Abstract
Haplo-insufficiency of the bHLH (basic helix-loop-helix) transcription factor single-minded 1 (SIM1) causes severe obesity in mice and humans. We hypothesized that common genetic variations in/near SIM1 could exert more subtle effects on its function and associate with human adiposity. First, SIM1 coding regions were sequenced in severely obese subjects, and two common nonsynonymous single-nucleotide polymorphisms (nsSNPs) in complete linkage disequilibrium (LD) were identified: Pro352Thr (rs3734354) and Ala371Val (rs3734355). We next carried out a SNP association study of five adiposity traits (BMI, % body fat, abdominal visceral and subcutaneous fat, and leptin concentrations) in 1,699 whites and 1,173 blacks. TagSNPs covering SIM1 and nearby conserved regions, and the only common nsSNP in SIM1's binding partner aryl-hydrocarbon receptor nuclear translocator 2 (ARNT2) (Gly679Ser/rs4072568), were investigated. The effects of rs3734355/4 on SIM1 activity were tested using an in vitro reporter assay. We replicated previous observations that homozygosity for the 371Val allele was associated with higher BMI in white males (P = 0.003). Together with previous findings in white males (combined n = 3,479), BMI was increased by 1.10 kg/m2 in 371Val homozygotes (95% confidence interval (CI): 0.25–1.95 kg/m2, P = 0.01). In vitro, the 352Thr-371Val haplotype impaired SIM1 transcriptional activity by 22% (P < 0.0001). TagSNP analysis of SIM1 revealed two SNPs in the 3′ region (rs9390322 and rs7746743) and another in intron 5 (rs3734353) to be significantly associated with various adiposity measures in ethnicity- and sex-specific manners after multiple testing correction. In white males, rs4072568 in ARNT2 was also associated with BMI (P = 9 × 10−4) and % body fat (P = 0.001). Our findings implicate heritable defects of the SIM1-ARNT2 axis in the predisposition to human obesity.
INTRODUCTION
Single-minded 1 (SIM1), a bHLH/PAS (basic helix-loop-helix/Per-Arnt-Sim) transcription factor, is essential for the terminal differentiation of second order hypothalamic paraventricular nucleus (PVN) neurons involved in the central regulation of food intake and energy homeostasis (1,2). SIM1 heterodimerizes with another bHLH/PAS factor, aryl-hydrocarbon receptor nuclear translocator 2 (ARNT2) to activate transcription (3). While Sim1 homozygous null mice die shortly after birth, heterozygotes are viable and develop hyperphagia and obesity (4,5). In humans, rare cases of severe, early onset obesity have been linked to haplo-insufficiency at the SIM1 locus (6–9). Given the extreme obesity phenotype observed when SIM1 function is reduced by half, it was of interest to determine the phenotype of individuals with common mutations in SIM1 that may exert more subtle effects on its function.
Homozygosity for a common SIM1 haplotype encompassing two linked nonsynonymous single-nucleotide polymorphisms (nsSNPs) within exon 9, Pro352Thr (rs3734354) and Ala371Val (rs3734355), has been previously associated with a higher BMI in white men (10). However, this association was not replicated in either of two subsequent studies of SIM1, one of 6,194 full- and mixed-heritage Pima Indians (11) and another of 1,275 obese children/adults and 1,395 lean controls from France (12). In Pima Indians, the most significant results for association between SIM1 SNPs and BMI were obtained within a block of noncoding SNPs extending from the 5′ region of SIM1 to intron 8 (11). Differences in mean BMI between homozygotes for the major allele and homozygotes for the minor allele were ~2.2 kg/m2. Two SNPs displaying the strongest associations with BMI in Pima Indians (rs3734353 and rs3213541) were not associated with BMI in French Caucasians, suggesting the presence of genetic heterogeneity. Further investigation of SIM1 in French Caucasians (12) did not yield any significant evidence for association with obesity after correction for multiple hypothesis testing. Moreover, neither (11) nor (12) reported sex-specific associations between SIM1 SNPs and adiposity, and neither study assumed a recessive mode of inheritance.
Here, we provide evidence to support the initial association between the Pro352Thr-Ala371Val haplotype and adiposity in white males (10), and demonstrate its biological relevance by showing that it impairs SIM1 activity in vitro. Moreover, by analyzing tagSNPs covering SIM1 and its flanking regions, we establish that the 352Thr-371Val haplotype, as well as another SNP (rs9390332, in the 3′ noncoding region) in moderate linkage disequilibrium (LD) with these two variants, provides the strongest association with adiposity in white males. Other SNPs in/near SIM1 were also associated with measures of adiposity in black subjects. Finally, we also find an association between the only common nsSNP in ARNT2 (Gly679Ser/rs4072568) and adiposity in white males, but there was no evidence for an interaction between the nsSNPs in SIM1 and ARNT2. Together, these results support the hypothesis that dysregulation of the SIM1-ARNT2 axis contributes to human obesity.
METHODS AND PROCEDURES
Subjects for SIM1 variant identification
Identification of nonsynonymous SIM1 variants was initially performed in severely obese subjects from an ongoing UCSF (University of California, San Francisco) study (13). The UCSF Committee on Human Research approved the protocols, and informed written consent was obtained from all participants.
Subjects and phenotype measurement in the Health, Aging, and Body Composition (Health ABC) Study
The Health ABC study is a population-based prospective study of 3,075 men and women (48.5% male; 41.7% black) aged 70–79 years, from Pittsburgh, PA and Memphis, TN. All participants were physically active at the time of entry into the study and provided informed consent. The study protocol was approved by the institutional review boards at the University of Pittsburgh (Pittsburgh, PA) and the University of Tennessee (Memphis, TN). The present study used data obtained from the baseline examination, during 1997–1998. Measurement of adiposity traits in these subjects has been described previously (14,15). Measures of global adiposity included BMI, the percentage of total body fat (% body fat, assessed by dual-emission X-ray absorptiometry) and fasting leptin (measured using the Sensitive Human Leptin RIA Kit from Linco Research, St Charles, MO). Regional adiposity, including abdominal visceral fat area (visceral fat, in cm2) and abdominal subcutaneous fat area (subcutaneous fat, in cm2), was assessed using computed tomography.
Sequencing of the SIM1 coding regions and genotyping of rs3734355
Genomic DNA was extracted from buffy coats using a Puregene DNA Purification Kit (Gentra Systems, Minneapolis, MN). SIM1 exons and splice junctions were amplified using the primers listed in Supplementary Table S1 online. PCR products were bidirectionally sequenced on an ABI 3700 sequencer (Applied Biosystems, Foster City, CA). Sequences were analyzed using Sequencher software (Gene Codes, Ann Arbor, MI), using the SIM1 GenBank sequence (accession number NM_005068.2) as a reference. The Ala371Val SNP (rs3734355) was genotyped using fluorescently labeled allele-specific primer extension assayed by fluorescence polarization template-directed dye incorporation (16). Quality control criteria are described in Supplementary Methods and Procedures online.
Selection and genotyping of tagSNPs and rs3734354 in SIM1 and rs4072568 in ARNT2
SNPs were selected using HapMap data Phase II (release 20) and genotyped using the Golden Gate Assay (Illumina, San Diego, CA). By considering conservation between human and mouse genomes at ≥75% identity for sequences of ≥100 bp using VISTA (17), the program Tagger (18) was used to select tagSNPs (r2 ≥ 0.8, minor allele frequency (MAF) ≥ 0.05) in a region spanning 45 kb upstream of the 5′ end to 60 kb downstream of the 3′ end of SIM1. SNP rs3734354, which is in complete LD with rs3734355, was included in all tagSNP selections and was used to validate the accuracy of genotypes for rs3734355. The tagSNPs selected in the CEU (European) population were forced to be included while picking tagSNPs in the YRI (Yoruban) population, and all (87) of these tagSNPs were genotyped in 2,982 subjects from the Health ABC cohort. Of the 87 genotyped SNPs, 77 SNPs in whites and 68 SNPs in blacks passed quality control (QC) criteria (see Supplementary Methods and Procedures online). Ethnicity-specific tagSNPs were then selected (r2 ≥ 0.8) among genotyped SNPs passing QC criteria using the genotype data from the Health ABC Study population. For SIM1, 38 SNPs (37 tagSNPs and rs3734355) in whites and 55 SNPs (54 tagSNPs and rs3734355) in blacks were tested for association. The only common nsSNP in ARNT2 (rs4072568), based on the dbSNP (Build ID: 126 and 130) and HapMap (release 20) databases, was also genotyped.
