Summary
Neurodegenerative phenotypes reflect complex, time-dependent molecular processes, whose elucidation may reveal neuronal class-specific therapeutic targets. The current focus in neurodegeneration has been on individual genes and pathways. In contrast, we assembled a genome-wide regulatory model (henceforth, interactome), whose unbiased interrogation revealed 23 candidate causal master regulators of neurodegeneration in an in vitro model of amyotrophic lateral sclerosis (ALS), characterized by a loss of spinal motor neurons (MNs). Of these, eight were confirmed as specific MN death drivers in our model of familial ALS, including NF-κB, which has long been considered a pro-survival factor. Through an extensive array of molecular, pharmacological and biochemical approaches we have confirmed that neuronal NF-κB drives the degeneration of MNs in both familial and sporadic models of ALS, thus providing proof-of-principle that regulatory network analysis is a valuable tool for studying cell-specific mechanisms of neurodegeneration.
Introduction
The contribution of non-neuronal cells such as astrocytes to the demise of neighboring neurons in a variety of neurodegenerative disorders (Garden and La Spada, 2012), including amyotrophic lateral sclerosis (ALS), is increasingly recognized. Specifically, we previously reported that astrocytes carrying a mutation in the superoxide dismutase-1 (SOD1) gene, which account for ∼12% of familial ALS cases (Renton et al., 2014), selectively kill mouse primary spinal motor neurons (MNs) and embryonic stem cell-derived MNs (ES-MNs) (Nagai et al., 2007; Re et al., 2014). This spontaneous neurodegenerative phenotype was observed either when MNs were cultured in the presence of mSOD1-expressing astrocytes or when they were exposed to medium conditioned by mutant astrocytes (Nagai et al., 2007; Re et al., 2014). Mirroring these findings from mouse, we showed that astrocytes derived from postmortem CNS samples of human sporadic ALS patients were also toxic to human ES-MNs (Re et al., 2014). Furthermore, it has been reported that mSOD1-expressing glial-restricted precursor cells grafted into spinal cords of wild-type rats cause MN loss in living animals (Papadeas et al., 2011), and that reduction of mSOD1 levels selectively in astrocytes prolongs survival in SOD1G37R transgenic mice (Yamanaka et al., 2008). Taken together, these observations suggest that astrocyte-triggered neurodegeneration is a general phenomenon in ALS, and is not restricted to in vitro systems, to mouse cells, or to mSOD1-linked ALS.
Phenotypic changes effected by pathological events are now routinely captured by gene expression profile (GEP) measurements, determining mRNA abundance on a genome-wide scale in a cellular population (Klein et al., 2003; Tothill et al., 2008). We have shown that analysis of large GEP datasets, using algorithms such as ARACNe, can produce accurate and comprehensive repertoires of regulatory interactions between transcription factors (TFs) and their transcriptional targets, known as regulatory networks or interactomes (Basso et al., 2005; Margolin et al., 2006). Interactome interrogation using the MAster Regulator INference algorithm (MARINa) has been especially effective in identifying TFs that could be experimentally validated as causal master regulators (MRs), i.e. genes that are both necessary and sufficient for implementation of phenotypes (Aytes et al., 2014; Carro et al., 2010; Lefebvre et al., 2010). However, such methods have never been used to identify drivers of neurodegenerative phenotypes.
In this paper, we show that computational assembly and interrogation of neural tissue specific interactomes using GEP signatures derived from purified ES-MNs, following exposure to mSOD1 astrocyte conditioned medium (ACM) for 3 days, identified 23 TFs as candidate MRs (i.e. drivers) of neurodegeneration. Of the 22 for which shRNA-mediated silencing reagents were available, 14 (∼64%) were shown to modulate MN loss. Specifically, silencing these genes in MNs decreased neurodegeneration, and of these, 11 displayed mSOD1 astrocyte-specific effects. Validated MRs included a key subunit of the nuclear factor kappa B (NF-κB) complex, whose role as effector of mSOD1 astrocyte-induced MN death was further confirmed by genetic and pharmacological approaches. Taken together, these data demonstrate the value of interactome analysis, using appropriate in vitro models, as an effective methodology to generate insights into the mechanisms of neurodegeneration.
Results
A model of non-cell autonomous neurodegeneration in ALS
Our earlier studies (Nagai et al., 2007) required co-culturing MNs with primary astrocytes or using mixed ES-MNs populations, which prevented generation of MN-specific GEP signatures. Thus, to generate signatures representative of early changes in MNs, following exposure to toxic mSOD1-expressing astrocytes, we had to first develop an in vitro model system yielding sufficiently homogeneous ES-MNs.
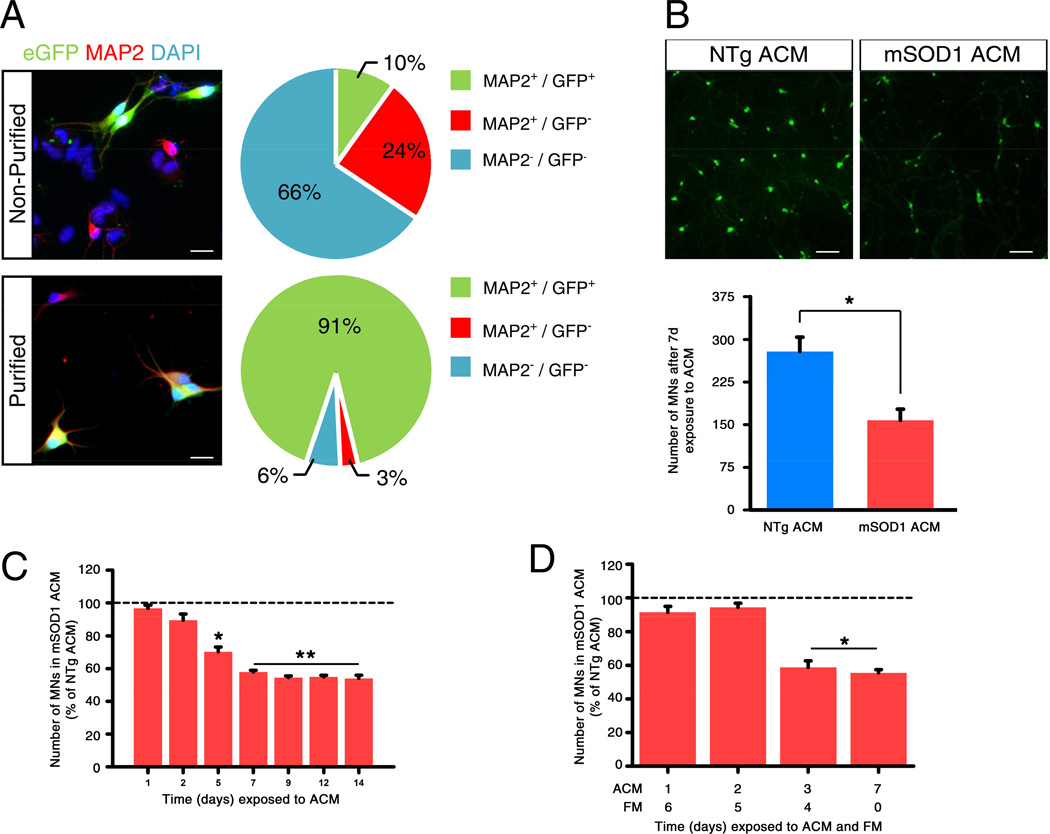
Accordingly, we started by substituting the glial cell monolayer with ACM (Nagai et al., 2007), thus avoiding signature contamination by co-cultured astrocytes. Then, to enrich ES-MN purity, we tested two distinct purification methods: fluorescence-activated cell sorting (FACS) and magnetic-activated cell sorting (MACS). For the latter we used a murine ES Hb9::GFP reporter cell line stably transduced with a viral vector expressing the cell surface receptor CD2 (cluster of differentiation 2), under the control of the MN-specific promoter Hb9. Both FACS-and MACS-purification processes gave ∼91% cell purity, following ES-MN differentiation. Yet total yield of purified ES-MNs using MACS was ∼5-fold greater than with FACS. MACS-purified ES-MNs (Figure 1A) remained susceptible to mSOD1 ACM exposure for 7 days (Figure 1B), thus eliminating the possibility that such process might select for non-representative ES-MN subpopulations or that CD2 expression might affect ES-MN properties.
Figure 1. Purified ES-MNs exposed to mSOD1 ACM is a potent model to study non-cell autonomous MN degeneration in ALS.
(A) Immunocytochemical illustrations and pie charts of cultures before and after MACS® enrichment. Scale bars: 20 µm.
(B) Immunocytochemical illustrations and bar graphs of purified ES-MN numbers after 7 days exposure to non-toxic (NTg ACM) or toxic mSOD1 ACM. Scale bars: 60 µm. *Student’s t test: t[8] = 3.78, p = 0.0054.
(C) Time course of purified ES-MN loss upon exposure to ACM. ES-MNs are counted at the indicated exposure days. Lower than mSOD1 ACM at day 1 and NTg ACM at all tested days (*p < 0.05, **p < 0.01; Newman-Keuls post-hoc).
(D) At 2 days post-plating, purified ES-MNs are exposed to ACM for 1, 2, 3 or 7 days, after which medium is replaced with fresh medium and cells are cultured for an additional 6, 5, 4 or 0 day, respectively. ES-MN numbers are determined after 7 days exposures to ACM + fresh medium (FM). Bars show the number of viable ES-MNs treated with mSOD1 ACM relative to the ones treated with NTg ACM (*p < 0.01; Newman-Keuls post-hoc).
