LaFlam et al. identify a novel and highly conserved noncoding DNA element, ACNS1, essential for Aire expression and immune tolerance regulation in thymic epithelial cells. They show that ACNS1 is an NF-κB–responsive element and that its loss results in development of spontaneous autoimmunity in mice.
Abstract
Thymic central tolerance is essential to preventing autoimmunity. In medullary thymic epithelial cells (mTECs), the Autoimmune regulator (Aire) gene plays an essential role in this process by driving the expression of a diverse set of tissue-specific antigens (TSAs), which are presented and help tolerize self-reactive thymocytes. Interestingly, Aire has a highly tissue-restricted pattern of expression, with only mTECs and peripheral extrathymic Aire-expressing cells (eTACs) known to express detectable levels in adults. Despite this high level of tissue specificity, the cis-regulatory elements that control Aire expression have remained obscure. Here, we identify a highly conserved noncoding DNA element that is essential for Aire expression. This element shows enrichment of enhancer-associated histone marks in mTECs and also has characteristics of being an NF-κB-responsive element. Finally, we find that this element is essential for Aire expression in vivo and necessary to prevent spontaneous autoimmunity, reflecting the importance of this regulatory DNA element in promoting immune tolerance.
The establishment and maintenance of immune tolerance requires the successful detection and control of self-reactive T cells in the thymus and the periphery. In the thymus, medullary thymic epithelial cells (mTECs) play a crucial role in this process, presenting a varied repertoire of antigens and thereby eliminating self-reactive T cells or driving them to a regulatory T cell fate (Derbinski et al., 2001; Malchow et al., 2013). Along with ubiquitous self-antigens, mTECs present thousands of tissue-specific antigens (TSAs), and expression of many of these TSAs depends on the Autoimmune regulator (Aire) gene (Anderson et al., 2002). Humans and mice with mutations in Aire develop an organ-specific autoimmune syndrome, underscoring the critical role of Aire in immune tolerance (Aaltonen et al., 1997; Nagamine et al., 1997; Anderson et al., 2002).
Interestingly, Aire expression is highly tissue restricted. It is expressed broadly early in embryogenesis but then restricted to a subset of mature mTECs and rare extrathymic Aire-expressing cells (eTACs), a DC-lineage–derived population present in secondary lymphoid tissues (Gardner et al., 2008, 2013). Previous research has demonstrated the importance of a few TNF receptor superfamily members, particularly Tnfrsf11a (RANK), in the development and maintenance of Aire-expressing mTECs (Rossi et al., 2007; Akiyama et al., 2008; Hikosaka et al., 2008; Khan et al., 2014). Likewise, noncanonical NF-κB family members, which can be activated via RANK signaling, are also essential for the development of Aire-expressing mTECs (Heino et al., 2000; Zhu et al., 2006). Despite this knowledge of factors that help promote expression of Aire, the DNA regulatory elements that help coordinate this process have not been clearly delineated. We sought to identify cis-regulatory elements (CREs) important for regulating Aire expression.
In vertebrates, gene regulation involves both proximal CREs—the promoter—and distal CREs, including enhancers and silencers. The sequences of such elements are often conserved (Noonan and McCallion, 2010). In addition, several epigenetic markers, including p300 binding and the histone modification acetylated lysine 27 of histone 3 (H3K27ac), often mark active enhancers (Visel et al., 2009; Creyghton et al., 2010). Here, we searched for candidate Aire-regulating elements through the use of both sequence conservation and chromatin immunoprecipitation and high-throughput sequencing (ChIP-seq). This focused our investigation on a highly conserved region ∼3 kb upstream of Aire that is an NF-κB-responsive element and is essential for Aire expression and immune tolerance.
