DNAM-1–mediated active biochemical signals initiated by a conserved ITT-like motif are essential for its capacity to enhance NK cell cytotoxicity and cytokine production. Enhancement of these signals may underlie the therapeutic impact of blocking anti-TIGIT antibodies for treatment of cancer and viral infections.
Abstract
DNAM-1 (CD226) is an activating receptor expressed on natural killer (NK) cells, CD8+ T cells, and other immune cells. Upon recognition of its ligands, CD155 and CD112, DNAM-1 promotes NK cell–mediated elimination of transformed and virus-infected cells. It also has a key role in expansion and maintenance of virus-specific memory NK cells. Herein, the mechanism by which DNAM-1 controls NK cell–mediated cytotoxicity and cytokine production was elucidated. Cytotoxicity and cytokine production triggered by DNAM-1 were mediated via a conserved tyrosine- and asparagine-based motif in the cytoplasmic domain of DNAM-1. Upon phosphorylation by Src kinases, this motif enabled binding of DNAM-1 to adaptor Grb2, leading to activation of enzymes Vav-1, phosphatidylinositol 3′ kinase, and phospholipase C-γ1. It also promoted activation of kinases Erk and Akt, and calcium fluxes. Although, as reported, DNAM-1 promoted adhesion, this function was signal-independent and insufficient to promote cytotoxicity. DNAM-1 signaling was also required to enhance cytotoxicity, by increasing actin polymerization and granule polarization. We propose that DNAM-1 promotes NK cell activation via an immunoreceptor tyrosine tail (ITT)–like motif coupling DNAM-1 to Grb2 and other downstream effectors.
DNAX accessory molecule-1 (DNAM-1), also known as CD226, is a receptor expressed on natural killer (NK) cells, CD8+ T cells, some CD4+ T cells, and some myeloid cells (Shibuya et al., 1996; Long et al., 2013; de Andrade et al., 2014; Martinet and Smyth, 2015). Although it is part of the Ig superfamily, DNAM-1 is rather unique. This uniqueness is especially evident in the cytoplasmic domain, which shares little or no homology with other Ig superfamily members. DNAM-1 recognizes CD155 (poliovirus receptor) and CD112 (nectin-2) as ligands, which are expressed on a broad range of cells, including transformed cells and virus-infected cells (Bottino et al., 2003). CD155, CD112, or both are also ligands for the inhibitory receptors TIGIT and CD96 (tactile), which are also expressed on immune cells. As loss of DNAM-1 enhances the availability of CD155 and CD112 for TIGIT and CD96, this feature complicates interpretation of phenotypes found in mice lacking DNAM-1.
DNAM-1 was initially identified as a molecule promoting cytotoxicity and cytokine secretion by NK cells and CD8+ T cells (Shibuya et al., 1996). Subsequent work revealed that DNAM-1 was important for NK cell–mediated killing of tumor cells such as melanoma cells, rhabdomyosarcoma cells, and Ewing’s sarcoma cells (Verhoeven et al., 2008; Lakshmikanth et al., 2009; Cho et al., 2010). Accordingly, DNAM-1–deficient mice were more susceptible to carcinogen-induced fibrosarcoma and papilloma in vivo (Gilfillan et al., 2008; Iguchi-Manaka et al., 2008). DNAM-1 was also implicated in NK cell–mediated elimination of HIV-infected CD4+ T cells and human cytomegalovirus (HCMV)-infected DCs (Magri et al., 2011; Matusali et al., 2012). Moreover, in mice, it played a critical role in expansion and survival of virus-specific memory NK cells in mouse cytomegalovirus (MCMV)–infected mice (Nabekura et al., 2014). A key role of DNAM-1 was also documented in CD8+ T cells. DNAM-1 was critical for the ability of CD8+ T cells to eliminate tumor cells and virus-infected cells during blocking antibody therapy targeting the inhibitory receptor TIGIT (Johnston et al., 2014). Likewise, DNAM-1 was necessary for the ability of CD8+ T cells to mediate acute graft-versus-host disease in mice (Nabekura et al., 2010).
Early studies suggested that human DNAM-1 promotes NK cell activation, at least in part, by acting as an adhesion receptor, which stabilizes physical contacts between NK cells and target cells (Shibuya et al., 1996, 1999). This function was reportedly dependent on the ability of DNAM-1 to bind in cis to integrin LFA-1. DNAM-1 was also shown to undergo phosphorylation at a conserved tyrosine (Y) in its cytoplasmic domain (Y319 in mouse and Y322 in human; Shibuya et al., 1999). This phosphorylation was reported to be mediated by Src family kinase Fyn. Although the precise role of Y319 was not elucidated (for simplification, mouse numbering will be used in this report), the ability of DNAM-1 to stimulate accumulation of virus-induced memory-like NK cells in mice was dependent on Y319 (Nabekura et al., 2014). Human DNAM-1 was also described to undergo phosphorylation at serine 326 (S326) in its cytoplasmic domain, as a result of the action of protein kinase C (Shibuya et al., 1998, 1999). This phosphorylation was reported to promote the DNAM-1–LFA-1 association and, in mice, to be critical for accumulation of memory-like NK cells in virus-infected mice (Shibuya et al., 1999; Nabekura et al., 2014).
Various reports have examined the possibility that DNAM-1 transduces intrinsic biochemical signals. In some studies, engagement of human DNAM-1 by anti–DNAM-1 antibodies failed to trigger activation of kinases Erk and Akt or calcium fluxes (Bryceson et al., 2006; Chen et al., 2007). In other studies, human DNAM-1 synergized with 2B4 to enhance tyrosine phosphorylation of adaptor SLP-76 and exchange factor Vav-1 (Kim and Long, 2012). This effect correlated with increased NK cell–mediated cytotoxicity and cytokine production. However, the mechanism by which DNAM-1 influenced these signals was not determined.
Whether DNAM-1 functions as an adhesion receptor, a signal transduction molecule or both during NK cell activation needs to be resolved. The structural basis for the ability of DNAM-1 to mediate possible signals, as well as the effectors of these signals, also requires clarification. Lastly, the cellular mechanism by which DNAM-1 promotes NK cell activation, i.e., enhanced cellular adhesion or other mechanisms, requires clarification.
Herein, we addressed these issues using a combination of genetic, biochemical, molecular, and imaging approaches. These studies showed that engagement of DNAM-1 triggered active signals, which were required for the ability to promote NK cell activation. These signals were initiated by Y319, but not S326, of DNAM-1. They also necessitated an asparagine at position 321 (N321), which cooperated with phosphorylated Y319 to bind adaptor Grb2. Binding of DNAM-1 to Grb2 enabled activation of exchange factor Vav-1, phosphatidylinositol (PI) 3′ kinase, and phospholipase C (PLC)-γ1. DNAM-1 signaling did not enhance cytotoxicity by promoting adhesion. Rather, it acted by enhancing actin polymerization and polarization of lytic granules toward target cells.
RESULTS
DNAM-1 triggers signals involving activation of Erk, Akt, and calcium fluxes
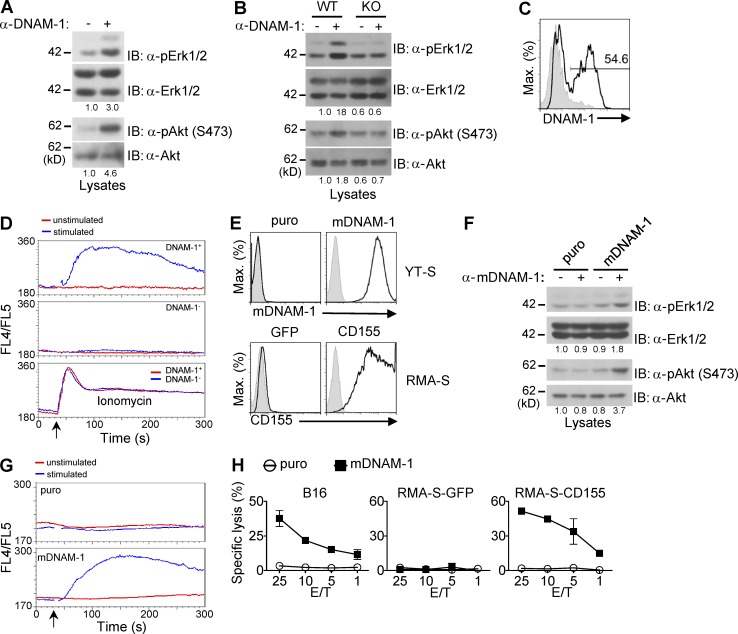
To resolve the mechanism by which DNAM-1 promotes NK cell activation, the ability of DNAM-1 to trigger intracellular signals in the absence of engagement of other activating receptors was examined (Fig. 1). Stimulation of normal mouse NK cells with anti–DNAM-1 antibodies (anti–DNAM-1) resulted in activation of kinases Erk and Akt (Fig. 1 A). This was not a result of triggering of CD16, an activating Fc receptor on NK cells, as these effects were lost in NK cells from DNAM-1–deficient mice (Fig. 1 B). We also examined the ability of DNAM-1 to trigger calcium fluxes (Fig. 1, C and D). Normal mouse NK cells contained both DNAM-1+ and DNAM-1− subsets, as previously reported (Tahara-Hanaoka et al., 2005; Fig. 1 C). Stimulation with anti–DNAM-1 induced calcium fluxes in DNAM-1+, but not in DNAM-1−, NK cells (Fig. 1 D).
Figure 1.
Impact of engagement of DNAM-1 on activation of Erk, Akt, and calcium fluxes in NK cells. (A) IL-2–activated NK cells from WT mice were stimulated for 2 min with anti–mouse DNAM-1 (480.2), followed by secondary antibodies (+) or secondary antibodies alone (-). Activation of Erk and Akt was assessed by immunoblotting with antibodies against phosphorylated Erk (α-pErk) or Akt (α-pAkt). Quantitation of relative phosphorylation is shown at the bottom. Representative of at least n = 3. (B) Same as A, except that WT and DNAM-1–deficient (KO) mice were analyzed. Representative of at least n = 3. (C) Expression of DNAM-1 on splenic NK cells (defined as NK1.1+CD3− cells) was analyzed by flow cytometry. Isotype control is shown as filled histograms. Percentage of DNAM-1+ NK cells is indicated. Representative of at least n = 5. (D) IL-2–activated NK cells were labeled with FITC-conjugated anti–mouse DNAM-1 and loaded with Indo-1, followed by secondary antibodies (stimulated) or not (unstimulated). The changes of intracellular calcium (monitored as FL4/FL5 ratio) in DNAM-1+ (top) and DNAM-1− (middle) NK cells were analyzed. Cells were treated with ionomycin (bottom) as positive control. Arrow indicates when the secondary antibodies or ionomycin was added. Representative of n = 5. (E) Expression of mouse DNAM-1 (top) and mouse CD155 (bottom) on transfected or retrovirus-transduced YT-S cells and RMA-S cells, respectively, was determined by flow cytometry. Cells expressing the puromycin resistance marker (puro) or GFP alone were used as control. Isotype control is shown as filled histogram. Representative of at least n = 3. (F) YT-S cells expressing mouse DNAM-1 (mDNAM-1) or puromycin-resistant marker (puro) alone were stimulated with anti–DNAM-1, and activation of Erk and Akt was assessed, as detailed for A. Representative of n = 5. (G) YT-S cells expressing mouse DNAM-1 (mDNAM-1) or puromycin-resistant marker (puro) alone were loaded with Indo-1. They were then stimulated with anti–mouse DNAM-1, followed by secondary antibodies (stimulated), or secondary antibodies alone (unstimulated). Changes of intracellular calcium were monitored as FL4/FL5 ratio. Arrow indicates when the secondary antibodies or ionomycin was added. Representative of n = 3. (H) Natural cytotoxicity of YT-S cells expressing mouse DNAM-1 (mDNAM-1) or puromycin resistant marker (puro) alone was tested in a 51Cr-release assay at indicated effector-to-target (E/T) ratio. The target cells used were B16 or RMA-S expressing or not expressing mouse CD155. SDs of duplicate values are depicted by error bars. Representative of n = 3.
