Background: The initiation of replication in φ29 mainly occurs opposite the 3′ penultimate nucleotide of the template.
Results: Mutations in the TP residue Phe-230 changed the specificity for the templating nucleotide.
Conclusion: The results suggest a role for Phe-230 in determining the templating nucleotide.
Significance: The results shed light on the positioning of the template nucleotide in organisms that initiate with protein-priming mechanism.
Keywords: bacteriophage, DNA polymerase, DNA replication, nucleic acid, viral polymerase
Abstract
Bacteriophage φ29 from Bacillus subtilis starts replication of its terminal protein (TP)-DNA by a protein-priming mechanism. To start replication, the DNA polymerase forms a heterodimer with a free TP that recognizes the replication origins, placed at both 5′ ends of the linear chromosome, and initiates replication using as primer the OH-group of Ser-232 of the TP. The initiation of φ29 TP-DNA replication mainly occurs opposite the second nucleotide at the 3′ end of the template. Earlier analyses of the template position that directs the initiation reaction were performed using single-stranded and double-stranded oligonucleotides containing the replication origin sequence without the parental TP. Here, we show that the parental TP has no influence in the determination of the nucleotide used as template in the initiation reaction. Previous studies showed that the priming domain of the primer TP determines the template position used for initiation. The results obtained here using mutant TPs at the priming loop where Ser-232 is located indicate that the aromatic residue Phe-230 is one of the determinants that allows the positioning of the penultimate nucleotide at the polymerization active site to direct insertion of the initiator dAMP during the initiation reaction. The role of Phe-230 in limiting the internalization of the template strand in the polymerization active site is discussed.
Introduction
DNA polymerases synthesize DNA exclusively in the 5′-3′ direction and are unable to start de novo DNA synthesis, compelling the use of a priming 3′ OH-group generally provided by a short RNA or DNA molecule. These features of the DNA polymerases create the so-called end replication problem of linear chromosomes according to which, after successive rounds of replication, the short region of unreplicated ssDNA at the end of the chromosome would lead to a progressive shortening of the daughter DNA (1). Many mechanisms have evolved to maintain the DNA ends of linear chromosomes through subsequent replications rounds, most of them making use of the presence of repetitive sequences at the ends that allow the formation of long concatemers, as in phage T7 (2), to circularize, as in phage λ (3), or to form hairpin loops to fill the incomplete 5′ ends, as in poxvirus (4–6) or in the linear plasmid of Borrelia (7).
Several prokaryotic and eukaryotic viruses, as well as linear plasmids from bacteria, fungi, higher plants, and Streptomyces spp. have solved the end replication problem by using a protein, called terminal protein (TP),6 to prime DNA synthesis from the end of their linear genomes (TP-DNA) (8). Thus, a specific amino acid of the TP (serine, threonine, or tyrosine) provides the OH-group to prime initiation of DNA replication, the TP becoming covalently linked to the 5′ ends of the nascent DNA (parental TP).
Bacillus subtilis phage φ29 initiates replication by a protein-priming mechanism and has become the paradigm in the study of TP-DNA replication. Phage φ29 has a linear dsDNA 19285 base pairs (bp) long with a 6-bp inverted terminal repeat (3′-TTTCAT) that, together with the terminal 12-bp and with the phage-encoded TP covalently attached at each 5′ DNA end, assemble a minimal replication origin (9). The DNA polymerase and a free TP molecule (primer TP) form a heterodimer (10) that recognizes the replication origin. Then, the DNA polymerase catalyzes the incorporation of the initiating dAMP to the OH-group of the TP residue Ser-232 (8), a reaction directed by the 3′ penultimate dTMP of the template strand. Once the initiation product (TP-dAMP) has been formed, it translocates backwards one position to recover the information of the 3′ terminal T (11). This so-called sliding-back mechanism requires a terminal repetition of at least two nucleotides and provides a way to prevent mutations at the φ29 DNA ends during initiation, as the 3′-5′ exonuclease activity of the φ29 DNA polymerase is not able to proofread the TP-dAMP initiation product (12). Once initiation has taken place, there is a transition stage in which the DNA polymerase synthesizes the first nine nucleotides. When the 10th nucleotide is inserted, the DNA polymerase dissociates from the TP (13) and the same DNA polymerase molecule catalyzes processive chain elongation via a strand displacement mechanism to fulfill TP-DNA replication (14). These two features of φ29 DNA polymerase, processivity and strand displacement capacity, enable the enzyme to replicate the viral genome without requiring the assistance of processivity and unwinding factors, unlike most replicases.
φ29 DNA polymerase consists of a N-terminal exonuclease domain (residues 1–189) and a C-terminal polymerization domain (residues 190–572) that, like in other DNA polymerases, is subdivided into the universally conserved subdomains: palm (containing the catalytic and DNA ligand residues), fingers (containing the dNTP ligands), and thumb (which confers stability to the primer) (15). Additionally, the polymerization domain has two insertions called terminal protein regions 1 and 2 (TPR1 and TPR2), specifically present in the protein-primed DNA polymerases subgroup (16, 17). The arch-like structure formed by the TPR2 insertion and the thumb subdomain confers a toroidal shape to the polymerization domain that encircles the nascent DNA, being responsible for the high processivity of the enzyme. In addition, the TPR2 subdomain together with the fingers subdomain and the exonuclease domain wrap the template strand, acting as a “molecular wedge” to unwind the parental DNA strands during replication (18). Despite the physical separation between the exonuclease and polymerization domains, these two opposite activities must act in concert to achieve a productive and accurate replication reaction.
