Abstract
Potato virus X (PVX) is a single-stranded RNA plant virus, historically investigated in light of the detrimental effects on potato, the world’s fourth most important food commodity. The study of the interactions with cells, and more generally with the plant, both locally and systemically, significantly contributed to unveil the mechanisms underlying gene silencing, fundamental not only in plant virology but also in the study of gene expression regulation. Unraveling the molecular events of PVX infection paved the way for the development of different viral expression vectors and consequential applications in functional genomics and in the biosynthesis of heterologous proteins in plants. Apart from that, the ease of manipulation and the knowledge of the virus structure (particle dimensions, shape and physicochemical features) are inspiring novel applications, mainly focused on nanobiotechnology. This review will lead the reader in this area, spanning from fundamental to applied research, embracing fields from plant pathology to vaccine and drug-targeted delivery, imaging and material sciences. Due to the versatile moods, PVX holds promise to become an interesting nanomaterial, in view to create the widest possible arsenal of new “bio-inspired” devices to face evolving issues in biomedicine and beyond.
Keywords: potato virus X, nanotechnology, plant virus, nanomedicine, nanoparticles
Introduction to Potato Virus X Molecular Pathology
Potato virus X (PVX) belongs to the genus Potexvirus of the Flexiviridae family that groups viruses phylogenetically related by replication mechanisms, structural proteins and genome type and organization (Martelli et al., 2007). PVX is mechanically transmitted and its main hosts are herbaceous plants, especially Solanaceae. It has been included among the pathogens with important economic impact and its effects can be worsened by co-infection with other viruses, in particular Potato virus Y (PVY; a phenomenon also known as “synergism”; Syller, 2012).
Potato virus X has a simple filamentous flexible structure of about 500 nm in length and 15 nm in diameter (Figure 1A). The virion has helical symmetry and a deeply grooved, highly hydrated surface (Parker et al., 2002). It is made of a single-stranded positive-sense RNA genome of approximately 6.4 kb (Huisman et al., 1988; Skryabin et al., 1988), wrapped in approximately 1300 units of a single coat protein (CP) type, with 8.9 CP units per helix turn (Kendall et al., 2008).
FIGURE 1.
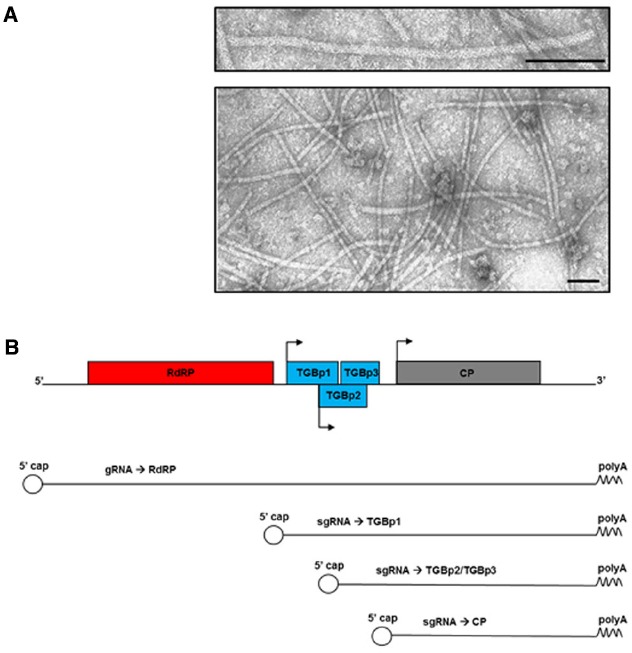
(A) Transmission electron micrographs of purified PVX nanoparticles (personal unpublished data). Bars: 100 nm. (B) Schematic diagram of PVX genome, genomic and subgenomic RNAs (gRNA and sgRNA, respectively), and the proteins they encode. RdRP: RNA-dependent RNA Polymerase; TGBp1/p2/p3: Triple Gene Block proteins; CP: Coat Protein. Arrows represent the subgenomic promoters. The 5′ cap and the polyA tail are indicated.
The genome is capped at the 5′-end and poly-adenylated at the 3′-terminus. It contains five open reading frames (ORFs) encoding five proteins: the RNA-dependent RNA Polymerase (RdRP), the movement proteins encoded by three overlapping ORFs that form the Triple Gene Block module (TGBp1, TGBp2, and TGBp3), and the CP (Figure 1B; Sonenberg et al., 1978; Morozov et al., 1987, 1991). The majority of these proteins are multifunctional and involved in several different steps of the infection process, together with numerous host factors. The RdRP, for example, contains a methyl transferase type 1 domain involved in viral RNAs capping (Rozanov et al., 1992), a helicase domain fundamental for viral replication (Davenport and Baulcombe, 1997), and a replicase domain (Longstaff et al., 1993); while TGBp1 has RNA-binding, ATPase (Leshchiner et al., 2006) and RNA-helicase (Kalinina et al., 2002) activities. Moreover, TGBp1 contributes to: (a) increase the size exclusion limit of the plasmodesmata (PD; Angell et al., 1996; Howard et al., 2004), (b) translationally activate the progeny virions (Karpova et al., 2006), and (c) counteract gene silencing (Voinnet et al., 2000), the plant innate mechanism of immune resistance.
When PVX enters plant cells, the particles unpack thanks to the phosphorylation events mediated by cellular enzymes on Ser/Thr residues located at the solvent-exposed N-terminus of the CP (Atabekov et al., 2001). The RdRP is directly synthetized from the viral RNA genome, exploiting the host translational machinery, to subsequently give rise to subgenomic RNAs and viral proteins (Batten et al., 2003). Later on, within infected cells, the replication complexes become part of structures called X-bodies that are formed next to the nucleus (Tilsner et al., 2012). X-bodies are isolated environments delimited by remodeled cellular membranes and organized in “compartments” that incorporate both viral and host components, coordinating the different phases of infection (replication, protein synthesis, assembly, and movement). The primary function of X-bodies is to protect and allow viral replication through the anchoring of RdRP in arrays to membranes, yet being sites where ribonucleoprotein (RNP) complexes (involved in cell-to-cell transfer of infection) and complete virus particles are assembled. TGBp1 has a central role in the formation of X-bodies as it participates to the remodeling of host actin filaments, endoplasmic reticulum (ER) and Golgi membranes, and to the recruitment of the viral TGBp2 and TGBp3. Within X-bodies, aggregates of TGBp1 and actin microfilaments wrap in spirals to form the typical “beaded sheets,” electron microscopy-visible structures in PVX infected cells (Linnik et al., 2013). The spirals are surrounded by naked viral RNAs and fine membrane hoops/granular vesicles that contain TGBp2, TGBp3, ribosomes, and viral replicase, probably representing replication sites. The TGBp2 and TGBp3 membrane-spanning proteins contribute individually and/or synergistically to the formation of these vesicles by remodeling the ER membranes. At the periphery of X-bodies, assembled viral particles accumulate.
