 6B). Furthermore, NMR analyses of cell culture medium from HepG2 cells incubated with Nam and NA and transiently overexpressing NAPRT revealed the accumulation of NAR irrespective of the presence of FK866 (Fig. 6C). However, the levels of the produced riboside were rather low (∼1.2–1.5 μm). To test whether such low concentrations would be sufficient to support cell survival, we incubated HepG2 cells in the presence of FK866 and low concentrations of NR or NAR (10−9–10−4 m). As can be inferred from Fig. 6D, low micromolar concentrations of either riboside were sufficient to maintain cell viability. Interestingly, NAR seemed to be sufficient at ∼1 μm concentrations, whereas about 10 times more NR was required to maintain viability. Thus, the NAPRT deficiency of HepG2 cells permitted demonstration of their capability to convert NA to NAR and to provide the riboside as NAD precursor to neighboring cells unable to metabolize NA.
FIGURE 6.
Nicotinic acid riboside released from HepG2 cells is taken up and used as NAD precursor by neighboring cells. HepG2 cells were cultivated in the presence of NA, NR, or NAR as indicated. Nam utilization was inhibited by FK866 addition as indicated. A, proposed scheme of nucleoside NR and/or NAR release from intrinsically NAPRT-deficient HepG2 cells overexpressing NAPRT-FLAG. B, extracellular nucleosides NR and NAR support NAD synthesis and cell viability in the presence of FK866. Cell viability was measured by MTT assay 7 days after the treatment. Viability of untreated cells (control) was taken as 100%. Data are presented as mean ± S.D. (error bars) (n = 3). C, 700-MHz 1H NMR spectra of cell culture medium obtained from control (transfected with empty vector) cells or from cells transiently transfected with vector encoding NAPRT. Cell culture medium was analyzed by NMR 7 days after transfection. D, low micromolar concentrations of nucleosides NR and NAR were sufficient to maintain viability of FK866-treated cells. Cell viability was measured by MTT assay 7 days after the treatment. Viability of untreated cells (control) was taken as 100%. Data are presented as mean ± S.D. (error bars) (n = 3). NAAD, nicotinic acid adenine dinucleotide.
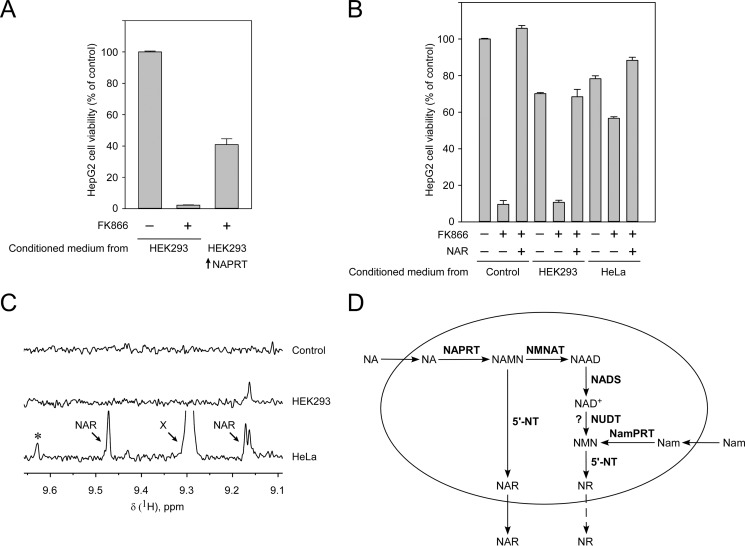
To analyze whether untransfected cells could produce sufficient amounts of NAR to support NAPRT-deficient HepG2 cells, HEK293 or HeLa cells were cultured in the presence of Nam and NA for 4 days. These preconditioned media were then used to culture HepG2 cells in the presence of FK866. To verify the suitability of this approach, we first used preconditioned medium from HEK293 cells transiently overexpressing NAPRT. As shown in Fig. 7A, the preconditioned medium from these cells led to a considerable survival of HepG2 cells grown in the presence of FK866. A similar experiment was then conducted using conditioned medium from untransfected HEK293 or HeLa cells. Although the medium from the HEK293 cells did not support viability, the medium from HeLa cells greatly increased survival of HepG2 cells grown in the presence of FK866 (Fig. 7B). As expected from these results, HeLa, but not HEK293, cells released appreciable amounts of NAR (Fig. 7C). Moreover, most likely, the HeLa cells also released NR (Fig. 7C, indicated by an asterisk). However, the observed signal was at the detection limit.