Mouse DNAM-1 was also ectopically expressed in the human NK cell line YT-S, which lacks CD16 (Fig. 1, E and H; and not depicted). Triggering of mouse DNAM-1 on YT-S cells resulted in activation of Erk and Akt (Fig. 1, E and F). Additionally, it triggered calcium fluxes (Fig. 1 G). Importantly, expression of mouse DNAM-1 on YT-S also enabled killing of B16, a mouse melanoma cell line expressing ligands for DNAM-1 (Dong et al., 2009), as well as RMA-S mouse lymphoma cells ectopically expressing mouse CD155 (Fig. 1, E and H). No killing was seen toward RMA-S cells lacking CD155, or when YT-S cells lacked mouse DNAM-1. Consequently, in this experimental system, engagement of DNAM-1 was strictly needed for target cell killing.
These data showed that engagement of DNAM-1 on NK cells triggered active biochemical signals involving activation of Erk, Akt, and calcium fluxes. These signals correlated with enhanced natural cytotoxicity.
Y319, but not S326, is critical for DNAM-1–mediated signaling and cytotoxicity
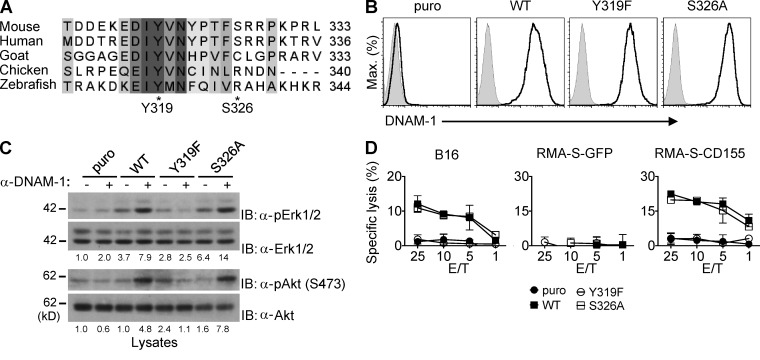
To determine the mechanism by which DNAM-1 coupled to these signals, we focused on the cytoplasmic domain. Although the sequence in this region is quite divergent across species, there is a stretch of 5 aa, D/EIYV/MN (where D is aspartic acid, E is glutamic acid, V is valine, M is methionine, and N is asparagine), that is highly conserved in all species, including mouse and human (Fig. 2 A). This sequence encompasses the aforementioned site of phosphorylation, Y319 (Shibuya et al., 1999). The other proposed phosphorylation site, S326, is located outside of this motif (Shibuya et al., 1998). Moreover, S326 is present in mouse and human, but not in other species.
Figure 2.
Y319, but not S326, is required for DNAM-1–dependent natural cytotoxicity. (A) Alignment of the C-terminal sequence of DNAM-1 from various species. Amino acids that are strictly conserved are highlighted in dark gray. Partially conserved residues are shown in lighter gray. The positions of tyrosine 319 (Y319) and serine 326 (S326) in the mouse sequence (indicated by asterisks) are shown. (B) Expression of DNAM-1 on YT-S cells transfected with cDNAs encoding WT mouse DNAM-1 or mouse DNAM-1 mutants carrying a tyrosine 319-to-phenylalanine 319 (Y319F) or serine 326-to-alanine 326 (S326A) substitution was determined by flow cytometry. Cells expressing the puromycin resistant marker (puro) alone are included as control. Isotype controls are shown as filled histograms. Representative of n = 4. (C) The YT-S cells described in B were stimulated with anti–DNAM-1 and activation of Erk and Akt was assessed, as detailed for Fig. 1 F. Quantitation of relative phosphorylation is shown at the bottom. Representative of n = 3. (D) The YT-S cells described in B were tested for natural cytotoxicity, as detailed for Fig. 1 H. SDs of duplicate values are depicted by error bars. Representative of n = 4.
To examine if these phosphorylation sites were critical for the ability of DNAM-1 to trigger signals and promote cytotoxicity, mutants of mouse DNAM-1 carrying a tyrosine 319-to-phenylalanine 319 (Y319F) or a serine 326-to-alanine 326 (S326A) mutation, two conserved substitutions aimed at preventing phosphorylation without affecting structure, were introduced in YT-S cells. Both mutants were expressed in amounts analogous to those of WT DNAM-1 (Fig. 2 B). The Y319F mutation fully abrogated the capacity of DNAM-1 to trigger activation of Erk and Akt (Fig. 2 C). In contrast, the S326A substitution had no effect. Likewise, Y319F, but not S326A, eliminated the capacity of DNAM-1 to mediate cytotoxicity toward B16 cells or RMA-S cells expressing CD155 (Fig. 2 D).
Hence, the highly conserved Y319, but not the less conserved S326, was needed for the ability of DNAM-1 to stimulate intracellular signals and cytotoxicity in YT-S cells.
Generation and analyses of DNAM-1 Y319F knock-in (KI) mouse
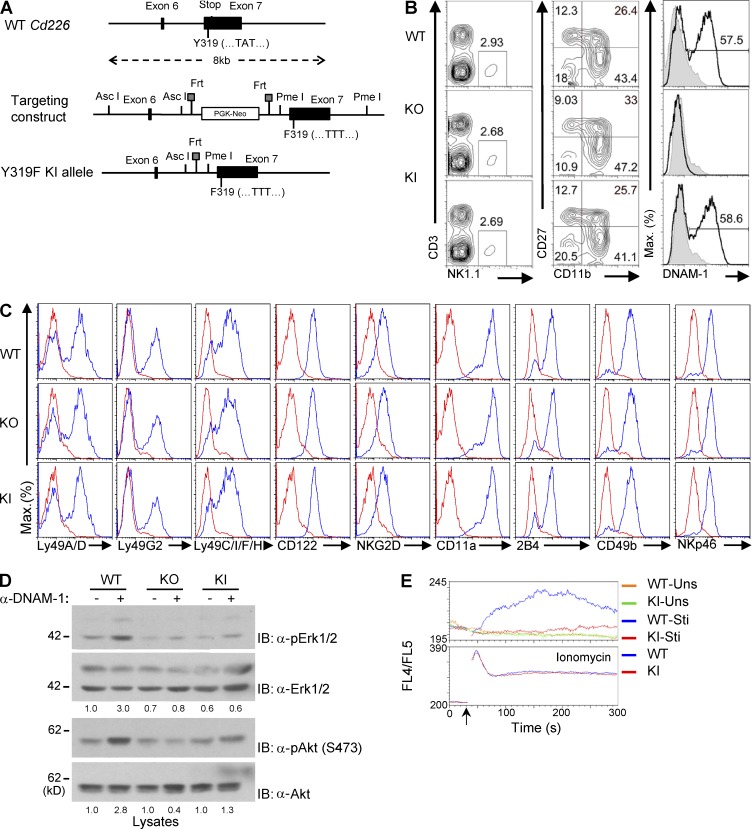
To establish the physiological relevance of this mechanism, a KI mouse, in which Y319 was changed to phenylalanine in the germline (Cd226Y319F mouse), was created (Fig. 3). The construct depicted in Fig. 3 A was used to introduce the mutation in embryonic stem (ES) cells from C57BL/6 mice. After germline transmission of the mutation, breeding of mutant mice with animals expressing the Flpe recombinase eliminated the neo selection cassette. As reported for DNAM-1–deficient mice (Gilfillan et al., 2008; Iguchi-Manaka et al., 2008), DNAM-1 Y319F mice exhibited no alterations of NK cell numbers, development, or repertoire (Fig. 3, B and C). These mice also displayed unaltered levels of DNAM-1 on NK cells. The proportion of DNAM-1+ NK cells was also not affected.
Figure 3.
Generation and signaling analyses of mice expressing DNAM-1 Y319F. (A) The genomic structure of the DNAM-1–encoding gene in the mouse, Cd226, is shown at the top. Tyrosine 319 (Y319) is encoded by exon 7, which also contains the stop codon. The targeting construct is shown in the middle. In this construct, the WT codon for Y319, TAT, was replaced by a codon for phenylalanine, TTT. The final Y319F knock-in (KI) Cd226 allele carrying the mutated codon is shown at the bottom. (B) Splenocytes from WT, DNAM-1–deficient (KO), or DNAM-1 Y319F (KI) mice were analyzed by flow cytometry. Proportions of NK cells (NK1.1+CD3− cells) are indicated (left). Expression of CD11b, CD27, and DNAM-1 on NK cells was also analyzed (middle and right). Percentages of NK cells in each quadrant (middle), as well as of DNAM-1+ NK cells (right), are shown. Isotype controls are depicted as filled histogram. Representative of n = 4. (C) Expression of various receptors was analyzed on NK1.1+CD3− splenocytes from WT, KI, or KO mice, using flow cytometry. Isotype controls are shown as the red line. Representative of n = 2. (D) Same as Fig. 1 A, except that WT, DNAM-1 KO, and DNAM-1 KI NK cells were analyzed. Quantitation of relative phosphorylation is shown at the bottom. Representative of n = 3. (E) Same as Fig. 1 D, except that only DNAM-1+ NK cells were analyzed. Arrow indicates when the secondary antibodies or ionomycin was added. Uns, unstimulated; Sti, stimulated. Representative of n = 3.
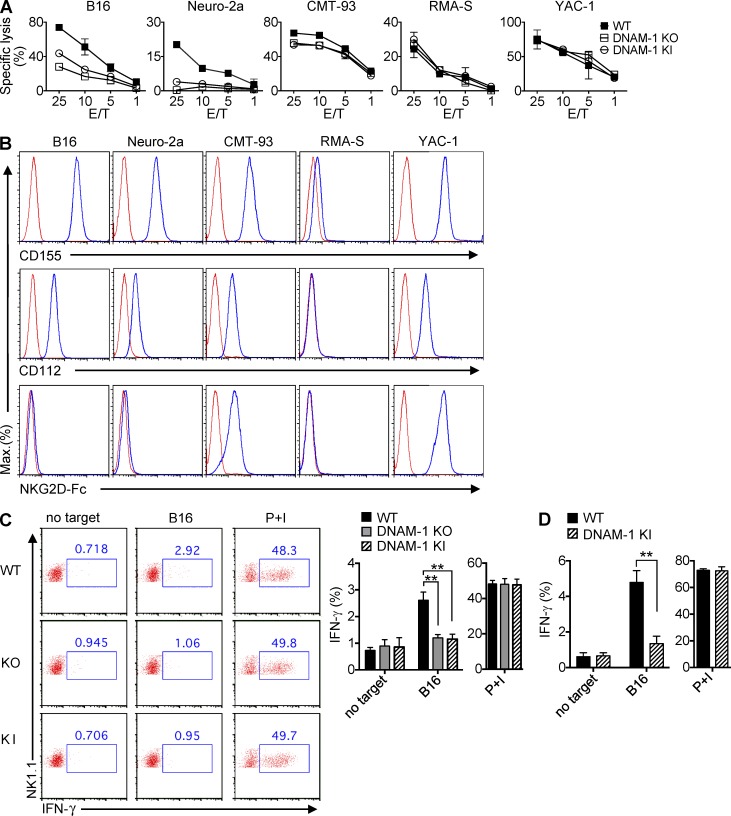
As was the case for NK cells lacking DNAM-1, NK cells expressing DNAM-1 Y319F displayed compromised activation of Erk and Akt in response to anti–DNAM-1 (Fig. 3 D). A markedly reduced ability to evoke calcium fluxes was also observed (Fig. 3 E). Furthermore, like DNAM-1–deficient NK cells, NK cells expressing DNAM-1 Y319F showed a reduced ability to kill the nonhematopoietic target cells B16 and Neuro-2a (neuroblastoma), which express ligands for DNAM-1, but not for NKG2D (Dong et al., 2009; Fig. 4, A and B). A defect was also seen toward CMT-93 (rectal carcinoma), which expresses ligands for DNAM-1 and NKG2D. Notably, with these three targets, the defects observed with DNAM-1 Y319F NK cells were slightly less severe than those noted with DNAM-1–deficient NK cells. The possible significance of this finding will be discussed below. No defect was seen in cytotoxicity toward RMA-S or YAC-1, either with DNAM-1–deficient or with DNAM-1 Y319F NK cells.
