Abstract
The Drosophila embryo at the mid-blastula transition (MBT) concurrently experiences a receding first wave of zygotic transcription and the surge of a massive second wave. It is not well understood how genes in the first wave become turned off transcriptionally and how their precise timing may impact embryonic development. Here we perturb the timing of the shutdown of Bicoid (Bcd)-dependent hunchback (hb) transcription in the embryo through the use of a Bcd mutant that has heightened activating potency. A delayed shutdown specifically increases Bcd-activated hb levels, and this alters spatial characteristics of the patterning outcome and causes developmental defects. Our study thus documents a specific participation of maternal activator input strength in the timing of molecular events in precise accordance with MBT morphological progression.
KEY WORDS: Bicoid, Drosophila, Transcription dynamics, Mid-blastula transition, Morphogen
Summary: Two waves of zygotic transcription occur during the mid-blastula transition in Drosophila. Shutdown of the first wave involves post-translational modulation of the maternal factor Bicoid.
INTRODUCTION
The transfer of developmental control from maternally deposited products to zygotically expressed products is a process that all animal embryos must experience during development (Tadros and Lipshitz, 2009). In Drosophila, a set of cellular events, such as cell cycle lengthening and cellularization, marks a specific stage within this period and is referred to as the mid-blastula transition (MBT) (Tadros and Lipshitz, 2009; Langley et al., 2014; Gebelein and Ma, 2015). During this time there is a massive wave of zygotic gene activation (Pritchard and Schubiger, 1996; De Renzis et al., 2007), which coincides with the receding of an earlier wave of transcription of zygotic genes, such as nullo, Serendipity-α (Sry-α), Sex-lethal (Sxl) and hunchback (hb) (Rose and Wieschaus, 1992; ten Bosch et al., 2006; Erickson and Quintero, 2007; Liu and Ma, 2013b). The mitotic phase is known to be disruptive to transcription prior to nuclear cycle (nc) 14 (Shermoen and O'Farrell, 1991). But during nc14, transcription of these early-expressing genes is turned off well before the mitotic phase (Rose and Wieschaus, 1992; Erickson and Quintero, 2007; Liu and Ma, 2013b). It remains poorly understood how this is controlled. Since maternally derived products become degraded while zygotic products accumulate during this time (Bushati et al., 2008; Lu et al., 2009; Lee et al., 2014), we can envision two simple mechanisms. It is possible that the observed transcriptional shutdown is reflective of a mere loss (decay) of maternally derived transcriptional activators or, alternatively, of repression by zygotically derived transcriptional repressors that have accumulated to sufficient levels.
Here we investigate these possibilities through the use of hb, a gap gene that is required for the formation of head and thoracic structures (Lehmann and Nusslein-Volhard, 1987). An important feature of hb transcription prior to its shutdown is that the maternally derived morphogenetic protein Bicoid (Bcd) acts as the primary input (He et al., 2008, 2015; Liu et al., 2011; Liu and Ma, 2011; Cheung et al., 2014). This feature contrasts with other early-expressing genes that are activated by sets of either complex or currently uncharacterized inputs (Salz and Erickson, 2010). We show that a reporter gene that is driven by synthetic Bcd binding sites also exhibits a characteristic shutdown phase of transcription at nc14, suggesting that properties intrinsic to Bcd as a transcriptional activator might be directly involved in the shutdown process. Through the use of a Bcd mutant lacking its primary sumoylation site (Liu and Ma, 2012), we show that the shutdown timing of Bcd-activated hb transcription at nc14 can be perturbed and that such a perturbation can lead to both molecular and phenotypic consequences. Our results do not support either of the two simple models envisioned above. Instead, they suggest that the shutdown of Bcd-activated transcription at nc14 is reflective of a general or global mechanism that specifically involves maternal activator input strength. Our results also underscore the importance of the shutdown phase of transcription dynamics during MBT in safeguarding patterning trajectories toward a normal outcome.
RESULTS
Evaluating hb as a tool for monitoring transcription shutdown during the MBT
To identify genes that are shut down during nc14, we analyzed the RNA-seq dataset previously generated from Drosophila embryos separated into four temporal groups of nc14 (Lott et al., 2011). Our analysis identified 194 genes that exhibit characteristics of being shut down (Table S1; see also Fig. S1 for dynamic profiles of expression levels of selected candidate genes and those that do not exhibit shutdown properties). The functions of these genes are highly enriched in such categories as ‘developmental proteins’, ‘embryonic morphogenesis’ and ‘cell fate commitment’ (Table S2), suggesting that this group of genes plays diverse and important roles in early embryo development. The Bcd target gene hb is among the list in Table S1, suggesting that mechanistic studies of hb as a representative of this class of genes have general significance.
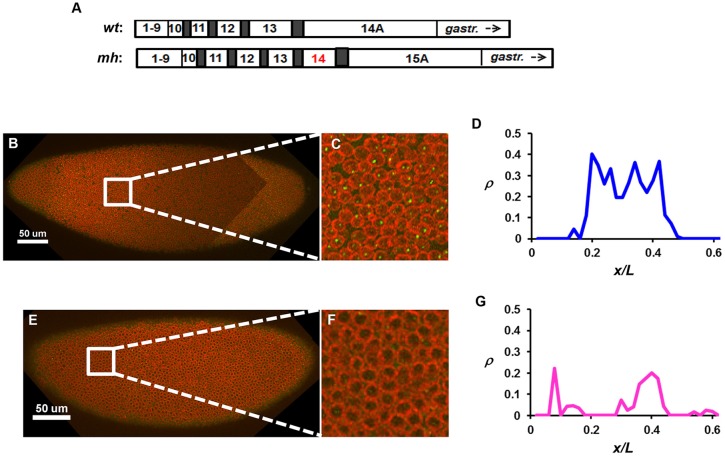
To determine whether the shutdown of hb transcription at nc14 is a property associated with the MBT, we took advantage of haploid embryos derived from homozygous maternal haploid (mh1) females (referred to as mh embryos) (Gans et al., 1975; Zalokar et al., 1975). The mh embryo undergoes an extra cleavage cycle, postponing the MBT to nc15 (Fig. 1A). We reasoned that, if shutdown of hb transcription is associated with the MBT, this event would be correspondingly postponed to nc15 in mh embryos, but if this event is associated with a specific time of development, then it would take place at nc14 in mh embryos (Lu et al., 2009). Importantly, the shutdown of Bcd-activated hb transcription is a very rapid event in wild-type embryos, taking place within a few minutes upon entering nc14 interphase, and it does not become reactivated after this shutdown event (Liu and Ma, 2013b). Thus, according to the MBT hypothesis, hb is expected to be transcriptionally active at early but not late nc15 in mh embryos and, according to the developmental time hypothesis, hb is expected to be transcriptionally inactive even at early nc15 in mh embryos.
Fig. 1.
hb transcription is shut down at nc15 in the mh Drosophila embryo. (A) Schematic showing an extra cleavage cycle (14) in mh embryos (Edgar et al., 1986; Rose and Wieschaus, 1992; Lu et al., 2009). gastr., gastrulation. (B,C) Image captured from an mh embryo at early nc15, with intron dots stained in green and the nuclear envelope in red. C is a magnified view of the image. (D) The ρ profile quantified from the image in B. The anterior part of the embryo is shown. (E-G) Images and ρ profile from an mh embryo at late nc15. A peak of the ρ profile (at a location of ∼0.4) in this embryo is likely to be reflective of Bcd-independent transcription at the PS4 position (see also Fig. 7 for further discussions).
We monitored active hb transcription through the use of an intronic probe that exclusively detects the nascent transcripts as discrete fluorescence dots inside the nucleus, referred to as intron dots (He et al., 2011, 2012; Liu and Ma, 2013b). This method is highly sensitive for evaluating the dynamic properties of active transcription because the detected signals are exclusively from nascent transcripts prior to the removal of intronic mRNA by splicing (Bothma et al., 2011; Liu and Ma, 2013b). For the endogenous hb, the intron dots accurately record the transcriptional status of the Bcd-responsive P2 promoter of individual copies of the gene (He et al., 2011, 2012; Liu and Ma, 2013b). Fig. 1B-E shows confocal images of two mh embryos at nc15. Whereas the early embryo exhibits robust active transcription as evidenced by the number of intron dots per nucleus (ρ), the late embryo has a deficit in ρ (Fig. 1F,G). These results support the hypothesis that hb shutdown is a property associated with the MBT.
