Abstract
Treatment of solid and liquid tobacco wastes with high nicotine content remains a longstanding challenge. Here, we explored an environmentally friendly approach to replace tobacco waste disposal with resource recovery by genetically engineering Pseudomonas putida. The biosynthesis of 3-succinoyl-pyridine (SP), a precursor in the production of hypotensive agents, from the tobacco waste was developed using whole cells of the engineered Pseudomonas strain, S16dspm. Under optimal conditions in fed-batch biotransformation, the final concentrations of product SP reached 9.8 g/L and 8.9 g/L from aqueous nicotine solution and crude suspension of the tobacco waste, respectively. In addition, the crystal compound SP produced from aqueous nicotine of the tobacco waste in batch biotransformation was of high purity and its isolation yield on nicotine was 54.2%. This study shows a promising route for processing environmental wastes as raw materials in order to produce valuable compounds.
Tobacco (Nicotiana, of the Solanaceae family), cash crop with a long history, is widely cultivated in USA, China, India, Brazil, and Cuba. China is the largest producer and consumer of tobacco worldwide, accounting for about a third of the total global consumption1. However, parts of tobacco leaves that can not be used in cigarette production are discarded because their nicotine content is in a high range of 3–6% (w/w)2. As an N-heterocyclic alkaloid, nicotine is the main toxic organic compound in solid or liquid tobacco wastes. The lethal dose of pure nicotine for adults is reported to be 60 mg, an oral LD50 of 0.8 mg/kg, or less (30–60 mg)3. In addition, nicotine released into the environment as a result of the tobacco industry has been designated as a Toxic Release Inventory (TRI) chemical by the United States Environmental Protection Agency4. Due to its high solubility in aqueous solutions, nicotine leached from tobacco wastes can be easily transported into ground water. In previous reports, nicotine has been increasingly detected in seepage and leakage water from landfill wastes5,6.
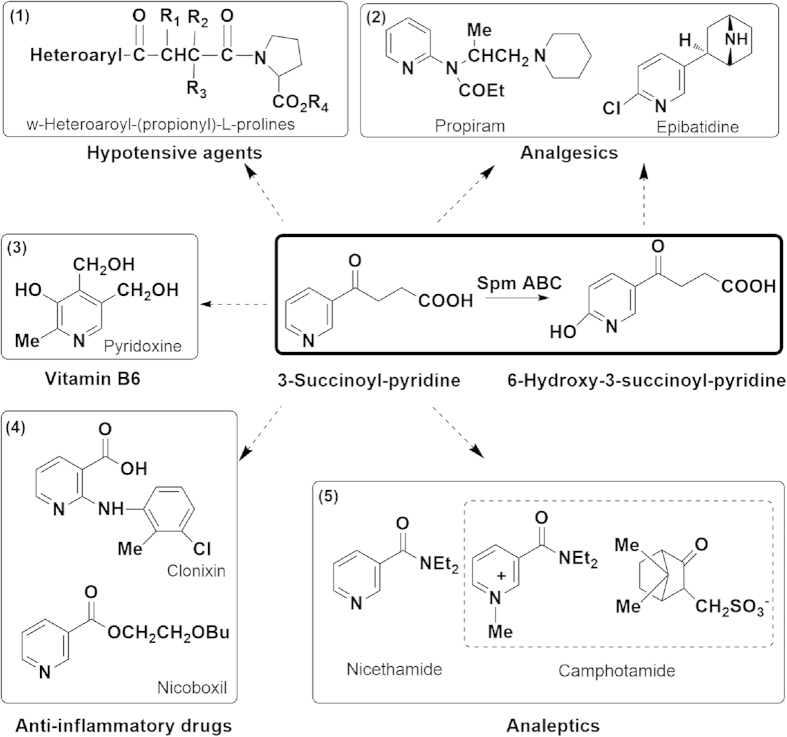
The development of methods to clean up and remove nicotine present in large amounts of tobacco wastes is imperative. Under methanogenic or aerobic conditions, a series of effective bioremediation processes focusing on nicotine reduction in tobacco wastes have been developed7,8,9. However, a variety of microorganisms play important roles in these biological methods. Bacteria of the genera Pseudomonas and Arthrobacter represent the two major types of bacterial species among these nicotinophilic microorganisms10. Because of their ability to degrade nicotine with high efficiency, certain microbes could be used to dispose of tobacco wastes11. Replacing waste disposal with resource recovery has been viewed as a potential environment-friendly and resource-saving future possibility12. In earlier times, to increase the value of tobacco wastes, such as low-grade tobacco leaves, processes were carried out to recover nicotine from these wastes and the extracted nicotine or nicotine sulfate was used as insecticide13,14,15. In addition, nicotine has potential application as a precursor molecule for the synthesis of valuable chemicals, especially many functional pyridines, which are difficult to prepare via chemical methods16,17. A number of pyridine derivatives exhibit bioactivity and medicinal value, such as vitamin B6 (pyridoxine), analgesics (propiram, epibatidine), analeptics (nicethamide, camphotamide) and anti-inflammaroty drugs (clonixin, nicoboxil) (Fig. 1)17,18,19. For example, 3-succinoyl-pyridine (SP) (also called γ-oxo-3-pyridinebutanoic acid) is a nicotine derivative and simple molecular pyridine compound and can be transformed into mammals hypotensive agents (ω-heteroaroyl-(propionyl)-L-prolines)20. The mechanism of nicotine degradation in species of the Arthrobacter genus has been reported in detail21. Moreover, the nicotine degradation pathway in Pseudomonas spp. has also been determined, particularly in Pseudomonas putida strain S1622,23,24,25. Based on these scientific developments, metabolic engineering could be a new approach for realizing customized multistep microbial synthesis of valuable compounds26. In previous studies, we have shown that strain S16 and its derivative have the ability to produce 6-hydroxy-3-succinoyl-pyridine (HSP)13,27, and these results verify that these species of Pseudomonas have the potential to utilize the nicotine in tobacco wastes and form a variety of valuable metabolites.
Figure 1. The hypotensive agents and typical pyridine drugs that may be derived from 3-succinoyl-pyridine (SP).