Figure 4.
Cytotoxicity and cytokine production by DNAM-1 Y319F mouse NK cells. (A) Natural cytotoxicity of NK cells from WT, DNAM-1 KO, or DNAM-1 KI mice was tested in a 51Cr-release assay, using as targets B16, Neuro-2a, CMT-93, RMA-S, or YAC-1. SDs of duplicate values are depicted by error bars. Representative of n = 5. (B) Expression of ligands for DNAM-1 and NKG2D on target cells. Expression of the DNAM-1 ligands, CD155 and CD112, and NKG2D ligands (detected using an NKG2D-Fc fusion protein) was assessed by flow cytometry. Isotype controls are shown as the red line. Representative of n = 3. (C and D) Splenocytes from poly(I:C)-primed mice were incubated with B16 for 6 h. Production of IFN-γ in total NK cells (NK1.1+CD3−; C) or DNAM-1+ NK cells (NK1.1+CD3−DNAM-1+; D) was analyzed by intracellular staining. Splenocytes without any target or splenocytes stimulated with phorbol myristate acetate and ionomycin (PMA/iono) served as controls. Representative dot plots are shown in C (left). Average values with SDs of triplicates from one representative experiment are depicted in C (right) and D. Unpaired Student’s t tests were performed. **, P < 0.01. Representative of n = 3 (C and D).
We also tested the impact of the Y319F mutation on production of IFN-γ (Fig. 4 C). Splenic NK cells were incubated or not with B16, and IFN-γ production was determined by intracellular staining. As was the case of lack of DNAM-1, mutation of Y319F abrogated the ability of DNAM-1 to promote IFN-γ production in response to B16. No difference was seen when cells were activated with phorbol myristate acetate (PMA) and ionomycin, which trigger cytokine production by bypassing activating receptors. Because only ∼50% of mouse NK cells express DNAM-1, we also compared production of IFN-γ by DNAM-1+ NK cells from WT or DNAM-1 KI mice (Fig. 4 D). Compared with WT NK cells, DNAM-1 KI NK cells displayed a markedly attenuated IFN-γ production in the presence of B16.
Thus, as was the case in YT-S cells, mutation of Y319 severely attenuated the ability of DNAM-1 to mediate signals, as well as to promote cytotoxicity and cytokine production in mouse NK cells.
DNAM-1 promotes cytotoxicity via a tyrosine- and asparagine-based motif that couples to Grb2
We postulated that, when phosphorylated, Y319 was coupling DNAM-1 to a specific set of intracellular signaling molecules promoting NK cell activation. To identify these effectors, we opted to use primarily the YT-S system, for the following reasons. First, it was easier to generate large numbers of YT-S cells for biochemical studies. Second, our derivatives of YT-S cells uniformly expressed mouse DNAM-1, whereas only ∼50% of mouse NK cells expressed DNAM-1 and this fraction was further diminished when NK cells were expanded in IL-2 (unpublished data). Third, stimulation of mouse DNAM-1 on YT-S cells generated signals (activation of Erk, Akt, and calcium fluxes) analogous to those seen in normal mouse NK cells. Moreover, in both systems, these signals were dependent on Y319. And fourth, YT-S cells were ideal for structure–function analyses of DNAM-1, as expression of mouse DNAM-1 in these cells was absolutely required for killing of mouse target cells expressing DNAM-1 ligands.
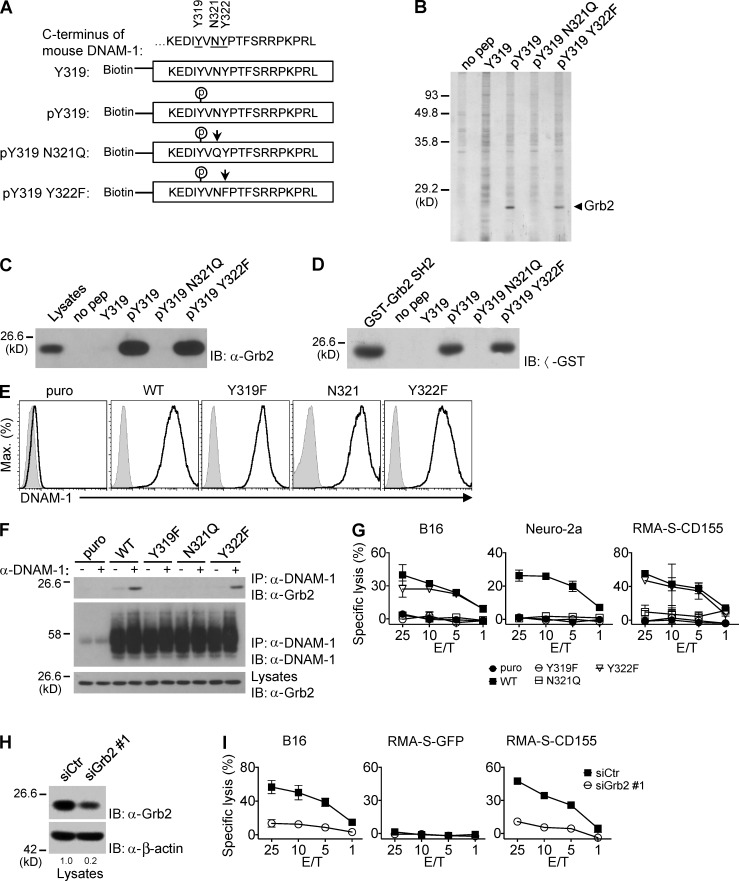
To identify effectors that directly interacted with DNAM-1, peptides corresponding to aa 315–333 in the cytoplasmic domain of mouse DNAM-1 were synthesized, phosphorylated or not at Y319 (Fig. 5 A). After immobilization onto agarose beads, peptides were used to capture binding proteins from lysates of YT-S cells (Fig. 5 B). Bound proteins were identified by silver staining of protein gels. A single polypeptide of ∼25 kD (p25) was bound to the peptide phosphorylated at Y319, but not to the unphosphorylated peptide. Mass spectrometry revealed that p25 was Grb2, an intracellular adaptor composed of one Src homology 2 (SH2) domain flanked by two Src homology 3 (SH3) domains (Lowenstein et al., 1992; Pawson and Scott, 1997). The ability of the phosphorylated DNAM-1 peptide to bind Grb2 was confirmed by immunoblotting of peptide-associated proteins with anti-Grb2 antibodies (Fig. 5 C).
Figure 5.
DNAM-1 promotes NK cell activation via a tyrosine- and asparagine-based motif that recruits Grb2. (A) Biotinylated peptides corresponding to the carboxyl-terminus of mouse DNAM-1 were synthesized, with (pY319) or without (Y319) phosphorylation at tyrosine 319. In some peptides, additional mutations were introduced: N321Q, in which asparagine 321 was mutated to glutamine and Y322F, in which tyrosine 322 was mutated to phenylalanine. The mutated residues are indicated by arrows. (B) DNAM-1 peptides were coupled to agarose beads and used to capture binding proteins from lysates of YT-S cells. Bound proteins were identified by silver staining of protein gels. Agarose beads alone (no pep) served as control. The migration of molecular mass markers is indicated on the left. Representative of n = 4. (C) DNAM-1 peptides were incubated with lysates of YT-S cells and bound proteins were probed by immunoblotting with anti-Grb2 antibodies. Representative of n = 4. (D) DNAM-1 peptides were incubated with lysates of bacteria expressing the indicated glutathione-S-transferase (GST) fusion proteins. Nonpurified GST-Grb2 SH2 fusion proteins served as control. Representative of n = 2. (E and F) YT-S cells were transfected with cDNAs encoding WT, Y319F, N321Q, or Y322F mouse DNAM-1, or the puromycin resistant marker (puro) alone. Expression of mouse DNAM-1 was tested by flow cytometry. Isotype controls are shown as filled histogram (E). Representative of n = 4. They were then stimulated (+) or not (−) with anti–DNAM-1 antibodies and the relevant secondary antibody (F). The association of DNAM-1 with Grb2 was detected by immunoblotting of DNAM-1 immunoprecipitates with anti-Grb2 antibodies. Migration of heavy chain of IgG is shown on the left. Representative of n = 3. (G) The YT-S cells described in Fig. 5 E were tested for natural cytotoxicity, as detailed for Fig. 1 H. SDs of duplicate values are depicted by error bars. Representative of n = 3. (H and I) YT-S cells expressing WT mouse DNAM-1 were transfected with Grb2-specific siRNAs (siGrb2 #1) or irrelevant siRNAs (siCtr) as control. Down-regulation of Grb2 expression was assessed by immunoblotting with anti-Grb2 (H). Cells were also tested for natural cytotoxicity, as detailed for Fig. 1 H (I). SDs of duplicate values are depicted by error bars. Quantitation of relative protein expression is shown at the bottom (H). Representative of n = 4.
Phosphorylated Y319 (pY319) is nested in the sequence pYVNY, which is compatible with the consensus binding motif for the SH2 domain of Grb2, pYxNx (where x is any residue; Songyang et al., 1994). Accordingly, the phosphorylated DNAM-1 peptide bound to recombinant Grb2 SH2 domains in vitro (Fig. 5 D). Moreover, the association between the phosphorylated DNAM-1 peptide was eliminated when the highly conserved asparagine at the +2 position was mutated to glutamine (N321Q mutation; Fig. 5, C and D). However, it was not affected when the less conserved residue at +3 position (Y322) was altered (Y322F mutation).
To show that DNAM-1 associated with Grb2 via the pYxNx motif in cells, YT-S cells expressing WT, Y319F, N321Q, or Y322F DNAM-1 were created and stimulated with anti–DNAM-1 (Fig. 5, E and F). The association of DNAM-1 with Grb2 was detected by immunoblotting of anti–DNAM-1 immunoprecipitates with anti-Grb2 (Fig. 5 F). WT DNAM-1 coimmunoprecipitated with Grb2 in cells stimulated with anti–DNAM-1 antibodies. Minimal association was seen in unstimulated cells. As shown in the aforementioned in vitro binding assays, this association was abolished by the Y319F or N321Q, but not the Y322F, mutation. All mutants were expressed in amounts analogous to those of WT DNAM-1 (Fig. 5 E).
To ascertain if Grb2 binding was critical for the activating function of DNAM-1, we first tested the ability of these various DNAM-1 mutants to promote cytotoxicity (Fig. 5 G). As was the case for mutation of Y319, the N321Q mutation eliminated the ability of DNAM-1 to enhance cytotoxicity toward B16, Neuro-2a, and CD155-expressing RMA-S. In contrast, the Y322F mutation had no effect.
Next, we ascertained the impact of reduced Grb2 expression on DNAM-1 function. Unfortunately, these experiments could not be performed with NK cells from Grb2-deficient mice, as lack of Grb2 interfered with proper NK cell development in mice (unpublished data). Hence, we down-regulated expression of Grb2 in YT-S cells, using siRNAs (Fig. 5, H and I; and not depicted). Two different Grb2-specific siRNAs, but not control siRNAs, induced an ∼40–70% diminution of Grb2 expression in YT-S cells (Fig. 5 H; and not depicted). Importantly, they also caused a reduction of cytotoxicity toward B16 and RMA-S cells expressing CD155 (Fig. 5 I; and not depicted). As the ability of YT-S cells to kill RMA-S cells expressing CD155 was strictly dependent on expression of DNAM-1 on YT-S cells (Fig. 1 H), the latter finding supported the notion that Grb2 was directly implicated in the activating function of DNAM-1.
Combined, these results showed that phosphorylation of the highly conserved tyrosine- and asparagine-based motif of DNAM-1 resulted in recruitment of Grb2, which was needed for the ability of DNAM-1 to promote NK cell–mediated cytotoxicity.