In the initial/middle phases of the infection, TGBp2 and TGBp3 exploit the dynamic ER/actin network to direct the transfer of RNP complexes, made of viral RNA and CP, from the perinuclear to the cortical ER (Tilsner et al., 2012). It has been recently suggested that later on the TGBp2 and TGBp3 remodeled ER-membranes form caps of the PD orifice and that these caps, harboring the RdRP and viral RNA, become the viral replication sites. According to this “co-replication model,” as soon as they are synthetized, RNP complexes are inserted by the TGBp1 into the PD for cell-to-cell transfer (Tilsner et al., 2013).
As for viral spread throughout the plant, the CP is necessary both in cell-to-cell and in systemic movement, but recently published data indicate that virus particle assembly is dispensable for local while essential for long-distance transfer (Santa Cruz et al., 1998; Betti et al., 2012). The CP has a pivotal role in virus particle morphology and stability, and strongly influences the efficiency of infection spreading (Lico et al., 2006). Unfortunately, atomic-resolution data defining its folding are not available. A recent revision of the protein model indicates that it contains seven α-helices and six β-strands and confirms that the N-terminus is exposed on the viral surface (Baratova et al., 2004; Lukashina et al., 2009, 2012).
Potato virus X has been used as model to study plant-virus interactions and to elucidate the plant-defense mechanisms and viral countermeasures such as suppression of silencing (Ruiz et al., 1998; Ratcliff et al., 1999). The plant is not a passive host during viral infection and viral RNA sensing induces the activation of defense responses that involve a complex set of proteins. The post-transcriptional RNA gene-silencing (PTGS) is a homology-dependent gene inactivation strategy that is triggered by the unusual presence in the plant cell of double stranded (ds) RNAs (Tenllado and Díaz-Ruíz, 2001). In the case of a viral infection, the PTGS response is activated by highly structured regions in the viral genomes or in the viral replication intermediates. These abnormal RNA structures are digested by a plant cell dsRNA-specific RNase, named Dicer, into 21–24 nucleotides RNA species (small interfering RNAs, siRNAs; Hamilton and Baulcombe, 1999). The siRNAs are then exploited through a sequence complementarity mechanism to recruit the homologous RNA into an RNA-induced silencing complex (RISC), where it is digested (Voinnet, 2005).
Also viral proteins contribute to trigger plant innate defense. In particular, the CP has been shown to work as pathogen avirulence effector of the Rx protein-mediated resistance (Bendahmane et al., 1995). Folding of the Rx protein is modified by CP binding and consequently nucleotide-binding site leucine-rich domains are exposed thus determining the activation of the signaling cascade and cell death (Moffett et al., 2002; Rairdan and Moffett, 2006).
Like other plant viruses, PVX has evolved mechanisms to counteract plant innate defense through one of its multifunctional protein, the TGBp1. The protein inhibits the formation of the effector complexes involved in RNA silencing (Bayne et al., 2005) by inducing, through the proteasome pathway, the degradation of the Argonaute 1 nuclease, one of the RISC components (Chiu et al., 2010).
Also the PVX-Potyviruses synergistic interaction that results in the enhancement of pathogenicity of PVX in Nicotiana benthamiana plants is somehow related to virus-induced gene silencing through the involvement of the Potyviruses P1/Helper component proteinase (HC-Pro; González-Jara et al., 2005; Pacheco et al., 2012).
To bring over these fundamental studies aimed to clarify the complex mechanisms underlying the plant-virus interaction, a number of viral expression vectors have been developed (Lico et al., 2008; Hefferon, 2012; Gleba et al., 2014; Peyret and Lomonossoff, 2015). First generation viral expression vectors usually harbor the cDNA form of the complete viral genome, sometimes engineered to easily insert foreign sequences as additional ORFs, or in substitution of a viral one, in association to a strong promoter (Chapman et al., 1992). These vectors can be directly used to infect the plant hosts, when the viral genome is under the control of a plant specific promoter, or used as template to generate in vitro an infectious transcript. In second-generation vectors, viral components are separately inserted into binary vectors and used to transform independently different Agrobacterium cells. Bacterial cells are then mixed together and used to co-infiltrate plant leaves (Gleba et al., 2014). Most of these vectors have found application in different technological fields.
In functional genomics studies, they are used to vehicle an endogenous gene fragment, triggering the specific suppression of the corresponding sequence in the genome (Ruiz et al., 1998; Baulcombe, 1999). This reverse genetics technique known as virus-induced gene silencing, is a high-throughput approach to the analysis of plant gene functions.
In “molecular farming,” PVX-based vectors are currently being used to vehicle and induce the expression in plants of foreign genes encoding high added-value biomolecules (Komarova et al., 2010).
Last, but only in the order of appearance list, these vectors appear extremely appealing also for nanotechnologies (Lico et al., 2013), as described in the following paragraphs.
PVX Nanoparticles for Subunit Vaccine Delivery
Subunit vaccines are formulations based on isolated pathogens components (proteins or peptides) that allow the activation of highly specific and protective immune responses. Nowadays, these vaccines are typically produced by recombinant DNA technologies. Compared to traditional inactivated or attenuated vaccines, subunit vaccines guarantee selectivity, specificity, low toxicity, stability and reduced risk of undesired side effects (Rappuoli, 2007). Main limitation of these vaccines is that isolated proteins or peptides being small (<10 nm) are per se unable to stimulate complete immune responses (innate, antibody, and cell-mediated). Indeed, it has been established that the efficiency of antigen uptake by antigen presenting cells (APCs) is strictly related to antigen sizes, and the larger surfaces of particulate antigens improve the interaction with APCs (Bachman and Jennings, 2010). To increase subunit vaccine immunogenicity, it is thus important to arrange isolated antigens in larger particles (20–200 nm). This is possible by entrapping them with adjuvants or by favoring self-assembly in supramolecular structures, such as in the case of virus-like particles generated by the self-assembly of viral capsid proteins (Rosenthal et al., 2014). A further possibility consists in delivering the antigen in association to nanoparticles, such as liposomes.
Many attempts have also been made to increase subunit vaccines immunogenicity and stability using genetically engineered plant virus nanoparticles as carriers for their delivery (Lico et al., 2007, 2013; Lebel et al., 2015; Streatfield et al., 2015). As for PVX, this provides for the fusion of the sequence encoding the immunogenic peptides or proteins in frame to the 5′-end of the gene encoding the CP in viral expression vectors. The N-terminus of the protein has indeed been demonstrated to be exposed on the external surface of the virion (Baratova et al., 1992). The modified viral expression vectors are then used to induce infection onset in plants and produce on large scale chimeric virus particles (CVPs) displaying on each CP subunit (approx. 1300 per virion) the (poly)peptide of interest (Figure 2). CVPs are then extracted from plant tissues and purified to be used in immunization trials.
FIGURE 2.