RESULTS AND DISCUSSION
Identification of candidate Aire cis-regulatory elements
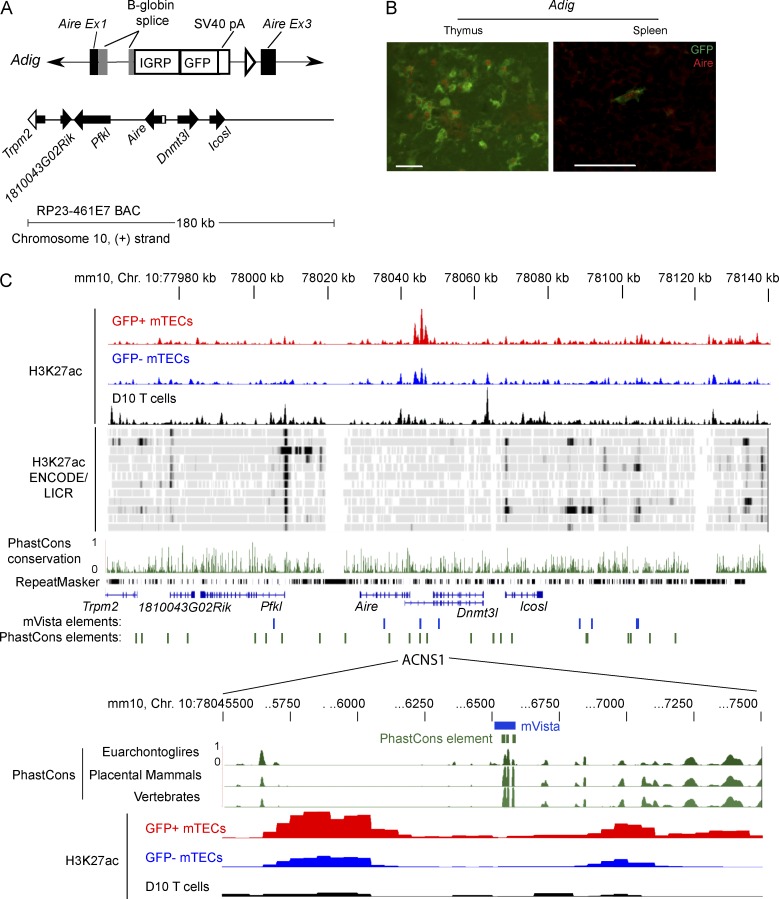
One challenge in identifying distal CREs for a particular gene is that they may be located tens or even hundreds of kilobases away. Fortuitously, however, our previously generated Aire reporter (Adig) mouse greatly narrowed our search window. The Adig mouse shows faithful recapitulation of Aire expression by means of a bacterial artificial chromosome (BAC) transgene in which the RP-23 461E7 BAC was modified by replacement of several exons of Aire with an IGRP-GFP fusion protein (Fig. 1, A and B; Gardner et al., 2008). This indicated that the essential Aire cis-regulatory elements are located within the 180-kb span of this BAC.
Figure 1.
Identification of candidate Aire cis-regulatory elements. (A) Schematic of Adig transgene: IGRP-GFP cassette replaces the coding portion of Aire exon 1, all of exon 2, and part of exon 3 in the RP23 461E7 BAC. (B) Immunofluorescent staining of GFP (green) and Aire (red) in frozen sections from Adig mice. Bars, 50 µm. (C) Alignment of conservation and ChIP-seq for the region spanned by the 461E7 BAC. The top three rows show unique reads from H3K27ac ChIP-seq of GFP+ and GFP− mTECs from Adig mice and the D10 T cell line. Below are selected H3K27ac ENCODE/LICR signal tracks, via UCSC Genome Browser: (top to bottom) brain, bone marrow, brown adipose tissue, heart, kidney, limb, liver, placenta, small intestine, spleen, testis, and thymus. These tracks, aligned to the mm9 genome, are juxtaposed here with H3K27ac reads aligned to mm10, as this 180-kb region differs between mm9 and mm10 at a nucleotide. Below are PhastCons placental mammal track and the RepeatMasker track from UCSC Genome Browser, Refseq genes, conserved noncoding sequences (P < 0.01) identified using mVista, and nonexon PhastCons conserved elements. The expanded region below shows a particular mVista-identified CNS and three PhastCons elements and H3K27ac ChIP-seq. H3K27ac mTEC tracks are representative of three samples, each composed of pooled mTECs from 6–12 mice, analyzed in the same ChIP-seq experiment as the D10 sample.