FIGURE 7.
Generation and release of NAR and NR by human cells. A, HEK293 cells with or without transient overexpression of NAPRT (as indicated) were cultured for 4 days in the presence of Nam and NA. The medium was then mixed 2:1 with fresh medium and used to culture HepG2 in the presence or absence of FK866 as indicated. The preconditioned medium was renewed every 24 h. Viability of the HepG2 cells was assessed after 7 days of culture. B, the experiment was conducted as in A except untransfected HEK293 or HeLa cells were used to generate the preconditioned medium. As a control, NAR was added to the medium as indicated. The data in A and B are represented as mean ± S.D. (error bars) (n = 3). C, NMR analyses of media from untransfected HEK293 and HeLa cells indicate release of NAR and likely NR (asterisk) from HeLa, but not HEK293, cells. X designates an unidentified peak. D, proposed mechanism of riboside generation from NA and Nam and their release from human cells. NAR can be generated from NAMN by human cytosolic 5′-nucleotidases and released from cells to the culture medium. A similar mechanism is proposed for the generation and release of NR. NUDT, Nudix hydrolase; NMNAT, NMN adenylyltransferase; NAAD, nicotinic acid adenine dinucleotide; NADS, NAD synthetase.
Discussion
The present study establishes NR and NAR as physiological intermediates of human NAD metabolism. Our results support the conclusion that cytosolic 5′-NTs, in particular CN-II and CN-III, are capable of dephosphorylating the corresponding mononucleotides NMN and NAMN. These mononucleotides are produced from the bases Nam and NA that are abundantly present in food sources (Fig. 7D). Because it is well documented that NR and NAR can be taken up and metabolized to NAD, the demonstration of their generation in human cells suggests a permanent turnover, similar to purine and pyrimidine nucleosides. That is, cells are likely to maintain and regulate NR and NAR pools to support NAD biosynthesis when needed.
Another key observation of this study is the capacity of human cells to release NAR at least under conditions of increased NAMN production. The generation and release of riboside precursors of NAD synthesis indicate that they may be important not only for intracellular NAD metabolism but also for the organism as a whole. This notion deserves attention because the released NAR, even at submicromolar concentrations, was sufficient to maintain viability of neighboring cells that were unable to use other NAD precursors. Such a high sensitivity and efficiency to use minute amounts of NAR (and somewhat higher concentrations of NR) indicates that these nucleosides could become an essential source for cells starved for other NAD precursors. Considering the positive effects of systemic NR application, for example, in mouse models of pathologies (13–17), it seems conceivable that generation or utilization of NAR or NR may be affected in these conditions, further pointing to an important physiological role of these processes.
Studying the physiology of NR and NAR metabolism has remained difficult because of the rather low concentrations. For example, in glioma cells, the intracellular concentration of these nucleosides has been estimated to be only submicromolar (30). The 1H NMR method applied in this study permitted the detection of such low concentrations in cell culture medium because the ribosides exhibited signals with chemical shifts in a range well separated from other components of the culture medium. Therefore, this method is convenient for these measurements as it does not require any preceding separation or purification steps other than the enrichment of soluble metabolites. Unfortunately, the low sensitivity necessitates acquisition of spectra over rather long periods of time. However, this drawback appears to be compensated by the possibility to simultaneously detect multiple metabolites not limited to NAD precursors.