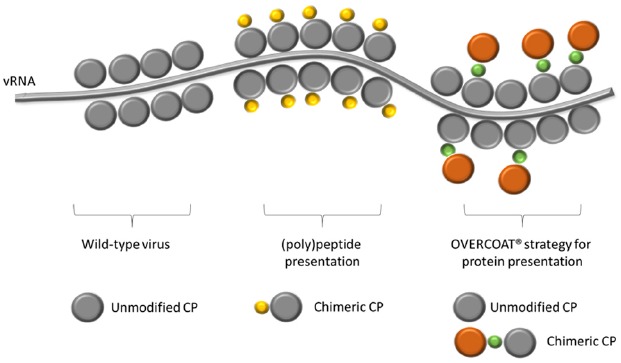
Schematic representation of a PVX nanoparticle, showing the viral RNA (vRNA) and the shell composition of: a wild-type virion; a chimeric virus particle exposing on every single CP unit (in gray) a copy of the heterologous (poly)peptide (in yellow); a partially chimeric virus particle in which only some of the CP units are fused to an entire protein (in orange) through the OVERCOAT® procedure. In green the FMDV 2A peptide.
Because the CP is involved in mediating the interaction with the plant environment, substantial modifications are not always tolerated in that they may interfere with both viral encapsidation and/or spread. These issues have been narrowly explored in the past years and some basic rules have been defined, whose compliance increases the chance of getting genetically stable infectious CVPs. Clearly, the shorter the exogenous (poly)peptide, the higher the probability that CP folding is preserved and compatible with efficient assembly. But length is not the unique parameter to be considered. Studies performed by fusing peptides (variable in length and amino acid composition) to a N-terminally truncated CP (a natural PVX mutant lacking the first 21 amino acids) have indicated that also isoelectric point, Ser/Thr and Trp content in the heterologous sequence may affect viral fitness (Lico et al., 2006). Recently, the insertion of a heterologous sequence between amino acids 23 and 24 at the N-terminus of the CP has also been reported (Vaculik et al., 2015). Nonetheless, up to now no CP domains other than the N-terminus have been identified that can harbor fusion peptides/proteins. Notably, the fusion to the C-terminus does not give rise to CVPs able to move throughout the plant (Cerovska et al., 2012). In fact, the C-terminus of the protein is crucial for the interaction with the viral RNA, hence for virion assembly (Nemykh et al., 2008).
On these bases, PVX particles displaying peptides of immunological relevance have been successfully produced in plants and their efficacy verified in animal models. PVX CVPs displaying the linear 2F5 epitope (2F5e) of the gp41 protein of Human Immunodeficiency Virus type 1 (HIV-1) envelope are able to induce the production of epitope-specific IgA and IgG, when administered intranasally or intraperitoneally in normal mice without the need to co-administer adjuvants. Notably, human dendritic cells pulsed with these CVPs are able to activate a 2F5e-specific human primary antibody response in severe combined immunodeficient mice engrafted with human lymphocytes. Finally, the antibodies induced by immunization with the CVPs are able to neutralize HIV-1 particles in viral neutralization assays (Marusic et al., 2001).
Potato virus X CVPs displaying a Major Histocompatibility Complex class I-restricted peptide of the nucleoprotein of influenza virus have also been engineered, taking into account mechanisms and limitations imposed by both plant virus biology and immune response. The synthetic DNA fragment encoding the influenza peptide was designed to be translated in an amino acid sequence optimized to: (i) obtain assembly of CVPs able to move systemically; (ii) minimize the risk of occurrence of unexpected mutations during viral replication; (iii) avoid detrimental effects of N-terminal modifications (such as acetylation) and improper C-terminal context on peptide processing by APCs. These rationally designed CVPs resulted to be genetically stable and triggered epitope-specific CD8+ T cells without the need of adjuvant co-delivery (Lico et al., 2009).
Overall, these data indicate that appropriately designed PVX CVPs are excellent epitope carriers endowed of adjuvant properties, probably not only because of the complex particulate structure but also because of the presence of the viral RNA that may trigger Toll-Like Receptor 7 on APCs (Jobsri et al., 2015). Altogether, these features account for the inherent ability of PVX to trigger a whole immune response, ideal condition to achieve complete protection from pathogens through vaccination.
Because one of the factors that may limit the construction of PVX CVPs is the length of the fused (poly)peptide, a “smart” procedure based on the OVERCOAT® PVX vector has also been devised to avoid possible steric hindrance of chimeric CPs during particle assembly (Santa Cruz et al., 1996). To this aim, a 16 amino acid linker peptide (2A) derived from the Foot and Mouth Disease Virus (FMDV) has been inserted between the foreign sequence and the CP. During translation, this sequence may induce a ribosomal skip with a frequency that is dependent on the nature of the 18–30 amino acids in the region immediately upstream the 2A cleavage site (Minskaia and Ryan, 2013; Minskaia et al., 2013). The incomplete FMDV 2A-mediated “cleavage” results in the production of a heterogeneous population of CPs including original and engineered version, both contributing to the assembly of fully functional CVPs able to spread throughout the plant (Figure 2). By this strategy, PVX particles were constructed displaying on the surface different (poly)peptides of immunological interest. In the case of the rotavirus major inner capsid protein (VP6; O’Brien et al., 2000) and the tuberculosis ESAT-6 antigen (Zelada et al., 2006), data demonstrated only the structural stability of the CVPs and the correct display of the foreign sequences. Subsequent papers demonstrated also the ability of these PVX CVPs to induce antibody responses in animal models (rabbits and mice). This is the case of the CVPs tailored to display the D2 peptide from Staphylococcus aureus fibronectin-binding protein (Brennan et al., 1999), two peptides from the E2 glycoprotein of the classical swine fever virus (Marconi et al., 2006) or the R9 peptide from the hypervariable region I of hepatitis C virus (Uhde-Holzem et al., 2010).
Alternative approaches to the construction of PVX CVPs, even if at higher costs, imply standard chemical protein conjugation procedures. In this case, the success depends on the presence of free amine groups (Lys residues) and carboxylate groups (Asp and Glu residues). These amino acids are all present in PVX CP. Different bioconjugation protocols have been tested to functionalize PVX, revealing that the carboxylates groups are either not solvent-exposed or not accessible. On the other hand, N-Hydroxysuccinimide (NHS)-ester based and Cu(I)-catalyzed azide-alkyne 1,3-dipolar cycloaddition (“CLICK”) reactions on amine groups are both feasible (Steinmetz et al., 2010).
Potato virus X particles displaying a peptide derived from the extracellular domain of the Human epidermal growth factor receptor 2 (HER2) have been constructed by conjugation to Lys on the CP surface through a two-step reaction via a heterofunctional NHS-(poly)ethyleneglycole (PEG)4-maleimide linker (Shukla et al., 2014a). HER2 is an antigen usually expressed on breast cancer cells and known to induce the activation of B lymphocytes and antibody production. The chemically derivatized PVX particles were tested in mice and demonstrated to be effective in inducing the overcoming of HER2 immunological tolerance by Enzyme Linked ImmunoSorbent Assay and confocal microscopy using HER2-positive human cancer cells.
