Abstract
Background:
Triple-negative breast cancer (TNBC) accounts for 15–20% of all breast cancer in women globally. This subtype often has early and high recurrence rates resulting in poor survival, partially due to lack of targeted therapies. Therefore, there is an urgent need to identify TNBC-specific biomarkers for early diagnosis and treatment monitoring, and to develop more effective targeted therapy.
Methods:
By using miRCURY LNA array platform, we compared the differential miRNA expressions in plasma of patient with TNBC (n=5) and non-TNBC (n=5), as well as healthy controls (n=5). Potential miRNAs were then validated in a large cohort of patients by real-time PCR.
Results:
Ten putative miRNAs from the microarray data that differentially expressed between non-TNBC and healthy controls were identified. In the screening phase (n=90), we selected five miRNAs (miR-92a-3p, miR-342-3p, miR-16, miR-21 and miR-199a-5p) that could discriminate TNBC from non-TNBC for further validation. Results showed that miR-16, miR-21 and miR-199a-5p were underexpressed in TNBC when compared with non-TNBC, and were further validated in a large cohort (n=252). In addition, post-operative plasma levels of miR-16, miR-21 and miR-199a-5p were significantly restored when compared with pre-operative plasma of TNBC. Plasma miR-199a-5p expression in TNBC had significant difference when compared with non-TNBC and healthy controls, the receiver-operator characteristics curve analysis revealed the highest area under curve (AUC=0.8838) among all. The expression levels were associated with TNM stage and tumour subtypes.
Conclusions:
Our data suggest that miR-199a-5p could be a TNBC-specific marker with diagnostic value and provide insights into targeted therapy in the treatment of TNBC.
Keywords: miR-199a-5p, triple-negative breast cancer, biomarker, circulating cell-free miRNA
Breast cancer is the most common cause of cancer mortality in women worldwide (GLOBOCAN 2012, http://globocan.iarc.fr/Pages/fact_sheets_population.aspx). Triple negative-breast cancer (TNBC) is characterised by lack of oestrogen receptor (ER), progesterone receptor (PR) and human epidermal growth factor receptor 2 (HER2). This subtype accounts for 15–20% of all breast cancers (Boyle, 2012). It is a heterogeneous disease that partake distinct histopathological features and clinical behaviour. Epidemiological studies revealed that TNBC is associated with young age (<40) and more frequently in African–American and black ethnicity (Boyle, 2012). TNBC tumours represent the most aggressive phenotype with relatively high recurrence rates (Dent et al, 2007). Patients with TNBC do not respond to endocrine therapy or HER2 targeted therapy, and treatment are based on a combination of commonly used breast cancer therapies including surgery, radiation and chemotherapy regimens.
Clinical trials conducted on TNBC using cisplatin-based regimes as neoadjuvant therapy found that the response rate to single-agent cisplatin was only 21%. An improved response rate was seen using a combination of paclitaxel/fluorouracil, doxorubicin and cyclophosphamide but this was still only 45% (Silver et al, 2010). TNBC and BRCA mutations associated tumour share common pathologies. Poly (ADP-ribose) polymerase (PARP) inhibitor is a class of anti-tumour agents which targets tumours with BRCA mutations but could only benefit a small group of patients (Metzger-Filho et al, 2012). Combining inhibitors of PARP and PI3K were shown to be effective in primary tumour xenografts where tumours had no BRCA mutations, and the clinical outcome of this trial is awaiting (Ibrahim et al, 2012). To date, the current treatment options for TNBC do not demonstrate promising clinical outcomes and often results in poor treatment response and overall survival rate. This is also partly due to the lack of biomarkers to envisage which groups of patients are likely to respond to specific chemotherapeutic and targeted therapies nor are there reliable biomarkers which have been identified to be used as a screening marker. Hence, there is a substantial need for discovering a predictive biomarker for diagnosis that could identify patients with this aggressive and fast growing tumour at an earlier stage to improve the prognosis. With limited targeted therapy currently available for TNBC, it is also important to have a marker for disease monitoring and evaluate the need to change therapy if the treatment response is poor.
MicroRNA (miRNA) has a critical regulatory role in gene expression and is involved in breast carcinogenesis (Serpico et al, 2014). Currently, there are over 1800 miRNAs identified in human (miRBase release 20) and these regulate >50% of the genes. Several lines of evidence have shown that miRNAs are differentially expressed in cancerous tissues when compared with adjacent non-tumour counterparts (Calin and Croce, 2006; Volinia et al, 2006). Expression profiles of miRNAs have been reported to form more defined cluster with similar tumour types than mRNA expression. Numerous studies suggested that miRNAs are highly stable and can be detected in the circulation (Mitchell et al, 2008). It has also been reported that miRNA patterns are unique in the cancers of breast (Ng et al, 2013), colon (Ng et al, 2009), gastric (Tsujiura et al, 2010), ovary (Shapira et al, 2014) and prostate (Brase et al, 2011). The recognition of miRNAs in the circulation marks the milestone in cancer diagnosis and therapeutic application. Detection of circulating miRNAs can serve as non-invasive and cost-effective markers to identify high-risk patients, which are beneficial in clinical setting in terms of diagnosis, prognosis and treatment response. Published data reported that the levels of miR-30a were lower in patients with breast cancer, and showed improved sensitivity and specificity compared with conventional circulating tumour markers (CEA and CA153) (Zeng et al, 2013). Furthermore, high level of miR-155 was associated with better clinical outcome in TNBC patients (Gasparini et al, 2014).
Our group has established a robust platform to profile miRNA in various diseases (Ng et al, 2009; Lai et al, 2013; Ng et al, 2013), and have identified a panel of breast cancer-associated miRNAs from plasma, which can potentially serve as screening tool for breast cancer patients (Ng et al, 2013). In this study, we aim to uncover TNBC-associated miRNAs in the circulation based on our established platform. The profiles of miRNAs in TNBC were compared with those in non-TNBC, as well as healthy controls. Candidate miRNAs were selected based on the differential expression between these groups, validated in an independent cohort, and correlated with clinical outcomes.
