Background: Integrins undergo large conformational changes when ligand-bound.
Results: Using nanodisc technology and EM, we observed Mn2+-activated αIIbβ3 integrin both alone and fibrin-bound.
Conclusion: MnCl2-activated αIIbβ3 integrin alone has a compact conformation, becoming fully upright and open when fibrin-bound.
Significance: The first structure of membrane-embedded integrins bound to physiological substrate reveals the importance of integrin extension in macromolecular ligand binding.
Keywords: cell adhesion, electron microscopy (EM), fibrin, platelet, signal transduction, αIIbβ3 integrin, nanodisc
Abstract
Increased ligand binding to integrin (“activation”) underpins many biological processes, such as leukocyte trafficking, cell migration, host-pathogen interaction, and hemostasis. Integrins exist in several conformations, ranging from compact and bent to extended and open. However, the exact conformation of membrane-embedded, full-length integrin bound to its physiological macromolecular ligand is still unclear. Integrin αIIbβ3, the most abundant integrin in platelets, has been a prototype for integrin activation studies. Using negative stain electron microscopy and nanodisc-embedding to provide a membrane-like environment, we visualized the conformation of full-length αIIbβ3 in both a Mn2+-activated, ligand-free state and a Mn2+-activated, fibrin-bound state. Activated but ligand-free integrins exist mainly in the compact conformation, whereas fibrin-bound αIIbβ3 predominantly exists in a fully extended, headpiece open conformation. Our results show that membrane-embedded, full-length integrin adopts an extended and open conformation when bound to its physiological macromolecular ligand.
Introduction
Integrins are non-covalent heterodimers containing α- and β-subunits, both of which have a large extracellular (ectodomain), a single transmembrane (TM)3 α-helix and a small cytoplasmic domain (1). Integrins respond to cytoplasmic signals by increasing the affinity of their ectodomains for ligands through an allosteric rearrangement that requires the binding of talin to the β-subunit cytoplasmic domain, in a process termed “integrin activation.” Integrin activation plays an important role in development, immune responses, hemostasis, and cancer (2). Integrin αIIbβ3, the most abundant surface protein in platelets, mediates platelet aggregation and is critical in hemostasis, thrombosis, and inflammation (3). Mutations of αIIbβ3 are known to cause diseases such as Glanzmann thrombasthenia, a disease of abnormal hemostasis (4). The affinity of integrin αIIbβ3 must be precisely regulated to enable hemostasis responses upon platelet stimulation while preventing pathological thrombosis due to uncontrolled platelet aggregation. Thus, αIIbβ3 has been a prototype for studies of integrin activation in health and disease (5–8).
In integrins without an α-A domain, which includes αIIbβ3, the α-subunit β-propeller domain, and the β-subunit β-A domain, jointly form a ligand binding pocket (see Fig. 1B) (9–11). The ligand binding pocket, the thigh domain of the α-subunit, the hybrid domain, the PSI domain, and I-EGF1 of the β-subunit are referred to as the headpiece and can be studied as a separate fragment (11, 12). The rest of the ectodomain including the calf-1 and calf-2 domains from the α-subunit, I-EGF2–4, and β-tail domains from the β-subunit comprise the leg pieces (Fig. 1B). Integrin conformation has been studied by x-ray crystallography, electron microscopy (EM), NMR, and molecular dynamics simulation of integrin fragments and detergent-solubilized integrin (1, 13–16). The reported conformations include a compact shape with the headpiece pointing toward the membrane (Fig. 1A) (compact, head down) (9, 10), a compact form with the headpiece pointing away from the membrane (compact, head up) (17), an extended form with hybrid and calf-1 domain juxtaposed (extended, headpiece closed) (18), and an extended form with the hybrid domain swung out (Fig. 1, B and C) (extended, headpiece open) (12, 18, 19). Despite these efforts, membrane-embedded, full-length integrin bound to its physiological ligand has not been directly visualized, and its precise conformation is not clear.
FIGURE 1.
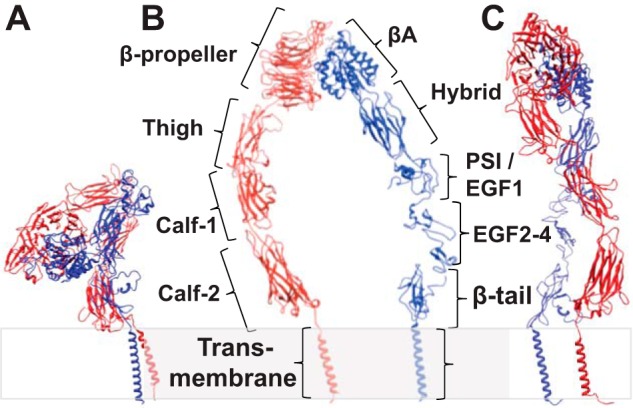
Schematic structure of integrin αIIbβ3. In the inhibited state (A), the integrin adopts a bent-over conformation, whereas when integrin is in the active state (B and C), it adopts an up-right, extended conformation. When the leg densities are in the same plane, they appear to be separate (B). However, if the view is from the side, the leg densities could appear crossed-over (C) even though they come from the same structure with widely separated legs when viewed from a different direction. Because there is no high resolution structure of full-length integrin αIIbβ3, details of the domain interaction and placement are imprecise. Panel A was generated using PDB structures 1JV2 (10) and 2K9J (51); panels B and C were generated using PDB structures 2VDM (49), 2P28 (52), 2K1A (53), and 2MRZ (53).
The controversy in the current literature necessitates the present focus on the conformation of membrane-embedded, full-length integrin bound to its physiological ligand fibrin. Activated αvβ3 integrin ectodomain in the compact, head-down conformation bound stably to a fragment of fibronectin (20), whereas in a comparable study using α5β1 integrin only the extended, headpiece-open conformation was observed when bound to the same protein ligand (12). In a similar controversy, crystals of αvβ3 integrin ectodomain bound to a cyclic RGD peptide, which typically activates integrins, showed the compact, head-down conformation (21), whereas in an EM study utilizing both αvβ3 and αIIbβ3 only the extended, headpiece-open conformation was observed in the presence of an RGD peptide (22). Adding to this controversy are the reports that both the compact and the extended-open conformation of integrin ectodomain fragments can be in the activated form (9, 20, 22–25).
