DNA sequencing uncovers the entire architecture of below-ground plant–fungus networks.
Keywords: plant communities, fungi, Ecology, network, mycorrhizae, mutualism, community stability, DNA barcoding, soil ecology, symbiosis
Abstract
In nature, plants and their pollinating and/or seed-dispersing animals form complex interaction networks. The commonly observed pattern of links between specialists and generalists in these networks has been predicted to promote species coexistence. Plants also build highly species-rich mutualistic networks below ground with root-associated fungi, and the structure of these plant–fungus networks may also affect terrestrial community processes. By compiling high-throughput DNA sequencing data sets of the symbiosis of plants and their root-associated fungi from three localities along a latitudinal gradient, we uncovered the entire network architecture of these interactions under contrasting environmental conditions. Each network included more than 30 plant species and hundreds of mycorrhizal and endophytic fungi belonging to diverse phylogenetic groups. The results were consistent with the notion that processes shaping host-plant specialization of fungal species generate a unique linkage pattern that strongly contrasts with the pattern of above-ground plant–partner networks. Specifically, plant–fungus networks lacked a “nested” architecture, which has been considered to promote species coexistence in plant–partner networks. Rather, the below-ground networks had a conspicuous “antinested” topology. Our findings lead to the working hypothesis that terrestrial plant community dynamics are likely determined by the balance between above-ground and below-ground webs of interspecific interactions.
INTRODUCTION
Ever since its introduction to ecology, network theory has repeatedly reorganized our understanding of the laws and processes that drive ecological community dynamics (1–3). The architecture of ecological networks and its consequences for community stability have been intensively investigated in mutualistic interactions involving plants and their pollinating or seed-dispersing animal partners (4–7). These plant–animal interactions commonly exhibit a “nested” network architecture (3, 8), in which specialists (that is, species with narrow partner ranges) mainly interact with subsets of the partners of generalists (4, 8) (Fig. 1A). Because nestedness is so prevalent in plant–animal mutualistic networks (3, 8), understanding its impact on plant community processes is the key to understanding how biotic environmental changes (for example, extinction of indigenous partners or introduction of alien partners) can alter plant community structure (9).
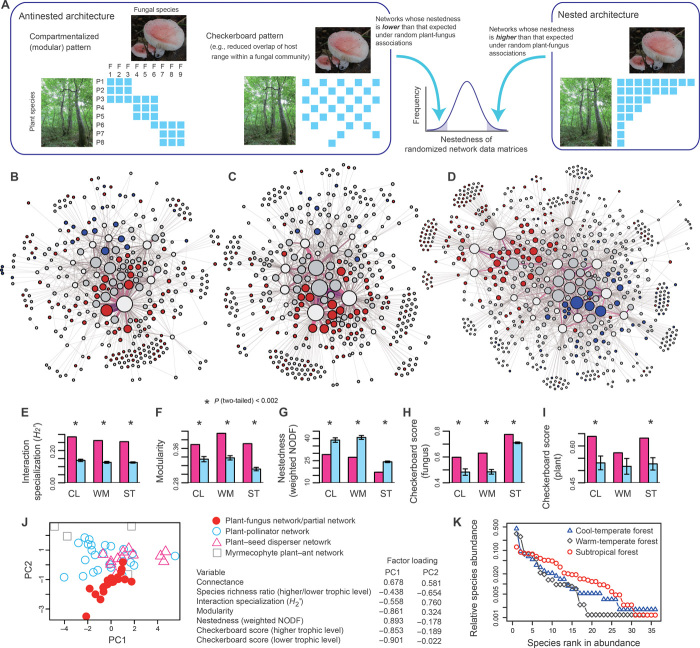
Fig. 1. Plant–fungus network architecture.
(A) Schematic example of nested and antinested plant–fungus associations. In a nested network, specialists (that is, species with narrow partner ranges) interact with subsets of the partners of generalists (that is, species with broad partner ranges). Networks whose nestedness estimates are higher/lower than that expected by chance are regarded as nested/antinested. Antinestedness can result from compartmentalized and checkerboard network patterns. (B to K) Observed network structure. In each network of (B) cool-temperate (CL) (36 plants and 278 fungi), (C) warm-temperate (WM) (33 plants and 343 fungi), and (D) subtropical (ST) (36 plants and 580 fungi) forests, ectomycorrhizal fungi (red), arbuscular mycorrhizal fungi (blue), and fungi with unknown functions (gray) are linked with their host plants (white). The size of circles represents the relative abundances of fungi or plants in each network. The network-level interaction specialization (E), modularity (F), and nestedness (G) of each network (red bar) are shown with those calculated for randomized networks (blue bar; ±SD). Asterisks represent significant deviations from randomized index values. In addition, checkerboard scores representing how the overlap of host/symbiont plants is avoided in fungal (H) or plant (I) communities are shown. With the use of those network indices, a principal component analysis was also performed (J). The examined ecological networks are plotted on the surface defined by principal components axes (PC1, principal component 1; PC2, principal component 2). For the networks of plants and their root-associated fungi, each index was calculated also for the partial networks representing associations between particular functional or taxonomic groups of fungi and their host plants. The “rank abundance” curve representing the compositional evenness/unevenness of each plant community (K) is also shown.
Theory predicts that a nested network architecture can promote plant species coexistence by offsetting among-plant competition, increasing persistence against random extinctions, and promoting facilitation (3, 4, 9). Moreover, a recent study (2) argued that a nested architecture can enhance species coexistence in mutualistic networks by increasing structural stability, which is mathematically defined as the range of parameter values that realize both feasible and dynamically stable equilibria. Thus, knowledge of such a potential link between network nestedness and species coexistence is crucial to preventing further plant biodiversity loss and consequential degrading of terrestrial ecosystem services worldwide.
Despite the potential importance of network architecture to plant species coexistence and the advances promoted by the study of plant–animal mutualism, we remain ignorant of the network structural features of most major forms of plant–partner interactions. In terrestrial ecosystems, plant species mutualistically interact not only with pollinators and seed dispersers but also with functionally and taxonomically diverse root-associated fungi (10, 11). Mycorrhizal symbiosis ecologically differs from interactions between plants and most of their pollinating and seed-dispersing agents because mycorrhizal fungi form physiologically intimate associations with host plants and some can simultaneously interact with more than one host individual (11, 12). Furthermore, because the symbiosis of plants and root-associated fungi originated early in the history of land plants, it is a ubiquitous and major component of all terrestrial ecosystems (10, 13, 14) involving broader taxonomic ranges of plant species than pollination and seed dispersal interactions (15–17). In below-ground plant–fungus symbiosis, not only mycorrhizal fungi, but also various clades of endophytic fungi supply host plants with soil nutrients and receive photosynthetic carbohydrates in return (10, 11). Among the major groups of root-associated fungi, ectomycorrhizal fungi display a relatively high host specificity and are considered to promote the dominance of plants in specific families such as Fagaceae, Pinaceae, and Dipterocarpaceae (11, 18). In contrast, arbuscular mycorrhizal and root-endophytic fungi generally have broad host ranges (11, 19), potentially working as interaction network hubs (5, 20) in plant–fungus networks and thereby connecting otherwise isolated groups of species (5). In addition to these mutualistic fungi, potentially commensalistic or antagonistic fungi may also be involved in interaction webs with plants (15–17). Consequently, analyses of how these symbiotic networks embrace multiple functional groups of fungi and their host plants provide pivotal opportunities for examining relationships between network architecture and species coexistence in ecologically complex interactions (1, 3, 4).
