Abstract
Among the four cultivated cotton species, G. hirsutum (allotetraploid) presently holds a primary place in cultivation. Efforts to further improve this primary cotton face the constraints of its narrow genetic base due to repeated selective breeding and hence demands enrichment of diversity in the gene pool. G. arboreum (diploid species) is an invaluable genetic resource with great potential in this direction. Based on the dispersal and domestication in different directions from Indus valley, different races of G. arboreum have evolved, each having certain traits like drought and disease resistance, which the tetraploid cotton lack. Due to lack of systematic, race wise characterization of G. arboreum germplasm, it has not been explored fully. During the present study, 100 polymorphic SSR loci were used to genotype 95 accessions belonging to 6 races of G. arboreum producing 246 polymorphic alleles; mean number of effective alleles was 1.505. AMOVA showed 14 % of molecular variance among population groups, 34 % among individuals and remaining 52 % within individuals. UPGMA dendrogram, based on Nei’s genetic distance, distributed the six populations in two major clusters of 3 populations each; race ‘bengalense’ was found more close to ‘cernuum’ than the others. The clustering of 95 genotypes by UPGMA tree generation as well as PCoA analysis clustered ‘bengalense’ genotypes into one group along with some genotypes of ‘cernuum’, while rest of the genotypes made separate clusters. Outcomes of this research should be helpful in identifying the genotypes for their further utilization in hybridization program to obtain high level of germplasm diversity.
Electronic supplementary material
The online version of this article (doi:10.1007/s12298-015-0326-y) contains supplementary material, which is available to authorized users.
Keywords: Asiatic cotton, Polymorphism information content, Genetic distance, SSR, UPGMA, PCoA
Introduction
Cotton is the world leading natural fiber crop on which the textile industries worldwide are largely based on. The genus Gossypium has 45–50 species, 40–45 are diploid (2n = 2× = 26) while 5 are allotetraploid (2n = 4× = 52). Spinnable fibers are obtained from two allotetraploid (G. hirsutum and G. barbedense) and two diploid (G. herbaceum and G. arboreum) species. Presently, tetraploid cotton (dominantly G. hirsutum) occupies a major fraction (>90 %) of world cotton cultivation because of superior fiber quality and has achieved the status of primary cotton; diploid species being cultivated only in traditional cotton growing areas of India, Pakistan, China, Bangladesh and Iran (Kulkarni et al. 2009). Efforts to further improve the plant and fiber traits of primary cotton face the constraints of narrow genetic base due to continuous selective breeding and selection. Enrichment of gene pool with genetic diversity is strongly needed for future gains in fiber industry (Abdalla et al. 2001). Transfer of allelic variation from diverse cotton germplasm resources to the primary cotton breeding gene pools by intraspecific and interspecific hybridization would be an important step in this direction.
G. arboreum (also known as Asiatic cotton) germplasm collection is an important genetic resource for tetraploid cotton improvement. G. arboreum has certain inherent qualities, which the tetraploids lack, like the ability to withstand drought and salinity (Maqbool et al. 2010; Tahir et al. 2011) and remarkable tolerance to several pests and disease, including bollworms (Dhawan et al. 1991), aphids and leafhoppers (Nibouche et al. 2008), rust, fungal (Wheeler et al. 1999) and viral (Mehetre et al. 2004; Akhtar et al. 2010) diseases. Natural G. arboreum fibers display various colours (e.g. white, off-white and tan) also and some of the accessions produce fibers with high strength (Mehetre et al. 2003). Some efforts have been put for introgressive breeding using G. arboreum as donor species to improve tetraploid cotton, especially for disease resistance and insect tolerance (Ansingkar et al. 2004; Kulkarni 2002), though the achievement was limited. A major problem in such efforts is the poor understanding of G. arboreum germplasm at molecular level.
A huge collection of G. arboreum germplasm is maintained at different centres worldwide (Kulkarni et al. 2009). Domestication of G. arboreum initiated during Indus valley civilization (3300–1300 BCE) (Hutchinson 1954) and from there it spread to different direction worldwide. During this dispersal it became adapted to diverse climate and soil conditions by developing distinct genetic and morphological features, based on which six different races were classified viz. ‘indicum’, ‘burmanicum’, ‘sinense’, ‘soudanense’, ‘bengalense’, ‘cernuum’ (Silow 1944; Hutchinson 1954; Brubaker et al. 1999). Each race has its own characteristics traits like race ‘indicum’ (cultivated in west India and coastal Tanzania) yield long fibers, race ‘cernuum’ (cultivated in North-east India) bear big bolls, higher lint% is observed with race ‘bengalense’ (cultivated in North and central India) while race ‘soudanense’ is well adapted to dry climatic conditions of Egypt and North Africa. ‘Sinense’ and ‘burmanicum’ are annual forms domesticated and cultivated popularly in China and Myanmar respectively with some cultivation in North-eastern regions of India also. Earlier the races were sown year by year by local farmers and diversity was dynamically maintained. During modern agriculture, new varieties have been introduced, often originating from crosses among elite inbred lines. Because of higher yields, the new varieties have largely replaced the old races. Therefore, analysis of genetic variation between and within races of G. arboreum is prerequisite for exploiting this germplasm in modern cotton improvement programs.
