Ladang et al. report that Elp3, a subunit of the Elongator complex, is induced by Wnt signaling and is required to initiate colon cancer development through the regulation of Sox9 translation. They also show that this mechanism is relevant in radiation-induced intestinal regeneration.
Abstract
Tumor initiation in the intestine can rapidly occur from Lgr5+ crypt columnar stem cells. Dclk1 is a marker of differentiated Tuft cells and, when coexpressed with Lgr5, also marks intestinal cancer stem cells. Here, we show that Elp3, the catalytic subunit of the Elongator complex, is required for Wnt-driven intestinal tumor initiation and radiation-induced regeneration by maintaining a subpool of Lgr5+/Dclk1+/Sox9+ cells. Elp3 deficiency dramatically delayed tumor appearance in Apc-mutated intestinal epithelia and greatly prolonged mice survival without affecting the normal epithelium. Specific ablation of Elp3 in Lgr5+ cells resulted in marked reduction of polyp formation upon Apc inactivation, in part due to a decreased number of Lgr5+/Dclk1+/Sox9+ cells. Mechanistically, Elp3 is induced by Wnt signaling and promotes Sox9 translation, which is needed to maintain the subpool of Lgr5+/Dclk1+ cancer stem cells. Consequently, Elp3 or Sox9 depletion led to similar defects in Dclk1+ cancer stem cells in ex vivo organoids. Finally, Elp3 deficiency strongly impaired radiation-induced intestinal regeneration, in part because of decreased Sox9 protein levels. Together, our data demonstrate the crucial role of Elp3 in maintaining a subpopulation of Lgr5-derived and Sox9-expressing cells needed to trigger Wnt-driven tumor initiation in the intestine.
The intestinal epithelium is characterized by a repetitive architecture made of crypt–villus units and is seen as a powerful experimental model to study adult stem cells in health and diseases (Clevers, 2013; Barker, 2014). Each villus is covered by a single layer of postmitotic cells and is surrounded at its base by multiple epithelial invaginations, referred to as crypts of Lieberkühn. Each crypt–villus unit is composed of six differentiated epithelial cell types. These are lysozyme and defensin-secreting Paneth cells located at the bottom of the crypts, absorptive enterocytes, Goblet and enteroendocrine cells that secrete mucus or hormones, microfold (M) cells that play essential roles in mucosal immunity, and rare post-mitotic Tuft cells (also referred to as Brush cells) whose biological functions remain to be defined (Clevers, 2013).
Tuft cells originate from poorly characterized tuft cell progenitors, are enriched in acetylated α-tubulin, and show characteristic microtubule and actin bundles located at the cell apex exposed to the luminal environment (Gerbe et al., 2011, 2012). They are distinct from intestinal secretory cells, as transcription factors such as Neurog3, Sox9, and Spdef are dispensable for their generation (Gerbe et al., 2011; Bjerknes et al., 2012). Tuft cells specifically express Doublecortin-like kinase 1 (Dclk1; also referred to as Dcamkl-1) as well as the transcription factor Gfi1B (Bjerknes et al., 2012; Gerbe et al., 2012). Sox9 is also expressed in Tuft cells, but is not seen as a specific tuft cell marker because of its strong expression in Paneth cells (Bastide et al., 2007; Mori-Akiyama et al., 2007). Identifying molecular determinants for the specification and differentiation of Tuft cells is therefore critical to shed more light on their poorly understood biological functions.
The intense self-renewal kinetics of the intestinal epithelium relies on crypt base columnar (CBC) stem cells located at the bottom of intestinal crypts together with Paneth cells. Cycling CBC cells express the Wnt target gene Leucine-rich repeat containing G protein-coupled receptor 5 (Lgr5; Barker et al., 2007). A second pool of quiescent stem cells has also been described in the intestine. Indeed, DNA label–retaining cells (LRCs), also referred to as +4 stem cells, are located above Paneth cells and express various markers such as Bmi1, Hopx, mTERT, and Lrig1 (Potten et al., 1978; Sangiorgi and Capecchi, 2008; Montgomery et al., 2011; Takeda et al., 2011; Powell et al., 2012). Importantly, a high level of plasticity occurs between Lgr5+ and LRC stem cells as Hopx-expressing +4 cells can generate Lgr5+ cells, whereas the latter can also express +4 stem cell markers (Takeda et al., 2011; Muñoz et al., 2012). Similarly, Lgr5+ cells mark a heterogeneous population as Lgr5high stem cells are mitotically active, whereas other Lgr5low progenitors exit the cell cycle (Basak et al., 2014).
Wnt signaling fuels the stem cell compartment and also critically controls tumor development in the intestine by promoting crypt proliferation through transcriptional activation of Tcf target genes (Clevers and Nusse, 2012). Although it is not totally clear how cancer stem cells are controlled by Wnt signaling, it has nevertheless been demonstrated that Lgr5+ stem cells critically drive tumor initiation (van de Wetering et al., 2002; Barker et al., 2009; Schepers et al., 2012; Clevers, 2013). The tuft cell marker Dclk1 also appears to distinguish cancer versus normal stem cells (Nakanishi et al., 2013). Dclk1+ cells have been demonstrated to sustain tumor growth in the intestine, but it is currently unclear how Dclk1 expression is regulated (Westphalen et al., 2014). A better characterization of proteins acting downstream of the oncogenic Wnt signaling pathway and required for the maintenance of intestinal cancer stem cells is therefore required.
The intestine, as a proliferative and self-renewing organ, is highly sensitive to DNA-damaging agents, but regenerates when subjected to high doses of radiation. Indeed, intestinal stem cells that escaped from massive p53-mediated apoptosis undergo cell proliferation and repopulate the intestine within a few days, a process that critically relies on Wnt signaling (Withers and Elkind, 1970; Merritt et al., 1994; Ashton et al., 2010; Cordero and Sansom, 2012). The mitotically active Lgr5+ cells, although dispensable for intestinal homeostasis, are critical for intestinal regeneration upon damage. Notably, upon caloric restriction or expression of the Wnt agonist R-spondin1, the increased number of Lgr5+ cells promotes protection against intestinal damage (Bhanja et al., 2009; Tian et al., 2011; Yilmaz et al., 2012; Metcalfe et al., 2013; Zhou et al., 2013). Similarly, a population of quiescent LRCs, defined as Paneth cell precursors that express Paneth and +4 markers, can revert back into proliferating Lgr5+ cells upon radiation (Buczacki et al., 2013). Another study also defined an intestinal Dclk1+ quiescent cell population required for regeneration as the genetic ablation of Dclk1 led to defects in recovery after intestinal injury (Westphalen et al., 2014). Moreover, the transcription factor Sox9 is also critical for epithelial regeneration after high-dose irradiation highlighted (Roche et al., 2015). Still, the molecular mechanism governing stem cell plasticity during radiation-induced intestinal regeneration remains poorly characterized.
Elp3 is the catalytic subunit of Elongator (Elp1-Elp6), a protein complex initially identified as a component of a hyperphosphorylated RNA polymerase II holoenzyme isolated from budding yeast chromatin. Elp3 sequence harbors motifs found in the GNAT family of histone acetyltransferases (HATs; Otero et al., 1999; Wittschieben et al., 1999; Kim et al., 2002) and a radical S-adenosylmethionine (SAM) domain at the N-terminal region that includes a FeS cluster critical for Elongator integrity (Greenwood et al., 2009). Elongator promotes transcriptional elongation in the nucleus through histone H3 acetylation but also, as a cytoplasmic complex, in translational efficiency by adding 5-methoxycarbonylmethyl (mcm5) and 5-carbamoylmethyl (ncm5) groups on uridines at the wobble position of some tRNAs (Kristjuhan et al., 2002; Winkler et al., 2002; Huang et al., 2005; Close et al., 2006; Esberg et al., 2006; Bauer et al., 2012; Lin et al., 2013). Elongator promotes cell migration in a variety of primary and transformed cells (Close et al., 2006, 2012; Creppe et al., 2009; Lee et al., 2009). However, it is unclear whether Elongator is involved in tumor development in vivo.
We show here that Elp3 expression is induced by Wnt signaling, promotes Tuft cell differentiation, and is essential for Wnt-driven tumor development in the intestine, as well as for radiation-induced intestinal regeneration. Mechanistically, our data support a model in which Lgr5+ cells rely on Elp3 to initiate tumor development upon constitutive Wnt signaling at least through the maintenance of a pool of Lgr5+/Dclk1+/Sox9+ cells. We show that Elp3 regulates Sox9 protein levels through tRNA modification. Therefore, we define Elp3 as a downstream effector of Wnt signaling that controls Sox9 translation and maintains a pool of Lgr5+/Dclk1+/Sox9+ cells to drive tumor initiation in the intestine.
RESULTS
ELP1 and ELP3 are Wnt target genes
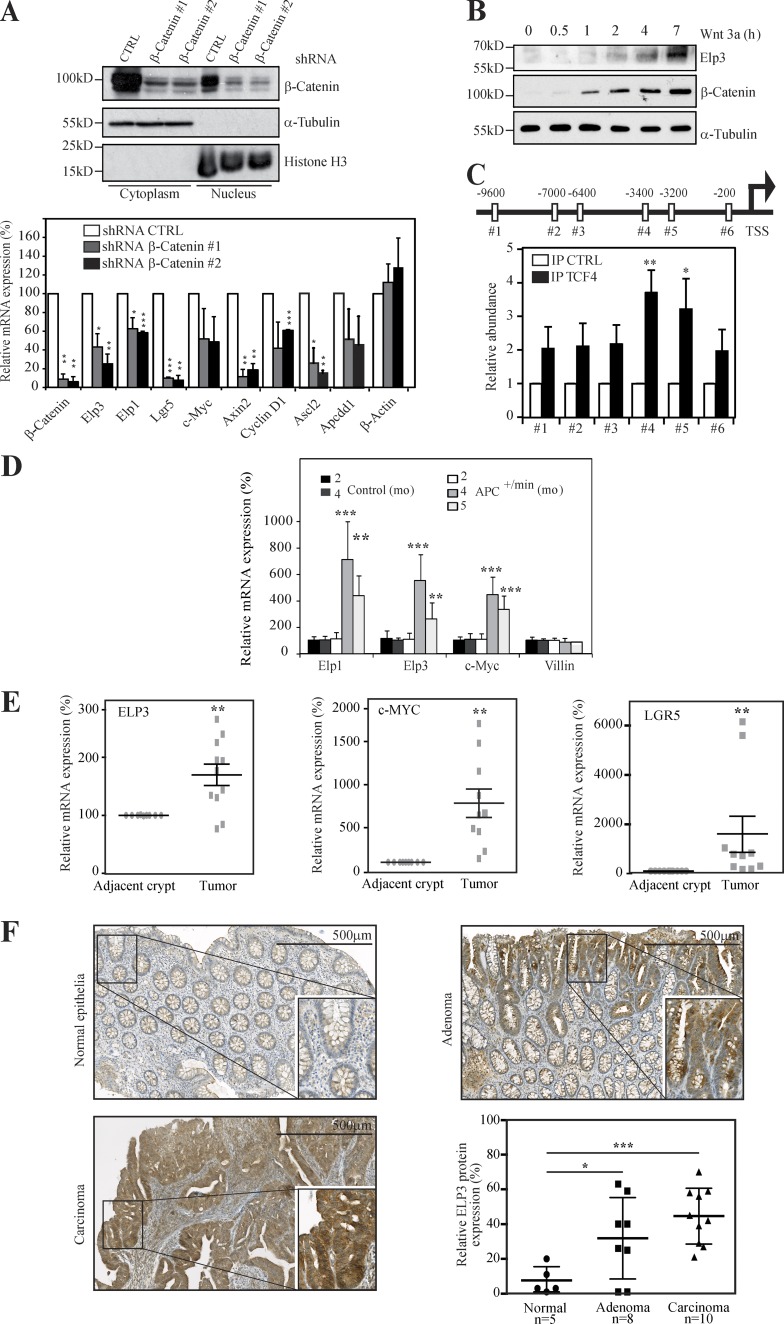
Elongator-deficient melanoma cells fail to form colonies in soft agar (Close et al., 2012). The anchorage-independent growth of two colon cancer–derived cell lines, HCT116 and HT29, is also strongly impaired after Elp3 depletion (unpublished data). Both HCT116 and HT29 cells harbor enhanced Wnt signaling as a result of β-catenin or APC mutations, respectively (Morin et al., 1997; Wang et al., 2003), suggesting a crucial role of Elongator in Wnt-driven tumorigenesis. As β-catenin critically drives colony formation in soft agar (Verma et al., 2003), we next investigated whether Elongator is functionally connected to Wnt- and β-catenin–dependent signaling pathways. β-catenin nuclear levels, as well as expression of described Wnt target genes, remained unchanged upon ELP3 deficiency in HT29 cells, indicating that Elongator is dispensable for Wnt signaling activation (unpublished data). We then impaired the Wnt pathway by generating β-catenin–depleted HT29 cells and noticed that ELP1 and ELP3 mRNA levels decreased upon β-catenin deficiency, similarly to other Wnt target genes, such as Lgr5, Axin 2, Cyclin D1, and c-Myc (Fig. 1 A). Conversely, Wnt activation in RKO cells, which have low intrinsic Wnt signaling,increased Elp3 protein levels (Fig. 1 B), suggesting that ELP3 expression is β-catenin-dependent in colon cancer–derived cells. The Elp3 promoter harbors multiple TCF4-binding sites, and we detected a specific recruitment of TCF4 on two sites located 3,400 and 3,200 bp upstream from the transcription start site by ChIP assays (Fig. 1 C). To investigate whether this finding is also relevant in vivo, we assessed Elp1 and Elp3 mRNA levels in control versus Apc+/min mice, which spontaneously develop adenocarcinomas as a result of constitutive Wnt signaling (Su et al., 1992). Similarly to c-Myc, Elp1 and Elp3 mRNAs dramatically increased over time in intestinal epithelial cells (IECs) from Apc+/min, but not from control mice (Fig. 1 D). ELP3 expression was also higher in human intestinal carcinomas than in normal adjacent tissues, similar to c-MYC (Fig. 1 E). Consistently, in human samples of colon cancer, ELP3 protein expression was enriched in parts enclosing adenomas and carcinomas compared with normal intestinal epithelia, as shown by anti-ELP3 immunohistochemistry analyses (Fig. 1 F). Collectively, our data indicate that Elp3 expression is Wnt and β-catenin dependent in the intestine and is enhanced upon constitutive Wnt signaling.
Figure 1.
