Vibrational imaging reveals vitamin E distribution on mouse skin in vivo and captures human breast cancer tissues in situ.
Keywords: Raman spectroscopy, label-free microscopy, molecular vibration
Abstract
In vivo vibrational spectroscopic imaging is inhibited by relatively slow spectral acquisition on the second scale and low photon collection efficiency for a highly scattering system. Recently developed multiplex coherent anti-Stokes Raman scattering and stimulated Raman scattering techniques have improved the spectral acquisition time down to microsecond scale. These methods using a spectrometer setting are not suitable for turbid systems in which nearly all photons are scattered. We demonstrate vibrational imaging by spatial frequency multiplexing of incident photons and single photodiode detection of a stimulated Raman spectrum within 60 μs. Compared to the spectrometer setting, our method improved the photon collection efficiency by two orders of magnitude for highly scattering specimens. We demonstrated in vivo imaging of vitamin E distribution on mouse skin and in situ imaging of human breast cancerous tissues. The reported work opens new opportunities for spectroscopic imaging in a surgical room and for development of deep-tissue Raman spectroscopy toward molecular level diagnosis.
INTRODUCTION
In vivo vibrational spectroscopic imaging is opening a new window on cellular machinery by visualizing the spatiotemporal dynamics of target molecules or intracellular organelles. However, in vivo spectroscopic imaging is not a simple combination of spectroscopy and microscopy. Two grand challenges need to be overcome to realize spectroscopic imaging of a living system. One is decreasing the spectral acquisition time to microsecond scale per pixel to allow real-time imaging of a highly dynamic living system. The other is extracting the spectral information from highly scattered photons, for which the conventional dispersion approach is no longer efficient.
To increase the imaging speed, single- or multicolor coherent anti-Stokes Raman scattering (CARS) and stimulated Raman scattering (SRS) have been used to map known species (1–4), which have reached video rate speed (5, 6). To resolve overlapping Raman bands, various frame-by-frame hyperspectral imaging platforms by Raman shift sweeping (7–12) have been reported. In these methods, the CARS or SRS spectra are derived from the whole image stack acquired on millisecond to second scale, which is insufficient for imaging living systems. Broadband CARS (13–15) and multiplex SRS (16–18) have been developed to allow spectral acquisition at each pixel. Using a tuned amplifier array, microsecond-scale multiplex SRS imaging has been demonstrated (19). Yet, these methods using a spectrometer setting are not suitable for turbid systems in which nearly all photons are scattered.
The spectrometer, conceptually demonstrated by Newton in 1672 and first built by Fraunhofer in 1814, uses a prism or grating to disperse the collimated signal light on a detector array, which requires a slit to reject scattered photons to achieve the desired spectral resolution. For highly scattered photons, this scheme becomes inefficient because most of the photons are rejected by the slit. An elegant way to overcome this difficulty is to code the excitation photons, collect the photons with a single detector, and then decode the spectral signal. To this end, spectrally tailored SRS using pulse shaping technology has been developed to map desired chemicals (20). Three-color SRS by modulation-multiplexing at kilohertz rate has been demonstrated with an acousto-optical tunable filter (21). Coherent control (22) and frequency comb (23) techniques allowed acquisition of a CARS spectrum at each pixel. These two techniques required tens of milliseconds or longer for each spectrum, which is not sufficient for in vivo imaging purposes.
Here, we demonstrate a spectrometer-free, microsecond-scale spectroscopic imaging scheme by using a single large-area photodiode to collect highly scattered photons within tens of microseconds for each pixel. We first modulate the intensity (Ip) of a spectrally dispersed broadband pump beam with distinct megahertz frequencies (Fig. 1A). The broadband laser is then combined with a narrow-band Stokes beam (IS) and focused onto a sample. Through the stimulated Raman gain (SRG) process, ΔI = σIpIS, where σ is the Raman scattering cross section, a Raman spectrum is coded into the intensity of the Stokes beam, with each Raman shift corresponding to a distinct modulation frequency. Then, a large-area detector collects the Stokes beam intensity in the time domain, and a Fourier transformation is performed to retrieve the SRG spectrum.
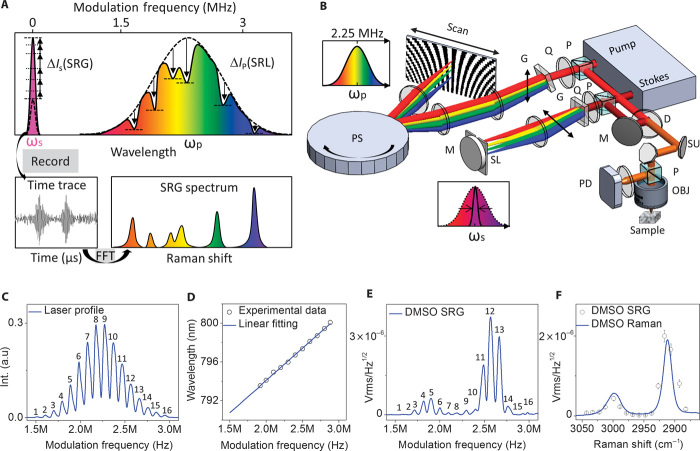
Fig. 1. Stimulated Raman spectroscopic imaging by spatial frequency multiplexing and single photodiode detection.