A lacZ reporter gene driven by Bcd binding sites and without hb cis-regulatory elements is sufficient to be shut down in embryos at nc14
It is well documented that sequence-specific transcriptional repressors play important roles in regulating the spatial and temporal behavior of gene transcription (Jaeger et al., 2004, 2012). The shutdown of hb transcription at nc14 could thus reflect the accumulation and action of such repressors (Little et al., 2013); nc14 is a time during which regulatory mechanisms among gap gene products begin to intensify (Surkova et al., 2008; Manu et al., 2009). To evaluate this possible mechanism of shutdown for Bcd-activated transcription, we generated a transgenic fly line with a lacZ reporter gene. This reporter gene, bcd6-lacZ, contains six synthetic Bcd binding sites upstream of a different core promoter (hsp70) other than the P2 promoter used in Bcd-dependent hb transcription. Bcd is known to be capable of recognizing multiple binding sites in the DNA in a highly cooperative manner (Ma et al., 1996; Burz et al., 1998).
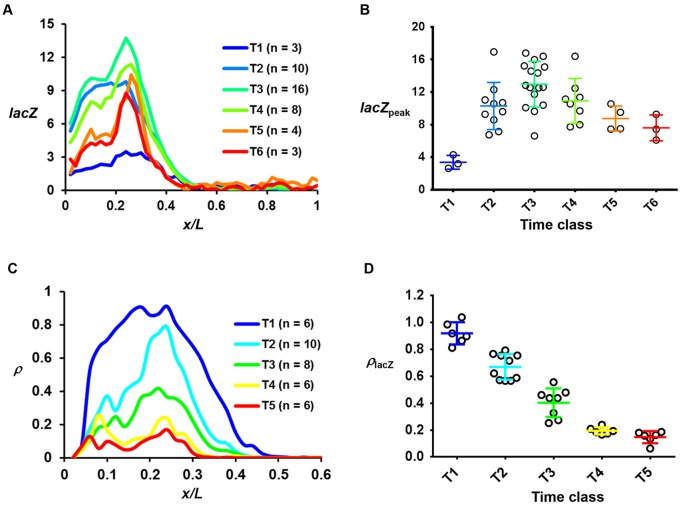
We performed quantitative fluorescence in situ hybridization (FISH) to quantify the product of this reporter gene in embryos grouped into temporal classes of ∼4 min intervals (T1, T2, T3, etc.). Fig. 2A shows the mean profiles of FISH intensities detecting lacZ mRNA at the indicated time classes (see Fig. S2A-F for individual profiles). The intensities extracted from the peak region in individual embryos (lacZpeak) are shown in Fig. 2B. Upon reaching its highest level at T3, lacZpeak as a group mean begins to descend as a function of time. This decrease, although less severe than that of hb mRNA (Liu and Ma, 2013b) and consistent with the longer lifetime of lacZ mRNA (Monsma et al., 1988), is supportive of the possibility that, similar to the endogenous hb gene, the bcd6-lacZ reporter also becomes turned off as embryos progress into nc14.
Fig. 2.
Transcription dynamics of the bcd6-lacZ reporter gene. (A) The mean lacZ FISH intensity profile (in arbitrary units, a.u.) in the indicated time classes. (B) The lacZpeak value in individual embryos that were grouped into the time classes. The mean and s.d. for each time class are shown (3.37±0.85, 10.29±2.91, 12.96±2.81, 10.92±2.75, 8.75±1.54 and 7.60±1.59 for T1 to T6, respectively). Each circle represents an individual embryo. The lacZpeak value for the first two time classes was derived from the plateau region owing to a lack of an apparent peak. (C,D) The mean ρ profile and ρlacZ for the indicated time classes. Similar to lacZpeak, the ρlacZ value at T1 was derived from the plateau region. The mean ρlacZ values are 0.92±0.08, 0.67±0.09, 0.40±0.11, 0.19±0.03 and 0.15±0.04 for T1 to T5, respectively. Time classes are ∼4 min intervals into nc14.
To test this possibility directly, we analyzed the temporal profile of the active sites of reporter gene transcription detected as discrete fluorescence dots inside the nucleus (Fig. S2G,H). Fig. 2C shows a plot of mean ρ as a function of relative anteroposterior (AP) position x/L in the different time classes (see Fig. S2J-M for individual profiles). The ρ profiles exhibit an overall resemblance to those of the lacZ mRNA FISH intensities (Fig. 2A), supportive of the suggestion that the probability of active transcription of gene copies at a location in the embryo is directly related to the levels of mature transcripts (He et al., 2011, 2012). An important aspect of these profiles that is particularly relevant to our current study is that they show a decrease in ρ as a function of time, illustrating a reduction in the probability of active transcription of this reporter gene during nc14. Specifically, the mean ρ at the peak region (ρlacZ) peaks at T1 and progressively decreases as a function of time (Fig. 2D). This kinetic profile of reporter gene shutdown is broadly comparable to that of the endogenous hb gene (Liu and Ma, 2013b). Since our bcd6-lacZ reporter gene is transcribed from a different core promoter and is driven by synthetic Bcd binding sites, our results show that cis-regulatory elements that are specific to hb are not required to mediate the shutdown of Bcd-activated transcription at nc14 (see Discussion for additional information).
A Bcd mutant with elevated activating potency forms a normal gradient in the embryo
Our bcd6-lacZ results suggest that the shutdown of Bcd-dependent hb transcription at nc14 might be reflective of the molecular properties of Bcd during this time. To test this possibility directly (see below), we took advantage of a Bcd mutant, BcdK308A, which has an elevated ability to activate transcription in Drosophila cells (Liu and Ma, 2012). We generated transgenic flies with ∼18 kb genomic fragments containing the bcd coding sequences for either wt Bcd or BcdK308A (referred to as bcdwt and bcdK308A transgenes, respectively). To ensure an identical genomic location of the two transgenes for our quantitative studies, we employed the ΦC31-mediated transgenesis approach and inserted them at a landing site on the second chromosome. We used embryos from bcdE1 mothers with a copy of either the bcdwt or bcdK308A transgene for technical convenience (referred to as wt and mutant embryos, respectively). It has been documented that bcd gene copy number only affects where hb is expressed in the embryo (i.e. its boundary position), not its level of expression (i.e. its amplitude) (Liu and Ma, 2013a). In addition, the hb expression amplitude is insensitive to whether Bcd is derived from an endogenous bcd locus or a transgenic copy (Fig. S3A-E).