The focus of more recent studies (17, 25, 26) has been on how well these observations made with ectodomain or ectodomain fragments correspond to the conformation of integrins embedded in the membrane. Observations such as a surprising compact, head-up conformation for inactive αIIbβ3 integrin in nanodiscs (17), a stable integrin αLβ2 conformation with intermediate affinity in cells (26), and conflicting inferences on the conformation of different integrin isoforms in the cell membrane drawn from fluorescence studies (25, 27–29) further complicate this issue.
We have previously reported that the talin head domain (THD), the integrin-activating fragment of talin, more than doubles the percentage of extended conformation of full-length integrin αIIbβ3 embedded in islands of phospholipid bilayers formed in nanodiscs (30). However, we were unable to determine the conformation of activated integrin bound to macromolecular ligands such as fibrin because we could not observe the THD in the proposed talin-integrin-fibrin complex. Because of these uncertainties, we lacked independent verification of the activation state of each individual integrin, i.e. we were unable to pinpoint the activation state of the individual integrin in the complex without using the extended conformation as the proxy. An alternative way of ensuring uniform and high percentage of integrin activation is Mn2+ treatment (31), which is often used as a positive control for integrin activation. Mn2+ replaces all Ca2+ and occupies 100% of the integrin metal ion sites (21), thus producing a homogeneously activated integrin population.
Here we reconstituted purified full-length αIIbβ3 integrin into phospholipid nanodiscs, activated these αIIbβ3 nanodiscs with Mn2+, and visualized them in complex with fibrin by negative stain EM. Our EM results show that fibrin-free, Mn2+-activated integrin adopts a compact conformation. However, integrins bound to fibrin, a physiological macromolecular ligand, predominantly adopt a fully extended, headpiece-open conformation.
Experimental Procedures
Preparation of Integrin Nanodiscs and Two-dimensional Fibrin Complexes
Full-length αIIbβ3 integrin was purified from outdated blood platelets following published procedures (30, 32, 33). Integrin nanodiscs were assembled using a protocol adapted previously (34, 35). Briefly, detergent-solubilized 1,2-dimyristoyl-sn-glycero-3-phosphocholine, 1,2-dimyristoyl-sn-glycero-3-phospho-(1′-rac-glycerol), membrane scaffold protein, integrin, and other solutes were combined, and the detergent was removed with SM-2 Biobeads. The assembled integrin nanodiscs were then purified with a size exclusion column.
The lipid monolayer technique previously described was employed to produce fibrin-integrin complexes suitable for EM (30). We used a rigid lipid monolayer consisting of dioctadecyldimethyl ammonium bromide and dipalmitoyl phosphatidylcholine. This lipid combination is in a gel phase, i.e. it is not fluid, at 4 °C, thereby preventing in-plane diffusion of the integrin nanodiscs after they bind the lipid. In the present paper, MnCl2 instead of THD was used to activate the integrin. We preferred Mn2+ over THD as the activator because it will activate all of the integrins, whereas THD at achievable concentrations in vitro will not (30). Preformed integrin nanodiscs were activated by incubation for 2 h at room temperature in 20 mm Tris, 50 mm NaCl, 50 mm MnCl2, pH 7.6. After injection under the monolayer the final Mn2+ concentration was 5 mm. Between 1.0 and 1.5 μl of the activated integrin solution was injected by Hamilton syringe into the buffer phase under the fibrin-lipid monolayer samples, all done at 4 °C. After a 24-h incubation, the lipid monolayer samples were evaluated by negative stain EM to verify the presence of an adequate concentration of integrin bound to the fibrin before data collection.
Electron Microscopy
Samples were stained with 2% uranyl acetate following previous procedures (30). Projection images were collected using low dose protocols on an FEI CM120 TEM (FEI, Hillsboro, OR) at 120 keV using a TVIPS F224 2K × 2K CCD camera (Tietz; TVIPS GmbH, Germany). The magnification was 60,000×; the defocus was ∼2.5 μm. The final pixel size was 0.222 nm. A total of 600 micrographs were collected.
Data Processing
Most image processing was performed with the I3 package (36), which utilizes modulation analysis and hierarchical ascendant classification.
Integrin nanodiscs could be readily identified because of their distinctive shape and size. Images with good density of both free and fibrin-bound integrin nanodiscs were picked manually for further processing. A total of 1515 fibrin-free integrin nanodisc particles were normalized, corrected for the contrast transfer function by Wiener filtering, and low pass-filtered to 1.78-nm resolution. For each of the selected particles, the nanodisc density was placed in the center and first aligned by translation only. A global average of all extracted particles was used as the initial reference for translational alignment. For the free integrin nanodiscs, rotational alignment was used to place the integrin density at the top of the nanodisc density. The procedure comprised the following: a randomly picked particle with clear integrin density was used as the initial reference, and subsequent cycles used the class averages as the reference for alignment of all the raw particles. When the integrin density was aligned well at the top of the nanodisc, multivariate data analysis and multi-reference alignment using only class averages, known as alignment by classification (37), were used to focus further alignment on the integrin density.
For the fibrin-bound integrin nanodiscs, we selected a total of 4,656 particles. Once the nanodisc density was centered well with the integrin density positioned in the bottom half of the image, a raw particle with clear, straight horizontal fibrin density at the bottom of the image was chosen as the initial reference. Rotational alignment was then applied to all raw particles to make the fibrin density horizontal as the second point of reference. In this way the integrin density was concentrated between the centered nanodisc and the horizontal fibrin. A trapezoidal shaped Boolean mask covering the space between the nanodisc and the fibrin was used for focused classification to separate fibrin-bound integrin conformations. This procedure minimizes the possibility of reference bias in the integrin class averages because no alignment is done on the integrin itself; all the alignment was done to center the nanodisc density and to place the fibrin fiber at the bottom.
The majority of class averages showed well defined nanodisc and fibrin density, but the integrin density was uninterpretable or ambiguous (more than one interpretation for leg pieces and head piece). We then examined montages of the class membership systematically rejecting class members of poor quality based on visibility of headpiece and legs (i.e. preservation in negative stain) thereby reducing the number of fibrin-bound integrin nanodiscs to 1,057.
