Background: Calcium-activated, family C2 cysteine peptidases (calpains) are rare among prokaryotes, and none have been characterized.
Results: Tpr peptidase from Porphyromonas gingivalis requires calcium for stability, autoprocessing, and activity.
Conclusion: Tpr is the calcium-regulated peptidase well adapted to function on the P. gingivalis cell surface.
Significance: Tpr is the first characterized bacteria-derived calpain-like peptidase.
Keywords: calcium, calpain, enzyme processing, periodontal disease, virulence factor, calcium-dependent activity
Abstract
Porphyromonas gingivalis is a peptide-fermenting asaccharolytic periodontal pathogen. Its genome contains several genes encoding cysteine peptidases other than gingipains. One of these genes (PG1055) encodes a protein called Tpr (thiol protease) that has sequence similarity to cysteine peptidases of the papain and calpain families. In this study we biochemically characterize Tpr. We found that the 55-kDa Tpr inactive zymogen proteolytically processes itself into active forms of 48, 37, and 33 kDa via sequential truncations at the N terminus. These processed molecular forms of Tpr are associated with the bacterial outer membrane where they are likely responsible for the generation of metabolic peptides required for survival of the pathogen. Both autoprocessing and activity were dependent on calcium concentrations >1 mm, consistent with the protein's activity within the intestinal and inflammatory milieus. Calcium also stabilized the Tpr structure and rendered the protein fully resistant to proteolytic degradation by gingipains. Together, our findings suggest that Tpr is an example of a bacterial calpain, a calcium-responsive peptidase that may generate substrates required for the peptide-fermenting metabolism of P. gingivalis. Aside from nutrient generation, Tpr may also be involved in evasion of host immune response through degradation of the antimicrobial peptide LL-37 and complement proteins C3, C4, and C5. Taken together, these results indicate that Tpr likely represents an important pathogenesis factor for P. gingivalis.
Introduction
Periodontitis is one of the most prevalent infectious inflammatory conditions that leads to damage of tooth-supporting structures (1). A severe form of the disease affects up to 2% of the population, although mild-to-moderate periodontitis is observed in the majority of adults (2). Results from clinical and epidemiologic studies suggest that periodontitis is a causal factor in the initiation and maintenance of the autoimmune inflammatory response in rheumatoid arthritis (3). Furthermore, periodontitis is also implicated in the development of cardiovascular diseases (4, 5) and diabetes (6).
Periodontitis is initiated by the colonization of the subgingival sulcus by pathogenic bacteria including Porphyromonas gingivalis, which is considered to be the major causative factor (7). This bacterium constitutes a consortium together with Tannerella forsythia and Treponema denticola, described as the “red complex,” which is strongly associated with the progression and severity of periodontitis (8). A common feature of these pathogens is secretion of peptidases that are required for pathogenesis. In fact, peptidases released by P. gingivalis and T. denticola function as important virulence factors (9, 10).
In comparison to other periodontal pathogens, the extracellular proteolytic system of P. gingivalis is very complex and encompasses a large number of specific peptidases. The most well characterized are cysteine peptidases referred to as Arg- and Lys-gingipains, which have endo- and exo-proteolytic activity selective for Arg-Xaa and Lys-Xaa peptide bonds, respectively (11–14). By contrast, very little is known about other peptidases, although they may also function as virulence factors and represent viable targets for drug design (15).
Bioinformatic analysis of the P. gingivalis genome revealed the presence of several genes of putative peptidases (16). One of these genes (PG1055) encodes a cysteine peptidase referred to as Tpr4 (thiol protease) (17). To date Tpr has only been expressed at very low yield in Escherichia coli and superficially characterized (18, 19). Part of the Tpr sequence shares significant sequence similarity with the catalytic domain of μ-calpain; therefore, it was assigned to clan CA, family C2 of the cysteine peptidases (MEROPS database ID number C02.022). The expression of the tpr gene is significantly elevated under nutrient-limited conditions (20), suggesting that Tpr participates in the generation of metabolic peptides assimilated by asaccharolytic P. gingivalis during these conditions, in which amino acids are used as the sole source of energy and carbon. Location of Tpr in the outer membrane fraction is consistent with this predicted function of Tpr (18). In this study we cloned, expressed, and purified a proform of Tpr (pro-Tpr). In addition this zymogen is autoprocessed and activated by the presence of high levels of calcium, suggesting that it may use calcium binding as a trigger to prevent premature activation of the peptidase.
Experimental Procedures
Reagents
Restriction enzymes and ligase T4 were obtained from Thermo Scientific (Waltham, MA). Oligonucleotides were acquired from Genomed (Warsaw, Poland). AccuPrimeTM Pfx DNA Polymerase was purchased from Invitrogen. Plasmid pGEX-6P-1, glutathione-SepharoseTM, PreScissionTM Protease, and SDS-PAGE High-Range Rainbow Marker were purchased from GE Healthcare. DNA cleanup, plasmid purification, and site-directed mutagenesis kits were from Stratagene (La Jolla, CA). Peptide substrates were purchased from Bachem (Bubendorf, Switzerland). All inhibitors (leupeptin: N-acetyl-l-leucyl-l-leucyl-l-argininal hemisulfate salt; E-64: trans-epoxy-succinyl-l-leucyl-amido (4-guanidino) butane; pepstatin A; IAA: iodoacetic acid; Pefabloc), isopropyl-1-thio-β-d-galactopyranoside, EDTA, and Bz-Arg-pNA were purchased from Sigma. Casein, human fibrinogen, and fibronectin were obtained from Sigma. Human complement proteins C3, C4, and C5 were purchased from Complement Technology (Tyler, TX). Synthetic LL-37 (LLGDFFRKSKEKIGKEFKRIVQRIKDFLRNLVPRTES) was obtained from GenScript (Piscataway, NJ).
Bacterial Strains and Culture Conditions
P. gingivalis W83 was grown anaerobically in Scheadler Broth supplemented with hemin (final concentration: 5 μg/ml), vitamin K (final concentration: 1 μg/ml), and cysteine-HCl (final concentration: 0.5 mg/ml). Cultures were incubated in an anaerobic chamber in a 5% CO2, 10% H2, 85% N2 atmosphere at 37 °C. E. coli was grown in Luria-Bertani (LB) broth supplemented with ampicillin (final concentration, 100 μg/ml) at 37 °C. E. coli DH5α was used as the host for plasmid construction and amplification, and E. coli BL21 was used for expression.
Construction of the WT and Mutant Protein Vectors
Genomic DNA of P. gingivalis was extracted from strain W83. The entire gene, PG1055, was amplified by PCR using the forward primer 5′-CCGGGATCCATGGAAAAGAAATTAGTACCGCAA-3′ (TPRnewFP) and the reverse primer 5′-CCGGGAATCCTTAATACATATATGCGATGTACCAGAA-3′ (TPRnewRP), which introduce BamHI and EcoRI restriction sites, respectively (underlined). The PCR product was purified and cloned into the pGEX-6P-1, which provides the sequence for an N-terminal glutathione S-transferase tag (GST) and inserts five additional residues (Gly-Pro-Leu-Gly-Ser) in the N terminus of Tpr. The WT (wild type) construct was used to produce C229A and C229S variants (TprC229A and TprC229S) in which the mutations substituted active site Cys-229 with alanine or serine. The following sets of primers were used: C229A-F, 5′-GGAATCGCTGGCGATGCTTACATGCTTGCAG-3′, and C229A-R, 5′-GCTGCAAGCATGTAAGCATCGCCAGCGATTCC-3′; C229S-F, 5′-GGAATCGCTGGCGATTCTTACATGCTTGCAGC-3′, and C229S-R, 5′-GCTGCAAGCATGTATCTATCGCCAGCGATTCC-3′. The mutations were verified by double-stranded DNA sequencing, and constructs were transformed into the electrocompetent E. coli BL21 strain using a Bio-Rad gene pulser (2 kV).
Recombinant Protein Expression and Purification
Protein production and purification procedures were essentially the same for WT and mutant Tpr. Routinely, E. coli transformed with expression constructs were grown at 37 °C in 1 liter of LB medium to an optical density (A600 nm) of 0.7. Isopropyl-1-thio-β-d-galactopyranoside was added into the culture to a final concentration of 0.5 mm to induce protein expression, and incubation was continued for 3 h at 37 °C. Cells were harvested, washed, and resuspended in 30 ml of PBS (140 mm NaCl, 2.7 mm KCl, 10 mm Na2HPO4, and 1.8 mm KH2PO4, pH 7.4) supplemented with 10 mm EDTA. The pellet was disrupted by ultrasonication on ice using a broad tip at an amplitude of 60% for 5 min (pulse = 30 s). After centrifugation (18,000 × g), the supernatant was collected, filtered through a 0.45-μm membrane, and applied onto a glutathione-Sepharose column (5 ml bed volume) equilibrated with PBS supplemented with 10 mm EDTA. The column was washed with 100 ml of the same buffer to elute unbound proteins. Bound proteins were eluted stepwise with elution buffer (50 mm Tris-HCl, pH 8.0, and 20 mm l-glutathione) and dialyzed against PreScission Protease cleavage buffer (50 mm Tris-HCl, 150 mm NaCl, 1 mm EDTA, and 1 mm dithiothreitol, pH 7.0) followed by the addition of 20 μl of PreScission Protease stock solution (2000 units/ml). The cleavage reaction was carried out for 16 h at 4 °C after which the sample was dialyzed against PBS containing 10 mm EDTA and reloaded onto the glutathione-Sepharose column to remove GST. The recombinant protein was collected in the flow-through. The final purification of recombinant protein was accomplished by gel filtration on Superdex 75 (16/60 gel filtration column) pre-equilibrated with 50 mm Tris-HCl, 150 mm NaCl, and 10 mm EDTA, pH 7.5. The total amount of purified protein was determined by absorbance at a 280-nm measurement using a Spectrophotometer NanoDrop 1000 (Thermo Scientific). The GST-free Tpr forms were routinely purified with a yield of ∼1 mg per liter of culture.