Statistical analysis
In the severely obese subjects from the UCSF study, differences in BMI values by genotype were examined using the Wilcoxon rank sum test. For the Health ABC Study participants, genetic association analysis of adiposity traits was performed using linear regression analysis. For analyses of BMI, % body fat, and leptin, the effects of age, sex, recruitment site, diabetes status, weekly levels of physical activity, smoking and drinking habits, and education levels were adjusted for in the model. Percentage body fat was included as a covariate in the analysis of leptin, and baseline height and weight were included as covariates for analyses of abdominal subcutaneous and visceral fat. Leptin, visceral fat, and subcutaneous fat were transformed by taking the square-root to approximate normal distributions. For case–control analysis of obesity within Health ABC participants, BMI at the baseline examination was used to define obese cases as BMI ≥ 30 kg/m2 and nonobese controls as BMI < 30 kg/m2. The effects of the same covariates used in linear regression of BMI were adjusted for in logistic regression models.
To determine the likely mode of inheritance, mean values of adiposity traits stratified by genotype were examined. All SNPs were modeled with an additive mode of inheritance, except for rs3734355 and rs3734354, which were modeled as recessive. To minimize the effects of population stratification, all analyses were stratified by ethnicity. Analyses were also stratified by sex in order to replicate previously reported results (10). To correct for multiple hypothesis testing, empirical P values were obtained by permutation testing using the min-P procedure with 100,000 replicates (19).
Analysis of the overall effect of rs3734355 on BMI was performed using inverse variance weighting for pooling of effect estimates from two population-based cohorts (10) and the Health ABC population-based cohort. Heterogeneity between studies was assessed using the I2 statistic and the P value from the Q-test. The overall effect size and P value was determined using a fixed effect model.
Values from the in vitro transcription assay were removed if the coefficient of variation between two reads was > 0.05. The ratio of the normalized fluorescence of the 352Thr-371Val double mutant SIM1 and its matched wild-type control was determined for each experiment, and the significance was assessed using a one-sample two-sided t test. All statistical analyses were performed using R software (http://www.r-project.org/).
In vitro transcription assay
The SIM1 coding sequence was amplified from human fetal kidney cDNA and cloned into pCDNA3.1/V5-His-TOPO (Invitrogen, Carlsbad, CA). Mutations were introduced into the vector using QuickChange (Stratagene, La Jolla, CA). Human ARNT2 was cloned from cDNA from HEK293 cells. The toll 4 CME site, consisting of three repeats of the sequence GGAGCATGCGGTACCAGAT(CTAGAAAT TTGTACGTGCCACAGA)3GGATCCGTG (20) was cloned into the KpnI/BglII sites of pLuc-MCS (Invitrogen), which contains a minimal basal promoter followed by the coding sequence for luciferase.
HEK 293 cells were cultured in α-minimum essential medium supplemented with 10% calf serum (Hyclone, Logan, UT), 0.002 mmol/l l-glutamine, nonessential amino acids, and penicillin/streptomycin in an incubator maintained at 37 °C and 5% CO2. Six hours before transfection, cells were seeded in 24-well plates at a density of 100,000 cells/well. Using Effectene reagent (Qiagen, Valencia, CA), each well was then transfected with pLuc-MCS or the pLuc-MCS containing SIM1 response element (120 ng), 80 ng of SIM1/ARNT2, as well as the plasmid pRL-RSV (Promega, Madison, WI), encoding Renilla luciferase, to control for transfection efficiency. Cells were lysed 48 h later, and firefly and Renilla luciferase activities were measured in lysates using the Dual Luciferase Reporter Assay System (Promega), according to the manufacturer's standard protocol.
RESULTS
All 11 exons of the human SIM1 were sequenced in 445 severely obese subjects (74% whites, mean BMI ± s.e.m: 48.0 ± 0.5 kg/m 2)) (13). Five nsSNPs were identified uniquely in five different patients (Thr361Ile, Ser622Pro, His644Arg, Arg665His, Asp707His) as well as two common nsSNPs in complete LD, Pro352Thr and Ala371Val. The rare Thr361Ile and Arg665His variants have been reported previously (11). The common Ala371Val (rs3734355) and Pro352Thr (rs3734354) variants are located in or near the SIM1 nuclear localization signal, respectively (21). Obese males homozygous for the 371Val allele had a higher BMI than obese males of the other two genotypes (P = 0.036) (Table 1).
Table 1.
BMI and genotype frequencies for single-minded 1 (SIM1) Ala371Val (rs3734355) in severely obese cases and lean controls
| Genotype |
|||||
|---|---|---|---|---|---|
| Ala/Ala |
Ala/Val |
Val/Val |
|||
| BMI (kg/m2) (n) | BMI (kg/m2) (n) | BMI (kg/m2) (n) | P | Freq (Val) | |
| All casesa | 48.9 ± 0.6 (297) | 47.5 ± 0.8 (98) | 51.6 ± 3.0 (14) | NS | 0.15 |
| Male cases | 48.5 ± 1.1 (83) | 44.5 ± 1.1 (26) | 54.2 ± 7.4 (2) | 0.036 | 0.14 |
| Female cases | 49.0 ± 0.7 (214) | 48.6 ± 0.9 (72) | 51.2 ± 3.4 (12) | NS | 0.16 |
| All controlsb | 22.9 ± 0.1 (379) | 23.0 ± 0.1 (166) | 22.6 ± 0.4 (19) | NS | 0.18 |
| Male controls | 23.3 ± 0.1 (126) | 23.3 ± 0.1 (47) | 23.4 ± 0.4 (8) | NS | 0.17 |
| Female controls | 22.7 ± 0.1 (253) | 22.8 ± 0.1 (119) | 22.1 ± 0.5 (11) | NS | 0.18 |
Cases had a BMI ±40 kg/m2 and were obtained from an ongoing study at UCSF (13). BMI is shown as mean ± s.e.m.
Sex- and age-matched lean controls (BMI ≤25 kg/m2) were obtained from the same study.
Considering previous findings (10), we investigated the potential relationship between rs3734355 and adiposity in a large population-based cohort, the Health ABC Study. Among whites, those homozygous for the minor (Val) allele at rs3734355 had greater adiposity than individuals with the other two genotypes (Table 2). SNP rs3734355 was more common in whites (MAF = 0.14) than blacks (MAF = 0.03). Sex-stratified analysis showed that in white males but not white females, homozygosity for the 371Val allele was associated with higher BMI (P = 0.003) and % body fat (P = 0.04) (Table 3). To validate our results, we also genotyped rs3734354 (see Methods and Procedures section), a SNP previously reported to be in complete LD with rs3734355 in whites (10). Among the white Health ABC subjects, these two SNPs were in nearly perfect LD (r2 = 0.97), and the association between rs3734354 and adiposity was nearly identical to that of rs3734355 (see Supplementary Table S2 online). A case–control analysis of white males from the Health ABC Study confirmed that homozygosity for the 371Val allele of rs3734355 was significantly associated with obesity (BMI ≥ 30 kg/m2) (odds ratio = 2.8, 95% confidence interval (CI): 1.0–7.9, P = 0.05).
Table 2.
