Abstract
Tibetan pigs live between 2500 and 4300 m above sea level on the Tibetan Plateau, and are better adapted to hypoxia than lowland pigs. MicroRNAs (miRNAs) are involved in a wide variety of cellular processes; however, their regulatory role in hypoxia adaptation remains unclear. In this study, miRNA-seq was used to identify differentially expressed miRNAs (DE miRNAs) in the cardiac muscle of Tibetan and Yorkshire pigs, which were both raised in high elevation environments. We obtained 108 M clean reads and 372 unique miRNAs, which included 210 known porcine miRNAs, 136 conserved in other mammals, and 26 novel pre-miRNAs. In addition, 20 DE miRNAs, including 10 up-regulated and 10 down-regulated miRNAs, were also found after comparison between Tibetan and Yorkshire pigs. We predicted miRNA targets based on differential expression and abundance in the two populations. Furthermore, the results of a Kyoto Encyclopedia of Genes and Genomes pathway analysis suggested that DE miRNAs in Tibetan and Yorkshire pigs are involved in hypoxia-related signaling pathways such as the mitogen-activated protein kinase, which is the mechanistic target of rapamycin, and the vascular endothelial growth factor, as well as cancer-related signaling pathways. Five DE miRNAs were randomly selected to validate the results of miRNA-seq using real-time polymerase chain reaction, and the results corresponded to those from the miRNA-seq, confirming that deep-sequencing methods are feasible and efficient. In our study, we identified various previously unknown hypoxia-related miRNAs in pigs, and the data obtained suggest that hypoxia-related miRNA expression patterns are significantly altered in the Tibetan pig compared to other species. Therefore, DE miRNAs may play an important role in organisms that have adapted to hypoxic environments.
Introduction
MicroRNAs (miRNAs) are small non-coding RNA molecules found in plants, animals, and viruses, and are widely believed to repress gene expression by binding to specific mRNA sequences [1–3]. Furthermore, miRNAs can be transferred to the nucleus [4] and guide the remodeling of chromatin and silencing of gene transcription [5], resulting in de novo DNA methylation [6]. Since miRNAs play different roles in multiple aspects of cellular function, it is not surprising that they are involved in hypoxia-related gene regulation. Kulshreshtha et al. [7] reported that experiencing hypoxia changes miRNA profiles in various cell types, and is affects the hypoxia-inducible factor (HIF) signal pathway. Moreover, several hypoxia-regulated miRNAs play roles in cell survival in hypoxic environments and have been implicated in the regulation of both upstream and downstream HIF signaling pathways, e.g., miR-20b and miR-17-92 clusters, while miR-199a regulates HIF-1α under hypoxic conditions [8–10], and miR-107, miR-210, miR-373, miR-23, miR-24, and miR-26 are induced by HIFs [7, 11, 12].
The Tibetan pig inhabits high-altitude regions (2500–4300 m) on the Qinghai-Tibet Plateau in southwestern China, and is well adapted to extreme elevations [13–15]. It is an ideal animal model for investigating the molecular mechanisms of hypoxia adaptation. Hypoxia could causes myocardial hypolasia, cardiomyopathy and reduced heart rate [16, 17]; therefore, identifying the regulatory mechanism of miRNAs in Tibetan pig cardiac muscle would elucidate the animals’ responses and molecular adaptation to hypoxic conditions, as well as enable us to determine not only the genes involved but also understand the regulation of specific hypoxia-related miRNAs. In this study, we conducted a comprehensive miRNA expression profile using miRNA-seq in the cardiac muscle of Tibetan and Yorkshire pigs raised in upland environments, established an overview of differential miRNA expression, and identified key miRNAs involved in hypoxia adaptation.
Materials and Methods
Sample preparation and RNA isolation
Sixteen 6-month-old castrated boars from populations of Tibetan pigs (TPs, n = 8) and Yorkshire pigs (YPs, n = 8) that were raised at the experimental farm of the Tibet Agriculture and Animal Husbandry College (Linzhi, 3000 m above sea level) were slaughtered and sampled. Cardiac tissue samples were immediately frozen in liquid nitrogen and stored at -80°C until RNA extraction. Total RNA for miRNA sequencing was extracted using TRIzol reagent (Invitrogen, San Diego, CA, USA) according to the manufacturer’s protocol. Extract quality was checked using a NanoDrop™ Biophotometer 2000 (Thermo Fisher Scientific Inc., West Palm Beach, FL, USA); a 260/280 nm absorbance ratio of 1.8–2.0 indicated a pure RNA sample. Equal quantities of the RNA extracted from the cardiac tissue of four pigs in each population were pooled into one. Thus, we had two duplicate samples in Tibetan pigs, two duplicate samples in Yorkshire pigs. Specific details are provided in S1 Table. The experiments were approved by the animal welfare committee of the State Key Laboratory for Agro-Biotechnology of the China Agricultural University (Approval number XK257). Pig farming at Linzhi was permitted, and the field study did not involve endangered or protected species.
Library construction and sequencing
Three micrograms of total RNA per sample was used for the construction of a small RNA library. Sequencing libraries were generated using NEBNext® Multiplex Small RNA Library Prep Set for Illumina® (NEB Ltd., UK) following the manufacturer’s recommendations, and index codes were added to attribute sequences to each sample. Polymerase chain reaction (PCR) products were purified on an 8% polyacrylamide gel (100 V, 80 min). DNA fragments corresponding to 140–160 bp (the length of small noncoding RNA plus 3′ and 5′ adaptors) were recovered and dissolved in 8 μL of elution buffer. The quality of the library was assessed using an Agilent Bioanalyzer 2100 system with DNA high-sensitivity chips. Clustering of the index-coded samples was performed on a cBot Cluster Generation System using a TruSeq SR Cluster Kit v3-cBot-HS (Illumina Inc.), according to the manufacturer’s protocol. After cluster generation, the library preparations were sequenced on a HiSeq™ 2000 platform and 50-bp paired-end reads were generated. The microRNA sequencing profile data were deposited in the Gene Expression Omnibus with the accession number GSE71550.