Materials and Methods
Study design and patients samples
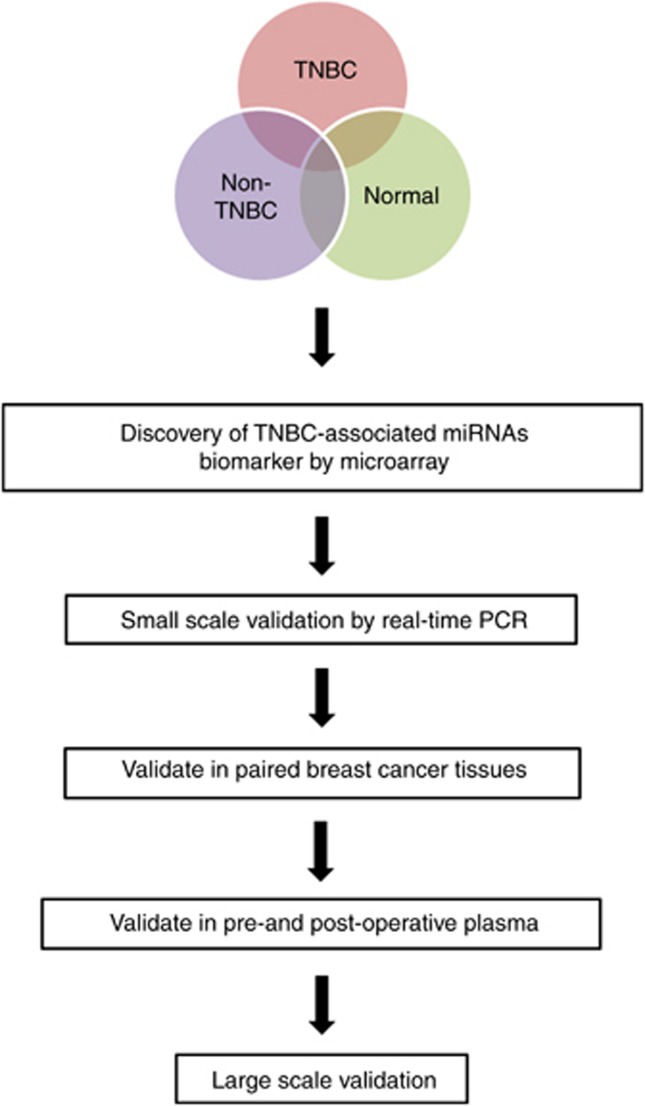
In the marker discovery phase, miRNA expression from plasma of TNBC (n=5), non-TNBC (n=5) and healthy controls (n=5) were profiled. In the validation phase, candidate miRNAs were selected and validated in a small scale independent cohort of breast cancer patients (n=90). Those miRNAs were then verified in primary breast tissues and pre- and post-operative plasma samples. Potential miRNAs were then validated in a large cohort of patients (n=252). The workflow of the study design is shown in Figure 1. A total of 67 TNBC female patients and 95 non-TNBC female patients were recruited with informed consent through the Queen Mary Hospital, the Tung Wah Hospital and the Hong Kong Sanatorium and Hospital and through the Hong Kong Hereditary Breast Cancer Family Registry. We also recruited 90 healthy controls from the Queen Mary Hospital and the Tung Wah Hospital who were confirmed not to have breast cancer and have no personal history of other cancers. The study was approved by the Institutional Review Board of the University of Hong Kong and the Hong Kong Sanatorium and Hospital. Patients' demographic and clinical outcomes were listed in Table 1.
Figure 1.
Workflow of the study design.
Table 1. Clinical characteristics of TNBC and non-TNBC breast cancer patients in validation set.
| TNBC (n=67) | Non-TNBC (n=95) | P-value | |
|---|---|---|---|
| Age (years; mean (s.d.)) | 55.7 (12.6) | 55.1 (11.8) | 0.4753 |
| Histological type | 0.1166 | ||
| DCIS | 0 | 3 | |
| IDC | 59 | 85 | |
| ILC | 1 | 3 | |
| Mixed (ILC+IDC) | 2 | 1 | |
| Others | 5 | 3 | |
| Histological tumour grade | 0.7058 | ||
| 1 | 1 | 11 | |
| 2 | 14 | 41 | |
| 3 | 41 | 28 | |
| NA | 11 | 12 | |
| Bilateral cancer (at the same time) | 1 | 1 | 1.0000 |
| Operation | 0.1695 | ||
| No | 2 | 0 | |
| Yes | 65 | 95 | |
| Lymph-node (N) | 0.7833 | ||
| N0 | 43 | 53 | |
| N1 | 11 | 27 | |
| N2 | 3 | 8 | |
| N3 | 8 | 7 | |
| NX | 2 | 0 | |
|
Distant metastasis | |||
| Before surgery | |||
| No | 65 | 91 | 1.0000 |
| Yes | 2 | 4 | |
| After surgery | |||
| No | 57 | 93 | 0.0040 |
| Yes | 10 | 2 | |
| Stage | 0.8016 | ||
| 0 | 1 | 4 | |
| I | 26 | 35 | |
| II | 25 | 39 | |
| III | 11 | 13 | |
| IV | 2 | 4 | |
| NA | 2 | 0 | |
| Tumour stage | 0.0921 | ||
| T0 | 2 | 5 | |
| T1 | 34 | 49 | |
| T2 | 22 | 39 | |
| T3 | 5 | 0 | |
| T4 | 2 | 0 | |
| TX | 2 | 2 | |
Abbreviations: DCIS=ductal carcinoma in situ; IDC=invasive ductal carcinoma; ILC=invasive lobular carcinoma; NA=not applicable. Bold represents statistical significance.
Samples processing and miRNA extraction
Total RNA were extracted from tissues and paraffin blocks using the TRIzol reagent (Invitrogen, Carlsbad, CA, USA) following the manufacturer's instructions. Blood samples were centrifuged at 1600 g for 10 min at 4 °C, and plasma was carefully transferred into new tubes followed by further centrifugation at 1600 g for 10 min at 4 °C. Total RNA containing small RNA using Trizol LS reagent and miRNeasy Mini Kit (Qiagen, Hilden, Germany) according to the manufacturer's suggestion protocol with the following modifications: 1 ml Trizol LS reagent was added to 500 μl plasma samples. After phase separation, 1.5 volume of 100% ethanol was added to the aqueous phase and the mixture was loaded into the miRNeasy column (Qiagen) according to the manufacturer's instructions. The concentration of all miRNA samples were quantified by NanoDrop 1000 (Thermo Scientific, Wilmington, DE, USA).
