Heart rhythm of fruit flies at different developmental stages can be noninvasively controlled by light using optogenetics.
Keywords: Optogenetics, Label-free imaging, Optical coherence microscopy, Optical coherence tomography, Drosophila, Heart development
Abstract
Electrical stimulation is currently the gold standard for cardiac pacing. However, it is invasive and nonspecific for cardiac tissues. We recently developed a noninvasive cardiac pacing technique using optogenetic tools, which are widely used in neuroscience. Optogenetic pacing of the heart provides high spatial and temporal precisions, is specific for cardiac tissues, avoids artifacts associated with electrical stimulation, and therefore promises to be a powerful tool in basic cardiac research. We demonstrated optogenetic control of heart rhythm in a well-established model organism, Drosophila melanogaster. We developed transgenic flies expressing a light-gated cation channel, channelrhodopsin-2 (ChR2), specifically in their hearts and demonstrated successful optogenetic pacing of ChR2-expressing Drosophila at different developmental stages, including the larva, pupa, and adult stages. A high-speed and ultrahigh-resolution optical coherence microscopy imaging system that is capable of providing images at a rate of 130 frames/s with axial and transverse resolutions of 1.5 and 3.9 μm, respectively, was used to noninvasively monitor Drosophila cardiac function and its response to pacing stimulation. The development of a noninvasive integrated optical pacing and imaging system provides a novel platform for performing research studies in developmental cardiology.
INTRODUCTION
For more than a century, electrical stimulation techniques have been used for pacing excitable tissues such as neurons and cardiomyocytes. During electrical stimulation, low-energy shocks are delivered through electrodes to initiate action potential in target tissues. Electrical stimulation of the heart has been widely used to investigate the pathogenesis and underlying mechanisms of various cardiovascular diseases such as ventricular tachycardia (1), cardiac arrest (2), and other rhythm disorders (3, 4). Although electrical pacing has been highly effective in controlling cardiac function and has provided a wealth of information on cardiac physiology, it has several limitations such as (i) requirement of invasive procedures for placement of electrodes, (ii) lack of specificity for cardiac tissues, and (iii) production of inhomogeneous areas of depolarization (5). Moreover, electrical shocks administered during the procedure may generate toxic gases, alter pH level, and cause tissue damage (6). These factors make electrical pacing nonoptimal for in vivo longitudinal studies and situations requiring pacing for longer durations.
Optical pacing is a promising alternative to electrical pacing techniques for basic cardiovascular research. In comparison to electrical pacing, noninvasive stimulation of cardiac tissues with light allows control of heart function in vivo with high spatial and temporal precisions. Pulsed infrared laser light has been used to pace intact embryonic quail hearts in vivo (7) and excised adult rabbit hearts (8). Moreover, optogenetic tools can be used to achieve optical pacing of the heart at lower power levels and with improved specificity for cardiac tissues. Over the last decade, optogenetic tools have been widely used in neuroscience to control neuronal function (9, 10). This is achieved by incorporating light-sensitive proteins, known as rhodopsins (10, 11), into target cell populations. One of these rhodopsins, named channelrhodopsin-2 (ChR2), is a light-gated cation channel that, when illuminated with blue light, allows positively charged ions to pass into cells (12). ChR2-expressing neurons depolarize upon illumination with blue light, resulting in an action potential. Complementing the optogenetic tools used for neuronal excitation, other light-activated ion pumps, such as halorhodopsins [for example, halorhodopsin from Natronomonas (NpHR)] and archaerhodopsins, can be used for neuronal inhibition (10, 13). Thus, optogenetics offers the capability to control well-defined neuronal activities in specific neuronal populations with unprecedented spatial and temporal precisions (13). Recent optogenetic studies in animal models have helped researchers to gain insights into the nature of neurological disorders, such as Parkinson’s disease (14) and depression (15), and to understand the course of action of therapeutic interventions (16).
Although optogenetics has been widely used in neuroscience, optogenetic pacing of the heart has been limited to date. In 2010, two separate studies demonstrated optogenetic pacing of the heart in zebrafish and mice models (17, 18). In the first study, ChR2-expressing cardiac tissues were used for light-induced stimulation of heart muscle in vitro and in mice undergoing open-chest preparation (17). In the other study, a genetically encoded, optically controlled pacemaker was created by expressing NpHR and ChR2 in zebrafish hearts (18). The cardiac pacemaker was located using patterned illumination, and optogenetic pacing was used to simulate tachycardia, bradycardia, atrioventricular blocks, and cardiac arrest by activating ChR2- and NpHR-expressing cardiomyocytes. The ability to noninvasively pace the heart using optogenetic tools offers an opportunity to investigate the underlying mechanisms of cardiac arrhythmias and to develop new treatment strategies (19–21).
Here, we demonstrated optogenetic pacing of the heart in a well-established and powerful model organism, Drosophila melanogaster. Drosophila shares many similarities with vertebrates at the early stages of heart development (22) and in underlying genetic mechanisms regulating cardiac function (23, 24). About 75% of disease-causing genes in humans have been estimated to have functional orthologs in Drosophila (25). Hence, Drosophila has been extensively used to investigate mechanisms associated with human diseases, including cardiac disorders (26–29). Given its wide-ranging possibilities for genetic manipulation, rapid life cycle, and low maintenance costs, Drosophila is an extremely valuable model organism for understanding cardiac arrhythmia mechanisms and for developing new therapeutic strategies. To monitor and capture the response of the Drosophila heart to stimulation pulses, we developed an ultrahigh-resolution optical coherence microscopy (OCM) system (30–33) that allows real-time, label-free, and nondestructive imaging of the Drosophila heart at micrometer-scale resolution. The heart tube of Drosophila is located on the dorsal side of its abdomen, usually within 300 μm of the tissue surface (34). Hence, it is possible to analyze the structure and function of the Drosophila heart at different developmental stages in vivo and noninvasively using OCM. Here, we developed an integrated optical pacing and imaging system to synchronously control and monitor Drosophila cardiac function in vivo. By performing noninvasive pacing experiments using this system, we quantitatively determined cardiac physiological parameters, such as refractory period (RP) and contraction time (CT), of the Drosophila heart at different stages of its life cycle.