During the last two decades, molecular markers have been extensively used for studying genetic diversity as well as genetic relationship among genotypes across species including cotton species, though much focus has been on the tetraploid cotton germplasm (Abdalla et al. 2001; Lu and Myers 2002; Han et al. 2006, Abdurakhmonov et al. 2008, 2009; Azmat and Khan 2010; Noormohammadi et al. 2011; Surgun et al. 2012; Dahab et al. 2013). Efforts have also been made for genetic diversity analysis of selected G. arboreum germplasm using different markers like RAPD (Rana and Bhat 2004; Mahmood et al. 2009; Mandaliya et al. 2010; Deosarkar et al. 2010; Dongre et al. 2011), ISSR (Dongre et al. 2007; Khandagale et al. 2007; Bardak and Bolek 2012) and microsatellites or simple sequence repeats (SSRs) (Guo et al. 2006; Liu et al. 2006; Dongre et al. 2007; Kantartzi et al. 2009; Deosarkar et al. 2010; Dongre et al. 2011; Noormohammadi et al. 2013), though no study explored the polymorphism among the six races of G. arboreum. So, the present work was designed to study genetic diversity among elite genotypes of six different races of G. arboreum using microsatellite markers, since microsatellites markers have edge over other marker system in cultivar fingerprinting and diversity studies,
Materials and methods
Plant materials and DNA extraction
Ninety five cotton genotypes belonging to six races of G. arboreum, as described in Table 1, were selected for the present study. The cotton plants were cultivated in two rows of 6 m length with 30 cm interplant distance in the experimental field of Central Institute of Cotton Research (CICR), regional station, Sirsa, Haryana, India, in a completely randomized design (CRD) with 3 replications. Single plant, having fresh and young leaves, was selected randomly from any of the three replicates of each genotype. Fresh and young leaves of selected plants were subjected to total genomic DNA extraction using CTAB method (Saghai-Maroof et al. 1984). Quality and quantity of extracted DNA was examined by running on 0.8 % agarose gel as well as by UV-Spectrophotometer method.
Table 1.
The genotypes, belonging to six different races of G. arboreum, selected for the present work
| No. | Accession | Population Group & source of collection | No. | Accession | Population group & source of collection |
|---|---|---|---|---|---|
| 1 | CISA-6-187 | Population group 1 (Bengalense race) Collected from C.I.C.R., Regional Station, Sirsa (Haryana) India |
49 | AKA-0106 | Population group 1 |
| 2 | CISA-6-123 | 50 | CINA-369 | ||
| 3 | CISA-6-209 | 51 | CAN-1006 | ||
| 4 | CISA-6-214 | 52 | HD-485 | ||
| 5 | CISA-6-256 | 53 | GAM-150 | ||
| 6 | CISA-6-295 | 54 | JTAPTI-007 | ||
| 7 | CISA-6-350 | 55 | CCA-8 | ||
| 8 | CISA-614 | 56 | LD-694 | ||
| 9 | CISA-6 | 57 | RG-8 | ||
| 10 | CISA-7 | 58 | HD-123 | ||
| 11 | CISA-8 | 59 | PA-255 | ||
| 12 | CISA-9 | 60 | LD-987 | ||
| 13 | CISA-10 | 61 | RG-579 | ||
| 14 | CISA-294 | 62 | LD-919 | ||
| 15 | CISA-64 | 63 | LD-936 | ||
| 16 | CISA-310 | 64 | LD-1010 | ||
| 17 | LD-327 | 65 | RG-595 | ||
| 18 | LD-733 | 66 | C- 1 | Population group 2 (Cernuum race) Collected from Genbank, C.I.C.R. Nagpur (Maharashtra) |
|
| 19 | ARBAS-105 | 67 | C- 2 | ||
| 20 | TKA-9102/03 | 68 | C-3 | ||
| 21 | MDL-2617 | 69 | C-4 | ||
| 22 | GBaV-107 | 70 | C-5 | ||
| 23 | PA-532 | 71 | C-6 | ||
| 24 | PA-686 | 72 | C-7 | ||
| 25 | RG-526 | 73 | C-8 | ||
| 26 | RG-540 | 74 | C-9 | ||
| 27 | RG-541 | 75 | C-10 | ||
| 28 | RG-514 | 76 | Id-1 | Population group 3 (Indicum race) Collected from Genbank, C.I.C.R. Nagpur (Maharashtra) |
|
| 29 | FDK-118 | 77 | Id-2 | ||
| 30 | TKA-9102 | 78 | Id-3 | ||
| 31 | KWP-902 | 79 | Id-4 | ||
| 32 | DLSA-17 | 80 | Id-5 | ||
| 33 | DLSA-1005 | 81 | S-1 | Population group 4 (Soudanese race) Collected from Genbank, C.I.C.R. Nagpur (Maharashtra) |
|
| 34 | DLSA-1006 | 82 | S-2 | ||
| 35 | LD-960 | 83 | S-3 | ||
| 36 | LD-909 | 84 | S-4 | ||
| 37 | FDK-124 | 85 | S-5 | ||
| 38 | PAIG-8/1 | 86 | Sin-1 | Population group 5 (Sinense race) Collected from Genbank, C.I.C.R. Nagpur (Maharashtra) |
|
| 39 | DAS-802 | 87 | Sin-2 | ||
| 40 | CCA-4 | 88 | Sin-3 | ||
| 41 | RAAS-931 | 89 | Sin-4 | ||
| 42 | GBaV-105 | 90 | Sin-5 | ||
| 43 | GBaV-120 | 91 | Bur-1 | Population group 6 (Burmanicum race) Collected from Genbank, C.I.C.R. Nagpur (Maharashtra) |
|
| 44 | ARBHA-0853 | 92 | Bur-2 | ||
| 45 | ARBAS-104 | 93 | Bur-3 | ||
| 46 | RAAS-36 | 94 | Bur-4 | ||
| 47 | RAAS-8 | 95 | Bur-5 | ||
| 48 | GAM-158 |
Microsatellite analysis
One hundred thirty microsatellite primer pairs were obtained from BNL (Brookhaven National Laboratory), NAU (Nanjing Agricultural University) and MUSS (M- Microsatellite, U- Last name of Principal Investigator, SS- Simple Sequences), sources for initial screening. Out of these, only 100 primers produced polymorphic and reproducible band pattern and hence these were selected for present study (Table 2). The sequence information of these primers is available at http://www.cottonmarker.org.
Table 2.
