Abstract
Background:
The overall survival for patients with squamous cell carcinoma of the tongue is low and the search for early diagnostic and prognostic markers is thus essential. MicroRNAs have been suggested as potential prognostic and diagnostic candidates in squamous cell carcinoma of head and neck in general.
Methods:
On the basis of the known differences between sub-sites within the oral cavity, we investigated the expression and role of microRNA-424 in squamous cell carcinoma arising in tongue. MicroRNA levels were measured by qRT–PCR in both tissue and plasma samples.
Results:
Levels of microRNA-424 were upregulated in tongue squamous cell carcinoma, but not in tumours originating from gingiva or floor of the mouth. Interestingly, microRNA-424 was downregulated in clinically normal tongue tissue next to tumour compared with completely healthy tongue, indicating that microRNA-424 could be a marker of field cancerisation in this tumour type. However, expression of microRNA-424 in a tongue-derived epithelial cell line revealed no significant changes in the expression profile of proteins and genes.
Conclusions:
Our patient data show that microRNA-424 alterations are a marker of field cancerisation specific for tongue tumourigenesis, which also could have a role in development of tongue squamous cell carcinoma.
Keywords: miRNA, tongue SCC, miRNA-424, field cancerisation
Squamous cell carcinoma of the head and neck (SCCHN), a collective term for malignant tumours arising in epithelial tissue in the upper aero-digestive tract, is among the 10 most common malignancies in the world. The SCCHN group comprises tumours in the oral cavity, nasal cavity, pharynx and larynx. Smoking and alcohol abuse are the two most well-known risk factors for developing the malignancy (Jefferies and Foulkes, 2001). In the subgroup of tumours occurring in oropharynx, a connection to human papilloma virus (HPV) has been demonstrated (Syrjanen, 2004), whereas the presence of HPV in other locations within the oral cavity is significantly lower (Kreimer et al, 2005).
Despite increasing knowledge about the development and the molecular mechanisms behind SCCHN the 5-year overall survival for the entire population of patients with SCCHN is still only around 50% (Sano and Myers, 2007; Mehanna et al, 2011). One reason for this is that tumours often are diagnosed at a late stage and thus have spread to cervical lymph nodes. Another reason is the development of loco-regional recurrences and secondary tumours, both contributing to a worse prognosis. Not only the tumours, but also the surrounding tissue close to the tumour shows genetic changes related to the neoplastic process. For intra-oral squamous cell carcinoma (SCC), such genetically changed fields can be detected within 7 cm from the tumour. The area of epithelial cells with genetic alterations is defined as field cancerisation (Braakhuis et al, 2004).
Several biomarkers have been studied in the search for new diagnostic and prognostic factors for SCCHN but none has so far turned out to be suitable for routine use in the clinic. One potential group of molecules suggested to be useful is microRNA (miRNA).
One of the first reports about miRNA came in 1993 (Lee et al, 1993) and today several thousands of miRNAs have been reported in miRBase. miRNAs are small (19–22 nucleotides) non-coding RNA molecules regulating processes like development, proliferation, differentiation, apoptosis and stress responses (Kloosterman and Plasterk, 2006). miRNAs act to inhibit the expression of their target proteins by complementary binding of the miRNA to the target mRNA sequence, leading to either degradation of the mRNA or inhibition of translation. It has been estimated that ∼30% of all genes are regulated by at least one miRNA (Bartel, 2004) and miRNA expression has been mapped in various cancers where they can function both as oncogenes and tumour suppressors (Ruan et al, 2009). For SCCHN, many studies have shown differentially expressed miRNAs although many of the miRNA profiling studies tend to have a poor agreement among them (Chen et al, 2013).
In tongue SCC, a miRNA profiling study of formalin-fixed paraffin-embedded samples showed miRNA-424 to be the highest upregulated miRNA (Rentoft et al, 2011). To further investigate the expression and function of miRNA-424 in these tumours we here analysed its expression in normal tongue, clinically normal tongue adjacent to SCC, tongue SCC and also in blood from patients with tongue SCC. Expression of miRNA-424 was further analysed in tumours from different sub-sites within the oral cavity. To study its function in more detail, we used an in vitro model and Illumina whole-genome array as well as a targeted multiplex protein assay. Our data show dysregulation of miRNA-424 in tongue SCC but not in other oral sites and we further suggest that lower levels of miRNA-424 can be used as an indicator of field cancerisation in the tongue.
