ABSTRACT
Cell surface adhesion receptors play diverse functions in multicellular development. In Dictyostelium, two immunoglobulin-like adhesion proteins, TgrB1 and TgrC1, are essential components with dual roles in morphogenesis and allorecognition during development. TgrB1 and TgrC1 form a heterophilic adhesion complex during cell contact and mediate intercellular communication. The underlying signaling pathways, however, have not been characterized. Here, we report on a mutation that suppresses the tgrB–tgrC1-defective developmental arrest. The mutated gene alg9 encodes a putative mannosyl transferase that participates in N-linked protein glycosylation. We show that alteration in N-linked glycosylation, caused by an alg9 mutation with a plasmid insertion (alg9ins) or tunicamycin treatment, can partially suppress the developmental phenotypes caused by tgrC1 deletion or replacement with an incompatible allele. The alg9ins mutation also preferentially primed cells toward a stalk-cell fate. Despite its effect on development, we found that altered N-linked glycosylation had no discernable effect on TgrB1-TgrC1-mediated allorecognition. Our results show that N-linked protein glycosylation can modulate developmental processes without disturbing cell-cell recognition, suggesting that tgrB1 and tgrC1 have distinct effects in the two processes.
KEY WORDS: Allorecognition, Development, N-glycosylation, Genetic suppression, Dictyostelium
Summary: In Dictyostelium, compromised N-glycosylation, but not ER stress, caused by tunicamycin treatment or alg9 mutation, partially suppresses the developmental arrest in tgrB1-tgrC1-defective mutants.
INTRODUCTION
Dictyostelium development begins when individual cells starve. Upon starvation, the amoebae aggregate and form loose mounds of ∼105 cells, followed by a transition into coherent tight mounds. Cell-type-specific genes are initially induced during late aggregation (Barklis and Lodish, 1983; Chisholm et al., 1984; Mehdy et al., 1983), as cells within mounds adopt either a prestalk or a prespore fate. The tight mounds subsequently elongate to form migrating slugs and culminate into fruiting bodies consisting of spore-containing sori held aloft by cellular stalks. The tgrB1 and tgrC1 genes encode two immunoglobulin-like cell adhesion molecules (Ig-like CAMs) that form a heterophilic cell-cell adhesion complex (Chen et al., 2013, 2014) and are crucial for the initiation of cell differentiation. Cells with disrupted tgrB1 or tgrC1 genes fail to express cell-type-specific genes and are unable to progress beyond the loose mound stage (Dynes et al., 1994; Iranfar et al., 2006). Genetic analyses indicate that TgrC1 plays a role in intercellular signaling during cell contact (Kibler et al., 2003) and interacts in a feed-forward loop with GBF, a G-box binding protein, to regulate the transcription of post-aggregation genes (Iranfar et al., 2006). The signaling pathways that mediate these activities, however, have not been well characterized.
In addition to their roles in developmental signaling, TgrB1 and TgrC1 mediate discrimination of kin and non-kin (allorecognition) through attractive cell-cell adhesion (Benabentos et al., 2009; Hirose et al., 2011). During aggregative development, Dictyostelium cells preferentially co-aggregate and form developing structures with genetically similar individuals (Ostrowski et al., 2008). The kin recognition process is dependent on the polymorphic tgrB1 and tgrC1 genes, and their sequence dissimilarity predicts the degree of segregation when two divergent strains are developed in chimera (Benabentos et al., 2009). Kin discrimination in Dictyostelium directs cooperative behavior toward relatives and can protect cells against non-kin cheaters (Ho et al., 2013). In this recognition system, the tgrB1 and tgrC1 genes of a given strain function as a matching allelic pair that is necessary and sufficient to mediate allorecognition (Hirose et al., 2011). Non-matching alleles taken from divergent strains fail to function as a functional pair. For example, cells that carry the laboratory wild-type (AX4) tgrB1 allele and a foreign tgrC1 allele from the wild isolate QS38 (hereafter called tgrC1QS38 cells) display impaired kin recognition and severe developmental defects, similar to the phenotypes of the tgrB1-null or tgrC1-null cells. Therefore, both kin recognition and cell-type differentiation are regulated by allele-specific heterophilic interactions between TgrB1 and TgrC1.
We hypothesized that TgrB1 and TgrC1 regulate these functions through signal-transduction pathways, and we wanted to analyze these pathways by searching for genetic modifiers of the tgrB1–tgrC1-defective phenotype. Here, we performed a genetic screen to identify suppressors of tgrC1. We found that a plasmid insertion on the alg9 gene, which is involved in the biosynthesis of N-linked glycans, partially suppresses the developmental defects, but not the allorecognition defects. These results suggest that the TgrB1–TgrC1 system has dual functions that are mediated through linked, but distinct, mechanisms.