Bold-solid lined box: SP is converted to 6-hydroxy-3-succinoyl-pyridine (HSP) by SP monoxygenase (SpmABC) in P. putida S1625. Solid lined box: (1) The hypotensive agent derived from SP, ω-heteroaroyl-(propionyl)-L-prolines20. (2) Analgesics derived from SP or HSP, propiram and epibatidine17,18. (3) Bioactivity pyridine derivative: vitamin B6 (pyridoxine)19. (4) Typical pyridine drugs: anti-inflammaroty drugs (clonixin, nicoboxil) and, (5) analeptics (nicethamide, camphotamide (dash line box))18.
In this study, we expected to introduce a promising “green” method of reusing and reducing the toxicity of tobacco wastes to efficiently synthesize SP using genetically engineering Pseudomonas putida. Moreover, this SP production could be initiated with aqueous nicotine solution extracted from the low-grade tobacco leaves and even begun directly with crude suspension of tobacco leaf powder. Therefore, this green strategy makes it possible to convert nicotine from tobacco wastes with high nicotine content into commercially valuable compounds.
Results
Detection of nicotine in discarded tobacco leaves
To determine the actual nicotine content of discarded tobacco leaves (the tobacco waste), linear standard curves of nicotine and 3-succinoyl-pyridine (SP) were drawn according to the data of high performance liquid chromatography (HPLC) analysis. Using the standard curve, the nicotine content in these tobacco leaves was calculated as a percentage, yielding results of 3.09% ± 0.02% (w/w) (Table 1). This value was used to determine the total amount of nicotine in the tobacco leaves.
Table 1. Determination of nicotine content in discarded tobacco leaves.
| Sample No | 1 | 2 | 3 |
|---|---|---|---|
| Mass of tobacco powder (g) | 0.528 | 0.507 | 0.508 |
| Nicotine content of tobacco extraction (g/L) | 0.162 | 0.157 | 0.158 |
| Nicotine content of tobacco leaves (%) | 3.07 | 3.10 | 3.11 |
| Average content of nicotine in tobacco leaves (%) | 3.09 ± 0.02 |
Separation of nicotine from the tobacco leaves
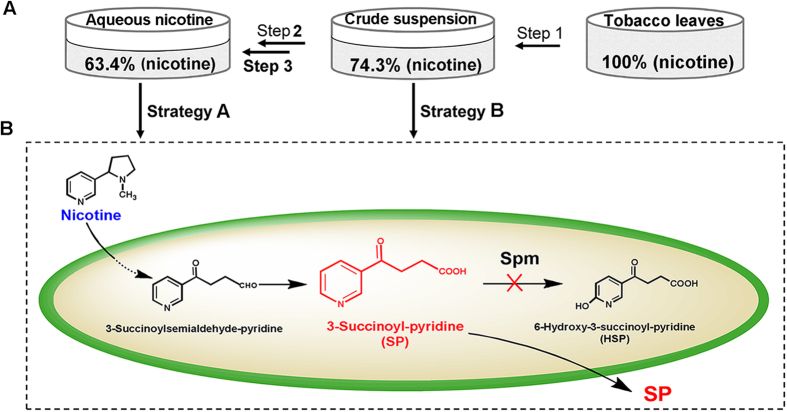
After the first step from 50 g tobacco leaves powder, 500 mL leach liquor was obtained and the nicotine content was calculated to be 2.3 g/L. The nicotine yield of step 1 was ~74.3% after immersion in water (Fig. 2). The second step was chloroform extraction, and the thin layer chromatography (TLC) results showed that there was little nicotine residue in aqueous phase after extraction. Finally, nicotine was separated into the aqueous sulfuric acid solution, resulting in 20 ml aqueous solution with 49.0 g/L nicotine content and a 63.4% yield by weight of the total nicotine (Fig. 2).
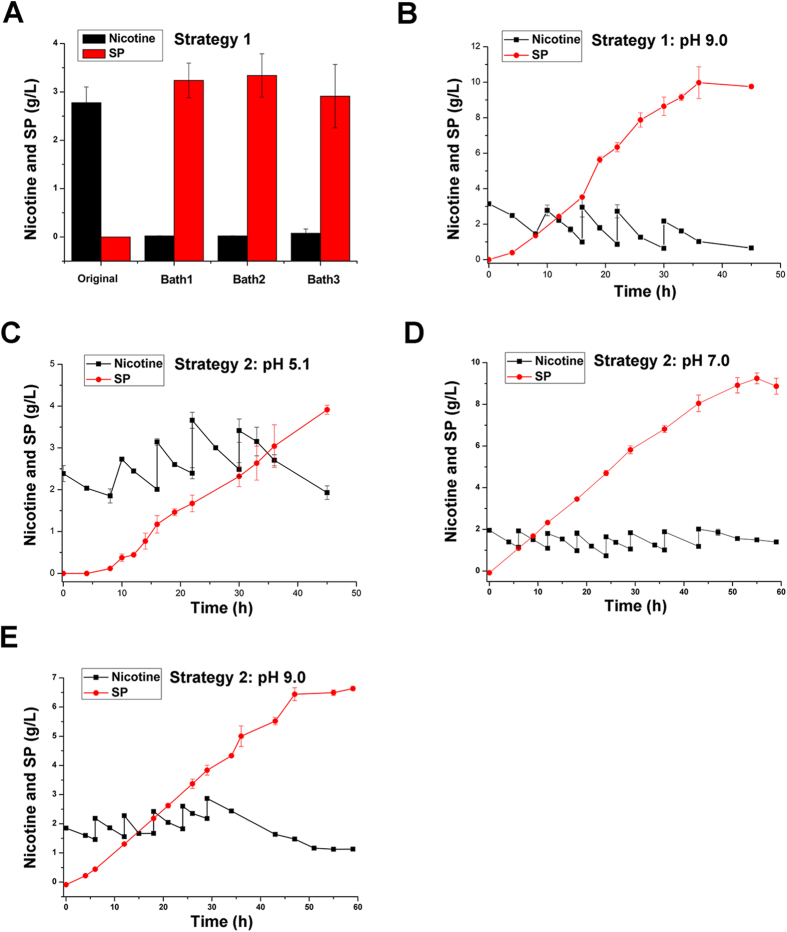
Figure 2. Strategies of SP production from the tobacco leaves using P. putida S16dspm.
