Shakya et al. identify the transcription factor Oct1 and its cofactor OCA-B as central mediators for generating memory T cell responses in mice.
Abstract
Epigenetic changes are crucial for the generation of immunological memory. Failure to generate or maintain these changes will result in poor memory responses. Similarly, augmenting or stabilizing the correct epigenetic states offers a potential method of enhancing memory. Yet the transcription factors that regulate these processes are poorly defined. We find that the transcription factor Oct1 and its cofactor OCA-B are selectively required for the in vivo generation of CD4+ memory T cells. More importantly, the memory cells that are formed do not respond properly to antigen reencounter. In vitro, both proteins are required to maintain a poised state at the Il2 target locus in resting but previously stimulated CD4+ T cells. OCA-B is also required for the robust reexpression of multiple other genes including Ifng. ChIPseq identifies ∼50 differentially expressed direct Oct1 and OCA-B targets. We identify an underlying mechanism involving OCA-B recruitment of the histone lysine demethylase Jmjd1a to targets such as Il2, Ifng, and Zbtb32. The findings pinpoint Oct1 and OCA-B as central mediators of CD4+ T cell memory.
Memory immune responses provide critical protection against viral, microbial, and parasitic pathogens. Understanding memory generation is therefore an area of paramount importance for vaccine development, but one in which our understanding remains inadequate. CD4+ memory T cells persist for long periods and provide protection as both central/circulating and tissue-resident cells (Hammarlund et al., 2003; Darrah et al., 2007; Williams et al., 2008; Pepper et al., 2011; Iijima and Iwasaki, 2014; Zens and Farber, 2015). Although transcription regulators such as T-bet, Eomes, Blimp-1, Bcl-6, STAT3, and Foxo1 have been implicated in the generation of T cell memory, mostly in CD8+ cells (Ichii et al., 2002, 2007; Intlekofer et al., 2007; Joshi et al., 2007; Kallies et al., 2009; Rutishauser et al., 2009; Banerjee et al., 2010; Cui et al., 2011; Rao et al., 2012; Hess Michelini et al., 2013), little is known regarding the mechanism, in particular with respect to CD4 memory. Furthermore, factors selectively regulating memory cell populations are not well described.
OCA-B/OBF-1/Bob.1 (gene symbol Pou2af1) is a transcriptional coactivator widely expressed along the developing B lymphocyte axis, but not during T cell development or in naive CD4+ T cells. Despite the fact that it is induced after T cell activation (Sauter and Matthias, 1997; Zwilling et al., 1997; Mueller et al., 2013), loss of OCA-B only minimally affects initial cytokine production at short time points after initial activation (Kim et al., 1996; Schubart et al., 1996; Brunner et al., 2007; Yosef et al., 2013). OCA-B localizes to target DNA by interacting with the sequence-specific DNA-binding transcription factors Oct1 (Pou2f1) and Oct2 (Pou2f2). Oct1 is constitutively expressed in CD4+ T cells (Garrity et al., 1994). Il2 is a T cell–specific Oct1 target (Ullman et al., 1991). This key cytokine is induced in naive CD4+ T cells after activation but expressed more robustly upon restimulation of previously stimulated T cells (Murayama et al., 2006). In vitro, poising of Il2 for later robust expression requires Oct1 (Shakya et al., 2011). To maintain a poised state, Oct1 recruits Jmjd1a/KDM3A to the Il2 promoter. Jmjd1a is a histone lysine demethylase that catalyzes the removal of histone H3K9me1 and -me2 marks (Yamane et al., 2006). Jmjd1a does not associate with Oct1 at Il2 in naive cells but rapidly associates after T cell activation. The MEK-ERK arm of the MAPK signaling pathway is required for initial association. In rested cells, Jmjd1a remains associated in the absence of MAPK activity (Shakya et al., 2011). This result suggested that another activity localizes Jmjd1a to Oct1 at the Il2 promoter at long time points.
Here we show that OCA-B is required for Jmjd1a association with Il2 specifically in resting but previously stimulated CD4+ T cells. Restimulation of OCA-B–deficient cells results in defective Il2 expression. Furthermore, we show that OCA-B is required for robust activity of multiple Oct1/OCA-B target genes in the restimulated condition. Using pathogen infection models, we show that Oct1 and OCA-B are both required for robust memory responses in vivo. These results identify Oct1 and its cofactor OCA-B as fundamental determinants of CD4 T cell memory, identify the relevant targets, and delineate a mechanism involving removal of negative epigenetic marks.
RESULTS
OCA-B is induced after naive CD4+ T cell activation and localizes Jmjd1a to Il2
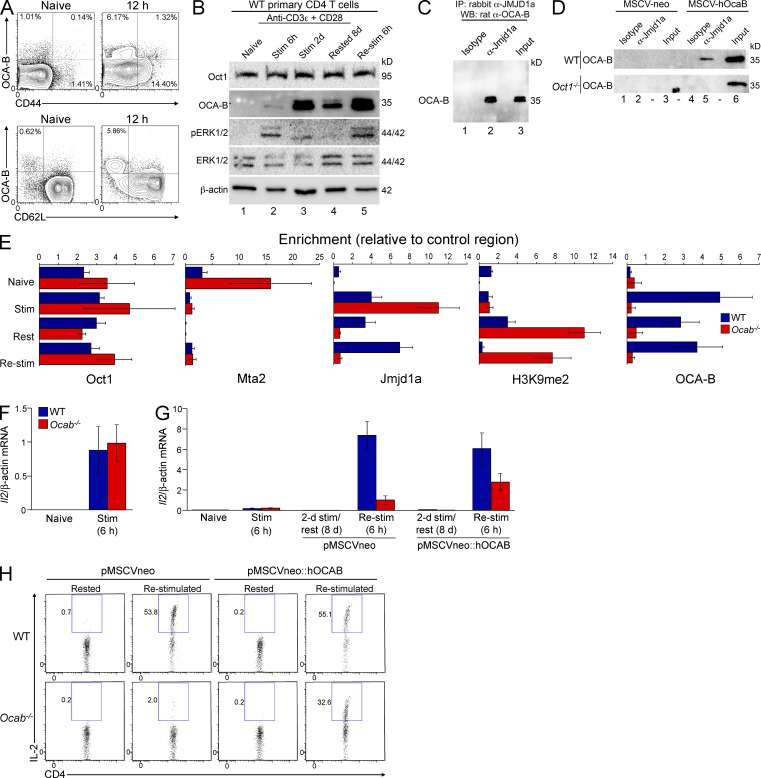
OCA-B is induced after T cell activation (Sauter and Matthias, 1997; Zwilling et al., 1997; Mueller et al., 2013). We corroborated the OCA-B induction using intracellular staining and Western blotting with primary naive mouse CD4+ T cells (CD8aneg,CD11bneg,CD45Rneg,DX5neg,Ter-119neg,CD44lo) stimulated with CD3ε and CD28 antibodies. OCA-B is not expressed in naive cells but becomes expressed around 6 h (Fig. 1, A and B). OCA-B persists in cells stimulated for 2 d and rested for an additional 8 d (Fig. 1 B, lane 4). We also used phospho-ERK1/2 antibodies to confirm the absence of MAPK activity in resting cells (Fig. 1 B).
Figure 1.
OCA-B localizes Jmjd1a to the Il2 promoter at long time points after T cell activation and is required for robust Il2 expression in rested but previously stimulated primary T cells. (A) Naive mouse splenic CD4+ T cells were stimulated in vitro for 12 h and stained for CD44, CD62L, and intracellular OCA-B. Naive cells are shown as a control. (B) Western blots showing the time course of naive helper T cell polyclonal activation. OCA-B induction is shown, as is phospho-ERK1/2 status. Oct1, ERK1/2, and β-actin are shown as controls. (C) 100 µg of total primary T cell extract in RIPA buffer was used for IP with anti-Jmjd1a antibody or isotype control. OCA-B Western blot (WB) is shown. Endogenous proteins were used. 2.5% input is shown in lane 3. (D) Similar to C except 100 µg of total WT or Oct1−/− 3T3 MEF extract in RIPA buffer was used. Human OCA-B was introduced by viral transduction. (E) ChIP-qPCR was performed at the Il2 promoter using purified naive T cells (Naive), 6-h-stimulated cells (Stim), cells stimulated for 2 d and cultured for 8 d in the absence of stimulus (Rested), or the same cells stimulated for 6 h (Re-stim). Antibodies specific to Oct1, Jmjd1a, Mta2 (NuRD), OCA-B, and H3K9me2 were used. Enrichment was calculated relative to a control genomic region, isotype control antibody, and standard input DNA. Values depict mean ± SD of three biological replicates. Differences in absolute levels of enrichment reflect variability in antibody properties. (F) WT and Ocab−/− cells were stimulated for 6 h, and Il2 mRNA expression was assessed using TaqMan RT-qPCR. mRNA levels were normalized to β-actin. Triplicate results are shown ± SD. (G) Il2 mRNA expression was measured in Naive, Stim, Rested, or Re-stim WT and Ocab−/− cells as in F. Expanding cells were additionally infected using MSCV (empty vector or encoding human OCA-B). Cells were not drug selected. Naive and Stim mRNA expression data are identical to F, except plotted on a different y axis. (H) Cells stimulated as in G were subjected to intracellular cytokine staining using antibodies against IL-2 and assessed by flow cytometry.
OCA-B interacts with Jmjd1a in coimmunoprecipitation (co-IP) assays using 2-d-stimulated WT naive CD4+ T cells (Fig. 1 C). To determine whether this interaction requires Oct1, we used Oct1-deficient MEFs (Tantin et al., 2005) stably transduced with murine stem cell virus (MSCV) retroviruses encoding human OCA-B or empty vector controls. Jmjd1a efficiently coimmunoprecipitated ectopically expressed OCA-B in WT but not Oct1-deficient fibroblasts (Fig. 1 D), indicating that Oct1 helps mediate the OCA-B/Jmjd1a interaction.
In naive T cells, Oct1 associates with an inhibitory chromatin remodeling complex known as nucleosome-remodeling deacetylase (NuRD) at the Il2 promoter, but rapidly switches to Jmjd1a after stimulation. Switching requires the MEK-ERK arm of the MAPK signaling pathway (Shakya et al., 2011). After removal of the stimulus, Jmjd1a is retained even in the absence of MAPK signals. To test whether OCA-B was the activity responsible for maintaining Jmjd1a at Il2, we performed chromatin IP (ChIP)–quantitative PCR (qPCR) using primary naive splenic WT or Pou2af1−/− (hereafter called Ocab−/−) CD4+ T cells. Cells were taken directly ex vivo (Naive), stimulated for 6 h (Stim), stimulated for 2 d and allowed to rest for 8 d (Rested), or rested and restimulated for 6 h (Re-stim). Oct1 constitutively associates with Il2 (Fig. 1 E). After T cell stimulation, MTA2 (a subunit of NuRD) dissociates from Il2 and Jmjd1a associates. Jmjd1a association was independent of OCA-B in Stim cells, as expected because ERK1/2 mediates the Oct1–Jmjd1a interaction at this time point (Shakya et al., 2011). Rested normal T cells maintain Jmjd1a association with Il2 simultaneously with the presence of H3K9me2, consistent with prior findings (Shakya et al., 2011). T cell stimulation may therefore augment Jmjd1a activity and/or decrease the activity of one or more H3K9me2 methyltransferases. In contrast to normal cells, Ocab−/− cells do not maintain association with Jmjd1a (Fig. 1 E). OCA-B is therefore required to localize Jmjd1a to Il2 selectively at long time points. OCA-B is also required to recruit Jmjd1a in Re-stim cells. Failure to recruit Jmjd1a is associated with H3K9me2 accumulation specifically at long time points in the OCA-B–deficient condition (Fig. 1 E).
