Abstract
Hepatitis C virus (HCV) causes persistent infection in 75% of cases and is a major public health problem worldwide. More than 92% of intravenous drug users (IDU) infected by human immunodeficiency virus type 1 (HIV-1) are seropositive for HCV, and it is conceivable that some HIV-1-infected IDU who remain uninfected by HCV may be genetically resistant.Here we conducted a case-control study to identify mutations in HCV entry coreceptors in HIV-infected IDU who remained uninfected by HCV. We recruited 138 patients, comprising 22 HIV+ HCV- case IDU and 116 HIV+ HCV+ control IDU. We focused on coreceptors in which point mutations are known to abolish HCV infectivity in vitro. Our previous study of the Claudin-1 gene revealed no specific variants in the same case population. Here we performed direct genomic sequencing of the Claudin-6, Claudin-9, Occludin and Scavenger receptor-B1 (SCARB1) gene coding regions. Most HIV+ HCV- IDU had no mutations in HCV coreceptors. However, two HIV+ HCV- patients harbored a total of four specific mutations/variants of HCV entry factors that were not found in the HIV+ HCV+ controls. One case patient harbored heterozygous variants of both Claudin-6 and Occludin, and the other case patient harbored two heterozygous variants of SCARB1. This suggests that HCV resistance might involve complex genetic events and factors other than coreceptors, a situation similar to that reported for HIV-1 resistance.
Introduction
Hepatitis C virus (HCV) infects more than 170 million individuals worldwide and is a major public health problem. HCV infection becomes persistent in 75% of cases. Chronic carriers frequently develop fibrosis, cirrhosis and, in some cases, hepatocellular carcinoma, especially if untreated. Because human immunodeficiency virus (HIV) and HCV share common routes of infection, coinfection with HCV is frequent among HIV-1-infected patients: in Europe, about one-third of all HIV-1-infected individuals have anti-HCV antibodies [1, 2]. However, the seroprevalence of HCV among HIV-1-infected individuals in France ranges from 8% when HIV-1 is transmitted via heterosexual contact to 41.7% among hemophiliacs and/or transfusion recipients and more than 92% among intravenous drug users (IDU) [3]. Thus, individuals who acquired HIV-1 via intravenous drug use but remain uninfected by HCV are rare.
HCV entry into hepatocytes is complex and not fully understood [4, 5]. It is a multistep process involving several host factors and coreceptors, including heparan sulfate [6], the tetraspanin CD81 [7, 8], the scavenger receptor B1 (SCARB1) [9], the tight-junction proteins claudin-1 (CLDN1) [10] 6 and 9 (CLDN6 and CLDN9) [11] and occludin (OCLN) [12]. The EGF and EphrinA2 receptors, the Niemann-Pick C1-like 1 cholesterol absorption receptor [13, 14] and, more recently, transferrin receptor 1 (TfR1) and cell-death-inducing DFFA like effector B (CIDEB) have all been reported to play a role in HCV entry [5]. Infectious virions complete their binding, endocytosis and fusion processes through sequential interactions with SR-B1 and CD81 earlier in the entry pathway, whereas the two tight junction proteins (claudin-1 and occludin) play important roles during a post-binding step [15]. Claudin-1 has also been shown to play a key role in cell-to-cell virus transmission [16]: no evidence of claudin-1-independent HCV entry has so far been reported [10, 17].
The aim of this case-control study was to identify mutations in HCV entry coreceptors in long-term HIV-infected intravenous drug users who remained uninfected by HCV. As previously described [18], we recruited 138 patients having acquired HIV-1 infection before 1995 through intravenous drug use. Cases (n = 22) were HIV-1-infected IDU who remained negative for anti-HCV antibodies and HCV RNA despite at least five years of intravenous drug use. Controls (n = 116) were HIV-1-infected IDU coinfected with HCV (anti-HCV antibody-positive).
We focused on coreceptors in which point mutations are known to prevent HCV infection in vitro [19–22]. In a previous case-control study of the CLDN1 gene in the same HIV-infected IDU population, we identified no specific coding variants in the HCV-uninfected case population [18]. Here we focused on the CLDN6, CLDN9, OCLN and SCARB1 coding regions.
Materials and Methods
Study participants
We conducted a case-control study of HIV-infected individuals followed in four Parisian clinical centres [18]. The objective was to compare the sequences of HCV entry factor genes between HIV-1-infected, HCV-uninfected IDU and HIV/HCV-coinfected IDU. Cases were defined as: (i) having acquired HIV-1 infection before 1995 through intravenous drug use, (ii) having injected illicit drugs for at least 5 years, and (iii) being negative for anti-HCV antibodies and HCV-RNA. Controls were defined as: (i) having acquired HIV-1 infection before 1995 through intravenous drug use, (ii) having injected illicit drugs for at least 5 years, and (iii) being positive for anti-HCV antibodies and HCV-RNA before any anti-HCV treatment. The case-control ratio was 1:5.
Patients 18 years of age or older were eligible for participation in this study if they had acquired HIV-1 through intravenous drug use before 1995, i.e. before substitution programs with methadone and buprenorphine were launched in France. Data on the HIV-1 transmission group and date of HIV-1 diagnosis were collected from the databases of the four clinical centres. The clinical charts of potentially eligible patients were then reviewed by the same investigator to check for injection drug history (including the date of first i.v. drug use). The study was restricted to individuals who did not have HIV-1-seropositive sexual partners at the time of HIV diagnosis and who were not transfusion recipients.