Next, to select the most relevant time points for GEP signature generation, we studied the kinetics of purified ES-MN loss in response to mSOD1 ACM. For up to 2 days, no difference could be detected in the number of ES-MNs exposed to mSOD1 or to non-transgenic (NTg) ACM (Figure 1C). Thereafter, the number of ES-MNs exposed to mSOD1 ACM declined monotonically until 7 days, reaching 50 ± 4% (n=5, mean ± s.e.m) of the value in control cultures (Figure 1C). Between 7 and 14 days exposure, there was no further reduction in the number of ES-MNs exposed to mSOD1 ACM as compared to those exposed to NTg ACM (Figure 1C). The kinetics and magnitude of ES-MN loss were comparable to those reported for mouse primary spinal MNs (Nagai et al., 2007) and for human ES-MNs exposed to sporadic human astrocytes (Re et al., 2014).
Finally, we assumed that activity of MN death drivers would likely occur before ES-MNs become irreversibly committed to death. To test for the existence of such a “point of no return”, ES-MNs were exposed to mSOD1 ACM for varying times, prior to switching to fresh MN medium for a combined time of 7 days (Figure 1D). Exposure to mSOD1 ACM for ≤ 2 days did not induce MN loss compared to controls (Figure 1D). In contrast, exposure to mSOD1 ACM for ≥ 3 days was sufficient to trigger loss of ∼50% of MNs at 7 days (Figure 1D). Thus, susceptible ES-MNs become committed to death by 3 days exposure to mSOD1 ACM. Taken together, these observations suggest that purified ES-MNs exposed to mSOD1 ACM provide a reliable non-cell autonomous model of progressive neurodegeneration. Moreover, their expandable nature, relevance to human disease, and synchronicity of death commitment by susceptible MNs make this a robust in vitro model to explore the molecular basis of neurodegeneration in ALS.
Gene expression signature of MN death
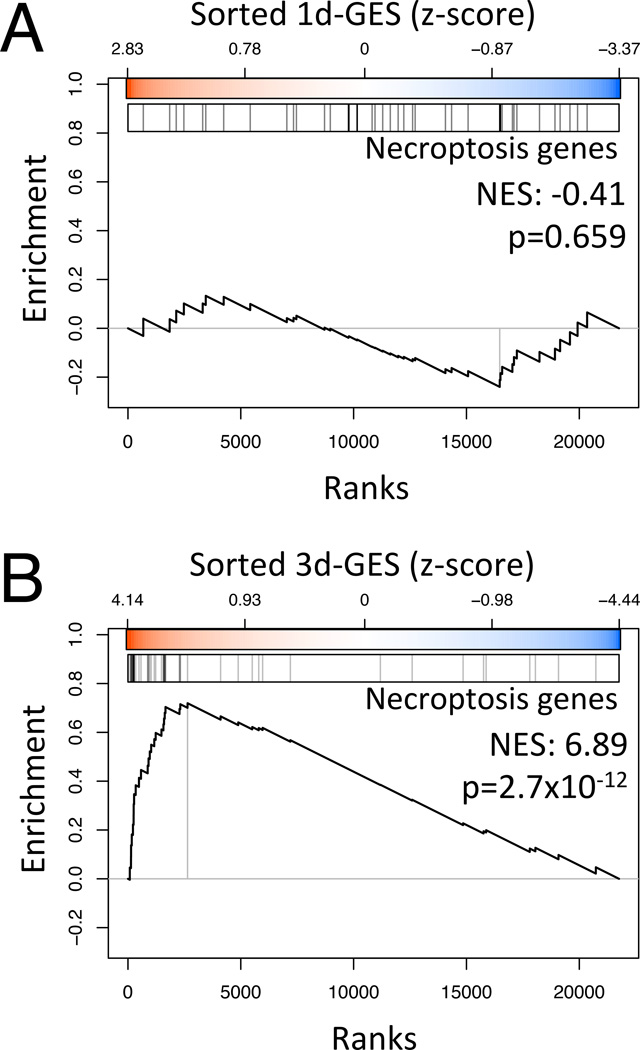
To identify molecular events correlated with neurodegeneration, we profiled gene expression by ES-MNs after 1 day and 3 days of exposure to either mSOD1 ACM, NTg ACM, or culture medium conditioned by astrocytes from transgenic mice overexpressing wild-type SOD1 (wtSOD1 ACM). Like NTg ACM, the latter is not toxic to ES-MNs (Nagai et al., 2007), but controls for any effects SOD1 overexpression might have on the GEP signatures. After correcting for potential GEP batch effects using a linear model that accounts for both treatment and batch variance, we estimated statistical significance by a moderated Student’s t-test (see Experimental Procedures). At 1 day following exposure, there were only 79 differentially expressed genes (false discovery rate, FDR < 1%) between ES-MNs exposed to either mSOD1 or control ACMs. However, at 3 days of exposure there were 620 differentially expressed genes (FDR < 1%; see Table S1). All profiled genes were rank-sorted from the most under- to the most over-expressed, based on the moderated t-statistic (see Experimental Procedures), to produce a 3-day gene expression signature (3d-GES).
To support the relevance of the generated GESs to the death mechanisms of MNs, we first performed a gene set enrichment analysis between our data and a set of 40 neuron-specific necroptosis-associated genes, previously characterized through a loss-of-function screen in zVAD-fmk-treated mouse fibrosarcoma cells (Hitomi et al., 2008). This specific analysis was prompted by our demonstration that both mouse mSOD1 familial-ALS and human sporadic ALS astrocytes kill MNs by necroptosis (Re et al., 2014), a caspase-independent form of programmed cell death (Pasparakis and Vandenabeele, 2015). Strikingly, the necroptosis-associated genes that were inferred to be expressed in normal neurons were highly enriched in the 3d-GES, but not in the 1d-GES (Figure 2 and Table S1). These findings suggest that the 3d-GES may be particularly suitable to dissect the molecular drivers of MN death in our ALS models.
Figure 2. Enrichment of the necroptosis program in the mSOD1-induced GES.
(A) Gene Set Enrichment Analysis for 40 neuron-specific necroptosis genes on the 1d-GES
(B) Same at 3d-GES.
The maximum value of the curve determines the enrichment score. The vertical lines indicate the projection of the neuron-specific necroptosis genes on the sorted GES (indicated by the color scale in z-score units). The normalized enrichment score (NES) and associated p-value are shown on each plot. See also Table S1.
Regulatory mechanisms controlling MN death in response to mSOD1 ACM
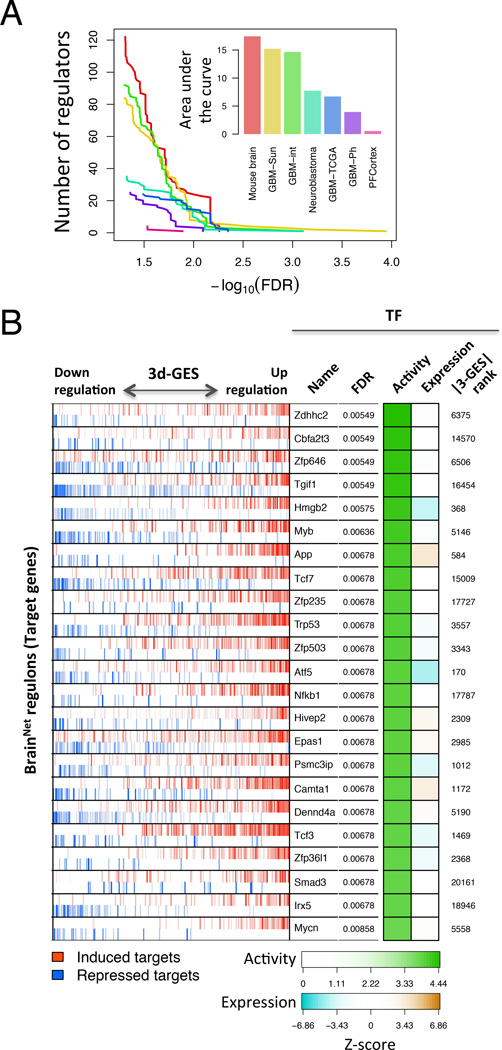
To identify the genes driving the 3d-GES and thus, being the causal determinants of MN degeneration, we utilized the previously validated MARINa algorithm (Carro et al., 2010; Lefebvre et al., 2010). A critical requirement for utilization of the algorithm is the availability of a context-specific interactome, representing a genome-wide map of regulatory interactions (i.e., the repertoire of transcriptional targets that are activated or repressed by each TF in the cellular context of interest). Assembly of such an interactome using the ARACNe algorithm requires >100 GEPs from independent samples, representative of a particular cellular context of interest (Margolin et al., 2006). Since a MN-specific interactome is not currently available, we evaluated whether other ARACNe-inferred interactomes (Basso et al., 2005), assembled from lineage-related tissues, could be used instead (see Supplemental Experimental Procedures). To select an optimal interactome, we first assessed whether statistical significance of MARINa-inferred MRs may represent a good indication of the interactome’s ability to represent a specific cellular context. This was confirmed by both increasingly degrading interactomes (in silico) and by using non-tissue-matched interactomes, which, as expected, substantially reduced the statistical significance of inferred MRs (Figure S1 and Experimental Procedures). MARINa analysis of the 3d-GES using seven distinct ARACNe-inferred interactomes revealed that a murine whole-brain interactome (BrainNet) out-performed all others, including those obtained from human glioma, neuroblastoma, and prefrontal cortex, as the most appropriate model for the murine ES-MN context (Figure 3A). The BrainNet was assembled by ARACNe analysis of 437 mouse brain biopsies, obtained from seven distinct brain regions of 20 inbred mouse strains, profiled by Nadler et al. (Gene Expression Omnibus GSE10415).