OCA-B promotes robust secondary Il2 responses
We tested Il2 expression in Naive, Stim, Rested, or Re-stim OCA-B–deficient CD4+ T cells. For these experiments. we transduced stimulated Ocab−/− T cells with MSCV retroviruses encoding OCA-B or empty vector controls. 6-h-stimulated Ocab−/− cells displayed no difference in Il2 expression compared with controls (Fig. 1 F). Cells of both genotypes also expanded equivalently after initial stimulation (not depicted). Re-stim WT cells displayed a >20-fold increase in Il2 expression over the prior stimulation, as expected. In contrast, Ocab−/− Re-stim cells display a severe defect, behaving similarly to naive cells (Fig. 1 G). The expression defect was partially rescued by transduction (∼40% efficiency) with MSCV-hOcab, indicating that the OCA-B requirement is cell intrinsic. Flow cytometry analysis produced similar results for IL-2 production (Fig. 1 H). These findings indicate that OCA-B is required for robust Il2 expression specifically in previously stimulated CD4+ T cells.
Oct1 and OCA-B coordinate poised memory gene expression programs in previously stimulated T cells
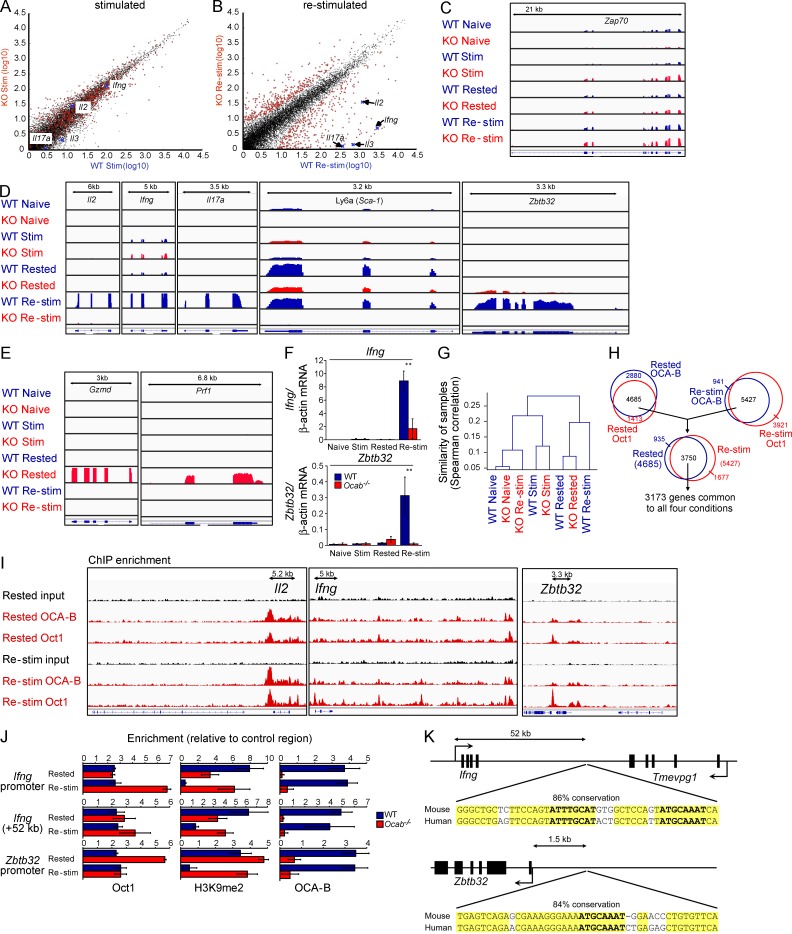
To test whether OCA-B similarly regulates additional genes, we performed RNAseq. Between 25 and 30 million sequence reads were generated for each sample, ∼50% of which aligned perfectly to the mouse reference genome. After initial activation, few gene expression changes were noted between stimulated WT and Ocab−/− cells (Fig. 2 A). In contrast, 110 genes normally induced more than twofold upon Re-stim showed a more than twofold diminution in Ocab−/− cells compared with WT (Fig. 2 B, Table 1, and Table S2). This set includes Il2 (#12), as well as Il9 (#3), Ifng (#5), Il17a (#9), and Il6 (#21). In addition, the stem cell marker Ly6a (Sca-1) was poorly expressed, as was Zbtb32 (Plzp). Zbtb32 encodes a transcriptional repressor of Prdm1/Blimp1 (Beaulieu et al., 2014), a known modulator of T cell memory (Rutishauser et al., 2009). RNAseq read density tracks for Zap70, a component of the T cell receptor signaling complex, are shown as a control in Fig. 2 C. Il2, Ifng, Il17a, Ly6a, and Zbtb32 are shown in Fig. 2 D. In addition, a small group of genes encoding CTL effector molecules was inappropriately expressed in Rested Ocab−/− cells, including Gzmd and Prf1 (Fig. 2 E) and the linked loci Gzme and Gzmg (not depicted). We independently validated differential expression of several genes including Il2 (not depicted), Ifng, and Zbtb32 using RT-qPCR (Fig. 2 F). Transcriptome clustering indicated that Naive, Stim, and Rested WT and Ocab−/− cells were most similar to each other, whereas Re-stim Ocab−/− cells clustered with naive cells (Fig. 2 G).
Figure 2.
OCA-B coordinates the expression of multiple direct target genes in Re-stim primary CD4+ T cells. (A) Mouse WT and Ocab−/− primary CD4+ T cells were isolated, stimulated for 6 h, and prepared for RNAseq. Gene expression scatter plot is shown from the two cell types based on cufflinks. Il17a, Il2, Il3, and Ifng are highlighted for purposes of comparison with B. Significantly changed genes in B are shown in red. Displayed values are log10 (fragments per kilobase per million + 1). (B) Similar data (as in A) using Re-stim cells. Differentially expressed genes are marked in red. (C) RNAseq read densities across mouse Zap70 were plotted as custom tracks using the integrated genome viewer (IGV), confirming RNAseq quality and normalization. Naive, Stim, Rested, and Re-stim cells are shown ± OCA-B. (D) Read densities for Il2, Ifng, Il17a, Ly6a, and Zbtb32. (E) Gzmd and Prf1. (F) Ifng and Zbtb32 mRNA levels were assessed by RT-qPCR using primary T cells stimulated as in A and B in biological triplicate. Error bars depict ±SD. **, Student’s t test p-value ≤0.005. (G) Spearman correlation values were determined for the entire alignable RNAseq dataset and plotted as a dendrogram. (H) Overlapping target genes identified by ChIPseq using antibodies against Oct1 and OCA-B in Rested and Re-stim T cells. 3,750 enriched regions corresponding to 3,173 nearby genes were identified in the intersection between the two conditions (Rested and Re-stim T cells) and two antibodies (Oct1 and OCA-B). (I) IGV images of Oct1 and OCA-B ChIPseq read densities in Rested and Re-stim T cells. (J) ChIP-qPCR using the mouse Il2, Ifng, and Zbtb32 promoter regions and an area downstream of Ifng. Enrichment over background for Oct1, H3K9me2, and OCA-B is shown in Rested and Re-stim WT and Ocab−/− primary CD4+ T cells. Values depict mean ± SD of three biological replicates. (K) Schematic and DNA sequence of two regions enriched by ChIPseq. (top) Element identified between Ifng and the Tmevpg1 lncRNA containing a dyad octamer motif conserved to humans. (bottom) Conserved element upstream of Zbtb32. Yellow highlighting indicates conserved nucleotides. Bold indicates canonical octamer motif.
Table 1. Top differentially down-regulated genes in Re-stim OCA-B/Pou2af1-deficient T cells relative to WT.
| No. | Gene | Description | Fold restimulation Ocab−/−/WT (log2) | Target in combined ChIPseq?a |
|---|---|---|---|---|
| 1 | CCL1 | chemokine (C-C motif) ligand 1 | −9.01 | |
| 2 | GZMB | granzyme B | −8.92 | |
| 3 | IL9 | interleukin 9 | −8.57 | |
| 4 | IFNG | interferon gamma | −8.49 | Yes |
| 5 | PENK | Preproenkephalin | −8.38 | |
| 6 | IL3 | interleukin 3 | −7.19 | |
| 7 | IL5 | interleukin 5 | −7.04 | Yes |
| 8 | IL22 | interleukin 22 | −6.43 | |
| 9 | IL17A | interleukin 17A | −6.33 | |
| 10 | CCL4 | chemokine (C-C motif) ligand 4 | −5.63 | |
| 11 | INHBA | inhibin beta-A | −5.30 | |
| 12 | IL2 | interleukin 2 | −5.00 | Yes |
| 13 | CCL3 | chemokine (C-C motif) ligand 3 | −4.91 | Yes |
| 14 | IL13 | interleukin 13 | −4.49 | |
| 15 | IL24 | interleukin 24 | −4.49 | |
| 16 | IL10 | interleukin 10 | −4.45 | Yes |
| 17 | LAG3 | lymphocyte activation gene 3 | −4.34 | Yes |
| 18 | ZBTB32 | zinc finger and BTB domain containing 32 | −4.16 | Yes |
| 19 | CXCL2 | chemokine (C-X-C motif) ligand 2 | −4.16 | |
| 20 | CRABP2 | cellular retinoic acid binding protein II | −4.09 | Yes |
| 21 | IL6 | interleukin 6 | −4.06 | |
| 22 | TIGIT | T cell immunoreceptor with Ig and ITIM domains | −3.70 | |
| 23 | TIMP1 | tissue inhibitor of metalloproteinase 1 | −3.65 | |
| 24 | RGS1 | regulator of G-protein signaling 1 | −3.56 | Yes |
| 25 | ERO1L | ERO1-like (Saccharomyces cerevisiae) | −3.56 | |
| 26 | 0610010O12RIK | RIKEN cDNA 0610010O12 gene | −3.52 | |
| 27 | SERPINB6B | serine (or cysteine) peptidase inhibitor, clade B, member 6b | −3.50 | Yes |
| 28 | CSF2 | colony stimulating factor 2 (granulocyte-macrophage) | −3.34 | Yes |
| 29 | IL4 | interleukin 4 | −3.33 | |
| 30 | TNFRSF4 (OX40) | tumor necrosis factor receptor superfamily, member 4 | −3.22 | Yes |
| 31 | SLAMF7 | SLAM family member 7 | −3.21 | Yes |
| 32 | CCL7 | chemokine (C-C motif) ligand 7 | −3.20 | |
| 33 | IL21 | interleukin 21 | −3.18 | Yes |
| 34 | EGLN3 | EGL nine homolog 3 (Caenorhabditis elegans) | −3.17 | Yes |
| 35 | CCL20 | chemokine (C-C motif) ligand 20 | −3.12 | |
| 36 | CISH | cytokine inducible SH2 containing protein | −3.06 | Yes |
| 37 | IL17F | interleukin 17F | −3.06 | |
| 38 | IL31 | interleukin 31 | −3.00 | Yes |
| 39 | IL2RA (CD25) | interleukin 2 receptor, alpha chain | −2.96 | Yes |
| 40 | SERPINB9 | serine (or cysteine) peptidase inhibitor, clade B, member 9 | −2.95 | Yes |
| 41 | OSM | oncostatin M | −2.93 | Yes |
| 42 | BATF | basic leucine zipper transcription factor, ATF-like | −2.89 | Yes |
| 43 | FAM132A | family with sequence similarity 132, member A | −2.77 | |
| 44 | ASNS | asparagine synthetase | −2.73 | |
| 45 | DDT | d-dopachrome tautomerase | −2.71 | |
| 46 | WISP2 | WNT1 inducible signaling pathway protein 2 | −2.69 | |
| 47 | CASP6 | caspase 6 | −2.65 | |
| 48 | TNFSF8 | tumor necrosis factor (ligand) superfamily, member 8 | −2.59 | Yes |
| 49 | CXCL3 | chemokine (C-X-C motif) ligand 3 | −2.56 | |
| 50 | IL27 | interleukin 27 | −2.52 | |
| 51 | HEMK1 | HemK methyltransferase family member 1 | −2.49 | Yes |
| 52 | CD200 | CD200 antigen | −2.42 | Yes |
| 53 | BC048355 | cDNA sequence BC048355 | −2.43 | |
| 54 | NRN1 | neuritin 1 | −2.42 | Yes |
| 55 | CTLA4 (CD152) | Cytotoxic T lymphocyte associated protein 4 | −2.39 | Yes |
| 56 | TBX21 (TBET) | T-box 21 | −2.37 | Yes |