Gel Electrophoresis
SDS-PAGE was used to verify homogeneity of purified protein and to monitor autocatalytic processing and degradation of physiological substrates and processing of TprC229S by active Tpr and other peptidases. To this end the Tris-HCl/Tricine buffer system was used with 12.5% gels (21). Gels were stained with 0.1% Coomassie Brilliant Blue R-250 in 10% acetic acid and destained with 10% acetic acid, 30% methanol solution.
N-terminal Sequence and MALDI-TOF Analysis
WT Tpr and the TprC229S mutein as well as low molecular mass forms of Tpr were separated by SDS-PAGE and electroblotted on polyvinylidene difluoride membrane. Membranes were stained with 0.5% Coomassie Blue, and protein bands of interest were subjected to automated Edman degradation on a Procise 494-HT protein sequencer (Proteomics and Bioinformatics Facility from Autonomous University of Barcelona, Barcelona, Spain). MALDI-TOF analysis was performed on an UltrafleXtrem MALDI-TOR/TOF (Bruker Daltonics and Proteomics and Bioinformatics Facility from Autonomous University of Barcelona, Barcelona, Spain).
Liquid Chromatography Tandem Mass Spectrometry (LC-MS/MS)
Samples were analyzed by nanoflow LC-MS/MS using an EASY-nLC II system (Thermo Scientific) connected to a TripleTOF 5600 mass spectrometer (AB Sciex). The Tpr-digested samples were dissolved in 0.1% formic acid and loaded onto a 0.1 × 20-mm C18 trap column and a 0.075 × 150-mm C18 analytical column. The peptides were eluted and electrosprayed directly into the mass spectrometer using a 20-min gradient from 5% to 35% solvent B (0.1% formic acid in 90% acetonitrile) with a flow of 250 nl/min. An information-dependent acquisition method was employed to automatically run experiments acquiring up to 25 MS/MS spectra per cycle using 0.9-s cycle times and an exclusion window of 6 s. The raw data were converted to MGF files and searched against Swiss-Prot mammals database (Version 2015_07) using an in-house Mascot search engine. No enzyme was specified, and oxidation of methionine was included as a variable modification. The peptide mass tolerance and fragment mass tolerance were set at 10 ppm and 0.2 Da respectively, and the significance threshold (p) was set at 0.01.
Production of Tpr-specific Antiserum
Recombinant protein Tpr55/Tpr48 was used to raise Tpr-specific antiserum by Capra Science Antibodies AB. A total of 400 μg of antigen in Freund's Complete Adjuvant was injected into one New Zealand rabbit, and immunization was carried out using a standard 12-week program including four immunizations (100 μg of antigen in Freund's Incomplete Adjuvant) at 2-week intervals and 3 bleeds. Antiserum from bleeds was analyzed by enzyme-linked immunosorbent assay using unconjugated peptide as the absorbed antigen.
Cellular Fractionation Procedure and Tpr Sublocalization
Cellular fractions of P. gingivalis W83 were prepared as described previously (22). All steps were carried out at 4 °C. Purity of the various fractions was verified by Western blotting using P. gingivalis anti-gingipains antibodies. The cellular fractions were mixed with sample buffer, boiled, and separated by reducing SDS-PAGE; resolved proteins were transferred onto polyvinylidene membrane. The membrane was blocked overnight with 3% BSA in TBS-Tween buffer (10 mm Tris-HCl, 150 mm NaCl, and 0.05% Tween 20, pH 7.5) at 4 °C and incubated with 10 μl of Tpr antiserum for 2 h at room temperature. Next, the membrane was washed with TBS-Tween buffer and incubated with goat anti-rabbit HRP-conjugated antibodies (Thermo Scientific Pierce) for 1 h at room temperature. The signals were detected by enhanced chemiluminescence (ECL) and visualized using ChemiDocTM MP System.
Autoprocessing of Tpr and DCG-04 Probing
Tpr was incubated at 37 °C and at room temperature in 50 mm Tris-HCl, 150 mm NaCl, 5 mm CaCl2, and 1 mm DTT, pH 7.5, for up to 24 h. At specific time intervals samples were withdrawn and subjected to DCG-04 (2 μm) active-site probing. Samples prepared as described above were separated by 12.5% SDS-PAGE and transferred to the PVDF membrane. The membrane was blocked overnight with 3% BSA in TBS-Tween buffer at 4 °C and then incubated with 1 μg/ml of streptavidin-horseradish peroxidase (Sigma) for 1 h at room temperature. DCG-04-reactive polypeptides were detected by ECL and visualized using ChemiDocTM MP System.
Proteolytic Activity Assays and Biochemical Characterization
Activity was determined using various fluorescent substrates (Bz-Arg-pNA, Z-Tyr-Val-Ala-Asp-7-amino-4-trifluorometylocumarin (AMC), Ac-Asp-Glu-Val-Asp-AFC, Ac-Leu-Glu-His-Asp-AFC, Z-Ala-Ala-Asn-AMC, H-Gly-Phe-AMC, Boc-Val-Leu-Lys-AMC, Boc-Ala-Ala-Gly-pNA, Suc-Leu-Leu-Val-Tyr-pNA, Z-Phe-Arg-pNA, H-Leu-Gly-pNA, Suc-Ala-Ala-Ala-pNa, Suc-Ala-Ala-Pro-Phe-pNA, N-carbobenzyloxy-Arg-pNA, Suc-Leu-Leu-Val-Tyr-AMC, Z-Arg-Arg-AMC, Z-Phe-Arg-AMC, DQ-gelatin (fluorescein conjugate gelatin), azocoll, and azocasein). Routine assays were performed at 37 °C using 250 nm titrated enzyme in 50 mm Tris-HCl, 5 mm CaCl2, 150 mm NaCl, and 0.2 mm sodium azide, pH 7.5, freshly supplemented with 1 mm DTT, with a substrate concentration of 5 μm. Before substrate addition, the enzyme was preincubated in assay buffer for 30 min. Formation of fluorescent degradation product was monitored continuously at 355-nm excitation and 460-nm emission for AMC derivatives and at 444-nm excitation and 538-nm emission for AFC and DQ-gelatin using a micro-titer fluorescent plate reader (Spectra Max Gemini, Molecular Devices). The activity on p-nitroanilide substrates was monitored at 405 nm. Proteolysis of azocoll and azocasein was assayed at 545 and 366 nm, respectively.
Human plasma fibrinogen and fibronectin were incubated with Tpr33 at a substrate/enzyme ratio of 100:1 and 10:1 by weight, respectively, at 37 °C in 50 mm Tris-HCl, 150 mm NaCl, 5 mm CaCl2, and 0.2 mm sodium azide, pH 7.5, and samples withdrawn at specific time points were subjected to SDS-PAGE. Proteins incubated with TprC229S and Tpr33 inhibited with E-64 served as controls. The same procedure was used for LL-37, casein, and human complement components C3, C4, and C5 (concentrations are indicated under “Results”).
Suc-Leu-Leu-Val-Tyr-AMC (5 μm) was used as substrate to determine the enzyme pH optimum, E-64 titration, and effect of inhibitors on proteolytic activity of Tpr. To test the inhibitory effect, the enzyme was preincubated for 1 h with different inhibitors (concentrations are indicated under “Results”) at room temperature, and residual activity was determined. The effect of Ca2+ ions on Tpr activity was determined by measuring the rate of hydrolysis of Suc-Leu-Leu-Val-Tyr-AMC after different incubation times in the assay buffer supplemented with increasing concentrations of Ca2+ (0, 10 μm, 100 μm, 0.5 mm, 1 mm, 2 mm, 3 mm, 5 mm, 7 mm, and 10 mm). The pH optimum was detected using the following buffers at 50 mm concentrations: MES (pH 5.5, 6.0, and 6.3), MOPS (pH 6.5, 7.0, and 7.6), HEPES (pH 7.0 and 7.5), Tris-HCl (pH 7.5, 8.0, and 8.5), and CAPS (pH 8.5, 9.0, and 10.0). All buffers contained 5 mm CaCl2, 150 mm NaCl, and 1 mm DTT. The Michaelis-Menten constant (Km) for enzymatic hydrolysis of Z-Phe-Arg-AMC and Suc-Leu-Leu-Val-Tyr-AMC was determined by plotting the experimentally determined reaction velocity (1/V) against the reciprocal of substrate concentration (1/[S]). Linear regression analysis was performed using GraphPad Prism 3.0, and software-fitting was employed to produce the Lineweaver-Burk plots. All kinetic measurements were performed in duplicate.
Processing of Pro-TprC229S by WT Tpr and Other Cysteine Peptidases
Five micrograms of TprC229S in 50 mm Tris-HCl, 150 mm NaCl, 5 mm CaCl2, and 1 mm DTT, pH 7.5, were incubated at 37 °C for up to 24 h with WT Tpr, and samples taken at specific time intervals were subjected to SDS-PAGE analysis. A total of 5 μg of the TprC229S in TNCT buffer (100 mm Tris-HCl, 150 mm NaCl, 0.05% Tween, and 20 mm l-cysteine, pH 7.6) in the presence or absence of 5 mm CaCl2 was incubated at 37 °C for 2 h with activated Kgp and RgpB (enzyme:substrate ratio is indicated under “Results”). Samples were separated by 12.5% SDS-PAGE.
Differential Scanning Fluorimetry
Differential scanning fluorimetry was used to identify the influence of Ca2+ ion concentration on TprC229S thermal stability (23). TprC229S (0.1 mg/ml) was dissolved in buffer (10 mm HEPES, pH 7.5) in the presence or absence of Ca2+ (0.1, 0.5, 1, 2, 3, 5, 7, and 10 mm) with 1:1000-fold diluted SYPRO Orange dye (Invitrogen) in a final reaction volume of 35 μl in a PCR plate. Samples were equilibrated at 25 °C for 5 min and then heated from 25 °C to 95 °C at a rate of 1 °C per min. Protein unfolding was monitored by measuring the fluorescence (at 490-nm excitation and 625-nm emission) intensity using an Mx3005P quantitative PCR system (Stratagene). Analysis of the results was performed using OriginPro 8.5, and the Boltzmann distribution curve was used to calculate the protein melting temperature (Tm).