Figure 3. Optimal interactome and inferred master regulators of the ES-MN molecular phenotype induced by 3 days of exposure to mSOD1 ACM.
(A) Number of regulators (y-axis) inferred as differentially active by MARINa for the mSOD1 ACM treated ES-MN 3-day GES, using seven alternative interactomes (indicated in the plot insert) for different false discovery rate (FDR) levels (x-axis). The plot insert shows the quantification of MARINa performance on seven alternative interactomes (x-axis) as the area under the curves shown in (A). See Supplemental Experimental Procedures for interactome details.
(B) Statistically significant MRs inferred by MARINa analysis of the BrainNet, using the 3-day signature of MNs exposed to mSOD1 ACM. The x-axis represents all genes, sorted from that with the largest decrease to the one with the largest increase in expression. For each MR, activated and repressed ARACNe-inferred targets are shown as red and blue bars, respectively, to indicate that MARINa considers them separately to compute MR significance, see Experimental Procedures for details. Due to the uncertainty in assigning directionality to a MR (see text), all significant MRs are included, comprising both those predicted as positive (Zdhhc2, Cbfa2t3, App, Zfp235, Hivep2, Epas1, Camta1, Dennd4a, Smad3 and Mycn) and negative (Zfp646, Tgif1, Hmgb2, Myb, Tcf7, Trp53, Zfp503, Atf5, Nfkb1, Psmc3ip, Tcf3, Zfp36l1 and Irx5) MRs. To improve the Figure readability by showing a concordant enrichment of activated and repressed targets, the x-axis is inverted for MRs predicted to be negative regulators of mSOD1 mediated toxicity. This transformation affects only the display of the results but not their analysis and interpretation. TF names and False discovery rate (FDR) significance for differential MR activity, as computed by MARINa, are shown to the right of the ‘bar-code’ plot. The first and second columns of the heatmap show a color code representative of differential TF activity (absolute normalized enrichment score) and differential TF expression (z-score), respectively. Numbers on the right of the heatmap represent the TF rank among differentially expressed genes (e.g., Zdhhc2 is the 6,375th most differentially expressed gene). See also Figure S1 and Table S2.
MARINa-based interrogation of the BrainNet using the 3d-GES identified 23 candidate MRs (FDR < 1%) as potential drivers of mSOD1 ACM-induced ES-MN death (Figure 3B and Table S2). As we have observed in other systems, despite robust changes in expression of downstream genes in response to the 3-day exposure to ACM, only three of the candidate MRs (Atf5, Hmgb2 and App) were differentially expressed between mSOD1 and control ACM-treated ES-MNs (FDR < 1%), and none of them was even in the top 100 most differentially-expressed genes (ranking at positions 170, 368 and 584, respectively; Figure 3B).
Experimental validation of MARINa-inferred Master Regulators
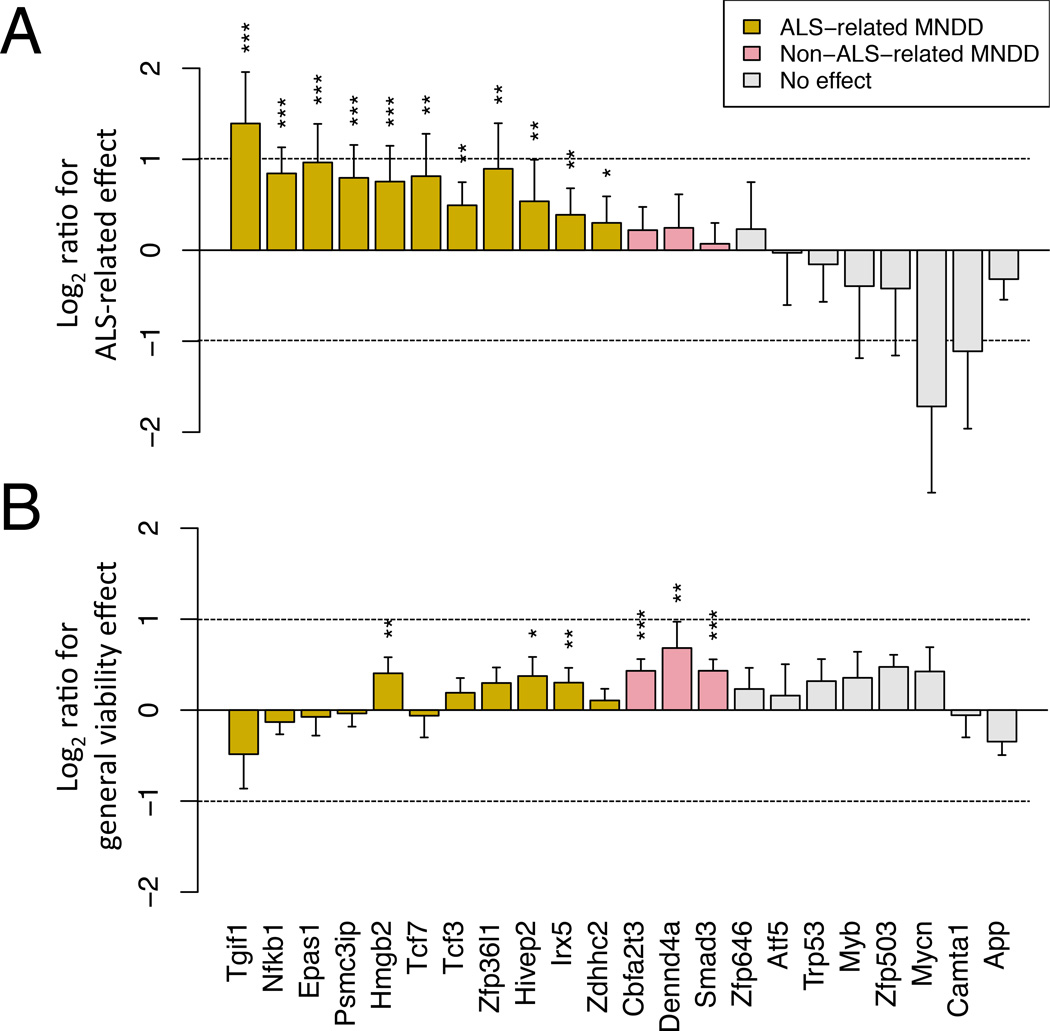
To validate these candidate MRs, we tested their role in astrocyte-induced MN death using lentiviral shRNA silencing in primary embryonic MNs grown on mSOD1 or NTg astrocyte monolayers (AML); Zfp235 could not be tested due to shRNA unavailability. For each of the 22 tested genes, two distinct, validated MISSION® shRNA hairpins were utilized. Thus, 66 shRNA-expressing viral vectors were individually used to infect primary MNs in suspension, prior to seeding them on an astrocyte monolayer (AML). Each was confirmed to produce ≥60% reduction in targeted MR expression at 2 days post-transduction, as assessed by qRT-PCR (Figure S2A). Primary MNs were transduced with the validated shRNAs, plated on mSOD1 AML, and counted after 7 days (Figure S2C). To reduce the risk of changes due to off-target effects, only consistent results reproduced by two distinct shRNA hairpins were considered. We first analyzed the degree to which MR silencing improved the ratio of the number of MNs grown on mSOD1 AML divided by the number of MNs grown on NTg AML. Once normalized for minor changes induced by empty vector control (Figure S2C), we found that the silencing 11 of the 22 MRs increased the mSOD1 AML:NTg AML MN ratio (Figure 4A; yellow bars, FDR < 0.1, 1-tail Student t-test); hence, we termed these 11 genes as “ALS-related MN death drivers”. Of these, only eight (Tgif1, Nfkb1, Epas1, Psmc3ip, Tcf7, Tcf3, Zfp36l1, Zdhhc2) appeared as mSOD1 astrocyte specific MN death drivers since their silencing increased the MN ratio without altering the number of MN grown on NTg AML (Figure 4B; yellow bars without asterisks) while the silencing of the three remaining (Hmgb2, Hivep2, Irx5) did increase both the ratio and the number of MN grown on NTg AML (Figure 4B; yellow bars with asterisks).
Figure 4. Candidate MR validation.
(A) ALS-related effect of shRNA mediated TF silencing on surviving MNs. The bars show the mean ± s.e.m for the ratio of the number of surviving ES-MN on mSOD1 vs. NTg AML following TF silencing, divided by the same ratio when the cells were transduced with the control empty vector (EV).
(B) General viability effect of each TF silencing. The bars show the mean ± s.e.m for the ratio of the number of surviving ES-MNs following shRNA mediated silencing of a specific TF vs. EV control, when cultured on an NTg astrocyte monolayer. * FDR < 0.1, ** FDR < 0.05, *** FDR < 0.01, Student’s t-test. The bar colors indicate the functional group of the driver gene, either ALS-related MN death driver (ALS-related MNDD) or non-ALS-related death driver (Non-ALS-related MNDD). See also Figure S2.
Furthermore, the silencing of three additional genes (Cbfa2t3, Dennd4a, Smad3) improved the number of MN grown on NTg AML but did not alter the mSOD1 AML:NTg AML MN ratio. As such, these were classified as “non-ALS-related MN death drivers” (Figure 4B, pink bars).