The Scientific Review Board of Bicêtre Hospital approved the study and all the patients gave their written informed consent to participate.
Sequencing of the CLDN6, CLDN9, OCLN and SCARB1 genes
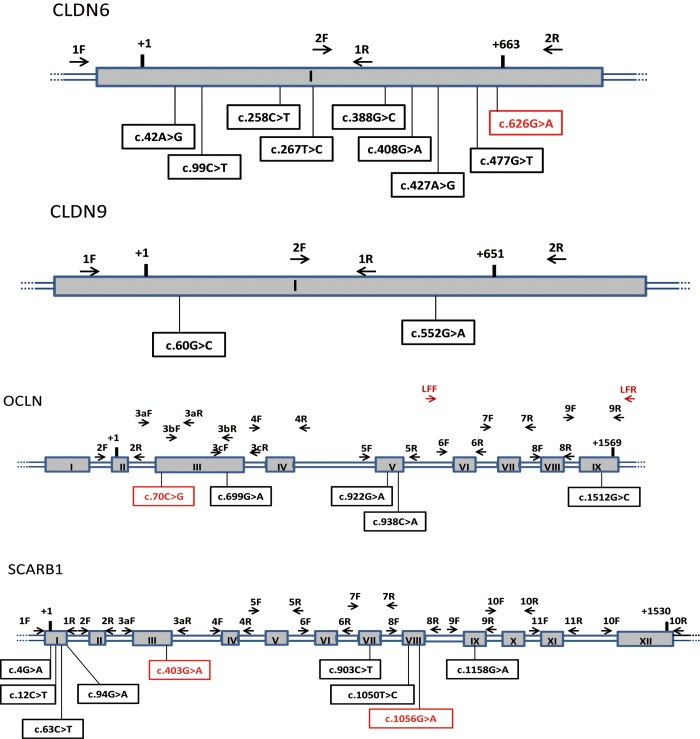
DNA was extracted from peripheral blood mononuclear cells with the Blood and Cell Culture DNA maxi kit (Qiagen). Forward (F) and reverse (R) primer pairs were designed to amplify all the exons and intron-exon junctions of each gene (Table 1, Table 2 and Fig 1). Each exon was amplified with the corresponding F/R primer pair in classical polymerase chain reaction (PCR) conditions: 5 min at 94°C followed by 30 cycles of 30 s at 95°C, the hybridization temperature being adjusted to each exon (Table 1 and Table 2), followed by a final elongation step of 8 min at 72°C.
Table 1. Primers used to sequence the CLDN6, CLDN9 and OCLN genes.
| Primer | exon | Sequence 5'-3' | Hybridation T°C | amplicon size bp |
|---|---|---|---|---|
| CLDN6 gene | ||||
| 1F | 1 | CTTGTTGTGCTTCTGTCCCA | 56 | 439 |
| 1R | 1 | AGGACCCCTGAGATGACAAA | 56 | |
| 2F | 1 | CTTGCTGGTCTACCTTGCTG | 56 | 467 |
| 2R | 1 | AAAAGGTACGAACCCATCCC | 56 | |
| CLDN9 gene | ||||
| 1F | 2 | CACACCAGACACACCCTCTG | 58 | 478 |
| 1R | 2 | AGCACACAGGGATGAGCAC | 58 | |
| 2F | 2 | GTGTACCACGTGTGTGGAGG | 58 | 440 |
| 2R | 2 | AGGAGGTTGTGATGGAGCAG | 58 | |
| OCLN gene | ||||
| 2F | 2 | GCAGTGAGCTGTGATTGGA | 59 | 340 |
| 2R | 2 | GCAAACACTTAAAGTTTCAACC | 59 | |
| 3Fa | 3 | CCAAATAAGTTGTGTTCTTTCTGC | 54 | 356 |
| 3Ra | 3 | CCAAAGCCACTTCCTCCATA | 54 | |
| 3Fb | 3 | TCTCCTCCAGGAGTGATTCG | 56 | 373 |
| 3Rb | 3 | ATGCCCAGGATAGCACTCAC | 56 | |
| 3Fc | 3 | CGCGTTGGTGATCTTTGTTA | 56 | 328 |
| 3Rc | 3 | TTGAAGGCCTCTGCTAAGGA | 56 | |
| 4F | 4 | CATTAGGCATTTTCTGAGGATTG | 60 | 586 |
| 4R | 4 | AACCATTTCCACTTAGCCCATC | 60 | |
| 5F | 5 | TGTGGGCGTGAGATAATGAGACCA | 56 | 595 |
| 5R | 5 | CCAGCTTTTGTGTGCACTGCTGG | 56 | |
| LRF | 6–9 | GGTTTGGTGAAGCATTTGCCTGTGAAG | 68 | 23153 |
| LRR | 6–9 | AACGACTAACCAGCACAGCATCCAAAG | 68 | |
| 6F | 6 | TGGTGTTTATTATGGCTGTGC | 62 | 372 |
| 6R | 6 | CAACACCTGGTTGGTCTCCT | 62 | |
| 7F | 7 | TCTCCATACCCAACCAGCTT | 62 | 337 |
| 7R | 7 | AGGATGCTGTACCTCCACA | 62 | |
| 8F | 8 | CCTTCAGACCTTCCTGCTGA | 62 | 311 |
| 8R | 8 | GAAAAGCTCTTCCTCCAGATG | 62 | |
| 9F | 9 | CAGGCACCTTGCGTATTTTAC | 62 | 352 |
| 9R | 9 | GCTCACAGAGGTTTGGCTTC | 62 |
Table 2. Primers used to sequence the SCARB1 gene.
