Abstract
Ligands possessing different physico-chemical structures productively interact with G protein-coupled receptors generating distinct downstream signaling events due to their abilities to activate/select idiosyncratic receptor entities (‘receptorsomes’) from the full spectrum of potential receptor partners. We have employed multiple novel informatic approaches to identify and characterize the in vivo transcriptomic signature of an arrestin-signaling biased ligand, [D-Trp12,Tyr34]-bPTH(7-34), acting at the parathyroid hormone type 1 receptor (PTH1R), across six different murine tissues after chronic drug exposure. We are able to demonstrate that [D-Trp12,Tyr34]-bPTH(7-34) elicits a distinctive arrestin-signaling focused transcriptomic response that is more coherently regulated, in an arrestin signaling-dependent manner, across more tissues than that of the pluripotent endogenous PTH1R ligand, hPTH(1-34). This arrestin-focused response signature is strongly linked with the transcriptional regulation of cell growth and development. Our informatic deconvolution of a conserved arrestin-dependent transcriptomic signature from wild type mice demonstrates a conceptual framework within which the in vivo outcomes of biased receptor signaling may be further investigated or predicted.
Keywords: G protein-coupled receptor, signaling bias, arrestin, informatic, transcriptome, in vivo
1. INTRODUCTION
The therapeutic targeting of heptahelical G protein-coupled receptors (GPCRs) has proved enormously successful as nearly half of the current pharmacopeia is composed of GPCR-regulating ligands of nearly every physico-chemical type [1]. Even with this success the functional impact of GPCR research upon therapeutic design is currently at a stage of enormous potential expansion, due to the enhanced appreciation of ‘pluridimensional’ signaling efficacy, and ligand signaling ‘bias’. The discovery of additional, non-G protein-dependent signaling modalities for GPCRs has necessitated the understanding that the functional actions of GPCR signaling may be best appreciated not in a low-dimensionality manner, typically with simple unitary output measurement of G protein or direct effector (e.g. adenylyl cyclase), but with high-dimensionality mechanisms and workflows that facilitate the gestalt monitoring of non-G protein-dependent activity [2, 3]. The prototypic non-G protein mediators of GPCR signaling are the beta-arrestins [4]. While productive GPCR engagement with G proteins appears to be relatively transient, GPCR association with arrestin molecules, as well as other subsequently described non-G protein accessory signaling factors [5], involves the creation of more complex, stable, higher-order multi-protein signaling structures termed ‘receptorsomes’. Thus, the spectrum of structurally-diverse receptorsomes present within cells/tissues facilitates the creation of pluridimensional GPCR efficacy profiles that have been revealed in recent years with the coordinated implementation of high-dimensionality data analysis techniques (transcriptomics and proteomics) [6, 7]. With the capacity for diverse and discrete signal transduction outcomes from GPCR activation, the existence of differential ligand-mediated signaling functions, induced by structurally divergent ligands, is evident. Hence chemical analogs of the endogenous cognate ligand, for a given receptor, are unlikely to possess the capacity to reproduce the comprehensive spectrum of signaling effects (via a balanced activation of diverse receptorsomes) employed to maintain standard physiology and thus will invariably demonstrate bias toward a subset of the physiological signaling spectrum [8–12]. The resultant downstream effects of biased ligands are likely to be a non-linear sum of both positive and negative actions across multiple efficacy dimensions – however as the majority of currently-existing therapeutics are analogs of endogenous ligands, the high-dimensionality elucidation and/or prediction of the physiological effects of biased agents may be the prime goal of future pharmacological research.
Our previous research has proven that [D-Trp12,Tyr34]-bPTH(7-34) [bPTH(7-34)] demonstrates arrestin pathway-selective biased agonism of the parathyroid hormone type 1 receptor (PTH1R). Using easily controlled systems, e.g. within in vitro settings, bPTH[7-34] exhibits classical efficacy reversal compared to the endogenous ligand, acting as an inverse PTH1R agonist for Gαs coupling and an agonist for arrestin-dependent signaling, e.g. ERK1/2 signaling, cell migration and anti-apoptotic signaling [13–15]. In more complex, less well-controlled environments, i.e. in vivo, the intermittent injection of bPTH(7-34) increases multiple bone density/integrity indices without stimulating osteoclast proliferation and bone resorption that are induced in the same conditions by the endogenous agonist counterpart, hPTH(1-34) [14]. Even with the multiple difficulties induced by long-term whole animal experimentation a strong mechanistic signaling diversity, at the transcriptomic response level in bone, between bPTH(7-34) and hPTH(1-34) was still evident [16]. These findings therefore suggest that indeed complex high-dimensionality responses can recapitulate the distinct signaling mechanisms entrained rapidly in simpler in vitro cell systems. Therefore it is likely that the biased behavior of pharmacotherapeutics can be connected to differential therapeutic outcomes. Hence controlled and predictable biased agonism presents itself as a facile mechanism to tailor the specific efficacy profile required to remediate, via support of palliative and diminution of detrimental signaling activities [12, 17], complex high-dimensionality pathophysiological disease profiles, such as those presented in multifactorial disorders such as diabetes or dementia. While biased agonism presents a promising future for therapeutic diversity and intelligently-engineered multifactorial efficacy profiles, our ability to identify novel and discrete signaling subsets, e.g. for biased arrestin-dependent signaling, from the full comprehensive endogenous spectrum remains limited [18]. Here, we have attempted to demonstrate that via multiple combinatorial informatic mechanisms the deconvolution, from a standard wild-type comprehensive signaling organismal paradigm, of selective biased agonism is possible and statistically measurable. Our approach employs highly complex physiological data, across multiple tissues, from animals chronically exposed either bPTH(7-34) and hPTH(1-34). The signaling deconvolution mechanisms described here for the definition of arrestin-biased signaling in vivo may represent a step towards formalizing discovery/refinement mechanisms for the creation of intelligently-designed tailored efficacy pharmacotherapeutics.
2. MATERIALS AND METHODS
2.1 Animals and drug treatment
Eleven-week-old male C57BL/6J mice were employed as control mice for the administration of either human PTH(1–34) (hPTH(1-34) (40 μg/kg.d), bovine (D-Trp12, Tyr34)-PTH(7-34) (bPTH(7-34) (40 μg/kg.d), or PBS vehicle for 28 days via Alzet osmotic minipumps (Model #1004, Durect Corp., Cuperteino, CA). The differential treatment protocols for hPTH(1-34) or bPTH(7-34) have been described previously [19]. Minipumps were implanted subcutaneously in the upper back of anesthetized mice. At the end of the infusion period, animals were sacrificed and target tissues, calvarial bone, heart, lung, liver, kidney and aorta, were harvested and stored at −80°C until mRNA isolation.
2.2 RNA Extraction and Oligonucleotide Microarray Hybridization
RNA isolation from at least three individual animals in each experimental group was carried out using a Qiagen RNeasy mini kit (Qiagen, Inc., Valencia, CA), as described previously [20]. RNA conversion to cDNA and subsequent hybridization with Sentrix MouseRef-8 Expression BeadChips (Illumina, San Diego) was performed as described previously [20]. Microarray data were analyzed using DIANE 6.0, a spreadsheet-based microarray analysis program based on the SAS JMP7.0 system. Raw microarray data were subjected to filtering and z normalization and tested for significant changes as described previously [20]. Initial filtering identified genes with a z ratio of ≥1.50. The z ratio is derived from the difference between the averages of the observed gene z scores, divided by the standard deviation of all of the differences for that particular comparison. Genes were then refined by calculating the false discovery rate, which controls for the expected proportion of falsely rejected hypotheses. Only those genes with false discovery rate <0.05 were included for analysis. These data were further analyzed using analysis of variance with significance set at p≤0.05. This allowed us to identify transcripts that differed in their intensity across all of the animal replicates and the various experimental conditions of the mice employed in this study. We have deposited the raw data at GEO/ArrayExpress under accession number GSE GSE64485, and we can confirm all details are MIAME-compliant.
2.3 Bioinformatic Analyses and Signaling Bias Deconvolution
Our primary high-dimensionality data stream for this study is the previously-described significant transcriptomic data generated using Illumina Sentrix MouseRef-8 Expression BeadChips (GSE64485) [19]. The transcriptomic data was differentially created using vehicle or hPTH(1-34)/bPTH(7-34) treatment of wild-type mice and analyzed in-depth as outlined in the following methodological sections.
2.3.1 Venn diagram separation of multi-tissue data
Array-derived transcript lists were analyzed using multiple forms of Venn analysis and functional annotation clustering. Separation of transcriptomic data gathered from six different tissues was performed using the Edwards Venn diagram application VENNTURE [21]. VENNTURE is a novel C++-based Venn diagram-generating application that can accommodate the input of up to 6 distinct data sets from a standard Excel spreadsheet. VENNTURE is freely-available (with additional instructional PDF) either at National Institutes of Health - National Institutes on Aging (http://www.irp.nia.nih.gov/branches/lci/nia_bioinformatics_software.html) or through Omicstools (http://omictools.com/vennture-s6317.html). VENNTURE allows Venn diagram set image generation with additional intersection data representation viewing modes. The Venn segment contents can either be viewed in the VENNTURE Venn diagram image itself or they can be exported to an annotated Excel spreadsheet.