Data analysis
Clean reads were obtained from raw data after strictly eliminating low-quality reads, trimming adaptor sequences, and mapping (allowing two end-nucleotide mismatches) to reference pig genomic sequences (S scrofa10.2.72). The matched reads ranging from 15 to 35 bp were used as a query against non-coding RNA data (rRNA, snoRNA, tRNA, etc.). After excluding reads matching non-miRNA databases, the remaining sequences were regarded as potential miRNA reads. Precursors of candidate miRNAs that had been assembled from the short-read library were then searched against the pig genome to determine their location on the chromosome. Known pig miRNAs were identified by mapping the matched mature miRNAs in miRBase20.0 to pig genome sequences. All of the unannotated reads that matched the pig genomic sequences were analyzed using miRDeep2 to predict novel miRNAs. In addition, all candidate novel miRNAs were classified as conserved or pig-specific, according to their sequence conservation among the candidate species.
The microRNA read counts were normalized by the trimmed mean of TPM (transcript per million) normalization method in the edgeR package [18, 19]. P-values to compare miRNA expression levels were calculated using DEGseq software. Fold change = log2 (normalized read counts in TP/normalized read counts in YP).
Bioinformatic analyses
For the primary analysis, the retained reads (clean reads) were mapped to the pig genome using the custom mapping tools including the Bowtie 0.12.5 package [20] and miRDeep2 (version mirdeep2_0_0_2) [21]. The pig genome sequence was obtained from ftp://ftp.ensembl.org/pub/release-72/fasta/sus_scrofa/dna/Sus_scrofa.Sscrofa10.2.72.dna.toplevel.fa.gz. Clean reads were initially mapped to miRBase20.0 (www.mirbase.org/) [22] to identify known miRNAs and miRNA hairpins previously characterized [23–25]. Unmappable reads were annotated and classified by referencing noncoding RNAs in the NONCODE (version3.0) [26] and Rfam 11.0 databases. Since there is no complete pig miRNA dataset in miRBase, we obtained known mature miRNA for pigs, humans, mice, cows, and sheep (326, 257, 1908, 783, and 153, respectively) from miRBase 20.0 for use in miRDeep (mirdeep2_0_0_2) [21] to predict novel pig miRNA. Venn diagrams were prepared using the Venn diagram function in R, based on lists of novel and known miRNAs identified in each group. DEGseq was used to identify DE miRNAs in TPs and YPs. A volcano plot was generated based on log2 normalized read counts and -log10 (P-value). The August 2010 release of the microRNA target detection software miRanda [27] was used for target gene prediction. Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway [28] annotations of the miRNA targets were found using the DAVID (Database for Annotation, Visualization, and Integrated Discovery) gene annotation tool[29].
Stem-loop reverse-transcription quantitative PCR (RT-qPCR) to validate miRNA expression
A stem-loop RT-qPCR assay was conducted to measure the quantity of specific mature miRNA expression and validate the DE miRNAs [30]. Briefly, a miRcute miRNA First-Strand cDNA Synthesis Kit (KR201, Tiangen Biotech Co. Ltd., Beijing, China) was used for reverse transcription and real-time PCR according to the manufacturer’s instructions. Pig 5S and U6 snRNA was used as an internal control. All of the reactions were run in triplicate, and a mixture of every cDNA sample was used for calibration. Primer sequences for miRNA amplification are listed in S2 Table. A BioRad CFX96 (Bio-Rad, CA, USA) was used to perform the RT-qPCR with a SYBR® Green PCR Master Mix (FP401, Tiangen Biotech Co. Ltd.). Eight biological samples consisted of 8 single individuals for each group in the measurement. Relative miRNA expression levels were calculated using the 2-ΔΔCT method [31].
Results
Overview of miRNA transcriptome profiles in pig cardiac muscle
We obtained 21.2 M to 32.0 M clean reads from each of the four small RNA libraries (S3 Table). The number of reads with 20–24 nt was greater than that of those with shorter or longer sequences (S1 Fig). Approximately half of the sequences were 22 nt in length, which coincided with the known specifity of Dicer processing and characteristics of mature miRNAs [32, 33]. Approximately 87% of the clean reads could be mapped to the pig genome (Sscrofa10.2.72, S3 Table).
Identification of pig miRNAs
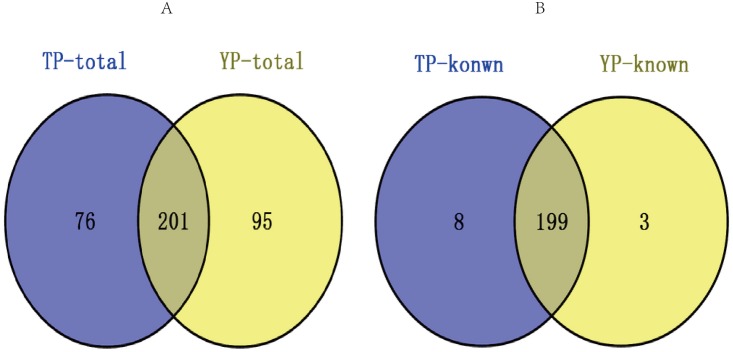
The annotated sequences were analyzed according to the data from miRBase20.0 (containing 326 mature miRNAs and 280 hairpin precursors). According to the results, 192–202 mature miRNAs and 230–234 hairpin precursors were identified (S3 Table). The remaining annotated sequences were compared using the Rfam and ncRNA databases after removal of the cellular structural RNAs, such as rRNAs, snoRNAs, snRNAs, scRNAs, and tRNAs. This revealed that nearly all remaining sequences were known miRNAs (S2 Fig). After filtering these data, 372 unique miRNAs, comprising 210 known porcine miRNAs (S4 Table) and 162 predicted candidate pre-miRNAs (S5 Table), were identified. According to this program for miRDeep2 software we identified the miRNAs that satisfy the requirements to be classified as novel, and the secondary structure of potential precursor and the mature miRNA, star sequences and loops were reliable. The TP and YP both expressed 201 miRNAs and in which 199 were known porcine miRNAs (Fig 1). In the 162 candidate miRNAs, 136 were conserved in other mammals (human, mouse, cow, and sheep) while the other 26 were considered as novel pre-miRNAs (S5 Table). The chromosomal locations of known and novel pre-miRNAs were determined using the pig reference genomic sequence. All miRNAs were aligned against autosomes or the X chromosome (S4 and S5 Tables).