| Primer | exon | Sequence 5'-3' | Hybridation T°C | amplicon size bp |
|---|---|---|---|---|
| SCARB1 gene | ||||
| 1F | 1 | ATGGCGGGGCTTGTCTTGGC | 64 | 617 |
| 1R | 1 | CCTGGCCTCCCTCGTGCTCT | 64 | |
| 2F | 2 | CCACCACCTCCTATCCCAAG | 64 | 366 |
| 2R | 2 | CCCCATCCCGTCCACTCTGA | 64 | |
| 3F | 3 | GTGTTGGGTGGGGGAGAGC | 64 | 527 |
| 3R | 3 | GACAGCACAGGGCCGAAAGC | 64 | |
| 4F | 4 | AGAGGGTGGTTCTGGTGTCC | 64 | 521 |
| 4R | 4 | AAGCCGGTTTGAGTCAGGTTC | 64 | |
| 5F | 5 | CTCAGCCCAGAATGTTCAGAC | 64 | 381 |
| 5R | 5 | CACTAACCCCACCTGCCCC | 64 | |
| 6F | 6 | AGCCTGCCCTCTTCCCAC | 60 | 374 |
| 6R | 6 | GCTACTGAGTCAAATCCACGA | 60 | |
| 7F | 7 | TGGGTGGGGAGGCAGAGTC | 60 | 460 |
| 7Rb | 7 | GCCAGAGATTAAGCAGACAGC | 60 | |
| 8F | 8 | TCCTGCCTCACCCCTTCTCT | 60 | 440 |
| 8R | 8 | CTTCCCACCACCCCAGCC | 60 | |
| 9F | 9 | GACGCCCACCCTCTTGACTG | 60 | 240 |
| 9R | 9 | GGACCACTGGAGCACTGAGC | 60 | |
| 10F | 10 | GGTGAGGGTTTAGTGTGTGC | 60 | 327 |
| 10R | 10 | AGGGTGAAGTTTCTGATACGC | 60 | |
| 11F | 11 | AGGCGGGCACAGAGGAAGG | 60 | 449 |
| 11R | 11 | CAGGCAGAGTAGTGGCAACG | 60 | |
| 12F | 12 | ATCGTTGAGGGTTGTTGGAC | 60 | 362 |
| 12R | 12 | GCTGAAGGAATGAGCAGGAC | 60 |
Fig 1. Schematic representation of of CLDN6, CLDN9, OCLN and SCARB1 genes.
Exons are represented by numbered gray rectangles and introns or non coding regions by double blue lines. Positions of primers pairs used for direct genomic sequencing are shown. Reference sequences of the transcripts are NM_021195.4 (CLDN6), NM_020982.3 (CLDN9), NM_002538.3 (OCLN) and NM_005505.4 (SCARB1). The numbering starts at the first base of the initiation codon ATG, and stops at the first base of the termination codon of the corresponding transcripts. The sequences of corresponding primers are shown in Table 1 and Table 2. F: Forward primer, R: Reverse primer, LR: Long Range PCR primer. SNPs identified by direct genomic sequencing are indicated, in red SNPs specific of the case population HIV+ HCV-.
To avoid amplification of the pseudogene for exons 6 to 9 of the OCLN gene, long-range (LR) PCR (Qiagen LongRange PCR Kit) was used as an intermediate step before PCR amplification of the exon. LR PCR amplification consisted of 10 min at 94°C followed by 30 cycles of 20 s at 94°C, 1 min at 68°C and 5 min at 72°C, and a final 10-min elongation step at 72°C. The amplicon was purified and used as a matrix to amplify OCLN exons 6 to 9, as described above.
Sequencing was performed on an ABI 3100 automated sequencer (Applied Biosystems, Foster City, CA, USA) according to the manufacturer’s instructions. Rare variants were confirmed by resequencing independent PCR products.
As a predicted splice site due to c.626G>A mutation in the CLDN6 gene (NM_021195.4) was detected, peripheral white blood cells were collected in a PAXgen Blood RNA collection tube (Qiagen) from the patient concerned, and RNA was extracted with a nucleic acid purification kit (PAXgen Blood RNA Kit, Qiagen). Then RT PCR was applied to the corresponding cDNAs using the RETROscript reverse transcription kit (Ambion, Lifes Technologies). PCR products were sequenced with primers corresponding to the C terminal part of CLDN6. All variants were verified by double-strand sequencing of two independent PCR products. The c.70C>G mutation in the OCLN gene (NM_002538.3) creates a new restriction site for BseX1. This mutation was confirmed by BseX1 (Fermentas, Thermo Scientific) restriction analysis of a PCR product.
