A sulfated peptide activates a rice immune receptor.
Keywords: Rice, immune receptors, Bacteria
Abstract
Surveillance of the extracellular environment by immune receptors is of central importance to eukaryotic survival. The rice receptor kinase XA21, which confers robust resistance to most strains of the Gram-negative bacterium Xanthomonas oryzae pv. oryzae (Xoo), is representative of a large class of cell surface immune receptors in plants and animals. We report the identification of a previously undescribed Xoo protein, called RaxX, which is required for activation of XA21-mediated immunity. Xoo strains that lack RaxX, or carry mutations in the single RaxX tyrosine residue (Y41), are able to evade XA21-mediated immunity. Y41 of RaxX is sulfated by the prokaryotic tyrosine sulfotransferase RaxST. Sulfated, but not nonsulfated, RaxX triggers hallmarks of the plant immune response in an XA21-dependent manner. A sulfated, 21–amino acid synthetic RaxX peptide (RaxX21-sY) is sufficient for this activity. Xoo field isolates that overcome XA21-mediated immunity encode an alternate raxX allele, suggesting that coevolutionary interactions between host and pathogen contribute to RaxX diversification. RaxX is highly conserved in many plant pathogenic Xanthomonas species. The new insights gained from the discovery and characterization of the sulfated protein, RaxX, can be applied to the development of resistant crop varieties and therapeutic reagents that have the potential to block microbial infection of both plants and animals.
INTRODUCTION
Plasma membrane–localized receptors are critical components of the innate immune responses of animals and plants (1–4). Many of these receptors recognize and respond to the presence of conserved microbial molecules and are often referred to as pattern recognition receptors (PRRs). In animals, this recognition is carried out, in part, by Toll-like receptors (TLRs) (3). Humans have 10 characterized TLRs that recognize conserved microbial molecules such as lipopolysaccharide or flagellin (5, 6).
In plants, cell surface receptor kinases (RKs) and receptor-like proteins (RLPs) recognize microbial molecules in the apoplast (7–13). Well-characterized leucine-rich repeat (LRR)–RKs include Arabidopsis FLS2 that detects flagellin, or its peptide epitope flg22, and the elongation factor Tu receptor (EFR) that detects the bacterial elongation factor Tu, or its peptide epitope elf18 (10, 14). Lacking an adaptive immune system, plants have an extended array of innate immune receptors encoded in their genome. Rice, for example, has more than 300 RKs predicted to serve as innate immune receptors based on the presence of a “non-RD” kinase domain, which lack the arginine-aspartate motif characteristic of most kinases (15, 16). Of the few non-RD RKs characterized to date, all have a role in innate immunity or symbiosis (4, 17).
The rice XA21 RK, one of the first innate immune receptors isolated, mediates recognition of the Gram-negative bacterium Xanthomonas oryzae pv. oryzae (Xoo), the causal agent of an agronomically important disease of rice (7, 18). Previous efforts to identify the microbial molecule that activates the XA21-mediated immune response led to the identification of a number of Xoo genes that are required for activation of XA21 (rax genes). These genes encode a tyrosine sulfotransferase, RaxST, and three components of a predicted type 1 secretion system (T1SS): a membrane fusion protein, RaxA; an adenosine triphosphate (ATP)–binding cassette (ABC) transporter, RaxB; and an outer membrane protein, RaxC. raxST, raxA, and raxB are located in a single operon (raxSTAB) (19). On the basis of these findings, we hypothesized that the activator of XA21-mediated immunity is a tyrosine-sulfated, type 1–secreted protein (19). However, the identity of this molecule has remained elusive (20–22). Here, we report the identification of the tyrosine-sulfated protein RaxX as the activator of XA21-mediated immunity.
RESULTS
RaxX is required for activation of XA21-mediated immunity
RaxB belongs to a subgroup of T1SS ABC transporters that contain a C39 peptidase domain (19). In other bacterial species, substrates for this class of secretion systems are often encoded in or near the T1SS operon (23). We therefore hypothesized that the RaxST/A/B substrate may be genetically linked to the raxSTAB operon. To test this hypothesis, we generated mutants in the Xoo strain PXO99 (24) with either a 1-kb deletion ~0.4 kb upstream from raxST (PXO99Δ1.0Sp) or a 1.7-kb deletion immediately after raxB (PXO99Δ1.7Sp) (Fig. 1A). The mutants were used to inoculate Taipei 309 (TP309), a rice variety lacking XA21, and XA21-TP309, a transgenic derivative expressing XA21 under control of its native promoter (7). On TP309, all strains formed long, water-soaked lesions typical of the disease (Fig. 1B). PXO99Δ1.7Sp formed short lesions on XA21-TP309 similar to the control strain PXO99. In contrast, PXO99Δ1.0Sp formed long lesions on XA21-TP309. These results suggested that the region upstream to the raxSTAB operon contains an element that is required for activation of XA21-mediated immunity.
Fig. 1. raxX, a small ORF located upstream of the raxSTAB operon, is required for activation of XA21-mediated immunity.
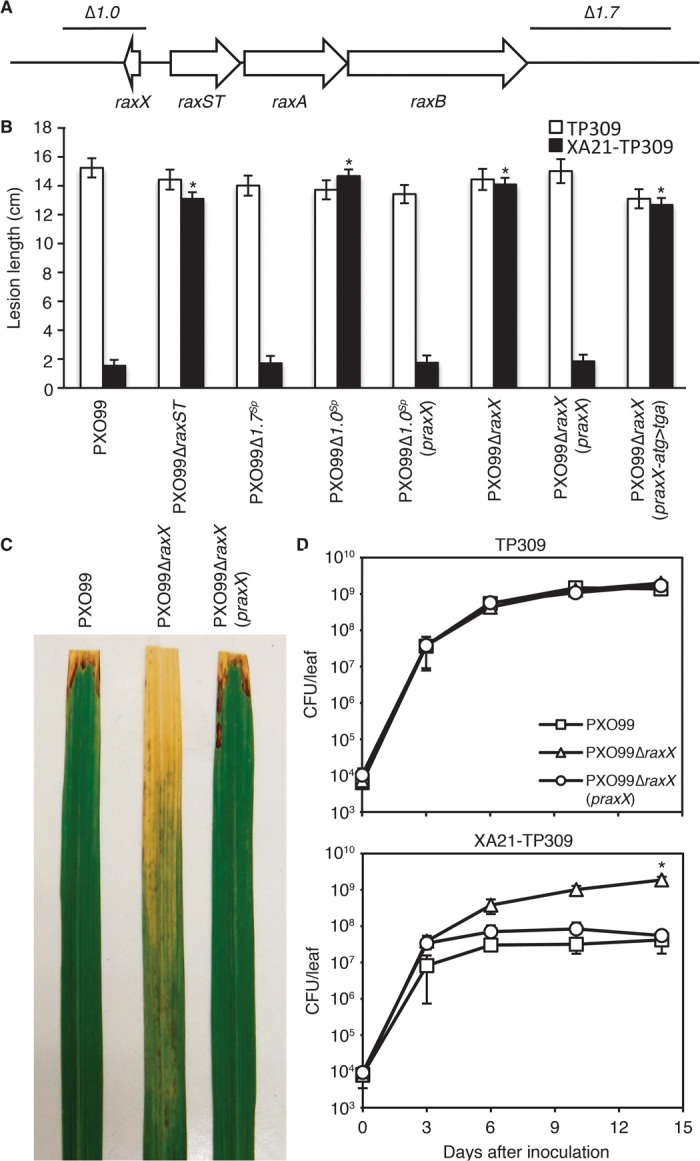
(A) raxST, raxA, and raxB are encoded in a single operon. A 1.0-kb region upstream of raxST and a 1.7-kb region downstream of raxB were deleted in PXO99Δ1.0Sp and PXO99Δ1.7Sp, respectively. The raxX ORF is located ~0.4 kb upstream of raxST and in the opposite orientation. (B) TP309 (open bars) or XA21-TP309 (black bars) were inoculated by clipping with scissors dipped in Xoo suspensions. Bars indicate the mean lesion length ± SE measured 14 days after inoculation (n ≥ 14). The “*” indicates statistically significant difference from PXO99 using Dunnett’s test (α = 0.01). No statistical differences in lesion length were observed on TP309 inoculated with the different strains. The experiment was repeated at least three times with similar results. (C) XA21-TP309 rice leaves display water-soaked lesions 2 weeks after inoculation with the indicated strains. (D) Growth of PXO99 (□), PXO99ΔraxX (△), and PXO99ΔraxX(praxX) (○) in rice leaves inoculated as in (B). In planta bacterial growth analysis was carried out as described (22). Bacterial quantification was determined as the number of colony-forming units (CFU) extracted per inoculated leaf. For the final data point, “*” indicates statistically significant difference from PXO99 using Dunnett’s test (α = 0.01, n = 4). The experiment was repeated twice with similar results.
Sequence analysis of this 1-kb region revealed the presence of a 183-nucleotide open reading frame (ORF) (covering the PXO99 genomic region 1,282,744–1,282,926, NC_010717). The ORF, which we named raxX (Fig. 1A), was not annotated in the original PXO99 genome (24), but has been recently noted as PXO_RS05990. We found that raxX is conserved among all sequenced Xanthomonas strains that encode the raxSTAB operon (fig. S1), but is not present in Xanthomonas species that lack the raxSTAB operon. RaxX shares no homology with other microbial proteins of known function.
To further investigate the role of RaxX, we transformed PXO99Δ1.0Sp with a broad-range host vector expressing raxX under control of its native promoter (praxX). PXO99Δ1.0Sp(praxX) regained the ability to activate XA21-mediated immunity and formed short lesions on XA21-TP309 (Fig. 1B). To confirm the importance of raxX in activation of XA21-mediated immunity, we generated a marker-free PXO99 mutant with the raxX ORF deleted (PXO99ΔraxX). As observed for PXO99Δ1.0Sp, PXO99ΔraxX evades the XA21-mediated immune response, forming long lesions on XA21-TP309 (Fig. 1, B and C). PXO99ΔraxX(praxX) also regained the ability to activate XA21-mediated immunity (Fig. 1, B and C). Similar results were obtained when these strains were inoculated on rice plants overexpressing XA21 in the Kitaake genetic background (Ubi::XA21) (fig. S2) (25). A variant of praxX carrying a mutation in the predicted start codon (praxX-atg>tga) failed to complement PXO99ΔraxX (Fig. 1B). PXO99ΔraxX bacterial populations accumulated to more than 30-fold higher levels in inoculated XA21-TP309 rice leaves than those of strains PXO99 and PXO99ΔraxX(praxX) (Fig. 1D). These results indicate that RaxX is required for activation of XA21-mediated immunity.