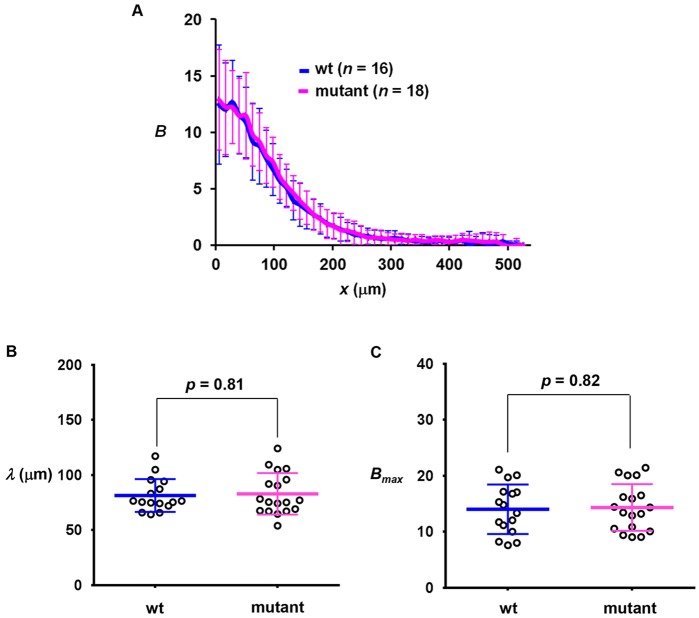
As further detailed below, despite an increased amplitude of hb expression in mutant embryos, the hb expression boundary does not exhibit a posterior shift, suggesting that mutating the primary sumoylation site of Bcd impacts its transcription activation function as opposed to the formation of a normal gradient. To directly evaluate the gradient profile of the mutant Bcd, we performed anti-Bcd immunostaining in embryos. Fig. 3A shows that the mean intensity profiles from wt and mutant embryos exhibit an overall similarity (see Fig. S3F,G for individual profiles). To compare the Bcd gradient profiles quantitatively, we measured their length constants (λ) and amplitudes (using the maximal Bcd intensities, Bmax, as proxies). Fig. 3B shows that Bcd gradient profiles from the wt and mutant embryos have a similar λ [81.42±14.87 µm (n=16) and 82.87±18.87 µm (n=18), respectively; P=0.81]. In addition, they have a similar Bmax [14.01±4.42 (n=16) and 14.34±4.17 (n=18), respectively; P=0.82; Fig. 3C]. These results document quantitatively that the sumoylation-defective Bcd mutant forms a normal concentration gradient (Grimm and Wieschaus, 2010), suggesting that its elevated activating function in the embryo is not associated with any alterations in Bcd protein stability.
Fig. 3.
wt and mutant embryos have similar Bcd gradient profiles. (A) The mean profile of Bcd intensities (a.u.) from wt and mutant embryos. (B,C) Measured λ (µm) and Bmax (a.u.) from individual wt and mutant embryos. Mean and s.d. are shown. P-value was calculated by Student's t-test.
The duration of active hb transcription is elongated in the mutant embryo
To evaluate specifically the relationship between the activating potency of Bcd and the timing of hb shutdown at nc14, we performed hb intron staining in embryos. To permit close evaluation of the temporal dynamics of hb transcription, which takes place rapidly at nc14, we grouped embryos into time classes (t1, t2, t3, etc.) that have a mean time interval of ∼1 min (Liu and Ma, 2013a) (see also Fig. 4 legend for additional information about time scale and other technical details). Fig. 4A,B shows the profiles of mean ρ as function of x/L in the different time classes of wt and mutant embryos (see Fig. S4 for the individual profiles).
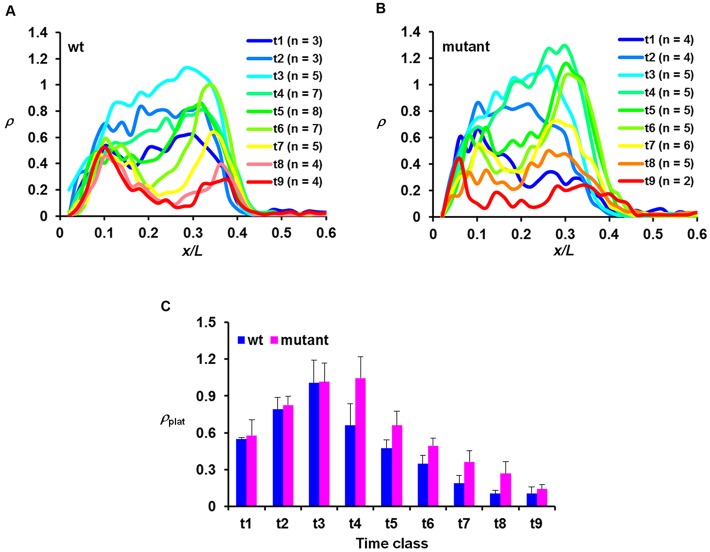
Fig. 4.
hb shutdown is delayed in the mutant embryo. (A,B) Mean ρ profiles for the indicated time classes from wt and mutant embryos. (C) Comparison of the mean ρplat values (with s.d.) for the indicated time classes between wt and mutant embryos. Time classes are ∼1 min intervals into nc14. The mean ρplat values are 0.55±0.01, 0.79±0.10, 1.01±0.18, 0.66±0.17, 0.47±0.07, 0.35±0.07, 0.19±0.07, 0.10±0.03 and 0.11±0.05 for wt and 0.58±0.13, 0.83±0.07, 1.02±0.15, 1.05±0.17, 0.66±0.12, 0.49±0.07, 0.36±0.09, 0.27±0.10 and 0.14±0.04 for mutant, for t1 to t9, respectively. P-values between wt and mutant embryos are 0.73, 0.63, 0.94, 3.4×10−3, 3.9×10−3, 4.2×10−3, 5.7×10−3, 1.4×10−2 and 0.46 for t1 to t9, respectively. Note the difference between the time scales in this figure (∼1 min) and Fig. 2 (∼4 min). In addition, the active copies of the endogenous hb gene shown in this figure were detected by an intronic probe, whereas those of the intronless lacZ reporter transgene were detected by a coding sequence probe. As discussed recently (Liu and Ma, 2013a), unlike intronic probes, the use of coding sequence probes can cause the detected active sites to ‘linger’ for a period of time after transcription initiation/elongation near the promoter has been shut off.
Fig. 4C shows the mean ρ at the plateau region (ρplat) for wt and mutant embryos in the indicated time classes (with s.d.). Consistent with the temporal dynamics of active hb transcription under control of the endogenous Bcd protein (Liu and Ma, 2013b), ρplat in our transgenic wt embryos (Fig. 3C, blue) also exhibits a rapid ascent upon entering nc14 interphase to reach its peak at t3. This is followed by a rapid descent, which represents the shutdown phase of active hb transcription at nc14. Similar to the wt embryos, ρplat in mutant embryos (Fig. 4C, pink) exhibits a similar rapid ascent, also reaching a peak at t3 and attaining a similar peak level (1.01±0.18 and 1.02±0.15 in wt and mutant embryos, respectively; P=0.96). However, in contrast to an immediate descent in wt embryos, ρplat in mutant embryos stays at its peak level until t4, and thereafter the quick descent begins. These results show evidence for an elongated duration of active hb transcription in the mutant embryo, resulting from a postponement of the shutdown phase, as opposed to an accelerated onset phase (see also Fig. S5 for data generated from independent transgenic lines).
A delayed shutdown phase of active hb transcription at nc14 leads to an increased level of hb mRNA
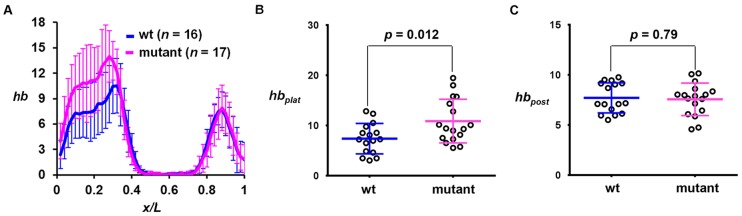
To investigate how an elongated duration of active transcription affects hb gene product accumulation, we performed quantitative FISH to detect mature hb mRNA in embryos. Fig. 5A shows the mean hb intensity profiles as a function of x/L, revealing a higher transcript level at the anterior domain in the mutant embryo (see Fig. S6A,B for individual profiles). The mean intensities at the plateau region (hbplat) for wt embryos were significantly lower than those in the mutant [Fig. 5B; 7.39±3.03 (n=16) and 10.89±4.35 (n=17), respectively, P=0.012]. Importantly, the peak level of hb intensity of the Bcd-independent posterior stripe (Fig. 5C; referred to as hbpost) is similar between wt and mutant embryos (P=0.79). In addition, the expression level of another gap gene, Kruppel (Kr), which is regulated primarily by cross-regulatory mechanisms (Jaeger et al., 2012), also remains similar in wt and mutant embryos (Fig. S6C-F). Together, these results document a specific increase in the amplitude of Bcd-activated hb expression in mutant embryos (see also Fig. S6G-K for hb amplitude increases detected in independent transgenic lines).