(A) Principle. Every color of the pump laser was modulated at a specific megahertz frequency. Through the SRG process, the modulation frequency transferred to the Stokes beam. A time trace was recorded, from which fast Fourier transform was performed to retrieve an SRG spectrum. (B) A lab-built multiplex-modulation SRS microscope. D, dichroic mirror; G, grating; SU, scanning unit; M, mirror; OBJ, objective; P, polarizing beam splitter; PD, photodiode; PS, polygon scanner; Q, quarter waveplate; SL, slit. (C) Modulated pump laser intensity as a function of modulation frequency. (D) Linear relation between wavelength and modulation frequency. (E) SRG intensity as a function of modulation frequency. (F) Retrieved SRG spectrum (circles) and spontaneous Raman spectrum (solid line).
RESULTS
Quantification of photon collection efficiency by ray tracing simulation
We quantitatively evaluated the photon collection efficiency by ray tracing simulations in nonsequential mode using Zemax. In brief, 107 rays were focused by a lens (f = 2 mm) into a 25-μm spot on the surface of a scattering medium. A Henyey-Greenstein scattering phase function was used to approximate the angular scattering dependence (24). The reflected rays were collected by the same lens and passed through a pinhole of 50-μm diameter on the focal plane (fig. S1A). The percentage of rays entering the spectrometer was 0.01% for a 2.0-mm-thick dermis tissue with scattering parameters measured by Tuchin et al. (25). In comparison, a detector (10 × 10 mm2) placed right after the lens collected many more reflected rays (fig. S1B). On the basis of our simulation result, the detection efficiency can be improved 200 times with a single detector to directly collect modulated photons and retrieve the spectral information through demodulation.
Single-photodiode detection of SRS spectra by spatial frequency multiplexing
We used femtosecond pulse shaping technology to implement spatial frequency multiplexing at megahertz level. A pulse shaper narrowed down the spectral width of the Stokes beam to 15 cm−1. The pump beam was dispersed vertically and then scanned horizontally by a polygon scanner onto a vertically aligned reflective pattern with 16 different densities, so that the spectrally dispersed laser source was modulated at 16 frequencies ranging from 1.5 to 3.0 MHz. After combining and focusing the pump and Stokes beams onto the sample, the Stokes beam, as the local oscillator, was collected by a large-area photodiode and amplified by a resonant circuit with a 2.25-MHz central frequency and a 1.5-MHz bandwidth. A data acquisition board recorded a 60-μs time trace of the SRG signal, and fast Fourier transformation demodulated the 16 frequency components to 16 Raman shifts. A schematic of our experimental setup is shown in Fig. 1B (detailed in Materials and Methods). Figure 1C shows the 16 distinct frequency components of the broadband pump beam, each with a 70-kHz bandwidth. The frequency difference between two adjacent components was 100 kHz, which prevented crosstalk between adjacent channels. Moreover, a frequency range of 1.5 to 3.0 MHz prevented crosstalk caused by harmonic frequencies. The pump laser intensity profile (Fig. 1C) was used for normalization of the SRG signal on each spectral channel. By relating the wavelengths of the pump laser to the modulation frequencies, a linear relationship was found between Raman shifts and modulation frequencies (Fig. 1D). We further acquired the SRG spectrum of 100% dimethyl sulfoxide (DMSO) solution. Figure 1E shows SRG intensity as a function of modulation frequency. After laser intensity normalization and Raman shift calibration, an SRG spectrum was retrieved, which reproduced the spontaneous Raman spectrum (Fig. 1F).
We note that collecting all dispersed photons by a single detector increases the level of shot noise shared among all spectral channels (26). To evaluate the noise level, we measured the SDs of 16 spectroscopic images as a function of laser power on the photodiode (fig. S2A). After normalizing the gain of each spectral channel (fig. S2B), we compared the noise levels with electronic noise and theoretical shot noise (fig. S2C). Under a laser power of 10 mW on the photodiode, the theoretical shot noise and the measured noise were 1.5 × 10−8 and 1.9 × 10−8 Vrms/Hz1/2, respectively, which exceeded the level of electronic noise (2.0 × 10−9 Vrms/Hz1/2). We further determined our detection sensitivity by recording the 2912 cm−1 Raman peak of diluted DMSO solutions. The SRG spectrum of 1.0% DMSO produced a signal level of 2.5 × 10−8 Vrms/Hz1/2 (fig. S2D), corresponding to a detection sensitivity of 3.2 × 10−6 dI/I modulation depth at 60-μs time constant.
