Abstract
miRNA-20b has been shown to be aberrantly expressed in several tumor types. However, the clinical significance of miRNA-20b in the prognosis of patients with hepatocellular carcinoma (HCC) is poorly understood, and the exact role of miRNA-20b in HCC remains unclear. The aim of the present study was to investigate the association of the expression of miR-20b with clinicopathological characteristics and overall survival of HCC patients analyzed by Kaplan-Meier analysis and Cox proportional hazards regression models. Meanwhile, the HIF-1α and VEGF targets of miR-20b have been confirmed. We found not only miR-20b regulation of HIF-1α and VEGF in normal but also regulation of miR-20b in hypoxia. This mechanism would help the tumor cells adapt to the different environments thus promoting the tumor invasion and development. The whole study suggests that miR-20b, HIF-1α, and VEGF serve as a potential therapeutic agent for hepatocellular carcinoma.
1. Introduction
Hepatocellular carcinoma (HCC) represents an extremely poor prognostic cancer that remains one of the third causes of cancer-related death and aggressive human malignancies represented worldwide [1, 2]. The dismal outcome has been attributed to the major hallmarks of HCC, intrahepatic metastases or postsurgical recurrence [3]. With much progress made in surgery and other treatments, the prognosis of HCC patients is still unsatisfactory due to the high rate of recurrence and metastasis. Thus, it is crucial to improve our understanding of the molecular mechanisms underlying HCC which will be critical for the improvement of therapeutic strategies for HCC patients [4]. However, tumor metastasis is considered to be one of the most complex cell activities because it is a multistep process of cascade involving cell invasion and intramedullary pin into the blood and lymph vessels, survival and arrests in the vascular system, the proliferation of extravasation.
MicroRNAs (miRNAs) are a class of small noncoding RNA molecules that regulate gene expression by binding to partially complementary recognition sequences of target mRNAs, either repressing miRNA translation or cleaving target miRNA and RNAs that are highly conserved between species [5–8]. In recent years, increasing studies indicate that miRNAs have crucial functions in specific cellular processes such as differentiation, morphogenesis, and tumorigenesis and they are also considered as oncogenes and tumor suppressors [9–12]. Recently, it has been manifested that the deregulation or dysfunction of miRNAs is involved in cancer development and related to clinical outcomes of cancer patients including HCC [13]. In HCC, miRNAs have been discovered to be aberrantly expressed and some of them are functionally involved in HCC carcinogenesis, progression, and metastasis [14, 15]. However, the roles of a large number of miRNAs are still unexplored in HCC [4].
Hypoxia inducible factor-1 (HIF-1) is the first identified mediator of cell response to hypoxia in mammalian cells cultured under reduced oxygen tension [16]. The transcription factor HIF-1 consists of HIF-1α and HIF-1β and is a key regulator responsible for the induction of genes that facilitate adaptation and survival of tumor cells from hypoxic microenvironment and confer on the tumor a worse malignant phenotype [17, 18]. The overexpression of HIF-1α was found in various types of cancers of both human and mouse [17, 19]. The HIF-1α complex acts as a transcription factor for many target genes in several aspects of cancer progression including angiogenesis, glucose metabolism, cell proliferation, and apoptosis [20, 21]. Vascular endothelial growth factor (VEGF) is one of the major target genes for HIF-1α that directly participates in angiogenesis and a recognized therapeutic target [22, 23]. VEGF, the most potent angiogenic molecule, participates specifically in promoting vascular endothelial cell division, proliferation, and migration [24].
Some studies have shown that miRNA-20b is deregulated in several types of cancers [25–29] and upregulation of miRNA-20b correlates with worse prognosis; all these studies indicated that miRNA-20b acts as a tumor promoter [28, 30–34]. Some studies have shown that miRNA-20b modulates HIF-1α and VEGF to keep tumor adapting to different environment and promoting cell division, proliferation, and migration [27, 33, 34]. At present, the clinical significance of miRNA-20b in the prognosis of patients with HCC is poorly understood, and the exact role of miRNA-20b in HCC remains unclear. Here, we investigated the association between miRNA-20b expression and clinicopathological parameters and assessed the effect of miRNA-20b modulating HIF-1α and VEGF on biological behaviours including cell proliferation, apoptosis, and migration of HepG2 cells.
2. Materials and Methods
2.1. Tissue Samples
A total of 76 cases of HCCs tissues were obtained from patients collected by the Institute of General Surgical Research, Second Affiliated Hospital, Yangzhou University. In addition, 76 normal liver tissues were used as controls. Clinicopathological characteristics parameters are shown in Table 1. All of the HCC patients have not received previous treatments like local ablation, radiation therapy, chemoembolization, or chemotherapy.
Table 1.
Association between miR-20b expression and clinicopathological features of HCC.
| Variables | miRNA-20b expression high (n = 45) |
miRNA-20b expression low (n = 31) |
P |
|---|---|---|---|
| Age (years) | |||
| ≤57 | 20 | 18 | 0.243 |
| >57 | 25 | 13 | |
| Gender | |||
| Male | 32 | 25 | 0.356 |
| Female | 13 | 6 | |
| Tumor size (cm) | |||
| ≥5 | 27 | 15 | 0.317 |
| <5 | 18 | 16 | |
| AFP (ng/mL) | |||
| ≥400 | 19 | 17 | 0.279 |
| <400 | 26 | 14 | |
| Metastasis | |||
| Yes | 37 | 10 | 0.000∗ |
| No | 8 | 21 | |
| Tumor grade | |||
| G1 | 12 | 6 | 0.147 |
| G2 | 17 | 7 | |
| G3 | 16 | 18 | |
| TNM stage | |||
| I-II | 14 | 26 | 0.000∗ |
| III-IV | 31 | 5 | |
| Tumor recurrence | |||
| Yes | 32 | 14 | 0.023∗ |
| No | 13 | 17 | |
| Microvascular invasion | |||
| Yes | 29 | 5 | 0.016∗ |
| No | 16 | 26 |
∗ P < 0.05.