Wnt signaling induces ELP3 expression. (A) HT29 cells were infected with the indicated lentiviral constructs and protein extracts from the resulting cells were subjected to Western blot (WB) analyses. α-tubulin and Histone H3 are used as controls for the cytoplasmic and nuclear fractions, respectively (top). At the bottom, control (shRNA CTRL) or β-catenin–depleted HT29 cells (shRNA β-Catenin #1 or #2) were subjected to quantitative real-time PCR analysis to monitor Wnt target genes, as well as ELP1 and ELP3 mRNA levels. Data from three independent experiments (mean values ± SD; Student’s t test; *, P < 0.05; **, P < 0.01; ***, P < 0.001) are shown. (B) Anti-ELP3, –β-Catenin, and –α-tubulin WB analyses with whole-cell lysates from RKO cells treated or not with Wnt 3a (100 ng/ml) for the indicated periods of time were performed. (C) TCF4-binding sites were identified on the Elp3 promoter (positions relative to the transcription start site are indicated). ChIP assays were conducted using extracts from HT29 cells and an anti-TCF4 or a nonrelevant (CTRL) antibody. Associated DNA was analyzed by quantitative real-time PCR using primers spanning the indicated TCF4-binding sites. Signals are expressed as IP/Input ratio for each primer pair normalized to control. Data from three experiments performed in triplicate are shown (mean values ± SD; Student’s t test; *, P < 0.05; **, P < 0.01). (D) Total mRNAs from IEC of control or Apc+/min mice were subjected to quantitative real-time PCR analysis to assess Elp3 and Elp1 expression. Abundance of Villin transcripts was used as control. Error bars denote SD. (**, P < 0.01; ***, P < 0.001; Mann-Whitney test; n ≥ 5). (E) Total mRNAs from nine intestinal carcinomas or from adjacent normal tissues were subjected to quantitative real-time PCR to assess ELP3, MYC, and LGR5 levels. The figure shows the mRNA expression levels of all transcripts in adenomas relative to their levels in normal adjacent tissues after normalization with β-2 microglobulin. (n = 9; **, P < 0.01; Wilcoxon-matched paired test). (F) Anti-ELP3 immunohistochemistry (IHC) was conducted in human cases of intestinal malignancies, as well as in normal intestinal epithelia. Representative images are shown. The average Elp3-specific signal has been quantified and plotted (mean values ± SD; Student’s t test; *, P < 0.05; ***, P < 0.001).
Elp3 promotes Tuft cell differentiation in the intestine
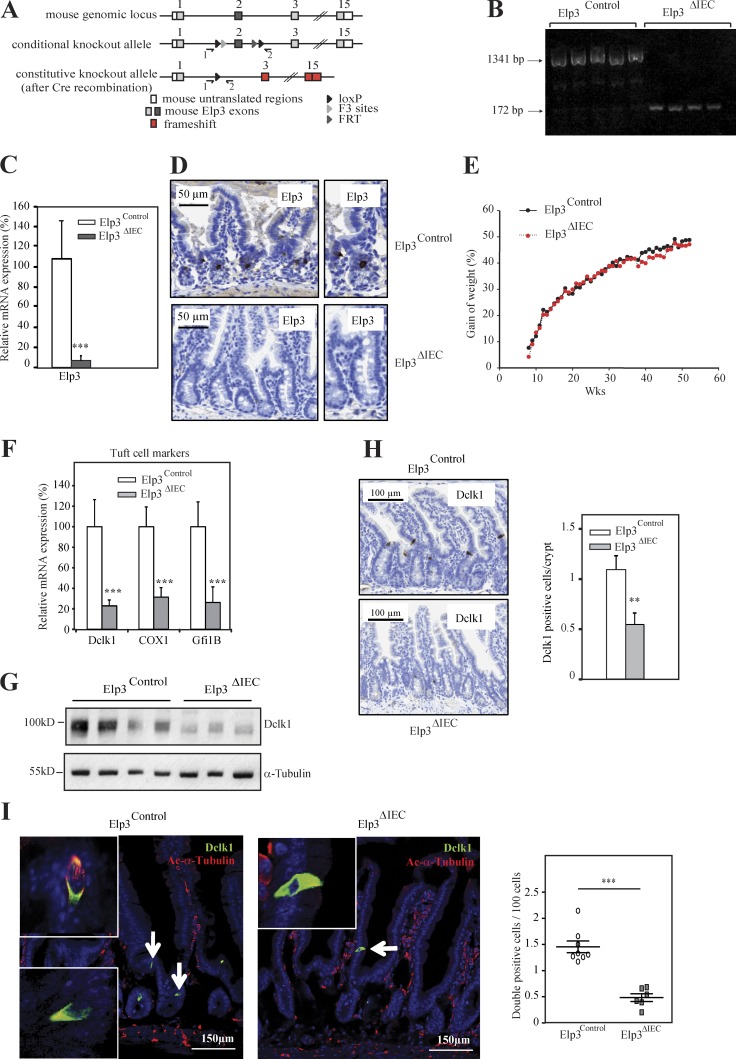
Having defined Elp3 as a Wnt target, we next assessed its role in intestinal homeostasis. We first generated a genetically engineered mouse model in which the exon 2 of the Elp3 gene was flanked with LoxP sites to create the conditional knockout (KO) allele (referred hereafter to as Elp3Control). Deletion of exon 2 upon Cre expression results in Elp3 loss of function by generating a frameshift to all downstream exons (Fig. 2 A). The constitutive knockout alleles generated by crossing our Elp3Control strain with the Villin-Cre mouse (referred hereafter to as Elp3ΔIEC) was indeed identified in IECs through PCR analysis (Fig. 2 B). As a result, Elp3 expression was severely decreased in IECs of Elp3ΔIEC mice (Fig. 2, C and D). The absence of Elp3 in IEC did not impact on the overall gain of weight of mice over lifetime (Fig. 2 E). Moreover, no obvious defect in the intestinal architecture was observed (unpublished data). The number of cycling cells in the transient amplifying compartment, as well as cell death at the top of intestinal villi were intact upon Elp3 deficiency, as judged by Ki67 and cleaved Caspase 3 staining performed in 4-mo-old control or Elp3ΔIEC mice (unpublished data). Moreover, mRNA levels of markers of CBC cells Olfm4, Lgr5, and Achaete Scute-like 2 (Ascl2; van der Flier et al., 2009) remained unchanged (unpublished data). Quiescent, Paneth, and Goblet cell markers were also properly expressed upon Elp3 deficiency, as were Atonal homologue 1 (Atoh1) and hairy/enhancer of split 1 (Hes1), which control the balance between secretory or absorptive lineages (Jensen et al., 2000; Yang et al., 2001; and unpublished data). Of note, mRNA levels of Chromogranin A, an enteroendocrine cell marker, slightly decreased upon Elp3 deficiency (unpublished data). Surprisingly, all tested markers of Tuft cell, namely Dclk1, Cox1, and Gfi1B (Gerbe et al., 2011, 2012; Bjerknes et al., 2012), were decreased in their mRNA levels upon Elp3 deficiency (Fig. 2 F). As a consequence, intestinal Dclk1 protein levels were strikingly reduced (Fig. 2 G). The number of Dclk1+ cells actually decreased in Elp3-deficient intestinal crypts, as judged by IHC analyses (Fig. 2 H), suggesting that Elp3 is critical for Tuft cell differentiation. IF analyses further showed that the number of Tuft cells, defined as Dclk1+/acetylated α-tubulin+ cells (Saqui-Salces et al., 2011), was significantly lower upon Elp3 deficiency in intestinal epithelia (Fig. 2 I). Thus, Elp3 specifically promotes the differentiation of Tuft cells in healthy crypts but does not affect the overall homeostasis of the intestine.
Figure 2.
Elp3 promotes Tuft cell differentiation. (A) LoxP sites flanking exon 2 of the Elp3 gene were inserted. Deletion of exon 2 upon Cre expression results in loss of function of Elp3 by generating a frameshift to all downstream exons (red boxes). Primers used for detection of the wild-type or mutated alleles are depicted by arrows. (B) Genomic DNA was extracted from IECs of Elp3lox/lox (Elp3Control) or Elp3lox/lox VillinCre/+ (Elp3ΔIEC) mice and subjected to PCR analysis using primers 1 and 2 to amplify the wild-type or recombinant allele (1,341- or 172-bp fragments, respectively). (C) mRNAs from Elp3Control and Elp3ΔIEC mice were subjected to quantitative real-time PCR analysis to assess Elp3 mRNA levels (mean values ± SD; Student’s t test; ***, P < 0.001). (D) Anti-Elp3 IHC analyses were performed with intestinal crypts from 120-d-old Elp3Control and Elp3ΔIEC mice. (E) The weight of five male Elp3Control and Elp3ΔIEC mice (8–55 wk old) was quantified and plotted. Weights were normalized at 8 wk of age for each genotype. (F) Intestinal epithelial cells (IECs) were isolated from 120-d-old Elp3Control and Elp3ΔIEC mice, and the resulting total mRNAs were subjected to quantitative real-time PCR analysis to assess mRNA levels of the indicated candidates (mean values ± SD; Student’s t test; ***, P < 0.001; n = 6). (G) Anti-Dclk1 and α-tubulin (loading control) WBs were performed on cell extracts from IECs of Elp3Control and Elp3ΔIEC 120-d-old mice. (H; left) Anti-Dclk1 IHC in intestinal crypts from the indicated genotypes. (right) Quantification of Dclk1-positive cells per crypt showing significant decrease of Dclk1 upon Elp3 deficiency in the intestine (mean values ± SD; Student’s t test; **, P < 0.01; n = 4). (I) Anti-Dclk1 and acetylated α-tubulin immunofluorescence (IF) analyses were performed to visualize Tuft cells in Elp3Control and Elp3ΔIEC mice, as indicated. The graph represents the quantification of the Dclk1/acetylated-α-tubulin double-positive cells in both genotypes (mean values ± SD; Student’s t test; ***, P < 0.001).
Wnt-driven tumor initiation requires Elp3
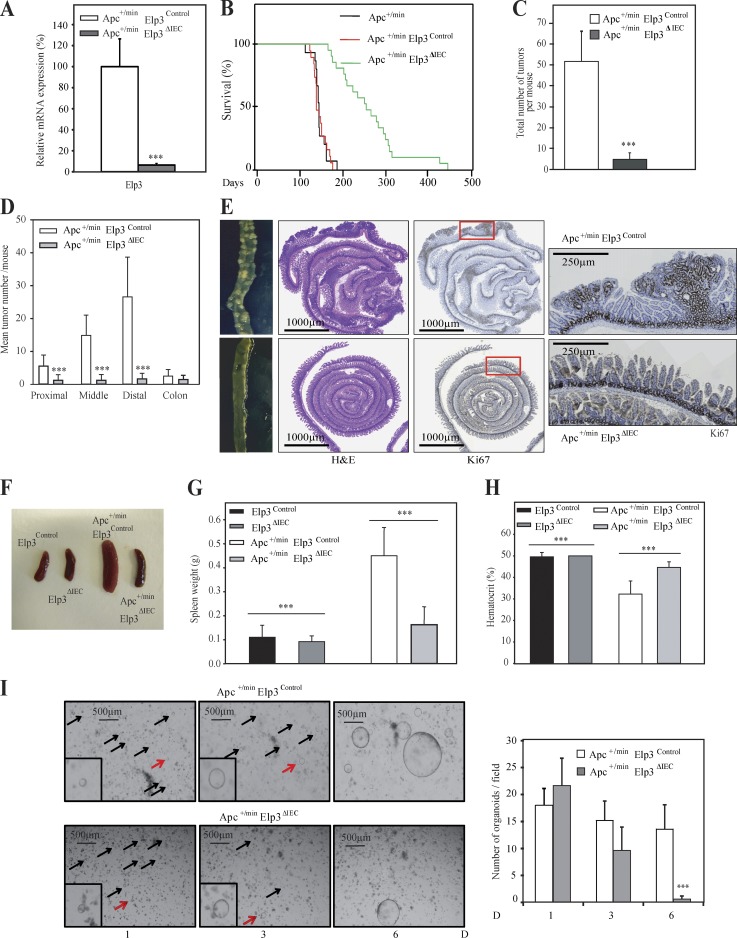
Because Elp3 is required for colony formation in colon cancer cells showing constitutive Wnt signaling, and because Wnt signaling promotes Elp3 expression in the intestine, we next explored the putative role of Elp3 in Wnt-driven intestinal tumor development by crossing Elp3Control or Elp3ΔIEC mice with the Apc+/min strain. As expected, Elp3 expression was severely impaired in intestinal crypts of the resulting mouse model (Apc+/min Elp3ΔIEC), as confirmed by real-time PCR analysis (Fig. 3 A). Strikingly, Elp3 deficiency in Apc+/min mice dramatically extended their life span due to a severe decrease in tumor number in proximal, middle, and distal intestines (Fig. 3, B–E). Splenomegaly, a typical feature of Apc+/min mice (Lane et al., 2010), did not occur upon Elp3 deficiency, as spleen weight of Apc+/min Elp3ΔIEC mice were almost identical to those seen in Elp3Control or in Elp3ΔIEC mice (Fig. 3, F and G). Moreover, decreased hematocrit levels, another feature of Apc+/min mice, was not observed upon Elp3 deficiency (Fig. 3 H). Additionally, Elp3 deletion strongly impaired spheroid structure formation from ex vivo culture of Apc-mutated intestinal crypts (Fig. 3 I). Together, these data demonstrate that Elp3 promotes Wnt-driven tumor initiation in the intestine.
Figure 3.
Elp3 is required for tumor development in a mouse model of colon cancer. (A) IECs were isolated from Apc+/min Elp3Control and Apc+/min Elp3ΔIEC 120-d-old mice and the resulting total mRNAs were subjected to quantitative real-time PCR analysis to assess Elp3 mRNA levels (mean values ± SD; Student’s t test; ***, P < 0.001; n = 4). (B) A Kaplan–Meyer curve (P < 0.001; n ≥ 16) was established with the indicated genotypes. (C and D) The histograms show the total number of adenomas in the entire intestine (C) or in the indicated areas (D) of 120-d-old control (Apc+/min Elp3Control) and Elp3-deficient mice (Apc+/min Elp3ΔIEC; mean values ± SD; Student’s t test; ***, P < 0.001; n ≥ 11). (E) Representative pictures of jejunal section of 120-d-old Apc+/min Elp3Control and Apc+/min Elp3ΔIEC mice. H&E and K67 staining are also illustrated. (F) Representative pictures of the spleen from the indicated genotypes. (G) Quantification of the spleen weight of 120-d-old mice from the indicated genotypes (mean values ± SD; Student’s t test; ***, P < 0.001; n ≥ 7). (H) Hematocrit levels in the indicated genotypes of 3-mo-old mice (Elp3Control, n = 4; Apc+/min Elp3Control, n = 9; Elp3ΔIEC, n = 3) and Apc+/min Elp3ΔIEC (n = 9) was assessed and plotted (mean values ± SD; Student’s t test; ***, P < 0.001). (I) Representative pictures of ex vivo organoid cultures using intestinal crypts from 120-d-old Apc+/min Elp3Control and Apc+/min Elp3ΔIEC mice are shown (left). Quantification of the total number of spheroid structures per well for each genotype is illustrated (mean values ± SD; Student’s t test; ***, P < 0.001). Red and black arrows depict cells that will give rise to spheroid structures or not, respectively.