| 57 | AGPAT4 | 1-acylglycerol-3-phosphate O-acyltransferase 4 (lysophosphatidic acid acyltransferase, delta) | −2.21 | |
| 58 | LRMP | lymphoid-restricted membrane protein | −2.21 | Yes |
| 59 | CD70 (TNFSF7) | CD70 antigen | −2.18 | |
| 60 | XCL1 | chemokine (C motif) ligand 1 | −2.17 | |
| 61 | 0610007P14RIK | RIKEN cDNA 0610007P14 gene | −2.13 | Yes |
| 62 | BATF2 | basic leucine zipper transcription factor, ATF-like 2 | −2.12 | |
| 63 | ACOT7 | acyl-CoA thioesterase 7 | −2.08 | Yes |
| 64 | ARHGDIG | Rho GDP dissociation inhibitor (GDI) gamma | −1.99 | |
| 65 | CCR8 | chemokine (C-C motif) receptor 8 | −1.97 | Yes |
| 66 | ABHD14A | abhydrolase domain containing 14A | −1.96 | |
| 67 | TNFSF14 | tumor necrosis factor (ligand) superfamily, member 14 | −1.94 | |
| 68 | HBEGF | heparin-binding EGF-like growth factor | −1.91 | |
| 69 | STC2 | stanniocalcin 2 | −1.89 | |
| 70 | ZEB2 | zinc finger E-box binding homeobox 2 | −1.86 | |
| 71 | PLA2G12A | phospholipase A2, group XIIA | −1.86 | |
| 72 | TNIP3 | TNFAIP3 interacting protein 3 | −1.78 | |
| 73 | ACY1 | aminoacylase 1 | −1.75 | |
| 74 | GSTT2 | glutathione S-transferase, theta 2 | −1.74 | Yes |
| 75 | SPRR2E | small proline rich protein 2E | −1.72 | |
| 76 | STAT5A | signal transducer and activator of transcription 5A | −1.70 | Yes |
| 77 | GALK1 | galactokinase 1 | −1.68 | Yes |
| 78 | SDF4 | stromal cell–derived factor 4 | −1.68 | Yes |
| 79 | MRPS2 | mitochondrial ribosomal protein S2 | −1.65 | |
| 80 | TIMM10 | Translocase of inner mitochondrial membrane 10 | −1.62 | |
| 81 | IL12RB1 | interleukin 12 receptor, beta 1 | −1.60 | Yes |
| 82 | SLC13A3 | solute carrier family 13 (sodium-dependent dicarboxylate transporter), member 3 | −1.60 | |
| 83 | ST6GALNAC4 | ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 4 | −1.58 | |
| 84 | RBPSUH-RS3 | recombining binding protein suppressor of hairless (Drosophila), related sequence 3 | −1.56 | |
| 85 | WISP1 | WNT1 inducible signaling pathway protein 1 | −1.56 | Yes |
| 86 | ETV6 | ets variant gene 6 (TEL oncogene) | −1.55 | Yes |
| 87 | POLR3G | polymerase (RNA) III (DNA directed) polypeptide G | −1.55 | |
| 88 | AREG | amphiregulin | −1.54 | |
| 89 | HIRIP3 | HIRA interacting protein 3 | −1.52 | Yes |
| 90 | TLCD1 | TLC domain containing 1 | −1.52 | Yes |
| 91 | PLAGL2 | pleomorphic adenoma gene-like 2 | −1.51 | Yes |
| 92 | NDRG1 | N-myc downstream regulated gene 1 | −1.50 | |
| 93 | ICOS | inducible T cell co-stimulator | −1.50 | Yes |
| 94 | MYC | myelocytomatosis oncogene | −1.38 | Yes |
| 95 | CCND2 | cyclin D2 | −1.37 | Yes |
| 96 | RBPJ | recombination signal binding protein for immunoglobulin kappa J region | −1.36 | Yes |
| 97 | MIF | macrophage migration inhibitory factor | −1.29 | |
| 98 | ELL2 | elongation factor RNA polymerase II 2 | −1.28 | |
| 99 | PPA1 | pyrophosphatase (inorganic) 1 | −1.24 | |
| 100 | COQ4 | coenzyme Q4 homolog (yeast) | −1.17 | |
| 101 | LAP3 | leucine aminopeptidase 3 | −1.17 | |
| 201 | GADD45B | growth arrest and DNA damage-inducible 45 beta | −1.13 | Yes |
| 103 | ECSCR | endothelial cell-specific chemotaxis regulator | −1.13 | |
| 104 | CHCHD4 | coiled-coil-helix-coiled-coil-helix domain containing 4 | −1.09 | Yes |
| 105 | ATF3 | activating transcription factor 3 | −1.07 | Yes |
| 106 | LAD1 | ladinin | −1.04 | |
| 107 | TMEM97 | transmembrane protein 97 | −1.03 | |
| 108 | HSPA5 | heat shock protein 5 | −1.02 | Yes |
| 109 | YWHAG | tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, gamma polypeptide | −1.02 | Yes |
| 110 | 1810029B16RIK | RIKEN cDNA 1810029B16 gene | −1.01 |
Included are genes normally induced by twofold or greater upon restimulation, showing twofold or greater down-regulation in the absence of OCA-B. See Table S2 for full dataset. Empty cells in the last column indicate “No,” although some genes may have just missed the arbitrary threshold and would be false negatives.
Merged data for ChIPseq binding of Oct1 and OCA-B in rested and restimulated T cells.
In T cells, Oct1 has been functionally associated with multiple target cytokine genes including Il2, Il4, Il5, Il8, Il12b, and Il13 (Kaushansky et al., 1994; Pfeuffer et al., 1994; Wu et al., 1997; Bert et al., 2000; Cron et al., 2001; Zhou et al., 2007; Kiesler et al., 2009). Oct1 additionally associates with widely expressed genes such as those encoding histones, TBP-associated factors (TAFs), and RNA polymerase II subunits (Sive et al., 1986; Fletcher et al., 1987; Kang et al., 2009). Oct1 is dispensable for expression of these genes and instead buffers them against inappropriate repression by external stress (Kang et al., 2009; Shakya et al., 2011). Finally, Oct1 also associates with cell type–specific genes in other lineages. Examples include immunoglobulins (Falkner and Zachau, 1984; Mason et al., 1985; Wirth et al., 1987) and Cdx2 (Jin and Li, 2001; Shakya et al., 2011). To study genome-wide Oct1 and OCA-B DNA binding, we performed ChIPseq. Because OCA-B ChIPseq was poor using formaldehyde cross-linking, we added a longer cross-linker to the reactions (Fig. S1). Peak calling from Rested or Re-stim cells enriched using Oct1 or OCA-B antibodies identified ∼5,000–7,000 enriched genomic regions, the majority of which (3,570) were common between the four conditions. The 3,570 regions corresponded to 3,173 nearby genes (Fig. 2 H and Table S3). Gene Ontogeny enrichments for the set of genes bound in all four conditions identified normal and deregulated immune function as the most significantly enriched components (Table S4). The 3,173 genes included 50 of the 110 differentially expressed genes identified by RNAseq (45%) as direct targets (Table 1). This set included Il2, Ifng, and Zbtb32 (Fig. 2 I). ChIP-qPCR at a site in the Ifng promoter (Penix et al., 1993) and newly identified sites in Zbtb32 and downstream of Ifng confirmed binding of Oct1 and OCA-B in Resting and Re-stim cells and extended these results to the H3K9me2 mark (Fig. 2 J).
Inspection of the Ifng and Zbtb32 regions revealed strong binding motifs conserved to human. The signal identified 52 kb downstream of Ifng was positioned between Ifng and Tmevpg1 (NeST), a long noncoding RNA (lncRNA) that supports Ifng expression (Vigneau et al., 2003; Collier et al., 2012; Gomez et al., 2013). This element contains a pair of conserved perfect consensus octamer motifs (Fig. 2 K). Interestingly, there were other spatially linked, coregulated direct target pairs, including Il2/Il21, Icos/Ctla4, Serpinb6b/SerpinB9, Tnfrsf4 (Ox40)/Sdf4, Hemk1/Cish, Wisp1/Ndrg1, and Chchcd4/Tmem97 (Table 1). Zbtb32 also contained a conserved perfect octamer motif ∼1.5 kb upstream (Fig. 2 K). There were many other targets with no expression changes, including housekeeping genes such as histones, Polr2a and Taf12 (Fig. 3). In contrast, genes expressed in other lineages such as immunoglobulins and Cdx2 were not bound, despite the fact that they contain perfect binding sites. Inappropriately expressed genes in resting Ocab−/− cells such as Gzmd and Prf1 (Fig. 2 E) were also not bound (not depicted).
Figure 3.
Oct1 and OCA-B associate with cell type–nonspecific genes. (A) IGV images of ChIPseq output for the Hist1h histone gene cluster. Peak calling is shown on the right. (B) Polr2a (encoding the RNA Pol II large subunit). (C) Taf12 (encoding a component of the TFIID transcription complex).
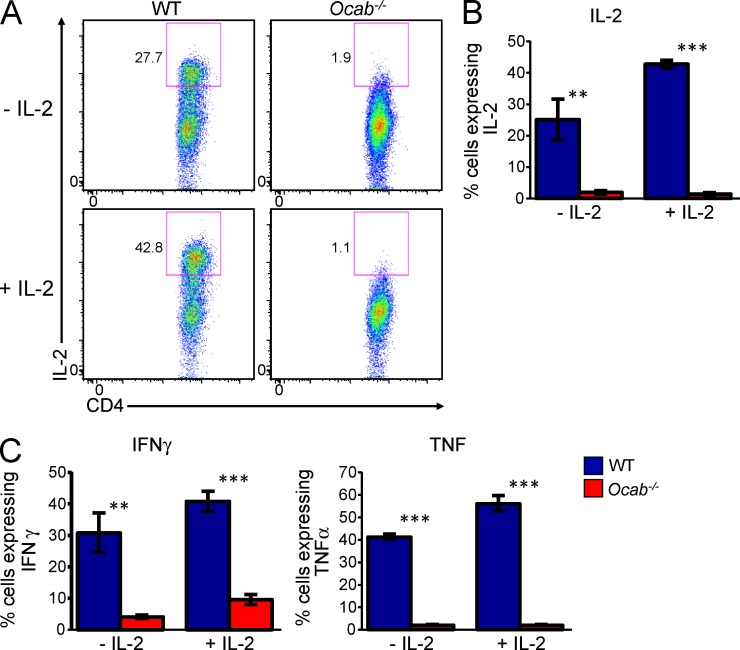
Despite the fact that OCA-B associates with multiple differentially expressed immunomodulatory genes, it was possible that defective IL-2 production contributes to defective expression of the other genes. To test this possibility, we provided exogenous IL-2 to WT and Ocab−/− cells after in vitro stimulation and then rested and restimulated as before. In the absence of IL-2, Ocab−/− cells produced significantly reduced quantities of IL-2, IFNγ, and TNF, as expected (Fig. 4). Supplemented IL-2 induced T cell expansion and increased the proportion of live cells in the culture regardless of genotype (not depicted). Exogenous IL-2 also significantly increased endogenous IL-2, IFNγ, and TNF cytokine production in WT cells. However, IL-2 failed to correct defects in cytokine production in Re-stim Ocab−/− cells (Fig. 4). Consistent with the ChIPseq results, these findings provide functional evidence that coordinate OCA-B regulation of multiple target genes contributes to the overall phenotype.
