Abstract
Pancreatic cancer (PC) is a highly malignant tumor derived from pancreas tissue and is one of the leading causes of death from cancer. Its molecular mechanism has been partially revealed by validating its oncogenes and tumor suppressor genes; however, the available data remain insufficient for medical workers to design effective treatments. Large-scale identification of PC-related genes can promote studies on PC. In this study, we propose a computational method for mining new candidate PC-related genes. A large network was constructed using protein-protein interaction information, and a shortest path approach was applied to mine new candidate genes based on validated PC-related genes. In addition, a permutation test was adopted to further select key candidate genes. Finally, for all discovered candidate genes, the likelihood that the genes are novel PC-related genes is discussed based on their currently known functions.
1. Introduction
The pancreas is a significant versatile organ of the digestive system and endocrine system; it is significant because it assists in digestion and maintains hormonal balance via digestive enzymes and specific hormones, respectively. Pancreatic cancer (PC) originates from the pancreas, has a high rate of metastasis, and is a highly malignant tumor derived from pancreas tissue. Among various types of PC, pancreatic ductal adenocarcinoma (PDAC) accounts for more than 90% of all pancreatic tumors. PDAC is a malignancy with a poor prognosis, which is demonstrated through its one-year survival rate of approximately 18% for all stages of the disease [1]. In the Western world, pancreatic cancer is one of the top killers for human beings [2]. In 2012 alone, it resulted in 33000 deaths all over the world. In the Western world, pancreatic cancer is the fourth leading cause of death from cancer with a poor prognosis (5-year survival in less than 5% of cases according to most reports). Such a high fatality rate is attributed to the low rate of diagnosis at early age. Only a minority of patients can receive proper treatment with a 5-year survival rate up to 22%. Therefore, it is significant and crucial to study this severe disease. Similar to most types of tumors, PC is induced by both environmental and hereditary elements. Extrinsic factors such as age, gender, race, cigarette smoking, and obesity are all factors that may contribute to tumor initiation [3–5]. Further, certain chronic pancreas-associated diseases, such as diabetes mellitus and chronic pancreatitis, are also related to PC [6, 7].
Over the last decade, the genetic background for PC, especially pancreatic adenocarcinoma (PAC) that comprises most cases, has been revealed through validating a list of oncogenes and tumor suppressor genes for PC. Based on hereditary features, mutations in pancreatic adenocarcinoma can be divided into two clusters defined as common somatic mutations and germline mutations. A somatic mutation, KRAS, is regarded as the earliest and key mutation in nonfamilial PAC initiation, and it aids in maintaining invasion status and tumor progression [8]. In addition to tumor development, more mutated genes contribute to malignant phenotypes. The tumor suppressor genes p16/INK4A are significant somatic mutations and are downregulated in pancreatic adenocarcinoma [9, 10]. In many types of tumors, excessive activation of the TGF-β pathway is a mechanism of tumor progression and invasion [11]. Another pair of tumor suppressor genes, SMAD/DPC4, is involved in PAC through regulating the TGF-β pathway and is critical to advanced tumors [12].
Further, heredofamilial pancreatic adenocarcinoma is associated with certain other significant genes with more complex mechanisms. As indicated by the available literature, most such genes participate in the DNA repair process, such as MSH1/2, PMS1/2, and BRCA1/2, which may participate in nonspecific tumor induction [13–15]. However, in several inherited familial conditions, specific known mutations have not been identified, which may hint at the complexity of carcinogenic mechanisms and the potential oncogenes as well as tumor suppressor genes [16].
To predict more potential tumor-related genes, we proposed a new method that considers protein interactions from STRING (Search Tool for the Retrieval of Interacting Genes/Proteins) [17] and mines potential PC-related genes. STRING is a database with massive amounts of information on physical and functional associations between different proteins. With an established score system, STRING enables a user to search and browse the protein interactions data as well as simultaneously quantify the statistic cooccurrence in the background [17]. Here, depending on the method and using a comprehensive analysis of the protein interaction network, we fully utilized the database containing reported PC-related genes and predicted potential genes involved in PC.
2. Materials and Methods
2.1. Materials
PC-related genes were retrieved from KEGG PATHWAY, which is one of the main databases in KEGG (Kyoto Encyclopedia of Genes and Genomes) [18]. By examining the pathway hsa05212, pancreatic cancer (http://www.genome.jp/dbget-bin/www_bget?hsa05212, accessed in December 2014), we obtained 65 PC-related genes, which comprise the gene set S. Detailed information on these genes is listed in Supplementary Material I available online at http://dx.doi.org/10.1155/2015/623121.
2.2. Method for Mining New Candidate Genes in a Protein-Protein Interaction Network
Protein-protein interaction (PPI) information is useful for investigating protein-related and gene-related problems [19–25]. Most methods for predicting protein attributes are based on the notion that two proteins that interact always share similar attributes [19–23]. Because PC-related genes must share common features related to PC, it is reasonable to use PPI information to identify whether a gene is related to PC. Here, we adopted the PPI information reported in STRING (version 9.1, http://www.string-db.org/) [17], which is a large online database that reports known and predicted protein interactions. The PPIs reported in STRING are derived from the following four sources: (1) genomic context, (2) high-throughput experiments, (3) (conserved) coexpression, and (4) previous knowledge, which imply that the physical and functional associations of the proteins were measured. To extract PPI information for humans, we downloaded the file “protein.links.v9.1.txt.gz” and selected protein interactions beginning with “9606.”, which produced 2,425,314 human PPIs. Each PPI includes two proteins and one score, which measures the strength of the interaction in the range between 150 and 999. For the formulation, let us denote the score of an interaction between proteins p 1 and p 2 as Q i(p 1, p 2).
The aforementioned PPIs were used to construct a large network by taking proteins as nodes. Two nodes are adjacent if and only if the corresponding proteins comprise an interaction that is a member of the 2,425,314 PPIs. Furthermore, the interaction score should also be added to the constructed network. To generate compatibility between our network and a shortest path approach, each edge e was assigned a weight w(e) as follows: w(e) = 1000 − Q i(p 1, p 2), where p 1 and p 2 are corresponding proteins of the endpoints of e.
Two proteins that can interact with each other always share similar attributes [19–23]. Further, considering the interaction score, this notion can be generalized as follows: two proteins in an interaction with a high score are more likely to share similar attributes than those with a low score. Moreover, if we consider a series of proteins p 1, p 2,…, p n such that two consecutive proteins can comprise an interaction with a high score, then these proteins may all share some common attributes. By mapping these proteins onto the constructed network, they may comprise a shortest path connecting p 1 and p n, which is a path connecting p 1 and p n such that the summation of the weights of edges on the path is minimum. Thus, we searched all shortest paths connecting any two PC-related genes. For two consecutive nodes in each of these paths, their corresponding proteins can comprise a PPI with a high score because they lie on a shortest path. As mentioned above, they can share similar functions. For a specific shortest path p 1, p 2,…, p n, where p 1 and p n are encoded by PC-related genes, p 2 shares similar functions with p 1; that is, p 2 may be encoded by an invalidated or known PC-related gene; p 3 shares similar functions with p 2; thus p 3 may also share similar functions with p 1 and may be encoded by an invalidated or known PC-related gene. This can be induced to p 4, p 5,…, p n−1. Thus, we extracted genes in these shortest paths and excluded those that were members of 65 PC-related genes. These genes were referred to as shortest path genes and deemed to have special relationships with PC. The similar scheme has been applied to extract novel genes or chemicals related to other diseases or some biological processes [26–29]. In addition, to distinguish those genes, each shortest path gene was assigned a value, which is referred to as betweenness and defined as the number of shortest paths that contain the gene. The betweenness indicates the direct and indirect associations between shortest path genes and PC-related genes [30].
2.3. Further Selection
By executing the method mentioned in Section 2.2, certain shortest path genes can be extracted from the constructed network. However, certain such genes may be false positives. To exclude this type of gene, the following method was adopted.
Certain nodes in the constructed network are general hubs; their corresponding genes may always receive a high betweenness value even if we randomly selected certain genes as PC-related genes, but the genes exhibit few relationships with PC. To exclude this type of gene among shortest path genes, we randomly produced 1,000 gene sets with the same size as S and compared the betweenness of the shortest path genes in these sets to S. The detailed procedures are described as follows.
-
(I)
Randomly produce 1,000 gene sets, such as S 1, S 2,…, S 1000, each of which with the same size as S, the set consisting of PC-related genes.
-
(II)
The method described in Section 2.2 was executed for 1,000 rounds. For the ith round, the PC-related genes in S were replaced with the genes in S i, and the shortest paths connecting any pair of genes in S i were searched, thereby counting the betweenness of each shortest path gene based on these shortest paths.
-
(III)
For each shortest path gene, there were 1,000 betweenness on randomly produced gene sets and one betweenness on S. After comparing them, we generated a measurement referred to as permutation FDR, which defines the proportion of randomly produced gene sets on which the betweenness was larger than that on S.
-
(IV)
To exclude shortest path genes with general hubs in the network and few relationships with PC, we excluded shortest path genes with permutation FDR values equal to or larger than 0.05. The remaining shortest path genes are referred to as candidate genes.