2.3.2 Functional enrichment annotation of primary transcriptomic data
To generate a higher-order functional interpretation of the primary transcriptomic data we applied multiple forms of pathway- and ontology-based annotation. Thus we applied Gene Ontology (GO: http://geneontology.org/) enrichment, canonical signaling pathway analysis (including both metabolic and cell signaling modules) with Ingenuity Pathway Analysis (IPA: http://www.ingenuity.com/), KEGG (Kyoto Encyclopedia of Genes and Genomes: http://www.genome.jp/kegg/) pathway enrichment analysis, Wikipathways enrichment (http://www.wikipathways.org/index.php/WikiPathways), transcription factor (TF) target enrichment analysis (www.broadinstitute.org/gsea/msigdb/geneset_page.jsp) and micro-RNA (miRNA) target enrichment analysis (www.mirbase.org/cgi-bin/mirna_summary). Parametric Geneset enrichment (PAGE) analyses such as GO, KEGG or MSigDB (Molecular Signatures Database: http://www.broadinstitute.org/gsea/msigdb/index.jsp)-PAGE are performed using either raw, or pre-filtered significant (to enhance profundity of output analysis), datasets to test significance of enrichment at a group/collection level rather than at the individual gene significance level [22]. GO, KEGG, TF and miRNA enrichment analysis was performed using Mus musculus Official Gene Symbols (http://www.ncbi.nlm.nih.gov/gene) input to the web-based application, WebGestalt (WEB-based Gene Set Analysis Toolkit: http://bioinfo.vanderbilt.edu/webgestalt/). To perform enrichment analysis for these different forms of annotation the whole mouse genome reference set was employed to generate estimates of expected random transcript frequency within a dataset the size of the experimentally-generated transcriptomes. Using the differences between expected and observed frequencies of transcript occurrence within the input dataset compared to the background enrichment of multiple annotation types can be made. For enrichment probabilities a standard hypergeometric test of significance is applied via WebGestalt: in our current workflows we applied the standard of accepted significant probability at the p≤0.05 level. For each of these parametric enrichment analytical workflows we also employed a cut-off of at least two significantly-regulated transcripts (from the original filtered/analysis of variance geneset) needing to be present within the specific pathway/ontology term to fully populate a particular, GO term group, KEGG pathway, TF or miRNA target. We employed similar enrichment criteria, i.e. enrichment probability of p≤0.05 created using at least two different transcripts populating the specific pathway or term group, for the IPA-based canonical signaling pathway analysis. IPA canonical signaling pathway additionally allows the inclusion of numerical z ratio qualifiers for each transcript, allowing the generation of potential pathway ‘activation’ z-scores. The activation z-score makes a prediction about the potential polarity of modulation of the specific enriched pathway using a cumulative score analysis of the different populating transcripts.
2.3.3 Protein interaction network analyses
Functional protein/gene network interaction analysis was performed using STRING version 10 (http://string-db.org/). STRING essentially generates predicted patterns of protein-protein interactions between input factors, either as individual proteins (using Official Gene Symbols) or as a batch of proteins, using multiple forms of datamining. STRING employs a curated database of known and predicted protein-protein interactions. The curated set of interactions include direct (physical) and indirect (functional) associations. The protein-protein associations are drawn from four source domains: ‘Genomic Context’; ‘High-throughput Experiments’; ‘Conserved Co-expression’ data; ‘Previous Knowledge’. STRING quantitatively integrates interaction data from these sources for a large number of organisms, and transfers information between these organisms where applicable. From the last reported update, the STRING database currently covers over 9×106 proteins from over two thousand organisms. For our present analysis specific transcript lists from the primary murine data were uploaded using the ‘multiple names’ batch input mode. For further analysis the specific species was set as Mus musculus to ensure the greatest degree of network coverage. In addition to the species settings the ‘highest confidence’ level (0.9) of network integrity was used. STRING enables the observation of evidence-based, confidence-based and action-based networks, using the same set of input data. Here we employed the most informative network, i.e. the evidence-based network. STRING evidence-based networks display forms of protein-protein interaction based on the following forms of empirical or informatic evidence: (genetic) Neighborhood; Gene Fusion; Co-occurrence; Co-expression; (empirical) Experiments; (curated) Databases; Textmining. STRING version 10 allows the generation of protein-protein interaction enrichment (with probability scoring), within the input dataset compared to the species-specific whole genome background dataset, using similar algorithms to PAGE. In addition the predicted level of protein-protein enrichment, generated by STRING we also mathematically calculated the number of protein-protein interactions (numbers of evidence-based scores) per input transcript in the network, as we all as the actual number of evidence-based scores for each protein in the network.
2.3.4 Informatic Keystone analysis and natural language processing informatics
Among a series of proteins within a functional network, e.g. in response to selective drug receptor activation modalities, there are likely to be proteins that help link subsets of molecular signaling pathways, thus effectively reducing the complexity of interactions required to coordinate complex multifactorial processes. These factors that facilitate communication across several signaling subsets are often termed ‘keystones’. To identify novel protein factors that may act as keystones, we performed combinatorial latent semantic indexing (LSI)-based analysis using multiple KEGG pathways significantly populated by hPTH(1-34) or bPTH(7-34) transcriptomic datasets. Thus we employed the text contents of our previously identified, significantly-populated KEGG pathways for the two drug transcriptome datasets, as the input interrogator terms for cross-pathway LSI with a curated genome-wide set of gene-word documents extracted from over 2×106 PubMed Abstracts using GeneIndexer (Computable Genomix: https://computablegenomix.com/geneindexer [23]). This process yields lists of proteins that possess a quantitative LSI cosine similarity-based correlation score associated with the input text term, i.e. the functional KEGG pathway, enriched in the ligand treatment datasets. The possible LSI cosine similarity correlation scores for a protein to be associated with an input interrogation term range from 0 to 1, with the stronger correlation scores approaching 1. Our minimal cosine similarity cut-off score criteria for data analysis of the generated protein lists was set at >0.1, which is accepted as demonstration of at least an implicit association between the user-generated input term and the associated output protein. To identify potentially multidimensional keystone factors in drug response transcriptomic datasets we combined the LSI correlation results using input KEGG terms significantly populated by hPTH(1-34) or bPTH(7-34) datasets into color-coded heatmap diagrams. To generate these text-protein correlation heatmaps we rejected correlations that only occurred between a specific protein and only one input pathway term. After taking together the total number of proteins demonstrating multiple (>2) KEGG term correlations, for each ligand transcriptome dataset, and performing a group statistical analysis (GraphPad Prism version 5.0), the numbers of multidimensionally pathway-linked proteins existing outside the 95% or 99% percentile for the number of multiple-correlated protein-pathway combinations was calculated.
2.3.5 Theoretical dataset creation
In the absence of extensive and comprehensive high-dimensionality datasets (transcriptomic or proteomic) that empirically delineate the qualitative nature of selective GPCR signaling modalities we intended to create un-biased theoretical signal-selective datasets to compare our empirical datasets with. To this end we employed biomedical text-based LSI to generate protein lists bearing at least an implicit association (cosine similarity >0.1) with multiple synonyms (to enhance correlated protein data efficiency) associated with beta-arrestin or G proteins. These lists were created using GeneIndexer (Computable Genomix) whole-genome interrogation with the beta-arrestin or G protein synonyms. In addition to these specific datasets, a generic set of proteins associated with multiple ‘cell signaling’ text synonyms was also created (also with cosine similarity >0.1). The intersecting proteins between the ‘cell signaling’ dataset and the selective beta-arrestin or G protein theoretical datasets would therefore create a set union forming either an ‘arrestin signaling’ or ‘G protein signaling’ theoretical dataset. These datasets, created in a simple, un-biased manner, we then employed to identify the potential degree of similarity of these theoretical signaling datasets with our empirically-generated high-dimensionality hPTH(1-34) or bPTH(7-34) transcriptomic datasets.
2.3.6 Natural language processing informatics
In addition to our application of the LSI-based GeneIndexer for multidimensional keystone analysis and theoretical dataset creation, we also applied additional informatic mechanisms based upon scientific natural language processing (NLP) algorithms to our high-dimensionality drug-response data. To provide a gene/protein-to-word biomedical semantic correlation, inverse to that generated using GeneIndexer, we used both Genes2WordCloud (http://www.maayanlab.net/G2W/help.php) and our own NLP-based platform Textrous! [24] (http://textrous.irp.nia.nih.gov/). Textrous! was specifically developed as an advanced rational web-based framework for the extraction of biomedical semantic meaning from a given input data set of arbitrary length. Textrous! simultaneously applies multiple NLP techniques including LSI, sentence splitting, word tokenization, parts-of-speech tagging, and noun-phrase chunking, to mine MEDLINE abstracts (http://www.nlm.nih.gov/bsd/pmresources.html), PubMed Central articles (http://www.ncbi.nlm.nih.gov/pmc/), articles from the Online Mendelian Inheritance in Man (OMIM: http://www.omim.org/), and Mammalian Phenotype annotation obtained from Jackson Laboratories (http://www.informatics.jax.org/phenotypes.shtml). Textrous! has the ability to generate meaningful output data, including both scientifically-relevant nouns as well as extended noun-phrases associated with these nouns, with even very small input datasets. Textrous! also generates multiple types of text extraction methodologies (collective and individual) for the selecting, ranking, clustering, and visualization of English words obtained from the user data. Textrous!, therefore, is able to facilitate the output of quantitatively significant and easily appreciable semantic words and phrases linked to both individual gene/protein and batch genomic/proteomic data. Using these multiple NLP-based processors we attempted to generate a more nuanced de novo interpretation of the high-dimensionality profile of bias GPCR agonist signaling. Complex bPTH(7-34)-based wordclouds were generated, using Wordle (http://www.wordle.net/), from Textrous! collective and/or individual processing outputs, including both nouns and associated noun-phrases. To create the input word sets for these clouds extracted noun-phrases were broken back down to individual words and then added to the original output nouns. These large semantically-associated word lists were then used as the direct input for Wordle-based cloud generation. Wordle-based clouds demonstrate the relative word frequencies from the input data by representing high occurrence frequencies with increased font size. Eventual word frequency scores were assessed from the input list using the online WriteWords (http://www.writewords.org.uk/word_count.asp) application.