MiRNA expression profiling
In the marker discovery phase, we profiled 5 age- and sex- matched individuals from plasma of patients with TNBC, non-TNBC and healthy controls using a miRCURY LNA Array V3.R (Exiqon, Vedbaek, Denmark), which contained 752 human miRNAs. This system is a real-time PCR-based array containing a panel of 384 well-established mature miRNA assays. The kit contains all reagents and primers, reverse transcription and qPCR. In brief, a poly-A tail is added to the mature miRNA template and then synthesised to cDNA by a poly-T primer with a 3′ degenerate anchor and 5′ universal tag. The cDNA is amplified by miRNA-specific and LNA-enhanced forward and reverse primers (Exiqon). SYBR Green PCR will be performed in LC480 Real-time PCR system (Roche, Basel, Switzerland).
MiRNA validation by real-time RT–PCR
MiRNAs were polyadenylated and reverse transcribed into cDNA by using miScript Reverse Transcription Kit (Qiagen) according to the manufacturer's instructions. Real-time qPCR was performed using LightCycler 480 SYBR Green I Master (Roche) in Roche LC480 system. The miRNA-specific forward primers sequences were designed based on the miRNA sequences obtained from the miRBase database (release 20; http://mirbase.org/). The miRNA-specific primer sequences for qPCR were listed in Supplementary Table 1. Each sample was run in duplicates for analysis. The expression levels of miRNAs were normalised to miR-484. Fold change of miRNA was calculated by the equation 2−ΔΔCt. ΔCt was calculated by subtracting the Ct values of miR-484 from the Ct values of the miRNA-of-interest. ΔΔCt was then calculated by subtracting ΔCt of the control from ΔCt of cancer.
Statistical analysis
The significance of plasma miRNA levels was determined by Mann–Whitney test, Wilcoxon test, χ2-test or Kruskal–Wallis test where appropriate. The sensitivity and specificity were calculated according to the standard formulas. Multivariate logistic regression model was established and leave-one-out cross validation to find the best logistic model. Receiver-operator characteristics (ROC) curves were established for discriminating patients with or without breast cancer. The optimal sensitivity and specificity from ROC curves were determined by a commonly used method. All P-values are two-sided and P<0.05 was considered statistically significant. All statistical calculations were performed by GraphPad PRISM 5 software (GraphPad Software, La Jolla, CA, USA).
Results
Discovery of TNBC-associated miRNAs
We profiled miRNA expression in plasma from patients with TNBC (n=5), non-TNBC (n=5) and age-matched healthy controls (n=5) using miRCURY LNA Array. With a cutoff value of 2-fold difference, there were 10 differentially expressed miRNAs in TNBC. As shown in Table 2, there were three upregulated (miR-342-3p, miR-23b-3p and miR-92a-3p) and seven downregulated miRNAs (miR-21 miR-16, miR-143, miR-185, miR-199a-3p, miR-199a-5p and miR-221) in TNBC when compared with non-TNBC and healthy controls.
Table 2. Dysregulated miRs in TNBC when compared with normal and non-TNBC.
| Downregulated miRs | TNBC/normal | TNBC/non-TNBC |
|---|---|---|
| miR-21 | 0.14 | 0.02 |
| miR-16 | 0.16 | 0.03 |
| miR-143 | 0.06 | 0.03 |
| miR-185 | 0.16 | 0.03 |
| miR-199a-3p | 0.12 | 0.05 |
| miR-199a-5p | 0.1 | 0.05 |
| miR-221 | 0.12 | 0.05 |
|
Upregulated miRs | ||
| miR-92a-3p | 6.1 | 2.03 |
| miR-23b-3p | 1.15 | 18.53 |
| miR-342-3p | 6.68 | 23.81 |
Abbreviations: miR=miRNA; TNBC=triple-negative breast cancer.
Selection of potential markers
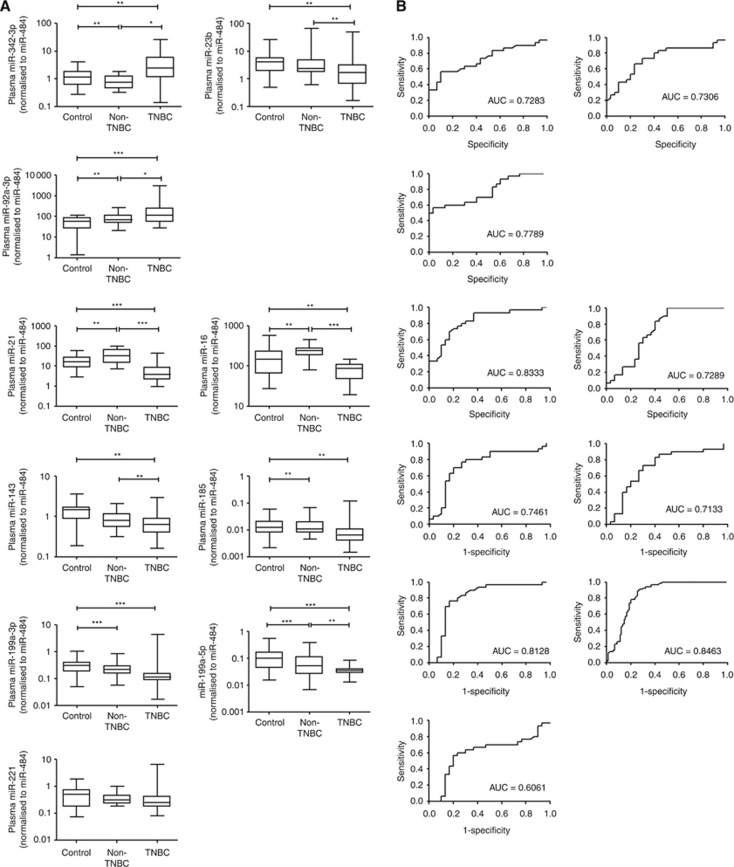
Ten putative miRNAs derived from microarray were validated in plasma of 30 breast cancer patients and 30 healthy controls by real-time PCR. The expression of these miRNAs was shown in Figure 2A, and we selected a marker that could differentiate TNBC from non-TNBC. Results showed that the expression levels of miR-16, miR-21, miR-199a-5p, miR-92a-3p and miR-342-3p were significantly different between TNBC, non-TNBC and healthy controls and was consistent with the array data. Receiver-operator characteristics curve analysis of miR-16 (area under curve (AUC)=0.7289, 95% CI: 0.60–0.86, P=0.002), miR-21 (AUC=0.8333, 95% CI: 0.73–0.94, P<0.0001) and miR-199a-5p (AUC=0.8463, 95% CI: 0.78–0.91, P<0.0001), miR-92a-3p (AUC=0.7789, 95% CI: 0.66–0.90, P=0.0002), miR-342-3p (AUC=0.7283, 95% CI: 0.60–0.86, P=0.002) (Figure 2B). Therefore, these five miRNAs were chosen for further validation.