The crystallographic structure of φ29 TP in complex with the DNA polymerase (19) showed that it consists of a disordered N-terminal domain (residues 1–73) responsible for DNA binding and TP nucleoid localization (20), an intermediate domain (residues 74–172) that confers specificity to the interaction with the DNA polymerase, connected through a hinge region to the C-terminal priming domain (residues 173–266) bearing the primer Ser-232 in the priming loop. The TP priming loop comprehends residues 227 to 233 and is located close to the end of the priming domain and, although its structure has not been solved, it should be located at the polymerization active site to allow the OH-group of Ser-232 to prime the initiation step of replication.
Previous studies performed with chimerical TPs allowed to propose that the priming domain is responsible for placing the internal 3′ nucleotide used as template during initiation at the polymerization active site (21). The results presented in this paper indicate that although the parental TP is not playing a significant role in the recognition of the template nucleotide used to direct the initiation reaction in φ29 TP-DNA replication, the correct positioning of the 3′ terminus of the template strand at the polymerase active site relies on residue Phe-230 of the TP priming loop, allowing the penultimate T to direct the insertion of the initiator dAMP.
Experimental Procedures
Nucleotides and DNAs
Unlabeled nucleotides were supplied by Amersham Biosciences Pharmacia. [α-32P]dATP (3000 Ci/mmol) and [α-32P]dTTP (3000 Ci/mmol) were supplied by PerkinElmer Life Sciences. Oligonucleotides were obtained from Invitrogen.
Site-directed Mutagenesis of φ29 TP
TP mutants were obtained using the QuikChange site-directed mutagenesis kit provided by Stratagene, using as template the plasmid pT7-3 that contains the viral gene 3 coding for the TP (22). The presence of the desired mutations, as well as the absence of additional ones was determined by sequencing the entire gene.
Proteins
TP mutants were expressed in Escherichia coli BL21(DE3) cells harboring gene 3 cloned into plasmid pT7-3 and further purified, essentially as described for the wild-type TP (23). Both, the wild-type φ29 DNA polymerase and the exonuclease-deficient mutant D12A/D66A (24) were expressed in E. coli BL21(DE3) cells harboring gene 2 cloned into plasmid pJLPM (a derivative of pT7–4w2 containing the wild-type gene 2 that encodes the φ29 DNA polymerase and its ribosomal binding site; the nucleotide T was changed into G in the Shine-Dalgarno sequence) and further purified essentially as described (25).
Preparation of Reconstituted TP-DNA
The 3′ ends of TP-DNA (6 nm) were partially degraded by the 3′-5′ exonuclease activity of Vent DNA polymerase in its reaction buffer during 15 min at 75 °C. Then, the degradation product was hybridized in an incubation mixture that contained 20 nm of the indicated 29-mer single-stranded oligonucleotide, 1.8 mm EDTA, 150 mm NaCl, and 0.02% Tween 20 during 5 min at 75 °C. The temperature was allowed to decrease to 55 °C during 99 min and remain for 5 min at 55 °C. Then, the temperature was lowered until 4 °C during 15 min.
Protein-primed Initiation Assay
The ability to carry out the initiation step of TP-DNA replication was analyzed as described (26). The incubation mixture contained, in 25 μl, 50 mm Tris-HCl, pH 7.5, 10 mm MgCl2, 20 mm ammonium sulfate, 1 mm dithiothreitol (DTT), 4% (v/v) glycerol, 1.6 nm φ29 TP-DNA as template, 0.1 mg/ml of bovine serum albumin (BSA), 0.2 μm dATP (1 μCi of [α-32P]dATP), 13 nm either wild-type or mutant φ29 TP, and 13 nm φ29 DNA polymerase. Samples were incubated for 10 min at 30 °C. Reactions were stopped by adding 10 mm EDTA, 0.1% SDS, and the samples were filtered through Sephadex G-50 spin columns in the presence of 0.1% SDS and further analyzed by SDS-PAGE in 12% polyacrylamide gels. Quantification was done by densitometric analysis of the labeled band corresponding to the TP-dAMP complex detected by autoradiography.
For the reconstituted DNA the reaction was carried out essentially as described above for the TP-DNA but in the presence of 13 nm of either wild-type or mutant TP, 13 nm D12A/D66A exonuclease-deficient φ29 DNA polymerase, 0.4 nm reconstituted DNA, 1 mm MnCl2, and 0.2 μm dATP (1 μCi of [α-32P]dATP). Samples were incubated for 30 min at 30 °C.
In the case of the TP-dAMP or TP-dTMP formation using as template 29-mer ssDNA instead of TP-DNA the reaction was carried out essentially as described above for the TP-DNA but in the presence of 2 μm of the indicated oligonucleotide, 130 nm of either wild-type or mutant TP, 60 nm of the D12A/D66A exonuclease-deficient φ29 DNA polymerase, 1 mm MnCl2, and either 0.2 μm dATP (1 μCi of [α-32P]dATP) or 0.2 μm dTTP (1 μCi of [α-32P]dTTP). Samples were incubated for 1 h at 12 °C.
TP-DNA Replication Assay
The replication assay was performed as described (26). The incubation mixture contained, in 25 μl, 50 mm Tris-HCl, pH 7.5, 10 mm MgCl2, 20 mm ammonium sulfate, 1 mm DTT, 4% (v/v) glycerol, 0.1 mg/ml of BSA, 20 μm each dNTP and [α-32P]dATP (1 μCi), 13 nm of either wild-type or mutant TP, 13 nm DNA polymerase, and 1.6 nm φ29 TP-DNA. After 10 min at 30 °C, the reaction was stopped by adding 10 mm EDTA, 0.1% SDS, and the samples were filtered through Sephadex G-50 spin columns in the presence of 0.1% SDS. Quantification of the DNA synthesized was carried out from the amount of radioactivity (Cerenkov radiation) corresponding to the excluded volume. The labeled DNA was denatured by treatment with 0.7 m NaOH and subjected to electrophoresis in alkaline 0.7% agarose gels, as described (27). After electrophoresis, the position of unit-length φ29 DNA (19285 nucleotides) was detected by ethidium bromide staining, and then the gels were dried and autoradiographed.