Alignment on only the nanodisc and fibrin for this subset of images generated interpretable class averages that incorporated only 23 integrin nanodiscs, suggesting that the integrins were broadly distributed across the gap between the nanodisc and fibrin as well as conformationally variable. Consequently, we did further alignment based on the integrin position. For the first cycle we did a single reference alignment of raw particles against a class average having the integrin facing vertically down. After this first cycle, we used multireference alignment by classification but masked the images on the integrin density. In this way we obtained four additional interpretable class averages incorporating a further 245 raw nanodisc images.
We then took the remaining integrin images supplemented with previously rejected images, a total of 4,388 particles, and repeated the analysis twice using a conventional single particle approach with alignment on the integrin nanodisc, only with the fibrin masked out. In each cycle we rejected 20% of particles with poor correlation. Both single and multireference alignment strategies were used reducing the particle count to 2,384 images. However, no new conformations were identified, and those that were found were substantially inferior in quality to the ones shown. Finally, we repeated the analysis using the EMAN software package (38). We computed 40 class averages, but they were no more interpretable than those produced using I3 and suggested no new conformations. We conclude from this that our unconventional image analysis was better suited than conventional methods to the analysis of these micrographs.
Model Building
A density map of a bent-over integrin was generated using the crystal structure of the ectodomain of integrin αIIbβ3 with the TM domain (PDB code 4CAK; Ref. 17) (Fig. 1A). The density map of active integrin (shown in Fig. 1, B and C) was generated mainly using the crystal structure of integrin αIIbβ3 with the open headpiece (PDB code 3FCU; Ref. 9). Projections of the electron density volume were generated using the Spider software package (39). Projections of these maps were generated in a grid covering the three Euler angles in increments of 10°.
Results
Structure of Fibrin-free Integrin Nanodiscs
Inactive integrin is believed to adopt a bent-over (referred to here as compact, head-down) conformation as shown by crystal structures of the αvβ3 ectodomain (9, 10). This structure has been confirmed for the α-I integrin αxβ2 (40). However, whether activated integrin is extended before engaging its ligand is still not clear. To study integrin conformation in a homogeneously activated integrin population, we investigated Mn2+-activated full-length integrin αIIbβ3 embedded in an island of membrane formed by our previously developed αIIbβ3 nanodiscs (30, 35). We picked, classified, and averaged 1,515 particles of integrin nanodiscs unbound by ligand from images of samples where we added integrin nanodiscs to preformed fibrin (Fig. 2). The fibrin-free integrin nanodiscs lie in the background of the fibrin-bound integrin nanodiscs so that the solution conditions and other variables are identical for both populations. The general shape of the unbound integrin nanodiscs had two distinctive but connected densities: one large and circular and the other smaller and irregular (Fig. 3). The large moiety at 12∼13-nm diameter was close to the size of nanodiscs predicted from the length of the membrane scaffold protein and observed in our previous study (30). Thus, the large moiety is assigned to the nanodisc density. The nanodiscs lay nearly exclusively “en face” on the surface of the lipid monolayer, an orientation that is likely to be due to the negative charge of the phospholipids and the positive charge of the lipid monolayer.
FIGURE 2.
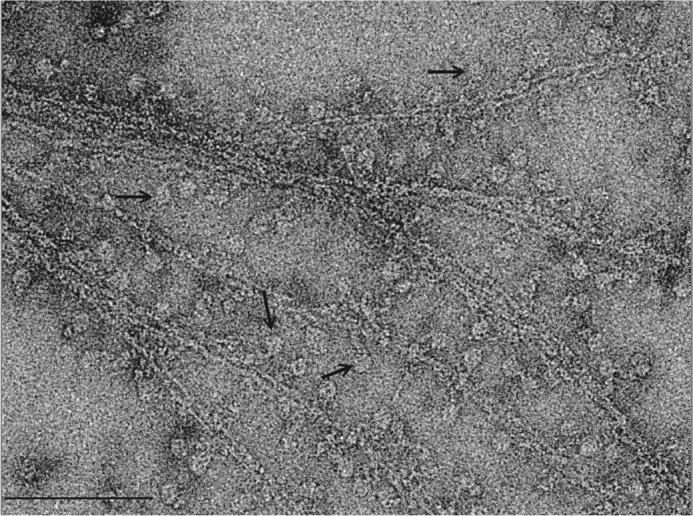
Fibrin rafts decorated with αIIbβ3 integrin nanodiscs. Electron micrograph of fibrin rafts formed on a positively charged lipid monolayer and was treated with αIIbβ3 integrin nanodiscs previously treated with Mn2+ to achieve an activated state with high affinity for fibrin. In this sample the nanodiscs density is disk-shaped; the filamentous structure is fibrin. The density connecting the nanodiscs with the fibrin is the integrin. The pointers locate some fibrin-bound integrin nanodiscs. Bar, 100 nm.
FIGURE 3.
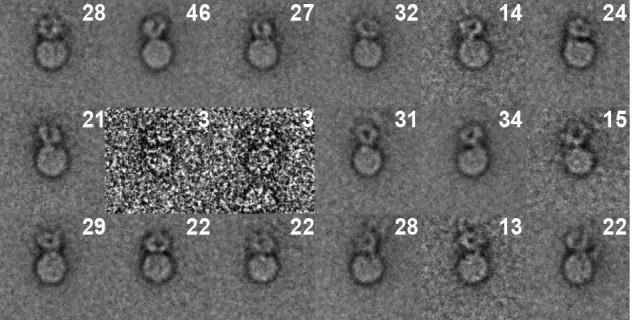
Class averages of Mn2+-activated, fibrin-free integrin nanodiscs. These class averages come from 414 of 1515 raw ligand-free integrin nanodiscs with the best morphological detail. Numbers in the upper right hand corner are the number of class members within each class average.
The small moiety is around 9∼10 nm in length in its longest dimension. Its size and shape resemble integrin density observed previously (30) and is assigned to the integrin ectodomain. The size of the integrin density is consistent with previous studies (17, 33, 41). The size is slightly smaller than previously observed for detergent-solubilized integrin (41), which is about 13 nm. However, the TM and cytoplasmic domains and the surrounding detergent micelle may increase the observed size in detergent. In our preparations using integrin nanodiscs, the ectodomain nearly always lay over the edge of the nanodiscs. The shape and length of the ectodomain is consistent with a bent-over conformation or other non-extended conformation (17, 33, 41). Computation of class averages and visual inspection of class members as well as raw micrographs revealed only a small fraction of fibrin-free integrin nanodiscs in the extended conformation. Classes with integrins clearly in the compact conformation comprised 1,394 of 1,515 fibrin-free nanodiscs (57 of 60 classes). The remaining three classes (121 nanodiscs) lacked a clearly defined ectodomain and so were classified visually. The ratio between bent-over and extended is close to 19:1. Thus, our data confirm that Mn2+-activated but ligand-free integrin αIIbβ3 predominantly adopts a compact conformation in a membrane environment (33). Some variability in the appearance of the active but ligand-free integrin nanodiscs was observed in the class averages, which probably represent different views of the non-extended integrin. However, they all showed a compact conformation. The αIIb and β3 leg pieces were not discernible in any of the views, strongly suggesting that the legs are juxtaposed.