RESULTS
The alg9ins mutation partially suppresses the tgrC1− phenotype
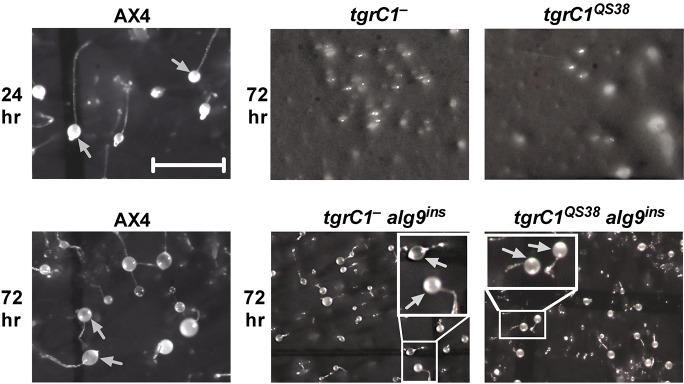
We performed a restriction-enzyme-mediated integration (REMI) screen for Dictyostelium discoideum genes that, once mutated, can mitigate the loose aggregate arrest of the tgrC1− and tgrC1QS38 cells (Fig. 1). In this screen, we identified a suppressor mutation with a plasmid insertion 19 bases upstream of the predicted start codon of the mannosyl transferase gene, alg9 (Fig. S1A). To test whether this insertion was the causative mutation, we excised the insertion plasmid along with flanking sequences from the original REMI mutant, using EcoRV as the restriction enzyme (Kuspa and Loomis, 1992). We then linearized the plasmid with EcoRV and used it to recapitulate the insertion event (alg9ins) by homologous recombination in fresh host cells (Fig. S1B). In the tgrC1− and tgrC1QS38 cells, recapitulation of the alg9ins partially suppressed the developmental arrest, resulting in the formation of small fruiting bodies at 72 h of development (Fig. 1). Therefore, alg9ins is most likely the mutation that suppressed the original developmental phenotype. We note that alg9ins had almost no effect on development in the wild-type AX4 background (Figs 1 and 2A).
Fig. 1.
alg9ins mutation partially suppresses the developmental arrest of tgrC1− and tgrC1QS38 cells. We developed cells on dark nitrocellulose filters for the indicated times and photographed multicellular structures from above. The relevant genotypes are indicated above each panel. Gray arrows indicate sori. Scale bar=1 mm. Insets are magnified 2×.
Fig. 2.

Sporulation efficiency of alg9ins mutants. We developed cells on dark filters for 72 h. Sporulation efficiency is the percentage of cells that formed spores. We used two clones of each strain carrying the recapitulated alg9ins mutation (c1 and c2) in different genetic backgrounds as indicated. Bars show mean and s.e.m. of independent replications. (A) AX4 cells and the related alg9ins mutants (n=8). (B) tgrC1− cells and the related alg9ins mutants. Sup. #9 is the original REMI mutant generated in the tgrC1− background (n=3–4). (C) tgrC1QS38 cells and the related alg9ins mutants (n=3–4). (D) tgrB1−/C1−cells and the related alg9ins mutants (n=3–4). Comparisons between bars (unpaired t-test): ns, not significant (P>0.05); **P≤0.01; ***P≤0.001. Notice that the y-axes are not the same between the different panels.
To quantify the effect of the alg9ins mutation on development, we measured the sporulation efficiency of the relevant strains. We tested two independent clones with the alg9ins mutation in both tgrC1− and tgrC1QS38 cells and observed significantly increased sporulation as compared with the respective parental strains (Fig. 2B and C). The original REMI suppressor (Sup. #9) found in the tgrC1− background showed a higher sporulation efficiency than the recapitulated strains (Fig. 2B). This observation is possibly due to uncharacterized secondary mutations. The alg9ins mutation in the AX4 background did not cause significant changes in sporulation efficiency compared with the parental AX4 cells (Fig. 2A). This observation indicates that an intact alg9 gene is not necessary for development under our test conditions. Fig. 2D shows that the tgrB1−/tgrC1− double knockout cells sporulated at a level similar to the tgrC1− alg9ins cells (Fig. 2B). Moreover, the alg9ins mutation did not improve the sporulation level of the tgrB1−/tgrC1− cells (Fig. 2D). These results suggest that modification of either tgrB1 or alg9 function might act through a common mechanism to suppress the sporulation defect in the tgrC1− background.
alg9ins mutation is associated with altered TgrB1 and TgrC1 proteins
alg9 encodes a mannosyl transferase that mediates the biosynthesis of N-glycan precursors in the endoplasmic reticulum (ER) (Fig. 3A). To test whether the pBSR1 insertion in the alg9 promoter affected the transcription of the alg9 gene, we examined the alg9 mRNA during development. We found that the tgrC1QS38 alg9ins cells produced a read-through transcript from the inserted pBSR1 plasmid through to the spliced alg9 gene (Fig. S1C). Using quantitative RT-PCR, we found that the expression level of the fusion pBSR1-alg9 transcript was significantly increased at 4, 8 and 12 h of development in the tgrC1QS38 alg9ins cells compared with the expression of the resident alg9 transcript in the parental tgrC1QS38 cells (Fig. S1D).
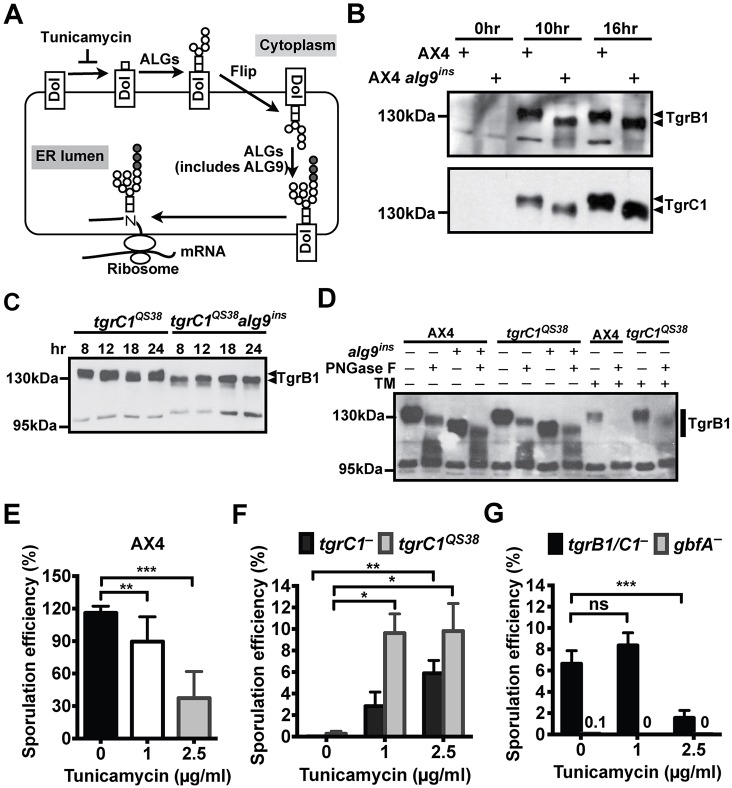
Fig. 3.