2.4 Protein Expression Analysis
Microarray results were validated by selective Western blots. Briefly, tissues except calvarial bone, were homogenized using sonication followed by fractionation using a Qiagen Q-proteome kit according to the manufacturer’s instructions (Qiagen, Inc., Valencia, CA). For all experiments, cytoplasmic fractions were used. Calvarial bones were pulverized on dry ice in a glass Dounce homogenizer and proteins were extracted for 30 min on wet ice in 200 μl of protein extraction buffer (2% sodium dodecyl sulfate; 2 M urea; 10 mM Tris-HCl (pH 6.8); 1 mM phenylmethlysulfonylfluoride). Homogenates were clarified by microcentrifugation for 10 min at 15,000 rpm, and the buffer composition was adjusted to 10% glycerol, 10 mM dithiothreitol, and 0.0025% bromophenol blue for SDS-PAGE. Each tissue homogenate was loaded onto a BisTris 4–12% polyacrylamide gel (Life Technologies) before electrotransfer to a PVDF membrane (Thermo Scientific, Rockford, IL). Proteins were identified using primary antisera at 1:1000 dilutions, followed by species-specific alkaline phosphatase-conjugated secondary antibodies (Sigma-Aldrich) at a 1:7000 dilution. Primary antibodies for Gapdh (glyceraldehyde 3-phosphate dehydrogenase), Bace2 (beta-site APP-cleaving enzyme 2) and RNaseK (RNAase K) were obtained from Santa Cruz (Santa Cruz, CA), AbCam (Cambridge, MA) and Abgent (San Diego, CA) respectively. PVDF-bound immune complexes were identified using enzyme-linked chemifluorescence and quantified using a Typhoon 9410 Phosphorimager (GE Healthcare).
2.5 Statistical Analyses
In each histogram, data represent the means ± S.E. Statistical analyses (Student’s t-test: paired or non-paired) were performed using GraphPad Prism (GraphPad Software, San Diego). p ≤ 0.05 (*), p ≤ 0.01 (**) and p ≤ 0.001 (***) were considered statistically significant.
3. RESULTS
3.1 PTH-ligand mediated transcriptomic activity in wild-type mice
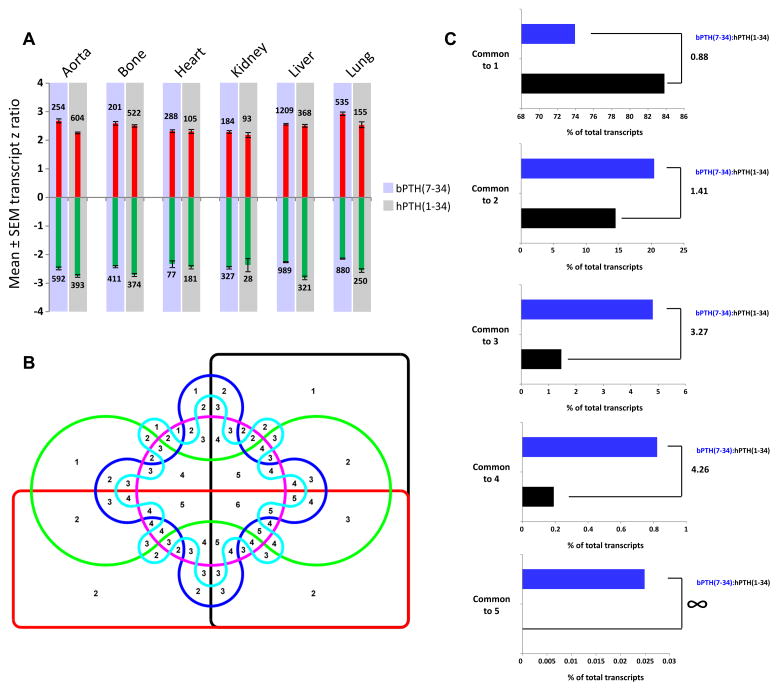
Transcriptomic data for each ligand treatment and tissue has been previously reported and validated [19]. Chronic infusion of both PTH1 receptor (PTH1R)-targeting ligands (hPTH(1-34) and bPTH(7-34)) mediated the complex regulation of multiple gene transcripts across all the six different tissues studied (Fig. 1A). While significant transcript modulation across the divergent tissues was stimulated, bPTH(7-34) ligand signaling resulted in a more tissue-coherent transcriptomic response across the six tissues at the level of transcript identity, using a 6-way Edwards Venn diagram analyzer (Fig. 1B). Transcripts significantly-regulated by bPTH(7-34) were more reproducibly modulated across the tissue range. A consistent greater percentage of the total transcripts significantly modulated by bPTH(7-34), compared to hPTH(1-34), was found across 2–6 different tissues (Fig. 1C).
Fig. 1.
bPTH(7-34) significantly regulates gene transcripts in a more cross-tissue conserved manner than the endogenous hPTH(1-34) ligand. (A) Depiction of mean total transcript z ratios (upregulated transcripts – red bars; downregulated transcripts – green bars). The total number of up- or down-regulated transcripts per ligand in each tissue is indicated next to the corresponding bars. (B) Edwards 6-way Venn diagram employed to separate the patterns of cross tissue significant transcript regulation induced by the two PTH1R-stimulating ligands. The numbers in the intersections indicate the number of set commonalities (1 to 6). (C) Percentage distribution of hPTH(1-34) (black bars) or bPTH(7-34) (blue bars) of significantly regulated transcripts across Edwards Venn sectors common to 1, 2, 3, 4 or 5 tissues (no transcripts were found commonly regulated across all six tissues studied: aorta, bone, heart, kidney, liver, lung). The ratio of the percentage sector occupation between bPTH(7-34) and hPTH(1-34) is indicated next to the appropriate histogram.
3.2 bPTH(7-34) regulates transcripts in a more cross-tissue polarity-coherent manner
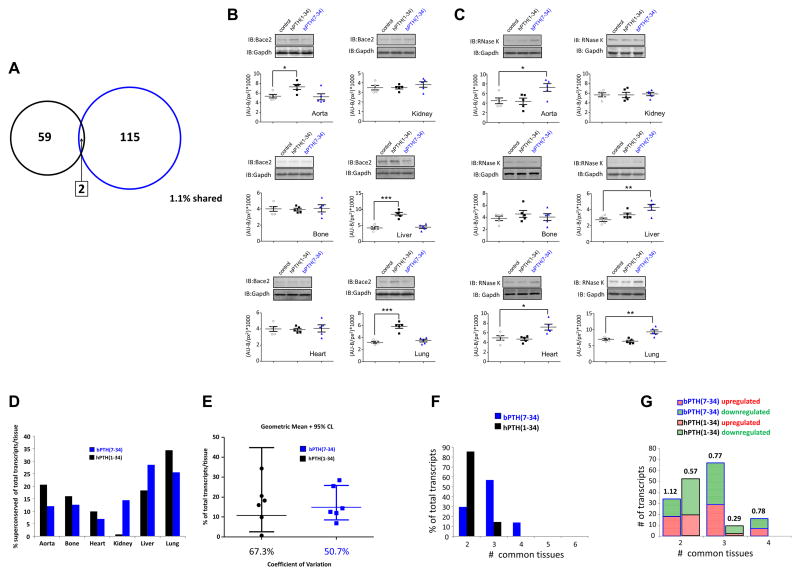
Long-term treatment of mice with hPTH(1-34) significantly modulated the expression of 2590 individual transcripts at a global level (across all the six tissues measured), while bPTH(7-34) significantly modulated 4016 individual transcripts across the six tissues. We previously demonstrated that bPTH(7-34) transcriptomic effects, at the significant gene identity level, show a greater degree of conservance across more tissues than hPTH(1-34) [19]. However, our previous analysis did not take into account ligand-controlled expression polarity coherence across multiple tissues. To assess this from our primary transcriptomic data we excluded the analysis of any transcripts that: i) were only found to be significantly regulated by hPTH(1-34)/bPTH(7-34) in only one tissue and, ii) were found to possess divergent expression polarities (either up- or down-regulation versus tissues from vehicle-treated mice) across between 2–6 tissues. Applying these criteria to our multiple tissue datasets we found that 117 bPTH(7-34)-controlled transcripts fulfilled these criteria (Table S1) while only 61 hPTH(1-34)-controlled transcripts also fulfilled these criteria (Table S2). We termed these highly-conserved, coherently-regulated datasets the ‘superconserved’ hPTH(1-34) or bPTH(7-34) datasets. These two datasets are highly likely to represent the tissue-independent functional signaling core of these two ligands. The superconserved datasets were virtually unique for both ligands (Fig. 2A), with the only 1.1% transcript identity being Krüppel-like Factor 2 (Klf2) and Acyl-CoA dehydrogenase (Acadl) – that were down-regulated by both ligands. To validate the polarity regulation pattern for bPTH(7-34) or hPTH(1-34)-controlled superconserved transcripts we randomly chose two independent transcripts, RNaseK (bPTH(7-34)-regulated) and Bace2 (hPTH(1-34)-regulated) to assess using selective Western blot analysis. We found the ligand-induced tissue expression pattern regulation, at the protein level, corroborated our primary transcriptomic data (Fig. 2B, C: Tables S1/S2).