Lgr5+ cells require Elp3 to promote Wnt-dependent tumor initiation in the intestine
Given the critical role of Lgr5+ cells in tumor initiation (Barker et al., 2009; Schepers et al., 2012), we next wondered whether the genetic inactivation of Elp3 in Lgr5+ cells would also impact on Wnt-driven tumor initiation. To address this issue, we first crossed the stem cell–specific Lgr5-EGFP-IRES-creERT2 knock-in mouse with our Elp3lox/lox strain and subsequently deleted Elp3 in the resulting mice through tamoxifen administration (referred to as Lgr5-EGFP-Elp3ΔCBC). Tamoxifen was also administered in the Lgr5-EGFP-IRES-creERT2 strain as control (referred to as Lgr5-EGFP-Elp3Control). Notably, Elp3 expression was enriched in cycling Lgr5+ (EGFP+) cells, which provides an additional demonstration for Wnt signaling acting as a driver of Elp3 expression (unpublished data). Impaired Elp3 expression was specifically observed in sorted EGFP+ cells upon tamoxifen injection (unpublished data). Importantly, Elp3 deficiency did not impact on the number of Lgr5+ cells (unpublished data). Therefore, Elp3 is dispensable for the maintenance of Lgr5+ stem cells in healthy crypts.
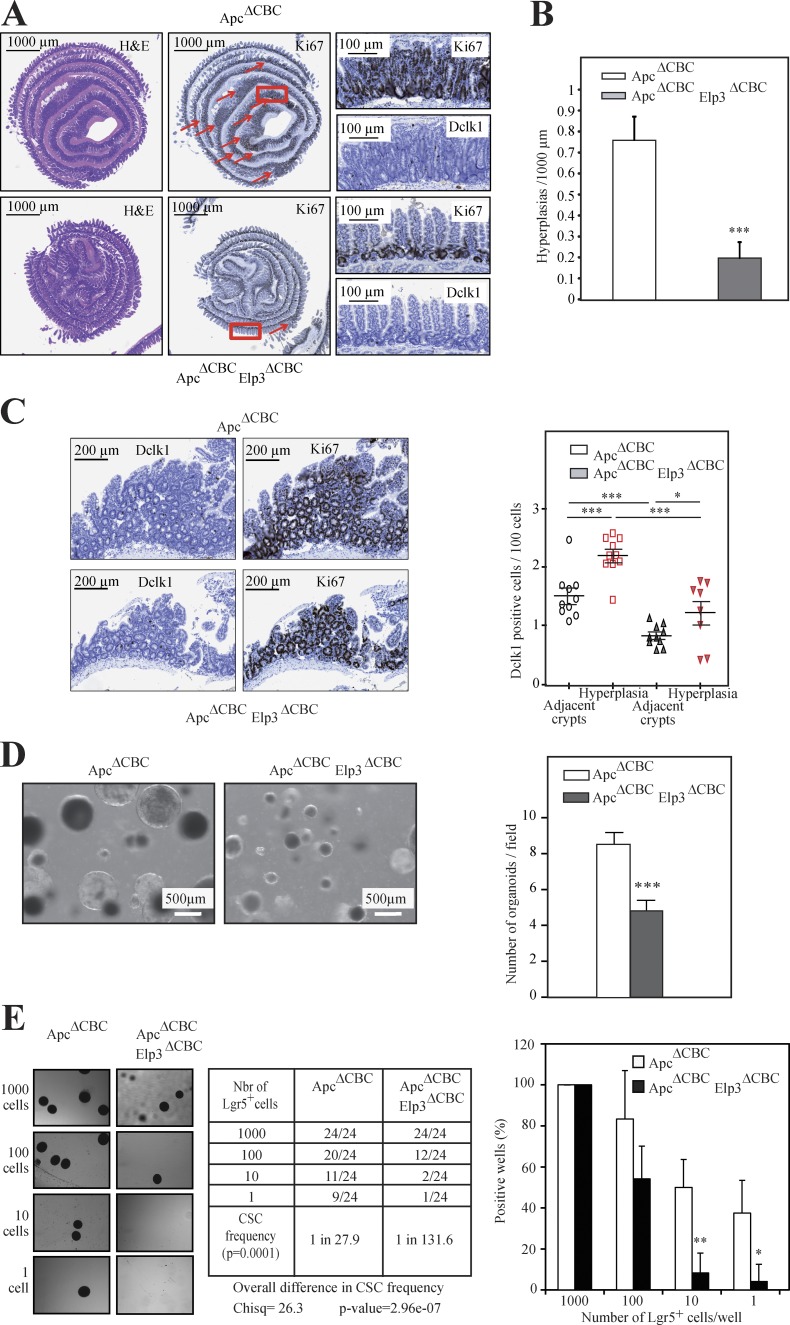
We next crossed the Apclox/lox strain with the Lgr5-EGFP-IRES-creERT2 knock-in mouse and with our Elp3lox/lox strain. Both Apc and Elp3 were simultaneously inactivated in Lgr5+ cells of the resulting mouse through tamoxifen administration (referred to as ApcΔCBC Elp3ΔCBC). Tamoxifen was also administered in mice resulting from the breeding of Apclox/lox mice with the Lgr5-EGFP-IRES-creERT2 strain (referred to as ApcΔCBC). As expected, numerous hyperplasias that resulted from constitutive Wnt signaling in Lgr5+ cells were observed 15 d after tamoxifen administration in ApcΔCBC mice, as demonstrated by H&E and Ki67 IHC analyses (Fig. 4, A and B). Those areas of hyperplasia also showed elevated numbers of Dclk1+ cells (Fig. 4 C). Elp3 deficiency in Lgr5+ and Apc-deleted cells did not impact the total number of Lgr5+ cells, but severely impaired tumor initiation, as judged by decreased numbers of hyperplastic foci (not depicted and Fig. 4, A and B, respectively). The number of Dclk1+ cells was also decreased in the rare Elp3-deficient hyperplastic foci (Fig. 4 C). Importantly, Dclk1+ cells found in these hyperplastic foci were largely negative for acetylated-α-tubulin staining, as compared with adjacent crypts, which supports the notion that they are not Tuft cells (unpublished data). Therefore, Elp3 expression in Lgr5+ cells is required for Wnt-dependent intestinal tumor initiation. Intestinal crypts extracted from ApcΔCBC Elp3ΔCBC mice also failed to efficiently generate spheroid structures ex vivo (Fig. 4 D). To further demonstrate that Elp3 expression is required for the self-renewal capacity of Lgr5+ cancer stem cells, FACS-sorted Lgr5+ cells from both ApcΔCBC and ApcΔCBC Elp3ΔCBC mice were subjected to an in vitro limiting dilution assay. Elp3 ablation strongly affected the Lgr5+ cells cancer stem cell potential (Fig. 4 E). Therefore, the cancer stem cell potential of Lgr5+ cells relies on Elp3 in the intestine.
Figure 4.
Elp3 deficiency in Lgr5+ cells impairs Wnt-driven tumor initiation. (A) ApcΔCBC and ApcΔCBC Elp3ΔCBC mice were intraperitoneally injected with tamoxifen and intestinal crypts, isolated 15 d later, and then subjected to H&E, anti-Ki67, and anti-Dclk1 IHCs, as indicated. Arrows highlight hyperplasias. (B) Quantification of the number of hyperplasias/1,000 µm in each genotype is illustrated (mean values ± SD; Student’s t test; ***, P < 0.001; n = 4). (C) The number of Dclk1+ cells per 100 cells in healthy areas and in hyperplasias was quantified for both genotypes (mean values ± SD; Student’s t test; *, P < 0.05; ***, P < 0.001; n = 10; right). Representative Dclk1 and Ki67 IHC analyses from the indicated genotypes are illustrated (left). (D) Representative pictures of ex vivo organoid cultures using intestinal crypts from ApcΔCBC and ApcΔCBC Elp3ΔCBC mice are shown. Quantification of the number of spheroid structures per field (mean values ± SD; Student’s t test; ***, P < 0.001). (E) Lgr5+ cells FACS-sorted from ApcΔCBC or ApcΔCBC Elp3ΔCBC mice (n = 4), 15 d after tamoxifen administration were serial diluted to generate ex vivo spheroid structures (n = 4). 7 d after seeding, the number of wells containing spheroid structures was quantified. The cancer stem cell (CSC) potential was calculated for both genotypes, using the ELDA software (see Materials and methods for details; Chisq = χ2 test). (right) Graph showing the percentage of positive wells per genotype normalized on 1,000-cell dilution conditions (mean values ± SD; Student’s t test; *, P < 0.05; **, P < 0.01). (left) A summary table of organoid-positive wells (absolute numbers) in all experimental conditions is shown with representative images of spheroid structures.
Elp3 maintains the pool of Dclk1+ cells in the intestine
We next explored the mechanisms underlying the role of Elp3 in Wnt-driven intestinal tumor initiation. Nuclear β-catenin protein levels, as well as mRNA levels of Wnt target genes, such as c-Myc, EphB2, and Tcf-1, were unchanged in 4-mo-old Apc+/min Elp3ΔIEC mice (unpublished data) as compared with Apc+/min Elp3Control mice. Moreover, crypt cell proliferation and cell apoptosis at the top of the villi remained unchanged upon Elp3 deficiency in Apc+/min mice (unpublished data), as were the number of goblet cells, apical enterocytes, Paneth, and enteroendocrine cells (unpublished data). Of note, Atoh1 and Hes1 mRNA levels were also unchanged in Elp3-deficient Apc+/min mice, as were several markers of cell differentiation, namely Krt20, Anpep, and Prss7 (unpublished data). In agreement with a dispensable role of Elp3 in the maintenance of Lgr5+ cells in ApcΔCBC mice, Lgr5, but also Ascl2, mRNA levels did not change upon Elp3 deficiency in intestinal tumors of Apc+/min mice (Fig. 5 A). Interestingly, in line with the fact that Dclk1+ cells in tumors are distinct from tuft cells, both Dclk1 and Gfi1B, but not COX1, were decreased at the mRNA level upon Elp3 deficiency in the Apc+/min tumors (Fig. 5 B). Dclk1, whose protein levels were higher in cystic lesions than in adenocarcinomas (Fig. 5 C), was also less expressed in IECs extracted from Apc+/min Elp3ΔIEC mice, as shown by IHC and WB analyses (Fig. 5, D and E). Consistently, the number of Dclk1+ cells in both tumors and adjacent regions significantly decreased in the intestine of Apc+/min Elp3ΔIEC mice (Fig. 5 F). Importantly, although the number of Tuft cells (Dclk1+ and acetylated α-tubulin+ cells) slightly decreased, the number of Dclk1+/acetylated α-tubulin− cells dramatically decreased in Apc+/min Elp3ΔIEC mice (Fig. 5 G). Importantly, the depletion of Dclk1 in ex vivo organoid cultures with intestinal crypts extracted from ApcΔCBC mice did not impact on Elp3 expression, but interfered with the generation of spheroid structures, similar to Elp3 deficiency (Fig. 5 H). Therefore, Elp3 promotes Wnt-driven tumor initiation in the intestine, at least by maintaining the pool of Dclk1+ cells.
Figure 5.
Elp3 deficiency in intestinal epithelial cells of Apc+/min mice impairs Dclk1 expression. (A and B) Tumors were isolated from 120-d-old Apc+/min Elp3Control and Apc+/min Elp3ΔIEC mice and the resulting total mRNAs were subjected to quantitative real-time PCR analysis to assess mRNA levels of the indicated candidates. Tumors efficiently recombined for Elp3 were selected. The mean value of mRNA levels in Apc+/min Elp3Control of each candidate was set to 100% and Apc+/min Elp3ΔIEC were expressed relative to that after normalization with GAPDH (mean values ± SD; Student’s t test; *, P < 0.05; n ≥ 4). (C) Dclk1 protein expression was assessed by IHC in intestinal cystic lesions and adenomas of 120-d-old Apc+/min mice. (D) Anti-Dclk1 IHC analyses were conducted with intestinal crypts of 120-d-old Apc+/min Elp3Control and Apc+/min Elp3ΔIEC mice. The number of Dclk1+ cells per intestinal crypt was quantified in each indicated genotype (mean values ± SD; Student’s t test; **, P < 0.01; n = 5). (E) Protein extracts from IECs of 120-d-old mice of the indicated genotypes were subjected to anti-Dclk1 and α-tubulin (loading control) WB analysis. (F) The number of Dclk1+ cells per 100 cells was quantified in the intestine from the indicated genotypes (mean values ± SD; Student’s t test; *, P < 0.05; ***, P < 0.001; n ≥ 4). (G) Anti-Dclk1 and acetylated α-tubulin IF analyses were performed to distinguish and quantify Tuft cells versus Dclk1+ cancer stem cells in Apc+/min Elp3Control and Apc+/min Elp3ΔIEC mice (mean values ± SD; Student’s t test; *, P < 0.05; **, P < 0.01; ***, P < 0.001; n ≥ 10 tumors). (H) ApcΔCBC organoids were infected with the indicated shRNA construct and their growth was assessed. Western blot analyses carried out with extracts from the resulting organoids are illustrated.The number of organoids per well was quantified. The histogram shows the data from three different mice (mean values ± SD; Student’s t test; **, P < 0.01).