Truncated Elongation Assay
The assay was performed essentially as described for the TP-DNA replication assay. For the analysis of the transition products, 160 nm wild-type or mutant TP, 60 nm exonuclease-deficient DNA polymerase mutant D12A/D66A, and 1.6 nm TP-DNA were incubated in the presence of 5 μm each dATP and dGTP, and 500 μm ddTTP for 30 min at 30 °C. The reaction was stopped by adding 10 mm EDTA, 0.1% SDS, and the samples were filtered through Sephadex G-50 spin columns in the presence of 0.1% SDS. The samples were analyzed by electrophoresis in SDS-12% polyacrylamide gels (360 × 280 × 0.5 mm) to obtain enough resolution to distinguish the TP bound to the first elongation products.
Results
Effect of the φ29 Parental TP in the Positioning of the First Templating Nucleotide
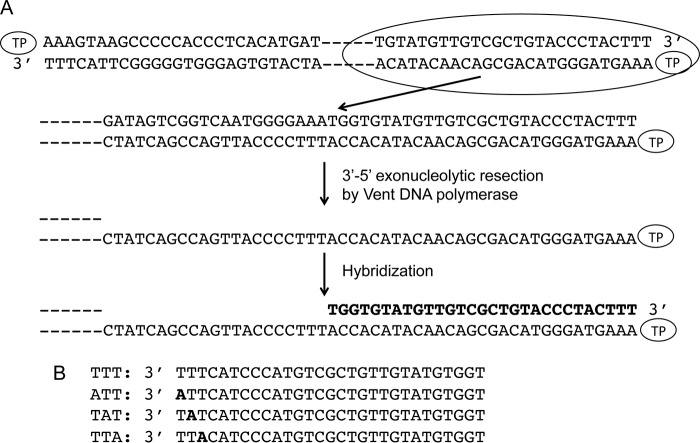
By using single-stranded synthetic oligonucleotides with the sequence of the replication origins (11) and TP-free dsDNA fragments containing the φ29 DNA terminal sequences (28) it was concluded that initiation of replication occurs mainly opposite the second nucleotide at the 3′ end of the template. However, the use of TP-free DNA ends precluded analysis of the influence of the parental TP in positioning the templating nucleotide during the initiation reaction. To address this question, full TP-DNA was partially digested with the 3′-5′ exonuclease activity of Vent DNA polymerase giving rise to TP-ssDNA regions (see scheme in Fig. 1). Replication origins were reconstituted after hybridization of 29-mer oligonucleotides containing either the wild-type sequence corresponding to the right replication origin (TTT) or single substitutions of the first (ATT), second (TAT), or third (TTA) thymine for adenine (see Fig. 1). These hybrid molecules were further used as substrate for the initiation reaction. Fig. 2A shows that, as expected, in the presence of the initiator [α-32P]dATP, the single-stranded 29-mer oligonucleotides TTT, ATT, and TTA supported formation of the initiation product TP-dAMP by the exonuclease-deficient φ29 DNA polymerase D12A/D66A (24) (the use of this mutant prevents the degradation of the template ssDNA). The small amount of TP-dAMP complex obtained with the template containing the TAT sequence is due to some initiation reaction directed by the third T (11). Essentially the same result was obtained with the reconstituted origins (Fig. 2B). The origins containing the TTT, ATT, and TTA sequences gave rise to the formation of the TP-dAMP initiation product, the TAT origin being the one that supported the lowest initiation reaction. The initiation levels observed with the reconstituted origins were not due either to potentially non-hybridized oligonucleotides (lane O) or to non-degraded TP-DNA (lane D). The results obtained indicate that the parental TP is not playing a significant role in determining the internal nucleotide used as template of the initiation reaction.
FIGURE 1.
A, schematic representation of reconstituted TP-DNA. φ29 TP-DNA is represented with a TP linked at each 5′ end. At the first step, the 3′-5′ exonuclease activity of Vent DNA polymerase degrades the 3′ ends of the TP-DNA. The resulting ssDNA region is further hybridized to 29-mer single-stranded oligonucleotides containing the φ29 right replication origin sequence and variants in the 3′ terminal thymines. B, 29-mer oligonucleotides containing wild-type and variants of the φ29 right replication origin sequence. The thymines changed into adenine are indicated in black letters.
FIGURE 2.
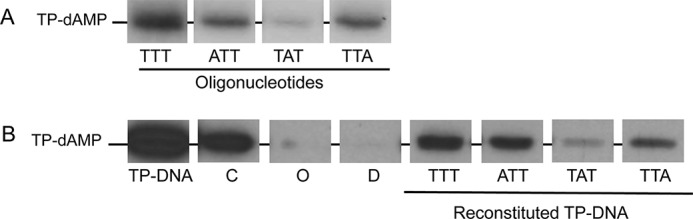
Formation of the TP-dAMP initiation complex using as template ssDNA (A) and reconstituted TP-DNA (B). The assays were carried out essentially as described under “Experimental Procedures” in the presence of 1 mm MnCl2. The position of the TP-dAMP is indicated. TP-DNA, initiation reaction using as template intact TP-DNA; C (control), initiation reaction using as template TP-DNA incubated at the same temperature and in the same reaction mixture as reconstituted DNA but in the absence of Vent polymerase and oligonucleotide; O (oligonucleotide), initiation reaction using as template the amount of oligonucleotide used in the hybridization but without TP-DNA; D (degraded), initiation reaction using as template TP-DNA with the 3′ ends degraded but not hybridized to the oligonucleotide.