(A) Step 1: pure water was used as solvent to obtain crude suspension of the tobacco leaves with nicotine recovery yield of 74.3%. Steps 2 and 3: chloroform extraction and acidic aqueous back-extraction were performed to obtain aqueous nicotine solution with nicotine recovery yield of 64.3%. (B) Nicotine degradation pathway in P. putida S16dspm was blocked at SP due to inactivation of Spm enzyme. Whole cells of S16dspm were used as biocatalysts for SP production following two strategies: the biotransformation to SP from aqueous nicotine solution (strategy A) and from crude suspension of the tobacco leaves (strategy B).
Preparation of the S16dspm biocatalyst
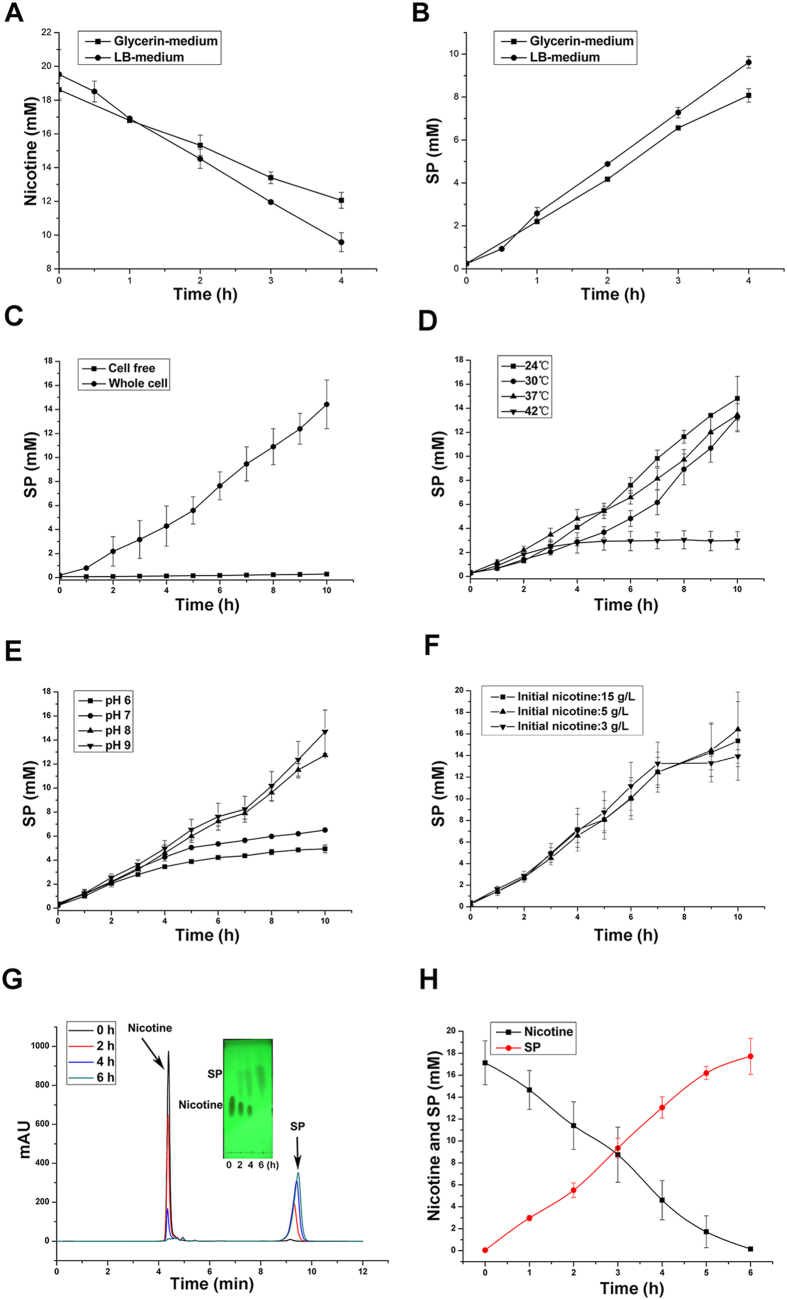
Due to the deletion of the spmA gene, the S16dspm strain has lost the ability to further degrade SP and cannot grow in nicotine medium25. Resting-cell reactions of S16dspm were investigated in glycerin and lysogeny borth (LB) medium, respectively. The resting cells obtained from culture in glycerin and LB medium showed no significant difference in nicotine transformation ability (Figs 3A,B). These data suggest that there were no differences between glycerin and LB medium for biocatalyst preparation. However, the whole-cell reactions showed low nicotine cell transforming activity without nicotine in either glycerin or LB medium, which indicated that the nicotine in the prepared media was necessary to improve the bioconversion ability of whole cells as an inducer.
Figure 3. Biotransformation to SP by strain S16dspm, preparation of biocatalyst, and optimal conditions for biotransformation.
Preparation of biocatalysts using glycerin (■) and LB (●) medium, respectively. (A) Nicotine degradation in the initial stage of resting-cell reactions with glycerin and LB medium. (B) SP formation in the initial stage of resting-cell reactions with glycerin and LB medium. Effects of reaction conditions for SP production. (C) SP formation by strain S16dspm under whole-cell condition (●) and cell-free condition (■). (D) SP formation by strain S16dspm at various temperatures, 24 °C (■), 30 °C (●), 37 °C (▲), 42 °C (▼). (E) SP formation by strain S16dspm at various pH values, pH6 (■), pH7 (●), pH8 (▲), pH9 (▼). (F) SP formation by strain S16dspm with different initial nicotine contents, 15 g/L (■), 5 g/L (▲), 3 g/L (▼). Analysis of the biotransformation results at the optimal conditions using resting cells of S16dapm. (G) TLC and HPLC qualitative analysis of nicotine degradation and SP formation. (H) The content changes of nicotine and SP during the biotransformation process detected by HPLC quantitative analysis. Nicotine (■); SP (●).
Optimal reaction conditions for synthesis of SP
Cell integrity was necessary for biotransformation of nicotine to SP because no SP synthesis occurred under cell-free condition (Fig. 3C). Moreover, the nicotine content was detected after 10 h under the cell-free condition, and the nicotine content remained unchanged. Therefore, the biocatalytic reaction should be conducted in the whole-cell system, and cell integrity is necessary for the biotransformation of nicotine.