Elp3 promotes Sox9 expression through tRNA modifications
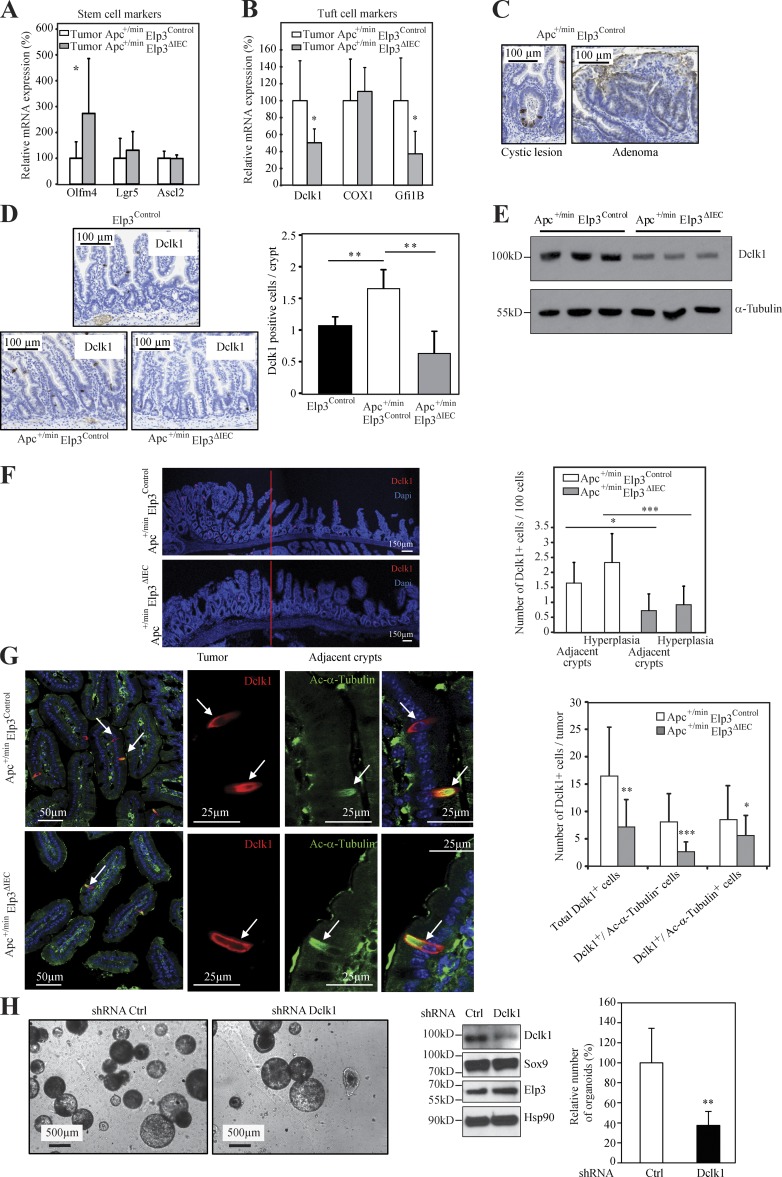
Constitutive Wnt signaling induces significant changes in gene and protein expression to promote tumor development (Sansom et al., 2007). Sox9 is required downstream of Wnt/β-catenin signaling for the long term self-renewal of oncogene-expressing cells in a genetic mouse model of basal cell carcinoma (Larsimont et al., 2015). Moreover, Sox9 is expressed in intestinal crypts as a Wnt target gene and is consequently overexpressed upon constitutive Wnt signaling (Blache et al., 2004). Therefore, we hypothesized that the blockage of Wnt-driven tumor development upon Elp3 deficiency may be due, at least in part, to a defective Sox9 expression. As expected, Sox9 levels were robustly increased in Apc+/min intestinal crypts as a result of constitutive Wnt signaling (Fig. 6 A). Importantly, its expression was severely decreased in Apc+/min Elp3ΔIEC mice to reach similar levels as in normal intestinal crypts (Fig. 6 A). Consistently, Sox9 protein levels dramatically decreased in Apc+/min Elp3ΔIEC versus Apc+/min Elp3Control mice (Fig. 6 B). Igfbp4, a stem cell marker whose expression is Sox9-dependent in the intestine (Merlos-Suárez et al., 2011; Shi et al., 2013), was consequently less expressed upon Elp3 deficiency in intestinal crypts from Apc+/min mice (Fig. 6 C). Thus, Elp3 is critical for the expression of Sox9 seen upon Apc loss. This conclusion was further supported by the fact that although Wnt 3a increased ELP3 and SOX9 expression, both at the mRNA and protein levels through β-catenin stabilization in control RKO cells, it failed to induce SOX9 protein expression upon ELP3 deficiency (Fig. 6 D and not depicted). Importantly, the number of ex vivo spheroid structures generated from ApcΔCBC mice similarly decreased upon Sox9 or Elp3 depletion, suggesting that Elp3 promotes intestinal Wnt-driven tumor initiation at least by promoting Sox9 expression (Fig. 6 E).
Figure 6.
Elp3 drives Sox9 expression. (A; left) Anti-Sox9 IHC analyses were conducted with intestinal crypts of 120-d-old mice of the indicated genotypes. (right) The number of Sox9+ cells per intestinal crypt was quantified in each genotype (mean values ± SD; Student’s t test; *, P < 0.05; ***, P < 0.001; n ≥ 4). (B) Protein extracts from IECs of 120-d-old mice of the indicated genotypes were subjected to anti-Sox9 and -Hsp90 (loading control) WB analyses. (C) IECs were isolated from 120-d-old mice of the indicated genotypes and the resulting total mRNAs were subjected to quantitative real-time PCR analysis to assess Sox9 and Igfbp4 mRNA levels (mean values ± SD; Student’s t test; **, P < 0.01; n = 5). (D) RKO cells were untreated or stimulated with Wnt 3a for the indicated periods of time and the resulting cell extracts were subjected to WB analyses using the indicated antibodies. (E) ApcΔCBC organoids were infected with the indicated shRNA construct and their growth was assessed. (right) the number of organoids per well was quantified. The histogram shows the data from three different mice (mean values ± SD; Student’s t test; *, P < 0.05; **, P < 0.01). (F) Total protein extracts from fission yeast cells of the indicated genotypes expressing Sox9 and containing either a plasmid overexpressing the tRNAlysUUU (K) or the corresponding empty vector (/) were subjected to anti-Sox9 and anti–α-tubulin (loading control) WB analyses. (G) A construct generating wild-type Sox9 or a Sox9 mutant in which all lysine residues (K) were mutated into arginines (R; Sox9ΔAAA) was expressed in yeast cells of the indicated genotypes and the resulting cell extracts were subjected to WB analyses.
Intriguingly, Sox9 mRNA levels remained unchanged upon Elp3 deficiency in the Apc+/min model, as shown by quantitative real-time PCR and in situ hybridization (Fig. 6 C and not depicted, respectively). Moreover, Sox9 transcription was properly induced upon Wnt 3a stimulation in RKO cells (unpublished data), suggesting that Elp3 regulates Sox9 expression at the posttranscriptional level. Of note, mRNA levels of various proteins known to regulate the acetylation or sumoylation of Sox9 (Tip60, Ubc9, Pias1, Pias4, and Senp2, respectively; Hattori et al., 2006, 2008) were properly expressed upon Elp3 deficiency in Apc+/min mice (unpublished data), suggesting that Sox9 post-translational modifications known to interfere with its stability are not responsible for the decreased protein levels seen upon Elp3 deficiency. In yeast, Elp3 promotes translational efficiency through tRNA modification (Huang et al., 2005). To understand if Sox9 decrease upon Elp3 deficiency is due to translational defects, we expressed a Sox9 cDNA in wild-type and elp3-deleted fission yeast cells (Fig. 6 F). Strikingly, the absence of Elp3 impeded Sox9 expression, which was restored upon overexpression of the unmodified tRNAlysUUU, a critical target of Elongator. As Elp3 promotes the translation of AAA-enriched polypeptides in yeast (Bauer et al., 2012), we next generated a Sox9 expression construct in which all three AAA codons (K167, 242, and 249) found in the Sox9 murin sequence were mutated into their cognate Elp3-insensitive AAG codons (Sox9ΔAAA). We then assessed Sox9ΔAAA expression in control versus Elp3 mutant yeast strain. Interestingly, Sox9ΔAAA but not wild-type Sox9 was properly expressed in yeast Elp3 mutant strain (Fig. 6 G). Therefore, Elp3-mediated tRNA modification is critical for Sox9 proper translation.
Elp3 expression is required to maintain a pool of Lgr5+/Dclk1+/Sox9+ cells in the intestine
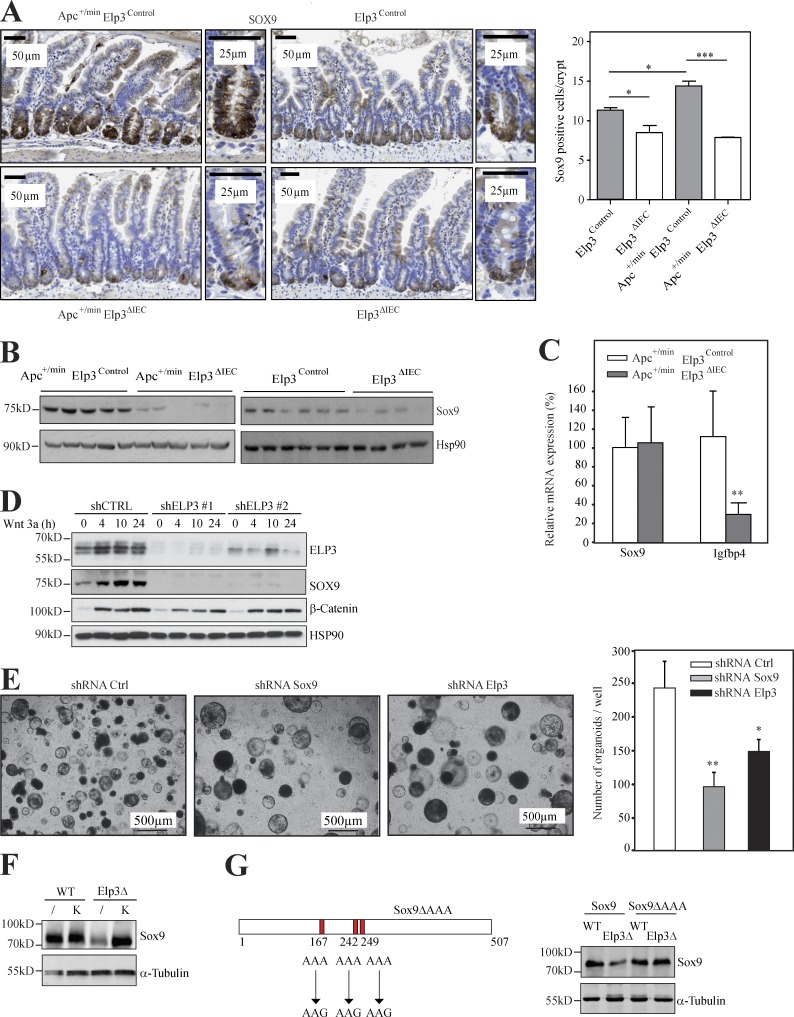
To better understand how the Elp3-dependent translation of Sox9 expression impacts on the number of Dclk1+ cells, we assessed Elp3, Sox9 and Dclk1 levels in ex vivo organoid cultures generated from ApcΔCBC mice and depleted for Elp3 or Sox9. As expected, Elp3 deficiency decreased both Dclk1 and Sox9 protein levels (Fig. 7 A). In agreement with our previous data, Sox9 mRNA levels did not decrease in Elp3-depleted organoids (unpublished data). Yet, both Dclk1 and Gfi1B, but not Cox1, mRNA levels were decreased upon Elp3 depletion, supporting the notion that Elp3 controls the maintenance of a subpopulation of Dclk1+ cells which differ from Tuft cells upon constitutive Wnt activation (Fig. 7 A). Importantly, Sox9 depletion in ex vivo organoids also largely decreased Dclk1 mRNA and protein levels, suggesting that a pool of Dclk1+ cells relies on both Sox9 and Elp3 expression for its maintenance. Finally, Dclk1 depletion in Apc+/min ex vivo organoids, did not affect Sox9 or Elp3 mRNA and protein levels, but decreased the number of spheroid structures as seen for Sox9 or Elp3 depletion (Fig. 5 H), strengthening the notion that both Elp3 and Sox9 are required to maintain Dclk1+ cells.
Figure 7.
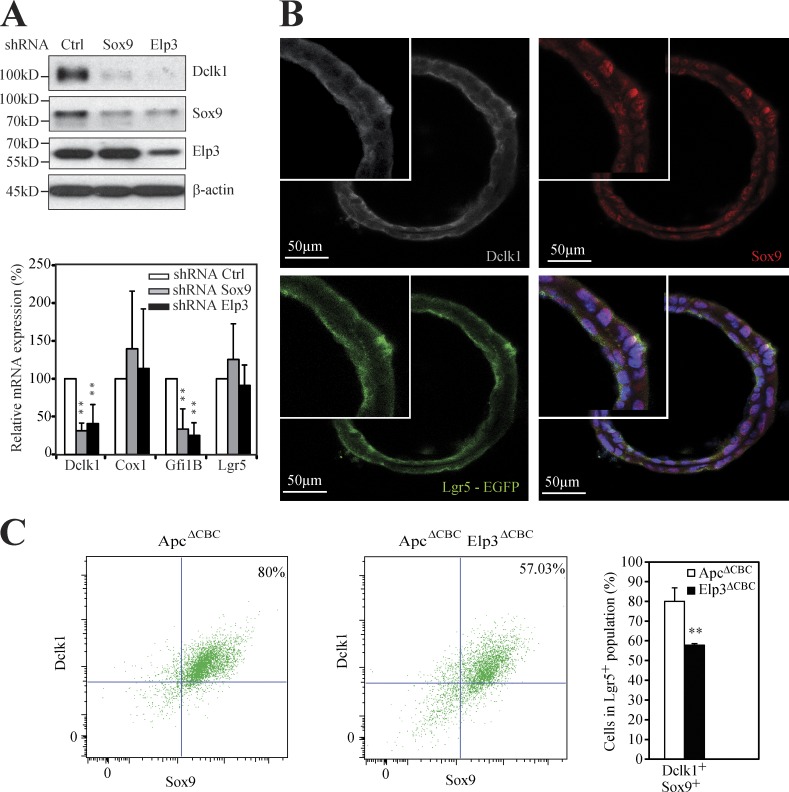
Elp3 expression is required to maintain a pool of Lgr5+/Dclk1+/Sox9+ cells upon constitutive Wnt signaling. (A) ApcΔCBC organoids were infected with a pool of lentiviral constructs delivering shRNA targeting Sox9, Elp3 or an irrelevant sequence used as negative control (shRNA Sox9, shRNA Elp3 and shRNA Ctrl, respectively). (top) Protein extracts from resulting ApcΔCBC organoids were subjected to anti-Dclk1, -Sox9, -Elp3 and β-Actin (loading control) WB analyses. (bottom) Total mRNAs from those organoids were subjected to quantitative real-time PCR analysis to monitor expression levels of the indicated transcripts. The abundance of transcripts in shRNA Ctrl condition was set to 100% and levels of the other transcripts were expressed relative to that after normalization to GAPDH. Data with organoids derived from three different mice are shown (mean values ± SD; Student’s t test; **, P < 0.01). (B) Anti-Dclk1 (gray), anti-Sox9 (red), and anti-EGFP (green) IF analyses were carried out to visualize the Lgr5+/Sox9+/Dclk1+ cell population in ApcΔCBC organoids. (C) Single-cell suspensions of digested organoids from ApcΔCBC or ApcΔCBC Elp3ΔCBC mice were stained for Sox9 and Dclk1 and were subjected to FACS analysis. Lgr5-EGFP+ cells were selected. (left) A representative Dot plot of Lgr5-EGFP+ cells stained for Sox9 and Dclk1 is illustrated. 10,000 events per mice were analyzed. (right) The histogram shows a quantification of the Dclk1+/Sox9+ cells in the Lgr5+ population (n ≥ 3 for each genotype; mean values ± SD; Student’s t test; **, P < 0.01).