PVX Nanoparticles for Imaging, Drug Delivery, and Diagnosis
Pioneering works in vaccine technology exploiting plant viruses such as PVX have worked as trailblazer in the field of nanotechnology. Indeed, the encouraging results prompted scientists in nanomedicine to broaden the exploration and exploitation of PVX particles as tools for targeted drug delivery or for diagnosis. In this perspective, the evaluation of the “biocompatibility” of viral particles, their natural tropism and biodistribution are fundamental to enter clinical trials. Indeed, the intrinsic toxicity cannot be taken for granted even if, for the animal cell, plant viruses are expected to behave as unreplicative and biologically safe nano-objects.
In vitro and in vivo experiments have been recently carried out to evaluate the toxicity and teratogenicity potential of unmodified PVX and Tomato Bushy Stunt Virus (TBSV), the type member of another viral genus and family, different in shape and dimensions (Blandino et al., 2015). Haemolysis assay was performed in vitro to determine detrimental effect of virus nanoparticles on human erythrocytes while toxicity and teratogenicity were evaluated in vivo through the early embryo assay (EEA). Chicken embryo is considered an excellent model to perform this kind of studies even if the reliability of the results depends on critical factors such as the choice of adequate controls, number of eggs per dose, and volume and route of delivery. Overall, the results of this study demonstrated that both plant viruses have neither cytotoxic effects on human erythrocytes nor toxic/teratogenic effects on chicken embryos. These data represent a good prerequisite for safety evaluation in view of forthcoming use of these nanoparticles as functionalized nanocarriers.
Biodistribution studies in vivo have been performed with PVX particles conjugated to various fluorescent dyes and to PEG with different chain lengths. PEG is frequently used to create a shield around biopharmaceuticals to increase solubility and stability and reduce immunogenicity even if, as a side effect, this masking reduces the interactions with cells, such as in the case of some plant virus nanoparticles (Raja et al., 2003; Schlick et al., 2005; Steinmetz and Manchester, 2009).
Preliminary studies in vitro, with human cervical cancer (HeLa) cells and murine fibroblasts (BalbCl7), highlight somewhat contradictory effects of PEGylation on PVX. In fact, this procedure effectively prevents interaction with cells of particles labeled with OregonGreen 488 (O488), while it promotes interaction if particles are labeled with Alexafluor 647 (A647; Steinmetz et al., 2010), and this is probably due to some intrinsic feature of the dye.
The analysis of the biodistribution and clearance of A647-labeled and PEGylated PVX in mice evidences that the viral nanoparticles remain mainly trapped by the mononuclear phagocyte system (MPS) that speeds up their removal from the circulation and from tissues. Nonetheless, the flexible filamentous PVX has enhanced tumor homing and tissue penetration, as compared to the icosahedral Cowpea Mosaic virus (Shukla et al., 2013, 2014b). Similarly, PVX CVPs constructed by the OVERCOAT® procedure and carrying the mCherry fluorescent protein but not conjugated to PEG, were shown to accumulate in the liver and to co-localize with macrophages for final clearance from tissues via MPS, 7 days post-administration in mice (Shukla et al., 2014c). It has been recently shown that it is possible to significantly improve PVX half-life in the serum reducing tissue accumulation, renal clearance as well as uptake by immune cells through derivatization of the viral surface with a PEG chain tailored in length and conformation (brush vs. mushroom; Lee et al., 2015).
Beside these early studies aimed to gain insights on PVX nanoparticles-based delivery, advanced studies have already started to exploit the virus as scaffold for the conjugation of chemical moieties, constructing nano-bullets for molecular imaging and targeted drug delivery.
A647-labeled PVX nanoparticles, bioconjugated with a 12 amino acid peptide with affinity to the epidermal growth factor receptor (EGFR), are effective in detection and imaging of carcinoma cell lines that upregulate EGFR, and prefer partitioning to cancer cells rather than to macrophages (Chariou et al., 2015). Furthermore, PVX particles conjugated to the Herceptin (Trastuzumab) monoclonal antibody specific for the peptide HER2 have been demonstrated to be able to induce the apoptosis of HER2 positive cell lines (Esfandiari et al., 2015).
Other PVX Nanoparticles Applications
Genetically or chemically modified PVX nanoparticles can be usefully applied also in fields other than vaccine and drug delivery or in vivo imaging.
A diagnostic kit for the early detection of the Sjogren’s Syndrome (SjS) has been recently developed (patent application No. 102015000020005 Italian Patent Office), based on PVX CVPs displaying a SjS diagnostic peptide. It has indeed been observed that serum antibodies recognize much less efficiently this soluble peptide if compared to the same peptide displayed on the viral scaffold so that the sensitivity of the assay is noticeably increased, preserving the specificity.
Potato virus X CVPs, displaying an antimicrobial decapeptide derived from an anti-idiotypic antibody that acts as a functional internal image of a microbicidal broad-spectrum yeast killer toxin, were active against Staphylococcus aureus and Candida albicans, as well as against the plant pathogens Erwinia carotovora, Botrytis cinerea, and Fusarium oxysporum. Moreover, they effectively protected plants from Pseudomonas syringae attacks (Donini et al., 2005).
Potato virus X CVPs carrying CPs fused to a single chain antibody specific to Diuron herbicide were purified from plant extracts by Diuron-based immune-capture, demonstrating their potential as tools for in situ applications including remediation of contaminated soils and waterways (Smolenska et al., 1998).
The fusion of GFP or mCherry protein to the PVX CP through the 2A peptide strategy allowed to follow infection progression throughout the plant, providing a non-invasive tool to the study of viral multiplication and spread (Santa Cruz et al., 1996, 1998; Roberts et al., 1997; Tilsner et al., 2013; Shukla et al., 2014c).
Potato virus X provides also opportunities for nanochemistry applications such as the construction of catalytic systems operating like metabolons in nature. A proof of concept has been published, in which the CVPs were constructed through the OVERCOAT® system to display Lipase B, an enzyme derived from Candida antarctica known to be an efficient enantiospecific catalyst for chemical hydrolysis. The virus-anchored lipase molecules retained their catalytic activity toward the substrate p-nitrophenyl caproate in solution (Carette et al., 2007). Unlike manmade supported catalysts, these PVX-based biocatalysts were easily produced using plant machinery and, because the virus particle is made of many proteins, they intrinsically possessed multiple catalytic sites.
Recently, PVX nanoparticles have also been investigated as scaffold for the attachment of metal atoms in an attempt to develop new bioinorganic materials for nanoelectronics. Platinum atom clusters (Pt nanoparticles, Pt-NPs) were built selectively at one end of the virus helical structure by chemical reduction, yielding virions that resemble a push-pin with a platinum head and a virus needle (Drygin et al., 2013). These results highlight the specificity of the coordinate metal interaction with a complex polyvalent biological structure, thus providing insights on the mechanisms of Pt-NPs formation, important to understand the interaction of viral particles with ionic metals. Chimeric PVX particles have also been constructed displaying multiple copies of a peptide that is known to induce the formation of SiO2. By the conditions adopted for synthesis, isolated SiO2 spots of about 10 nm were formed along the surface of the chimeric virus. The not uniform SiO2 coating allowed additional immunogold labeling of the particles. The possibility to combine silicification with immunolabeling provides a method to develop triple-hybrid complex structures, with controllable composition and functional properties (Van Rijn et al., 2015).