The polymorphic 100 SSR markersused in present study with number of alleles, size range and PIC values
| S. No | Primer name | No. of alleles | Size range (bp) | PIC | S. No | Primer name | No. of alleles | Size range (bp) | PIC |
|---|---|---|---|---|---|---|---|---|---|
| 1 | NAU-1067 | 3 | 160–156 | 0.528 | 51 | MUSS-243 | 2 | 180–176 | 0.375 |
| 2 | NAU-3911 | 2 | 194–190 | 0.040 | 52 | BNL-1395 | 2 | 250–245 | 0.021 |
| 3 | NAU-2083 | 2 | 224–220 | 0.040 | 53 | BNL-1694 | 4 | 289–272 | 0.695 |
| 4 | MUSS-422 | 2 | 265–260 | 0.040 | 54 | BNL-1604 | 2 | 120–115 | 0.021 |
| 5 | BNL-3580 | 4 | 197–190 | 0.517 | 55 | BNL-1531 | 2 | 239–235 | 0.021 |
| 6 | BNL-3888 | 2 | 130–126 | 0.021 | 56 | NAU-2432 | 2 | 207–204 | 0.040 |
| 7 | BNL-3090 | 2 | 240–234 | 0.040 | 57 | NAU-2308 | 3 | 154–146 | 0.567 |
| 8 | NAU-2095 | 2 | 219–215 | 0.040 | 58 | NAU-3590 | 2 | 223–220 | 0.021 |
| 9 | BNL-3424 | 2 | 197–192 | 0.040 | 59 | NAU-3793 | 3 | 262–255 | 0.574 |
| 10 | MUSS-73 | 2 | 208–205 | 0.041 | 60 | BNL-3792 | 2 | 243–240 | 0.021 |
| 11 | MUSS-599 | 2 | 230–227 | 0.021 | 61 | BNL-3257 | 2 | 130–127 | 0.021 |
| 12 | NAU-5383 | 4 | 162–110 | 0.666 | 62 | BNL-1017 | 2 | 205–202 | 0.040 |
| 13 | BNL-3971 | 2 | 200–196 | 0.021 | 63 | NAU-2407 | 2 | 105–102 | 0.021 |
| 14 | BNL-1897 | 2 | 190–186 | 0.040 | 64 | NAU-920 | 2 | 155–150 | 0.021 |
| 15 | BNL-1434 | 3 | 148–137 | 0.554 | 65 | MUSS-189 | 2 | 216–210 | 0.021 |
| 16 | NAU-5499 | 3 | 145–154 | 0.581 | 66 | BNL-686 | 3 | 210–202 | 0.581 |
| 17 | BNL-3259 | 2 | 105–102 | 0.021 | 67 | BNL-1162 | 2 | 134–130 | 0.021 |
| 18 | MUSS-207 | 2 | 224–220 | 0.021 | 68 | BNL-1672 | 2 | 180–177 | 0.021 |
| 19 | MUSS-192 | 2 | 220–217 | 0.040 | 69 | NAU-2322 | 2 | 240–236 | 0.040 |
| 20 | MUSS-172 | 2 | 219–215 | 0.021 | 70 | NAU-3052 | 2 | 190–185 | 0.021 |
| 21 | NAU-1070 | 2 | 250–245 | 0.021 | 71 | NAU-1009 | 2 | 125–122 | 0.021 |
| 22 | BNL-226 | 2 | 244–240 | 0.021 | 72 | NAU-1046 | 2 | 200–197 | 0.021 |
| 23 | NAU-1190 | 2 | 190–185 | 0.021 | 73 | BNL-4053 | 2 | 220–215 | 0.021 |
| 24 | BNL-3441 | 2 | 180–175 | 0.021 | 74 | NAU-3467 | 2 | 224–220 | 0.021 |
| 25 | BNL-2443 | 2 | 196–190 | 0.021 | 75 | MUSS-68 | 2 | 205–203 | 0.021 |
| 26 | NAU-1167 | 2 | 190–186 | 0.021 | 76 | NAU-3454 | 2 | 234–230 | 0.021 |
| 27 | NAU-3083 | 2 | 145–140 | 0.021 | 77 | BNL-2530 | 2 | 180–176 | 0.021 |
| 28 | NAU-2363 | 3 | 192–180 | 0.593 | 78 | BNL-256 | 2 | 230–226 | 0.040 |
| 29 | BNL-4047 | 3 | 183–177 | 0.557 | 79 | BNL-2631 | 2 | 219–215 | 0.021 |
| 30 | BNL-530 | 2 | 120–116 | 0.040 | 80 | BNL-3895 | 2 | 240–236 | 0.372 |
| 31 | NAU-3093 | 2 | 170–167 | 0.021 | 81 | NAU-1182 | 2 | 206–200 | 0.021 |
| 32 | BNL-4049 | 4 | 143–127 | 0.692 | 82 | MUSS-123 | 2 | 200–195 | 0.021 |
| 33 | BNL-2572 | 4 | 207–170 | 0.586 | 83 | BNL-1231 | 2 | 191–188 | 0.021 |
| 34 | NAU-2865 | 2 | 204–200 | 0.040 | 84 | BNL-1066 | 4 | 206–198 | 0.593 |
| 35 | NAU-2000 | 4 | 195–180 | 0.700 | 85 | NAU-1162 | 2 | 235–229 | 0.021 |
| 36 | MUSS-99 | 3 | 320–310 | 0.577 | 86 | BNL-1404 | 2 | 225–219 | 0.021 |
| 37 | BNL-3995 | 2 | 174–170 | 0.021 | 87 | BNL-3147 | 2 | 107–104 | 0.021 |
| 38 | BNL-3992 | 3 | 205–196 | 0.570 | 88 | NAU-3377 | 2 | 237–234 | 0.021 |
| 39 | BNL-542 | 3 | 204–195 | 0.590 | 89 | MUSS-26 | 2 | 200–196 | 0.021 |
| 40 | BNL-3241 | 6 | 230–170 | 0.800 | 90 | NAU-3426 | 4 | 260–220 | 0.700 |
| 41 | NAU-934 | 3 | 152–147 | 0.589 | 91 | NAU-4047 | 2 | 190–186 | 0.021 |
| 42 | BNL-3359 | 2 | 205–202 | 0.021 | 92 | BNL-3261 | 2 | 149–146 | 0.021 |
| 43 | BNL-2569 | 2 | 175–170 | 0.021 | 93 | NAU-1278 | 2 | 215–209 | 0.021 |
| 44 | BNL-1440 | 2 | 201–197 | 0.021 | 94 | BNL-1673 | 2 | 234–229 | 0.021 |
| 45 | NAU-1151 | 4 | 233–220 | 0.659 | 95 | BNL-1679 | 2 | 211–208 | 0.021 |
| 46 | NAU-2580 | 2 | 205–202 | 0.040 | 96 | NAU-2038 | 3 | 210–200 | 0.500 |
| 47 | NAU-3206 | 3 | 247–240 | 0.564 | 97 | NAU-1141 | 2 | 156–150 | 0.021 |
| 48 | NAU-3427 | 3 | 197–190 | 0.579 | 98 | BNL-2652 | 2 | 175–170 | 0.021 |
| 49 | NAU-933 | 3 | 173–165 | 0.554 | 99 | BNL-4029 | 2 | 220–217 | 0.021 |
| 50 | NAU-4030 | 2 | 159–155 | 0.021 | 100 | BNL-1707 | 4 | 180–140 | 0.702 |
The bold names are the SSR primers which exhibited PIC value greater than or equal to 0.5