Recently, Choi et al. (17) reported that full-length integrin αIIbβ3 in nanodiscs adopts a compact, head-up conformation with the headpiece pointed away from the membrane and not toward the membrane as is commonly thought. We, therefore, generated density maps from three compact structures, PDB codes 3IJE, 3FCS, and 4CAK (9, 25). We then computed projection maps covering the whole of Euler space and aligned each with 18 class averages displaying the best-defined integrin density (Fig. 3). The projections from the Choi et al. (17) model (PDB code 4CAK) appeared to be most similar to our class averages (Fig. 4) due to the fact that the membrane-proximal part of the integrin molecule was always adjacent to the nanodiscs. The best alignment of the other two structures (PDB codes 3IJE and 3FCS) placed the membrane-proximal domains adjacent to the nanodiscs <50% of the time. Such a placement is unlikely in the integrin nanodiscs specimen. The difference in correlation coefficient between our map and the three-modeled maps was small (∼20%), but the Choi (17) model produced consistently better correlation. The significant difference between this study and that of Choi et al. (17) is the shape of the nanodisc in projection. In our experiment we used a positively charged lipid monolayer to support the nanodiscs, whereas Choi et al. (17) used glow-discharged carbon-coated grids, a process that makes the carbon film generally negatively charged if done in air. The negative phospholipid in the nanodiscs would be attracted to the positively charged lipid monolayer making the nanodiscs orient en face, thereby presenting a circular shape. This would not be the case with glow discharging, which is likely to bind the integrin to the carbon film rather than the lipids in the nanodiscs. Choi et al. (17) found that the orientation of the nanodiscs varies considerably, thus presenting ellipsoidal shapes rather than orienting en face as in our case, consistent with this interpretation.
FIGURE 4.
Modeled projections from three compact integrin atomic structures. A, PDB code 3IJE (25). B, PDB code 3FCS (9). C, PDB code 4CAK (17). White arrows point to the membrane proximal part of the integrin head when wrongly oriented relative to the nanodisc, located at the bottom as shown in Fig. 3, after cross correlation. Lack of an arrow indicates that the membrane proximal region is correctly positioned at the bottom adjacent to the nanodisc. The corresponding projections in each series of projections have the highest correlation coefficient with the class-averages shown in Fig. 3. In A and B the membrane proximal densities of the integrin in some projections are not positioned at the bottom where they must be located if the TM domain is to be located within the nanodiscs. Conversely, the projections in C computed from PDB code 4CAK are always positioned with the membrane proximal density at the bottom adjacent to the nanodisc, as required from the known domain topology.
Structure of Fibrin-bound Integrin Nanodiscs
To facilitate the visualization of αIIbβ3-ligand interaction, we chose fibrin, the product of fibrinogen polymerization after thrombin cleavage, as the ligand for αIIbβ3. Fibrin fibers were formed on a lipid monolayer to enforce a two-dimensional arrangement of the sample, which simplifies interpretation (42, 43). Fibrin and fibrinogen, the physiological ligands of αIIbβ3, have been used in numerous studies of platelet and integrin activation (1, 3, 44–49). The binding of these ligands has been regarded as a measure for integrin activation. Furthermore, no direct observation of the molecular structure of αIIbβ3 in complex with fibrin has been reported previous to this study.
In micrographs of the negatively stained fibrin complexes (Fig. 2), the fibrin fibers exist as a two-dimensional bundle of varying width. Decoration of αIIbβ3 nanodiscs along the edge of the fibrin filaments can be clearly seen in the form of round disks generally at a uniform distance from the filament and connected to it by integrin densities. Because ligand binding is the measure of integrin activation, these αIIbβ3 nanodiscs decorating the fibrin filaments are, by definition, the activated form. We processed images of the fibrin-attached integrin nanodiscs, as we have done with ligand-free integrin nanodiscs. Starting with 4,656 single particle images, the nanodisc density was placed in the center of the boxed image, which was large enough to include both the integrin and a few strands of the fibrin raft. During the processing, the fibrin filament density was rotated to be oriented parallel to the bottom of the box as much as possible. This gave a frame of reference consisting of the centered integrin nanodiscs and the horizontally oriented fibrin strands, which made it easier to visualize the variable shape of integrin and fibrin. We could also focus the multivariate data analysis on the smaller area between the nanodiscs and the fibrin fiber where the integrin is presumed to lie. Because of the complicated geometry and variable size of the fibrin bundles, we were unable to obtain a unique structure of fibrin; each averaged fibrin filament is essentially featureless. Single, double, and even triple strands of fibrin were found in the class averages. To further reduce the heterogeneity problem, the extracted fibrin-bound integrin nanodiscs were further separated based on the number of fibrin strands within the boxed area. Because the angle between integrin and fibrin filament in the raw images shows considerable variation (Fig. 2), classification by multivariate data analysis grouped particles with similar conformation and similar integrin/fibrin angle into separate classes, resulting in considerably smaller population per class. Most of these classes were uninterpretable, necessitating visual editing of the class membership according to image quality, thereby reducing the number of usable single particle images by ∼75%. At the end of the processing, only four classes incorporating a total of 23 raw particles of the remaining 1,057 images showed both distinct headpiece and leg densities (Fig. 5A). Although few in number, these classes represent the most objective structures as no alignment has been done on the integrin itself. Based on the class averages, we measured the length of the extended integrin to be 20 ± 1 nm, consistent with that calculated from extended molecular models and that observed with full-length integrin αIIbβ3 in detergent (41).
FIGURE 5.