Of note, 8 of 22 genes (36%) were validated as mSOD1 astrocyte specific MN death drivers, which represents a proportion much higher than the one expected by chance (p = 4.4×10−13, Binomial test), assuming a true positive rate (TPR) <1% as reported in screens for genes affecting viability in mutation-specific manner (Beronja et al., 2013; Rosenbluh et al., 2012). Overall, 14 of 22 candidate MRs (64%) were validated as MN death drivers irrespective of the astrocyte genotypes, which is also highly significant (p = 8.6×10−11), assuming a TPR ≤ 10%, as previously observed for non-specific viability assays (Beronja et al., 2013).
NF-κB is activated in response to mSOD1 ACM and mediates ES-MN cell death
Among the 14 validated MRs, silencing of NF-κB subunit encoded by the Nfkb1 gene – as evidenced by ≥75% reduction of both the mRNA and protein levels (Figure S2A and B) – was associated with the most statistically significant reduction in mSOD1 AML-induced MN loss (Figure 4A), without any effect on MN number exposed to NTg astrocytes (Figure 4B). While NF-κB subunit p65 overexpression in spinal MNs from patients with ALS has been reported (Jiang et al., 2005) and NF-κB activation in microglia has been linked to MN degeneration in ALS models (Frakes et al., 2014; Swarup et al., 2011), its role as a causal driver of MN death remains to be established. Thus, to provide additional validation of our unbiased approach as a methodology for the identification of causal functional determinants of MN neurodegeneration, we decided to further investigate the role of NF-κB in MN degeneration.
NF-κB is a heterodimeric protein complex composed of different subunits of the Rel TF family, which upon activation, translocates from the cytosol to the nucleus, where it binds to the κB motif of target genes to regulate their expression (Ghosh et al., 2012). Thus, to confirm NF-κB activation and to identify the subunits involved, we performed a NF-κB DNA-binding ELISA. This assay revealed a strong but transient increase in DNA binding for the p50 and p65 subunits in nuclear extracts of purified ES-MNs exposed to mSOD1 ACM (Figure 5A). Conversely, no change in DNA binding was observed for NF-κB subunits p52 and RelB (Figure S3A). Given the robust ELISA results obtained for the p65 subunit (Figure 5A), we also examined its subcellular partition by Western blot as an additional sign of NF-κB canonical pathway activation in our ALS model. We found that the nuclear, but not the cytosolic, content of p65 was increased in MNs exposed to mSOD1 but not NTg ACM (Figure 5B and Figure S3B). In light of these results, we returned to our 3d-GES data (Table S1) and found that none of the NF-κB subunits was differentially expressed at the mRNA level, but that p50 and p65 encoded by Nfkb1 and RelA were significantly dysregulated at the protein activity level (FDR < 0.05) in MNs exposed to mSOD1 vs. NTg ACM, as inferred by the MARINa analysis (Table S2). Thus, collectively, these results support the notion that mSOD1 astrocyte-induced MN toxicity is associated with NF-κB activation via its canonical (p65-p50 complex) rather than non-canonical (p52-RelB complex) pathway. Critically, such a molecular event was successfully detected by the interactome-based unbiased approach.
Figure 5. NF-κB pathway is activated in ES-MNs exposed to mSOD1 ACM.
(A) DNA-binding ELISA assay for the canonical NF-κB pathway subunits p65 and p50 in nuclear extracts from purified ES-MNs exposed to either mSOD1 ACM or NTg ACM for 0 (just before adding the ACM) to 7 days. The o.d. values are normalized to the o.d. of ES-MNs exposed to NTg ACM for 0 days. *p < 0.05, **p < 0.01, ***p < 0.001; Newman-Keuls post-hoc.
(B) Western blot analysis for NF-κB p65 subunit content in the nuclear fractions from purified ES-MNs exposed for 1 to 4 days to either mSOD1 or NTg ACM. Histone H3 is used as a nuclear housekeeping marker. NF-κB control (Ctrl) is a cell extract provided from Cell Signaling.
(C) Western blot analysis of phosphorylated IκB (p-IκB) and total IκB from purified ES-MNs exposed to either mSOD1 or NTg ACM for 1, 2, 3 or 7 days. β-actin is the loading control. p-IκB protein content is normalized by the corresponding amount of total IκB. See also Figure S3.
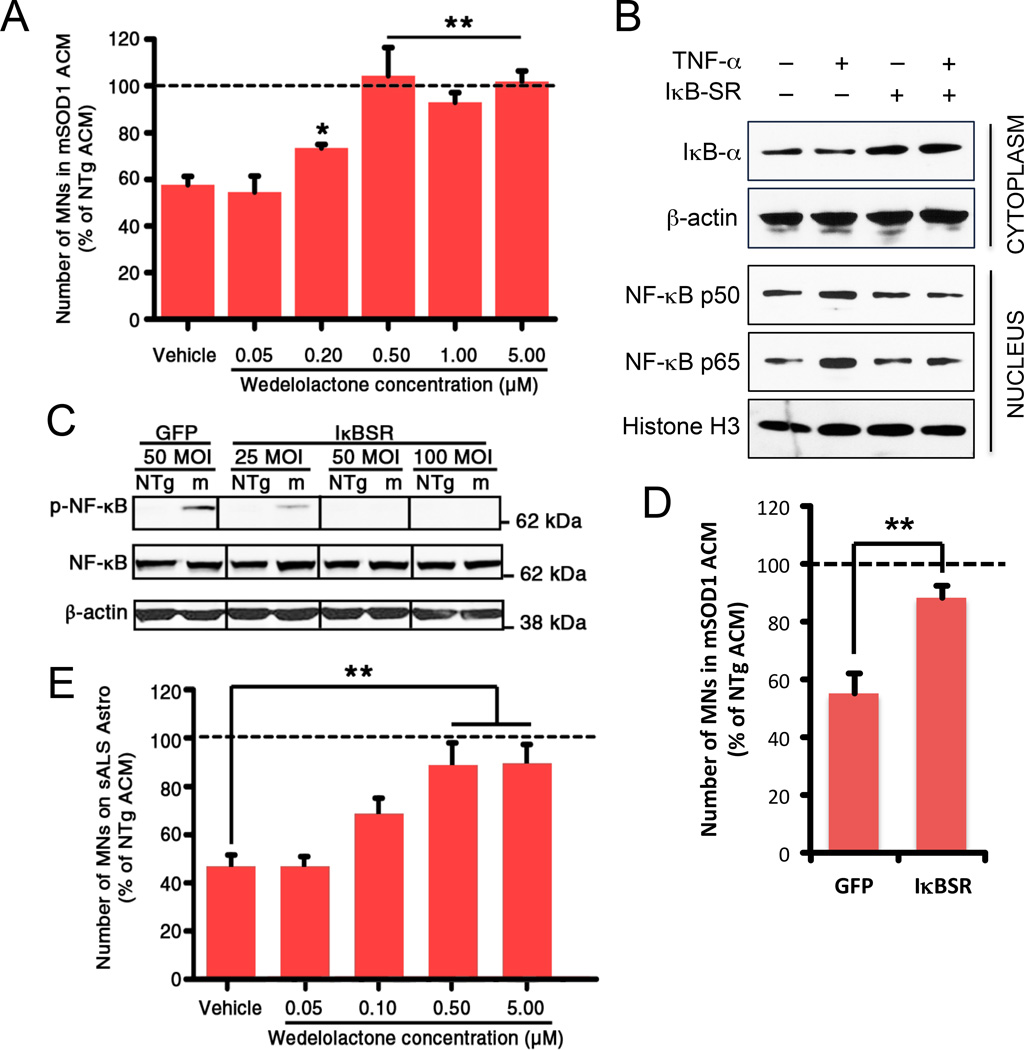
Signals that induce NF-κB activity trigger the inactivation, by phosphorylation, of its endogenous inhibitor, IκB (Ghosh et al., 2012) and we observed an increase in cytosolic phosphorylated IκB in MNs exposed to mSOD1 ACM that paralleled the nuclear accumulation of p65 (Figure 5C and 5B). Given this finding, we next decided to assess the effect of NF-κB inhibition on ACM-induced MN death by using wedelolactone, a natural compound that prevents the phosphorylation and ensuing degradation of IκB (Kobori et al., 2004). Consistent with its known mechanism of action, wedelolactone stabilized IκB in cultured MNs (Figure S4A) and protected against ES-MN loss caused by mSOD1 ACM (Figure 6A). This effect was specific to NF-κB-mediated cell death, since when we exposed primary MNs to two other stressors known to kill MNs via non-NF-κB mechanisms – Fas ligand (Raoul et al., 2002) and domoic acid (Xu et al., 2008) – wedelolactone conferred no protection (Figure S4B). These results were corroborated by a different NF-κB antagonist, namely BMS-345541, which also fully protected MNs demise in response to mSOD1 astrocytes (Figure S4C).
Figure 6. Inhibition of NF-κB activity rescues ES-MNs exposed to mSOD1 ACM.
(A) At 2 day post-plating, purified ES-MNs exposed to either mSOD1 or NTg ACM are treated with Wedelolactone at concentrations ranging from 0.05 to 5 µM. After seven days, the MNs are fixed and their survival is assessed by immunostaining. *p < 0.05, **p < 0.01; Newman-Keuls post-hoc.