PCR amplification was performed in a volume of 20 μl containing 2 μl of DNA (50 ng/μl), 0.5 μM of each primer (Sigma-Aldrich), 200 μM of dNTPs (Sigma-Aldrich), 0.5 U Taq polymerase (Sigma-Aldrich) and 1X PCR buffer (Sigma-Aldrich). Thirty five cycles, each consisting of 1 min denaturation at 95 °C, 1 min at annealing temperature (optimized separately for each primer pair, generally Tm-5 °C) and 2 min polymerization at 72 °C, were performed in a thermo cycler (Bio-Rad, USA). The PCR products were separated by electrophoresis in a horizontal gel system at 100 V for 4 h in 4 % metaphor gel and polymorphism was visualized by staining with ethidium bromide. Finally the gel was photographed under Gel Documentation system (Bio-Rad, USA).
Data analysis
The profiles revealed by SSR markers were scored as present (1) or absent (0) for each of the SSR loci. Genetic diversity was calculated at each locus by means of allelic polymorphism information content (PIC) (Anderson et al. 1993), with program CERVUS version 3.0 based on allelic frequencies among all 95 genotypes. PIC values for each locus were calculated as: PICj = 1-∑p2lj, plj is the frequency if the lth allele for locus j and is summed over its L alleles. Markers were classified as informative when PIC ≥0.5.
Several other genetic diversity parameters were determined viz. number of SSR locus (N), number of different allele (Na), effective number of allele (Ne), Shannon’s index (I), observed heterozygosity (Ho) and expected heterozygosity (He). The fixation index (F) which is equal to (Hexp-Ho)/Hexp, was also computed for all the loci and population being studied. This was accompanied by Analysis of Molecular Variance (AMOVA) in order to reveal significant difference between various genotypes and population groups. UPGMA (Unweighted Paired Group using Mean Average) dendrogram of 6 population groups was drawn based on Nei’s genetic distance, modified from Neighbour procedure of PHYLIP ver. 3.5. Similarity matrices were generated among the cultivars studied using ‘Simqual’ subprogram of software NTSYS and used for grouping of the genotypes by UPGMA clustering method. Ordination based on principle coordinate analysis (PCoA) was also done. All computations for determination of genetic parameters, clustering, AMOVA and PCoA analysis was done using softwares- NTSYS ver 2.02, POPGENE ver. 3.2 and GenAlex 6.5.
Results and discussion
Microsatellite diversity
The hundred selected microsatellite primer pairs, when used to amplify genomic DNA of selected 95 genotypes of G. arboreum, yielded a total of 240 alleles (all polymorphic), quite distinct on metaphor gels (Supplementary Fig. 1). The mean number of alleles obtained per locus was 2.4 while the number of alleles per locus varied from 2 to 6. The PIC values ranged from 0.021 to 0.80 (average 0.206) (Table 2). The average PIC obtained during the present study was less to that obtained by Kantartzi et al. (2009) (average PIC 0.42), while analyzing genetic diversity in G. arboreum cultivars using microsatellites, though the range obtained by them was also different (0.00 to 0.68). Liu et al. (2006) also reported high average PIC (0.31), compared to that obtained in present study, and the average PIC value obtained by Lacape et al. (2007) was also higher (average 0.55) than our value. This variation can be attributed to selection of different genotypes and primers for the study.
Of the 100 selected SSRs loci, 27 loci were found highly informative as they yielded PIC value of ≥0.5. As our selection of SSR loci was biased towards di-nucleotides, so it was difficult to correlate the polymorphism with the repeat type of SSR loci. SSR polymorphism has, however, sometimes been correlated with repeat length, and dinucleotide AT-rich repeats have been found more polymorphic than other kinds of repeats by some groups like Cavagnaro et al. (2010) and Kantartzi et al. (2009). Guo et al. (2007) found the polymorphic rate of tetranucleotide and dinucleotide repeat types slightly higher than that of trinucleotide repeat types.
Analysis of genetic diversity over all loci showed the mean number of effective alleles (Ne) = 1.505, mean value of Shannon’s information index (I) = 0.343 and a mean value of 0.203 expected heterozygosity (He) (Table 3). The highest value of Ne (1.595) was observed for population group 1, while highest value of I (0.359) and He (0.209) occurred in population group 5. Highest observed heterozygosity (Ho) value (0.144) was found in population group 6 whereas lowest value for the same parameters occurred in population group 3.
Table 3.