Figure 2.
Small scale validation of miRNAs in TNBC, non-TNBC and healthy controls (n=90). (A) Box plot of plasma levels of three upregulated (miR-342-3p, miR-23b-3p and miR-92a-3p) and seven downregulated miRNAs (miR-21 miR-16, miR-143, miR-185, miR-199a-3p, miR-199a-5p and miR-221) in TNBC patients. The expression of miRNAs was normalised to miR-484. The lines inside the boxes represent the medians. The boxes mark the interval between the 25th and 75th percentiles. The whiskers denote the interval between the 10th and 90th percentiles. Statistically significant differences were determined using Mann–Whitney tests. (B) Receiver-operator characteristics (ROC) curve analysis of miRNAs to discriminate TNBC patients from healthy controls. *P<0.05, **P<0.01, ***P<0.001.
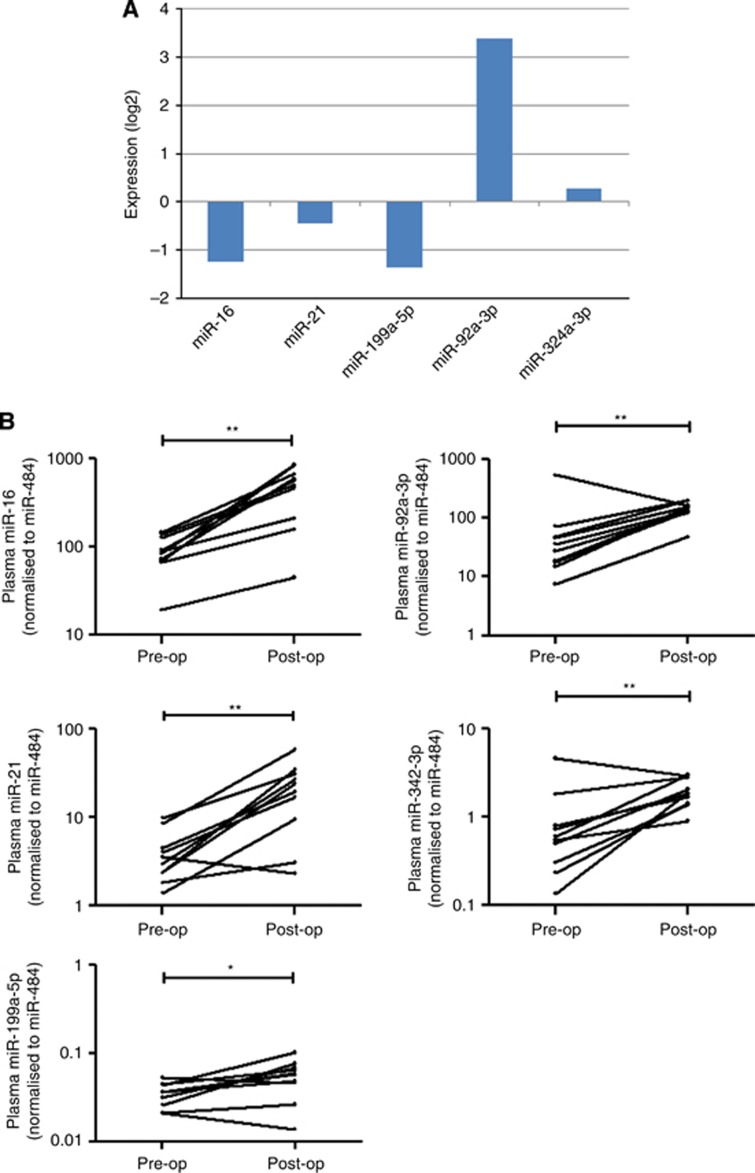
To examine whether these miRNAs were associated with tumour load, we compared the expression of these miRNAs in paired primary breast tissues, as well as pre- and post-operative plasma samples. The expression levels of miR-92a-3p and miR-324-3p were higher in primary breast cancer tumour tissues (n=11) when compared with the adjacent non-tumour counterparts (Figure 3A). On the other hand, miR-16, miR-21 and miR-199a-5p had a lower expression in tumour tissues, which was in line with the data in plasma. Moreover, we compared the expression levels of miR-16, miR-21, miR-199a-5p, miR-92a-3p and miR-342-3p between pre-operative (pre) and post-operative (post) plasma (n=10). Results showed that the expression levels of miR-16, miR-21 and miR-199a-5p were reversed in the post-operative plasma, however, reversal of expression levels were not seen in miR-92a-3p and miR-342-3p (Figure 3B). These data confirmed that miR-16, miR-21 and miR-199a-5p were associated with tumour load, which may potentially be used as a diagnostic marker for TNBC.
Figure 3.
Selection and validation of potential miRNAs. (A) Fold change of miRNAs in primary breast tumour/non-tumour adjacent tissues. (B) Comparison of miRNAs levels in plasma of TNBC patients (n=10) before (pre-operative) and 14 days after (post-operative) removal of the breast tumour. Expression levels of the miRNAs are normalised to miR-484. Statistically significant differences were determined using Wilcoxon tests. *P<0.05, **P<0.01.