(B) Western blot analysis of IκB-a protein levels in the cytoplasmic fractions, and NF-κB subunits p50 and p65 in the nuclear fractions, from ES-MNs transduced with adenoviral vectors carrying GFP or super repressive IκB (IκB-SR) coding sequences (MOI 50). NF-κB translocation to the nucleous was induced by TNF-a (10 ng/ml).
(C) Western blot illustrating the effect of adenoviral-mediated expression of super repressive IκB (IκBSR) on phosphorylated p65 subunit (p-NF-κB) in extracts from purified ES-MNs exposed to either mSOD1 or NTg ACM for 1 day in comparison to ES-MNs transduced with adenovirus expressing green fluorescent protein (GFP) only. β-actin is the loading control.
(D) Effect of ES-MNs transduction by IκBSR (50 MOI) against mSOD1 ACM ES-MN toxicity. **p < 0.01; Newman-Keuls post-hoc.
(E) Dose-dependent protection of wedelolactone of human ES-MNs exposed to AML sporadic ALS or non-disease control astrocytes. *p < 0.05, **p < 0.01; Newman-Keuls post-hoc. See also Figure S4.
Because small molecules like wedelolactone or BMS-345541 are often fraught with off-target effects, we also assessed the role of NF-κB in mSOD1-mediated MN loss by genetic means using an adenoviral vector (AV) expressing a dominant-negative IκB mutant. Consistent with the expected inhibition of NF-κB by the IκB mutant, we observed increased levels of IκB directly associated with the multiplicity of infection (MOI) used to transduce ES-MNs (Figure S4D), a consequent decreased in nuclear p65 and p50 (Figure 6B), and decreased levels of phosphorylated p65 (Figure 6C). More importantly, ES-MNs transduced with MOI ≥50 not only showed near complete absence of p65 phosphorylation (Figure 6C), but also were resistant to mSOD1 ACM-induced toxicity (Figure 6D).
Lastly, we targeted NF-κB in our fully humanized co-culture model composed of astrocytes from adult human sporadic ALS cases and human ES cell-derived MNs (Re et al., 2014). As in the mouse mSOD1 model, wedelolactone reduced the death of human ES-MNs exposed to human sporadic ALS astrocytes in a dose-dependent manner (Figure 6E). Thus, upon exposure to familial or sporadic ALS astrocytes, neuronal NF-κB, in contrast to its roles in many other settings and cell types, is instrumental in driving MN death.
Discussion
Interrogation of cell context-specific interactomes assembled de novo from large molecular profiles allows the identification of key genetic modules and causal MRs underlying physiologic and pathologic phenotypes (Aytes et al., 2014; Carro et al., 2010; Lefebvre et al., 2010), as well as of causal driver mutations responsible for the aberrant activity of these regulators (Chen et al., 2014). However, interactome analysis for neurobiological phenotypes remains poorly developed, with only a handful of relevant examples. For instance, integrative analysis of a co-expression network using both gene expression and GWAS data was used to rank-order genes that may play a role in late-onset Alzheimer’s disease (Zhang et al., 2013), and to elucidate networks that may play roles in autism and schizophrenia (Chang et al., 2015; Gilman et al., 2012). Despite these examples, we are not aware of attempts made to interrogate causal interactomes as a way of systematically elucidating and experimentally validating MRs of neurological disease. This is particularly vital in adult-onset neurodegenerative disorders, where identification of potential therapeutic targets is strongly dependent on establishing more direct clues about the specific mechanism of neuronal demise.
In the present manuscript, we report an unbiased, systems biology analysis of gene expression signatures from an in vitro ALS model, using an interactome that had been reverse-engineered from mouse whole-brain gene expression profiles. The proposed interactome-based analysis was successful in identifying a comprehensive set of regulatory genes driving MN degeneration in a highly relevant cell model of ALS, including 14 causal effector genes as well as their transcriptional programs. We were able to experimentally confirm these inferences by showing that lentiviral-mediated shRNA silencing of 11 of these MRs in spinal MNs reduced mSOD1 astrocyte toxicity. Of the 23 candidate MRs predicted to regulate MN transcriptional response following mSOD1 ACM exposure, only three were significantly differentially expressed and none was in the top 100 most differentially expressed. This finding is consistent with previous reports (Carro et al., 2010; Lefebvre et al., 2010) and the fact that changes in MR activity are likely post-translationally driven. Therefore, the proposed methodology represents a significant progress over more conventional GEP analyses, as it reliably and systematically identified causal functional determinants of the degenerative phenotype that were not differentially expressed.
Among the MRs whose silencing mitigated the MN death phenotypes, eight (Tgif1, Nfkb1, Epas1, Psmc3ip, Tcf7, Tcf3, Zfp36l1, Zdhhc2) reduced neuronal death following exposure to mSOD1 astrocytes, without significantly altering the number of surviving MNs following exposure to NTg astrocytes (Figure 4). This phenotypic pattern is consistent with their specific role as key effectors of the lethal programs responsible for killing MNs in response to mSOD1 astrocytes. Each of these MRs has been linked to regulation of cell survival/death especially during development and, according to the Allen Spinal Cord Atlas, they are all expressed in the spinal cord of the mouse. However, except for NF-κB (Mattson and Camandola, 2001), none of these MRs has so far been associated to adult-onset chronic neurodegenerative disorders. It will be thus exciting to link the developmental role of these MRs to their function in neurodegeneration.
While in most instances NF-κB activation is known for its pro-survival role, several lines of evidence suggest a pathogenic role of this TF in ALS (Akizuki et al., 2013; Jiang et al., 2005; Maruyama et al., 2010; Swarup et al., 2011). Yet, NF-κB inactivation in MNs is not known to abrogate mSOD1 astrocyte-mediated MN death. Furthermore, among all the other identified MRs, we realized that high-quality reagents were available for the study of NF-κB. Thus, NF-κB provided the most direct and concrete proof-of-principle that our unbiased approach can elucidate potentially meaningful neurodegenerative determinants. Accordingly, we performed a series of in-depth functional studies on NF-κB in our in vitro models of ALS. These investigations revealed that NF-κB canonical pathway was activated in MNs exposed to mSOD1 ACM and that its inhibition protects these neurons against mouse mSOD1-expressing and human sporadic ALS astrocytes. Our results thus confirm that NF-κB is a driver of MN death and that this action is conserved between mouse and human and shared between familial and sporadic ALS models. Our findings also imply that MN death is mediated by NF-κB activation within MNs and not glial cells, since astrocyte specific NF-κB inhibition does not attenuate MN death in transgenic mSOD1 mice (Crosio et al., 2011) and our in vitro ALS models are essentially devoid of microglia (Nagai et al., 2007; Re et al., 2014) which were shown to kill cultured MNs by conventional NF-κB-dependent neuroinflammation (Frakes et al., 2014).
Our demonstration that inferred neuron-specific necroptosis genes (Hitomi et al., 2008) are enriched in the 3d-GES may be regarded as an independent confirmation of our recent report that, in both mouse and human in vitro models of ALS, MNs die by necroptosis (Re et al., 2014). However, we noted two intriguing facts with respect to our findings and necroptosis. First, MN degeneration is driven by activation and not repression of NF-κB, as reported in other necroptosis contexts (Pasparakis and Vandenabeele, 2015). For instance, in response to inflammatory stimuli in other cell types, NF-κB activation is promoted by polyubiquitination of receptor-interacting protein-1 (RIP1) whereas necroptosis is triggered by RIP1 deubiquitination and phosphorylation (Pasparakis and Vandenabeele, 2015), two processes that would appear mutually exclusive. Furthermore, it has been reported that NF-κB can upregulate c-FLIPL, a caspase-like molecule that forms a heterodimer with caspase-8 and FADD (Micheau et al., 2001), and that the c-FLIPL-caspase-8-FADD complex inhibits RIP3 activation and, consequently, necroptosis (Dillon et al., 2012). A possible explanation to this paradox may lie in the timing of the two events: upon mSOD1 ACM exposure, p65 and p50 DNA binding peak at 1 day (Figure 5A) whereas overt MN death occurs later (Figure 1C) suggesting that NF-κB activation precedes rater then coincides with the induction of necroptosis. Second, our analysis is restricted to genes annotated as transcriptional regulators. Of the 40 inferred neuron-specific necroptosis genes (Hitomi et al., 2008), only three are represented as transcriptional regulators in our interactome (Zfp521, Zmym3 and Sall2), and could have thus been inferred by our analysis. Since these genes were identified by siRNA screen of zVAD-fmk-exposed fibrosarcoma cells followed by neuron gene-expression based filtering – and not from ACM-exposed MNs – the probability that none of these three genes is a MN-specific regulator of necroptosis is quite large (p = 0.38, based on a 27.5% overlap of the 40 gene signature with MR targets). As a result, while the full signature may be enriched in bona fide neuronal necroptosis regulators, there is no statistically significant expectation that any of these three genes may regulate necroptosis in MNs. However, while none of these genes were identified as MRs of MN death (Figure 3), the targets (i.e., regulons) of four of the 11 validated MRs showed statistically significant overlap with the 40 inferred neuron-necroptosis genes (Epas1, Hivep2, Tgif1 and Tcf7; FDR < 0.1, Fisher’s exact test), suggesting a potential role of these MRs as drivers of necroptosis in MNs. Clearly, future studies designed to assess the necroptosis regulatory network, specifically in MNs, are warranted to link ALS astrocyte-induced MN toxicity to this particular form of programmed cell death. Nonetheless, the current knowledge of necroptosis signaling (Pasparakis and Vandenabeele, 2015) taken together with the data demonstrating the mutual involvement of RIP1 and NF-κB in our ALS models, suggest that a member of the tumor necrosis receptor superfamily (Tnfrsf) most likely transduces the astrocyte-mediated, extracellular toxic signal. Since Tnfrsf 1, 6, and 16 have previously been excluded (Nagai et al., 2007; Re et al., 2014) and transcripts for Tnfrsf 4, 8, 9, 11, 13, 14, 17, and 25 were not detected in our ES-MNs (data not shown), this list can be narrowed considerably to only a handful of potential candidates, whose role in engaging necroptosis in MNs deserves further investigation.