Figure 4.
Supplemental IL-2 does not complement OCA-B deficiency. (A) WT and Ocab−/− naive CD4+ T cells were stimulated with CD3ε and CD28 antibodies for 2 d. The cells were rested for 8 d either in the presence or absence of 30 IU/ml exogenously added IL-2. Cells were restimulated with the same antibodies for 6 h. Re-stim cells were stained for CD4 and intracellular cytokines. Intracellular expression of IL-2 is shown. (B) Mean of IL-2 expression from four sets of T cells independently purified from separate mice. Percentage of cytokine producing cells is plotted. Mean of quadruplicate results is shown ± SD. **, Student’s t test p-value ≤0.005; ***, p-value ≤0.0001. (C) Similar quantification (as in B) of IFNγ and TNF expression.
OCA-B–deficient T cells manifest poor memory recall responses in vivo
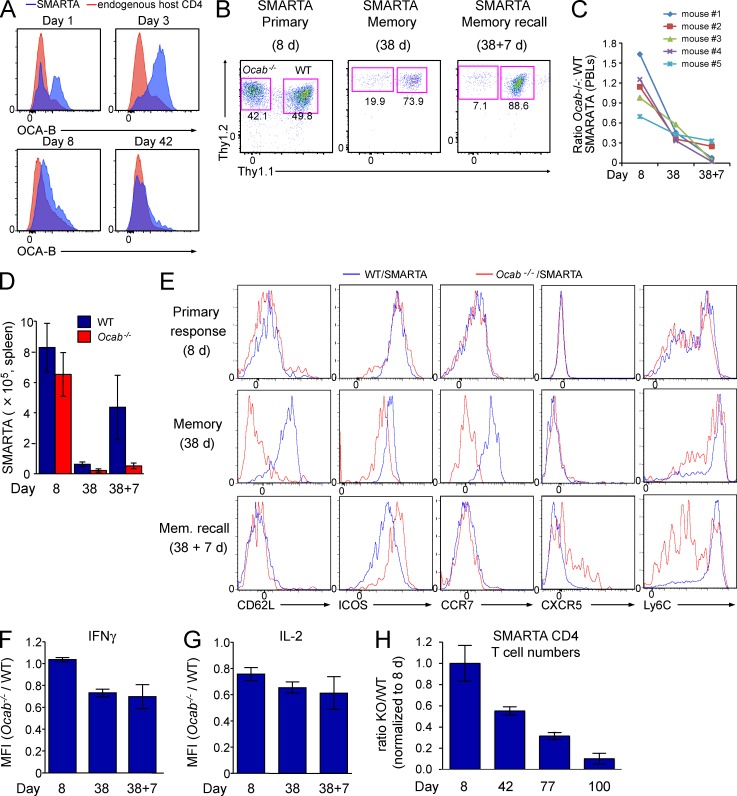
To test OCA-B induction during pathogen responses, we used lymphocytic choriomeningitis virus (LCMV) infection in conjunction with SMARTA T cell receptor transgenic mice, which are specific for the immunodominant CD4 epitope of the LCMV glycoprotein (GP61–80) and generate strong Th1 CD4 responses (Oxenius et al., 1998). We adoptively transferred 5 × 105 WT Thy1.1+ SMARTA transgenic naive T cells from spleens and lymph nodes (purity >95%; not depicted) into Thy1.2+ recipients. 1 d later, recipient mice were infected with LCMV, after which OCA-B expression was tested at 1, 3, 8, and 42 d after infection using intracellular antibody staining. Even without secondary antibody amplification, OCA-B protein could be detected over background in SMARTA T cell populations activated in vivo (Fig. 5 A).
Figure 5.
OCA-B is required for robust memory CD4+ T cell function in vivo. (A) WT SMARTA (Thy1.1+) T cells were transferred into Thy1.2+ mice, which were infected with LCMV 1 d later. Splenocytes were harvested at the indicated time points after infection. Cells were stained with OCA-B antibodies and subjected to flow cytometry. Enrichment relative to host T cells is shown. (B) Ocab−/− SMARTA (Thy1.2+Ly5.2+) and WT control SMARTA (Thy1.1+Thy1.2+Ly5.2+) T cells were co-transferred into Ly5.1+ mice, which were infected with LCMV. Frequencies of each population were tracked at the indicated time points. Representative plots, gated on donor SMARTA cells, indicate the relative distribution of WT and Ocab−/− SMARTA T cells in the spleen at the peak of the primary response (Primary, 8 d after infection), after the establishment of memory (Memory, 38 d after infection), or 7 d after rechallenge with Lm-gp61 (Memory recall). (C) For each time point, the ratio of Ocab−/− to WT SMARTA blood cells was calculated for each of five mice. (D) For each time point, total splenic CD4+ Ocab−/− and WT SMARTA T cell numbers were averaged and plotted. Three animals were sacrificed at days 8 and 38, and four animals at day 38 + 7. Error bars depict ±SD. (E) Cells isolated as in D were stained using the indicated antibodies and analyzed by flow cytometry. Cells were gated for viability and CD4 expression. (F) The ratio of IFNγ mean fluorescence intensities (MFIs) in WT and Ocab−/− SMARTA T cells was calculated from each of five mice and plotted as mean ± SD. Cells were stimulated with peptide/brefeldin-A ex vivo before intracellular cytokine staining. (G) Similar calculation (as in F) using IL-2 MFIs. (H) WT (Thy1.1+) and Ocab−/− (Thy1.1+Thy1.2+) SMARTA cells were combined and adoptively transferred into Thy1.2 recipients. After LCMV infection, cohorts of four mice were sacrificed at 8, 42, 77, and 100 d. T cell numbers at peak response were averaged. For each genotype, T cell counts were divided by the number at peak response and averaged. Error bars depict ± SD.
The failure to robustly induce Il2 and other target genes in vitro selectively at long time points suggested that OCA-B–deficient T cells might display impaired memory responses in vivo. We therefore tested whether OCA-B promotes T cell memory. We combined 104 Ocab−/− Ly5.2+Thy1.2+ SMARTA T cells 1:1 with Thy1.1+Thy1.2+Ly5.2+ WT SMARTA transgenic cells and adoptively transferred them into Ly5.1+5.2+ recipients. This approach allows simultaneous tracking of WT and Ocab−/− SMARTA cells within the same host. At peak response (8 d after infection), Ocab−/− SMARTA CD4+ T cell expansion was equivalent to WT (Fig. 5 B). By 38 d after infection, fewer OCA-B–deficient SMARTA cells populated the memory compartment. The numbers ranged between 3- and 50-fold fewer in different mice (Fig. 5 C).
The most important functional test of a memory T cell is its response to a secondary challenge. We rechallenged mice with recombinant Listeria monocytogenes (Lm)–expressing LCMV GP61–80 (Lm-gp61). Although WT SMARTA cells responded robustly, OCA-B–deficient SMARTA cells failed to generate recall responses 7 d after rechallenge (Fig. 5 B, 38 + 7 d). Similar findings were made using five mice analyzing either peripheral blood (Fig. 5 C) or total numbers in the spleen (Fig. 5 D). Ocab−/− and WT SMARTA cells also differed in their ability to adopt CD62Lhi and CCR7hi central memory phenotypes (Fig. 5 E), indicative of retention of effector properties. In addition, upon rechallenge, expression of the differentiation marker Ly6C was decreased specifically in Ocab−/− cells, whereas CXCR5 was increased, suggesting a failure to form Th1 effector cells (Fig. 5 E). The remaining T cells produced only slightly less IL-2 and IFNγ (Fig. 5, F and G), indicating a difference between the in vitro data and the SMARTA transgenic system. The difference between the in vivo and in vitro findings may be attributable to the more complex in vivo environment, which includes signals missing from the in vitro system. IL-2, for example, is essential for in vitro T cell expansion but is not required for primary expansion in vivo (Williams et al., 2006). A second reason may be the fixed specificity of the SMARTA transgenic T cells. Cumulatively, these findings indicate a crucial role for OCA-B in the in vivo generation of functionally responsive Th1 memory cells. Because the aforementioned experiments were performed using Ocab−/− and control cells in the same mouse, they also indicate that the observed defects are intrinsic to T cells.
The aforementioned data represent early memory time points. To investigate memory at longer time points and to investigate polyclonal responses, we tested OCA-B–deficient mice and matched WT controls. WT and OCA-B–deficient mice were infected with LCMV and sacrificed at 8, 42, and 77 d for memory time points and at 42 + 7 and 77 + 7 d for Lm-gp61 rechallenge. Ocab−/− mice are viable and fertile and show no known defects in hematopoiesis but show B cell–intrinsic defects after activation and lack germinal centers (Kim et al., 1996; Schubart et al., 1996; Qin et al., 1998). We noted that the peak CD4+ T cell response to LCMV in whole body OCA-B knockouts was reduced compared with controls. However, superimposed on this defect was an additional memory formation and recall response defect (not depicted). Because B cells are thought to regulate T cell memory formation (Whitmire et al., 2009; Mollo et al., 2013), this defect could be an indirect consequence of B cell immunodeficiency in Ocab−/− mice. To circumvent problems associated with B cell defects in whole body knockouts, we challenged mice adoptively transferred with WT and OCA-B–deficient SMARTA transgenic T cells with LCMV and monitored mice for 100 d without rechallenge. Ocab−/− Thy1.1+Thy1.2+ and WT Thy1.1+ SMARTA transgenic naive T cells were adoptively transferred into Thy1.2+ recipients. After LCMV infection, T cell numbers for each genotype were monitored as a fraction of peak response. WT numbers declined slightly as a function of time, as expected (not depicted); however, beyond this decline, OCA-B deficiency resulted in severe and progressive decreases in memory T cell numbers (Fig. 5 H). These results indicate that the OCA-B deficiency reduces the ability of cells to survive in the absence of antigenic stimulation.
Oct1-deficient T cells show selective defects in memory establishment and recall response
We tested whether Oct1 deficiency also selectively controls CD4 memory. Germline Oct1−/− mice die before birth (Sebastiano et al., 2010). A slightly less severe germline-deficient allele (Wang et al., 2004) survives long enough to perform fetal liver transplants. We generated bone marrow chimeras by transplanting Oct1-deficient, Thy1.1+1.2+ SMARTA transgenic fetal liver cells into irradiated Rag1−/− recipients. Parallel Oct1-sufficient SMARTA radiation chimeras were also made using bone marrow from a Thy1.1+ SMARTA transgenic mouse. The chimeras were rested for 10 wk to allow engraftment, after which naive WT and Oct1-deficient SMARTA splenic T cells were harvested, combined 1:1, and adoptively co-transferred into Thy1.2+ recipients. These mice were infected with LCMV 1 d later. By the peak effector response (day 8 after infection), both WT and Oct1-deficient splenic SMARTA CD4+ T cells expanded robustly (Fig. 6 A). Both WT and Oct1-deficient effector SMARTA T cells also initially adopted memory phenotypes with similar efficiencies, as comparable numbers of CD62L+ memory cells were present at day 8 (not depicted). However, by 42 d after infection, approximately threefold fewer Oct1-deficient SMARTA cells populated the memory compartment compared with WT in the same host (Fig. 6 A, 42 d). The difference in T cell numbers ranged between 2.5- and 6.5-fold (Fig. 6 B). WT splenic SMARTA cells responded robustly to rechallenge with Lm-gp61, whereas Oct1-deficient SMARTA cells failed to generate robust recall responses (Fig. 6 A, 42 + 7 d). Approximately 10-fold fewer Oct1-deficient T cells were present in every mouse (Fig. 6 B). Similar findings were made in liver and mesenteric lymph node (not depicted). Relative to OCA-B deficiency, the slightly less robust phenotype is likely attributable to the hypomorphic Oct1 allele (Wang et al., 2004), the presence of low amounts of Oct2 in CD4+ T cells, or incomplete deletion (see Fig. 7 G below). As with Ocab−/− SMARTA T cells, the few memory cells that were produced expressed near normal IL-2 and IFNγ (Fig. 6 C).
