Abstract
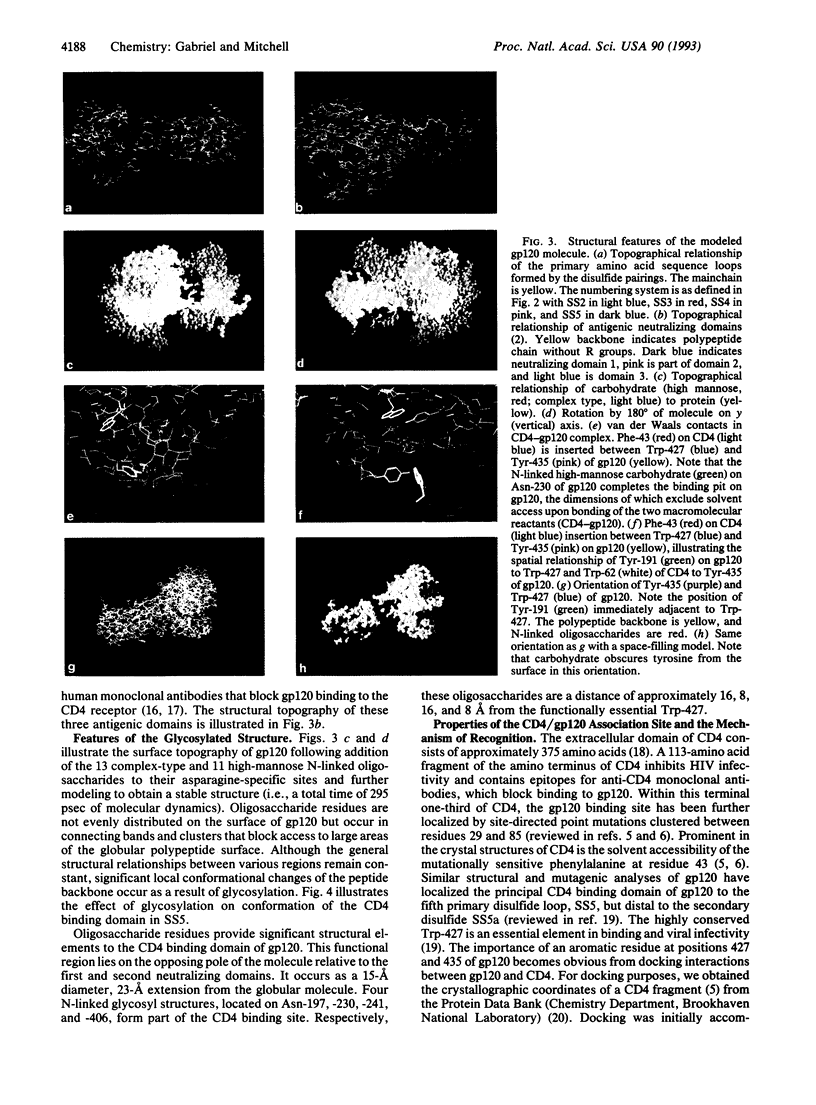
The atomic structure of a truncated glycoprotein gp120 from human immunodeficiency virus 1 (HIV-1) that contains the principal neutralizing antigenic sites and the CD4 binding domain has been derived by molecular dynamics and calculation of potential energy using the DREIDING force field. The resultant N-glycosylated molecular model is consistent with known properties of gp120 and docks with CD4 with a substantial reduction in the sum of the internal potential energies of the individual proteins (delta E = -200 kcal/mol). The primary mechanism of recognition and binding is the insertion of the solvent-accessible Phe-43 of CD4 into a gp120 solvent-accessible acceptor pit formed by Trp-427, Tyr-435, and the high-mannose oligosaccharide N-linked to Asn-230. delta E for the nonglycosylated complex is reduced significantly (-75 kcal/mol). Binding is by pi-pi* interactions of the aromatic groups forming a hydrophobic, thermodynamically stable environment for these functional noncovalent bonding participants. This model for gp120 provides a theoretical basis for the evaluation of HIV molecular pathogenesis involving the env proteins, the analysis of conformation on functional immune response of the host, and the design of nonproteinaceous inhibitors specific for the CD4 binding site on gp120.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- ANFINSEN C. B., HABER E., SELA M., WHITE F. H., Jr The kinetics of formation of native ribonuclease during oxidation of the reduced polypeptide chain. Proc Natl Acad Sci U S A. 1961 Sep 15;47:1309–1314. doi: 10.1073/pnas.47.9.1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anfinsen C. B. Principles that govern the folding of protein chains. Science. 1973 Jul 20;181(4096):223–230. doi: 10.1126/science.181.4096.223. [DOI] [PubMed] [Google Scholar]
- Bernstein F. C., Koetzle T. F., Williams G. J., Meyer E. F., Jr, Brice M. D., Rodgers J. R., Kennard O., Shimanouchi T., Tasumi M. The Protein Data Bank: a computer-based archival file for macromolecular structures. J Mol Biol. 1977 May 25;112(3):535–542. doi: 10.1016/s0022-2836(77)80200-3. [DOI] [PubMed] [Google Scholar]
- Burley S. K., Petsko G. A. Aromatic-aromatic interaction: a mechanism of protein structure stabilization. Science. 1985 Jul 5;229(4708):23–28. doi: 10.1126/science.3892686. [DOI] [PubMed] [Google Scholar]
- Chao B. H., Costopoulos D. S., Curiel T., Bertonis J. M., Chisholm P., Williams C., Schooley R. T., Rosa J. J., Fisher R. A., Maraganore J. M. A 113-amino acid fragment of CD4 produced in Escherichia coli blocks human immunodeficiency virus-induced cell fusion. J Biol Chem. 1989 Apr 5;264(10):5812–5817. [PubMed] [Google Scholar]
- Cordonnier A., Montagnier L., Emerman M. Single amino-acid changes in HIV envelope affect viral tropism and receptor binding. Nature. 1989 Aug 17;340(6234):571–574. doi: 10.1038/340571a0. [DOI] [PubMed] [Google Scholar]