In a previous study, we found that a large network of 33 plant species and their root-associated fungi in a warm-temperate forest (35°02′N, 135°47′E) had unique properties, including the absence of nestedness (21). In this study, we examine the generality of those architectural features in plant–fungus networks by compiling additional large next-generation sequencing (NGS) data sets of the symbiosis of plants and their root-associated fungi in two contrasting forests in Japan [cool-temperate (42°40′N, 141°36′E) (17) and subtropical (30°26′N, 130°30′E) (15) sites] (fig. S1 and data file S1). Incorporation of these additional communities, together with the use of NGS, allows not only a comparative analysis of variation in the architectural properties of these networks of whole plant and fungal communities along a latitudinal gradient but also a deeper analysis of the phylogenetic and functional complexity of ecological networks than has been previously possible. We evaluate how these results differ in some important respects from patterns found in other kinds of ecological networks.
RESULTS
Network architecture
All three plant–fungus networks shared some architectural properties with other kinds of species networks (Fig. 1, figs. S2 and S3, and tables S1 and S2). Plants and fungi in each forest were associated with a narrower range of partners than expected under null models that assumed random associations between hosts and symbionts (Fig. 1E and table S1). Moreover, the networks were compartmentalized into modules (5) of closely associated plants and fungi (Fig. 1F and fig. S4), although the estimated modularity was as low as that observed in other ecological networks (fig. S3).
Although these features are also commonly reported in other ecological networks (5, 22), the network structure of plant–fungus networks was idiosyncratic in terms of nestedness (Fig. 1J, fig. S3, and table S1). These plant–fungus networks consistently lacked a nested interaction structure (Fig. 1G and table S1) despite variation in climatic environments and local plant community structures among the three forests (Fig. 1K). Rather, nestedness in these networks was consistently lower than expected by chance, forming a linked pattern termed “antinestedness” (Fig. 1G and table S1) (23). Antinestedness is defined by the structure of networks whose nestedness is significantly lower than that observed in randomized network matrices (23). Therefore, nested linkage patterns, in which specialists interact with subsets of the partners of generalists, were significantly broken in plant–fungus networks. We used antinestedness as a measure of deviation from nestedness, which has been placed at the core of network architecture–species coexistence debates (1–4). We focused our subsequent analyses on the continuum spanning from nested to antinested network patterns, examining whether an antinested pattern per se can be a consequence of other community-assembly patterns such as network compartmentalization or highly differentiated partner ranges (23) (Fig. 1A) because nested networks can simultaneously have a compartmentalized (modular) architecture (24).
Partial networks
Because our plant–fungus networks included functionally and taxonomically more diverse species than previously investigated plant–animal networks (fig. S1), each of the examined networks could represent a network of semi-independent networks (that is, a highly compartmentalized network), resulting in antinestedness (Fig. 1A) (23). If so, we would expect antinestedness not to characterize “partial” networks, each of which is composed of a single functional or taxonomic group of fungi and their host plants (Fig. 2). Contrary to this expectation, an antinested architecture persisted in 9 of 15 examined “partial” networks (Fig. 3 and table S1). In addition, even for each network module of closely connected plants and fungi (fig. S4, A to C), interactions were nonnested or even antinested (fig. S4, D to I). Albeit rare, the presence of such an antinested architecture has been previously reported in small plant–fungus networks with a few plant species (25) and in networks of host plants and protective ants (26). Our results show that antinestedness persists in large networks of interacting plants and fungi. Given these past studies and our present results, an antinested architecture, rather than a nested architecture, could be a predominant property of below-ground plant–fungus networks.
Fig. 2. Partial networks.
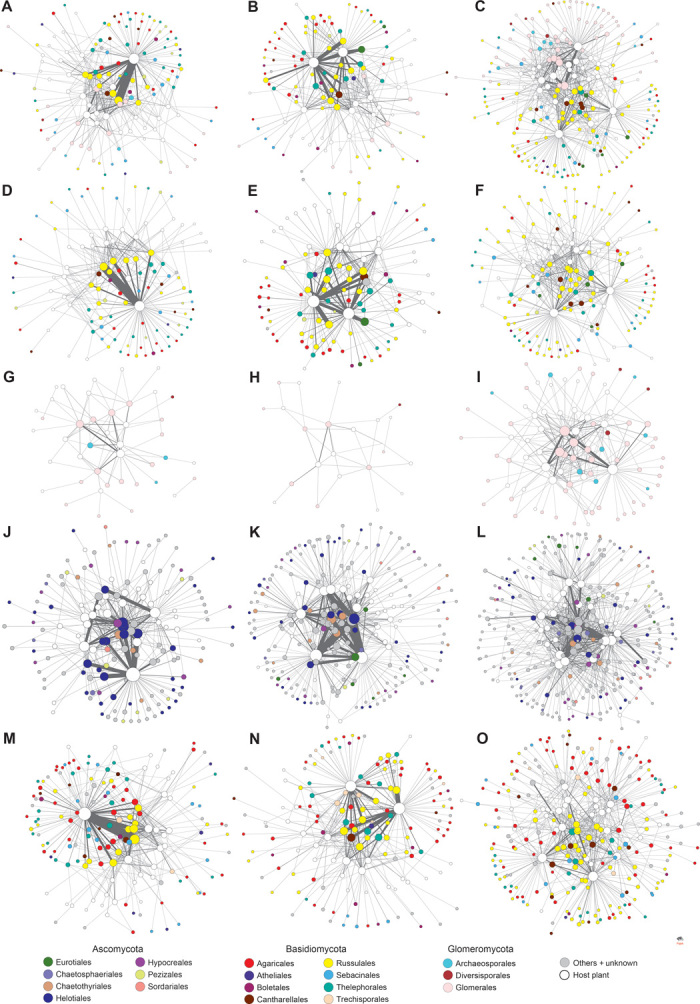
The architecture of partial networks, including each functional or taxonomic group of fungi and their host plants, is shown for the cool-temperate (left), warm-temperate (middle), and subtropical (right) forests. (A to C) Mycorrhizal partial networks including both ectomycorrhizal and arbuscular mycorrhizal fungi. (D to F) Ectomycorrhizal partial networks. (G to I) Arbuscular mycorrhizal partial networks. (J to L) Ascomycete partial networks. (M to O) Basidiomycete partial networks.
Fig. 3. Topological properties of partial networks.
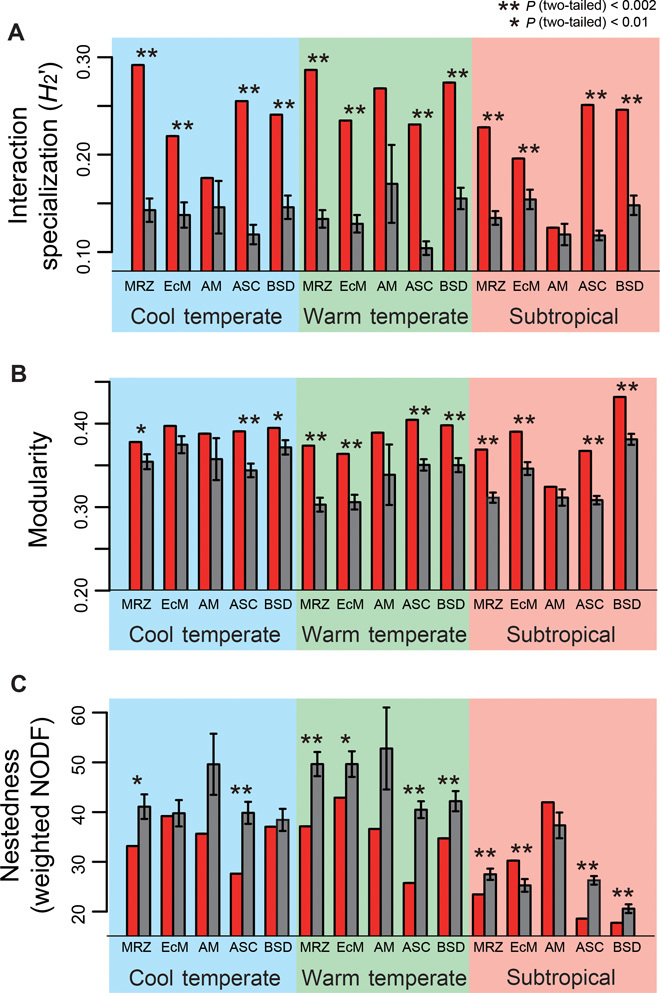
(A to C) The network-level interaction specialization (A), modularity (B), and nestedness (C) of each partial network (red bar) are shown with those calculated for randomized networks (gray bar; ±SD). Asterisks represent significant deviations from randomized index values. MRZ, mycorrhizal partial network; EcM, ectomycorrhizal partial network; AM, arbuscular mycorrhizal partial network; ASC, ascomycete partial network; BSD, basidiomycete partial network.