Adiposity-related traits in the Health ABC cohort by SNP genotype and ethnicity
| Traitsa | n | Mean ± s.d. | n | Mean ± s.d. | n | Mean ± s.d. | n | Mean ± s.d. | n | Mean ± s.d. | n | Mean ± s.d. |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| rs3734355 (SIM1) |
||||||||||||
| Whites |
Blacks |
|||||||||||
| Ala/Ala | Ala/Val | Val/Val | Ala/Ala | Ala/Val | Val/Val | |||||||
| BMI (kg/m2) | 1,308 | 26.5 ± 4.1 | 418 | 26.6 ± 4.2 | 36 | 28.3 ± 3.8 | 1,161 | 28.7 ± 5.4 | 78 | 28.3 ± 5.4 | 1 | 31.9 |
| % Body fat | 1,260 | 34.6 ± 7.2 | 398 | 34.9 ± 7.1 | 34 | 37.6 ± 6.2 | 1,125 | 35.4 ± 8.6 | 75 | 35.5 ± 8.4 | 1 | 49.0 |
| Leptin (ng/ml) | 1,294 | 12.5 ± 11.3 | 413 | 13.0 ± 11.5 | 35 | 16.2 ± 12.9 | 1,140 | 17.1 ± 13.9 | 76 | 16.4 ± 13.3 | 1 | 38.0 |
| VAT (cm2) | 1,255 | 152.7 ± 69.9 | 405 | 147.8 ± 67.1 | 35 | 182.9 ± 73.8 | 1,116 | 130.0 ± 61.7 | 76 | 128.1 ± 57.5 | 0 | – b |
| SAT (cm2) | 1,223 | 263.4 ± 101.6 | 402 | 266.6 ± 103.7 | 35 | 303.3 ± 80.6 | 1,066 | 314.3 ± 138.7 | 72 | 321.7 ± 143.6 | 0 | – b |
| rs9390332 (SIM1) |
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Whites |
Blacks |
|||||||||||
| A/A | A/T | T/T | A/A | A/T | T/T | |||||||
| BMI (kg/m2) | 1,004 | 26.3 ± 4.1 | 559 | 26.8 ± 4.2 | 90 | 27.6 ± 4.1 | 1,050 | 28.7 ± 5.5 | 122 | 28.5 ± 5.2 | 2 | 29.1 ± 4.0 |
| % Body fat | 974 | 34.3 ± 7.2 | 528 | 35.3 ± 7.2 | 88 | 36.1 ± 6.8 | 1,016 | 35.4 ± 8.6 | 117 | 36.3 ± 8.7 | 2 | 38.8 ± 14.4 |
| Leptin (ng/ml) | 994 | 12.4 ± 11.4 | 551 | 13.4 ± 11.6 | 88 | 14.3 ± 13.4 | 1,030 | 17.2 ± 13.8 | 119 | 18.2 ± 14.8 | 2 | 24.9 ± 18.6 |
| VAT (cm2) | 962 | 149.9 ± 67.2 | 540 | 153.6 ± 71.7 | 86 | 166.4 ± 72.2 | 1,007 | 129.8 ± 61.7 | 119 | 134.7 ± 60.3 | 1 | 236.3 |
| SAT (cm2) | 942 | 260.6 ± 102.8 | 529 | 271.9 ± 102.7 | 85 | 285.6 ± 99.3 | 961 | 314.8 ± 140.0 | 111 | 323.3 ± 138.7 | 1 | 205.3 |
| rs3734353 (SIM1) |
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Whites |
Blacks |
|||||||||||
| A/A | A/C | C/C | A/A | A/C | C/C | |||||||
| BMI (kg/m2) | 831 | 26.6 ± 4.1 | 686 | 26.5 ± 4.1 | 134 | 26.5 ± 4.5 | 626 | 28.7 ± 5.3 | 465 | 28.7 ± 5.7 | 81 | 28.9 ± 5.2 |
| % Body fat | 798 | 34.6 ± 7.4 | 660 | 34.9 ± 6.8 | 131 | 34.8 ± 7.4 | 606 | 35.5 ± 8.7 | 447 | 35.3 ± 8.6 | 80 | 36.9 ± 8.2 |
| Leptin (ng/ml) | 821 | 12.6 ± 11.8 | 679 | 13.1 ± 11.4 | 131 | 12.7 ± 10.9 | 615 | 16.8 ± 13.8 | 456 | 17.9 ± 14.2 | 78 | 18.3 ± 13.6 |
| VAT (cm2) | 796 | 153.5 ± 71.6 | 664 | 150.2 ± 67.0 | 128 | 152.9 ± 63.6 | 604 | 130.8 ± 62.5 | 442 | 130.7 ± 60.9 | 79 | 125.4 ± 60.0 |
| SAT (cm2) | 777 | 265.4 ± 102.2 | 655 | 265.1 ± 102.5 | 124 | 271.2 ± 107.3 | 575 | 312.6 ± 135.9 | 424 | 317.8 ± 146.0 | 72 | 325.8 ± 134.3 |
| rs7746743 (SIM1) |
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Whites |
Blacks |
|||||||||||
| A/A | A/T | T/T | A/A | A/T | T/T | |||||||
| BMI (kg/m2) | 324 | 26.4 ± 4.4 | 839 | 26.6 ± 4.2 | 491 | 26.6 ± 3.9 | 373 | 29.3 ± 5.6 | 568 | 28.6 ± 5.4 | 234 | 27.8 ± 5.4 |
| % Body fat | 310 | 34.8 ± 7.5 | 809 | 34.9 ± 7.2 | 472 | 34.5 ± 7.0 | 362 | 36.0 ± 8.9 | 549 | 35.4 ± 8.7 | 225 | 35.0 ± 8.0 |
| Leptin (ng/ml) | 322 | 13.1 ± 12.0 | 828 | 12.9 ± 11.6 | 484 | 12.5 ± 11.3 | 360 | 17.7 ± 13.8 | 559 | 17.0 ± 14.0 | 233 | 17.4 ± 14.0 |
| VAT (cm2) | 312 | 147.1 ± 64.8 | 804 | 153.8 ± 71.4 | 473 | 152.4 ± 67.6 | 353 | 131.4 ± 58.2 | 552 | 131.0 ± 62.0 | 223 | 127.3 ± 65.9 |
| SAT (cm2) | 307 | 265.3 ± 107.8 | 786 | 267.7 ± 103.1 | 464 | 262.6 ± 98.7 | 336 | 329.0 ± 145.0 | 521 | 315.5 ± 138.8 | 217 | 295.0 ± 131.6 |
| rs4072568 (ARNT2) |
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Whites |
Blacks |
|||||||||||
| Gly/Gly | Gly/Ser | Ser/Ser | Gly/Gly | Gly/Ser | Ser/Ser | |||||||
| BMI (kg/m2) | 1,043 | 26.8 ± 4.1 | 549 | 26.2 ± 4.2 | 61 | 25.6 ± 3.5 | 877 | 28.6 ± 5.5 | 274 | 28.8 ± 5.5 | 22 | 29.4 ± 4.2 |
| % Body fat | 1,003 | 34.9 ± 7.0 | 526 | 34.4 ± 7.5 | 61 | 34.2 ± 7.1 | 844 | 35.5 ± 8.5 | 269 | 35.3 ± 9.0 | 21 | 37.6 ± 8.5 |
| Leptin (ng/ml) | 1,030 | 13.0 ± 11.5 | 542 | 12.7 ± 11.8 | 61 | 10.8 ± 10.0 | 859 | 17.4 ± 13.8 | 269 | 16.9 ± 14.2 | 22 | 19.8 ± 16.0 |
| VAT (cm2) | 1,005 | 155.7 ± 71.0 | 528 | 146.3 ± 65.5 | 55 | 142.3 ± 60.5 | 842 | 129.8 ± 61.3 | 262 | 133.1 ± 64.0 | 22 | 122.6 ± 42.7 |
| SAT (cm2) | 983 | 270.0 ± 100.6 | 518 | 258.3 ± 107.2 | 55 | 259.2 ± 94.3 | 797 | 313.5 ± 137.3 | 254 | 319.9 ± 145.4 | 21 | 347.3 ± 167.2 |
None of the trait values are transformed or adjusted for covariates.
The sample size was zero (measures of regional adiposity were not available for this participant).
ARNT2, aryl-hydrocarbon receptor nuclear translocator 2; SAT, abdominal subcutaneous adipose tissue; SIM1, single-minded 1; SNP, single-nucleotide polymorphism; VAT, abdominal visceral adipose tissue.
Table 3.