Materials and methods
Patient material
After obtaining informed consent, tumour biopsies were collected from consecutive patients diagnosed with tongue SCC. A biopsy was also taken from clinically normal tissue on the contralateral side of the tongue. In addition, a control group of tongue biopsies from healthy individuals was collected. Tissue samples were snap-frozen in liquid nitrogen and stored at −80 °C until RNA was extracted. For analysis of miRNA-424 in tongue tissue 22 tongue tumours, 16 samples from clinically normal tongue tissue and 14 normal tongue samples from healthy individuals were included. Tongue SCC included only tumours of the mobile tongue, and were based on specific localisation further subdivided into: tongue, border of the tongue and tongue with overgrowth to floor of the mouth. When analysing miRNA-424 in different locations within the oral cavity, paired samples of tumour and clinically normal tissue adjacent to the tumour from the same patient from the tongue (n=15), gingiva (n=8) and floor of the mouth (n=4) were analysed as previously described (Boldrup et al, 2012). Blood samples were collected from seven patients with tongue tumours at the time of surgical treatment, and five healthy controls. The project was approved by the local Ethical Committee (dnr 08-003M). For patient data see Table 1.
Table 1. Patient information.
| Sample ID | Age | Sex | Location | TNM | Samples included in only tongue miRNA-424 analysis | Samples included in different location analysis of miRNA-424a | Plasma samples Exiqon panel/qRT–PCR |
|---|---|---|---|---|---|---|---|
| N1 | 32 | F | 2 | Healthy | C | ||
| N2 | 49 | F | 2 | Healthy | C | ||
| N3 | 25 | F | 2 | Healthy | C | ||
| N4 | 30 | M | 2 | Healthy | C | ||
| N5 | 27 | M | 2 | Healthy | C | ||
| N6 | 42 | F | 2 | Healthy | C | ||
| N7 | 32 | F | 2 | Healthy | C | ||
| N8 | 41 | F | 2 | Healthy | C | ||
| N9 | 35 | F | 2 | Healthy | C | ||
| N10 | 57 | M | 2 | Healthy | C | ||
| N11 | 45 | M | 2 | Healthy | C | ||
| N12 | 37 | M | 2 | Healthy | C | ||
| N13 | 48 | F | 2 | Healthy | C | ||
| N14 | 59 | F | 2 | Healthy | C | ||
| T42 | 69 | F | 1 | T2N0M0 | N | ||
| T68 | 62 | M | 1 | T2N0M0 | T | P | |
| T82 | 19 | F | 1 | T4aN0M0 | T | P | |
| T51 | 74 | M | 1 | T2N0M0 | T/N | T/N | |
| T53 | 55 | M | 1 | T1N0M0 | T/N | T/N | |
| T59 | 68 | F | 1 | T2N0M0 | T/N | T/N | |
| T76 | 59 | M | 1 | T4aN0M0 | T/N | T/N | |
| T85 | 88 | F | 1 | T2N0M0 | T/N | T/N | |
| T14 | 78 | F | 2 | T2N1M0 | T | ||
| T29 | 65 | F | 2 | T2N0M0 | T | ||
| T70 | 71 | M | 2 | T1N0M0 | T | ||
| T92 | 63 | F | 2 | T2N0M0 | T | ||
| T11 | 79 | M | 2 | T2N0M0 | T/N | T/N | |
| T35 | 24 | F | 2 | T2N0M0 | T/N | T/N | |
| T79 | 61 | M | 2 | T1N0M0 | T/N | T/N | |
| T24 | 65 | M | 3 | T1N0M0 | T | ||
| T40 | 81 | F | 3 | T4N2bM0 | T/N | T/N | |
| T49 | 52 | F | 3 | T4N2cM0 | T/N | T/N | |
| T56 | 41 | F | 3 | T2N2bM0 | T/N | T/N | |
| T58 | 62 | M | 3 | T1N0M0 | T/N | T/N | |
| T61 | 70 | M | 3 | T4aN0M0 | T/N | T/N | |
| T65 | 81 | F | 3 | T2N0M0 | T/N | T/N | P |
| T73 | 81 | M | 3 | T4aN0M0 | T/N | T/N | |
| T83 | 64 | F | 2 | T1N0M0 | P | ||
| T105 | 64 | M | 2 | T1N0M0 | P | ||
| T111 | 31 | F | 2 | T1N0M0 | P | ||
| T98 | 31 | M | 3 | T2N0M0 | P | ||
| T62 | 75 | M | 4 | T4aN2bM0 | T/N | ||
| T63 | 62 | M | 4 | T1N0M0 | T/N | ||
| T64 | 57 | M | 4 | T4N0M0 | T/N | ||
| T69 | 39 | M | 4 | T2N0M0 | T/N | ||
| T33 | 85 | M | 5a | T4N0M0 | T/N | ||
| T36 | 63 | M | 5a | T4N0M0 | T/N | ||
| T41 | 83 | F | 5a | T4N2bM0 | T/N | ||