Results
Expression and Purification
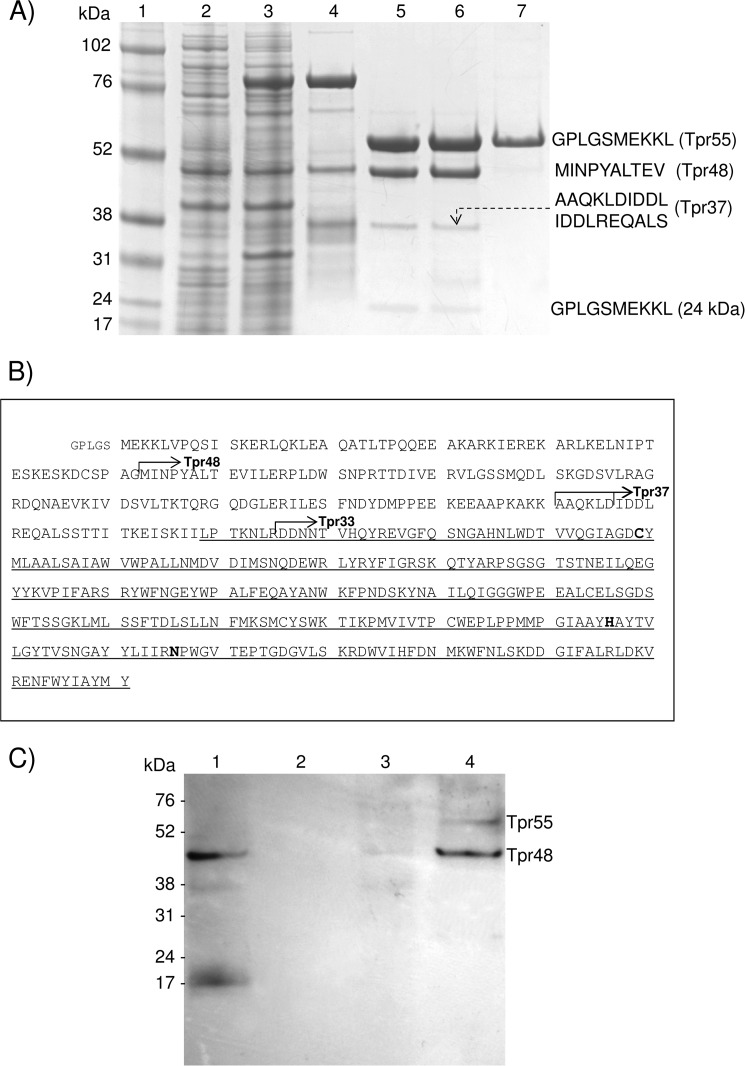
Recombinant, WT, full-length Tpr, and a mutated form of the enzyme in which the putative catalytic cysteine residue was substituted with serine (TprC229S) were expressed at high yield (>2 mg/liter) in E. coli as 84-kDa fusion proteins with an N-terminal glutathione S-transferase domain (GST-Tpr). Of note, only a negligible amount (<0.25 mg/5 liters of culture) of recombinant TprC229A was obtained in the same system. The recombinant proteins were isolated from bacterial extracts by affinity chromatography on glutathione-Sepharose, and the GST tag (29 kDa) was removed by digestion with PreScission Protease. The recombinant Tpr protein was further purified by gel filtration. TprC229S was obtained as a homogenous 55-kDa protein (Fig. 1A, lane 7), whereas purification of the WT enzyme yielded three protein species of 55, 48, and 37 kDa (as determined by MALDI-TOF mass spectrometry) (Fig. 1A, lane 5). These same processed species were obtained even in the presence of a protease inhibitor mixture to prevent proteolysis. Notably, the 55-kDa form of the WT enzyme (Tpr55) (Fig. 1A, lane 6) could only be obtained if EDTA was present in all buffers used for enzyme purification. N-terminal amino acid sequence analysis revealed that the 48-kDa form (Tpr48) is derived from full-length Tpr by hydrolysis of the Gly-62–Met-63 peptide bond (Fig. 1B), whereas trace amounts of the 37-kDa protein (Tpr37) were generated by proteolysis of the Lys-160–Ala-161 and Asp-166–Ile-167 peptide bonds. These results suggest that full-length Tpr must undergo proteolysis within the E. coli cytoplasm and/or during an initial step of the purification procedure, precluding successful separation of the homogenous 55-kDa form of the active enzyme.
FIGURE 1.
Expression of WT and catalytic mutant Tpr and assessment of their autoprocessing activities. A, both WT (Tpr) and the active-site cysteine-to-serine mutant (TprC229S) were expressed in E. coli and purified. Lane 1, molecular mass marker (kDa); lanes 2 and 3, cell extracts of E. coli before and 3 h after induction of GST-Tpr expression with isopropyl-1-thio-β-d-galactopyranoside, respectively; lane 4, a fusion protein GST-Tpr (84 kDa) obtained from E. coli extract by affinity chromatography on glutathione-Sepharose; lane 5, Tpr55, Tpr48, and Tpr37 after GST was cleaved off by PreScission Protease and removed on glutathione-Sepharose; lane 6, Tpr55, Tpr48, and Tpr37 purified by gel filtration; lane 7, TprC229S (55 kDa) expressed and purified in the same way as WT Tpr. N-terminal sequences of Trp55, TprC229S, Tpr48, and Tpr37, determined by Edman degradation, are shown on the right side. B, the primary structure of Tpr with identified proteolytic cleavage sites (arrows) that generated the Tpr48 (Gly-62–Met-63), Tpr37 (Lys-160–Ala-161 and Asp-166–Ile-167), and Tpr33 (Arg-195–Asp-196) forms of the peptidase. Catalytic residues (Cys-229, His-406, and Asn-426), identified by alignment of the Tpr sequence with papain- and calpain-like peptidases, are in bold font. A domain similar to papain is indicated by a thin underline, starting from Leu-189. Of note, Tpr is encoded without a signal peptide. A pentapeptide (small font) preceding Met-1 is derived from the plasmid. C, Tpr55 and Tpr48 were localized in P. gingivalis W83 outer membrane and cellular extract. Lane 1, whole cellular extract; lane 2, cytoplasmic/periplasmic fraction; lane 3, inner membrane fraction; lane 4, sarkosyl-insoluble outer membrane fraction. Western blot analysis of cell fractions of P. gingivalis were probed with Tpr-specific rabbit antiserum.
By subjecting a sarkosyl-insoluble outer membrane preparation to Western blot analysis with a Tpr-specific antiserum, we were able to confirm that the two primary Tpr forms (55 and 48 kDa) are localized on the outer membrane of P. gingivalis W83 (Fig. 1C). Significantly, the Tpr protein was also found in whole cellular extract but not in cytoplasmic and periplasmic fractions. These results confirm the outer membrane localization of Tpr (18).
Tpr Autoprocessing and Activity Assessment of Different Molecular Forms of Tpr
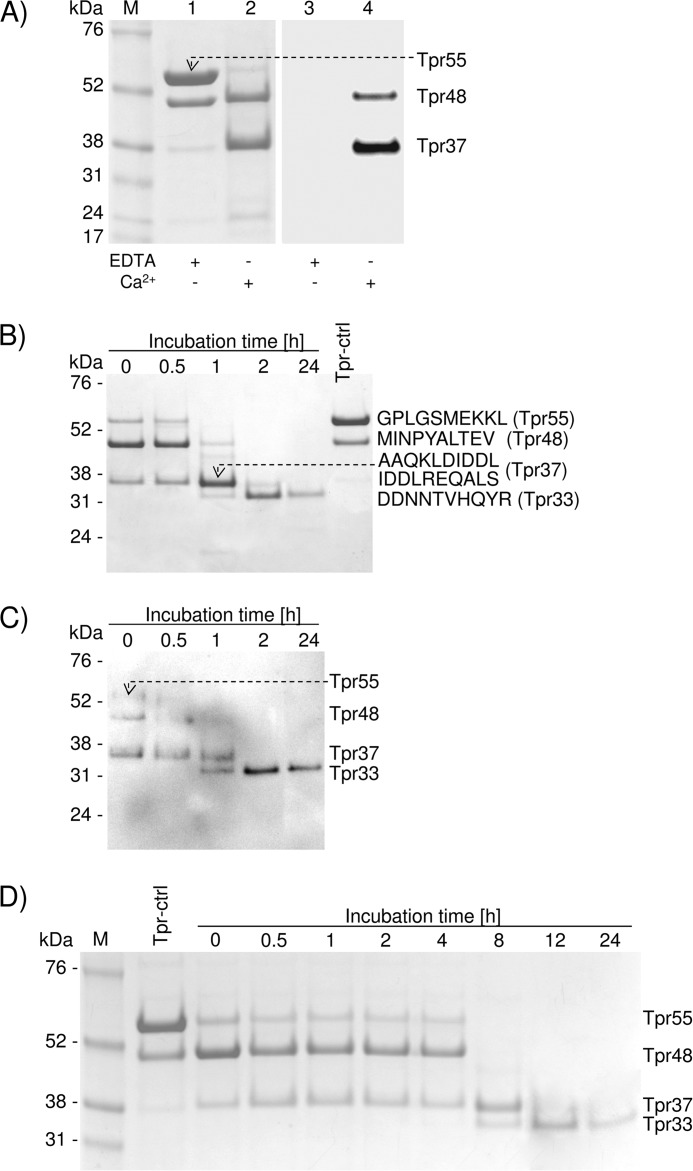
A large prodomain precedes the Tpr peptidase domain and may contribute to latency. However, in the case of recombinant Tpr, the level of latency appeared to be rather low because even in the presence of inhibitors, the full-length pro-Tpr55 was processed to Tpr48 by the autoproteolytic cleavage of the Gly-62–Met-63 peptide bond. To determine whether these forms of Tpr are proteolytically active, we applied a biotinylated derivative of E-64 (DCG-04), an activity-based probe that covalently binds to the active-site cysteine residue of clan CA/CB cysteine peptidases (24). We pretreated purified Tpr55/Tpr48 with DCG-04 in the activity assay buffer and in a buffer in which Ca2+ was chelated by EDTA. We found that in the presence of Ca2+, Tpr55 was converted to Tpr48 and Tpr37 (Fig. 2A, lane 2), both of which were labeled by DCG-04, indicating that they represent active forms of Tpr (Fig. 2A, lane 4). By contrast, EDTA prevented not only autoproteolytic processing of Tpr (Fig. 2A, lane 1) but also labeling by DCG-04 (Fig. 2A, lane 3). Together, these results confirm the essential role of calcium ions in activity and autoprocessing of pro-Tpr to the mature, active forms of the enzyme.