Tunicamycin treatment mimics the suppression effect of alg9ins. (A) An illustration of N-linked glycan biosynthesis: precursor N-linked glycan is initially assembled on dolichol (Dol) on the ER membrane before being transferred to a nascent polypeptide. The lipid-bound N-glycan precursor is extended by adding sugar residues one at a time by individual asparagine-linked glycosylation enzymes (ALGs). The process begins on the cytoplasmic face of the ER and then the lipid-bound N-glycan intermediate flips into the ER lumen. ALG proteins in the ER lumen, including ALG9, finish the synthesis of the ER N-glycan precursor. Addition of the first N-acetylgalactosamine to dolichol can be inhibited by tunicamycin. Sugar monomers: N-acetylgalactosamine (open rectangles), mannose (open circles) and glucose (filled circles). (B-C) We developed cells (AX4, tgrC1QS38 and their respective alg9ins mutants) on dark nitrocellulose filters for various times, as indicated. Cell lysates were analyzed by western blotting with anti-TgrB1 or anti-TgrC1 antibodies. The positions of TgrB1 and TgrC1 protein bands are indicated on the right of the panels. Molecular weights (kDa) are indicated on the left of the panels. (D) We developed cells (AX4, tgrC1QS38 and their respective alg9ins mutant) on dark nitrocellulose filters for 8 h with (+) or without (–) 2.5 µg/ml of tunicamycin (TM). Cell lysates were prepared and incubated with (+) or without (–) PNGase F to remove N-glycans on glycoproteins. Molecular weights and TgrB1 protein bands are indicated on the left and right of the panel, respectively. (E-G) We developed cells on dark nitrocellulose filters for 72 h with the addition of the indicated concentrations of tunicamycin. Sporulation efficiency is the percentage of cells that formed spores. Bars show mean and s.e.m. (E) AX4 cells (n=8). (F) tgrC1− (n=6) and tgrC1QS38 cells (n=7). (G) tgrB1−/C1− (n=7) and gbfA− cells (n=3). Numbers above the x-axis indicate values that are too low to be viewed as bars. Comparisons between bars (one-way ANOVA followed by Tukey's post hoc-test): ns, not significant (P>0.05); *P≤0.05; **P≤0.01; ***P≤0.001. Notice that the y-axes are not the same between the different panels.
Both disruption and overexpression of alg9 lead to altered ER N-glycan profiles and to increased relative abundance of smaller oligosaccharides (Frank and Aebi, 2005; Hykollari et al., 2013). To test whether the alg9ins mutation altered N-linked glycosylation, we performed western blot analysis to evaluate the size of the TgrB1 and TgrC1 proteins, which have 19 and 22 putative N-glycosylation sites, respectively. Fig. 3B shows that, in AX4 alg9ins cells, the TgrB1 and TgrC1 proteins indeed had a reduced size compared with the parental AX4 cells at 10 and 16 h of development (these proteins are not expressed at 0 h of development). A reduced molecular mass of TgrB1 was also observed in the tgrC1QS38 alg9ins cells at 8, 12, 18 and 24 h of development (Fig. 3C). These results suggest that the alg9ins mutation may compromise N-glycan biosynthesis. To test that possibility, we used PNGase F, which is an endoglycosidic enzyme that cleaves N-glycans lacking α(1→3)-fucose residues from glycoproteins. If the difference in protein size were solely due to change in N-linked glycosylation, we would expect the molecular weights of the proteins to be identical after removing N-glycans by PNGase F. Fig. 3D shows that after PNGase F treatment, the TgrB1 protein in the alg9ins mutant was smaller than its counterpart in the parental strain, suggesting that the alg9ins mutation resulted in alteration in both N-linked glycosylation and in other, as yet unidentified posttranslational modifications on TgrB1.
Tunicamycin treatment mimics the effects of the alg9ins mutation
To further test whether the phenotypic suppression was associated with alteration in the biosynthesis of N-glycans, we used a pharmacological approach to inhibit glycosylation with tunicamycin (Fig. 3A). First, we found that AX4 cells developed in the presence of 1 µg/ml or 2.5 µg/ml tunicamycin showed significantly decreased sporulation efficiency (Fig. 3E), indicating that the drug was effective at these concentrations. We then found that tunicamycin treatment mimicked the suppression caused by the alg9ins mutation in tgrC1− and tgrC1QS38 cells. In both cases, sporulation efficiency was significantly increased following drug treatment (Fig. 3F), thereby suggesting that compromising N-linked glycosylation can suppress the tgrC1-defective phenotype. These results also suggest that the suppression effect of the alg9ins mutation was possibly due to compromised N-linked glycosylation. Moreover, tunicamycin treatment (2.5 µl/ml) reduced the expression level but not the molecular weight of the TgrB1 protein (Fig. 3D), suggesting that the remaining TgrB1 protein had intact posttranslational modifications. Finally, drug treatment did not improve development in tgrB1−/tgrC1− cells (Fig. 3E), consistent with the lack of suppression by the alg9ins mutation in that genetic background (Fig. 2D).
GBF is a DNA binding protein that participates in a feed-forward loop with TgrC1 to regulate the expression of post-aggregation genes. gbfA− and tgrC1− single knockout cells have similar transcriptional profiles and developmental phenotypes, as both arrest at the loose aggregate stage (Iranfar et al., 2006). Therefore, we tested whether tunicamycin treatment could suppress the sporulation defect of the gbfA− cells. Fig. 3F shows that gbfA− cells failed to form spores in the presence of tunicamycin, suggesting that intact GBF function is necessary for the tunicamycin-mediated phenotypic suppression of tgrC1−.