The central tyrosine of RaxX is sulfated by RaxST
In addition to raxST, we have previously identified two other rax genes involved in microbial sulfation. These genes, raxP and raxQ, encode an ATP sulfurylase and an adenosine 5′-phosphosulfate kinase, and work in concert to produce the universal sulfuryl group donor 3′-phosphoadenosine 5′-phosphosulfate (PAPS) (26). The requirement of these three genes for activation of XA21-mediated immunity by Xoo suggests that tyrosine sulfation plays a key functional role in this process.
To further investigate this possibility, we transformed a raxST mutant strain (PXO99ΔraxSTSp), which forms long lesions on XA21-TP309, with a plasmid expressing raxST under control of its native promoter (praxST). PXO99ΔraxSTSp(praxST) regained the ability to activate XA21-mediated immunity (Fig. 2A). RaxST carries a predicted PAPS binding motif conserved in mammalian sulfotransferases including the human tyrosine sulfotransferases TPST1 and TPST2 (fig. S3) (19). In TPST2, mutation of the conserved arginine in the PAPS binding motif impairs enzymatic activity (27). We generated a similar mutation in raxST and tested if the expression of this mutant variant on a plasmid (praxST-R35A) could complement the PXO99ΔraxSTSp infection phenotype on XA21-expressing rice plants. The strain PXO99ΔraxSTSp (praxST-R35A) failed to activate XA21-mediated immunity (Fig. 2A), indicating that the sulfotransferase activity of RaxST is critical for its function.
Fig. 2. RaxST sulfates RaxX on tyrosine 41.
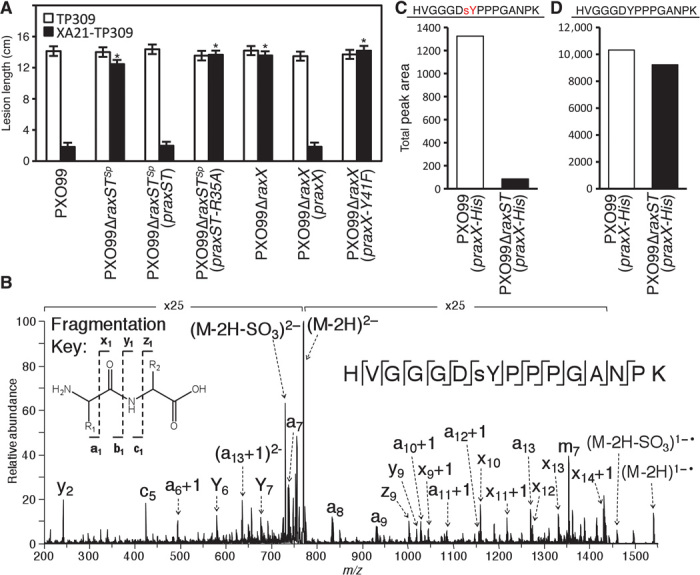
(A) The predicted PAPS binding residue R35 of RaxST and Y41 of RaxX are required for activation of XA21-mediated immunity. TP309 (open bars) and XA21-TP309 (black bars) were inoculated with the indicated Xoo strains, and lesion lengths were measured 14 days later as described in Fig. 1B. Bars indicate the mean lesion length ± SE (n ≥ 14). The “*” indicates statistically significant difference from PXO99 using Dunnett’s test (α = 0.01). (B) Ultraviolet photodissociation (UVPD) mass spectrum of a tyrosine-sulfated peptide (HVGGGDsYPPPGANPK, 2–, m/z 770) from trypsin digestion of in vitro sulfated RaxX39. After incubation with Escherichia coli–expressed and purified His-RaxST, RaxX39 was digested with trypsin and analyzed by LC-UVPD-MS/MS in the negative nanoelectrospray mode to generate a, c, x, y, and z product ions, which are defined in the fragmentation key in the upper left corner of the spectrum. SO3 is retained on all product ions, allowing the sulfate modification to be localized to Y41. Neutral losses of SO3 from the precursor ion and charge reduced radical precursor ion are denoted as (M-2H-SO3)2− and (M-2H-SO3)1−•, respectively, in the spectrum. The ion labeled “m7” refers to the neutral loss of the sulfotyrosine side chain without additional fragmentation of the peptide backbone. Examination of extracted ion chromatograms of the sulfated and nonsulfated peptides suggests that the sulfated peptide is 100× lower in abundance than the nonsulfated peptide (see fig. S9). (C and D) RaxX-His proteins purified from PXO99(praxX-His) and PXO99ΔraxST(praxX-His) were analyzed by selected reaction monitoring-MS (SRM-MS) (fig. S8). Total peak areas (arbitrary units) were quantitated for sulfated (C) and nonsulfated (D) tryptic RaxX peptides covering Y41.
On the basis of the genetic association of raxX with the raxSTAB operon, the importance of tyrosine sulfation for activation of the XA21-mediated immune response, and the presence of a single tyrosine residue in PXO99 RaxX (Y41) that is conserved among all available RaxX sequences (fig. S1), we hypothesized that RaxX Y41 is sulfated by RaxST and that this sulfation is required for RaxX function. To test this hypothesis, we transformed PXO99ΔraxX with a plasmid carrying a derivative of RaxX with tyrosine 41 mutated to phenylalanine [PXO99ΔraxX(praxX-Y41F)]. PXO99ΔraxX(praxX-Y41F) failed to activate XA21-mediated immunity in XA21-TP309 (Fig. 2A and fig. S4). We also demonstrated that sulfated RaxX peptides, but not peptides carrying an Y41 to F substitution, are immunogenic on XA21-expressing rice plants (fig. S5 and next section). These results support the hypothesis that sulfation of RaxX Y41 is required for its activation of XA21-mediated immunity.
To determine whether RaxX Y41 is sulfated by RaxST in vitro, we incubated a chemically synthesized peptide covering the C-terminal region of RaxX (RaxX39, amino acids 22 to 60) with purified His-RaxST (fig. S6) in the presence of PAPS. Trypsin-digested peptides were analyzed by liquid chromatography–tandem mass spectrometry (LC-MS/MS) in both negative and positive nanoelectrospray modes with ultraviolet photodissociation (UVPD) to generate informative a, b, c, x, y, and z product ions from cleavage of the peptide backbone. This method has previously been shown to facilitate the characterization of sulfated tyrosine residues within peptides by MS/MS (28, 29). The lability of the sulfation modification has proven problematic for other MS/MS methods. In negative ion mode, the sulfate group is retained on all product ions, thus allowing the sulfate modification to be unequivocally localized to Y41 of RaxX. MS/MS data showed fragment ions that account for 93% sequence coverage of peptide HVGGGDsYPPPGANPK (Fig. 2B). The high-resolution verification of the peptide mass in the negative mode MS1 (HVGGGDsYPPPGANPK) is displayed in fig. S7. The extracted ion chromatograms of the peptides of interest and positive mode UVPD mass spectrum are shown in figs. S8 and S9, respectively.
We next tested if RaxX is sulfated in vivo. Using selected reaction monitoring-MS (SRM-MS), we observed sulfation on tryptic peptides covering Y41 derived from RaxX-His purified from PXO99(praxX-His) (Fig. 2C and figs. S10 and S11). The sulfated version of the tryptic peptide covering Y41 of RaxX-His purified from PXO99ΔraxST(praxX-His) was not detectable with multiple SRM transitions above the background noise level (~50 to 100 cycles per second) (Fig. 2C and fig. S11). In contrast, we did detect the corresponding nonsulfated peptide covering Y41 at high levels and with high confidence for RaxX-His isolated from both PXO99 (praxX-His) and PXO99ΔraxST(praxX-His) (Fig. 2D and fig. S11). In combination, these results demonstrate that RaxST sulfates RaxX on Y41 in vivo and that sulfation of RaxX is required for its immunogenic activity on XA21-expressing rice plants.
RaxX is sufficient for activation of XA21-mediated immunity
Infection assays using bacterial mutants clearly demonstrate that RaxX is required for activation of XA21-mediated immunity. We next sought to determine whether sulfated RaxX can trigger XA21-mediated defense responses in the absence of Xoo. For this purpose, we produced full-length sulfated recombinant RaxX (RaxX60, 1 to 60 amino acids) using an expanded genetic code approach (30, 31). We heterologously expressed RaxX in E. coli together with an engineered aminoacyl-tRNA (transfer RNA) synthetase specific for sulfotyrosine, a cognate engineered amber suppressor tRNA, and the nonstandard amino acid sulfotyrosine (sY) (32). Nonsulfated RaxX was also expressed in E. coli without sulfotyrosine. We confirmed purity and tyrosine sulfation status of RaxX60 by gel-based assays and SRM-MS analysis (fig. S12). We tested if the resulting highly purified, full-length, sulfated RaxX60-sY and nonsulfated RaxX60-Y proteins could trigger defense gene expression in leaves of rice plants overexpressing XA21 (Ubi::XA21). As shown in fig. S13, sulfated RaxX60-sY, but not the nonsulfated form RaxX60-Y, triggered strong up-regulation of defense marker genes (table S1) in detached leaves of Ubi::XA21. Leaves from Kitaake rice plants, which lack the XA21 immune receptor, are insensitive to RaxX60-Y and RaxX60-sY. These results demonstrate that sulfated RaxX60-sY is sufficient to activate XA21-mediated defense gene expression in rice.