We used the online tool mVista (Frazer et al., 2004; Loots and Ovcharenko, 2004) to align regions homologous to the Aire-recapitulating BAC from seven placental mammals; the accompanying RankVista algorithm identified CNSs based on this alignment. We compared these CNSs to the nonexon conserved elements already identified by the PhastCons algorithm, which are publicly available through the UCSC Genome Browser (Fig. 1 C). Our attention was caught by the sole mVista-identified CNS to overlap with PhastCons elements, a region ∼3 kb upstream of Aire that we called Aire CNS1 (ACNS1).
Because H3K27ac is enriched at enhancers in the tissues in which the enhancers are active (Creyghton et al., 2010), we performed anti-H3K27ac ChIP-seq on highly purified GFP+ and GFP− mTECs sorted from the Adig (Aire-GFP reporter) mouse (Gardner et al., 2008). We also used the Th2-skewed T cell line D10 as a non-Aire-expressing control. In both mTEC subsets, but not D10 cells, H3K27ac was enriched near ACNS1 (Fig. 1 C). H3K27ac is similarly not enriched at this site in a diverse set of non-Aire-expressing tissues for which ChIP-seq data are publicly available (Fig. 1 C).
CNS1 is an NF-κB–responsive element
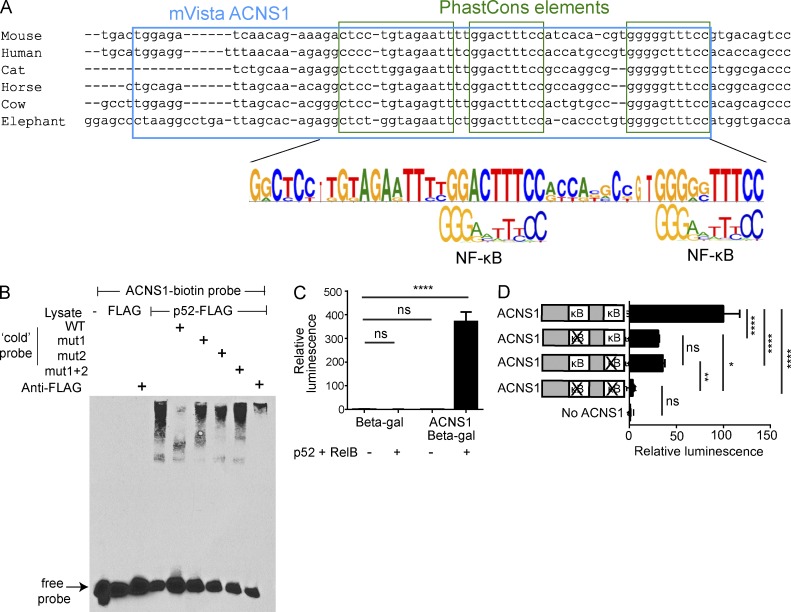
We next examined the sequence of ACNS1 and found it contained two highly conserved putative NF-κB binding sites (Fig. 2 A). Using electrophoretic mobility shift assays (EMSAs), we found binding of p52-FLAG–transfected nuclear lysates to ACNS1 (Fig. 2 B). Abrogation of this shift by an excess of WT but not mutant ACNS1 probe and supershift in the presence of anti-FLAG antibody demonstrate the specificity of this interaction. p52 is the active form of p100, encoded by Nfkb2, which is essential for the normal generation of Aire-expressing mTECs (Zhu et al., 2006). We then investigated whether ACNS1 can enhance transcription in an NF-κB-dependent manner. We inserted ACNS1 into a thymidine kinase minimal promoter (TK) β-galactosidase (β-gal) reporter plasmid and transfected this into 293T cells with or without p52 and RelB. We found substantial β-gal expression only when ACNS1 was present and p52 and RelB were co-transfected (Fig. 2 C). We also mutated each NF-κB–binding site alone or in combination and found that each site had an equal and approximately additive effect on reporter expression (Fig. 2 D). The finding that ACNS1 can act as an enhancer in response to transcription factors known to be critical in mTECs, together with its sequence conservation and the nearby H3K27ac accumulation, encouraged our hypothesis that ACNS1 regulates Aire expression.