3. Results and Discussion
3.1. Shortest Path Genes
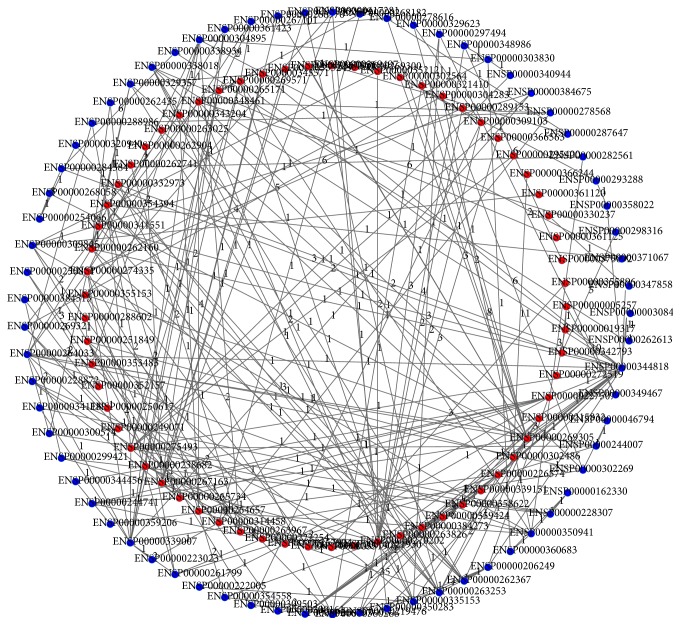
Based on the method described in Section 2.2, we searched all shortest paths connecting any pair of PC-related genes and produced 2,080 shortest paths (each pair of PC-related genes can be connected by a shortest path), which are provided in Supplementary Material II. A graph with these 2,080 shortest paths is shown in Figure 1. The detailed information of edges in this graph is provided in Supplementary Material III. These 2,080 paths involved 134 Ensembl gene IDs. We excluded 65 Ensembl IDs for PC-related genes, resulting in 69 shortest path genes, which are listed in Supplementary Material IV. In addition, we counted the betweenness of each shortest path gene, which is also provided in Supplementary Material IV.
Figure 1.
A graph consisting of 2,080 shortest paths. The nodes on the inner circle (red nodes) represent 65 PC-related genes, while the nodes on the outer circle (blue nodes) represent 69 shortest path genes. The numbers on the edges represent the weights of the edges.
3.2. Candidate Genes
As mentioned in Section 3.1, several shortest path genes were retrieved. According to the principle underlying the method in Section 2.2, these genes may have special relationships with PC. However, certain such genes may be false positives and have few relationships with PC. Thus, we performed a permutation test to control for this type of genes. After calculating the permutation FDR for each shortest path gene, which are listed in Supplementary Material IV, we discarded the genes with a permutation FDR greater than or equal to 0.05, thereby generating thirteen candidate genes, which are listed in Table 1.
Table 1.
Detailed information on the thirteen candidate genes.
| Row number | Ensembl ID | Gene symbol | Full name | Betweenness | Permutation FDR |
|---|---|---|---|---|---|
| 1 | ENSP00000216797 | NFKBIA | Nuclear Factor of Kappa Light Polypeptide Gene Enhancer in B-Cells Inhibitor, Alpha | 73 | 0.002 |
| 2 | ENSP00000269321 | ARHGDIA | Rho GDP Dissociation Inhibitor (GDI) Alpha | 94 | 0.012 |
| 3 | ENSP00000347858 | XIAP | X-Linked Inhibitor of Apoptosis, E3 Ubiquitin Protein Ligase | 64 | 0.006 |
| 4 | ENSP00000250894 | MAPK8IP3 | Mitogen-Activated Protein Kinase 8 Interacting Protein 3 | 64 | <0.001 |
| 5 | ENSP00000262613 | SLC9A3R1 | Solute Carrier Family 9, Subfamily A (NHE3, Cation Proton Antiporter 3), Member 3 Regulator 1 | 128 | 0.031 |
| 6 | ENSP00000384515 | PARVB | Parvin, Beta | 64 | <0.001 |
| 7 | ENSP00000254066 | RARA | Retinoic Acid Receptor, Alpha | 24 | 0.005 |
| 8 | ENSP00000299421 | ILK1 | Integrin-linked kinase 1 | 64 | 0.03 |
| 9 | ENSP00000338934 | EZR | Ezrin | 128 | 0.026 |
| 10 | ENSP00000309845 | HRAS | Harvey Rat Sarcoma Viral Oncogene Homolog | 113 | 0.033 |
| 11 | ENSP00000335153 | HSP90AA1 | Heat Shock Protein 90 kDa Alpha (Cytosolic), Class A Member 1 | 230 | 0.007 |
| 12 | ENSP00000348986 | INS-IGF2 | INS-IGF2 Readthrough | 64 | 0.031 |
| 13 | ENSP00000284384 | PRKCA | Protein Kinase C, Alpha | 175 | <0.001 |
3.3. Analysis of Significant Candidate Genes
Using our method, we predicted thirteen genes that may participate in PC. Based on the principle underlying our method, the candidate genes are specifically connected with PC-related genes. Moreover, such genes have all been reported as genes that are relevant to PC and may exhibit diverse functions in tumor initiation and invasion, especially in PC.
Among the candidate genes, three are inhibitory genes that may participate in PC through their respective mechanisms. The first candidate gene is NFKBIA (see row 1 of Table 1, with betweenness 73 and permutation FDR 0.002), which is expressed in pancreatic tissues [31]. NFKBIA encodes a member of the NF-kappa-B inhibitor family, which has been confirmed to further participate in interactions with REL dimers to inhibit the NF-kappa-B pathway in processes of inflammation immune response and tumorigenesis [32, 33]. Targeted by a specific microRNA, miR196a, NFKBIA has been proved to be associated with PC, especially in the metastasis process [31]. As we have mentioned above, in most tumors including pancreatic cancer, NF-kappa-B pathway has been widely reported to be overactivated and has a close connection with the patients' prognosis, indicating the underlying relationship between NFKBIA and the pancreatic cancer [34–37]. Mutations have been widely reported to contribute to specific functional alteration of crucial proteins including NFKBIA in diseases especially in various cancer subtypes like pancreatic cancer [38–41]. What is more, as one of the crucial inhibitory components of NF-kappa-B pathway, NFKBIA has been reported to be downregulated in cancer [42, 43]. The overexpression of NFKBIA has also been reported to be associated with a better prognosis of various treatment methods in different human tumor subtypes, especially for the prognosis of patients that have taken alpha 1-adrenoceptor antagonists and radiotherapy [44, 45]. Further, considering the inflammation associated function of such gene, such mutations and expression alteration may contribute to the process of tumorigenesis through two individual regulation mechanisms: proliferation associated pathways that involve NF-kappa-B and specific immune response associated pathways in tumor microenvironment [46]. In addition to the NF-kappa-B pathway, the JNK pathway is also a specific pathway in tumor initiation and progression, including PC [47, 48]. A specific regulator of Rho protein exchange reactions which is crucial for JNK pathway, ARHGDIA (see row 2 of Table 1, with betweenness 94 and permutation FDR 0.012), was also identified using our method and functions in several types of tumors [49, 50]. In addition, this apoptosis inhibitory protein has been confirmed to control Rho protein homeostasis and participate in the initiation and progression of PC through the apoptosis associated GDP/GTP exchange reaction via RhoA-Rho pathway [51]. Apoptosis inhibition widely exists in tumor tissues and is an effective way to induce malignant cells, implying the potential relationship between ARHGDIA and pancreatic cancer [52, 53]. As we all know, pathways that contribute to the proliferation and invasion of cancer cells (JNK-STAT, RhoA-Rho signaling pathway, etc.) have all been confirmed to involve phosphorylation and dephosphorylation process which may be regulated by the survival associated GDP/GTP exchange reaction, further validating the potential relationship between ARHGDIA and pancreatic cancer [54–58]. What is more, regarded as a therapeutic target of pancreatic cancer, ARHGDIA (also known as RhoGDI) has been actually proved to be associated with the proliferation and invasion process of the pancreatic tumorigenesis which may further affect the prognosis of various patients [51]. The expression quantity of such gene, ARHGDIA, as the specific negative regulator of Rho protein exchange reactions, has been widely confirmed to be downregulated in tumors, which further induce the activation of survival associated Rho GTPases and promote the progression of various tumors [55–57]. As for specific mutations that may affect the biological function of such gene, coincidentally, specific triple Y156F/S101A/S174A-RhoGDI mutation has been confirmed to be related to the progression of pancreatic cancer [58]. Furthermore, we also obtained the gene XIAP as one of the candidate genes in our list (see row 3 of Table 1, with betweenness 64 and permutation FDR 0.006) that encodes a functional apoptosis suppressor protein. As the most potent apoptosis suppressor, XIAP interacts with several caspases directly and prevents the apoptosis process [59–61]. In PC, XIAP has been proved to be critical for the progression and prognosis of this disease via several classical pathways (Erk, PTEN/PI3K/AKT, etc.) [62–64]. What is more, the overexpression of XIAP has been confirmed to be associated with poor prognosis of pancreatic cancer. Therefore, such protein has already been applied as a useful laboratory test parameter and a functional therapeutic target of pancreatic cancer [64, 65]. As an immune associated functional gene, the polymorphism of XIAP may contribute to the complex and personalized immune response to pancreatic cancers. Some of the functional variants have already been proved to be directly associated with the prognosis of various subtypes of pancreatic cancer, indicating the potential significant role of XIAP in pancreatic cancer [62, 66].