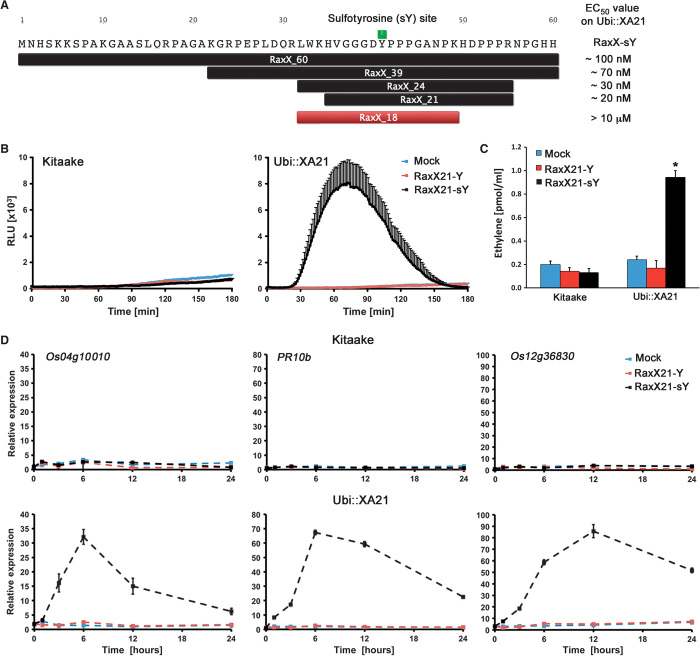
To identify a “minimal” epitope of RaxX that is sufficient to trigger these responses, we took biochemical and rational design approaches. We first tested whether chemically synthesized RaxX39 is sufficient to trigger XA21-dependent defense gene expression (Fig. 3A and fig. S14). We found that sulfated RaxX39-sY triggers defense gene expression in an XA21-dependent manner, whereas nonsulfated RaxX39 does not (fig. S14). To further narrow down the active region, we subjected RaxX39 to digestion with four site-specific proteases (GluC, ArgC, AspN, and trypsin). The predicted digestion patterns were confirmed by gel-based assays and by SRM-MS analysis for ArgC, AspN, and trypsin digests (fig. S15). We tested the resulting RaxX fragments for their ability to activate XA21-dependent signaling (fig. S16A). Only RaxX39-sY digestion products resulting from GluC and ArgC treatments retained activity on Ubi::XA21 plants. The ability of the ArgC fragment (32 to 55 amino acids) to activate XA21-mediated immunity was confirmed with chemically synthesized RaxX24-sY (32 to 55 amino acids) peptides (Fig. 3A and fig. S16B). Next, we tested N- and C-terminal truncated versions of RaxX24-sY peptides. Chemically synthesized RaxX21-sY (35 to 55 amino acids) retained the ability to induce XA21-dependent signaling, whereas RaxX18-sY (32 to 49 amino acids) was compromised in this activity (Fig. 3A and fig. S16B). These results indicate that a chemically synthesized tyrosine-sulfated 21–amino acid derivative of RaxX, named RaxX21-sY, is sufficient to activate XA21-mediated defense responses.
Fig. 3. Sulfated RaxX triggers XA21-mediated defense responses.
(A) Amino acid sequence of RaxX from PXO99. RaxX derivative peptides of varying lengths were tested for their ability to activate XA21-dependent signaling. Sulfated peptides shown in black trigger XA21-mediated defense responses, whereas nonsulfated peptides and the sulfated peptide RaxX_18 shown in red do not (figs. S13 to S21). “EC50 value on Ubi::XA21” refers to the EC50 values determined by monitoring total ROS production over 3 hours after application of the RaxX protein and peptide derivatives (fig. S20) at different concentrations on Kitaake rice expressing XA21 under the control of the ubiquitin promoter (Ubi::XA21) (n = 6). (B) ROS production in leaves of Kitaake and Ubi::XA21 rice plants treated with H2O (mock), RaxX21-Y, or RaxX21-sY (250 nM) (n = 6). RLU, relative light units. (C) Ethylene production in leaves of Kitaake and Ubi::XA21 rice plants after 4 hours of treatment with H2O (mock), RaxX21-Y, or RaxX21-sY (1 μM). The “*” indicates statistically significant difference from mock treatment using Dunnett’s test (α = 0.01, n = 3). (D) Temporal changes in defense marker gene (Os04g10010, PR10b, and Os12g36830) expression in leaves of Kitaake and Ubi::XA21 rice plants treated with H2O (mock), RaxX21-Y, or RaxX21-sY (500 nM, n = 3). All data points depict means ± SE. These experiments were repeated at least three times with similar results.
In addition to activation of defense marker gene expression, the activation of PRR-triggered immunity in plants often involves the production of ethylene and reactive oxygen species (ROS) (11, 33). These responses are known to contribute to the final disease outcome in many plant pathogen interactions (11, 33). We therefore tested if RaxX21-sY can trigger these hallmarks of plant innate immune signaling in XA21-expressing rice leaves (Fig. 3, B to D, and figs. S17 to S19). As shown in Fig. 3 (B to D), only sulfated RaxX21-sY, but not RaxX21-Y, was able to activate defense marker gene expression and the production of ROS and ethylene in an XA21-dependent manner. These responses were most pronounced in rice plants overexpressing XA21 (Ubi::XA21). Nonetheless, we also observed significant RaxX21-sY–mediated up-regulation of defense gene expression and ROS production in Kitaake lines expressing XA21 from its native promoter (XA21::XA21) (figs. S18 and S19), with the highest levels of ROS production observed in rice leaves at later developmental stages (fig. S19).
To assess the sensitivity of XA21-expressing rice plants toward the different RaxX epitopes, we established the RaxX concentration needed to trigger the half maximal response (often referred to as the EC50 value) using the fast and quantitative ROS assay. The EC50 value in Ubi::XA21-expressing rice plants ranges from ~100 nM for RaxX60-sY, ~70 nM for RaxX39-sY, ~30 nM for RaxX24-sY, to ~20 nM for RaxX21-sY (Fig. 3A and fig. S20). In contrast, the further truncated peptide RaxX18-sY was completely inactive, even at high peptide concentrations (fig. S20). As an additional control, we tested an unrelated tyrosine-sulfated peptide called axY(S)22, derived from a PXO99 outer membrane protein (ACD57244), for its ability to activate XA21-mediated immunity (20, 22). Rice leaves overexpressing XA21 were insensitive to axY(S)22 at high peptide concentrations (1 μM) under conditions in which RaxX21-sY induced ROS production and defense gene expression changes (fig. S21). In summary, our data demonstrate that XA21-mediated immune signaling in rice is specifically induced by RaxX60-sY, and sulfated epitopes derived from the full RaxX60 protein, at low nanomolar concentrations.
A RaxX variant evades XA21-mediated immunity
When XA21 was first discovered, it was demonstrated to confer resistance to most tested strains of Xoo (18, 34). Since that time, several field strains that evade XA21-mediated immunity have been identified (35, 36). For most of these strains, the molecular basis of immune evasion is unknown. However, we previously demonstrated that two of these Xoo strains, DY89031 and CK89021, carry mutations in raxA and raxST, respectively (19). We hypothesized that other strains may evade XA21-mediated immunity because they encode variant raxX alleles. Indeed, we found that RaxX from the Indian field isolate IXO685, which evades XA21-mediated immunity (35), differs in nine amino acids as compared with RaxX from PXO99 (Fig. 4A). Four amino acid substitutions are located in the RaxX21 sequence.
Fig. 4. Comparative genomics and mutational analyses identify key amino acids required for RaxX activity.
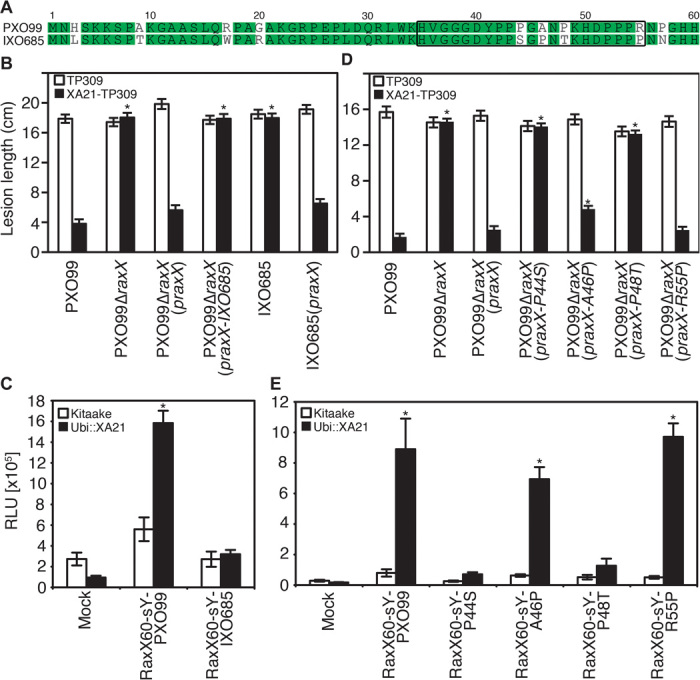
(A) Alignment of amino acid sequences of RaxX from PXO99 and IXO685. The region corresponding to RaxX21 is boxed. (B and C) RaxX from the field strain IXO685 does not trigger XA21-mediated immune response. (B) TP309 (open bars) and XA21-TP309 (black bars) were inoculated with the indicated Xoo strains, and lesion lengths were measured 14 days later as described in Fig. 1B. Bars indicate the mean lesion length ± SE (n ≥ 14). (C) ROS production in leaves of Kitaake (open bars) and Ubi::XA21 (black bars) rice plants treated with the indicated peptides (250 nM) or mock treatment. Bars depict mean RLU over 3 hours ± SE (n = 6). (D and E) RaxX point mutation analysis reveals that P44 and P48 are required for activation of XA21-mediated immune response. (D) TP309 (open bars) and XA21-TP309 (black bars) were inoculated with the indicated Xoo strains, and lesions were measured 14 days later as described in Fig. 1B. Bars indicate the mean lesion length ± SE (n ≥ 14). (E) ROS production in leaves of Kitaake (open bars) and Ubi::XA21 (black bars) rice plants treated with the indicated peptides (250 nM) or mock treatment. Bars depict average RLU ± SE (n = 6). For (B) to (D), the “*” indicates statistically significant difference from mock treatments using Dunnett’s test (α = 0.01). These experiments were repeated at least three times with similar results.