An alternative explanation for the phenotypic suppression observed with alg9ins or tunicamycin treatment could be that ER stress caused suppression of the developmental defects. We therefore tested whether two ER stress inducers, thapsigargin and 17-N-allylamino-17-demethoxygeldanamycin, could suppress the tgrC1− defective phenotype. Fig. 4 shows that inducing ER stress did not improve sporulation of the tgrC1− and tgrC1QS38 cells. In that same figure, we show that the drug concentrations we used had negative effects on the sporulation efficiency of the wild type (AX4; Fig. 4A) but that no significant sporulation was observed in the tgrC1− and tgrC1QS38 cells treated with the same drug concentrations (Fig. 4B and C). These results suggest that altered N-glycosylation is the most likely cause of the developmental suppression observed with tunicamycin.
Fig. 4.
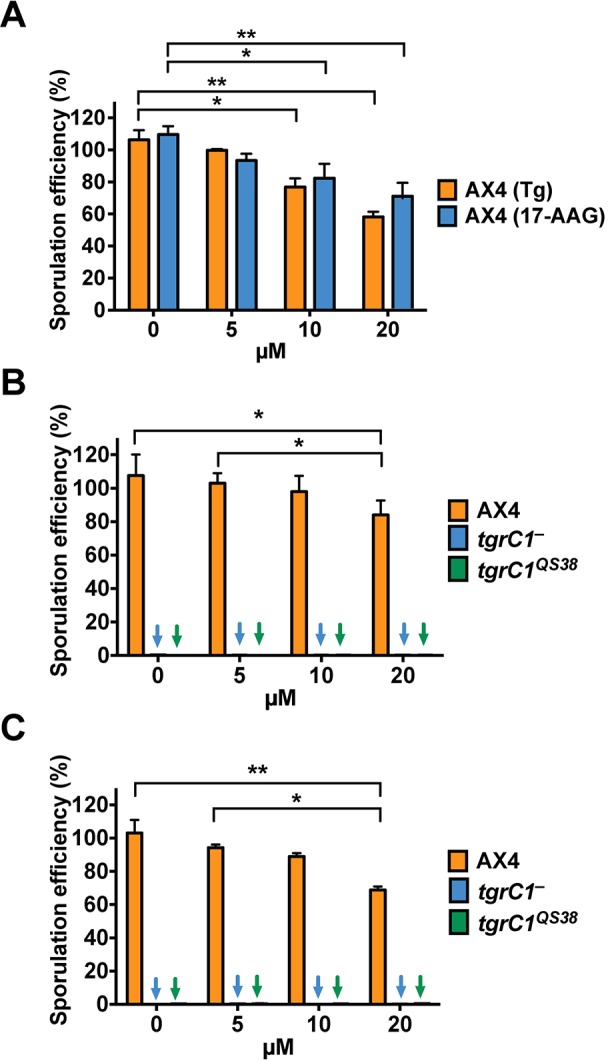
ER stress does not suppress the tgrC1− and the tgrC1QS38 phenotypes. We developed cells on dark nitrocellulose filters in the presence of one of the ER stress inducers, thapsigargin (Tg) or 17-N-Allylamino-17-demethoxygeldanamycin (17-AAG). Sporulation efficiency is the percentage of the total number of cells that formed spores. Bars show mean and s.e.m. (A) Treatment of wild-type (AX4) cells with the indicated concentrations of Tg or 17-AAG for 30 h (n=3). (B) 17-AAG treatment of AX4, tgrC1− and tgrC1QS38 cells for 72 h (n=3). (C) Thapsigargin treatment of AX4, tgrC1− and tgrC1QS38 cells for 72 h (n=3). Colored arrows (blue, tgrC1−; green, tgrC1QS38) indicate bars with values below the limit of detection. Comparisons between bars (one-way ANOVA followed by Tukey's post hoc-test): *P≤0.05; **P≤0.01.