Fig. 2.
Superconserved transcript analysis reveals strong distinctions between bPTH(7-34) and hPTH(1-34) signaling patterns: (A) Venn diagram indicating the degree of transcript identity between superconserved transcript datasets. Superconserved datasets represent transcripts coherently-regulated (i.e. consistent expression polarity changes compared to vehicle-treated controls) across at least two different experimental tissues that were significantly controlled by either hPTH(1-34) (black) or bPTH(7-34) blue. (B–C) Superconserved dataset Western blot validation of cross-tissue significant expression variation for hPTH(1-34) modulation of Bace2 expression (B) and bPTH(7-34) modulation of RnaseK expression (C). Western blot band intensities (measured as arbitrary absorbance units-background fluorescence per square pixel (AU-B/px2) indicate the mean ± SEM for n=5 datapoints. (D) Relative tissue distribution patterns of significantly-regulated superconserved dataset transcripts. (E) Degree of cross-tissue expression variation for transcripts comprising the superconserved datasets for hPTH(1-34) (black) or bPTH(7-34) (blue). Data is represented as the geometric mean ± 95% confidence limits. (F) Tissue commonality levels of superconserved hPTH(1-34) or bPTH(7-34) significantly-regulated transcripts. (G) Expression polarity (shaded red – upregulated; shaded green – downregulated) modulation balance for constituents of the superconserved datasets for hPTH(1-34) (black outlined bars) and bPTH(7-34) (blue outlined bars).
Investigating the multi-tissue distribution of the superconserved dataset transcripts we found that the bPTH(7-34) superconserved dataset was more evenly-distributed across the six tissues studied, suggesting a more coherent and conserved systemic signaling functionality for bPTH(7-34) compared to hPTH(1-34) (Fig. 2D,E). In addition to a more systemic tissue balance of the bPTH(7-34) signaling repertoire (compared to hPTH(1-34)), the bPTH(7-34) superconserved dataset comprised more transcripts common to a greater number of tissues than for hPTH(1-34) (Fig. 2F). We also found that the relative balance between the number of up- or down-regulated transcripts between either hPTH(1-34) and bPTH(7-34) superconserved datasets was distinct, i.e. the ratio between up- and down-regulated transcripts was near unity for bPTH(7-34) (mean 0.89) and was considerably lower for hPTH(1-34) (mean 0.43) (Fig. 2G).
3.3 Superconserved bPTH(7-34) transcripts represent a more coherent functional interactive signaling network than hPTH(1-34)
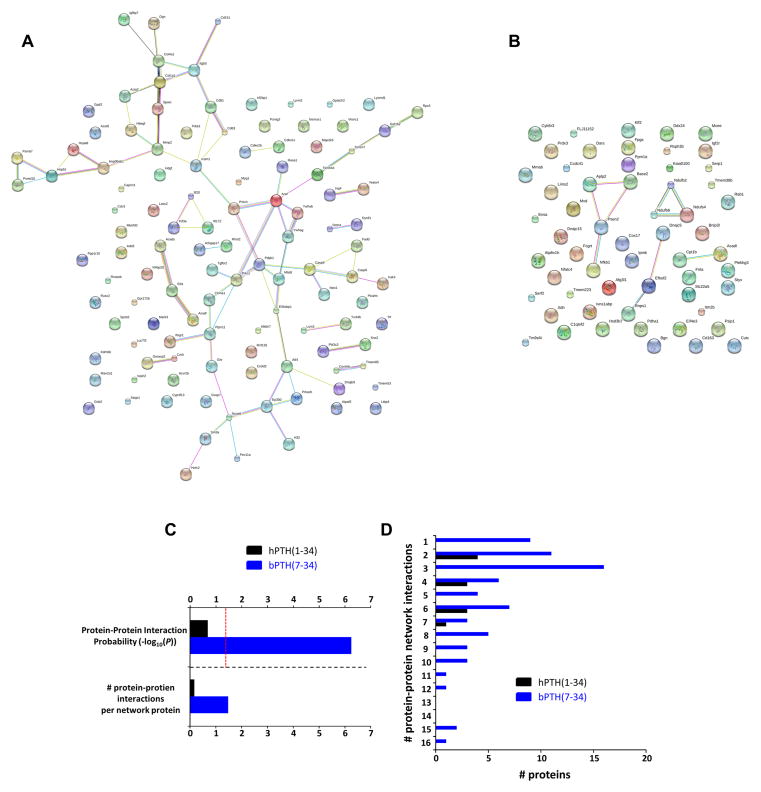
Applying multidimensional evidence-based protein-protein connectivity analysis (EMBL-STRING), the significantly regulated factors in the bPTH(7-34) superconserved dataset (Fig. 3A–B) demonstrated a greater degree of interactivity compared to the factors comprising the hPTH(1-34) superconserved dataset. We found that the bPTH(7-34) superconserved dataset demonstrated a significant protein-protein interaction probability (p= 5.57e-7), while the hPTH(1-34) superconserved dataset did not present any significant interaction enrichment (Fig. 3C). In addition to containing a greater number of more-connected superconserved factors, the number of protein-protein interactions per superconserved factor was considerably greater for bPTH(7-34) compared to hPTH(1-34) (Fig. 3C). For the bPTH(7-34)-derived interaction network there were also a demonstrably greater number of highly-connected factors than for the hPTH(1-34)-derived network (Fig. 3D).
Fig. 3.
Network interaction predictions for superconserved transcript datasets: (A) Evidence-based STRING interaction network for the superconserved transcript dataset induced by bPTH(7-34). (B) Evidence-based STRING interaction network for the superconserved transcript dataset induced by hPTH(1-34). Lines connecting nodes in the network indicate inferred or demonstrated functional interactions. (C) Protein-protein interaction metrics for the bPTH(7-34) (blue bars) or hPTH(1-34) (black bars) superconserved networks. The bPTH(7-34) superconserved network demonstrates a significant enrichment of protein-protein interactions, no significant enrichment occurs for the superconserved hPTH(1-34) dataset. (D) The bPTH(7- 34) superconserved functional network possess a greater number of more-interconnected factors than the network of superconserved hPTH(1-34)-regulated transcripts.
3.4 Signaling pathway distinctiveness of superconserved bPTH(7-34) and hPTH(1-34) datasets
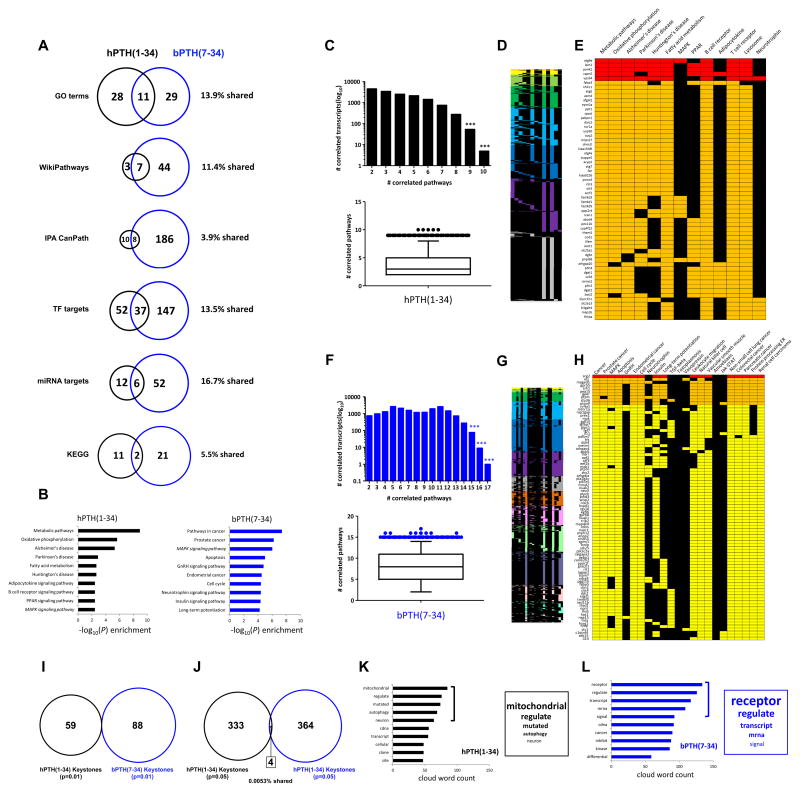
While it is clear that at the transcript identity level the arrestin-biased ligand bPTH(7-34) appears to create a largely unique response pattern, we also assessed a higher level of functional appreciation of these datasets, i.e. enriched pathway and functional analyses. Hence employing the superconserved hPTH(1-34)/bPTH(7-34) datasets we performed Gene Ontology (GO) clustering analysis, signaling pathway enrichment (KEGG (hPTH(1-34) Table S3, bPTH(7-34) Table S4), WikiPathways (hPTH(1-34) Table S5, bPTH(7-34) Table S6), IPA – Canonical Signaling Pathways (hPTH(1-34) Table S7, bPTH(7-34) Table S8), Transcription Factor regulation predictions (TF: hPTH(1-34) Table S9, bPTH(7-34) Table S10) and miRNA target analysis (hPTH(1-34) Table S11, bPTH(7-34) Table S12) (Fig. 4A). Inspecting these differential modes of investigation of the coordinated higher transcript organization and function we found again that the biased bPTH(7-34) ligand generated a primarily unique signaling effect (3.9–16.7% commonality). Demonstrating the strong functional distinction between hPTH(1-34) and bPTH(7-34) the representation of the top 10 highest probability enriched KEGG pathways reveals that the superconserved bPTH(7-34) signaling profile is linked with processes that control cell cycle regulation as well as neuro- and insulinotropic pathways (Fig. 4B). In contrast, using the same criteria hPTH(1-34) superconserved signaling appears to be linked to metabolic functions and inflammatory pathways. We have previously demonstrated the utility of our novel combinatorial informatics methodology in the discovery of signaling keystone factors [25] using a KEGG pathway-LSI hybrid pipeline. Keystone proteins represent a special trophic subset of signaling factors that connect multiple diverse predicted signaling paradigms significantly generated by primary datasets (either transcriptomic or proteomic). Our ability therefore to generate a significantly enriched (p<0.01) ‘keystone’ dataset also represents a mechanism by which higher-order signaling functions of GPCR ligands can be compared in an un-biased manner. To this end we applied keystone analysis to the superconserved datasets for hPTH(1-34) and bPTH(7-34). Using the significantly enriched KEGG pathways generated from the respective superconserved datasets (p<0.05: hPTH(1-34) – 10 pathways (Fig. 4C–E); bPTH(7-34) – 17 pathways (Fig. 4F–H)) as input interrogation terms in GeneIndexer we were able to identify genelists demonstrating consistent (correlating to >2 KEGG pathways) implicit associations (Cosine Similarity Score >0.1) with the KEGG pathways (Fig 4C–E, F–H: Table S13). Using 99% percentile cut-offs for the most stringent investigation we were able to identify a specific ‘keystone’ subset created from the two ligand-induced superconserved datasets. We found that the identity of these functional keystones (99% percentile cut-off) were completely unique to the two ligand superconserved datasets (Fig. 4I). Even with a lowering of the keystone inclusion cut-off to the 95% percentile (resulting in a 4.2 and 5.7 fold increase in included genes for bPTH(7-34) and hPTH(1-34) respectively: Table S13) the