Having established a link between Sox9 expression and Dclk1+ cells, we next explored whether a Dclk1+/Sox9+ cell population can be detected in Lgr5-expressing cells and if so, whether Elp3 plays any role in its maintenance. Lgr5+/Dclk1+/Sox9+ cells were indeed identified by IF in ex vivo organoids from ApcΔCBC mice (Fig. 7 B). To address the role of Elp3 in their maintenance, we sorted EGFP-Lgr5+ cells from single-cell suspension of digested organoids generated from ApcΔCBC or ApcΔCBC Elp3ΔCBC mice by FACS analysis (unpublished data). Although Lgr5+ cell number did not change upon Elp3 deficiency, the amount of Dclk1+/Sox9+ cells in the Lgr5+ population strongly decreased upon Elp3 deficiency in ex vivo organoids from ApcΔCBC mice (unpublished data and Fig. 7 C, respectively). Therefore, Elp3 promotes Wnt-driven tumor initiation, at least in part, by maintaining the pool of Lgr5+/Dclk1+/Sox9+ cells in the intestine.
Intestinal crypt regeneration upon irradiation requires Elp3
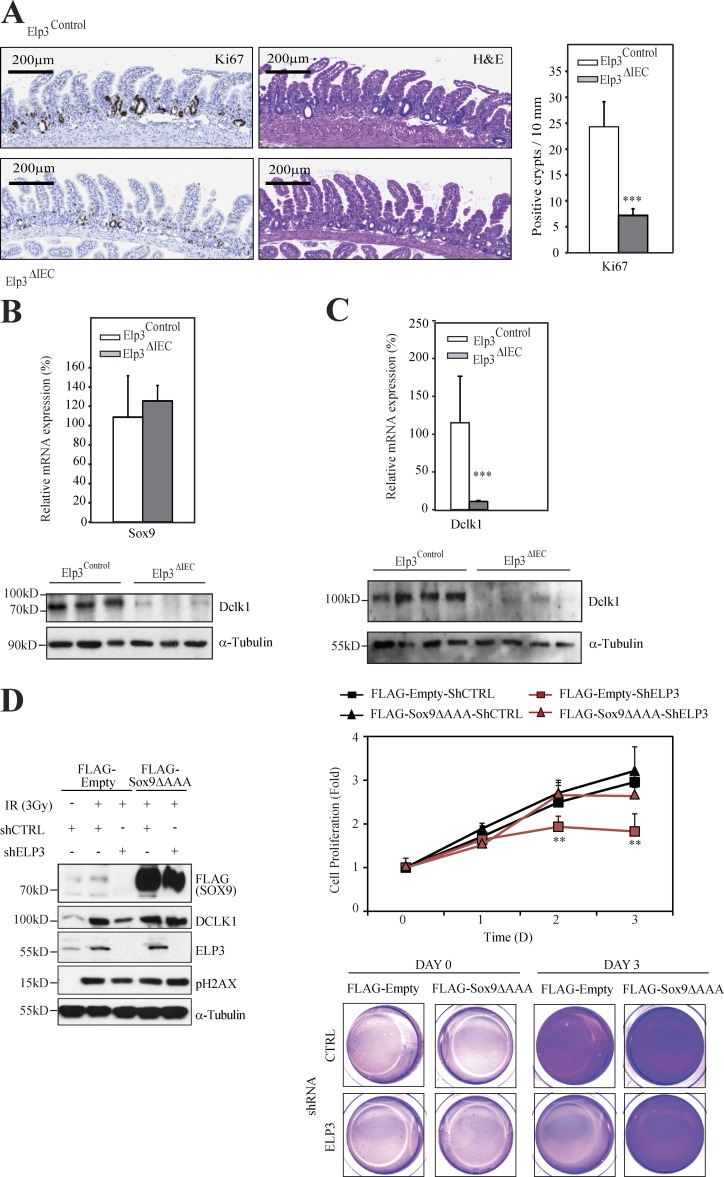
Because Wnt signaling critically promotes intestinal regeneration upon DNA damage (Ashton et al., 2010), and because Dclk1 expression has been shown to be required for survival after radiation injury (May et al., 2014), we next explored whether Elp3 deficiency had any consequence on radiation-induced intestinal regeneration. Elp3Control or Elp3ΔIEC mice were subjected to 14Gy irradiation and cell proliferation was assessed in intestinal crypts. The number of regenerating crypts dramatically decreased upon Elp3 deficiency, as assessed by Ki67 staining performed 3 d after irradiation (Fig. 8 A). Sox9 protein, but not mRNA, levels severely decreased upon Elp3 deficiency in regenerating crypts from irradiated mice (Fig. 8 B). Importantly, mRNA and protein levels of Dclk1 were also dramatically impaired in IECs of irradiated Elp3-deficient mice (Fig. 8 C). To further explore whether Elp3 promotes radiation-induced intestinal regeneration through Sox9 expression, we expressed the Elp3 insensitive Sox9ΔAAA mutant into control or Elp3-deficient RKO cells and assessed cell proliferation post-irradiation (Fig. 8 D). Importantly, Elp3-deficient RKO cells in which Sox9 expression was expectedly defective failed to efficiently proliferate post-irradiation. On the other hand, Elp3-deficient RKO cells in which the Sox9ΔAAA mutant was ectopically expressed proliferated similarly to control RKO cells and showed normalized levels of Dclk1 (Fig. 8 D). Collectively, our data demonstrate that Elp3 mediates regeneration after irradiation by maintaining Sox9 expression.
Figure 8.
Elp3 promotes radiation-induced intestinal regeneration. (A; left) Representative pictures of Ki67 and H&E staining of intestinal crypts from Elp3Control and Elp3ΔIEC mice 3 d after 14 Gy irradiation. (right) The number of regenerating crypts according to Ki67+ cells per mm was quantified for the indicated genotypes (mean values ± SD; Student’s t test; ***, P < 0.001; n = 5). (B) Protein and mRNA extracts from IECs from Elp3Control and Elp3ΔIEC mice 3 d after irradiation were subjected to anti-Sox9 and Hsp90 (loading control) WB or to quantitative real-time PCR analyses (n = 4). (C) Protein and mRNA extracts from jejunal epithelial cells from Elp3Control and Elp3ΔIEC mice 3 d after irradiation were subjected to anti-Dclk1 and α-tubulin (loading control) WB or to quantitative Real time PCR analyses (mean values ± SD; Student’s t test; ***, P < 0.001; n = 4). (D) Control or Elp3-depleted RKO cells were transfected with an empty vector or with the Sox9ΔAAA mutant, as indicated and were subsequently untreated or irradiated with 3 Gy/min (Day 0). (left) Cell extracts were subjected to WB analyses using the indicated antibodies. Cells showing DNA damage are positive for pH2AX. (right) Cell proliferation in all experimental conditions was measured by Crystal Violet at the indicated time points. The mean of two independent experiments performed in duplicate is illustrated (mean values ± SD; Student’s t test; **, P < 0.01). At the bottom, representative images of cells stained with Crystal Violet are shown.
DISCUSSION
We defined here Elp3 as a target of Wnt signaling dispensable for intestinal homeostasis but essential for Tuft cell differentiation, Wnt-driven tumor initiation, and radiation-induced regeneration in the intestine. Although nuclear levels of β-catenin and constitutive Wnt-dependent gene transcription do not require Elp3 in the intestine, our data demonstrate that the translation of Sox9, a protein induced by Wnt, is controlled by Elp3 to maintain a pool of Lgr5+/Dclk1+/Sox9+ cancer stem cells.
Tuft cells derive from Lgr5+ crypt columnar stem cells but their differentiation and biological roles remain poorly understood (Gerbe et al., 2011; Bjerknes et al., 2012). We found here that Elp3 is critical for Tuft cell differentiation, as its absence leads to dramatic reduction of Dclk1+ Tuft cells in the intestinal epithelium. The reduction of differentiated Tuft cells in Elp3-deficient mice did not lead to any detectable functional defect or to significant change in intestinal architecture, indicating that Tuft cells are dispensable for global intestinal homeostasis. Mechanistically, it is unclear how Elp3 promotes Tuft cell differentiation. Levels of the Atoh1/Math1 transcription factor, which appear to be critical for secretory cell lineage commitment and for Tuft cell differentiation, are normally expressed in Elp3-deficient intestinal crypts (Shroyer et al., 2007; Gerbe et al., 2011). Therefore, Elp3 expression may control a late step of the epithelial cell lineage commitment. A gene network between Atoh1, Hes1, and Gfi1B was previously demonstrated to be critical for epithelial lineage commitment in the intestine (Bjerknes et al., 2012). High concentrations of Gfi1B in daughter cells of short-lived progenitors commit them to Tuft cell differentiation. Our data support this model, as Gfi1B mRNA levels are severely decreased in Elp3-deficient intestinal crypts. Therefore, Elp3 may promote Tuft cell differentiation through Gfi1B.
Cell migration of multiple primary and transformed cells requires Elongator (Close et al., 2006; Johansen et al., 2008; Creppe et al., 2009; Lee et al., 2009). Cell invasion of glioblastoma-, neuroblastoma-, and melanoma-derived cells also relies on Elp3 (Close et al., 2006, 2012). Further, our results show that colonosphere formation from colon cancer–derived cells was greatly dependent on Elp3. It remains to be seen whether the inactivation of Elp3 in ex vivo organoid cultures generated from human cases of intestinal cancer also interferes with their growth. We also show here that Elp3 is essential in initial steps of tumor development in vivo using a mouse model of intestinal Wnt-dependent tumor initiation. Our data demonstrate that Elp3 expression increased upon constitutive Wnt activation. Therefore, we highlight a positive correlation between Elp3 expression and the activation of a major oncogenic pathway. This suggests that Elp3 may also be critically involved in the development of other epithelial malignancies harboring deregulated Wnt signaling (Michaelson and Leder, 2001; Ayyanan et al., 2006). Future studies will be dedicated to the identification of all oncogenic pathways that regulate Elp3 expression.
Dclk1+ cells are differentiated Tuft cells in normal intestinal epithelium but they were also defined as cancer stem cells upon Apc loss (Nakanishi et al., 2013). A subpopulation of long-lived and quiescent Dclk1+ cells acting as tumor-initiating cells was also identified (Westphalen et al., 2014). Surprisingly, loss of Apc in these Dclk1+ cells did not cause nuclear translocation of β-catenin and, consequently, did not trigger adenoma formation. These quiescent Dclk1+ cells could nevertheless be reactivated and act as tumor-initiating cells only when Apc loss was combined with an inflammatory stimulus such as dextran sodium sulfate (DSS) colitis (Westphalen et al., 2014). We show that Elp3 deficiency in Apc-mutated Lgr5+ intestinal stem cells blocks tumor initiation by limiting the number of Lgr5+/Dclk1+/Sox9+ cancer stem cells. Because a bidirectional conversion between tumor initiating and differentiated cells that relies on inflammatory signals has been established (Schwitalla et al., 2013), it remains to be tested whether fully differentiated Dclk1+ Tuft cells can undergo dedifferentiation to convert into so-called Dclk1+ tumor initiating cells.
Our data defined Elp3 as a critical protein for Sox9 translation in both normal and Apc-mutated intestinal crypts. Sox9 is expressed in several differentiated epithelial lineages, such as Tuft and Paneth cells. Moreover, two distinct Sox9-expressing stem cell populations were also defined using EGFP-expressing transgenic mice, namely Sox9-EGFPlow and Sox9-EGFPhigh populations (Van Landeghem et al., 2012). Sox9-EGFPlow cells are Lgr5-enriched stem cells, whereas Sox9-EGFPhigh cells express enteroendocrine and +4 stem cell markers. The decreased Sox9 expression seen upon Elp3 deficiency does not result from a lower number of Paneth cells, as they appear to be functional, at least based on intact lysozyme levels seen in Elp3-deficient intestinal crypts. Similarly, the enteroendocrine and +4 stem cells populations are also intact upon Elp3 deficiency. A study concluded that Sox9 is dispensable for Tuft cell differentiation (Gerbe et al., 2011). Whether Elp3 promotes Tuft cell differentiation through Sox9 regulation remains unclear, and it is likely that other candidates whose Wnt-dependent protein translation relies on Elp3 contribute to Tuft cell differentiation.
The role of Sox9 in Wnt-driven tumor development in vivo remains controversial. Indeed, Sox9 inactivation appears to increase tumor burden in the Apc+/min model, which appears to be consistent with the fact that Sox9 acts as a Wnt inhibitor (Akiyama et al., 2004; Shi et al., 2013). On the other hand, Insulin Receptor Substrate-1 (IRS-1) deficiency also limited tumor development in Apc+/min mice, at least because of impaired Sox9 expression (Ramocki et al., 2008). These data, combined with our study, rather support the hypothesis that a defective Sox9 expression in cancer stem cells impairs Wnt-driven cancer development and intestinal regeneration. Indeed, our results show that Sox9 deficiency also impacts on the growth of Apc-mutated ex vivo organoids. Our data are actually in agreement with the recently demonstrated key role of Sox9 in promoting the self-renewal of oncogene-expressing cells downstream of Wnt signaling in a genetic model of basal cell carcinoma (Larsimont et al., 2015). This study, combined with our current work, may reveal a role of Elp3 in any cancer model in which Sox9 critically maintains the self-renewal capacity of cancer stem cells.