Network architectural properties correlated with nestedness/antinestedness
We next explored how specific network structural factors may correlate with antinestedness. Although highly specialized and highly compartmentalized plant–fungus networks and partial networks were strongly antinested (Fig. 4, A and B), there was a clearer relationship between antinestedness and reduced host range overlap (Fig. 1H) in fungal communities (Fig. 4C and fig. S5). That is, strong antinested patterns were observed in the networks and partial networks in which host-plant overlap is reduced, potentially because of competition among fungal species or other factors such as habitat preferences (Fig. 4C). Plant communities, on the other hand, behaved differently. That is, variation in partner overlap was lower in plant communities (Fig. 4D) than in fungal communities (Fig. 4C) and no correlation was observed between the range of symbiont overlap among plant species and network (or partial network) antinestedness (Fig. 4D). A multiple regression analysis further indicated that reduced host range overlap significantly explained the variation found in the nestedness/antinestedness of plant–fungus networks (Fig. 4E).
Fig. 4. Network properties explaining variation in (anti)nestedness.
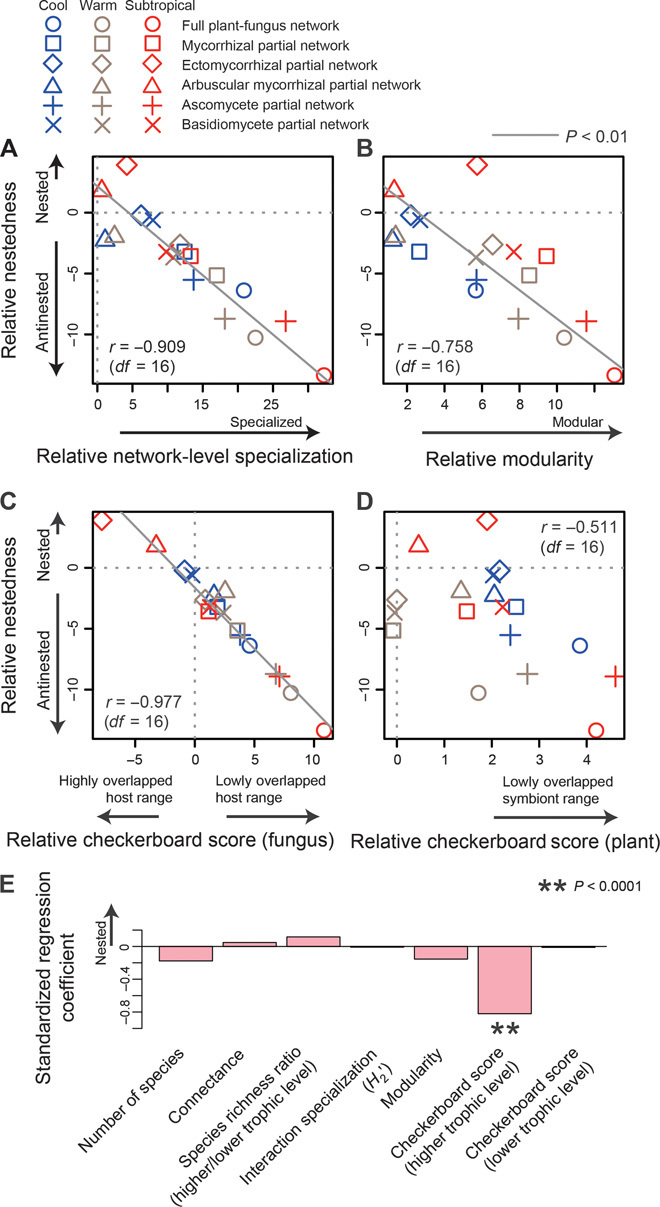
(A to D) Weighted NODF nestedness (vertical axes) and other network indices (horizontal axes) were calculated for each plant–fungus network or partial network. Positive/negative values indicate that network index estimates of observed plant–fungus association matrices are larger/smaller than those of randomized matrices (that is, relative nestedness, etc.). There was a significant correlation between nestedness and network-level specialization (A) or modularity (B). Further conspicuously, stronger antinestedness was observed for the networks and partial networks with less host-plant overlap in fungal communities (C). The extent of the within-plant-community overlap of fungal symbionts displayed no relationship with network nestedness (D). (E) Multiple regression analysis (df = 10) further indicated that variation in relative nestedness was solely explained by the extent of host range overlap among fungal species.
Roles of each fungal species
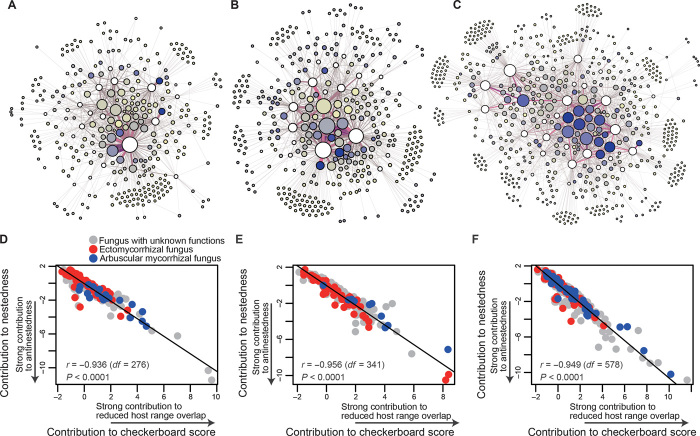
The inferred strong relationship between reduced host range overlap and antinestedness was also supported by a correlational analysis evaluating the roles of each fungal species in plant–fungus networks (Fig. 5 and fig. S6). In each network, fungal species that reduced host range overlap in fungal communities also increased antinestedness (Fig. 5). Competition for space in host root systems (27, 28) may select for fungi that avoid host range overlap and may be a potential process leading to an antinested architecture in plant–fungus networks. This proposition leads to the working hypothesis that competition with earlier fungal colonizers plays key roles in the emergence of antinestedness through below-ground community assembly processes.
Fig. 5. Contribution of each fungal species.
(A to C) Fungal species with strong effects on network antinestedness. In each of the cool-temperate (A), warm-temperate (B), and subtropical (C) forests, fungal species that strongly contribute to decreasing network nestedness are indicated in dark blue. White circles represent plant species. (D to F) Relationship between the contribution of each fungal species to network nestedness (vertical axes) and the contribution of each fungal species to reduced host range overlap in a fungal community (horizontal axes). Fungal species that more conspicuously avoided the overlap of host-plant species with others more strongly contributed to the emergence of network antinestedness.