In addition to inhibitory genes, a group of scaffold genes was also among the thirteen candidate genes. MAPK8IP3 (see row 4 of Table 1, with betweenness 64 and permutation FDR <0.001), which is also known as JIP3, is a scaffold protein involved in the JNK pathway [67]. Associated with the MAP kinase cascade, it may be related to PC via potential downstream mechanisms [67–69]. With high tissue specificity, such gene has been reported to actually induce the excessive proliferation of pancreatic cells, which may further initiate the biological process of pancreatic cancer [70]. MAPK8IP3 is also one of the crucial components of JNK pathway which has been overactivated in tumors including pancreatic tumor [71, 72]. During the initiation and invasion stages of pancreatic cancer, MAPK8IP3 has been reported to be overexpressed which may further contribute to the excessive proliferation of pancreatic cells in clinical cases [70]. Mutations have also been identified in MAPK8IP3 which has been reported to partially interfere with its own functions under pathological conditions [73, 74]. Some of the specific mutations of MAPK8IP3 have been reported to influence the adhesion and invasion process of various cancer cells including the pancreatic cancer cells, validating the crucial role of MAPK8IP3 in pancreatic cancer [75, 76]. As another functional scaffold protein that participates in the interaction between the cell membrane and cytoskeleton, our candidate gene SLC9A3R1 (see row 5 of Table 1, with betweenness 128 and permutation FDR 0.031) regulates the subcellular location and function of SLC9A3 and plays a specific role in the Wnt pathway [77–79]. Known as NHERF1, SLC9A3R1 acts as a scaffold protein in many types of tumors depending on its specific role in Wnt pathway, especially in PC [78, 80, 81]. During the course of pancreatic cancer raising, SLC9A3R1 has been confirmed to be overexpressed and involve the poor prognosis of such disease [81, 82]. Just like other candidates encoding scaffold proteins, SLC9A3R1 has also been proved to contain specific nonsynonymous mutations in various cancer subtypes, especially in pancreatic cancer [83, 84]. Coding an adaptor protein, the candidate gene PARVB (see row 6 of Table 1, with betweenness 64 and permutation FDR <0.001) is critical to certain specific biological processes in cancer, such as ERK signaling pathway and focal adhesion which both have been identified in pancreatic cancer [85, 86]. Therefore, this gene is critical for tumorigenesis and may have a specific role in PC [87]. As a crucial adaptor protein, the overexpression of PARVB has been proved to be associated with high cell proliferation rate which indicates worsening clinical prognosis of tumors, including pancreatic cancer [86, 88]. The polymerase of such gene, PARVB, has also been identified and confirmed to be associated with a group of severe diseases including cancers [87]. Such abnormal expression level and high polymerase in tumor cells suggest that PARVB may play a crucial role in the specific pathogenic process of pancreatic cancer.
Six remaining genes may also be associated with PC in their respective way. RARA (see row 7 of Table 1, with betweenness 24 and permutation FDR 0.005) encodes a specific receptor for retinoic acid, which may further regulate germ cell development during spermatogenesis and the expression of specific genes through recruiting the chromatin complex (KMT2E, MLL5, etc.) [89–91]. As we all know, the stem-cell-like features of tumor cells may be associated with the potential invasion and proliferation ability of such tumor cells [92–94]. In pancreatic cancer, it has been confirmed that the more “stem-like” the cancer cell is, the more malignant the cancer is [95]. Similar to its specific function in germ cells, RARA is related to the regulation of stem- cell-like features in PC, which implies that it may further affect the prognosis of such a severe malignancy [96]. The overexpression of RARA has been reported to be related to specific biological behaviors of tumors such as EMT (epithelial-to-mesenchymal) and invasion, which indicates poor prognosis of pancreatic cancers in clinical cases [97]. Although few mutations have been identified in RARA, crucial fusion gene variants have been widely identified in cancers which may be analyzed later. As we have mentioned above, SLC9A3R1 has been reported to regulate cellular morphology as a scaffold protein [78]. Similarly, the candidate gene ILK1 (see row 8 of Table 1, with betweenness 64 and permutation FDR 0.005) also mediates cell architecture, signaling transduction and cell adhesion via integrin-mediated signaling transduction, especially in tumor tissues [98, 99]. Although studies have not explained clearly how ILK1 actually contributes to the process of pancreatic cancer, it has been shown to participate in several types of tumors and may be associated with several unique molecules (GSK, AKT, PTEN, etc.) that all have been confirmed to be critical to PC [98–103]. Recently, such gene has been reported to be associated with the expression of TGF-α, which may partially explain the underlying carcinogenic mechanism of such gene [104]. What is more, such gene has been confirmed to be overexpressed during the whole biological process of tumorigenesis including the proliferation, migration, and invasion of pancreatic cancers [104–106]. Further, specific mutations have also been identified in such gene. Integrin-linked kinase 1 (ILK1) and HSP90 (HSP90AA1, which is also in our list and will be analyzed later) have both been confirmed to contain quite a lot of tumor-associated variants. Such mutations of ILK1 have been reported to be functional pancreatic cancer biomarkers [107–109]. All in all, ILK1 has been proved to regulate the tumorigenesis process of PC through a specific TGF-α-associated mechanism. As a functional SLC9A3R1-associated protein, a specific protein ezrin is encoded by one of our candidate genes EZR (see row 9 of Table 1, with betweenness 128 and permutation FDR 0.026) [110]. Crucial for cytoplasmic peripheral membrane, EZR is associated with the actin cytoskeleton and regulates the surface expression of actin [111, 112]. It has been widely reported that ezrin is critical for the proliferation, metastasis, and invasion process of several tumor subtypes, especially of pancreatic cancer [113, 114]. The overexpression of EZR has been widely reported to be associated with the initiation, proliferation, and metastasis processes of tumor, which strongly affect the patients' prognosis especially in pancreatic cancer [115–117]. EZR, as the crucial scaffold protein which may interact with SLC9A3R1 that we have mentioned above, has also been reported to contain specific mutations that may contribute to the tumorigenesis process of various cancer subtypes including pancreatic cancer [118]. All in all, EZR may be a potential pancreatic-cancer-associated gene and may contribute to similar pathways with SL9A3R1 as we have mentioned above [110].
Further, we also predicted one of the most famous oncogenes HRAS (see row 10 of Table 1, with betweenness 113 and permutation FDR 0.033), which binds GDP/GTP and exhibits intrinsic GTPase activity [119]. Associated with several tumors and similar to its homologues (r-RAS, k-Ras, etc.), HRAS is a powerful oncogene that can initiate tumors through inducing excessive growth factor activation in cells and promoting malignant proliferation of tumor cells, especially in tumors in situ [120–122]. In PC, such gene has also been regarded as a main oncogene and is widely reported to be overexpressed during the entire clinical course of pancreatic cancer, validating our prediction of the potential pancreatic cancer-associated genes [123]. Several genetic alterations in HRAS (mutations, CNVs, etc.) have been identified and reported to be associated with such a severe type of cancer [124–126]. Recently, some of the crucial mutations have already been confirmed to directly contribute to the tumorigenesis of pancreatic cancer, indicating the underlying relationship between mutations of candidate genes and the process of tumorigenesis [127]. Heat shock proteins compose a unique group of proteins produced by cells to resist harmful and stressful conditions, including the tumor microenvironment, which contains less oxygen and a lower pH [128–130]. This cluster of proteins has been confirmed to be functional in tumor microenvironment and promote the progression of various tumor subtypes [131, 132]. One of the candidate genes, HSP90AA1 (see row 11 of Table 1, with betweenness 230 and permutation FDR 0.007), also encodes a specific heat shock protein, heat shock protein 90 kDa alpha, which has been reported to be expressed in the cytoplasm. Several types of heat shock proteins (such as HSP20, HSP70, and HSP90), including our predicted protein, are not only related to tumor genesis but specifically contribute to PC [133–135]. Heat shock proteins have also been confirmed to be overexpressed in pancreatic cancer, including HSP90 which is encoded by our predicted gene [136–140]. The overexpression of heat shock proteins has been confirmed to actually contribute to the tumorigenesis process of pancreatic cancer which validates our predication of HSP90AA1 as a functional candidate gene [133]. As a hormone-associated gene, our candidate gene INS-IGF2 (see row 12 of Table 1, with betweenness 64 and permutation FDR 0.031) is a readthrough gene of INS and IGF2. This gene may act as a posttranscriptional regulatory factor for the two genes INS and IGF2, which has been shown to be associated with several pancreatic-associated diseases, such as diabetes [141–143]. As a specific fusion gene, INS-IGF2 participates in several cancer types, especially in prostate cancer [144–146]. The fusion of a tumor-associated gene and a pancreas-related gene, INS-IGF2, probably participates in PC progression and invasion specifically. Recently, such fusion gene INS-IGF2 is reported to be specifically expressed in pancreatic islets especially in patients with autoimmune diseases under pathological conditions [147, 148]. Since immune response is quite significant for tumor surveillance, INS-IGF2 is definitely associated with the unique tumor-associated immune response and may further contribute to the prognosis of pancreatic cancer [148]. Although genes like RARA (mostly identified as FIP1L1/PML-RARA fusion gene) (we have mentioned above) and INS-IGF2 have not been reported to contain specific mutations that may be associated with pancreatic cancer, such fusion genes have both been confirmed to have abnormal gene functions which may directly contribute to the tumorigenesis of various cancer subtypes, including pancreatic cancer [149, 150]. The last candidate gene PRKCA (see row 13 of Table 1, with betweenness 175 and permutation FDR <0.001) encodes a specific kinase that has been reported in several types of tumors [151, 152]. As a widely reported oncogene, PRKCA contributes to the phosphorylation process of crucial tumor-associated proteins [153]. The overexpression of PRKCA has been proved to enhance the transformed proliferation and invasion process of pancreatic cancer which further definitely influences the prognosis of such disease [154, 155]. While at the same time, genes like PRKCA which encodes functional kinase may be strongly affected by genetic alterations especially by some specific nonsynonymous mutations [75, 156]. Therefore, the polymerase of gene PRKCA may definitely affect the function of protein kinase C which has also been identified in pancreatic cancer [156, 157]. However, recent studies show that, at least in vitro, PRKCA has a specific function in tumor suppression [156]. The complex role of PKC in cancer may reflect the complicated interactive relationship between PRKCA and PC [156, 158]. All in all, our predicted gene PRKCA has been confirmed to be definitely associated with pancreatic cancer, while, at the same time, the various functions of PRKCA imply the complex role of such gene in pancreatic cancer.
Consequently, all candidate genes obtained using our method play a critical role in tumor initiation and progression. Because the detailed regulation mechanisms of only few genes remain unclear, those candidate genes may be genes that are critical to PC. PC features genetic heterogeneity, and many familial hereditary related genes remain ambiguous [16]. Therefore, in addition to the confirmed genes, such as KRAS and p16/INK41 [8, 9], more genes must be screened and validated to clearly demonstrate the mechanisms of pancreatic adenocarcinoma. In conclusion, based on reported PC-related genes, the proposed method was effective at predicting candidate tumor-associated genes in PC.