FIGURE 2.
Tpr55 is a zymogen requiring Ca2+ for sequential autoproteolytic processing to generate active forms of the enzyme: first Tpr48, then Tpr37, and finally stable Tpr33. A, purified Tpr55/Tpr48 (in buffer containing EDTA) was incubated in assay buffer without calcium (EDTA +/Ca2+ −) or with CaCl2 (EDTA −/Ca2+ +), and samples were treated with DCG-04. After a 15-min incubation at 37 °C, the reaction was terminated by boiling in denaturing sample buffer. Proteins were resolved by SDS-PAGE and subjected to Western blot analysis. Lanes 1 and 2, protein staining; lanes 3 and 4, detection of probe labeled active Tpr. B and C, purified Tpr55/Tpr48 was supplemented with CaCl2 and treated with DCG-04 (at time 0 or after incubation at 37 °C for specific time intervals). A sample incubated in buffer with EDTA (Tpr-ctrl) served as a control. Proteins were resolved by SDS-PAGE, and gels were stained for protein (B) or subjected to Western blot analysis to detect probe labeled proteins (C). SDS-PAGE-separated proteins were subjected to N-terminal sequencing analysis. D, purified Tpr55/Tpr48 was incubated in the assay buffer with Ca2+ at 21 °C for the indicated periods of time, and samples were analyzed by SDS-PAGE.
To investigate the time-dependent autoprocessing and activity of processed forms of Tpr in more detail, we activated Tpr55/Tpr48 with calcium and then treated with E-64 (Fig. 2B) or DCG-04 at different time intervals during incubation at 37 °C (Fig. 2C). Inhibitors added simultaneously with CaCl2 (incubation time = 0 h) failed to prevent autoproteolysis of Tpr55, which was processed to the low molecular mass forms (Tpr48 and Tpr37). Tpr55 was first cleaved to Tpr48, which was almost completely converted to Tpr37 within 1 h of incubation. Finally, Tpr37 was further truncated to Tpr33, a form of the enzyme that was stable for at least 24 h (Fig. 2B). N-terminal sequence analysis revealed sequential proteolysis events, first at Gly-62–Met-63 (Trp48), then at Lys-160–Ala-161 and Asp-166–Ile-167 (Tpr37), and finally at Arg-195–Asp-196 (Tpr33), leading to sequential generation of Tpr48, Tpr37, and Tpr33 from the high molecular weight precursor. Autoprocessing of Tpr also occurred at 21 °C. Moreover, although the initial cleavage happened at the same rate as observed at 37 °C, the subsequent processing of Tpr48 occurred at a much slower rate (Fig. 2D). Significantly, DCG-04 did not label Tpr55, whereas the intensity of labeling of Tpr48 was visibly weaker than that of Tpr37 and Tpr33 (Fig. 2C) despite the presence of more Trp48 protein at incubation times of 0 and 30 min.
Activity and Inhibition Profile of Tpr
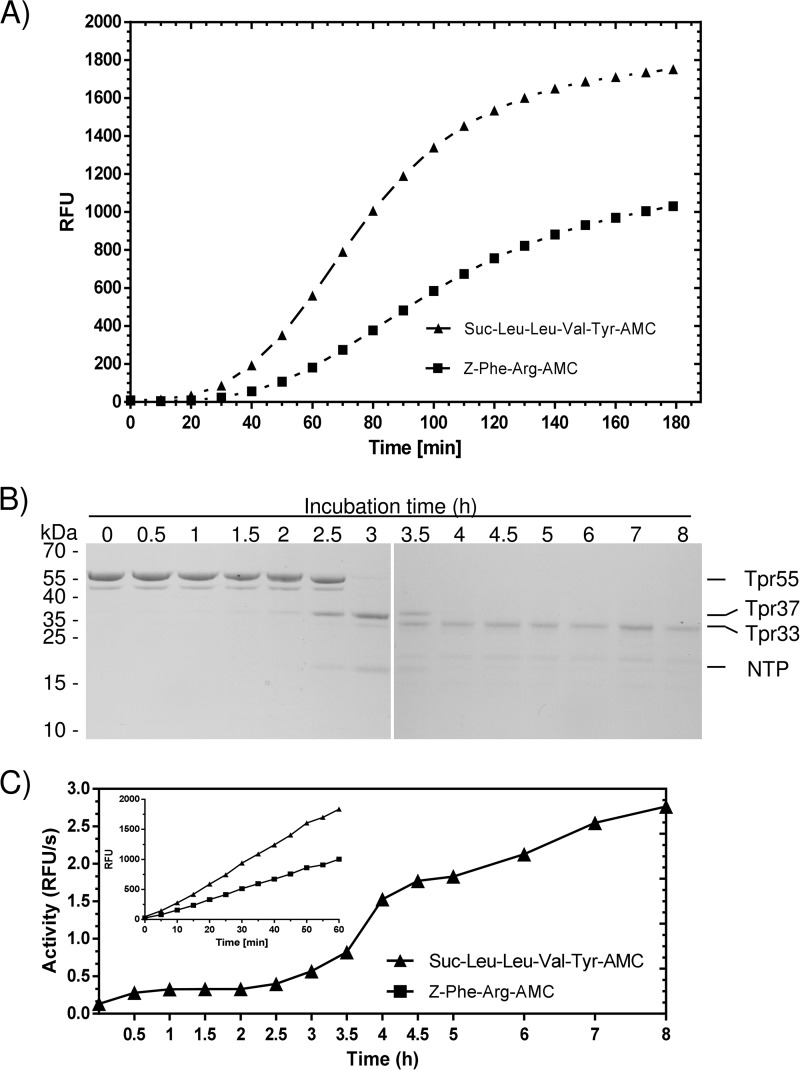
To confirm the enzymatic activity of the Tpr peptidase, we used a panel of substrates including Bz-Arg-pNA, Z-Tyr-Val-Ala-Asp-AFC, Ac-Asp-Glu-Val-Asp-AFC, Ac-Leu-Glu-His-Asp-AFC, Z-Ala-Ala-Asn-AMC, H-Gly-Phe-AMC, Boc-Val-Leu-Lys-AMC, Suc-Leu-Leu-Val-Tyr-pNA, Z-Phe-Arg-pNA, Boc-Ala-Ala-Gly-pNA, H-Leu-Gly-pNA, Suc-Ala-Ala-Ala-pNa, Suc-Ala-Ala-Pro-Phe-pNA, N-carbobenzyloxy-Arg-pNA, Suc-Leu-Leu-Val-Tyr-AMC, Z-Arg-Arg-AMC, Z-Phe-Arg-AMC, DQ-gelatin, azocoll, and azocasein. Of all of these chromogenic or fluorogenic synthetic peptide substrates, only Suc-Leu-Leu-Val-Tyr-AMC and Z-Phe-Arg-AMC were hydrolyzed efficiently (Fig. 3A).
FIGURE 3.
Tpr hydrolyzes Suc-Leu-Leu-Val-Tyr-AMC and Z-Phe-Arg-AMC with a lag-phase (A), which is apparently due to the enzyme inhibition by the N-terminal remaining non-covalently associated with Tpr33 (B and C). Recombinant Tpr (Tpr55/Tpr48) was preincubated in the assay buffer for 1 h before the substrate was added, and then fluorescence was recorded (λex = 355 nm and λem = 460 nm) for 3 h (A). Tpr processing was activated by calcium (2.5 mm final concentration), and at specific time points aliquots were withdrawn and subjected to the SDS-PAGE analysis (B). Simultaneously, the activity was measured employing Suc-Leu-Leu-Val-Tyr-AMC as the substrate (C). The inset shows substrates hydrolysis by Tpr preincubated in the presence of 2.5 mm CaCl2 for 4 h. RFU, relative fluorescence units.
Interestingly a 40-min lag phase in substrate hydrolysis was observed for both substrates (Fig. 3A). To assess the lag phenomenon, we monitored time-dependent Tpr autoprocessing at 2.5 mm concentration of Ca2+, which is equal to the physiological calcium concentration in gingival crevicular fluid and human serum. To this end the Tpr55/48 sample in EDTA was extensively dialyzed against 50 mm Tris, 150 mm NaCl, 0.2 mm sodium azide, pH 7.5 buffer. The autoprocessing reaction was started by the addition of CaCl2, and aliquots taken at different time points were subjected to SDS-PAGE (Fig. 3B) and assayed for enzyme activity employing Suc-Leu-Leu-Val-Tyr-AMC (Fig. 3C). In such conditions Tpr55 was converted directly to Tpr37 (without forming Tpr48) and the intact N-terminal profragment (NTP) was clearly, but transiently (time points 2.5, 3, and 3.5 h) visible in the SDS-PAGE (Fig. 3B). We observed the dramatic increase of activity after 3 h of incubation (Fig. 3C), which correlated with release of Tpr33 and degradation of the NTP (Fig. 3B). These results suggest that the lag phase is an effect of latency maintained by the NTP. Indeed the Tpr55 enzyme preincubated for 4 h when the NPT is already degraded (Fig. 3B) hydrolyzed substrates without a lag phase (Fig. 3C, inset). To further confirm that the NTP could form a noncovalent complex with Tpr37, TprC229S was subjected to limited proteolysis by trypsin, resulting in removal of the NTP and generation of TprC229S37. The resulting forms of Tpr were separated by size exclusion chromatography, and a single peak with molecular mass equivalent to the complex (55 kDa) was eluted. The SDS-PAGE analysis of the purified protein confirmed a stoichiometric amount of both the NTP and TprC229S37 (data not shown).