Fig. 5.
hb mRNA level is increased in mutant embryos. (A) Mean profiles of hb intensities (a.u.) from wt and mutant embryos. (B,C) Measured hbplat and hbpost values in individual wt or mutant embryos. The mean hbplat values are shown in the text and the mean hbpost values are 7.71±1.50 and 7.59±2.45 in wt and mutant embryos, respectively. There is a significant difference (Student's t-test) between wt and mutant embryos in hbplat but not in hbpost.
The Bcd mutant alters the spatial characteristics of AP patterning and causes developmental defects
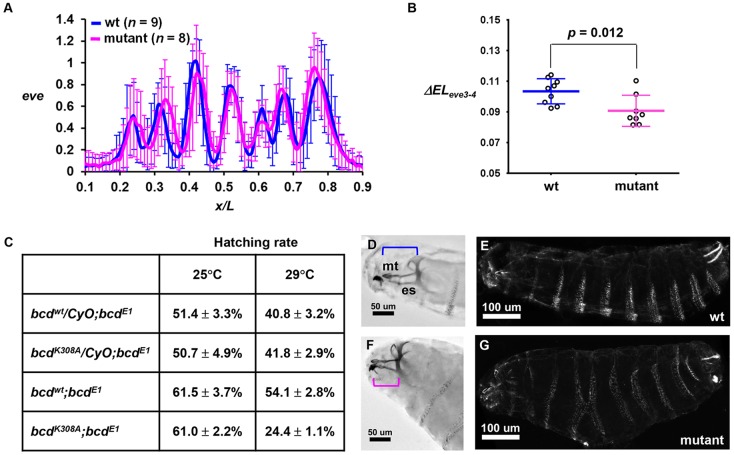
To determine whether an elevated amplitude of hb expression caused by a stronger activating potency of Bcd can be propagated downstream of the AP patterning network, we measured even-skipped (eve) expression profiles in embryos. Fig. 6A shows that the mean normalized intensity profiles from the wt and mutant embryos have different spatial characteristics (see Fig. S7A,B for individual profiles). Here we focused specifically on the distance between the anterior boundaries of eve stripes 3 and 4 (referred to as ΔELeve3-4), a spatial feature known to be directly sensitive to hb gene dose (Payankaulam and Arnosti, 2008; Yu and Small, 2008). We recorded ΔELeve3-4 values of 10.34±1.59 and 9.07±1.01 (percentage egg length, mean±s.d.) in wt (n=9) and mutant (n=8) embryos, respectively (Fig. 6B; P=0.012). These findings document that the impact of an increased activating strength of Bcd on hb expression can be propagated downstream of the AP patterning network.
Fig. 6.
Developmental defects exhibited by mutant embryos. (A) Normalized eve intensity profile from wt and mutant embryos. (B) ΔELeve3-4 value in individual wt and mutant embryos. (C) The Bcd mutant reduces the hatching rate in a gene dosage- and temperature-dependent manner (29°C versus 25°C for two versus one copy of the mutant transgene). (D,E) The cuticle pattern of an embryo from the bcdwt;bcdE1 mother at 29°C showing (D) the head structure from Nomarski microscopy and (E) the denticle pattern of the whole body under dark-field microscopy. (F,G) Cuticle patterns of an embryo from the bcdK308A;bcdE1 mother at 29°C. Note a shortening in mediate tooth (mt) and epistomal sclerite (es) and a denticle fusion in this embryo. Brackets indicate the spans from mt to the end of es.
The altered transcription patterns in mutant embryos led to developmental defects. First, we compared the hatching rates of the embryos from mothers containing different copy numbers of the transgenes at different temperatures. Our results (Fig. 6C) show a hatching rate reduction between embryos from bcdE1 mothers with two copies of bcdwt versus bcdK308A transgenes (referred to as bcdwt;bcdE1 and bcdK308A;bcdE1 embryos, respectively) at 29°C but not at 25°C. Embryos from bcdE1 mothers with only one copy of the wt or mutant transgenes did not exhibit a difference in hatching rate. These results suggest a hatching rate reduction caused by the Bcd mutant in a gene dosage- and temperature-dependent manner. In addition, we examined the cuticle patterns of the bcdwt;bcdE1 and bcdK308A;bcdE1 embryos at a temperature (29°C) that caused a hatching rate difference. We detected mutant embryos that exhibited head structure defects (Fig. 6D,E), including a shortening in median tooth (mt) and epistomal sclerite (es). Among the morphologically defective embryos, a fusion between denticle bands could also be observed (Fig. 6F,G).
The impact of enhanced activating potency of Bcd is highly specific
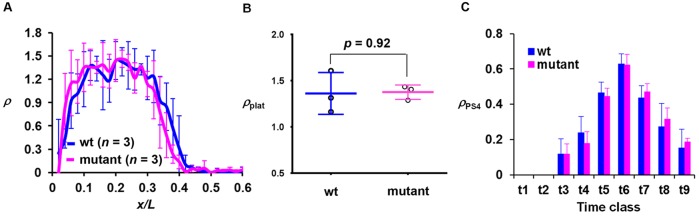
As shown above, an altered shutdown phase, but not onset, of hb transcription at nc14 in embryos containing the sumoylation-defective Bcd mutant reveals a temporal specificity (i.e. time dependence) in the effect caused by this mutant protein. This argues against a model in which the mutant protein leads to a temporally non-specific increase in hb transcription probability during nc14. To further evaluate the temporal specificity of the effect of the Bcd mutant on the time scale of nuclear cycles we analyzed intron staining images of embryos at nc13. Fig. 7A shows the mean ρ profiles of wt and mutant embryos (see Fig. S7C,D for individual profiles). Importantly, the wt and mutant embryos have similar intron dot numbers in the plateau region: ρplat=1.36±0.23 (n=3) and 1.38±0.08 (n=3), respectively (Fig. 7B; P=0.92). These results further argue against a temporally non-specific increase of hb transcription probability caused by this mutant protein. Furthermore, unlike nc14, the amplitude of hb expression at nc13 is unaffected by the sumoylation mutation (Fig. S7E-H; see also Fig. S7I-L for data from two independent transgenic lines).
Fig. 7.
The regulation of hb transcription shutdown is stage and input dependent. (A) The mean ρ profile at nc13 from wt and mutant embryos. (B) The ρplat value from individual wt and mutant embryos at nc13. (C) The mean ρPS4 value at different time classes of nc14 was compared between wt and mutant embryos. PS4-specific transcription is not detectable at t1 and t2. The mean ρPS4 values (with s.d.) are 0.12±0.08, 0.24±0.09, 0.46±0.06, 0.63±0.06, 0.44±0.07, 0.27±0.13 and 0.15±0.11 for wt and 0.12±0.06, 0.18±0.07, 0.44±0.05, 0.62±0.06, 0.47±0.05, 0.32±0.06 and 0.19±0.02 for mutant for t3 to t9, respectively. P-values between wt and mutant embryos are 0.95, 0.08, 0.44, 0.88, 0.45, 0.31 and 0.32 for t3 to t9, respectively (Student's t-test).
To further evaluate the specificity of the effect of the Bcd mutation, we analyzed active hb transcription at a location (PS4) known to be independent of Bcd as a direct input (Perry et al., 2012; Chen et al., 2013). The intron dot number at PS4 (ρPS4) exhibits similar profiles in wt and mutant embryos at nc14 (Fig. 7C; see also Fig. S5P for data from two independent transgenic flies). These findings document that, in contrast to a delayed shutdown of Bcd-dependent hb transcription, the temporal behavior of active hb transcription at the PS4 location is unaffected.