- Creighton T. E. Experimental studies of protein folding and unfolding. Prog Biophys Mol Biol. 1978;33(3):231–297. doi: 10.1016/0079-6107(79)90030-0. [DOI] [PubMed] [Google Scholar]
- Creighton T. E., Goldenberg D. P. Kinetic role of a meta-stable native-like two-disulphide species in the folding transition of bovine pancreatic trypsin inhibitor. J Mol Biol. 1984 Nov 5;179(3):497–526. doi: 10.1016/0022-2836(84)90077-9. [DOI] [PubMed] [Google Scholar]
- Devereux J., Haeberli P., Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 1):387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earl P. L., Moss B., Doms R. W. Folding, interaction with GRP78-BiP, assembly, and transport of the human immunodeficiency virus type 1 envelope protein. J Virol. 1991 Apr;65(4):2047–2055. doi: 10.1128/jvi.65.4.2047-2055.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haigwood N. L., Nara P. L., Brooks E., Van Nest G. A., Ott G., Higgins K. W., Dunlop N., Scandella C. J., Eichberg J. W., Steimer K. S. Native but not denatured recombinant human immunodeficiency virus type 1 gp120 generates broad-spectrum neutralizing antibodies in baboons. J Virol. 1992 Jan;66(1):172–182. doi: 10.1128/jvi.66.1.172-182.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho D. D., McKeating J. A., Li X. L., Moudgil T., Daar E. S., Sun N. C., Robinson J. E. Conformational epitope on gp120 important in CD4 binding and human immunodeficiency virus type 1 neutralization identified by a human monoclonal antibody. J Virol. 1991 Jan;65(1):489–493. doi: 10.1128/jvi.65.1.489-493.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang C. Y., Nara P., Chamat S., Caralli V., Ryskamp T., Haigwood N., Newman R., Köhler H. Evidence for non-V3-specific neutralizing antibodies that interfere with gp120/CD4 binding in human immunodeficiency virus 1-infected humans. Proc Natl Acad Sci U S A. 1991 Jul 15;88(14):6171–6175. doi: 10.1073/pnas.88.14.6171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonard C. K., Spellman M. W., Riddle L., Harris R. J., Thomas J. N., Gregory T. J. Assignment of intrachain disulfide bonds and characterization of potential glycosylation sites of the type 1 recombinant human immunodeficiency virus envelope glycoprotein (gp120) expressed in Chinese hamster ovary cells. J Biol Chem. 1990 Jun 25;265(18):10373–10382. [PubMed] [Google Scholar]
- Modrow S., Hahn B. H., Shaw G. M., Gallo R. C., Wong-Staal F., Wolf H. Computer-assisted analysis of envelope protein sequences of seven human immunodeficiency virus isolates: prediction of antigenic epitopes in conserved and variable regions. J Virol. 1987 Feb;61(2):570–578. doi: 10.1128/jvi.61.2.570-578.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montefiori D. C., Robinson W. E., Jr, Mitchell W. M. Role of protein N-glycosylation in pathogenesis of human immunodeficiency virus type 1. Proc Natl Acad Sci U S A. 1988 Dec;85(23):9248–9252. doi: 10.1073/pnas.85.23.9248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muesing M. A., Smith D. H., Cabradilla C. D., Benton C. V., Lasky L. A., Capon D. J. Nucleic acid structure and expression of the human AIDS/lymphadenopathy retrovirus. Nature. 1985 Feb 7;313(6002):450–458. doi: 10.1038/313450a0. [DOI] [PubMed] [Google Scholar]