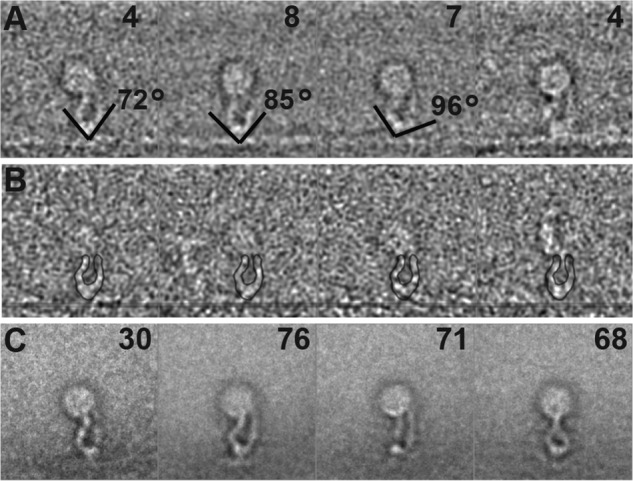
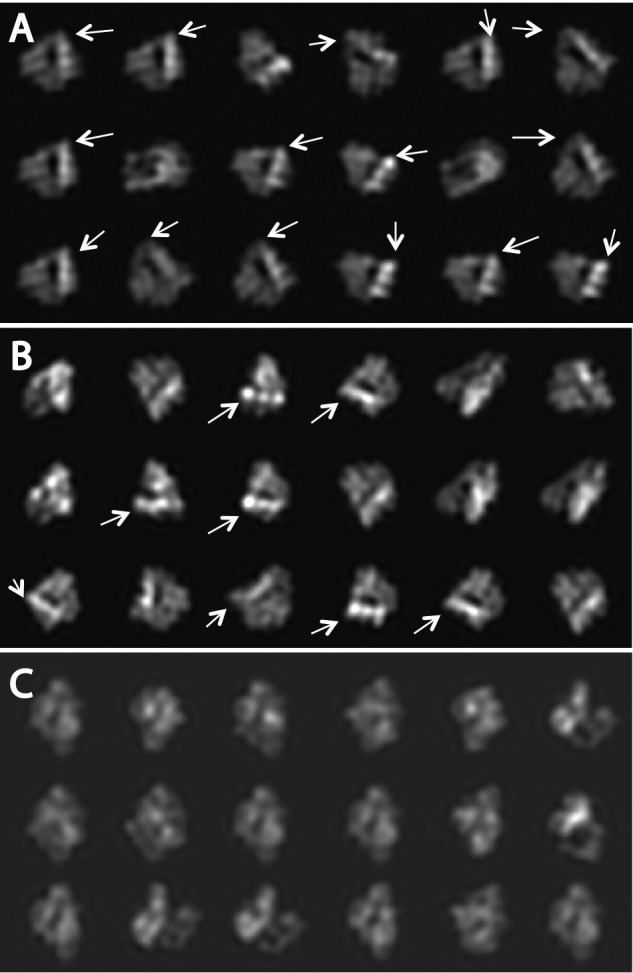
Class-averages of extended αIIbβ3 integrin nanodiscs bound to fibrin. A, montage of four class averages from fibrin-bound integrin nanodiscs from the alignment that excluded the integrin representing 23 of 1,057 images. The number of the members in each class is in the top right corner. The lines show the direction of the adjacent leg densities of the headpiece. The angle between line pairs ranges from 72° to 96°. B, montage of the members in the top left panel class average. The position of the fibrin fiber has been marked for clarity with a translucent line. The outline of the class average is superimposed on the class member for clarity. C, montage of class averages representing 245 of 1,034 integrin nanodisc images from panel A obtained from an alignment on the integrin instead of the fibrin and nanodisc density. The number of class members is noted in the upper right-hand corner of each panel. Note that although all of these integrins are bound to the fibrin, the fibrin orientation with respect to the integrin is highly variable.
Both class averages and the raw images in the class clearly show that integrin head pieces are in an open conformation and the two leg pieces are discernible and separated (Fig. 5, A and B). They generally do not appear to be crossed as seen in talin-activated but ligand-free αIIbβ3 nanodiscs (30). The leg densities from both chains at the edge of the nanodiscs remain separated but not by much. The largest leg separation is about 4.5 nm. The angle between the hybrid domain and thigh domain is 84° ± 12°, consistent with a hybrid domain in a swung-out position after ligand engagement.
Using the remaining 1,034 raw particles, we did another alignment this time using the integrin density. We obtained an additional 4 class averages encompassing 245 integrin nanodiscs that showed clear integrin headpiece and leg density, but in this case the fibrin density was blurred out (Fig. 5C). Thus, if the fibrin was well aligned, few of the integrins were well aligned, and when the integrins were aligned, very little of the fibrin was aligned. The other class averages either did not show both the headpiece and leg-piece densities clearly or were ambiguous with more than a pair of legs or no clear separation between headpiece and fibrin. However, this additional procedure increased the number of particles averaged by >10-fold. Three of these integrin-aligned class averages resemble the first four classes from the fibrin alignment. The fourth is different, resembling mirrored images of extended integrin nanodiscs activated by THD (see Fig. 7C of Ref. 30) in that the leg densities appear to cross each other near the edge of the nanodisc. The integrin height obtained in the integrin-aligned class averages is 18 ± 1 nm, slightly smaller than the fibrin-aligned class averages.
After removing those raw images contained in the best class averages, we subjected all the remaining 4,388 fibrin-bound integrin nanodisc images to conventional single particle processing using a high rate of particle rejection. This effort produced no new conformations, and those that were similar were of inferior quality to those reported above.
Comparison of Fibrin-free and Fibrin-bound Integrin
The class averages clearly showed that in the fibrin-bound state, the integrins are extended, the leg pieces of α- and β-subunits are well separated (although they sometimes cross near the edge of the nanodisc), and the head-piece is open, whereas in the Mn2+-activated but ligand-free state, the integrins are bent over, and their leg pieces are juxtaposed and indiscernible. To directly show that ligand engagement induces the extension of integrin, a typical class average of activated but ligand-free integrin nanodiscs was superimposed with ligand-bound integrin nanodiscs by bringing their nanodiscs densities into register (Fig. 6). This superimposed image clearly demonstrates that the length of activated but fibrin-free integrin is much shorter than the fibrin-bound integrin. Furthermore, the distance between the leg pieces is larger in the fibrin-bound integrin than that in the ligand-free integrin.
FIGURE 6.