2.2. Cell Culture
The human HCC-derived cell lines HepG2 were provided from the Medical Academy of Yangzhou University and cells were cultured in Dulbecco's modified essential medium (DMEM, Invitrogen Corp., USA). Both media were supplemented with 10% heat-inactivated fetal bovine serum (Zhejiang Tianhang Biotechnology Co., Ltd., China), penicillin, and streptomycin, at 37°C in a humidified incubator with 5% CO2. Cellular hypoxia environments were stimulated with 300 mM CoCl2 (Sigma-Aldrich Co. LLC., USA) for 24 h.
2.3. RT-qPCR
Total RNA from cell lines or tissue samples was extracted using a mirVana miRNA Isolation Kit (Ambion, Austin, TX, USA) following the manufacturer's instructions. The purity and concentration of RNA samples were assessed by standard spectrophotometric methods 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA). Briefly, 5 ng of RNA was added to RT reaction, and then the cDNA served as the template for amplification of PCR with sequence-specific primers (Sangon Biotech, Shanghai, China) using SYBR PrimeScript miRNA RT-PCR kit (Takara Biotechnology Co. Ltd., Dalian, China). All reactions were run in triplicate on the iCycler iQ Multicolor Real-Time PCR Detection System (BioRad, Hercules, CA, USA). Small nucleolar RNA GAPDH was used as an internal standard for normalization. Each sample was run in duplicate for analysis. The change for mRNA in HCC tissues relative to the matched normal liver tissues was calculated using the 2−ΔΔCt method, where ΔΔCt = ΔCt HCC/normal liver tissues and ΔCt = CtmiR-20b − CtGAPDH.
For miR-20b, the primers were as follows: forward, 5′-TGTCAACGATACGCTACGA-3′ and reverse, 5′-GCTCATAGTGCAGGTAGA-3′; GAPDH forward, 5′-GTGGTCCAGGGTTTCTTACT-3′ and reverse, 5′-GTTGTCTCCTGCGACTTCA-3′; HIF-1α forward, 5′-AACGACAAGAAAAAGATAAGTTCT-3′ and reverse, 5′-GTTTGGTGTGGTTACATA-3′; VEGF forward, 5′-CAGGAACAAGGGCCTCTGTCT-3′ and reverse, 5′-TGTCCCTCTGACAATGTGCCATC-3′.
2.4. Transfection of miRNA
Transfection was performed using Lipofectamine 2000 (Invitrogen Life Technologies), in accordance with the manufacturer's instructions. For miRNA-20b functional analysis, the HepG2 cells were transfected with the scrambled miRNA as a negative control, miRNA-20b mimics, or miRNA-20b inhibitor (Ambion, Life Technologies, Grand Island, USA). For HIF-1α or VEGF functional analysis, the HepG2 cells were transfected with HIF-1α or VEGF-specific small interfering (si)RNA or pcDNA3.1-HIF-1α plasmid (Sangon Biotech, Shanghai, China). The transfection assay was performed as described in study [27].
2.5. Luciferase Reporter Gene Assay
The mRNA sequence targeted by the miRNA was predicted using TargetScan, miRanda, and NBmiRTar. The fragment was designated as HIF-1α 3′-UTR and inserted into pMIR-REPORTTM luciferase reporter vector (Sac I and Hind III restriction enzyme sites; Ambion, Life Technologies, Grand Island, USA). Another expressing vector was also constructed by the insertion of a mutated HIF-1α 3′-UTR using QuikChangeH Site-Directed Mutagenesis Kit (Stratagene, Santa Clara, CA). Then, the recombinant reporter vectors with normal and mutated HIF-1α 3′-UTR were cotransfected with miR-20b into HepG2 cells, respectively, using TransMessenger Transfection Reagent (Tiangen Biochemical Technology (Beijing) Co., Ltd., Beijing, China). The luciferase assay was performed according to the manufacturer's instructions.
2.6. Western Blot Analysis
Protein concentration was determined by BCA Protein Assay Kit (Santa Cruz, USA). About 30 μg protein extracts were electrophoresed on 10% sodium dodecyl sulfate-polyacrylamide gel (SDS-PAGE), transferred onto PVDF membranes (BioRad Laboratories, Hercules, CA, USA), and incubated for 1 h in TBS containing 5% nonfat milk and 0.1% Tween-20 incubated overnight at 4°C with the following primary antibodies: anti-HIF-1α (1 : 1000, BD Transduction Laboratories, USA). After washing in TBS with 0.1% Tween-20, they were incubated for 1 h at room temperature with HRP-conjugated anti-rabbit antibody (1 : 1,000, Santa Cruz Biotechnology Inc., USA). Immunoreactivity was detected by enhanced chemiluminescence (ECL kit, Santa Cruz Biotechnology Inc., USA) and visualized by autoradiography. The level of β-actin (1 : 1,000, Santa Cruz Biotechnology Inc., USA) was used as a control of the amount of protein loaded into each lane and the optical density of each band was measured using ImageJ.
2.7. Viability Assay
For the cell viability assay, cells were seeded into a 96-well plate in quintuplicate; the cell growth was measured by CellTiter 96 AQueous One Solution Cell Proliferation Assay (MTS; Promega (Beijing) Biotechnology Co., Ltd., Beijing, China) after the indicated periods. Absorbance was measured at 540 nm using Versamax microplate reader (Molecular Devices, Sunnyvale, CA).
2.8. Annexin V-FITC/PI Determination of Apoptosis
Add precooled 70% ethanol to fixed cells. Take 1 mL of the cell suspension and add it to a centrifuge tube, centrifuge (4°C, 1000 r/min, 5 min). Use 1x binding buffer to adjust 1 × 106/mL; take 100 μL cell suspension to detect cell apoptosis, according to the Annexin V-FITC/PI kit (Nanjing Kaiji Biological Technology Development Co., Ltd., Nanjing, China) specification method steps.
2.9. Statistical Analysis
Data are presented as the mean ± standard deviation (SD). The significance of differences was evaluated by t-test. Differences with P values <0.05 were considered statistically significant. The postoperative survival rate was analyzed with the Kaplan-Meier method, and differences in survival rates were assessed with the log-rank test. All statistical analyses were performed using GraphPad Prism 6 software (GraphPad Software, San Diego, CA, USA).