| T50 | 73 | F | 5a | T4N2bM0 | T/N | ||
| T32 | 80 | F | 5b | T4N2bM0 | T/N | ||
| T34 | 72 | M | 5b | T4NxMx | T/N | ||
| T55 | 79 | F | 5b | T4N2bM0 | T/N | ||
| T67 | 72 | M | 5b | T4N0M0 | T/N | ||
| C25 | 53 | F | Healthy | P | |||
| C26 | 50 | F | Healthy | P | |||
| C27 | 47 | F | Healthy | P | |||
| C28 | 67 | F | Healthy | P | |||
| C29 | 45 | F | Healthy | P |
Abbreviations: C=control; F=female; M=male; N=normal; P=plasma; T=tumour; T/N=tumour and normal; TNM=primary tumour; regional lymph nodes; distant metastasis.
1=tongue; 2=border of the tongue; 3=border of the tongue and tongue with overgrowth to floor of mouth; 4=floor of the mouth; 5=gingiva (a=upper jaw, b=lower jaw).
Previously used in Boldrup et al, 2012.
RNA extraction and qRT–PCR of tissue samples
Biopsies were homogenised in either trizol (samples from a previous study) (Boldrup et al, 2012) or lysis buffer from RNA/protein purification kit (to extract both RNA and protein from the same biopsy) using a Precellys (Bertin Technologies, Artigues Pres Bordeaux, France) and total RNA including miRNA was extracted using either chloroform or an RNA/protein purification kit (Norgen, Thorold, ON, Canada). After dilution in water, no difference in RNA quality or yield was observed between the different methods according to measurement with nano-drop and bioanalyzer. For cDNA synthesis, 20 ng of total RNA was used with the mercury LNA Universal cDNA synthesis kit according to the manufacturer's protocol (Exiqon, Vedbaek, Denmark). The total volume of each reaction was 20 μl. Expression levels of miRNA-424 and the reference gene SNORD48 were analysed with real-time PCR amplification for individual assays using LNA primer sets for miRNA (Exiqon).
Blood analysis
RNA was prepared from plasma samples of seven patients with tongue SCC and five healthy individuals using the miRCURY RNA isolation kit for biofluids (Exiqon) according to the manufacturer's protocol. In brief, 200 μl plasma was mixed with 60 μl lysis solution, thereafter 20 μl protein precipitation solution was added. After centrifugation the clear supernatant was transferred to a new tube and 270 μl isopropanol was added. Samples were then placed in a mini spin column for purification of RNA. After several washing steps RNA was eluted in 50 μl water. cDNA was prepared from 2 μl RNA in a 10 μl cDNA reaction using the same universal cDNA synthesis kit from Exiqon.
Cell transfections
PE/CA, a human cell line derived from a patient with tongue SCC (ECACC), was transfected with miRNA-424 inhibitor (Qiagen) using HiPerFect (Qiagen). Levels of miRNA-424 in PE/CA cells are in the same range as in tongue SCC, detected at Ct 24–25 in qRT–PCR. As negative control Allstars siRNA was included (Qiagen). Cells were harvested 24 and 48 h after transfection.
Illumina array and proseek
RNA and protein was extracted from cell lines transfected with miRNA-424 inhibitor using the RNA and protein purification kit (Norgen). RNA was labelled with TargetAmp-Nano Labeling Kit for illumina Expression BeadChip (Epicenter, Madison, WI, USA) to produce cRNA which was hybridised to illumina HumanHT-12 v4 bead chip and analysed with an iScan system. cDNA was synthesised from RNA with a miscript II RT kit (Qiagen) and expression of miRNA-424 was measured with miscript primer assay and sybrgreen (Qiagen). Protein extracts were analysed by Olink (Uppsala, Sweden) with the proseek multiplex oncology I, a protein panel of 92 well-characterised markers for tumorigenesis.