Further validation of markers
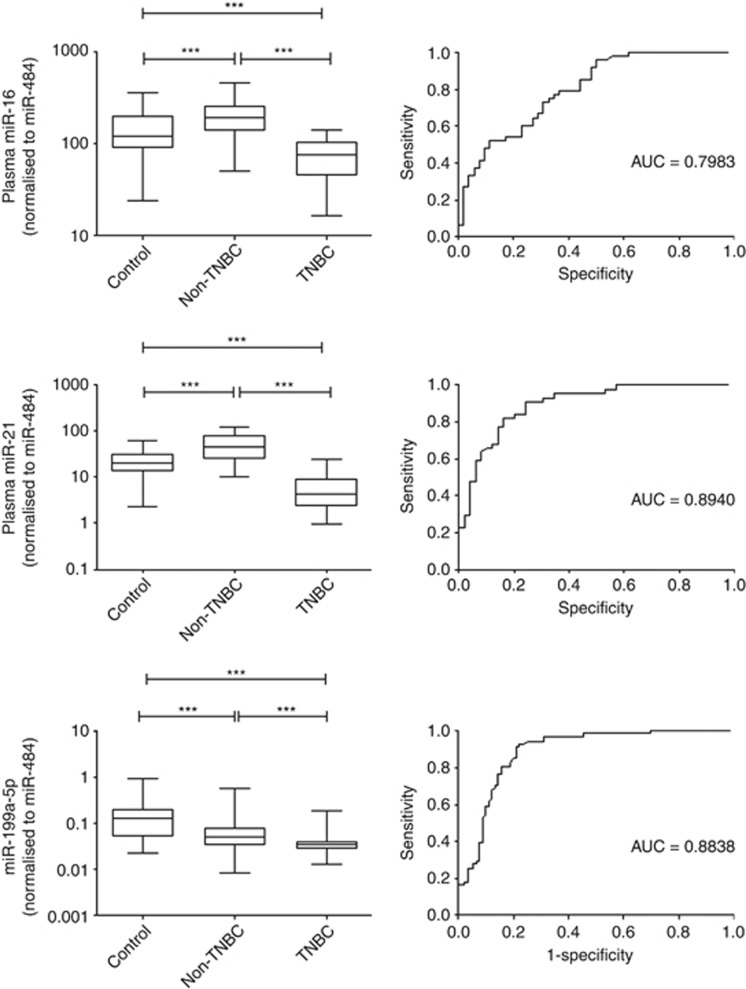
Mir-16, miR-21 and miR-199a-5p were then further validated in a large cohort of patients containing 67 TNBC (stage 0 & I, n=27; stage II, n=25; stage III & IV, n=13), 95 non-TNBC (stage 0 & I, n=39; stage II, n=39; stage III & IV, n=17) and 90 healthy controls. Results showed that all three miRNAs showed a significant lower expression in TNBC than non-TNBC (Figure 4). Receiver-operator characteristics curve analyses revealed that miR-16 (AUC=0.7983, 95% CI: 0.71–0.88, P<0.0001), miR-21 (AUC=0.8740, 95% CI: 0.81–0.94, P<0.0001), miR-199a-5p (AUC=0.8838, 95% CI: 0.83–0.94, P<0.0001) were differentiating TNBC from healthy controls and non-TNBC patients. Results showed that miR-199a-5p with the highest ROC value maybe a potential biomarker to discriminate TNBC from non-TNBC.
Figure 4.
Large scale validation of three miRNAs in a cohort of 252 patients. Box plot of plasma levels of miR-16 miR-21 and miR-199a-5p in TNBC patients. The expression of miRNAs was normalised to miR-484. The lines inside the boxes represent the medians. The boxes mark the interval between the 25th and 75th percentiles. The whiskers denote the interval between the 10th and 90th percentiles. Statistically significant differences were determined using Mann–Whitney tests. Receiver-operator characteristics (ROC) curve analysis of miRNAs to discriminate TNBC patients from healthy controls. ***P<0.001.
Correlation with clinical outcomes
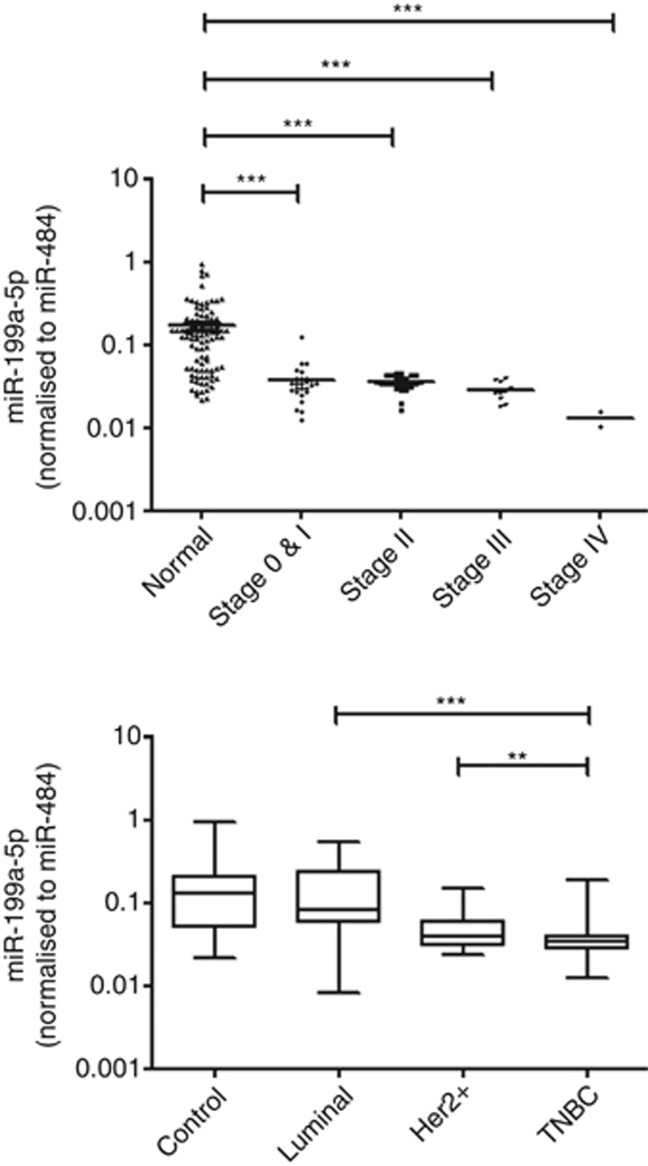
To correlate if the plasma level of miR-199a-5p is associated with tumour stage, patients were then stratified into TNM stages. Plasma miR-199a-5p expression in TNBC patients had significant difference when compared with healthy controls (stage 0 & I, P<0.001; stage II, P<0.001; stage III, P<0.001; stage IV, P<0.001) (Figure 5). In addition, there was a trend of decrease in the expression level of miR-199a-5p with tumour stage, in which patients with stage IV breast cancer had a significantly lower expression than those with stage 0 & I disease (P=0.028).