Comparative analysis of factors associated with nestedness
Intratrophic level patterns related to network nestedness/antinestedness may be common not only in plant–fungus networks but also in other categories of plant–partner networks (Fig. 5). In a multiple regression analysis of a meta-dataset including plant–fungus, plant–pollinator, plant–seed disperser, and plant–protective ant networks (Fig. 1J), we found that the extent of host-plant overlap among partner (that is, higher trophic level) species explained variation in nestedness better than other indices of network architecture (Fig. 6 and table S3). We also found that the effects of reduced host range overlap were highly significant even after controlling for the effects of other network properties, such as interaction category, species richness, connectance, species richness ratio, and symbiont range overlap among plants (table S3). These results suggest that plant–partner network architecture, in general, depends on the within-trophic-level assembly rules of animals or fungi.
Fig. 6. Comparative analysis of factors associated with nestedness.
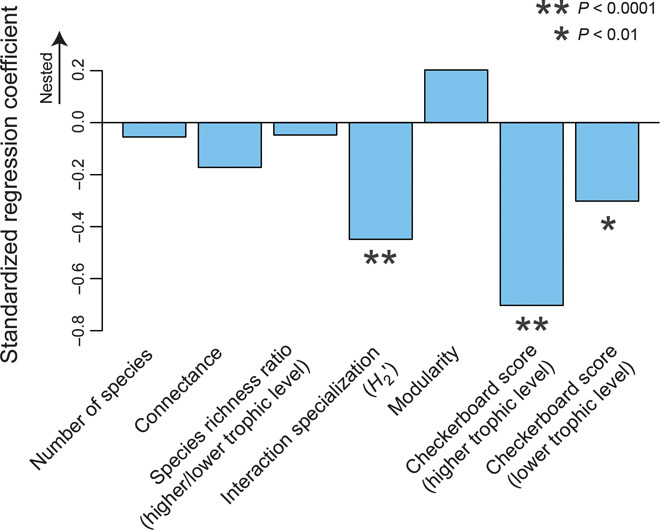
Across the metadata of the 59 ecological networks analyzed in Fig. 1J, a multiple regression of nestedness (weighted NODF) was performed with seven network indices (df = 51). Smaller values indicate stronger contribution to antinestedness.
DISCUSSION
This study is the first to provide a picture of whole kingdom-to-kingdom ecological networks at three sites along a latitudinal gradient by exploring how phylogenetically and functionally diverse fungi constitute structured (that is, nonrandom or nested or antinested) interaction networks with plant communities. Our results build on studies that have provided insights into plant–mycorrhizal fungus networks based on DNA sequencing data sets (16, 29–31). Previous studies of plant–fungus networks, however, have focused on either arbuscular mycorrhizal fungi (29–32) or ectomycorrhizal fungi (25) partly because these two fungal functional groups had been thought to considerably differ in their host specificity (11). Recent high-throughput sequencing analyses have altered the conventional view by showing that both arbuscular mycorrhizal and ectomycorrhizal fungal clades include host-specific and generalist fungi (16, 17, 33). Moreover, a plant individual can be simultaneously colonized not only by arbuscular mycorrhizal and ectomycorrhizal fungi (34) but also by diverse endophytic fungi (15, 28). By targeting both arbuscular mycorrhizal and ectomycorrhizal fungi as well as endophytic fungi, we have shown how functionally and phylogenetically diverse fungi constituted networks with the whole plant communities. This study therefore provides a basis for contemplating how plant community structure is organized by the entire root-associated fungal community in light of findings in above-ground plant–partner networks.
Although relatively comprehensive, our study is nevertheless limited in its ability to test the mechanisms that we broadly propose or extrapolate to other systems at larger scales. Even so, it provides several testable hypotheses that future research in plant–microbe networks will be more able to address with more resources and rapidly advancing methods. Our study lacks replicate habitats along the latitudinal gradient. We can therefore observe patterns that suggest mechanism but not readily confirm them. Furthermore, there are continuing challenges in applying operational taxonomic units (OTUs) in biologically realistic ways in molecular microbial community research (21, 35). Thus, although we have more fully characterized the relationship between network architecture and species interactions than previous studies and have uncovered a diversity of intriguing network patterns, our findings primarily provide a more solid basis for future experimental studies and theoretical models.
Our strongest finding is that, although most plant–pollinator and plant–seed disperser networks have nested architectures, the species-rich below-ground networks between plants and fungi are consistently antinested. There are several potential biological explanations for why the below-ground networks differ in structure from the above-ground plant–partner networks. These plant–fungus networks are composed of symbiotic interactions that could ecologically range from mutualism to antagonism, depending on environmental conditions (36). Generally, networks of symbiotic interactions are often more compartmentalized and less nested than networks of interaction among free-living species (26), and antagonistic networks are sometimes less nested than mutualistic networks (3, 8). In addition, below-ground plant–fungus symbioses are unique in that some fungi are potentially able to establish symbiotic interactions with two or more host individuals (37, 38). Finally, the potentially competitive aspects of the interactions identified here may themselves contribute to antinestedness. Collectively, these ecological aspects of plant root–associated fungus interactions may partially or fully explain the observed patterns, but studies partitioning these effects are needed.
The observed antinestedness in plant–fungus networks suggests that the relationship between network architecture and ecological community processes [for example, relationship between nestedness and species coexistence (2–4)] is more complex and variable than has previously been anticipated based on the studies of above-ground plant–pollinator and plant–seed disperser networks. Network architectural or compositional properties other than nestedness might be major determinants of community persistence in below-ground plant–fungus symbiosis. Recent theoretical studies have shown, for example, that the existence of antagonists in predominantly mutualistic interaction networks (39) could greatly affect community stability.
The observed antinested architecture also highlights the hypothesis that below-ground plant–fungus symbiosis may restrict, rather than support, the species coexistence of plants in terrestrial ecosystems [sensu (40)]. Ectomycorrhizal fungi, for instance, can promote the monodominance of their specific host-plant species in otherwise highly species-rich tropical forests (18). Thus, plants are simultaneously involved in several above-ground and below-ground networks, of which some (for example, plant–pollinator and plant–seed disperser networks) may promote the coexistence of their species, whereas others may counteract the facilitative effects. Accordingly, a comprehensive understanding of plant community structure and dynamics can be achieved only when we consider both above-ground and below-ground processes of plant–partner interactions.
Although our results provide insights into how some plant–fungus networks are organized, our present data are limited in their ability to extrapolate the potential mechanisms by which plant community structure is organized by below-ground fungal communities worldwide. This study included no replicate sites at each climatic region, and the present observational approach precluded explicit tests of the relationship between plant–fungus network architecture and community persistence. In addition, more rare fungi will be detected by deeper sequencing in future analyses, and use of OTUs in microbial diversity analyses in itself deserves continuing methodological attention (21, 35). The influence of spatial autocorrelations in sampling plots on the estimated plant–fungus network architecture also deserves further theoretical analysis (21, 25). Consequently, our present study primarily provides a basis for important working hypotheses on the linkage between above-ground and below-ground community dynamics, instead of giving an opportunity for strict hypothesis testing. Future theoretical and experimental studies will deepen our understanding of how plant communities are organized by both above-ground and below-ground partners.
MATERIALS AND METHODS
Data
We used the data sets of three community ecological studies that used NGS (454 pyrosequencing) to thoroughly uncover associations between plants and their functionally and phylogenetically diverse root-associated fungi (15–17). In general, DNA sequencing–based studies allow not only the high-throughput investigation of interspecific interactions but also the detection of a number of novel interactions that were unobservable with conventional methods based on direct observations (41, 42). Thus, NGS is promising in further accelerating those DNA sequencing–based approaches of ecological network analyses. Such data sets likely include not only mutually beneficial host–symbiont links but also a wide range of neutral, commensalistic, and potentially antagonistic interactions (43, 44). Thus, while NGS-based DNA barcoding approaches enable a simultaneous analysis of a greater number of taxa, obtained host–symbiont data sets can involve multiple types of ecological interaction.