Our results identify CN-II and CN-III, but not CN-I, as being able to generate NR and NAR in vitro. Moreover, overexpression of these enzymes considerably enhances NAR release from cells overexpressing NAPRT. The yeast 5′-NT Sdt1 had a similar effect. Importantly, Sdt1 has previously been shown to mediate physiological NR generation in yeast (18). It is further worth noting that the affinity of Sdt1 toward NMN and NAMN is very close to those found for the human cytosolic 5′-NTs, which are in the millimolar range. This rather low affinity indicates that only under conditions of increased mononucleotide (NMN or NAMN) availability will their cleavage become significant. This notion is in line with the detectable release of NAR upon NAPRT overexpression.
The intracellular concentrations of NR and NAR seem to be rather low (30). Overexpression of NAPRT in HEK293 cells resulted in detectable amounts of NAR in the medium, which was obviously a consequence of a considerable increase in NA utilization. Interestingly, even though often referred to as rate-limiting, overexpression of NamPRT hardly increased cellular NAD levels. Likewise, it did not increase NR production and release to detectable levels. These observations are in accordance with the suggestion that NamPRT is inhibited by physiological NAD concentrations (22). Still, even overexpression of 5′-NTs, which cleave NMN and thereby counteract NAD synthesis, did not lead to NR accumulation in the medium beyond the detection limit. Besides NAMN being preferred over NMN as substrates by these 5′-NTs, it could be that the system mediating the transport of these nucleosides across the plasma membrane has a lower affinity toward NR compared with NAR. The observations that HeLa cells release NAR rather than NR are in line with these suggestions. The generation and release of at least NAR by HeLa cells, in quantities that suffice to maintain NAD biosynthesis and cell survival of NAPRT-deficient HepG2 cells, indicates a potential role of these ribosides for intercellular exchange of NAD precursors.
Taken together, our data document the first mechanism of NR and NAR generation in human cells, namely through NMN and NAMN dephosphorylation by known cytosolic 5′-NTs. Consequently, turnover of these ribosides represents an integral part of human NAD metabolism. Unexpectedly, low micromolar concentrations of NAR or NR as sole NAD precursors are sufficient to maintain cell viability. The demonstration that at least NAR can be produced and delivered by cells in these amounts indicates the possibility that different cell types could preferentially use different NAD precursors and deliver them to one another.
Author Contributions
A. N. and M. Z. conceived and coordinated the study. V. K., K. S., K. N., C. D., M. N., A. Y., and A. N. conducted experiments. P. R. and M. E. M. synthesized NAR and NR. All authors analyzed data. M. K. contributed to the setup of NMR experiments and the drafting of the manuscript. A. N., M. Z., and V. K. wrote the manuscript. All authors contributed to the final version of the paper.
Acknowledgments
We thank Prof. Beverly S. Mitchell (Stanford University School of Medicine, Stanford, CA) for the plasmid encoding CN-IA protein. This work was carried out using scientific equipment of the Center of Shared Usage “The analytical center of nano- and biotechnologies of Peter the Great St. Petersburg Polytechnic University.”
This work was supported by Russian Foundation for Basic Research Grants 14-04-01765 a and 14-04-32117 mol_a and by the Norwegian Research Council. The authors declare that they have no conflicts of interest with the contents of this article.
- Nam
- nicotinamide
- NA
- nicotinic acid
- NR
- nicotinamide riboside
- NAR
- nicotinic acid riboside
- 5′-NT
- 5′-nucleotidase
- NMN
- nicotinamide mononucleotide
- NAMN
- nicotinic acid mononucleotide
- MTT
- 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide
- CN
- cytosolic 5′-nucleotidase
- NamPRT
- Nam phosphoribosyltransferase
- NAPRT
- NA phosphoribosyltransferase.