Grb2 couples DNAM-1 to Vav-1, PI3′K, and PLC-γ1
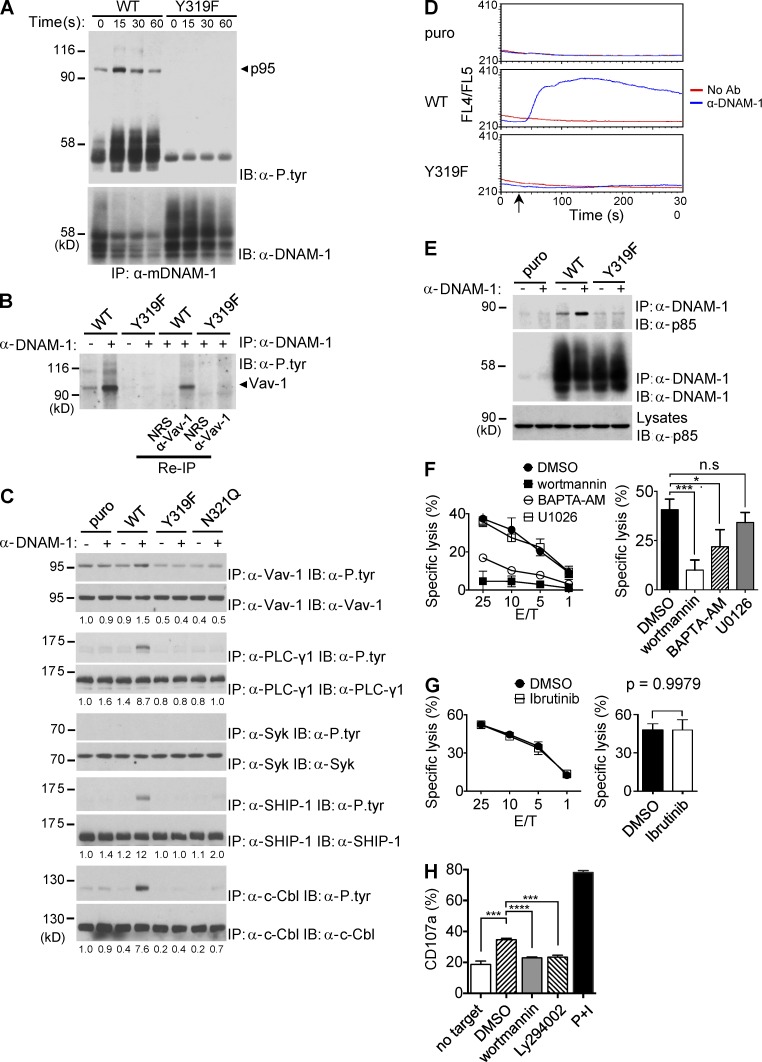
Next, we postulated that, whereas the SH2 domain of Grb2 interacted with tyrosine-phosphorylated DNAM-1, one or both of its SH3 domains were involved in recruitment of downstream effectors. To identify these effectors, several approaches were taken. First, DNAM-1 was immunoprecipitated from YT-S cells expressing WT DNAM-1 or DNAM-1 Y319F, stimulated or not with anti–DNAM-1. DNAM-1–associated proteins were then detected by immunoblotting with antiphosphotyrosine antibodies (Fig. 6 A). Immunoprecipitates of WT DNAM-1 from stimulated cells contained tyrosine phosphorylated proteins of ∼45–50 kD (p45-50) and ∼95 kD (p95). Both products were less prominently detectable in unstimulated cells and were absent in cells bearing DNAM-1 Y319F.
Figure 6.
The tyrosine- and asparagine-based motif couples DNAM-1 to multiple effectors. (A) YT-S cells expressing WT or Y319F mouse DNAM-1 were stimulated for the indicated times with anti–DNAM-1 and the relevant secondary antibody. DNAM-1 was then immunoprecipitated from cell lysates, and DNAM-1–associated proteins were detected by immunoblotting with anti-phosphotyrosine antibodies (α-P.tyr). The positions of DNAM-1 and p95 are indicated on the right. Migration of prestained molecular weight markers and heavy chain of IgG is shown on the left. It is notable that the amount of DNAM-1 recovered from cells expressing WT DNAM-1, but not DNAM-1 Y319F, was diminished upon anti–DNAM-1 stimulation. This finding likely indicated that signaling-competent DNAM-1 became detergent-insoluble upon stimulation. Representative of n = 4. (B) YT-S cells expressing the indicated variants of DNAM-1 were stimulated for 15 s with anti–DNAM-1 antibodies and the relevant secondary antibody. After extensive washing, eluates of DNAM-1 immunoprecipitates were reimmunoprecipitated with anti-Vav-1 or normal rabbit serum (NRS). Tyrosine phosphorylated proteins were detected by anti-phosphotyrosine immunoblotting. Representative of n = 3. (C) YT-S cells expressing the indicated variants of DNAM-1 or the puromycin resistance marker (puro) alone were stimulated with anti–DNAM-1 followed by the relevant secondary antibodies. Lysates were then immunoprecipitated with antibodies against the specified substrates and tyrosine phosphorylation was analyzed by immunoblotting with α-P.tyr. Quantitation of relative phosphorylation is shown at the bottom. Representative of at least n = 3. (D) YT-S cells expressing the indicated variants of DNAM-1 or the puromycin resistance marker (puro) alone were stimulated with anti–DNAM-1 antibodies and the relevant secondary antibody. Changes in intracellular calcium were analyzed as described for Fig. 1 G. Representative of n = 3. (E) YT-S cells expressing the indicated variants of DNAM-1 or the puromycin resistance marker (puro) alone were stimulated for 15 s with anti–DNAM-1 antibodies and the relevant secondary antibody. DNAM-1 was then immunoprecipitated and its association with p85 was detected by immunoblotting with anti-p85. Representative of n = 3. (F and G) YT-S cells expressing WT mouse DNAM-1 were treated with wortmannin (0.1 µM), 1,2-bis(2-aminophenoxy)ethane-N,N,N′,N′-tetraacetic acid tetrakis(acetoxymethyl ester; BAPTA-AM; 40 µM), U0126 (1 µM), ibrutinib (10 nM), or dimethysulfoxide (DMSO) alone as control, and their capacity to kill RMA-S-CD155 cells was assessed in a 51Cr-release assay (left). Average values at the 25:1 E/T ratio from 5 independent experiments are shown on the right. Representative of n = 5 (F) or n = 3 (G). SDs of duplicate values are depicted by error bars. Unpaired Student’s t tests were performed. n.s., not statistically significant. *, P < 0.05; ***, P < 0.001. (H) Splenocytes from WT mice were stimulated with B16 cells in the presence of the indicated compounds, as detailed for H. Expression of CD107a on DNAM-1+ NK cells (NK1.1+CD3−DNAM-1+) was analyzed by flow cytometry. Splenocytes alone or splenocytes stimulated with phorbol myristate acetate and ionomycin (PMA/iono) served as controls. Average values with SDs of triplicate values from one representative experiment are depicted. Unpaired Student’s t tests were performed. ***, P < 0.001; ****, P < 0.0001. Representative of n = 3.
Whereas p45-50 was likely DNAM-1 itself, p95 was suggestive of Vav-1, an exchange factor known to associate with Grb2 SH3 domains (Ramos-Morales et al., 1994; Bustelo, 2001). To address if p95 was Vav-1, denatured eluates of DNAM-1 immunoprecipitates were reimmunoprecipitated with anti–Vav-1 antibodies. Tyrosine-phosphorylated proteins were then detected by antiphosphotyrosine immunoblotting (Fig. 6 B). Tyrosine-phosphorylated p95 was recovered by reimmunoprecipitation with anti–Vav-1 antibodies, but not with normal rabbit serum. This experiment confirmed that the DNAM-1–associated p95 was Vav-1.
We then evaluated whether other known regulators of NK cell activation were tyrosine phosphorylated in response to DNAM-1 engagement by immunoprecipitating total cell lysates with substrate-specific antibodies, followed by antiphosphotyrosine immunoblotting (Fig. 6 C). Engagement of WT DNAM-1 triggered tyrosine phosphorylation not only of Vav-1, but also of phospholipase (PLC)-γ1, ubiquitin ligase c-Cbl, and lipid phosphatase SHIP-1. There was no impact on tyrosine phosphorylation of kinase Syk or adaptor SLP-76 (Fig. 6 C; not depicted). These signals were absent in cells containing DNAM-1 Y319F or DNAM-1 N312Q. Mutation of Y319F also abolished the ability of DNAM-1 to increase intracellular calcium, which is a downstream effector of PLC-γ1 (Fig. 6 D). It should be noted that PLC-γ1, c-Cbl, and SHIP-1 were not detectably associated with DNAM-1 (unpublished data).
Next, activation of Akt in response to DNAM-1 (Fig. 1) raised the possibility that DNAM-1 also stimulated PI3′K, a key upstream regulator of Akt. Because the Grb2 SH3 domains can bind to the PI3′K regulatory subunit p85 (Wang et al., 1995), we examined whether engagement of DNAM-1 triggered recruitment of p85 to DNAM-1 (Fig. 6 E). Triggering of WT DNAM-1, but not DNAM-1 Y319F, induced coimmunoprecipitation of DNAM-1 with p85. This effect was absent in cells lacking DNAM-1.
Finally, to help identify the signals playing key roles in DNAM-1–mediated cytotoxicity, YT-S cells expressing mouse DNAM-1 were treated with various pharmacological inhibitors, and the impact on cytotoxicity toward RMA-S cells expressing CD155 was determined (Fig. 6, F and G). This system was chosen because killing of the target cell variant was strictly dependent on DNAM-1 function in YT-S cells (Fig. 1 H). Wortmannin, a PI3′K inhibitor, caused a profound suppression of cytotoxicity mediated by DNAM-1 (Fig. 6 F and not depicted). A less prominent effect was seen with 1,2-bis(2-aminophenoxy)ethane-N,N,N′,N′-tetraacetic acid tetrakis(acetoxymethyl ester; BAPTA-AM), a calcium chelator. However, no impact was observed with U0126, an inhibitor of the Erk pathway, or ibrutinib, an inhibitor of the kinase Btk (Fig. 6, F and G). These various compounds had no deleterious effect on cell viability (unpublished data).
To provide evidence that PI3′K was also involved in DNAM-1–dependent degranulation in primary NK cells, we tested the effect of two PI3′K inhibitors, wortmannin and Ly294002, on the ability of primary mouse NK cells to expose CD107a, a marker of granule exocytosis, in response to B16 melanoma cells (Fig. 6 H). Treatment with either inhibitor resulted in significant inhibition on the increased CD107a exposure in response to B16 cells.
As Vav-1, c-Cbl, SHIP-1, and p85 bind the Grb2 SH3 domains (Ramos-Morales et al., 1994; Odai et al., 1995; Kavanaugh et al., 1996), these findings supported that idea that Grb2 was responsible for coupling DNAM-1 to these effectors. Our data with pharmacological inhibitors also suggested that activation of PI3′K and, to a lesser extent, calcium fluxes (a PLC-γ1 effector) were especially important for DNAM-1–dependent cytotoxicity.