Figure 6.
Germline Oct1 loss impairs CD4 memory. (A) Naive CD4+ T cells were isolated from spleens of Rag1−/− mice engrafted with Oct1−/− SMARTA (Thy1.1+Thy1.2+) fetal liver or WT SMARTA control (Thy1.1+). Isolated cells were combined 1:1 and adoptively transferred into Thy1.2+ recipients, which were then infected with LCMV. Flow cytometry plots are shown of splenic CD4+ T cells at peak response (8 d), after the establishment of memory (42 d), and after rechallenge with Lm-gp61 (42 + 7 d). Data from a single mouse representative of five examples are shown. For each plot (gated on CD4), host T cells (Thy1.2+), WT transferred T cells (Thy1.1+), and Oct1-deficient transferred T cells (Thy1.1/1.2+) are shown. (B) Quantification of averaged relative T cell numbers from five mice. Error bars depict ±SD. (C) Same calculation as in B except using IFNγ/IL-2 double producers.
Figure 7.
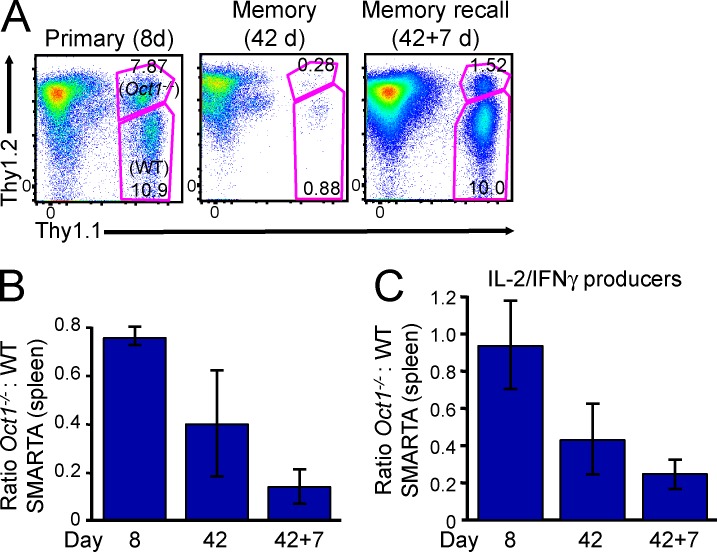
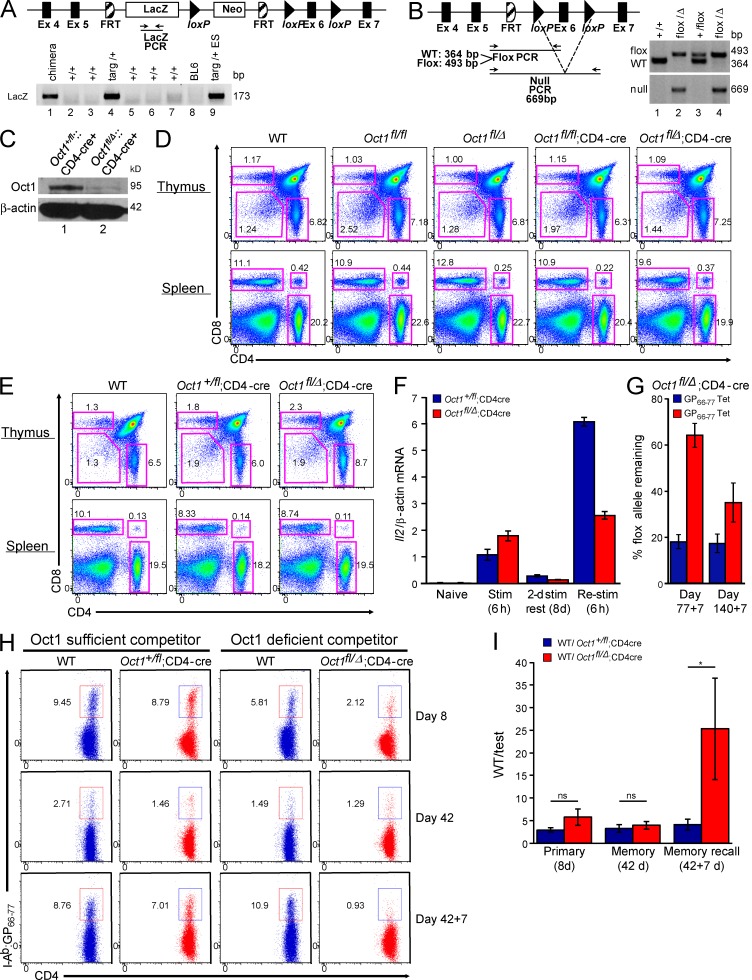
Conditional loss of Oct1 in T cells impairs CD4 memory. (A) Generation of Oct1 (Pou2f1) conditional mice. (top) Oct1 targeting event. (bottom) Agarose gel showing PCR data from chimera generated by ESC blastocyst injection (lane 1). Breeding generated a germline transmittant founder (lane 4). Targeted ESCs are shown as a control (lane 9). (B, left) After deletion of the LacZ reporter by breeding onto a germline FLIP recombinase expressing mouse, a conditional allele was generated in which loxP sites flank Oct1 exon 6, which encodes part of the DNA binding domain. (right) Genotyping of representative WT, floxed, and null alleles. (C) Oct1 protein levels were measured in CD4+ T cells purified from Oct1fl/Δ;CD4-cre and control Oct1+/fl;CD4-cre mice by Western blot. Purity was ∼90%. (D, top) CD4+/CD8+ thymocyte profiles from 8-wk-old Oct1 conditional-sufficient or -deficient mice and WT control. (bottom) CD4+/CD8+ splenic T cell profiles from the same mice. (E) Similar data (as in D) from 35-wk-old mice. (F) Oct1fl/Δ;CD4-cre and control Oct1+/fl;CD4-cre splenic naive CD4+ T cells were stimulated in vitro, rested, and restimulated. Il2 mRNA levels were monitored by TaqMan RT-qPCR. Means of replicates from T cells taken from three individual mice of each genotype are shown ± SD. (G) Oct1fl/Δ;CD4-cre mice were infected with LCMV, rested for either 77 or 140 d, and subsequently rechallenged using Lm-gp61. For each of the two rechallenge time points, splenic CD4+ cells were isolated by magnetic separation, stained with I-Ab/gp66–77 tetramers, and FACS sorted, and the conditional (floxed) allele was detected by qPCR. Loss was measured relative to tail DNA from the same mouse. Means of three mice ± SD are shown. (H) LCMV-reactive CD4+ T cells were assessed using mixed radiation chimeras in which lethally irradiated Thy1.1 host mice were repopulated with Ly5.2;Thy1.2 bone marrow from either Oct1+/fl or Oct1fl/Δ mice. Bone marrow was mixed 1:1 with WT Ly5.1;Thy1.2 control bone marrow. Oct1fl/Δ and control Oct1+/fl T cells are shown after infection with LCMV (8 and 42 d) or rechallenge with Lm-gp61 (42 + 7 d) next to the WT competitor cells from the same mouse. (I) For each mouse (n = 4–5), the ratio of I-Ab/gp66–77 tetramer–positive cells for each genotype (WT/test) was calculated. Mean of ratios ± SD is plotted. The Mann–Whitney test was used to compare WT and test populations at each time point (p-values: 8 d = 0.3429; 42 d = 0.4206; *, 42 + 7 d = 0.0159).
To study the role of Oct1 in polyclonal T cells responses, we generated Oct1 conditional mice (Fig. 7, A and B). Analysis of litters from Pou2f1fl/Δ (hereafter called Oct1fl/Δ, Δ will be used to distinguish this allele from the germline allele used above) or Oct1fl/+ intercrosses confirmed that the null allele was embryonic lethal and the floxed allele viable in the homozygous condition (Table S1). We crossed these mice to CD4-cre mice (Lee et al., 2001), which resulted in deletion of Oct1 in isolated splenic CD4+ T cells (Fig. 7 C). These mice showed normal thymocyte and splenocyte ratios (Fig. 7, D and E) and numbers (not depicted), and normal CD4+Foxp3+Helios+ T reg numbers (not depicted). We isolated Oct1fl/Δ;CD4-cre and control Oct1+/fl;CD4-cre splenic naive CD4+ T cells and stimulated, rested, and restimulated them in vitro. As before, there were significant defects in Il2 mRNA expression, specifically in Re-stim cells (Fig. 7 F).
LCMV was administered to male Oct1fl/Δ;CD4-cre and control Oct1+/fl;CD4-cre mice, and specific T cell responses were monitored by tetramer staining. Unexpectedly, little difference in memory formation or recall response was observed (not depicted). To determine whether the floxed allele was efficiently deleted in these experiments, we performed qPCR of the floxed and null alleles of sorted tetramer-negative and -positive Oct1fl/Δ;CD4-cre CD4+ T cells, assessing amplification relative to tail DNA from the same mouse. We observed a large increase in the presence of the floxed allele in tetramer-positive cells (Fig. 7 G), indicating a selection bias against loss of the floxed allele in cells entering the memory pool.
If Oct1-sufficient cells preferentially form memory cells, we would expect that in a mixed population of Oct1fl/Δ;CD4-cre cells and control cells, Oct1 deficiency would confer a competitive disadvantage, specifically in the memory compartment. To test this hypothesis, we generated mixed radiation chimeras in which Oct1fl/Δ;CD4-cre;Ly5.2 or Oct1fl/+;CD4-cre;Ly5.2 bone marrow hematopoietic progenitors were coinjected with Oct1+/+;Ly5.1 progenitors into lethally irradiated Thy1.1+ recipients. 8 wk after reconstitution, mice were infected with LCMV. Oct1fl/+ CD4+ T cells mounted normal polyclonal primary responses, entered into the memory pool, and generated robust recall responses as measured by tetramer staining (Fig. 7 H). Ratios of absolute cell numbers are shown in Fig. 7 I. Competing cells also formed memory and responded to rechallenge. Oct1fl/Δ;CD4-cre cells also generated normal primary responses and contributed to the memory pool, but upon rechallenge showed a 10-fold defect in proliferative response relative to competing WT cells (Fig. 7, H and I). The few cells that entered the memory pool and responded to rechallenge produced IL-2 and IFNγ upon peptide restimulation (not depicted).
DISCUSSION
Cells must maintain key genes in a silent state while simultaneously keeping them poised for later expression. This property is critical for memory lymphocytes; however, the mechanisms are unclear. Our findings pinpoint OCA-B, a transcriptional coactivator, together with Oct1, a transcription factor with which it docks, as activities that prevent stable repression of multiple target genes, to help maintain silent but poised transcriptional states associated with immunological memory. We definitively identify ∼50 direct targets poised for later expression by Oct1/OCA-B. We also show that OCA-B and Oct1 regulate memory T cell states in vivo. Oct1 is constitutively expressed in T cells, whereas OCA-B is induced after T cell activation. Although neither protein specifically marks memory cells, both proteins are functionally specific for CD4 memory. Notably, deletion of either OCA-B or Oct1 has no visible effects on T cell development or homeostasis.