Elp3 promotes Sox9 translation through tRNA modification in fission yeast. A study in this model suggested that proteins whose translation requires Elp3 show enriched representation of AAA codons (Bauer et al., 2012). Surprisingly, Sox9 coding sequence is not enriched in AAA codons, yet tRNAlysUUU efficiently restored Sox9 expression in elp3-deleted yeast, and a Sox9 mutant in which all 3 AAA codons were mutated is still properly translated in elp3-deficient yeast cells. Therefore, our data further strengthen the notion that AAA codons rely on properly modified tRNAs to be efficiently translated and also define Sox9 as a Wnt-induced protein whose expression is critically controlled at the translation level.
The identity of intestinal cells required for radiation-induced intestinal regeneration has been the subject of multiple studies. These cells are Sox9+ and reexpress Lgr5 to undergo proliferation in regenerating intestinal crypts (Van Landeghem et al., 2012; Metcalfe et al., 2013). Consistently, Sox9 expression is required to maintain reserve stem cells to promote regeneration upon radiation (Roche et al., 2015). We show that Elp3 is required in radiation-induced regeneration in the intestine, at least by promoting Sox9 expression at the protein level. ELP3-deficient cells in which a Sox9 mutant whose translation is ELP3-independent is ectopically expressed efficiently proliferate after irradiation. Dclk1 expression was also severely decreased upon Elp3 deficiency in the regenerating intestinal crypts. Although a study had shown that Dclk1 inactivation did not impair the number of surviving crypts, but decreased survival of irradiated mice as a result of disrupted tight junctions (May et al., 2014), another study highlighted a critical role for a subpopulation of Dclk1+ cells in radiation-induced intestinal regeneration (Westphalen et al., 2014). Our results suggest that intestinal regeneration upon radiation may rely on a specific Lgr5+/Dclk1+/Sox9+ pool of cells whose maintenance critically relies on Elp3. Our study defines a key role for Elp3 in the DNA damage response in the intestine. An increased sensitivity of Elp3 mutants to DNA damage was previously reported in yeast, which demonstrates that Elp3 critically controls the DNA damage response in multiple organisms (Li et al., 2009; Chen et al., 2011).
The molecular mechanisms underlying Elp3 functions remain poorly understood. It is currently believed that Elp3 promotes the acetylation of multiple substrates in distinct cell compartments to regulate transcriptional elongation in the nucleus and protein translation in the cytoplasm. Such hypothesis was recently experimentally validated while addressing the role of Elp3 in meiosis during spermatogenesis (Lin et al., 2013). We showed here that Elp3 acts downstream of Wnt, and as such, may contribute to Wnt-dependent protein acetylation, transcriptional elongation and/or to Wnt-dependent protein translation through tRNA modifications to promote tumor initiation and intestinal regeneration. Because Elp3 has been recently defined as a tRNA acetyltransferase (Selvadurai et al., 2014), our future studies will be dedicated to the elucidation of the Wnt- and Elp3-dependent protein signature involved in tumor development.
Elp3 deficiency in Apc-mutated intestinal crypts shares many features with the genetic inactivation of Myc in those crypts (Sansom et al., 2007). Whereas our data disclose a role of Elp3 in the transcription of Myc, both Elp3 and Myc may control the same pool of cancer stem cells to drive tumor development in the intestine. Interestingly, the genetic inactivation of Raptor, an essential component of mTORC1, also interferes with Wnt-dependent tumor growth but not with normal gut homeostasis (Faller et al., 2014). mTORC1 is required for cell proliferation in Apc-mutated intestinal crypts through S6K activation by increasing the rate of translational elongation of yet to be defined polypeptides. Therefore, it is tempting to speculate that Elp3 may act in this pathway by promoting protein translation of those polypeptides through tRNA modifications.
As cancer stem cells critically rely on Elp3 to promote Wnt-dependent cancer development, and given the fact that Elp3 deficiency in normal intestine did not interfere with intestinal homeostasis, targeting Elp3 activity may represent a promising therapeutic approach to treat colon cancer. Future studies in human samples of intestinal cancers will be performed to generalize our conclusions and to validate the hypothesis that Elp3 is a suitable target to treat intestinal cancers showing constitutive Wnt signaling.
MATERIALS AND METHODS
Cell lines and reagents
HT29 and RKO cells were cultured in McCoy’s 5A medium supplemented with 10% FBS, 1% glutamine, and 1% penicillin/streptomycin. HEK 293 Lentix cells were cultured in DMEM medium supplemented with 10% FBS, 1% glutamine, and 1% penicillin/streptomycin.
Anti–β-catenin, -Elp1, and -Hsp90 antibodies used in Western blot analyses were obtained from Santa Cruz Biotechnology, Inc. The anti-Elp3 antibody was provided by Cell Signaling Technology. Anti–α-tubulin and –acetyl-α-tubulin antibodies were purchased from Sigma-Aldrich. Anti-Histone H3, -pH2AX, and -Dclk1 antibodies were obtained from Abcam, and anti-Sox9 and Ephrin B2 antibodies were purchased from EMD Millipore and R&D Systems, respectively.
Generating the conditional Elp3 KO mouse
The Elp3 gene (Ensembl gene ID, mouse ENSMUSG00000022031) is located on chromosome 14. Elp3 exon 1 harbors the translation initiation codon. The targeting strategy was designed to generate the conditional KO (cKO), as well as the constitutive KO alleles of the Elp3 gene (Taconic; Fig. 2 A). The targeting vector was generated using BAC clones from the C57BL/6J RPCIB-731 BAC library (clones RP23-233H18, RP23-220M13, RP23-236C3, RP23-435F17, and RP23-450B15) and transfected into the C57BL/6N Tac ES cell line (Taconic). Exon 2 was flanked by loxP sites, and the positive selection marker was flanked by F3 (Puromycin resistance [PuroR]) sites and inserted into intron 1. Homologous recombinant clones were isolated using positive (PuroR) selection. Mutant Elp3 mice were generated by Taconic. In brief, E3.5 blastocysts from superovulated BALB/cbfemales were injected with targeted C57BL/6 N.tac ES cells and transferred to pseudopregnant NMR1 females. Chimerism was determined according to the black/white coat color which reflects the contribution of ES cells to the BALB/c host. Highly chimeric mice were bred to C57BL/6 females and germline transmission was identified by C57BL/6 (black) in offspring was performed by Southern blot analysis. Crossbreeding of chimeric mice with Flp Deleter mice for in vivo selection marker deletion was performed to generate mice heterozygous for the conditional KO allele. The constitutive KO allele was obtained after Cre-mediated recombination. Deletion of exon 2 results in the loss of function of Elp3 by generating a frameshift to all downstream exons. For genotyping analysis, the floxed allele, defined as the conditional allele, was identified by PCR conducted on DNAs extracted from tails (Fig. 2 A). Primer sequences are available upon request. To inactivate Elp3 in the intestine (Elp3ΔIEC), the Elp3loxp/loxp mouse was crossed with the VillinCre/+ strain. PCR analysis using primers 1 and 2 were conducted on DNAs extracted from tissues of control or Elp3ΔIEC mice to amplify 1,341- and 172-bp products from the conditional versus the constitutive knockout allele, respectively (Fig. 2, A and B).
Transgenic mouse models, tamoxifen injections, and exposures to radiation
The Apc+/min strain (C57BL/6J-ApcMin/J), as well as the Lgr5-EGFP-IRES-creERT2 (B6.129P2-Lgr5tm1(cre/ESR1)Cle/J) heterozygote knock-in mouse were purchased from The Jackson Laboratory, whereas the VillinCre/+ strain was purchased from Genoway. The Apclox/lox strain was previously described (Sansom et al., 2007). All mice were in the C57BL/6 background, excepted for Apclox/lox mice that were three parts C57BL6 and one part Sv129. All experiments were approved by the local ethical committee (University of Liège, Liège, Belgium). For conditional inactivation of Elp3 in Lgr5+ cells, four daily injections of 2 mg tamoxifen in sunflower oil were performed intraperitoneally to the stem cell–specific Lgr5-EGFP-IRES-creERT2 knock-in mice as control (Lgr5-EGFP-Elp3Control) or to mice resulting from the breeding between Lgr5-EGFP-IRES-creERT2 mice and the Elp3Control strain to generate the Lgr5-EGFP-Elp3ΔCBCstrain. For the simultaneous inactivation of Elp3 and Apc in Lgr5+ cells, four daily injections of 2 mg tamoxifen in sunflower oil were performed intraperitoneally to mice resulting from the cross of Apclox/lox mice with the Lgr5-EGFP-IRES-creERT2 strain (ApcΔCBC) as control or to mice resulting from the cross of Apclox/lox mice with the Lgr5-EGFP-IRES-creERT2 and the Elp3Control strain (ApcΔCBC Elp3ΔCBC). The sequence of all primers used for genotyping purposes are available upon request. To assess radiation-induced intestinal regeneration, 8–10-wk-old mice were exposed to 14Gy from a 137Cs source, as previously described (Ashton et al., 2010). Mice were given buprenorphine twice a day and were sacrificed 72 h after radiation.
Intestinal epithelial cells extraction and ex vivo organoid cultures
Intestines and colons were extracted from mice and cecum was removed. Intestine was divided into three parts of the same size and defined as the proximal, middle, and distal intestine. Bowels were subsequently cut in half longitudinally. Half of the tissue was rolled and used for histological purposes and the other half was used for intestinal epithelial cells extraction. In brief, bowels were washed for 10 min at 37°C in a PBS-DTT (1 mM) buffer, and then incubated for 15 min at 37°C in a HBSS-EDTA buffer (30 mM). Cells were harvested, washed twice in PBS, and flash frozen.
Ex vivo organoid cultures were performed as previously described (Sato et al., 2009). In brief, small pieces of intestine were incubated in 2 mM EDTA-PBS for 30 min at 4°C. Crypts were extracted, washed twice in PBS and cultured at same density in Matrigel (BD). DMEM/F12 supplemented with EGF (20 ng/ml), Noggin (100 ng/ml), and R-Spondin (500 ng/ml) was added every 2 d. Apc-mutated organoids were cultured in DMEM/F12 supplemented with EGF (20 ng/ml) and Noggin (100 ng/ml) without R-Spondin. For the generation of organoids from ApcΔCBC and ApcΔCBC Elp3ΔCBC mice, animals were treated with tamoxifen.
Limiting dilution assay
Crypts from ApcΔCBC and ApcΔCBC Elp3ΔCBC mice treated with tamoxifen were extracted as described above. To generate a unicellular suspension of the extracted crypts, we digested them for 30 min at 37°C with 10× Trypsin and centrifuged them at 150 rcf for 5 min. The pellet was resuspended in 1%BSA/1× PBS and filtered in a 70-µm cell strainer to be centrifuged at 150 rcf for 5 min. Finally, the pellet was resuspended in 1 ml 1%BSA/1× PBS and EGFP-Lgr5+ cells were sorted using the FACSAria (BD). EGFP-Lgr5+ sorted cells were counted and seeded in Matrigel as indicated in the figure. DMEM/F12 supplemented with EGF (20 ng/ml) and Noggin (100 ng/ml) was added every 2 d. Wells positive for organoid growth were quantified after 7 d. Statistical analysis was performed by as previously described (Hu and Smyth, 2009).
Flow cytometry
Crypts were extracted after 30 min of incubation in 2 mM EDTA-PBS at 4°C, and then digested into single cells using the trypLE express solution (Life Technologies) containing 200 U of DNase for 30 min at 37°C. Cells were washed in 1% BSA/PBS. Resuspended cells were sorted on the BD FACSAria. For phenotyping, organoids were collected, washed in PBS, and digested for 20 min using the trypLE express solution. The resulting single-cell solution was fixed with 4% paraformaldehyde solution, permeabilized with Perm-Wash solution (BD), and stained with anti-Sox9 and -Dclk1 antibodies (Abcam) for 30 min at 4°C. Corresponding secondary antibodies (Life Technologies) were incubated for 30 min at 4°C. The FlowJo software was used to analyze data.
Total RNA extraction and real-time PCR
Total RNA from cell lines was extracted using the total RNA kit (Omega Biotek) according to manufacturer’s instructions. For mice samples, total RNA was extracted using the TriPure Isolation Reagent (Roche). All samples were subsequently treated with the Recombinant DNase (Roche). cDNAs were synthesized using the Revert Aid H Minus First Strand cDNA Synthesis kit (Fermentas). Subsequent PCRs were performed using the Power SYBR Green PCR Master kit (Takara Bio Inc.) on the LightCycler 480 (Roche). The mean value of mRNA levels in control mice was set to 100% and levels in mutant mice were expressed relative to that after normalization with glyceraldehyde-3-phosphate dehydrogenase GAPDH. Data are plotted as mean values ± SD. All primer sequences used are available upon request.
Lentiviral infections
The control or shRNAs targeting Dclk1, ELP3, β-catenin, or Sox9 were purchased from Sigma-Aldrich. Lentiviral infections in RKO orHT29 cells were performed as previously described (Creppe et al., 2009). Lentiviral infections in ex vivo organoids were performed as previously described (Koo et al., 2012).
Chromatin immunoprecipitation assays
Cells were fixed in formaldehyde 1% for 10 min at room temperature, quenched in 1.25 M glycine and lysed in SDS 1% lysis buffer. Lysates were sonicated 15 min using the Bioruptor (Diagenode), cleared by centrifugation and used for immunoprecipitation. Extracts were precleared by 1 h incubation with protein G/Herring sperm DNA/BSA, and immunoprecipitation was performed by incubating overnight at 4°C with the anti-TCF4 antibody, using IgG antibody as negative control, and then 1 h with protein G/Herring sperm DNA/BSA. Protein–DNA complexes were washed as per standard ChIP techniques. After elution, proteinase K treatment, and reversal of cross-links, DNA fragments were analyzed by quantitative real-time PCR using SYBr Green for detection. Input DNA was analyzed simultaneously and used as normalization. Primers used are available upon request.
Immunofluorescence and immunohistochemistry analyses
For immunohistochemistry analyses, samples were fixed overnight in 4% formalin solution and embedded in paraffin. In brief, 3-µm-thick sections of FFPE tumors were deparaffinized and antigen retrieval was performed by boiling the sections in citrate buffer at pH 6.0 or EDTA at pH 9.0 for 20 min. Primary antibodies used were as follows: Dclk1 (1:50; pH 6.0; Abcam), Sox9 (1:50; pH 6.0; Millipore), Ki67 (1:50; pH 6.0; Cell Marque), corresponding secondary antibody detection kits for reduced background on murine tissue were used (Histofine Simple Stain Mouse MAX PO; Medac) and stained on an automated stainer (LabVision Autostainer 480S; Thermo Fisher Scientific). All slides were scanned with a Panoramic 250 slide scanner (3D Histech.com).