Each of the three NGS studies was conducted at a 30 m × 30 m plot in a cool-temperate forest (42°40′N, 141°36′E) (17), a 59 m × 15 m plot in a warm-temperate forest [35°02′N, 135°47′E; examined in a previous network study (21)] (16), or a 29 m × 39 m plot in a subtropical forest (30°26′N, 130°30′E) (15) in Japan. Plant species in the family Fagaceae were the most abundant in all of the three forests. However, the heterogeneity of plant community composition [that is, patterns observed in relative species abundance (45)] substantially varied among the forests in the different climatic regions. That is, the cool-temperate forest was dominated by a hybrid population of two Quercus species (17) and the warm-temperate forest included two dominant Quercus species (16), whereas the subtropical forest consisted of various taxonomic groups of plants with comparable abundances (15).
In each forest, 2-cm fragments of terminal roots were sampled at 1-m horizontal intervals (15–17). The root samples were then subjected to highly parallelized 454 pyrosequencing, wherein the host-plant and fungal symbionts of each root sample were determined based on chloroplast rbcL and nuclear internal transcribed spacer (ITS) sequences, respectively (15–17). In total, both rbcL and ITS sequences were successfully obtained from 577, 834, and 849 root samples in the cool-temperate, warm-temperate, and subtropical forests, respectively (15–17). A preliminary study of the warm-temperate forest (21) indicated that about 500 root samples collected within a 1-m interval forest plot were sufficient for characterizing the architectural properties of plant root–associated fungus networks. In addition, the previous study also showed that the estimates of network indices did not greatly change with increasing mean distance among sampling positions in a study site (21).
As the sampling was indiscriminate in terms of root morphology and mycorrhizal type, the data included not only ectomycorrhizal or arbuscular mycorrhizal fungi but also diverse taxonomic clades of possibly root-endophytic and parasitic fungi (fig. S1 and data file S1). Meanwhile, this sampling method allowed the estimation of the relative frequency of plant–fungus associations in each forest (16, 43). As expected from the abovementioned geographic variation in plant community structure, the compositional evenness of plant species across the randomly collected root samples was low in the cool-temperate (Simpson’s diversity index = 0.78) and warm-temperate (Simpson’s diversity index = 0.81) forests but was high in the subtropical forest [Simpson’s diversity index = 0.94; see also Fig. 1K for species rank abundance (45) curves].
The obtained plant–fungus association data sets consisted of two types of matrices (data file S1): “sample-level” and “species-level” matrices. In sample-level matrices, rows represent samples with their host-plant information, columns represent fungal OTUs, and each cell entry indicates the presence or absence of a fungal species in a root sample; hereafter, we use the term “fungal species” instead of fungal OTUs for simplicity, keeping in mind that molecular OTUs do not necessarily correspond to species units defined with the biological species concept. In species-level matrices, rows represent plant species, columns represent fungal species, and each cell entry indicates the number of root samples in which focal plant–fungus association is detected. The latter matrices were used as plant–fungus association matrices for the analyses of network architecture, whereas the former matrices were used to obtain randomized plant–fungus matrices in null-model analyses. The species representing the ITS sequences that were not identified as belonging to the kingdom Fungi were excluded from the original data sets. When fungal species were defined with a cutoff ITS sequence similarity of 95%, the compiled data sets of cool-temperate, warm-temperate, and subtropical forests included 36 plant and 287 fungal species, 33 plant and 343 fungal species, and 36 plant and 580 fungal species, respectively. A previous study (21) indicated that varying cutoff fungal ITS sequence similarities from 81% to 97% did not qualitatively alter the statistical results of the analyses of below-ground plant–fungus network architecture.
Each local network data set included diverse taxonomic and functional groups of fungi (fig. S1): not only mycorrhizal fungi but also possibly endophytic or parasitic fungi were observed (15–17). Moreover, the data sets contained plant–fungus associations that did not fall within the conventional classification of mycorrhizal symbiosis [for example, nontypical associations between Fagaceae plants and arbuscular mycorrhizal fungi (15–17)]. Although recent mycological studies have shown that such nontypical plant–fungus associations could be more common than have been previously recognized (34, 46), those associations might include fungi with no ecological impact on host plants (44). Therefore, although many of the plant–fungus associations in our data sets could represent mutualistic ecological interactions, each network link may be more conservatively described in a potential continuum spanning from mutualism to commensalism and antagonism (36, 47). Such variation in ecological outcomes of interactions is most likely common in many ecological networks that have been analyzed as mutualistic networks: for example, large observational data of flower visitors usually involve associations between plants and noneffective pollinators or nectar robbers (48).
Local network architecture
The plant–fungus network architecture of each forest was visualized based on the “ForceAtlas2” layout algorithm (49) using the program Gephi (50). The H2′ metric (23) of network-level interaction specialization, Barber’s metric (51) of network modularity, the weighted NODF metric (52) of nestedness, and checkerboard scores (53) representing the avoidance of partner overlap in plant–fungus communities were examined in randomization tests to reveal the network architectural features of each forest. Note that checkerboard scores were standardized to vary between 0 (the lowest possible level of niche segregation) and 1 (the highest possible level of niche segregation) in our study (54).
In the randomization analysis, we randomized the label of plant species in a sample-level matrix and converted the randomized sample-level matrices into randomized species-level matrices [shuffle-sample model (model A); 1000 permutations]. We confirmed the results of this model by comparing the outcomes to two types of alternative null models. In additional analyses, species-level matrices were directly randomized with the “r2dtable” (54) (model B) and “vaznull” (55) (model C) null model algorithms, as implemented in the R 3.0.2 bipartite 2.04 package (54). The latter kept the species richness, marginal totals (that is, column and row sums), and connectance observed in the original matrices, whereas the former changed the connectance of randomized matrices. Qualitatively consistent results were obtained by the three alternative null models (tables S1 and S2). Calculations of H2′, weighted NODF, and modularity were performed using the R bipartite package and the MODULAR program (56) for simulated annealing–based estimation of network modularity (57), respectively. Using these programs, we further investigated the architectural features of local plant–fungus networks by evaluating interaction evenness, standard NODF nestedness (58) for binary data formats, Newman and Girvan’s metric (59) of modularity, and niche overlap metric (54) based on Raup-Crick β-diversity (60) (fig. S2). The degree distribution of each local network is available in fig. S2 (F to K).
Comparative analysis of network architecture
We examined the similarity of plant root–associated fungus networks to other forms of interaction by compiling data sets from other studies (data file S1). The compiled data set incorporated 25 plant–seed disperser, 51 plant–pollinator, 4 myrmecophyte plant–ant, 3 anemone–anemone fish, 7 fish–invertebrate parasite, and 20 plant–herbivore (5 inner tissue feeding and 15 browsing herbivore systems) networks, as well as 30 food webs. Our plant–fungus network data sets included functionally and phylogenetically more diverse organisms (fig. S1) than most previous ecological network studies. This difference among studies in species-level basal complexity might affect the results of the comparative analysis regardless of potential difference in assembly processes. Therefore, we examined not only the network architecture of the entire plant–root fungus communities but also that of the “partial networks” consisting of the respective functional or taxonomic groups of fungi (25, 31) and their host plants. The examined partial networks were categorized as follows: “mycorrhizal partial networks” (that is, ectomycorrhizal + arbuscular mycorrhizal fungi), “ectomycorrhizal partial networks” (ectomycorrhizal fungi), “arbuscular partial networks” (arbuscular mycorrhizal fungi), “ascomycete partial networks” (Ascomycota fungi), and “basidiomycete partial networks” (Basidiomycota fungi). We also added two published data sets of arbuscular mycorrhizal symbiosis (61, 62) to the meta-dataset.