Figure 5.
Association of miR-199a-5p expression with tumour stage. Plasma miR-199a-5p expression in TNBC patients were stratified by TMN stage and had statistical significance with healthy controls. **P<0.01, ***P<0.001.
More importantly, the plasma levels of miR-199a-5p were categorised into tumour subtypes, that is, ER/PR+ (luminal), HER2+ and TNBC. Result showed that tumours bearing hormone receptors (luminal and HER2+) had a higher expression of miR-199a-5p than in TNBC, implicating hormone receptors may have a role in the regulation of miR-199-5p in breast cancer.
Discussion
Triple-negative breast cancer represents an aggressive phenotype of breast cancer. It has been well-documented that TNBC is associated with advanced stage, family history and occurs in patients who are BRCA1 mutation carriers (Lee et al, 2011). Epidermal growth factor receptor (EGFR) was one of the first anti-cancer agents which was used to treat TNBC. However, <20% of the patients responded to anti-EGFR agents in the clinical setting, suggesting other signal pathways were prevailing in these group of patients (Carey et al, 2012). Others suggest that anti-angiogenesis-based drugs can result in a better response in TNBC in the neoadjuvant setting but whether survival would be affected is still very much unknown (Sikov et al, 2015; von Minckwitz et al, 2014). Lack of available targeted therapy is one of the obstacles in treating TNBC, which could partially explain the poor prognosis in patients with TNBC compared with other breast cancer subtypes (Foulkes et al, 2010). MiRNA profiling could help to understand the pathogenesis of TNBC, can be implicated in the clinical application as biomarkers for diagnosis and prognosis. In the present study, we used our established platform to examine the miRNAs that can discriminate TNBC from other breast cancer subtypes.
We have listed several differentially expressed miRNAs in TNBC and compared their expression in pre- and post-operative plasma (Table 2 and Figure 3B). We have demonstrated that miR-16, miR-21 and miR-199a-5p had a significant lower expression in TNBC than in non-TNBC, in which only miR-199a-5p was associated with tumour stage (Figure 5). In our previous study, miR-16 and miR-21 were upregulated in primary tissues and blood samples in non-TNBC cases (Ng et al, 2013). Interestingly, we found that these miRNAs were downregulated in the plasma, as well as in cancerous tissues of TNBC patients (Figure 4). Piles of evidence showed that miR-21 was one of the ER-responsive miRNAs in cancers (Terao et al, 2011; Tilghman et al, 2012), which could partly explain the low expression of miR-21 in TNBC. Recently, a panel of miRNAs (including miR-16) has been identified to be used as a predictive marker for overall survival in TNBC cases (Cascione et al, 2013). Furthermore, a four-miRNA signature (miR-16, miR-25, miR-222 and miR-324-3p) has been associated with increased risk of breast cancer (Hu et al, 2012). However, the exact functions of miR-16 in breast cancer have not been clearly defined. The tumour suppressive role of miR-16 was linked to cyclin E regulation in breast cancer (Rivas et al, 2012). In the case of hepatitis B virus-related hepatocellular carcinoma, downregulation of miR-15 and miR-16 was observed in the viral RNA (Wang et al, 2013). These studies revealed the tumour suppressor function of miR-16, which reverberated our data on the low expression of miR-16 in more aggressive subtype of breast cancer.
The action of miR-199a-5p in the development of cancers has not been studied until recently. Higher expression of miR-199a-5p was found in cancerous tissues than non-tumour counterparts and was associated with lymph-node metastasis in patients with gastric cancer (He et al, 2014). Forced expression of miR-199a-5p has been shown to stimulate EMT process and metastasis in gastric cancer through serum response factor (Zhao et al, 2014). On the other hand, the anti-tumorigenic activity of miR-199a-5p was seen in multiple myeloma cells by targeting endothelial-cell migration and adhesion molecules (VCAM-1 and ICAM-1) (Raimondi et al, 2014). Recently, a three-miRNA plasma panel with low expression of miR-199a-5p could discriminate patients with COPD and squamous cell carcinomas. This early-response miRNA has contributed to the 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone (NNK)-induced pulmonary carcinogenesis (Kalscheuer et al, 2008). There has only been one study which demonstrated that miR-199a-5p is a regulator of autophagy and sensitised breast cancer cells to irradiation (Yi et al, 2013). Consistent with our current findings, miR-199a-5p was downregulated in cancers of prostate, colon and bladder, in which it could suppress surface-exposed glucose-related protein (GRP78) and enhance chemosensitivity to chemotherapy in cancer cells (Su et al, 2013).
It is widely believed that circulating miRNAs are released from the tumour cells, and yet the level of specific miRNAs would be altered before and after removal of tumours. There is a possibility of hormonal influence, which regulates the miRNA biogenesis and the subsequent cellular responses in breast carcinogenesis. These hypotheses warrant further investigation and provide the basis for the understanding of the molecular mechanisms of specific miRNAs. Though the results were promising, our study was limited by small sample number of TNBC patients and those with stage IV disease. The evaluation of the association between metastasis and expression of miR-199a-5p would be useful and is awaited to be proven with a large sample size. Nevertheless, results of this study provide a non-invasive screening tool to identify patients with TNBC phenotype, which offers a more convenient and sensitive method when compared with immunohistochemistry.
To the best of our knowledge, this is the first study to unravel low expression of miR-199a-5p in TNBC subtype when compared with other breast cancer subtypes in the circulation. With the inclusion of non-TNBC subtypes in this study, we could have a clearer picture of the changes in expression levels in TNBC subtypes, which was not common in most of the studies. The expression of miR-199a-5p was associated with a disease stage, which could be potentially useful as a predictive marker for early diagnosis for TNBC in the clinical settings. This study provides the basis for drug development, which may serve as a target for treating TNBC patients.