Acquisition of stimulated Raman spectra from diffuse photons
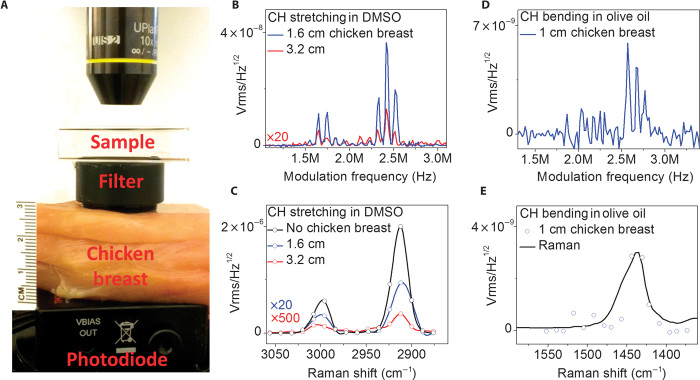
To show that our technique is capable of extracting a Raman spectrum from highly diffuse photons, we focused the pump and Stokes light with a 10× objective into a cuvette filled with a specimen (Fig. 2A). After passing through the filter, the Stokes laser beam propagated through a chicken breast tissue and was received by a photodiode with 60-μs integration time. We first recorded the SRG spectra of DMSO in the spectral window of C-H stretching vibration 2800 to 3000 cm−1). The raw modulation spectra of DMSO reached a signal-to-noise ratio (SNR) of ~60 and ~6 for 1.6- and 3.2-cm chicken breast tissues, respectively (Fig. 2B). The calibrated spectra are shown in Fig. 2C. We then recorded the SRG spectra of olive oil in the C-H bending vibration (~1450 cm−1). With 1.0-cm chicken breast tissues before the photodiode, the raw modulation spectrum showed an SNR of ~3 (Fig. 2D), and the calibrated SRG spectrum matched the spontaneous Raman spectrum (Fig. 2E). These results collectively demonstrated microsecond-scale spectrum extraction from diffuse photons in both C-H stretching and fingerprint vibration regions.
Fig. 2. Microsecond-scale acquisition of SRG spectra from completely diffused photons.
(A) Experiment configuration. (B) SRG intensity of DMSO as a function of modulation frequency with 1.6- or 3.2-cm chicken breast tissues before photodiode. (C) Calibrated SRG spectra of DMSO. (D) SRG intensity of olive oil as a function of modulation frequency with 1.0-cm chicken breast tissues before photodiode. (E) Calibrated SRG spectra of olive oil. The integration time was 60 μs.
Following drug diffusion through mouse skin tissue in vivo
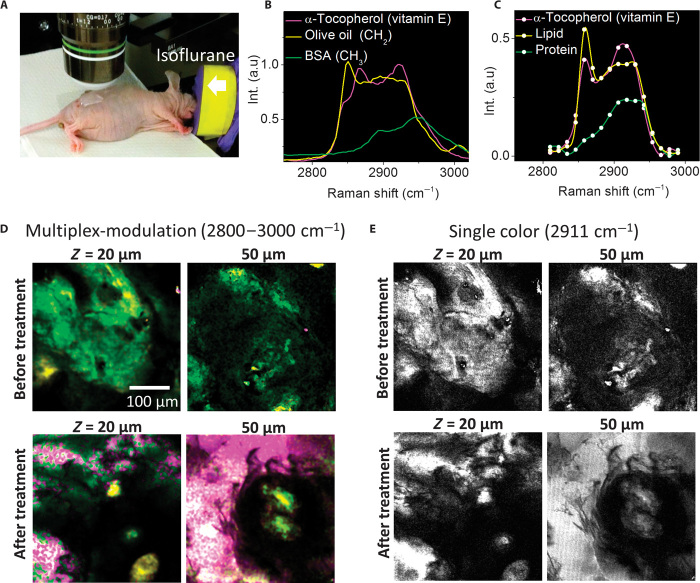
Our technique allowed in vivo monitoring of drug delivery into a mouse skin. Topical drug delivery is traditionally analyzed by tape-stripping or microdialysis techniques. Single-color CARS and SRS microscopy have been used to map deuterated drug in the skin using the isolated C-D stretching vibration (27, 28). Here, we demonstrate in vivo mapping of vitamin E, in the form of α-tocopherol, on the skin of a live mouse (Fig. 3A). Vitamin E is a fat-soluble antioxidant widely used for the maintenance of healthy skin. This molecule exhibits a spectral profile overlapping with the skin tissue in the C-H stretching vibration (Fig. 3B). To distinguish vitamin E from the skin tissue, we acquired an SRG spectrum at each pixel and performed multivariate curve resolution (MCR) analysis of 40,000 SRG spectra. The SRG image was decomposed into vitamin E (in pink), lipid (in yellow), and protein (in green) as shown in Fig. 3C. Their concentration distributions in the epidermis layer (~20 μm below the surface) and around the sebaceous gland (~50 μm below the surface) are shown in Fig. 3D. Before the treatment, only lipid and protein components were found. After 5 min of treatment, vitamin E molecules were found (in pink) in the epidermis layer and enriched in the sebaceous gland (Fig. 3D). In contrast, selective imaging of vitamin E molecules was impossible with single-color SRS microscopy (Fig. 3E). Together, these data showed that our technique is capable of mapping a specific chemical in a spectrally overlapped tissue in vivo.
Fig. 3. In vivo monitoring of vitamin E distribution on mouse skin.