Role of the Priming Loop Aromatic Residue Phe-230 in Positioning the Internal 3′ Nucleotide Used as Template
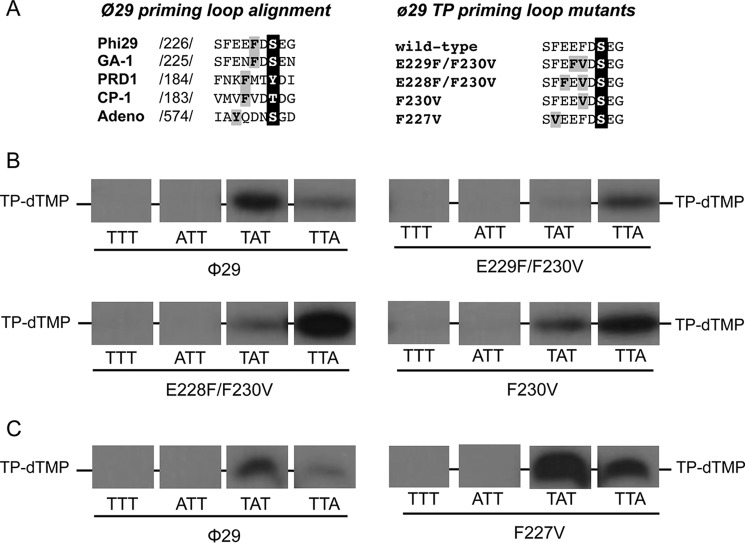
Previous assays with chimerical TPs indicated that the priming domain is responsible for placing the templating nucleotide at the polymerization active site during initiation (21). The TP priming domain (residues 173–266) is comprised of a four-helix bundle structure, the priming loop (residues 227–233) being located between the last and penultimate α helixes of the domain (19). Although the structure of this priming loop is disordered in the crystallographic structure of the φ29 DNA polymerase-TP complex, it should be positioned at the polymerization active site in this heterodimer to allow Ser-232 to prime the formation of the initiation product by the DNA polymerase. As shown in Fig. 3A the alignment of the corresponding priming loop of the TPs from different viruses that initiate the replication of their genomes with a TP shows the presence of aromatic residues close to the initiating (priming) residue. Interestingly, there seems to be a correlation between the distance of the aromatic residue to the priming amino acid and the initiation position. Accordingly, in phages φ29 and GA-1 that initiate opposite the second 3′ T (11, 29) the aromatic residue is located two positions before the priming residue. In Cp-1 and PRD1 an aromatic residue is three positions away from the initiator amino acid and the initiation reaction is directed by the third (30) and fourth (31) 3′ nucleotide, respectively. In adenovirus the replication occurs opposite the fourth nucleotide at the 3′ end of the template (32) and the aromatic residue is four positions away from the initiator residue. Thus, it is tempting to speculate that the aromatic residue could be playing some role in the selection of the templating nucleotide.
FIGURE 3.
A, alignment of the amino acid sequence of the priming loop of different viruses that initiate replication with a terminal protein. Numbers between slashes indicate the amino acid position relative to the N-terminal end of each TP. The priming residue of the different viruses is in white letters over black background. The aromatic residues are in black letters over gray background. φ29 TP priming loop mutants. The priming-loop of the wild-type and mutant TPs is shown. The primer serine is in white letters over black background and the residues changed are in black letters over gray background. B and C, ssDNA-dependent in vitro formation of the initiation complex with φ29 TP mutants of the priming loop aromatic residues Phe-230 (B) and Phe-227 (C). The assay was performed essentially as described under “Experimental Procedures” in the presence of 1 mm MnCl2. The 3′ end of the 29-mer oligonucleotide used is indicated. [α-32P]dTTP was used as initiator nucleotide. Samples were incubated for 1 h at 12 °C and then stopped, processed, and analyzed by SDS-PAGE and autoradiography. The position of the TP-dTMP complex is indicated. Panels corresponding to mutants E229F/F230V and F230V were 2-fold overexposed to better detect the initiation signal.
To study the role of the aromatic residue in positioning the template nucleotide we made site-directed mutagenesis in the priming loop of φ29 TP obtaining three mutants: E229F/F230V, E228F/F230V, and F230V (see Fig. 3A). In the single mutant F230V the aromatic residue Phe-230 of the priming loop is removed, whereas in the double mutants the loop is moved one (E229F/F230V) or two (E228F/F230V) positions N terminally. In the case of the mutant E229F/F230V the aromatic residue is in a position similar to that occupied in the TPs from PRD1 and Cp-1 and in the case of the mutant E228F/F230V the aromatic residue was moved N terminally to a position similar to that occupied in the TP from adenovirus.
To determine whether the introduced mutations affected the template nucleotide that directs the initiation reaction, the 29-mer single-stranded oligonucleotides TTT, ATT, TAT, and TTA were used as template. To detect the formation of TP-dTMP when the templating nucleotide is an adenine, [α-32P]dTTP was used as the initiator nucleotide. As it can be observed in Fig. 3B, the wild-type TP primes initiation mainly opposite the second 3′ nucleotide of the template (TAT) although some initiation reaction also takes place opposite the third 3′ nucleotide, as described (11). In contrast, with mutant TPs F230V, E229F/F230V, and E228F/F230 formation of TP-dTMP was mainly directed by the third nucleotide of the template strand (TTA). These results indicated that when the aromatic residue Phe-230 was removed or the distance to the priming residue was increased one (E229F/F230V) or two (E228F/F230V) positions the initiation occurred in a more internal position (third instead of second 3′ terminal nucleotide), suggesting that the location of aromatic residue Phe-230 within the priming loop could be a determinant to place the templating nucleotide used to direct the φ29 protein-primed initiation reaction.