Compromised N-linked glycosylation does not affect allorecognition
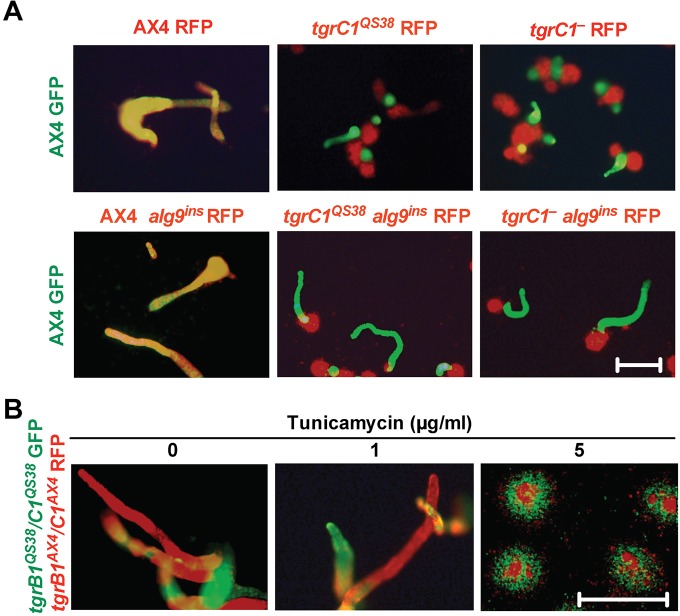
N-linked glycosylation can influence the binding kinetics of cell-cell adhesion (Langer et al., 2012). We tested whether the specific interaction of TgrB1 and TgrC1 is dependent on N-linked glycosylation by performing a segregation assay (Benabentos et al., 2009). We developed two strains labeled with different fluorescence markers in chimera and followed their segregation and development patterns (Fig. 5). AX4 cells labeled with GFP mixed well with compatible AX4 cells labeled with RFP, but they segregated from the RFP-labeled tgrC1− and tgrC1QS38 cells (Fig. 5A, top row). Identical results were observed in the respective alg9ins strains: GFP-labeled AX4 cells mixed well with RFP-labeled AX4 alg9ins cells but segregated from the RFP-labeled tgrC1− alg9ins and tgrC1QS38 alg9ins cells (Fig. 5A, bottom row). These findings suggest that N-linked glycans do not influence the function of TgrB1 and TgrC1 in allorecognition. To further test this conclusion, we observed the effect of tunicamycin treatment on allorecognition. We mixed two incompatible gene replacement strains (Hirose et al., 2011), GFP-labeled tgrB1QS38tgrC1QS38 and RFP-labeled tgrB1AX4tgrC1AX4, in the presence of various concentrations of tunicamycin and followed their development and segregation patterns (Fig. 5B). We found that, in the absence of tunicamycin, the two strains segregated into distinct slugs, as previously reported (Hirose et al., 2011). Similar results were observed in the presence of 1 µg/ml tunicamycin, suggesting that a drug concentration that restores partial sporulation in the tgrC1-defective strains (Fig. 3F) does not significantly compromise segregation (Fig. 5B). Using a higher drug concentration (5 µg/ml tunicamycin), we found that development was arrested at the loose aggregate stage, but that cells with distinct allotypes still segregated. We conclude that reduction of N-linked glycosylation does not have an overt effect on the allele-specific interactions of TgrB1 and TgrC1.
Fig. 5.
Allorecognition is not modified by the alg9ins mutation nor by tunicamycin treatment. We developed the GFP- and RFP-labeled strains in chimera on non-nutrient agar. We took GFP and RFP images at 14–17 h of development and merged the images such that yellow indicates mixing of GFP- and RFP-labeled cells whereas green or red indicates segregated clonal populations. The relevant genotypes of the mixed strains are indicated. (A) GFP-labeled AX4 cells were developed in chimera with RFP-labeled strains as indicated above each panel. Scale bar=0.5 mm. (B) tgrB1QS38/C1QS38 GFP cells and tgrB1AX4/C1AX4 RFP cells were developed in chimera with the addition of tunicamycin at the indicated concentrations. Scale bar=0.5 mm.
The alg9ins mutation primes cells for prestalk differentiation
Glycan structures on the cell surface change temporally and spatially during cell differentiation and morphogenesis, and different cell types have unique signatures of N-linked and O-linked glycans (Haltiwanger and Lowe, 2004). We tested whether changes in N-glycan structures in the alg9ins mutants may have led to altered spore and stalk differentiation. We developed cells with or without the alg9ins mutation clonally and in chimera, and we tested the sporulation efficiencies and cell-type preferences of the strains. We found that alg9+ cells contributed the majority of the spores when developed in chimera with their respective alg9ins mutants. In Fig. 6A we show that pure populations of AX4 (I) and of AX4 alg9ins (II) cells had the expected ∼90% sporulation efficiency. When developed in a chimera at equal proportions (I+II), the AX4 spores were more abundant (55.6% of the total number of spores) than the AX4 alg9ins spores. In Fig. 6B we show that a pure population of tgrC1QS38 alg9ins cells was able to form spores at ∼17% efficiency (IV) whereas a pure population of tgrC1QS38 cells failed to form spores altogether (III). In the respective chimera (III+IV), the tgrC1QS38 cells made up the vast majority of the spores (88.6%). These findings suggest that the alg9ins mutation has a non-cell-autonomous component in its suppressor effect on spore differentiation. Moreover, in chimeric slugs of AX4 and its respective alg9ins mutant, the alg9ins cells were enriched in the prestalk O region (Fig. 6C). These findings suggest that alg9ins cells have a bias toward prestalk O differentiation.
Fig. 6.

Suppression by alg9 inactivation is non-cell-autonomous and costly. We developed cells either clonally or in chimera at a 1:1 ratio on dark nitrocellulose filters for 72 h. Sporulation efficiency is the percentage of cells that formed spores. Bars show mean and s.e.m. (A) AX4 cells (I, black bars), AX4 alg9ins (II, white bars) and the respective mix (I+II). AX4 made up 55.6% of the spores in the chimera (n=3). (B) tgrC1QS38 cells (III, white bars), tgrC1QS38 alg9ins cells (IV, gray bars) and the respective mix (III+IV). tgrC1QS38 alg9ins cells made up 88.6% of the spores in the chimera (n=3). Notice that the y-axes are not the same in the different panels. (C) AX4 GFP and AX4 alg9ins RFP cells were developed in chimera on non-nutrient agar for 16 h. Fluorescence images of a representative slug are shown. The prestalk O region is indicated by gray arrows. Left: GFP channel; Middle: merged image channel; Right, RFP channel. (D) A model, suggesting that suppression of the tgrC1-defective phenotype by either tgrB1 deletion, alg9ins mutation or tunicamycin treatment share a similar mechanism that is dependent on GBF activity.