(A) Experiment configuration. The nude mouse was anesthetized with isoflurane. (B)Spontaneous Raman spectra of vitamin E, olive oil (rich in CH2), and bovine serum albumin (rich in CH3). (C) MCR output spectra of three components assigned to lipid, protein, and vitamin E. (D) MCR concentration maps of lipid (in yellow) and protein (in green) before and after the treatment. (E) SRG images at 2911 cm−1. The pixel dwell time was 60 μs. Scale bar, 100 μm.
Spectroscopic imaging of human breast cancerous tissue in situ
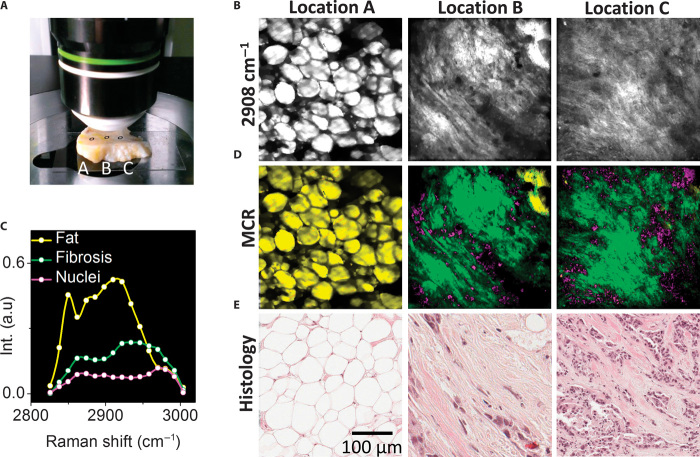
We further demonstrated the capability of our scheme for imaging human breast cancer and stroma in situ. Current medical diagnosis of malignancies relies on in vitro hematoxylin and eosin (H&E) staining and histological examination of biopsies. Two-color SRS allowed selective imaging of lipids and proteins in brain tissue on the basis of CH2 vibration (2845 cm−1) and CH3 vibration (2930 cm−1) (29). Broadband CARS (15) and wavelength-sweeping SRS microscopy (9) were used to map the chemical composition of thin tissue sections. Here, we demonstrate epi-detected stimulated Raman spectroscopic imaging of intact human breast cancerous tissues (Fig. 4A). SRG images at 2908 cm−1 showed the morphology of the cancerous tissue at three different locations (Fig. 4B). MCR analysis decomposed the SRG spectra into three components (Fig. 4C). The spectral profile with a peak around 2850 cm−1 was assigned to the adipose breast tissue. The peak at 2940 cm−1 was assigned to fibrosis, and the 2960 cm−1 peak was assigned to cell nuclei. The corresponding MCR concentration maps (Fig. 4D) closely matched the histological examination of sliced tissues of the same area (Fig. 4E).
Fig. 4. In situ mapping of human patient breast cancer and stroma.
(A) Digital picture of a 5-mm-thick human breast cancerous tissue. (B) SRG images of locations indicated in (A) at 2908 cm−1. (C) MCR output spectra of three components assigned to fat, fibrosis, and cell nuclei. (D) MCR concentration maps of fat (in yellow), fibrosis (in green), and cell nuclei (in pink). (E) Histological analysis (H&E staining) of the same tissue. The pixel dwell time was 600 μs. Scale bar, 100 μm.
DISCUSSION
The current work demonstrated a platform for acquisition of a Raman spectrum from highly scattered photons at 60-μs integration time. Our method is conceptually different from Fourier-transform Raman spectroscopy, which sends the collimated signal light to a Michelson interferometer and retrieves the spectrum encoded in the Fourier domain (30). A greater potential of our technology can be realized by placing a ring-shaped detector between the focusing lens and the specimen (6). In this way, up to 40% of diffuse photons can be collected to generate a vibrational spectrum in a spectrometer-free manner. In another direction toward minimally invasive molecular-level diagnosis, the combination of our spatial frequency multiplexing scheme with techniques that enable focusing of light through a turbid tissue, such as time reversal of ultrasound-encoded light (31) or digital phase conjugation with dynamic wavefront shaping (32), promises a new avenue to implement deep-tissue vibrational spectroscopy for minimally invasive in vivo diagnosis.
It is interesting to compare the detection sensitivity between our approach and single-color excitation SRS. For single-color SRS microscopy, the detection sensitivity of a 50 μM retinol or 5 mM methanol solution was reported by Freudiger et al. (33) with 1-min integration time. The 5 mM concentration limit for cholesterol reported by Freudiger et al. was achieved at 1-s integration time. Here, we achieved a 141 mM DMSO solution sensitivity at 60-μs integration time (fig.S2D). At 1-s integration time, our SNR would be improved 129 times in theory, which corresponds to a detection sensitivity of 1.1 mM. Thus, the detection sensitivity of our setup is on the same level as single-color SRS. This sensitivity is partly attributed to shot-noise limited detection and partly to spectral analysis of the whole data set.
It is important to note that acquiring a spectrum at each pixel allows for separation of the SRS signal from the unwanted cross-phase modulation. Because cross-phase modulation is largely wavelength-independent, by using spectral analysis such as MCR, we can separate the Raman signal from this nonresonant background.