Fig 1. Venn diagrams demonstrating relationships among miRNA in Tibetan and Yorkshire pigs.
(A) Venn diagram for total miRNAs (contained novel miRNAs and known miRNAs). (B) Venn diagram for known porcine miRNAs. TP and YP miRNAs marked in blue and yellow cycle, respectively. Tibetan pigs (TP) and Yorkshire pigs (YP).
DE miRNAs in TPs and YPs
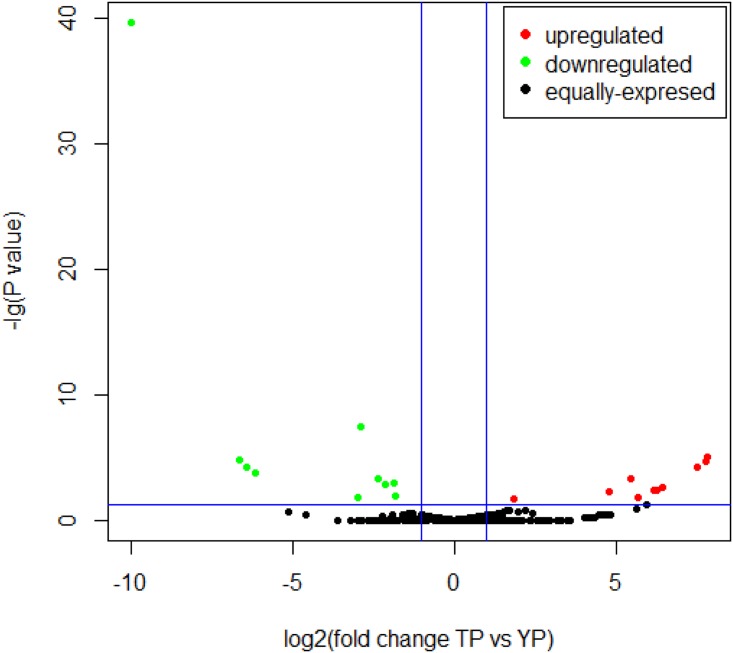
Based on the criteria of a fold change either ≥2 or ≤0.5 and P ≤ 0.05, 20 DE miRNAs were identified when comparing 372 miRNAs between TP and YP (Fig 2 and S6 Table). Of these, 10 miRNAs (ssc-miR-210, ssc-miR-1343, 12_3058, ssc-miR-676-5p, GL894044.2_23796, 1_4279, 13_5125, ssc-miR-194b-5p, ssc-miR-142-5p, and ssc-miR-421-5p) were up-regulated and 10 (ssc-miR-101, 1_1126, 4_13655, GL892805.1_27591, ssc-miR-320, ssc-miR-7136-5p, ssc-miR-214, ssc-miR-10b, 7_17790, and ssc-miR-206) were downregulated in the TP relative to the YP.
Fig 2. Volcano plot displaying differentially expressed miRNAs identified using miRNA-seq in Tibetan and Yorkshire pigs.
The y-axis represents the mean expression value of log10 (P-value) and the x-axis displays the log2-fold change value. Up-regulated and downregulated miRNAs are shown in red and green, respectively. Black dots indicate genes with no significant change in expression.
Functional analysis of DE miRNAs
Using miRanda software and the Ensemble database, 2389 and 2295 target genes were predicted from the 10 up-regulated and 10 downregulated miRNAs, respectively (S7A and S7B Table). The predicted target genes were classified in order to identify pathways that were actively regulated by miRNAs, according to the KEGG functional annotations made using DAVID (S8 Table, Tables 1 and 2). Thirteen putative target genes of up-regulated miRNAs enriched the mammalian target of the rapamycin (mTOR) signaling pathway (S3 Fig), which can be induced to enhance angiogenesis in response to hypoxia [34]. The mitogen-activated protein kinase (MAPK) signaling pathway was enriched according to 47 putative target genes of the up-regulated miRNAs and 53 putative target genes of the downregulated miRNAs (S4 Fig). The MAPK signaling pathway participates in the activation of HIFs and is involved in various cellular functions, including cell proliferation, differentiation, and migration [35].
Table 1. The Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways enriched for targets of the 10 up-regulated miRNAs in Tibetan pigs.
| Signaling pathway term | Count | P-value | Benjamini |
|---|---|---|---|
| Phosphatidylinositol signaling system | 22 | 7.16E-04 | 0.128 |
| Endocytosis | 37 | 1.66E-02 | 0.799 |
| Chondroitin sulfate biosynthesis | 8 | 2.40E-02 | 0.787 |
| Inositol phosphate metabolism | 14 | 2.87E-02 | 0.751 |
| O-Glycan biosynthesis | 9 | 4.54E-02 | 0.831 |
| mTOR signaling pathway | 13 | 4.67E-02 | 0.782 |
| Ubiquitin mediated proteolysis | 27 | 5.27E-02 | 0.772 |
| MAPK signaling pathway | 47 | 5.95E-02 | 0.769 |
| Pancreatic cancer | 16 | 6.32E-02 | 0.750 |
| RNA polymerase | 8 | 8.04E-02 | 0.798 |
| Apoptosis | 18 | 8.31E-02 | 0.778 |
| B cell receptor signaling pathway | 16 | 8.47E-02 | 0.756 |
| Cell cycle | 24 | 8.67E-02 | 0.736 |
| N-Glycan biosynthesis | 11 | 9.24E-02 | 0.734 |
| Renal cell carcinoma | 15 | 9.36E-02 | 0.714 |
Table 2. Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways enriched for targets of the 10 downregulated miRNAs in Tibetan pigs.