Taking into account the presence of an additional aromatic residue in the φ29 TP priming loop (Phe-227) we analyzed whether it was playing any role in the initiation reaction. To address this question, φ29 TP residue Phe-227 was changed into valine (F227V mutant) and further evaluated in the initiation reaction described above. As it can be observed in Fig. 3C, when this mutant was used as primer, the initiation mainly occurred opposite the second nucleotide of the template and to some extent opposite the third nucleotide, like with the wild-type TP, indicating that the aromatic residue Phe-227 does not seem to contribute to the placement of the 3′ terminus of the template strand at the polymerization active site.
To study if the residues changed in the mutant TPs could be playing a role in further steps of φ29 TP-DNA replication, the formation of the TP-dAMP initiation complex and its further elongation was evaluated using as template φ29 TP-DNA. As shown in Fig. 4A and Table 1, when the DNA polymerase used the TP mutants as primer the formation of the TP-dAMP was impaired, especially when residue Phe-230 was removed or moved away from the priming residue. As mentioned in the Introduction, once formation of the TP-dAMP product was catalyzed, the same DNA polymerase elongates it via strand displacement to yield full-length φ29 TP-DNA. To ascertain to what extent the mutations introduced at the TP affect the replication process, assays were carried out using a minimal replication system based on φ29 TP-DNA, DNA polymerase, and TP (14). As observed in Fig. 4B and Table 1, when mutant TPs were used as primers, defects in the replication reaction were even more pronounced that during the initiation, mutant TPs E229F/F230V, E228F/F230V, F230V, and F227V being 11-, 100-, 27-, and 4-fold were less efficient in supporting DNA replication than the wild-type protein.
FIGURE 4.
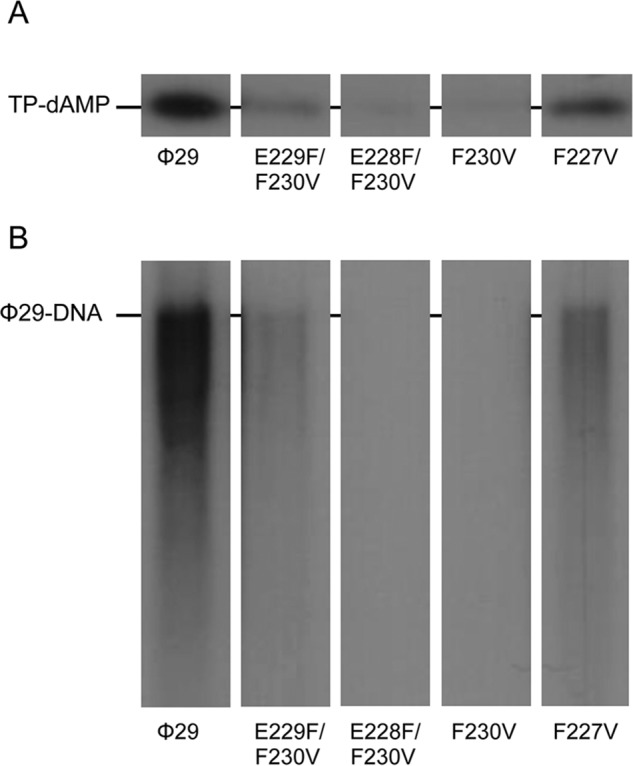
φ29 TP-DNA initiation and replication with φ29 TP mutants at the priming loop. The assays were carried out as described in “Experimental Procedures” using as template TP-DNA in the presence of 10 mm MgCl2. A, in vitro protein-primed initiation in the presence of TP-DNA. After 10 min of incubation at 30 °C, the reactions were stopped, processed, and analyzed by SDS-12% polyacrylamide gels and autoradiography. The position of the TP-dAMP complex is indicated. B, TP-DNA replication. After incubation for 10 min at 30 °C, the length of the synthesized DNA was analyzed by alkaline agarose gel electrophoresis. The migration position of unit length φ29 DNA is indicated.
TABLE 1.
Activity of wild-type and mutant φ29 TPs
Data represent the mean ± S.D. obtained from at least three independent experiments. The activity for the wild-type DNA polymerase when the wild-type TP was used as primer in the replication reaction was 1.6 nmol of nucleotide incorporated per minute and in the initiation reaction 0.5 fmol of nucleotide incorporated per minute.
| φ29 TP | TP-dAMP formationa | TP-DNA replicationb |
|---|---|---|
| Wild-type | 100 | 100 |
| E229F/F230V | 26.5 ± 9.7 | 9.1 ± 3.5 |
| E228F/F230V | 11.8 ± 6.5 | 1 ± 0.5 |
| F230V | 15.1 ± 7.3 | 3.7 ± 1.9 |
| F227V | 53.2 ± 18.4 | 28.2 ± 4.7 |
a TP-dAMP formation using ϕ29 TP-DNA as template.
b Numbers indicate the activity (in percentage) of mutant TPs with respect to the wild-type TP.
Furthermore, we investigated whether the heterodimer formed by the mutant TPs and DNA polymerase was able to recover the first and second 3′ nucleotides. To this end truncated elongation assays were performed in the presence of TP-DNA, dATP, dGTP, and ddTTP, to allow elongation to take place only up to the first A of the template. As mentioned above, initiation with the wild-type TP occurs using as template the penultimate 3′ T and then the TP-dAMP product translocates one position backwards to recover the information of the 3′ terminal T in a mechanism called sliding-back (11). Then, when the wild-type TP was used as primer, the elongation continued up to the incorporation of the ddTMP opposite the fifth nucleotide (Fig. 5), indicating that all the positions of the template, including the first nucleotide, were replicated. As it can be observed, 40% of the elongated initiation products stopped at the fourth position, most likely due to the inefficiency displayed by φ29 DNA polymerase to insert ddNMPs. When mutant F227V was used as primer, the DNA polymerase gave rise to a 5-nucleotide long product (60%), indicating a recovery of the first 3′ terminal nucleotide as it occurred with the wild-type TP. In contrast, when aromatic residue Phe-230 was removed (F230V) or the distance was increased one (E229F/F230V) or two (E228F/F230V) positions the elongation mainly stopped opposite the fourth nucleotide (70, 55, and 70%, respectively). This suggests that after the initiation opposite the third nucleotide, most of the initiation products performed only a sliding-back step between the third and second positions. A possible explanation is that initiation at the third 3′ nucleotide may affect the performance of the additional sliding-back step between the second and the first position that would be required to recover the 3′ end information.