DISCUSSION
Our finding that tunicamycin treatment recapitulates the phenotype caused by the alg9ins mutation suggests the alg9ins mutation may result in imbalance in the N-linked glycosylation process. Both the genetic and pharmacological approaches partially suppressed the loose mound phenotype of the tgrC1− and tgrC1QS38, suggesting that N-glycans play a role in the regulation of developmental progression past the loose mound stage. Interestingly, previous studies have shown that developmental progression and cell differentiation in Dictyostelium are associated with changes in the N-glycan profile (Lam and Siu, 1981; Schiller et al., 2009). Dictyostelium cells form two major cell types during development – stalk cells and spores. Prespore cells are more active in N-glycan biosynthesis than prestalk cells (Riley et al., 1993). In contrast, the deglycosylation enzyme, DdPNGase, is more abundant in prestalk cells and is required for stalk differentiation (Gosain et al., 2012). Moreover, the glycosylation mutant modB preferentially forms stalk cells when developed in chimera with wild-type cells (Houle et al., 1989). Therefore, our finding that the alg9ins mutants preferentially contributed to the stalk population in chimera suggests that the alg9ins mutation resulted in compromised N-linked glycosylation. In sum, our results and the previous studies support the conclusion that temporal control of glycosylation plays regulatory roles in cell differentiation and developmental progression in Dictyostelium.
Altered N-linked glycosylation alleviates the tgrC1− phenotype, but the mechanism is not obvious. There can be hundreds of glycoproteins whose stability and activity are affected by N-linked glycosylation. Therefore, the phenotypic suppression can be a result of the collective outputs of some or all of the affected biological processes. Using TgrB1 and TgrC1 as indicators of posttranslational modification, we noticed a decrease in the molecular weights of the proteins in the alg9ins mutants. However, by using PNGase F to remove the N-linked glycans, we found that modifications other than N-linked glycosylation are probably at play as well. Since we did not observe the same change in protein size in the tunicamycin-treated cells, the results argue that other mechanisms might be involved. Finally, our studies cannot determine whether the altered modification on TgrB1 and TgrC1 contributed to the phenotypic suppression. Specifically, our results show that the sporulation defect of tgrC1− cells can be partially mitigated by either tunicamycin treatment or the alg9ins mutation. It is known that tgrB1 deletion in the tgrC1− background also allows cells to sporulate at a reduced efficiency (Hirose et al., 2011). We found no additive effect in suppression when tgrB1 deletion was combined with tunicamycin treatment or the alg9ins mutation. These results suggest that the three manipulations act through a common pathway. We also found that GBF, which participates in a developmental feed-forward loop with TgrC1 (Iranfar et al., 2006), is required for the tunicamycin-induced phenotypic suppression. In sum, these results suggest that tunicamycin treatment and alg9ins mutation may act directly or indirectly on the TgrB1-related pathway and depend on GBF activity (Fig. 6D).
We were initially concerned that ER stress, which could be caused by abnormal N-linked glycosylation, might contribute to the observed developmental suppression. ER-stress-inducing drugs that do not affect N-linked glycosylation did not suppress the sporulation defect, suggesting that this possibility is rather unlikely. The TgrB1 and TgrC1 cell adhesion system of Dictyostelium is an essential component of development, with roles in morphogenesis and allorecognition (Dynes et al., 1994; Hirose et al., 2011). Although the precise signaling pathways that mediate these roles are uncertain, it is clear that allorecognition is mediated by allele-specific interactions between TgrB1 molecules on the outer membrane of one cell and matching TgrC1 molecules on the outer membrane of a neighboring cell (Chen et al., 2013, 2014; Hirose et al., 2011). It was therefore surprising to find that allorecognition, which is mediated by TgrB1 and TgrC1, was not affected by compromised N-linked glycosylation. The binding specificity of TgrB1 and TgrC1 may thus be independent of N-linked glycosylation. It is possible that TgrB1 and TgrC1 interactions elicit two signaling pathways within the cells, one of which is dedicated to development and dependent on N-linked glycosylation and another that participates in allorecognition in an N-linked glycosylation independent manner.
In summary, our results suggest that kin discrimination and developmental signaling in the TgrB1–TgrC1 adhesion system have independent components. We propose that N-linked glycosylation plays more important roles in the developmental processes of Dictyostelium and less so in the allorecognition mediated by the TgrB1–TgrC1 adhesion system. Ultimately, this study uses the tgrB1–tgrC1-defective backgrounds to provide new insights into the roles of N-linked glycosylation on developmental progression.
MATERIALS AND METHODS
Strains and cell growth
All the Dictyostelium discoideum strains used here are described in Table S1. Cells were grown in shaking suspension in HL5 medium supplemented with 50 U/ml penicillin and 50 µg/ml streptomycin at 22°C (Miranda et al., 2013). In drug-resistant strains, 5 µg/ml Blasticidin-S or 10 µg/ml G418 was added as necessary. Prior to any experiment, cells were grown in HL5 without Blasticidin-S or G418 for at least 24 h.
Development and sporulation efficiency
Cells were grown to mid-log phase, washed once with KK2 buffer (16.3 mM KH2PO4, 3.7 mM K2HPO4) and resuspended in PDF buffer (9.2 mM K2HPO4, 13.2 mM KH2PO4, 20 mM KCl, 1 mM CaCl2, 2.5 mM MgSO4, pH 6.4) containing 500 µg/ml streptomycin. The cells were deposited on dark nitrocellulose filters at a density of 4×106 cells/cm2. The filters were placed on a paper filter soaked in PDF buffer and incubated in a humid chamber at 22°C. To measure sporulation efficiency, we harvested spores by detergent treatment (KK2 buffer containing 0.1% NP-40 and 10 mM EDTA) after 30 or 72 h of development. Sporulation efficiency was calculated as the fraction (%) of the number of spores formed relative to the number of cells developed.
Drug treatment
Tunicamycin, thapsigargin and 17-N-Allylamino-17-demethoxygeldanamycin (all purchased from Sigma-Aldrich) were prepared as stock solutions in DMSO and added at the indicated concentrations into the media at the onset of development.