The biotransformation reactions were performed at different temperatures and pH values in deionized water. The SP synthesis activity was optimal at 24 °C, and the relatively high temperature of 42 °C deteriorated the efficiency of SP formation (Fig. 3D). The acidic condition was found to hinder SP formation, and the optimum pH value was 9.0 (Fig. 3E). According to an earlier report, a nicotine content of 6 g/L or more in culture was poisonous to the growth of bacterium S1622. However, under the catalytic reaction condition containing 5.6 mg/ml (dry cell weight, DCW) of resting cells (OD620 ~ 10), the biotransformation could be conducted even with an initial nicotine of 15 g/L. Moreover, the SP formation rate was not obviously effected when the initial nicotine content was increased (Fig. 3F). The results suggest that a nicotine content of 3 g/L is an appropriate initial nicotine concentration, and increased substrate concentration could improve the final yield of SP but not the rate of SP formation.
Then, the whole-cell reaction was performed with all of the optimal conditions mentioned above, and the results of TLC/HPLC analysis are shown in Fig. 3G. After incubation for 6 h, almost all of the nicotine has been converted into metabolite SP, and the transformation was an equimolar reaction (Fig. 3H).
Isolation and identification of the SP compound
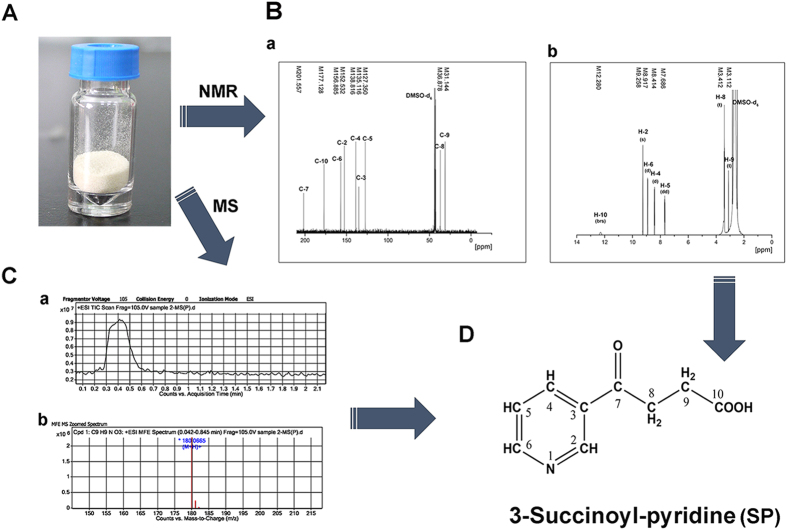
To isolate the metabolite SP, the methods used were performed as previous described and the powder was purified by recrystallization (Fig. 4A)13. The crystals showed no impurities when analyzed by HPLC. Finally, about 0.72 g of the (4.02 mmol) metabolite SP was obtained from 1.20 g (7.41 mmol) nicotine and the isolation yield of crystal SP was 54.2%. The structure of metabolite SP was detected and further analyzed by means of mass spectrometry (MS) (m/z 180.0665 [M + H]+) (Fig. 4C) and 1H nuclear magnetic resonance (NMR) (DMSO-d6, 400 MHz, δ: 12.28, 9.26, 8.92, 8.41, 7.69, 3.41, 3.11)/13C NMR (DMSO-d6, 100 MHz, δ: 201.56, 177.13, 156.89, 152.53, 138.82, 135.12, 127.35, 36.88, 31.14) spectrometry (Fig. 4B). The 13C spectrum showed that there are nine carbon atoms in the SP compound, and each type of carbon atom in the molecule is represented by a peak (Fig. 4D). The NMR data for SP are consistent with those in a previous report by Wang et al.23.
Figure 4. Identification of product SP by ESI-MS and NMR.
(A) The crystal compound SP got in this study. (B) 13C-NMR (B-a) and 1H-NMR (B-b) spectra of the product SP. Measurements were performed in DMSO-d6. C. The analysis results by ESI-MS. The total ionization chromatography (C-a) and the ion mass of [M+H]+ (C-b) of the product SP. (D) Molecular structural formula of SP with the positions of carbon (C) and hydrogen (H) atoms detected by NMR.
Batch and fed-batch biotransformations to SP
For batch biotransformation of nicotine to SP, the biocatalyst was reused three times, and each time, the initial nicotine concentration was 3.0 g/L. The SP concentrations with the conversion yields on nicotine were 3.2 ± 0.35 g/L (97.0%), 3.3 ± 0.45 g/L (100.0%), and 2.9 ± 0.65 (87.9%), respectively. The results showed that whole cells of the strain S16dspm could be reused and that, in these subsequent uses, they had almost the same efficient catalytic capabilities as in their first use (Fig. 5A). For fed-batch biotransformation of strategy A, the substrate nicotine was fed intermittently to maintain a substrate concentration above 1 g/L. The time courses of nicotine feeding and SP production are shown in Fig. 5B. SP product was efficiently accumulated after a reaction time of 20 h; the final concentration of SP transformed from nicotine was 9.8 ± 0.11 g/L, with a conversion yield of 83.8% for 45 h.
Figure 5. Time course of batch and fed-batch biotransformations by P. putida S16dspm.
(A) For batch biotransformation of aqueous nicotine solution, the biocatalysts were reused three times. Nicotine (black) and SP (red) concentration were analyzed by HPLC at the end of each batch reaction. (B) Time course of fed-batch biotransformation from aqueous nicotine solution by P. putida S16dspm at pH 9.0, 24 °C, and 5.6 mg/ml DCW resting cells. Nicotine (■); SP (●). (C–E) Time course of fed-batch biotransformation by P. putida S16dspm from crude tobacco extract at pH 5.1 (C), pH 7.0 (D), and pH 9.0 (E). The fed-batch biotransformation from crude tobacco extract were performed at different pH conditions. Nicotine (■); SP (●).