3. Result
3.1. miR-20b Is Upregulated in HCC Tissues
From the qRT-PCR result, we found that miR-20b expression levels were significantly upregulated in HCC cancer tissues compared to those in the normal control (P = 0.000, Figure 1).
Figure 1.
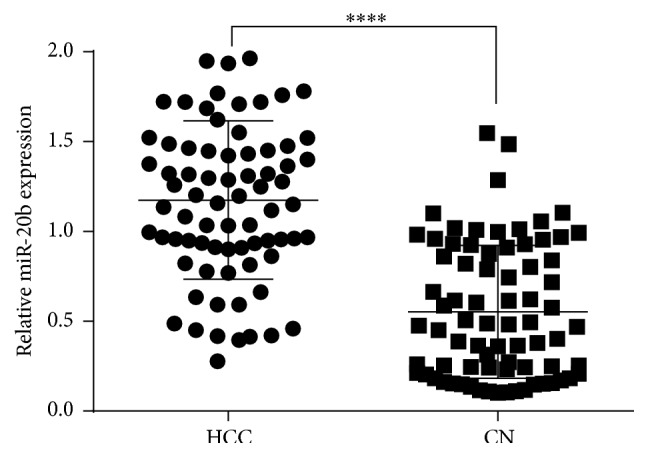
Comparison of miR-20b expression levels between HCC tissues and normal tissues (P = 0.000).
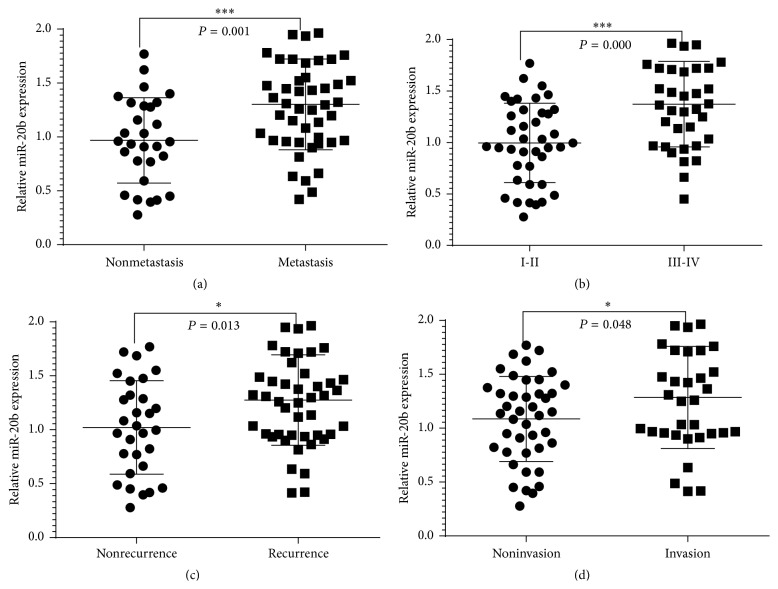
Correlation between levels of miR-20b expression and clinicopathological characteristics of HCC patients was observed.
The miR-20b expression levels were classified as high or low in relation to the median value. Meanwhile, we found the expression levels of miR-20b in HCC patients had no significant correlation with gender, age, tumor size, AFP (ng/mL), and tumor grade (P > 0.05; Table 1). But when comparing miR-20b expression in tumor metastasis, TNM stage, tumor recurrence, and microvascular invasion we found a significant difference (Figure 2).
Figure 2.
The correlation between levels of miR-20b and tumor metastasis, TNM stage, tumor recurrence, and microvascular invasion. (a) Tumor metastasis (P = 0.001), TNM stage (P = 0.000), tumor recurrence (P = 0.013), and microvascular invasion (P = 0.048).
3.2. miR-20b Expression and Postoperative Survival
Kaplan-Meier survival curve and log-rank test for 76 patients with HCC with high expression or low expression of miRNA-20b in tumor tissue were analyzed. The overall survival (OS) rate of HCC patients was significantly lower with high miR-20b mRNA expression than that in those with low expression; it means that high miR-20b expression might be correlated with poor prognosis of HCC patients (P = 0.01; Figure 3). We also use univariate and multivariate Cox model analyses which were performed to determine the correlation of miR-20b expression with overall survival of HCC patients. In univariate analysis the found metastasis, TNM stage, tumor recurrence, microvascular invasion, and miR-20b were statistically significant prognosis factors (Table 2). A multivariate analysis confirmed that metastasis, TNM stage, microvascular invasion, and miR-20b expression were significant independent predictors of poor survival of HCC (Table 3).
Figure 3.
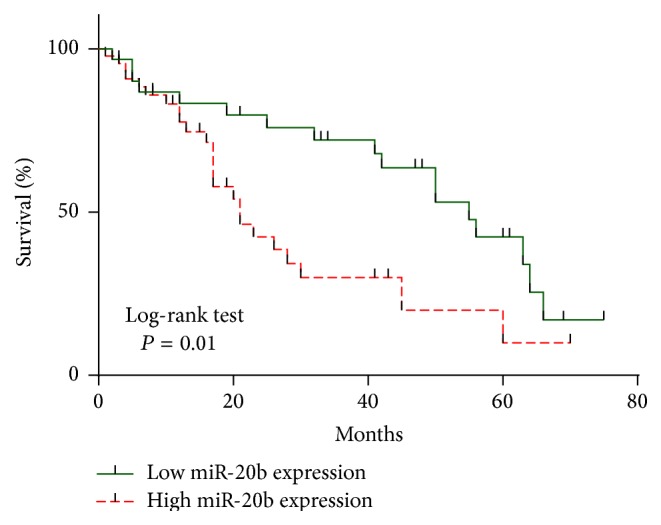
Kaplan-Meier survival curve of HCC patients. Patients in the high expression group had significantly poorer prognosis than those in low expression group, analyzed by log-rank tests which are indicated (P = 0.01).
Table 2.