RESULTS AND DISCUSSION
Optogenetic pacing in Drosophila
To noninvasively monitor Drosophila heart function in real time in response to optical stimulations, we developed an integrated optical pacing and ultrahigh-resolution OCM imaging system (Fig. 1A). We also developed transgenic Drosophila models expressing ChR2 specifically in their hearts (24B-GAL4;UAS-H134R-ChR2) for the pacing experiments (Fig. 1B). When the heart tube of ChR2-expressing flies was illuminated with blue-light pulses, each pulse induced depolarization in pacemaker cells, triggering a heart contraction. Figure 1C shows successful optical pacing of an adult fly expressing ChR2 at three different pacing frequencies. The fly with a resting heart rate (RHR) of 6 Hz was successfully paced at 8, 9, and 10 Hz, respectively. The fly heart consistently followed different pacing frequencies and returned to its RHR as soon as stimulation pulses were turned off. In comparison, stimulation pulses did not have any influence on the heart rate (HR) of control fly (24B-GAL4/+; Fig. 1C). Figure 1C demonstrates the feasibility of noninvasively applying optogenetic pacing to tune HR in an adult Drosophila by varying the frequency of stimulation pulses.
Fig. 1. Optogenetic pacing of the Drosophila heart.
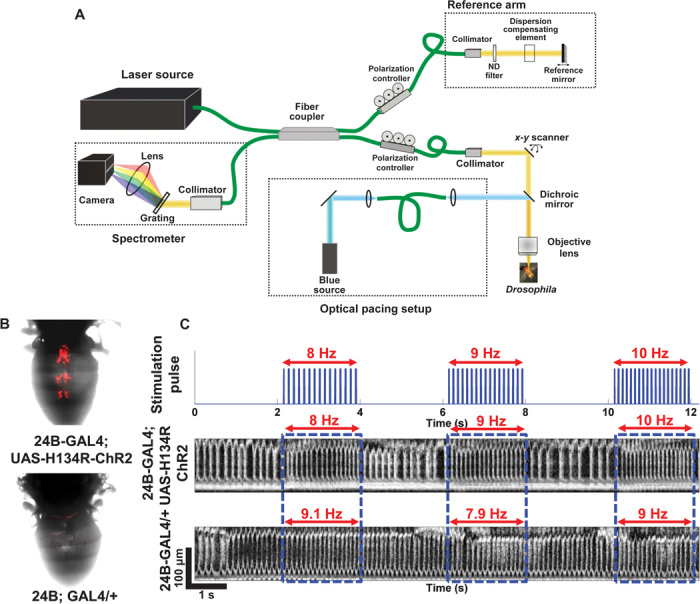
(A) Schematic of the integrated OCM imaging and pacing system. The optogenetic excitation beam was coupled with the sample arm of the spectral domain OCM system using a dichroic beam splitter. (B) Comparison of cardiac-specific mCherry fluorescence expression between ChR2-expressing transgenic flies (24B-GAL4;UAS-H134R-ChR2) and control flies (24B-GAL4/+). (C) M-mode images showing optogenetic pacing in ChR2 and control adult flies. The ChR2 fly heart with an RHR of 6 Hz was successfully paced at three different frequencies: 8, 9, and 10 Hz. In comparison, the control fly heart was not responsive to optical pacing stimulations.
Characterization of optogenetic pacing
Optimal power levels and pulse widths required to achieve successful pacing at different developmental stages were determined and are shown in Fig. 2 (A and B) and fig. S1. Among the flies used for threshold measurements, the mean RHR at the larva, pupa, and adult stages was 4.5, 2.6, and 7.1 Hz, respectively (n = 10 at each stage). The pacing frequency for threshold measurements was set slightly above the RHR at each stage as 5, 3.5, and 10 Hz for larval, pupal, and adult flies, respectively. For threshold power measurements, 20-ms pulses were used. One hundred percent pacing at the larva and pupa stages was achieved using 16-mW pulses (Fig. 2A). However, optimal power level tends to vary in adult flies. With 16-mW pulses, only ~60% of adult flies followed the pacing frequency. For threshold pulse-width measurements using 16-mW pulses, 100% pacing for pulse widths >10 ms was achieved at both larva and pupa stages (Fig. 2B). During the adult stage, 20-ms pulses stimulated the highest number of flies (~83%) to follow the pacing frequency. When the length of excitation pulses in adult flies was extended to 40 ms, additional heart contractions were often observed between excitation pulses, which resulted in a deviation of the measured HR from the pacing frequency. This could be related to the faster cardiac dynamics observed in adult flies (see results in Fig. 4). On the basis of these experiments, 16-mW and 20-ms light pulses were determined to be optimal for pacing experiments at different developmental stages. This corresponded to an irradiance of ~35.5 mW/mm2 for larval and pupal flies and ~12 mW/mm2 for adult flies. The irradiance level used in the current study was considerably higher than that used for cardiac optogenetic pacing in mouse and rat hearts through direct illumination of blue-light pulses (17, 21). However, we noninvasively performed cardiac pacing through intact fly cuticle. Hence, the higher irradiance level used in this study, which is still below the American National Standards Institute (ANSI) safety limit, was expected to achieve reliable pacing.
Fig. 2. Characterization of cardiac optogenetic pacing.