When crude tobacco extract was used as the substrate in fed-batch biotransformation, the optimal pH for the reaction was 7.0 and the final SP concentration was 8.9 ± 0.38 g/L with a conversion yield of 89.9% (Fig. 5D). Compared to the concentrations produced when using a pH of 7.0 or 9.0, the SP concentration was lowest at pH 5.1 (initial pH value of crude tobacco leaf powder extract): 3.9 ± 0.11 g/L with a yield on nicotine of 54.9% for 46 h (Fig. 5C). The SP concentration obtained using crude tobacco extract was 6.6 ± 0.07 g/L with a yield on nicotine of 85.7% for 59 h (Fig. 5E), which was lower than that obtained using aqueous nicotine solution at pH 9.0 (Fig. 5B).
Function of the pps_3984 gene on nicotine degradation in strain S16
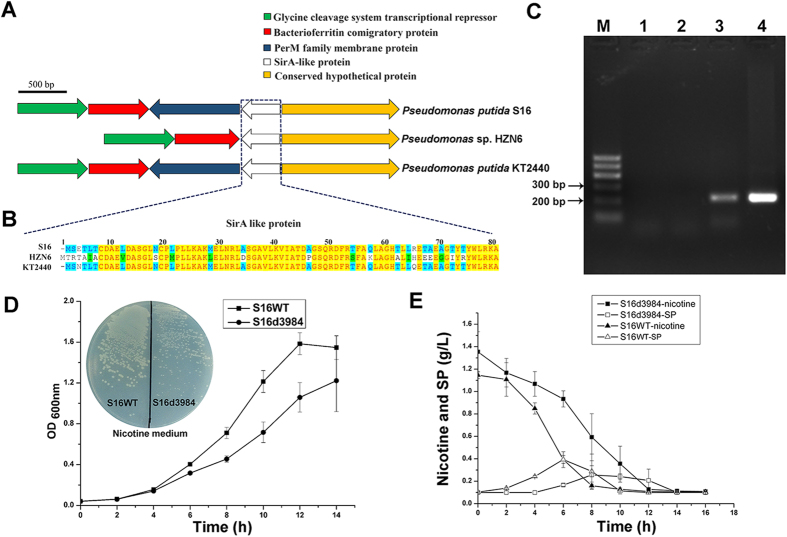
The biocatalyst S16dspm in this work lost its ability to degrade metabolite SP25. Interestingly, Qiu et al. reported that when the sirA2 (sirA like) gene was disrupted in Pseudomonas sp. HZN6, the sirA2-disrupted mutant lost the ability to degrade SP28. In P. putida S16, a sirA-like gene (pps_3984 gene) may also exist, located in a similar gene cluster to that of sirA2/sirA in Pseudomonas sp. HZN6 and P. putida KT2440 (Fig. 6A). The pps_3984 protein shows 76% and 97% similarity in amino acid sequence identity to SirA2 protein in Pseudomonas sp. HZN6 and SirA protein in P. putida KT2440, respectively (Fig. 6B). Semi-quantitative reverse transcription polymerase chain reaction (RT-PCR) analysis of pps_3984 showed that this gene was transcribed when strain S16 was grown in nicotine medium (Fig. 6C). However, the mutant of strain S16 with pps_3984 gene deletion, S16d3984, could also grow in nicotine medium (Fig. 6D). This mutant strain did not lose the ability of nicotine and SP degradation and had no ability to accumulate metabolite SP (Fig. 6E). These results show that pps_3984 gene is not crucial in the nicotine degradation of strain S16.
Figure 6. Characteristics of pps_3984 gene in P. putida S16.
(A) The sirA like gene clusters in P. putida S16 (GenBank no. NC_015733), Pseudomonas sp. HZN6 (GenBank no. HQ832741), and P. putida KT2440 (GenBank no. NC_002947). Green–glycine cleavage system transcriptional repressor. Red–bacterioferritin comigratory protein. Blue–PerM family membrane protein. White–SirA-like protein. Yellow–conserved hypothetical protein. (B) Multiply sequence alignment of SirA like proteins from P. putida S16 (GenBank no. WP_004375233), Pseudomonas sp. HZN6 (GenBank no. AEK25019), and P. putida KT2440 (GenBank no. NP_743393) using Vector NTI software. Identical conservative sites are highlighted in yellow, comparatively conservative sites are highlighted in blue and green. (C) Semi-quantitative RT-PCR analysis of pps_3984 gene in P. putida S16. Pure water (lane 1), total RNA (lane 2), total cDNA (lane 3), and genome (lane 4) from S16 cells grown in nicotine medium were used as templates for PCR with oligonucleotide pairs of pps_3984 gene. Lane M was DNA marker. (D) Growth curves of wild type P. putida S16 (S16WT) (■) and the pps_3984 gene disrupted mutant S16d3984 (●) in nicotine medium with nicotine as the sole carbon and nitrogen source, and S16WT and S16d3984 grown on nicotine plate. (E) HPLC analysis of changes of nicotine and SP concentration by culturing S16WT and S16d3984 in nicotine medium. Nicotine degradation by S16d3984 (■). SP metabolism by S16d3984 (□). Nicotine degradation by S16WT (▲). SP metabolism by S16WT (Δ).
Discussion
Tobacco wastes containing alkaloids are intractable wastes. In recent years, the nicotine metabolism of Pseudomonas strains has been studied in depth, especially in P. putida S16 and Pseudomonas sp. HZN622,28. These strains, with high nicotine degrading efficiency, have potential applications in tobacco waste treatment. Meanwhile, biotransformation by means of engineered microbial cells makes a possibility in resource recovery of tobacco wastes with high nicotine content12. In early times, 6-hydroxy-(S)-nicotine (productivity, 30 g/L) and 6-hydroxy-3-succinoylpyridine (productivity, 15 g/L) (HSP) were produced by fermentation processes using Arthrobacter oxydans NRRL-B-3603 and Pseudomonas sp. DSM8653, respectively16. In addition, P. putida S16 and its derivative P-HSP were developed for the production of HSP in our previous work, and the productivity of HSP could reach 16.3 g/L13,27. In this paper, genetically engineered strain S16dspm was used to produce a new nicotine analogue 3-succinoyl-pyridine (SP) with a productivity of 9.8 g/L. Over 7000 exciting drugs are pyridine derivatives in the pharmaceutical industry, and the selective functionalization of the pyridine ring is difficult to control by chemical means17,18,19. So the production of simple molecular pyridine compounds, such as SP, HSP or 2,5-dihydroxypyridine (2,5-DHP), by engineered biocatalyst will provide abundant starting materials for the synthesis of bioactivity pyridine drugs.