Figure 2.
ACNS1 is an NF-κB-responsive element. (A) Comparison of consensus ACNS1 sequence to κB sequence motif. (B) EMSA using nuclear lysates from 293T cells transfected with p52-FLAG, incubated with biotinylated ACNS1 probe. Untagged WT and mutant probes (first, second, or both κB sites, respectively), and anti-FLAG antibody included in specified lanes. (C) Relative luminescence after chemiluminescent detection of β-gal in cellular lysates of 293T cells 48 h after transfection with TK-β-gal or CNS1-TK-β-gal plasmid ± RelB-FLAG and p52-FLAG. (D) Relative luminescence after chemiluminescent detection of β-gal in cellular lysates of 293T cells, 48 h after transfection with RelB-FLAG and p52-FLAG and WT or mutated CNS1-TK-β-gal. In C and D, normalized for transfection efficiency using pRL-CMV. All data are representative of at least three independent experiments. Statistical analysis using Student’s t test: ns, not significant; *, P < 0.05; **, P < 0.01; ****, P < 0.0001.
A small region upstream of Aire can support Aire-recapitulating expression
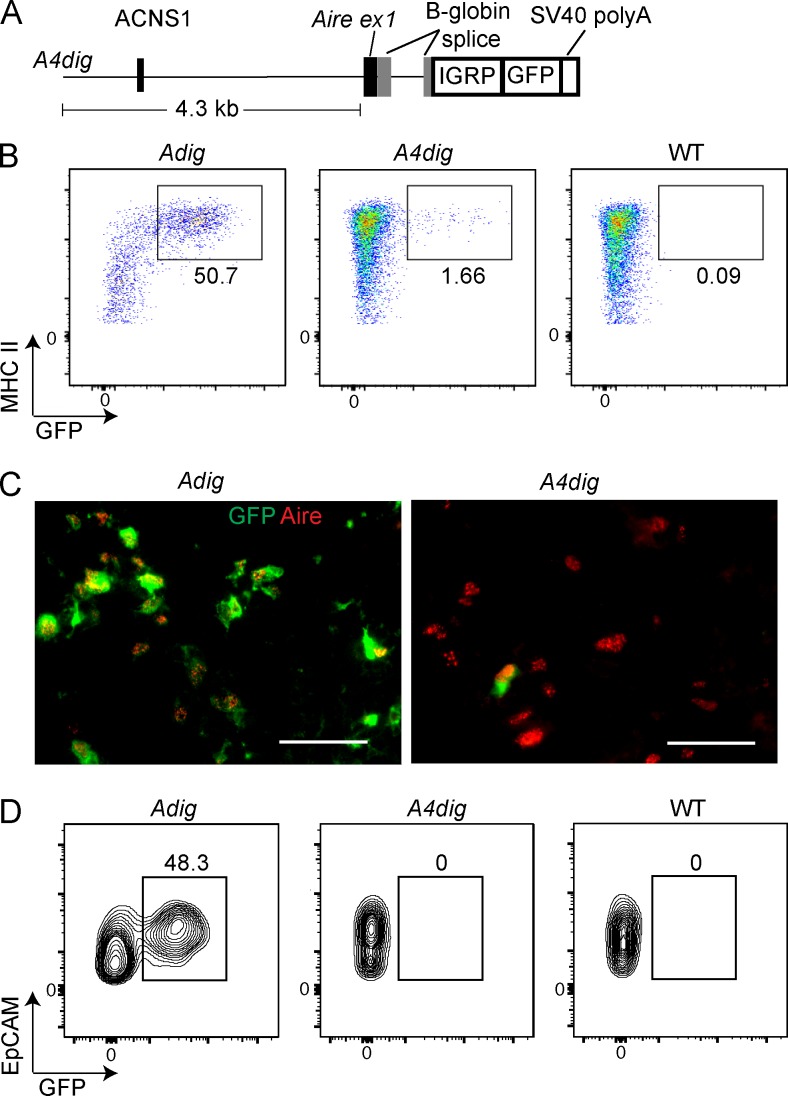
We began in vivo functional assays by creating a small transgene, consisting of ∼4.3 kb of sequence present immediately upstream of Aire, followed by an IGRP–GFP fusion protein cassette (the A4dig mouse; Fig. 3 A). We generated eight founders, six of which we analyzed at the F0 generation and two at F1 and later. We found four lines had variegated thymic GFP expression (the other four had no thymic GFP expression). Remarkably, in all four cases, this expression was restricted solely to Aire+ mTECs (Fig. 3, B and C). However, unlike in the BAC reporter mouse, only a minority of Aire+ mTECs were GFP+. In addition, there were no GFP+ eTACs in any of the A4dig lines (Fig. 3 D). The frequency of GFP+ cells did not significantly correlate with transgene copy number (unpublished data).