FIGURE 5.
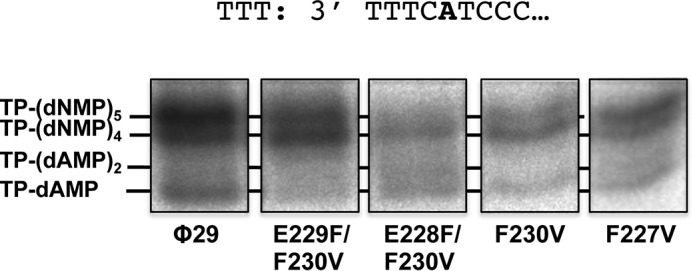
φ29 TP-DNA truncated elongation with the TP mutants (transition assay). The assay was performed as described under “Experimental Procedures” in the presence of dATP, dGTP, and ddTTP and 10 mm MgCl2, using TP-DNA as template of the reaction. After incubation for 30 min at 30 °C, the different transition products were analyzed by high resolution SDS-12% PAGE.
Discussion
The first step of φ29 DNA replication requires the formation of a heterodimer between the DNA polymerase and the primer TP that further recognizes the replication origins located at both ends of the linear genome. The DNA polymerase catalyzes the covalent linkage of the initial dAMP to the hydroxyl group of Ser-232 of the TP and its further elongation (8). Similarly, TP-DNA replication of bacteriophages PRD1 (31, 33), Cp-1 (30), and the φ29-related phages GA-1 (22, 29) and Nf (22, 34) was also shown to occur by a protein-primed mechanism, involving its corresponding DNA polymerase and TP. Initiation of φ29 DNA replication occurs at the second 3′ nucleotide of the template. The first nucleotide is then recovered by sliding-back of the initiation product TP-dAMP (11). After incorporation of the 10th nucleotide there is a dissociation of the DNA polymerase (13) that continues replication via a strand displacement mechanism to fulfill TP-DNA replication (14).
The complex between the replicative DNA polymerase and a free TP molecule interacts with the replication origins by specific recognition of the parental TP and DNA sequences. The presence of the parental TP, which is covalently linked to the 5′ ends of the non-template strand by a previous cycle of replication, is the main signal to be recognized by the polymerase-TP complex for the initiation of replication, because when terminal DNA fragments lacking parental TP were used as templates, the initiation reaction decreased 6–10-fold with respect to the activity obtained with TP-DNA (34, 35). Due to its important role in recruiting the φ29 DNA polymerase/TP heterodimer to the origins we analyzed whether the parental TP was conditioning the correct placement of the heterodimer with respect to the 3′ ends of the molecule used as template. In this work we used reconstituted dsDNA that contains the parental TP and the φ29 replication origin sequence or variants of the first, second, or third nucleotide. This template allowed us to analyze the role of the parental TP in determining the template position during the initiation reaction. When reconstituted DNA was used as template of the initiation reaction the result was the same as obtained by using single- or double-stranded oligonucleotides containing φ29 replication origins and variants. Therefore, the parental TP does not seem to be involved in the determination of the nucleotide that directs the initiation reaction.
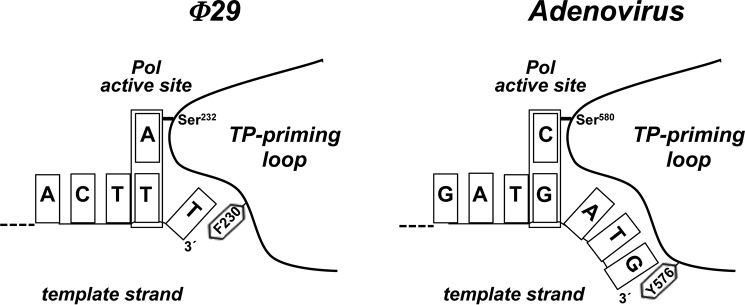
Previous assays showed that the priming domain is the main structural determinant that dictates the internal 3′ nucleotide used as template during initiation (21). Here, we have analyzed the role of the aromatic residue Phe-230 of the TP priming loop, proximal to the priming Ser-232. The results showed that mutations in the aromatic residue Phe-230 changed the specificity for the templating nucleotide, as initiation occurred mainly opposite the third 3′ T instead of the second one. An appealing hypothesis would be that this aromatic residue acts as a structural barrier that limits the internalization of the 3′ end of the template strand, conditioning the placement at the polymerization active site of the correct templating nucleotide. Fig. 6 shows a schematic of the priming-loop of the φ29 and adenovirus TPs, with the corresponding aromatic residues Phe-230 and Tyr-576, respectively, and how they could be interacting with the 3′ terminal base. Such a contact would block further internalization, allowing the second and fourth position in φ29 and adenovirus, respectively, to be placed at the polymerization active site.
FIGURE 6.
Schematic representation of the placement of the 3′ end of the template strand at the active site of φ29 and adenovirus DNA polymerases. The templating nucleotide and the aromatic residue of the priming loop are indicated. See text for details.