Transformation
Plasmids were transformed into cells by electroporation as described (Kuspa and Loomis, 1992; Ostrowski et al., 2008). In brief, plasmids were electroporated into 0.5 to 1×107 cells by pulsing twice at 0.95 kV and 25 µF. Cells were allowed to recover for one day in submerged culture with HL5 before the addition of Blasticidin-S (5 µg/ml) or G418 (5 µg/ml) as necessary. To recapitulate the alg9 insertion, the rescued plasmid was linearized with EcoRV and transformed into cells (Fig. S1A). Clones were screened for the correct integration event by PCR with the following primers: F, 5′-CACATCCTTGCTGTTTCAC-3′; R, 5′-TGGACTGTATTCCCATGTTTG-3′; R1, 5′-GCCATCCCATGAATACTG-3′; F1, 5′-TACCGATGAAGAACTCTCAC-3′; T7, 5′-TAATACGACTCACTATAGGG-3′ (Fig. S1).
REMI mutagenesis and suppressor screen
We performed REMI mutagenesis as described (Kuspa and Loomis, 1992; Shaulsky et al., 1996). The parental strains were tgrC1− and tgrC1QS38 cells that arrest at the loose aggregate stage during development. 10 µg BamHI-linearized pBSR1 and 30 U DpnII were transformed into 1×107 cells. Pools of 100–250 REMI mutants were grown on 10-cm SM-agar plates in association with Klebsiella aerogenes at a density of 0.1 to 1×105 cells per plate. After 7–10 days, all cells were collected, selected for spores by detergent treatment (KK2 buffer containing 0.1% NP-40 and 10 mM EDTA) and washed once with KK2 buffer. Serial dilutions of spores were germinated on bacterial lawns on SM-agar plates to allow the formation of single-cell-derived colonies. Plates were screened for developmental suppressors that form fruiting bodies.
RNA extraction and quantitative RT-PCR
We developed cells, harvested 5×107 cells at 0, 4, 8, 12, 18 or 24 h of development, and resuspended them in 1 ml Trizol reagent (Invitrogen). Total RNA was extracted according to the manufacturer's recommendations. cDNA was synthesized by using SuperScript II reverse transcriptase (ThermoFisher #18064-014) and quantified by quantitative RT-PCR. Two independent biological samples of each strain were collected for cDNA synthesis. The alg9 expression level was quantified and normalized to the expression level of Ig7. Primers for alg9: 5′-GGCATTGTTACCAGTTATTG-3′ and 5′-AGGAATCTTTCTTCTTTATGTG-3′. Primers for Ig7: 5′-TTACATTTATTAGACCCGAAACCAAGCG-3′ and 5′-TTCCCTTTAGACCTATGGACCTTAGCG-3′.
Segregation
GFP- and RFP-tagged cells were grown in pure populations. We washed the cells once with KK2, mixed at a 1:1 ratio in PDF buffer and deposited on non-nutrient agars (1.5% KK2 agar). Cells were developed for the indicated period of time before being photographed under an epifluorescence microscope. Merged images of GFP and RFP channels are shown.
Western blotting
We developed cells, harvested 5×107 cells at various time points of development and lysed them in 300 µl lysis buffer [20 mM Tris-HCl (pH 8.0), 120 mM NaCl, 0.1 mM PMSF, 2 mM EDTA, 1 mM sodium orthovanadate, 0.5% NP-40] for 10 min at 4°C. After centrifugation, the supernatants were denatured with 1× glycoprotein denaturing buffer (0.5% SDS, 40 mM DTT) at 100°C for 10 min and used for SDS-PAGE. For PNGase F digestion (NEB #0704), 10 µl of denatured cell lysate were treated with 1000 units of PNGase F at 37°C overnight. After SDS-PAGE, proteins were transferred to a nitrocellulose membrane, blocked with 10% non-fat milk and blotted with anti-TgrB1 or anti-TgrC1 multi-clonal antibodies. Target proteins were detected by using horseradish peroxidase-conjugated secondary antibodies, followed by the enhanced chemiluminescence detection kit (GE Healthcare).
Acknowledgements
We thank Y. Wang for insightful discussions, and C. Dinh and M. Kurasawa for helpful advice.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
C.-L.F.L. and G.S. conceived the study. C.-L.F.L. conducted the experiments and analyzed the data. G.C. performed the western blotting shown in Fig. 3B–D. A.N.W. performed the segregation assays shown in Fig. 5B. G.S. provided feedback on the manuscript. All authors read and approved the final manuscript.
Funding
This work was supported by the National Institutes of Health [grant number R01 GM098276]. C.-L.F.L. was partially funded by the Burroughs Wellcome Fund through the Houston Laboratory and Population Sciences Training Program in Gene-Environment Interaction. Deposited in PMC for release after 12 months.