Acknowledgments
We are thankful to Dr Ellen Li Charitable Foundation, Kerry Kuok Foundation, United Ally Research Limited and Hong Kong Hereditary Breast Cancer Family Registry for their support. This work was partially supported by the General Research Fund (HKU775911M), ITF-University-Industry Collaboration Programme (ITF-UICP) (UIM/205) and the Seed Funding from Committee on Research and Conference Grants, The University of Hong Kong (201402159002).
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on British Journal of Cancer website (http://www.nature.com/bjc)
This work is published under the standard license to publish agreement. After 12 months the work will become freely available and the license terms will switch to a Creative Commons Attribution-NonCommercial-Share Alike 4.0 Unported License.
Supplementary Material
References
- Boyle P (2012) Triple-negative breast cancer: epidemiological considerations and recommendations. Ann Oncol 23(Suppl 6): vi7–12. [DOI] [PubMed] [Google Scholar]
- Brase JC, Johannes M, Schlomm T, Falth M, Haese A, Steuber T, Beissbarth T, Kuner R, Sultmann H (2011) Circulating miRNAs are correlated with tumor progression in prostate cancer. Int J Cancer 128(3): 608–616. [DOI] [PubMed] [Google Scholar]
- Calin GA, Croce CM (2006) MicroRNA signatures in human cancers. Nat Rev Cancer 6(11): 857–866. [DOI] [PubMed] [Google Scholar]
- Carey LA, Rugo HS, Marcom PK, Mayer EL, Esteva FJ, Ma CX, Liu MC, Storniolo AM, Rimawi MF, Forero-Torres A, Wolff AC, Hobday TJ, Ivanova A, Chiu WK, Ferraro M, Burrows E, Bernard PS, Hoadley KA, Perou CM, Winer EP (2012) TBCRC 001: randomized phase II study of cetuximab in combination with carboplatin in stage IV triple-negative breast cancer. J Clin Oncol 30(21): 2615–2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cascione L, Gasparini P, Lovat F, Carasi S, Pulvirenti A, Ferro A, Alder H, He G, Vecchione A, Croce CM, Shapiro CL, Huebner K (2013) Integrated microRNA and mRNA signatures associated with survival in triple negative breast cancer. PLoS One 8(2): e55910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dent R, Trudeau M, Pritchard KI, Hanna WM, Kahn HK, Sawka CA, Lickley LA, Rawlinson E, Sun P, Narod SA (2007) Triple-negative breast cancer: clinical features and patterns of recurrence. Clin Cancer Res 13(15 Pt 1): 4429–4434. [DOI] [PubMed] [Google Scholar]
- Foulkes WD, Smith IE, Reis-Filho JS (2010) Triple-negative breast cancer. N Engl J Med 363(20): 1938–1948. [DOI] [PubMed] [Google Scholar]
- Gasparini P, Lovat F, Fassan M, Casadei L, Cascione L, Jacob NK, Carasi S, Palmieri D, Costinean S, Shapiro CL, Huebner K, Croce CM (2014) Protective role of miR-155 in breast cancer through RAD51 targeting impairs homologous recombination after irradiation. Proc Natl Acad Sci USA 111(12): 4536–4541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He XJ, Ma YY, Yu S, Jiang XT, Lu YD, Tao L, Wang HP, Hu ZM, Tao HQ (2014) Up-regulated miR-199a-5p in gastric cancer functions as an oncogene and targets klotho. BMC Cancer 14: 218. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Hu Z, Dong J, Wang LE, Ma H, Liu J, Zhao Y, Tang J, Chen X, Dai J, Wei Q, Zhang C, Shen H (2012) Serum microRNA profiling and breast cancer risk: the use of miR-484/191 as endogenous controls. Carcinogenesis 33(4): 828–834. [DOI] [PubMed] [Google Scholar]
- Ibrahim YH, Garcia-Garcia C, Serra V, He L, Torres-Lockhart K, Prat A, Anton P, Cozar P, Guzman M, Grueso J, Rodriguez O, Calvo MT, Aura C, Diez O, Rubio IT, Perez J, Rodon J, Cortes J, Ellisen LW, Scaltriti M, Baselga J (2012) PI3K inhibition impairs BRCA1/2 expression and sensitizes BRCA-proficient triple-negative breast cancer to PARP inhibition. Cancer Discov 2(11): 1036–1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalscheuer S, Zhang X, Zeng Y, Upadhyaya P (2008) Differential expression of microRNAs in early-stage neoplastic transformation in the lungs of F344 rats chronically treated with the tobacco carcinogen 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone. Carcinogenesis 29(12): 2394–2399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai CT, Ng EK, Chow PC, Kwong A, Cheung YF (2013) Circulating microRNA expression profile and systemic right ventricular function in adults after atrial switch operation for complete transposition of the great arteries. BMC Cardiovasc Disord 13: 73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee E, McKean-Cowdin R, Ma H, Spicer DV, Van Den Berg D, Bernstein L, Ursin G (2011) Characteristics of triple-negative breast cancer in patients with a BRCA1 mutation: results from a population-based study of young women. J Clin Oncol 29(33): 4373–4380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metzger-Filho O, Tutt A, de Azambuja E, Saini KS, Viale G, Loi S, Bradbury I, Bliss JM, Azim HA Jr, Ellis P, Di Leo A, Baselga J, Sotiriou C, Piccart-Gebhart M (2012) Dissecting the heterogeneity of triple-negative breast cancer. J Clin Oncol 30(15): 1879–1887. [DOI] [PubMed] [Google Scholar]
- Mitchell PS, Parkin RK, Kroh EM, Fritz BR, Wyman SK, Pogosova-Agadjanyan EL, Peterson A, Noteboom J, O'Briant KC, Allen A, Lin DW, Urban N, Drescher CW, Knudsen BS, Stirewalt DL, Gentleman R, Vessella RL, Nelson PS, Martin DB, Tewari M (2008) Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci USA 105(30): 10513–10518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng EK, Chong WW, Jin H, Lam EK, Shin VY, Yu J, Poon TC, Ng SS, Sung JJ (2009) Differential expression of microRNAs in plasma of patients with colorectal cancer: a potential marker for colorectal cancer screening. Gut 58(10): 1375–1381. [DOI] [PubMed] [Google Scholar]