Univariate analysis of clinicopathological factors for overall survival.
| Variable | n | Hazard ratio | 95% CI | P value |
|---|---|---|---|---|
| Age (years) | ||||
| ≤57 | 38 | 1 | 0.866–1.417 | 0.350 |
| >57 | 28 | 0.728 | ||
| Gender | ||||
| Male | 57 | 1 | 0.912–3.749 | 0.088 |
| Female | 19 | 1.849 | ||
| Tumor size (cm) | ||||
| ≥5 | 42 | 1 | 0.264–1.116 | 0.097 |
| <5 | 34 | 0.543 | ||
| AFP (ng/mL) | ||||
| ≥400 | 36 | 1 | 0.387–1.506 | 0.436 |
| <400 | 40 | 0.434 | ||
| Metastasis | ||||
| Yes | 47 | 1 | 1.396–5.647 | 0.004 ∗ |
| No | 29 | 2.808 | ||
| Tumor grade | ||||
| G1 | 18 | 1 | 0.710–2.751 | 0.333 |
| G2 | 24 | 1.398 | ||
| G3 | 34 | |||
| TNM stage | ||||
| I-II | 40 | 1 | 1.626–6.946 | 0.001 ∗ |
| III-IV | 36 | 3.360 | ||
| Tumor recurrence | ||||
| Yes | 46 | 1 | 0.171–0.657 | 0.007 ∗ |
| No | 30 | 0.360 | ||
| Microvascular invasion | ||||
| Yes | 34 | 1 | 1.187–5.931 | 0.017 ∗ |
| No | 42 | 2.654 | ||
| miR-20b | ||||
| High | 45 | 1 | 0.129–0.528 | 0.000 ∗ |
| Low | 31 | 0.261 |
∗ P < 0.05.
Table 3.
Multivariate analysis of clinicopathological factors for overall survival.
| Variable | n | Hazard ratio | 95% CI | P value |
|---|---|---|---|---|
| Metastasis | ||||
| Yes | 47 | 0.295 | 0.124–0.696 | 0.005 ∗ |
| No | 29 | |||
| TNM stage | ||||
| I-II | 40 | 5.031 | 1.627–15.558 | 0.005 ∗ |
| III-IV | 36 | |||
| Tumor recurrence | ||||
| Yes | 46 | 1.418 | 0.522–3.854 | 0.493 |
| No | 30 | |||
| Microvascular invasion | ||||
| Yes | 34 | 0.215 | 0.088–0.527 | 0.001 ∗ |
| No | 42 | |||
| miR-20b | ||||
| High | 45 | 5.018 | 2.325–11.223 | 0.000 ∗ |
| Low | 31 |
∗ P < 0.05.
3.3. Inverse Level of miR-20b and HIF-1α, VEGF in HepG2 Cells under Normal or Hypoxia-Mimetic Conditions
To further investigate the role of miR-20b in the regulation of HIF-1α, VEGF expression in normal and hypoxia-mimetic conditions was analyzed. We used CoCl2 to mimic hypoxia conditions, the cells treatment of CoCl2 for 24 h. The levels of miR-20b and HIF-1α, VEGF expression in normal and hypoxic conditions were analyzed (Figure 4). Then the HepG2 cells were transfected with scrambled miRNA, miR-20b mimics, and miR-20b inhibitor, respectively. The transfection efficiency is as shown in Figure 5.
Figure 4.
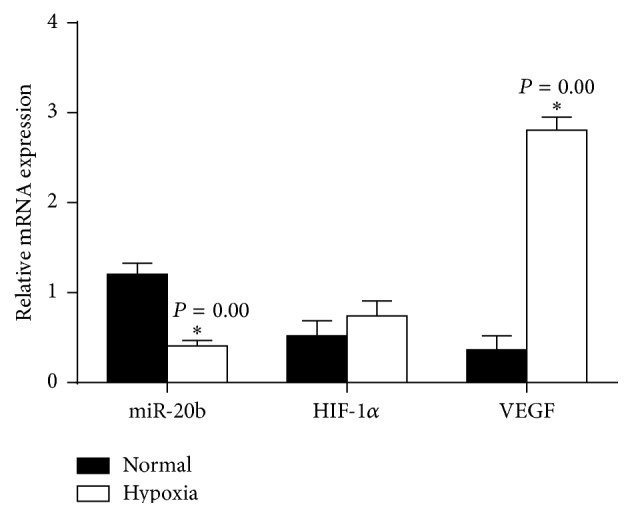
The levels of miR-20b and HIF-1α, VEGF expression after treatment of CoCl2 for 24 h. ∗∗ P < 0.01 compared with normal group.
Figure 5.
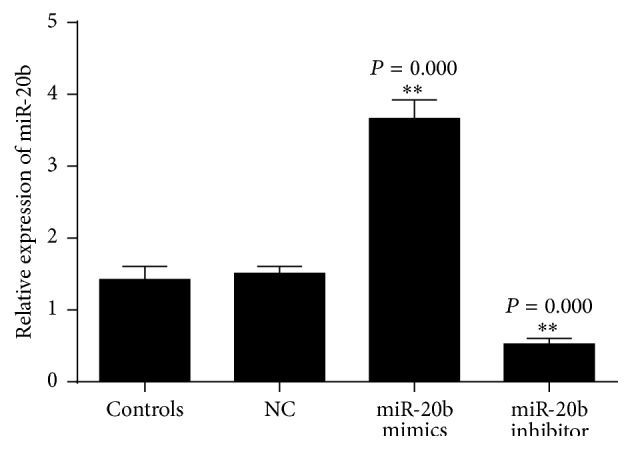
The transcription efficiency was performed to determine the levels of miR-20b transfected with scrambled miRNA (NC), miR-20b mimics, and miR-20b inhibitor. ∗∗ P < 0.05 compared with control.