Identification of miRNA-424-binding sites and miRNA-424 targets
In the search for transcription factor-binding sites within the miRNA-424 promoter the bioinformatic tool LASAGNA-Search 2.0 (Lee and Huang, 2013) was used. The search was restricted to 1000 bp upstream of the transcription start site.
Looking at potential targets for miRNA-424 TargetScan was used.
Statistical analysis
Differences in miRNA-424 expression were assessed by Mann–Whitney tests and for comparison of miRNA-424 expression in different locations a Wilcoxon rank test was used. Array data was analysed with the software Chipster (an open source project, http://chipster.csc.fi/) and the P-value was set to 0.05 allowing for false discovery rate correction. The protein data from proseek was analysed with the Mann–Whitney test. SPSS version 22 (IBM, Armonk, NY, USA) was used for all statistical analyses.
Results
Expression of miRNA-424 in tongue tumours
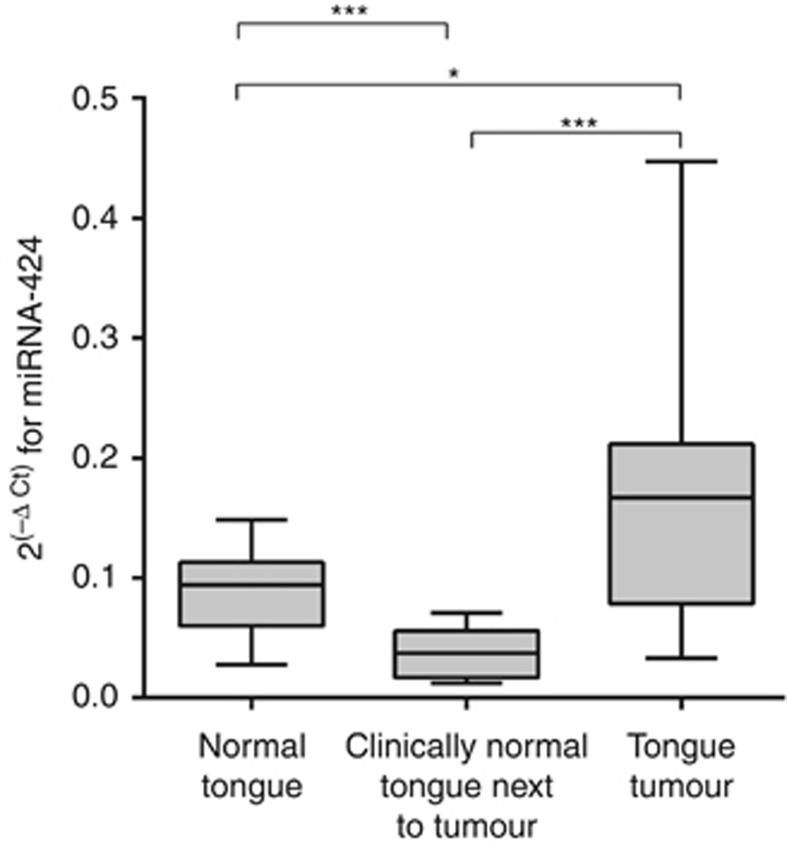
Measuring the expression of miRNA-424 in tongue tissue showed that levels were highest in tongue tumours followed by normal tongue tissue from healthy individuals, whereas the lowest levels were observed in clinically normal tissue adjacent to tongue SCC. There was a significant difference (P<0.001) comparing clinically normal tongue next to tumour either with healthy normal tongue or with tongue tumour. Also comparing tongue tumours with healthy normal tongue showed significant difference in expression (P<0.05) (Figure 1). No correlation between miRNA-424 levels and age or sex could be seen.
Figure 1.
Analysis of miRNA-424 expression in tongue SCC by qRT–PCR. Comparing miRNA-424 levels between 14 normal tongue samples from healthy individuals with 16 samples of clinically normal tongue tissue next to tumour, and 22 tongue tumour samples showed a significant difference between all groups. *P<0.05, ***P<0.001.