DISCUSSION
A fundamental feature of animal development is the control of gene expression to achieve specific spatial and temporal patterns in a highly coordinated way (Gebelein and Ma, 2015). There are two aspects of the temporal dynamics of a gene's transcription in a developmental system: onset and duration (Garcia et al., 2013; Liu and Ma, 2013b; Lucas et al., 2013). Proper control of the temporal dynamics of transcription is particularly important for developmental systems that progress rapidly, such as the early Drosophila embryo (Foe and Alberts, 1983; Guilgur et al., 2014). It is well known that alterations in the transcriptional activation of early zygotic genes can cause morphological defects in the Drosophila embryo (Bushati et al., 2008; Liang et al., 2008; Lagha et al., 2013; Sung et al., 2013). Our results show that the timing of hb transcription shutdown is associated with the MBT and can be perturbed specifically in embryos containing a Bcd mutant defective in sumoylation (Fig. 4). A postponement of hb shutdown at nc14 can elongate the duration of active transcription, leading to increased levels of hb gene products and patterning defects (Fig. 6). These results show that the precise timing of the shutdown phase of Bcd-activated hb transcription at nc14 is important for normal development. The effects of the Bcd sumoylation mutant on hb shutdown are highly specific, and they are restricted to the shutdown phase (without affecting the onset phase) only at nc14 and only on Bcd-activated hb transcription (Fig. 7). It should be noted that our current results do not show that Bcd sumoylation is a temporally regulated event during the MBT, although it represents an attractive possibility that remains to be tested in the future. Temporally regulated Bcd sumoylation could directly account for the temporally restricted effects of the Bcd mutant, although ‘constitutive’ Bcd sumoylation can also exert time-dependent actions in association with the global events of the system (e.g. mitotic cycles and morphological progression of the MBT).
Our results provide new insights into how Bcd-activated hb transcription becomes shut down at nc14. Our evaluations of the bcd6-lacZ reporter gene demonstrate that neither the P2 promoter of hb nor any of its cis-regulatory elements are required for the shutdown of Bcd-activated transcription at nc14. Importantly, hb shutdown takes place at a time when the Bcd concentration gradient remains intact (Liu and Ma, 2013b). It has been suggested that specific pathways can become activated at the MBT to cause rapid degradation of maternal proteins such as Twine (Di Talia et al., 2013). If hb shutdown were to merely reflect a decaying Bcd gradient at nc14, we would have expected a shutdown process that initiates near mid-embryo (where the Bcd concentration is low) and ‘spreads' toward the anterior (with increasing Bcd concentrations). But our results do not support this prediction (Fig. S8A,B; see also Liu and Ma, 2013a). In addition, neither the length constant nor the amplitude of the Bcd gradient profile is affected by the Bcd mutation (Fig. 3). Thus, our results show that the timing of Bcd-activated transcription during nc14 requires neither a physical disappearance of this maternal activator nor the accumulating activities of sequence-specific zygotic repressors. Instead, it is the functional potency of the maternal activator Bcd that is part of the mechanism of timing the molecular events in accordance with MZT morphological progression. Importantly, the potency of the Bcd activator can be either strengthened (this study) or weakened (Liu and Ma, 2013a) to tune – in opposite directions – the hb shutdown timing and hb expression level. We note that our bcd6-lacZ reporter results do not formally exclude the possibility that the shutdown of Bcd-activated transcription at nc14 involves a zygotic repressor(s) that operates by competing with Bcd binding to its DNA sites. But we currently do not favor this possibility because the position independence and rapidity of the shutdown event would likely require any such zygotic repressor not only to have the same/overlapping Bcd binding specificity but also to accumulate in a spatially non-restricted (i.e. covering the entire hb expression domain) and temporally abrupt way at nc14.
As part of the receding of the first wave of zygotic transcription in association with the MBT (Rose and Wieschaus, 1992; Erickson and Quintero, 2007), hb is among a group of genes that exhibit a shutdown phase at nc14. These genes play key roles in different processes that are ongoing during the MBT, suggesting the possibility that a global or general mechanism might regulate the shutdown events of transcription in a coordinated manner. Our study shows that the timing of the shutdown can be postponed by elevated Bcd activating potency, but only to a degree. For genes that are activated by combinatorial sets of maternal inputs, it remains to be determined whether activator functions, as opposed to protein availability, are also regulated during transcription shutdown at the MBT. Genes that are transcriptionally active prior to the MBT tend to share promoter features that are distinct from those of genes that become activated during MBT (Chen et al., 2013). An intriguing possibility is that the MBT might be associated with a systematic change in the composition of the transcription machinery (Phillips and Pitt, 1985). But the fact that a synthetic reporter containing a different core promoter also exhibits a shutdown phase at nc14 indicates that the hb P2 promoter is not required.
A recent study reveals an interplay between zygotic transcription and DNA replication at the MBT (Blythe and Wieschaus, 2015). It has been proposed that euchromatin DNA is replicated within a few minutes into nc14 interphase (Shermoen et al., 2010). This DNA replication time coincides broadly with the time of hb shutdown, raising the question of whether DNA replication at the hb locus might trigger its transcription shutdown. Fig. S8C,D shows an embryo that we were able to capture in which nuclei contain more than two intron dots – a positive indicator of DNA replication at the hb gene locus. The strong intron staining detectable at the anterior of the embryo indicates that Bcd-activated hb transcription has not yet been turned off (nuclear height measurements suggest that this embryo belongs to time class t2). These results thus suggest that DNA replication at the hb locus does not directly trigger its transcription shutdown at nc14.
Sumoylation is a post-translational modification that regulates a variety of biological processes through mechanisms that may involve protein-protein interactions, subcellular localization and protein stability (Gareau and Lima, 2010). From the perspective of developmental biology, many transcriptional activators with important developmental roles are substrates of sumoylation (Geiss-Friedlander and Melchior, 2007; Nie et al., 2009). It has been reported that sumoylation of Medea (Med), an intracellular transducer of the Drosophila morphogen Decapentaplegic (Dpp), triggers Med nuclear export and, therefore, restricts the range of Dpp signaling (Miles et al., 2008). The lengthening of the duration of Bcd-activated hb transcription caused by the Bcd sumoylation mutation increases the amplitude of hb expression without extending its expression boundary. Thus, sumoylation of proteins involved in morphogen functions can alter either the action range (in space) or the output level (due to action time). In yeast, sumoylation has been suggested to play a role in terminating inducible activation events by evicting activator molecules from promoters. For example, disruption of Gcn4 sumoylation can extend its promoter association and increase the expression level of the target gene ARG1 (Rosonina et al., 2010). Whether sumoylation of Bcd plays a mechanistically equivalent role in evicting Bcd molecules from the hb enhancer at nc14 remains an open question and speculative possibility.
MATERIALS AND METHODS
Identification of genes subject to transcription shutdown at nc14
We used a published RNA-seq dataset (Lott et al., 2011) to identify genes that become shut down during nc14. This dataset consists of normalized read counts (RPKM) per gene for individual embryos that have been temporally ranked and grouped into four time classes in nc14 interphase (referred to as 14A, 14B, 14C and 14D, from young to old). We performed two analyses aimed at systematically identifying genes that exhibit characteristics of transcription shutdown at nc14. First, we selected transcripts that could be positively identified as zygotic products expressed from paternal gene copies [marked with single-nucleotide polymorphisms (SNPs)] (Lott et al., 2011). Among the transcripts that have an RPKM value >2, we calculated the RPKM ratios between two adjacent time classes: 14A to 14B, 14B to 14C, or 14C to 14D. We defined a gene to possess a shutdown property during nc14 if any of these three ratios is greater than 1.5. This first analysis led to the identification of 156 shutdown genes from a total of 3401 genes analyzed. Second, based on the list of genes that were categorized as zygotic (Lott et al., 2011), we selected 1658 genes that do not overlap with the 3401 genes used in the first analysis. This second analysis led to the identification of an additional 38 shutdown genes using the same criteria defined above. The two gene lists combined gave a total of 194 shutdown genes (Table S1). The Database for Annotation, Visualization and Integrated Discovery (DAVID) was then used for the assessment of functional enrichment in the identified shutdown genes.
Plasmid construction and Drosophila transgenesis
Two copies of oligonucleotide 5′-GGCGCGTCGACAGGTTCTAATCCCGGTCTAATCCCTCGAGTCTAATCCCATGAGTCGACGGTCGACCCCCG-3′ were cloned (SalI sites in italics) in tandem into the SalI site of plasmid pCZ3005 (Zhao et al., 2002) to yield plasmid ljb3058. The DNA fragment containing six Bcd binding sites (TAATCC, underlined) was amplified from ljb3058 and cloned into the XbaI site of the vector placZattB (Bischof et al., 2007) to yield the bcd6-lacZ reporter plasmid ljb5003. Using a commercial service (Rainbow Transgenic Flies), ljb5003 was inserted into the docking site VK00037 of fly line 24872 through φC31 integrase-mediated transformation to generate the bcd6-lacZ transgenic flies.