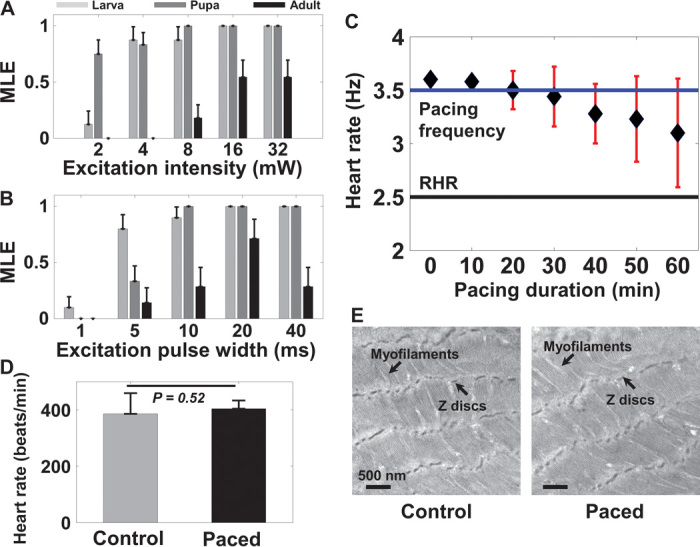
(A and B) Different stimulation power levels (A) and pulse widths (B) were used to determine optimal stimulation parameters to achieve successful pacing at different developmental stages (n = 10). On the basis of acquired data, the MLE to achieve successful pacing at different power levels and pulse widths was determined. Results are presented as mean ± SEM. (C) HR measurements in early pupae continuously paced for 1 hour at 3.5 Hz (n = 10). About 50% of early pupae successfully followed the pacing frequency for 1 hour. Results are presented as mean ± SD. (D) No significant differences in the HR of adult flies were observed between flies that were paced for 1 hour during the early pupa stage and control flies (n = 10). The average HR for paced and control flies was 404 ± 23.9 and 386 ± 73.4 beats/min, respectively (P = 0.52). Results are presented as mean ± SD. (E) TEM images of adult fly heart obtained from around the A1 segment of paced and control flies. TEM images show a normal myofibril structure and continuous Z discs in both groups. Results in (D) and (E) demonstrate that optogenetic pacing was safe and that pacing for 1 hour during the early pupa stage did not adversely affect heart development in Drosophila.
Fig. 4. Analysis of cardiac dynamics at different developmental stages.
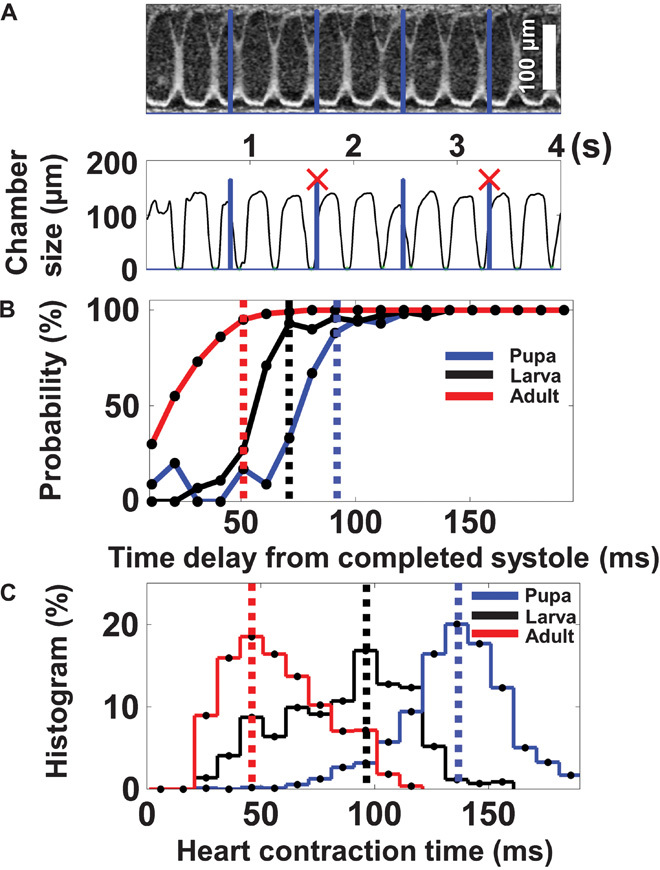
(A) M-mode image showing that pacing pulses incident during RP could not initiate a heart contraction (pulses marked with X). (B) The probability that each stimulation pulse would initiate a heart contraction was characterized at different time delays with respect to previously completed systole. RP was estimated as the time delay required after the preceding systole for achieving a 90% probability of successful pacing. It was shortest at the adult stage, intermediate at the larva stage, and longest at the pupa stage. (C) Comparison of heart CT at different developmental stages. CT had a similar trend as RP: shortest at the adult stage, intermediate at the larva stage, and slowest at the pupa stage.
A group of 10 early pupae with an average RHR of 2.5 Hz were paced continuously at 3.5 Hz for 1 hour to explore the safety of extended optogenetic pacing (Fig. 2C). More than 80% of the pupae followed the pacing frequency for the first 30 min. Some of the early pupae started to deviate from the pacing frequency after 30 min, which may be related to intermittent periods of cardiac inactivity during the pupa stage in Drosophila (35, 36). For example, a wild-type pupal heart is only active for about 60% of the time at 8 hours after puparium formation and for about 30% of the time at 16 hours after puparium formation (36). Hence, continuous pacing of early pupae at ~140% of their RHR for 1 hour may have imposed significant stress on the developing heart, which may result in failure to follow the pacing frequency in some of the specimens. However, ~50% of pupae were still following the pacing frequency even after continuous pacing for 1 hour. All of these pupae continued to develop normally and emerged as adult flies. On the first day of their adult stage, the HR of these paced specimens did not show any significant difference from that of age-matched control flies (Fig. 2D). Moreover, transmission electron microscopy (TEM) images of the heart tube from paced and control flies showed normal myofibril structures with regular myofilament arrays and continuous Z discs (Fig. 2E). These results suggested that 1 hour of optogenetic pacing at the early pupa stage was safe and did not have any observable long-term adverse effects on the development of the Drosophila heart.