Conclusion
Overall, the knowledge of PVX, type-member of the Potexvirus genus, has evolved from basic molecular pathology to applications in biomedicine and material science.
The knowledge of plant-pathogen interactions and related silencing mechanisms has opened the way to the development of novel genetically-controlled strategies of plant infection, and through these to functional genomics and molecular farming.
By virtue of the innate versatility, PVX is now in the spotlight as a promising nanoobject for applications in vaccinology, drug targeting and delivery, diagnostic kits development and also as imaging reporter tool.
Last but not least, it is investigated as scaffold for enzymatic reactions or nanoelectronics.
For a long time, other plant viruses, mainly spherical or rigid rod-shaped, have been preferred to PVX for this kind of applications, but the data reviewed here clearly demonstrate that the flexuous mood of this nanoparticle is not a constraint rather a positive trait to be adequately exploited to respond to specific needs. Although many of the illustrated applications are still in their infancy, there is no reason to suppose they will not progress rapidly.
The shift of PVX from foe to friend has been made possible because this virus has been largely investigated in its innermost mechanisms. This cultural trail clearly witnesses how basic research can trigger a virtuous cycle of knowledge, removing existing interdisciplinary boundaries and opening up unimagined prospects, thus fuelling the development of innovative technologies.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
- Angell S. M., Davies C., Baulcombe D. C. (1996). Cell-to-cell movement of Potato virus X is associated with a change in the size-exclusion limit of plasmodesmata in tricomere cells of Nicotiana clevelandii. Virology 216, 197–201. 10.1006/viro.1996.0046 [DOI] [PubMed] [Google Scholar]
- Atabekov J. G., Rodionova N. P., Karpova O. V., Kozlovsky S. V., Novikov V. K., Arkhipenko M. V. (2001). Translational activation of encapsidated potato virus X RNA by coat protein phosphorylation. Virology 286, 466–474. 10.1006/viro.2001.1013 [DOI] [PubMed] [Google Scholar]
- Bachman M. F., Jennings G. T. (2010). Vaccine delivery: a matter of size, geometry, kinetics and molecular patterns. Nat. Rev. Immunol. 10, 787–796. 10.1038/nri2868 [DOI] [PubMed] [Google Scholar]
- Baratova L. A., Grebenshchikov N. I., Dobrov E. N., Gedrovich A. V., Kashirin I. A., Shishkov A. V., et al. (1992). The organization of potato virus X coat proteins in virus particles studied by tritium planigraphy and model building. Virology 188, 175–180. 10.1016/0042-6822(92)90747-D [DOI] [PubMed] [Google Scholar]
- Baratova L. A., Fedorova N. V., Dobrov E. N., Lukashina E. V., Kharlanov A. N., Nasonov V. V., et al. (2004). N-Terminal segment of potato virus X coat protein subunits in glycosylated and mediates formation of a bound water shell on the virion surface. Eur. J. Biochem. 271, 3136–3145. 10.1111/j.1432-1033.2004.04243.x [DOI] [PubMed] [Google Scholar]
- Batten J. S., Yoshinari S., Hemenway C. (2003). Potato virus X: a model system for virus replication, movement and gene expression. Mol. Plant Pathol. 4, 125–131. 10.1046/j.1364-3703.2003.00156.x [DOI] [PubMed] [Google Scholar]
- Baulcombe D. C. (1999). Fast forward genetics based on virus-induced gene silencing. Curr. Opin. Plant Biol. 2, 109–113. 10.1016/S1369-5266(99)80022-3 [DOI] [PubMed] [Google Scholar]
- Bayne E. H., Rakitina D. V., Morozov S. Y., Baulcombe D. C. (2005). Cell-to-cell movement of Potato Potexvirus X is dependent on suppression of RNA silencing. Plant J. 44, 471–482. 10.1111/j.1365-313X.2005.02539.x [DOI] [PubMed] [Google Scholar]
- Bendahmane A., Köhn B. A., Dedi C., Baulcombe D. C. (1995). The coat protein of potato virus X is a strain-specific elicitor of Rx1-mediated virus resistance in potato. Plant J. 8, 933–941. 10.1046/j.1365-313X.1995.8060933.x [DOI] [PubMed] [Google Scholar]
- Betti C., Lico C., Maffi D., D’Angeli S., Altamura M. M., Benvenuto E., et al. (2012). Potato virus X movement in Nicotiana benthamiana: new details revealed by chimeric coat protein variants. Mol. Plant Pathol. 13, 198–203. 10.1111/j.1364-3703.2011.00739.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blandino A., Lico C., Baschieri S., Barberini L., Cirotto C., Blasi P., et al. (2015). In vitro and in vivo toxicity evaluation of plant virus nanocarriers. Colloids Surf. B Biointerfaces 129, 130–136. 10.1016/j.colsurfb.2015.03.039 [DOI] [PubMed] [Google Scholar]
- Brennan F. R., Jones T. D., Longstaff M., Chapman S., Bellaby T., Smith H., et al. (1999). Immunogenicity of peptides derived from a fibronectin-binding protein of S. aureus expressed on two different plant viruses. Vaccine 17, 1846–1857. 10.1016/S0264-410X(98)00485-X [DOI] [PubMed] [Google Scholar]
- Carette N., Engelkamp H., Akpa E., Pierre S. J., Cameron N. R., Christianen P. C., et al. (2007). A virus-based biocatalyst. Nat. Nanotechnol. 2, 226–229. 10.1038/nnano.2007.76 [DOI] [PubMed] [Google Scholar]
- Cerovska N., Hoffmeisterova H., Moravec T., Plchova H., Folwarczna J., Synkova H., et al. (2012). Transient expression of Human papillomavirus type 16 L2 epitope fused to N- and C-terminus of coat protein of Potato virus X in plants. J. Biosci. 37, 125–133. 10.1007/s12038-011-9177-z [DOI] [PubMed] [Google Scholar]
- Chapman S., Kavanagh T., Baulcombe D. (1992). Potato virus X as a vector for gene expression in plants. Plant J. 2, 549–557. [DOI] [PubMed] [Google Scholar]
- Chariou P. L., Lee K. L., Wen A. M., Gulati N. M., Stewart P. L., Steinmetz N. F. (2015). Detection and imaging of aggressive cancer cells using an epidermal growth factor receptor (EGFR)-targeted filamentous plant virus-based nanoparticle. Bioconjug. Chem. 26, 262–269. 10.1021/bc500545z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu M. H., Chen I. H., Baulcombe D. C., Tsai C. H. (2010). The silencing suppressor P25 of Potato virus X interacts with Argonaute1 and mediates its degradation through the proteasome pathway. Mol. Plant Pathol. 11, 641–649. 10.1111/j.1364-3703.2010.00634.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davenport G. F., Baulcombe D. C. (1997). Mutation of the GKS motif of the RNA-dependent RNA polymerase from potato virus X disables or eliminates virus replication. J. Gen. Virol. 78, 1247–1251. 10.1099/0022-1317-78-6-1247 [DOI] [PubMed] [Google Scholar]