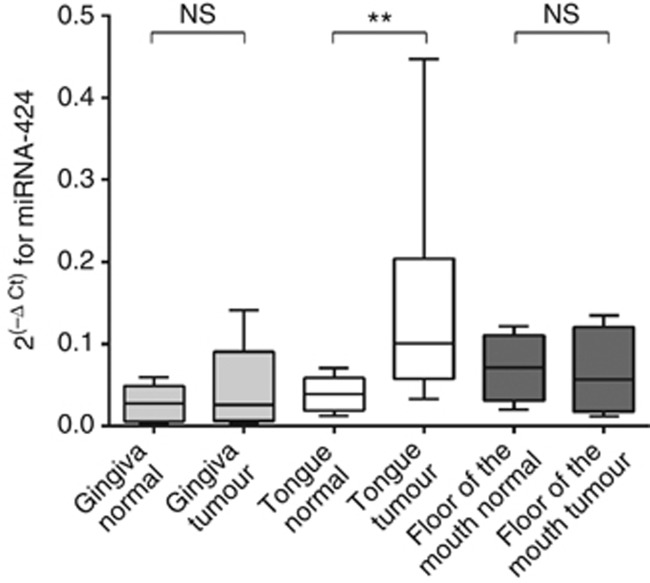
Significant difference in miRNA-424 expression in tongue SCC but not in gingival and floor of the mouth tumours
Our previous study showed that different areas within the oral cavity vary in their miRNA expression (Boldrup et al, 2012). Therefore tumours with corresponding normal tissue from gingiva, tongue and floor of the mouth (Boldrup et al, 2012) were analysed also for expression of miRNA-424. Results showed a significant difference between tumour and corresponding clinically normal tissue in tongue (P<0.01), whereas no significant difference was seen between gingival tumours or tumours in the floor of the mouth and their corresponding clinically looking normal tissue (Figure 2).
Figure 2.
Comparison of miRNA-424 expression in tumours of different locations in the oral cavity. Paired samples of clinically normal tissue next to tumour and tumour samples from three different locations, gingiva, tongue and floor of the mouth were investigated for miRNA-424 expression. Results showed significantly different expression only between tongue tumours and clinically normal tongue controls (P=0.001). Neither gingiva nor floor of the mouth tumours showed any significant difference in miRNA-424 expression compared with their corresponding clinically normal tissue, NSP>0.05, **P<0.01.
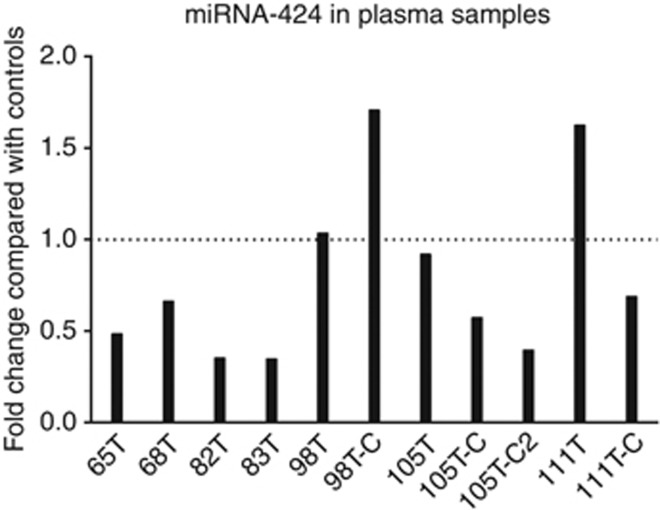
Analysis of miRNA-424 expression in plasma samples
Several miRNAs have been reported to be detectable in blood (Lawrie et al, 2008; Weber et al, 2010) but no reports, to our knowledge, have identified miRNA-424 in blood. Blood samples from patients with tongue SCC were analysed for miRNA-424 expression and compared with a group of healthy controls. miRNA-424 could be detected in plasma, however, levels were very low, close to the assay limits for detection, and also showed highly variable levels between individuals, both tumour patients and healthy controls (Figure 3). No significant difference between tumour patients and healthy controls could be found.
Figure 3.
miRNA-424 expression in plasma from patients with tongue SCC. Analysis of miRNA-424 levels in plasma samples from seven patients with tongue SCC compared with plasma from healthy individuals showing interindividual variation. The graph shows fold change when comparing tumour patients with the group of controls. All values were normalised against a geometric mean of miRNA-93, miRNA-103, miRNA-191 and miRNA-423. One follow-up sample was collected 3 months after treatment from two patients, and two follow-up samples were available from one patient, 3 and 6 months after treatment. These samples were labelled −C and −C2. The mean 2−(ΔCt) for plasma from patients with tumour was 0.105 and for healthy controls 0.144, P=0.343.