For each of the 160 compiled network (and partial network) matrices, we estimated connectance and network modularity. In the modularity analysis, Newman and Girvan’s metric was used because the compiled data sets included not only bipartite but also unipartite forms of network matrices. In addition, for 59 bipartite-format networks with interaction frequency information (data file S1), H2′ interaction specialization and weighted NODF nestedness were also estimated. We also estimated checkerboard scores for the 130 binary bipartite networks with information on trophic levels. Moreover, for the 59 networks for which all of the above estimates were available, their architectural features were further compared in a principal component analysis. In this analysis, a correlation matrix was obtained based on connectance, the ratio of species of higher trophic level to species of lower trophic level (species richness ratio), H2′, modularity (Barber’s metric for bipartite matrices), weighted NODF, and checkerboard scores (fig. S3 and tables S1 and S2).
Network modules and nestedness
For each plant–fungus network data set, we inferred the modules of closely associated plant and fungal species. Although simulated annealing–based optimization of modularity (57) has been recently implemented for quantitative (interaction frequency) data sets (63), our plant–fungus data sets were too large to be analyzed by this approach. Among other existing module detection algorithms, the Infomap method, which uses the “code length” necessary to describe random walkers’ movements (64), detects network modules the most accurately (65). For each plant–fungus quantitative network data set, we performed 1000 Infomap runs and estimated network modules based on the consensus clustering approach (66). The five largest modules in each local plant–fungus data set, with their compositions of included fungal functional groups, are indicated in fig. S4.
We next tested whether the links in the inferred modules display a nested architecture. Specifically, randomization tests of weighted NODF nestedness were conducted for each module encompassing more than three plant species. Because the data sets used for this analysis were species-level matrices, randomized matrices were obtained based on the r2dtable (model B) and vaznull (model C) null models.
Network architectural properties correlated with nestedness/antinestedness
We conducted a comparative analysis to infer how an antinested network architecture was organized in plant–fungus networks. We first evaluated how observed network index estimates deviated from those expected under null models by standardizing each network index as
where Iobserved is a network index value of an observed network matrix, and mean(Irandomized) and SD(Irandomized) are the average and standard deviation of the network index values of randomized matrices, respectively. For each plant–fungus network or partial network of the three forests, the relative index values were calculated based on weighted NODF nestedness, H2′ interaction specialization, Barber’s modularity, and checkerboard scores under the three null models mentioned above. We then examined how relative nestedness was correlated with relative interaction specialization or relative modularity (Pearson’s correlation tests) across 18 plant–fungus networks and partial networks (that is, 3 full networks + 3 × 5 partial networks). In addition, relative checkerboard metrics were used in the correlation tests to examine the potential effects of partner overlap in plant–fungus communities on network (anti)nestedness. The results with checkerboard scores were confirmed by an additional test using niche overlap scores (fig. S5).
We examined the relative contributions of various network properties on nestedness/antinestedness by conducting a multiple regression analysis, in which relative nestedness was regressed on the number of species, connectance, species richness ratio (fungus to plant), relative interaction specialization, relative modularity, and relative checkerboard scores of plant–fungus networks/partial networks. In the model, all of the response and explanatory variables were z-standardized (zero mean and unit variance) to obtain the standardized regression coefficients of the explanatory variables.
Contribution of each fungal species to network architecture
In further examining the potential relationship between antinestedness and the patterns observed in partner overlap in fungal communities (Fig. 2, we evaluated how each fungal species contributed to network (anti)nestedness based on the extent to which it overlapped with other species in the use of host plants. We first evaluated the contribution of each fungus to network architecture as proposed in a previous study (67)
where Iobserved is a network index value of an observed network matrix, and mean(Irandomized, i) and SD(Irandomized, i) are the average and standard deviation of the network index values of randomized matrices, respectively, in which only the links of a fungal species i are randomized. Because our network data sets were quantitative (that is, with interaction frequency information), links of a fungal species i were randomized in sample-level binary matrices depicting the presence/absence of each fungal species in each root sample (100 permutations for each fungal species). The randomized sample-level matrices were then converted into species-level quantitative matrices representing the frequency of each plant–fungus association.
By this approach, the contribution of each fungal species to network architecture was evaluated in terms of weighted NODF nestedness, as well as checkerboard scores and niche overlap scores for host-plant overlap in fungal communities. On the basis of Pearson’s correlation tests of contribution to nestedness and each of the remaining contribution parameters, we examined whether fungal species substantially decreasing host-plant overlap in fungal communities also contributed greatly to the emergence of an antinested network architecture.
Comparative analysis of factors associated with nestedness
Across the 59 bipartite-format networks with interaction frequency information (see data mentioned previously), we constructed an analysis of variance model to examine the factors determining the nestedness (weighted NODF) of plant–partner networks. The explanatory variables examined were interaction category (plant–fungus, plant–pollinator, plant–seed disperser, or plant–protective ant network), number of species, network connectance, species richness ratio, interaction specialization, modularity, and checkerboard scores representing the extent of host/symbiont range overlap avoidance. In addition, the relative contribution of various network indices to variation in nestedness was examined in a multiple regression model. In the model, all of the response and explanatory variables were z-standardized (zero mean and unit variance) to obtain standardized regression coefficients.
Acknowledgments
We thank the anonymous reviewers and S. Naeem for their productive comments on the manuscript. Funding: This work was financially supported by the Hakubi Center for Advanced Research, Kyoto University, Japan Society for the Promotion of Science Grants-in-Aid for Scientific Research (KAKENHI; grant 26711026), and the Funding Program for Next-Generation World-Leading Researchers of Cabinet Office, Government of Japan (grant GS014) to H.T. P.R.G. was supported by Fundação de Amparo à Pesquisa do Estado de São Paulo (grant 2009/54422-8), J.M.O. was supported by the Danish Science Research Council (grant 1323-00278), and J.N.T. was supported by NSF (grant DEB-1048333). Author contributions: H.T. designed the research and obtained funding. H.T. performed statistical analyses based on discussions with P.R.G., J.M.O., and J.N.T. All of the authors wrote the paper. Competing interests: The authors declare that they have no competing interests. Data and materials availability: Correspondence and requests for materials should be addressed to H.T. (hiro.toju@gmail.com).
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/1/9/e1500291/DC1
Data File S1. Data sets of the three plant–fungus networks and the list of data sets used in the comparative analysis.
Fig. S1. Diversity in local networks.
Fig. S2. Additional analyses of the architectural properties of local plant–fungus networks.
Fig. S3. Comparison of network architecture between plant–fungus networks and other types of ecological networks.
Fig. S4. Network modules and nestedness.
Fig. S5. Relationship between (anti)nestedness and other network architectural properties (r2dtable and vaznull models).
Fig. S6. Contribution of each fungal species to (anti)nested network architecture.
Table S1. Randomization tests of network architectural indices with three types of null models.
Table S2. Randomization tests of checkerboard patterns.
Table S3. Comparative analysis of factors associated with nestedness (analysis of variance).