Collectively, our data suggest that Tpr55 is a precursor zymogen whose autoprocessing is initiated by calcium and proceeds through the NTP degradation exerted by residual proteolytic activity of Tpr37 in a complex with the NTP. In vivo, however, it is likely that other proteolytic enzymes abundant on the P. gingivalis surface also degrade the NTP. The released NTP, however, remains non-covalently associated with the enzyme, in which activity is released only after the NTP is degraded.
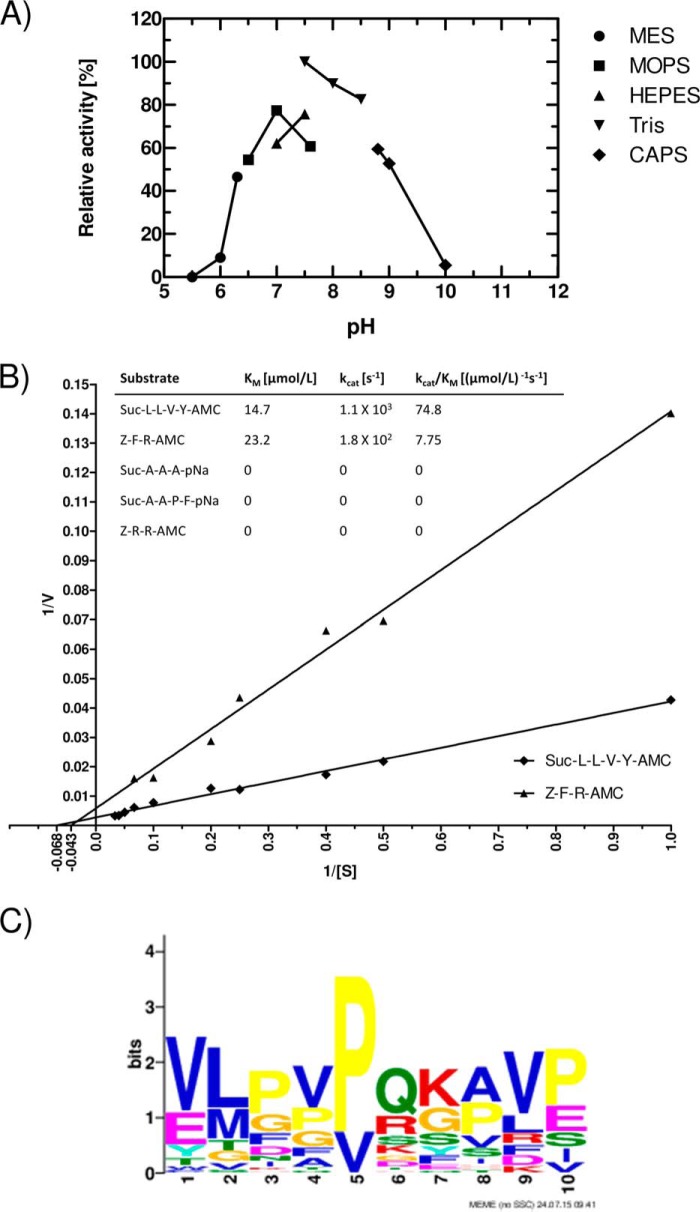
The recombinant enzyme was also active over a rather broad pH range, from 6.5 to 9.0 (Fig. 4A), with a pH optimum of 7.5. The activity was dependent on buffer composition; for all subsequent investigations, 50 mm Tris-HCl and 150 mm NaCl, pH 7.5, supplemented with 5 mm CaCl2 and 1 mm DTT was used as an assay buffer.
FIGURE 4.
Tpr hydrolyzes Suc-Leu-Leu-Val-Tyr-AMC at a pH in the range from 6.5 to 9.0 (A), obeying Michaelis-Menten kinetics (B), and the enzyme has a broad specificity (C). The pH profile was determined by assaying the activity in buffers of different pH (all containing 5 mm CaCl2 and 1 mm DTT), with substrate added after 30 min of preincubation. Activity (relative fluorescence units/min at the linear phase of fluorescence increase) at 50 mm Tris-HCl, 5 mm CaCl2, and 1 mm DTT, pH 7.5, was arbitrarily taken as 100% (A). Km values for Suc-Leu-Leu-Val-Tyr-AMC and Z-Phe-Arg-AMC were determined from the initial velocity of hydrolysis of substrates at concentrations in the range from 0.5 μm to 30 μm (B). A Tpr cleavage consensus sequence motif was derived from digestion of bovine casein and human fibrinogen by Tpr33 (C). Residues at positions 6 and 7 are equivalent to P1-P1′. The motif was discovered using MEME (Multiple Em for Motif Elicitation) with default parameters except for site distribution = 0 or 1 set for the presence or absence, respectively, of a given residue.
We further characterized Tpr33 activity against the Z-Phe-Arg-AMC and Suc-Leu-Leu-Val-Tyr-AMC substrates. For both substrates, the enzymatic reaction obeyed the Michaelis-Menten kinetics, and Km and kcat values could be calculated from a Lineweaver-Burk plot (Fig. 4B). The Km values of 14.7 μm and 23.2 μm and kcat values of 1.1 × 103 s−1 and 1.8 × 102 s−1 were obtained for the Suc-Leu-Leu-Val-Tyr-AMC and Z-Phe-Arg-AMC, respectively. Of note, no activity was observed for structurally similar substrates (Fig. 4B, inset table), and among the several tested proteinaceous substrates, Tpr degraded only DQ-gelatin (267.12 relative fluorescence units/s/μg of protein) while exhibiting no evidence of proteolysis of azocoll or azocasein. Furthermore, regardless of molecular form (55, 48, 37, or 33 kDa), Tpr did not degrade unlabeled gelatin and casein as assessed by zymography (data not shown), contradicting a previous report (17).
To further define Tpr cleavage specificity, the enzyme was incubated with bovine casein and human fibrinogen for a set time where no fragments higher than 10 kDa were visible on SDS-PAGE gels. Peptides in the digests were identified by mass spectrometry, allowing identification of the specific sites hydrolyzed within the polypeptide chains of the substrates. Based on these results, we were able to map the observed sequence specificity using a web logo method (Fig. 4C). Like the human calpains (Merops id C02.001, C02.002), Tpr has an overall broad specificity with some preferences for Val/Glu, Leu/Met, Val, and Pro/Glu at positions P6, P5, P3′, and P4′, respectively. We also observed a slight preference for positively charged amino acids and Gln at the P1 position. Interestingly, we also found almost exclusive preference for proline and valine at the P2 position. Regardless of this result for the P2 position, we did not observe any clear consensus sequential motif for the protease.
Significant sequence identity of Tpr, especially around the catalytic residues, to cysteine peptidases of the papain (C1) and calpain (C2) families clearly indicated that Cys-229 is the catalytic residue. This was confirmed by the observation that TprC299S did not exhibit any proteolytic activity. Consistent with its similarity to C1 and C2 clan peptidases, Tpr activity was inhibited by E-64, a diagnostic cysteine peptidase inhibitor. Inhibition by E-64 was stoichiometric and allowed active-site titration of Tpr (data not shown). Using this inhibitor, we found that the concentration of active Tpr in different preparations varied from 60 to 70% of total protein content. Apart from E-64, leupeptin and iodoacetic acid also quantitatively inhibited Tpr activity, whereas the aspartic peptidase inhibitor, pepstatin A, had no effect (Table 1). Conversely, Tpr was completely inhibited by TLCK, an inhibitor of serine and cysteine peptidases, but only partially by Pefabloc, a serine protease-specific inhibitor (Table 1).
TABLE 1.
Effect of inhibitors and activators on the proteolytic activity of Tpr
Activity was determined at 37 °C using Suc-LLVY-AMC as a substrate. Activity in buffer (50 mm Tris-HCl and 150 mm NaCl, pH 7.5, supplemented with 5 mm CaCl2) alone was taken as 100%. Sample treated with 5 mm and 10 mm EDTA was saturated with Ca2+ (15 mm and 25 mm, respectively), and residual activity was determined.
| Inhibitors and metal ions | Concentration | % of control |
|---|---|---|
| E-64 | 50 μm | 0 |
| 100 μm | 0 | |
| Leupeptin | 50 μm | 0 |
| 100 μm | 0 | |
| Iodoacetic acid | 1 mm | 0 |
| 5 mm | 0 | |
| Pepstatin A | 50 μm | 73 |
| 100 μm | 60 | |
| Orthophenanthroline | 50 μm | 101 |
| 100 μm | 104 | |
| TLCK | 50 μm | 0 |
| 100 μm | 0 | |
| Pefabloc | 50 μm | 67 |
| 100 μm | 58 | |
| EDTA | 5 mm | 0 |
| 10 mm | 0 | |
| Ca2+ | – | 0 |
| 10 μm | 0 | |
| 100 μm | 0 | |
| 0.5 mm | 0 | |
| 1 mm | 0 | |
| 2 mm | 25 | |
| 3 mm | 54 | |
| 5 mm | 100 | |
| 7 mm | 35 | |
| 10 mm | 31 | |
| DTT | 1 mm | 109 |
| l-Cysteine | 1 mm | 99 |
In accordance with its similarity to the calpains, Tpr required calcium ions for enzymatic activity. Detectable hydrolysis of fluorogenic substrates was observed only at a Ca2+ concentration higher than 1 mm, with maximal activity at a Ca2+ concentration of 5 mm. Of note, Trp activity inhibition by higher calcium concentration is typical for cysteine proteases and is apparently due to propensity of a thiol group of the active site cysteine to chelate divalent cations. In line with this calcium dependence, the calcium chelator EDTA effectively inhibited Tpr activity, and the effect was fully reversed by the addition of excess CaCl2 (Table 1). In contrast to calcium, supplementation of the assay buffer with reducing agents (DTT and l-cysteine) only slightly increased activity of the enzyme. Finally, o-phenanthroline, a Zn2+-chelating agent and inhibitor of Zn2+-dependent metallopeptidases, had no effect on Tpr activity.