| Signaling pathway term | Count | P-value | Benjamini |
|---|---|---|---|
| Long-term depression | 23 | 6.11E-05 | 0.011 |
| Endometrial cancer | 16 | 2.76E-03 | 0.229 |
| Pathways in cancer | 63 | 2.86E-03 | 0.164 |
| MAPK signaling pathway | 53 | 3.27E-03 | 0.143 |
| Vascular smooth muscle contraction | 27 | 3.32E-03 | 0.117 |
| Focal adhesion | 42 | 3.71E-03 | 0.110 |
| Non-small cell lung cancer | 16 | 4.12E-03 | 0.105 |
| ErbB signaling pathway | 22 | 4.91E-03 | 0.109 |
| Phosphatidylinositol signaling system | 19 | 8.15E-03 | 0.157 |
| VEGF signaling pathway | 19 | 9.42E-03 | 0.163 |
| Chronic myeloid leukemia | 19 | 9.42E-03 | 0.163 |
| B cell receptor signaling pathway | 19 | 9.42E-03 | 0.163 |
| GnRH signaling pathway | 23 | 9.96E-03 | 0.157 |
| Renal cell carcinoma | 18 | 1.02E-02 | 0.148 |
| Inositol phosphate metabolism | 15 | 1.05E-02 | 0.142 |
| Melanoma | 18 | 1.17E-02 | 0.146 |
| Insulin signaling pathway | 29 | 1.21E-02 | 0.141 |
| Pancreatic cancer | 18 | 1.35E-02 | 0.147 |
| Toll-like receptor signaling pathway | 23 | 1.42E-02 | 0.146 |
| Protein export | 5 | 1.47E-02 | 0.144 |
| Glioma | 16 | 1.82E-02 | 0.166 |
| Endocytosis | 36 | 2.04E-02 | 0.176 |
| Apoptosis | 20 | 2.11E-02 | 0.174 |
| Melanogenesis | 22 | 2.17E-02 | 0.171 |
| Chemokine signaling pathway | 36 | 2.56E-02 | 0.191 |
| Gap junction | 20 | 2.64E-02 | 0.189 |
| Fc epsilon RI signaling pathway | 18 | 2.87E-02 | 0.197 |
| Colorectal cancer | 19 | 2.92E-02 | 0.193 |
| Wnt signaling pathway | 30 | 2.92E-02 | 0.187 |
| Fc gamma R-mediated phagocytosis | 20 | 4.84E-02 | 0.283 |
| Prostate cancer | 19 | 4.88E-02 | 0.277 |
| Adherens junction | 17 | 4.92E-02 | 0.271 |
| T cell receptor signaling pathway | 22 | 5.13E-02 | 0.274 |
| Small cell lung cancer | 18 | 5.42E-02 | 0.279 |
| Basal cell carcinoma | 13 | 5.98E-02 | 0.296 |
| Progesterone-mediated oocyte maturation | 18 | 6.54E-02 | 0.312 |
| Vibrio cholerae infection | 13 | 6.72E-02 | 0.312 |
| Epithelial cell signaling in Helicobacter pylori infection | 15 | 6.74E-02 | 0.305 |
| Sphingolipid metabolism | 10 | 7.13E-02 | 0.313 |
| Notch signaling pathway | 11 | 9.34E-02 | 0.384 |
| Axon guidance | 24 | 9.60E-02 | 0.385 |
Interestingly, the renal cell carcinoma pathway was enriched by the identification of 15 putative target genes of the downregulated miRNAs (S8 Table and S5 Fig). In this case EPAS1 (Endothelial Per-Arnt-Sim (PAS) domain protein 1), EGLN3 (EGL-nine homolog-3), VEGFC (vascular endothelial growth factor C), and EGLN1 (EGL-nine homolog-1) have been reported to have strong high-altitude selective signatures in the TP [36, 37]. It was noteworthy that 63 target genes of the downregulated miRNAs in the TP belonged to cancer-related pathways (S6 Fig). These pathways are involved in sustained angiogenesis, proliferation, genomic damage, and inhibition of differentiation, all of which are closely related to cancer growth and development under hypoxic conditions [38].
Validation of DE miRNAs
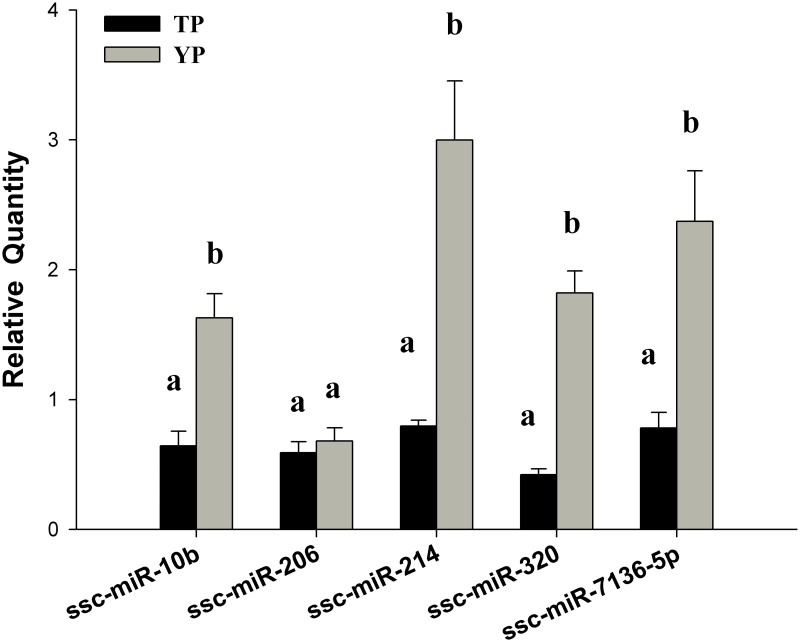
Expression levels of five miRNAs (ssc-miR-10b, ssc-miR-206, ssc-miR-214, ssc-miR-320, and ssc-miR-7136-5p) in the cardiac tissues of TPs and YPs were assessed by stem-loop qPCR. The results showed that four of the five miRNAs had significantly higher expression levels in TPs than in YPs (Fig 3), which was consistent with the miRNA-seq data. The different expression trends observed in the miRNA-seq data and from the RT-qPCR were uniform for five miRNAs in the TPs and YPs (S9 Table).