REFERENCES AND NOTES
- 1.Allesina S., Tang S., Stability criteria for complex ecosystems. Nature 483, 205–208 (2012). [DOI] [PubMed] [Google Scholar]
- 2.Rohr R. P., Saavedra S., Bascompte J., On the structural stability of mutualistic systems. Science 345, 1253497 (2014). [DOI] [PubMed] [Google Scholar]
- 3.Thébault E., Fontaine C., Stability of ecological communities and the architecture of mutualistic and trophic networks. Science 329, 853–856 (2010). [DOI] [PubMed] [Google Scholar]
- 4.Bastolla U., Fortuna M. A., Pascual-García A., Ferrera A., Luque B., Bascompte J., The architecture of mutualistic networks minimizes competition and increases biodiversity. Nature 458, 1018–1020 (2009). [DOI] [PubMed] [Google Scholar]
- 5.Olesen J. M., Bascompte J., Dupont Y. L., Jordano P., The modularity of pollination networks. Proc. Natl. Acad. Sci. U.S.A. 104, 19891–19896 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Suweis S., Simini F., Banavar J. R., Maritan A., Emergence of structural and dynamical properties of ecological mutualistic networks. Nature 500, 449–452 (2013). [DOI] [PubMed] [Google Scholar]
- 7.Staniczenko P. P. A., Kopp J. C., Allesina S., The ghost of nestedness in ecological networks. Nat. Commun. 4, 1391 (2013). [DOI] [PubMed] [Google Scholar]
- 8.Bascompte J., Jordano P., Melián C. J., Olesen J. M., The nested assembly of plant-animal mutualistic networks. Proc. Natl. Acad. Sci. U.S.A. 100, 9383–9387 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lever J. J., van Nes E. H., Scheffer M., Bascompte J., The sudden collapse of pollinator communities. Ecol. Lett. 17, 350–359 (2014). [DOI] [PubMed] [Google Scholar]
- 10.R. D. Bardgett, D. A. Wardle, Aboveground–Belowground Linkages: Biotic Interactions, Ecosystem Processes, and Global Change (Oxford Univ. Press, Oxford, UK, 2010). [Google Scholar]
- 11.S. E. Smith, D. J. Read, Mycorrhizal Symbiosis (Elsevier, New York, NY, ed. 3, 2008). [Google Scholar]
- 12.J. N. Thompson, The Geographic Mosaic of Coevolution (The University of Chicago Press, Chicago, IL, 2005). [Google Scholar]
- 13.Bever J. D., Soil community feedback and the coexistence of competitors: Conceptual frameworks and empirical tests. New Phytol. 157, 465–473 (2003). [DOI] [PubMed] [Google Scholar]
- 14.Wardle D. A., Bardgett R. D., Klironomos J. N., Setälä H., van der Putten W. H., Wall D. H., Ecological linkages between aboveground and belowground biota. Science 304, 1629–1633 (2004). [DOI] [PubMed] [Google Scholar]
- 15.Toju H., Sato H., Tanabe A. S., Diversity and spatial structure of belowground plant-fungal symbiosis in a mixed subtropical forest of ectomycorrhizal and arbuscular mycorrhizal plants. PLOS One 9, e86566 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Toju H., Sato H., Yamamoto S., Kadowaki K., Tanabe A. S., Yazawa S., Nishimura O., Agata K., How are plant and fungal communities linked to each other in belowground ecosystems? A massively parallel pyrosequencing analysis of the association specificity of root-associated fungi and their host plants. Ecol. Evol. 3, 3112–3124 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Toju H., Yamamoto S., Sato H., Tanabe A. S., Sharing of diverse mycorrhizal and root-endophytic fungi among plant species in an oak-dominated cool-temperate forest. PLOS One 8, e78248 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McGuire K. L., Common ectomycorrhizal networks may maintain monodominance in a tropical rain forest. Ecology 88, 567–574 (2007). [DOI] [PubMed] [Google Scholar]
- 19.Newsham K. K., A meta-analysis of plant responses to dark septate root endophytes. New Phytol. 190, 783–793 (2011). [DOI] [PubMed] [Google Scholar]
- 20.Guimarães P. R. Jr, Jordano P., Thompson J. N., Evolution and coevolution in mutualistic networks. Ecol. Lett. 14, 877–885 (2011). [DOI] [PubMed] [Google Scholar]
- 21.Toju H., Guimarães P. R. Jr, Olesen J. M., Thompson J. N., Assembly of complex plant-fungus networks. Nat. Commun. 5, 5273 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Blüthgen N., Menzel F., Hovestadt T., Fiala B., Specialization, constraints, and conflicting interests in mutualistic networks. Curr. Biol. 17, 341–346 (2007). [DOI] [PubMed] [Google Scholar]
- 23.Almeida-Neto M., Guimarães P. R. Jr, Lewinsohn T. M., On nestedness analyses: Rethinking matrix temperature and anti-nestedness. Oikos 116, 716–722 (2007). [Google Scholar]
- 24.Fortuna M. A., Stouffer D. B., Olesen J. M., Jordano P., Mouillot D., Krasnov B. R., Poulin R., Bascompte J., Nestedness versus modularity in ecological networks: Two sides of the same coin? J. Anim. Ecol. 79, 811–817 (2010). [DOI] [PubMed] [Google Scholar]
- 25.Bahram M., Harend H., Tedersoo L., Network perspectives of ectomycorrhizal associations. Fungal Ecol. 7, 70–77 (2014). [Google Scholar]
- 26.Guimarães P. R. Jr, Rico-Gray V., Oliveira P. S., Izzo T. J., dos Reis S. F., Thompson J. N., Interaction intimacy affects structure and coevolutionary dynamics in mutualistic networks. Curr. Biol. 17, 1797–1803 (2007). [DOI] [PubMed] [Google Scholar]
- 27.Kennedy P., Ectomycorrhizal fungi and interspecific competition: Species interactions, community structure, coexistence mechanisms, and future research directions. New Phytol. 187, 895–910 (2010). [DOI] [PubMed] [Google Scholar]
- 28.Yamamoto S., Sato H., Tanabe A. S., Hidaka A., Kadowaki K., Toju H., Spatial segregation and aggregation of ectomycorrhizal and root-endophytic fungi in the seedlings of two Quercus species. PLOS One 9, e96363 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bennett A. E., Daniell T. J., Öpik M., Davison J., Moora M., Zobel M., Selosse M. A., Evans D., Arbuscular mycorrhizal fungal networks vary throughout the growing season and between successional stages. PLOS One 8, e83241 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chagnon P.-L., Bradley R. L., Klironomos J. N., Using ecological network theory to evaluate the causes and consequences of arbuscular mycorrhizal community structure. New Phytol. 194, 307–312 (2012). [DOI] [PubMed] [Google Scholar]
- 31.Montesinos-Navarro A., Segarra-Moragues J. G., Valiente-Banuet A., Verdú M., The network structure of plant–arbuscular mycorrhizal fungi. New Phytol. 194, 536–547 (2012). [DOI] [PubMed] [Google Scholar]
- 32.Veresoglou S. D., Rillig M. C., Do closely related plants host similar arbuscular mycorrhizal fungal communities? A meta-analysis. Plant Soil 377, 395–406 (2014). [Google Scholar]
- 33.Peay K. G., Russo S. E., McGuire K. L., Lim Z., Chan J. P., Tan S., Davies S. J., Lack of host specificity leads to independent assortment of dipterocarps and ectomycorrhizal fungi across a soil fertility gradient. Ecol. Lett. 18, 807–816 (2015). [DOI] [PubMed] [Google Scholar]
- 34.Dickie I. A., Koide R. T., Fayish A. C., Vesicular–arbuscular mycorrhizal infection of Quercus rubra seedlings. New Phytol. 151, 257–264 (2001). [DOI] [PubMed] [Google Scholar]
- 35.Nilsson R. H., Kristiansson E., Ryberg M., Hallenberg N., Larsson K. H., Intraspecific ITS variability in the kingdom Fungi as expressed in the international sequence databases and its implications for molecular species identification. Evol. Bioinform. 4, 193–201 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kiers E. T., Denison R. F., Sanctions, cooperation, and the stability of plant-rhizosphere mutualisms. Ann. Rev. Ecol. Evol. Syst. 39, 215–236 (2008). [Google Scholar]