Figure 3.
A small transgene is able to reproduce the cell type specificity of Aire expression. (A) Schematic of A4dig transgene: 4.3-kb region extending upstream from the translation start site of Aire followed by an IGRP-GFP cassette. (B) Flow cytometry of mTECs, showing frequency (%) of GFP+ cells in Adig, A4dig, and WT mice. (C) Immunofluorescent staining of frozen thymic sections, with staining for GFP (green) and Aire (red). Bars, 50 µm. (D) Flow cytometry of eTACs showing frequency (%) of GFP+ cells in Adig, A4dig, and WT mice. Data in B and D are representative of eight distinct A4dig founder lines analyzed across three experiments. Data in C are representative of two independent experiments with two to three mice per genotype.
Although the frequency of reporter-expressing mTECs in these A4dig mice was far lower than that seen in our standard Adig reporter mouse, the MFI of GFP in GFP+ cells was similar (Fig. 3 B). This suggests a stochastic process in which robust transgene expression occurs once a particular regulatory threshold is cleared. This sort of specific, variegated expression has been observed in other transgenes—and, in fact, in endogenous genes upon deletion of certain regulatory regions (Ronai et al., 1999). As such, we interpret the inconsistency of transgene expression in the A4dig mouse as indicating that the A4dig transgene lacks one or more important regulatory elements, located elsewhere in the Aire-recapitulating BAC, which normally stabilize expression. We were nevertheless struck by how this relatively small transgene was able to produce a highly specific pattern of expression, restricted even to the point of not being expressed in Aire− mTECs.