Genetic diversity parameters in six population group of G. arboreum
| Pop | N | Na | Ne | I | Ho | He | F | |
|---|---|---|---|---|---|---|---|---|
| Pop 1 | Mean | 65.000 | 1.860 | 1.595 | 0.351 | 0.127 | 0.201 | 0.507 |
| SE | 0.000 | 0.122 | 0.099 | 0.052 | 0.032 | 0.030 | 0.067 | |
| Pop 2 | Mean | 10.000 | 1.830 | 1.544 | 0.355 | 0.134 | 0.204 | 0.564 |
| SE | 0.000 | 0.109 | 0.087 | 0.048 | 0.033 | 0.027 | 0.065 | |
| Pop 3 | Mean | 5.000 | 1.650 | 1.447 | 0.311 | 0.120 | 0.188 | 0.509 |
| SE | 0.000 | 0.097 | 0.075 | 0.043 | 0.032 | 0.025 | 0.071 | |
| Pop 4 | Mean | 5.000 | 1.710 | 1.470 | 0.334 | 0.126 | 0.203 | 0.510 |
| SE | 0.000 | 0.101 | 0.072 | 0.044 | 0.032 | 0.026 | 0.072 | |
| Pop 5 | Mean | 5.000 | 1.770 | 1.499 | 0.359 | 0.136 | 0.215 | 0.527 |
| SE | 0.000 | 0.101 | 0.076 | 0.044 | 0.034 | 0.025 | 0.073 | |
| Pop 6 | Mean | 5.000 | 1.720 | 1.475 | 0.344 | 0.144 | 0.209 | 0.469 |
| SE | 0.000 | 0.096 | 0.070 | 0.043 | 0.035 | 0.025 | 0.079 | |
| Total | Mean | 15.822 | 1.757 | 1.505 | 0.343 | 0.131 | 0.203 | 0.515 |
| SE | 0.000 | 0.043 | 0.033 | 0.019 | 0.013 | 0.011 | 0.029 |
Na Number of different allele, Ne Effective number of allele, I Shannon’s index, Ho Observed heterozygosity, He Expected heterozygosity, F Fixation index
AMOVA analysis (after 999 numbers of permutations) was performed among populations, within population group and within individuals. The analysis indicated that 14 % of molecular variance is due to 6 population groups, 34 % is due to genetic variations among accessions in each population group and remaining 52 % is observed within individuals (Table 4). Present analysis showed significant difference among population groups, among individuals of a group and within individuals (p = 0.001). In previous similar study by Wang et al. (2011), 92 % of total variation was found confined to within population variation whereas only 8 % of total variation was due to among population variation, as analyzed by AMOVA. Noormohammadi et al. (2013), in diploid cotton genotypes after analysis by AMOVA, found 2 % of total variation due to population groups and 98 % due to genetic variations among accessions in each population group; such a low polymorphism among population groups could be due to inclusion of inter-specific hybrids, second backcross progenies and F5 plants of the same cross.
Table 4.
Parameters obtained by Analysis of molecular variance (AMOVA), during present study
| Source | df | SS | MS | Est. Var. | % Molecular variance |
|---|---|---|---|---|---|
| Among Population | 5 | 249.02 | 49.80 | 1.80 | 14 % |
| Among individuals (in each population group) | 89 | 1319.95 | 14.83 | 4.21 | 34 % |
| Within individuals | 95 | 609 | 6.41 | 6.41 | 52 % |
| Total | 189 | 2177.97 | 12.42 | 100 % |
df Degree of freedom, SS Sum of squares, MS Mean Square
UPGMA dendrogram based on Nei’s genetic distance distributed 6 population groups into 2 main clusters. Population group 1 (race ‘bengalense’), 2 (race “cernuum’) and 6 (race ‘burmanicum’) formed the first main cluster whereas population group 3 (race ‘indicum’), 4 (race ‘soudanense’) and 5 (race ‘sinense’) formed the second main cluster (Fig. 1). The genetic distance coefficient among 6 population groups ranged from 0.046–0.094 (Table 5). Evolutionary studies indicate race ‘indicum’ as most primitive perennial form in western India, dispersal of which to various regions evolved other races like ‘burmanicum’, ‘soudanense’, ‘sinense’ and ‘bengalense’. In the present study, indicum showed maximum genetic similarity to race sinense (0.9508) followed by soudanense (0.9273), bangalense (0.9165) and burmanicum (0.9142). ‘Cernuum’ and ‘indicum’ were found most distant (0.094). Evolutionary studies indicate ‘Cernuum’ to have evolved independently in the Assam hills of North-East India and Chittagang hills of Bangladesh (Kulkarni et al. 2009). However, during the present investigation, cernuum did not appear as independent group but exhibited remarkable similarities with bengalense and burmanicum, together forming one main cluster in the dendrogram while the rest three formed another main cluster (Fig. 1). Since, the evolution of these races is not very primitive, the loci used during present investigation may not be polymorphic enough for accurate grouping.
Fig. 1.
A phylogenetic UPGMA tree of six G. arboreum populations, based on Nei’s genetic distance and generated by POPGENE ver 3.2 software
Table 5.