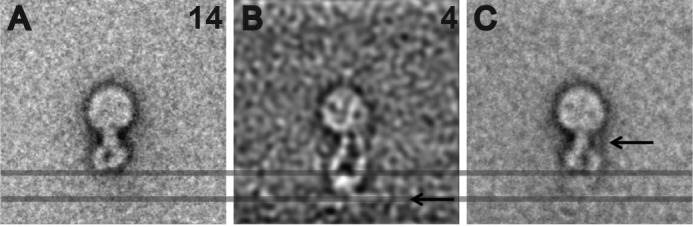
Superposition of free and fibrin-bound integrin nanodiscs. A, fibrin-free integrin nanodiscs. B, fibrin-bound integrin nanodiscs. C, overlap of fibrin-free and fibrin-bound integrin nanodiscs. The arrow in B indicates the location of the fibrin density, which is distant from the integrin density when it is bent-over. Another significant change locates to the lower leg densities (arrow in C), which are clearly separated in the fibrin-bound integrin nanodiscs (B).
Discussion
Our previous papers showed no dramatic change on the length of the ectodomain when the full-length αIIbβ3 integrin was activated by 1 mm Mn2+ when incorporated into small unilamellar vesicles (33). The average length is around 11 nm, which means there is no large conformational change when activated by Mn2+ alone. The presence of a compact conformation in detergent after Mn2+ activation was later confirmed (41). Here we further confirmed that integrin in a membrane bilayer environment after Mn2+ activation adopts primarily a compact conformation with no dramatic length change. In Eng et al. (41), 60% of the integrin population adopted a compact head-down conformation after Mn2+ activation; even after the addition of another ligand mimetic L-739758, 20% of the integrins remain in the compact head-down conformation. In our result the ratio of compact to extended conformation for the unbound integrin nanodisc is close to 19:1, which is much higher than the 2.3:1 observed in the above-mentioned experiment. The higher ratio in our result could be due to the membrane environment, in which the integrin transmembrane domain interactions are stabilized by formation of inner and outer membrane clasps (50, 51), thereby maintaining the compact conformation.
Because the fibrin-free integrin nanodiscs were quantified in the presence of ligand, it could be argued that the low percentage of activated integrins in the extended but ligand-free state compared with the 40% predicted by previous studies in detergent (41) was due to fibrin removing the upright, activated integrins from the pool of activated integrins. The fact the Mn2+-treated nanodiscs were incubated with the fibrin for 24 h before EM specimens were made argues against this possibility. If fibrin indeed removed all the extended integrins, it seems likely that the prolonged incubation would have allowed the remaining fibrin-free nanodiscs to reach a new equilibrium even at 4 °C. Furthermore, the rigid characteristic of the lipid monolayer used would have prevented in-plane diffusion of the lipid-bound nanodiscs and limited the possibility that fibrin may concentrate around itself the active, extended integrin nanodiscs among the integrin nanodiscs attached to lipid monolayer. Integrins in the extended or bent conformation, if they did not happen to hit the fibrin when first absorbed onto lipid monolayer, will therefore not “see” fibrin during the entire experiment due to nonfluidity of the underlying monolayer. These integrin nanodiscs away from fibrin would presumably remain bound in whatever conformation they were in and stay at a locus when they first contacted the monolayer. Thus, we feel that the internal control of ligand-free nanodiscs provides a reasonable measure of the proportion of extended integrins in solution.
Integrin αIIbβ3 extends to 18–20 nm in length upon binding to fibrin. This length is very close to the 23-nm maximum length of intact integrin αIIbβ3 in detergent (41). Considering the TM and cytoplasmic domains are visible in detergent-solubilized integrins but invisible in the negatively stained integrin nanodiscs, the length of the ectodomain in our study is reasonable. Although we cannot identify the TM domain directly, it is reasonable to expect that separation of the TM domains of the α- and β-subunits has occurred based on the degree of separation between the adjacent lower leg densities. The moderate size of the lower leg separation, 4.5 nm, is consistent with the recent observation that TM domains from the two subunits are still close to each other after headpiece opening (41), although the size of the nanodiscs may have constrained the degree of separation. The fibrin-bound integrin adopts an extended, headpiece-open conformation as confirmed by the angle between the adjacent leg densities.
Therefore, full-length αIIbβ3 integrin embedded in a phospholipid bilayer is in an extended, headpiece-open conformation when bound to its macromolecular ligand. Our results can be reconciled with previous reports on both sides of the controversy (12, 19, 20, 22). It is possible that the presence of a lipid bilayer would restrict a macromolecular ligand or protein ligand from accessing the membrane-facing ligand pocket in a bent integrin conformer even if it is activated. Thus, truncating the transmembrane domain and removal of the lipid bilayer would enable such an interaction with an activated but bent integrin conformer, as seen previously (20).
Recently, Choi et al. (17) reported that the headpiece of inactive integrin αIIbβ3 points away from the membrane (compact, head-up) rather than toward it as depicted in most current models. We, therefore, tested three models for the compact conformation. The model from Choi et al. (17) was the better fit but largely in terms of consistently providing a realistic orientation of the membrane proximal domains with respect to the nanodiscs. Nevertheless, due to the preferred orientation and possible flattening due to drying, our projections themselves are insufficient to conclude that the headpiece points away from the membrane.
To summarize, we have investigated the conformation of integrin in a physiological-like condition. Our study used full-length integrin αIIbβ3 incorporated into nanodiscs to mimic the interaction between the TM domain and the membrane environment. Mn2+-activated but ligand-free integrin in a lipid bilayer remains in the non-extended conformation, but integrin nanodiscs predominantly adopt an extended, headpiece-open conformation when bound to its macromolecular ligand. We captured the images of both conformations and presented the first direct observation of global conformational change of membrane-embedded integrin upon binding to its physiological ligand.
Author Contributions
A. D. performed the data collection and image analysis, F. Y. prepared and characterized the integrin nanodiscs, D. W. T. prepared the specimens used for electron microscopy, G. H. carried out preliminary studies investigating feasibility, A. D., F. Y., D. W. T., M. H. G., and K. A. T. wrote the paper, and M. H. G. and K. A. T. supervised the experimental work in their respective laboratories.
Acknowledgment
We utilized the instruments at the Biological Science Imaging Resource supported by Florida State University for this study.