Fyn and LFA-1 are not critical for DNAM-1 signaling
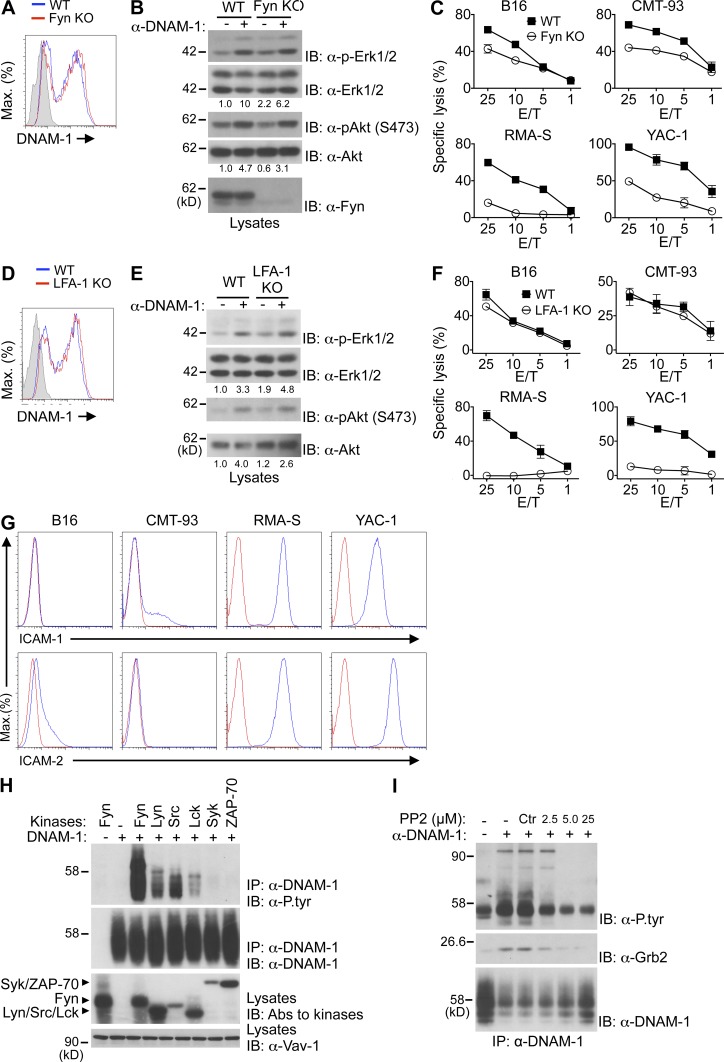
Previous studies suggested that Fyn was critical for phosphorylation of DNAM-1 at Y319 and, thus, for DNAM-1 function (Shibuya et al., 1999). To assess if Fyn was needed for DNAM-1 signaling, the ability of DNAM-1 to mediate signals and enhance cytotoxicity was examined in NK cells from Fyn-deficient mice (Fig. 7, A–C). Fyn deficiency had no impact on DNAM-1 expression on NK cells (Fig. 7 A). More importantly, it had little or no effect on the capacity of DNAM-1 to trigger activation of Erk and Akt (Fig. 7 B). Nonetheless, as previously reported, loss of Fyn reduced the ability of NK cells to kill a broad range of targets (Bloch-Queyrat et al., 2005; Dong et al., 2012; Fig. 7 C). This effect was seen whether target cells expressed or not DNAM-1 ligands. These findings indicated that Fyn was not absolutely required for the signaling capacity of DNAM-1. However, it was needed for NK cell–mediated cytotoxicity, presumably by receptors other than DNAM-1.
Figure 7.
Role of Fyn and LFA-1 in DNAM-1 signaling. (A) Expression of DNAM-1 was analyzed on NK cells (NK1.1+CD3− splenocytes) from WT and Fyn-deficient (KO) mice. Isotype control is shown as filled histogram. Representative of n = 4. (B) NK cells from the indicated mice were stimulated for 2 min with anti–DNAM-1 antibodies and the relevant secondary antibody. Phosphorylation of Erk and Akt was assessed as detailed for Fig. 1 A. Quantitation of relative phosphorylation is shown at the bottom. Representative of n = 2. (C) Natural cytotoxicity of NK cells toward a variety of targets was tested in a 51Cr-release assay at indicated effector-to-target (E/T) ratio. SDs of duplicate values are depicted by error bars. Representative of n = 4. (D–F) Same as A–C, except that WT and LFA-1–deficient (KO) mice were analyzed. Representative of n = 3. (G) Expression of LFA-1 ligands on the indicated targets was determined by flow cytometry. Red line, isotype control. Representative of n = 2. (H) Mouse DNAM-1 was coexpressed with the indicated protein tyrosine kinases (PTKs) in Cos-1 cells. DNAM-1 was then immunoprecipitated and probed by immunoblotting with anti-phosphotyrosine (α-P.tyr) antibodies. Expression of PTKs was detected by probing total cell lysates with a cocktail of antibodies against the various PTKs. Representative of n = 3. (I) YT-S cells expressing mouse DNAM-1 were stimulated for 15 s with anti–DNAM-1 and the relevant secondary antibodies, in the absence or in the presence of the indicated concentrations of the Src family kinase inhibitor, PP2. DNAM-1 then was recovered from cell lysates by immunoprecipitation and probed by immunoblotting with anti-phosphotyrosine (α-P.tyr) or anti-Grb2 antibodies. Treatment with DMSO alone served as control (Ctr). The migration of molecular mass markers is indicated on the left. Representative of n = 3.
It was also proposed that LFA-1 was critical for the capacity of DNAM-1 to promote NK cell functions (Shibuya et al., 1999). To examine if LFA-1 was required for DNAM-1 signaling, the function of DNAM-1 was examined in NK cells from LFA-1–deficient mice (Fig. 7, D–F). Lack of LFA-1 had no impact on DNAM-1 expression (Fig. 7 D). Surprisingly, it also had no effect on the capacity of DNAM-1 to activate Erk and Akt (Fig. 7 E). However, lack of LFA-1 reduced the ability of NK cells to kill some, but not all, target cells. As was the case for Fyn deficiency, this effect was seen whether target cells did or did not express DNAM-1 ligands. In particular, an effect was seen on RMA-S, which expressed ICAM-1 and ICAM-2, but did not express DNAM-1 ligands (Fig. 7 F). In contrast, there was no impact on B16 and CMT-93, which had little or no ICAM-1 and ICAM-2 (Fig. 7 G), but were killed in a DNAM-1–dependent manner (Fig. 4). These findings implied that LFA-1 was not strictly needed for the signaling capabilities and activating function of DNAM-1.
Next, we determined which classes of PTKs were responsible for phosphorylation of Y319 of DNAM-1 (Fig. 7, H and I). Co-transfection experiments in Cos-1 cells revealed that Src family kinases Fyn and, to a lesser extent, Lyn, Src, and Lck, were able to trigger tyrosine phosphorylation of DNAM-1 (Fig. 7 H). No phosphorylation was induced by Syk and ZAP-70, which belong to a distinct PTK family. In the light of these results, we examined the impact of Src family kinase inhibitor PP2 on tyrosine phosphorylation of DNAM-1 in NK cells (Fig. 7 I). Treatment of YT-S cells with PP2 fully prevented the ability of anti–DNAM-1 to induce tyrosine phosphorylation of DNAM-1 and the associated Vav-1. It also suppressed the ability of anti–DNAM-1 to trigger the association of DNAM-1 with Grb2.
Thus, although Fyn and LFA-1 played a key role in activation via a wide range of activating receptors, they were not strictly required for DNAM-1 signaling. Furthermore, multiple Src family kinases, including not only Fyn but also Lyn, Src, and Lck, were able to trigger DNAM-1 tyrosine phosphorylation.
DNAM-1 signaling promotes actin polymerization and granule polarization
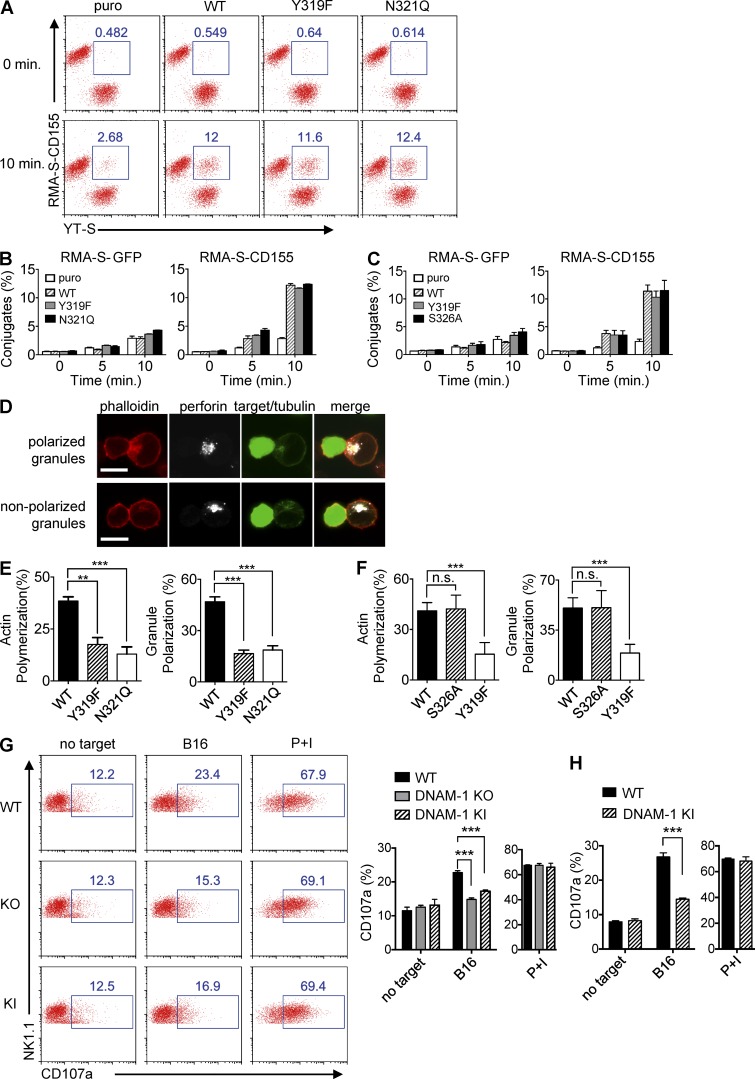
DNAM-1 was shown to serve at least in part as an adhesion receptor, which promotes NK cell activation by stabilizing conjugate formation between NK cells and target cells, one of the key steps of NK cell synapse formation (Shibuya et al., 1996; Orange, 2008). To assess if this function required DNAM-1 signaling and was sufficient to promote NK cell activation, we evaluated the impact of DNAM-1 and its signaling motif on various steps of NK cell–mediated cytotoxicity (Fig. 8). First, conjugate formation between NK cells and target cells was analyzed, using YT-S cells expressing various forms of mouse DNAM-1 and a flow cytometry–based assay (Fig. 8, A–C). WT DNAM-1 markedly enhanced the ability of NK cells to form stable conjugates with RMA-S cells expressing mouse CD155 (Fig. 8 A). Surprisingly, however, the signaling-defective DNAM-1 Y319F and N321Q mutants also increased conjugate formation, even though they did not promote cytotoxicity. A time-course analysis and a quantitation are depicted in Fig. 8 B. The signaling-competent DNAM-1 S326A mutant behaved like WT DNAM-1 in this assay (Fig. 8 C).
Figure 8.
Impact of DNAM-1 signaling on conjugate formation, actin polymerization, and granule polarization. (A–C) The indicated derivatives of YT-S cells (labeled with Brilliant Violet 421-conjugated anti–human NKp46) were incubated with RMA-S-GFP or RMA-S-CD155 (labeled with Alexa Fluor 647–conjugated anti–mouse SLAM). Formation of conjugates (identified as NKp46+SLAM+) at the indicated times was examined by flow cytometry. A representative experiment is shown in A. The percentages of conjugates formed are indicated at the top. Statistical analysis of one representative experiment with SD of duplicate values is shown in B and C. Representative of at least n = 3. (D–F) Derivatives of YT-S cells and CellTrace violet-loaded RMA-S-CD155 cells were incubated for 30 min, loaded on poly-L-lysine–coated slides, and stained with phalloidin, anti-perforin, and anti-tubulin. Conjugates were analyzed by confocal microscopy. Staining of representative conjugates in which cytotoxic granules of YT-S cells were polarized or nonpolarized toward the synapse are shown in D. Bars, 10 µm. Percentages of conjugates in which YT-S cells displayed polymerized actin near the synapse or polarized granules are shown in E and F. For each YT-S derivative, >100 conjugates were analyzed. Averages of four values for each cell type pooled from two independent experiments are depicted in E and F. SDs are shown by error bars. Unpaired Student’s t tests were performed. n.s., non significant. **, P < 0.01; ***, P < 0.001. (G and H) Splenocytes from the indicated poly(I:C)-primed mice were incubated for 6 h with B16 cells. Expression of CD107a on NK cells (NK1.1+CD3−; G) or DNAM-1+ NK cells (NK1.1+CD3−DNAM-1+; H) was analyzed by flow cytometry. Splenocytes alone or splenocytes stimulated with phorbol myristate acetate and ionomycin (PMA/iono) served as controls. Representative dot plots are shown in (G, left). Percentages of CD107a-positive NK cells are shown above the blue boxes. Average values with SDs of triplicate values from one representative experiment are depicted in G (right; H). Unpaired Student’s t tests were performed. ***, P < 0.001. Representative of n = 4 (G and H).