- Donini M., Lico C., Baschieri S., Conti S., Magliani W., Polonelli L., et al. (2005). Production of an engineered killer peptide in Nicotiana benthamiana by using a potato virus X expression system. Appl. Environ. Microbiol. 71, 6360–6367. 10.1128/AEM.71.10.6360-6367.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drygin Y., Kondakova O., Atabekov J. (2013). Production of platinum atom nanoclusters at one end of helical plant viruses. Adv. Virol. 2013:746796. 10.1155/2013/746796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esfandiari N., Arzanani M. K., Soleimani M., Kohi-Habibi M., Svendsen W. E. (2015). A new application of plant virus nanoparticles as drug delivery in breast cancer. Tumour. Biol. 10.1007/s13277-015-3867-3[Epub ahead of print]. [DOI] [PubMed] [Google Scholar]
- Gleba Y. Y., Tusé D., Giritch A. (2014). Plant viral vectors for delivery by Agrobacterium. Curr. Top. Microbiol. Immunol. 375, 155–192. 10.1007/82_2013_352 [DOI] [PubMed] [Google Scholar]
- González-Jara P., Atencio H. A., Martínez-García B., Barajas D., Tenllado F., Díaz-Ruiz J. R. (2005). A single amino acid mutation in the Plum pox virus helper component-proteinase gene abolishes both synergistic and RNA silencing suppression activities. Phytopathology 95, 894–901. 10.1094/PHYTO-95-0894 [DOI] [PubMed] [Google Scholar]
- Hamilton A. J., Baulcombe D. C. (1999). A species of small antisense RNA in posttranscriptional gene silencing in plants. Science 286, 950–952. 10.1126/science.286.5441.950 [DOI] [PubMed] [Google Scholar]
- Hefferon K. L. (2012). Plant virus expression vectors set the stage as production platforms for biopharmaceutical proteins. Virology 433, 1–6. 10.1016/j.virol.2012.06.012 [DOI] [PubMed] [Google Scholar]
- Howard A. R., Hepffer M. L., Ju H. J., Krishnamurthy K., Payton M. E., Verchot-Lubicz J. (2004). Potato virus X TGBp1 induces plasmodesmata gating and moves between cells in several host species whereas CP moves only in N. benthamiana leaves. Virology 328, 185–197. 10.1016/j.virol.2004.06.039 [DOI] [PubMed] [Google Scholar]
- Huisman M. J., Linthorst H. J., Bol J. F., Cornelissen J. C. (1988). The complete nucleotide sequence of potato virus X and its homologies at the amino acid level with various plus-stranded RNA viruses. J. Gen. Virol. 69, 1789–1798. 10.1099/0022-1317-69-8-1789 [DOI] [PubMed] [Google Scholar]
- Jobsri J., Allen A., Rajagopal D., Shipton M., Kanyuka K., Lomonossoff G. P., et al. (2015). Plant virus particles carrying tumour antigen activate TLR7 and Induce high levels of protective antibody. PLoS ONE 10:e0118096. 10.1371/journal.pone.0118096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalinina N. O., Rakitina D. A., Soloyev A. G., Schiemann J., Morozov S. Y. (2002). RNA helicase activity of the plant virus movement proteins encoded by the first gene of the triple gene block. Virology 296, 321–329. 10.1006/viro.2001.1328 [DOI] [PubMed] [Google Scholar]
- Karpova O. V., Zayakina O. V., Arkhipenko M. V., Sheval E. V., Kiselyova O. I., Poljakov V. Y., et al. (2006). Potato virus X RNA-mediated assembly of single-tailed ternary ‘coat protein-RNA-movement protein’ complexes. J. Gen. Virol. 87, 2731–2740. 10.1099/vir.0.81993-0 [DOI] [PubMed] [Google Scholar]
- Kendall A., McDonald M., Bian W., Bowles T., Baumgarten S. C., Shi J., et al. (2008). Structure of flexible filamentous plant viruses. J. Virol. 82, 9546–9554. 10.1128/JVI.00895-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komarova T. V., Baschieri S., Donini M., Marusic C., Benvenuto E., Dorokhov Y. L. (2010). Transient expression systems for plant-derived biopharmaceuticals. Expert. Rev. Vaccines 9, 859–876. 10.1586/erv.10.85 [DOI] [PubMed] [Google Scholar]
- Lebel M. E., Chartrand K., Leclerc D., Lamarre A. (2015). Plant viruses as nanoparticle-based vaccines and adjuvants. Vaccines 3, 620–637. 10.3390/vaccines3030620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K. L., Shukla S., Wu M., Ayat N. R., El Sanadi C. E., Wen A. M., et al. (2015). Stealth filaments: polymer chain length and conformation affect the in vivo fate of PEGylated potato virus X. Acta Biomater. 19, 166–179. 10.1016/j.actbio.2015.03.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leshchiner A. D., Solovyev A. G., Morozov S. Y., Kalinina N. O. (2006). A minimal region in the NTPase/helicase domain of the TGBp1 plant virus movement protein is responsible for ATPase activity and cooperative RNA binding. J. Gen. Virol. 87, 3087–3095. 10.1099/vir.0.81971-0 [DOI] [PubMed] [Google Scholar]
- Lico C., Baschieri S., Marusic C., Benvenuto E. (2007). “Molecular farming for antigen (vaccine) production in plants,” in Improvement of Crop Plants for Industrial End Uses, ed. Ranalli P. (Dordrecht, Netherland: Springer; ), 417–433. [Google Scholar]
- Lico C., Capuano F., Renzone G., Donini M., Marusic C., Scaloni A., et al. (2006). Peptide display on Potato virus X: molecular features of the coat protein-fused peptide affecting cell-to-cell and phloem movement of chimeric virus particles. J. Gen. Virol. 87, 3103–3112. 10.1099/vir.0.82097-0 [DOI] [PubMed] [Google Scholar]
- Lico C., Chen Q., Santi L. (2008). Viral vectors for production of recombinant proteins in plants. J. Cell. Physiol. 216, 366–377. 10.1002/jcp.21423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lico C., Mancini C., Italiani P., Betti C., Boraschi D., Benvenuto E., et al. (2009). Plant-produced potato virus X chimeric particles displaying an influenza virus-derived peptide activate specific CD8+ T cells in mice. Vaccine 27, 5069–5076. 10.1016/j.vaccine.2009.06.045 [DOI] [PubMed] [Google Scholar]
- Lico C., Schoubben A., Baschieri S., Blasi P., Santi L. (2013). Nanoparticles in biomedicine: new insights from plant viruses. Curr. Med. Chem. 20, 3471–3487. 10.2174/09298673113209990035 [DOI] [PubMed] [Google Scholar]