References
- 1.Nikiforov A., Kulikova V., and Ziegler M. (2015) The human NAD metabolome: functions, metabolism and compartmentalization. Crit. Rev. Biochem. Mol. Biol. 50, 284–297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Houtkooper R. H., Cantó C., Wanders R. J., and Auwerx J. (2010) The secret life of NAD+: an old metabolite controlling new metabolic signaling pathways. Endocr. Rev. 31, 194–223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Haigis M. C., and Sinclair D. A. (2010) Mammalian sirtuins: biological insights and disease relevance. Annu. Rev. Pathol. 5, 253–295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hassa P. O., and Hottiger M. O. (2008) The diverse biological roles of mammalian PARPS, a small but powerful family of poly-ADP-ribose polymerases. Front. Biosci. 13, 3046–3082 [DOI] [PubMed] [Google Scholar]
- 5.Koch-Nolte F., Kernstock S., Mueller-Dieckmann C., Weiss M. S., and Haag F. (2008) Mammalian ADP-ribosyltransferases and ADP-ribosylhydrolases. Front. Biosci. 13, 6716–6729 [DOI] [PubMed] [Google Scholar]
- 6.Fliegert R., Gasser A., and Guse A. H. (2007) Regulation of calcium signalling by adenine-based second messengers. Biochem. Soc. Trans. 35, 109–114 [DOI] [PubMed] [Google Scholar]
- 7.Belenky P., Bogan K. L., and Brenner C. (2007) NAD+ metabolism in health and disease. Trends Biochem. Sci. 32, 12–19 [DOI] [PubMed] [Google Scholar]
- 8.Magni G., Orsomando G., Raffelli N., and Ruggieri S. (2008) Enzymology of mammalian NAD metabolism in health and disease. Front. Biosci. 13, 6135–6154 [DOI] [PubMed] [Google Scholar]
- 9.Chiarugi A., Dölle C., Felici R., and Ziegler M. (2012) The NAD metabolome—a key determinant of cancer cell biology. Nat. Rev. Cancer 12, 741–752 [DOI] [PubMed] [Google Scholar]
- 10.Dölle C., Skoge R. H., Vanlinden M. R., and Ziegler M. (2013) NAD biosynthesis in humans—enzymes, metabolites and therapeutic aspects. Curr. Top. Med. Chem. 13, 2907–2917 [DOI] [PubMed] [Google Scholar]
- 11.Magni G., Di Stefano M., Orsomando G., Raffaelli N., and Ruggieri S. (2009) NAD(P) biosynthesis enzymes as potential targets for selective drug design. Curr. Med. Chem. 16, 1372–1390 [DOI] [PubMed] [Google Scholar]
- 12.Nikiforov A., Dölle C., Niere M., and Ziegler M. (2011) Pathways and subcellular compartmentation of NAD biosynthesis in human cells: from entry of extracellular precursors to mitochondrial NAD generation. J. Biol. Chem. 286, 21767–21778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gong B., Pan Y., Vempati P., Zhao W., Knable L., Ho L., Wang J., Sastre M., Ono K., Sauve A. A., and Pasinetti G. M. (2013) Nicotinamide riboside restores cognition through an upregulation of proliferator-activated receptor-γ coactivator 1alpha regulated β-secretase 1 degradation and mitochondrial gene expression in Alzheimer's mouse models. Neurobiol. Aging 34, 1581–1588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brown K. D., Maqsood S., Huang J. Y., Pan Y., Harkcom W., Li W., Sauve A., Verdin E., and Jaffrey S. R. (2014) Activation of SIRT3 by the NAD+ precursor nicotinamide riboside protects from noise-induced hearing loss. Cell Metab. 20, 1059–1068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cantó C., Houtkooper R. H., Pirinen E., Youn D. Y., Oosterveer M. H., Cen Y., Fernandez-Marcos P. J., Yamamoto H., Andreux P. A., Cettour-Rose P., Gademann K., Rinsch C., Schoonjans K., Sauve A. A., and Auwerx J. (2012) The NAD+ precursor nicotinamide riboside enhances oxidative metabolism and protects against high-fat diet-induced obesity. Cell Metab. 