This observation implied that the ability of DNAM-1 to enhance adhesion was not sufficient to promote cytotoxicity. In this light, we examined the impact of DNAM-1 on subsequent events of NK cell activation, in particular actin polymerization and polarization of lytic granules, using confocal microscopy (Orange, 2008; Fig. 8, D–F). YT-S cells expressing WT DNAM-1, DNAM-1 Y319F, DNAM-1 N321Q, or DNAM-1 S326A were incubated with RMA-S cells expressing mouse CD155 for a fixed amount of time. They were then stained with fluorescent phalloidin, to detect polymerized actin, and antibodies against perforin and tubulin to detect lytic granules and microtubule-organizing centers, respectively. Target cells were also distinguished from NK cells, by labeling the former with CellTrace violet. Examples of conjugates in which NK cells displayed polarized or nonpolarized granules are depicted in Fig. 8 D.
In keeping with the aforementioned results, conjugates between NK cells and target cells were more abundant when YT-S cells expressed any of the DNAM-1 variants, compared with cells lacking DNAM-1 (unpublished data). However, among these conjugates, NK cells expressing WT DNAM-1 and DNAM-1 S326A exhibited more frequent actin polymerization and polarized granules, in comparison to cells expressing the signaling-defective DNAM-1 Y319F and N322Q (Fig. 8, E and F).
Primary mouse NK cells are difficult to analyze by confocal microscopy. To show that DNAM-1 signaling also enhanced granule polarization in mouse NK cells, we evaluated externalization of CD107a in NK cells from WT, DNAM-1–deficient, or DNAM-1 Y319F mice (Fig. 8, G and H). Compared with WT NK cells, NK cells from DNAM-1–deficient or DNAM-1 Y319F mice exhibited diminished CD107a exposure in response to B16 targets (Fig. 8 G). No difference was seen in response to PMA and ionomycin. Similar results were obtained when only DNAM-1+ NK cells from WT and DNAM-1 Y319F mice were compared (Fig. 8 H).
Therefore, augmentation of NK cell adhesion by DNAM-1 was signaling-independent and insufficient to promote cytotoxicity. DNAM-1 signaling enhanced NK cell cytotoxicity by favoring actin polymerization and lytic granule polarization, rather than by enhancing conjugate formation.
DISCUSSION
Herein, we examined whether DNAM-1 mediates intrinsic signals and whether these signals are critical for DNAM-1 to promote NK cell activation. Our initial studies showed that engagement of mouse DNAM-1 on NK cells by anti–DNAM-1 antibodies resulted in activation of Erk, Akt and calcium fluxes, three positive regulators of NK cell activation. This effect was seen in normal mouse NK cells and the human NK cell line YT-S. It was strictly dependent on DNAM-1 expression on NK cells, implying that it was truly mediated by engagement of DNAM-1, rather than by nonspecific triggering of other receptors such as Fc receptors. It is possible that the inability to detect these signals in an earlier study using a human NK cell line was due to differences in stimulation protocols or in the NK cell populations studied (Chen et al., 2007).
Structure–function analyses provided strong evidence that DNAM-1–mediated signals were crucial for the capacity to promote NK cell activation. In particular, mutation of Y319, a site of tyrosine phosphorylation in the cytoplasmic domain of DNAM-1, completely abrogated the ability of DNAM-1 to signal and promote cytotoxicity in YT-S cells. Interestingly, similar effects were seen when an asparagine located at the +2 position vis-à-vis Y319, N321, was mutated. In contrast, no effect was seen when a less conserved site of phosphorylation, S326, was mutated. Thus, the tyrosine- and asparagine-containing motif, but not S326, was necessary for the ability of DNAM-1 to signal and promote NK cell activation. Both Y319 and N321 are highly conserved across species.
Introduction of the DNAM-1 Y319F mutation in the mouse germline also dramatically reduced the capacity of DNAM-1 to trigger signals, natural cytotoxicity, and cytokine production in mouse NK cells. This observation validated the findings made in YT-S cells. It should be mentioned, however, that the Y319F mutation did not completely eliminate all functions of DNAM-1 in mouse NK cells. A small residual activity seemingly remained. In comparison to NK cells lacking DNAM-1, cells expressing DNAM-1 Y319F had a slightly less compromised ability to mediate cytotoxicity toward B16 and Neuro-2a. Furthermore, DNAM-1 Y319F was capable of triggering a small degree of calcium flux, although this response was much weaker than that evoked by WT DNAM-1. Thus, in mouse NK cells, the Y319F mutant was capable of mediating some signals, albeit much more weakly than WT DNAM-1. It is possible that this residual activity related to the existence of an additional minor signaling motif, which might be active in mouse NK cells but not in YT-S cells. Future studies will be needed to address this possibility.
The molecular mechanism by which the tyrosine- and asparagine-containing motif triggered signals was next characterized. Immunoprecipitation experiments in YT-S cells showed that engagement of DNAM-1 triggered tyrosine phosphorylation of DNAM-1. This phosphorylation was abolished when Y319 was mutated, implying that it occurred at Y319. Engagement of DNAM-1 also induced tyrosine phosphorylation of Vav-1, PLC-γ1, c-Cbl, and SHIP-1. All these events were eliminated when either Y319 or N321 was mutated.
Several lines of evidence indicated that the tyrosine- and asparagine-based motif was promoting NK cell activation by linking DNAM-1 to Grb2. First, the tyrosine- and asparagine-based motif is a typical consensus binding sequence for the Grb2 SH2 domain (pYxNx; Songyang et al., 1994). Second, Grb2 was the sole polypeptide recovered with a peptide encompassing phosphorylated Y319 and N321 of DNAM-1 in pull-down assays. Third, mutation of Y319 or N321 eliminated the ability of DNAM-1 to bind Grb2 and, in parallel, abolished its capacity to signal and enhance cytotoxicity in a system in which cytotoxicity was strictly dependent on DNAM-1. Fourth, depletion of Grb2 using two different siRNAs reduced the ability of DNAM-1 to mediate killing by YT-S cells, once again in a setting where killing was absolutely DNAM-1 dependent. Fifth, other effectors of DNAM-1 signaling, namely Vav-1, c-Cbl, SHIP-1, and p85 of PI3′K are known to bind the SH3 domains of Grb2 (Ramos-Morales et al., 1994; Odai et al., 1995; Wang et al., 1995; Kavanaugh et al., 1996).
Although the role of these various effectors in DNAM-1–mediated signals and cytotoxicity remains to be clarified, we speculate that Vav-1, PI3′K, and PLC-γ1 were likely responsible for DNAM-1–elicited activation of Erk, Akt, and calcium fluxes, as well as for induction of effector functions. Our experiments with pharmacological inhibitors suggested that PI3′K and, to a lesser extent, calcium fluxes were especially important for induction of cytotoxicity. Conversely, c-Cbl and SHIP-1 might be part of a negative feedback mechanism aimed at restricting the activating impact of DNAM-1.
Early studies showed that human DNAM-1 promoted adhesion between NK cells and target cells (Shibuya et al., 1996). This finding was confirmed in the present study using mouse DNAM-1. However, we also observed that, although the signal-defective DNAM-1 mutants Y319F and N321Q promoted conjugate formation, they failed to enhance cytotoxicity. This observation implied that enhanced adhesion was not sufficient for the activating function of DNAM-1. Along these lines, confocal microscopy studies in YT-S cells showed that WT DNAM-1 was also able to augment actin polymerization and polarization of lytic granules toward target cells. Importantly, these effects were absent with the signal-defective Y319F and N321Q DNAM-1 variants. The ability of DNAM-1 to promote granule exocytosis was also eliminated when the Y319F mutation was introduced in mouse NK cells. On these bases, we propose that DNAM-1–induced adhesion is likely to be important, but is not sufficient, for enhancing NK cell activation. In addition, the ability of DNAM-1, at least mouse DNAM-1, to stimulate actin polymerization and granule polarization, which require DNAM-1 signaling, is required. Although it remains to be established that an analogous mechanism exists for human DNAM-1, it seems likely to be the case as the signaling motif of DNAM-1 is strictly conserved in human DNAM-1.
Fyn and LFA-1 were previously suggested to play key roles in the function of human DNAM-1 (Shibuya et al., 1999). However, using NK cells from Fyn-deficient or LFA-1–deficient mice, we observed that Fyn and LFA-1 were not strictly needed for DNAM-1 signaling. Nonetheless, as reported, both were necessary for NK cell activation in response to various targets, whether targets did or did not express ligands for DNAM-1 (Matsumoto et al., 1998; Bloch-Queyrat et al., 2005; Dong et al., 2012). This finding implied that Fyn and LFA-1 were needed for the function of activating receptors other than DNAM-1. Based on transfection assays in Cos-1 cells, it is probable that, in addition to Fyn, other members of the Src family, such as Lyn, Src, and Lck, participated in DNAM-1 signaling, at least in the mouse. Likewise, it is likely that, in addition to LFA-1, other integrins were implicated in DNAM-1–dependent killing. This is especially true for targets such as B16 and CMT-93, which were killed in a DNAM-1–dependent manner but lacked ligands for LFA-1.
Although our data showed that the highly conserved Y319, but not the less conserved S326, was needed for the capacity of mouse DNAM-1 to mediate signals and promote NK cell cytotoxicity, Nabekura et al. (2014) recently showed that both Y319 and S326 were critical for mouse DNAM-1–dependent accumulation of memory NK cells in MCMV-infected mice. One possible explanation for these divergent results is that signals emanating from S326 are not needed for natural cytotoxicity, but are required for expansion of memory-like NK cells. It is also plausible that these different findings reflected distinctions in experimental conditions or systems. We mutated S326 to alanine (a conserved substitution) and tested the function of this mutant in the human YT-S cell line. Alternatively, Nabekura et al. (2014) replaced S326 by phenylalanine and studied the activity of this mutant in a competitive assay using bone marrow–reconstituted mice.
Although DNAM-1 may be strictly speaking the sole member of its family, other activating and co-stimulatory receptors use Grb2 as their immediate effector, by way of a sequence christened immunoglobulin tyrosine tail (ITT) motif (Engels et al., 2009; Engels and Wienands, 2011). These ITT motif-containing receptors include NKG2D, CD28, and membrane-bound IgG. Whereas DNAM-1 interacted with Grb2 by way of the sequence pYVNY (critical residues are underlined; this study), NKG2D interacts with Grb2 via the motif pYINM found in the NKG2D-associated adaptor DAP-10 (Engels and Wienands, 2011). Conversely, CD28 interacts with Grb2 through the sequence pYMNM, whereas membrane-bound IgG binds Grb2 via the sequence pYRNM. In this manner, DNAM-1 is analogous to ITT motif-bearing receptors. However, in the case of NKG2D, CD28, and surface IgG, the ITT motif also directly interacts with the p85 subunit of PI3′K. This interaction is dependent on the methionine at the +3 position of their ITT motif. Although DNAM-1 does not carry a methionine at the +3 position, it also coupled to PI3′K, seemingly by an indirect mechanism involving binding of Grb2 to p85. Thus, it would appear that DNAM-1 also signals via an ITT-like motif.
One of the concerns in elucidating the functions of DNAM-1 using DNAM-1–deficient mice is that loss of DNAM-1 not only obliterates DNAM-1–dependent signals, but also frees DNAM-1 ligands for potential binding to inhibitory receptors TIGIT and CD96 (Martinet and Smyth, 2015). Thus, enhanced triggering of TIGIT and CD96, rather than loss of DNAM-1–dependent signals, might explain functional changes in DNAM-1–deficient immune cells. Our finding that mutation of Y319 of DNAM-1 markedly reduced DNAM-1–dependent cytotoxicity and cytokine production in mouse NK cells, without compromising DNAM-1 expression, firmly established that these functions are intrinsic activities of DNAM-1. Given the functional interplay between DNAM-1, CD96, and TIGIT in anti-tumor immunity in vivo, it will be interesting in the future to examine if tumor rejection and mouse survival differs between animals lacking DNAM-1 (in which DNAM-1 function is ablated, and DNAM-1 ligands are more available for CD96 and TIGIT) and mice expressing DNAM-1 Y319F (in which DNAM-1 signaling is compromised, but DNAM-1 ligands are presumably not more available for CD96 and TIGIT).