3.4. HIF-1α, VEGF Are Direct Targets of miR-20b
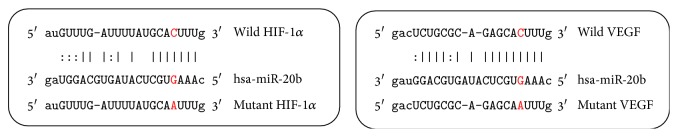
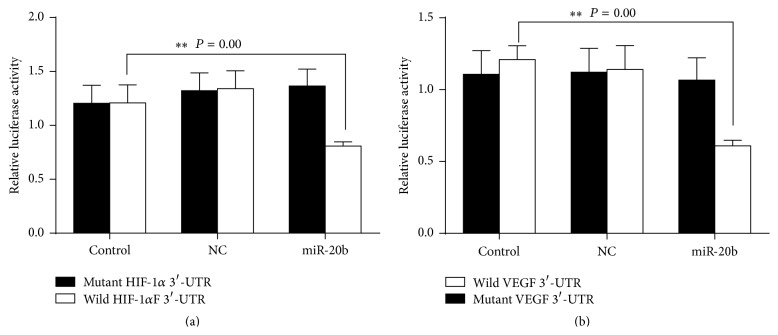
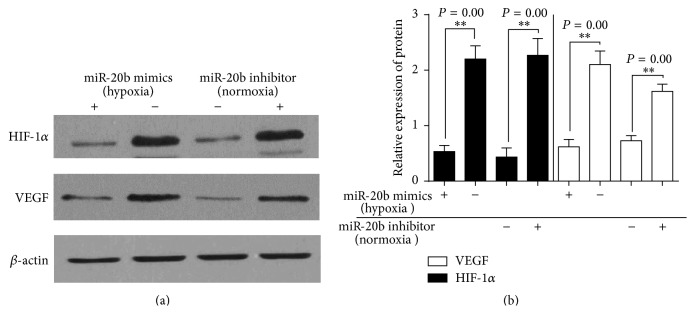
Bioinformatical predication was performed using TargetScan, miRanda, and NBmiRTar to predict targets of miR-20b. We found miR-20b at the 3′-UTR of HIF-1α, VEGF which were highly conserved (Figure 6). To verify whether HIF-1α, VEGF were direct targets of miR-20b, the wild and mutant types of HIF-1α, VEGF 3′-UTR were generated. The dual-luciferase reporter assay was subsequently performed in hepatocellular carcinoma HepG2 cells. As shown in Figures 7(a) and 7(b), the luciferase activity was significantly reduced in HepG2 cells cotransfected with the wild-type 3′-UTR of HIF-1α, VEGF and miR-20b mimics but unchanged in HepG2 cells cotransfected with the mutant HIF-1α, VEGF 3′-UTR and miR-20b mimics, indicating that miR-20b directly binds to the 3′-UTR of HIF-1α, VEGF in HepG2 cells. We transfected miR-20b mimics and miR-20b inhibitor to HepG2 cells. The miR-20b inhibitor was transfected into normoxic cells; miR-20b mimics were transfected into hypoxic cells and the HIF-1α, VEGF expression detected by Western blot (Figure 8).
Figure 6.
miR-20b targeted 3′-UTR of HIF-1α, VEGF mRNA.
Figure 7.
Luciferase reporter assay in HepG2 cells. Cells were cotransfection of HepG2 cells with miR-20b mimics and wild-type HIF-1α, VEGF 3′-UTR led to a marked decrease in luciferase activity and cotransfection with miR-20b and mutant HIF-1α, VEGF 3′-UTR had no effect on luciferase activity, and cotransfection with NC miRNA and wild-type HIF-1α, VEGF 3′-UTR or mutant HIF-1α, VEGF 3′-UTR also showed no difference, ∗∗ P < 0.05 compared with negative control.
Figure 8.
In hypoxia group showing HIF-1α, VEGF decreased after miR-20b treated transfected. In normal group miR-20b inhibitor increased HIF-1α, VEGF protein by Western blot (a). Use ImageJ to analyze relative expression of proteins (b). ∗∗ P < 0.05 compared with each control group.
3.5. miR-20b Enhances HepG2 Cell Proliferation
The MTS assay was used to investigate the miR-20b effects cell viability. We transfected miR-20b mimics into cells; the cell viabilities significantly increased after transfected miR-20b mimics for 24 h in normoxia comparison with that of negative control (miR-NC) (Figure 9; ∗∗ P < 0.01). We also transfected miR-20b inhibitor into cells in normoxia; the cell viabilities were dramatically restrained compared with that of negative control (inhibitor-NC) transfected cells (Figure 9; ∗∗ P < 0.01), to verify the ways miR-20b affects tumor cells growth. Furthermore we also transfected inhibitor + si-HIF-1α or inhibitor + si-VEGF into cells. When the cells were cotransfected with miR-20b inhibitor and HIF-1α-siRNA or VEGF-siRNA the cell viabilities significantly increased compared with only transfected miR-20b inhibitor (Figure 9; ∗∗ P < 0.01).
Figure 9.
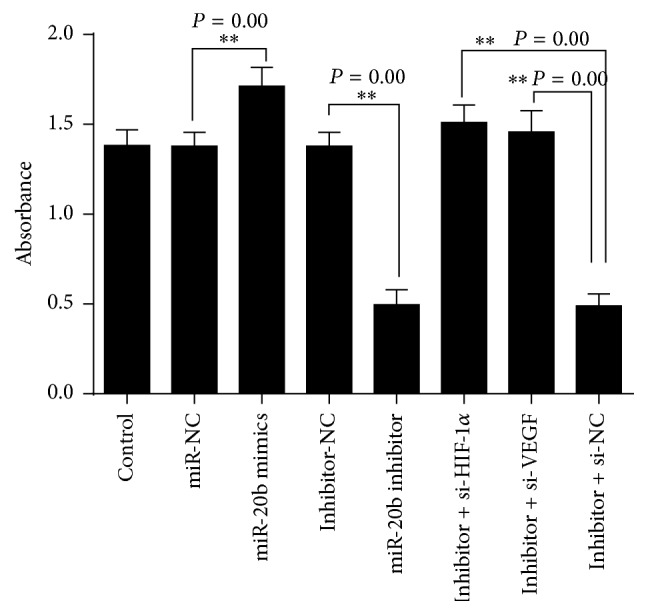
The cells viability detected by MTS assays. HepG2 cells were transfected with miR-20b mimics, miR-20b inhibitor, inhibitor + si-HIF-1α, and inhibitor + si-VEGF. Inhibitor + si-HIF-1α: miR-20b inhibitor + HIF-1α-siRNA; inhibitor + si-VEGF: miR-20b inhibitor + VEGF-siRNA. ∗∗ P < 0.01 compared with control.