Fig 3. Five cardiac tissue differentially expressed (DE) miRNAs validated by reverse-transcription quantitative polymerase chain reaction.
Relative expression levels of DE miRNAs. Upper letters (a, b) on bars denote significantly different expression levels in the same miRNA (P < 0.05).
Discussion
High-altitude populations of pigs have evolved genetic adaptations that allow for survival in extremely hypoxic environments. Previous reports have demonstrated that TPs in particular exhibit a distinct suite of phenotypic and physiological traits, including thin-walled pulmonary vascular structures and high blood flow [13, 14, 39], which are shaped by natural and artificial selection, allowing them to adapt to high-altitude environments [40]. The relatively recently identified miRNAs constitute a novel class of master regulators that control gene expression, and are responsible for a variety of normal and pathological cellular processes. High-throughput sequencing has generated new insights into global gene expression, and provided evidence for the complexity of the mammalian transcriptome and allowed for the development of miRNAomics [41].
In this study, miRNA profiles in the cardiac tissues of TPs and YPs, both adapted to a high-altitude environment, were obtained using next-generation sequencing. We identified 20 DE miRNAs in the two pig breeds, of which ssc-miR-214 downregulated expression in the TP. Previous studies have demonstrated that alcohol depresses glutathione reductase (GSR) and P450 oxidoreductase (POR) gene expression by the up-regulation of miR-214 that induces oxidative stress, which plays an important role in responses to hypoxia [42, 43].
The most notably up-regulated miRNA in the TP (FC = 230), ssc-miR-210, is induced by HIF1α under hypoxic conditions in mice [11, 12]. miR-210 is located on an intron of a noncoding RNA, transcribed from AK123483 on the human chromosome 11p15.5, and its expression correlates with VEGF regulation and angiogenesis in breast cancer patients [44]. Using bioinformatics analyses, miR-210-regulated factors have been found to be implicated in DNA repair pathways [11], and has been found to play roles in modulating the expression of proteins involved in the homology-dependent repair and nucleotide-excision repair pathways, and reverses cellular DNA damage during hypoxia. Our results demonstrate that miR-210 is expressed more in the TP than in the YP, which suggests that miR-210 could modulate key factors that are related to cellular or organic hypoxia adaptation pathways in the TP.
Based on these functional pathways, 100 putative target genes (including 47 up-regulated miRNAs and 53 downregulated miRNAs) were enriched in the MAPK signaling pathway (S4 Fig). The MAPK pathway may up-regulate HIF activity, and plays an essential role in tumor growth and transformation that depends on angiogenesis and changes in glucose metabolism [35]. The expression of a gene that is involved in cancer and MAPK signaling pathways, FOS (FBJ murine osteosarcoma viral oncogene homolog) (S7B Table), is increased by miR-101 under hypoxic conditions [45, 46]. Furthermore, FOS may be a potential target of ssc-miR-101, which was downregulated in the TP in this study.
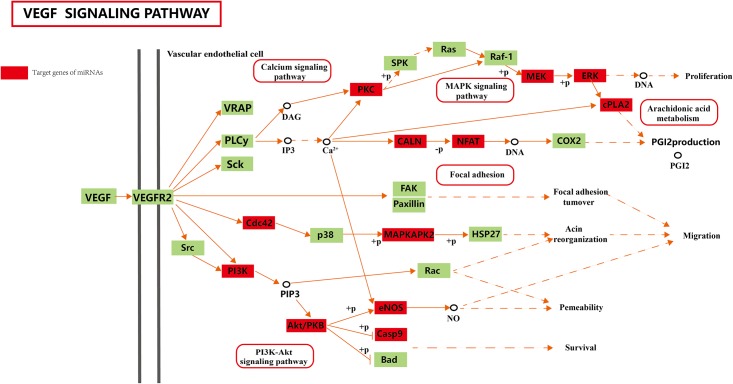
The VEGF signaling pathway regulates angiogenesis, endothelial cell growth, proliferation, and migration, affects the permeability of blood vessels [47, 48], and plays important roles in hypoxia adaptation. In our study, 19 putative target genes that are regulated by downregulated miRNAs (ssc-miR-101, ssc-miR-7136-5p, ssc-miR-214, ssc-miR-10b, ssc-miR-206, and ssc-miR-320) were found to be involved in the VEGF signaling pathway (Fig 4). VEGF up-regulation by HIF is accompanied by the stability of its mRNA and an increase in translation, which are essential for hypoxia-related angiogenesis. Another important pathway that was targeted by downregulated miRNAs in cardiac tissue is involved in vascular smooth muscle contraction. This pathway is associated with energy metabolism, including soluble guanylyl cyclase [49] and potassium and calcium channels (such as sarcoplasmic reticulum calcium ATPase) [50, 51].
Fig 4. The VEGF signaling pathway enriched by 19 putative target genes of downregulated miRNAs.
Red boxes represent the target genes of miRNAs.