We tested if raxX from IXO685 expressed under control of the native promoter of PXO99 raxX was able to complement the PXO99ΔraxX mutant phenotype on XA21-expressing rice plants. We found that PXO99ΔraxX(praxX-IXO685) was unable to activate XA21-mediated immunity (Fig. 4B). Likewise, recombinant, highly purified RaxX60-sY–IXO685 produced using our expanded genetic code system was not able to trigger XA21-mediated ROS production (Fig. 4C and fig. S22). Expression of the PXO99 raxX allele in the IXO685 strain conferred the strain with the ability to activate XA21-mediated immunity (Fig. 4B). These results indicate that IXO685 can overcome XA21-mediated immunity because it has acquired several nonsynonymous substitutions in the raxX ORF. Two other Indian field isolates (IXO651 and IXO1221), which are also able to evade XA21-mediated immunity (35) and share the identical raxX sequence with IXO685, can also be complemented with praxX (fig. S23).
We carried out mutational analyses of PXO99 RaxX to determine if any of the four identified amino acid substitutions within the RaxX21 epitope of the XA21-evading strains are essential for XA21 activation. The four residues were mutated individually in praxX, and the mutated alleles were tested for functional complementation of the PXO99ΔraxX mutant by inoculation of XA21-expressing rice plants (Fig. 4D). PXO99ΔraxX(praxX-P44S) and PXO99ΔraxX(praxX-P48T) formed long lesions on XA21-TP309, PXO99ΔraxX(praxX-A46P) displayed an intermediate phenotype, and PXO99ΔraxX(praxX-R55P) formed short lesions. Consistent with this result, highly purified recombinant RaxX60-sY–P44S and RaxX60-sY–P48T proteins, produced using our expanded genetic code system, are unable to induce ROS production in Kitaake Ubi::XA21 (Fig. 4E and fig. S22). Thus, P44 and P48 are required for activation of XA21-mediated immunity. That is, the alterations of these specific amino acids in RaxX of IXO685, IXO651, and IXO1221 enable these strains to evade XA21-mediated immunity.
DISCUSSION
Our results indicate that the presence or absence of sulfation and the residues near Y41 are decisive for the ability of RaxX to trigger XA21-mediated immunity and that Y41 is a substrate for RaxST-mediated sulfation. We have identified RaxX in at least eight Xanthomonas species that infect maize, cassava, sugar cane, tomatoes, peppers, wheat, alfalfa, onions, banana, and citrus (fig. S1). In all of these strains, raxX is encoded upstream of raxST, raxA, and raxB. The colocalization of the rax genes suggests that they function in a common biological process and that the RaxX proteins in these species are also substrates for RaxST-mediated sulfation.
Although tyrosine sulfation has not been demonstrated in other prokaryotic species, it is a common posttranslational modification of eukaryotic proteins and plays important roles in regulating plant development and in mediating the interactions of plants and animals with microbes. For example, in humans, sulfation of the C-C chemokine receptor type 5 (CCR5) is critical for binding of the envelope glycoprotein gp120 by the HIV (37). In plants, the sulfated peptide signaling factors PSK (phytosulfokine) and PSY1 (plant peptide containing sulfated tyrosine 1) have been implicated in xylem trachea development, root growth, suppression of plant defense, and promotion of plant health (38, 39).
Although RaxX shares no homology with microbial proteins of known function, we have noted a remarkable similarity between the sulfated, secreted Arabidopsis 18–amino acid peptide signaling factor PSY1, four predicted rice PSY1 orthologs, and RaxX residues 40 to 55 (fig. S24). These similarities suggest the exciting possibility that in the absence of XA21, Xoo and other Xanthomonads use sulfated RaxX to mimic PSY1-like peptides to suppress host defense responses, facilitate infection, or enhance plant health, maintenance of which is critically important for propagation and survival of biotrophic pathogens (39–41).
The robust immunity conferred by XA21 to most Xoo strains is a highly valued agronomic trait, and therefore, XA21 has been introgressed into diverse rice varieties grown by rice farmers (42–47). The discovery of raxX and allelic variants that can evade XA21-mediated recognition can be used to screen Xoo field populations for the presence of potentially virulent variants. Such an anticipatory breeding approach would alert farmers of the need to plant different varieties or mixtures before the breakdown of XA21-mediated immunity.
The knowledge gained from these studies can also be used to expand the spectrum of XA21-mediated immunity to additional strains of Xoo. For example, new synthetic receptors of XA21 with novel engineered recognition specificities such as those that can recognize RaxX from IXO685 would be of great interest. Because Xanthomonas species affect virtually all crop plants, it is expected that future studies will lead to new strategies for engineering resistance to non-rice pathogens. For example, X. axonopodis pv. citrumelo, X. euvesicatoria, and X. oryzae pv. oryzicola, which cause serious diseases on citrus, tomato, and rice, carry alternate raxX alleles (fig. S1). This knowledge provides an opportunity to engineer new XA21 variants that recognize these alleles and use the novel variants for engineering plants resistant to Xanthomonas infections that cannot be controlled by more conventional genetic methods.
The sulfated RaxX/XA21 interaction represents a new biological system for studies of the interaction of sulfated epitopes and their receptors. A recent report of the generation of a potent new antiviral drug (48), which was based on more than 10 years of work on a single family of sulfated peptides, indicates the clinical relevance of studies of sulfopeptides. Thus, the studies reported here are applicable to generation of therapeutic reagents with the potential to block microbial infection of both plants and animals.
MATERIALS AND METHODS
Bacterial culture and growth conditions
Lists of plasmids and bacterial strains used in this study are provided in tables S2 and S3. Xoo was cultured on peptone sucrose agar (PSA) plates at 28°C with the appropriate antibiotic(s). YEB medium [yeast extract (5 g/liter), tryptone (10 g/liter), NaCl (5 g/liter), sucrose (5 g/liter), MgSO4 (0.5 g/liter), pH 7.3] was used for liquid culture at 28°C with shaking at 230 rpm. E. coli was cultured at 37°C and 230 rpm in lysogenic broth (LB) for cloning or M9T (per 1 liter: M9 plus 10 g of tryptone, 5 g of NaCl; after autoclaving, 1 ml of 1 M MgSO4 and 1 ml of 0.02% biotin were added). Kanamycin was used at 50 μg/ml, carbenicillin at 50 μg/ml, spectinomycin at 50 μg/ml, and cephalexin at 20 μg/ml.
Xoo mutation and complementation
Xoo mutants were generated in Philippine race 6 strain PXO99Az, a derivative of strain PXO99 (referred to simply as PXO99 in this manuscript) (24, 49). PXO99ΔraxSTSp, PXO99Δ1.0Sp, and PXO99Δ1.7Sp were generated by marker exchange mutagenesis using the suicide vector pJP5603 (50). About 500–bp (base pair) sequences flanking both sides of the region to be deleted were sequentially cloned into the multiple cloning site using the restriction enzyme sites Kpn I/Bam HI and Bam HI/Sal I. A spectinomycin resistance cassette was ligated between the two flanking sequences using the Bam HI site. PXO99-competent cells were transformed with the suicide plasmids by electroporation and plated to PSA with spectinomycin (50 μg/ml). Colonies with the expected antibiotic resistance phenotype for double crossover (spectinomycin-resistant, kanamycin-sensitive) were validated by polymerase chain reaction (PCR).
Marker-free raxX (PXO99ΔraxX) and raxST (PXO99ΔraxST) mutants were generated using the vector pUFR80 (51). About 500-bp sequences flanking were sequentially cloned into the multiple cloning site of pUFR80 using the sites Eco RI/Bam HI and Bam HI/Hind III. praxX-KO-MF and praxST-KO-MF were transformed into PXO99-competent cells by electroporation, and the cells were plated to nutrient agar (NA) (CM0001, OXOID) with kanamycin (50 μg/ml). Transformants were grown in nutrient broth without kanamycin and plated to NA with 5% sucrose to select for a second crossover event. Colonies were analyzed by PCR to identify deletion mutants.
Strains were transformed using derivatives of the broad host range vector pVSP61 (52). The raxX and raxST sequences along with their predicted promoter region (300 bp upstream of the predicted start codons) were cloned into pVSP61 using the Eco RI/Hind III sites. Point mutations were generated by QuikChange mutagenesis. For praxX-IXO685, the raxX promoter from PXO99 was first introduced into pVSP61 using the sites Eco RI/Bam HI to generate praxXprom. The raxX sequence from IXO685 was amplified from genomic DNA and cloned into praxXprom behind the PXO99 raxX promoter using the restriction sites Bam HI/Hind III.
Rice and rice growth conditions for inoculations
Oryza sativa ssp. japonica rice varieties TP309, a 106-17–derived transgenic line of TP309 carrying XA21 driven by its own promoter (XA21-TP309) (7), Kitaake, and Ubi::Myc::XA21-Kitaake line 7-A-8 (Ubi::XA21) (25) were used for rice inoculations. Native TP309 and Kitaake do not contain XA21. Seeds were germinated in distilled water at 28°C for 1 week and then transplanted into sandy soil (80% sand, 20% peat from Redi-Gro) in 5.5-inch square pots (two seedlings per pot). Plants were grown in tubs in a greenhouse. The plants were top-watered daily with fertilizer water [N, 58 ppm (parts per million); P, 15 ppm; K, 55 ppm; Ca, 20 ppm; Mg, 13 ppm; S, 49 ppm; Fe, 1 ppm; Cu, 0.06 ppm; Mn, 0.4 ppm; Mo, 0.02 ppm; Zn, 0.1 ppm; B, 0.4 ppm] for 4 weeks, followed by water for 2 weeks. Six weeks after planting, the rice was transferred to a growth chamber set to 28°C/24°C, 80%/85% humidity, and 14/10-hour lighting for the day/night cycle.
Plants were inoculated 2 to 3 days after transfer using the scissors clipping method (53). Bacteria for inoculation were taken from PSA plates and resuspended in water at a density of 108 CFU/ml. Water-soaked lesions were measured 14 days after inoculation. In planta bacterial growth analysis was carried out as described (22). Four biological replicates were measured for each data point. Each biological replicate was the mean number of colonies from four technical replicates.