Replicated SIM1 nsSNP association results in white subjects from the Health ABC Study
| Dependent variable per SNPa | Whites (n = 1,613–1,711)b |
White males (n = 854–907)b |
White females (n = 759–804)b |
|||
|---|---|---|---|---|---|---|
| β ± s.e. | P | β ± s.e. | P | β ± s.e. | P | |
| rs3734355 | ||||||
| BMI (kg/m2) | 1.31 ± 0.67 | 0.05 | 2.69 ± 0.91 | 0.003 | 0.29 ± 0.99 | NS |
| % Body fat | 1.71 ± 0.87 | 0.05 | 2.42 ± 1.20 | 0.04 | 1.08 ± 1.26 | NS |
| Leptin (ng/ml) | 0.35 ± 0.21 | NS | 0.47 ± 0.26 | NS | 0.26 ± 0.34 | NS |
| Leptin (ng/ml)c | 0.06 ± 0.16 | NS | 0.03 ± 0.20 | NS | 0.11 ± 0.24 | NS |
| VAT (cm2) | 0.46 ± 0.33 | NS | 0.16 ± 0.50 | NS | 0.78 ± 0.42 | NS |
| SAT (cm2) | –0.09 ± 0.27 | NS | 0.03 ± 0.38 | NS | –0.06 ± 0.37 | NS |
NS, nonsignificant; SAT, abdominal subcutaneous adipose tissue; SIM1, single-minded 1; nsSNP, nonsynonymous single-nucleotide polymorphism; VAT, abdominal visceral adipose tissue.
rs3734355 was studied assuming a recessive mode of inheritance.
Sample size range for 6 models, sample sizes for each model are found in Supplementary Table S2 online.
Leptin outcome was adjusted for percentage of body fat.
A previous report utilizing two population-based cohorts (Ely and EPIC-Norfolk) found the rs3734354/rs3734355 haplotype to be associated with higher BMI among white males using a recessive model (10). Combining these results with those from the Health ABC study (total n = 3,479 white males) revealed that homozygosity for the 371Val allele of rs3734355 was associated with an increase in BMI of 1.10 kg/m2 (95% CI: 0.25–1.95, P = 0.01). There was no evidence for heterogeneity between the studies. Sex-specific association results were not provided in the other two reports investigating the relationship between SIM1 and adiposity (11,12).
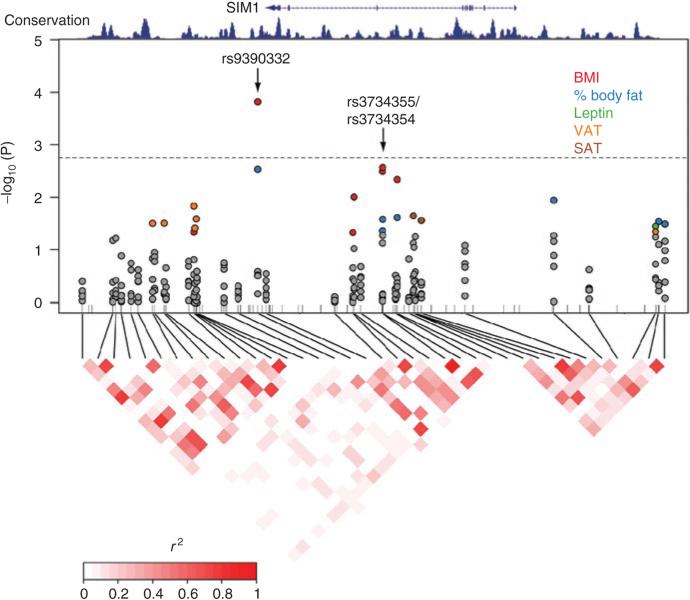
Next, common genetic variation in the rest of the SIM1 locus and the surrounding noncoding conserved regions was surveyed using ethnicity-specific tagSNPs (see Supplementary Figure S1 online, Figure 1). Using Health ABC genotype data from SNPs passing QC criteria, 38 SNPs (37 tagSNPs and rs3734355) in whites and 55 SNPs (54 tagSNPs and rs3734355) in blacks were selected and tested for association (see Supplementary Table S2 online). In addition to the nominally significant associations between rs3734355/4 and adiposity (Table 3), we found three tagSNPs in Health ABC subjects that remained significantly associated with adiposity after correction for multiple testing. The first of these, rs9390332, is located 6.5 kb downstream from the end of the SIM1 mRNA transcript and is in moderate LD with rs3734355 (r2 = 0.54, see Supplementary Figure S1 online). rs9390332 was significantly associated with BMI and % body fat in the entire group of white subjects and in white males (Figure 1, Table 4). After correction for multiple testing, rs9390332 remained significantly associated with BMI in white males (Punadjusted = 2 × 10−4, Pempirical = 0.005) under an additive genetic model. The MAF for rs9390332 was 0.22 in whites but only 0.05 in blacks, thus limiting the power to detect an association in the latter group.
Figure 1.
Association analysis and linkage disequilibrium plot for the 38 tagSNPs genotyped in SIM1 and flanking regions in white males from the Health ABC Study. The upper plot shows the genomic organization of SIM1 and the degree of conservation among mammalian genomic sequences, with nominally significant P values for associations between various adiposity traits (BMI, % body fat, leptin, VAT, and SAT) at each of 38 tagSNPs shown below. Nonsignificant association P values at the 0.05 level are filled gray, and the colors of the significant (P < 0.05) association P values correspond to the colors in the legend for the traits with which they are associated. The dashed line indicates the nominal P value cutoff that results in a significant empirical P value from the permutation test for BMI in white males. Association models assumed an additive mode of inheritance, except for rs3734355/rs3734354, which are modeled using a recessive mode of inheritance. The ticks along the X-axis mark the positions of the 78 genotyped SNPs that passed QC and had an MAF ≥0.05 among whites in the Health ABC Study. The lower plot is of linkage disequilibrium (r2 units) across SIM1 in white subjects from the Health ABC Study. MAF, minor allele frequency; SAT, abdominal subcutaneous adipose tissue; SIM1, single-minded 1; SNPs, single-nucleotide polymorphisms; VAT, abdominal visceral adipose tissue.
Table 4.
SIM1 tagSNP association results passing multiple test correction
| Dependent variable per SNPa | Whites (n = 1,509–1,603)b |
White males (n = 801–854)b |
White females (n = 708–749)b |
Blacks (n = 1,017–1,115)b |
Black males (n = 435–473)b |
Black females (n = 582–642)b |
||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| β ± s.e. | P | β ± s.e. | P | β ± s.e. | P | β ± s.e. | P | β ± s.e. | P | β ± s.e. | P | |
| rs9390332 | ||||||||||||
| BMI (kg/m2) | 0.43 ± 0.16 | 0.009 | 0.78 ± 0.21 | 2 × 10–4* | 0.01 ± 0.26 | NS | 0.10 ± 0.48 | NS | 0.54 ± 0.62 | NS | –0.26 ± 0.71 | NS |
| % Body fat | 0.49 ± 0.21 | 0.02 | 0.80 ± 0.27 | 0.003 | 0.19 ± 0.33 | NS | 1.05 ± 0.54 | 0.05 | 1.30 ± 0.77 | NS | 0.86 ± 0.74 | NS |
| Leptin (ng/ml) | 0.05 ± 0.05 | NS | 0.07 ± 0.06 | NS | 0.04 ± 0.09 | NS | 0.21 ± 0.12 | NS | 0.22 ± 0.16 | NS | 0.17 ± 0.18 | NS |
| Leptin (ng/ml)c | –0.02 ± 0.04 | NS | –0.05 ± 0.04 | NS | 0.01 ± 0.06 | NS | 0.06 ± 0.09 | NS | 0.09 ± 0.12 | NS | 0.00 ± 0.14 | NS |
| VAT (cm2) | 0.01 ± 0.08 | NS | 0.12 ± 0.12 | NS | –0.09 ± 0.11 | NS | 0.45 ± 0.20 | 0.02 | 0.73 ± 0.31 | 0.02 | 0.18 ± 0.26 | NS |
| SAT (cm2) | –0.05 ± 0.07 | NS | –0.04 ± 0.09 | NS | 0.00 ± 0.10 | NS | 0.35 ± 0.17 | 0.05 | 0.28 ± 0.26 | NS | 0.41 ± 0.24 | NS |
| rs3734353 | ||||||||||||