Hydrolysis of Physiological Substrates
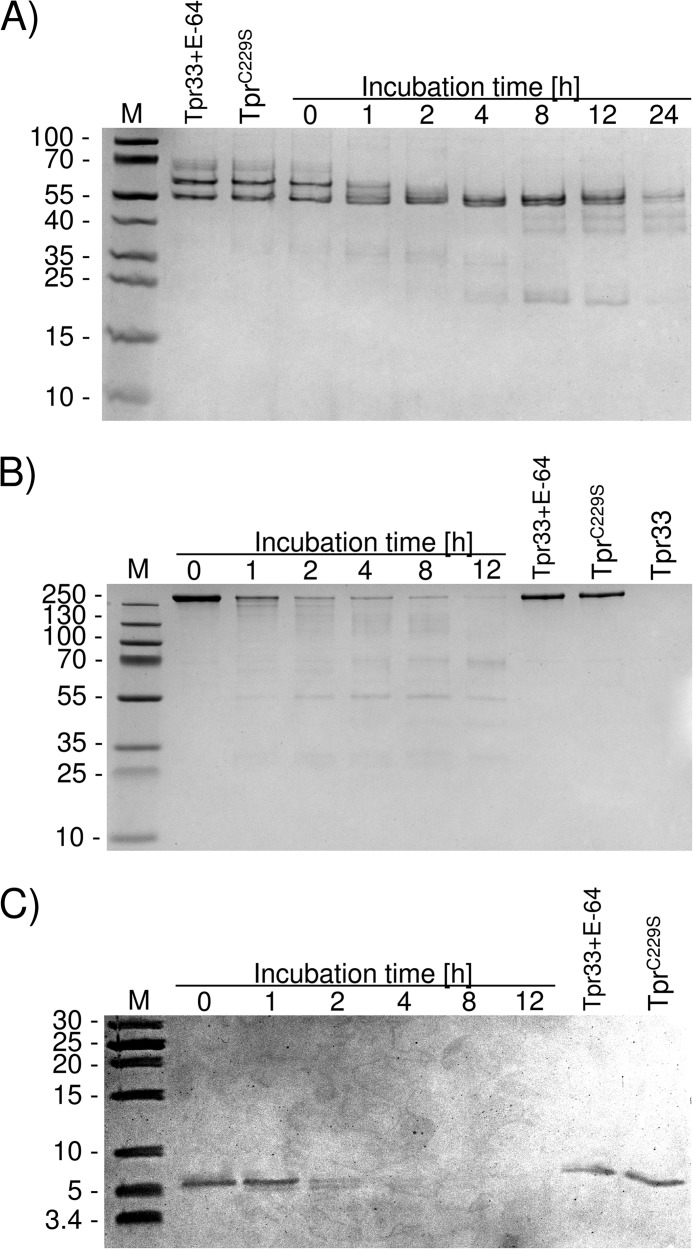
To determine whether Tpr could be functioning as a potential virulence factor for P. gingivalis, we assessed its ability to degrade the human proteins myoglobin, hemoglobin, fibrinogen, fibronectin, human complement components, and LL-37, an antimicrobial peptide from the cathelicidin family. Tpr33 efficiently degraded fibrinogen and fibronectin. At a 100:1 (substrate:enzyme) weight ratio, nearly 50% of fibrinogen was cleaved within 1 h (Fig. 5A). In the case of fibronectin, the substrate was rapidly degraded by Tpr33 (Fig. 5B). Tpr33 also efficiently degraded LL-37 (Fig. 5C) at the 10:1 weight ratio and digested human complement compounds, C3, C4, and C5 (Fig. 6, A–C) at a molar ratio of 5:1 (substrate:enzyme). Myoglobin and hemoglobin were not cleaved by Tpr33.
FIGURE 5.
Tpr33 degrades the fibrinogen (A), fibronectin (B), and LL-37 (C). Fibrinogen (100 μg), fibronectin (50 μg), and LL-37 (10 μg) were incubated at 37 °C in 100 μl of 100 mm Tris-HCl, 5 mm CaCl2, pH 7.5. At the indicated time points, aliquots (10 μl) were withdrawn from the reaction mixture, mixed with 10 μl of reducing SDS-PAGE sample buffer, and denatured at 95 °C for 5 min to stop the reaction. Samples (20 μl) were subjected to SDS-PAGE. Substrates incubated with Tpr33 inhibited with E-64 and TprC229S served as non-digested protein control.
FIGURE 6.
Tpr33 hydrolyzes the complement proteins C3 (A), C4 (B), and C5 (C). Complement compounds (100 μg) were incubated at 37 °C in 100 μl of 100 mm Tris-HCl, 5 mm CaCl2, pH 7.5, up to 12 h at a 5:1 substrate/protease molar ratio. At the indicated time points, aliquots (10 μl) were withdrawn from the reaction mixture, mixed with 10 μl of reducing SDS-PAGE sample buffer, and denatured at 95 °C for 5 min to stop the reaction. Samples (20 μl) were subjected to SDS-PAGE. Substrates incubated with Tpr33 inhibited with E-64 and TprC229S served as non-digested protein control.
Processing of Pro-TprC229S by WT Tpr and Other Cysteine Peptidases
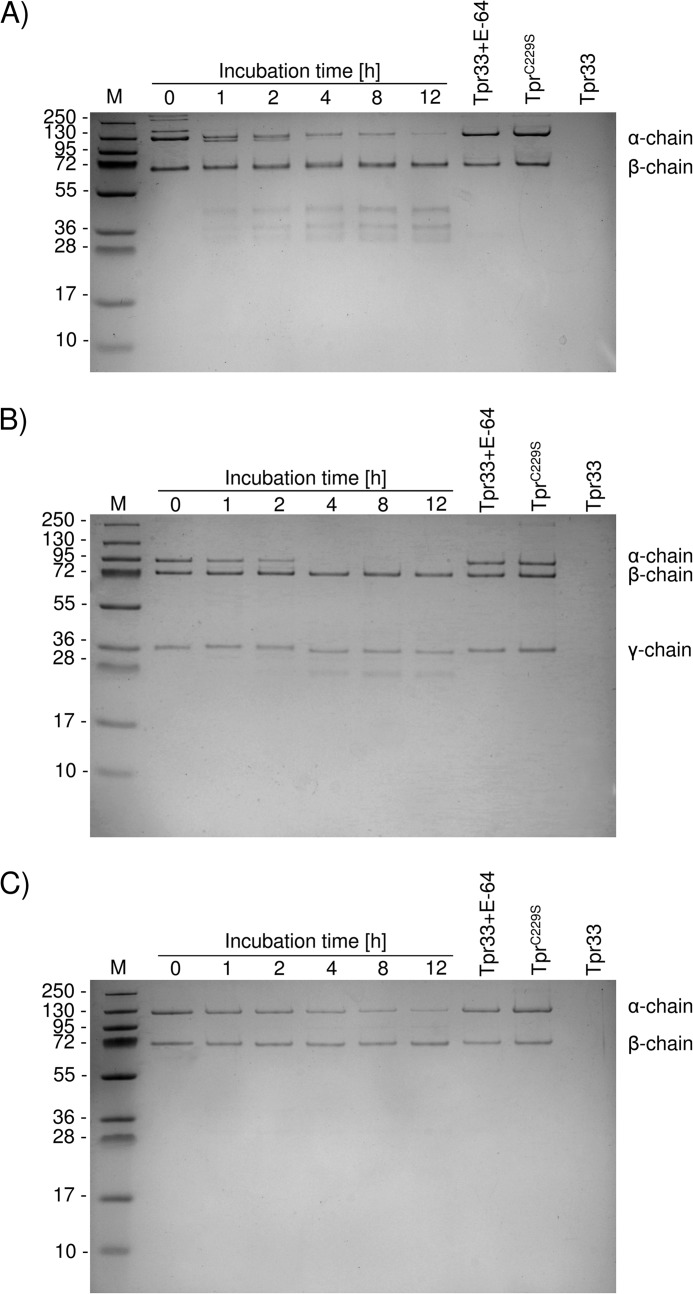
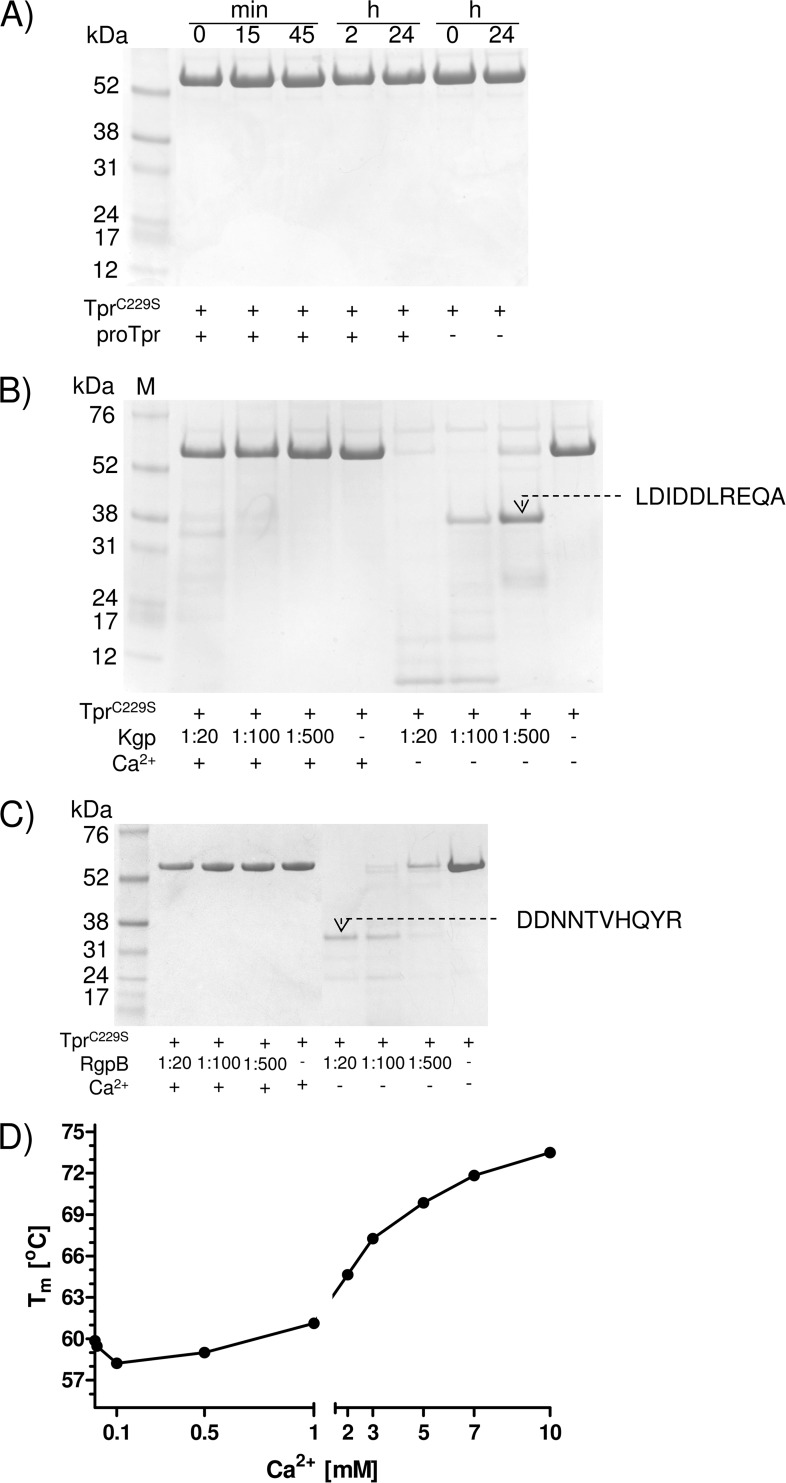
P. gingivalis is highly proteolytic as a result of gingipain expression; thus, the finding of calcium-induced autoactivation of pro-Tpr55 does not preclude involvement of other peptidases in processing of the Tpr55 zymogen. To determine whether other peptidases are capable of processing Tpr55 in trans, we performed experiments using the inactive variant of pro-Tpr (TprC229S). We initially incubated this inactive mutant with different proteolytic enzymes, including WT Tpr, Kgp, and Rgp, and assessed proteolysis by SDS-PAGE. At a molar ratio 1:100 of active Tpr to TprC229S, cleavage of TprC229S did not occur even after 24 h of incubation regardless of the presence or absence of calcium (Fig. 7A). This finding indicates that calcium-dependent activation is an intramolecular event.