Generation of RaxX peptides
Sulfated (sY) and nonsulfated (Y) versions of RaxX39 (KGRPEPLDQRLWKHVGGGDYPPPGANPKHDPPPRNPGHH), RaxX24 (LWKHVGGGDYPPPGANPKHDPPPR), RaxX21 (HVGGGDYPPPGANPKHDPPPR), RaxX18 (LWKHVGGGDYPPPGANPK), and ax22(S)Y (20) were ordered from Pacific Immunology. The peptides were checked for purity by high-performance liquid chromatography and MS analysis by the company. All peptides were resuspended in ddH2O. No other solvents were required to dissolve the peptide.
For the expanded genetic code approach, full-length RaxX (RaxX60) from PXO99 was expressed as MBP-3C–RaxX-His fusion protein in E. coli C321.ΔA.exp (31). E. coli C321.ΔA.exp was cotransformed with SYP29_pULTRA_SY, made by cloning the sY-specific aminoacyl-tRNA synthetase from (32) into the pULTRA system (54) and pBAD/MBP-3C–RaxX-His (Amber) to generate sulfated RaxX or pBAD/MBP-3C–RaxX-His to generate nonsulfated RaxX. Transformed bacteria were grown in 0.5 liter of M9T medium. For expression of sulfated RaxX, sulfotyrosine (sY), synthesized as described in (32), was added to a final concentration of 3 mM. Cultures were grown at 37°C with shaking at 230 rpm. Expression was induced at an optical density of 0.6 to 0.7 by addition of 1 mM isopropyl-β-D-thiogalactopyranoside and 0.25% (w/v) arabinose for 5 hours. MBP-3C–RaxX-His was purified from intracellular total protein extracts over a 5-ml Ni-NTA column by fast protein liquid chromatography (FPLC) (ÄKTA, GE Healthcare). The MBP-tag was removed by overnight digestion with 3C protease at a concentration of 1:200 (w/w) at 4°C. The MBP was separated from full-length RaxX-His over a 1-ml SP-XL column, a strong cation exchanger, on an FPLC (ÄKTA, GE Healthcare). Nonsulfated RaxX was expressed and purified in the same way, except pBAD/MBP-3C–RaxX-His plasmids without Amber mutations were used, and no sulfotyrosine was added to the cultures. The yield for sulfated and nonsulfated highly purified RaxX-His was approximately 1 mg of protein per 1 liter of culture. All RaxX allelic variants were prepared according to the same protocol.
In vitro sulfation of RaxX by His-RaxST
RaxST was expressed as a full-length fusion protein with an N-terminal His-tag in BL21(DE3) cells and purified using Ni-NTA agarose beads (Qiagen). His-RaxST expression was confirmed by SDS–polyacrylamide gel electrophoresis with Coomassie blue staining and Western blot analysis with Penta-His antibody (Qiagen) (fig. S4). The in vitro sulfation reaction was carried out as described by Han et al. (28). Purified RaxST (20 μg) was incubated with 170 μg of RaxX39 in the presence of sulfation buffer [25 mM tris (pH 7.5), 150 mM NaCl, 20 mM MgCl2, 480 μM PAPS] for 4 hours at 30°C.
Nano-LC-UVPD-MS
Proteins from in vitro sulfation reactions were buffer-exchanged into 50 mM tris-HCl (pH 8.5), 1 M urea using a 3-kD molecular weight cutoff (MWCO) filter and digested with trypsin overnight at 37°C using a 1:25 enzyme to substrate ratio. After digestion, samples were passed through a 10K MWCO filter to separate RaxX39 peptides from trypsin.
Tryptic peptides of RaxX39 were separated by reversed-phase LC at 300 nl/min using a Dionex Ultimate 3000 NSLC system configured for preconcentration. Mobile phases A and B were water and acetonitrile, respectively, both containing 0.1% formic acid. A solution of 2% acetonitrile and 0.1% formic acid was used for sample loading onto a New Objective IntegraFrit 100-μm × 3-cm trap column packed in-house with 5-μm, 100-Å Michrom Magic C18 AQ. After 5 min of preconcentration at 7 μl/min, the trap column was switched in-line with a New Objective PicoFrit analytical column (75 μm × 15 cm) packed with Waters 3.5-μm, 130-Å XBridge BEH C18. A 23-min linear gradient starting at 2% B and ending at 40% B was used for separation.
LC-MS/MS analysis was performed on a Thermo Scientific Velos Pro mass spectrometer equipped with a Coherent ExciStar XS Excimer laser operated at 193 nm with a pulse frequency of 500 Hz (55). Data acquisition was carried out by first performing a full MS1 scan from mass/charge ratio (m/z) 300 to 2000 followed by three MS/MS UVPD events to activate the doubly charged sulfopeptide of interest, HVGGGDsYPPPGANPK, at m/z 769.8 in negative mode and m/z 771.8 in positive mode. A single laser pulse was used for ion activation in negative mode during a 2.0-ms activation period. Two laser pulses were used for ion activation in the positive mode. In both polarities, the isolation width was 3 daltons and the activation q value was 0.10. MS/MS fragmentation patterns were manually interpreted. For the UVPD mass spectrum shown in Fig. 2B, the neutral loss of SO3 and CO2 from the precursor ion and/or charge reduced precursor ion are denoted as “–SO3” and “–CO2,” respectively, in the spectrum. Product ions a + 1 and x + 1 correspond to homolytic cleavage of the Cα-carbonyl bond of the peptide backbone without subsequent transfer of hydrogen radicals to produce even electron a and x ions, which are the diagnostic N- and C-terminal ions used to identify the peptide.
High-resolution mass measurement of the diagnostic tryptic peptide corresponding to HVGGGDsYPPPGANPK from the RaxX39 sample sulfated by RaxST in vitro was undertaken on a Thermo Fisher Scientific Instruments Orbitrap Elite mass spectrometer operated at an FT resolution of 120,000 at m/z 400.
Isolation of RaxX from PXO99 for SRM-MS
Briefly, RaxX-His proteins were purified from Xoo strains and their sulfation was analyzed by SRM-MS. PXO99(praxX-His) and PXO99ΔraxST(praxX-His) were cultured in 1 liter of XVM2 (56) liquid medium for 40 hours at 28°C. The cells were harvested by centrifugation at 10,000 rpm for 20 min at 4°C. RaxX-His was purified from intracellular total protein extract over a 5-ml Ni-NTA column on an FPLC (ÄKTA, GE Healthcare). Isolation of Rax-His was confirmed by Coomassie blue staining and Western blot analysis with Penta-His antibody (Qiagen) (fig. S6).
Selected reaction monitoring–mass spectrometry
Purified RaxX-His was digested with 1 μg of trypsin in the presence of 5 mM dithiothreitol (DTT). Peptide digests were analyzed using an Agilent 1260 LC system operating in normal flow mode at 400 μl/min coupled to an Agilent 6460QQQ Mass Spectrometer. Peptides were separated on an Ascentis Express Peptide C18 column [2.7-μm particle size, 160-Å pore size, 5-cm length × 2.1-mm inside diameter (ID), coupled to a 5-mm × 2.1-mm ID guard column with similar particle and pore size, operating at 60°C; Sigma-Aldrich] using a 43-min method with the following gradient: initial conditions were 98% buffer A (0.1% formic acid), 2% buffer B (99.9% acetonitrile, 0.1% formic acid), after which B was increased to 35% over 35.70 min, followed by an increase to 90% B in 18 s, where it was held for 2 min. Buffer B was then decreased to 5% in 30 s, where it was held for 2.5 min, then further decreased to 2%, and held for 2 min to reequilibrate the column.
The eluted peptides were ionized via an Agilent Jet Stream ESI source operating in positive ion mode with the following source parameters: gas temperature = 250°C, gas flow = 13 liters/min, nebulizer pressure = 35 psi, sheath gas temperature = 250°C, sheath gas flow = 11 liters/min, capillary voltage = 3500 V, nozzle voltage = 0 V. The data were acquired using Agilent MassHunter version B.06.00. Data were acquired in MRM mode with transitions generated via Skyline (57) software (version 2.6). In silico SRM transitions were selected with 2+ and 3+ charge states, no missed tryptic cleavages, sulfated and nonsulfated tyrosine modifications, and singly charged fragment ions from both the y and b series. The Skyline-generated method was refined to three transitions per peptide and set to Agilent 6460QQQ for instrument-specific collision energies. The final method consists of 36 transitions, with a 25-ms dwell time per transition and MS1 and MS2 resolution set to unit. Acquired SRM data were imported into Skyline where confident peptide quantification was based on multiple concurrent SRM transitions above background signal and peak areas were refined with the mProphet algorithm (58).
Rice and rice growth conditions for plant defense assays
Kitaake, Ubi::Myc::XA21-Kitaake line 7-A-8 (Ubi::XA21) (25), and XA21::XA21-Kitaake (25) were grown in growth chambers at 24° or 28°C with a 14-hour/10-hour light-dark cycle at 80% humidity. Rice plants were grown in hydroponic system using A-OK Starter Plugs (Grodan) and watered with Hoagland’s solution twice a week.
Assays for ROS
Leaves of 4- to 6-week-old hydroponically grown rice plants were cut longitudinally along the mid vein and then transversely into 1- to 1.5-mm-thick leaf pieces. These leaf pieces were floated on autoclaved ddH2O overnight. The next morning, two leaf pieces were transferred into one well of a 96-well white plate containing 100 μl of excitation solution [5 to 20 mM L-012 (Wako) and horseradish peroxidase (0.5 to 2 mg/ml; Sigma)]. Leaf pieces were treated with the indicated concentration of peptides or mock-treated (ddH2O or a peptide buffer solution). Chemiluminescence was measured for 3 hours with 0.5 s per reading with a high-sensitivity plate reader (TriStar, Berthold).