| BMI (kg/m2) | 0.07 ± 0.15 | NS | 0.15 ± 0.19 | NS | –0.02 ± 0.25 | NS | 0.07 ± 0.24 | NS | 0.37 ± 0.33 | NS | –0.17 ± 0.35 | NS |
| % Body fat | 0.09 ± 0.20 | NS | 0.42 ± 0.25 | NS | –0.29 ± 0.31 | NS | 0.04 ± 0.27 | NS | 0.09 ± 0.40 | NS | –0.03 ± 0.37 | NS |
| Leptin (ng/ml) | 0.04 ± 0.05 | NS | 0.10 ± 0.05 | NS | –0.02 ± 0.08 | NS | 0.08 ± 0.06 | NS | 0.25 ± 0.08 | 0.003 | –0.04 ± 0.09 | NS |
| Leptin (ng/ml)c | 0.02 ± 0.04 | NS | 0.02 ± 0.04 | NS | 0.04 ± 0.06 | NS | 0.07 ± 0.05 | NS | 0.21 ± 0.06 | 4 × 10–4* | –0.03 ± 0.07 | NS |
| VAT (cm2) | 0.05 ± 0.08 | NS | 0.13 ± 0.11 | NS | –0.04 ± 0.11 | NS | 0.01 ± 0.10 | NS | –0.14 ± 0.16 | NS | 0.09 ± 0.13 | NS |
| SAT (cm2) | 0.05 ± 0.06 | NS | 0.12 ± 0.08 | NS | –0.01 ± 0.09 | NS | 0.07 ± 0.09 | NS | –0.04 ± 0.14 | NS | 0.16 ± 0.11 | NS |
| rs7746743 | ||||||||||||
| BMI (kg/m2) | –0.04 ± 0.14 | NS | 0.07 ± 0.18 | NS | –0.17 ± 0.23 | NS | –0.83 ± 0.21 | 1 × 10–4* | –0.43 ± 0.28 | NS | –1.06 ± 0.31 | 7 × 10–4* |
| % Body fat | –0.04 ± 0.18 | NS | 0.09 ± 0.23 | NS | –0.14 ± 0.28 | NS | –1.09 ± 0.24 | 5 × 10–6* | –0.66 ± 0.34 | NS | –1.34 ± 0.33 | 5 × 10–5* |
| Leptin (ng/ml) | 0.00 ± 0.04 | NS | –0.02 ± 0.05 | NS | 0.03 ± 0.08 | NS | –0.13 ± 0.06 | 0.02 | –0.08 ± 0.07 | NS | –0.14 ± 0.08 | NS |
| Leptin (ng/ml)c | 0.01 ± 0.03 | NS | –0.03 ± 0.04 | NS | 0.06 ± 0.06 | NS | 0.01 ± 0.04 | NS | 0.01 ± 0.05 | NS | 0.03 ± 0.06 | NS |
| VAT (cm2) | 0.11 ± 0.07 | NS | 0.21 ± 0.10 | 0.03 | –0.05 ± 0.10 | NS | 0.11 ± 0.09 | NS | 0.01 ± 0.14 | NS | 0.15 ± 0.11 | NS |
| SAT (cm2) | 0.05 ± 0.06 | NS | 0.04 ± 0.08 | NS | 0.08 ± 0.09 | NS | –0.17 ± 0.08 | 0.02 | –0.29 ± 0.12 | 0.01 | –0.10 ± 0.10 | NS |
NS, nonsignificant; SAT, abdominal subcutaneous adipose tissue; SIM1, single-minded 1; SNP single-nucleotide polymorphism; VAT, abdominal visceral adipose tissue.
rs9390332, rs3734353, and rs7746743 were studied assuming an additive mode of inheritance.
Sample size range for 6 models, sample sizes for each model are found in Supplementary Table S2 online.
Leptin outcome was adjusted for percentage of body fat.
Pempirical < 0.05.
The remaining two SNPs were significantly associated with adiposity in black Health ABC subjects after correction for multiple testing. rs3734353, a SNP in intron 5 that was previously associated with BMI in Pima Indians (11), was common in Health ABC whites and blacks (see Supplementary Table S2 online) and was associated with leptin concentrations in black males (Punadjusted = 0.003). This association became even more significant when leptin concentrations were adjusted for % body fat (Punadjusted = 4 × 10−4, Pempirical = 0.02) (Table 4). rs7746743, located 35 kb downstream of the SIM1 polyadenylation signal, was also common in both whites and blacks (see Supplementary Table S2 online) and was significantly associated with BMI (Punadjusted = 1 × 10−4, Pempirical = 0.004) and % body fat (Punadjusted = 5 × 10−6, Pempirical = 3 × 10−4) after correction for multiple testing (Table 4). When analyzed by sex, these associations were only significant in black females (for BMI: Punadjusted = 7 × 10−4, Pempirical = 0.03; for % body fat: Punadjusted = 5 × 10−5, Pempirical = 0.002) (Table 4).
We extended our study by investigating the only common nsSNP, rs4072568, in SIM1's heterodimerization partner, ARNT2. rs4072568 was associated with BMI, % body fat, and leptin concentrations in the entire group of white subjects (Table 5). Sex-specific analyses indicated that the association was confined to white males (for BMI: P = 9 × 10−4; for % body fat: P = 0.001; for leptin concentrations: P = 0.04). There was no evidence for a statistical interaction between rs3734355 in SIM1 and rs4072568 in ARNT2 in the association with BMI, however.
Table 5.
ARNT2 nsSNP association results
| Dependent variable per SNPa | Whites (n = 1,509–1,602)b |
White males (n = 802–854)b |
White females (n = 707–748)b |
Blacks (n = 1,018–1,113)b |
Black males (n = 435–472)b |
Black females (n = 583-641)b |
||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| β ± s.e. | P | β ± s.e. | P | β ± s.e. | P | β ± s.e. | P | β ± s.e. | P | β ± s.e. | P | |
| rs4072568 | ||||||||||||
| BMI (kg/m2) | –0.51 ± 0.18 | 0.004 | –0.73 ± 0.22 | 9 × 10–4 | –0.21 ± 0.28 | NS | 0.00 ± 0.31 | NS | –0.47 ± 0.41 | NS | 0.39 ± 0.46 | NS |
| % Body fat | –0.61 ± 0.22 | 0.007 | –0.92 ± 0.28 | 0.001 | –0.17 ± 0.35 | NS | –0.04 ± 0.35 | NS | –0.76 ± 0.50 | NS | 0.59 ± 0.48 | NS |
| Leptin (ng/ml) | –0.13 ± 0.06 | 0.02 | –0.13 ± 0.06 | 0.04 | –0.13 ± 0.10 | NS | –0.08 ± 0.08 | NS | –0.27 ± 0.11 | 0.01 | 0.09 ± 0.12 | NS |
| Leptin (ng/ml)c | –0.03 ± 0.04 | NS | 0.01 ± 0.05 | NS | –0.10 ± 0.07 | NS | –0.04 ± 0.06 | NS | –0.12 ± 0.07 | NS | 0.02 ± 0.09 | NS |
| VAT (cm2) | 0.00 ± 0.09 | NS | 0.05 ± 0.13 | NS | –0.02 ± 0.12 | NS | –0.03 ± 0.13 | NS | 0.08 ± 0.20 | NS | –0.09 ± 0.16 | NS |
| SAT (cm2) | –0.07 ± 0.07 | NS | –0.08 ± 0.10 | NS | –0.06 ± 0.11 | NS | 0.09 ± 0.11 | NS | –0.09 ± 0.17 | NS | 0.24 ± 0.15 | NS |
ARNT2, aryl-hydrocarbon receptor nuclear translocator 2; NS, nonsignificant; nsSNP, nonsynonymous single-nucleotide polymorphism; SAT, abdominal subcutaneous adipose tissue; VAT, abdominal visceral adipose tissue.
rs4072568 was studied assuming an additive mode of inheritance.
Sample size range for 6 models, sample sizes for each model are found in Supplementary Table S2 online.
Leptin outcome adjusted for percentage of body fat.
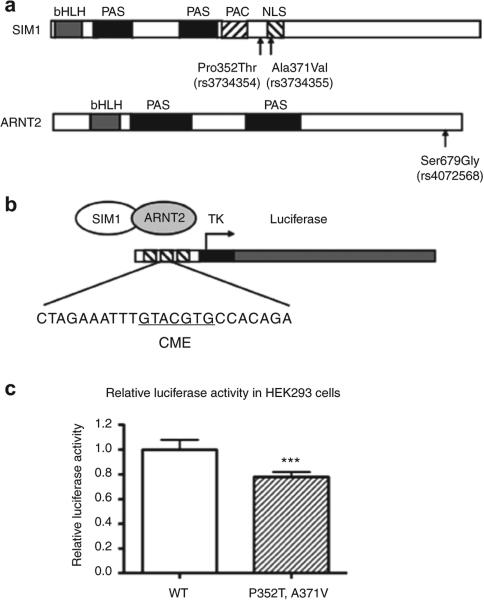
Finally, we investigated the functional effects of the 352Thr-371Val haplotype on SIM1 activity in an in vitro transcriptional assay. When transfected with wild-type ARNT2 into HEK293 cells, the 352Thr-371Val SIM1 double mutant displayed reduced transcriptional activity relative to the wild-type SIM1 (to 78% of wild type, 95% CI: 75–82%, P < 0.0001) (Figure 2).