Nei’s genetic distance (below diagonal) and Nei’s genetic identity (above diagonal), among six population groups
| Pop1 | Pop2 | Pop3 | Pop4 | Pop5 | Pop6 | |
|---|---|---|---|---|---|---|
| Pop1 | 0.9542 | 0.9165 | 0.9383 | 0.9265 | 0.9243 | |
| Pop 2 | 0.0469 | 0.9100 | 0.9273 | 0.9392 | 0.9421 | |
| Pop 3 | 0.0871 | 0.0943 | 0.9289 | 0.9508 | 0.9142 | |
| Pop 4 | 0.0636 | 0.0755 | 0.0737 | 0.9389 | 0.9183 | |
| Pop 5 | 0.0763 | 0.0628 | 0.0504 | 0.0631 | 0.9380 | |
| Pop 6 | 0.0787 | 0.0596 | 0.0897 | 0.0853 | 0.0640 |
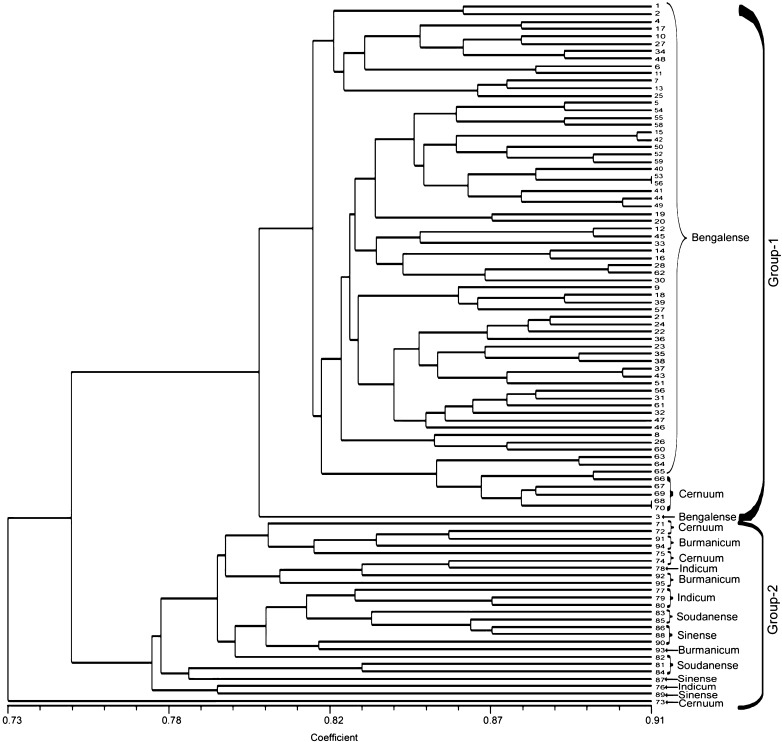
UPGMA tree based on similarity matrix of the 95 G. arboreum accessions, using 100 times bootstrapping, depicted cophenetic correlation value of r = 0.82. The cluster tree analysis distributed the genotypes into two major groups (Fig. 2). Group 1 consists of all the accessions belonging to race ‘bengalense’ and 5 accessions of race ‘cernuum’. Group 2 consists of other 4 races (5 genotypes each) and 5 genotypes of race ‘cernuum’ which are distributed randomly, not indicating a clear differentiation of subgroups. The result was similar to dendrogram drawn on the basis of Nei’s genetic distance. The similarity coefficient among 95 accessions ranged from 0.73–0.91. The maximum similarity of 0.91 has been observed between genotype 53 and 56 (both belong to race bengalense) and genotype 68 and 70 (both of race cernuum), followed by similarity coefficient of 0.905 between genotype 15 and 42 (race bengalense). The two major groups shared similarity coefficient of 0.75.
Fig. 2.
A UPGMA tree of 95 genotypes of G. arboreum generated by NTSYSpc2.02 software
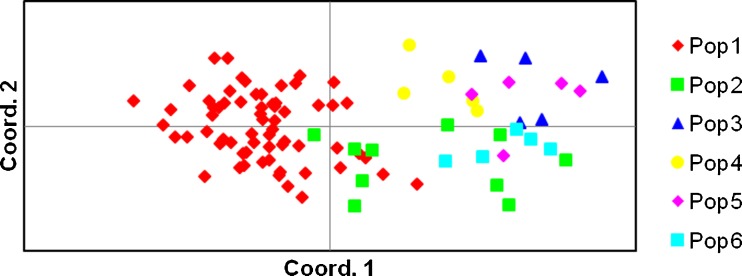
Principal coordinate analysis (PCoA) is a technique which highlights the similarities and differences in the given data by reducing the number of dimensions without much loss of information. The PCoA plot of cotton genotypes after 999 reiterations (Fig. 3) supported the grouping obtained by clustering by UPGMA methods. The PCoA plot exhibited one major and distinct group constituting all the genotypes of bengalense along with five accessions of cernuum, while a second diffused group of rest of genotypes of all the races (Fig. 3).
Fig. 3.
Clustering of 95 genotypes of G. arboreum obtained by ordination based on Principal Coordinate Analysis (PCoA)
A narrow genetic base has been reported in Gossypium species (Iqbal et al. 1997; Abdalla et al. 2001). Plant breeders desire to use Gossypium arboreum as an invaluable genetic resource for improving both diploid and tetraploid cotton production. No study till date has been reported for genetic diversity and population structure characterization of all six races of G. arboreum. The comprehensive molecular characterization of selected cotton germplasm collections, during the present study gives insights regarding the level and distribution of genetic diversity in existing resources and provides insights into genetic subdivisions within each race.
Electronic supplementary material
(DOC 189 kb)
Acknowledgments
Financial support was provided by the University Grants Commission (UGC), Ministry of Human Resource Development, Government of India, New Delhi, India in the form of Major Research Project to Priyanka Siwach (Principal Investigator) and Surender Kumar Verma (Co-Principal Investigator). The authors are grateful to the Director, CICR, Sirsa and Director CICR, Nagpur for providing the germplasm. We also acknowledge the infrastructural facilities provided by Chaudhary Devi Lal University, Sirsa, Haryana, India for all the lab work. Acknowledgement is also extended to CICR, Sirsa for allowing the use of experimental field facility and polyhouse facility.
Compliance with Ethical Standards
Conflict of Interest
The authors declare that they have no conflict of interest.