Optogenetic pacing of Drosophila at different developmental stages
To demonstrate the feasibility of optogenetic pacing at different developmental stages, we paced the Drosophila heart at different stages of its life cycle, including the larva, pupa, and adult stages. The Drosophila heart exhibits significant structural and functional variations during its life cycle (36, 37). During the development of Drosophila from a larva to an adult fly, its heart transforms from a tubular structure with eight abdominal segments to a conical structure with five abdominal segments (34, 37). Alongside structural changes, HR reduces significantly at the early pupa stage, with heartbeat completely stopping on the second day of the pupa stage. Subsequently, the heart begins beating again on the third day of the pupa stage, and HR further increases to reach its maximum during the adult stage (36). Because of significant structural and functional variations during its life cycle, Drosophila can provide insights into distinct developmental and physiological processes.
Considering the differences in cardiac structure at different developmental stages, we further optimized excitation beam location and spot size to pace the Drosophila heart at different developmental stages (Fig. 3, A, D, and G). Representative M-mode OCM images demonstrating successful optogenetic pacing of larval, pupal, and adult flies are shown in Fig. 3 (B, E, and H, respectively). The 1:1 correspondence between excitation pulses and heart contractions can be clearly observed in these images. Heart rhythm was optically tuned to different frequencies at each developmental stage to determine the range for reliable optogenetic pacing (n = 6 at each stage). Pacing rates with frequencies 25 to 50% above the average RHR was consistently achieved at different developmental stages (Fig. 3, C, F, and I, and videos S1 to S3), although cardiac pacing in adult flies showed larger variations. This could be partly attributed to inherently larger variations in RHR among adult flies and more prominent neuromuscular coupling at this stage. Unlike the larva and early pupa stages, heart contractions in adult flies are not completely myogenic because the cardiac muscle layer is extensively innervated during metamorphosis (38). The fly heart failed to follow the pacing rhythm when the pacing frequency was too high. These results are consistent with previous observations in ChR2-expressing zebrafish hearts, where the zebrafish hearts could not be paced at frequencies outside the physiological range (18). For pacing frequencies lower than the RHR, spontaneous heart contractions could occur between stimulation pulses, causing the effective HR to be higher than the pacing frequency (Fig. 4A). These spontaneous heart contractions can be stopped by expressing inhibitory hyperpolarizing opsins, such as NpHR, in the Drosophila heart. Illumination with orange light has been shown to instantaneously block zebrafish heart contractions and could induce cardiac arrest and atrioventricular blocks of varying severity in NpHR-expressing hearts (18). Hence, one could potentially develop double-transgenic fly models simultaneously expressing ChR2 and NpHR in the heart to achieve reliable underpacing. Consequently, spontaneous contractions of the heart could be inhibited by hyperpolarizing the cardiac pacemaker using green or orange light between blue stimulation pulses, thereby increasing the frequency range over which reliable pacing can be achieved. Moreover, double-transgenic fly models would enable studies to identify the exact location of the anterior and posterior pacemakers in an adult fly and to assess their relative contributions to the cardiac function of an adult fly. Pacing efficiency (PE) for different pacing frequencies at each developmental stage was also determined to further characterize the effectiveness of optogenetic pacing (fig. S2).
Fig. 3. Optogenetic pacing of the Drosophila heart at different development stages.
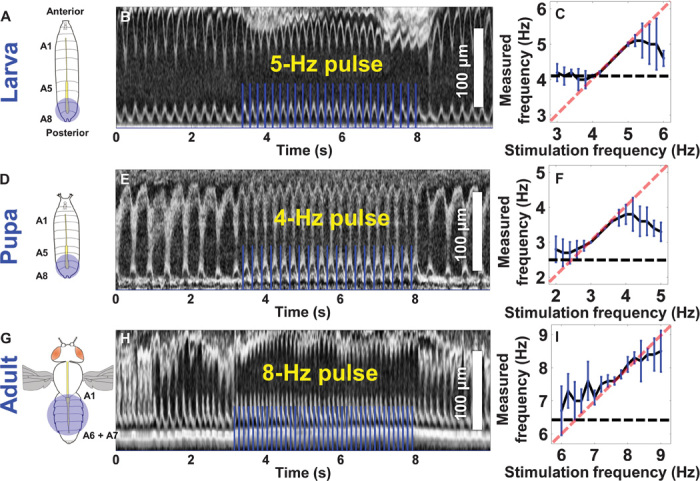
(A, D, and G) Schematic showing pacing stimulation location and spot size used at the larva, pupa, and adult stages. (B, E, and H) M-mode images showing optogenetic pacing at the larva, pupa, and adult stages. The 1:1 correspondence between pacing pulse and heart contraction can be clearly observed. (C, F, and I) Frequency tuning curves showing the reliable range for optogenetic pacing at different stages (n = 6 at each stage). HR was tuned around the average RHR at the respective stages. In larvae, the fly heart linearly followed the pacing frequency from 4 to 5.2 Hz, where the average RHR was 4.1 Hz. In early pupae, the average RHR was 2.5 Hz, and the heart linearly followed the pacing frequency in the range between 2.8 and 4 Hz. During the adult stage (with a higher average RHR of 6.4 Hz), HR and pacing frequency showed a linear relationship between 7 and 8.6 Hz. The fly heart reliably followed the pacing frequency up to ~25, ~50, and ~35% above the RHR at the larva, pupa, and adult stages, respectively. Results are presented as mean ± SD.
Quantification of parameters of cardiac dynamics
We analyzed the relative timing between stimulation pulses and heart contractions to gain further insights into the cardiac dynamics and physiology of Drosophila at different developmental stages. We observed that stimulation pulses incident during RP after each heart contraction failed to initiate an additional heart contraction (Fig. 4A). The data for successful and failed pulses at different time delays with respect to previous systole at different developmental stages are summarized in fig. S3. The probability that a stimulation pulse would initiate a successful heart contraction depended on the time delay between the completed systole and the onset of the stimulation pulse (Fig. 4B). From these results, we estimated RP as the time delay at which there is ~90% probability that a stimulation pulse would initiate a heart contraction. We observed that RP was longest during the pupa stage (~90 ms), intermediate during the larva stage (~70 ms), and shortest during the adult stage (~50 ms). Thus, RP has been determined to depend on the developmental stage and to vary alongside HR, with RP getting longer during less active periods such as the pupa stage. In addition to RP, heart CT (defined as the time delay between the onset of a successful optical stimulation pulse and the complete systole of the resultant heart contraction) was also quantified (Fig. 4C). CT was found to be shortest at the adult stage (~40 ms), intermediate at the larva stage (~90 ms), and longest at the pupa stage (~130 ms). These quantitative analyses of stage-dependent cardiac dynamics provided additional insights into the cardiac physiology of Drosophila at different developmental stages.