Bioinformatics analyses
Polyphen and SIFT (Sort Intolerant From Tolerant) are sequence homology based tools that predicts whether an amino acid substitution in a protein will have a phenotypic effect. These prediction softwares are based on the principle that protein evolution is correlated with protein function. Positions important for function should be conserved in an alignment of the protein family, whereas unimportant positions should appear diverse in an alignment. This software will predict which mutants may have a phenotypic effect.
SIFT takes a query sequence or a SNP and uses multiple alignment information to predict tolerated (meaning no predicted effect on structure or protein function) and deleterious (that can render the resulting protein nonfunctional or defective) substitutions for every position of the query sequence or for the SNP. From a query sequence SIFT searches for similar sequences, chooses closely related sequences that may share similar function to the query sequence, obtains the alignment of these chosen sequences, and calculates normalized probabilities for all possible substitutions from the alignment. Positions with normalized probabilities less than 0.05 are predicted to be deleterious, those greater than or equal to 0.05 are predicted to be tolerated (http://sift.jcvi.org/).
Polyphen-2 (http://genetics.bwh.harvard.edu/pph2/) predicts the functional significance of an allele replacement from its individual features using HumDIv and HumVar datasets (compiled human disease-causing mutations from the UniProtKB database). For a mutation, PolyPhen-2 calculates probability that this mutation is damaging and reports estimates of false positive rate (the chance that the mutation is classified as damaging when it is in fact non-damaging) and true positive rate (the chance that the mutation is classified as damaging when it is indeed damaging), mutation is also appraised qualitatively, as benign (correspond to tolerated for SIFT), possibly damaging, or probably damaging (damaging corresponds to deleterious for SIFT). Higher the score is higher the mutation is predicted damaging. The prediction confidence depends on rates of false and true positive and allows to qualify the mutation of possibly, probably or benign.
Results
A total of 138 Caucasian patients were enrolled between February and December 2008, comprising 22 cases (20 men) and 116 controls (93 men). They were all followed in four Parisian clinical centers managing patients infected with HIV-1. Their median age was 46 years. The median year of first documented HIV-1 seropositivity was 1989 (range 1984–1994). They all reported using intravenous drug use for more than five years before 1995.
In the control patients (n = 116), the estimated median year of HCV acquisition (corresponding to the first year of intravenous drug use) was 1992 (range 1982–1994). The distribution of the main HCV genotypes was as follows: 60% genotype 1, 20% genotype 3, 10% genotype 4 and 7% genotype 2. CD4 T cell count did not differ between the case and the control populations at the time of first diagnosis of HIV infection.
In our previous study of the CLDN1 gene, we identified no specific coding variants in the case population [18]. Here, direct genomic sequencing of the whole coding region and adjacent intron-exon junctions of the CLDN6, CLDN9, OCLN and SCARB1 genes in the case and control populations enabled us to identify several new mutations and single nucleotide polymorphisms (SNPs).
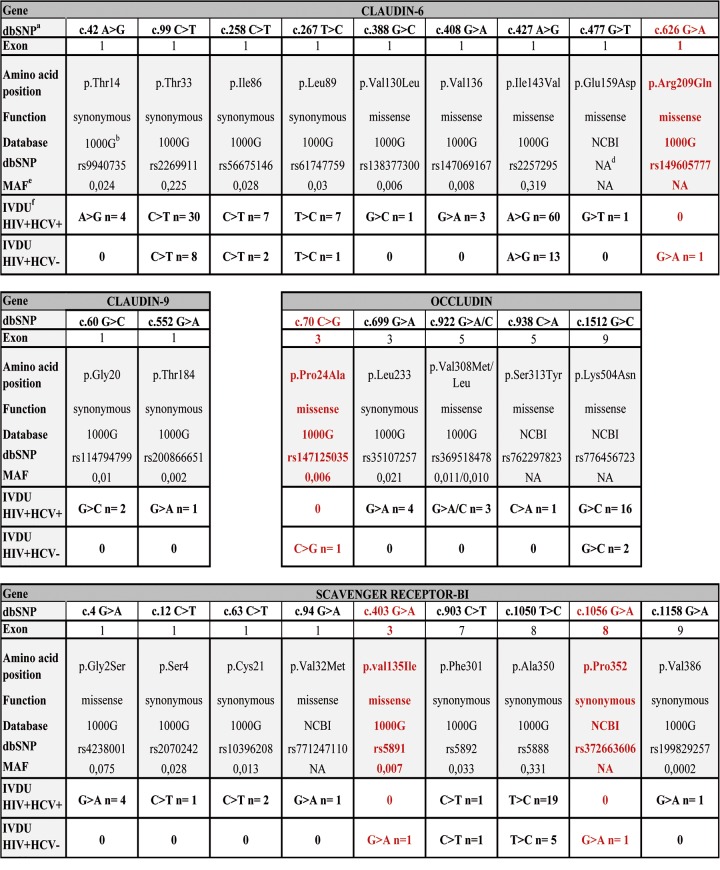
In the CLDN6 gene, we found SNPs in both populations, corresponding to frequent polymorphisms described in conventional databases (NCBI and 1000 Genomes databases) (Fig 2). However, one SNP, rs149605777 (Fig 2), corresponding to a c.626G>A heterozygous coding variant, was found only in the HIV+HCV- case population (G>A n = 1). This variant leads to a missense Arg209Gln (R209Q) change in the C-terminal cytosolic tail of CLDN6, with a non-attributed MAF (minor allele frequency) in the NCBI database. Prediction tools (PolyPhen 2 and SIFT) showed that this mutation is possibly damaging or tolerated (scores of 0.533 with PolyPhen and 0.43 with SIFT). Human Splicing Finder (HSF) software showed that this c.626G>A substitution potentially creates a splice acceptor site. However, sequencing of cDNAs derived from the patient’s blood cell mRNA showed no evidence of splicing in vivo (see Materials and Methods).