Oct1 has an extensive history in the immunological literature, beginning with the immunoglobulin and interleukin-2 loci (Falkner and Zachau, 1984; Mason et al., 1985; Wirth et al., 1987; Ullman et al., 1991) and including multiple cytokine targets such as Ifng and IL-3, -4, -5, -8, -12b, and -13 (Penix et al., 1993; Kaushansky et al., 1994; Pfeuffer et al., 1994; Duncliffe et al., 1997; Wu et al., 1997; Cron et al., 2001; Zhou et al., 2007; Kiesler et al., 2009). More recently Oct1 has been implicated in interchromosomal communication between the Th2 and Th17 loci and IFNγ, mediated via an interaction with CTCF, at early stages of T cell activation and differentiation (Kim et al., 2014). OCA-B has been intensively studied in B cells but less so in T cells. In B cells, OCA-B has also been implicated in communication between distantly bound sites at the IgH gene locus via interactions with GTF2I/TFII-I (Ren et al., 2011).
Rested but previously stimulated Oct1-deficient T cells fail to recruit the histone lysine demethylase Jmjd1a to the Il2 promoter and manifest poor secondary Il2 responses in resting but previously stimulated T cells (Shakya et al., 2011). MEK is required for initial recruitment of Jmjd1a after T cell activation; however, these findings do not explain how Jmjd1a remains stably associated with Il2 in resting, previously stimulated T cells. We show that OCA-B docks with Oct1 at Il2 and is required to localize Jmjd1a to Il2 at long time points after T cell activation. OCA-B is also required to localize Jmjd1a to Il2 in Re-stim T cells, possibly because CD4+ memory T cells have reduced ERK activity after stimulation (Hussain et al., 2002; Adachi and Davis, 2011). Provision of exogenous IL-2 to cultures increases T cell survival in vitro regardless of genotype and boosts cytokine production in WT cells, but fails to correct cytokine expression defects in Ocab−/− cells. This result strongly suggests that there are other critical OCA-B targets. Consistent with this supposition, OCA-B is required for robust expression of multiple genes specifically in Re-stim T cells. Using ChIPseq, we confirm Oct1 and OCA-B binding at many of these differentially regulated genes. In the case of Ifng, binding occurs at a downstream site containing two conserved perfect octamer sequences. This site is located between Ifng and its regulatory lncRNA Tmevpg1/NeST (Vigneau et al., 2003; Collier et al., 2012; Gomez et al., 2013) in a configuration similar to Igf2 and the H19 lncRNA, where a similar dyad octamer site is present between the two (Hori et al., 2002).
In the absence of IL-2, CD8+ memory T cells form normally but are impaired in response to secondary challenge (Williams et al., 2006; Mitchell et al., 2010). The failure to robustly induce IL-2 and other cytokines in vitro suggested that Ocab−/− T cells may display impaired memory responses in vivo. We show that OCA-B deficiency results in poor memory T cell numbers and failure to generate robust secondary responses. Unlike the in vitro situation, IL-2 and IFNγ production on a per cell basis is largely normal. This difference is most likely attributable to the more complex in vivo environment. As one example, IL-2 is essential for in vitro T cell expansion but is not required for primary expansion in vivo (D’Souza and Lefrançois, 2003; Williams et al., 2006). Another reason may be the fixed antigen specificity of SMARTA transgenic T cells.
If the in vivo phenotype associated with OCA-B deficiency is a product of docking with Oct1 at critical target genes, Oct1 deficiency should also manifest selective memory T cell defects. We tested this prediction using adoptive transfer of CD4+ SMARTA T cells lacking Oct1 from Rag1−/− mice engrafted with Oct1-deficient SMARTA fetal liver hematopoietic progenitor cells. Separately, we used a conditional Oct1 allele bred onto CD4-cre. In both cases profound and selective memory cell formation and recall response defects were observed, similar to OCA-B deficiency. These findings suggest that OCA-B and Oct1 act through a common transcriptional pathway to promote CD4+ memory T cell function. Collectively, the findings indicate that the epigenetic memory established by Jmjd1a localization by OCA-B and Oct1 to critical targets constitutes a fundamental determinant of physiological CD4+ T cell memory. Control of transcriptional states by OCA-B/Oct1 partnerships may also represent a more general mechanism applying to CD8+ T cell or B cell memory and hematopoietic/leukemic stem cell maintenance, although these remain to be tested.
MATERIALS AND METHODS
T cell isolation and in vitro stimulation
Naive helper T cells were isolated and simulated as described previously (Shakya et al., 2011). The C57BL/6J strain background was used for all experiments. LCMV was administered to male knockout and transplant recipient mice at 8 wk of age.
Flow cytometry
For OCA-B T cell intracellular staining, 8 × 105 cells were washed in 200 µl PBS plus 3% FBS and fixed/permeabilized in 100 µl Cytofix/Cytoperm (BD). Cells were vortexed and incubated for 30 min at 4°C. Subsequently, 100 µl Perm/Wash buffer (BD) was added, and cells were collected by centrifugation. Cells were resuspended in 200 µl Perm/Wash buffer plus 15 µl anti–OCA-B–PE (Santa Cruz Biotechnology, Inc.), briefly vortexed, and incubated for 30 min at 4°C. After addition of 100 µl Perm/Wash buffer and collection of cells, the stained cell pellet was resuspended in 200 µl PBS + 3% FBS for analysis by flow cytometry. Peptide restimulation, APC-tetramer staining, and antibody staining for flow cytometry were conducted as described previously (Williams et al., 2008). For cytokine staining, brefeldin A (Golgi-plug, 1 mg/µl; BD) was used. Data were collected using a FACSCanto and LSR Fortessa (both BD) and analyzed using FlowJo software (Tree Star).
ChIP
ChIP-qPCR was performed as described previously (Tantin et al., 2013). Antibodies used were OCA-B (Santa Cruz Biotechnology, Inc.), Oct1 (Bethyl Laboratories, Inc.), Jmjd1a (Abcam), and H3K9me2 (EMD Millipore). Mouse Ifng promoter primers were Ch-MmIFNg-F, 5′-CCCCCACCTATCTGTCACCATC-3′ and Ch-MmIFNg-R, 5′-AGAGGGTCCAAAGGATCAGC-3′. Ifng downstream dyad octamer primers were Ch-MmIFNgregion2-F, 5′-CATGACTATGGTGTGAGCTG-3′ and Ch-MmIFNgregion2-R, 5′-AAAATGTTGACCAGACAAGC-3′. Zbtb32 primers were Ch-mZbtb32-F, 5′-GAAGGGTTAGTAGGGTGGAG-3′ and Ch-mZbtb32-R, 5′-GGAAGGCTTGTATCTGTTTG-3′.
RT-qPCR
Mouse Il2 mRNA levels were measured as described previously (Shakya et al., 2011). Primer pairs for mouse Ifng and Zbtb32 were taken from Huffaker et al. (2012) and Yoon et al. (2012), respectively.
RNAseq
Naive CD4+ T cells (CD8aneg,CD11bneg,CD45Rneg,DX5neg,Ter-119neg,CD44lo) purified from four WT and six Ocab−/− spleens were pooled. For each time point, cells were seeded into 5 individual wells of a 6-well plate. Cells in each well were independently stimulated as described previously (Shakya et al., 2011). RNA was extracted using a kit (RNeasy; QIAGEN) and combined for processing and RNAseq. RNA concentration was determined using a Quant-iT RNA assay kit and a Qubit fluorometer (Invitrogen).
ChIPseq
Oct1 and OCA-B ChIPseq used antibodies from Santa Cruz Biotechnology, Inc. IP and library preparation were performed identically to a recent study (Shakya et al., 2015) with the following exceptions. For cross-linking, 5 mM disuccinimidyl glutarate (DSG; Thermo Fisher Scientific) was included with formaldehyde. Cross-linking was performed at room temperature for 30 min. During cross-linking with DSG, 1% formaldehyde was added for the final 10 min. ChIP DNA concentration was determined using a Quant-iT dsDNA HS assay kit (Invitrogen) and a Qubit fluorometer. Data were collected using MiSeq and HiSeq 2000 instruments (Illumina) and combined. Sequenced reads were aligned to the reference mouse genome (mm9/NCBI Build 37) using previously published methodologies (Ram et al., 2011). Because the longer DSG cross-linker generated wider enrichments compared with formaldehyde alone, a higher 2-kb cutoff was applied to filter spurious peaks (Fig. S1). This method produced maximum enrichment around transcription start sites using a control H3K4me3 antibody (Fig. S1). Intersecting regions (physically overlapping) in all four conditions (OCA-B– and Oct1-enriched material from Rested and Re-stim T cells) were generated using BEDTools (Quinlan and Hall, 2010). Nearest gene analysis and GO enrichment were performed with the genomic regions enrichment of annotations tool (GREAT; McLean et al., 2010) using a 100,000-bp cutoff.
Oct1 (Pou2f1) conditional mice
C57BL/6 mouse embryonic stem cells (ESCs) carrying an agouti mutation and a targeted Pou2f1 allele were obtained from EUCOMM. ESCs were injected into blastocysts of C57BL/6 mice at the University of Utah gene targeting facility. Chimeric mice were crossed with C57BL/6 mice to produce founders. Founders were identified by genotyping using primers spanning the LacZ gene. Founders were bred with B6.ACTB-FLPe mice to remove the neomycin cassette and generate conditional mice. The conditional mice were bred with HPRT-Cre transgenic mice to generate germline null alleles. The flox and null alleles were crossed to CD4-cre (gift of J. Round, University of Utah, Salt Lake City, UT) to generate mice with CD4+ T cells null for Oct1. Genotyping was performed using the following primers: LacZ forward, 5′-ACTATCCCGACCGCCTTACTG-3′; LacZ reverse, 5′-GTAGCGGCTGATGTTGAACTGG-3′; Frtdel forward, 5′-GACTTGTCTTCTGATCTCCACATGC-3′; Frtdel reverse, 5′-AAACTTAGGGCCTCTGCAAGAAG-3′; HPRTcre forward, 5′-CCCTTCATAGAGACAAGGAATGTGTCC-3′; HPRTcre reverse, 5′-AGTGCTTAATGTAATCCAGCAGGTCA-3′; CD4cre forward, 5′-CGATGCAACGAGTGATGAGG-3′; and CD4cre reverse, 5′-GCATTGCTGTCACTTGGTCGT-3′. The presence of WT and floxed alleles was determined by PCR using Oct1-3′-WT and Oct1-5′-flox primers, whereas the presence of the null allele was determined using Oct1-3′-WT and Oct1-5′-null primers. Oct1-3′WT, 5′-ACAGATGCTCGCATTTTAGTTG-3′; Oct1-5′flox, 5′-ACACACACAAACAGCAATTATC-3′; and Oct1-5′null, 5′-TTGCCCAAGTCTTGTCTATCTG-3′.
T cell retroviral transduction
T cells were transduced with MSCV retrovirus by spin infection. Cells were centrifuged with 293T retroviral supernatant at 1,500 g for 1.5 h at 37°C in the presence of 4 µg/ml polybrene. The MSCVneo-hOcab plasmid (Casellas et al., 2002) was gift of R. Roeder (The Rockefeller University, New York, NY).
OCA-B and Oct1 radiation chimeras, LCMV, and Lm-gp61 infection
Chimeras were generated as described previously (Williams et al., 2008). In brief, 104 WT and either Oct1- or OCA-B–deficient naive SMARTA transgenic T cells were isolated and adoptively transferred (i.v.) into recipient mice. Combinations of Thy1 and Ly5 alleles were used to discriminate transplanted and host T cell subsets, as indicated. For viral infection, mice were inoculated with LCMV Armstrong 53b (2 × 105 PFU, IP). For bacterial infection, mice were inoculated with Lm-gp61 (106 CFU, i.v.).
qPCR genotyping and calculation of loss of floxed allele
For each indicated time point, splenic CD4+ cells from Oct1fl/Δ;CD4-cre mice were isolated by magnetic separation and stained with gp66-77 tetramers. Tetramer-positive and -negative cells were isolated using a FACSAria (BD). Isolated DNA was used to detect the conditional (floxed) allele by qPCR. Loss was measured relative to tail DNA from the same mouse. The PCR primer pair also amplifies the WT Oct1 allele; however, no WT allele was present in these experiments. Specifically, the fold loss of the floxed allele was calculated by generating a deviation of T cell DNA from tail DNA from the same mouse for the floxed allele:
and a deviation for the null allele:
To normalize for amounts of tail DNA and T cell DNA for an individual mouse, ΔΔCp was calculated:
Because loss of the floxed allele also causes gain of the null allele, to calculate loss of the null allele ΔΔCp was halved:
where x is the fold change in the floxed allele.