percentage of transcripts shared between the two keystone lists was only 0.0053% (Fig. 4J). While the scrutiny of keystone identities demonstrates a strong divergence between the potential high-order trophic regulation of the two ligand signaling profiles, we also gained a more gestalt functional appreciation of these datasets using another web-based platform capable of a reverse gene-word association capacity compared to GeneIndexer, i.e. Genes2WordCloud (http://www.maayanlab.net/G2W/help.php: [26]). Using a similar curated PubMed Abstract reference database we were able to generate correlated wordlists associated with the input keystones from hPTH(1-34) and bPTH(7-34) analysis (Table S14). Ranking the words with highest counts created by the input keystone lists we again demonstrated a clear diversity between hPTH(1-34)- and bPTH(7-34)-derived data (Fig. 4K,L respectively). Interestingly, the top five scoring keystone-derived words from bPTH(7-34) superconserved data essentially provided a textual reconstruction (receptor-regulate-transcript-mRNA-signal) of the major functional modality of GPCR-based β-arrestin signaling [14, 16, 18], i.e. arrestin-mediated control of transcriptional responses from stimulated GPCRs [18].
Fig. 4.
High-dimensionality signaling distinction between bPTH(7-34) and hPTH(1-34). (A) Employing multiple forms of informatics functional annotation for the superconserved datasets of hPTH(1-34) (black circles) or bPTH(7-34) (blue circles) a minimal degree of shared function is evident (the % shared identity is indicated for each analytical platform). (B) Divergence of most-significantly populated KEGG signaling pathways between superconserved transcripts from hPTH(1-34)- (black bars) or bPTH(7-34) (blue bars)-treated mice. For each dataset the top 10 highest probability-scoring KEGG pathways are illustrated. KEGG pathways denoted in italics are common between the two ligand datasets. (C) hPTH(1-34) superconserved keystone factor analysis (employing significantly-regulated KEGG pathways to search for commonly-associated transcripts between the greatest number of predicted signaling functions – 10 pathways employed). Genes identified to possess a degree of correlation > 99% percentile are indicated in the lower panel. (D) Full gene-KEGG pathway interrogation term matrix. (E) Expanded section of full matrix (D) depicting the genes possessing a 99% percentile level of cross pathway commonality. The color coding of the gene blocks indicates the relative number correlations with the input signaling pathways at the top of the matrix. (F) bPTH(7-34) superconserved keystone factor analysis (employing significantly-regulated KEGG pathways to search for commonly-associated transcripts between the greatest number of predicted signaling functions – 10 pathways employed). Genes identified to possess a degree of correlation > 99% percentile are indicated in the lower panel. (G) Full gene-KEGG pathway interrogation term matrix. (H) Expanded section of full matrix (G) depicting the genes possessing a 99% percentile level of cross pathway commonality. The color coding of the gene blocks indicates the relative number correlations with the input signaling pathways at the top of the matrix. (I) Degree of commonality between identified keystone factors between hPTH(1-34) and bPTH(7-34) superconserved datasets at the 99% percentile confidence limit (I) and at the 95% percentile confidence limit (J). (K) Natural language semantic analysis (Genes2WordCloud: Top 5 scoring word frequencies) of the 99% percentile keystone factors for the hPTH(1-34) (K) or bPTH(7-34) (L) superconserved datasets.
3.5 Arrestin-based bias of bPTH(7-34) signaling in a native in vivo background
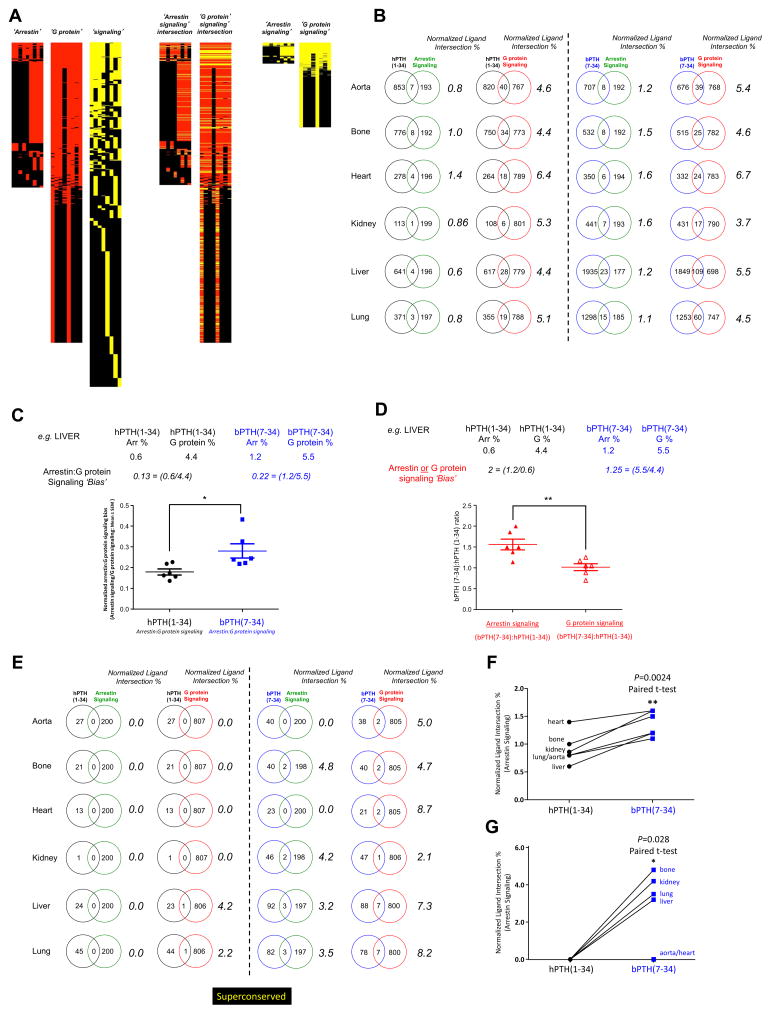
We have previously reported that the bone tissue phenotype associated with bPTH(7-34) signaling is lost in a β-arrestin 2-null background [16], suggesting that its in vivo activity, like its known in vitro effects [13–15], are β-arrestin 2-dependent. We next employed bioinformatic approaches to determine whether the complex transcriptomic effects of bPTH(7-34) were also, in our native wild-type (WT) murine in vivo system, strongly linked to β-arrestin. To determine the extent to which the ligand-specific transcriptomic signatures were β-arrestin 2-dependent we employed a series of combinatorial informatics approaches. We first intended to determine the correlation between the observed WT tissue responses to each ligand and a qualitative library of transcripts semantically-linked to arrestin-dependent signaling that was generated with the entire PubMed Scientific Abstract database (www.ncbi.org). To extract arrestin-selective ligand signaling information from WT mouse transcriptomes and probe for GPCR signaling bias, theoretical transcriptomic signaling datasets were constructed and then impartially compared to the empirically-derived bPTH(7-34) and hPTH(1-34) transcriptomic datasets. Using biomedical database LSI (GeneIndexer, ComputableGenomix Inc.: [23]), we extracted transcripts demonstrating statistically-based associations with input ‘interrogation’ terms used to create the theoretical datasets: ‘arrestin’, ‘G protein’, and ‘cell signaling’ (Table S15). Using this PubMed-based database of more than 2×106 abstracts we generated lists of genes from the whole murine genomic background set associated (with Cosine Similarity Score > 0.1 – representing at least an implicit association) with the input interrogation terms semantically-linked to ‘arrestin’ (Table S16), ‘G protein’ (Table S17) or ‘cell signaling’ (Table S18). Heatmap representations of these ‘arrestin’ or ‘G protein’ associated genelists are shown in Fig. (5A). Horizontal red-colored rows represent implicitly-correlating genes and vertical columns represent the specific input interrogation terms employed (Table S15). To impartially create the resultant ‘arrestin signaling’ or ‘G protein signaling’ theoretical datasets, the intersection between the ‘arrestin’ or ’G protein’ sets and the ‘cellular signaling’ set was identified and extracted (Fig. 5A: Table S19 – ‘arrestin signaling’; Table S20 – ‘G protein signaling’). Using these theoretical arrestin- and G protein-signaling datasets, we compared the degree of intersection between these diverse theoretical ‘signaling’ sets and our experimental transcriptomic data (total significantly-regulated transcripts) from the six tissues treated with bPTH(7-34) or hPTH(1-34) (Fig. 5B). Compared to the hPTH(1-34) transcriptomes, it was evident that bPTH(7-34) exhibited a greater percentage of normalized dataset intersection with the ‘arrestin-signaling’ set than with the ‘G protein signaling’ set. Using the normalized dataset intersection values for all six tissues displayed, we found that the ‘arrestin-signaling’ to ‘G protein-signaling’ ratio for bPTH(7-34) transcriptomic activity was significantly greater (non-paired t-test) than that of hPTH(1-34) (Fig. 5C; p<0.05, liver tissue set values are shown for illustration). When the ligands were compared directly for their arrestin or G protein signaling bias [bPTH(7-34) arrestin signaling: hPTH(1-34) arrestin signaling versus bPTH(7-34) G protein signaling: hPTH(1-34) G protein signaling] we found that bPTH(7-34) transcriptomic responses showed a significantly greater tendency for ‘arrestin-signaling’ compared to ‘G protein-signaling’ (Fig. 5D; p<0.01, liver tissue set values are shown for illustration). We next sought to apply a similar arrestin-dependence of signaling analysis for the superconserved dataset across the six tissues (Fig. 5E). We found that the bPTH(7-34) superconserved dataset demonstrated a greater degree of overlap with the ‘arrestin signaling’ set compared to the hPTH(1-34) superconserved dataset. Using a paired t-test applied across the different tissues this degree of normalized Venn set overlap was significantly greater for bPTH(7-34) than hPTH(7-34) data at both the whole significant transcriptome level (Fig. 5F) and at the superconserved level (Fig. 5G).