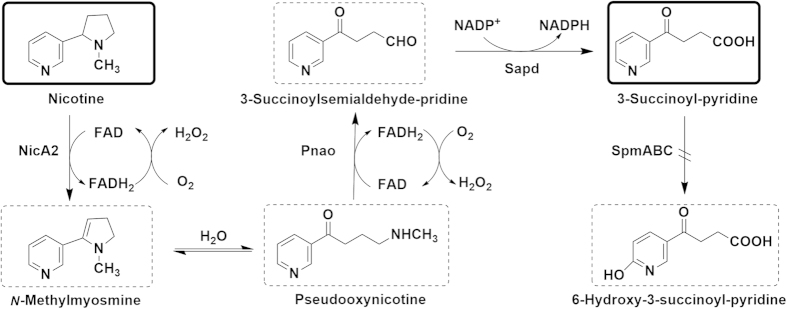
The advantage of whole-cell biocatalysts is that energy can be provided and cofactors can be recycled when redox reactions are involved17,29. The biotransformation from nicotine to SP in S16dspm needs multi-enzymatic reactions, and the putative cofactors involved were shown in Fig. 7 30,31. The reaction from nicotine to SP didn’t consume energy, and the putative cofactors FAD/FADH2 involved in the reaction from nicotine to 3-succinoylsemialdehyde-pyridine were self-regeneration under aerobic conditions29. The last step of reaction from 3-succinoylsemialdehyde-pyridine to the product SP was catalyzed by enzyme Sapd with cofactors NADP+/NADPH. However, the Sapd was not essential for nicotine metabolism in S16 because the mutant strain S16dsapd with the deletion of sapd gene could still use nicotine to grow. The most reasonable explanation was that 3-succinoylsemialdehyde-pyridine could be easily oxidized to SP by other aldehyde dehydrogenases in the whole cell biocatalysts of mutant strain25,31. Thus, these facts could explain why the biotransformation in this work using the whole cells S16dspm as biocatalysts was sustainable.
Figure 7. Intermediates and putative cofactors between nicotine and the product SP in P. putida S16dspm.
Biotransformation from nicotine to SP was catalyzed by multi-enzymes: NicA2, nicotine oxido-reductase; Pnao, pseudooxynicotine amine oxidase; Sapd, 3-succinoylsemialdehyde-pyridine dehydrogenase; SpmABC, SP monoxygenase. Cofactors involved in the reaction between nicotine and pseudooxynicotine were suggested to be FAD/FADH2 according to reaction catalyzed by 6-hydroxy-L-nicotine oxidase30. Cofactors involved in the reaction from pseudooxynicotine to SP were suggested to be FAD/FADH2 and NADP+/NADPH according to similar reactions catalyzed by PNAO and SAPD in Pseudomonas sp. HZN631.
The engineered strain S16dspm achieved high accumulation of SP from both aqueous nicotine and crude tobacco powder extract at the optimal conditions. To test the biocatalytic capability of S16dspm, we explored two strategies. Strategy A: the aqueous nicotine solution extracted from the tobacco leaves was used as substrate (Fig. 2 left). Compared with crude suspension of the tobacco leaves, the reaction solution was simple when the reaction substrate was aqueous nicotine solution, and the optimal pH value for SP production was 9.0 (Fig. 3E), the same as that for HSP production27. When the bioconversion reaction system is uncomplicated, the alkaline condition is conducive to SP formation due to its property as a weak acid. Strategy B: the biotransformation could be performed directly in crude tobacco extract in an aqueous solution (Fig. 2 right). However, the initial pH value of crude tobacco extract was 5.1, and as shown in Fig. 5C, this condition was not suitable for SP production. Intriguingly, the SP concentration obtained at pH 9.0 was not the maximum concentration, and it was lower than that obtained at pH 7.0 (Fig. 5D,E) when crude tobacco extract was used as the substrate. This may be due to the fact that crude tobacco extract consists of complex components, including pectins, tannins and alkaloids which can make the reaction mixture become muddy under the alkaline condition and this could inhibit biocatalyst activity. Moreover, SP was isolated from the reaction solution easily in strategy A due to its simple component reaction system. However, when the transformation was carried out in crude tobacco extract using pure water as a solvent in strategy B, the isolation and purification SP-compound crystals remained to be investigated, and would provide a greener approach of SP production than strategy A due to its use of an organic solvent.
In summary, the efficient and specific biocatalyst could be easily prepared using the reusable whole cells of S16dspm. Under the optimal biotransformation process, the isolation yield of crystal compound SP on nicotine was 54.2%. Additionally, this production process for SP reveals that spmA gene is crucial in SP degradation of strain S16 rather than the pps_3984 gene, a sirA-like gene. Resource recovery of tobacco waste remains a long-term challenge, and this study thus offers a useful strategy to accomplish the recovery of discarded tobacco wastes.
Methods
Chemicals
l-(−)-Nicotine was obtained from Fluka Chemie GmbH (Switzerland). SP was obtained from Toronto Research Chemicals (Canada). All other reagents used were of analytical grade.
Bacterial strains, plasmids, and culture conditions
Pseudomonas putida S16 was isolated from soil and identified as a nicotine-degrading strain as previously decribed22. Strain S16 was deposited at Deutsche Sammlung von Mikroorganismen und Zellkulturen with in Gottingen (Germany) under DSM No. 28022. The mutant strain P. putida S16dspm was derived from strain S16 in our previous sutudy25. The bacterium S16dspm was cultivated in LB medium or glycerin medium containing 1 g/L nicotine at 30 °C.
Construction of mutant strain S16d3984 and semi-quantitative RT-PCR analysis of pps_3984 gene
The disruption of gene pps_3984 was performed using suicide plasmid pK18mob according to the previous methods25,27. The primers pps_3984F (5′- AGCTAAGCTTCTGTGACGCCGAACTGGAC-3′, HindIII) and pps_3984R (5′- GCGAATTCGTGTAGGTACCGGCCTCGGC-3′, EcoRI) were used in this work. Semi-quantitative RT-PCR analysis of pps_3984 gene was performed as previously described32.