Online supplemental material
Fig. S1 shows the ChIPseq analysis workflow. Table S1 shows numbers of offspring with the indicated genotype from Oct1−/+ and Oct1fl/+ intercrosses. Table S2, included as a separate Excel file, shows normalized RNAseq gene expression changes. Table S3, included as a separate Excel file, shows Oct1/OCA-B target gene identification using ChIPseq. Table S4, included as a separate Excel file, shows Gene Ontology enrichments for ChIPseq data. Online supplemental material is available at http://www.jem.org/cgi/content/full/jem.20150363/DC1.
Supplementary Material
ACKNOWLEDGMENTS
OCA-B–deficient mice and the MSCVneo-hOcab viral construct encoding human OCA-B were provided by R. Roeder. We thank S. Tamowski and J. Kaplan for assistance generating Oct1 conditional mice and J. Round for providing CD4-cre mice.
A. Shalek was supported by the Searle Scholars Program. This work was supported by a grant from National Institutes of Health/National Institute of Allergy and Infectious Diseases (R01AI100873 to D. Tantin).
The authors declare no competing financial interests.
Footnotes
Abbreviations used:
- ChIP
- chromatin IP
- DSG
- disuccinimidyl glutarate
- ESC
- embryonic stem cell
- IGV
- integrated genome viewer
- IP
- immunoprecipitation
- LCMV
- lymphocytic choriomeningitis virus
- Lm
- Listeria monocytogenes
- lncRNA
- long noncoding RNA
- MSCV
- murine stem cell virus
- NuRD
- nucleosome-remodeling deacetylase
- qPCR
- quantitative PCR
References
- Adachi K., and Davis M.M.. 2011. T-cell receptor ligation induces distinct signaling pathways in naive vs. antigen-experienced T cells. Proc. Natl. Acad. Sci. USA. 108:1549–1554. 10.1073/pnas.1017340108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee A., Gordon S.M., Intlekofer A.M., Paley M.A., Mooney E.C., Lindsten T., Wherry E.J., and Reiner S.L.. 2010. Cutting edge: The transcription factor eomesodermin enables CD8+ T cells to compete for the memory cell niche. J. Immunol. 185:4988–4992. 10.4049/jimmunol.1002042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaulieu A.M., Zawislak C.L., Nakayama T., and Sun J.C.. 2014. The transcription factor Zbtb32 controls the proliferative burst of virus-specific natural killer cells responding to infection. Nat. Immunol. 15:546–553. 10.1038/ni.2876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bert A.G., Burrows J., Hawwari A., Vadas M.A., and Cockerill P.N.. 2000. Reconstitution of T cell-specific transcription directed by composite NFAT/Oct elements. J. Immunol. 165:5646–5655. 10.4049/jimmunol.165.10.5646 [DOI] [PubMed] [Google Scholar]
- Brunner C., Sindrilaru A., Girkontaite I., Fischer K.D., Sunderkötter C., and Wirth T.. 2007. BOB.1/OBF.1 controls the balance of TH1 and TH2 immune responses. EMBO J. 26:3191–3202. 10.1038/sj.emboj.7601742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casellas R., Jankovic M., Meyer G., Gazumyan A., Luo Y., Roeder R., and Nussenzweig M.. 2002. OcaB is required for normal transcription and V(D)J recombination of a subset of immunoglobulin κ genes. Cell. 110:575–585. 10.1016/S0092-8674(02)00911-X [DOI] [PubMed] [Google Scholar]
- Collier S.P., Collins P.L., Williams C.L., Boothby M.R., and Aune T.M.. 2012. Cutting edge: influence of Tmevpg1, a long intergenic noncoding RNA, on the expression of Ifng by Th1 cells. J. Immunol. 189:2084–2088. 10.4049/jimmunol.1200774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cron R.Q., Zhou B., Brunvand M.W., and Lewis D.B.. 2001. Octamer proteins inhibit IL-4 gene transcription in normal human CD4 T cells. Genes Immun. 2:464–468. 10.1038/sj.gene.6363811 [DOI] [PubMed] [Google Scholar]
- Cui W., Liu Y., Weinstein J.S., Craft J., and Kaech S.M.. 2011. An interleukin-21-interleukin-10-STAT3 pathway is critical for functional maturation of memory CD8+ T cells. Immunity. 35:792–805. 10.1016/j.immuni.2011.09.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Souza W.N., and Lefrançois L.. 2003. IL-2 is not required for the initiation of CD8 T cell cycling but sustains expansion. J. Immunol. 171:5727–5735. 10.4049/jimmunol.171.11.5727 [DOI] [PubMed] [Google Scholar]
- Darrah P.A., Patel D.T., De Luca P.M., Lindsay R.W., Davey D.F., Flynn B.J., Hoff S.T., Andersen P., Reed S.G., Morris S.L., et al. . 2007. Multifunctional TH1 cells define a correlate of vaccine-mediated protection against Leishmania major. Nat. Med. 13:843–850. 10.1038/nm1592 [DOI] [PubMed] [Google Scholar]
- Duncliffe K.N., Bert A.G., Vadas M.A., and Cockerill P.N.. 1997. A T cell-specific enhancer in the interleukin-3 locus is activated cooperatively by Oct and NFAT elements within a DNase I-hypersensitive site. Immunity. 6:175–185. 10.1016/S1074-7613(00)80424-0 [DOI] [PubMed] [Google Scholar]
- Falkner F.G., and Zachau H.G.. 1984. Correct transcription of an immunoglobulin κ gene requires an upstream fragment containing conserved sequence elements. Nature. 310:71–74. 10.1038/310071a0 [DOI] [PubMed] [Google Scholar]
- Fletcher C., Heintz N., and Roeder R.G.. 1987. Purification and characterization of OTF-1, a transcription factor regulating cell cycle expression of a human histone H2b gene. Cell. 51:773–781. 10.1016/0092-8674(87)90100-0 [DOI] [PubMed] [Google Scholar]
- Garrity P.A., Chen D., Rothenberg E.V., and Wold B.J.. 1994. Interleukin-2 transcription is regulated in vivo at the level of coordinated binding of both constitutive and regulated factors. Mol. Cell. Biol. 14:2159–2169. 10.1128/MCB.14.3.2159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez J.A., Wapinski O.L., Yang Y.W., Bureau J.F., Gopinath S., Monack D.M., Chang H.Y., Brahic M., and Kirkegaard K.. 2013. The NeST long ncRNA controls microbial susceptibility and epigenetic activation of the interferon-γ locus. Cell. 152:743–754. 10.1016/j.cell.2013.01.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammarlund E., Lewis M.W., Hansen S.G., Strelow L.I., Nelson J.A., Sexton G.J., Hanifin J.M., and Slifka M.K.. 2003. Duration of antiviral immunity after smallpox vaccination. Nat. Med. 9:1131–1137. 10.1038/nm917 [DOI] [PubMed] [Google Scholar]
- Hess Michelini R., Doedens A.L., Goldrath A.W., and Hedrick S.M.. 2013. Differentiation of CD8 memory T cells depends on Foxo1. J. Exp. Med. 210:1189–1200. 10.1084/jem.20130392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hori N., Nakano H., Takeuchi T., Kato H., Hamaguchi S., Oshimura M., and Sato K.. 2002. A dyad oct-binding sequence functions as a maintenance sequence for the unmethylated state within the H19/Igf2-imprinted control region. J. Biol. Chem. 277:27960–27967. 10.1074/jbc.M202280200 [DOI] [PubMed] [Google Scholar]
- Huffaker T.B., Hu R., Runtsch M.C., Bake E., Chen X., Zhao J., Round J.L., Baltimore D., and O’Connell R.M.. 2012. Epistasis between microRNAs 155 and 146a during T cell-mediated antitumor immunity. Cell Reports. 2:1697–1709. 10.1016/j.celrep.2012.10.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussain S.F., Anderson C.F., and Farber D.L.. 2002. Differential SLP-76 expression and TCR-mediated signaling in effector and memory CD4 T cells. J. Immunol. 168:1557–1565. 10.4049/jimmunol.168.4.1557 [DOI] [PubMed] [Google Scholar]
- Ichii H., Sakamoto A., Hatano M., Okada S., Toyama H., Taki S., Arima M., Kuroda Y., and Tokuhisa T.. 2002. Role for Bcl-6 in the generation and maintenance of memory CD8+ T cells. Nat. Immunol. 3:558–563. 10.1038/ni802 [DOI] [PubMed] [Google Scholar]
- Ichii H., Sakamoto A., Arima M., Hatano M., Kuroda Y., and Tokuhisa T.. 2007. Bcl6 is essential for the generation of long-term memory CD4+ T cells. Int. Immunol. 19:427–433. 10.1093/intimm/dxm007 [DOI] [PubMed] [Google Scholar]
- Iijima N., and Iwasaki A.. 2014. A local macrophage chemokine network sustains protective tissue-resident memory CD4 T cells. Science. 346:93–98. 10.1126/science.1257530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Intlekofer A.M., Takemoto N., Kao C., Banerjee A., Schambach F., Northrop J.K., Shen H., Wherry E.J., and Reiner S.L.. 2007. Requirement for T-bet in the aberrant differentiation of unhelped memory CD8+ T cells. J. Exp. Med. 204:2015–2021. 10.1084/jem.20070841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin T., and Li H.. 2001. Pou homeodomain protein OCT1 is implicated in the expression of the caudal-related homeobox gene Cdx-2. J. Biol. Chem. 276:14752–14758. 10.1074/jbc.M008277200 [DOI] [PubMed] [Google Scholar]
- Joshi N.S., Cui W., Chandele A., Lee H.K., Urso D.R., Hagman J., Gapin L., and Kaech S.M.. 2007. Inflammation directs memory precursor and short-lived effector CD8+ T cell fates via the graded expression of T-bet transcription factor. Immunity. 27:281–295. 10.1016/j.immuni.2007.07.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kallies A., Xin A., Belz G.T., and Nutt S.L.. 2009. Blimp-1 transcription factor is required for the differentiation of effector CD8+ T cells and memory responses. Immunity. 31:283–295. 10.1016/j.immuni.2009.06.021 [DOI] [PubMed] [Google Scholar]
- Kang J., Gemberling M., Nakamura M., Whitby F.G., Handa H., Fairbrother W.G., and Tantin D.. 2009. A general mechanism for transcription regulation by Oct1 and Oct4 in response to genotoxic and oxidative stress. Genes Dev. 23:208–222. 10.1101/gad.1750709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaushansky K., Shoemaker S.G., O’Rork C.A., and McCarty J.M.. 1994. Coordinate regulation of multiple human lymphokine genes by Oct-1 and potentially novel 45 and 43 kDa polypeptides. J. Immunol. 152:1812–1820. [PubMed] [Google Scholar]
- Kiesler P., Shakya A., Tantin D., and Vercelli D.. 2009. An allergy-associated polymorphism in a novel regulatory element enhances IL13 expression. Hum. Mol. Genet. 18:4513–4520. 10.1093/hmg/ddp411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim L.K., Esplugues E., Zorca C.E., Parisi F., Kluger Y., Kim T.H., Galjart N.J., and Flavell R.A.. 2014. Oct-1 regulates IL-17 expression by directing interchromosomal associations in conjunction with CTCF in T cells. Mol. Cell. 54:56–66. 10.1016/j.molcel.2014.02.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim U., Qin X.F., Gong S., Stevens S., Luo Y., Nussenzweig M., and Roeder R.G.. 1996. The B-cell-specific transcription coactivator OCA-B/OBF-1/Bob-1 is essential for normal production of immunoglobulin isotypes. Nature. 383:542–547. 10.1038/383542a0 [DOI] [PubMed] [Google Scholar]
- Lee P.P., Fitzpatrick D.R., Beard C., Jessup H.K., Lehar S., Makar K.W., Pérez-Melgosa M., Sweetser M.T., Schlissel M.S., Nguyen S., et al. . 2001. A critical role for Dnmt1 and DNA methylation in T cell development, function, and survival. Immunity. 15:763–774. 10.1016/S1074-7613(01)00227-8 [DOI] [PubMed] [Google Scholar]