ACNS1 is necessary for Aire expression
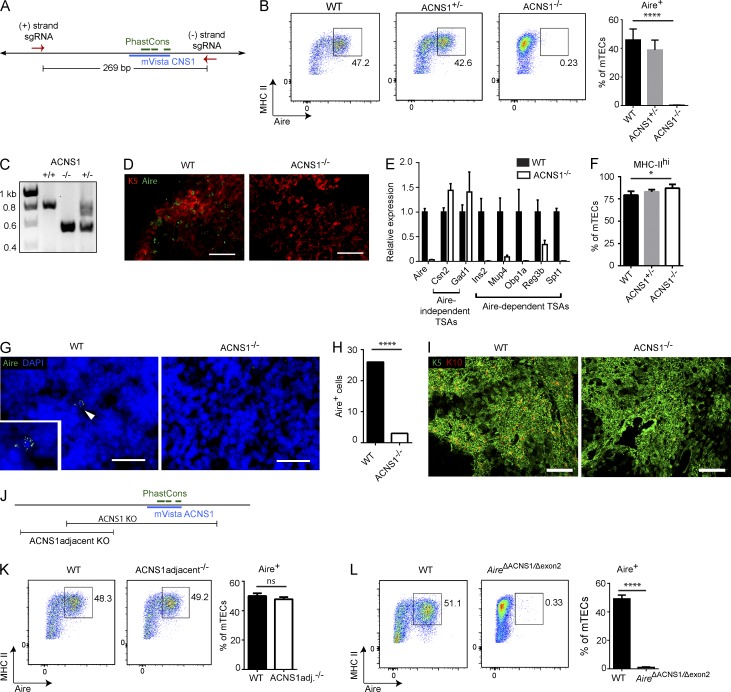
We also investigated whether ACNS1 is required for Aire expression. To do so, we took advantage of the recently developed genome editing technology CRISPR-Cas9 (Ran et al., 2013). A single guide RNA is usually sufficient to knock out a coding gene via a frameshift and the resulting premature stop codon. As noncoding functional elements are relatively more resistant to small alterations, we sought to delete all of ACNS1, as well as some flanking sequence (Fig. 4 A). Two guide RNAs and Cas9 mRNA were injected into fertilized B6 oocytes. The resulting pups were screened by PCR and Sanger sequenced. We found that approximately one in seven pups had the desired deletion, in each case in heterozygosity (unpublished data).
Figure 4.
ACNS1 is required for Aire expression. (A) Schematic of the CRISPR-Cas9–mediated germline deletion of ACNS1. Arrows show the sites targeted by guide RNAs. (B) Flow cytometry of mTECs, showing frequency (%) of Aire+ cells. Bar graph (right) shows mean ± SD frequencies of Aire+ cells among mTECs from WT, ACNS1+/−, and ACNS1−/− mice. (C) Gel electrophoresis of products from ACNS1 genotyping PCR. Broad upper band is consistently seen in heterozygotes. (D) Immunofluorescent staining for cytokeratin-5 (red) and Aire (green) in frozen thymic sections from WT and ACNS1−/− mice. Bars, 50 µm. (E) Quantitative PCR analysis of Aire and select TSA genes, comparing RNA from WT and ACNS1−/− mTECs, showing mean ± SD (technical replicates), normalized to Actb. (F) Bar graph shows mean ± SD frequencies of MHChi cells among mTECs from WT, ACNS1+/−, and ACNS1−/− mice. (G) Immunofluorescent staining for Aire (green) with DAPI counterstain in frozen lymph node sections from WT and ACNS1−/− mice. Bars, 25 µm. Inset in WT image shows Aire+ cell indicated by arrowhead. (H) Graph summarizes the results in G, showing the number of Aire+ cells observed in 80 lymph node sections from WT and ACNS1−/− mice. (I) Immunofluorescent staining for cytokeratin-5 (green) and cytokeratin-10 (red) in frozen thymic sections from WT and ACNS1−/− mice. Bars, 250 µm. (J) Schematic showing ACNS1adjacent deletion and ACNS1 deletion. (K) Flow cytometry of mTECs, showing frequency (%) of Aire+ cells. Bar graph (right) shows mean ± SD frequencies of Aire+ cells among mTECs from WT and ACNS1adjacent−/− mice. (L) Flow cytometry of mTECs, showing frequency (%) of Aire+ cells. Bar graph (right) shows mean ± SD frequencies of Aire+ cells among mTECs from WT and AireΔACNS1/Δexon2 (compound heterozygote) mice. Data in B, F, K, and L each summarize three independent experiments with one or more mice per group and totaling at least five mice per group. Data in E are representative of two independent experiments with three replicates each. Data in D and G are representative of two independent experiments with at least three mice per group. Data in I are representative of two independent experiments with at least two mice per group. B, K, and L were analyzed by Student’s t test, and H was analyzed by Garwood method of Poisson distribution confidence interval: ns, not significant; *, P < 0.05; and ****, P < 0.0001.