For immunofluorescence, frozen sections were incubated in the target retrieval solution (Dako) for 7 min. Primary antibodies are listed in the Cell lines and reagents section. Slides were analyzed using a sp5 confocal microscope (Leica).
Expression of Sox9 in fission yeast and tRNA overexpression
The pLVX-Sox9-EGFP expression construct, a gift from S.T. Magness (University of North Carolina at Chapel Hill, Chapel Hill, NC), was used as template to generate the Sox9ΔAAA mutant by site-directed mutagenesis, using the protocol provided by the manufacturer (Agilent Technologies). Wild-type Sox9 and the Sox9ΔAAA cDNA mutant were amplified by PCR and cloned in the fission yeast ura4-based pREP4 that allows expression from the nmt1 promoter, which is repressed by thiamine. Wild-type and elp3-deleted strains were transformed with the pREP4-Sox9 plasmid and selected in EMM medium in the absence of uracil. The tRNAlysUUU cloned in a leu2-based vector as previously described (Bauer et al., 2012), or an empty vector, were also transformed and co-selected with the pREP4-sox9 on medium lacking both leucine and uracil. The expression of Sox9 was induced by growing cells for 22 h in the absence of thiamine. Cells were harvested at OD 0.5 (10 ml) and lysed in the presence of zirconium beads, as previously described (Fersht et al., 2007). Total protein extracts were subjected to anti-Sox9 and α-tubulin (loading control) Western blot analyses.
Statistical analysis
Student’s t test was performed to test statistical relevance, unless otherwise specified. For quantification of Elp3 immunohistochemistry of human biopsies, total H-DAB positive areas of each slide were calculated using the ImageJ software. Specifically, H-DAB positivity was quantified from the transformed images by ImageJ IHC Toolbox. Measurements were calculated in pixels, and the analysis was done blindly. Data are expressed as percentage of H-DAB–positive pixels on total pixels per slide.
ACKNOWLEDGMENTS
We thank S.T. Magness (University of North Carolina at Chapel Hill, Chapel Hill, NC) for the gift of the pLVX-Sox9-EGFP expression construct, as well as the GIGA Imaging and Flow Cytometry Facility.
This study was supported by grants from the Belgian National Funds for Scientific Research (FNRS), TELEVIE, the Belgian Federation against cancer, the University of Liege (Concerted Research Action Program [BIO-ACET]) and Fonds Spéciaux [C-11/03]), the Centre Anti-Cancéreux, the Leon Fredericq Fundation (ULg), as well as by the Walloon Excellence in Life Sciences and Biotechnology (WELBIO). P. Close and A. Chariot are Research Associate and Senior Research Associate at the FNRS, respectively.
The authors declare no competing financial interests.
Footnotes
Abbreviations used:
- CBC
- Crypt base columnar
- Dclk1
- Doublecortin-like kinase 1
- Hes1
- Hairy/enhancer of split1
- LRCs
- label-retaining cells
- Mcm5
- 5-methoxycarbonylmethyl
- SAM
- S-adenosylmethionine
References
- Akiyama H., Lyons J.P., Mori-Akiyama Y., Yang X., Zhang R., Zhang Z., Deng J.M., Taketo M.M., Nakamura T., Behringer R.R., et al. 2004. Interactions between Sox9 and beta-catenin control chondrocyte differentiation. Genes Dev. 18:1072–1087. 10.1101/gad.1171104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashton G.H., Morton J.P., Myant K., Phesse T.J., Ridgway R.A., Marsh V., Wilkins J.A., Athineos D., Muncan V., Kemp R., et al. 2010. Focal adhesion kinase is required for intestinal regeneration and tumorigenesis downstream of Wnt/c-Myc signaling. Dev. Cell. 19:259–269. 10.1016/j.devcel.2010.07.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayyanan A., Civenni G., Ciarloni L., Morel C., Mueller N., Lefort K., Mandinova A., Raffoul W., Fiche M., Dotto G.P., and Brisken C.. 2006. Increased Wnt signaling triggers oncogenic conversion of human breast epithelial cells by a Notch-dependent mechanism. Proc. Natl. Acad. Sci. USA. 103:3799–3804. 10.1073/pnas.0600065103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker N. 2014. Adult intestinal stem cells: critical drivers of epithelial homeostasis and regeneration. Nat. Rev. Mol. Cell Biol. 15:19–33. 10.1038/nrm3721 [DOI] [PubMed] [Google Scholar]
- Barker N., van Es J.H., Kuipers J., Kujala P., van den Born M., Cozijnsen M., Haegebarth A., Korving J., Begthel H., Peters P.J., and Clevers H.. 2007. Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature. 449:1003–1007. 10.1038/nature06196 [DOI] [PubMed] [Google Scholar]
- Barker N., Ridgway R.A., van Es J.H., van de Wetering M., Begthel H., van den Born M., Danenberg E., Clarke A.R., Sansom O.J., and Clevers H.. 2009. Crypt stem cells as the cells-of-origin of intestinal cancer. Nature. 457:608–611. 10.1038/nature07602 [DOI] [PubMed] [Google Scholar]
- Basak O., van de Born M., Korving J., Beumer J., van der Elst S., van Es J.H., and Clevers H.. 2014. Mapping early fate determination in Lgr5+ crypt stem cells using a novel Ki67-RFP allele. EMBO J. 33:2057–2068. 10.15252/embj.201488017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastide P., Darido C., Pannequin J., Kist R., Robine S., Marty-Double C., Bibeau F., Scherer G., Joubert D., Hollande F., et al. 2007. Sox9 regulates cell proliferation and is required for Paneth cell differentiation in the intestinal epithelium. J. Cell Biol. 178:635–648. 10.1083/jcb.200704152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer F., Matsuyama A., Candiracci J., Dieu M., Scheliga J., Wolf D.A., Yoshida M., and Hermand D.. 2012. Translational control of cell division by Elongator. Cell Reports. 1:424–433. 10.1016/j.celrep.2012.04.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhanja P., Saha S., Kabarriti R., Liu L., Roy-Chowdhury N., Roy-Chowdhury J., Sellers R.S., Alfieri A.A., and Guha C.. 2009. Protective role of R-spondin1, an intestinal stem cell growth factor, against radiation-induced gastrointestinal syndrome in mice. PLoS One. 4:e8014 10.1371/journal.pone.0008014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjerknes M., Khandanpour C., Möröy T., Fujiyama T., Hoshino M., Klisch T.J., Ding Q., Gan L., Wang J., Martín M.G., and Cheng H.. 2012. Origin of the brush cell lineage in the mouse intestinal epithelium. Dev. Biol. 362:194–218. 10.1016/j.ydbio.2011.12.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blache P., van de Wetering M., Duluc I., Domon C., Berta P., Freund J.N., Clevers H., and Jay P.. 2004. SOX9 is an intestine crypt transcription factor, is regulated by the Wnt pathway, and represses the CDX2 and MUC2 genes. J. Cell Biol. 166:37–47. 10.1083/jcb.200311021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buczacki S.J., Zecchini H.I., Nicholson A.M., Russell R., Vermeulen L., Kemp R., and Winton D.J.. 2013. Intestinal label-retaining cells are secretory precursors expressing Lgr5. Nature. 495:65–69. 10.1038/nature11965 [DOI] [PubMed] [Google Scholar]
- Chen C., Huang B., Eliasson M., Rydén P., and Byström A.S.. 2011. Elongator complex influences telomeric gene silencing and DNA damage response by its role in wobble uridine tRNA modification. PLoS Genet. 7:e1002258 10.1371/journal.pgen.1002258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clevers H. 2013. The intestinal crypt, a prototype stem cell compartment. Cell. 154:274–284. 10.1016/j.cell.2013.07.004 [DOI] [PubMed] [Google Scholar]
- Clevers H., and Nusse R.. 2012. Wnt/β-catenin signaling and disease. Cell. 149:1192–1205. 10.1016/j.cell.2012.05.012 [DOI] [PubMed] [Google Scholar]
- Close P., Hawkes N., Cornez I., Creppe C., Lambert C.A., Rogister B., Siebenlist U., Merville M.P., Slaugenhaupt S.A., Bours V., et al. 2006. Transcription impairment and cell migration defects in elongator-depleted cells: implication for familial dysautonomia. Mol. Cell. 22:521–531. 10.1016/j.molcel.2006.04.017 [DOI] [PubMed] [Google Scholar]
- Close P., Gillard M., Ladang A., Jiang Z., Papuga J., Hawkes N., Nguyen L., Chapelle J.P., Bouillenne F., Svejstrup J., et al. 2012. DERP6 (ELP5) and C3ORF75 (ELP6) regulate tumorigenicity and migration of melanoma cells as subunits of Elongator. J. Biol. Chem. 287:32535–32545. 10.1074/jbc.M112.402727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordero J.B., and Sansom O.J.. 2012. Wnt signalling and its role in stem cell-driven intestinal regeneration and hyperplasia. Acta Physiol. (Oxf.). 204:137–143. 10.1111/j.1748-1716.2011.02288.x [DOI] [PubMed] [Google Scholar]
- Creppe C., Malinouskaya L., Volvert M.L., Gillard M., Close P., Malaise O., Laguesse S., Cornez I., Rahmouni S., Ormenese S., et al. 2009. Elongator controls the migration and differentiation of cortical neurons through acetylation of alpha-tubulin. Cell. 136:551–564. 10.1016/j.cell.2008.11.043 [DOI] [PubMed] [Google Scholar]
- Esberg A., Huang B., Johansson M.J., and Byström A.S.. 2006. Elevated levels of two tRNA species bypass the requirement for elongator complex in transcription and exocytosis. Mol. Cell. 24:139–148. 10.1016/j.molcel.2006.07.031 [DOI] [PubMed] [Google Scholar]
- Faller W.J., Jackson T.J., Knight J.R., Ridgway R.A., Jamieson T., Karim S.A., Jones C., Radulescu S., Huels D.J., Myant K.B., et al. 2014. mTORC1-mediated translational elongation limits intestinal tumour initiation and growth. Nature. 517:497–500. 10.1038/nature13896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fersht N., Hermand D., Hayles J., and Nurse P.. 2007. Cdc18/CDC6 activates the Rad3-dependent checkpoint in the fission yeast. Nucleic Acids Res. 35:5323–5337. 10.1093/nar/gkm527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerbe F., van Es J.H., Makrini L., Brulin B., Mellitzer G., Robine S., Romagnolo B., Shroyer N.F., Bourgaux J.F., Pignodel C., et al. 2011. Distinct ATOH1 and Neurog3 requirements define tuft cells as a new secretory cell type in the intestinal epithelium. J. Cell Biol. 192:767–780. 10.1083/jcb.201010127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerbe F., Legraverend C., and Jay P.. 2012. The intestinal epithelium tuft cells: specification and function. Cell. Mol. Life Sci. 69:2907–2917. 10.1007/s00018-012-0984-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenwood C., Selth L.A., Dirac-Svejstrup A.B., and Svejstrup J.Q.. 2009. An iron-sulfur cluster domain in Elp3 important for the structural integrity of elongator. J. Biol. Chem. 284:141–149. 10.1074/jbc.M805312200 [DOI] [PubMed] [Google Scholar]
- Hattori T., Eberspaecher H., Lu J., Zhang R., Nishida T., Kahyo T., Yasuda H., and de Crombrugghe B.. 2006. Interactions between PIAS proteins and SOX9 result in an increase in the cellular concentrations of SOX9. J. Biol. Chem. 281:14417–14428. 10.1074/jbc.M511330200 [DOI] [PubMed] [Google Scholar]
- Hattori T., Coustry F., Stephens S., Eberspaecher H., Takigawa M., Yasuda H., and de Crombrugghe B.. 2008. Transcriptional regulation of chondrogenesis by coactivator Tip60 via chromatin association with Sox9 and Sox5. Nucleic Acids Res. 36:3011–3024. 10.1093/nar/gkn150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y., and Smyth G.K.. 2009. ELDA: extreme limiting dilution analysis for comparing depleted and enriched populations in stem cell and other assays. J. Immunol. Methods. 347:70–78. 10.1016/j.jim.2009.06.008 [DOI] [PubMed] [Google Scholar]
- Huang B., Johansson M.J., and Byström A.S.. 2005. An early step in wobble uridine tRNA modification requires the Elongator complex. RNA. 11:424–436. 10.1261/rna.7247705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen J., Pedersen E.E., Galante P., Hald J., Heller R.S., Ishibashi M., Kageyama R., Guillemot F., Serup P., and Madsen O.D.. 2000. Control of endodermal endocrine development by Hes-1. Nat. Genet. 24:36–44. 10.1038/71657 [DOI] [PubMed] [Google Scholar]
- Johansen L.D., Naumanen T., Knudsen A., Westerlund N., Gromova I., Junttila M., Nielsen C., Bøttzauw T., Tolkovsky A., Westermarck J., et al. 2008. IKAP localizes to membrane ruffles with filamin A and regulates actin cytoskeleton organization and cell migration. J. Cell Sci. 121:854–864. 10.1242/jcs.013722 [DOI] [PubMed] [Google Scholar]
- Kim J.H., Lane W.S., and Reinberg D.. 2002. Human Elongator facilitates RNA polymerase II transcription through chromatin. Proc. Natl. Acad. Sci. USA. 99:1241–1246. 10.1073/pnas.251672198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koo B.K., Stange D.E., Sato T., Karthaus W., Farin H.F., Huch M., van Es J.H., and Clevers H.. 2012. Controlled gene expression in primary Lgr5 organoid cultures. Nat. Methods. 9:81–83. 10.1038/nmeth.1802 [DOI] [PubMed] [Google Scholar]