Four DE miRNAs (ssc-miR-210, ssc-miR-1343, ssc-miR-142-5p, and ssc-miR-421-5p) that are involved in the renal cell carcinoma pathway were upregulated in TPs, and the corresponding target genes were EPAS1, EGLN3, EGLN1, and VEGFC. Both EPAS1 and EGLN1 are closely associated with the high-altitude adaptation of Tibetan populations [37], and are involved in hypoxia pathways as key regulators during chronic hypoxia [52]. Genetic variation in EPAS1 and EGLN1 is associated with Hb levels and high-altitude adaptation in Tibetan populations [36, 53, 54]. MiRNA-pathway-enrichment analysis revealed that the miRNAs might contribute to high-altitude adaptation by participating in signaling pathway in TPs. VEGFC is involved in two signaling pathways: renal cell carcinoma and mTOR. Hypoxia signaling (in particular HIF-1α) regulates the expression of VEGFC, which is one of the key lymphangiogenic factors [55]. Liang et al. [56] reported an associative correlation between HIF-1α and VEGFC in cancer. Here, four DE miRNAs related to hypoxia were found to have high expression levels in TPs, suggesting that miRNAs and target-gene regulation enabled the TP to adapt to a hypoxic environment.
In conclusion, this study significantly increased the number of hypoxia-related miRNAs known in the pig, and identified miRNAs with significantly altered in the TP. Using miRNA-seq, 372 miRNAs were found in the cardiac tissues of pigs living at high altitudes. Twenty DE miRNAs were identified and subjected to bioinformatics functional analyses. The results suggest that 20 miRNAs involved in the mTOR signaling pathway, the MAPK signaling pathway, the renal cell carcinoma pathway, various cancer pathways, the VEGF signaling pathway, and the vascular smooth muscle contraction pathway play regulatory roles in hypoxia adaptation in the TP. This study provides new insights that will advance the study adaptation to hypoxia in humans and other mammals living at high altitude. The next step, the functional mechanism of the DE miRNA regulating the hypoxia adaptation in the Tibetan pig was to be investigated.
Supporting Information
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
Acknowledgments
We would like to thank Wenyou Gou, Qian Zhang, and Xiangxiang Li for assistance with sample collection.
Data Availability
All analysis results are in the paper and its Supporting Information files. The microRNA sequencing profile data is deposited in the Gene Expression Omnibus with accession number GSE71550.
Funding Statement
This work was supported by the National Natural Science Foundation of China (31160441), the National Major Special Project on New Varieties Cultivation for Transgenic Organisms (2014ZX08009-003-006), and the National Key Technology Research and Development Program (2012BAD03B03), to HZ. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Rajewsky N. microRNA target predictions in animals. Nat Genet. 2006; 38: S8–S13. 10.1038/ng1798 [DOI] [PubMed] [Google Scholar]
- 2. Flynt AS, Lai EC. Biological principles of microRNA-mediated regulation: shared themes amid diversity. Nat Rev Genet. 2008; 9: 831–842. 10.1038/nrg2455 PMCID: PMC2729318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Pasquinelli AE. MicroRNAs and their targets: recognition, regulation and an emerging reciprocal relationship. Nat Rev Genet. 2012; 13: 271–282. 10.1038/nrg3162 [DOI] [PubMed] [Google Scholar]
- 4. Liao D, Johnson RS. Hypoxia: a key regulator of angiogenesis in cancer. Cancer Metast Rev. 2007; 26: 281–290. [DOI] [PubMed] [Google Scholar]
- 5. Zardo G, Ciolfi A, Vian L, Starnes LM, Billi M, Racanicchi S, et al. Polycombs and microRNA-223 regulate human granulopoiesis by transcriptional control of target gene expression. Blood. 2012; 119: 4034–4046. 10.1182/blood-2011-08-371344 [DOI] [PubMed] [Google Scholar]
- 6. Sinkkonen L, Hugenschmidt T, Berninger P, Gaidatzis D, Mohn F, Artus-Revel CG, et al. MicroRNAs control de novo DNA methylation through regulation of transcriptional repressors in mouse embryonic stem cells. Nat Struct Mol Biol. 2008; 15: 259–267. 10.1038/nsmb.1391 [DOI] [PubMed] [Google Scholar]
- 7. Kulshreshtha R, Ferracin M, Wojcik SE, Garzon R, Alder H, Agosto-Perez FJ, et al. A microRNA signature of hypoxia. Mol Cell Biol. 2007; 27: 1859–1867. 10.1128/MCB.01395-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Taguchi A, Yanagisawa K, Tanaka M, Cao K, Matsuyama Y, Goto H, et al. Identification of hypoxia-inducible factor-1α as a novel target for miR-17-92 microRNA cluster. Cancer Res. 2008; 68: 5540–5545. 10.1158/0008-5472.CAN-07-6460 [DOI] [PubMed] [Google Scholar]
- 9. Lei Z, Li B, Yang Z, Fang H, Zhang G, Feng Z, et al. Regulation of HIF-1and VEGF by miR-20b tunes tumor cells to adapt to the alteration of oxygen concentration. PloS one. 2009; 4: e7629 10.1371/journal.pone.0007629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Rane S, He M, Sayed D, Vashistha H, Malhotra A, Sadoshima J, et al. Downregulation of miR-199a derepresses hypoxia-inducible factor-1α and Sirtuin 1 and recapitulates hypoxia preconditioning in cardiac myocytes. Circ Res. 2009; 104: 879–886. 10.1161/CIRCRESAHA.108.193102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Crosby ME, Kulshreshtha R, Ivan M, Glazer PM. MicroRNA regulation of DNA repair gene expression in hypoxic stress. Cancer Res. 2009; 69: 1221–1229. 10.1158/0008-5472.CAN-08-2516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Huang X, Ding L, Bennewith KL, Tong RT, Welford SM, Ang KK, et al. Hypoxia-inducible mir-210 regulates normoxic gene expression involved in tumor initiation. Mol Cell. 2009; 35: 856–867. 10.1016/j.molcel.2009.09.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Pan PW, Zhao SH, Yu M, Liu B, Xiong TA, Li K. Identification of Differentially Expressed Genes in the Longissimus Dorsi Muscle Tissue between Duroc and Erhualian Pig by mRNA Differential Display. Asan Autral J Anim. 2003; 16: 1066–1070. 10.5713/ajas.2003.1066 [DOI] [Google Scholar]