The role of miRNA-424 in tumour cell line
To investigate the role and function of miRNA-424 in tongue SCC, a tongue tumour cell line, PE/CA, was transfected with miRNA-424 inhibitor. Transfection efficiency was demonstrated with qRT–PCR, showing a 92-fold reduction in miRNA-424 levels at 24 h and a 75 fold reduction 48 h after transfection (Supplementary Figure). To evaluate if miRNA-424 had any effect on specific mRNA expression, samples were analysed with whole-genome RNA expression analysis. No genes were, however, significantly differentially expressed when comparing cells transfected with miRNA-424 inhibitor to mock-transfected cells. In the next step we investigated whether the effect of miRNA-424 could be at protein level rather than at RNA level using a multiplex protein assay. A panel of 92 proteins connected to tumorigenesis were analysed in the cell line transfected with miRNA-424 inhibitor compared with mock-transfected cells. Results showed 42 out of the 92 proteins to be detectable in the cell extracts. None of these 42 proteins were, however, significantly differentially expressed between cells transfected with miRNA-424 inhibitor and control cells.
miRNA-424-binding sites and target analysis
To identify transcription factors with at least theoretical ability to bind to and thus also potentially involved in regulation of miRNA-424, the promoter was analysed by LASAGNA-Search 2.0. Of the top 20 candidates summarised in the Supplementary Table several, such as HSF1 and MEIS1, have a known connection to SCC (Ishiwata et al, 2012; Mitra et al, 2014).
In the search for potential targets of miRNA-424, Targetscan was used and 20 top candidates identified based on total context score (Garcia et al, 2011). Results which are summarised in the Supplementary Table also here include genes with a known connection to SCC, for example FGF2 and CCNE1 (Freier et al, 2010; Marshall et al, 2011).
Discussion
A major problem with tongue SCC is the poor prognosis which has not improved over the past 25–30 years. In the search for factors to improve prognosis, including early detection, much recent focus has been upon miRNAs, which also have been suggested for use as diagnostic tools for a wide variety of human diseases (De Guire et al, 2013). Our previous studies have shown miRNA-21 to be upregulated and miRNA-125b and miRNA-203 both to be downregulated in tongue SCC compared with clinically normal tissue adjacent to the tumours (Boldrup et al, 2012). Furthermore, our array data has shown miRNA-424 to be the highest upregulated miRNA in tongue SCC (Rentoft et al, 2011). In the present study the significant upregulation of miRNA-424 in tongue SCC was confirmed, both compared with clinically normal tongue adjacent to the tumours as well as to completely normal tongue in healthy individuals. An interesting finding was that levels of miRNA-424 were significantly lower in clinically normal tongue adjacent to tongue SCC compared with normal tongue in healthy donors. This implies that decreased levels of miRNA-424 serve as a marker of field cancerisation. Furthermore, the downregulation of this miRNA in normal tongue adjacent to tumour could indicate that it is an important player under normal conditions which is repressed under conditions of tumour formation. The significant increase in tumours could in turn be an effect of the complete lack of normal regulation seen in tumour tissue.
The use of blood samples for analysis of factors important in diagnostics and especially in follow-up of patients treated for tongue SCC is advantageous, as sampling is a non-invasive method compared with the taking of tissue biopsies. Previous studies have shown the possibility to use plasma for detection of miRNA in patients with SCCHN (Hsu et al, 2012), and a correlation between levels in plasma and tissue has also been shown for several tumour types (Wang et al, 2010; Brase et al, 2011). The present results showed two out of three patients with follow-up samples to have decreased miRNA-424 expression and one to have increased expression at the first follow-up after treatment. This finding, in combination with the high variation in levels of this miRNA seen in plasma from both controls and patients with tongue SCC does not make it an ideal blood marker for early detection of tongue SCC or tumour recurrence.