Figure 2.
In vitro transcriptional assay for SIM1 activity. (a) Human SIM1 and ARNT2 cDNAs were cloned into the mammalian expression vector pCDNA3.1/V5-His-TOPO (Invitrogen, Carlsbad, CA) and the nsSNPs rs3734354 and rs3734355 were introduced into SIM1 by site-directed mutagenesis. Functional domains in SIM1 and ARNT2 (and the only common SNP in ARNT2, rs4072568/Gly679Ser) are shown. (b) The SIM1 (either WT or 352Thr-371Val) and ARNT2 plasmids were transfected into HEK293 cells using Effectene reagent, along with the pLuc-MCS containing the SIM1 response element (CME) as well as the plasmid pRL-RSV (Promega, Madison, WI), which encodes Renilla luciferase, to control for transfection efficiency. (c) Relative to the wild-type SIM1, the 352Thr-371Val SIM1 double mutant displayed reduced transcriptional activity (to 78% of WT, 95% CI: 0.75–0.82, P < 0.0001) in HEK293 cells. Results shown are the average of seven independent experiments. ARNT2, aryl-hydrocarbon receptor nuclear translocator 2; bHLH, basic helix-loop-helix; CI, confidence interval; CME, CNS midline enhancer; NLS, nuclear localization signal; nsSNP, nonsynonymous single-nucleotide polymorphism; PAC, conserved domain occurring C-terminal to a PAS domain; PAS, Per-Arnt-Sim domain; SIM1, single-minded 1; WT, wild type.
DISCUSSION
We have confirmed previous findings (10) that homozygosity for a common SIM1 haplotype consisting of two amino acid changes (Pro352Val/rs3734354 and Ala371Val/rs3734355) is associated with an increase in BMI in white males. We extended these findings in several ways. First, we obtained nominal evidence for an association between homozygosity for this haplotype and a more informative measure of adiposity (% body fat). Second, by systematically scanning the SIM1 region (from 45 kb upstream of the 5′ end to 60 kb downstream of the 3′ end), we demonstrated that the most significant association with adiposity in white males was at these two nsSNPs, and at a related SNP, rs9390332, in the 3′ flanking region. rs9390332 is located in/near regions of high mammalian conservation 6.5 kb downstream of the SIM1 polyadenylation signal, and is in moderate LD (r2 = 0.54) with rs3734355/4. Lastly, we strengthened the biological relevance of this association by showing that the 352Thr-371Val haplotype impairs SIM1's ability to activate target gene transcription. Future studies will be required to determine whether this occurs via impaired nuclear localization (as rs3734355 is located in the SIM1 nuclear localization signal (21)), and whether rs9390332 alters SIM1 function in an as yet unidentified manner. It is also unknown whether SIM1 352Thr-371Val homozygotes recapitulate the other phenotypes of hyperphagia, hyperinsulinemia, or increased linear growth observed in SIM1 haploinsufficient mice (5); although milder effects on these phenotypes would be expected based on our functional results. Interestingly, a comparable impairment of SIM1 in vitro transcriptional activity (to 67% of wild-type levels, P = 0.093) by the 352Thr-371Val haplotype was observed in another recent report (12). The additional association between the only common nsSNP in SIM1's heterodimerization partner ARNT2 and BMI in white men further highlights the role of this axis in human body weight regulation.
The greater significance of the association between the 352Thr-371Val haplotype and BMI in the present study (P = 0.002) compared to the initial report (P = 0.04) (10) was in spite of a smaller sample size (907 white men vs. 2,166 white men in the EPIC-Norfolk Study) and was not due to the use of different covariates. It is possible that the effects of this haplotype on adiposity may be age-dependent, as one of the recent studies that did not find an association between rs3734354/5 and adiposity (12) examined subjects who were ~10 years younger than the subjects studied in (10) and ~30 years younger than the subjects in the present study. In addition to its critical role for hypothalamic development, Sim1 is also known to influence feeding behavior in adult mice (22,23). One interpretation of the available data is that the subtle defect in SIM1 function conferred by the 352Thr-371Val haplotype causes a slight shift toward positive energy balance throughout life. Therefore, it is conceivable that significant differences in adiposity between genotype groups may not become apparent until well into middle age. This possibility remains to be investigated in longitudinal studies.
Genome-wide association studies (GWAS) for obesity in whites (24–26) have not reported significant associations for BMI at rs3734355, rs3734354, or rs9390332. This could be partly due to the absence of these SNPs from many of the genotyping panels used for the discovery stages of recent GWAS, although their genotypes could be imputed. Moreover, these recent GWAS assumed an additive mode of inheritance for all SNPs, whereas we found a much more significant association between rs3734355 and BMI in white males when using a recessive model (Padd = 0.04 compared to Prec = 0.003). The closest positive result obtained in a GWAS for BMI was obtained at rs2572016, about 1.3 million bp centromeric to rs3734355, (P = 0.05 in a meta-analysis of 16,876 individuals) (24). However, this SNP is separated from rs3734354/rs3734355 (and from rs7746743 and rs9390332) by several known human genes (MCHR2, PRDM13, CCNC, USP45, SFRS18, and COQ3). Finally, our results suggest that the effect of rs3734355/rs3734354 on BMI is stronger in males, yet none of these GWAS have performed separate analyses by sex.
We scanned the SIM1 region with as many or more SNPs than used in current genotyping platforms. In whites from the Health ABC Study, 78 genotyped SNPs passed QC, yielding a density of one genotyped SNP every 2.3 kb. In blacks, 69 of the 88 genotyped SNPs passed QC, yielding a density of one genotyped SNP every 2.7 kb. For this same genomic region, the Affymetrix 500K, 5.0, and 6.0, and the Illumina Infinium HumanHap 370K, 610K, and 1M arrays genotype 37, 34, 62, 27, 46, and 69 SNPs, respectively.
The most plausible mechanism by which genetic variation in SIM1 might contribute to adiposity is via the regulation of feeding behavior. An individual with a balanced translocation interrupting SIM1 displayed early onset hyperphagic obesity but no obvious defect in energy expenditure (6). Sim1+/− mice exhibit a partial failure of the terminal migration and differentiation of hypothalamic neurons in the PVN, supraoptic, and anterior periventricular nuclei (1,4). Sim1 is also expressed postdevelopmentally in the PVN, basomedial amygdala, and lateral hypothalamus (27). Adenovirally mediated overexpression or silencing of Sim1 expression in the PVN of weaned mice inhibits and stimulates food intake, respectively (22).
SIM1's inhibitory effects on feeding are likely to involve a reduction in the expression of specific peptides in the PVN. Secretory PVN neurons produce a variety of peptides, including somatostatin, arginine vasopressin, oxytocin, corticotropin-releasing hormone (CRH) and thyrotropin-releasing hormone. In the hypothalamus of a Sim1+/− mouse strain, the mRNA expression levels of all of these peptides are reduced by 20–80%, relative to wild-type mice (2). In particular, normal oscillations in Crh and oxytocin (oxt) mRNA expression in response to feeding and fasting were either markedly attenuated (Crh) or completely abolished (oxt) in Sim1+/− mice. Crh and oxt mediate the anorectic effects of leptin in the central nervous system (28,29). In particular, oxytocin is thought to facilitate the termination of feeding behavior, as intracerebroventricular infusion of oxytocin produces a dose-dependent reduction in feeding in rats fed ad libitum as well as those fed again after a period of food deprivation (30,31). Oxytocin is implicated in the hyperphagia of Sim1+/− mice, as intracerebroventricular administration of an oxytocin antagonist increased food intake in Sim1+/− mice but not wild-type mice, while central oxytocin administration abolished hyperphagia in Sim1+/− mice but had no effect in wild-type mice (2). Intriguingly, oxytocin's effects on feeding cessation may be nutrient-specific, as oxytocin-deficient mice have an increased preference for ingesting excessive amounts of carbohydrates (32). Accordingly, the reduced transcriptional activity of the SIM1 352Thr-371Val haplotype could promote weight gain in homozygous individuals through a defect in feeding cessation and/or a change in macronutrient preference. This remains to be investigated in future studies.