Gene expression assays
Rice leaf tissue was treated with peptides as described previously with slight adaptations (59). Leaves of 4-week-old hydroponically grown rice plants were cut into 2-cm-long strips and incubated for at least 12 hours in ddH2O to reduce residual wound signal. Leaf strips were treated with the indicated peptides and for the indicated time. Leaf tissue was snap-frozen in liquid nitrogen and processed appropriately. PR10b and Os04g10010 were previously described as defense marker genes in rice (59). Other defense marker genes, Os12g36830 and Os06g37224, were identified in RNAseq experiments as up-regulated genes after elf18 treatment of rice leaves expressing EFR::XA21::GFP (60) (http://genome.jgi.doe.gov/pages/dynamicOrganismDownload.jsf?organism=OrysatspilotREDO). For quantitative reverse transcription PCRs (qRT-PCRs), the Bio-Rad SsoFast EvaGreen Supermix was used. The genes and qRT-PCR primer pairs are listed in table S1. qRT-PCRs were run for 40 cycles with annealing and amplification at 62°C for 5 s and denaturation at 95°C for 5 s. The expression levels of all defense marker genes were normalized to the actin gene expression level and to the respective mock-treated control at 0 or 1 hour.
Ethylene production
Leaves of 3- to 4-week-old hydroponically grown rice plants were cut in small horizontal strips (~0.3 to 0.5 mm) and floated on water in petri dishes overnight at room temperature. Ethylene biosynthesis was assayed by placing leaf samples in 6-ml tubes with 500 μl of water or the indicated peptide at a concentration of 1 μM as indicated. Tubes were sealed with rubber caps, and ethylene accumulating in the headspace within 4 hours of incubation was determined by gas chromatography as described (61).
RaxX39 peptide digestion
RaxX39-Y or RaxX39-sY (10 μg) was digested in buffer D (50 mM tris-HCl, 1 mM DTT) with 1 μg of one of the following site-specific proteases: GluC, ArgC, AspN, or trypsin. Digestions or mock reactions were performed at 25°C for 16 hours.
Statistical analysis
Statistical analyses were performed using JMP software (SAS Institute Inc.).
Acknowledgments
We thank W. Sun for sharing the vectors pVSP61 and pUFR80. We also thank M. G. Anil and D. Mishra for helpful discussions. Funding: Funded by NIH GM59962 and the US Israel Binational Science Foundation grant #2 2011062. This work was also conducted in part by the Joint BioEnergy Institute and was supported by the Office of Science, Office of Biological and Environmental Research, of the U.S. Department of Energy under contract no. DE-AC02-05CH11231. B.S. was supported by a European Molecular Biology Organization long-term fellowship (ALTF 1290-2011) and by a Human Frontiers Science Program long-term postdoctoral fellowship (LT000674/2012). The laboratories of P.B.P. and R.V.S. are supported by “Plant-Microbe and Soil Interactions” project (BSC0117) of the Council of Scientific and Industrial Research. J.S.B. and M.R.R. were supported by NIH R21 GM099028 (J.S.B.) and Welch Foundation F1155 (J.S.B.). D.C. was supported by Monsanto’s Beachell-Borlaug International Scholars Program fellowship. Author contributions: R.N.P., B.S., A.J., and P.C.R. conceived and planned the work. R.N.P., B.S., A.J., N.T., F.L., M.A., M.R.R., L.J.G.C., D.D.L., H.C., O.B., A.D., D.D.V., D.C., W.Z., X.Z., J.L.H., D.R., D.M., M.C., and C.J.P. performed experiments. X.L., S.M., P.B.P., R.V.S., H.K., C.C.L., and P.C.R. provided critical reagents and information. R.N.P., B.S., A.J., M.R.R., J.S.B., and P.C.R. wrote the manuscript with assistance and feedback from other authors. Competing interests: R.N.P., B.S., A.J., W.Z., and P.C.R. have filed a patent describing the isolation and expression of RaxX and its application for engineering resistance in plants (U.S. provisional patent application no. 62/013,709).
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/1/6/e1500245/DC1
Fig. S1. Sequence alignment of 15 raxX alleles.
Fig. S2. raxX is required for activation of XA21-mediated immunity in the Ubi::XA21 Kitaake (Ubi::XA21) genetic background.
Fig. S3. Alignment of the PAPS binding motif of RaxST from PXO99, TPST1 from human, and TPST2 from human.
Fig. S4. RaxX Y41 is required for activation of XA21-mediated immunity.
Fig. S5. RaxX Y41F does not trigger the XA21-mediated immune response.
Fig. S6. Purification of the full-length RaxST protein carrying an N-terminal His-tag.
Fig. S7. Expanded MS1 spectrum of HVGGGDsYPPPGANPK from trypsin digestion of in vitro sulfated RaxX39.
Fig. S8. Extracted ion chromatograms (XIC) showing the difference in ion abundances for the sulfated and nonsulfated peptide HVGGGDYPPPGANPK (2− charge state).
Fig. S9. Positive UVPD mass spectrum of HVGGGDsYPPPGANPK from trypsin digestion of in vitro sulfated RaxX39.
Fig. S10. RaxX-His proteins were purified from PXO99(praxX-His) and PXO99ΔraxST(praxX-His).
Fig. S11. SRM-MS analysis of the tryptic RaxX peptide HVGGGDYPPPGANPK from PXO99(praxX-His) and PXO99ΔraxST(praxX-His).
Fig. S12. Expression and sulfation of RaxX60-Y and RaxX60-sY.
Fig. S13. Heterologously expressed, full-length RaxX60-sY activates XA21-mediated defense gene expression.
Fig. S14. Chemically synthesized RaxX39-sY activates XA21-mediated defense gene expression.
Fig. S15. Digestion of RaxX39-sY by the four site-specific proteases GluC, trypsin, ArgC, and ApsN.
Fig. S16. A 21–amino acid sulfated peptide derived from RaxX is sufficient to activate XA21-mediated defense gene expression and the production of ROS.
Fig. S17. Chemically synthesized RaxX21-sY is sufficient to activate XA21-mediated defense gene expression.
Fig. S18. Chemically synthesized RaxX21-sY activates gene expression in Kitaake lines expressing XA21 from its native promoter (XA21:XA21).
Fig. S19. Chemically synthesized RaxX21-sY activates the production of ROS in Kitaake lines expressing XA21 from its native promoter (XA21:XA21).
Fig. S20. ROS production dose-response curves in Ubi::XA21 leaves treated with chemically synthesized RaxX sulfated peptides.
Fig. S21. The tyrosine-sulfated peptide axY(S)22, derived from Ax21, does not trigger XA21-mediated immune responses in rice.
Fig. S22. Expression and purification of different RaxX alleles.
Fig. S23. Complementation of Xoo isolates IXO651, IXO685, and IXO1221 with praxX confers the ability to activate XA21-mediated immunity.
Fig. S24. RaxX is similar to the Arabidopsis thaliana peptide signaling factors PSY1 and to rice PSY1 homologs.
Table S1. Marker genes and primers used for qPCR.
Table S2. Plasmids used in this study.
Table S3. Bacterial strains used in this study.
REFERENCES AND NOTES
- 1.Jones J. D., Dangl J. L., The plant immune system. Nature 444, 323–329 (2006). [DOI] [PubMed] [Google Scholar]
- 2.Ronald P. C., Beutler B., Plant and animal sensors of conserved microbial signatures. Science 330, 1061–1064 (2010). [DOI] [PubMed] [Google Scholar]
- 3.Kawai T., Akira S., The role of pattern-recognition receptors in innate immunity: Update on Toll-like receptors. Nat. Immunol. 11, 373–384 (2010). [DOI] [PubMed] [Google Scholar]
- 4.Schwessinger B., Ronald P. C., Plant innate immunity: Perception of conserved microbial signatures. Annu. Rev. Plant Biol. 63, 451–482 (2012). [DOI] [PubMed] [Google Scholar]
- 5.Poltorak A., He X., Smirnova I., Liu M. Y., Van Huffel C., Du X., Birdwell D., Alejos E., Silva M., Galanos C., Freudenberg M., Ricciardi-Castagnoli P., Layton B., Beutler B., Defective LPS signaling in C3H/HeJ and C57BL/10ScCr mice: Mutations in Tlr4 gene. Science 282, 2085–2088 (1998). [DOI] [PubMed] [Google Scholar]
- 6.Hayashi F., Smith K. D., Ozinsky A., Hawn T. R., Yi E. C., Goodlett D. R., Eng J. K., Akira S., Underhill D. M., Aderem A., The innate immune response to bacterial flagellin is mediated by Toll-like receptor 5. Nature 410, 1099–1103 (1998). [DOI] [PubMed] [Google Scholar]
- 7.Song W.-Y., Wang G.-L., Chen L.-L., Kim H.-S., Pi L.-Y., Holsten T., Gardner J., Wang B., Zhai W.-X., Zhu L.-H., Fauquet C., Ronald P., A receptor kinase-like protein encoded by the rice disease resistance gene, Xa21. Science 270, 1804–1806 (1998). [DOI] [PubMed] [Google Scholar]
- 8.Chen X., Shang J., Chen D., Lei C., Zou Y., Zhai W., Liu G., Xu J., Ling Z., Cao G., Ma B., Wang Y., Zhao X., Li S., Zhu L., A B-lectin receptor kinase gene conferring rice blast resistance. Plant J. 46, 794–804 (2006). [DOI] [PubMed] [Google Scholar]
- 9.Sun X., Cao Y., Yang Z., Xu C., Li X., Wang S., Zhang Q., Xa26, a gene conferring resistance to Xanthomonas oryzae pv. oryzae in rice, encodes an LRR receptor kinase-like protein. Plant J. 37, 517–527 (2004). [DOI] [PubMed] [Google Scholar]
- 10.Gomez-Gomez L., Boller T., FLS2: An LRR receptor–like kinase involved in the perception of the bacterial elicitor flagellin in Arabidopsis. Mol. Cell 5, 1003–1011 (2000). [DOI] [PubMed] [Google Scholar]
- 11.Macho A. P., Zipfel C., Plant PRRs and the activation of innate immune signaling. Mol. Cell 54, 263–272 (2014). [DOI] [PubMed] [Google Scholar]