In order to learn more about its role and mechanism of action in tongue SCC, we inhibited miRNA-424 in a tongue SCC cell line and looked at the effects at global mRNA levels. Surprisingly, no significant mRNA changes were seen. As miRNAs can exert their effect through either mRNA degradation or inhibition of mRNA translation to influence protein levels (Selbach et al, 2008), the lack of effects on mRNA levels using global expression profiling arrays indicate that the effects of miRNA-424 are likely to be at the level of mRNA translational inhibition. We therefore analysed levels in a panel of oncology related proteins and found about half of the 92 proteins to be detectable in the transfected cell line. None of these proteins were, however, significantly differently expressed between treated and untreated cells. As this panel only represents a small group of proteins with a connection to tumours in general, it cannot be excluded that there are translational targets for miRNA-424 outside this panel. However, it is also possible that miRNA-424 serves as tumour marker and has limited impact on tumour formation. In addition, in the in vitro experiment only miRNA-424 was targeted for inhibition, whereas in vivo, several of the members in the miRNA-424/miRNA-503 cluster might be needed working together to have a function. Another possibility is that miRNA-424 is regulated through methylation since it contains a CPG island.
We also searched for transcription factors that might bind to miRNA-424 as well as potential targets of miRNA-424 in silico and found many candidate regulators and targets of miRNA-424 of which many already have a known connection to SCC. Their interaction with and correlation to miRNA-424 in vivo was, however, not possible to clarify based on the present material.
Taken together our data clearly show that expression of miRNA-424 differs between tumours of different locations within the oral cavity, and that levels are significantly upregulated in squamous cell carcinomas of the tongue. One important finding is that miRNA-424 is a potential marker of field cancerisation changes in the tongue, as it is downregulated in apparently normal tissue surrounding the tumours compared with tongue tissue from healthy individuals. These results support the use of miRNA-424 as a marker for tumour presence in tongue, either as an indicator of remaining or relapsing tumour in patients treated for tongue SCC or as a marker of suspect tumour growth in patients with symptoms but still no clinical signs of a tongue tumour. Further studies of the effects of miRNA-424 in tongue SCC and the cancerisation field will also be valuable to improve our understanding of these processes and provide reliable markers for clinical use.
Acknowledgments
This investigation was supported by grants from Lion's Cancer Research Foundation, Umeå University, the Swedish Cancer Society Contract number 14 0752 and Västerbotten County Council.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on British Journal of Cancer website (http://www.nature.com/bjc)
This work is published under the standard license to publish agreement. After 12 months the work will become freely available and the license terms will switch to a Creative Commons Attribution-NonCommercial-Share Alike 4.0 Unported License.
Supplementary Material
References
- Bartel DP (2004) MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116: 281–297. [DOI] [PubMed] [Google Scholar]
- Boldrup L, Coates PJ, Wahlgren M, Laurell G, Nylander K (2012) Subsite-based alterations in miR-21, miR-125b, and miR-203 in squamous cell carcinoma of the oral cavity and correlation to important target proteins. J Carcinog 11: 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braakhuis BJ, Leemans CR, Brakenhoff RH (2004) A genetic progression model of oral cancer: current evidence and clinical implications. J Oral Pathol Med 33: 317–322. [DOI] [PubMed] [Google Scholar]
- Brase JC, Johannes M, Schlomm T, Falth M, Haese A, Steuber T, Beissbarth T, Kuner R, Sultmann H (2011) Circulating miRNAs are correlated with tumor progression in prostate cancer. Int J Cancer 128: 608–616. [DOI] [PubMed] [Google Scholar]
- Chen D, Cabay RJ, Jin Y, Wang A, Lu Y, Shah-Khan M, Zhou X (2013) MicroRNA deregulations in head and neck squamous cell carcinomas. J Oral Maxillofac Res 4: e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Guire V, Robitaille R, Tetreault N, Guerin R, Menard C, Bambace N, Sapieha P (2013) Circulating miRNAs as sensitive and specific biomarkers for the diagnosis and monitoring of human diseases: promises and challenges. Clin Biochem 46: 846–860. [DOI] [PubMed] [Google Scholar]