Fig 2. Variants of HCV entry factors found in HIV-infected, HCV-uninfected patients but not in HIV/HCV-coinfected controls.
a Database single nucleotide polymorphism. b 1000 Genomes database. c refSNP reference identification number of single nucleotide polymorphism. d Not attributed. e Minor allele frequency. f Intravenous drug users.
In the CLDN9 gene, we identified SNPs already described in databases (Fig 2). There was no statistically significant difference between the control and case populations.
In the OCLN gene, we identified several SNPs and also one rare heterozygous variant, exclusively in the case population (Fig 2). This rare rs147125035 variant was present in only one HIV+HCV- patient and corresponded to a missense c.70C>G substitution with a MAF frequency of 0.006. It leads to a Pro24Ala (P24A) change (Fig 2) in the N-terminus of OCLN. This variant is predicted to be potentially damaging or deleterious (scores of 1 with PolyPhen and 0.12 with SIFT). Interestingly, this rare variant was found in the case patient harboring the CLDN6 mutation described above. HSF software identified no splice site for this variant. Both mutated residues in CLDN6 and OCLN are highly conserved in different species.
We also identified two variants of the SCARB1 gene, both in another case patient (Fig 2): rs5891 (MAF: 0.007) and rs372663606 (no attributed MAF in NCBI), corresponding to heterozygous c.403G>A missense (leading to a p.Val135Ile change) and c.1056G>A synonymous (p.Pro352) substitutions, respectively. HSF software identified no splice site for this variant. The c.403G>A missense substitution is predicted to be benign/tolerated (scores of 0.322 with PolyPhen and 0.5 with SIFT).
Thus, we found that 2 of the 22 HIV+HCV- IDU case patients displayed specific mutations or variants in HCV cellular receptors. Interestingly, one patient harbored two variants of different cellular factors required for HCV entry. The affected residues are highly conserved in different species and are likely damaging. One mutation has no frequency described in databases, while the other variant is rare and was not found in the control population.
Discussion
The overall seroprevalence of HCV in HIV-infected patients is around 24% [23], but it can reach 92.8% in HIV-1-infected IDU (2). As observed in the Urban Health Study, patients who acquire HIV-1 through intravenous drug use but remain uninfected by HCV are rare [24]. The latter study involved 25 Caucasian HIV+HCV- IDU patients. Here, we selected individuals with prolonged exposure to HCV (at least five years of IDU) and in whom HIV-1 infection was diagnosed before 1995, in order to avoid false resistance to HCV infection. We therefore considered that the HCV-negative patients were very likely resistant, as most or all of them would have been exposed to HCV for several years.
To explore the possibility that variations in HCV cell entry factors might be involved in resistance to HCV infection, we sequenced the genes of four entry factors in which point mutations are known to abolish HCV entry in vitro [19–22]. We found variants in CLDN6, OCLN and SCARB1, solely in the cases (2/22, 18%). The corresponding mutations affected residues that are highly conserved in various species and are thus likely to be damaging. Two mutations had no reported database frequency, while the other two variants were rare and not found in the control population. Interestingly, the same case patient harbored one heterozygous mutation in CLDN6, together with one rare heterozygous mutation in OCLN, both of which were likely damaging and involved residues highly conserved in different species. The probability of finding both variants in the same case patient was extremely low. Another case patient harbored two mutations in SCARB1, but prediction tools suggested that they were unlikely to have functional consequences. Once again, the likelihood of the two variants occurring in the same patient was extremely low. The combinations of new mutations or rare variants specifically present in HIV-infected but HCV-uninfected patients but not in HIV and HCV-infected patients suggest that HCV resistance might be a multigenic phenomenon.
While many studies have explored the role of genetic variants of chemokine receptors in the susceptibility and progression of HIV disease, the full coding regions of several coreceptor genes have rarely been sequenced in the same sexually exposed but HIV-uninfected population [25–36]. In keeping with our results, a large proportion of individuals who remain HIV-uninfected despite repeated sexual exposure harbor no mutations in HIV entry coreceptors; only 10–18% of these individuals have heterozygous mutations and as few as 1–2.8% have homozygous mutations in CCR5 (mostly CCR5Δ32) [25, 26, 37–41]. This suggests that host resistance to HIV may involve other mechanisms besides coreceptor mutations, especially as heterozygous CCR5Δ32 mutations simply slow HIV disease progression, without preventing infection [25, 26, 42]. The full coding regions of several coreceptor genes have never previously been sequenced in an HIV-resistant IDU population [25, 26, 28, 29, 32, 40, 43, 44]. Only known specific variants of CCR5, CCR2, SD F1 RANTES have so far been studied. The only study of HCV-infected, HIV-uninfected patients (HCV infection being a marker of intravenous exposure to HIV) revealed a strong protective effect of a high copy number of the CCL3L1 gene, which encodes a CCR5 ligand [25, 45].