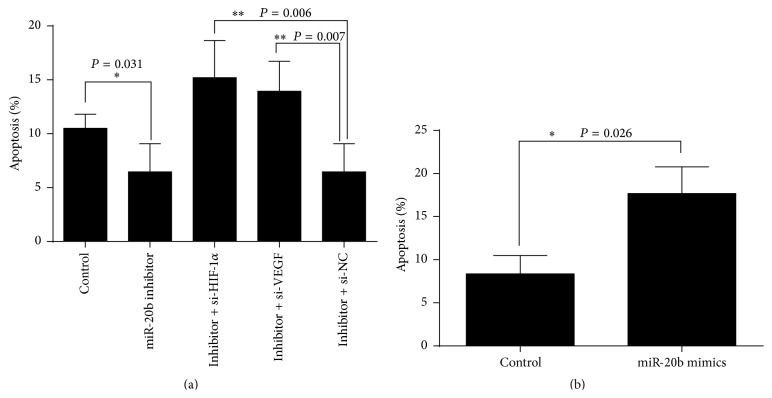
3.6. Downregulation of miR-20b Enhanced the Resistance to Apoptosis
The Annexin V-FITC/PI assay was used to investigate the cell apoptosis effects of miR-20b. We transfected miR-20b inhibitor into cells; the cell apoptosis significantly decreased in comparison with that of control in normal environment (Figure 10(a); ∗ P < 0.05). Furthermore we also transfected inhibitor + si-HIF-1α or inhibitor + si-VEGF into cells for 24 h in normoxia. When the cells were cotransfected with miR-20b inhibitor and HIF-1α-siRNA or VEGF-siRNA the cell apoptosis significantly increased compared with only transfected control group (Figure 10(a); ∗∗ P < 0.01). By the way, we transfected miR-20b mimics for 24 h and used CoCl2 to mimetic hypoxia conditions; the cells treatment of CoCl2 for 24 h was then detected by flow cytometry. Comparison with control group transfected miR-20b mimics the cells apoptosis significantly increased (Figure 10(b); ∗ P < 0.05).
Figure 10.
The cells apoptosis detected by Annexin V-FITC/PI assay. HepG2 cells were transfected with miR-20b inhibitor, inhibitor + si-HIF-1α, and inhibitor + si-VEGF. Inhibitor + si-HIF-1α: miR-20b inhibitor + HIF-1α-siRNA; inhibitor + si-VEGF: miR-20b inhibitor + VEGF-siRNA. ∗∗ P < 0.01 compared with control in normoxia (a). Transfected miR-20b mimics for 24 h in mimetic hypoxia conditions; apoptosis significantly increased in hypoxia conditions (b).
4. Discussion
In the present study, many miRNAs have confirmed contribution to the initiation and progression of HCC. With further in-depth studies more and more miRNAs have shown that take part in the regulation of hepatocellular carcinoma development. Furthermore, it has been shown that miRNAs can function as tumor suppressors or oncogenes and repress the expression of important cancer-related genes and might prove useful biomarkers in the diagnosis and treatment of cancers [35]. The important role in hepatocellular carcinoma tells us that understanding of miRNA function will provide us with broad prospects to understand and overcome tumor in the future.
miR-20b belongs to the miR-106a-363 cluster, which together with miR-17-92 and miR-106b-25 clusters forms a large family of highly similar miRNAs called the miR-17 family [28]. In the present study, the high expression levels of miR-20b often promote tumor development so the miR-20b suggested can serve as a potential oncogene. In our study we found that miR-20b expression levels were significantly upregulated in HCC cancer tissues compared to those in the normal control (P = 0.000, Figure 1). Furthermore, miR-20b expression showed significant association with tumor metastasis, TNM stage, tumor recurrence, and microvascular invasion comparison of clinicopathological factors which is an important clinical determinant for the prognosis of HCC patients. In Kaplan-Meier survival curve analysis, OS rates of HCC patients rates indicated that with high miR-20b expression there was significantly poorer survival in comparison with low miR-20b expression. In a multivariate Cox model, we found that high miR-20b expression was an independent factor for predicting the 5-year OS of HCC patients. From clinicopathological characteristics study we found that the expression of miR-20b was upregulated in HCC. High expression of miR-20b was significantly associated with tumor progression and decreased OS in patients of HCC indicating that it might play critical roles in HCC progression and development.
Based on our clinicopathological characteristics study, we further investigated the function and possible mechanisms of miR-20b in regulating some biological properties of HepG2 cells. We confirmed that miR-20b binds with the HIF-1α and VEGF 3′-UTR by luciferase reporter assay. Under normoxic conditions, posttranslational HIF-1α is rapidly degraded by the proteasome and usually not detectable and HIF-1α is a mediator of cell response to hypoxia in mammalian cells cultured under reduced oxygen tension [24]. Among the target genes of HIF-1α, vascular endothelial growth factor (VEGF) is one of the major target genes for HIF-1α that directly participates in angiogenesis [36]. We used CoCl2 in mimetic hypoxia conditions and found inverse level of miR-20b and HIF-1α, VEGF in HepG2 cells under normal or hypoxia-mimetic conditions. From the result we found that in hypoxia group miR-20b expression significantly decreased and VEGF mRNA significantly increased; the hypoxia conditions are not to change HIF-1α mRNA expression but significantly increase HIF-1α protein. To verify relationship between miR-20b and HIF-1α, VEGF expression, we also transfected miR-20b mimics and miR-20b inhibitor to HepG2 cells. The miR-20b inhibitor was transfected into normoxic cells; miR-20b mimics were transfected into hypoxic cells and the HIF-1α, VEGF expression detected by Western blot. As a result we found HIF-1α and VEGF are targets by miR-20b. In normal condition HepG2 highly expresses miR-20b but reduces HIF-1α, VEGF. Interestingly, once we put the cells in hypoxia-mimetic conditions the cell response to hypoxia would highly express HIF-1α, VEGF and then reduced levels of miR-20b.