- Linnik O., Liesche J., Tilsner J., Oparka K. J. (2013). Unraveling the structure of viral replication complexes at super-resolution. Front. Plant Sci. 4:6. 10.3389/fpls.2013.00006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longstaff M., Brigneti G., Boccard F., Chapman S., Baulcombe D. (1993). Extreme resistance to potato virus X infection in plants expressing a modified component of the putative viral replicase. EMBO J. 12, 379–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukashina E., Badun G., Fedorova N., Ksenofontov A., Nemykh M., Serebryakova M., et al. (2009). Tritium planigraphy study of structural alterations in the coat protein of Potato virus X induced by binding of its triple gene block 1 protein to virions. FEBS J. 276, 7006–7015. 10.1111/j.1742-4658.2009.07408.x [DOI] [PubMed] [Google Scholar]
- Lukashina E., Ksenofontov A., Fedorova N., Badun G., Mukhamedzhanova A., Karpova O., et al. (2012). Analysis of the role of the coat protein N-terminal segment in Potato virus X virion stability and functional activity. Mol. Plant Pathol. 13, 38–45. 10.1111/j.1364-3703.2011.00725.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marconi G., Albertini E., Barone P., De Marchis F., Lico C., Marusic C., et al. (2006). In planta production of two peptides of the Classical Swine Fever Virus (CSFV) E2 glycoprotein fused to the coat protein of potato virus X. BMC Biotechnol. 6: 29. 10.1186/1472-6750-6-29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martelli G. P., Adams M. J., Kreuze J. F., Dolja V. V. (2007). Family Flexiviridae: a case study in virion and genome plasticity. Ann. Rev. Phytopathol. 45, 73–100. 10.1146/annurev.phyto.45.062806.094401 [DOI] [PubMed] [Google Scholar]
- Marusic C., Rizza P., Lattanzi L., Mancini C., Spada M., Belardelli F., et al. (2001). Chimeric plant virus particles as immunogens for inducing murine and human immune responses against human immunodeficiency virus type 1. J. Virol. 75, 8434–8439. 10.1128/JVI.75.18.8434-8439.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minskaia E., Nicholson J., Ryan M. D. (2013). Optimisation of the foot-and-mouth disease virus 2A co-expression system for biomedical applications. BMC Biotechnol. 13:67. 10.1186/1472-6750-13-67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minskaia E., Ryan M. D. (2013). Protein coexpression using FMDV 2A: effect of “linker” residues. Biomed. Res. Int. 2013:291730. 10.1155/2013/291730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moffett P., Farnham G., Peart J., Baulcombe D. C. (2002). Interaction between domains of a plant NBS-LRR protein in disease resistance-related cell death. EMBO J. 21, 4511–4519. 10.1093/emboj/cdf453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morozov S. Y., Lukasheva L. I., Chernov B. K., Skryabin K. G., Atabekov J. G. (1987). Nucleotide sequence of the open reading frames adjacent to the coat protein cistron in the potato virus X genome. FEBS Lett. 213, 438–442. 10.1016/0014-5793(87)81538-7 [DOI] [Google Scholar]
- Morozov S. Y., Miroshnichenko N. A., Solovyev A. G., Fedorkin O. N., Zelenina D. A., Lukasheva L. I., et al. (1991). Expression strategy of the potato virus X triple gene block. J. Gen. Virol. 72, 2039–2042. 10.1099/0022-1317-72-8-2039 [DOI] [PubMed] [Google Scholar]
- Nemykh M. A., Efimov A. V., Novikov V. K., Orlov V. N., Arutyunyan A. M., Drachev V. A., et al. (2008). One more probable structural transition in potato virus X virions and a revised model of the virus coat protein structure. Virology 373, 61–71. 10.1016/j.virol.2007.11.024 [DOI] [PubMed] [Google Scholar]
- O’Brien G. J., Bryant C. J., Voogd C., Greenberg H. B., Gardner R. C., Bellamy A. R. (2000). Rotavirus VP6 expressed by PVX vectors in Nicotiana benthamiana coats PVX rods and also assembles into virus-like particles. Virology 270, 444–453. 10.1006/viro.2000.0314 [DOI] [PubMed] [Google Scholar]
- Pacheco R., García-Marcos A., Barajas D., Martiáñez J., Tenllado F. (2012). PVX-potyvirus synergistic infections differentially alter microRNA accumulation in Nicotiana benthamiana. Virus Res. 165, 231–235. 10.1016/j.virusres.2012.02.012 [DOI] [PubMed] [Google Scholar]
- Parker L., Kendall A., Stubbs G. (2002). Surface features of potato virus X from fiber diffraction. Virology 300, 291–295. 10.1006/viro.2002.1483 [DOI] [PubMed] [Google Scholar]
- Peyret H., Lomonossoff G. P. (2015). When plant virology met Agrobacterium: the rise of the deconstructed clones. Plant Biotechnol. J. 13, 1121–1135. 10.1111/pbi.12412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rairdan G. J., Moffett P. (2006). Distinct domains in the ARC region of the potato resistance protein Rx mediate LRR binding and inhibition of activation. Plant Cell 18, 2082–2093. 10.1105/tpc.106.042747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raja K. S., Wang Q., Gonzalez M. J., Manchester M., Johnson J. E., Finn M. G. (2003). Hybrid virus-polymer materials. 1. Synthesis and properties of PEG-decorated cowpea mosaic virus. Biomacromolecules 4, 472–476. 10.1021/bm025740+ [DOI] [PubMed] [Google Scholar]
- Rappuoli R. (2007). Bridging the knowledge gaps in vaccine design. Nat. Biotechnol. 25, 1361–1366. 10.1038/nbt1207-1361 [DOI] [PubMed] [Google Scholar]
- Ratcliff F. G., MacFarlane S. A., Baulcombe D. C. (1999). Gene silencing without DNA. rna-mediated cross-protection between viruses. Plant Cell 11, 1207–1216. 10.1105/tpc.11.7.1207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts A. G., Cruz S. S., Roberts I. M., Prior D., Turgeon R., Oparka K. J. (1997). Phloem unloading in sink leaves of Nicotiana benthamiana: comparison of a fluorescent solute with a fluorescent virus. Plant Cell 9, 1381–1396. 10.1105/tpc.9.8.1381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenthal J. A., Chen L., Baker J. L., Putnam D., DeLisa M. P. (2014). Pathogen-like particles: biomimetic vaccine carriers engineered at the nanoscale. Curr. Opin. Biotechnol. 28, 51–58. 10.1016/j.copbio.2013.11.005 [DOI] [PubMed] [Google Scholar]
- Rozanov M. N., Koonin E. V., Gorbalenya A. E. (1992). Conservation of the putative methyltransferase domain: a hallmark of the ‘Sindbis-like’ supergroup of positive-strand RNA viruses. J. Gen. Virol. 73, 2129–2134. 10.1099/0022-1317-73-8-2129 [DOI] [PubMed] [Google Scholar]