4. Conclusions
In this study, we proposed a computational method to mine new candidate genes related to pancreatic cancer, which utilized protein-protein interaction information. The analyses of the obtained candidate genes indicate that they may be novel PC-related genes. Hopefully, this contribution will promote studies on pancreatic cancer and provide new hope for designing effective treatments.
Supplementary Material
Supplementary Material I: lists 65 PC-related genes and their ensembl IDs.
Supplementary Material II: lists the detailed information of 2,080 shortest paths.
Supplementary Material III: lists edges in a graph consisting of shortest paths connecting any two PC-related genes.
Supplementary Material IV: lists 69 shortest path genes and their betweenness and permutation FDRs.
Acknowledgments
This study was supported by the National Basic Research Program of China (2011CB510101 and 2011CB510102), the National Natural Science Foundation of China (31371335), and the Innovation Program of Shanghai Municipal Education Commission (12ZZ087).
Conflict of Interests
The authors declare that there is no conflict of interests regarding the publication of this paper.
References
- 1.Hidalgo M., Cascinu S., Kleeff J., et al. Addressing the challenges of pancreatic cancer: future directions for improving outcomes. Pancreatology. 2015;15(1):8–18. doi: 10.1016/j.pan.2014.10.001. [DOI] [PubMed] [Google Scholar]
- 2.Verbeke C., Löhr M., Severin Karlsson J., Del Chiaro M. Pathology reporting of pancreatic cancer following neoadjuvant therapy: challenges and uncertainties. Cancer Treatment Reviews. 2015;41(1):17–26. doi: 10.1016/j.ctrv.2014.11.002. [DOI] [PubMed] [Google Scholar]
- 3.Freilich J., Strom T., Springett G., et al. Age and resected pancreatic cancer outcomes. International Journal of Radiation Oncology Biology Physics. 2014;90(1, supplement):p. S356. doi: 10.1016/j.ijrobp.2014.05.1157. [DOI] [Google Scholar]
- 4.Mizuno S., Nakai Y., Isayama H., et al. Smoking, family history of cancer, and diabetes mellitus are associated with the age of onset of pancreatic cancer in japanese patients. Pancreas. 2014;43(7):1014–1017. doi: 10.1097/MPA.0000000000000158. [DOI] [PubMed] [Google Scholar]
- 5.Preziosi G., Oben J. A., Fusai G. Obesity and pancreatic cancer. Surgical Oncology. 2014;23(2):61–71. doi: 10.1016/j.suronc.2014.02.003. [DOI] [PubMed] [Google Scholar]
- 6.Deng S., Zhu S., Wang B., et al. Chronic pancreatitis and pancreatic cancer demonstrate active epithelial-mesenchymal transition profile, regulated by miR-217-SIRT1 pathway. Cancer Letters. 2014;355(2):184–191. doi: 10.1016/j.canlet.2014.08.007. [DOI] [PubMed] [Google Scholar]
- 7.Snima K. S., Nair R. S., Nair S. V., Kamath C. R., Lakshmanan V.-K. Combination of anti-diabetic drug metformin and boswellic acid nanoparticles: a novel strategy for pancreatic cancer therapy. Journal of Biomedical Nanotechnology. 2015;11(1):93–104. doi: 10.1166/jbn.2015.1877. [DOI] [PubMed] [Google Scholar]
- 8.Lüttges J., Reinecke-Lüthge A., Möllmann B., et al. Duct changes and K-ras mutations in the disease-free pancreas: analysis of type, age relation and spatial distribution. Virchows Archiv. 1999;435(5):461–468. doi: 10.1007/s004280050428. [DOI] [PubMed] [Google Scholar]
- 9.Georgiadou D., Sergentanis T. N., Sakellariou S., et al. Cyclin D1, p16INK4A and p27Kip1 in pancreatic adenocarcinoma: assessing prognostic implications through quantitative image analysis. Acta Pathologica, Microbiologica, Et Immunologica Scandinavica. 2014;122(12):1230–1239. doi: 10.1111/apm.12289. [DOI] [PubMed] [Google Scholar]
- 10.Wang Z., Ali S., Banerjee S., et al. Activated K-Ras and INK4a/Arf deficiency promote aggressiveness of pancreatic cancer by induction of EMT consistent with cancer stem cell phenotype. Journal of Cellular Physiology. 2013;228(3):556–562. doi: 10.1002/jcp.24162. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 11.Zhang L., Liu W., Qin Y., Wu R. Expression of TGF-β1 in Wilms' tumor was associated with invasiveness and disease progression. Journal of Pediatric Urology. 2014;10(5):962–968. doi: 10.1016/j.jpurol.2014.01.010. [DOI] [PubMed] [Google Scholar]
- 12.El-Gohary Y., Tulachan S., Guo P., et al. Smad signaling pathways regulate pancreatic endocrine development. Developmental Biology. 2013;378(2):83–93. doi: 10.1016/j.ydbio.2013.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Veenstra J. A. Immunocytochemical demonstration of a homology in peptidergic neurosecretory cells in the suboesophageal ganglion of a beetle and a locust with antisera to bovine pancreatic polypeptide, FMRFamide, vasopressin and α-MSH. Neuroscience Letters. 1984;48(2):185–190. doi: 10.1016/0304-3940(84)90017-x. [DOI] [PubMed] [Google Scholar]
- 14.Das Gupta R., Kolodner R. Identification of postmeiotic segregation (PMS) mutants as novel alleles of mismatch repair (MMR) genes. The FASEB Journal. 1997;11(9):p. 1971. [Google Scholar]
- 15.Golan T., Kanji Z. S., Epelbaum R., et al. Overall survival and clinical characteristics of pancreatic cancer in BRCA mutation carriers. British Journal of Cancer. 2014;111(6):1132–1138. doi: 10.1038/bjc.2014.418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Klein A. P., Brune K. A., Petersen G. M., et al. Prospective risk of pancreatic cancer in familial pancreatic cancer kindreds. Cancer Research. 2004;64(7):2634–2638. doi: 10.1158/0008-5472.can-03-3823. [DOI] [PubMed] [Google Scholar]
- 17.Szklarczyk D., Franceschini A., Wyder S., et al. STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Research. 2015;43(1):D447–D452. doi: 10.1093/nar/gku1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kanehisa M., Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Research. 2000;28(1):27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Deng M., Zhang K., Mehta S., Chen T., Sun F. Prediction of protein function using protein-protein interaction data. Journal of Computational Biology. 2003;10(6):947–960. doi: 10.1089/106652703322756168. [DOI] [PubMed] [Google Scholar]
- 20.Hu L., Huang T., Shi X., Lu W.-C., Cai Y.-D., Chou K.-C. Predicting functions of proteins in mouse based on weighted protein-protein interaction network and protein hybrid properties. PLoS ONE. 2011;6(1) doi: 10.1371/journal.pone.0014556.e14556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gao P., Wang Q.-P., Chen L., Huang T. Prediction of human genes regulatory functions based on proteinprotein interaction network. Protein and Peptide Letters. 2012;19(9):910–916. doi: 10.2174/092986612802084528. [DOI] [PubMed] [Google Scholar]
- 22.Ng K.-L., Ciou J.-S., Huang C.-H. Prediction of protein functions based on function-function correlation relations. Computers in Biology and Medicine. 2010;40(3):300–305. doi: 10.1016/j.compbiomed.2010.01.001. [DOI] [PubMed] [Google Scholar]
- 23.Sharan R., Ulitsky I., Shamir R. Network-based prediction of protein function. Molecular Systems Biology. 2007;3, article 88 doi: 10.1038/msb4100129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bogdanov P., Singh A. K. Molecular function prediction using neighborhood features. IEEE/ACM Transactions on Computational Biology and Bioinformatics. 2010;7(2):208–217. doi: 10.1109/tcbb.2009.81. [DOI] [PubMed] [Google Scholar]
- 25.AbdulHameed M. D. M., Tawa G. J., Kumar K., et al. Systems level analysis and identification of pathways and networks associated with liver fibrosis. PLoS ONE. 2014;9(11) doi: 10.1371/journal.pone.0112193.e112193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen L., Chu C., Lu J., Kong X., Huang T., Cai Y. D. A computational method for the identification of new candidate carcinogenic and non-carcinogenic chemicals. Molecular BioSystems. 2015;11(9):2541–2550. doi: 10.1039/c5mb00276a. [DOI] [PubMed] [Google Scholar]
- 27.Gui T., Dong X., Li R., Li Y., Wang Z. Identification of hepatocellular carcinoma-related genes with a machine learning and network analysis. Journal of Computational Biology. 2015;22(1):63–71. doi: 10.1089/cmb.2014.0122. [DOI] [PubMed] [Google Scholar]
- 28.Chen L., Chu C., Kong X., Huang G., Huang T., Cai Y. A hybrid computational method for the discovery of novel reproduction-related genes. PLoS ONE. 2015;10(3) doi: 10.1371/journal.pone.0117090.e0117090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li B.-Q., Zhang J., Huang T., Zhang L., Cai Y.-D. Identification of retinoblastoma related genes with shortest path in a protein-protein interaction network. Biochimie. 2012;94(9):1910–1917. doi: 10.1016/j.biochi.2012.05.005. [DOI] [PubMed] [Google Scholar]
- 30.Craven J. B. M. Markov Networks for Detecting Overlapping Elements in Sequence Data. MIT Press; 2005. [Google Scholar]