Fig. 5.
bPTH(7-34) in vivo transcriptomic activity is strongly associated with arrestin-biased signaling. (A) Generation of arrestin, G protein and cellular signaling theoretical datasets using latent semantic indexing. A PubMed-based database of >2×106 indexed abstracts was used to generate lists of genes, from the whole murine genomic background set associated with the input interrogation terms, semantically-linked to ‘arrestin’, ‘G protein’ or ‘cell signaling’-related text terms. The intersections between the ‘arrestin’ or ‘G protein’ datasets with the ‘cellular signaling’ dataset were used to create the resultant theoretical ‘arrestin signaling’ and ‘G protein signaling’ datasets. (B) Percentage intersection scoring (normalized for different dataset sizes) for both hPTH(1-34) and bPTH(7-34) transcriptomic response datasets (full significantly-regulated datasets) with either the ‘arrestin signaling’ or ‘G protein signaling’ theoretical datasets. (C) Arrestin:G protein signaling bias analysis for hPTH(1-34) and bPTH(7-34). An example calculation from the intersection analysis results for liver data is depicted. (D) Arrestin or G protein signaling bias analysis for hPTH(1-34) and bPTH(7-34). An example calculation from the intersection analysis results for liver is depicted. (E) Percentage intersection scoring (normalized for different dataset sizes) for both hPTH(1-34) and bPTH(7-34) superconserved transcriptomic response datasets with either the ‘arrestin signaling’ or ‘G protein signaling’ theoretical datasets. (F) Percentage intersection analysis (paired t-test across tissues) for the whole significant transcriptome data revealed a significantly greater (p=0.0024) degree of intersection between bPTH(7-34) empirical data with the ‘arrestin signaling’ theoretical dataset compared to hPTH(1-34) data. (G) Percentage intersection analysis (paired t-test across tissues) for the superconserved transcriptome data again revealed a significantly greater (p=0.028) degree of intersection between bPTH(7-34) empirical data with the ‘arrestin signaling’ theoretical dataset compared to hPTH(1-34) data.
3.6 Semantic analysis of the molecular signature of bPTH(7-34) arrestin-dependent signaling in vivo
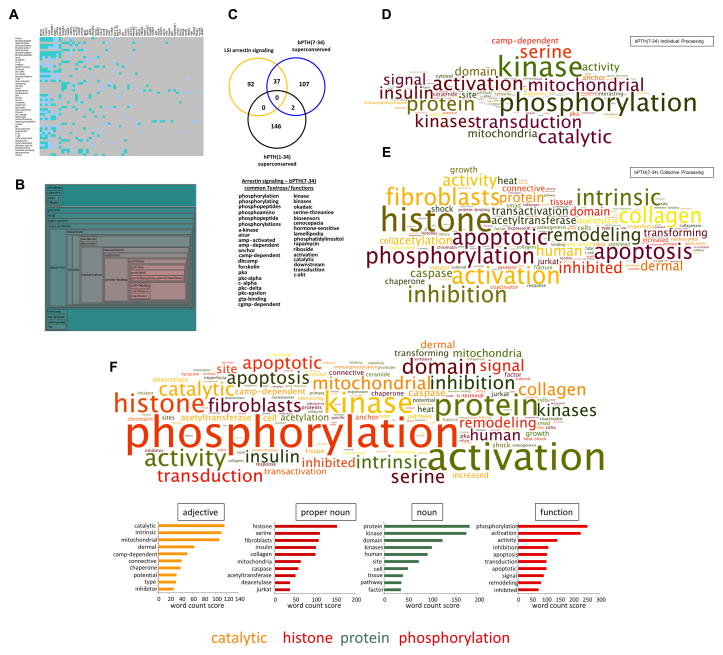
To develop a comprehensive description of the functional relevance of the most conserved core activities of bPTH(7-34) across multiple tissues we employed our novel reverse LSI application Textrous! [24]. Using the superconserved bPTH(7-34) dataset (Table S1) we performed both individual (Fig. 6A) and collective (Fig. 6B) textual processing. Individual transcript processing with Textrous! investigates the strongest individual links between scientifically-relevant words (curated from PubMed (http://www.ncbi.nlm.nih.gov/pubmed/), OMIM (Online Mendelian Inheritance in Man - http://www.omim.org/) and the Jackson Laboratories Mammalian Phenotypes Database (http://www.informatics.jax.org/)) and individual transcripts, while collective processing attempts to generate a hierarchical wordcloud indicating the most strongly associated word groups with the specific entire input dataset, in this case the superconserved bPTH(7-34) dataset (Table S21 – collective processing metrics). Using individual processing of the superconserved bPTH(7-34) dataset we found a strong clustering of terms involved in protein phosphorylation and non-receptor tyrosine kinase (Src (proto-oncogene c-Src), Abl (Abelson murine leukemia viral oncogene homolog 1)) activity – both functions canonically associated with GPCR-arrestin-dependent signaling [4, 27]. With collective processing, a distinct transcriptionally-related post-translational modifying phenotype of the bPTH(7-34) superconserved dataset was revealed (Fig. 6B). We next performed Textrous! semantic analysis for the hPTH(1-34) superconserved dataset as well as for the transcripts we previously identified in the LSI-derived ‘arrestin signaling’ set (Table S19). Upon analysis of the similarity between these three text outputs (‘arrestin signaling’ dataset, bPTH(7-34) superconserved and hPTH(1-34) superconserved: adding both collective and individual word outputs together (Table S22) we found a strikingly higher degree of overlap between the Textrous! outputs for the ‘arrestin signaling’ theoretical LSI dataset and the bPTH(7-34) superconserved set, compared to the hPTH(1-34) superconserved set (37 common words for ‘arrestin signaling and bPTH(7-34) superconserved compared to 0 for arrestin signaling and hPTH(1-34): Fig. 6C). Thus again the most conserved cross-tissue activity of bPTH(7-34) is demonstrably more arrestin-based than hPTH(1-34). To crystallize the potential ‘signature’ of this arrestin-specific receptor signaling we extracted strongly-associated noun-phrases for the bPTH(7-34) dataset using Textrous! (collective processing Noun-Phrases – Table S23; individual processing Noun-Phrases – Table S24), and then (after dismantling of all noun-phrases to render simpler single word input lists) generated clustered wordclouds for individual transcript processing (Fig. 6D: word count results – Table S25) and for collective transcript processing (Fig. 6E: word count results – Table S26). Interestingly it appears that when considering the more singular activities of genes (i.e. individual processing) in the bPTH(7-34) superconserved set, kinase signaling activity seems the most prominent, while when assessing the collective activity of the bPTH(7-34) superconserved set (i.e. collective processing) we see a bias towards cellular growth, remodeling, histone regulation and cell cycle control. Therefore when assessing the collective transcriptional activity of bPTH(7-34) across multiple tissues we revealed a profound activation of cell growth and development pathways when the arrestin-linked genomic factors strongly interact with each other. The more individual functions of this arrestin signaling mode by bPTH(7-34) seem to control signaling events such as kinase activation and phosphorylation. As the complex biological effects of bPTH(7-34) across multiple tissues are likely caused by both a combination of both individualistic and collective activity of downstream factors we also generated an in-depth semantic appreciation of this full gestalt activity. We therefore created a wordcloud using Textrous!-extracted nouns and the associated noun-phrases linked to those LSI-associated nouns from both individual and collective processing streams (Fig. 6F: word count results obtained with WriteWords (http://www.writewords.org.uk/word_count.asp) – Table S27). We next grouped the highest frequency words (top 100 scoring words) according to their syntactic nature (Table S28) to synthesize the most representative functional description of bPTH(7-34) superconserved signaling, i.e. ‘catalytic histone protein phosphorylation’. This novel method of high-dimensionality data analysis thus attempts to generate a simple prosaic interpretation of complex high-dimensionality data using PubMed/OMIM/JaxInformatics-derived naturalistic language elements. It is highly interesting to note that recent research has indeed demonstrated the functional relevance of histone phosphorylation to transcriptional signaling events that regulate transcription and cell cycle control [28] - events that form the core of arrestin-dependent signaling activity of bPTH(7-34) both in vitro [14] and in vivo [16].