In conclusion, early diagnosis and evaluation of efficacy in clinical trials for all neurodegenerative disorders are hindered by the absence of robust biomarkers for the first phase of the disease. In addition, there is a chronic lack of validated therapeutic targets to allow for pharmacological intervention at early stages of disease. Systems biology analysis of a cell culture system that models non-cell-autonomous elements of neurodegeneration allowed us to identify a statistically significant and numerically limited set of potential MRs of MN degeneration. These drivers – including NF-κB for which we provide proof-of-principle validation – may in the future serve both as biomarkers and as therapeutic targets for ALS. Furthermore, an extension of our approach holds the promise of shedding comparable light on the regulatory networks that are affected in other neurodegenerative diseases, which constitute a major challenge to society over the coming decades. Indeed, regardless of whether these genes will be confirmed as key effectors of human ALS phenotype, these results show that MR analysis is extremely effective in identifying driver genes responsible for the implementation of neurodegenerative phenotypes. In a field where discovery of key mediators of disease phenotype has proven both complex and inefficient, the ability to elucidate and validate more than a dozen causal modulators of a death phenotype suggests that this kind of analyses may significantly complement more traditional assays for the study of individual genes. Thus, further availability of human-derived ALS signature will allow to either confirm the identity of the currently reported genes or to discover their human counterpart.
Experimental Procedures
Primary cultures of astrocytes and preparation of astrocyte conditioned media
ACMs were prepared from astrocyte cultures of different genotypes, collected and frozen in several aliquots as described before (Nagai et al., 2007; Re et al., 2014). On the day of the assay, ACM was supplemented with 4.5 mg ml−1 D-glucose (final concentration), 100 U ml−1 penicillin and 100 µg ml−1 streptomycin (penicillin/streptomycin, Invitrogen), and trophic factors (0.5 ng ml−1 glial cell line-derived neurotrophic factor, 1 ng ml−1 brain-derived neurotrophic factor, and 10 ng ml−1 ciliary neurotrophic factor [trophic factor cocktail, R&D Systems]). The ACM was then filtered (0.22 µm cellulose acetate membrane filter, Nalgene) before being added to the MN cultures.
Primary culture of human astrocytes
Human astrocyte cultures were made following the protocol previously described (Re et al., 2014) using fresh human autopsied tissues obtained within 11 h of death from Columbia Medical Center Morgue and the National Disease Research Interchange (NDRI, PA, USA). Produced astrocytes were frozen in RecoveryTM Cell Culture Freezing Medium (Invitrogen) and stored in liquid nitrogen until used. All of the human astrocytes used in this study were passaged for a maximum of 5.
Production of primary MN cultures
Spinal neuronal cultures were performed from embryonic 12.5 days Hlxb9-GFP1Tmj transgenic rodents as previously described (Nagai et al., 2007). The culture medium was either ACM or MN medium (neurobasal medium; Invitrogen) containing 2% horse serum (heat inactivated; Invitrogen), 2% B27 supplement, 0.5 mM glutamine (Invitrogen), 25 µM 2-mercaptoethanol, and penicillin/streptomycin supplemented with the cocktail of trophic factors described above.
Production of ES-MN cultures and purification methods
Both human and mouse ES-MNs were produced as previously described (Nagai et al., 2007; Re et al., 2014). For purification of the mouse ES-MNs (see below), ES derived from transgenic Hlxb9-GFP1Tmj mice (Wichterle et al., 2002) were engineered to expressed eGFP and CD2 driven by the mouse Hb9 promoter. The produced cell suspensions were washed with the MN medium described above, resuspended (∼20 million cells ml−1) in PBS containing 1% BSA, and processed by fluorescence-activated cell sorting (FACS) or by magnetic-activated cell sorting (MACS).
Cell sorting was performed using a Becton Dickinson FACS Aria cytometer (BD Biosciences) at the Flow Cytometry Core Facility of Columbia University (http://www.ccc.columbia.edu/FCCF/flow_cytometry.html). Forward scatter (FS) and side scatter (SSC) were collected through a filter. The GFP signal was collected in the FL1 channel. A light scatter gate was drawn in the SSC versus FS plot to exclude debris and include the viable cells. Cells in the gate were displayed in a single-parameter histogram for GFP and final gate settings determined to collect the GFP+ cells. Post-sorting, the cells were collected in MN medium supplemented with 3% fetal bovine serum. Before plating, the purified MN suspensions were collected by centrifugation (300xg, 5 min) and resuspended in MN medium.
As for MACS, upon dissociation and wash with L15 medium (Sigma) containing 50 µM EDTA, 4% BSA (Sigma), 25 mM glucose, 2% horse serum (Sigma), 40 µg ml−1 DNAse, 500 µg ml−1 Insulin (Sigma), 0.01M Putrecine (Sigma), 10 mg ml−1 conalbumin (Sigma), 30 µM Na2SeO3 (Sigma), cell suspensions were incubated for 20 min at 4°C in 80 µl of L15 medium containing 2.1 µg of anti-rat CD2 antibody (Invitrogen) per 15 million dissociated cells. After washing with L15 medium, cells were incubated for 20 min at 4°C with an anti-mouse antibody conjugated to magnetic microbeads (Miltenyi Biotec). Finally, cells were passed through a magnetic column to separate CD2-GFP+ cells from the other types. Post-sorting, the cells were collected in MN medium and were plated for culture for 2 days in MN medium before being exposed to ACM.
Immunocytochemistry
For immunocytochemistry, cells were processed as we previously described (Nagai et al., 2007; Re et al., 2014). Primary antibodies used were: rabbit polyclonal anti–eGFP (1:1000; Molecular Probes) and anti–HB9 (1:1000; Abcam), and mouse monoclonal anti–MAP2 (1:1000; Chemicon).
Gene expression profile analysis of ES-MNs
Total mRNA was isolated at 1 and 3 days (see Supplemental Experimental Procedures for details on strategies to reduce potential batch effects) using TRI Reagent Solution (Ambion) and MagMAX-96 total RNA Isolation kit (Ambion). A minimum of 20 million, 100 nucleotides single end reads were obtained by a HiSeq2000 platform (Illumina), which were mapped to the mouse genome (UCSC-mm9) with Bowtie (Langmead et al., 2009) and counted at the gene level using GenomicFeatures R-system package (Bioconductor (Gentleman et al., 2004)). The expression data was then normalized to account for different library sizes and the variance was stabilized based on the negative binomial distribution as implemented in the DESeq R-system package (Bioconductor). The raw and normalized data are available from the Gene Expression Omnibus (GSE49023) and figshare (http://dx.doi.org/10.6084/m9.figshare.809573), respectively. To account for batch effects, differential gene expression analysis was performed by fitting a two-factor, linear mixed-effects model to the normalized data, independently estimating the variance due to treatment and MN culture conditions (batch effect, see Supplemental Experimental Procedures for details). Statistical significance was estimated by a moderated Student’s t-test implemented in the limma package (Dudoit et al., 2003) for the R-system (Bioconductor).
Enrichment of the 1-GES and 3-GES on a set of 40 neuron-specific necroptosis associated genes (Hitomi et al., 2008) was performed by Gene Set Enrichment Analysis as described in the original publication (Subramanian et al., 2005).
Master Regulators analysis
Interactomes were reverse engineered by ARACNe (Margolin et al., 2006) from six different GEP data sets, for 1,857 human and 1,507 mouse transcription factors (see Supplemental Experimental Procedures for details on the datasets and algorithm). Human interactome genes were mapped to their mouse orthologs by the HomoloGene database (http://www.ncbi.nlm.nih.gov/homologene). All the interactomes used in this publication, as well as a sweave file including the source code for the analysis workflow, are available for download from figshare (http://dx.doi.org/10.6084/m9.figshare.809573).
We used an improved version of the MARINa original algorithm (Carro et al., 2010; Lefebvre et al., 2010) with the following modifications: (1) it tests for a global shift in the position of the regulons when projected on the GES instead of the Kolmorogov-Smirnov test statistics; (2) it test the enrichment of each tail, or globally, according to the regulator mode of action, either activator, repressor or mixed, respectively; (3) takes into account pleiotropic regulation of each target gene by multiple regulators when computing the enrichment. Statistical significance, including p-value and normalized enrichment score (NES), is estimated by comparison to a null model generated by permuting the samples uniformly at random 1,000 times. The MARINa algorithm is implemented in the “ssmarina” R-package available for download from figshare (http://dx.doi.org/10.6084/m9.figshare.785718).
Gene silencing in primary MNs using shRNA lentiviral particles
All lentiviral particle-packaged shRNAs in pLKO.1-puro plasmids were obtained from Sigma Mission (see Supplemental Experimental Procedures). As a control, MISSION® pLKO.1-puro Empty Vector Control Transduction Particles (#SHC001V; EV) was used in all of the experiments since during our pilot work we found no difference between EV and MISSION® pLKO.1-puro Non-Mammalian shRNA Control Transduction Particles which targets no known mammalian genes (#SHC002V).