- Nygren A., Bergman T., Matthews T., Jörnvall H., Wigzell H. 95- and 25-kDa fragments of the human immunodeficiency virus envelope glycoprotein gp120 bind to the CD4 receptor. Proc Natl Acad Sci U S A. 1988 Sep;85(17):6543–6546. doi: 10.1073/pnas.85.17.6543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson W. E., Jr, Mitchell W. M. Neutralization and enhancement of in vitro and in vivo HIV and simian immunodeficiency virus infections. AIDS. 1990;4 (Suppl 1):S151–S162. [PubMed] [Google Scholar]
- Robinson W. E., Jr, Montefiori D. C., Mitchell W. M. Evidence that mannosyl residues are involved in human immunodeficiency virus type 1 (HIV-1) pathogenesis. AIDS Res Hum Retroviruses. 1987 Fall;3(3):265–282. doi: 10.1089/aid.1987.3.265. [DOI] [PubMed] [Google Scholar]
- Ryu S. E., Kwong P. D., Truneh A., Porter T. G., Arthos J., Rosenberg M., Dai X. P., Xuong N. H., Axel R., Sweet R. W. Crystal structure of an HIV-binding recombinant fragment of human CD4. Nature. 1990 Nov 29;348(6300):419–426. doi: 10.1038/348419a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sodroski J., Goh W. C., Rosen C., Campbell K., Haseltine W. A. Role of the HTLV-III/LAV envelope in syncytium formation and cytopathicity. 1986 Jul 31-Aug 6Nature. 322(6078):470–474. doi: 10.1038/322470a0. [DOI] [PubMed] [Google Scholar]
- Thali M., Olshevsky U., Furman C., Gabuzda D., Posner M., Sodroski J. Characterization of a discontinuous human immunodeficiency virus type 1 gp120 epitope recognized by a broadly reactive neutralizing human monoclonal antibody. J Virol. 1991 Nov;65(11):6188–6193. doi: 10.1128/jvi.65.11.6188-6193.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veronese F. D., DeVico A. L., Copeland T. D., Oroszlan S., Gallo R. C., Sarngadharan M. G. Characterization of gp41 as the transmembrane protein coded by the HTLV-III/LAV envelope gene. Science. 1985 Sep 27;229(4720):1402–1405. doi: 10.1126/science.2994223. [DOI] [PubMed] [Google Scholar]
- Wang J. H., Yan Y. W., Garrett T. P., Liu J. H., Rodgers D. W., Garlick R. L., Tarr G. E., Husain Y., Reinherz E. L., Harrison S. C. Atomic structure of a fragment of human CD4 containing two immunoglobulin-like domains. Nature. 1990 Nov 29;348(6300):411–418. doi: 10.1038/348411a0. [DOI] [PubMed] [Google Scholar]
- Weissman J. S., Kim P. S. Reexamination of the folding of BPTI: predominance of native intermediates. Science. 1991 Sep 20;253(5026):1386–1393. doi: 10.1126/science.1716783. [DOI] [PubMed] [Google Scholar]
- Willey R. L., Smith D. H., Lasky L. A., Theodore T. S., Earl P. L., Moss B., Capon D. J., Martin M. A. In vitro mutagenesis identifies a region within the envelope gene of the human immunodeficiency virus that is critical for infectivity. J Virol. 1988 Jan;62(1):139–147. doi: 10.1128/jvi.62.1.139-147.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]