- Ng EK, Li R, Shin VY, Jin HC, Leung CP, Ma ES, Pang R, Chua D, Chu KM, Law WL, Law SY, Poon RT, Kwong A (2013) Circulating microRNAs as specific biomarkers for breast cancer detection. PLoS One 8(1): e53141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raimondi L, Amodio N, Di Martino MT, Altomare E, Leotta M, Caracciolo D, Gulla A, Neri A, Taverna S, D'Aquila P, Alessandro R, Giordano A, Tagliaferri P, Tassone P (2014) Targeting of multiple myeloma-related angiogenesis by miR-199a-5p mimics: in vitro and in vivo anti-tumor activity. Oncotarget 5(10): 3039–3054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivas MA, Venturutti L, Huang YW, Schillaci R, Huang TH, Elizalde PV (2012) Downregulation of the tumor-suppressor miR-16 via progestin-mediated oncogenic signaling contributes to breast cancer development. Breast Cancer Res 14(3): R77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serpico D, Molino L, Di Cosimo S (2014) microRNAs in breast cancer development and treatment. Cancer Treat Rev 40(5): 595–604. [DOI] [PubMed] [Google Scholar]
- Shapira I, Oswald M, Lovecchio J, Khalili H, Menzin A, Whyte J, Dos Santos L, Liang S, Bhuiya T, Keogh M, Mason C, Sultan K, Budman D, Gregersen PK, Lee AT (2014) Circulating biomarkers for detection of ovarian cancer and predicting cancer outcomes. Br J Cancer 110(4): 976–983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikov WM, Berry DA, Perou CM, Singh B, Cirrincione CT, Tolaney SM, Kuzma CS, Pluard TJ, Somlo G, Port ER, Golshan M, Bellon JR, Collyar D, Hahn OM, Carey LA, Hudis CA, Winer EP (2015) Impact of the addition of carboplatin and/or bevacizumab to neoadjuvant once-per-week paclitaxel followed by dose-dense doxorubicin and cyclophosphamide on pathologic complete response rates in stage II to III triple-negative breast cancer: CALGB 40603 (Alliance). J Clin Oncol 33(1): 13–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silver DP, Richardson AL, Eklund AC, Wang ZC, Szallasi Z, Li Q, Juul N, Leong CO, Calogrias D, Buraimoh A, Fatima A, Gelman RS, Ryan PD, Tung NM, De Nicolo A, Ganesan S, Miron A, Colin C, Sgroi DC, Ellisen LW, Winer EP, Garber JE (2010) Efficacy of neoadjuvant Cisplatin in triple-negative breast cancer. J Clin Oncol 28(7): 1145–1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su SF, Chang YW, Andreu-Vieyra C, Fang JY, Yang Z, Han B, Lee AS, Liang G (2013) miR-30d, miR-181a and miR-199a-5p cooperatively suppress the endoplasmic reticulum chaperone and signaling regulator GRP78 in cancer. Oncogene 32(39): 4694–4701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terao M, Fratelli M, Kurosaki M, Zanetti A, Guarnaccia V, Paroni G, Tsykin A, Lupi M, Gianni M, Goodall GJ, Garattini E (2011) Induction of miR-21 by retinoic acid in estrogen receptor-positive breast carcinoma cells: biological correlates and molecular targets. J Biol Chem 286(5): 4027–4042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tilghman SL, Bratton MR, Segar HC, Martin EC, Rhodes LV, Li M, McLachlan JA, Wiese TE, Nephew KP, Burow ME (2012) Endocrine disruptor regulation of microRNA expression in breast carcinoma cells. PLoS One 7(3): e32754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsujiura M, Ichikawa D, Komatsu S, Shiozaki A, Takeshita H, Kosuga T, Konishi H, Morimura R, Deguchi K, Fujiwara H, Okamoto K, Otsuji E (2010) Circulating microRNAs in plasma of patients with gastric cancers. Br J Cancer 102(7): 1174–1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volinia S, Calin GA, Liu CG, Ambs S, Cimmino A, Petrocca F, Visone R, Iorio M, Roldo C, Ferracin M, Prueitt RL, Yanaihara N, Lanza G, Scarpa A, Vecchione A, Negrini M, Harris CC, Croce CM (2006) A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci USA 103(7): 2257–2261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Minckwitz G, Loibl S, Untch M, Eidtmann H, Rezai M, Fasching PA, Tesch H, Eggemann H, Schrader I, Kittel K, Hanusch C, Huober J, Solbach C, Jackisch C, Kunz G, Blohmer JU, Hauschild M, Fehm T, Nekljudova V, Gerber B groups GA-Bs (2014) Survival after neoadjuvant chemotherapy with or without bevacizumab or everolimus for HER2-negative primary breast cancer (GBG 44-GeparQuinto) dagger. Ann Oncol 25(12): 2363–2372. [DOI] [PubMed] [Google Scholar]
- Wang Y, Jiang L, Ji X, Yang B, Zhang Y, Fu XD (2013) Hepatitis B viral RNA directly mediates down-regulation of the tumor suppressor microRNA miR-15a/miR-16-1 in hepatocytes. J Biol Chem 288(25): 18484–18493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi H, Liang B, Jia J, Liang N, Xu H, Ju G, Ma S, Liu X (2013) Differential roles of miR-199a-5p in radiation-induced autophagy in breast cancer cells. FEBS Lett 587(5): 436–443. [DOI] [PubMed] [Google Scholar]
- Zeng RC, Zhang W, Yan XQ, Ye ZQ, Chen ED, Huang DP, Zhang XH, Huang GL (2013) Down-regulation of miRNA-30a in human plasma is a novel marker for breast cancer. Med Oncol 30(1): 477. [DOI] [PubMed] [Google Scholar]
- Zhao X, He L, Li T, Lu Y, Miao Y, Liang S, Guo H, Bai M, Xie H, Luo G, Zhou L, Shen G, Guo C, Bai F, Sun S, Wu K, Nie Y, Fan D (2014) SRF expedites metastasis and modulates the epithelial to mesenchymal transition by regulating miR-199a-5p expression in human gastric cancer. Cell Death Differ 21(12): 1900–1913. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.