Freshly dissociated primary MNs were obtained, as described above, and exposed to lentiviral particles at a MOI of 15. Then cells in suspension were centrifuged (800xg, 30 min) before to be resuspended in fresh MN medium and cultured for 7 days on mSOD1 AML. Thereafter, transduced cells were fixed and counted following the protocols mentioned above. In each experiment, some transduced MNs were also cultured without AML to verify the efficiency of silencing following mRNA extraction. In addition, in each experiment, non-transduced MNs were plated, in a separate 24-well plate, on NTg versus mSOD1 AML to confirm the toxicity of mSOD1 AML on MNs.
Real-time quantitative PCR
Confirmation of knockdown was performed by qRT-PCR using RNA samples obtained from 24- or 96-well cell extraction (RNAqueous Micro-Kit, Ambion, Invitrogen). RNA (50–100 ng) was first reverse transcribed to cDNA (500 ng µl−1) using Superscript II three-step reverse transcription PCR (Invitrogen). A two-step qRT-PCR was carried out by Realplex 4 Mastercycler PCR System (Eppendorf) using FAM-conjugated gene specific primers (Applied Biosystems, Invitrogen) in triplicate for each sample. VIC-conjugated GAPDH (Applied Biosystems), a standard and accepted housekeeping gene, was used as a control against which results were normalized. Melt-curve analysis was done for all samples to exclude the possibility of primer-dimer formation. Fold change was based on ΔΔCT values (Applied Biosystems Guide to PCR).
Nuclear/cytoplasmic subcellular fractionation
Harvested cells were homogenized in buffer A (250 mM sucrose, 20 mM HEPES, 10 mM KCl, 1.5 mM MgCl2, 2 mM EDTA, pH 7.4) supplemented with a protease inhibitor cocktail (Complete mini, Roche Applied Science). For each preparation, an aliquot of cell homogenate was saved and the rest was centrifuged (15,000 rpm, 15 min). The pellet (P1) was used for nuclear purification, whereas the supernatant (S1) was used for the cytosolic purification. To purify nuclei, P1 was re-homogenized in buffer B (2 M sucrose, 50 mM Tris-HCl, 5 mM MgCl2, 1 mM DTT and 1 mM PMSF, pH 7.4) supplemented with the protease inhibitor cocktail and then centrifuged (15,000 rpm, 15 min) through a cushion of buffer B. The resulting pellet was enriched with nuclei and was designated as nuclear fraction. As S1, it was centrifuged (15,000 rpm, 15 min). The resulting supernatant was further centrifuged (15,000 rpm, 15 min). The supernatant of the latter centrifugation was designated as the cytosolic fraction. After each preparation and prior to using, the purities of the subcellular fractions were confirmed by Western blot for subcellular markers: histone H3 for nuclei and GAPDH for cytosol.
DNA-binding assay
To assess NF-κB activation, we quantified the DNA-bound NF-κB subunits p50, p52, p65, and RelB using a commercial DNA-binding ELISA kit (TransAM®, Active Motif). For this assay, 20 µg of nuclear extracts, prepared as outlined above, were added per well in a 96-well plate with 30 µl Complete Binding Buffer provided with the kit and was run as recommended by the manufacturer. The colorimetric reaction was read with a spectrophotometer (Dtx 880 Multimode Detector, Beckman Coulter) at 450 nm.
Western blots of total and phosphorylated proteins
At the indicated selected time points after exposure to ACM or infection with viral vector, MNs were harvested in lysis buffer containing sodium orthovanadate and sodium fluoride to avoid dephosphorylation. The following primary antibodies were used: rabbit anti-total NF-κB p65 antibody (Cell Signaling), anti-phosphorylated NF-κB p65 antibody (Cell Signaling), anti-GAPDH antibody (Cell Signaling), anti-histone H3 (Cell Signaling), anti-total IκB (Cell Signaling), anti-phosphorylated IκB (Cell Signaling), and mouse anti- β-actin (Sigma). The dilution used for all the antibodies was 1:1000, except for β-actin, for which it was 1:20,000. Western blots using antibodies raised against the cytosolic and nuclear markers cited above were used as loading controls. All bands were quantified by Odyssey Infrared Imaging system using IRDye dye-labeled secondary antibodies (LI-COR). Results were expressed as ratios of arbitrary fluorescence units for the protein of interest over arbitrary fluorescence units for loading controls or as ratios of phosphorylated protein of interest over total protein of interest.
Pharmacological treatments and AV transduction
Wedelolactone (Calbiochem; final concentrations 50 nM–5 µM) or BMS-345541 (Selleckchem.com; 5 µM) were dissolved in DMSO. TNFα (Sigma) was added to cultures at a concentration of 10 ng ml−1 for either 20 min or 1 hr. Fas ligand (Sigma) and domoic acid (Sigma) were added to MNs at concentrations 10 ng ml−1 (Raoul et al., 2002) and 1 µM (Xu et al., 2008), respectively, after five days in vitro of being in fresh medium. Cell survival was evaluated at 7 days in vitro by counting eGFP+ neurons as described below. MNs were transduced with AV expressing either CMV-GFP or CMV-IκB-SR (Aleyasin et al., 2004) with a MOI ranging from 25 to 100.
Statistics for cell counting and Western blot
Each experiment was performed in 2 to 3 technical replicates and data are the average of 3 to 5 biological replicates. Results for the cell counting were the average of 3–6 coverslips. Each coverslip was counted in its entirety under fluorescent examination. All values were expressed as mean ± s.e.m unless stated otherwise. Differences between means were analyzed using a two-tailed Student’s t-test. Differences among means were analyzed using a two- or three-way ANOVA. When ANOVA showed significant differences, pairwise comparisons between means were tested by Newman-Keuls post-hoc testing. In all analyses, the null hypothesis was rejected at the 0.05 level. All statistical analyses were performed using SigmaStat (Systat Software Inc., San Jose, CA) or R (http://www.R-project.org/).
Supplementary Material
Acknowledgements
The authors wish to thank Dr. Sankar Ghosh for his advice and guidance on the NF-κB studies, Mr. James Caicedo for his invaluable assistance with the mouse cell cultures, and Dr. David Park from University of Ottawa, Canada, for kindly providing us with the AV super-repressor IκB mutant. This study was supported by Target-ALS, the ALS Association, Project A.L.S., NIH/NINDS Grants NS072428-04, NS088009-02, NS078614-02, NS062055-03, NS042269-05A2, NS072182-01, the NIH Roadmap National Centers for Biomedical Computing (5U54CA121852-08), the US Department of Defense (Grant W81XWH-12-1-0431; SRI 5-21306; W81XWH-13-1-0416). BI is recipient of a Brunie Award for Stem Cell Research from Columbia University. DBR is recipient of a Career Development Award from the NIEHS Center of Northern Manhattan (NIEHS ES009089). VLV and AJ are recipients of an Award for exchange programs between France and the United States from the Philippe Foundation.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author Contributions
B.I., M.J.A., D.B.R, F.L., C.E.H., A. C. and S.P. designed experiments; B.I. D.B.R., V.L.V., K.P., F.L., Su. P., R.P., C.Y., G.F.C., A.J., and S.P. were responsible for the experimental part of the study; G.F.C. and C.E.H. designed, created and validated the GFP/CD2 ES-MNs; M. J.A. and A.C. were responsible for the computational part of the study; B.I., D.B.R., F.L. and S.P. analyzed all the data related to the experimental part of the study; and M.J.A., F.L., C.E.H., A.C. and S.P. wrote the manuscript.