It is unclear why the association between 352Thr-371Val haplotype and adiposity is confined to males; such sex-specific effects on adiposity are not observed in SIM1 haploinsufficient mice (5,27). PVN oxytocin expression is influenced by female sex hormones (33,34); however, the female subjects in the present study were aged 70–79 and therefore postmenopausal. CRH-producing neurons in the human PVN display strong androgen- and estrogen-receptor immunoreactivity (35,36), and expression of androgen receptor in combination with testosterone treatment represses CRH promoter activity in cultured neuroblastoma cells (35). Whether the sex-specific effects of the SIM1 352Thr-371Val haplotype involve a greater suppression of PVN oxytocin and/or CRH expression will require further investigation.
Notably, Sim1+/− mice are also resistant to the anorexigenic effects of a melanocortin agonist (37), and display a marked reduction in hypothalamic Mc4r mRNA expression (23). If the same is true in humans, this suggests that a subtle defect in SIM1 function could recapitulate, at least in part, obesity due to MC4R deficiency (38).
The lack of a significant association between SIM1 352Thr-371Val haplotype and adiposity in blacks was most likely due to the low MAF (0.03). However, our results obtained in these subjects do support a relationship between genetic variation in SIM1 and adiposity: the single black female homozygous for the rs3734355 371Val allele had a much higher BMI than the mean BMI for carriers of the 371Ala allele (Table 2), and we obtained independent positive associations at rs3734353 and rs7746743 for global measures of adiposity that remained significant after correction for multiple testing (Table 4). rs3734353 is located within intron 5, and rs7746743 is located 35 kb downstream of the SIM1 polyadenylation signal within a 41 bp conserved region that is 85.4% conserved between mice and humans (39). One could speculate that this region functions as a distal enhancer to regulate SIM1 expression, as the nearest refSeq gene to rs7746743 other than SIM1 is MCHR2, which is ~300 kb centromerically. The lack of an association between these two SNPs and adiposity in whites cannot be explained by differences in MAF (see Supplementary Table S2 online), and therefore indicates the presence of substantial genetic heterogeneity in addition to the sex-specific effects of SIM1 genotype on adiposity.
A recent investigation of SIM1 in Pima Indians (11) found that the most significant and reproducible associations with BMI were obtained for SNPs spanning a region from the 5′ untranslated region to intron 8, assuming an additive mode of inheritance. In Pima Indians, neither rs3734355 nor rs3734354 were associated with BMI, however, and attempts to replicate the most significant association results (rs3734353 and rs3213541 in both the full- and partial-heritage Pima Indians) in French whites were unsuccessful (11,12). Marked differences in allele frequency between populations were observed. For example, the frequencies of rs3734355 and rs3734354 were much higher in Pima Indians (MAF ~0.26) than in either the whites (0.14) or blacks (0.03) studied here. Consistent with the findings in French whites (12), we also failed to replicate the rs3734353 result among white Health ABC subjects, although we did obtain robust evidence for association at rs3734353 with leptin concentrations in black males (Table 4). We did not investigate rs3213541, as it was excluded due to violation of Hardy–Weinberg equilibrium (see Supplementary Methods and Procedures online). Lastly, the SNP rs7746743, where we obtained our most significant results for all black subjects (men and women) and black women, is outside of the region studied by Traurig et al. (11).
In conclusion, we have obtained evidence that dysregulation of the SIM1-ARNT2 axis, binding partners required for the differentiation of specific neurons involved in the regulation of appetite and body weight, contributes to adiposity in whites and blacks. This finding reinforces the view that human obesity is influenced by genetic variation in developmental factors (40), many of which are predominantly expressed in the central nervous system (26).
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by funding from the National Institutes of Health (K01 AG022782, R01 AG023692, R01 DK60540), the Intramural Research Program of the NIH, National Institute on Aging (N01-AG-6-2101, N01-AG-6-2103, N01-AG-6-2106), the American Diabetes Association (1-05-RA-54), the National Center for Research Resources (M01 RR-00079), and the UCSF Diabetes and Endocrinology Research Center (P30 DK63720). M.M.S. is supported by a National Health and Medical Research Council of Australia (NHMRC) Program Grant “Pathways to Diabetes Prevention.” D.S.E. is supported by NIH training grant T32 DK007418. M.I.V. is supported by a Postdoctoral Fellowship from the Spanish Ministerio de Educacion y Ciencia. I.M. is supported by Mentored Research Scientist Development Award from the National Institute of Diabetes and Digestive and Kidney Diseases (K01 DK083029). C.V. is supported by an Established Investigator Award from the American Heart Association. The authors would like to thank all subjects for their participation in the study.
Footnotes
SUPPLEMENTARY MATERIAL
Supplementary material is linked to the online version of the paper at http://www.nature.com/oby
DISCLOSURE
The authors declared no conflict of interest.
REFERENCES
- 1.Xu C, Fan CM. Allocation of paraventricular and supraoptic neurons requires SIM1 function: a role for a SIM1 downstream gene PlexinC1. Mol Endocrinol. 2007;21:1234–1245. doi: 10.1210/me.2007-0034. [DOI] [PubMed] [Google Scholar]
- 2.Kublaoui BM, Gemelli T, Tolson KP, Wang Y, Zinn AR. Oxytocin deficiency mediates hyperphagic obesity of Sim1 haploinsufficient mice. Mol Endocrinol. 2008;22:1723–1734. doi: 10.1210/me.2008-0067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Michaud JL, DeRossi C, May NR, Holdener BC, Fan CM. ARNT2 acts as the dimerization partner of SIM1 for the development of the hypothalamus. Mech Dev. 2000;90:253–261. doi: 10.1016/s0925-4773(99)00328-7. [DOI] [PubMed] [Google Scholar]
- 4.Michaud JL, Rosenquist T, May NR, Fan CM. Development of neuroendocrine lineages requires the bHLH-PAS transcription factor SIM1. Genes Dev. 1998;12:3264–3275. doi: 10.1101/gad.12.20.3264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Michaud JL, Boucher F, Melnyk A, et al. Sim1 haploinsufficiency causes hyperphagia, obesity and reduction of the paraventricular nucleus of the hypothalamus. Hum Mol Genet. 2001;10:1465–1473. doi: 10.1093/hmg/10.14.1465. [DOI] [PubMed] [Google Scholar]
- 6.Holder JL, Jr, Butte NF, Zinn AR. Profound obesity associated with a balanced translocation that disrupts the SIM1 gene. Hum Mol Genet. 2000;9:101–108. doi: 10.1093/hmg/9.1.101. [DOI] [PubMed] [Google Scholar]
- 7.Bonaglia MC, Ciccone R, Gimelli G, et al. Detailed phenotype-genotype study in five patients with chromosome 6q16 deletion: narrowing the critical region for Prader-Willi-like phenotype. Eur J Hum Genet. 2008;16:1443–1449. doi: 10.1038/ejhg.2008.119. [DOI] [PubMed] [Google Scholar]
- 8.Meyre D, Lecoeur C, Delplanque J, et al. A genome-wide scan for childhood obesity-associated traits in French families shows significant linkage on chromosome 6q22.31-q23.2. Diabetes. 2004;53:803–811. doi: 10.2337/diabetes.53.3.803. [DOI] [PubMed] [Google Scholar]
- 9.Wang JC, Turner L, Lomax B, Eydoux P. A 5-Mb microdeletion at 6q16.1-q16.3 with SIM gene deletion and obesity. Am J Med Genet A. 2008;146A:2975–2978. doi: 10.1002/ajmg.a.32555. [DOI] [PubMed] [Google Scholar]