In present studies reported, miR-20b expression could affect cell biological behaviour [28, 31, 32, 34, 37]. We used the loss-of-function and gain-of-function approaches which showed that miR-20b could impact the cell viability and apoptosis. The miR-20b mimics and miR-20b inhibitor were transfected into HepG2 cells in normoxic condition. In miR-20b mimics group, the cells viability significant increased compared with control group. While in miR-20b inhibitor group the cells viability decreased significantly, at some time we cotransfected miR-20b inhibitor and HIF-1α-siRNA or VEGF-siRNA; the cells proliferative capacity has been enhanced. The result tells us that miR-20b maintains tumor cell growth through its regulation of HIF-1α and VEGF in normal condition. The cells apoptosis result also suggests that high expression of HIF-1α and VEGF in normal condition would help cells to resist apoptosis. Considering the regulation of apoptosis genes by HIF-1α and VEGF [38, 39], when in hypoxia-mimetic conditions we transfected miR-20b mimics into cells; it is means that we also decreased levels of HIF-1α and VEGF which lead to cells apoptosis raised.
In conclusion, in the present study we report that miR-20b is associated with poor overall survival of HCC patients, suggesting its potential prognostic values in this disease type, and identified HIF-1α and VEGF as direct targets of miR20b in hepatocellular carcinoma cells. From our cells studies we found not only miR-20b regulation of HIF-1α and VEGF in normal but also HIF-1α and VEGF regulation of miR-20b in hypoxia. This mechanism would help the tumor cells adapt to the different environments thus promoting the tumor invasion and development.
Acknowledgments
This work was supported in part by grants from the Yangzhou Scientific and Technological Project (YZ2014064) and the Project Sponsored by the Institute of General Surgical Research, Second Affiliated Hospital, Yangzhou University.
Ethical Approval
The study was approved by the Institute of General Surgical Research, Second Affiliated Hospital, Yangzhou University, Jiangsu, China. All specimens were handled and made anonymous according to the ethical and legal standards.
Consent
The authors obtained written informed consent from all participants involved in their study.
Disclosure
Tong-min Xue, Li-de Tao, and Miao Zhang are listed as co-first authors with equal contribution to the paper.
Conflict of Interests
The authors have no conflict of interests to disclose.
References
- 1.Parkin D. M., Bray F., Ferlay J., Pisani P. Global cancer statistics, 2002. CA: A Cancer Journal for Clinicians. 2005;55(2):74–108. doi: 10.3322/canjclin.55.2.74. [DOI] [PubMed] [Google Scholar]
- 2.Thorgeirsson S. S., Grisham J. W. Molecular pathogenesis of human hepatocellular carcinoma. Nature Genetics. 2002;31(4):339–346. doi: 10.1038/ng0802-339. [DOI] [PubMed] [Google Scholar]
- 3.Budhu A., Jia H.-L., Forgues M., et al. Identification of metastasis-related microRNAs in hepatocellular carcinoma. Hepatology. 2008;47(3):897–907. doi: 10.1002/hep.22160. [DOI] [PubMed] [Google Scholar]
- 4.Chen P., Zhao X., Ma L. Downregulation of microRNA-100 correlates with tumor progression and poor prognosis in hepatocellular carcinoma. Molecular and Cellular Biochemistry. 2013;383(1-2):49–58. doi: 10.1007/s11010-013-1753-0. [DOI] [PubMed] [Google Scholar]
- 5.Bartel D. P. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116(2):281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 6.He L., Hannon G. J. MicroRNAs: small RNAs with a big role in gene regulation. Nature Reviews Genetics. 2004;5(7):522–531. doi: 10.1038/nrg1379. [DOI] [PubMed] [Google Scholar]
- 7.Nelson P., Kiriakidou M., Sharma A., Maniataki E., Mourelatos Z. The microRNA world: small is mighty. Trends in Biochemical Sciences. 2003;28(10):534–540. doi: 10.1016/j.tibs.2003.08.005. [DOI] [PubMed] [Google Scholar]
- 8.Chen P., Xi Q., Wang Q., Wei P. Downregulation of microRNA-100 correlates with tumor progression and poor prognosis in colorectal cancer. Medical Oncology. 2014;31, article 235 doi: 10.1007/s12032-014-0235-x. [DOI] [PubMed] [Google Scholar]
- 9.Ventura A., Jacks T. MicroRNAs and cancer: short RNAs go a long way. Cell. 2009;136(4):586–591. doi: 10.1016/j.cell.2009.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Iorio M. V., Croce C. M. MicroRNAs in cancer: small molecules with a huge impact. Journal of Clinical Oncology. 2009;27(34):5848–5856. doi: 10.1200/jco.2009.24.0317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Esquela-Kerscher A., Slack F. J. Oncomirs—microRNAs with a role in cancer. Nature Reviews Cancer. 2006;6(4):259–269. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- 12.Ambros V. The functions of animal microRNAs. Nature. 2004;431(7006):350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 13.Wang J., Li J., Shen J., Wang C., Yang L., Zhang X. MicroRNA-182 downregulates metastasis suppressor 1 and contributes to metastasis of hepatocellular carcinoma. BMC Cancer. 2012;12, article 227 doi: 10.1186/1471-2407-12-227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Braconi C., Henry J. C., Kogure T., Schmittgen T., Patel T. The role of microRNAs in human liver cancers. Seminars in Oncology. 2011;38(6):752–763. doi: 10.1053/j.seminoncol.2011.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Negrini M., Gramantieri L., Sabbioni S., Croce C. M. microRNA involvement in hepatocellular carcinoma. Anti-Cancer Agents in Medicinal Chemistry. 2011;11(6):500–521. doi: 10.2174/187152011796011037. [DOI] [PubMed] [Google Scholar]