- Mason J.O., Williams G.T., and Neuberger M.S.. 1985. Transcription cell type specificity is conferred by an immunoglobulin VH gene promoter that includes a functional consensus sequence. Cell. 41:479–487. 10.1016/S0092-8674(85)80021-0 [DOI] [PubMed] [Google Scholar]
- McLean C.Y., Bristor D., Hiller M., Clarke S.L., Schaar B.T., Lowe C.B., Wenger A.M., and Bejerano G.. 2010. GREAT improves functional interpretation of cis-regulatory regions. Nat. Biotechnol. 28:495–501. 10.1038/nbt.1630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell D.M., Ravkov E.V., and Williams M.A.. 2010. Distinct roles for IL-2 and IL-15 in the differentiation and survival of CD8+ effector and memory T cells. J. Immunol. 184:6719–6730. 10.4049/jimmunol.0904089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mollo S.B., Zajac A.J., and Harrington L.E.. 2013. Temporal requirements for B cells in the establishment of CD4 T cell memory. J. Immunol. 191:6052–6059. 10.4049/jimmunol.1302033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller K., Quandt J., Marienfeld R.B., Weihrich P., Fiedler K., Claussnitzer M., Laumen H., Vaeth M., Berberich-Siebelt F., Serfling E., et al. . 2013. Octamer-dependent transcription in T cells is mediated by NFAT and NF-κB. Nucleic Acids Res. 41:2138–2154. 10.1093/nar/gks1349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murayama A., Sakura K., Nakama M., Yasuzawa-Tanaka K., Fujita E., Tateishi Y., Wang Y., Ushijima T., Baba T., Shibuya K., et al. . 2006. A specific CpG site demethylation in the human interleukin 2 gene promoter is an epigenetic memory. EMBO J. 25:1081–1092. 10.1038/sj.emboj.7601012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oxenius A., Bachmann M.F., Zinkernagel R.M., and Hengartner H.. 1998. Virus-specific major MHC class II-restricted TCR-transgenic mice: effects on humoral and cellular immune responses after viral infection. Eur. J. Immunol. 28:390–400. [DOI] [PubMed] [Google Scholar]
- Penix L., Weaver W.M., Pang Y., Young H.A., and Wilson C.B.. 1993. Two essential regulatory elements in the human interferon gamma promoter confer activation specific expression in T cells. J. Exp. Med. 178:1483–1496. 10.1084/jem.178.5.1483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pepper M., Pagán A.J., Igyártó B.Z., Taylor J.J., and Jenkins M.K.. 2011. Opposing signals from the Bcl6 transcription factor and the interleukin-2 receptor generate T helper 1 central and effector memory cells. Immunity. 35:583–595. 10.1016/j.immuni.2011.09.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeuffer I., Klein-Hessling S., Heinfling A., Chuvpilo S., Escher C., Brabletz T., Hentsch B., Schwarzenbach H., Matthias P., and Serfling E.. 1994. Octamer factors exert a dual effect on the IL-2 and IL-4 promoters. J. Immunol. 153:5572–5585. [PubMed] [Google Scholar]
- Qin X.F., Reichlin A., Luo Y., Roeder R.G., and Nussenzweig M.C.. 1998. OCA-B integrates B cell antigen receptor-, CD40L- and IL 4-mediated signals for the germinal center pathway of B cell development. EMBO J. 17:5066–5075. 10.1093/emboj/17.17.5066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan A.R., and Hall I.M.. 2010. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 26:841–842. 10.1093/bioinformatics/btq033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ram O., Goren A., Amit I., Shoresh N., Yosef N., Ernst J., Kellis M., Gymrek M., Issner R., Coyne M., et al. . 2011. Combinatorial patterning of chromatin regulators uncovered by genome-wide location analysis in human cells. Cell. 147:1628–1639. 10.1016/j.cell.2011.09.057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao R.R., Li Q., Gubbels Bupp M.R., and Shrikant P.A.. 2012. Transcription factor Foxo1 represses T-bet-mediated effector functions and promotes memory CD8+ T cell differentiation. Immunity. 36:374–387. 10.1016/j.immuni.2012.01.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren X., Siegel R., Kim U., and Roeder R.G.. 2011. Direct interactions of OCA-B and TFII-I regulate immunoglobulin heavy-chain gene transcription by facilitating enhancer-promoter communication. Mol. Cell. 42:342–355. 10.1016/j.molcel.2011.04.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutishauser R.L., Martins G.A., Kalachikov S., Chandele A., Parish I.A., Meffre E., Jacob J., Calame K., and Kaech S.M.. 2009. Transcriptional repressor Blimp-1 promotes CD8(+) T cell terminal differentiation and represses the acquisition of central memory T cell properties. Immunity. 31:296–308. 10.1016/j.immuni.2009.05.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauter P., and Matthias P.. 1997. The B cell-specific coactivator OBF-1 (OCA-B, Bob-1) is inducible in T cells and its expression is dispensable for IL-2 gene induction. Immunobiology. 198:207–216. 10.1016/S0171-2985(97)80041-1 [DOI] [PubMed] [Google Scholar]
- Schubart D.B., Rolink A., Kosco-Vilbois M.H., Botteri F., and Matthias P.. 1996. B-cell-specific coactivator OBF-1/OCA-B/Bob1 required for immune response and germinal centre formation. Nature. 383:538–542. 10.1038/383538a0 [DOI] [PubMed] [Google Scholar]
- Sebastiano V., Dalvai M., Gentile L., Schubart K., Sutter J., Wu G.M., Tapia N., Esch D., Ju J.Y., Hübner K., et al. . 2010. Oct1 regulates trophoblast development during early mouse embryogenesis. Development. 137:3551–3560. 10.1242/dev.047027 [DOI] [PubMed] [Google Scholar]
- Shakya A., Kang J., Chumley J., Williams M.A., and Tantin D.. 2011. Oct1 is a switchable, bipotential stabilizer of repressed and inducible transcriptional states. J. Biol. Chem. 286:450–459. 10.1074/jbc.M110.174045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shakya A., Callister C., Goren A., Yosef N., Garg N., Khoddami V., Nix D., Regev A., and Tantin D.. 2015. Pluripotency transcription factor Oct4 mediates stepwise nucleosome demethylation and depletion. Mol. Cell. Biol. 35:1014–1025. 10.1128/MCB.01105-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sive H.L., Heintz N., and Roeder R.G.. 1986. Multiple sequence elements are required for maximal in vitro transcription of a human histone H2B gene. Mol. Cell. Biol. 6:3329–3340. 10.1128/MCB.6.10.3329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tantin D., Schild-Poulter C., Wang V., Haché R.J., and Sharp P.A.. 2005. The octamer binding transcription factor Oct-1 is a stress sensor. Cancer Res. 65:10750–10758. 10.1158/0008-5472.CAN-05-2399 [DOI] [PubMed] [Google Scholar]
- Tantin D., Voth W.P., and Shakya A.. 2013. Efficient chromatin immunoprecipitation using limiting amounts of biomass. J. Vis. Exp. 75:e50064 10.3791/50064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ullman K.S., Flanagan W.M., Edwards C.A., and Crabtree G.R.. 1991. Activation of early gene expression in T lymphocytes by Oct-1 and an inducible protein, OAP40. Science. 254:558–562. 10.1126/science.1683003 [DOI] [PubMed] [Google Scholar]
- Vigneau S., Rohrlich P.S., Brahic M., and Bureau J.F.. 2003. Tmevpg1, a candidate gene for the control of Theiler’s virus persistence, could be implicated in the regulation of gamma interferon. J. Virol. 77:5632–5638. 10.1128/JVI.77.10.5632-5638.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang V.E.H., Schmidt T., Chen J., Sharp P.A., and Tantin D.. 2004. Embryonic lethality, decreased erythropoiesis, and defective octamer-dependent promoter activation in Oct-1-deficient mice. Mol. Cell. Biol. 24:1022–1032. 10.1128/MCB.24.3.1022-1032.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitmire J.K., Asano M.S., Kaech S.M., Sarkar S., Hannum L.G., Shlomchik M.J., and Ahmed R.. 2009. Requirement of B cells for generating CD4+ T cell memory. J. Immunol. 182:1868–1876. 10.4049/jimmunol.0802501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams M.A., Tyznik A.J., and Bevan M.J.. 2006. Interleukin-2 signals during priming are required for secondary expansion of CD8+ memory T cells. Nature. 441:890–893. 10.1038/nature04790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams M.A., Ravkov E.V., and Bevan M.J.. 2008. Rapid culling of the CD4+ T cell repertoire in the transition from effector to memory. Immunity. 28:533–545. 10.1016/j.immuni.2008.02.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wirth T., Staudt L., and Baltimore D.. 1987. An octamer oligonucleotide upstream of a TATA motif is sufficient for lymphoid-specific promoter activity. Nature. 329:174–178. 10.1038/329174a0 [DOI] [PubMed] [Google Scholar]
- Wu G.D., Lai E.J., Huang N., and Wen X.. 1997. Oct-1 and CCAAT/enhancer-binding protein (C/EBP) bind to overlapping elements within the interleukin-8 promoter. The role of Oct-1 as a transcriptional repressor. J. Biol. Chem. 272:2396–2403. 10.1074/jbc.272.4.2396 [DOI] [PubMed] [Google Scholar]
- Yamane K., Toumazou C., Tsukada Y., Erdjument-Bromage H., Tempst P., Wong J., and Zhang Y.. 2006. JHDM2A, a JmjC-containing H3K9 demethylase, facilitates transcription activation by androgen receptor. Cell. 125:483–495. 10.1016/j.cell.2006.03.027 [DOI] [PubMed] [Google Scholar]
- Yoon H.S., Scharer C.D., Majumder P., Davis C.W., Butler R., Zinzow-Kramer W., Skountzou I., Koutsonanos D.G., Ahmed R., and Boss J.M.. 2012. ZBTB32 is an early repressor of the CIITA and MHC class II gene expression during B cell differentiation to plasma cells. J. Immunol. 189:2393–2403. 10.4049/jimmunol.1103371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yosef N., Shalek A.K., Gaublomme J.T., Jin H., Lee Y., Awasthi A., Wu C., Karwacz K., Xiao S., Jorgolli M., et al. . 2013. Dynamic regulatory network controlling TH17 cell differentiation. Nature. 496:461–468. 10.1038/nature11981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zens K.D., and Farber D.L.. 2015. Memory CD4 T cells in influenza. Curr. Top. Microbiol. Immunol. 386:399–421. 10.1007/82_2014_401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou L., Nazarian A.A., Xu J., Tantin D., Corcoran L.M., and Smale S.T.. 2007. An inducible enhancer required for Il12b promoter activity in an insulated chromatin environment. Mol. Cell. Biol. 27:2698–2712. 10.1128/MCB.00788-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwilling S., Dieckmann A., Pfisterer P., Angel P., and Wirth T.. 1997. Inducible expression and phosphorylation of coactivator BOB.1/OBF.1 in T cells. Science. 277:221–225. 10.1126/science.277.5323.221 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.