- 31.Huang F., Tang J., Zhuang X., et al. MiR-196a promotes pancreatic cancer progression by targeting nuclear factor kappa-B-inhibitor alpha. PLoS ONE. 2014;9(2) doi: 10.1371/journal.pone.0087897.e87897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chang P.-Y., Draheim K., Kelliher M. A., Miyamoto S. NFKB1 is a direct target of the TAL1 oncoprotein in human T leukemia cells. Cancer Research. 2006;66(12):6008–6013. doi: 10.1158/0008-5472.CAN-06-0194. [DOI] [PubMed] [Google Scholar]
- 33.Fullard N., Wilson C. L., Oakley F. Roles of c-Rel signalling in inflammation and disease. International Journal of Biochemistry and Cell Biology. 2012;44(6):851–860. doi: 10.1016/j.biocel.2012.02.017. [DOI] [PubMed] [Google Scholar]
- 34.Ito Y., Kikuchi E., Tanaka N., et al. Down-regulation of NF kappa B activation is an effective therapeutic modality in acquired platinum-resistant bladder cancer. BMC Cancer. 2015;15, article 324 doi: 10.1186/s12885-015-1315-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lessard L., Mes-Masson A.-M., Lamarre L., Wall L., Lattouf J.-B., Saad F. NF-κB nuclear localization and its prognostic significance in prostate cancer. BJU International. 2003;91(4):417–420. doi: 10.1046/j.1464-410x.2003.04104.x. [DOI] [PubMed] [Google Scholar]
- 36.Gilmore T., Gapuzan M.-E., Kalaitzidis D., Starczynowski D. Rel/NF-κB/IκB signal transduction in the generation and treatment of human cancer. Cancer Letters. 2002;181(1):1–9. doi: 10.1016/s0304-3835(01)00795-9. [DOI] [PubMed] [Google Scholar]
- 37.Haefner B. NF-kappa B: arresting a major culprit in cancer. Drug Discovery Today. 2002;7(12):653–663. doi: 10.1016/s1359-6446(02)02309-7. [DOI] [PubMed] [Google Scholar]
- 38.Kinugasa H., Nouso K., Miyahara K., et al. Detection of K-ras gene mutation by liquid biopsy in patients with pancreatic cancer. Cancer. 2015;121(13):2271–2280. doi: 10.1002/cncr.29364. [DOI] [PubMed] [Google Scholar]
- 39.Purohit A., Varney M., Rachagani S., Ouellette M., Batra S. K., Singh R. K. CXCR2 signaling axis- a helping hand to KRAS(G12D) mutation in pancreatic cancer initiation. Cancer Research. 2014;74, article 4874 doi: 10.1158/1538-7445.am2014-4874. [DOI] [Google Scholar]
- 40.Hayashi H., Ueno H., Sakamoto Y., et al. Gene mutation profile of pancreatic cancer in Japanese patients and its association with prognosis. Annals of Oncology. 2014;25 [Google Scholar]
- 41.Saito K., Nakai Y., Isayama H., et al. Associations between K-ras mutation, smoking, and prognosis of pancreatic cancer. Journal of Clinical Oncology. 2014;32(supplement 3, abstract 298) [Google Scholar]
- 42.Bredel M., Kim H., Nanda T. K., et al. Deletion of the tumor suppressor NFKBIA in triple-negative breast cancer. International Journal of Radiation Oncology, Biology, Physics. 2013;87(2):p. S98. doi: 10.1016/j.ijrobp.2013.06.254. [DOI] [Google Scholar]
- 43.Harsh G. R., Scholtens D. M., Yadav A. K., et al. NFKBIA in glioblastomas: tumor suppressor and potent predictor of outcome. Journal of Neurosurgery. 2011;115:p. A404. [Google Scholar]
- 44.Mukogawa T., Koyama F., Tachibana M., et al. Adenovirus-mediated gene transduction of truncated IκBα enhances radiosensitivity in human colon cancer cells. Cancer Science. 2003;94(8):745–750. doi: 10.1111/j.1349-7006.2003.tb01513.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Partin J. V., Anglin I. E., Kyprianou N. Quinazoline-based α1-adrenoceptor antagonists induce prostate cancer cell apoptosis via TGF-β signalling and IκBα induction. British Journal of Cancer. 2003;88(10):1615–1621. doi: 10.1038/sj.bjc.6600961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Reid-Lombardo K. M., Fridley B. L., Bamlet W. R., Cunningham J. M., Sarr M. G., Petersen G. M. Survival is associated with genetic variation in inflammatory pathway genes among patients with resected and unresected pancreatic cancer. Annals of Surgery. 2013;257(6):1096–1102. doi: 10.1097/SLA.0b013e318275b7e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Suzuki S., Okada M., Shibuya K., et al. JNK suppression of chemotherapeutic agents-induced ROS confers chemoresistance on pancreatic cancer stem cells. Oncotarget. 2015;6(1):458–470. doi: 10.18632/oncotarget.2693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yuan X. P., Dong M., Li X., Zhou J. P. GRP78 promotes the invasion of pancreatic cancer cells by FAK and JNK. Molecular and Cellular Biochemistry. 2014;398(1-2):55–62. doi: 10.1007/s11010-014-2204-2. [DOI] [PubMed] [Google Scholar]
- 49.Liang L., Li Q., Huang L. Y., et al. Loss of ARHGDIA expression is associated with poor prognosis in HCC and promotes invasion and metastasis of HCC cells. International Journal of Oncology. 2014;45(2):659–666. doi: 10.3892/ijo.2014.2451. [DOI] [PubMed] [Google Scholar]
- 50.Dal Bo M., Pozzo F., Bomben R., et al. ARHGDIA, a mutant TP53-associated Rho GDP dissociation inhibitor, is over-expressed in gene expression profiles of TP53 disrupted chronic lymphocytic leukaemia cells. British Journal of Haematology. 2013;161(4):596–599. doi: 10.1111/bjh.12258. [DOI] [PubMed] [Google Scholar]
- 51.Harding M. A., Theodorescu D. RhoGDI signaling provides targets for cancer therapy. European Journal of Cancer. 2010;46(7):1252–1259. doi: 10.1016/j.ejca.2010.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dixit D., Sharma V., Ghosh S., Mehta V. S., Sen E. Inhibition of Casein kinase-2 induces p53-dependent cell cycle arrest and sensitizes glioblastoma cells to tumor necrosis factor (TNFα)-induced apoptosis through SIRT1 inhibition. Cell Death and Disease. 2012;3(2, article e271) doi: 10.1038/cddis.2012.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Liu Q., Zhao X.-Y., Bai R.-Z., et al. Induction of tumor inhibition and apoptosis by a candidate tumor suppressor gene DRR1 on 3p21.1. Oncology Reports. 2009;22(5):1069–1075. doi: 10.3892/or_00000538. [DOI] [PubMed] [Google Scholar]
- 54.Marinissen M. J., Chiariello M., Tanos T., Bernard O., Narumiya S., Gutkind J. S. The small GTP-binding protein RhoA regulates c-jun by a ROCK-JNK signaling axis. Molecular Cell. 2004;14(1):29–41. doi: 10.1016/s1097-2765(04)00153-4. [DOI] [PubMed] [Google Scholar]
- 55.Bozza W. P., Zhang Y., Hallett K., Rivera Rosado L. A., Zhang B. RhoGDI deficiency induces constitutive activation of Rho GTPases and COX-2 pathways in association with breast cancer progression. Oncotarget. 2015 doi: 10.18632/oncotarget.5416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hooshmand S., Ghaderi A., Yusoff K., Thilakavathy K., Rosli R., Mojtahedi Z. Differentially expressed proteins in ER+ MCF7 and ER− MDA-MB-231 human breast cancer cells by RhoGDI-α silencing and overexpression. Asian Pacific Journal of Cancer Prevention. 2014;15(7):3311–3317. doi: 10.7314/apjcp.2014.15.7.3311. [DOI] [PubMed] [Google Scholar]
- 57.Zhang B. Rho GDP dissociation inhibitors as potential targets for anticancer treatment. Drug Resistance Updates. 2006;9(3):134–141. doi: 10.1016/j.drup.2006.06.001. [DOI] [PubMed] [Google Scholar]
- 58.Wang Z. X., Thurmond D. C. Differential phosphorylation of RhoGDI mediates the distinct cycling of Cdc42 and Rac1 to regulate second-phase insulin secretion. Journal of Biological Chemistry. 2010;285(9):6186–6197. doi: 10.1074/jbc.m109.072421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Jiao G., Guo W., Ren T., et al. BMPR2 inhibition induced apoptosis and autophagy via destabilization of XIAP in human chondrosarcoma cells. Cell Death and Disease. 2014;5(12) doi: 10.1038/cddis.2014.540.e1571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hu R., Yang Y., Liu Z., et al. The XIAP inhibitor Embelin enhances TRAIL-induced apoptosis in human leukemia cells by DR4 and DR5 upregulation. Tumor Biology. 2015;36(2):769–777. doi: 10.1007/s13277-014-2702-6. [DOI] [PubMed] [Google Scholar]
- 61.Cao Z., Li X., Li J., Luo W., Huang C., Chen J. X-linked inhibitor of apoptosis protein (XIAP) lacking RING domain localizes to the nuclear and promotes cancer cell anchorage-independent growth by targeting the E2F1/Cyclin E axis. Oncotarget. 2014;5(16):7126–7137. doi: 10.18632/oncotarget.2227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yi X. P., Han T., Li Y. X., Long X. Y., Li W. Z. Simultaneous silencing of XIAP and survivin causes partial mesenchymal-epithelial transition of human pancreatic cancer cells via the PTEN/PI3K/Akt pathway. Molecular Medicine Reports. 2015;12:601–608. doi: 10.3892/mmr.2015.3380. [DOI] [PubMed] [Google Scholar]
- 63.Cao L.-P., Song J.-L., Yi X.-P., Li Y.-X. Double inhibition of NF-κB and XIAP via RNAi enhances the sensitivity of pancreatic cancer cells to gemcitabine. Oncology Reports. 2013;29(4):1659–1665. doi: 10.3892/or.2013.2246. [DOI] [PubMed] [Google Scholar]
- 64.Wang Z.-H., Chen H., Guo H.-C., et al. Enhanced antitumor efficacy by the combination of emodin and gemcitabine against human pancreatic cancer cells via downregulation of the expression of XIAP in vitro and in vivo. International Journal of Oncology. 2011;39(5):1123–1131. doi: 10.3892/ijo.2011.1115. [DOI] [PubMed] [Google Scholar]