- 12.Cao Y., Liang Y., Tanaka K., Nguyen C. T., Jedrzejczak R. P., Joachimiak A., Stacey G., The kinase LYK5 is a major chitin receptor in Arabidopsis and forms a chitin-induced complex with related kinase CERK1. ELife 3, e03766 (2014) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hayafune M., Berisio R., Marchetti R., Silipo A., Kayama M., Desaki Y., Arima S., Squeglia F., Ruggiero A., Tokuyasu K., Molinaro A., Kaku H., Shibuya N., Chitin-induced activation of immune signaling by the rice receptor CEBiP relies on a unique sandwich-type dimerization. Proc. Natl. Acad. Sci. U.S.A. 111, E404–E413 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zipfel C., Kunze G., Chinchilla D., Caniard A., Jones J. D., Boller T., Felix G., Perception of the bacterial PAMP EF-Tu by the receptor EFR restricts Agrobacterium-mediated transformation. Cell 125, 749–760 (2006). [DOI] [PubMed] [Google Scholar]
- 15.Dardick C., Ronald P., Plant and animal pathogen recognition receptors signal through non-RD kinases. PLOS Pathog. 2, e2 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dardick C., Schwessinger B., Ronald P., Non-arginine-aspartate (non-RD) kinases are associated with innate immune receptors that recognize conserved microbial signatures. Curr. Opin. Plant Biol. 15, 358–366 (2012). [DOI] [PubMed] [Google Scholar]
- 17.Berrabah F., Bourcy M., Eschstruth A., Cayrel A., Guefrachi I., Mergaert P., Wen J., Jean V., Mysore K. S., Gourion B., Ratet P., A nonRD receptor-like kinase prevents nodule early senescence and defense-like reactions during symbiosis. New Phytol. 203, 1305–1314 (2014). [DOI] [PubMed] [Google Scholar]
- 18.Khush G. S., Bacalangco E., Ogawa T., A new gene for resistance to bacterial blight from O. longistaminata. Rice Genet. Newsl. 7, 121–122 (1990). [Google Scholar]
- 19.da Silva F. G., Shen Y., Dardick C., Burdman S., Yadav R. C., de Leon A. L., Ronald P. C., Bacterial genes involved in type I secretion and sulfation are required to elicit the rice Xa21-mediated innate immune response. Mol. Plant Microbe Interact. 17, 593–601 (2004). [DOI] [PubMed] [Google Scholar]
- 20.Lee S. W., Han S. W., Sririyanum M., Park C. J., Seo Y. S., Ronald P. C., Retraction. A type I–secreted, sulfated peptide triggers XA21-mediated innate immunity. Science 342, 191 (2013). [DOI] [PubMed] [Google Scholar]
- 21.Ronald P., Lab Life: The anatomy of a retraction. Sci. Am. (2013). [Google Scholar]
- 22.Bahar O., Pruitt R., Luu D. D., Schwessinger B., Daudi A., Liu F., Ruan R., Fontaine-Bodin L., Koebnik R., Ronald P., The Xanthomonas Ax21 protein is processed by the general secretory system and is secreted in association with outer membrane vesicles. PeerJ 2, e242 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Michiels J., Dirix G., Vanderleyden J., Xi C., Processing and export of peptide pheromones and bacteriocins in Gram-negative bacteria. Trends Microbiol. 9, 164–168 (2001). [DOI] [PubMed] [Google Scholar]
- 24.Salzberg S. L., Sommer D. D., Schatz M. C., Phillippy A. M., Rabinowicz P. D., Tsuge S., Furutani A., Ochiai H., Delcher A. L., Kelley D., Madupu R., Puiu D., Radune D., Shumway M., Trapnell C., Aparna G., Jha G., Pandey A., Patil P. B., Ishihara H., Meyer D. F., Szurek B., Verdier V., Koebnik R., Dow J. M., Ryan R. P., Hirata H., Tsuyumu S., Won Lee S., Seo Y. S., Sriariyanum M., Ronald P. C., Sonti R. V., Van Sluys M. A., Leach J. E., White F. F., Bogdanove A. J., Genome sequence and rapid evolution of the rice pathogen Xanthomonas oryzae pv. oryzae PXO99A. BMC Genomics 9, 204 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Park C. J., Lee S. W., Chern M., Sharma R., Canlas P. E., Song M. Y., Jeon J. S., Ronald P. C., Ectopic expression of rice Xa21 overcomes developmentally controlled resistance to Xanthomonas oryzae pv. oryzae. Plant Sci. 179, 466–471 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shen Y., Sharma P., da Silva F. G., Ronald P., The Xanthomonas oryzae pv. lozengeoryzae raxP and raxQ genes encode an ATP sulphurylase and adenosine-5′-phosphosulphate kinase that are required for AvrXa21 avirulence activity. Mol. Microbiol. 44, 37–48 (2002). [DOI] [PubMed] [Google Scholar]
- 27.Teramoto T., Fujikawa Y., Kawaguchi Y., Kurogi K., Soejima M., Adachi R., Nakanishi Y., Mishiro-Sato E., Liu M. C., Sakakibara Y., Suiko M., Kimura M., Kakuta Y., Crystal structure of human tyrosylprotein sulfotransferase-2 reveals the mechanism of protein tyrosine sulfation reaction. Nat. Commun. 4, 1572 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Han S.-W., Lee S.-W., Bahar O., Schwessinger B., Robinson M. R., Shaw J. B., Madsen J. A., Brodbelt J. S., Ronald P. C., Tyrosine sulfation in a Gram-negative bacterium. Nat. Commun. 3, 1153 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Robinson M. R., Moore K. L., Brodbelt J. S., Direct identification of tyrosine sulfation by using ultraviolet photodissociation mass spectrometry. J. Am. Soc. Mass Spectrom. 25, 1461–1471 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu C. C., Schultz P. G., Recombinant expression of selectively sulfated proteins in Escherichia coli. Nat. Biotechnol. 24, 1436–1440 (2006). [DOI] [PubMed] [Google Scholar]
- 31.Lajoie M. J., Rovner A. J., Goodman D. B., Aerni H.-R., Haimovich A. D., Kuznetsov G., Mercer J. A., Wang H. H., Carr P. A., Mosberg J. A., Rohland N., Schultz P. G., Jacobson J. M., Rinehart J., Genomically recoded organisms expand biological functions. Science 342, 357–360 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu C. C., Cellitti S. E., Geierstanger B. H., Schultz P. G., Efficient expression of tyrosine-sulfated proteins in E. coli using an expanded genetic code. Nat. Protoc. 4, 1784–1789 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Boller T., Felix G., A renaissance of elicitors: Perception of microbe-associated molecular patterns and danger signals by pattern-recognition receptors. Annu. Rev. Plant Biol. 60, 379–406 (2009). [DOI] [PubMed] [Google Scholar]
- 34.Wang G. L., Song W. Y., Ruan D. L., Sideris S., Ronald P. C., The cloned gene, Xa21, confers resistance to multiple Xanthomonas oryzae pv. oryzae isolates in transgenic plants. Mol. Plant Microbe Interact. 9, 850–855 (1996). [DOI] [PubMed] [Google Scholar]
- 35.Mishra D., Vishnupriya M. R., Anil M. G., Konda K., Raj Y., Sonti R. V., Pathotype and genetic diversity amongst Indian isolates of Xanthomonas oryzae pv. oryzae. PLOS One 8, e81996 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang C. L., Qin T. F., Yu H. M., Zhang X. P., Che J. Y., Gao Y., Zheng C. K., Yang B., Zhao K. J., The broad bacterial blight resistance of rice line CBB23 is triggered by a novel transcription activator-like (TAL) effector of Xanthomonas oryzae pv. oryzae. Mol. Plant Pathol. 15, 333–341 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Farzan M., Mirzabekov T., Kolchinsky P., Wyatt R., Cayabyab M., Gerard N. P., Gerard C., Sodroski J., Choe H., Tyrosine sulfation of the amino terminus of CCR5 facilitates HIV-1 entry. Cell 96, 667–676 (1999). [DOI] [PubMed] [Google Scholar]
- 38.Matsubayashi Y., Posttranslationally modified small-peptide signals in plants. Annu. Rev. Plant Biol. 65, 385–413 (2014). [DOI] [PubMed] [Google Scholar]
- 39.Mosher S., Seybold H., Rodriguez P., Stahl M., Davies K. A., Dayaratne S., Morillo S. A., Wierzba M., Favery B., Keller H., Tax F. E., Kemmerling B., The tyrosine-sulfated peptide receptors PSKR1 and PSY1R modify the immunity of Arabidopsis to biotrophic and necrotrophic pathogens in an antagonistic manner. Plant J. 73, 469–482 (2014). [DOI] [PubMed] [Google Scholar]
- 40.Amano Y., Tsubouchi H., Shinohara H., Ogawa M., Matsubayashi Y., Tyrosine-sulfated glycopeptide involved in cellular proliferation and expansion in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 104, 18333–18338 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mosher S., Kemmerling B., PSKR1 and PSY1R-mediated regulation of plant defense responses. Plant Signal. Behav. 8, e24119 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sundaram M., Vishnupriya M. R., Biradar S. K., Laha G. S., Reddy G. A., Rani N. S., Sarma N. P., Sonti R. V., Marker assisted introgression of bacterial blight resistance in Samba Mahsuri, an elite indica rice variety. Euphytica 160, 411–422 (2008). [Google Scholar]
- 43.Huang B., Xu J. Y., Hou M. S., Ali J., Mou T. M., Introgression of bacterial blight resistance genes Xa7, Xa21, Xa22 and Xa23 into hybrid rice restorer lines by molecular marker-assisted selection. Euphytica 187, 449–459 (2012). [Google Scholar]
- 44.Chen S., Lin X. H., Xu C. G., Zhang Q., Improvement of bacterial blight resistance of ‘Minghui 63’, an elite restorer line of hybrid rice, by molecular marker-assisted selection. Crop Sci. 40, 239–244 (2000). [Google Scholar]
- 45.Toenniessen G. H., O’Toole J. C., DeVries J., Advances in plant biotechnology and its adoption in developing countries. Curr. Opin. Plant Biol. 6, 191–198 (2003). [DOI] [PubMed] [Google Scholar]
- 46.Gopalakrishnan S., Sharma R. K., Rajkumar K. A., Joseph M., Singh V. P., Singh A. K., Bhat K. V., Singh N. K., Mohapatra T., Integrating marker assisted background analysis with foreground selection for identification of superior bacterial blight resistant recombinants in Basmati rice. Plant Breed. 127, 131–139 (2008). [Google Scholar]