Residues Phe-230 and Phe-227 could be also involved in the proper stabilization/orientation of the priming residue Ser-232 at the polymerization active site of DNA polymerase, explaining the lower levels of initiation and replication exhibited by the mutants, specially F230V. During the transition step, whereas the priming domain must move in response to the translocation of the DNA after nucleotide incorporation, the intermediate domain would be in a fixed orientation on the polymerase by means of stable contacts with the TPR1 subdomain (36). Some displacements of the priming domain appear possible without dissociation due to the flexibility of the hinge region that connects the intermediate and priming domains. After incorporation of 6–7 nucleotides the proximity of the priming Ser to the hinge region would impede a further priming domain rotation, promoting complex dissociation (19). The ability of mutant TPs F230V, E229F/F230V, and E228F/F230V to accomplish the first sliding-back step better than the second one, could indicate that the φ29 DNA polymerase/TP heterodimer is structurally adapted to perform mainly one backward movement of the initiation product. During the sliding-back a set of interactions between the TP priming domain and the DNA polymerase TPR2 and thumb subdomains should be broken and new contacts should be established. This fact could compel the polymerase to move forward, explaining why, although mutant TPs can prime initiation opposite the third T, the recovery of the terminal 3′ T is hindered.
Author Contributions
A. P. conducted the experiments, analyzed the results, and wrote most of the paper. J. M. L. performed the pJLPM plasmid. E. L. performed the TP mutant E228F/F230V. L. V. purified the TP mutants. M. S. and M. V. conceived the idea for the project, discussed the results, and wrote the paper with A. P.
This work was supported in part by the Spanish Ministry of Economy and Competitiveness Grants BFU2014-52656-P (to M. S.) and BFU 2014-53791P (to M. V.), Consolider-Ingenio from the Spanish Ministry of Science and Innovation Grant CSD2007–00015 (to M. S.), and a Institutional grant from the Fundación Ramón Areces to the Centro de Biología Molecular “Severo Ochoa.” The authors declare that they have no conflicts of interest with the content of this article.
- TP
- terminal protein
- TPR
- terminal protein region.
References
- 1.Kornberg A., and Baker T. A. (1992) DNA Replication, 2nd Ed., W.H. Freeman, New York [Google Scholar]
- 2.Schlegel R. A., and Thomas C. A. Jr. (1972) Some special structural features of intracellular bacteriophage T7 concatemers. J. Mol. Biol. 68, 319–345 [DOI] [PubMed] [Google Scholar]
- 3.Taylor K., and Wegrzyn G. (1995) Replication of coliphage λDNA. FEMS Microbiol. Rev. 17, 109–119 [DOI] [PubMed] [Google Scholar]
- 4.Geshelin P., and Berns K. I. (1974) Characterization and localization of the naturally occurring cross-links in vaccinia virus DNA. J. Mol. Biol. 88, 785–796 [DOI] [PubMed] [Google Scholar]
- 5.Baroudy B. M., Venkatesan S., and Moss B. (1982) Incompletely base-paired flip-flop terminal loops link the two DNA strands of the vaccinia virus genome into one uninterrupted polynucleotide chain. Cell 28, 315–324 [DOI] [PubMed] [Google Scholar]
- 6.Garon C. F., Barbosa E., and Moss B. (1978) Visualization of an inverted terminal repetition in vaccinia virus DNA. Proc. Natl. Acad. Sci. U.S.A. 75, 4863–4867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Barbour A. G., and Garon C. F. (1987) Linear plasmids of the bacterium Borrelia burgdorferi have covalently closed ends. Science 237, 409–411 [DOI] [PubMed] [Google Scholar]
- 8.Salas M. (1991) Protein-priming of DNA replication. Annu. Rev. Biochem. 60, 39–71 [DOI] [PubMed] [Google Scholar]
- 9.Gutiérrez J., Vinós J., Prieto I., Méndez E., Hermoso J. M., and Salas M. (1986) Signals in the φ29 DNA-terminal protein template for the initiation of phage φ29 DNA replication. Virology 155, 474–483 [DOI] [PubMed] [Google Scholar]
- 10.Blanco L., Prieto I., Gutiérrez J., Bernad A., Lázaro J. M., Hermoso J. M., and Salas M. (1987) Effect of NH4+ ions on φ29 DNA-protein p3 replication: formation of a complex between the terminal protein and the DNA polymerase. J. Virol. 61, 3983–3991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Méndez J., Blanco L., Esteban J. A., Bernad A., and Salas M. (1992) Initiation of φ29 DNA replication occurs at the second 3′ nucleotide of the linear template: a sliding-back mechanism for protein-primed DNA replication. Proc. Natl. Acad. Sci. U.S.A. 89, 9579–9583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Esteban J. A., Salas M., and Blanco L. (1993) Fidelity of φ29 DNA polymerase: comparison between protein-primed initiation and DNA polymerization. J. Biol. Chem. 268, 2719–2726 [PubMed] [Google Scholar]
- 13.Mendez J., Blanco L., and Salas M. (1997) Protein-primed DNA replication: a transition between two modes of priming by a unique DNA polymerase. EMBO J. 16, 2519–2527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Blanco L., Bernad A., Lázaro J. M., Martín G., Garmendia C., and Salas M. (1989) Highly efficient DNA synthesis by the phage φ29 DNA polymerase: symmetrical mode of DNA replication. J. Biol. Chem. 264, 8935–8940 [PubMed] [Google Scholar]
- 15.Kamtekar S., Berman A. J., Wang J., Lázaro J. M., de Vega M., Blanco L., Salas M., and Steitz T. A. (2004) Insights into strand displacement and processivity from the crystal structure of the protein-primed DNA polymerase of bacteriophage φ29. Mol Cell 16, 609–618 [DOI] [PubMed] [Google Scholar]