Analysis of the homologous gene cluster in P. putida S16, P. putida KT2440 and Pseudomonas sp. HZN6 and sequence similarity of amino acid were performed by Vector NTI software.
Determination of nicotine content in discarded tobacco leaves
A batch of discarded tobacco leaves (the tobacco waste) was obtained from Henan province, China. Dried, pulverized, and sieved tobacco powder was prepared for nicotine extraction. Exactly 100 ml of methanol-0.1 M NaOH (1:1, [vol/vol]) were added to 0.5 g tobacco powder, and then the mixture was sonicated for 1 h. After centrifugation at 12,000 rpm for 2 min, this suspension was diluted with twice volume of anhydrous ethanol and prepared for HPLC analysis.
Extraction of nicotine from discarded tobacco leaves
Strategy A (Fig. 2 right): in order to obtain aqueous nicotine solution, aqueous extracts of tobacco leaves (the tobacco waste) were obtained after step 1, and then the chloroform (1/5 volume) was added to the crude tobacco extract to obtain nicotine in organic phase. Ultimately, back-extraction was performed by dilute sulfuric acid (pH ≤ 4) to get nicotine into aqueous phase.
Strategy B (Fig. 2 left): in order to obtain crude suspension of the tobacco leaves, dried tobacco powder (1 part) was suspended in distilled water (12 parts), and the mixture was left at 60 °C temp with stirring for 200 minutes, then the filter residue was removed to get the crude tobacco extract.
Transformation of nicotine to SP using S16dspm resting cells
Mutant strain S16dspm was cultured at 30 °C in a rotary shaker at 200 rpm in LB medium or glycerin medium. In mid-exponential phase, cells were harvested by centrifugation at 4,500 rpm for 20 min at 4 °C, then washed with 0.1 M PBS for the first time and with distilled water for twice. The transformation reactions were carried out in a 500-ml flask containing 5.6 mg/ml DCW of resting cells (OD620nm ~ 10), 3–15 g/L nicotine and deionized water, with shaking at 120 rpm at 24–42 °C.
Culture temperature, pH, nicotine content and cellular integrity were studied to identify the optimal conditions for SP production. To identify the optimal temperature for the SP production, reactions were conducted in deionized water containing 3 g/L nicotine at 24 °C, 30 °C, 37 °C, and 42 °C, respectively, with the initial pH 8.0. The optimal pH was identified among pH values of 6.0, 7.0, 8.0 and 9.0. The pH was adjusted using 0.5 M NaOH. Under the optimal temperature and pH condition, the effect of initial nicotine content (3.0, 5.0, 10.0, or 15.0 g/L) was investigated. To evaluate whether the cellular integrity was necessary for the reaction, nicotine transformation was compared between whole-cell and cell-free conditions. The subsequent work was carried out under optimized conditions.
Batch and fed-batch biotransformations
The biotransformations to SP from aqueous nicotine solution or crude suspension of the leaves powder were conducted in a 1-L conical flask containing 300 ml reaction solution under optimized conditions. For batch transformation, the biocatalyst was collected after the reaction for 8 hours, and reused for another 8 hours. For fed-batch transformation, nicotine was fed at the desired time. Specifically, the reactions in crude leaves powder extract were carried out under different conditions of pH 5.1, pH 7.0, and pH 9.0, respectively.
Sample preparation and analytical methods
Samples of resting-cell reactions were obtained at desired time, and then twice volume of ethanol was added to stop the reaction. The supernatant obtained after centrifugation at 12,000 rpm for 2 min was filtered by 0.22 μm polyamide filter before quantitative analysis by HPLC. HPLC was performed as previously described13.
TLC was used to analyze the metabolite SP. The TLC experiment was performed on 0.2 ± 0.03 mm silica gel (HSGF 254, YJY, Yantai, China) with chloroform-ethanol-methanol-0.5 M NaOH (30:15:2:1.5 [vol/vol]) as mobile phase. The spots of nicotine and SP were shown under UV light (254 nm), or detected by incubating it in iodine vapor.
Isolation and identification of metabolite SP
After resting-cell incubation with nicotine for ~6 h, the reaction was stopped by removing cells via centrifugation at 8,000 rpm for 10 min at 4 °C. Then the supernatant was evaporated at 50 °C under reduced pressure. The residual liquid was adjusted to pH 3 and then filtration and drying were performed to obtain the white precipitate SP. To identify the structure of metabolite SP, electrospray ionization (ESI)-MS and NMR were performed as previous reports23.
Additional Information
How to cite this article: Wang, W. et al. Sustainable production of valuable compound 3-succinoyl-pyridine by genetically engineering Pseudomonas putida using the tobacco waste. Sci. Rep. 5, 16411; doi: 10.1038/srep16411 (2015).
Acknowledgments
This work was supported in part by grants from the Chinese National Natural Science Foundation (31230002 and 31270154), the Chinese National Science Foundation for Excellent Young Scholars (31422004), and partially from the National Basic Research Program of China (2013CB733901). We also acknowledge the “Chen Xing” project from Shanghai Jiaotong University.
Footnotes
Author Contributions P.X., W.W. and H.T. conceived and designed the project and experiments. W.W. performed the experiments. P.X. contributed reagents and materials. W.W. and H.T. wrote the paper. All authors reviewed the paper.