CONCLUSION
We developed a novel integrated optogenetic pacing and imaging platform to noninvasively control heart rhythm in a well-established cardiovascular model organism, D. melanogaster, at different stages of its development. In vivo pacing studies of Drosophila performed using this system allowed us to quantitatively characterize the parameters of cardiac dynamics (such as RP and CT) and the variations of these parameters at the different developmental stages of Drosophila. In addition, this noninvasive optogenetic pacing technique offers the possibility to perform in vivo longitudinal pacing studies of Drosophila without sacrificing the specimens, which can be a powerful research tool for studying cardiac disorders associated with heart development and functioning. This provides a great potential for developing a new class of experiments in developmental cardiology to understand arrhythmia mechanisms and to develop new therapeutic strategies for various cardiac disorders. Moreover, the ability to noninvasively pace the heart using optogenetic tools may, in the future, form the basis for a new generation of light-driven cardiac pacemakers and muscle actuators in larger animal models.
MATERIALS AND METHODS
Integrated pacing and OCM imaging system
An integrated optical pacing and OCM imaging system was designed and developed for this study (Fig. 1A). A portion of the white-light spectrum from a supercontinuum source (SC-400-4; Fianium Ltd.) with a central wavelength of ~800 nm and a bandwidth of ~220 nm was used as light source for the OCM system. Similar to the study of Liu et al. (39), the OCM system used a 45° rod mirror to generate an annular sample probe beam for obtaining an extended depth of focus. The OCM system was capable of providing axial and transverse resolutions of 1.5 and 3.9 μm, respectively. Light returning from the reference and sample arms was detected using a spectrometer with a transmission grating of 600 lines/mm (Wasatch Photonics) and a 2048-pixel line-scan camera operating at 20,000 A-scans/s (AViiVA EM4; e2v technologies plc). The spectral domain OCM system was capable of acquiring M-mode [two-dimensional (2D) plus time] images of the Drosophila heart at a frame rate of 130 Hz. The high imaging speed and ultrahigh resolution of the OCM system allowed distinct visualization of Drosophila heart structure and an accurate analysis of its function.
The optogenetic excitation source was a diode-pumped solid-state laser (Blue Ray DPSS; Laserlands Inc.) emitting at 475 nm. The frequency of the excitation pulse was controlled using a function generator (Agilent 33210A; Keysight Technologies) connected to the laser source. As shown in Fig. 1A, the optogenetic excitation pulse was coupled to the OCM sample arm using a dichroic beam splitter (Semrock Inc.). The positions of the pacing and imaging beams could be independently adjusted.
Transgenic fly models
The upstream activating sequence (UAS)–GAL4 system is one of the most powerful tools for targeted gene expression in which the yeast GAL4 transcription factor activates the transcription of its target genes by binding to UAS cis-regulatory sites (40). By mating the UAS transgenic fly with various tissue-, cell-, or developmental stage–specific GAL4 lines, the UAS-GAL4 system limits the ectopic expression or RNA interference silencing of a gene in specific tissues (for example, heart) or cell types (for example, muscle) or at various developmental stages (for example, larva and adult)—powerful strategies that are often necessary to avoid the lethal effects of changing the expression levels of a target protein, for instance, in the Drosophila heart (40, 41). 24B-GAL4 drives target gene expression early in the invaginating mesoderm; later, the expression becomes restricted to myogenic lineage, including cardioblasts and probably all myoblasts (42). For the pacing experiments, the UAS-GAL4 system was used to specifically overexpress ChR2 and to coexpress mCherry fluorescence in the Drosophila heart (24B-GAL4;UAS-H134R-ChR2). Wild-type flies with a heterozygous 24B driver (24B-GAL4/+) were used as control flies. For TEM analysis of heart ultrastructure, 10 hearts from 1-day-old paced and control adult flies were examined and imaged at around the A1 segment, as described previously, Ahsen et al. (29) and Zaffan et al. (43).
Fly food preparation
Flies were fed standard fly food mixed with 1 mM all-trans-retinal (ATR; Sigma-Aldrich Co.) to obtain the optimal response to pacing stimulation. Standard fly food is prepared by mixing Formula 4-24 (Instant Drosophila Medium; Carolina Biological Supply Company), water, and yeast. For preparation of 1 mM ATR food, 100 μl of 100 mM ATR in 100% ethanol was added for every 10 ml of standard fly food. Male and female parent flies were transferred into vials containing ATR food, and these vials were kept in the dark. Four to 5 days after mixing the parent flies, ChR2-expressing larvae were obtained from these vials. The parent flies were transferred out, and the larvae were used for the pacing experiments. With temperature maintained at 25°C, it usually takes between ~6 and ~10 days after egg formation for the flies to reach the early pupa and adult stages, respectively.
Fly preparation before pacing and imaging
Flies were paced at different developmental stages (such as the larva, pupa, and adults stages). During the larva stage, flies were mounted on a glass slide using a double-sided tape. Early pupae were directly mounted on the glass slide. Only those pupae <6 hours after puparium formation were used for pupa-stage pacing experiments. During the adult stage, flies were anesthetized with FlyNap (Carolina Biological Supply Company) for 2 to 3 min in a closed container (44). After anesthesia, flies were taped on a microscope cover slide by their wings, with the dorsal side facing up. OCM imaging and optogenetic pacing were conducted within 30 min before the effects of anesthesia wore off to minimize motion artifacts.