References
- Akizuki M, Yamashita H, Uemura K, Maruyama H, Kawakami H, Ito H, Takahashi R. Optineurin suppression causes neuronal cell death via NF-kappaB pathway. J Neurochem. 2013;126:699–704. doi: 10.1111/jnc.12326. [DOI] [PubMed] [Google Scholar]
- Aleyasin H, Cregan SP, Iyirhiaro G, O'Hare MJ, Callaghan SM, Slack RS, Park DS. Nuclear factor-(kappa)B modulates the p53 response in neurons exposed to DNA damage. J Neurosci. 2004;24:2963–2973. doi: 10.1523/JNEUROSCI.0155-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aytes A, Mitrofanova A, Lefebvre C, Alvarez MJ, Castillo-Martin M, Zheng T, Eastham JA, Gopalan A, Pienta KJ, Shen MM, et al. Cross-species regulatory network analysis identifies a synergistic interaction between FOXM1 and CENPF that drives prostate cancer malignancy. Cancer Cell. 2014;25:638–651. doi: 10.1016/j.ccr.2014.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basso K, Margolin AA, Stolovitzky G, Klein U, la-Favera R, Califano A. Reverse engineering of regulatory networks in human B cells. Nat Genet. 2005;37:382–390. doi: 10.1038/ng1532. [DOI] [PubMed] [Google Scholar]
- Beronja S, Janki P, Heller E, Lien WH, Keyes BE, Oshimori N, Fuchs E. RNAi screens in mice identify physiological regulators of oncogenic growth. Nature. 2013;501:185–190. doi: 10.1038/nature12464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carro MS, Lim WK, Alvarez MJ, Bollo RJ, Zhao X, Snyder EY, Sulman EP, Anne SL, Doetsch F, Colman H, et al. The transcriptional network for mesenchymal transformation of brain tumours. Nature. 2010;463:318–325. doi: 10.1038/nature08712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang J, Gilman SR, Chiang AH, Sanders SJ, Vitkup D. Genotype to phenotype relationships in autism spectrum disorders. Nat Neurosci. 2015;18:191–198. doi: 10.1038/nn.3907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen JC, Alvarez MJ, Talos F, Dhruv H, Rieckhof GE, Iyer A, Diefes KL, Aldape K, Berens M, Shen MM, et al. Identification of causal genetic drivers of human disease through systems-level analysis of regulatory networks. Cell. 2014;159:402–414. doi: 10.1016/j.cell.2014.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crosio C, Valle C, Casciati A, Iaccarino C, Carri MT. Astroglial inhibition of NF-kappaB does not ameliorate disease onset and progression in a mouse model for amyotrophic lateral sclerosis (ALS) PLoS One. 2011;6:e17187. doi: 10.1371/journal.pone.0017187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dillon CP, Oberst A, Weinlich R, Janke LJ, Kang TB, Ben-Moshe T, Mak TW, Wallach D, Green DR. Survival function of the FADD-CASPASE-8-cFLIP(L) complex. Cell reports. 2012;1:401–407. doi: 10.1016/j.celrep.2012.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudoit S, Gentleman RC, Quackenbush J. Open source software for the analysis of microarray data. Biotechniques. 2003;(Suppl):45–51. [PubMed] [Google Scholar]
- Frakes A, Ferraiuolo L, Haidet-Phillips AM, Schmelzer L, Braun L, Miranda CJ, Ladner KJ, Bevan AK, Foust KD, Godbout JP, et al. Microglia induce motor neuron death via the classical NF-κB pathway in amyotrophic lateral sclerosis. Neuron. 2014;81:1009–1023. doi: 10.1016/j.neuron.2014.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garden GA, La Spada AR. Intercellular (mis)communication in neurodegenerative disease. Neuron. 2012;73:886–901. doi: 10.1016/j.neuron.2012.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh G, Wang VY, Huang DB, Fusco A. NF-kappaB regulation: lessons from structures. Immunol Rev. 2012;246:36–58. doi: 10.1111/j.1600-065X.2012.01097.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilman SR, Chang J, Xu B, Bawa TS, Gogos JA, Karayiorgou M, Vitkup D. Diverse types of genetic variation converge on functional gene networks involved in schizophrenia. Nat Neurosci. 2012;15:1723–1728. doi: 10.1038/nn.3261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hitomi J, Christofferson DE, Ng A, Yao J, Degterev A, Xavier RJ, Yuan J. Identification of a molecular signaling network that regulates a cellular necrotic cell death pathway. Cell. 2008;135:1311–1323. doi: 10.1016/j.cell.2008.10.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang YM, Yamamoto M, Kobayashi Y, Yoshihara T, Liang Y, Terao S, Takeuchi H, Ishigaki S, Katsuno M, Adachi H, et al. Gene expression profile of spinal motor neurons in sporadic amyotrophic lateral sclerosis. Ann Neurol. 2005;57:236–251. doi: 10.1002/ana.20379. [DOI] [PubMed] [Google Scholar]
- Klein U, Tu Y, Stolovitzky GA, Keller JL, Haddad J, Jr, Miljkovic V, Cattoretti G, Califano A, Dalla-Favera R. Transcriptional analysis of the B cell germinal center reaction. Proc Natl Acad Sci USA. 2003;100:2639–2644. doi: 10.1073/pnas.0437996100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobori M, Yang Z, Gong D, Heissmeyer V, Zhu H, Jung YK, Gakidis MA, Rao A, Sekine T, Ikegami F, et al. Wedelolactone suppresses LPS-induced caspase-11 expression by directly inhibiting the IKK complex. Cell Death Differ. 2004;11:123–130. doi: 10.1038/sj.cdd.4401325. [DOI] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefebvre C, Rajbhandari P, Alvarez MJ, Bandaru P, Lim WK, Sato M, Wang K, Sumazin P, Kustagi M, Bisikirska BC, et al. A human B-cell interactome identifies MYB and FOXM1 as master regulators of proliferation in germinal centers. Mol Syst Biol. 2010;6:377. doi: 10.1038/msb.2010.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margolin AA, Nemenman I, Basso K, Wiggins C, Stolovitzky G, Dalla Favera R, Califano A. ARACNE: an algorithm for the reconstruction of gene regulatory networks in a mammalian cellular context. BMC Bioinformatics. 2006;(7 Suppl 1):S7. doi: 10.1186/1471-2105-7-S1-S7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruyama H, Morino H, Ito H, Izumi Y, Kato H, Watanabe Y, Kinoshita Y, Kamada M, Nodera H, Suzuki H, et al. Mutations of optineurin in amyotrophic lateral sclerosis. Nature. 2010;465:223–226. doi: 10.1038/nature08971. [DOI] [PubMed] [Google Scholar]
- Mattson MP, Camandola S. NF-kappaB in neuronal plasticity and neurodegenerative disorders. J Clin Invest. 2001;107:247–254. doi: 10.1172/JCI11916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Micheau O, Lens S, Gaide O, Alevizopoulos K, Tschopp J. NF-kappaB signals induce the expression of c-FLIP. Mol Cell Biol. 2001;21:5299–5305. doi: 10.1128/MCB.21.16.5299-5305.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagai M, Re DB, Nagata T, Chalazonitis A, Jessell TM, Wichterle H, Przedborski S. Astrocytes expressing ALS-linked mutated SOD1 release factors selectively toxic to motor neurons. Nat Neurosci. 2007;10:615–622. doi: 10.1038/nn1876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papadeas ST, Kraig SE, O'Banion C, Lepore AC, Maragakis NJ. Astrocytes carrying the superoxide dismutase 1 (SOD1G93A) mutation induce wild-type motor neuron degeneration in vivo. Proc Natl Acad Sci U S A. 2011;108:17803–17808. doi: 10.1073/pnas.1103141108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasparakis M, Vandenabeele P. Necroptosis and its role in inflammation. Nature. 2015;517:311–320. doi: 10.1038/nature14191. [DOI] [PubMed] [Google Scholar]
- Raoul C, Estevez A, Nishimune H, Cleveland D, deLapeyriere O, Henderson C, Haase G, Pettmann B. Motoneuron Death Triggered by a Specific Pathway Downstream of Fas. Potentiation by ALS-Linked SOD1 Mutations. Neuron. 2002;35:1067–1083. doi: 10.1016/s0896-6273(02)00905-4. [DOI] [PubMed] [Google Scholar]
- Re DB, Le Verche V, Yu C, Amoroso MW, Politi KA, Phani S, Ikiz B, Hoffmann L, Koolen M, Nagata T, et al. Necroptosis Drives Motor Neuron Death in Models of Both Sporadic and Familial ALS. Neuron. 2014;81:1001–1008. doi: 10.1016/j.neuron.2014.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renton AE, Chio A, Traynor BJ. State of play in amyotrophic lateral sclerosis genetics. Nat Neurosci. 2014;17:17–23. doi: 10.1038/nn.3584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenbluh J, Nijhawan D, Cox AG, Li X, Neal JT, Schafer EJ, Zack TI, Wang X, Tsherniak A, Schinzel AC, et al. beta-Catenin-driven cancers require a YAP1 transcriptional complex for survival and tumorigenesis. Cell. 2012;151:1457–1473. doi: 10.1016/j.cell.2012.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swarup V, Phaneuf D, Dupre N, Petri S, Strong M, Kriz J, Julien JP. Deregulation of TDP-43 in amyotrophic lateral sclerosis triggers nuclear factor kappaB-mediated pathogenic pathways. J Exp Med. 2011;208:2429–2447. doi: 10.1084/jem.20111313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tothill RW, Tinker AV, George J, Brown R, Fox SB, Lade S, Johnson DS, Trivett MK, Etemadmoghadam D, Locandro B, et al. Novel molecular subtypes of serous and endometrioid ovarian cancer linked to clinical outcome. Clin Cancer Res. 2008;14:5198–5208. doi: 10.1158/1078-0432.CCR-08-0196. [DOI] [PubMed] [Google Scholar]
- Wichterle H, Lieberam I, Porter JA, Jessell TM. Directed differentiation of embryonic stem cells into motor neurons. Cell. 2002;110:385–397. doi: 10.1016/s0092-8674(02)00835-8. [DOI] [PubMed] [Google Scholar]
- Xu R, Tao Y, Wu C, Yi J, Yang Y, Yang R, Hong D. Domoic acid induced spinal cord lesions in adult mice: evidence for the possible molecular pathways of excitatory amino acids in spinal cord lesions. Neurotoxicology. 2008;29:700–707. doi: 10.1016/j.neuro.2008.04.011. [DOI] [PubMed] [Google Scholar]
- Yamanaka K, Chun SJ, Boillee S, Fujimori-Tonou N, Yamashita H, Gutmann DH, Takahashi R, Misawa H, Cleveland DW. Astrocytes as determinants of disease progression in inherited amyotrophic lateral sclerosis. Nat Neurosci. 2008;11:251–253. doi: 10.1038/nn2047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B, Gaiteri C, Bodea LG, Wang Z, McElwee J, Podtelezhnikov AA, Zhang C, Xie T, Tran L, Dobrin R, et al. Integrated systems approach identifies genetic nodes and networks in late-onset Alzheimer's disease. Cell. 2013;153:707–720. doi: 10.1016/j.cell.2013.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.