- 65.Zai H., Yi X., Li Y., Jiang C., Lü X. Simultaneous inhibition of XIAP and survivin expression on EMT and invasion of human pancreatic cancer cells. Journal of Central South University. 2012;37(9):883–888. doi: 10.3969/j.issn.1672-7347.2012.09.004. [DOI] [PubMed] [Google Scholar]
- 66.Yu R., Albarenque S. M., Cool R. H., Quax W. J., Mohr A., Zwacka R. M. DR4 specific TRAIL variants are more efficacious than wild-type TRAIL in pancreatic cancer. Cancer Biology and Therapy. 2014;15(12):1658–1666. doi: 10.4161/15384047.2014.972183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sun T., Yu N., Zhai L.-K., et al. C-Jun NH2-terminal kinase (JNK)-interacting protein-3 (JIP3) regulates neuronal axon elongation in a kinesin- and JNK-dependent manner. Journal of Biological Chemistry. 2013;288(20):14531–14543. doi: 10.1074/jbc.m113.464453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Matsuura H., Nishitoh H., Takeda K., et al. Phosphorylation-dependent scaffolding role of JSAP1/JIP3 in the ASK1-JNK signaling pathway. A new mode of regulation of the MAP kinase cascade. The Journal of Biological Chemistry. 2002;277(43):40703–40709. doi: 10.1074/jbc.m202004200. [DOI] [PubMed] [Google Scholar]
- 69.Skrypek N., Vasseur R., Vincent A., Duchêne B., Van Seuningen I., Jonckheere N. The oncogenic receptor ErbB2 modulates gemcitabine and irinotecan/SN-38 chemoresistance of human pancreatic cancer cells via hCNT1 transporter and multidrug-resistance associated protein MRP-2. Oncotarget. 2015;6(13):10853–10867. doi: 10.18632/oncotarget.3414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Størling J., Allaman-Pillet N., Karlsen A. E., Billestrup N., Bonny C., Mandrup-Poulsen T. Antitumorigenic effect of proteasome inhibitors on insulinoma cells. Endocrinology. 2005;146(4):1718–1726. doi: 10.1210/en.2004-0963. [DOI] [PubMed] [Google Scholar]
- 71.Standen C. L., Kennedy N. J., Flavell R. A., Davis R. J. Signal transduction cross talk mediated by Jun N-terminal kinase-interacting protein and insulin receptor substrate scaffold protein complexes. Molecular and Cellular Biology. 2009;29(17):4831–4840. doi: 10.1128/mcb.00155-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ongusaha P. P., Qi H. H., Raj L., et al. Identification of ROCK1 as an upstream activator of the JIP-3 to JNK signaling axis in response to UVB damage. Science Signaling. 2008;1(47):p. ra14. doi: 10.1126/scisignal.1161938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Rao S. N., Balaji V. N. Molecular docking studies on JNK inhibitors at the allosteric JNK-JIP interaction site. Current Science. 2012;102(7):1009–1016. [Google Scholar]
- 74.Engström W., Granerus M. Expression of JNK-interacting protein JIP-1 and insulin-like growth factor II in Wilms tumour cell lines and primary Wilms tumours. Anticancer Research. 2009;29(7):2467–2472. [PubMed] [Google Scholar]
- 75.Takino T., Nakada M., Miyamori H., et al. JSAP1/JIP3 cooperates with focal adhesion kinase to regulate c-Jun N-terminal kinase and cell migration. The Journal of Biological Chemistry. 2005;280(45):37772–37781. doi: 10.1074/jbc.m505241200. [DOI] [PubMed] [Google Scholar]
- 76.Matsuguchi T., Masuda A., Sugimoto K., Nagai Y., Yoshikai Y. JNK-interacting protein 3 associates with Toll-like receptor 4 and is involved in LPS-mediated JNK activation. The EMBO Journal. 2003;22(17):4455–4464. doi: 10.1093/emboj/cdg438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Broere N., Chen M., Cinar A., et al. Defective jejunal and colonic salt absorption and altered Na+/H+ exchanger 3 (NHE3) activity in NHE regulatory factor 1 (NHERF1) adaptor protein-deficient mice. Pflügers Archiv—European Journal of Physiology. 2009;457(5):1079–1091. doi: 10.1007/s00424-008-0579-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Bellizzi A., Greco M. R., Rubino R., et al. The scaffolding protein NHERF1 sensitizes EGFR-dependent tumor growth, motility and invadopodia function to gefitinib treatment in breast cancer cells. International Journal of Oncology. 2015;46(3):1214–1224. doi: 10.3892/ijo.2014.2805. [DOI] [PubMed] [Google Scholar]
- 79.Hopwood B., Tsykin A., Findlay D. M., Fazzalari N. L. Microarray gene expression profiling of osteoarthritic bone suggests altered bone remodelling, WNT and transforming growth factor-β/bone morphogenic protein signalling. Arthritis Research and Therapy. 2007;9(5, article R100) doi: 10.1186/ar2301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Jiang Y., Wang S., Holcomb J., et al. Crystallographic analysis of NHERF1-PLCbeta3 interaction provides structural basis for CXCR2 signaling in pancreatic cancer. Biochemical and Biophysical Research Communications. 2014;446(2):638–643. doi: 10.1016/j.bbrc.2014.03.028. [DOI] [PubMed] [Google Scholar]
- 81.Wang S., Wu Y., Hou Y., et al. CXCR2 macromolecular complex in pancreatic cancer: a potential therapeutic target in tumor growth. Translational Oncology. 2013;6(2):216–225. doi: 10.1593/tlo.13133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Mangia A., Saponaro C., Malfettone A., et al. Involvement of nuclear NHERF1 in colorectal cancer progression. Oncology Reports. 2012;28(3):889–894. doi: 10.3892/or.2012.1895. [DOI] [PubMed] [Google Scholar]
- 83.Hayashi Y., Molina J. R., Hamilton S. R., Georgescu M.-M. NHERF1/EBP50 is a new marker in colorectal cancer. Neoplasia. 2010;12(12):1013–1022. doi: 10.1593/neo.10780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Jiang Y. Y., Wang S. O., Holcomb J., et al. Crystallographic analysis of NHERF1-PLCβ3 interaction provides structural basis for CXCR2 signaling in pancreatic cancer. Biochemical and Biophysical Research Communications. 2014;446(2):638–643. doi: 10.1016/j.bbrc.2014.03.028. [DOI] [PubMed] [Google Scholar]
- 85.Fukuda T., Guo L., Shi X., Wu C. CH-ILKBP regulates cell survival by facilitating the membrane translocation of protein kinase B/Akt. The Journal of Cell Biology. 2003;160(7):1001–1008. doi: 10.1083/jcb.200212113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Eslami A., Miyaguchi K., Mogushi K., et al. PARVB overexpression increases cell migration capability and defines high risk for endophytic growth and metastasis in tongue squamous cell carcinoma. British Journal of Cancer. 2015;112(2):338–344. doi: 10.1038/bjc.2014.590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Sepulveda J. L., Wu C. The parvins. Cellular and Molecular Life Sciences. 2006;63(1):25–35. doi: 10.1007/s00018-005-5355-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Rosenberger G., Jantke I., Gal A., Kutsche K. Interaction of αPIX (ARHGEF6) with β-parvin (PARVB) suggests an involvement of αPIX in integrin-mediated signaling. Human Molecular Genetics. 2003;12(2):155–167. doi: 10.1093/hmg/ddg019. [DOI] [PubMed] [Google Scholar]
- 89.Nejad N. A., Amidi F., Hoseini M. A., et al. Male germ-like cell differentiation potential of human umbilical cord Wharton's jelly-derived mesenchymal stem cells in co-culture with human placenta cells in presence of BMP4 and retinoic acid. Iranian Journal of Basic Medical Sciences. 2015;18(4):325–333. [PMC free article] [PubMed] [Google Scholar]
- 90.Chung S. S. W., Wang X., Roberts S. S., Griffey S. M., Reczek P. R., Wolgemuth D. J. Oral administration of a retinoic Acid receptor antagonist reversibly inhibits spermatogenesis in mice. Endocrinology. 2011;152(6):2492–2502. doi: 10.1210/en.2010-0941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Flajollet S., Lefebvre B., Cudejko C., Staels B., Lefebvre P. The core component of the mammalian SWI/SNF complex SMARCD3/BAF60c is a coactivator for the nuclear retinoic acid receptor. Molecular and Cellular Endocrinology. 2007;270(1-2):23–32. doi: 10.1016/j.mce.2007.02.004. [DOI] [PubMed] [Google Scholar]
- 92.Wu Q. H., Guo R. J., Lin M., Zhou B., Wang Y. F. MicroRNA-200a inhibits CD133/1+ ovarian cancer stem cells migration and invasion by targeting E-cadherin repressor ZEB2. Gynecologic Oncology. 2011;122(1):149–154. doi: 10.1016/j.ygyno.2011.03.026. [DOI] [PubMed] [Google Scholar]
- 93.Yu F., Li J., Chen H., et al. Kruppel-like factor 4 (KLF4) is required for maintenance of breast cancer stem cells and for cell migration and invasion. Oncogene. 2011;30(18):2161–2172. doi: 10.1038/onc.2010.591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Bae K. M., Parker N. N., Dai Y., Vieweg J., Siemann D. W. E-cadherin plasticity in prostate cancer stem cell invasion. American Journal of Cancer Research. 2011;1:71–84. [PMC free article] [PubMed] [Google Scholar]