- Ruiz M. T., Voinnet O., Baulcombe D. C. (1998). Initiation and maintenance of virus-induced gene silencing. Plant Cell 10, 937–946. 10.1105/tpc.10.6.937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santa Cruz S., Chapman S., Roberts A. G., Roberts I. M., Prior D. A., Oparka K. J. (1996). Assembly and movement of a plant virus carrying a green fluorescent protein overcoat. Proc. Natl. Acad. Sci. U.S.A. 93, 6286–6290. 10.1073/pnas.93.13.6286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santa Cruz S., Roberts A. G., Prior D. A., Chapman S., Oparka K. J. (1998). Cell-to-cell and phloem-mediated transport of potato virus X. The role of virions. Plant Cell 10, 495–510. 10.1105/tpc.10.4.495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlick T. L., Ding Z., Kovacs E. W., Francis M. B. (2005). Dual-surface modification of the tobacco mosaic virus. J. Am. Chem. Soc. 127, 3718–3723. 10.1021/ja046239n [DOI] [PubMed] [Google Scholar]
- Shukla S., Ablack A. L., Wen A. M., Lee K. L., Lewis J. D., Steinmetz N. F. (2013). Increased tumor homing and tissue penetration of the filamentous plant viral nanoparticle Potato virus X. Mol. Pharm. 10, 33–42. 10.1021/mp300240m [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shukla S., Wen A. M., Commandeur U., Steinmetz N. F. (2014a). Presentation of HER2 epitopes using a filamentous plant virus-based vaccination platform. J. Mater. Chem. B 2, 6249–6258. 10.1039/C4TB00749B [DOI] [PubMed] [Google Scholar]
- Shukla S., Wen A. M., Ayat N. R., Commandeur U., Gopalkrishnan R., Broome A. M., et al. (2014b). Biodistribution and clearance of a filamentous plant virus in healthy and tumor-bearing mice. Nanomedicine 9, 221–235. 10.2217/nnm.13.75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shukla S., Dickmeis C., Nagarajan A. S., Fischer R., Commandeur U., Steinmetz N. F. (2014c). Molecular farming of fluorescent virus-based nanoparticles for optical imaging in plants, human cells and mouse models. Biomater. Sci. 2, 784–797. 10.1039/c3bm60277j [DOI] [PubMed] [Google Scholar]
- Skryabin K. G., Kraev A. S., Morozov S. Y., Rozanov M. N., Chernov B. K., Lukasheva L. I., et al. (1988). The nucleotide sequence of potato virus X RNA. Nucleic Acids Res. 16, 10929–10930. 10.1093/nar/16.22.10929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smolenska L., Roberts I. M., Learmonth D., Porter A. J., Harris W. J., Wilson T. M., et al. (1998). Production of a functional single chain antibody attached to the surface of a plant virus. FEBS Lett. 441, 379–382. 10.1016/S0014-5793(98)01586-5 [DOI] [PubMed] [Google Scholar]
- Sonenberg N., Shatkin A. J., Ricciardi R. P., Rubin M., Goodman R. M. (1978). Analysis of terminal structures of RNA from potato virus X. Nucleic Acids Res. 5, 2501–2512. 10.1093/nar/5.7.2501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinmetz N. F., Manchester M. (2009). PEGylated viral nanoparticles for biomedicine: the impact of PEG chain length on VNP cell interactions in vitro and ex vivo. Biomacromolecules 10, 784–792. 10.1021/bm8012742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinmetz N. F., Mertens M. E., Taurog R. E., Johnson J. E., Commandeur U., Fischer R., et al. (2010). Potato virus X as a novel platform for potential biomedical applications. Nano Lett. 10, 305–312. 10.1021/nl9035753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Streatfield S. J., Kushnir N., Yusibov V. (2015). Plant-produced candidate countermeasures against emerging and reemerging infections and bioterror agents. Plant Biotechnol. J. 13, 1136–1159. 10.1111/pbi.12475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Syller J. (2012). Facilitative and antagonistic interactions between plant viruses in mixed infections. Mol. Plant Pathol. 13, 204–216. 10.1111/j.1364-3703.2011.00734.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenllado F., Díaz-Ruíz J. R. (2001). Double-stranded RNA-mediated interference with plant virus infection. J. Virol. 75, 12288–12297. 10.1128/JVI.75.24.12288-12297.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tilsner J., Linnik O., Wright K. M., Bell K., Roberts A. G., Lacomme C., et al. (2012). The TGB1 movement protein of Potato virus X reorganizes actin and endomembrane into the X-body, a viral replication factory. Plant Physiol. 158, 1359–1370. 10.1104/pp.111.189605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tilsner J., Linnik O., Louveaux M., Roberts I. M., Chapman S. N., Oparla K. J. (2013). Replication and trafficking of a plant virus are coupled at the entrances of plasmodesmata. J. Cell Biol. 201, 981–995. 10.1083/jcb.201304003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhde-Holzem K., Schlösser V., Viazov S., Fischer R., Commandeur U. (2010). Immunogenic properties of chimeric potato virus X particles displaying the hepatitis C virus hypervariable region I peptide R9. J. Virol. Methods 166, 12–20. 10.1016/j.jviromet.2010.01.017 [DOI] [PubMed] [Google Scholar]
- Vaculik P., Plchova H., Moravec T., Hoffmeisterova H., Cerovska N., Smahel M. (2015). Potato virus X displaying the E7 peptide derived from human papillomavirus type 16: a novel position for epitope presentation. Plant Cell Tiss. Organ Cult. 120, 671–680. 10.1007/s11240-014-0634-x [DOI] [Google Scholar]
- Van Rijn P., Van Bezouwen L. S., Fischer R., Boekema E. J., Böker A., Commandeur U. (2015). Virus-SiO2 and Virus-SiO2-Au hybrid particles with tunable morphology. Part. Part. Syst. Charact. 32, 43–47. 10.1002/ppsc.201400068 [DOI] [Google Scholar]
- Voinnet O. (2005). Induction and suppression of RNA silencing: insights from viral infections. Nat. Rev. Genet. 6, 206–220. 10.1038/nrg1555 [DOI] [PubMed] [Google Scholar]
- Voinnet O., Lederer C., Baulcombe D. C. (2000). A viral movement protein prevents spread of gene silencing signal in Nicotiana benthamiana. Cell 103, 157–167. 10.1016/S0092-8674(00)00095-7 [DOI] [PubMed] [Google Scholar]
- Zelada A. M., Calamante G., de la Paz Santangelo M., Bigi F., Verna F., Mentaberry A., et al. (2006). Expression of tuberculosis antigen ESAT-6 in Nicotiana tabacum using a potato virus X-based vector. Tuberculosis 86, 263–267. 10.1016/j.tube.2006.01.003 [DOI] [PubMed] [Google Scholar]