- Kristjuhan A., Walker J., Suka N., Grunstein M., Roberts D., Cairns B.R., and Svejstrup J.Q.. 2002. Transcriptional inhibition of genes with severe histone h3 hypoacetylation in the coding region. Mol. Cell. 10:925–933. 10.1016/S1097-2765(02)00647-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane S.W., Sykes S.M., Al-Shahrour F., Shterental S., Paktinat M., Lo Celso C., Jesneck J.L., Ebert B.L., Williams D.A., and Gilliland D.G.. 2010. The Apc(min) mouse has altered hematopoietic stem cell function and provides a model for MPD/MDS. Blood. 115:3489–3497. 10.1182/blood-2009-11-251728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsimont J.C., Youssef K.K., Sánchez-Danés A., Sukumaran V., Defrance M., Delatte B., Liagre M., Baatsen P., Marine J.C., Lippens S., et al. 2015. Sox9 Controls Self-Renewal of Oncogene Targeted Cells and Links Tumor Initiation and Invasion. Cell Stem Cell. 17:60–73. 10.1016/j.stem.2015.05.008 [DOI] [PubMed] [Google Scholar]
- Lee G., Papapetrou E.P., Kim H., Chambers S.M., Tomishima M.J., Fasano C.A., Ganat Y.M., Menon J., Shimizu F., Viale A., et al. 2009. Modelling pathogenesis and treatment of familial dysautonomia using patient-specific iPSCs. Nature. 461:402–406. 10.1038/nature08320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q., Fazly A.M., Zhou H., Huang S., Zhang Z., and Stillman B.. 2009. The elongator complex interacts with PCNA and modulates transcriptional silencing and sensitivity to DNA damage agents. PLoS Genet. 5:e1000684 10.1371/journal.pgen.1000684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin F.J., Shen L., Jang C.W., Falnes P.O., and Zhang Y.. 2013. Ikbkap/Elp1 deficiency causes male infertility by disrupting meiotic progression. PLoS Genet. 9:e1003516 10.1371/journal.pgen.1003516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- May R., Qu D., Weygant N., Chandrakesan P., Ali N., Lightfoot S.A., Li L., Sureban S.M., and Houchen C.W.. 2014. Brief report: Dclk1 deletion in tuft cells results in impaired epithelial repair after radiation injury. Stem Cells. 32:822–827. 10.1002/stem.1566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merlos-Suárez A., Barriga F.M., Jung P., Iglesias M., Céspedes M.V., Rossell D., Sevillano M., Hernando-Momblona X., da Silva-Diz V., Muñoz P., et al. 2011. The intestinal stem cell signature identifies colorectal cancer stem cells and predicts disease relapse. Cell Stem Cell. 8:511–524. 10.1016/j.stem.2011.02.020 [DOI] [PubMed] [Google Scholar]
- Merritt A.J., Potten C.S., Kemp C.J., Hickman J.A., Balmain A., Lane D.P., and Hall P.A.. 1994. The role of p53 in spontaneous and radiation-induced apoptosis in the gastrointestinal tract of normal and p53-deficient mice. Cancer Res. 54:614–617. [PubMed] [Google Scholar]
- Metcalfe C., Kljavin N.M., Ybarra R., and de Sauvage F.J.. 2013. Lgr5+ stem cells are indispensable for radiation-induced intestinal regeneration. Cell Stem Cell. 14:149–159. 10.1016/j.stem.2013.11.008 [DOI] [PubMed] [Google Scholar]
- Michaelson J.S., and Leder P.. 2001. beta-catenin is a downstream effector of Wnt-mediated tumorigenesis in the mammary gland. Oncogene. 20:5093–5099. 10.1038/sj.onc.1204586 [DOI] [PubMed] [Google Scholar]
- Montgomery R.K., Carlone D.L., Richmond C.A., Farilla L., Kranendonk M.E., Henderson D.E., Baffour-Awuah N.Y., Ambruzs D.M., Fogli L.K., Algra S., and Breault D.T.. 2011. Mouse telomerase reverse transcriptase (mTert) expression marks slowly cycling intestinal stem cells. Proc. Natl. Acad. Sci. USA. 108:179–184. 10.1073/pnas.1013004108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori-Akiyama Y., van den Born M., van Es J.H., Hamilton S.R., Adams H.P., Zhang J., Clevers H., and de Crombrugghe B.. 2007. SOX9 is required for the differentiation of paneth cells in the intestinal epithelium. Gastroenterology. 133:539–546. 10.1053/j.gastro.2007.05.020 [DOI] [PubMed] [Google Scholar]
- Morin P.J., Sparks A.B., Korinek V., Barker N., Clevers H., Vogelstein B., and Kinzler K.W.. 1997. Activation of beta-catenin-Tcf signaling in colon cancer by mutations in beta-catenin or APC. Science. 275:1787–1790. 10.1126/science.275.5307.1787 [DOI] [PubMed] [Google Scholar]
- Muñoz J., Stange D.E., Schepers A.G., van de Wetering M., Koo B.K., Itzkovitz S., Volckmann R., Kung K.S., Koster J., Radulescu S., et al. 2012. The Lgr5 intestinal stem cell signature: robust expression of proposed quiescent '+4′ cell markers. EMBO J. 31:3079–3091. 10.1038/emboj.2012.166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakanishi Y., Seno H., Fukuoka A., Ueo T., Yamaga Y., Maruno T., Nakanishi N., Kanda K., Komekado H., Kawada M., et al. 2013. Dclk1 distinguishes between tumor and normal stem cells in the intestine. Nat. Genet. 45:98–103. 10.1038/ng.2481 [DOI] [PubMed] [Google Scholar]
- Otero G., Fellows J., Li Y., de Bizemont T., Dirac A.M., Gustafsson C.M., Erdjument-Bromage H., Tempst P., and Svejstrup J.Q.. 1999. Elongator, a multisubunit component of a novel RNA polymerase II holoenzyme for transcriptional elongation. Mol. Cell. 3:109–118. 10.1016/S1097-2765(00)80179-3 [DOI] [PubMed] [Google Scholar]
- Potten C.S., Hume W.J., Reid P., and Cairns J.. 1978. The segregation of DNA in epithelial stem cells. Cell. 15:899–906. 10.1016/0092-8674(78)90274-X [DOI] [PubMed] [Google Scholar]
- Powell A.E., Wang Y., Li Y., Poulin E.J., Means A.L., Washington M.K., Higginbotham J.N., Juchheim A., Prasad N., Levy S.E., et al. 2012. The pan-ErbB negative regulator Lrig1 is an intestinal stem cell marker that functions as a tumor suppressor. Cell. 149:146–158. 10.1016/j.cell.2012.02.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramocki N.M., Wilkins H.R., Magness S.T., Simmons J.G., Scull B.P., Lee G.H., McNaughton K.K., and Lund P.K.. 2008. Insulin receptor substrate-1 deficiency promotes apoptosis in the putative intestinal crypt stem cell region, limits Apcmin/+ tumors, and regulates Sox9. Endocrinology. 149:261–267. 10.1210/en.2007-0869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roche K.C., Gracz A.D., Liu X.F., Newton V., Akiyama H., and Magness S.T.. 2015. SOX9 Maintains Reserve Stem Cells and Preserves Radioresistance in Mouse Small Intestine. Gastroenterology. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sangiorgi E., and Capecchi M.R.. 2008. Bmi1 is expressed in vivo in intestinal stem cells. Nat. Genet. 40:915–920. 10.1038/ng.165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sansom O.J., Meniel V.S., Muncan V., Phesse T.J., Wilkins J.A., Reed K.R., Vass J.K., Athineos D., Clevers H., and Clarke A.R.. 2007. Myc deletion rescues Apc deficiency in the small intestine. Nature. 446:676–679. 10.1038/nature05674 [DOI] [PubMed] [Google Scholar]
- Saqui-Salces M., Keeley T.M., Grosse A.S., Qiao X.T., El-Zaatari M., Gumucio D.L., Samuelson L.C., and Merchant J.L.. 2011. Gastric tuft cells express DCLK1 and are expanded in hyperplasia. Histochem. Cell Biol. 136:191–204. 10.1007/s00418-011-0831-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato T., Vries R.G., Snippert H.J., van de Wetering M., Barker N., Stange D.E., van Es J.H., Abo A., Kujala P., Peters P.J., and Clevers H.. 2009. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature. 459:262–265. 10.1038/nature07935 [DOI] [PubMed] [Google Scholar]
- Schepers A.G., Snippert H.J., Stange D.E., van den Born M., van Es J.H., van de Wetering M., and Clevers H.. 2012. Lineage tracing reveals Lgr5+ stem cell activity in mouse intestinal adenomas. Science. 337:730–735. 10.1126/science.1224676 [DOI] [PubMed] [Google Scholar]
- Schwitalla S., Fingerle A.A., Cammareri P., Nebelsiek T., Göktuna S.I., Ziegler P.K., Canli O., Heijmans J., Huels D.J., Moreaux G., et al. 2013. Intestinal tumorigenesis initiated by dedifferentiation and acquisition of stem-cell-like properties. Cell. 152:25–38. 10.1016/j.cell.2012.12.012 [DOI] [PubMed] [Google Scholar]
- Selvadurai K., Wang P., Seimetz J., and Huang R.H.. 2014. Archaeal Elp3 catalyzes tRNA wobble uridine modification at C5 via a radical mechanism. Nat. Chem. Biol. 10:810–812. 10.1038/nchembio.1610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Z., Chiang C.I., Mistretta T.A., Major A., and Mori-Akiyama Y.. 2013. SOX9 directly regulates IGFBP-4 in the intestinal epithelium. Am. J. Physiol. Gastrointest. Liver Physiol. 305:G74–G83. 10.1152/ajpgi.00086.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shroyer N.F., Helmrath M.A., Wang V.Y., Antalffy B., Henning S.J., and Zoghbi H.Y.. 2007. Intestine-specific ablation of mouse atonal homolog 1 (Math1) reveals a role in cellular homeostasis. Gastroenterology. 132:2478–2488. 10.1053/j.gastro.2007.03.047 [DOI] [PubMed] [Google Scholar]
- Su L.K., Kinzler K.W., Vogelstein B., Preisinger A.C., Moser A.R., Luongo C., Gould K.A., and Dove W.F.. 1992. Multiple intestinal neoplasia caused by a mutation in the murine homolog of the APC gene. Science. 256:668–670. 10.1126/science.1350108 [DOI] [PubMed] [Google Scholar]
- Takeda N., Jain R., LeBoeuf M.R., Wang Q., Lu M.M., and Epstein J.A.. 2011. Interconversion between intestinal stem cell populations in distinct niches. Science. 334:1420–1424. 10.1126/science.1213214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian H., Biehs B., Warming S., Leong K.G., Rangell L., Klein O.D., and de Sauvage F.J.. 2011. A reserve stem cell population in small intestine renders Lgr5-positive cells dispensable. Nature. 478:255–259. 10.1038/nature10408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Flier L.G., van Gijn M.E., Hatzis P., Kujala P., Haegebarth A., Stange D.E., Begthel H., van den Born M., Guryev V., Oving I., et al. 2009. Transcription factor achaete scute-like 2 controls intestinal stem cell fate. Cell. 136:903–912. 10.1016/j.cell.2009.01.031 [DOI] [PubMed] [Google Scholar]
- van de Wetering M., Sancho E., Verweij C., de Lau W., Oving I., Hurlstone A., van der Horn K., Batlle E., Coudreuse D., Haramis A.P., et al. 2002. The beta-catenin/TCF-4 complex imposes a crypt progenitor phenotype on colorectal cancer cells. Cell. 111:241–250. 10.1016/S0092-8674(02)01014-0 [DOI] [PubMed] [Google Scholar]
- Van Landeghem L., Santoro M.A., Krebs A.E., Mah A.T., Dehmer J.J., Gracz A.D., Scull B.P., McNaughton K., Magness S.T., and Lund P.K.. 2012. Activation of two distinct Sox9-EGFP-expressing intestinal stem cell populations during crypt regeneration after irradiation. Am. J. Physiol. Gastrointest. Liver Physiol. 302:G1111–G1132. 10.1152/ajpgi.00519.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma U.N., Surabhi R.M., Schmaltieg A., Becerra C., and Gaynor R.B.. 2003. Small interfering RNAs directed against beta-catenin inhibit the in vitro and in vivo growth of colon cancer cells. Clin. Cancer Res. 9:1291–1300. [PubMed] [Google Scholar]
- Wang Z., Vogelstein B., and Kinzler K.W.. 2003. Phosphorylation of beta-catenin at S33, S37, or T41 can occur in the absence of phosphorylation at T45 in colon cancer cells. Cancer Res. 63:5234–5235. [PubMed] [Google Scholar]
- Westphalen C.B., Asfaha S., Hayakawa Y., Takemoto Y., Lukin D.J., Nuber A.H., Brandtner A., Setlik W., Remotti H., Muley A., et al. 2014. Long-lived intestinal tuft cells serve as colon cancer-initiating cells. J. Clin. Invest. 124:1283–1295. 10.1172/JCI73434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkler G.S., Kristjuhan A., Erdjument-Bromage H., Tempst P., and Svejstrup J.Q.. 2002. Elongator is a histone H3 and H4 acetyltransferase important for normal histone acetylation levels in vivo. Proc. Natl. Acad. Sci. USA. 99:3517–3522. 10.1073/pnas.022042899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Withers H.R., and Elkind M.M.. 1970. Microcolony survival assay for cells of mouse intestinal mucosa exposed to radiation. Int. J. Radiat. Biol. Relat. Stud. Phys. Chem. Med. 17:261–267. 10.1080/09553007014550291 [DOI] [PubMed] [Google Scholar]
- Wittschieben B.O., Otero G., de Bizemont T., Fellows J., Erdjument-Bromage H., Ohba R., Li Y., Allis C.D., Tempst P., and Svejstrup J.Q.. 1999. A novel histone acetyltransferase is an integral subunit of elongating RNA polymerase II holoenzyme. Mol. Cell. 4:123–128. 10.1016/S1097-2765(00)80194-X [DOI] [PubMed] [Google Scholar]
- Yang Q., Bermingham N.A., Finegold M.J., and Zoghbi H.Y.. 2001. Requirement of Math1 for secretory cell lineage commitment in the mouse intestine. Science. 294:2155–2158. 10.1126/science.1065718 [DOI] [PubMed] [Google Scholar]
- Yilmaz O.H., Katajisto P., Lamming D.W., Gültekin Y., Bauer-Rowe K.E., Sengupta S., Birsoy K., Dursun A., Yilmaz V.O., Selig M., et al. 2012. mTORC1 in the Paneth cell niche couples intestinal stem-cell function to calorie intake. Nature. 486:490–495. 10.1038/nature11163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou W.J., Geng Z.H., Spence J.R., and Geng J.G.. 2013. Induction of intestinal stem cells by R-spondin 1 and Slit2 augments chemoradioprotection. Nature. 501:107–111. 10.1038/nature12416 [DOI] [PMC free article] [PubMed] [Google Scholar]