A ∼6.3 kb Drosophila genomic DNA fragment (located between BamHI and EcoRI sites), which contains the bcd coding sequence and flanking sequences, was cloned into the pCaSpeRBcdBgleII transgenic vector [a gift of Dr David Stein (Zhao et al., 2002)]. The resultant plasmids contained either the wt Bcd or BcdK308A coding sequences and were named ljb4056 and ljb4057, respectively. The bcdK308A mutation in ljb4057 was generated by PCR-based site-directed mutagenesis. ljb4057 was used to introduce the bcdk308A mutation into the CH322-100D18 plasmid using the recombineering-mediated gap repair method (Venken et al., 2006) to obtain plasmid ljb5006. An identical, parallel cloning experiment was carried out for ljb4056 to obtain plasmid ljb5005; thus, the two plasmids ljb5005 and ljb5006 differ only by the bcdk308A mutation. The CH322-100D18 plasmid was purchased from BACPAC Resources Center (BPRC) at Children's Hospital Oakland Research Institute (CHORI). It is a bacterial artificial chromosome (BAC) vector containing ∼18.5 kb bcd genomic DNA and the bacterial attachment (attB) site for site-specific integration. The resulting ljb5005 and ljb5006 were then inserted into the same docking site VK00001 of fly line 24861 through φC31 integrase-mediated transformation to generate transgenic flies (Rainbow Transgenic Flies). ljb4056 and ljb4057 were also used to generate transgenic flies through standard P-element-mediated transformation with the commercial service. The fly line 7130 with the mh1 mutation was obtained from the Bloomington Stock Center.
Embryo staining, imaging and data analysis
All embryos (0-4 h) used in quantitative FISH and immunostaining were collected at 25°C. Quantitative FISH to detect lacZ, hb or eve mRNA in embryos, imaging and data analysis were performed as previously described (He et al., 2008; Liu and Ma, 2011, 2013a). Briefly, quantitative FISH was performed on a side-by-side basis wherever data were used for direct comparisons and all images were captured within a linear range under identical settings during a single imaging cycle. All images focused on the midsagittal plane and, for each embryo, included one that captured the nuclei stained with DAPI. Nuclear cycle was determined by the number of nuclei on the dorsal side of the midsagittal image; typically, embryos at nc13 have ∼55-65 identifiable nuclei that are round in shape. Embryos at nc14 have ∼70-80 identifiable nuclei, which are initially round and become progressively elongated (along the apical-basal axis) as a function of developmental time (Liu and Ma, 2013b). In order to monitor the temporal dynamics of bcd6-lacZ transcription, embryos at nc14 were grouped into time classes (referred to as T1, T2, T3, etc.) based on the estimated nuclear height. The interval between the time classes is ∼4 min according to an established conversion (Fung et al., 1998). To extract data along the AP axis, each embryo was divided into 50 bins (He et al., 2008). The lacZpeak in an individual embryo was defined as the mean intensity of five consecutive bins with the peak position in the middle. Embryos that are ∼8-16 min into the interphase of nc14 (combination of time classes T3 and T4) were selected for comparisons of cytoplasmic hb mRNA levels. hbplat was calculated as follows. For a given embryo, five consecutive bins located 0.1 x/L (fractional embryo length) anterior to its hb expression boundary position (xhb) were grouped together and the mean intensity value of this group was defined as hbplat. Thus, hbplat is devoid of both PS4 and the variable anterior stripe (Tautz et al., 1987; Margolis et al., 1995). We defined xhb as the position at which hb intensity is half maximal. Embryo selection and boundary calculation for eve experiments were carried out as described previously (Liu and Ma, 2011, 2013a).
Quantitative FISH to detect nascent hb transcripts as intron dots in embryos was performed as follows. Digoxigenin (Roche)-labeled antisense RNA probes targeting the hb intron were prepared and FISH was performed (He et al., 2011). Wheat germ agglutinin (WGA) with Alexa Fluor 555 conjugate was used to stain the nuclear membrane to mark the nuclear boundary. To maximize the number of intron dots detected, we imaged embryos that had been flattened (He et al., 2011); a separate, midsagittal image for each embryo was also captured for measuring nuclear heights to allow the temporal ranking of embryos and their grouping into time classes. We generally took 8-10 z-sections (covering 4-5 µm along the z-axis) to capture all the hb intron dots in each nucleus (He et al., 2011). Threshold settings for hb intron dot detection were as follows (Porcher et al., 2010; Liu and Ma, 2013a,b). The pixel number threshold was set at ≥3 and the intensity threshold for an individual embryo was set to be the lower limit at which no nuclei in the broad anterior hb expression domain had more than two intron dots. Similar to the definition of hbplat, five consecutive bins located 0.1 x/L anterior to the hb expression boundary were grouped together as the plateau region. The mean ρ value from this region (denoted as ρplat) and the average intensity per intron dot within this group were calculated for individual embryos. Quantitative FISH experiments to detect nascent lacZ transcripts with an RNA probe targeting the full-length lacZ gene were performed in the same way as described above, and images with lacZ nascent transcripts captured were analyzed in the same way as for hb. ρlacZ was defined as the mean ρ value from five consecutive bins with the peak position in the middle.
Quantitative immunostaining for Bcd (anti-Bcd antibody, Santa Cruz Biotechnology, sc-66818; 1:100) in embryos, imaging, intensity measurements and calculation of length constant (λ) values were described previously (Liu and Ma, 2011). Specifically, the λ value for an individual embryo is obtained by fitting its Bcd intensity profile to the equation: B=A*e−x/λ+C, where B is Bcd intensity at location x, A is amplitude and C is background (both A and C are constants). The embryos used in the analysis were ∼8-16 min into the interphase of nc14 based on nuclear height.
Hatching rate measurement and cuticle pattern examination
Hatching rate was measured as follows. Freshly eclosed flies were transferred to a fresh vial and maintained at the specified temperature for 2 days and then allowed to lay eggs on a grape agar plate. After removing the flies, the number of embryos on the plate was counted and the plate was further incubated at the specified temperature. The larvae that emerged on the plate were counted and removed every day until no new emergence occurred. Hatching rate was calculated by dividing the total number of larvae by the number of embryos on the plate. To examine the cuticle pattern, embryos were collected on the grape agar plate overnight and allowed to age for 24-36 h at 29°C. Cuticles were then prepared according to the Hoyer's method (Ashburner, 1989) and images were captured by dark-field (whole body) and Nomarski (head) microscopy.
Statistical analysis
All experimental values are shown as the mean with s.d., with n being the number of independent samples. All image processing and statistical analyses, including Student's t-test (two-tailed) and curve fitting, were performed using Matlab software (MathWorks).
Acknowledgements
We thank members of our groups at CCHMC, in particular David Cheung, Feng He, Chuanxuan Wei and Honggang Wu, for discussions and assistance.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
J.L. and J.M. conceived and designed the study, interpreted the results, and wrote and approved the paper; J.L. performed all experiments, data acquisition and analysis, and generated all figures.
Funding
This work was supported in part by grants from the National Institutes of Health [1R01GM101373] and National Science Foundation [IOS-0843424] to J.M. Deposited in PMC for release after 12 months.