- 16.Semenza G. L., Wang G. L. A nuclear factor induced by hypoxia via de novo protein synthesis binds to the human erythropoietin gene enhancer at a site required for transcriptional activation. Molecular and Cellular Biology. 1992;12(12):5447–5454. doi: 10.1128/mcb.12.12.5447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Weidemann A., Johnson R. S. Biology of HIF-1alpha. Cell Death and Differentiation. 2008;15(4):621–627. doi: 10.1038/cdd.2008.12. [DOI] [PubMed] [Google Scholar]
- 18.Semenza G. L. Targeting HIF-1 for cancer therapy. Nature Reviews Cancer. 2003;3(10):721–732. doi: 10.1038/nrc1187. [DOI] [PubMed] [Google Scholar]
- 19.Ke Q., Costa M. Hypoxia-inducible factor-1 (HIF-1) Molecular Pharmacology. 2006;70(5):1469–1480. doi: 10.1124/mol.106.027029. [DOI] [PubMed] [Google Scholar]
- 20.Hong S., Lee H., Kim K. W. HIF-1α: a valid therapeutic target for tumor therapy. Cancer Research and Treatment. 2004;36(6):343–353. doi: 10.4143/crt.2004.36.6.343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Burroughs S. K., Kaluz S., Wang D., Wang K., Van Meir E. G., Wang B. Hypoxia inducible factor pathway inhibitors as anticancer therapeutics. Future Medicinal Chemistry. 2013;5(5):553–572. doi: 10.4155/fmc.13.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ferrara N., Kerbel R. S. Angiogenesis as a therapeutic target. Nature. 2005;438(7070):967–974. doi: 10.1038/nature04483. [DOI] [PubMed] [Google Scholar]
- 23.Hicklin D. J., Ellis L. M. Role of the vascular endothelial growth factor pathway in tumor growth and angiogenesis. Journal of Clinical Oncology. 2005;23(5):1011–1027. doi: 10.1200/JCO.2005.06.081. [DOI] [PubMed] [Google Scholar]
- 24.Luo D., Wang Z., Wu J., Jiang C., Wu J. The role of hypoxia inducible factor-1 in hepatocellular carcinoma. BioMed Research International. 2014;2014:11. doi: 10.1155/2014/409272.409272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cheung T.-H., Man K.-N. M., Yu M.-Y., et al. Dysregulated microRNAs in the pathogenesis and progression of cervical neoplasm. Cell Cycle. 2012;11(15):2876–2884. doi: 10.4161/cc.21278. [DOI] [PubMed] [Google Scholar]
- 26.Katada T., Ishiguro H., Kuwabara Y., et al. MicroRNA expression profile in undifferentiated gastric cancer. International Journal of Oncology. 2009;34(2):537–542. doi: 10.3892/ijo_00000179. [DOI] [PubMed] [Google Scholar]
- 27.Lei Z., Li B., Yang Z., et al. Regulation of HIF-1α and VEGF by miR-20b tunes tumor cells to adapt to the alteration of oxygen concentration. PLoS ONE. 2009;4(10) doi: 10.1371/journal.pone.0007629.e7629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhou W., Shi G., Zhang Q., Wu Q., Li B., Zhang Z. MicroRNA-20b promotes cell growth of breast cancer cells partly via targeting phosphatase and tensin homologue (PTEN) Cell & Bioscience. 2014;4, article 62 doi: 10.1186/2045-3701-4-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ingwersen J., Menge T., Wingerath B., et al. Natalizumab restores aberrant miRNA expression profile in multiple sclerosis and reveals a critical role for miR-20b. Annals of Clinical and Translational Neurology. 2015;2(1):43–55. doi: 10.1002/acn3.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yamaguchi T., Iijima T., Wakaume R., et al. Underexpression of miR-126 and miR-20b in hereditary and nonhereditary colorectal tumors. Oncology. 2014;87(1):58–66. doi: 10.1159/000363303. [DOI] [PubMed] [Google Scholar]
- 31.Zhu J., Chen L., Zou L., et al. MiR-20b, -21, and -130b inhibit PTEN expression resulting in B7-H1 over-expression in advanced colorectal cancer. Human Immunology. 2014;75(4):348–353. doi: 10.1016/j.humimm.2014.01.006. [DOI] [PubMed] [Google Scholar]
- 32.Saleiban A., Faxälv L., Claesson K., Jönsson J.-I., Osman A. miR-20b regulates expression of proteinase-activated receptor-1 (PAR-1) thrombin receptor in melanoma cells. Pigment Cell and Melanoma Research. 2014;27(3):431–441. doi: 10.1111/pcmr.12217. [DOI] [PubMed] [Google Scholar]
- 33.Li J.-Y., Zhang Y., Zhang W.-H., Jia S., Kang Y., Zhu X.-Y. Differential distribution of miR-20a and miR-20b may underly metastatic heterogeneity of breast cancers. Asian Pacific Journal of Cancer Prevention. 2012;13(5):1901–1906. doi: 10.7314/APJCP.2012.13.5.1901. [DOI] [PubMed] [Google Scholar]
- 34.Cascio S., D'Andrea A., Ferla R., et al. miR-20b modulates VEGF expression by targeting HIF-1 alpha and STAT3 in MCF-7 breast cancer cells. Journal of Cellular Physiology. 2010;224(1):242–249. doi: 10.1002/jcp.22126. [DOI] [PubMed] [Google Scholar]
- 35.Wang C.-J., Zhou Z.-G., Wang L., et al. Clinicopathological significance of microRNA-31, -143 and -145 expression in colorectal cancer. Disease Markers. 2009;26(1):27–34. doi: 10.3233/DMA-2009-0601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Martinez-Fernandez L., Pons Z., Margalef M., Arola-Arnal A., Muguerza B. Regulation of vascular endothelial genes by dietary flavonoids: structure-expression relationship studies and the role of the transcription factor KLF-2. Journal of Nutritional Biochemistry. 2015;26:277–284. doi: 10.1016/j.jnutbio.2014.11.003. [DOI] [PubMed] [Google Scholar]
- 37.Yu Y.-H., Wu D.-S., Huang F.-F., et al. MicroRNA-20b and ERK1/2 pathway independently regulate the expression of tissue factor in hematopoietic and trophoblastic differentiation of human embryonic stem cells. Stem Cell Research and Therapy. 2013;4, article 121 doi: 10.1186/scrt332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sasabe E., Tatemoto Y., Li D., Yamamoto T., Osaki T. Mechanism of HIF-1alpha-dependent suppression of hypoxia-induced apoptosis in squamous cell carcinoma cells. Cancer Science. 2005;96(7):394–402. doi: 10.1111/j.1349-7006.2005.00065.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xu M., Zheng Y.-L., Xie X.-Y., et al. Sorafenib blocks the HIF-1α/VEGFA pathway, inhibits tumor invasion, and induces apoptosis in hepatoma cells. DNA and Cell Biology. 2014;33(5):275–281. doi: 10.1089/dna.2013.2184. [DOI] [PubMed] [Google Scholar]