References
- Abdalla AM, Reddy OUK, El-Zik KM, Pepper AE. Genetic diversity and relationships of diploid and tetraploid cottons revealed using AFLP. Theor Appl Genet. 2001;102:222–229. doi: 10.1007/s001220051639. [DOI] [Google Scholar]
- Abdurakhmonov IY, Kohel RJ, Yu JZ, Pepper AE, Abdullaev AA, Kushanov FN, Salakhutdinov IB, Buriev ZT, Saha S, Scheffler BE, Jenkins JN, Abdukarimov A. Molecular diversity and association mapping of fiber quality traits in exotic G. hirsutum L. Germplasm. Genomics. 2008;92:478–487. doi: 10.1016/j.ygeno.2008.07.013. [DOI] [PubMed] [Google Scholar]
- Abdurakhmonov IY, Saha S, Jenkins JN, Buriev ZT, Shermatov SE, Scheffler BE, Pepper AE, Yu JZ, Kohel RJ, Abdukarimov A. Linkage disequilibrium based association mapping of fiber quality traits in G. hirsutum l. variety germplasm. Genetica. 2009;136:401–417. doi: 10.1007/s10709-008-9337-8. [DOI] [PubMed] [Google Scholar]
- Akhtar KP, Haidar S, Khan MKR, Ahmad M, Sarwar N. Evaluation of Gossypium species for resistance to cotton leaf curl burewela virus. Ann Appl Biol. 2010;157:135–147. doi: 10.1111/j.1744-7348.2010.00416.x. [DOI] [Google Scholar]
- Anderson JA, Churchill GA, Autrique JE, Tanksley SD, Sorells ME. Optimising parental selection for genetic linkage maps. Genome. 1993;36:181–186. doi: 10.1139/g93-024. [DOI] [PubMed] [Google Scholar]
- Ansingkar AS, Kadke PP, Borikar ST, Bhosle SS 2004 Altering G. hirsutum cotton at cellular level to impart multiple sucking pests resistance through interspecific hybridisation. In: Khadi B M, Kategeri I S, Patil S S, Vamadevaiah H M, Patil B R and Manjula S M (Eds), Proceedings of international symposium on “strategies for sustainable cotton production-a global vision”1, crop improvement, 23–25 November 2004, University of Agricultural Sciences, Dharwad, pp. 101–103.
- Azmat MA, Khan AA. Assessment of genetic diversity among the varieties of Gossypium arboreum and Gossypium hirsutum through random amplification of polymorphic DNA (RAPD) and simple sequence repeat (SSR) markers. Pak J Bot. 2010;42:3173–3181. [Google Scholar]
- Bardak A, Bolek Y. Genetic diversity of diploid and tetraploid cottons by SSR and ISSR markers. Turk J Field Crops. 2012;17:139–144. [Google Scholar]
- Brubaker CL, Bourland FM, Wendel JF. The origin and domestication of cotton. In: Smith C W, Cothren J T, editors. Cotton: origin, history, technology and production. New York: Wiley; 1999. pp. 3–31. [Google Scholar]
- Cavagnaro PF, Senalik DA, Yang L, Simon PW, Harkins TT, Kodira CD. Genome-wide characterization of simple sequence repeats in cucumber (Cucumis sativus L.) BMC Genomics. 2010;11(1):569. doi: 10.1186/1471-2164-11-569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahab AA, Saeed M, Mohamed BB, Ashraf MA, Puspito AN, Bajwa KS, Shahid AA, Husnain T. Genetic diversity assessment of cotton (Gossypium hirsutum L.) genotypes from Pakistan using simple sequence repeat markers. Aust J Crop Sci. 2013;7(2):261–267. [Google Scholar]
- Deosarkar VD, Gupta DB, Gaikwad DB. Genetic diversity studies in intra-specific desi cotton (G. arboreum) through DNA marker. J Cotton Res Dev. 2010;24:133–137. [Google Scholar]
- Dhawan AK, Simwat GS, Sidhu AS. Field reaction of some varieties of Asiatic cotton (Gossypium arboreum L.) to sucking and boll worm pests. J Res, Punjab Agricultural University, Ludhiana, Punjab, India. 1991;28:57–62. [Google Scholar]
- Dongre AB, Bhandarkar M, Banerjee S. Genetic diversity in tetraploid and diploid cotton (Gossypium spp.) using ISSR and microsatellite DNA markers. Indian J Biotechnol. 2007;6:349–353. [Google Scholar]
- Dongre AB, Ramteke DM, Bhandarkar MR, Raut MP, Meshram KJ. Genetic diversity analysis of Gossypium arboreum (diploid cotton) cultivars revealed by PCR based molecular markers. J Cotton Res Dev. 2011;25:1–8. [Google Scholar]
- Guo WZ, Zhou BL, Yang LM, Wang W, Zhang TZ. Genetic diversity of landraces in Gossypium arboreum L. race sinense assessed with simple sequence repeat markers. J Integr Plant Biol. 2006;48:1008–1017. doi: 10.1111/j.1744-7909.2006.00316.x. [DOI] [Google Scholar]
- Guo W, Cai C, Wang C, Han Z, Song X, Wang K, Niu X, Wang C, Lu K, Shi B, Zhang T. A microsatellite-based, gene-rich linkage map reveals genome structure, function and evolution in Gossypium. Genetics. 2007;176:527–541. doi: 10.1534/genetics.107.070375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Z, Wang C, Song X, Guo W, Gou J, Li C, Chen X, Zhang T. Characteristics development and mapping of Gossypium hirsutum derived EST-SSRs in allotetraploid cotton. Theor Appl Genet. 2006;112:430–439. doi: 10.1007/s00122-005-0142-9. [DOI] [PubMed] [Google Scholar]
- Hutchinson J B. New evidence on the origin of the old world cotton. Heredity. 1954;8:225–241. doi: 10.1038/hdy.1954.20. [DOI] [Google Scholar]
- Iqbal MJ, Aziz N, Saeed NA, Zafar Y, Malik KA. Genetic diversity evaluation of some elite cotton varieties by RAPD- analysis. Theor Appl Genet. 1997;94:139–144. doi: 10.1007/s001220050392. [DOI] [PubMed] [Google Scholar]
- Kantartzi SK, Ulloa M, Sacks E, Stewart JM. Assessing genetic diversity in Gossypium arboreum L. cultivars using genomic and EST-derived microsatellites. Genetica. 2009;136:141–147. doi: 10.1007/s10709-008-9327-x. [DOI] [PubMed] [Google Scholar]
- Khandagale GB, Dongre AB, Kalpande HV, Salunkhe SN 2007 Molecular evaluation of elite cotton cultivars using DNA markers. WCRC-4, Cotton: Nature’s High-Tech Fiber.