Fig. 6.
Defining in vivo arrestin-biased signaling using natural linguistic analysis: (A) Individual Textrous! processing of the superconserved multi-tissue transcriptomic dataset from bPTH(7-34)-treated animals. The heatmap blue-to-teal grid squares indicate a decreasing strength of statistical correlation between a specific gene and word: grey squares demonstrate a lack of correlation. (B) Collective Textrous! processing of the superconserved multi-tissue transcriptomic dataset from bPTH(7-34)-treated animals. The hierarchical wordcloud indicates the strength of correlation (size of text (larger = greater correlation) and green-to-red (red = high correlation) hue of the cloud). (C) The combined Textrous! analysis of the LSI-derived theoretical ‘arrestin signaling’ theoretical dataset (yellow circle) and the superconserved hPTH(1-34) (black circle) and bPTH(7-34) (blue circle) datasets was assessed for intersections using Venn analysis. The superconserved bPTH(7-34)-mediated transcriptomic responses demonstrate a considerably more arrestin-biased action compared to hPTH(1-34). The specific significantly-associated words, common between the output from the theoretical ‘arrestin signaling’ set and the empirical bPTH(7-34) transcriptomic superconserved set, are indicated in the lower panel. (D) Wordcloud interpretation of the individual Textrous! output (dismantled noun–phrases) from the bPTH(7-34) superconserved dataset. (E) Wordcloud interpretation of the collective Textrous! output (dismantled noun-phrases) from the bPTH(7-34) superconserved dataset. (F) Wordcloud interpretation of the collective + individual Textrous! outputs (dismantled noun–phrases) from the bPTH(7-34) superconserved dataset. Syntactic clustering of the highest word count elements in the lower panel (adjective, proper noun, noun, function) facilitates a coherent natural language-based specific interpretation of the high-dimensionality nature of the biased signaling effects of bPTH(7-34) at a systems-level.
4. DISCUSSION
We have employed multiple orthogonal informatic approaches to elucidate the profound distinctions between the complex signaling profiles, at a systemic level, of two GPCR-interacting ligands that target the same receptor target. Our results demonstrate that the diverse systems-level phenotypic effects of hPTH(1-34) and bPTH(7-34) [16, 19] are due to the largely distinct higher-order receptor functionality between a conventional pluripotent GPCR agonist and an arrestin pathway-selective agonist. As most GPCRs are expressed in multiple tissues in the body endogenous cognate ligands that act via these receptors, of-necessity possess a pluridimensional efficacy profile to ensure coordinated receptor signaling across diverse organs [29]. Essentially such pluridimensional efficacy profiles are effected by the capacity of the endogenous cognate ligand to productively activate the broadest spectrum of discrete functional receptorsome entities [11, 12]. In contrast to the endogenous cognate ligand for a given GPCR xenobiotic ligands, due to their divergent physico-chemical properties, tend to demonstrate bias towards specific signaling pathways linked to a smaller repertoire of pre-structured receptorsome entities. This limited signaling repertoire therefore may result in a more focused and consistent functional efficacy profile as the variation in downstream effects will be less varied across multiple tissues. This reduced functional spectrum therefore will likely yield an increased ability to predict the phenotype-level effect of pharmacotherapeutics across diverse cell types – thus representing a major breakthrough with regards to the design of selective efficacy agents. In line with this we found that the long-term, high-dimensionality transcriptomic effects of bPTH(7-34) were more consistently conserved across cells in diverse tissue backgrounds than the effects of hPTH(1-34) (Fig. 1). In addition to a whole transcriptome analysis we derived a superconserved subset of data, representing tissue conserved and coherent ligand-mediated transcript regulation, for the endogenous (hPTH(1-34)) and biased ligand (bPTH(7-34)). This superconserved profile again was more consistently maintained in diverse cell types (Fig. 2), was nearly completely divergent at the transcript identity level between the two ligands (Fig. 2) and demonstrated a significantly enriched level of predicted protein-protein interactions (Fig. 3). When investigated at a higher-order of functional interaction, i.e. pathway and miRNA/TF target analysis, again a profound functional divergence was evident for the superconserved properties of hPTH(1-34) and bPTH(7-34) (Fig. 4A). Employing our novel molecular keystone analysis [25], to impartially discover the proteins that coordinate the gestalt levels of ligand responses, we found that the network-regulatory factors associated with high-dimensionality hPTH(1-34) or bPTH(7-34) responses were again nearly completely distinct (Fig. 4D). Illustrating the relevance of our keystone analyses we found that natural language-based informatic interpretation of our impartial findings were in strong accordance with the empirically-identified (from in vitro experiments: [13, 14]) arrestin-dependent functions of biased GPCR ligands (Fig. 4E). Using publicly-available databases and LSI-based informatic applications we were able to statistically assess the presence of high-dimensionality arrestin signaling bias from our empirical in vivo data. Using unbiased comparisons between our physiological data (both total significant transcriptomic and significant superconserved transcriptomic) with formally-derived theoretical signaling datasets, we were able to demonstrate a statistically-significant arrestin bias of the complex signaling effects of bPTH(7-34), compared to hPTH(1-34), across multiple tissues (Fig. 5). To aid our impartial description and analysis of this novel systems-level biased ligand (bPTH(7-34)) response profiles we employed our previously-developed natural language processing, LSI-based text association platform Textrous! [24] (Fig. 6A–B). Using Textrous!-based investigation (generating extracted word lists from a complex multifactorial database generated using PubMed, OMIM and the Mammalian Phenotypes Database at Jackson Laboratories) we were able to confirm the validity of our identified arrestin-biased ligand effects and their close relationship to the theoretically-created arrestin-signaling dataset (Fig. 6C). Using the natural language output of Textrous! we were also able to identify the bifunctional features of the arrestin-biased ligand bPTH(7-34), i.e. simple signaling functional effects (kinase functionality) and complex ensemble effects (e.g. transcriptional effects controlling long-term phenotypic actions). Via the assembly of the natural language output products of the superconserved bPTH(7-34) signaling profile we were able to generate a rudimentary syntactic description of this complex systems-level high-dimensionality efficacy profile (Fig. 6F). This procedure potentially illustrates an important mechanism for future informatic data extraction and output refinement, i.e. the need for easily comprehensible natural language rendering of novel, non-canonical signaling pathways. Such an advance may represent an additional process by which complex, high-dimensionality data can be interpreted without the constraints of pre-determined signaling pathway classification.
5. CONCLUSION
The current state of investigation into conventional versus arrestin-selective agonist efficacy has been almost exclusively based on short-term in vitro assays of receptor conformation [30], effector coupling [31], second messenger generation [13–15, 31], or protein phosphorylation [32, 33]. In contrast we have demonstrated, using high-dimensionality transcriptomic data from multiple tissue types subjected to chronic ligand stimulation, that arrestin-biased ligands entrain a unique, complex and predictable efficacy profile that is distinct from endogenous pluridimensional ligands that is responsible for the specific phenotypic effects of the biased drug [19]. Our evidence, extracted using multiple orthogonal, classical and non-classical informatic platforms, suggests that as biased signals temporally and physically propagate, the intrinsic coherent nature of the distinct biased GPCR signaling event is not diluted, disrupted or lost during its complex functional impact on diverse physiological systems. Therefore it is likely that with the discovery of further biased ligands, their complex downstream phenotypic effects may be highly predictable, both in simple cellular systems as well as hyper-complex organismal level systems. Illustrating this point, in this and our previous study [19], the most conserved bPTH(7-34)-activated multi-tissue dataset was strongly associated with signaling pathways and biological processes related to cell cycle control, modulation of cell growth, somatic energy regulation and interleukin/cytokine-mediated signaling. Importantly, these results are consistent with our independent functional genomic and biological analysis of bPTH(7-34) actions in bone [14, 16] and with emerging data in the cancer field suggesting that arrestins are critical regulators of tumor cell proliferation, survival and metastasis [34–43].