- 14. Gong JJ, He ZP, Li ZQ, Lv XB, Ying SC, Chen XH. Investigation on fattening and carcass traits in Tibetan pig and its combinations. Southwest China Journal of Agricultural Sciences. 2007; 20: 1109–1112. [Google Scholar]
- 15. Qi S, Chen J, Guo R, Yu B, Chen D. β-defensins gene expression in tissues of the crossbred and Tibetan pigs. Livest Sci. 2009; 123: 161–168. 10.1016/j.livsci.2008.11.009 [DOI] [Google Scholar]
- 16. Ream M, Ray AM, Chandra R, Chikaraishi DM. Early fetal hypoxia leads to growth restriction and myocardial thinning. American Journal of Physiology-Regulatory, Integrative and Comparative Physiology. 2008; 295: R583–R595. 10.1152/ajpregu.00771.2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Tintu A, Rouwet E, Verlohren S, Brinkmann J, Ahmad S, Crispi F, et al. Hypoxia induces dilated cardiomyopathy in the chick embryo: mechanism, intervention, and long-term consequences. PLoS One. 2009; 4: e5155 10.1371/journal.pone.0005155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010; 26: 139–140. 10.1093/bioinformatics/btp616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Wang L, Feng Z, Wang X, Wang X, Zhang X. DEGseq: an R package for identifying differentially expressed genes from RNA-seq data. Bioinformatics. 2010; 26: 136–138. 10.1093/bioinformatics/btp612 [DOI] [PubMed] [Google Scholar]
- 20. Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009; 10: R25 10.1186/gb-2009-10-3-r25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Friedl A Nder MR, Chen W, Adamidi C, Maaskola J, Einspanier R, Knespel S, et al. Discovering microRNAs from deep sequencing data using miRDeep. Nat Biotechnol. 2008; 26: 407–415. 10.1038/nbt1394 [DOI] [PubMed] [Google Scholar]
- 22. Kozomara A, Griffiths-Jones S. miRBase: annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res. 2014; 42: 68–73. 10.1093/nar/gkt1181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Krawczynski K, Najmula J, Bauersachs S, Kaczmarek MM. MicroRNAome of Porcine Conceptuses and Trophoblasts: Expression Profile of microRNAs and Their Potential to Regulate Genes Crucial for Establishment of Pregnancy. Biol Reprod. 2015; 92: 21 10.1095/biolreprod.114.123588 [DOI] [PubMed] [Google Scholar]
- 24. Zhou S, Liu Y, Prater K, Zheng Y, Cai L. Roles of microRNAs in pressure overload-and ischemia-related myocardial remodeling. Life Sci. 2013; 93: 855–862. 10.1016/j.lfs.2013.08.023 [DOI] [PubMed] [Google Scholar]
- 25. Martini P, Sales G, Brugiolo M, Gandaglia A, Naso F, De Pitt A C, et al. Tissue-specific expression and regulatory networks of pig microRNAome. PloS one. 2014; 9: e89755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Bu D, Yu K, Sun S, Xie C, Skogerb O G, Miao R, et al. NONCODE v3. 0: integrative annotation of long noncoding RNAs. Nucleic Acids Res. 2012; 40: 210–215. 10.1093/nar/gkr1175 PMCID: PMC3245065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Betel D, Wilson M, Gabow A, Marks DS, Sander C. The microRNA.org resource: targets and expression. Nucleic Acids Res. 2008; 36: D149–153. 10.1093/nar/gkm995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kanehisa M, Goto S, Sato Y, Kawashima M, Furumichi M, Tanabe M. Data, information, knowledge and principle: back to metabolism in KEGG. Nucleic Acids Res. 2014; 42: D199–D205. 10.1093/nar/gkt1076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID Bioinformatics Resources. Nature Protoc. 2009; 4: 44–57. 10.1038/nprot.2008.211 [DOI] [PubMed] [Google Scholar]
- 30. Chen C, Ridzon DA, Broomer AJ, Zhou Z, Lee DH, Nguyen JT, et al. Real-time quantification of microRNAs by stem—loop RT—PCR. Nucleic Acids Res. 2005; 33: e179 10.1093/nar/gni178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Huang J, Ju Z, Li Q, Hou Q, Wang C, Li J, et al. Solexa sequencing of novel and differentially expressed microRNAs in testicular and ovarian tissues in Holstein cattle. Int J Biol Sci. 2011; 7: 1016–1026. 10.7150/ijbs.7.1016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lau NC, Lim LP, Weinstein EG, Bartel DP. An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science. 2001; 294: 858–862. 10.1126/science.1065062 [DOI] [PubMed] [Google Scholar]
- 33. Zhang B, Stellwag EJ, Pan X. Large-scale genome analysis reveals unique features of microRNAs. Gene. 2009; 443: 100–109. 10.1016/j.gene.2009.04.027 [DOI] [PubMed] [Google Scholar]
- 34. Wouters BG, Koritzinsky M. Hypoxia signalling through mTOR and the unfolded protein response in cancer. Nat Rev Cancer. 2008; 8: 851–864. 10.1038/nrc2501 [DOI] [PubMed] [Google Scholar]
- 35. Sang N, Stiehl DP, Bohensky J, Leshchinsky I, Srinivas V, Caro J. MAPK signaling up-regulates the activity of hypoxia-inducible factors by its effects on p300. J Biol Chem. 2003; 278: 14013–14019. 10.1074/jbc.M209702200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Simonson TS, Yang Y, Huff CD, Yun H, Qin G, Witherspoon DJ, et al. Genetic evidence for high-altitude adaptation in Tibet. Science. 2010; 329: 72–75. 10.1126/science.1189406 [DOI] [PubMed] [Google Scholar]