- Freier K, Knoepfle K, Flechtenmacher C, Pungs S, Devens F, Toedt G, Hofele C, Joos S, Lichter P, Radlwimmer B (2010) Recurrent copy number gain of transcription factor SOX2 and corresponding high protein expression in oral squamous cell carcinoma. Genes Chromosomes Cancer 49: 9–16. [DOI] [PubMed] [Google Scholar]
- Garcia DM, Baek D, Shin C, Bell GW, Grimson A, Bartel DP (2011) Weak seed-pairing stability and high target-site abundance decrease the proficiency of lsy-6 and other microRNAs. Nat Struct Mol Biol 18: 1139–1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu CM, Lin PM, Wang YM, Chen ZJ, Lin SF, Yang MY (2012) Circulating miRNA is a novel marker for head and neck squamous cell carcinoma. Tumour Biol 33: 1933–1942. [DOI] [PubMed] [Google Scholar]
- Ishiwata J, Kasamatsu A, Sakuma K, Iyoda M, Yamatoji M, Usukura K, Ishige S, Shimizu T, Yamano Y, Ogawara K, Shiiba M, Tanzawa H, Uzawa K (2012) State of heat shock factor 1 expression as a putative diagnostic marker for oral squamous cell carcinoma. Int J Oncol 40: 47–52. [DOI] [PubMed] [Google Scholar]
- Jefferies S, Foulkes WD (2001) Genetic mechanisms in squamous cell carcinoma of the head and neck. Oral Oncol 37: 115–126. [DOI] [PubMed] [Google Scholar]
- Kloosterman WP, Plasterk RH (2006) The diverse functions of microRNAs in animal development and disease. Dev Cell 11: 441–450. [DOI] [PubMed] [Google Scholar]
- Kreimer AR, Clifford GM, Boyle P, Franceschi S (2005) Human papillomavirus types in head and neck squamous cell carcinomas worldwide: a systematic review. Cancer Epidemiol Biomarkers Prev 14: 467–475. [DOI] [PubMed] [Google Scholar]
- Lawrie CH, Gal S, Dunlop HM, Pushkaran B, Liggins AP, Pulford K, Banham AH, Pezzella F, Boultwood J, Wainscoat JS, Hatton CS, Harris AL (2008) Detection of elevated levels of tumour-associated microRNAs in serum of patients with diffuse large B-cell lymphoma. Br J Haematol 141: 672–675. [DOI] [PubMed] [Google Scholar]
- Lee C, Huang CH (2013) LASAGNA-Search: an integrated web tool for transcription factor binding site search and visualization. Biotechniques 54: 141–153. [DOI] [PubMed] [Google Scholar]
- Lee RC, Feinbaum RL, Ambros V (1993) The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 75: 843–854. [DOI] [PubMed] [Google Scholar]
- Marshall ME, Hinz TK, Kono SA, Singleton KR, Bichon B, Ware KE, Marek L, Frederick BA, Raben D, Heasley LE (2011) Fibroblast growth factor receptors are components of autocrine signaling networks in head and neck squamous cell carcinoma cells. Clin Cancer Res 17: 5016–5025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehanna H, Paleri V, West CM, Nutting C (2011) Head and neck cancer-part 1: epidemiology, presentation, and preservation. Clin Otolaryngol 36: 65–68. [DOI] [PubMed] [Google Scholar]
- Mitra S, Mukherjee N, Das S, Das P, Panda CK, Chakrabarti J (2014) Anomalous altered expressions of downstream gene-targets in TP53-miRNA pathways in head and neck cancer. Sci Rep 4: 6280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rentoft M, Fahlen J, Coates PJ, Laurell G, Sjostrom B, Ryden P, Nylander K (2011) miRNA analysis of formalin-fixed squamous cell carcinomas of the tongue is affected by age of the samples. Int J Oncol 38: 61–69. [PubMed] [Google Scholar]
- Ruan K, Fang X, Ouyang G (2009) MicroRNAs: novel regulators in the hallmarks of human cancer. Cancer Lett 285: 116–126. [DOI] [PubMed] [Google Scholar]
- Sano D, Myers JN (2007) Metastasis of squamous cell carcinoma of the oral tongue. Cancer Metastasis Rev 26: 645–662. [DOI] [PubMed] [Google Scholar]
- Selbach M, Schwanhausser B, Thierfelder N, Fang Z, Khanin R, Rajewsky N (2008) Widespread changes in protein synthesis induced by microRNAs. Nature 455: 58–63. [DOI] [PubMed] [Google Scholar]
- Syrjanen S (2004) HPV infections and tonsillar carcinoma. J Clin Pathol 57: 449–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F, Zheng Z, Guo J, Ding X (2010) Correlation and quantitation of microRNA aberrant expression in tissues and sera from patients with breast tumor. Gynecol Oncol 119: 586–593. [DOI] [PubMed] [Google Scholar]
- Weber JA, Baxter DH, Zhang S, Huang DY, Huang KH, Lee MJ, Galas DJ, Wang K (2010) The microRNA spectrum in 12 body fluids. Clin Chem 56: 1733–1741. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.