In contrast to WT mice, in which ∼45% of mTECs are Aire+, in ACNS1−/− mice there is a complete absence of Aire+ mTECs (Fig. 4, B and D). Quantitative RT-PCR analysis of RNA from FACS-isolated mTECs showed a large decrease in expression of Aire (Fig. 4 E). We also observed far fewer Aire+ cells in lymph nodes (Fig. 4, G and H). Together, these results demonstrate that ACNS1 is essential for Aire expression in both the thymus and secondary lymphoid tissues.
In addition, we observed that a larger proportion of mTECs in ACNS1−/− mice are MHChi (Fig. 4 F). We also found that there were fewer cytokeratin 10 (K10)–expressing cells in the thymi of ACNS1−/− mice (Fig. 4 I). K10 expression is enriched in mature, post-Aire, involucrin+ mTECs (White et al., 2010). This result suggests that in the absence of ACNS1, maturation of mTECs from MHC-IIhi Aire+ to MHC-IIlo Aire− K10+ cells is impaired, a finding consistent with observation of this defect in Aire−/− mice (Wang et al., 2012; and unpublished data). In contrast to these changes in mTEC subset frequencies, we did not see a difference in the frequency of cTECs in ACNS1−/− mice (unpublished data).
When screening the CRISPR-targeted pups, we noticed one founder with a smaller, partially overlapping deletion that spares the conserved mVista-identified and overlapping PhastCons elements (Fig. 4 J). Mice homozygous for this ACNS1-adjacent deletion had the same frequency of Aire+ mTECs as WT mice (Fig. 4 K), implying that the essential regulatory region is smaller than the 269-bp region deleted in the ACNS1−/− mice.
CRISPR-Cas9–mediated genome editing carries the risk of undesired off-target mutations (Fu et al., 2013), raising the possibility that the observed phenotype is not a result of deletion of ACNS1. We took a few approaches to address this concern. First, analyses were performed on mice that were at least two generations removed from the founder. Given that mice used in breeding in each generation were chosen based on their ACNS1 genotype, it is unlikely that many of the analyzed ACNS1−/− mice would also be homozygous for another CRISPR-induced mutation unless it was strongly linked to ACNS1. Second, we found that mice homozygous for a second independently generated ACNS1 deletion, as well as compound heterozygotes bearing one copy of each of these two ACNS1 deletion alleles also lacked Aire+ mTECs (unpublished data). Finally, we crossed ACNS1+/− mice with traditionally targeted Aire+/− mice, in which deletion of exon 2 of Aire leads to a premature stop codon (Anderson et al., 2002), to generate compound heterozygous AireΔACNS1/Δexon2 mice. Again, there were no Aire+ cells in these mice, which lacked ACNS1 on one chromosome and lacked exon 2 of Aire on the other chromosome (Fig. 4 L). Collectively, these results argue that deletion of ACNS1, and not an off-target mutation, is responsible for the observed loss of Aire expression.