In summary, our results showed that DNAM-1 mediates active biochemical signals that are essential for its capacity to enhance NK cell cytotoxicity and cytokine production. These signals are initiated by a conserved ITT or ITT-like motif (pYxNx), which undergoes phosphorylation by Src family kinases and recruits the adaptor Grb2. In turn, Grb2 activates Vav-1, PI3′K, and PLC-γ1, which leads to activation of Erk, Akt, and calcium fluxes. Such a pathway promotes actin polymerization and granule polarization, thereby enhancing cytotoxicity. As DNAM-1 is expressed in other immune cells, including CD8+ T cells, it is reasonable to speculate that DNAM-1 mediates similar signals and effects in these cells. Likewise, it is likely that an enhancement of these signals, due to augmented triggering of DNAM-1 by its ligands, underlies the therapeutic impact of blocking anti-TIGIT antibodies for treatment of cancer and viral infections (Johnston et al., 2014).
MATERIALS AND METHODS
Mice
To generate a mouse strain in which Y319 of DNAM-1 was substituted by phenylalanine, the targeting vector depicted in Fig. 3 A was created. This construct contained a 5-kb 3′ arm harboring exon 7, in which the codon for Y319 (TAT) was replaced by the codon for phenylalanine (TTT). The construct was transfected in C57BL/6-derived Bruce 4 ES cells. Cells with successful homologous recombination were injected into blastocysts to generate chimeric mice. After germline transmission, mice were crossed with a transgenic mouse expressing the Flpe recombinase [B6.SJL-Tg(ACTFLPe)9205Dym/J; The Jackson Laboratory] to eliminate the neo cassette. Presence of the DNAM-1 Y319F mutation was confirmed by sequencing of mouse genomic DNA. DNAM-1–deficient mice in the C57BL/6 background were previously described (Gilfillan et al., 2008). LFA-1–deficient mice (B6.129S7-Itgaltm1Bll/J) were obtained from The Jackson Laboratory (Ding et al., 1999). Mice lacking Fyn were described previously (Dong et al., 2012). All mice were maintained in the C57BL/6 background. Littermates were used as controls in all experiments. Animal experimentation was approved by the Animal Care Committee of IRCM and performed as defined by the Canadian Council of Animal Care.
cDNAs, plasmids, and siRNAs
cDNAs encoding mouse DNAM-1 and CD155 were obtained from Thermo Fisher Scientific. Mutants of DNAM-1 (Y319F; S326A; N321Q; Y322F) were generated by PCR using the Site-Directed Mutagenesis Kit (QuickChange; Agilent Technologies), according to the instructions of the manufacturer. The resulting cDNAs were verified by sequencing (unpublished data). For expression in YT-S cells, cDNAs coding for DNAM-1 were cloned into the vector pSRα-Puro, which contains the puromycin resistance gene (puro). For expression in Cos-1 cells, cDNAs encoding DNAM-1 or various PTKs were cloned in the vector pXM139. For expression of CD155 in RMA-S cells, the cDNA encoding CD155 was cloned into the retroviral vector pFB-GFP. Grb2-specific siRNAs (SI00300328, SI02654750) and control scrambled siRNAs (1027280) were obtained from QIAGEN. Transfection of siRNAs into YT-S cells was performed as described elsewhere (Guo et al., 2015).
Cells
To obtain freshly isolated NK cells, mice were injected intraperitoneally with poly(I:C; 250 µg; Sigma-Aldrich). After 36 h, spleen NK cells were purified by positive selection (STEMCELL Technologies). For generation of IL-2–activated NK cells, spleen NK cells were enriched by negative selection (STEMCELL Technologies) and cultured for 5 d in the presence of 1,000 U/ml of mouse IL-2 (PeproTech). YT-S (human NK cell line), B16F10 (mouse melanoma), CMT-93 (mouse rectal carcinoma), RMA-S (mouse lymphoma), and YAC-1 (mouse thymoma) were described (Dong et al., 2009; Pérez-Quintero et al., 2014). Neuro-2a (neuroblastoma) was a gift from N. Seidah (Institut de recherches cliniques de Montréal, Montreal, Quebec, Canada). YT-S cells expressing various mutants of mouse DNAM-1 were generated by electroporation and selected in puromycin-containing medium. RMA-S cells expressing green fluorescent protein (GFP) alone or in combination with mouse CD155 were produced by retroviral infection, as detailed previously (Cruz-Munoz et al., 2009). Cos-1 (monkey kidney fibroblast) and its transfection were outlined previously (Guo et al., 2015).
Antibodies
A monoclonal antibody against mouse DNAM-1 (480.2) was previously described (Gilfillan et al., 2008). Polyclonal antibodies against mouse DNAM-1 were generated by immunizing rabbits with the mouse DNAM-1 cytoplasmic domain fused to glutathione-S-transferase (GST). Antibodies against Vav-1, SHIP-1, c-Cbl, Fyn, Lyn, Src, Lck, Syk, and ZAP-70 were produced in our laboratory. Monoclonal antibodies recognizing DNAM-1 (10E5), CD155 (TX56), CD3 (145-2C11), NK1.1 (PK136), CD11b (M1/70), CD27 (LG 7F9), IFN-γ (XMG1.2), CD107a (1D4B), Ly49A/D (11A8), Ly49G2 (LGL-1), Ly49C/I/F/H (14B11), CD122 (TMb1), NKG2D (CX5), CD11a (M17/4), 2B4 (244F4), CD49b (DX5), NKp46 and ICAM-1 (CD54; YN1/1.7.4), and matched isotype controls, were obtained from eBioscience. Antibodies against CD112 (MAB3387) were purchased from Abnova. Antibodies against ICAM-2 (CD102; 3C4), human NKp46 (9E2), and perforin (dG9) were obtained from BioLegend. NKG2D-Fc was obtained from R&D Systems. Polyclonal rabbit anti–rat IgG and goat anti–rat IgG antibodies were purchased from Jackson ImmunoResearch Laboratories. Antibodies against phosphorylated Erk (E10), p44/p42 MAPK (9102), phosphorylated Akt (9271), and Akt (9272) were obtained from Cell Signaling Technology. Antibodies to β-actin (C4), Grb2 (C-23), and PLC-γ1 (sc-81) were purchased from Santa Cruz Biotechnology, Inc. Antibodies against GST (AB3282), p85 (06–497), and phosphotyrosine (4G10) were from Millipore. Anti-tubulin (236–10501) and phalloidin (A22283) were purchased from Life Technologies. Antibodies against mouse SLAM (12F12) were produced in our laboratory.
In vitro NK cell assays
51Cr release assays were performed as outlined previously, using either YT-S cells or IL-2–activated NK cells (Dong et al., 2009). For measurement of IFN-γ production and CD107a exposure, splenocytes (1 × 106) from poly(I:C)-primed mice were incubated for 6 h with B16 cells (1 × 105), and assays were performed as previously described (Dong et al., 2009). Splenocytes stimulated with phorbol myristate acetate (50 ng/ml) plus ionomycin (1 µM) served as controls. For inhibition of PI3′K and the upstream activator of Erk, MEK, YT-S cells, or splenocytes were preincubated for 1 h with 0.1 µM wortmannin (EMD Millipore) or 1 µM U0126 (Sigma-Aldrich), respectively. For inhibition of calcium fluxes, YT-S cells were preincubated for 0.5 h with 40 µM BAPTA-AM (Sigma-Aldrich). For inhibition of Btk, YT-S cells were preincubated for 0.5 h with 10 nM ibrutinib (Sigma-Aldrich). For immunoblot analyses, YT-S cells or IL-2–activated mouse NK cells were incubated with anti–mouse DNAM-1 (480.2) for 10 min at room temperature. After being washed, cells were stimulated with the relevant secondary antibody at 37°C for the indicated time. Immunoprecipitations, reimmunoprecipitations, and immunoblots were performed as previously described (Yu et al., 2001; Dong et al., 2012). To study calcium fluxes in mouse NK cells, IL-2–activated NK cells were labeled with FITC-conjugated anti–DNAM-1 (10E5) and loaded with Indo-1 (Invitrogen) for 20 min at 37°C. After being washed, cells were loaded on a BD LSR cell analyzer and the relevant secondary antibody was added after 30 s. DNAM-1+ NK cells were gated and changes in intracellular calcium over time were measured as the Fluo-4 (FL4) to Fluo-5 (FL5) ratio. Cells without addition of secondary antibody or stimulated with ionomycin (0.2 µM) were used as control. Calcium fluxes in YT-S cells were measured in a similar way, except that YT-S cells were stimulated by anti–mouse DNAM-1 (480.2), followed by the relevant secondary antibody. Cells treated with only the secondary antibody served as control. Conjugate formation assay and confocal microscopy were previously described (Dong et al., 2009; Pérez-Quintero et al., 2014).
Peptide pull-down assay, silver staining, and mass spectrometry
Biotinylated peptides encompassing the 19 aa C-terminal sequence of mouse DNAM-1, phosphorylated or not at Y319, were synthesized by the W.M. Keck Facility (Yale University). Variants of this peptide in which individual residues were mutated were also created. Peptide pull-down assay was performed as described previously (Dong et al., 2009; Pérez-Quintero et al., 2014). Silver staining and mass spectrometry were performed at the Proteomic Mass Spectrometry Platform of IRCM.
Statistical analyses
Prism 6 (GraphPad Software) was used for unpaired Student’s t tests (two-tailed).
ACKNOWLEDGMENTS
We thank the members of the Veillette laboratory for discussions.
This work was supported by grants from the Canadian Institutes of Health Research (CIHR) and the Canadian Cancer Society Research Institute (CCSRI) to A. Veillette. N. Wu is recipient of a Fellowship from Fonds de la recherche du Québec – Santé (FRQS). A. Veillette holds the Canada Research Chair in Signaling in the Immune System.
The authors declare no competing financial interests.
Author contributions: Z. Zhang planned experiments, performed experiments, interpreted data, and wrote the manuscript. N. Wu, Y. Lu, and D. Davidson planned experiments, performed experiments, and interpreted data. M. Colonna provided critical reagents. A. Veillette planned experiments, generated reagents, interpreted data, wrote the manuscript, and obtained funding.