Supplementary information
Supplementary information available online at http://dev.biologists.org/lookup/suppl/doi:10.1242/dev.126946/-/DC1
References
- Ashburner M. (1989). Drosophila: A Laboratory Mannual. Cold spring Harbor, NY: Cold Spring Harbor Laboratory Press. [Google Scholar]
- Bischof J., Maeda R. K., Hediger M., Karch F. and Basler K. (2007). An optimized transgenesis system for Drosophila using germ-line-specific phiC31 integrases. Proc. Natl. Acad. Sci. USA 104, 3312-3317. 10.1073/pnas.0611511104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blythe S. A. and Wieschaus E. F. (2015). Zygotic genome activation triggers the DNA replication checkpoint at the midblastula transition. Cell 160, 1169-1181. 10.1016/j.cell.2015.01.050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bothma J. P., Magliocco J. and Levine M. (2011). The snail repressor inhibits release, not elongation, of paused Pol II in the Drosophila embryo. Curr. Biol. 21, 1571-1577. 10.1016/j.cub.2011.08.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burz D. S., Rivera-Pomar R., Jäckle H. and Hanes S. D. (1998). Cooperative DNA-binding by Bicoid provides a mechanism for threshold-dependent gene activation in the Drosophila embryo. EMBO J. 17, 5998-6009. 10.1093/emboj/17.20.5998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bushati N., Stark A., Brennecke J. and Cohen S. M. (2008). Temporal reciprocity of miRNAs and their targets during the maternal-to-zygotic transition in Drosophila. Curr. Biol. 18, 501-506. 10.1016/j.cub.2008.02.081 [DOI] [PubMed] [Google Scholar]
- Chen K., Johnston J., Shao W., Meier S., Staber C. and Zeitlinger J. (2013). A global change in RNA polymerase II pausing during the Drosophila midblastula transition. eLife 2, e00861 10.7554/eLife.00861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung D., Miles C., Kreitman M. and Ma J. (2014). Adaptation of the length scale and amplitude of the Bicoid gradient profile to achieve robust patterning in abnormally large Drosophila melanogaster embryos. Development 141, 124-135. 10.1242/dev.098640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Renzis S., Elemento O., Tavazoie S. and Wieschaus E. F. (2007). Unmasking activation of the zygotic genome using chromosomal deletions in the Drosophila embryo. PLoS Biol. 5, e117 10.1371/journal.pbio.0050117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Talia S., She R., Blythe S. A., Lu X., Zhang Q. F. and Wieschaus E. F. (2013). Posttranslational control of Cdc25 degradation terminates Drosophila's early cell-cycle program. Curr. Biol. 23, 127-132. 10.1016/j.cub.2012.11.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar B. A., Kiehle C. P. and Schubiger G. (1986). Cell cycle control by the nucleo-cytoplasmic ratio in early Drosophila development. Cell 44, 365-372. 10.1016/0092-8674(86)90771-3 [DOI] [PubMed] [Google Scholar]
- Erickson J. W. and Quintero J. J. (2007). Indirect effects of ploidy suggest X chromosome dose, not the X:A ratio, signals sex in Drosophila. PLoS Biol. 5, e332 10.1371/journal.pbio.0050332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foe V. E. and Alberts B. M. (1983). Studies of nuclear and cytoplasmic behaviour during the five mitotic cycles that precede gastrulation in Drosophila embryogenesis. J. Cell Sci. 61, 31-70. [DOI] [PubMed] [Google Scholar]
- Fung J. C., Marshall W. F., Dernburg A., Agard D. A. and Sedat J. W. (1998). Homologous chromosome pairing in Drosophila melanogaster proceeds through multiple independent initiations. J. Cell Biol. 141, 5-20. 10.1083/jcb.141.1.5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gans M., Audit C. and Masson M. (1975). Isolation and characterization of sex-linked female-sterile mutants in Drosophila melanogaster. Genetics 81, 683-704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia H. G., Tikhonov M., Lin A. and Gregor T. (2013). Quantitative imaging of transcription in living Drosophila embryos links polymerase activity to patterning. Curr. Biol. 23, 2140-2145. 10.1016/j.cub.2013.08.054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gareau J. R. and Lima C. D. (2010). The SUMO pathway: emerging mechanisms that shape specificity, conjugation and recognition. Nat. Rev. Mol. Cell Biol. 11, 861-871. 10.1038/nrm3011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gebelein B. and Ma J. (2015). Regulation in the early Drosophila embryo. Rev. Cell Biol. Mol. Med. (in press). [Google Scholar]
- Geiss-Friedlander R. and Melchior F. (2007). Concepts in sumoylation: a decade on. Nat. Rev. Mol. Cell Biol. 8, 947-956. 10.1038/nrm2293 [DOI] [PubMed] [Google Scholar]
- Grimm O. and Wieschaus E. (2010). The Bicoid gradient is shaped independently of nuclei. Development 137, 2857-2862. 10.1242/dev.052589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guilgur L. G., Prudêncio P., Sobral D., Liszekova D., Rosa A. and Martinho R. G. (2014). Requirement for highly efficient pre-mRNA splicing during Drosophila early embryonic development. eLife 3, e02181 10.7554/eLife.02181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He F., Wen Y., Deng J., Lin X., Lu L. J., Jiao R. and Ma J. (2008). Probing intrinsic properties of a robust morphogen gradient in Drosophila. Dev. Cell 15, 558-567. 10.1016/j.devcel.2008.09.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He F., Ren J., Wang W. and Ma J. (2011). A multiscale investigation of bicoid-dependent transcriptional events in Drosophila embryos. PLoS ONE 6, e19122 10.1371/journal.pone.0019122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He F., Ren J., Wang W. and Ma J. (2012). Evaluating the Drosophila Bicoid morphogen gradient system through dissecting the noise in transcriptional bursts. Bioinformatics 28, 970-975. 10.1093/bioinformatics/bts068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He F., Wei C., Wu H., Cheung D., Jiao R. and Ma J. (2015). Fundamental origins and limits for scaling a maternal morphogen gradient. Nat. Commun. 6, 6679 10.1038/ncomms7679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaeger J., Surkova S., Blagov M., Janssens H., Kosman D., Kozlov K. N., Manu K. N., Myasnikova E., Vanario-Alonso C. E., Samsonova M. et al. (2004). Dynamic control of positional information in the early Drosophila embryo. Nature 430, 368-371. 10.1038/nature02678 [DOI] [PubMed] [Google Scholar]
- Jaeger J., Manu J. and Reinitz J. (2012). Drosophila blastoderm patterning. Curr. Opin. Genet. Dev. 22, 533-541. 10.1016/j.gde.2012.10.005 [DOI] [PubMed] [Google Scholar]
- Lagha M., Bothma J. P., Esposito E., Ng S., Stefanik L., Tsui C., Johnston J., Chen K., Gilmour D. S., Zeitlinger J. et al. (2013). Paused Pol II coordinates tissue morphogenesis in the Drosophila embryo. Cell 153, 976-987. 10.1016/j.cell.2013.04.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langley A. R., Smith J. C., Stemple D. L. and Harvey S. A. (2014). New insights into the maternal to zygotic transition. Development 141, 3834-3841. 10.1242/dev.102368 [DOI] [PubMed] [Google Scholar]
- Lee M. T., Bonneau A. R. and Giraldez A. J. (2014). Zygotic genome activation during the maternal-to-zygotic transition. Annu. Rev. Cell Dev. Biol. 30, 581-613. 10.1146/annurev-cellbio-100913-013027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehmann R. and Nüsslein-Volhard C. (1987). hunchback, a gene required for segmentation of an anterior and posterior region of the Drosophila embryo. Dev. Biol. 119, 402-417. 10.1016/0012-1606(87)90045-5 [DOI] [PubMed] [Google Scholar]