Supplementary information
Supplementary information available online at http://jcs.biologists.org/lookup/suppl/doi:10.1242/jcs.172882/-/DC1
References
- Barklis E. and Lodish H. F. (1983). Regulation of dictyostelium discoideum mRNAs specific for prespore or prestalk cells. Cell 32, 1139-1148. 10.1016/0092-8674(83)90297-0 [DOI] [PubMed] [Google Scholar]
- Benabentos R., Hirose S., Sucgang R., Curk T., Katoh M., Ostrowski E. A., Strassmann J. E., Queller D. C., Zupan B., Shaulsky G. et al. (2009). Polymorphic members of the lag gene family mediate kin discrimination in Dictyostelium. Curr. Biol. 19, 567-572. 10.1016/j.cub.2009.02.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen G., Wang J., Xu X., Wu X., Piao R. and Siu C.-H. (2013). TgrC1 mediates cell–cell adhesion by interacting with TgrB1 via mutual IPT/TIG domains during development of Dictyostelium discoideum. Biochem. J. 452, 259-269. 10.1042/BJ20121674 [DOI] [PubMed] [Google Scholar]
- Chen G., Xu X., Wu X., Thomson A. and Siu C.-H. (2014). Assembly of the TgrB1–TgrC1 cell adhesion complex during Dictyostelium discoideum development. Biochem. J. 459, 241-249. 10.1042/BJ20131594 [DOI] [PubMed] [Google Scholar]
- Chisholm R. L., Barklis E. and Lodish H. F. (1984). Mechanism of sequential induction of cell-type specific mRNAs in Dictyostelium differentiation. Nature 310, 67-69. 10.1038/310067a0 [DOI] [PubMed] [Google Scholar]
- Dynes J. L., Clark A. M., Shaulsky G., Kuspa A., Loomis W. F. and Firtel R. A. (1994). LagC is required for cell-cell interactions that are essential for cell-type differentiation in Dictyostelium. Genes Dev. 8, 948-958. 10.1101/gad.8.8.948 [DOI] [PubMed] [Google Scholar]
- Frank C. G. and Aebi M. (2005). ALG9 mannosyltransferase is involved in two different steps of lipid-linked oligosaccharide biosynthesis. Glycobiology 15, 1156-1163. 10.1093/glycob/cwj002 [DOI] [PubMed] [Google Scholar]
- Gosain A., Lohia R., Shrivastava A. and Saran S. (2012). Identification and characterization of peptide: N- glycanase from Dictyostelium discoideum. BMC Biochem. 13, 9 10.1186/1471-2091-13-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haltiwanger R. S. and Lowe J. B. (2004). Role of glycosylation in development. Annu. Rev. Biochem. 73, 491-537. 10.1146/annurev.biochem.73.011303.074043 [DOI] [PubMed] [Google Scholar]
- Hirose S., Benabentos R., Ho H.-I., Kuspa A. and Shaulsky G. (2011). Self-recognition in social amoebae is mediated by allelic pairs of tiger genes. Science 333, 467-470. 10.1126/science.1203903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho H.-I., Hirose S., Kuspa A. and Shaulsky G. (2013). Kin recognition protects cooperators against cheaters. Curr. Biol. 23, 1590-1595. 10.1016/j.cub.2013.06.049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houle J., Balthazar J. and West C. M. (1989). A glycosylation mutation affects cell fate in chimeras of Dictyostelium discoideum. Proc. Natl. Acad. Sci. USA 86, 3679-3683. 10.1073/pnas.86.10.3679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hykollari A., Balog C. I. A., Rendić D., Braulke T., Wilson I. B. H. and Paschinger K. (2013). Mass spectrometric analysis of neutral and anionic N-glycans from a Dictyostelium discoideum model for human congenital disorder of glycosylation CDG IL. J. Proteome Res. 12, 1173-1187. 10.1021/pr300806b [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iranfar N., Fuller D. and Loomis W. F. (2006). Transcriptional regulation of post-aggregation genes in Dictyostelium by a feed-forward loop involving GBF and LagC. Dev. Biol. 290, 460-469. 10.1016/j.ydbio.2005.11.035 [DOI] [PubMed] [Google Scholar]
- Kibler K., Svetz J., Nguyen T.-L., Shaw C. and Shaulsky G. (2003). A cell-adhesion pathway regulates intercellular communication during Dictyostelium development. Dev. Biol. 264, 506-521. 10.1016/j.ydbio.2003.08.025 [DOI] [PubMed] [Google Scholar]
- Kuspa A. and Loomis W. F. (1992). Tagging developmental genes in Dictyostelium by restriction enzyme-mediated integration of plasmid DNA. Proc. Natl. Acad. Sci. USA 89, 8803-8807. 10.1073/pnas.89.18.8803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam T. Y. and Siu C.-H. (1981). Synthesis of stage-specific glycoproteins in Dictyostelium discoideum during development. Dev. Biol. 83, 127-137. 10.1016/S0012-1606(81)80015-2 [DOI] [PubMed] [Google Scholar]
- Langer M. D., Guo H., Shashikanth N., Pierce J. M. and Leckband D. E. (2012). N-glycosylation alters cadherin-mediated intercellular binding kinetics. J. Cell Sci. 125, 2478-2485. 10.1242/jcs.101147 [DOI] [PubMed] [Google Scholar]
- Mehdy M. C., Ratner D. and Firtel R. A. (1983). Induction and modulation of cell-type-specific gene expression in Dictyostelium. Cell 32, 763-771. 10.1016/0092-8674(83)90062-4 [DOI] [PubMed] [Google Scholar]
- Miranda E. R., Zhuchenko O., Toplak M., Santhanam B., Zupan B., Kuspa A. and Shaulsky G. (2013). ABC transporters in Dictyostelium discoideum development. PLoS ONE 8, e70040 10.1371/journal.pone.0070040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostrowski E. A., Katoh M., Shaulsky G., Queller D. C., Strassmann J. E. and Barton, N. H. (2008). Kin discrimination increases with genetic distance in a social amoeba. PLoS Biol. 6, e287 10.1371/journal.pbio.0060287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riley G. R., West C. M. and Henderson E. J. (1993). Cell differentiation in Dictyostelium discoideum controls assembly of protein-linked glycans. Glycobiology 3, 165-177. 10.1093/glycob/3.2.165 [DOI] [PubMed] [Google Scholar]
- Schiller B., Hykollari A., Voglmeir J., Pöltl G., Hummel K., Razzazi-Fazeli E., Geyer R. and Wilson I. B. H. (2009). Development of Dictyostelium discoideum is associated with alteration of fucosylated N-glycan structures. Biochem. J. 423, 41-52. 10.1042/BJ20090786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaulsky G., Escalante R. and Loomis W. F. (1996). Developmental signal transduction pathways uncovered by genetic suppressors. Proc. Natl. Acad. Sci. USA 93, 15260-15265. 10.1073/pnas.93.26.15260 [DOI] [PMC free article] [PubMed] [Google Scholar]