Pacing protocols
During the larva and early pupa stages, the cardiac pacemaker is located in the posterior region of the heart (A8). The larval heart and the pupal heart always beat in the anterograde direction (34). Hence, pacing pulses with a spot size of ~380 μm were used to excite the A8 region of the larval and pupal hearts (Fig. 3, A and D), and M-mode (2D plus time) OCM images from the corresponding region were acquired. During later stages, such as in late pupae and adults, heartbeat alternates between the anterograde direction and the retrograde direction. Hence, the adult heart is hypothesized to have two pacemakers: a retrograde pacemaker located around the A1 region and an anterograde pacemaker located around the A5-A6 junction (34). Blue-light pulses with a larger spot size (~650 μm) were used to simultaneously trigger both pacemakers in the adult heart (Fig. 3G). Considering the differences in excitation spot size, a 16-mW excitation intensity corresponded to an irradiance of ~35.5 mW/mm2 for larval and pupal flies and ~12 mW/mm2 for adult flies. This excitation intensity is well below the maximum permissible skin exposure recommended by the ANSI for safe use of lasers (ANSI Z136.1). In adult flies, M-mode OCM images were obtained from the A1 segment of the heart tube. OCM M-mode image acquisition was synchronized with pacing stimulation, and pulse trigger from the excitation laser was recorded.
For the pacing experiments, flies were prescreened before the actual pacing measurements to select flies showing optimal response to pacing stimulation. During the prescreening procedure, fly hearts were stimulated with blue pulses at three different frequencies (100, 120, and 140%) with respect to the RHR at the respective stages. Only those flies that followed at least two of the three pacing frequencies, indicating a stronger expression of ChR2, were used for the pacing experiments.
Data analysis
Transverse M-mode images obtained across the heart over a region covering 0.28 mm × 0.57 mm (x-z plane) were used to analyze the response of the Drosophila heart to pacing stimulation. M-mode images spanning ~6 s were acquired for threshold measurements. For frequency tuning measurements, alternate periods of stimulation at three different pacing frequencies were used in each individual M-mode data set (videos S1 to S3). After acquisition of OCM M-mode images, the heart region is segmented and the Fourier transform of variations in axial heart dimensions with time is used to calculate HR. For a more detailed analysis of the pacing measurements, M-mode OCM images were resliced to obtain a cross-sectional image showing variations of heart chamber size with time, similar to the OCM images shown in Fig. 1C. Subsequently, pacing pulses from the excitation laser were superimposed on this image to determine the correspondence between pacing pulses and heart contractions (Fig. 3, B, E, and H).
For differentiation of natural heart contractions and stimulation-induced heart contractions, the fly heart was considered as following the pacing frequency only when the heart followed the pacing stimulation for more than three consecutive pulses. In addition, for the pacing to be considered successful, the measured HR needs to be within ±0.1 Hz of the pacing frequency. At each pacing frequency, PE was estimated as the ratio between the number of pacing pulses causing a heart contraction and the total number of stimulation pulses. For threshold measurements, flies at different developmental stages (n = 10 at each stage) were used to characterize optogenetic pacing at different excitation intensities and pulse widths (Fig. 2, A and B). The pacing frequency was set slightly above the RHR at each stage. Among the 10 flies, the number of flies that followed the pacing frequency at the respective excitation intensities and pulse widths was determined. On the basis of the results of this experiment, the probability of achieving reliable pacing at a given power level and at a given pulse width was determined by calculating the maximum likelihood estimate (MLE). For a binomial distribution with n observations of which k are successes, the MLE of the probability of success equals k/n (45).
For a detailed analysis of time delays between pacing stimulation and heart contraction, a custom program written in MATLAB was used to estimate RP and CT at different developmental stages. A histogram of successful and failed pacing pulses at different time delays after the preceding systole indicates the minimum time delay required to initiate a heart contraction (fig. S3). RP was estimated as the time delay at which pacing success reached 90% (Fig. 4B). Furthermore, the average heart CT at different stages was determined as the time delay between the onset of a successful pacing pulse and the complete systole of the resultant heart contraction. Student’s t test was used for statistical analysis, and results were considered statistically significant at P < 0.05.
Acknowledgments
We would like to acknowledge M.-T. Wei and H. D. Ou-Yang (Department of Physics, Lehigh University) for their assistance with confocal fluorescence imaging. We would also like to acknowledge J. Jerwick for proofreading the English language of this manuscript. Funding: This work was supported by the Lehigh University Start-Up Fund, the NIH (R00EB010071 to C.Z., R15EB019704 to C.Z. and A.L., R03AR063271 to A.L., and R01AG014713 and R01MH060009 to R.E.T.), the NSF (1455613 to C.Z. and A.L.), the Cure Alzheimer’s Fund (to R.E.T.), and the Massachusetts General Hospital (Executive Committee on Research Award to A.L.). Author contributions: C.Z., A.A., and A.L. conceived and designed the experiments; A.L. and R.E.T. provided access to the fly facilities at the Massachusetts General Hospital; A.L. prepared the animal models; A.A., A.L., and C.Z. performed the experiments; A.A. and C.Z. analyzed the OCM data; and A.A., A.L., and C.Z. wrote the manuscript. All authors read the manuscript and provided constructive feedbacks. Competing interests: The authors declare that they have no competing interests. Data and materials availability: All relevant data to support the conclusions are within the paper and its Supporting Materials. The authors will make raw data available upon request.
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/1/9/e1500639/DC1
Fig. S1. Cardiac stimulation of control flies (24B-GAL4/+) at different developmental stages.
Fig. S2. Estimation of PE at different pacing frequencies.
Fig. S3. Histogram analysis of RP at different developmental stages.
Video S1. Tuning the HR of a Drosophila larva at 4, 5, and 6 Hz.
Video S2. Tuning the HR of a Drosophila early pupa at 3, 3.5, and 4 Hz.