Our imaging platform can be applied to monitor the spatiotemporal dynamics of target molecules in nontransparent living tissues from which the signal photons are completely scattered. In these systems, spectral acquisition by the conventional dispersion approach would lead to low photon collection efficiency. Because the animal motion and molecular diffusion are on the second scale, a microsecond-scale spectral acquisition speed is required to collect a spectroscopic image within seconds. Our technique meets this requirement by recording a 60-μs signal time trace from highly scattered photons and retrieving a spectrum that is encoded in the modulation frequency domain. We expect broad applications including in vivo assessment of local drug bioavailability in human skin. Although we focused on epi-detection, our technique is also applicable to forward detection of scattering samples. We performed spectroscopic imaging of live Caenorhabditis elegans and showed its capability to distinguish lipid- and protein-rich compartments (fig. S3).
Our technology opens an avenue toward in vivo intraoperative histological imaging. Current intraoperative tools for cancerous tissue assessment, including cytological examination, frozen section analysis, ultrasound, and radio-frequency spectroscopy (34–38), either are time-consuming or lack sufficient tumor specificity. Recently reported Raman spectroscopy technology for intraoperative tumor residual detection has shown the ability to differentiate cancer cells from normal brain tissue with a sensitivity of 93% and a specificity of 91% by point spectral measurement within 1 s (39). Our technique reported here improved the acquisition speed of a vibrational spectrum to microsecond scale, thus allowing real-time spectroscopic imaging. With the aid of the MCR analysis, we further generated chemical images of bulk human cancerous tissue, which reproduced the H&E staining and histological examination results. Future development of a mobile intraoperative vibrational spectroscopic imaging tool would enable generation of histological data from intact tissues in vivo to assess residual diseases in a surgical room.
The spectral coverage of our technology can be improved. The number of spectral channels (N) is determined as N = (fmax − fmin)/Δf, where fmax is the maximum modulation frequency, fmin is the minimum modulation frequency, and Δf is the bandwidth of each spectral channel. The spectrum acquisition time (T) is determined by T = 2/Δf. Thematically, by modulating an ultrafast pulse from 20 to 40 MHz with 70-kHz bandwidth, we will be able to generate 285 spectral channels within 29 μs. Moreover, with a 10-fs pulse, we will be able to cover the entire fingerprint region with a spectral window of 1470 cm−1.
In summary, our technique improved the Raman spectral acquisition speed to microsecond scale and at the same time allowed the retrieval of a vibrational spectrum from highly scattered photons. Using this approach, we demonstrated in situ imaging of cancerous human breast tissue and showed results that reproduced H&E staining and histological examination. Collectively, our technique has the potential of pushing Raman spectroscopy from a single point measurement tool toward in vivo clinical imaging applications.
MATERIALS AND METHODS
Spatial frequency multiplexing on an SRS microscope
A tunable 80-MHz pulsed laser (InSight, Spectra Physics) provided two synchronized outputs. The tunable pump beam provided up to 1.0-W power, 120-fs pulse duration, and a tuning range from 680 to 1300 nm. The fixed 1040-nm beam with 0.5-W power and ~200-fs pulse width served as the Stokes beam. The Stokes beam was sent to a diffraction grating (1200 grooves/mm), and a slit on the Fourier plane narrowed the bandwidth. The full width at half maximum was measured to be 3 ps by an autocorrelator, and the power was 100 mW. A broadband pump beam covering 180 cm−1 at 800 nm was dispersed by a diffraction grating (1200 grooves/mm) in the x direction and scanned by a 17-kHz polygon scanner (Lincoln Laser) in the y direction on a photolithography mask, which modulated the reflected intensities of each wavelength at 16 different frequencies ranging from 1.5 to 3.0 MHz. The photolithography mask was designed with 16 one-dimensional arrays of points in the x direction. Within each array, there were 88 to 176 periods in the y direction. These features were written by blue chrome, which exhibits an optical density of 3 and low reflectivity (less than 5%) at 800 nm, on a 4 inch × 4 inch × 0.060 inch piece of soda lime. We placed a mirror (E03, Thorlabs) after the photomask to reflect the modulated 800-nm laser beam. Therefore, the modulation depth was close to 95%. A similar scheme was used to modulate the beam in the kilohertz range in a previous study (26). For imaging applications, the modulated pump and Stokes beams were sent to a lab-built microscope. A 25× objective (XLPlan N 25×, Olympus) was used to focus the light on the sample. The pump and Stokes laser powers on the sample were 30 and 50 mW, respectively. The SRG signal was then collected by the same objective, and a polarizing beam splitter was used to reflect the polarization-scrambled SRG signal to a photodiode (s3994-01, Hamamatsu). This epi-detection scheme collected 3% of photons from the sample. For the forward-detected imaging scheme, the Stokes beam was collected by an oil condenser and sent to the photodiode. The induced photocurrent was amplified through a resonant amplifier circuit, which used the parasitic capacitance of the photodiode to create a central resonance frequency of 2.25 MHz and a bandwidth of 1.5 MHz. The amplified signal was sent to a high-speed data acquisition board with up to 140 MHz sampling rate (ATS 460, AlazarTech). The data acquisition was synchronized with the galvo mirrors in the lab-built microscope and the polygon scanner in the pulse shaper by LabVIEW. A 60-μs time trace of SRG signal with a sampling rate of 10 MHz was recorded for each pixel. Fast Fourier transformation was performed in LabVIEW to retrieve the SRG spectra in the modulation frequency domain. For each spectral channel, we integrated over the 70-kHz bandwidth. After the laser power normalization and Raman shift calibration, an SRG spectrum was retrieved. For an image with 200 × 200 pixels, it took 2.4 s to record a total of 40,000 time traces. The data transfer from the data acquisition board to the computer required ~3 s. The following Fourier transformation at each pixel and image reconstruction by LabVIEW required 1 min.