- Kulkarni VN 2002 Hirsutisation of G. arboreum cotton and genetic emendation of G. hirsutum for sucking pest resistance. Ph. D thesis submitted to University of Agricultural Sciences, Dharwad, India.
- Kulkarni VN, Khadi BM, Manjula S, Lalitadas M, Deshapande A, Narayanan SS 2009 The worldwide gene pools of Gossypium arboreum L. and G. herbaceum L. and their improvement, In: Paterson a H (ed.) Genetics and genomics of cotton, plant genetics and genomics: crops and models 3, pp 69–100.
- Lacape JM, Dessauw D, Rajab M, Noyer JL, Hau B. Microsatellite diversity in tetraploid Gossypium germplasm: assembling a highly informative genotyping set of cotton SSRs. Mol Breed. 2007;19(1):45–58. doi: 10.1007/s11032-006-9042-1. [DOI] [Google Scholar]
- Liu D, Guo X, Lin Z, Nie Y, Zhang X. Genetic diversity of Asian cotton (Gossypium arboreum L.) in China evaluated by microsatellite analysis. Genet Resour Crop Evol. 2006;53:1145–1152. doi: 10.1007/s10722-005-1304-y. [DOI] [Google Scholar]
- Lu J, Myers O. Genetic relationships and discrimination of ten influential upland cotton varieties using RAPD markers. Theor Appl Genet. 2002;105:325–331. doi: 10.1007/s00122-002-0947-8. [DOI] [PubMed] [Google Scholar]
- Mahmood Z, Raheel F, Dasti AA, Shahzadi S, Athar M, Qayyum M (2009) Genetic diversity analysis of the species of Gossypium by using RAPD markers. Afr J Biotechnol 8:3691–3697
- Mandaliya VB, Pandya RV, Thaker VS. Genetic diversity analysis of cotton (Gossypium) hybrids using RAPD markers. J Cotton Res Dev. 2010;24(2):127–132. [Google Scholar]
- Maqbool A, Abbas W, Rao A Q, Irfan M, Zahur M, et al. Gossypium arboreum GHSP26 enhances drought tolerance in Gossypium hirsutum. Biotechnol Prog. 2010;26:21–25. doi: 10.1002/btpr.306. [DOI] [PubMed] [Google Scholar]
- Mehetre SS, Aher AR, Gawande VL, Patil VR, Mokate AS. Induced polyploidy in gossypium: a tool to overcome interspecific compatibility of cultivated tetraploid and diploid cottons. Curr Sci. 2003;84:1510–1512. [Google Scholar]
- Mehetre SS, Shinde GC, Pardeshi SU 2004 Status and strategies of host plant resistance for biotic stress in cotton. In: Khadi B M, Kategeri I S, Patil S S, Vamadevaiah H M, Patil B R, Manjula S M (Eds) Proceedings of international symposium on “strategies for sustainable cotton production-a global vision”1, crop improvement, 23–25 November 2004, University of Agricultural Sciences, Dharwad, pp.31–47.
- Nibouche S, Brevault T, Klassou C, Dessauw D, Hau B. Assessment of the resistance of cotton germplasm (Gossypium spp.) to aphids (Homoptera, Aphididae) and leafhoppers (Homoptera : Cicadellidae, Typhlocybinae): methodology and genetic variability. Plant Breed. 2008;127:376–382. doi: 10.1111/j.1439-0523.2008.01499.x. [DOI] [Google Scholar]
- Noormohammadi Z, Shojaei-Jesvaghani F, Sheidai M, Farahani F, Alishah O. Inter simple sequence repeat (ISSR) and random amplified polymorphic DNA (RAPD) analyses of genetic diversity in Mehr cotton cultivar and its crossing progenies. Afr J Biotechnol. 2011;10:11839–11847. [Google Scholar]
- Noormohammadi Z, Rahnama A, Sheidai M. EST-SSR and SSR analyses of genetic diversity in diploid cotton genotypes from Iran. Nucleus. 2013;56(3):171–178. doi: 10.1007/s13237-013-0094-4. [DOI] [Google Scholar]
- Rana MK, Bhat KV. Analysis of molecular variance in cotton (Gossypium spp.) using RAPD markers. Indian J Genet. 2004;64:85–86. [Google Scholar]
- Saghai-Maroof MA, Soliman KM, Jorgensen RA, Allard RW. Ribosomal DNA spacer-length polymorphisms in barley: mendelian inheritance, chromosomal location and population dynamics. Proc Natl Acad Sci. 1984;81:8014–8018. doi: 10.1073/pnas.81.24.8014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silow RA. The genetics of species development in old world cottons. J Genet. 1944;46:62–77. doi: 10.1007/BF02986694. [DOI] [Google Scholar]
- Surgun Y, Col B, Burun B. Genetic diversity and identification of some Turkish cotton genotypes (Gossypium hirsutum L.) by RAPD-PCR analysis. Turk J Biol. 2012;36:143–150. [Google Scholar]
- Tahir MS, Khan NUI, Sajid UR. Development of an interspecific hybrid (triploid) by crossing Gossypium hirsutum and G. arboreum. Cytologia. 2011;76:193–199. doi: 10.1508/cytologia.76.193. [DOI] [Google Scholar]
- Wang XQ, Feng CH, Lin ZX, Zhang XL. Genetic diversity of sea-island cotton (Gossypium barbadense) revealed by mapped SSRs. Genet Mol Res. 2011;10(4):3620–3631. doi: 10.4238/2011.December.8.5. [DOI] [PubMed] [Google Scholar]
- Wheeler TA, Gannaway JR, Keating K. Identification of resistance to Thielaviopsis basicola in diploid cotton. Plant Dis. 1999;83:831–833. doi: 10.1094/PDIS.1999.83.9.831. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOC 189 kb)