- 95.Yasuda A., Sawai H., Takahashi H., et al. The stem cell factor/c-kit receptor pathway enhances proliferation and invasion of pancreatic cancer cells. Molecular Cancer. 2006;5, article 46 doi: 10.1186/1476-4598-5-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Herreros-Villanueva M., Er T. K., Bujanda L. Retinoic acid reduces stem cell-like features in pancreatic cancer cells. Pancreas. 2015;44(6):918–924. doi: 10.1097/mpa.0000000000000373. [DOI] [PubMed] [Google Scholar]
- 97.Doi A., Ishikawa K., Shibata N., et al. Enhanced expression of retinoic acid receptor alpha (RARA) induces epithelial-to-mesenchymal transition and disruption of mammary acinar structures. Molecular Oncology. 2015;9(2):355–364. doi: 10.1016/j.molonc.2014.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Yang H.-J., Zheng Y.-B., Ji T., et al. Overexpression of ILK1 in breast cancer associates with poor prognosis. Tumor Biology. 2013;34(6):3933–3938. doi: 10.1007/s13277-013-0981-y. [DOI] [PubMed] [Google Scholar]
- 99.Leung-Hagesteijn C., Mahendra A., Naruszewicz I., Hannigan G. E. Modulation of integrin signal transduction by ILKAP, a protein phosphatase 2C associating with the integrin-linked kinase, ILK1. The EMBO Journal. 2001;20(9):2160–2170. doi: 10.1093/emboj/20.9.2160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Sanghera J., Costello P., Kojic L., et al. Integrin-linked kinase 1 (ILK1): a ‘hot’ therapeutic target. Clinical Cancer Research. 2001;7:3766s–3767s. [Google Scholar]
- 101.Bang D., Wilson W., Ryan M., Yeh J. J., Baldwin A. S. GSK-3α promotes oncogenic KRAS function in pancreatic cancer via TAK1-TAB stabilization and regulation of noncanonical NF-κB. Cancer Discovery. 2013;3(6):690–703. doi: 10.1158/2159-8290.cd-12-0541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Shi P., Yin T., Zhou F., Cui P., Gou S., Wang C. Valproic acid sensitizes pancreatic cancer cells to natural killer cell-mediated lysis by upregulating MICA and MICB via the PI3K/Akt signaling pathway. BMC Cancer. 2014;14, article 370 doi: 10.1186/1471-2407-14-370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Chow J. Y. C., Ban M., Wu H. L., et al. TGF-β downregulates PTEN via activation of NF-κB in pancreatic cancer cells. American Journal of Physiology: Gastrointestinal and Liver Physiology. 2010;298(2):G275–G282. doi: 10.1152/ajpgi.00344.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Chang L.-H., Pan S.-L., Lai C.-Y., Tsai A.-C., Teng C.-M. Activated PAR-2 regulates pancreatic cancer progression through ILK/HIF-α-induced TGF-α expression and MEK/VEGF-A-mediated angiogenesis. The American Journal of Pathology. 2013;183(2):566–575. doi: 10.1016/j.ajpath.2013.04.022. [DOI] [PubMed] [Google Scholar]
- 105.Huang A. H., Pan S. H., Chang W. H., et al. PARVA promotes metastasis by modulating ILK signalling pathway in lung adenocarcinoma. PLoS ONE. 2015;10(3) doi: 10.1371/journal.pone.0118530.e0118530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Yan Z. P., Yin H. Z., Wang R., et al. Overexpression of integrin-linked kinase (ILK) promotes migration and invasion of colorectal cancer cells by inducing epithelial-mesenchymal transition via NF-κB signaling. Acta Histochemica. 2014;116(3):527–533. doi: 10.1016/j.acthis.2013.11.001. [DOI] [PubMed] [Google Scholar]
- 107.Traister A., Walsh M., Aafaqi S., et al. Mutation in integrin-linked kinase (ILKR211A) and heat-shock protein 70 comprise a broadly cardioprotective complex. PLoS ONE. 2013;8(11) doi: 10.1371/journal.pone.0077331.e77331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Gilbert J. A., Adhikari L. J., Lloyd R. V., Halfdanarson T. R., Muders M. H., Ames M. M. Molecular markers for novel therapeutic strategies in pancreatic endocrine tumors. Pancreas. 2013;42(3):411–421. doi: 10.1097/mpa.0b013e31826cb243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Bergmann F., Aulmann S., Sipos B., et al. Acinar cell carcinomas of the pancreas: a molecular analysis in a series of 57 cases. Virchows Archiv. 2014;465(6):661–672. doi: 10.1007/s00428-014-1657-8. [DOI] [PubMed] [Google Scholar]
- 110.Mery L., Strauss B., Dufour J. F., Krause K. H., Hoth M. The PDZ-interacting domain of TRPC4 controls its localization and surface expression in HEK293 cells. Journal of Cell Science. 2002;115(17):3497–3508. doi: 10.1242/jcs.115.17.3497. [DOI] [PubMed] [Google Scholar]
- 111.Del Giudice I., Messina M., Chiaretti S., et al. Behind the scenes of non-nodal MCL: downmodulation of genes involved in actin cytoskeleton organization, cell projection, cell adhesion, tumour invasion, TP53 pathway and mutated status of immunoglobulin heavy chain genes. British Journal of Haematology. 2012;156(5):601–611. doi: 10.1111/j.1365-2141.2011.08962.x. [DOI] [PubMed] [Google Scholar]
- 112.Li X. L., Lu X., Parvathaneni S., et al. Identification of RECQ1-regulated transcriptome uncovers a role of RECQ1 in regulation of cancer cell migration and invasion. Cell Cycle. 2014;13(15):2431–2445. doi: 10.4161/cc.29419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Pastor M. D., Nogal A., Molina-Pinelo S., et al. Identification of proteomic signatures associated with lung cancer and COPD. Journal of Proteomics. 2013;89:227–237. doi: 10.1016/j.jprot.2013.04.037. [DOI] [PubMed] [Google Scholar]
- 114.Figueroa J. D., Han S. S., Garcia-Closas M., et al. Genome-wide interaction study of smoking and bladder cancer risk. Carcinogenesis. 2014;35:1737–1744. doi: 10.1093/carcin/bgu064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Zhou J. H., Feng Y. J., Tao K. T., et al. The expression and phosphorylation of ezrin and merlin in human pancreatic cancer. International Journal of Oncology. 2014;45(6):2059–2067. doi: 10.3892/ijo.2014.2381. [DOI] [PubMed] [Google Scholar]
- 116.Meng Y. X., Lu Z. H., Yu S. N., Zhang Q. A., Ma Y. H., Chen J. Ezrin promotes invasion and metastasis of pancreatic cancer cells. Journal of Translational Medicine. 2010;8, article 61 doi: 10.1186/1479-5876-8-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Kocher H. M., Sandle J., Mirza T. A., Li N. F., Hart I. R. Ezrin interacts with cortactin to form podosomal rosettes in pancreatic cancer cells. Gut. 2009;58(2):271–284. doi: 10.1136/gut.2008.159871. [DOI] [PubMed] [Google Scholar]
- 118.Oda Y., Aishima S., Morimatsu K., et al. Differential ezrin and phosphorylated ezrin expression profiles between pancreatic intraepithelial neoplasia, intraductal papillary mucinous neoplasm, and invasive ductal carcinoma of the pancreas. Human Pathology. 2013;44(8):1487–1498. doi: 10.1016/j.humpath.2012.12.001. [DOI] [PubMed] [Google Scholar]
- 119.Thevathasan J. V., Tan E., Zheng H., et al. The small GTPase HRas shapes local PI3K signals through positive feedback and regulates persistent membrane extension in migrating fibroblasts. Molecular Biology of the Cell. 2013;24(14):2228–2237. doi: 10.1091/mbc.e12-12-0905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Fensterer H., Giehl K., Buchholz M., et al. Expression profiling of the influence of RAS mutants on the TGFB1-induced phenotype of the pancreatic cancer cell line PANC-1. Genes Chromosomes & Cancer. 2004;39(3):224–235. doi: 10.1002/gcc.20000. [DOI] [PubMed] [Google Scholar]
- 121.Schönleben F., Qiu W., Allendorf J. D., Chabot J. A., Remotti H. E., Su G. H. Molecular analysis of PIK3CA, BRAF, and RAS oncogenes in periampullary and ampullary adenomas and carcinomas. Journal of Gastrointestinal Surgery. 2009;13(8):1510–1516. doi: 10.1007/s11605-009-0917-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Sayyah J., Bartakova A., Nogal N., Quilliam L. A., Stupack D. G., Brown J. H. The Ras-related protein, Rap1A, mediates thrombin-stimulated, integrin-dependent glioblastoma cell proliferation and tumor growth. The Journal of Biological Chemistry. 2014;289(25):17689–17698. doi: 10.1074/jbc.m113.536227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Park J.-S., Lim K.-M., Park S. G., et al. Pancreatic cancer induced by in vivo electroporation-enhanced sleeping beauty transposon gene delivery system in mouse. Pancreas. 2014;43(4):614–618. doi: 10.1097/mpa.0000000000000102. [DOI] [PubMed] [Google Scholar]
- 124.Shichinohe T., Senmaru N., Furuuchi K., et al. Suppression of pancreatic cancer by the dominant negative ras mutant, N116Y. The Journal of Surgical Research. 1996;66(2):125–130. doi: 10.1006/jsre.1996.0383. [DOI] [PubMed] [Google Scholar]
- 125.Sirivatanauksorn V., Sirivatanauksorn Y., Lemoine N. R. Molecular pattern of ductal pancreatic cancer. Langenbeck's Archives of Surgery. 1998;383(2):105–115. doi: 10.1007/s004230050101. [DOI] [PubMed] [Google Scholar]
- 126.Nagy A., Kozma L., Kiss I., et al. Copy number of cancer genes predict tumor grade and survival of pancreatic cancer patients. Anticancer Research. 2001;21(2):1321–1326. [PubMed] [Google Scholar]
- 127.Seufferlein T., Van Lint J., Liptay S., Adler G., Schmid R. M. Transforming growth factor α activates Ha-Ras in human pancreatic cancer cells with Ki-ras mutations. Gastroenterology. 1999;116(6):1441–1452. doi: 10.1016/s0016-5085(99)70509-3. [DOI] [PubMed] [Google Scholar]