MCR analysis
MCR and alternative least squares fitting were used to decompose the SRG spectral images into chemical maps of major components in a specimen (11). MCR is a bilinear model (40) capable of decomposing a measured spectral data set D into concentration profiles and spectra of chemical components, represented by matrices C and ST, D = C·ST + E. Here, T is the transpose of matrix of S. E is the residual matrix or experimental error. The inputs to MCR are the data set D and the reference spectra of each component. S contains the output spectra of all fitted components. The output concentration of a chemical component at each pixel is expressed as a percentage relative to the intensity of the MCR-optimized spectrum. Given an initial estimate of pure spectra from either principal components analysis or prior knowledge, an alternating least squares algorithm calculates C and S iteratively until the results optimally fit the data matrix D. Nonnegativity on both concentration maps and spectral profiles is applied as a constraint during the alternating least squares iteration. To reduce ambiguity associated with MCR decomposition, a data augmentation matrix composed of repeating reference spectra can be added to the spectral data set D if needed. The enhanced weight on pure reference spectra ensures that the MCR algorithm selectively recovers concentration profiles for corresponding Raman bands from experimental data set D.
Mouse tissue preparation
The protocol for this animal study (no. 1111000114) was approved by the Purdue Animal Care and Use Committee. A male athymic nude mouse (6 weeks old) under inhalation anesthesia with isoflurane was used for in vivo imaging.
Human tissue materials and examination
The breast tumor samples were excised from a patient diagnosed with invasive ductal carcinoma and then fixed in 10% buffered formalin. The SRS imaging experiments followed an Institutional Review Board protocol (no. 0905008097) approved by Purdue University. After SRS imaging, the tissues were sectioned and stained with H&E for histological examination in the Purdue Histology and Phenotyping Laboratory.
Acknowledgments
We thank A. Weiner for helpful discussions. Funding: This work was supported by the Keck Foundation and NIH grant GM104681 to J.-X.C. Author contributions: C.-S.L. and Pu W. designed the experiment. G.E. designed the electronic circuits. C.-S.L., Pu W., Ping W., J.L., and H.J.L. performed the experiments. C.-S.L. carried out the data analysis. J.-X.C. conceived the study and provided overall guidance. All authors discussed the results and contributed to the writing of the manuscript. Competing interests: J.-X.C. has a financial interest in Vibonix Inc. Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and Supplementary Materials and will be made available by the authors upon request.
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/1/9/e1500738/DC1
Vibrational spectroscopic imaging of live C. elegans
Fig. S1. Comparison of photon collection efficiency by a spectrometer to that by a single detector.
Fig. S2. Detection sensitivity of spatial frequency multiplexing SRS microscopy.
Fig. S3. Spectroscopic imaging of a live wild-type C. elegans.
REFERENCES AND NOTES
- 1.Wei L., Hu F., Shen Y., Chen Z., Yu Y., Lin C.-C., Wang M. C., Min W., Live-cell imaging of alkyne-tagged small biomolecules by stimulated Raman scattering. Nat. Methods 11, 410–412 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lee H. J., Zhang W. D., Zhang D. L., Yang Y., Liu B., Barker E. L., Buhman K. K., Slipchenko L. V., Dai M., Cheng J.-X., Assessing cholesterol storage in live cells and C. elegans by stimulated Raman scattering imaging of phenyl-diyne cholesterol. Sci. Rep. 5, 7930 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hong S., Chen T., Zhu Y., Li A., Huang Y., Chen X., Live-cell stimulated Raman scattering imaging of alkyne-tagged biomolecules. Angew. Chem. Int. Ed. 53, 5827–5831 (2014). [DOI] [PubMed] [Google Scholar]
- 4.Kong L., Ji M., Holtom G. R., Fu D., Freudiger C. W., Xie X. S., Multicolor stimulated Raman scattering microscopy with a rapidly tunable optical parametric oscillator. Opt. Lett. 38, 145–147 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Evans C. L., Potma E. O., Puoris’haag M., Côté D., Lin C. P., Xie X. S., Chemical imaging of tissue in vivo with video-rate coherent anti-Stokes Raman scattering microscopy. Proc. Natl. Acad. Sci. U.S.A. 102, 16807–16812 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Saar B. G., Freudiger C. W., Reichman J., Stanley C. M., Holtom G. R., Xie X. S., Video-rate molecular imaging in vivo with stimulated Raman scattering. Science 330, 1368–1370 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lin C.-Y., Suhalim J. L., Nien C. L., Miljković M. D., Diem M., Jester J. V., Potma E. O., Picosecond spectral coherent anti-Stokes Raman scattering imaging with principal component analysis of meibomian glands. J. Biomed. Opt. 16, 021104 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Garbacik E. T., Herek J. L., Otto C., Offerhaus H. L., Rapid identification of heterogeneous mixture components with hyperspectral coherent anti-Stokes Raman scattering imaging. J. Raman Spectrosc. 43, 651–655 (2012). [Google Scholar]