This work was supported by National Institutes of Health Grants HL 078784, HL 31950, and HL 117807 (to M. H. G.) and GM 64346 (Cell Migration Consortium; to K. A. T. and M. H. G.). The authors declare that they have no competing conflicts of interest with the contents of this article.
- TM
- transmembrane
- THD
- talin head domain.
References
- 1.Hynes R. O. (2002) Integrins: bidirectional, allosteric signaling machines. Cell 110, 673–687 [DOI] [PubMed] [Google Scholar]
- 2.Hynes R. O. (2002) A reevaluation of integrins as regulators of angiogenesis. Nat. Med. 8, 918–921 [DOI] [PubMed] [Google Scholar]
- 3.Coller B. S., and Shattil S. J. (2008) The GPIIb/IIIa (integrin αIIbβ3) odyssey: a technology-driven saga of a receptor with twists, turns, and even a bend. Blood 112, 3011–3025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kato A. (1997) The biologic and clinical spectrum of Glanzmann's thrombasthenia: implications of integrin αIIbβ3 for its pathogenesis. Crit. Rev. Oncol. Hematol. 26, 1–23 [DOI] [PubMed] [Google Scholar]
- 5.Quinn M. J., Byzova T. V., Qin J., Topol E. J., and Plow E. F. (2003) Integrin αIIbβ3 and its antagonism. Arterioscler. Thromb. Vasc. Biol. 23, 945–952 [DOI] [PubMed] [Google Scholar]
- 6.Bledzka K., Smyth S. S., and Plow E. F. (2013) Integrin αIIbβ3: from discovery to efficacious therapeutic target. Circ. Res. 112, 1189–1200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shattil S. J., and Newman P. J. (2004) Integrins: dynamic scaffolds for adhesion and signaling in platelets. Blood 104, 1606–1615 [DOI] [PubMed] [Google Scholar]
- 8.Shattil S. J., Kim C., and Ginsberg M. H. (2010) The final steps of integrin activation: the end game. Nat. Rev. Mol. Cell Biol. 11, 288–300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhu J., Luo B. H., Xiao T., Zhang C., Nishida N., and Springer T. A. (2008) Structure of a complete integrin ectodomain in a physiologic resting state and activation and deactivation by applied forces. Mol. Cell 32, 849–861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xiong J. P., Stehle T., Diefenbach B., Zhang R., Dunker R., Scott D. L., Joachimiak A., Goodman S. L., and Arnaout M. A. (2001) Crystal structure of the extracellular segment of integrin αvβ3. Science 294, 339–345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Springer T. A., and Dustin M. L. (2012) Integrin inside-out signaling and the immunological synapse. Curr. Opin. Cell Biol. 24, 107–115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Takagi J., Strokovich K., Springer T. A., and Walz T. (2003) Structure of integrin α5β1 in complex with fibronectin. EMBO J. 22, 4607–4615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Arnaout M. A., Mahalingam B., and Xiong J. P. (2005) Integrin structure, allostery, and bidirectional signaling. Annu. Rev. Cell Dev. Biol. 21, 381–410 [DOI] [PubMed] [Google Scholar]
- 14.Luo B. H., Carman C. V., and Springer T. A. (2007) Structural basis of integrin regulation and signaling. Annu. Rev. Immunol. 25, 619–647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Campbell I. D., and Humphries M. J. (2011) Integrin structure, activation, and interactions. Cold Spring Harb. Perspect. Biol. 3, a004994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim C., Ye F., and Ginsberg M. H. (2011) Regulation of integrin activation. Annu. Rev. Cell Dev. Biol. 27, 321–345 [DOI] [PubMed] [Google Scholar]
- 17.Choi W. S., Rice W. J., Stokes D. L., and Coller B. S. (2013) Three-dimensional reconstruction of intact human integrin αIIbβ3: new implications for activation-dependent ligand binding. Blood 122, 4165–4171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen X., Xie C., Nishida N., Li Z., Walz T., and Springer T. A. (2010) Requirement of open headpiece conformation for activation of leukocyte integrin αxβ2. Proc. Natl. Acad. Sci. U.S.A. 107, 14727–14732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nishida N., Xie C., Shimaoka M., Cheng Y., Walz T., and Springer T. A. (2006) Activation of leukocyte β2 integrins by conversion from bent to extended conformations. Immunity 25, 583–594 [DOI] [PubMed] [Google Scholar]
- 20.Adair B. D., Xiong J. P., Maddock C., Goodman S. L., Arnaout M. A., and Yeager M. (2005) Three-dimensional EM structure of the ectodomain of integrin αvβ3 in a complex with fibronectin. J. Cell Biol. 168, 1109–1118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xiong J. P., Stehle T., Zhang R., Joachimiak A., Frech M., Goodman S. L., and Arnaout M. A. (2002) Crystal structure of the extracellular segment of integrin αvβ3 in complex with an Arg-Gly-Asp ligand. Science 296, 151–155 [DOI] [PubMed] [Google Scholar]
- 22.Takagi J., Petre B. M., Walz T., and Springer T. A. (2002) Global conformational rearrangements in integrin extracellular domains in outside-in and inside-out signaling. Cell 110, 599–511 [DOI] [PubMed] [Google Scholar]
- 23.Xiong J. P., Stehle T., Goodman S. L., and Arnaout M. A. (2003) New insights into the structural basis of integrin activation. Blood 102, 1155–1159 [DOI] [PubMed] [Google Scholar]
- 24.Zhu J., Boylan B., Luo B. H., Newman P. J., and Springer T. A. (2007) Tests of the extension and deadbolt models of integrin activation. J. Biol. Chem. 282, 11914–11920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xiong J. P., Mahalingham B., Alonso J. L., Borrelli L. A., Rui X., Anand S., Hyman B. T., Rysiok T., Müller-Pompalla D., Goodman S. L., and Arnaout M. A. (2009) Crystal structure of the complete integrin αvβ3 ectodomain plus an α/β transmembrane fragment. J. Cell Biol. 186, 589–600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lefort C. T., Rossaint J., Moser M., Petrich B. G., Zarbock A., Monkley S. J., Critchley D. R., Ginsberg M. H., Fässler R., and Ley K. (2012) Distinct roles for talin-1 and kindlin-3 in LFA-1 extension and affinity regulation. Blood 119, 4275–4282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chigaev A., Buranda T., Dwyer D. C., Prossnitz E. R., and Sklar L. A. (2003) FRET detection of cellular α4-integrin conformational activation. Biophys. J. 85, 3951–3962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Coutinho A., García C., González-Rodríguez J., and Lillo M. P. (2007) Conformational changes in human integrin αIIbβ3 after platelet activation, monitored by FRET. Biophys. Chem. 130, 76–87 [DOI] [PubMed] [Google Scholar]