- 16.Blasco M. A., Blanco L., Parés E., Salas M., and Bernad A. (1990) Structural and functional analysis of temperature-sensitive mutants of the phage φ29 DNA polymerase. Nucleic Acids Res. 18, 4763–4770 [PMC free article] [PubMed] [Google Scholar]
- 17.Dufour E., Méndez J., Lázaro J. M., de Vega M., Blanco L., and Salas M. (2000) An aspartic acid residue in TPR-1, a specific region of protein-priming DNA polymerases, is required for the functional interaction with primer terminal protein. J. Mol. Biol. 304, 289–300 [DOI] [PubMed] [Google Scholar]
- 18.Rodríguez I., Lázaro J. M., Blanco L., Kamtekar S., Berman A. J., Wang J., Steitz T. A., Salas M., and de Vega M. (2005) A specific subdomain in φ29 DNA polymerase confers both processivity and strand-displacement capacity. Proc. Natl. Acad. Sci. U.S.A. 102, 6407–641215845765 [Google Scholar]
- 19.Kamtekar S., Berman A. J., Wang J., Lázaro J. M., de Vega M., Blanco L., Salas M., and Steitz T. A. (2006) The φ29 DNA polymerase:protein-primer structure suggests a model for the initiation to elongation transition. EMBO J. 25, 1335–1343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Muñoz-Espín D., Holguera I., Ballesteros-Plaza D., Carballido-López R., and Salas M. (2010) Viral terminal protein directs early organization of phage DNA replication at the bacterial nucleoid. Proc. Natl. Acad. Sci. U.S.A. 107, 16548–16553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Longás E., Villar L., Lázaro J. M., de Vega M., and Salas M. (2008) Phage φ29 and Nf terminal protein-priming domain specifies the internal template nucleotide to initiate DNA replication. Proc. Natl. Acad. Sci. U.S.A. 105, 18290–18295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Longás E., de Vega M., Lázaro J. M., and Salas M. (2006) Functional characterization of highly processive protein-primed DNA polymerases from phages Nf and GA-1, endowed with a potent strand displacement capacity. Nucleic Acids Res. 34, 6051–6063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mencía M., Gella P., Camacho A., de Vega M., and Salas M. (2011) Terminal protein-primed amplification of heterologous DNA with a minimal replication system based on phage φ29. Proc. Natl. Acad. Sci. U.S.A. 108, 18655–18660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bernad A., Blanco L., Lázaro J. M., Martín G., and Salas M. (1989) A conserved 3′-5′ exonuclease active site in prokaryotic and eukaryotic DNA polymerases. Cell 59, 219–228 [DOI] [PubMed] [Google Scholar]
- 25.Lázaro J. M., Blanco L., and Salas M. (1995) Purification of bacteriophage φ29 DNA polymerase. Methods Enzymol. 262, 42–49 [DOI] [PubMed] [Google Scholar]
- 26.de Vega M., Blanco L., and Salas M. (1998) φ29 DNA polymerase residue Ser-122, a single-stranded DNA ligand for 3′-5′ exonucleolysis, is required to interact with the terminal protein. J. Biol. Chem. 273, 28966–28977 [DOI] [PubMed] [Google Scholar]
- 27.McDonell M. W., Simon M. N., and Studier F. W. (1977) Analysis of restriction fragments of T7 DNA and determination of molecular weights by electrophoresis in neutral and alkaline gels. J. Mol. Biol. 110, 119–146 [DOI] [PubMed] [Google Scholar]
- 28.Gonzalez-Huici V., Salas M., and Hermoso J. M. (2000) Sequence requirements for protein-primed initiation and elongation of phage φ29 DNA replication. J. Biol. Chem. 275, 40547–40553 [DOI] [PubMed] [Google Scholar]
- 29.Illana B., Blanco L., and Salas M. (1996) Functional characterization of the genes coding for the terminal protein and DNA polymerase from bacteriophage GA-1: evidence for a sliding-back mechanism during protein-primed GA-1 DNA replication. J. Mol. Biol. 264, 453–464 [DOI] [PubMed] [Google Scholar]
- 30.Martín A. C., Blanco L., García P., Salas M., and Méndez J. (1996) In vitro protein-primed initiation of pneumococcal phage Cp-1 DNA replication occurs at the third 3′ nucleotide of the linear template: a stepwise sliding-back mechanism. J. Mol. Biol. 260, 369–377 [DOI] [PubMed] [Google Scholar]
- 31.Caldentey J., Blanco L., Bamford D. H., and Salas M. (1993) In vitro replication of bacteriophage PRD1 DNA: characterization of the protein-primed initiation site. Nucleic Acids Res. 21, 3725–3730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.King A. J., and van der Vliet P. C. (1994) A precursor terminal protein-trinucleotide intermediate during initiation of adenovirus DNA replication: regeneration of molecular ends in vitro by a jumping back mechanism. EMBO J. 13, 5786–5792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Caldentey J., Blanco L., Savilahti H., Bamford D. H., and Salas M. (1992) In vitro replication of bacteriophage PRD1 DNA: metal activation of protein-primed initiation and DNA elongation. Nucleic Acids Res. 20, 3971–3976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gonzalez-Huici V., Lázaro J. M., Salas M., and Hermoso J. M. (2000) Specific recognition of parental terminal protein by DNA polymerase for initiation of protein-primed DNA replication. J. Biol. Chem. 275, 14678–14683 [DOI] [PubMed] [Google Scholar]
- 35.Gutiérrez J., García J. A., Blanco L., and Salas M. (1986) Cloning and template activity of the origins of replication of phage φ29 DNA. Gene 43, 1–11 [DOI] [PubMed] [Google Scholar]
- 36.Pérez-Arnaiz P., Longás E., Villar L., Lázaro J. M., Salas M., and de Vega M. (2007) Involvement of phage φ29 DNA polymerase and terminal protein subdomains in conferring specificity during initiation of protein-primed DNA replication. Nucleic Acids Res. 35, 7061–7073 [DOI] [PMC free article] [PubMed] [Google Scholar]