FIGURE 7.
TprC229S is impervious to proteolysis by active forms of Tpr (A) and in the presence of Ca2+ also blocks degradation by Kgp (B) and Rgp (C) because the protein is stabilized (D). Recombinant TprC229S was incubated with Tpr55/Tpr48 at 37 °C in the activity assay buffer at the molar ratio 100:1 for the indicated time periods (A) or with gingipains at different molar ratios in the presence or absence of Ca2+ for 1 h (B and C). Boiling in a denaturing sample buffer stopped the reaction, and samples were analyzed by SDS-PAGE followed by protein staining and analysis by N-terminal sequencing. The effect of increasing concentrations of calcium ions on the Tm of TprC229S was measured by differential scanning fluorimetry (D).
Furthermore, in the presence of calcium, pro-TprC229S was resistant to proteolysis by Kgp and RgpB. In the absence of calcium, both Kgp and RgpB degraded the TprC229S protein when applied at high concentrations (1:20 molar ratio). At lower concentrations (1:100 molar ratio), distinct fragments of 37 and 33 kDa were released from TprC229S by Kgp and RgpB, respectively (Fig. 7, B and C). N-terminal sequence analysis of these species revealed cleavages at Lys-154–Leu-155 (Kgp) and Arg-195–Asp-196 (RgpB) in the region where autoproteolytic processing occurs. Cumulatively, these results suggest that Ca2+ exerts a profound effect on the Tpr55 molecule, making it resistant to proteolysis by exogenous peptidases.
To further study the calcium-exerted stability of TprC229S, we subjected the protein to differential scanning fluorimetry in the presence of increasing concentrations of Ca2+ ions. Melting temperatures (Tm) were calculated to demonstrate the effect of Ca2+ ions on TprC229S stability. As shown in Fig. 7D, Tm values were not affected by sub-millimolar Ca2+ but drastically increased at concentrations higher than 1 mm, reaching the maximal Tm = 73.5 °C at 10 mm Ca2+. The Ca2+-induced stability profile is consistent with our findings of Tpr activation by calcium and the resistance of calcium-loaded TprC229S to degradation by gingipains.
Discussion
According to the MEROPS Peptidase Database, genes encoding calpain-like peptidases (family C2 of cysteine peptidases) are very rare among prokaryotes. These proteins are scattered among 55 species of 6 different phyla of Bacteria, with the largest number found in environmental species of Firmicutes (17 species), Cyanobacteria (14 species), and Proteobacteria (ten species) rather than in commensal gut microorganisms of the genus Bacteroides (six species). Thus far, P. gingivalis is the only human pathogen possessing a gene encoding a calpain-like cysteine peptidase. Except for the sequences surrounding its catalytic residues, Tpr shares very low overall primary structure similarity with other calpain- and papain-like peptidases of both eukaryotic and prokaryotic origin. The sequence apparently diverged from a common ancestor gene to fulfill a specific function in P. gingivalis. Here, we delineated the biochemical nature of Tpr, which is the first bacterial calpain-like peptidase characterized to date.
Tpr is produced as an inactive proenzyme of 55 kDa (Tpr55), which is prone to autoactivation by limited proteolysis (Fig. 2). The first cleavage occurs at the Gly-62–Met-63 peptide bond via intramolecular proteolysis, leading to the formation of the 48-kDa form of the enzyme (Trp48), which further processes itself to the 37- and 33-kDa forms of the active enzyme (Tpr37 and Tpr33). The presence of the Tpr55, Tpr48, and Tpr37 forms can be detected in P. gingivalis cell homogenate and in preparations from the outer membrane but not in the inner membrane or in cytoplasm/periplasm fractions (Fig. 1C). This finding is consistent with the cell-surface location of Tpr (18) and suggests that despite the absence of a typical signal peptide, Tpr is translocated across the inner membrane and is trafficked to the cell surface. The surface location of Tpr is essential for its suggested function in producing metabolic peptides by degradation of host proteins (17). In line with this suggestion, Tpr expression is strongly enhanced by nutrient limitation (20) and in P. gingivalis isogenic mutants deficient in Rgp activity (25).
Tpr autoprocessing and activity is dependent on the presence of calcium at millimolar concentrations (Table 1). Because intracellular calcium concentrations are in the low micromolar range (26, 27), the calcium dependence of Tpr activation provides P. gingivalis with an effective mechanism for controlling its enzymatic activity. Namely, Tpr is activated and exerts its proteolytic functions only after being secreted to the cell surface, where the extracellular calcium concentration in the growth medium or the gingival crevicular fluid exceeds 1 mm. Calcium-induced activation is apparently due to stabilization of the Tpr structure, as indicated by the drastic calcium-dependent increase in protein stability, as measured by differential scanning fluorimetry (Fig. 7D).
Although we cannot provide a direct structural explanation for the increased stability of Tpr in the presence of calcium, it is likely that the molecular mechanism of this phenomenon is similar to that described for the eukaryotic calpains. After binding of calcium to two subdomains (IIa and IIb) responsible for formation of the catalytic cleft, the calpain molecule undergoes significant conformational changes, which include opening of the active site cleft and movement of the IIa and IIb domains to produce a more compact enzyme (28, 29). Taking into account that the calcium binding sites within domains IIa and IIb are conserved for Tpr and calpains, similar rearrangements on binding of calcium would be predicted for Tpr. These changes may explain the observed increased thermostability of the peptidase in the presence of calcium. Moreover, calcium-loaded Tpr is completely resistant to proteolytic degradation by gingipains (Fig. 7, B and C). This is probably an important adaptation of Tpr to function in the highly proteolytic environment of the P. gingivalis cell surface, which is coated with gingipains or in GCF, where gingipains are present at concentrations of up to 100 μm (30). Another possible adaptation that allows activity to be maintained in the extracellular milieu is Tpr's unusual lack of requirement for reducing agents (Table 1). This may be especially important for P. gingivalis cells that enter into gingival tissue away from the reducing environment of the gingival crevicular fluid.
One would expect that a peptidase engaged in nutrient acquisition should have high activity as well as a broad specificity and pH optimum. As expected, this is the case for Tpr, which has both a broad pH optimum and substrate specificity (Fig. 4, A and C). However, our results for the substrate specificity of Tpr were derived only from the hydrolysis of only two proteins. Therefore, a more comprehensive analysis of diverse substrate libraries will be required to provide a clearer picture of the overall substrate specificity of Tpr.
Our results show that Tpr degrades several physiologically relevant proteins and peptides including complement proteins C3, C4, C5, and antimicrobial peptide LL-37 (Figs. 5 and 6). The complement system and LL-37 are crucial elements of the innate immune system within the human gingival crevice. Thus, degradation of these proteins may contribute to protection from immune clearance of P. gingivalis. Interestingly, similar to other proteases secreted by periodontopathogens, Tpr preferentially cleaved the α-chains of C3 and C5, which may result in generation of potent anaphylotoxins C3a and C5a, respectively. The high level of C3a and C5a at the site of infection may impair neutrophil functions and increase inflammation, which could result in additional periodontal tissue damage leading to progression of disease (31).
Interestingly, Tpr also efficiently degraded other human proteins (fibrinogen, fibronectin) abundant in gingival crevicular fluid, thus generating peptides (Fig. 5). Peptides are substrates for the fermentative metabolism of P. gingivalis, but only some support bacterial growth (32). In this context it is plausible that the cell-surface location of Tpr coupled with its specificity allows P. gingivalis to efficiently generate a limited spectrum of peptides in close proximity to the bacteria so that they are efficiently captured and transported inside the cell. Therefore, Tpr may be involved in regulating P. gingivalis virulence through both evasion of host defense and generation of nutrients.