In conclusion, in the first study of its type, we detected mutations in HCV entry factors in two (18%) of 22 HIV-infected patients who were highly exposed to HCV but remained uninfected. However, most such patients had no mutations in coreceptors in which single mutations are known to abolish HCV entry in vitro, highlighting the complexity of the mechanisms governing HCV resistance and entry in vivo. The observed combination of a new mutation with specific variants in two HCV-exposed but uninfected patients suggests that resistance to HCV may be associated with other, additive genetic factors, as also suggested for resistance to HIV. Whole-genome studies are now needed to identify the different cellular factors involved in HCV resistance.
Acknowledgments
We thank Olivier Alibeu for technical assistance. This work was supported by the French National Agency for Research on AIDS and Viral Hepatitis (ANRS) (grants ANRS AO-1-2008 and ANRS AO-2-2013) and by grants from Roche laboratories (Dr. Alexandrina Pinta) and Janssen laboratories (Dr. Rima Lahoulou). The funders had no role in the study design, data collection and analysis, decision to publish or preparation of the manuscript.
Data Availability
All relevant data are within the paper.
Funding Statement
This work was supported by the French National Agency for Research on AIDS and Viral Hepatitis (ANRS) (grants ANRS AO-1-2008 and ANRS AO-2-2013) and by grants from Roche laboratories (Dr. Alexandrina Pinta) and Janssen laboratories (Dr. Rima Lahoulou). The funders had no role in the study design, data collection and analysis, decision to publish or preparation of the manuscript.
References
- 1. Andreoni M, Giacometti A, Maida I, Meraviglia P, Ripamonti D, Sarmati L. HIV-HCV co-infection: epidemiology, pathogenesis and therapeutic implications. European review for medical and pharmacological sciences. 2012;16(11):1473–83. . [PubMed] [Google Scholar]
- 2. Soriano V, Vispo E, Labarga P, Medrano J, Barreiro P. Viral hepatitis and HIV co-infection. Antiviral Res. 2010;85(1):303–15. 10.1016/j.antiviral.2009.10.021 . [DOI] [PubMed] [Google Scholar]
- 3. Larsen C, Pialoux G, Salmon D, Antona D, Le Strat Y, Piroth L, et al. Prevalence of hepatitis C and hepatitis B infection in the HIV-infected population of France, 2004. Euro Surveill. 2008;13(22). . [PubMed] [Google Scholar]
- 4. Bartosch B, Dubuisson J. Recent advances in hepatitis C virus cell entry. Viruses. 2010;2(3):692–709. 10.3390/v2030692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ding Q, von Schaewen M, Ploss A. The impact of hepatitis C virus entry on viral tropism. Cell host & microbe. 2014;16(5):562–8. 10.1016/j.chom.2014.10.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Barth H, Schafer C, Adah MI, Zhang F, Linhardt RJ, Toyoda H, et al. Cellular binding of hepatitis C virus envelope glycoprotein E2 requires cell surface heparan sulfate. The Journal of biological chemistry. 2003;278(42):41003–12. 10.1074/jbc.M302267200 . [DOI] [PubMed] [Google Scholar]
- 7. Pileri P, Uematsu Y, Campagnoli S, Galli G, Falugi F, Petracca R, et al. Binding of hepatitis C virus to CD81. Science. 1998;282(5390):938–41. . [DOI] [PubMed] [Google Scholar]
- 8. Zona L, Tawar RG, Zeisel MB, Xiao F, Schuster C, Lupberger J, et al. CD81-receptor associations—impact for hepatitis C virus entry and antiviral therapies. Viruses. 2014;6(2):875–92. 10.3390/v6020875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Scarselli E, Ansuini H, Cerino R, Roccasecca RM, Acali S, Filocamo G, et al. The human scavenger receptor class B type I is a novel candidate receptor for the hepatitis C virus. Embo J. 2002;21(19):5017–25. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Evans MJ, von Hahn T, Tscherne DM, Syder AJ, Panis M, Wolk B, et al. Claudin-1 is a hepatitis C virus co-receptor required for a late step in entry. Nature. 2007;446(7137):801–5. . [DOI] [PubMed] [Google Scholar]
- 11. Zheng A, Yuan F, Li Y, Zhu F, Hou P, Li J, et al. Claudin-6 and claudin-9 function as additional coreceptors for hepatitis C virus. J Virol. 2007;81(22):12465–71. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ploss A, Evans MJ, Gaysinskaya VA, Panis M, You H, de Jong YP, et al. Human occludin is a hepatitis C virus entry factor required for infection of mouse cells. Nature. 2009;457(7231):882–6. 10.1038/nature07684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Sainz B Jr., Barretto N, Martin DN, Hiraga N, Imamura M, Hussain S, et al. Identification of the Niemann-Pick C1-like 1 cholesterol absorption receptor as a new hepatitis C virus entry factor. Nat Med. 2012;18(2):281–5. 10.1038/nm.2581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lupberger J, Zeisel MB, Xiao F, Thumann C, Fofana I, Zona L, et al. EGFR and EphA2 are host factors for hepatitis C virus entry and possible targets for antiviral therapy. Nat Med. 2011;17(5):589–95. 10.1038/nm.2341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Zeisel MB, Fofana I, Fafi-Kremer S, Baumert TF. Hepatitis C virus entry into hepatocytes: molecular mechanisms and targets for antiviral therapies. Journal of hepatology. 2011;54(3):566–76. 10.1016/j.jhep.2010.10.014 . [DOI] [PubMed] [Google Scholar]