- 9.Ozeki Y., Umemura W., Otsuka Y., Satoh S., Hashimoto H., Sumimura K., Nishizawa N., Fukui K., Itoh K., High-speed molecular spectral imaging of tissue with stimulated Raman scattering. Nat. Photonics 6, 844–851 (2012). [Google Scholar]
- 10.Fu D., Holtom G., Freudiger C., Zhang X., Xie X. S., Hyperspectral imaging with stimulated Raman scattering by chirped femtosecond lasers. J. Phys. Chem. B 117, 4634–4640 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang D., Wang P., Slipchenko M. N., Ben-Amotz D., Weiner A. M., Cheng J.-X., Quantitative vibrational imaging by hyperspectral stimulated Raman scattering microscopy and multivariate curve resolution analysis. Anal. Chem. 85, 98–106 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Masia F., Glen A., Stephens P., Borri P., Langbein W., Quantitative chemical imaging and unsupervised analysis using hyperspectral coherent anti-Stokes Raman scattering microscopy. Anal. Chem. 85, 10820–10828 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Müller M., Schins J. M., Imaging the thermodynamic state of lipid membranes with multiplex CARS microscopy. J. Phys. Chem. B 106, 3715–3723 (2002). [Google Scholar]
- 14.Kee T. W., Cicerone M. T., Simple approach to one-laser, broadband coherent anti-Stokes Raman scattering microscopy. Opt. Lett. 29, 2701–2703 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Camp C. H. Jr, Lee Y. J., Heddleston J. M., Hartshorn C. M., Walker A. R. H., Rich J. N., Lathia J. D., Cicerone M. T., High-speed coherent Raman fingerprint imaging of biological tissues. Nat. Photonics 8, 627–634 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rock W., Bonn M., Parekh S. H., Near shot-noise limited hyperspectral stimulated Raman scattering spectroscopy using low energy lasers and a fast CMOS array. Opt. Express 21, 15113–15120 (2013). [DOI] [PubMed] [Google Scholar]
- 17.Seto K., Okuda Y., Tokunaga E., Kobayashi T., Development of a multiplex stimulated Raman microscope for spectral imaging through multi-channel lock-in detection. Rev. Sci. Instrum. 84, 083705 (2013). [DOI] [PubMed] [Google Scholar]
- 18.Marx B., Czerwinski L., Light R., Somekh M., Gilch P., Multichannel detectors for femtosecond stimulated Raman microscopy—Ideal and real ones. J. Raman Spectrosc. 45, 521–527 (2014). [Google Scholar]
- 19.Liao C.-S., Slipchenko M. N., Wang P., Li J., Lee S.-Y., Oglesbee R. A., Cheng J.-X., Microsecond scale vibrational spectroscopic imaging by multiplex stimulated Raman scattering microscopy. Light Sci. Appl. 4, e265 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Freudiger C. W., Min W., Holtom G. R., Xu B. W., Dantus M., Xie X. S., Highly specific label-free molecular imaging with spectrally tailored excitation-stimulated Raman scattering (STE-SRS) microscopy. Nat. Photonics 5, 103–109 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fu D., Lu F.-K., Zhang X., Freudiger C., Pernik D. R., Holtom G., Xie X. S., Quantitative chemical imaging with multiplex stimulated Raman scattering microscopy. J. Am. Chem. Soc. 134, 3623–3626 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dudovich N., Oron D., Silberberg Y., Single-pulse coherently controlled nonlinear Raman spectroscopy and microscopy. Nature 418, 512–514 (2002). [DOI] [PubMed] [Google Scholar]
- 23.Ideguchi T., Holzner S., Bernhardt B., Guelachvili G., Picqué N., Hänsch T. W., Coherent Raman spectro-imaging with laser frequency combs. Nature 502, 355–358 (2013). [DOI] [PubMed] [Google Scholar]
- 24.Cheong W.-F., Prahl S. A., Welch A. J., A review of the optical-properties of biological tissues. IEEE J. Quantum Elect. 26, 2166–2185 (1990). [Google Scholar]
- 25.Tuchin V. V., Utz S. R., Yaroslavsky I. V., Tissue optics, light-distribution, and spectroscopy. Opt. Eng. 33, 3178–3188 (1994). [Google Scholar]