Video S3. Tuning the HR of an adult Drosophila at 8, 9, and 10 Hz.
REFERENCES AND NOTES
- 1.Hsieh C. H. C., Chia E.-M., Huang K., Lu J., Barry M., Pouliopoulos J., Ross D. L., Thomas S. P., Kovoor P., Evolution of ventricular tachycardia and its electrophysiological substrate early after myocardial infarction an ovine model. Circ. Arrhythm. Electrophysiol. 6, 1010–1017 (2013). [DOI] [PubMed] [Google Scholar]
- 2.Chen M.-H., Liu T.-W., Xie L., Song F.-Q., He T., Zeng Z.-Y., Mo S.-R., A simpler cardiac arrest model in rats. Am. J. Emerg. Med. 25, 623–630 (2007). [DOI] [PubMed] [Google Scholar]
- 3.Kim S. W., Kim H. W., Huang W., Okada M., Welge J. A., Wang Y., Ashraf M., Cardiac stem cells with electrical stimulation improve ischaemic heart function through regulation of connective tissue growth factor and miR-378. Cardiovasc. Res. 100, 241–251 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Akasaka T., Ocorr K., Drug discovery through functional screening in the Drosophila heart. Methods Mol. Biol. 577, 235–249 (2009). [DOI] [PubMed] [Google Scholar]
- 5.Knisley S. B., Trayanova N., Aguel F., Roles of electric field and fiber structure in cardiac electric stimulation. Biophys. J. 77, 1404–1417 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Merrill D. R., Bikson M., Jefferys J. G. R., Electrical stimulation of excitable tissue: Design of efficacious and safe protocols. J. Neurosci. Methods 141, 171–198 (2005). [DOI] [PubMed] [Google Scholar]
- 7.Jenkins M. W., Duke A. R., Gu S., Doughman Y., Chiel H. J., Fujioka H., Watanabe M., Jansen E. D., Rollins A. M., Optical pacing of the embryonic heart. Nat. Photonics 4, 623–626 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jenkins M. W., Wang Y. T., Doughman Y. Q., Watanabe M., Cheng Y., Rollins A. M., Optical pacing of the adult rabbit heart. Biomed. Opt. Express 4, 1626–1635 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Boyden E. S., Zhang F., Bamberg E., Nagel G., Deisseroth K., Millisecond-timescale, genetically targeted optical control of neural activity. Nat. Neurosci. 8, 1263–1268 (2005). [DOI] [PubMed] [Google Scholar]
- 10.Fenno L., Yizhar O., Deisseroth K., The development and application of optogenetics. Annu. Rev. Neurosci. 34, 389–412 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Boyden E. S., A history of optogenetics: The development of tools for controlling brain circuits with light. F1000 Biol. Rep. 3, 11 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nagel G., Szellas T., Huhn W., Kateriya S., Adeishvili N., Berthold P., Ollig D., Hegemann P., Bamberg E., Channelrhodopsin-2, a directly light-gated cation-selective membrane channel. Proc. Natl. Acad. Sci. U.S.A. 100, 13940–13945 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang F., Wang L.-P., Brauner M., Liewald J. F., Kay K., Watzke N., Wood P. G., Bamberg E., Nagel G., Gottschalk A., Deisseroth K., Multimodal fast optical interrogation of neural circuitry. Nature 446, 633–639 (2007). [DOI] [PubMed] [Google Scholar]
- 14.Gradinaru V., Mogri M., Thompson K. R., Henderson J. M., Deisseroth K., Optical deconstruction of parkinsonian neural circuitry. Science 324, 354–359 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chaudhury D., Walsh J. J., Friedman A. K., Juarez B., Ku S. M., Koo J. W., Ferguson D., Tsai H.-C., Pomeranz L., Christoffel D. J., Nectow A. R., Ekstrand M., Domingos A., Mazei-Robison M. S., Mouzon E., Lobo M. K., Neve R. L., Friedman J. M., Russo S. J., Deisseroth K., Nestler E. J., Han M.-H., Rapid regulation of depression-related behaviours by control of midbrain dopamine neurons. Nature 493, 532–536 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Busskamp V., Picaud S., Sahel J. A., Roska B., Optogenetic therapy for retinitis pigmentosa. Gene Ther. 19, 169–175 (2012). [DOI] [PubMed] [Google Scholar]
- 17.Bruegmann T., Malan D., Hesse M., Beiert T., Fuegemann C. J., Fleischmann B. K., Sasse P., Optogenetic control of heart muscle in vitro and in vivo. Nat. Methods 7, 897–900 (2010). [DOI] [PubMed] [Google Scholar]
- 18.Arrenberg A. B., Stainier D. Y. R., Baier H., Huisken J., Optogenetic control of cardiac function. Science 330, 971–974 (2010). [DOI] [PubMed] [Google Scholar]
- 19.Jia Z., Valiunas V., Lu Z., Bien H., Liu H., Wang H.-Z., Rosati B., Brink P. R., Cohen I. S., Entcheva E., Stimulating cardiac muscle by light: Cardiac optogenetics by cell delivery. Circ. Arrhythm. Electrophysiol. 4, 753–760 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Entcheva E., Cardiac optogenetics. Am. J. Physiol. Heart Circ. Physiol. 304, H1179–H1191 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nussinovitch U., Gepstein L., Optogenetics for in vivo cardiac pacing and resynchronization therapies. Nat. Biotechnol. 33, 750–754 (2015). [DOI] [PubMed] [Google Scholar]
- 22.Bodmer R., Venkatesh T. V., Heart development in Drosophila and vertebrates: Conservation of molecular mechanisms. Dev. Genet. 22, 181–186 (1998). [DOI] [PubMed] [Google Scholar]