In conclusion we have demonstrated that GPCR ligands that preferentially activate an arrestin-coupled form of a receptor generate a highly characteristic transcriptomic phenotype in wild-type mice that can be deconvoluted and identified, even after chronic drug treatment. Our data and methodology therefore suggest the potential presence of a conserved generic arrestin response mechanism for GPCRs. Eventual identification and screening for this arrestin-signature may facilitate the rational development of biased drugs that target arrestin signaling pathways. This creation of selective signaling bias holds great promise for the derivation of more effective therapeutics as positive signaling paradigms can be maximized, through rational informed design, contemporaneous with attenuation of unwanted signaling effects.
Supplementary Material
Acknowledgments
The study was in-part funded by the National Institutes of Health Grants AG000916-01 (SM) and the National Institutes of Health National Institute of General Medical Sciences [Grant R01-GM095497]; the National Institutes of Health National Institute of Diabetes, Digestive and Kidney Diseases [Grant R01-DK055524]; the National Institutes of Health National Institute of Child Health and Human Development [Grant T32-HD043446] (LML).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Maudsley S, Martin B, Luttrell LM. J Pharmacol Exp Ther. 2005;314:485–494. doi: 10.1124/jpet.105.083121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Galandrin S, Bouvier M. Mol Pharmacol. 2006;70:1575–1584. doi: 10.1124/mol.106.026716. [DOI] [PubMed] [Google Scholar]
- 3.Martin B, Chen H, Daimon CM, Chadwick W, Siddiqui S, et al. Front Physiol. 2013;4:190. doi: 10.3389/fphys.2013.00190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Luttrell LM, Ferguson SS, Daaka Y, Miller WE, Maudsley S, et al. Science. 1999;283:655–661. doi: 10.1126/science.283.5402.655. [DOI] [PubMed] [Google Scholar]
- 5.Maurice P, Guillaume JL, Benleulmi-Chaachoua A, Daulat AM, Kamal M, et al. Adv Pharmacol. 2011;62:349–380. doi: 10.1016/B978-0-12-385952-5.00001-4. [DOI] [PubMed] [Google Scholar]
- 6.Maudsley S, Chadwick W, Wang L, Zhou Y, Martin B, et al. Methods Mol Biol. 2011;756:99–130. doi: 10.1007/978-1-61779-160-4_5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Maudsley S, Siddiqui S, Martin B. Prog Mol Biol Transl Sci. 2013;118:431–467. doi: 10.1016/B978-0-12-394440-5.00017-6. [DOI] [PubMed] [Google Scholar]
- 8.Wei H, Ahn S, Shenoy SK, Karnik SS, Hunyady L, et al. Proc Natl Acad Sci USA. 2003;100:10782–10787. doi: 10.1073/pnas.1834556100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kenakin T. Mol Pharmacol. 2007;72:1393–1401. doi: 10.1124/mol.107.040352. [DOI] [PubMed] [Google Scholar]
- 10.Kenakin T, Miller LJ. Pharmacol Rev. 2010;62:265–304. doi: 10.1124/pr.108.000992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Luttrell LM, Kenakin TP. Methods Mol Biol. 2011;756:3–35. doi: 10.1007/978-1-61779-160-4_1. [DOI] [PubMed] [Google Scholar]
- 12.Maudsley S, Patel SA, Park SS, Luttrell LM, Martin B. Mini Rev Med Chem. 2012;12:831–840. doi: 10.2174/138955712800959071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gesty-Palmer D, Chen M, Reiter E, Ahn S, Nelson CD, et al. J Biol Chem. 2006;281:10856–10864. doi: 10.1074/jbc.M513380200. [DOI] [PubMed] [Google Scholar]
- 14.Gesty-Palmer D, Flannery P, Yuan L, Corsino L, Spurney R, et al. Sci Transl Med. 2009;1:1ra1. doi: 10.1126/scitranslmed.3000071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Appleton KM, Lee MH, Alele C, Alele C, Luttrell DK, et al. Methods Enzymol. 2013;522:229–262. doi: 10.1016/B978-0-12-407865-9.00013-3. [DOI] [PubMed] [Google Scholar]
- 16.Gesty-Palmer D, Yuan L, Martin B, Wood WH, 3rd, Lee MH, et al. Mol Endocrinol. 2013;27:296–314. doi: 10.1210/me.2012-1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Maudsley S. Mini Rev Med Chem. 2012;12:803. doi: 10.2174/138955712800959161. [DOI] [PubMed] [Google Scholar]
- 18.Luttrell LM, Gesty-Palmer D. Pharmacol Rev. 2010;62:305–330. doi: 10.1124/pr.109.002436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Maudsley S, Martin B, Gesty-Palmer D, Cheung H, Johnson C, et al. Mol Pharmacol. 2015;87:706–717. doi: 10.1124/mol.114.095224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cong WN, Chadwick W, Wang R, Daimon CM, Cai H, et al. J Biol Chem. 2015;290:2728–2743. doi: 10.1074/jbc.M114.588608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Martin B, Chadwick W, Yi T, Park SS, Lu D, et al. PLoS One. 2012;7:e36911. doi: 10.1371/journal.pone.0036911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mootha VK, Lindgren CM, Eriksson KF, Subramanian A, Sihag S, et al. Nat Genet. 2003;34:267–273. doi: 10.1038/ng1180. [DOI] [PubMed] [Google Scholar]
- 23.Martin B, Chadwick W, Cong WN, Pantaleo N, Daimon CM, et al. J Biol Chem. 2012;287:31766–31782. doi: 10.1074/jbc.M112.387316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen H, Martin B, Daimon CM, Siddiqui S, Luttrell LM, et al. PLoS One. 2013;8:e62665. doi: 10.1371/journal.pone.0062665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chadwick W, Martin B, Chapter MC, Park SS, Wang L, et al. PLoS One. 2012;7:e36975. doi: 10.1371/journal.pone.0036975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Desai J, Flatow JM, Song J, Zhu LJ, Du P, et al. Adv Exp Med Biol. 2010;680:709–715. doi: 10.1007/978-1-4419-5913-3_79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xiao K, Sun J, Kim J, Rajagopal S, Zhai B, et al. Proc Natl Acad Sci USA. 2010;107:15299–15304. doi: 10.1073/pnas.1008461107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Basnet H, Su XB, Tan Y, Meisenhelder J, Merkurjev D, et al. Nature. 2014;516:267–271. doi: 10.1038/nature13736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Janssens J, Etienne H, Idriss S, Azmi A, Martin B, et al. Front Endocrinol. 2014;5 doi: 10.3389/fendo.2014.00142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu JJ, Horst R, Katritch V, Stevens RC, Wuthrich K. Science. 2012;335:1106–1110. doi: 10.1126/science.1215802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sauliere A, Bellot M, Paris H, Denis C, Finana F, et al. Nat Chem Biol. 2012;8:622–630. doi: 10.1038/nchembio.961. [DOI] [PubMed] [Google Scholar]
- 32.Kendall RT, Strungs EG, Rachidi SM, Lee MH, El-Shewy HM, et al. J Biol Chem. 2011;286:19880–19891. doi: 10.1074/jbc.M111.233080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Christensen GL, Kelstrup CD, Lyngso C, Sarwar U, Bogebo R, et al. Mol Cell Proteomics. 2010;9:1540–1553. doi: 10.1074/mcp.M900550-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dasgupta P, Rastogi S, Pillai S, Ordonez-Ercan D, Morris M, et al. J Clin Invest. 2006;116:2208–2217. doi: 10.1172/JCI28164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Moussa O, Ashton AW, Fraig M, Garrett-Mayer E, Ghoneim MA, et al. Cancer Res. 2008;68:4097–4104. doi: 10.1158/0008-5472.CAN-07-6560. [DOI] [PubMed] [Google Scholar]
- 36.Chun KS, Lao HC, Trempus CS, Okada M, Langenbach R. Carcinogenesis. 2009;30:1620–1627. doi: 10.1093/carcin/bgp168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lakshmikanthan V, Zou L, Kim JI, Michal A, Nie Z, et al. Proc Natl Acad Sci USA. 2009;106:9379–9384. doi: 10.1073/pnas.0900258106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu H, Long J, Zhang PH, Li K, Tan JJ, et al. Int J Hematol. 2011;93:494–501. doi: 10.1007/s12185-011-0824-9. [DOI] [PubMed] [Google Scholar]
- 39.Fereshteh M, Ito T, Kovacs JJ, Zhao C, Kwon HY, et al. Proc Natl Acad Sci USA. 2012;109:12532–12537. doi: 10.1073/pnas.1209815109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Buchanan FG, Gorden DL, Matta P, Shi Q, Matrisian LM, et al. Proc Natl Acad Sci USA. 2006;103:1492–1497. doi: 10.1073/pnas.0510562103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li TT, Alemayehu M, Aziziyeh AI, Pape C, Pampillo M, et al. Mol Cancer Res. 2009;7:1064–1077. doi: 10.1158/1541-7786.MCR-08-0578. [DOI] [PubMed] [Google Scholar]
- 42.Rosano L, Cianfrocca R, Masi S, Spinella F, Di Castro V, et al. Proc Natl Acad Sci USA. 2009;106:2806–2811. doi: 10.1073/pnas.0807158106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bonnans C, Flaceliere M, Grillet F, Dantec C, Desvignes JP, et al. Proc Natl Acad Sci USA. 2012;109:3047–3052. doi: 10.1073/pnas.1109457109. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.