- 26.Howard S. S., Straub A., Horton N. G., Kobat D., Xu C., Frequency-multiplexed in vivo multiphoton phosphorescence lifetime microscopy. Nat. Photonics 7, 33–37 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Saar B. G., Contreras-Rojas L. R., Xie X. S., Guy R. H., Imaging drug delivery to skin with stimulated Raman scattering microscopy. Mol. Pharm. 8, 969–975 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen X., Grégoire S., Formanek F., Galey J.-B., Rigneault H., Quantitative 3D molecular cutaneous absorption in human skin using label free nonlinear microscopy. J. Control. Release 200, 78–86 (2015). [DOI] [PubMed] [Google Scholar]
- 29.Ji M., Orringer D. A., Freudiger C. W., Ramkissoon S., Liu X., Lau D., Golby A. J., Norton I., Hayashi M., Agar N. Y. R., Young G. S., Spino C., Santagata S., Camelo-Piragua S., Ligon K. L., Sagher O., Xie X. S., Rapid, label-free detection of brain tumors with stimulated Raman scattering microscopy. Sci. Transl. Med. 5, 201ra119 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hirschfeld T., Chase B., FT-Raman spectroscopy: Development and justification. Appl. Spectrosc. 40, 133–137 (1986). [Google Scholar]
- 31.Xu X., Liu H., Wang L. V., Time-reversed ultrasonically encoded optical focusing into scattering media. Nat. Photonics 5, 154–157 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang Y. M., Judkewitz B., DiMarzio C. A., Yang C., Deep-tissue focal fluorescence imaging with digitally time-reversed ultrasound-encoded light. Nat. Commun. 3, 928 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Freudiger C. W., Min W., Saar B. G., Lu S., Holtom G. R., He C., Tsai J. C., Kang J. X., Xie X. S., Label-free biomedical imaging with high sensitivity by stimulated Raman scattering microscopy. Science 322, 1857–1861 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Klimberg V. S., Westbrook K. C., Korourian S., Use of touch preps for diagnosis and evaluations of surgical margins in breast cancer. Ann. Surg. Oncol. 5, 220–226 (1998). [DOI] [PubMed] [Google Scholar]
- 35.Valdes E. K., Boolbol S. K., Ali I., Feldman S. M., Cohen J.-M., Intraoperative touch preparation cytology for margin assessment in breast-conservation surgery: Does it work for lobular carcinoma? Ann. Surg. Oncol. 14, 2940–2945 (2007). [DOI] [PubMed] [Google Scholar]
- 36.Olson T. P., Harter J., Muñoz A., Mahvi D. M., Breslin T. M., Frozen section analysis for intraoperative margin assessment during breast-conserving surgery results in low rates of re-excision and local recurrence. Ann. Surg. Oncol. 14, 2953–2960 (2007). [DOI] [PubMed] [Google Scholar]
- 37.Karni T., Pappo I., Sandbank J., Lavon O., Kent V., Spector R., Morgenstern S., Lelcuk S., A device for real-time, intraoperative margin assessment in breast-conservation surgery. Am. J. Surg. 194, 467–473 (2007). [DOI] [PubMed] [Google Scholar]
- 38.Doyle T. E., Factor R. E., Ellefson C. L., Sorensen K. M., Ambrose B. J., Goodrich J. B., Hart V. P., Jensen S. C., Patel H., Neumayer L. A., High-frequency ultrasound for intraoperative margin assessments in breast conservation surgery: A feasibility study. BMC Cancer 11, 444 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jermyn M., Mok K., Mercier J., Desroches J., Pichette J., Saint-Arnaud K., Bernstein L., Guiot M.-C., Petrecca K., Leblond F., Intraoperative brain cancer detection with Raman spectroscopy in humans. Sci. Transl. Med. 7, 274ra19 (2015). [DOI] [PubMed] [Google Scholar]
- 40.de Juan A., Tauler R., Multivariate curve resolution (MCR) from 2000: Progress in concepts and applications. Crit. Rev. Anal. Chem. 36, 163–176 (2006). [Google Scholar]
- 41.Hellerer T., Axäng C., Brackmann C., Hillertz P., Pilon M., Enejder A., Monitoring of lipid storage in Caenorhabditis elegans using coherent anti-Stokes Raman scattering (CARS) microscopy. Proc. Natl. Acad. Sci. U.S.A. 104, 14658–14663 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang M. C., Min W., Freudiger C. W., Ruvkun G., Xie X. S., RNAi screening for fat regulatory genes with SRS microscopy. Nat. Methods 8, 135–138 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang P., Liu B., Zhang D., Belew M. Y., Tissenbaum H. A., Cheng J.-X., Imaging lipid metabolism in live Caenorhabditis elegans using fingerprint vibrations. Angew. Chem. Int. Ed. 53, 11787–11792 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.O’Rourke E. J., Soukas A. A., Carr C. E., Ruvkun G., C. elegans major fats are stored in vesicles distinct from lysosome-related organelles. Cell Metab. 10, 430–435 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/1/9/e1500738/DC1
Vibrational spectroscopic imaging of live C. elegans
Fig. S1. Comparison of photon collection efficiency by a spectrometer to that by a single detector.
Fig. S2. Detection sensitivity of spatial frequency multiplexing SRS microscopy.
Fig. S3. Spectroscopic imaging of a live wild-type C. elegans.