15, 838–847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Khan N. A., Auranen M., Paetau I., Pirinen E., Euro L., Forsström S., Pasila L., Velagapudi V., Carroll C. J., Auwerx J., and Suomalainen A. (2014) Effective treatment of mitochondrial myopathy by nicotinamide riboside, a vitamin B3. EMBO Mol. Med. 6, 721–731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cerutti R., Pirinen E., Lamperti C., Marchet S., Sauve A. A., Li W., Leoni V., Schon E. A., Dantzer F., Auwerx J., Viscomi C., and Zeviani M. (2014) NAD+-dependent activation of Sirt1 corrects the phenotype in a mouse model of mitochondrial disease. Cell Metab. 19, 1042–1049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bogan K. L., Evans C., Belenky P., Song P., Burant C. F., Kennedy R., and Brenner C. (2009) Identification of Isn1 and Sdt1 as glucose- and vitamin-regulated nicotinamide mononucleotide and nicotinic acid mononucleotide [corrected] 5′-nucleotidases responsible for production of nicotinamide riboside and nicotinic acid riboside. J. Biol. Chem. 284, 34861–34869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lu S. P., and Lin S. J. (2011) Phosphate-responsive signaling pathway is a novel component of NAD+ metabolism in Saccharomyces cerevisiae. J. Biol. Chem. 286, 14271–14281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lu S. P., Kato M., and Lin S. J. (2009) Assimilation of endogenous nicotinamide riboside is essential for calorie restriction-mediated life span extension in Saccharomyces cerevisiae. J. Biol. Chem. 284, 17110–17119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Belenky P., Stebbins R., Bogan K. L., Evans C. R., and Brenner C. (2011) Nrt1 and Tna1-independent export of NAD+ precursor vitamins promotes NAD+ homeostasis and allows engineering of vitamin production. PLoS One 6, e19710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hara N., Yamada K., Shibata T., Osago H., Hashimoto T., and Tsuchiya M. (2007) Elevation of cellular NAD levels by nicotinic acid and involvement of nicotinic acid phosphoribosyltransferase in human cells. J. Biol. Chem. 282, 24574–24582 [DOI] [PubMed] [Google Scholar]
- 23.Ipata P. L., and Balestri F. (2013) The functional logic of cytosolic 5′-nucleotidases. Curr. Med. Chem. 20, 4205–4216 [DOI] [PubMed] [Google Scholar]
- 24.Hunsucker S. A., Mitchell B. S., and Spychala J. (2005) The 5′-nucleotidases as regulators of nucleotide and drug metabolism. Pharmacol. Ther. 107, 1–30 [DOI] [PubMed] [Google Scholar]
- 25.Hunsucker S. A., Spychala J., and Mitchell B. S. (2001) Human cytosolic 5′-nucleotidase I: characterization and role in nucleoside analog resistance. J. Biol. Chem. 276, 10498–10504 [DOI] [PubMed] [Google Scholar]
- 26.Ipata P. L., and Tozzi M. G. (2006) Recent advances in structure and function of cytosolic IMP-GMP specific 5′-nucleotidase II (cN-II). Purinergic Signal. 2, 669–675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Spychała J., Madrid-Marina V., and Fox I. H. (1988) High Km soluble 5′-nucleotidase from human placenta. Properties and allosteric regulation by IMP and ATP. J. Biol. Chem. 263, 18759–18765 [PubMed] [Google Scholar]
- 28.Amici A., and Magni G. (2002) Human erythrocyte pyrimidine 5′-nucleotidase, PN-I. Arch. Biochem. Biophys. 397, 184–190 [DOI] [PubMed] [Google Scholar]
- 29.Hasmann M., and Schemainda I. (2003) FK866, a highly specific noncompetitive inhibitor of nicotinamide phosphoribosyltransferase, represents a novel mechanism for induction of tumor cell apoptosis. Cancer Res. 63, 7436–7442 [PubMed] [Google Scholar]
- 30.Trammell S. A., and Brenner C. (2013) Targeted, LCMS-based metabolomics for quantitative measurement of NAD+ metabolites. Comput. Struct. Biotechnol. J. 4, e201301012. [DOI] [PMC free article] [PubMed] [Google Scholar]