- 29.Kim M., Carman C. V., and Springer T. A. (2003) Bidirectional transmembrane signaling by cytoplasmic domain separation in integrins. Science 301, 1720–1725 [DOI] [PubMed] [Google Scholar]
- 30.Ye F., Hu G., Taylor D., Ratnikov B., Bobkov A. A., McLean M. A., Sligar S. G., Taylor K. A., and Ginsberg M. H. (2010) Recreation of the terminal events in physiological integrin activation. J. Cell Biol. 188, 157–173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gailit J., and Ruoslahti E. (1988) Regulation of the fibronectin receptor affinity by divalent cations. J. Biol. Chem. 263, 12927–12932 [PubMed] [Google Scholar]
- 32.Adair B. D., and Yeager M. (2002) Three-dimensional model of the human platelet integrin αIIbβ3 based on electron cryomicroscopy and x-ray crystallography. Proc. Natl. Acad. Sci. U.S.A. 99, 14059–14064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ye F., Liu J., Winkler H., and Taylor K. A. (2008) Integrin αIIbβ3 in a membrane environment remains the same height after Mn2+ activation when observed by cryoelectron tomography. J. Mol. Biol. 378, 976–986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nath A., Atkins W. M., and Sligar S. G. (2007) Applications of phospholipid bilayer nanodiscs in the study of membranes and membrane proteins. Biochemistry 46, 2059–2069 [DOI] [PubMed] [Google Scholar]
- 35.Denisov I. G., Grinkova Y. V., Lazarides A. A., and Sligar S. G. (2004) Directed self-assembly of monodisperse phospholipid bilayer Nanodiscs with controlled size. J. Am. Chem. Soc. 126, 3477–3487 [DOI] [PubMed] [Google Scholar]
- 36.Winkler H., Zhu P., Liu J., Ye F., Roux K. H., and Taylor K. A. (2009) Tomographic subvolume alignment and subvolume classification applied to myosin V and SIV envelope spikes. J. Struct. Biol. 165, 64–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dube P., Tavares P., Lurz R., and van Heel M. (1993) The portal protein of bacteriophage SPP1: a DNA pump with 13-fold symmetry. EMBO J. 12, 1303–1309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tang G., Peng L., Baldwin P. R., Mann D. S., Jiang W., Rees I., and Ludtke S. J. (2007) EMAN2: an extensible image processing suite for electron microscopy. J. Struct. Biol. 157, 38–46 [DOI] [PubMed] [Google Scholar]
- 39.Frank J., Radermacher M., Penczek P., Zhu J., Li Y., Ladjadj M., and Leith A. (1996) SPIDER and WEB: processing and visualization of images in 3D electron microscopy and related fields. J. Struct. Biol. 116, 190–199 [DOI] [PubMed] [Google Scholar]
- 40.Xie C., Zhu J., Chen X., Mi L., Nishida N., and Springer T. A. (2010) Structure of an integrin with an αI domain, complement receptor type 4. EMBO J. 29, 666–679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Eng E. T., Smagghe B. J., Walz T., and Springer T. A. (2011) Intact αIIbβ3 integrin is extended after activation as measured by solution x-ray scattering and electron microscopy. J. Biol. Chem. 286, 35218–35226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Taylor D. W., Kelly D. F., Cheng A., and Taylor K. A. (2007) On the freezing and identification of lipid monolayer 2-D arrays for cryoelectron microscopy. J. Struct. Biol. 160, 305–312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Taylor K. A., and Taylor D. W. (1999) Structural studies of cytoskeletal protein arrays formed on lipid monolayers. J. Struct. Biol. 128, 75–81 [DOI] [PubMed] [Google Scholar]
- 44.Nachman R. L., and Leung L. L. (1982) Complex formation of platelet membrane glycoproteins IIb and IIIa with fibrinogen. J. Clin. Invest. 69, 263–269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Parise L. V., and Phillips D. R. (1985) Reconstitution of the purified platelet fibrinogen receptor: fibrinogen binding properties of the glycoprotein IIb-IIIa complex. J. Biol. Chem. 260, 10698–10707 [PubMed] [Google Scholar]
- 46.Müller B., Zerwes H. G., Tangemann K., Peter J., and Engel J. (1993) Two-step binding mechanism of fibrinogen to αIIbβ3 integrin reconstituted into planar lipid bilayers. J. Biol. Chem. 268, 6800–6808 [PubMed] [Google Scholar]
- 47.Xiao T., Takagi J., Coller B. S., Wang J. H., and Springer T. A. (2004) Structural basis for allostery in integrins and binding to fibrinogen-mimetic therapeutics. Nature 432, 59–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mosesson M. W. (2005) Fibrinogen and fibrin structure and functions. J. Thromb. Haemost. 3, 1894–1904 [DOI] [PubMed] [Google Scholar]
- 49.Springer T. A., Zhu J., and Xiao T. (2008) Structural basis for distinctive recognition of fibrinogen γC peptide by the platelet integrin αIIbβ3. J. Cell Biol. 182, 791–800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kim C., Lau T. L., Ulmer T. S., and Ginsberg M. H. (2009) Interactions of platelet integrin αIIb and β3 transmembrane domains in mammalian cell membranes and their role in integrin activation. Blood 113, 4747–4753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lau T. L., Kim C., Ginsberg M. H., and Ulmer T. S. (2009) The structure of the integrin αIIbβ3 transmembrane complex explains integrin transmembrane signalling. EMBO J. 28, 1351–1361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shi M., Foo S. Y., Tan S. M., Mitchell E. P., Law S. K., and Lescar J. (2007) A structural hypothesis for the transition between bent and extended conformations of the leukocyte β2 integrins. J. Biol. Chem. 282, 30198–30206 [DOI] [PubMed] [Google Scholar]
- 53.Lau T. L., Dua V., and Ulmer T. S. (2008) Structure of the integrin αIIb transmembrane segment. J. Biol. Chem. 283, 16162–16168 [DOI] [PMC free article] [PubMed] [Google Scholar]