Here we describe the expression and biochemical characterization of the first prokaryote-derived calpain-like peptidase with calcium-dependent autoprocessing and activity, These and other features, including cell-surface localization, resistance to proteolysis, and degradation of complement proteins and LL-37, seem to have evolved to allow this peptidase to not only fulfill the nutritional requirements of P. gingivalis but also to protect the bacterium against action of human innate immune system. Our group is now testing this attractive hypothesis.
Author Contributions
D. S. designed, performed, and analyzed all the experiments (except the results shown in Fig. 3, B and C) and wrote the paper. M. K. performed and analyzed the experiments shown in Fig. 3, B and C, and helped with designing the experiments shown in Figs. 5 and 6. I. B. T. and J. J. E. provided technical assistance and contributed to the preparation of Figs. 1, 2, and 4. A. S. and D. B. provided technical assistance, and A. S. designed the primers for construction of expression vector. M. B. designed part of the study and reviewed a draft of the manuscript. M. A. and J. P. designed and coordinated the study. All authors reviewed the results and approved the final version of the manuscript.
Acknowledgments
We are grateful to Anna-Cathrine Carlberg-Löfström for technical assistance. The Faculty of Biochemistry, Biophysics, and Biotechnology of Jagiellonian University is a partner of the Leading National Research Center (KNOW) supported by the Ministry of Science and Higher Education.
This work was supported in part by United States National Institutes of Health Grant DE 022597, the European Commission (FP7-PEOPLE-2011-ITN-290246 “RAPID” and FP7-HEALTH-F3–2012-306029 “TRIGGER”), the Polish Ministry of Science and Higher Education (Project 2975/7.PR/13/2014/2), and the National Science Center (2012/04/A/NZ1/00051, NCN, Krakow, Poland). The authors declare that they have no conflicts of interest with the contents of this article.
- Tpr
- thiol protease
- Bz-
- benzoyl
- pNA
- p-nitroanilide
- Z-
- benzyloxycarbonyl
- AMC
- 7-amino-4-trifluorometylocumarin
- CAPS
- 3-(cyclohexylamino)propanesulfonic acid
- NTP
- N-terminal profragment
- TLCK
- 1-chloro-3-tosylamide-7-amino-l-2-heptanone
- AFC
- aminofluoromethylcoumarin
- Boc-
- t-butyloxycarbonyl
- Suc-
- N-succinyl
- H-
- unblocked amino group.
References
- 1.Cobb C. M., Williams K. B., and Gerkovitch M. M. (2009) Is the prevalence of periodontitis in the U.S.A. in decline? Periodontol. 2000 50, 13–24 [DOI] [PubMed] [Google Scholar]
- 2.Burt B. (2005) Position paper: epidemiology of periodontal diseases. J. Periodontol. 76, 1406–1419 [DOI] [PubMed] [Google Scholar]
- 3.de Pablo P., Chapple I. L., Buckley C. D., and Dietrich T. (2009) Periodontitis in systemic rheumatic diseases. Nat. Rev. Rheumatol. 5, 218–224 [DOI] [PubMed] [Google Scholar]
- 4.Wegner N., Wait R., and Venables P. J. (2009) Evolutionarily conserved antigens in autoimmune disease: implications for an infective aetiology. Int. J. Biochem. Cell Biol. 41, 390–397 [DOI] [PubMed] [Google Scholar]
- 5.Friedewald V. E., Kornman K. S., Beck J. D., Genco R., Goldfine A., Libby P., Offenbacher S., Ridker P. M., Van Dyke T. E., and Roberts W. C. (2009) The American Journal of Cardiology and Journal of Periodontology Editors' Consensus: Periodontitis and Atherosclerotic Cardiovascular Disease. Am. J. Cardiol. 104, 59–68 [DOI] [PubMed] [Google Scholar]
- 6.Chee B., Park B., and Bartold P. M. (2013) Periodontitis and type II diabetes: a two-way relationship. Int. J. Evid. Based. Healthc. 11, 317–329 [DOI] [PubMed] [Google Scholar]
- 7.Darby I., and Curtis M. (2001) Microbiology of periodontal disease in children and young adults. Periodontol. 2000 26, 33–53 [DOI] [PubMed] [Google Scholar]
- 8.Socransky S. S., Haffajee A. D., Cugini M. A., Smith C., and Kent R. L. (1998) Microbial complexes in subgingival plaque. J. Clin. Periodontol. 25, 134–144 [DOI] [PubMed] [Google Scholar]
- 9.Travis J., Pike R., Imamura T., and Potempa J. (1997) Porphyromonas gingivalis proteinases as virulence factors in the development of periodontitis. J. Periodontal Res. 32, 120–125 [DOI] [PubMed] [Google Scholar]
- 10.Dashper S. G., Seers C. A., Tan K. H., and Reynolds E. C. (2011) Virulence factors of the oral spirochete Treponema denticola. J. Dent. Res. 90, 691–703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Imamura T. (2003) The role of gingipains in the pathogenesis of periodontal disease. J. Periodontol. 74, 111–118 [DOI] [PubMed] [Google Scholar]
- 12.Potempa J., Sroka A., Imamura T., and Travis J. (2003) Gingipains, the major cysteine proteinases and virulence factors of Porphyromonas gingivalis: structure, function, and assembly of multidomain protein complexes. Curr. Protein Pept. Sci. 4, 397–407 [DOI] [PubMed] [Google Scholar]
- 13.Fitzpatrick R. E., Aprico A., Wijeyewickrema L. C., Pagel C. N., Wong D. M., Potempa J., Mackie E. J., and Pike R. N. (2009) High molecular weight gingipains from Porphyromonas gingivalis induce cytokine responses from human macrophage-like cells via a nonproteolytic mechanism. J. Innate Immun. 1, 109–117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Veillard F., Potempa B., Poreba M., Drag M., and Potempa J. (2012) Gingipain aminopeptidase activities in Porphyromonas gingivalis. Biol. Chem. 393, 1471–1476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Walker C. B., and Bueno L. C. (1997) Antibiotic resistance in an oral isolate of Prevotella intermedia. Clin. Infect. Dis. 25, S281–S283 [DOI] [PubMed] [Google Scholar]
- 16.Potempa J., Pavloff N., and Travis J. (1995) Porphyromonas gingivalis: a proteinase/gene accounting audit. Trends Microbiol. 3, 430–434 [DOI] [PubMed] [Google Scholar]
- 17.Bourgeau G., Lapointe H., Péloquin P., and Mayrand D. (1992) Cloning, expression, and sequencing of a protease gene (tpr) from Porphyromonas gingivalis W83 in Escherichia coli. Infect. Immun. 60, 3186–3192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Park Y., Lu B., Mazur C., and McBride B. C. (1997) Inducible expression of a Porphyromonas gingivalis W83 membrane-associated protease. Infect. Immun. 65, 1101–1104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Park Y., and McBride B. C. (1993) Characterization of the tpr gene product and isolation of a specific protease-deficient mutant of Porphyromonas gingivalis W83. Infect. Immun. 61, 4139–4146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lu B., and McBride B. C. (1998) Expression of the tpr protease gene of Porphyromonas gingivalis is regulated by peptide nutrients. Infect. Immun. 66, 5147–5156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schägger H., and von Jagow G. (1987) Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal. Biochem. 166, 368–379 [DOI] [PubMed] [Google Scholar]
- 22.Nguyen K. A., Travis J., and Potempa J. (2007) Does the importance of the C-terminal residues in the maturation of RgpB from Porphyromonas gingivalis reveal a novel mechanism for protein export in a subgroup of gram-negative bacteria? J. Bacteriol. 189, 833–843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Niesen F. H., Berglund H., and Vedadi M. (2007) The use of differential scanning fluorimetry to detect ligand interactions that promote protein stability. Nat. Protoc. 2, 2212–2221 [DOI] [PubMed] [Google Scholar]
- 24.Greenbaum D., Baruch A., Hayrapetian L., Darula Z., Burlingame A., Medzihradszky K. F., and Bogyo M. (2002) Chemical approaches for functionally probing the proteome. Mol. Cell. Proteomics 1, 60–68 [DOI] [PubMed] [Google Scholar]
- 25.Tokuda M., Chen W., Karunakaran T., and Kuramitsu H. K. (1998) Regulation of protease expression in Porphyromonas gingivalis. Infect. Immun. 66, 5232–5237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dominguez D. C. (2004) Calcium signalling in bacteria. Mol. Microbiol. 54, 291–297 [DOI] [PubMed] [Google Scholar]
- 27.Jones H. E., Holland I. B., and Campbell A. K. (2002) Direct measurement of free Ca2+ shows different regulation of Ca2+ between the periplasm and the cytosol of Escherichia coli. Cell Calcium. 32, 183–192 [DOI] [PubMed] [Google Scholar]
- 28.Suzuki K., Hata S., Kawabata Y., and Sorimachi H. (2004) Structure, Activation, and Biology of Calpain. Diabetes 53, S12–S18 [DOI] [PubMed] [Google Scholar]
- 29.Hanna R. A., Campbell R. L., and Davies P. L. (2008) Calcium-bound structure of calpain and its mechanism of inhibition by calpastatin. Nature 456, 409–412 [DOI] [PubMed] [Google Scholar]
- 30.Guentsch A., Kramesberger M., Sroka A., Pfister W., Potempa J., and Eick S. (2011) Comparison of gingival crevicular fluid sampling methods in patients with severe chronic periodontitis. J. Periodontol. 82, 1051–1060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Potempa M., and Potempa J. (2012) Protease-dependent mechanisms of complement evasion by bacterial pathogens. Biol. Chem. 393, 873–888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wyss C. (1992) Growth of Porphyromonas gingivalis, Treponema denticola, T. pectinovorum, T. socranskii, and T. vincentii in a chemically defined medium. J. Clin. Microbiol. 30, 2225–2229 [DOI] [PMC free article] [PubMed] [Google Scholar]