- 16. Timpe JM, Stamataki Z, Jennings A, Hu K, Farquhar MJ, Harris HJ, et al. Hepatitis C virus cell-cell transmission in hepatoma cells in the presence of neutralizing antibodies. Hepatology. 2008;47(1):17–24. 10.1002/hep.21959 . [DOI] [PubMed] [Google Scholar]
- 17. Haid S, Windisch MP, Bartenschlager R, Pietschmann T. Mouse-specific residues of claudin-1 limit hepatitis C virus genotype 2a infection in a human hepatocyte cell line. J Virol. 2010;84(2):964–75. 10.1128/JVI.01504-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ghosn J, Fouquet B, Quertainmont Y, Salmon D, Sahali S, Rioux C, et al. Claudin-1 gene variants and susceptibility to hepatitis C infection in HIV-1 infected intravenous drug users (an ANRS case-control study). Journal of medical virology. 2015;87(4):619–24. 10.1002/jmv.24088 . [DOI] [PubMed] [Google Scholar]
- 19. Samreen B, Khaliq S, Ashfaq UA, Khan M, Afzal N, Shahzad MA, et al. Hepatitis C virus entry: role of host and viral factors. Infection, genetics and evolution: journal of molecular epidemiology and evolutionary genetics in infectious diseases. 2012;12(8):1699–709. 10.1016/j.meegid.2012.07.010 . [DOI] [PubMed] [Google Scholar]
- 20. Meredith LW, Wilson GK, Fletcher NF, McKeating JA. Hepatitis C virus entry: beyond receptors. Reviews in medical virology. 2012;22(3):182–93. 10.1002/rmv.723 . [DOI] [PubMed] [Google Scholar]
- 21. Belouzard S, Cocquerel L, Dubuisson J. Hepatitis C virus entry into the hepatocyte. Cent Eur J Biol. 2011;6(6):933–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Flint M, von Hahn T, Zhang J, Farquhar M, Jones CT, Balfe P, et al. Diverse CD81 proteins support hepatitis C virus infection. J Virol. 2006;80(22):11331–42. 10.1128/JVI.00104-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Loko MA, Salmon D, Carrieri P, Winnock M, Mora M, Merchadou L, et al. The French national prospective cohort of patients co-infected with HIV and HCV (ANRS CO13 HEPAVIH): early findings, 2006–2010. BMC infectious diseases. 2010;10:303 10.1186/1471-2334-10-303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Bekker V, Chanock SJ, Yeager M, Hutchinson AA, von Hahn T, Chen S, et al. Genetic variation in CLDN1 and susceptibility to hepatitis C virus infection. J Viral Hepat. 2010;17(3):192–200. 10.1111/j.1365-2893.2009.01166.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Chatterjee A, Rathore A, Vidyant S, Kakkar K, Dhole TN. Chemokines and chemokine receptors in susceptibility to HIV-1 infection and progression to AIDS. Disease markers. 2012;32(3):143–51. 10.3233/DMA-2011-0874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Restrepo C, Rallon NI, Carrillo J, Soriano V, Blanco J, Benito JM. Host factors involved in low susceptibility to HIV infection. AIDS reviews. 2011;13(1):30–40. . [PubMed] [Google Scholar]
- 27. McLaren PJ, Coulonges C, Ripke S, van den Berg L, Buchbinder S, Carrington M, et al. Association study of common genetic variants and HIV-1 acquisition in 6,300 infected cases and 7,200 controls. PLoS pathogens. 2013;9(7):e1003515 10.1371/journal.ppat.1003515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Capoulade-Metay C, Ma L, Truong LX, Dudoit Y, Versmisse P, Nguyen NV, et al. New CCR5 variants associated with reduced HIV coreceptor function in southeast Asia. Aids. 2004;18(17):2243–52. . [DOI] [PubMed] [Google Scholar]
- 29. Teixeira SL, Bastos FI, Hacker MA, Morgado MG. Distribution of CCR5 genotypes and HLA Class I B alleles in HIV-1 infected and uninfected injecting drug users from Rio de Janeiro, Brazil. Infection, genetics and evolution: journal of molecular epidemiology and evolutionary genetics in infectious diseases. 2009;9(4):638–42. 10.1016/j.meegid.2009.03.007 . [DOI] [PubMed] [Google Scholar]