Footnotes
Abbreviations used:
- DNAM-1
- DNAX accessory molecule-1
- ITT
- immunoreceptor tyrosine tail
- TIGIT
- T cell immunoreceptor with Ig and ITIM domains
References
- Bloch-Queyrat C., Fondanèche M.C., Chen R., Yin L., Relouzat F., Veillette A., Fischer A., and Latour S.. 2005. Regulation of natural cytotoxicity by the adaptor SAP and the Src-related kinase Fyn. J. Exp. Med. 202:181–192. 10.1084/jem.20050449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bottino C., Castriconi R., Pende D., Rivera P., Nanni M., Carnemolla B., Cantoni C., Grassi J., Marcenaro S., Reymond N., et al. . 2003. Identification of PVR (CD155) and Nectin-2 (CD112) as cell surface ligands for the human DNAM-1 (CD226) activating molecule. J. Exp. Med. 198:557–567. 10.1084/jem.20030788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryceson Y.T., March M.E., Ljunggren H.G., and Long E.O.. 2006. Synergy among receptors on resting NK cells for the activation of natural cytotoxicity and cytokine secretion. Blood. 107:159–166. 10.1182/blood-2005-04-1351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bustelo X.R. 2001. Vav proteins, adaptors and cell signaling. Oncogene. 20:6372–6381. 10.1038/sj.onc.1204780 [DOI] [PubMed] [Google Scholar]
- Chen X., Trivedi P.P., Ge B., Krzewski K., and Strominger J.L.. 2007. Many NK cell receptors activate ERK2 and JNK1 to trigger microtubule organizing center and granule polarization and cytotoxicity. Proc. Natl. Acad. Sci. USA. 104:6329–6334. 10.1073/pnas.0611655104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho D., Shook D.R., Shimasaki N., Chang Y.H., Fujisaki H., and Campana D.. 2010. Cytotoxicity of activated natural killer cells against pediatric solid tumors. Clin. Cancer Res. 16:3901–3909. 10.1158/1078-0432.CCR-10-0735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruz-Munoz M.E., Dong Z., Shi X., Zhang S., and Veillette A.. 2009. Influence of CRACC, a SLAM family receptor coupled to the adaptor EAT-2, on natural killer cell function. Nat. Immunol. 10:297–305. 10.1038/ni.1693 [DOI] [PubMed] [Google Scholar]
- de Andrade L.F., Smyth M.J., and Martinet L.. 2014. DNAM-1 control of natural killer cells functions through nectin and nectin-like proteins. Immunol. Cell Biol. 92:237–244. 10.1038/icb.2013.95 [DOI] [PubMed] [Google Scholar]
- Ding Z.M., Babensee J.E., Simon S.I., Lu H., Perrard J.L., Bullard D.C., Dai X.Y., Bromley S.K., Dustin M.L., Entman M.L., et al. . 1999. Relative contribution of LFA-1 and Mac-1 to neutrophil adhesion and migration. J. Immunol. 163:5029–5038. [PubMed] [Google Scholar]
- Dong Z., Cruz-Munoz M.E., Zhong M.C., Chen R., Latour S., and Veillette A.. 2009. Essential function for SAP family adaptors in the surveillance of hematopoietic cells by natural killer cells. Nat. Immunol. 10:973–980. 10.1038/ni.1763 [DOI] [PubMed] [Google Scholar]
- Dong Z., Davidson D., Pérez-Quintero L.A., Kurosaki T., Swat W., and Veillette A.. 2012. The adaptor SAP controls NK cell activation by regulating the enzymes Vav-1 and SHIP-1 and by enhancing conjugates with target cells. Immunity. 36:974–985. 10.1016/j.immuni.2012.03.023 [DOI] [PubMed] [Google Scholar]
- Engels N., and Wienands J.. 2011. The signaling tool box for tyrosine-based costimulation of lymphocytes. Curr. Opin. Immunol. 23:324–329. 10.1016/j.coi.2011.01.005 [DOI] [PubMed] [Google Scholar]
- Engels N., König L.M., Heemann C., Lutz J., Tsubata T., Griep S., Schrader V., and Wienands J.. 2009. Recruitment of the cytoplasmic adaptor Grb2 to surface IgG and IgE provides antigen receptor-intrinsic costimulation to class-switched B cells. Nat. Immunol. 10:1018–1025. 10.1038/ni.1764 [DOI] [PubMed] [Google Scholar]
- Gilfillan S., Chan C.J., Cella M., Haynes N.M., Rapaport A.S., Boles K.S., Andrews D.M., Smyth M.J., and Colonna M.. 2008. DNAM-1 promotes activation of cytotoxic lymphocytes by nonprofessional antigen-presenting cells and tumors. J. Exp. Med. 205:2965–2973. 10.1084/jem.20081752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo H., Cruz-Munoz M.E., Wu N., Robbins M., and Veillette A.. 2015. Immune cell inhibition by SLAMF7 is mediated by a mechanism requiring src kinases, CD45, and SHIP-1 that is defective in multiple myeloma cells. Mol. Cell. Biol. 35:41–51. 10.1128/MCB.01107-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iguchi-Manaka A., Kai H., Yamashita Y., Shibata K., Tahara-Hanaoka S., Honda S., Yasui T., Kikutani H., Shibuya K., and Shibuya A.. 2008. Accelerated tumor growth in mice deficient in DNAM-1 receptor. J. Exp. Med. 205:2959–2964. 10.1084/jem.20081611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston R.J., Comps-Agrar L., Hackney J., Yu X., Huseni M., Yang Y., Park S., Javinal V., Chiu H., Irving B., et al. . 2014. The immunoreceptor TIGIT regulates antitumor and antiviral CD8(+) T cell effector function. Cancer Cell. 26:923–937. 10.1016/j.ccell.2014.10.018 [DOI] [PubMed] [Google Scholar]
- Kavanaugh W.M., Pot D.A., Chin S.M., Deuter-Reinhard M., Jefferson A.B., Norris F.A., Masiarz F.R., Cousens L.S., Majerus P.W., and Williams L.T.. 1996. Multiple forms of an inositol polyphosphate 5-phosphatase form signaling complexes with Shc and Grb2. Curr. Biol. 6:438–445. 10.1016/S0960-9822(02)00511-0 [DOI] [PubMed] [Google Scholar]
- Kim H.S., and Long E.O.. 2012. Complementary phosphorylation sites in the adaptor protein SLP-76 promote synergistic activation of natural killer cells. Sci. Signal. 5:ra49 10.1126/scisignal.2002754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lakshmikanth T., Burke S., Ali T.H., Kimpfler S., Ursini F., Ruggeri L., Capanni M., Umansky V., Paschen A., Sucker A., et al. . 2009. NCRs and DNAM-1 mediate NK cell recognition and lysis of human and mouse melanoma cell lines in vitro and in vivo. J. Clin. Invest. 119:1251–1263. 10.1172/JCI36022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long E.O., Kim H.S., Liu D., Peterson M.E., and Rajagopalan S.. 2013. Controlling natural killer cell responses: integration of signals for activation and inhibition. Annu. Rev. Immunol. 31:227–258. 10.1146/annurev-immunol-020711-075005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowenstein E.J., Daly R.J., Batzer A.G., Li W., Margolis B., Lammers R., Ullrich A., Skolnik E.Y., Bar-Sagi D., and Schlessinger J.. 1992. The SH2 and SH3 domain-containing protein GRB2 links receptor tyrosine kinases to ras signaling. Cell. 70:431–442. 10.1016/0092-8674(92)90167-B [DOI] [PubMed] [Google Scholar]
- Magri G., Muntasell A., Romo N., Sáez-Borderías A., Pende D., Geraghty D.E., Hengel H., Angulo A., Moretta A., and López-Botet M.. 2011. NKp46 and DNAM-1 NK-cell receptors drive the response to human cytomegalovirus-infected myeloid dendritic cells overcoming viral immune evasion strategies. Blood. 117:848–856. 10.1182/blood-2010-08-301374 [DOI] [PubMed] [Google Scholar]
- Martinet L., and Smyth M.J.. 2015. Balancing natural killer cell activation through paired receptors. Nat. Rev. Immunol. 15:243–254. 10.1038/nri3799 [DOI] [PubMed] [Google Scholar]
- Matsumoto G., Nghiem M.P., Nozaki N., Schmits R., and Penninger J.M.. 1998. Cooperation between CD44 and LFA-1/CD11a adhesion receptors in lymphokine-activated killer cell cytotoxicity. J. Immunol. 160:5781–5789. [PubMed] [Google Scholar]
- Matusali G., Potestà M., Santoni A., Cerboni C., and Doria M.. 2012. The human immunodeficiency virus type 1 Nef and Vpu proteins downregulate the natural killer cell-activating ligand PVR. J. Virol. 86:4496–4504. 10.1128/JVI.05788-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nabekura T., Shibuya K., Takenaka E., Kai H., Shibata K., Yamashita Y., Harada K., Tahara-Hanaoka S., Honda S., and Shibuya A.. 2010. Critical role of DNAX accessory molecule-1 (DNAM-1) in the development of acute graft-versus-host disease in mice. Proc. Natl. Acad. Sci. USA. 107:18593–18598. 10.1073/pnas.1005582107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nabekura T., Kanaya M., Shibuya A., Fu G., Gascoigne N.R., and Lanier L.L.. 2014. Costimulatory molecule DNAM-1 is essential for optimal differentiation of memory natural killer cells during mouse cytomegalovirus infection. Immunity. 40:225–234. 10.1016/j.immuni.2013.12.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odai H., Sasaki K., Iwamatsu A., Hanazono Y., Tanaka T., Mitani K., Yazaki Y., and Hirai H.. 1995. The proto-oncogene product c-Cbl becomes tyrosine phosphorylated by stimulation with GM-CSF or Epo and constitutively binds to the SH3 domain of Grb2/Ash in human hematopoietic cells. J. Biol. Chem. 270:10800–10805. 10.1074/jbc.270.18.10800 [DOI] [PubMed] [Google Scholar]
- Orange J.S. 2008. Formation and function of the lytic NK-cell immunological synapse. Nat. Rev. Immunol. 8:713–725. 10.1038/nri2381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pawson T., and Scott J.D.. 1997. Signaling through scaffold, anchoring, and adaptor proteins. Science. 278:2075–2080. 10.1126/science.278.5346.2075 [DOI] [PubMed] [Google Scholar]
- Pérez-Quintero L.A., Roncagalli R., Guo H., Latour S., Davidson D., and Veillette A.. 2014. EAT-2, a SAP-like adaptor, controls NK cell activation through phospholipase Cγ, Ca++, and Erk, leading to granule polarization. J. Exp. Med. 211:727–742. 10.1084/jem.20132038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos-Morales F., Druker B.J., and Fischer S.. 1994. Vav binds to several SH2/SH3 containing proteins in activated lymphocytes. Oncogene. 9:1917–1923. [PubMed] [Google Scholar]
- Shibuya A., Campbell D., Hannum C., Yssel H., Franz-Bacon K., McClanahan T., Kitamura T., Nicholl J., Sutherland G.R., Lanier L.L., and Phillips J.H.. 1996. DNAM-1, a novel adhesion molecule involved in the cytolytic function of T lymphocytes. Immunity. 4:573–581. 10.1016/S1074-7613(00)70060-4 [DOI] [PubMed] [Google Scholar]
- Shibuya A., Lanier L.L., and Phillips J.H.. 1998. Protein kinase C is involved in the regulation of both signaling and adhesion mediated by DNAX accessory molecule-1 receptor. J. Immunol. 161:1671–1676. [PubMed] [Google Scholar]
- Shibuya K., Lanier L.L., Phillips J.H., Ochs H.D., Shimizu K., Nakayama E., Nakauchi H., and Shibuya A.. 1999. Physical and functional association of LFA-1 with DNAM-1 adhesion molecule. Immunity. 11:615–623. 10.1016/S1074-7613(00)80136-3 [DOI] [PubMed] [Google Scholar]
- Songyang Z., Shoelson S.E., McGlade J., Olivier P., Pawson T., Bustelo X.R., Barbacid M., Sabe H., Hanafusa H., Yi T., et al. . 1994. Specific motifs recognized by the SH2 domains of Csk, 3BP2, fps/fes, GRB-2, HCP, SHC, Syk, and Vav. Mol. Cell. Biol. 14:2777–2785. 10.1128/MCB.14.4.2777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tahara-Hanaoka S., Miyamoto A., Hara A., Honda S., Shibuya K., and Shibuya A.. 2005. Identification and characterization of murine DNAM-1 (CD226) and its poliovirus receptor family ligands. Biochem. Biophys. Res. Commun. 329:996–1000. 10.1016/j.bbrc.2005.02.067 [DOI] [PubMed] [Google Scholar]
- Verhoeven D.H., de Hooge A.S., Mooiman E.C., Santos S.J., ten Dam M.M., Gelderblom H., Melief C.J., Hogendoorn P.C., Egeler R.M., van Tol M.J., et al. . 2008. NK cells recognize and lyse Ewing sarcoma cells through NKG2D and DNAM-1 receptor dependent pathways. Mol. Immunol. 45:3917–3925. 10.1016/j.molimm.2008.06.016 [DOI] [PubMed] [Google Scholar]
- Wang J., Auger K.R., Jarvis L., Shi Y., and Roberts T.M.. 1995. Direct association of Grb2 with the p85 subunit of phosphatidylinositol 3-kinase. J. Biol. Chem. 270:12774–12780. 10.1074/jbc.270.21.12774 [DOI] [PubMed] [Google Scholar]
- Yu J., Riou C., Davidson D., Minhas R., Robson J.D., Julius M., Arnold R., Kiefer F., and Veillette A.. 2001. Synergistic regulation of immunoreceptor signaling by SLP-76-related adaptor Clnk and serine/threonine protein kinase HPK-1. Mol. Cell. Biol. 21:6102–6112. 10.1128/MCB.21.18.6102-6112.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]