- 37. Yi X, Liang Y, Huerta-Sanchez E, Jin X, Cuo ZXP, Pool JE, et al. Sequencing of 50 human exomes reveals adaptation to high altitude. Science. 2010; 329: 75–78. 10.1126/science.1190371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Shen C, Beroukhim R, Schumacher SE, Zhou J, Chang M, Signoretti S, et al. Genetic and functional studies implicate HIF1alpha as a 14q kidney cancer suppressor gene. Cancer Discov. 2011:222–235. 10.1158/2159-8290.CD-11-0098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Monge C, Leon-Velarde F. Physiological adaptation to high altitude: oxygen transport in mammals and birds. Physiol Rev. 1991; 71: 1135–1172. [DOI] [PubMed] [Google Scholar]
- 40. Yang S, Zhang H, Mao H, Yan D, Lu S, Lian L, et al. The local origin of the Tibetan pig and additional insights into the origin of Asian pigs. PloS one. 2011; 6: e28215 10.1371/journal.pone.0028215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Wilhelm BT, Landry JEE. RNA-Seq—quantitative measurement of expression through massively parallel RNA-sequencing. Methods. 2009; 48: 249–257. 10.1016/j.ymeth.2009.03.016 [DOI] [PubMed] [Google Scholar]
- 42. Zhou Y, Tang X, Song Q, Ji Y, Wang H, Jiao H, et al. Identification and characterization of pig embryo microRNAs by Solexa sequencing. Reprod Domest Anim. 2013; 48: 112–120. 10.1111/j.1439-0531.2012.02040.x [DOI] [PubMed] [Google Scholar]
- 43. Dong X, Liu H, Chen F, Li D, Zhao Y. MiR-214 Promotes the Alcohol-Induced Oxidative Stress via Down-Regulation of Glutathione Reductase and Cytochrome P450 Oxidoreductase in Liver Cells. Alcoholism: Clinical and Experimental Research. 2014; 38: 68–77. 10.1111/acer.12209 [DOI] [PubMed] [Google Scholar]
- 44. Foekens JA, Sieuwerts AM, Smid M, Look MP, de Weerd V, Boersma AW, et al. Four miRNAs associated with aggressiveness of lymph node-negative, estrogen receptor-positive human breast cancer. Proceedings of the National Academy of Sciences. 2008; 105: 13021–13026. 10.1073/pnas.0803304105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Rybnikova E, Glushchenko T, Tyulkova E, Baranova K, Samoilov M. Mild hypobaric hypoxia preconditioning up-regulates expression of transcription factors c-Fos and NGFI-A in rat neocortex and hippocampus. Neurosci Res. 2009; 65: 360–366. 10.1016/j.neures.2009.08.013 [DOI] [PubMed] [Google Scholar]
- 46. Yuen RK, Chen B, Blair JD, Robinson WP, Nelson DM. Hypoxia alters the epigenetic profile in cultured human placental trophoblasts. Epigenetics-US. 2013; 8: 192–202. 10.4161/epi.23400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Ray PS, Jia J, Yao P, Majumder M, Hatzoglou M, Fox PL. A stress-responsive RNA switch regulates VEGFA expression. Nature. 2009; 457: 915–919. 10.1038/nature07598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Oladipupo S, Hu S, Kovalski J, Yao J, Santeford A, Sohn RE, et al. VEGF is essential for hypoxia-inducible factor-mediated neovascularization but dispensable for endothelial sprouting. Proceedings of the National Academy of Sciences. 2011; 108: 13264–13269. 10.1073/pnas.1101321108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Gerassimou C, Kotanidou A, Zhou Z, Simoes D, Roussos C, Papapetropoulos A. Regulation of the expression of soluble guanylyl cyclase by reactive oxygen species. Brit J Pharmacol. 2007; 150: 1084–1091. 10.1038/sj.bjp.0707179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Magder S. Reactive oxygen species: toxic molecules or spark of life. Crit Care. 2006; 10: 208 10.1186/cc3992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Chan CK, Vanhoutte PM. Hypoxia, vascular smooth muscles and endothelium. Acta Pharmaceutica Sinica B. 2013; 3: 1–7. 10.1016/j.apsb.2012.12.007 [DOI] [Google Scholar]
- 52. De B, Huajun X, Cuihong Z, Jun Z, Xiaoyan D, Xiaopeng L. Systems biology approach to study the high altitude adaptation in tibetans. Braz Arch Biol Techn. 2013; 56: 53–60. 10.1590/S1516-89132013000100007 [DOI] [Google Scholar]
- 53. Beall CM, Cavalleri GL, Deng L, Elston RC, Gao Y, Knight J, et al. Natural selection on EPAS1 (HIF2α) associated with low hemoglobin concentration in Tibetan highlanders. Proceedings of the National Academy of Sciences. 2010; 107: 11459–11464. 10.1073/pnas.1002443107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Lou H, Lu Y, Lu D, Fu R, Wang X, Feng Q, et al. A 3.4-kb Copy-Number Deletion near EPAS1 Is Significantly Enriched in High-Altitude Tibetans but Absent from the Denisovan Sequence. AM. J. Hum. Genet. 2015; 97: 54–66. 10.1016/j.ajhg.2015.05.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Alitalo A, Detmar M. Interaction of tumor cells and lymphatic vessels in cancer progression. Oncogene. 2012; 31: 4499–4508. 10.1038/onc.2011.602 [DOI] [PubMed] [Google Scholar]
- 56. Liang X, Yang D, Hu J, Hao X, Gao J, Mao Z. Hypoxia inducible factor-1alpha expression correlates with vascular endothelial growth factor-C expression and lymphangiogenesis/angiogenesis in oral squamous cell carcinoma. Anticancer Res. 2008; 28: 1659–1666. . [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
Data Availability Statement
All analysis results are in the paper and its Supporting Information files. The microRNA sequencing profile data is deposited in the Gene Expression Omnibus with accession number GSE71550.