- 37.Beiler K. J., Durall D. M., Simard S. W., Maxwell S. A., Kretzer A. M., Architecture of the wood-wide web: Rhizopogon spp. genets link multiple Douglas-fir cohorts. New Phytol. 185, 543–553 (2010). [DOI] [PubMed] [Google Scholar]
- 38.Nara K., Ectomycorrhizal networks and seedling establishment during early primary succession. New Phytol. 169, 169–178 (2006). [DOI] [PubMed] [Google Scholar]
- 39.Mougi A., Kondoh M., Diversity of interaction types and ecological community stability. Science 337, 349–351 (2012). [DOI] [PubMed] [Google Scholar]
- 40.Fontaine C., Guimarães P. R. Jr, Kéfi S., Loeuille N., Memmott J., van der Putten W. H., van Veen F. J. F., Thébault E., The ecological and evolutionary implications of merging different types of networks. Ecol. Lett. 14, 1170–1181 (2011). [DOI] [PubMed] [Google Scholar]
- 41.Wirta H. K., Hebert P. D., Kaartinen R., Prosser S. W., Várkonyi G., Roslin T., Complementary molecular information changes our perception of food web structure. Proc. Natl. Acad. Sci. U.S.A. 111, 1885–1890 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pompanon F., Deagle B. E., Symondson W. O. C., Brown D. S., Jarman S. N., Taberlet P., Who is eating what: Diet assessment using next generation sequencing. Mol. Ecol. 21, 1931–1950 (2012). [DOI] [PubMed] [Google Scholar]
- 43.Toju H., Yamamoto S., Sato H., Tanabe A. S., Gilbert G. S., Kadowaki K., Community composition of root-associated fungi in a Quercus-dominated temperate forest: “Codominance” of mycorrhizal and root-endophytic fungi. Ecol. Evol. 3, 1281–1293 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Caruso T., Rillig M. C., Garlaschelli D., On the application of network theory to arbuscular mycorrhizal fungi–plant interactions: The importance of basic assumptions. New Phytol. 194, 891–894 (2012). [DOI] [PubMed] [Google Scholar]
- 45.S. P. Hubbell, The Unified Neutral Theory of Biodiversity and Biogeography (Princeton Univ. Press, Princeton, NJ, 2001). [Google Scholar]
- 46.Wagg C., Pautler M., Massicotte H. B., Peterson R. L., The co-occurrence of ectomycorrhizal, arbuscular mycorrhizal, and dark septate fungi in seedlings of four members of the Pinaceae. Mycorrhiza 18, 103–110 (2008). [DOI] [PubMed] [Google Scholar]
- 47.Kiers E. T., van der Heijden M. G. A., Mutualistic stability in the arbuscular mycorrhizal symbiosis: Exploring hypotheses of evolutionary cooperation. Ecology 87, 1627–1636 (2006). [DOI] [PubMed] [Google Scholar]
- 48.Kato M., Kakutani T., Inoue T., Itino T., Insect-flower relationship in the primary beech forest of Ashu, Kyoto: An overview of the flowering phenology and the seasonal pattern of insect visits. Contr. Biol. Lab. Kyoto Univ. 27, 309–376 (1990). [Google Scholar]
- 49.Jacomy M., Heymann S., Venturini T., Bastian M., ForceAtlas2, a continuous graph layout algorithm for handy network visualization designed for the Gephi software. PLOS One 9, e98679 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bastian M., Heymann S., Jacomy M., Gephi: An open source software for exploring and manipulating networks, ICWSM 8, 361–362 (2009). [Google Scholar]
- 51.Barber M. J., Modularity and community detection in bipartite networks. Phys. Rev. E 76, 066102 (2007). [DOI] [PubMed] [Google Scholar]
- 52.Almeida-Neto M., Ulrich W., A straightforward computational approach for measuring nestedness using quantitative matrices. Environ. Model. Software 26, 173–178 (2011). [Google Scholar]
- 53.Stone L., Roberts A., Competitive exclusion, or species aggregation? Oecologia 91, 419–424 (1992). [DOI] [PubMed] [Google Scholar]
- 54.Dormann C. F., Fründ J., Blüthgen N., Gruber B., Indices, graphs and null models: Analyzing bipartite ecological networks. Open Ecol. J. 2, 7–24 (2009). [Google Scholar]
- 55.Vázquez D. P., Melián C. J., Williams N. M., Blüthgen N., Krasnov B. R., Poulin R., Species abundance and asymmetric interaction strength in ecological networks. Oikos 116, 1120–1127 (2007). [Google Scholar]
- 56.Marquitti F. M. D., Guimarães P. R. Jr, Pires M. M., Bittencourt L. F., MODULAR: Software for the autonomous computation of modularity in large network sets. Ecography 37, 221–224 (2014). [Google Scholar]
- 57.Guimerà R., Amaral L. A. N., Cartography of complex networks: Modules and universal roles, J. Stat. Mech. Theor. Exp. 2005, P02001 ( 2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Almeida-Neto M., Guimarães P., Guimarães P. R. Jr, Loyola R. D., Ulrich W., A consistent metric for nestedness analysis in ecological systems: Reconciling concept and measurement. Oikos 117, 1227–1239 (2008). [Google Scholar]
- 59.Newman M. E. J., Girvan M., Finding and evaluating community structure in networks. Phys. Rev. E. 69, 026113 (2004). [DOI] [PubMed] [Google Scholar]
- 60.P. Legendre, L. Ledendre, Numerical Ecology. 2nd English Edition (Elsevier, Amsterdam, Netherlands, 1998). [Google Scholar]
- 61.Davison J., Öpik M., Daniell T. J., Moora M., Zobel M., Arbuscular mycorrhizal fungal communities in plant roots are not random assemblages. FEMS Microbial. Ecol. 78, 103–115 (2011). [DOI] [PubMed] [Google Scholar]
- 62.Montesinos-Navarro A., Segarra-Moragues J. G., Valiente-Banuet A., Verdú M., Plant facilitation occurs between species differing in their associated arbuscular mycorrhizal fungi. New Phytol. 196, 835–844 (2012). [DOI] [PubMed] [Google Scholar]
- 63.Schleuning M., Ingmann L., Strauß R., Fritz S. A., Dalsgaard B., Dehling D. M., Plein M., Saavedra F., Sandel B., Svenning J.-C., Böhning-Gaese K., Dormann C. F., Ecological, historical and evolutionary determinants of modularity in weighted seed-dispersal networks. Ecol. Lett. 17, 454–463 (2014). [DOI] [PubMed] [Google Scholar]
- 64.Rosvall M., Bergstrom C. T., Multilevel compression of random walks on networks reveals hierarchical organization in large integrated systems. PLOS One 6, e18209 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Orman G. K., Labatut V., Cherifi H., Comparative evaluation of community detection algorithms: A topological approach. J. Stat. Mech. Theor. Exp. 2012, P08001 ( 2012). [Google Scholar]
- 66.Lancichinetti A., Fortunato S., Consensus clustering in complex networks. Sci. Rep. 2, 336 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Saavedra S., Stouffer D. B., Uzzi B., Bascompte J., Strong contributors to network persistence are the most vulnerable to extinction. Nature 478, 233–235 (2011). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/1/9/e1500291/DC1
Data File S1. Data sets of the three plant–fungus networks and the list of data sets used in the comparative analysis.
Fig. S1. Diversity in local networks.
Fig. S2. Additional analyses of the architectural properties of local plant–fungus networks.
Fig. S3. Comparison of network architecture between plant–fungus networks and other types of ecological networks.
Fig. S4. Network modules and nestedness.
Fig. S5. Relationship between (anti)nestedness and other network architectural properties (r2dtable and vaznull models).
Fig. S6. Contribution of each fungal species to (anti)nested network architecture.
Table S1. Randomization tests of network architectural indices with three types of null models.
Table S2. Randomization tests of checkerboard patterns.
Table S3. Comparative analysis of factors associated with nestedness (analysis of variance).