- 47.Win K. M., Korinsak S., Jantaboon J., Siangliw M., Lanceras-Siangliw J., Sirithunya P., Vanavichit A., Pantuwan G., Jongdee B., Sidhiwong N., Toojinda T., Breeding the Thai jasmine rice variety kdml105 for non-age-related broad-spectrum resistance to bacterial blight disease based on combined marker-assisted and phenotypic selection. Field Crops Res. 137, 186–194 (2012). [Google Scholar]
- 48.Gardner M. R., Kattenhorn L. M., Kondur H. R., M von Schaewen, Dorfman T., Chiang J. J., Haworth K. G., Decker J. M., Alpert M. D., Bailey C. C., Neale E. S. Jr, Fellinger C. H., Joshi V. R., Fuchs S. P., Martinez-Navio J. M., Quinlan B. D., Yao A. Y., Mouquet H., Gorman J., Zhang B., Poignard P., Nussenzweig M. C., Burton D. R., Kwong P. D., Piatak M. Jr, Lifson J. D., Gao G., Desrosiers R. C., Evans D. T., Hahn B. H., Ploss A., Cannon P. M., Seaman M. S., Farzan M., AAV-expressed eCD4-Ig provides durable protection from multiple SHIV challenges. Nature 519, 87–91 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hopkins C. M., White F. F., Choi S. H., Guo A., Leach J. E., Identification of a family of avirulence genes from Xanthomonas oryzae pv. oryzae. Mol. Plant Microbe Interact. 5, 451–459 (1992). [DOI] [PubMed] [Google Scholar]
- 50.Penfold R. J., Pemberton J. M., An improved suicide vector for construction of chromosomal insertion mutations in bacteria. Gene 118, 145–146 (1992). [DOI] [PubMed] [Google Scholar]
- 51.Castañeda A., Reddy J. D., El-Yacoubi B., Gabriel D. W., Mutagenesis of all eight avr genes in Xanthomonas campestris pv. campestris had no detected effect on pathogenicity, but one avr gene affected race specificity. Mol. Plant Microbe Interact. 18, 1306–1317 (2005). [DOI] [PubMed] [Google Scholar]
- 52.Loper J. E., Lindow S. E., A biological sensor for iron available to bacteria in their habitats on plant surfaces. Appl. Environ. Microbiol. 60, 1934–1941 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kauffman H., Reddy A., Hsieh S., Merca S., Improved technique for evaluating resistance of rice varieties to Xanthomonas oryzae. Plant Dis. Rep. 57, 537–541 (1973). [Google Scholar]
- 54.Chatterjee A., Sun S. B., Furman J. L., Xiao H., Schultz P. G., A versatile platform for single- and multiple-unnatural amino acid mutagenesis in Escherichia coli. Biochemistry 52, 1828–1837 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Madsen J. A., Xu H., Robinson M. R., Horton A. P., Shaw J. B., Giles D. K., Kaoud T. S., Dalby K. N., Trent M. S., Brodbelt J. S., High-throughput database search and large-scale negative polarity liquid chromatography-tandem mass spectrometry with ultraviolet photodissociation for complex proteomic samples. Mol. Cell. Proteomics 12, 2604–2614 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wengelnik K., Bonas U., HrpXv, an AraC-type regulator, activates expression of five of the six loci in the hrp cluster of Xanthomonas campestris pv. vesicatoria. J. Bacteriol. 178, 3462–3469 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.MacLean B., Tomazela D. M., Shulman N., Chambers M., Finney G. L., Frewen B., Kern R., Tabb D. L., Liebler D. C., MacCoss M. J., Skyline: An open source document editor for creating and analyzing targeted proteomics experiments. Bioinformatics 26, 966–968 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Reiter L., Rinner O., Picotti P., Hüttenhain R., Beck M., Brusniak M. Y., Hengartner M. O., Aebersold R., mProphet: Automated data processing and statistical validation for large-scale SRM experiments. Nat. Methods 8, 430–435 (2011). [DOI] [PubMed] [Google Scholar]
- 59.Chen X., Zuo S., Schwessinger B., Chern M., Canlas P. E., Ruan D., Zhou X., Wang J., Daudi A., Petzold C. J., Heazlewood J. L., Ronald P. C., An XA21-associated kinase (OsSERK2) regulates immunity mediated by the XA21 and XA3 immune receptors. Mol. Plant 7, 874–892 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Schwessinger B., Bahar O., Thomas N., Holton N., Nekrasov V., Ruan D., Canlas P. E., Daudi A., Petzold C. J., Singan V. R., Kuo R., Chovatia M., Daum C., Heazlewood J. L., Zipfel C., Ronald P. C., Transgenic expression of the dicotyledonous pattern recognition receptor EFR in rice leads to ligand-dependent activation of defense responses. PLOS Pathog. 11, e1004809 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Felix G., Duran J. D., Volko S., Boller T., Plants have a sensitive perception system for the most conserved domain of bacterial flagellin. Plant J. 18, 265–276 (1999). [DOI] [PubMed] [Google Scholar]
- 62.Kearse M., Moir R., Wilson A., Stones-Havas S., Cheung M., Sturrock S., Buxton S., Cooper A., Markowitz S., Duran C., Thierer T., Ashton B., Meintjes P., Drummond A., Geneious Basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28, 1647–1649 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.McGee J. D., Hamer J. E., Hodges T. K., Characterization of a PR-10 pathogenesis-related gene family induced in rice during infection with Magnaporthe grisea. Mol. Plant Microbe Interact. 14, 877–886 (2001). [DOI] [PubMed] [Google Scholar]
- 64.Shimura K., Okada A., Okada K., Jikumaru Y., Ko K. W., Toyomasu T., Sassa T., Hasegawa M., Kodama O., Shibuya N., Koga J., Nojiri H., Yamane H., Identification of a biosynthetic gene cluster in rice for momilactones. J. Biol. Chem. 282, 34013–34018 (2007). [DOI] [PubMed] [Google Scholar]
- 65.Atawong A., Hasegawa M., Kodama O., Biosynthesis of rice phytoalexin: Enzymatic conversion of 3β-hydroxy-9β-pimara-7,15-dien-19,6β-olide to momilactone A. Biosci. Biotechnol. Biochem. 66, 566–570 (2002). [DOI] [PubMed] [Google Scholar]
- 66.Hashimoto M., Kisseleva L., Sawa S., Furukawa T., Komatsu S., Koshiba T., A novel rice PR10 protein, RSOsPR10, specifically induced in roots by biotic and abiotic stresses, possibly via the jasmonic acid signaling pathway. Plant Cell Physiol. 45, 550–559 (2004). [DOI] [PubMed] [Google Scholar]
- 67.Itoh H., Tatsumi T., Sakamoto T., Otomo K., Toyomasu T., Kitano H., Ashikari M., Ichihara S., Matsuoka M., A rice semi-dwarf gene, Tan-Ginbozu (D35), encodes the gibberellin biosynthesis enzyme, ent-kaurene oxidase. Plant Mol. Biol. 54, 533–547 (2004). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/1/6/e1500245/DC1
Fig. S1. Sequence alignment of 15 raxX alleles.
Fig. S2. raxX is required for activation of XA21-mediated immunity in the Ubi::XA21 Kitaake (Ubi::XA21) genetic background.
Fig. S3. Alignment of the PAPS binding motif of RaxST from PXO99, TPST1 from human, and TPST2 from human.
Fig. S4. RaxX Y41 is required for activation of XA21-mediated immunity.
Fig. S5. RaxX Y41F does not trigger the XA21-mediated immune response.
Fig. S6. Purification of the full-length RaxST protein carrying an N-terminal His-tag.
Fig. S7. Expanded MS1 spectrum of HVGGGDsYPPPGANPK from trypsin digestion of in vitro sulfated RaxX39.
Fig. S8. Extracted ion chromatograms (XIC) showing the difference in ion abundances for the sulfated and nonsulfated peptide HVGGGDYPPPGANPK (2− charge state).
Fig. S9. Positive UVPD mass spectrum of HVGGGDsYPPPGANPK from trypsin digestion of in vitro sulfated RaxX39.
Fig. S10. RaxX-His proteins were purified from PXO99(praxX-His) and PXO99ΔraxST(praxX-His).
Fig. S11. SRM-MS analysis of the tryptic RaxX peptide HVGGGDYPPPGANPK from PXO99(praxX-His) and PXO99ΔraxST(praxX-His).
Fig. S12. Expression and sulfation of RaxX60-Y and RaxX60-sY.
Fig. S13. Heterologously expressed, full-length RaxX60-sY activates XA21-mediated defense gene expression.
Fig. S14. Chemically synthesized RaxX39-sY activates XA21-mediated defense gene expression.
Fig. S15. Digestion of RaxX39-sY by the four site-specific proteases GluC, trypsin, ArgC, and ApsN.
Fig. S16. A 21–amino acid sulfated peptide derived from RaxX is sufficient to activate XA21-mediated defense gene expression and the production of ROS.
Fig. S17. Chemically synthesized RaxX21-sY is sufficient to activate XA21-mediated defense gene expression.
Fig. S18. Chemically synthesized RaxX21-sY activates gene expression in Kitaake lines expressing XA21 from its native promoter (XA21:XA21).
Fig. S19. Chemically synthesized RaxX21-sY activates the production of ROS in Kitaake lines expressing XA21 from its native promoter (XA21:XA21).
Fig. S20. ROS production dose-response curves in Ubi::XA21 leaves treated with chemically synthesized RaxX sulfated peptides.
Fig. S21. The tyrosine-sulfated peptide axY(S)22, derived from Ax21, does not trigger XA21-mediated immune responses in rice.
Fig. S22. Expression and purification of different RaxX alleles.
Fig. S23. Complementation of Xoo isolates IXO651, IXO685, and IXO1221 with praxX confers the ability to activate XA21-mediated immunity.
Fig. S24. RaxX is similar to the Arabidopsis thaliana peptide signaling factors PSY1 and to rice PSY1 homologs.
Table S1. Marker genes and primers used for qPCR.
Table S2. Plasmids used in this study.
Table S3. Bacterial strains used in this study.