- 128.Haggerty T. J., Dunn I. S., Rose L. B., Newton E. E., Pandolfi F., Kurnick J. T. Heat shock protein-90 inhibitors enhance antigen expression on melanomas and increase T cell recognition of tumor cells. PLoS ONE. 2014;9(12) doi: 10.1371/journal.pone.0114506.e114506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Zhou Y. J., Binder R. J. The heat shock protein-CD91 pathway mediates tumor immunosurveillance. OncoImmunology. 2014;3(4) doi: 10.4161/onci.28222.e28222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Dodd K., Nance S., Quezada M., et al. Tumor-derived inducible heat-shock protein 70 (HSP70) is an essential component of anti-tumor immunity. Oncogene. 2015;34:1312–1322. doi: 10.1038/onc.2014.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Qiao N., Zhu Y., Li H., Qu Z., Xiao Z. Expression of heat shock protein 20 inversely correlated with tumor progression in patients with ovarian cancer. European Journal of Gynaecological Oncology. 2014;35(5):576–579. doi: 10.12892/ejgot25602014. [DOI] [PubMed] [Google Scholar]
- 132.Ramp U., Mahotka C., Heikaus S., et al. Expression of heat shock protein 70 in renal cell carcinoma and its relation to tumor progression and prognosis. Histology and Histopathology. 2007;22(10–12):1099–1107. doi: 10.14670/HH-22.1099. [DOI] [PubMed] [Google Scholar]
- 133.Hyun J. J., Lee H. S., Keum B., et al. Expression of heat shock protein 70 modulates the chemoresponsiveness of pancreatic cancer. Gut and Liver. 2013;7(6):739–746. doi: 10.5009/gnl.2013.7.6.739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Kuramitsu Y., Wang Y., Taba K., et al. Heat-shock protein 27 plays the key role in gemcitabine-resistance of pancreatic cancer cells. Anticancer Research. 2012;32(6):2295–2299. [PubMed] [Google Scholar]
- 135.Zhang Z., Kawamura K., Jiang Y., et al. Heat-shock protein 90 inhibitors synergistically enhance melanoma differentiation-associated gene-7-mediated cell killing of human pancreatic carcinoma. Cancer Gene Therapy. 2013;20(12):663–670. doi: 10.1038/cgt.2013.66. [DOI] [PubMed] [Google Scholar]
- 136.MacKenzie T. N., Mujumdar N., Banerjee S., et al. Triptolide induces the expression of miR-142-3p: a negative regulator of heat shock protein 70 and pancreatic cancer cell proliferation. Molecular Cancer Therapeutics. 2013;12(7):1266–1275. doi: 10.1158/1535-7163.mct-12-1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Yu Y. K., Hamza A., Zhang T., et al. Withaferin A targets heat shock protein 90 in pancreatic cancer cells. Biochemical Pharmacology. 2010;79(4):542–551. doi: 10.1016/j.bcp.2009.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Mori-Iwamoto S., Taba K., Kuramitsu Y., et al. Interferon-γ down-regulates heat shock protein 27 of pancreatic cancer cells and helps in the cytotoxic effect of gemcitabine. Pancreas. 2009;38(2):224–226. doi: 10.1097/mpa.0b013e3181773970. [DOI] [PubMed] [Google Scholar]
- 139.Lang S. A., Moser C., Gaumann A., et al. Targeting heat shock protein 90 in pancreatic cancer impairs insulin-like growth factor-I receptor signaling, disrupts an interleukin-6/signal-transducer and activator of transcription 3/hypoxia-inducible factor-1α autocrine loop, and reduces orthotopic tumor growth. Clinical Cancer Research. 2007;13(21):6459–6468. doi: 10.1158/1078-0432.ccr-07-1104. [DOI] [PubMed] [Google Scholar]
- 140.Dudeja V., Phillips P., Sharif R., Zhan Y., Dawra R., Saluja A. Heat shock protein 70 (HSP70) confers resistance to apoptosts in pancreatic cancer cells by lysosomal stabilization. Pancreas. 2006;33(4):p. 457. doi: 10.1097/00006676-200611000-00073. [DOI] [Google Scholar]
- 141.Fendler W., Klich I., Cieślik-Heinrich A., Wyka K., Szadkowska A., Młynarski W. Increased risk of type 1 diabetes in Polish children—association with INS-IGF2 5′VNTR and lack of association with HLA haplotype. Endokrynologia Polska. 2011;62(5):436–442. [PubMed] [Google Scholar]
- 142.Sutherland G., Mellick G., Newman J., et al. Haplotype analysis of the IGF2-INS-TH gene cluster in Parkinson's disease. American Journal of Medical Genetics. Part B: Neuropsychiatric Genetics. 2008;147(4):495–499. doi: 10.1002/ajmg.b.30633. [DOI] [PubMed] [Google Scholar]
- 143.Polychronakos C., Kukuvitis A., Giannoukakis N., Colle E. Parental imprinting effect at the IPIS-IGF2 diabetes susceptibility locus. Diabetologia. 1995;38(6):715–719. doi: 10.1007/s001250050343. [DOI] [PubMed] [Google Scholar]
- 144.Ho G. Y. F., Melman A., Liu S.-M., et al. Polymorphism of the insulin gene is associated with increased prostate cancer risk. British Journal of Cancer. 2003;88(2):263–269. doi: 10.1038/sj.bjc.6600747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Eaves I. A., Bennett S. T., Forster P., et al. Transmission ratio distortion at the INS-IGF2 VNTR. Nature Genetics. 1999;22(4):324–325. doi: 10.1038/11890. [DOI] [PubMed] [Google Scholar]
- 146.Haiman C. A., Han Y., Feng Y., et al. Genome-wide testing of putative functional exonic variants in rlationship with breast and prostate cancer risk in a multiethnic population. PLoS Genetics. 2013;9(3) doi: 10.1371/journal.pgen.1003419.e1003419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Kanatsuna N., Delli A., Andersson C., et al. Doubly reactive INS-IGF2 autoantibodies in children with newly diagnosed autoimmune (type 1) diabetes. Scandinavian Journal of Immunology. 2015;82:361–369. doi: 10.1111/sji.12325. [DOI] [PubMed] [Google Scholar]
- 148.Kanatsuna N., Taneera J., Vaziri-Sani F., et al. Autoimmunity against INS-IGF2 protein expressed in human pancreatic islets. Journal of Biological Chemistry. 2013;288(40):29013–29023. doi: 10.1074/jbc.m113.478222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Paroni G., Fratelli M., Gardini G., et al. Synergistic antitumor activity of lapatinib and retinoids on a novel subtype of breast cancer with coamplification of ERBB2 and RARA. Oncogene. 2012;31(29):3431–3443. doi: 10.1038/onc.2011.506. [DOI] [PubMed] [Google Scholar]
- 150.Saumet A., Vetter G., Bouttier M., et al. Transcriptional repression of microRNA genes by PML-RARA increases expression of key cancer proteins in acute promyelocytic leukemia. Blood. 2009;113(2):412–421. doi: 10.1182/blood-2008-05-158139. [DOI] [PubMed] [Google Scholar]
- 151.Bailey T. A., Luan H., Tom E., et al. A kinase inhibitor screen reveals protein kinase C-dependent endocytic recycling of ErbB2 in breast cancer cells. The Journal of Biological Chemistry. 2014;289(44):30443–30458. doi: 10.1074/jbc.m114.608992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Qi H., Sun B., Zhao X., et al. Wnt5a promotes vasculogenic mimicry and epithelial-mesenchymal transition via protein kinase Cα in epithelial ovarian cancer. Oncology Reports. 2014;32(2):771–779. doi: 10.3892/or.2014.3229. [DOI] [PubMed] [Google Scholar]
- 153.Doller A., Winkler C., Azrilian I., et al. High-constitutive HuR phosphorylation at Ser 318 by PKC delta propagates tumor relevant functions in colon carcinoma cells. Carcinogenesis. 2011;32(5):676–685. doi: 10.1093/carcin/bgr024. [DOI] [PubMed] [Google Scholar]
- 154.Butler A. M., Scotti Buzhardt M. L. S., Li S., Smith K. E., Fields A. P., Murray N. R. Protein kinase C Zeta regulates human pancreatic cancer cell transformed growth and invasion through a STAT3-dependent mechanism. PLoS ONE. 2013;8(8) doi: 10.1371/journal.pone.0072061.e72061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Chen Y., Yu G. Z., Yu D. H., Zhu M. H. PKCα-induced drug resistance in pancreatic cancer cells is associated with transforming growth factor-β1. Journal of Experimental & Clinical Cancer Research. 2010;29(1, article 104) doi: 10.1186/1756-9966-29-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Antal C. E., Hudson A. M., Kang E., et al. Cancer-associated protein kinase C mutations reveal kinase's role as tumor suppressor. Cell. 2015;160(3):489–502. doi: 10.1016/j.cell.2015.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Linch M., Sanz-Garcia M., Soriano E., et al. A cancer-associated mutation in atypical protein kinase Cι occurs in a substrate-specific recruitment motif. Science Signaling. 2013;6(293, article ra82) doi: 10.1126/scisignal.2004068. [DOI] [PubMed] [Google Scholar]
- 158.Storz P. Targeting protein kinase C subtypes in pancreatic cancer. Expert Review of Anticancer Therapy. 2015;15(4):433–438. doi: 10.1586/14737140.2015.1003810. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Material I: lists 65 PC-related genes and their ensembl IDs.
Supplementary Material II: lists the detailed information of 2,080 shortest paths.
Supplementary Material III: lists edges in a graph consisting of shortest paths connecting any two PC-related genes.
Supplementary Material IV: lists 69 shortest path genes and their betweenness and permutation FDRs.