References
- Davis R. M., Wakefield M., Amos A. & Gupta P. C. The hitchhiker’s guide to tobacco control: a global assessment of harms, remedies, and controversies. Annu. Rev. Public Health 28, 171–194 (2007). [DOI] [PubMed] [Google Scholar]
- Zhang Y. D., Luo C. R., Wang H. L. & Lu G. F. Advances in microbial degradation of nicotine and its application. Tob. Sci. Technol. 197, 3–7 (2003). [Google Scholar]
- Mayer B. How much nicotine kills a human? Tracing back the generally accepted lethal dose to dubious self-experiments in the nineteenth century. Arch. Toxicol. 88, 5–7 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorgan C. A. Statistical record of the environment, 3rd ed. (Gale Research Incorporated, 1995). [Google Scholar]
- Slack R. J., Gronow J. R. & Voulvoulis N. Household hazardous waste in municipal landfills: contaminants in leachate. Sci. Total Eenviron. 337, 119–137 (2005). [DOI] [PubMed] [Google Scholar]
- Schwarzbauer J., Heim S., Brinker S. & Littke R. Occurrence and alteration of organic contaminants in seepage and leakage water from a waste deposit landfill. Water Res. 36, 2275–2287 (2002). [DOI] [PubMed] [Google Scholar]
- Meher K. K., Panchwagh A. M., Rangrass S. & Gollakota K. G. Biomethanation of tobacco waste. Environ. Pollut. 90, 199–202 (1995). [DOI] [PubMed] [Google Scholar]
- Briški F., Horgas N., Vuković M. & Gomzi Z. Aerobic composting of tobacco industry solid waste—simulation of the process. Clean Techn. Environ. Policy 5, 295–301 (2003). [Google Scholar]
- Piotrowska-Cyplik A., Olejnik A., Cyplik P., Dach J. & Czarnecki Z. The kinetics of nicotine degradation, enzyme activities and genotoxic potential in the characterization of tobacco waste composting. Bioresour. Technol. 100, 5037–5044 (2009). [DOI] [PubMed] [Google Scholar]
- Gurusamy R. & Natarajan S. Current status on biochemistry and molecular biology of microbial degradation of nicotine. Scientific World Journal 2013, e125385 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser J. P., Feng Y. C. & Bollag J. M. Microbial metabolism of pyridine, quinoline, acridine, and their derivatives under aerobic and anaerobic conditions. Microbiol Rev. 60, 483–498 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iranpour R. et al. Environmental engineering: energy value of replacing waste disposal with resource recovery. Science 285, 706–711 (1999). [DOI] [PubMed] [Google Scholar]
- Wang S. N. et al. “Green” route to 6-hydroxy-3-succinoyl-pyridine from (S)-nicotine of tobacco waste by whole cells of a Pseudomonas sp. Environ. Sci. Technol. 39, 6877–6880 (2005). [DOI] [PubMed] [Google Scholar]
- De Lucas A., Cañizares P., García M. A., Gomez J. & Rodriguez J. F. Recovery of nicotine from aqueous extracts of tobacco wastes by an H+-form strong-acid ion exchanger. Ind. Eng. Chem. Res. 37, 4783–4791 (1998). [Google Scholar]
- Rincon J. et al. Preliminary study on the supercritical carbon dioxide extraction of nicotine from tobacco wastes. Sep. Sci. Technol. 33, 411–423 (1998). [Google Scholar]
- Roduit J. P., Wellig A. & Kiener A. Renewable functionalized pyridines derived from microbial metabolites of the alkaloid (S)-nicotine. Heterocycles 46, 1687–1702 (1997). [Google Scholar]
- Schmid A. et al. Industrial biocatalysis today and tomorrow. Nature 409, 258–268 (2001). [DOI] [PubMed] [Google Scholar]
- Lukevits E. Pyridine derivatives in the drug arsenal (150 years of pyridine chemistry). Chem. Heterocycl. Compd. 31, 639–650 (1995). [Google Scholar]
- Henry G. D. De novo synthesis of substituted pyridines. Tetrahedron 60, 6043–6061 (2004). [Google Scholar]
- McEvoy F. J., Wright W. B., Birnberg G. H. & Albright J. D., inventors; American Cyanamid Company, assignee. ω-Heteroaroyl (propionyl or butyryl)-L-prolines. United States patent US 4, 299, 769. 1981. Nov 10. [Google Scholar]
- Brandsch R. Microbiology and biochemistry of nicotine degradation. Appl. Microbiol. Biotechnol. 69, 493–498 (2006). [DOI] [PubMed] [Google Scholar]
- Wang S. N. et al. Biodegradation and detoxification of nicotine in tobacco solid waste by a Pseudomonas sp. Biotechnol. Lett. 26, 1493–1496 (2004). [DOI] [PubMed] [Google Scholar]
- Wang S. N., Liu Z., Tang H. Z., Meng J. & Xu P. Characterization of environmentally friendly nicotine degradation by Pseudomonas putida biotype A strain S16. Microbiology 153, 1556–1565 (2007). [DOI] [PubMed] [Google Scholar]
- Tang H. Z. et al. Genomic analysis of Pseudomonas putida: genes in a genome island are crucial for nicotine degradation. Sci. Rep. 2, 377 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang H. Z. et al. Systematic unraveling of the unsolved pathway of nicotine degradation in Pseudomonas. PLoS Genet. 9, e1003923 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoemaker H. E., Mink D. & Wubbolts M. G. Dispelling the myths–biocatalysis in industrial synthesis. Science 299, 1694–1697 (2003). [DOI] [PubMed] [Google Scholar]
- Yu H., Tang H. Z. & Xu P. Green strategy from waste to value-added-chemical production: efficient biosynthesis of 6-hydroxy-3-succinoyl-pyridine by an engineered biocatalyst. Sci. Rep. 4, 5397 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu J. G. et al. A sirA-like gene, sirA2, is essential for 3-succinoyl-pyridine metabolism in the newly isolated nicotine-degrading Pseudomonas sp. HZN6 strain. Appl. Microbiol. Biotechnol. 92, 1023–1032 (2011). [DOI] [PubMed] [Google Scholar]
- Zhao H. M. & van der Donk W. A. Regeneration of cofactors for use in biocatalysis. Curr. Opin. Biotechnol. 14, 583–589 (2003). [DOI] [PubMed] [Google Scholar]
- Kachalova G., Decker K., Holt A. & Bartunik H. D. Crystallographic snapshots of the complete reaction cycle of nicotine degradation by an amine oxidase of the monoamine oxidase (MAO) family. Proc. Natl. Acad. Sci. USA 108, 4800–4805 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu J. G. et al. Functional identification of two novel genes from Pseudomonas sp. strain HZN6 involved in the catabolism of nicotine. Appl. Environ. Microbiol. 78, 2154–2160 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L. J., Tang H. Z., Yu H., Yao Y. X. & Xu P. An unusual repressor controls the expression of a crucial nicotine-degrading gene cluster in Pseudomonas putida S16. Mol. Microbiol. 91, 1252–1269 (2014). [DOI] [PubMed] [Google Scholar]