- 10.Hung CC, Luan J, Sims M, et al. Studies of the SIM1 gene in relation to human obesity and obesity-related traits. Int J Obes (Lond) 2007;31:429–434. doi: 10.1038/sj.ijo.0803443. [DOI] [PubMed] [Google Scholar]
- 11.Traurig M, Mack J, Hanson RL, et al. Common variation in SIM1 is reproducibly associated with BMI in Pima Indians. Diabetes. 2009;58:1682–1689. doi: 10.2337/db09-0028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ghoussaini M, Stutzmann F, Couturier C, et al. Analysis of the SIM1 contribution to polygenic obesity in the French population. Obesity (Silver Spring) 2010;18:1670–1675. doi: 10.1038/oby.2009.468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Swarbrick MM, Waldenmaier B, Pennacchio LA, et al. Lack of support for the association between GAD2 polymorphisms and severe human obesity. PLoS Biol. 2005;3:e315. doi: 10.1371/journal.pbio.0030315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Snijder MB, Visser M, Dekker JM, et al. The prediction of visceral fat by dual-energy X-ray absorptiometry in the elderly: a comparison with computed tomography and anthropometry. Int J Obes Relat Metab Disord. 2002;26:984–993. doi: 10.1038/sj.ijo.0801968. [DOI] [PubMed] [Google Scholar]
- 15.Ruhl CE, Everhart JE, Ding J, et al. Health, Aging, and Body Composition Study. Serum leptin concentrations and body adipose measures in older black and white adults. Am J Clin Nutr. 2004;80:576–583. doi: 10.1093/ajcn/80.3.576. [DOI] [PubMed] [Google Scholar]
- 16.Chen X, Levine L, Kwok PY. Fluorescence polarization in homogeneous nucleic acid analysis. Genome Res. 1999;9:492–498. [PMC free article] [PubMed] [Google Scholar]
- 17.Frazer KA, Pachter L, Poliakov A, Rubin EM, Dubchak I. VISTA: computational tools for comparative genomics. Nucleic Acids Res. 2004;32:W273–W279. doi: 10.1093/nar/gkh458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.de Bakker PI, Yelensky R, Pe'er I, et al. Efficiency and power in genetic association studies. Nat Genet. 2005;37:1217–1223. doi: 10.1038/ng1669. [DOI] [PubMed] [Google Scholar]
- 19.Westfall PH, Young SS. Resampling-Based Multiple Testing: Examples and Methods for P-value Adjustment. Wiley-Interscience; New York: 1993. [Google Scholar]
- 20.Moffett P, Pelletier J. Different transcriptional properties of mSim-1 and mSim-2. FEBS Lett. 2000;466:80–86. doi: 10.1016/s0014-5793(99)01750-0. [DOI] [PubMed] [Google Scholar]
- 21.Yamaki A, Kudoh J, Shimizu N, Shimizu Y. A novel nuclear localization signal in the human single-minded proteins SIM1 and SIM2. Biochem Biophys Res Commun. 2004;313:482–488. doi: 10.1016/j.bbrc.2003.11.168. [DOI] [PubMed] [Google Scholar]
- 22.Yang C, Gagnon D, Vachon P, et al. Adenoviral-mediated modulation of Sim1 expression in the paraventricular nucleus affects food intake. J Neurosci. 2006;26:7116–7120. doi: 10.1523/JNEUROSCI.0672-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tolson KP, Gemelli T, Gautron L, et al. Postnatal Sim1 deficiency causes hyperphagic obesity and reduced Mc4r and oxytocin expression. J Neurosci. 2010;30:3803–3812. doi: 10.1523/JNEUROSCI.5444-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Loos RJ, Lindgren CM, Li S, et al. Prostate, Lung, Colorectal, and Ovarian (PLCO) Cancer Screening Trial; KORA; Nurses’ Health Study; Diabetes Genetics Initiative; SardiNIA Study; Wellcome Trust Case Control Consortium; FUSION. Common variants near MC4R are associated with fat mass, weight and risk of obesity. Nat Genet. 2008;40:768–775. doi: 10.1038/ng.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Thorleifsson G, Walters GB, Gudbjartsson DF, et al. Genome-wide association yields new sequence variants at seven loci that associate with measures of obesity. Nat Genet. 2009;41:18–24. doi: 10.1038/ng.274. [DOI] [PubMed] [Google Scholar]
- 26.Willer CJ, Speliotes EK, Loos RJ, et al. Wellcome Trust Case Control Consortium; Genetic Investigation of ANthropometric Traits Consortium. Six new loci associated with body mass index highlight a neuronal influence on body weight regulation. Nat Genet. 2009;41:25–34. doi: 10.1038/ng.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Holder JL, Jr, Zhang L, Kublaoui BM, et al. Sim1 gene dosage modulates the homeostatic feeding response to increased dietary fat in mice. Am J Physiol Endocrinol Metab. 2004;287:E105–E113. doi: 10.1152/ajpendo.00446.2003. [DOI] [PubMed] [Google Scholar]
- 28.Masaki T, Yoshimichi G, Chiba S, et al. Corticotropin-releasing hormone-mediated pathway of leptin to regulate feeding, adiposity, and uncoupling protein expression in mice. Endocrinology. 2003;144:3547–3554. doi: 10.1210/en.2003-0301. [DOI] [PubMed] [Google Scholar]
- 29.Blevins JE, Schwartz MW, Baskin DG. Evidence that paraventricular nucleus oxytocin neurons link hypothalamic leptin action to caudal brain stem nuclei controlling meal size. Am J Physiol Regul Integr Comp Physiol. 2004;287:R87–R96. doi: 10.1152/ajpregu.00604.2003. [DOI] [PubMed] [Google Scholar]
- 30.Olson BR, Drutarosky MD, Chow MS, et al. Oxytocin and an oxytocin agonist administered centrally decrease food intake in rats. Peptides. 1991;12:113–118. doi: 10.1016/0196-9781(91)90176-p. [DOI] [PubMed] [Google Scholar]
- 31.Benelli A, Bertolini A, Arletti R. Oxytocin-induced inhibition of feeding and drinking: no sexual dimorphism in rats. Neuropeptides. 1991;20:57–62. doi: 10.1016/0143-4179(91)90040-p. [DOI] [PubMed] [Google Scholar]
- 32.Miedlar JA, Rinaman L, Vollmer RR, Amico JA. Oxytocin gene deletion mice overconsume palatable sucrose solution but not palatable lipid emulsions. Am J Physiol Regul Integr Comp Physiol. 2007;293:R1063–R1068. doi: 10.1152/ajpregu.00228.2007. [DOI] [PubMed] [Google Scholar]
- 33.Miller FD, Ozimek G, Milner RJ, Bloom FE. Regulation of neuronal oxytocin mRNA by ovarian steroids in the mature and developing hypothalamus. Proc Natl Acad Sci USA. 1989;86:2468–2472. doi: 10.1073/pnas.86.7.2468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Thomas A, Kim NB, Amico JA. Differential regulation of oxytocin and vasopressin messenger ribonucleic acid levels by gonadal steroids in postpartum rats. Brain Res. 1996;738:48–52. doi: 10.1016/0006-8993(96)00760-3. [DOI] [PubMed] [Google Scholar]
- 35.Bao AM, Fischer DF, Wu YH, et al. A direct androgenic involvement in the expression of human corticotropin-releasing hormone. Mol Psychiatry. 2006;11:567–576. doi: 10.1038/sj.mp.4001800. [DOI] [PubMed] [Google Scholar]
- 36.Bao AM, Hestiantoro A, Van Someren EJ, Swaab DF, Zhou JN. Colocalization of corticotropin-releasing hormone and oestrogen receptor-alpha in the paraventricular nucleus of the hypothalamus in mood disorders. Brain. 2005;128:1301–1313. doi: 10.1093/brain/awh448. [DOI] [PubMed] [Google Scholar]
- 37.Kublaoui BM, Holder JL, Jr, Gemelli T, Zinn AR. Sim1 haploinsufficiency impairs melanocortin-mediated anorexia and activation of paraventricular nucleus neurons. Mol Endocrinol. 2006;20:2483–2492. doi: 10.1210/me.2005-0483. [DOI] [PubMed] [Google Scholar]
- 38.Vaisse C, Clement K, Durand E, et al. Melanocortin-4 receptor mutations are a frequent and heterogeneous cause of morbid obesity. J Clin Invest. 2000;106:253–262. doi: 10.1172/JCI9238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.University of California SC. UCSC Genome Bioinformatics Site. 2010 < http://genome.ucsc.edu/>.
- 40.Levin BE. Why some of us get fat and what we can do about it. J Physiol (Lond) 2007;583:425–430. doi: 10.1113/jphysiol.2007.135434. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.