- Liang H.-L., Nien C.-Y., Liu H.-Y., Metzstein M. M., Kirov N. and Rushlow C. (2008). The zinc-finger protein Zelda is a key activator of the early zygotic genome in Drosophila. Nature 456, 400-403. 10.1038/nature07388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Little S. C., Tikhonov M. and Gregor T. (2013). Precise developmental gene expression arises from globally stochastic transcriptional activity. Cell 154, 789-800. 10.1016/j.cell.2013.07.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J. and Ma J. (2011). Fates-shifted is an F-box protein that targets Bicoid for degradation and regulates developmental fate determination in Drosophila embryos. Nat. Cell Biol. 13, 22-29. 10.1038/ncb2141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J. and Ma J. (2012). Drosophila Bicoid is a substrate of sumoylation and its activator function is subject to inhibition by this post-translational modification. FEBS Lett. 586, 1719-1723. 10.1016/j.febslet.2012.04.059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J. and Ma J. (2013a). Dampened regulates the activating potency of Bicoid and the embryonic patterning outcome in Drosophila. Nat. Commun. 4, 2968 10.1038/ncomms3968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J. and Ma J. (2013b). Uncovering a dynamic feature of the transcriptional regulatory network for anterior-posterior patterning in the Drosophila embryo. PLoS ONE 8, e62641 10.1371/journal.pone.0062641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., He F. and Ma J. (2011). Morphogen gradient formation and action: insights from studying Bicoid protein degradation. Fly 5, 242-246. 10.4161/fly.5.3.15837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lott S. E., Villalta J. E., Schroth G. P., Luo S., Tonkin L. A. and Eisen M. B. (2011). Noncanonical compensation of zygotic X transcription in early Drosophila melanogaster development revealed through single-embryo RNA-seq. PLoS Biol. 9, e1000590 10.1371/journal.pbio.1000590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu X., Li J. M., Elemento O., Tavazoie S. and Wieschaus E. F. (2009). Coupling of zygotic transcription to mitotic control at the Drosophila mid-blastula transition. Development 136, 2101-2110. 10.1242/dev.034421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucas T., Ferraro T., Roelens B., De Las Heras Chanes J., Walczak A. M., Coppey M. and Dostatni N. (2013). Live imaging of bicoid-dependent transcription in Drosophila embryos. Curr. Biol. 23, 2135-2139. 10.1016/j.cub.2013.08.053 [DOI] [PubMed] [Google Scholar]
- Ma X., Yuan D., Diepold K., Scarborough T. and Ma J. (1996). The Drosophila morphogenetic protein Bicoid binds DNA cooperatively. Development 122, 1195-1206. [DOI] [PubMed] [Google Scholar]
- Manu N., Surkova S., Spirov A. V., Gursky V. V., Janssens H., Kim A.-R., Radulescu O., Vanario-Alonso C. E., Sharp D. H., Samsonova M. et al. (2009). Canalization of gene expression in the Drosophila blastoderm by gap gene cross regulation. PLoS Biol. 7, e49 10.1371/journal.pbio.1000049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margolis J. S., Borowsky M. L., Steingrimsson E., Shim C. W., Lengyel J. A. and Posakony J. W. (1995). Posterior stripe expression of hunchback is driven from two promoters by a common enhancer element. Development 121, 3067-3077. [DOI] [PubMed] [Google Scholar]
- Miles W. O., Jaffray E., Campbell S. G., Takeda S., Bayston L. J., Basu S. P., Li M., Raftery L. A., Ashe M. P., Hay R. T. et al. (2008). Medea SUMOylation restricts the signaling range of the Dpp morphogen in the Drosophila embryo. Genes Dev. 22, 2578-2590. 10.1101/gad.494808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monsma S. A., Ard R., Lis J. T. and Wolfner M. F. (1988). Localized heat-shock induction in Drosophila melanogaster. J. Exp. Zool. 247, 279-284. 10.1002/jez.1402470312 [DOI] [PubMed] [Google Scholar]
- Nie M., Xie Y., Loo J. A. and Courey A. J. (2009). Genetic and proteomic evidence for roles of Drosophila SUMO in cell cycle control, Ras signaling, and early pattern formation. PLoS ONE 4, e5905 10.1371/journal.pone.0005905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payankaulam S. and Arnosti D. N. (2008). Gene regulation: boundaries within limits. Curr. Biol. 18, R653-R655. 10.1016/j.cub.2008.06.040 [DOI] [PubMed] [Google Scholar]
- Perry M. W., Bothma J. P., Luu R. D. and Levine M. (2012). Precision of hunchback expression in the Drosophila embryo. Curr. Biol. 22, 2247-2252. 10.1016/j.cub.2012.09.051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips J. P. and Pitt A. (1985). The RNA polymerases of Drosophila melanogaster during early development. Biochem. Genet. 23, 655-676. 10.1007/BF02399402 [DOI] [PubMed] [Google Scholar]
- Porcher A., Abu-Arish A., Huart S., Roelens B., Fradin C. and Dostatni N. (2010). The time to measure positional information: maternal hunchback is required for the synchrony of the Bicoid transcriptional response at the onset of zygotic transcription. Development 137, 2795-2804. 10.1242/dev.051300 [DOI] [PubMed] [Google Scholar]
- Pritchard D. K. and Schubiger G. (1996). Activation of transcription in Drosophila embryos is a gradual process mediated by the nucleocytoplasmic ratio. Genes Dev. 10, 1131-1142. 10.1101/gad.10.9.1131 [DOI] [PubMed] [Google Scholar]
- Rose L. S. and Wieschaus E. (1992). The Drosophila cellularization gene nullo produces a blastoderm-specific transcript whose levels respond to the nucleocytoplasmic ratio. Genes Dev. 6, 1255-1268. 10.1101/gad.6.7.1255 [DOI] [PubMed] [Google Scholar]
- Rosonina E., Duncan S. M. and Manley J. L. (2010). SUMO functions in constitutive transcription and during activation of inducible genes in yeast. Genes Dev. 24, 1242-1252. 10.1101/gad.1917910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salz H. K. and Erickson J. W. (2010). Sex determination in Drosophila: the view from the top. Fly 4, 60-70. 10.4161/fly.4.1.11277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shermoen A. W. and O'Farrell P. H. (1991). Progression of the cell cycle through mitosis leads to abortion of nascent transcripts. Cell 67, 303-310. 10.1016/0092-8674(91)90182-X [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shermoen A. W., McCleland M. L. and O'Farrell P. H. (2010). Developmental control of late replication and S phase length. Curr. Biol. 20, 2067-2077. 10.1016/j.cub.2010.10.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung H.-W., Spangenberg S., Vogt N. and Grosshans J. (2013). Number of nuclear divisions in the Drosophila blastoderm controlled by onset of zygotic transcription. Curr. Biol. 23, 133-138. 10.1016/j.cub.2012.12.013 [DOI] [PubMed] [Google Scholar]
- Surkova S., Kosman D., Kozlov K., Manu K., Myasnikova E., Samsonova A. A., Spirov A., Vanario-Alonso C. E., Samsonova M. and Reinitz J. (2008). Characterization of the Drosophila segment determination morphome. Dev. Biol. 313, 844-862. 10.1016/j.ydbio.2007.10.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tadros W. and Lipshitz H. D. (2009). The maternal-to-zygotic transition: a play in two acts. Development 136, 3033-3042. 10.1242/dev.033183 [DOI] [PubMed] [Google Scholar]
- Tautz D., Lehmann R., Schnürch H., Schuh R., Seifert E., Kienlin A., Jones K. and Jäckle H. (1987). Finger protein of novel structure encoded by hunchback, a second member of the gap class of drosophila segmentation genes. Nature 327, 383-389. 10.1038/327383a0 [DOI] [Google Scholar]
- ten Bosch J. R., Benavides J. A. and Cline T. W. (2006). The TAGteam DNA motif controls the timing of Drosophila pre-blastoderm transcription. Development 133, 1967-1977. 10.1242/dev.02373 [DOI] [PubMed] [Google Scholar]
- Venken K. J. T., He Y., Hoskins R. A. and Bellen H. J. (2006). P[acman]: a BAC transgenic platform for targeted insertion of large DNA fragments in D. melanogaster. Science 314, 1747-1751. 10.1126/science.1134426 [DOI] [PubMed] [Google Scholar]
- Yu D. and Small S. (2008). Precise registration of gene expression boundaries by a repressive morphogen in Drosophila. Curr. Biol. 18, 868-876. 10.1016/j.cub.2008.05.050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zalokar M., Audit C. and Erk I. (1975). Developmental defects of female-sterile mutants of Drosophila melanogaster. Dev. Biol. 47, 419-432. 10.1016/0012-1606(75)90295-X [DOI] [PubMed] [Google Scholar]
- Zhao C., York A., Yang F., Forsthoefel D. J., Dave V., Fu D., Zhang D., Corado M. S., Small S., Seeger M. A. et al. (2002). The activity of the Drosophila morphogenetic protein Bicoid is inhibited by a domain located outside its homeodomain. Development 129, 1669-1680. [DOI] [PubMed] [Google Scholar]