- 30. Nkenfou CN, Mekue LC, Nana CT, Kuiate JR. Distribution of CCR5-Delta32, CCR5 promoter 59029 A/G, CCR2-64I and SDF1-3'A genetic polymorphisms in HIV-1 infected and uninfected patients in the west region of Cameroon. BMC research notes. 2013;6:288 10.1186/1756-0500-6-288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Sarkar K, Das SS, Pal R, Bal B, Madhusudan P, Chakraborti S. HIV infection and host genetic mutation among injecting drug-users of northeastern states of India. Journal of health, population, and nutrition. 2010;28(2):130–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Wang FS, Hong WG, Cao Y, Liu MX, Jin L, Hu LP, et al. Population survey of CCR5 delta32, CCR5 m303, CCR2b 64I, and SDF1 3'A allele frequencies in indigenous Chinese healthy individuals, and in HIV-1-infected and HIV-1-uninfected individuals in HIV-1 risk groups. Journal of acquired immune deficiency syndromes. 2003;32(2):124–30. . [DOI] [PubMed] [Google Scholar]
- 33. Salem AH, Farid E, Fadel R, Abu-Hijleh M, Almawi W, Han K, et al. Distribution of four HIV type 1-resistance polymorphisms (CCR5-Delta32, CCR5-m303, CCR2-64I, and SDF1-3'A) in the Bahraini population. AIDS research and human retroviruses. 2009;25(10):973–7. 10.1089/aid.2009.0066 . [DOI] [PubMed] [Google Scholar]
- 34. Su B, Jin L, Hu F, Xiao J, Luo J, Lu D, et al. Distribution of two HIV-1-resistant polymorphisms (SDF1-3'A and CCR2-64I) in East Asian and world populations and its implication in AIDS epidemiology. American journal of human genetics. 1999;65(4):1047–53. 10.1086/302568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Zheng BJ, Zhao XY, Zhu NS, Chan CP, Wong KH, Chan KC, et al. Polymorphisms of CCR5 gene in a southern Chinese population and their effects on disease progression in HIV infections. Aids. 2002;16(18):2480–2. 10.1097/01.aids.0000042583.93174.96 . [DOI] [PubMed] [Google Scholar]
- 36. Martinson JJ, Hong L, Karanicolas R, Moore JP, Kostrikis LG. Global distribution of the CCR2-64I/CCR5-59653T HIV-1 disease-protective haplotype. Aids. 2000;14(5):483–9. . [DOI] [PubMed] [Google Scholar]
- 37. Dean M, Carrington M, Winkler C, Huttley GA, Smith MW, Allikmets R, et al. Genetic restriction of HIV-1 infection and progression to AIDS by a deletion allele of the CKR5 structural gene. Hemophilia Growth and Development Study, Multicenter AIDS Cohort Study, Multicenter Hemophilia Cohort Study, San Francisco City Cohort, ALIVE Study. Science. 1996;273(5283):1856–62. . [DOI] [PubMed] [Google Scholar]
- 38. Samson M, Libert F, Doranz BJ, Rucker J, Liesnard C, Farber CM, et al. Resistance to HIV-1 infection in caucasian individuals bearing mutant alleles of the CCR-5 chemokine receptor gene. Nature. 1996;382(6593):722–5. 10.1038/382722a0 . [DOI] [PubMed] [Google Scholar]
- 39. Zwolinska K, Knysz B, Rybka K, Pazgan-Simon M, Gasiorowski J, Sobczynski M, et al. Protective effect of CCR5-Delta32 against HIV infection by the heterosexual mode of transmission in a Polish population. AIDS research and human retroviruses. 2013;29(1):54–60. 10.1089/AID.2011.0362 . [DOI] [PubMed] [Google Scholar]
- 40. Marmor M, Sheppard HW, Donnell D, Bozeman S, Celum C, Buchbinder S, et al. Homozygous and heterozygous CCR5-Delta32 genotypes are associated with resistance to HIV infection. Journal of acquired immune deficiency syndromes. 2001;27(5):472–81. . [DOI] [PubMed] [Google Scholar]
- 41. Adojaan M, Molder T, Mannik A, Kivisild T, Villems R, Krispin T, et al. High prevalence of the CCR5Delta32 HIV-resistance mutation among Estonian HIV type 1-infected individuals. AIDS research and human retroviruses. 2007;23(2):193–7. 10.1089/aid.2006.0113 . [DOI] [PubMed] [Google Scholar]
- 42. Smith MW, Dean M, Carrington M, Winkler C, Huttley GA, Lomb DA, et al. Contrasting genetic influence of CCR2 and CCR5 variants on HIV-1 infection and disease progression. Hemophilia Growth and Development Study (HGDS), Multicenter AIDS Cohort Study (MACS), Multicenter Hemophilia Cohort Study (MHCS), San Francisco City Cohort (SFCC), ALIVE Study. Science. 1997;277(5328):959–65. . [DOI] [PubMed] [Google Scholar]
- 43. Alvarez V, Lopez-Larrea C, Coto E. Mutational analysis of the CCR5 and CXCR4 genes (HIV-1 co-receptors) in resistance to HIV-1 infection and AIDS development among intravenous drug users. Human genetics. 1998;102(4):483–6. . [DOI] [PubMed] [Google Scholar]
- 44. Li H, Liu TJ, Hong ZH. Gene polymorphisms in CCR5, CCR2, SDF1 and RANTES among Chinese Han population with HIV-1 infection. Infection, genetics and evolution: journal of molecular epidemiology and evolutionary genetics in infectious diseases. 2014;24:99–104. 10.1016/j.meegid.2014.03.009 . [DOI] [PubMed] [Google Scholar]
- 45. Huik K, Sadam M, Karki T, Avi R, Krispin T, Paap P, et al. CCL3L1 copy number is a strong genetic determinant of HIV seropositivity in Caucasian intravenous drug users. The Journal of infectious diseases. 2010;201(5):730–9. 10.1086/650491 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All relevant data are within the paper.