- 23.Piazza N., Wessells R. J., Drosophila models of cardiac disease. Prog. Mol. Biol. Transl. Sci. 100, 155–210 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pandey U. B., Nichols C. D., Human disease models in Drosophila melanogaster and the role of the fly in therapeutic drug discovery. Pharmacol. Rev. 63, 411–436 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Reiter L. T., Potocki L., Chien S., Gribskov M., Bier E., A systematic analysis of human disease-associated gene sequences in Drosophila melanogaster. Genome Res. 11, 1114–1125 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ocorr K., Reeves N. L., Wessells R. J., Fink M., Chen H.-S. V., Akasaka T., Yasuda S., Metzger J. M., Giles W., Posakony J. W., Bodmer R., KCNQ potassium channel mutations cause cardiac arrhythmias in Drosophila that mimic the effects of aging. Proc. Natl. Acad. Sci. U.S.A. 104, 3943–3948 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wolf M. J., Amrein H., Izatt J. A., Choma M. A., Reedy M. C., Rockman H. A., Drosophila as a model for the identification of genes causing adult human heart disease. Proc. Natl. Acad. Sci. U.S.A. 103, 1394–1399 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li A., Zhou C., Moore J., Zhang P., Tsai T. H., Lee H. C., Romano D. M., McKee M. L., Schoenfeld D. A., Serra M. J., Raygor K., Cantiello H. F., Fujimoto J. G., Tanzi R. E., Changes in the expression of the Alzheimer’s disease–associated presenilin gene in Drosophila heart leads to cardiac dysfunction. Curr. Alzheimer Res. 8, 313–322 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li A., Ahsen O. O., Liu J. J., Du C., McKee M. L., Yang Y., Wasco W., Newton-Cheh C. H., O’Donnell C. J., Fujimoto J. G., Zhou C., Tanzi R. E., Silencing of the Drosophila ortholog of SOX5 in heart leads to cardiac dysfunction as detected by optical coherence tomography. Hum. Mol. Genet. 22, 3798–3806 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huang D., Swanson E. A., Lin C. P., Schuman J. S., Stinson W. G., Chang W., Hee M. R., Flotte T., Gregory K., Puliafito C. A., Fujimoto J. G., Optical coherence tomography. Science 254, 1178–1181 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Izatt J. A., Hee M. R., Owen G. M., Swanson E. A., Fujimoto J. G., Optical coherence microscopy in scattering media. Opt. Lett. 19, 590–592 (1994). [DOI] [PubMed] [Google Scholar]
- 32.Zhou C., Cohen D. W., Wang Y., Lee H.-C., Mondelblatt A. E., Tsai T.-H., Aguirre A. D., Fujimoto J. G., Connolly J. L., Integrated optical coherence tomography and microscopy for ex vivo multiscale evaluation of human breast tissues. Cancer Res. 70, 10071–10079 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.W. Drexler, J. G. Fujimoto, Optical Coherence Tomography: Technology and Applications (Springer, Berlin, 2008), p. 1346. [Google Scholar]
- 34.T. Rizki, in The Genetics and Biology of Drosophila, M. Ashburner, T. Wright, Eds. (Academic Press, London, 1978), vol. 2b, pp. 397–452. [Google Scholar]
- 35.Sláma K., Farkas R., Heartbeat patterns during the postembryonic development of Drosophila melanogaster. J. Insect Physiol. 51, 489–503 (2005). [DOI] [PubMed] [Google Scholar]
- 36.Alex A., Li A., Zeng X., Tate R. E., McKee M. L., Capen D. E., Zhang Z., Tanzi R. E., Zhou C., A circadian clock gene, Cry, affects heart morphogenesis and function in Drosophila as revealed by optical coherence microscopy. PLOS One 10, e0137236 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Monier B., Astier M., Sémériva M., Perrin L., Steroid-dependent modification of Hox function drives myocyte reprogramming in the Drosophila heart. Development 132, 5283–5293 (2005). [DOI] [PubMed] [Google Scholar]
- 38.Dulcis D., Levine R. B., Glutamatergic innervation of the heart initiates retrograde contractions in adult Drosophila melanogaster. J. Neurosci. 25, 271–280 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu L., Gardecki J. A., Nadkarni S. K., Toussaint J. D., Yagi Y., Bouma B. E., Tearney G. J., Imaging the subcellular structure of human coronary atherosclerosis using micro-optical coherence tomography. Nat. Med. 17, 1010–1014 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Brand A. H., Perrimon N., Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development 118, 401–415 (1993). [DOI] [PubMed] [Google Scholar]
- 41.Fischer J. A., Giniger E., Maniatis T., Ptashne M., GAL4 activates transcription in Drosophila. Nature 332, 853–856 (1988). [DOI] [PubMed] [Google Scholar]
- 42.Zaffran S., Astier M., Gratecos D., Sémériva M., The held out wings (how) Drosophila gene encodes a putative RNA-binding protein involved in the control of muscular and cardiac activity. Development 124, 2087–2098 (1997). [DOI] [PubMed] [Google Scholar]
- 43.W. Sullivan, M. Ashburner, R. S. Hawley, Drosophila Protocols (Cold Spring Harbor Laboratory Press, New York, 2008), p. 724. [Google Scholar]
- 44.Paternostro G., Vignola C., Bartsch D.-U., Omens J. H., McCulloch A. D., Reed J. C., Age-associated cardiac dysfunction in Drosophila melanogaster. Circ. Res. 88, 1053–1058 (2001). [DOI] [PubMed] [Google Scholar]
- 45.B. Rosner, Fundamentals of Biostatistics (Brooks/Cole, Boston, MA, 2006), chap. 6, pp. 181–183. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/1/9/e1500639/DC1
Fig. S1. Cardiac stimulation of control flies (24B-GAL4/+) at different developmental stages.
Fig. S2. Estimation of PE at different pacing frequencies.
Fig. S3. Histogram analysis of RP at different developmental stages.
Video S1. Tuning the HR of a Drosophila larva at 4, 5, and 6 Hz.
Video S2. Tuning the HR of a Drosophila early pupa at 3, 3.5, and 4 Hz.
Video S3. Tuning the HR of an adult Drosophila at 8, 9, and 10 Hz.