Loss of ACNS1 leads to spontaneous autoimmunity
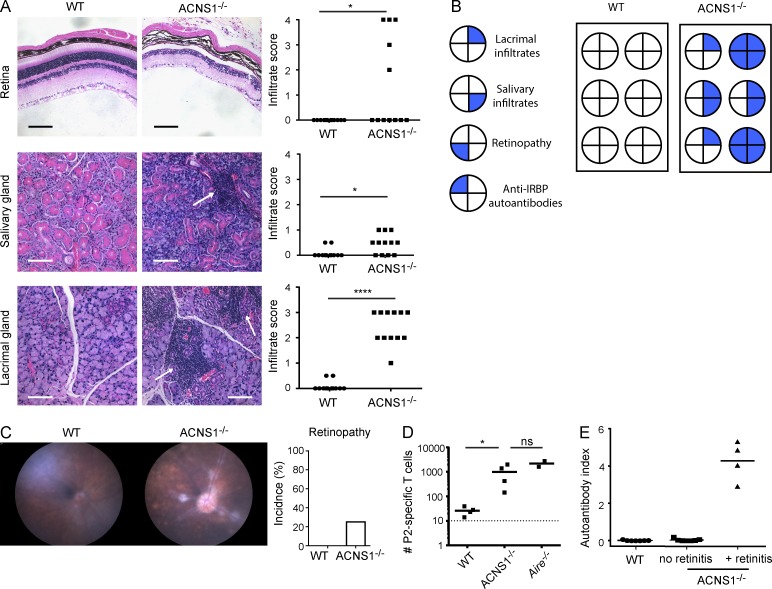
Consistent with their lack of Aire expression, we observed that ACNS1−/− mice develop spontaneous autoimmunity. Histological examination showed mononuclear infiltrates in lacrimal and salivary glands and retinal destruction in ACNS1−/− mice (Fig. 5, A and B). Funduscopic examination similarly revealed retinopathy in 10–15-wk-old ACNS1−/− mice (Fig. 5 C). In the absence of Aire, decreased expression of TSAs allows for the escape of T cells specific for these antigens, leading to autoimmunity (DeVoss et al., 2006). Quantitative RT-PCR of sorted mTECs revealed that ACNS1−/− mice showed substantially decreased expression of Aire-dependent, but not Aire-independent, TSAs (Fig. 4 E). Consistent with this decrease in TSA expression, ACNS1−/− mice show loss of tolerance to the Aire-dependent TSA IRBP (Rbp3); loss of thymic expression of this antigen in Aire−/− mice permits escape of IRBP-reactive T cells and leads to autoimmune retinitis (DeVoss et al., 2006). Specifically, after immunization with P2, an IRBP-derived peptide, ACNS1−/− mice had more P2-specific T cells compared with WT mice (Fig. 5 D). Also, at 10 wk of age, approximately one-third of ACNS1−/− mice had spontaneously generated autoantibodies against IRBP (Fig. 5 E), with an exact correspondence between the presence of autoantibodies at 10 wk of age and retinal damage, as determined by histology, at 14–16 wk of age. In sum, these results demonstrate that ACNS1−/− mice develop spontaneous autoimmunity in a similar pattern and penetrance to other Aire-deficient mice in the relatively autoimmune-resistant C57BL/6 background (Su et al., 2008; Hubert et al., 2009).
Figure 5.
Mice lacking ACNS1 develop spontaneous autoimmunity. (A) Representative hematoxylin and eosin–stained retinal, salivary, and lacrimal sections from 14–16-wk-old WT and ACNS1−/− mice; arrows indicate mononuclear infiltrates. Bars, 100 µm. Graphs (right) show disease severity scoring for each tissue; each dot is a single mouse and short horizontal lines show the means. (B) Correlation of four manifestations of autoimmunity in a cohort of WT and ACNS1−/− mice. (C) Representative funduscopic images of retinas of 10–15-wk-old WT and ACNS1−/− mice. Graph summarizes the incidence of retinopathy in each genotype. (D) WT, ACNS1−/−, and Aire−/− mice, the last serving as a positive control, were immunized with the IRBP P2 peptide and lymph nodes and spleen harvested 9–10 d later for P2-I-Ab tetramer-based quantitation of P2-specific T cells. Each dot on the graph represents an individual mouse, the short horizontal lines show the means, and the dotted line represents the limit of detection. (E) Radioligand binding assay of anti-IRBP antibodies in sera collected from 10-wk-old WT and ACNS1−/− mice. Autoantibody index of 1 is defined using a sample with 367 µg/ml of polyclonal rabbit anti-IRBP antibody. Data in A are from two independent cohorts, each with six WT and six ACNS1−/− mice; B summarizes one of these cohorts. Data in C are pooled from several experiments, totaling 22 WT and 8 ACNS1−/− mice. Data in D are representative of two independent experiments, each with four WT, four ACNS1−/−, and two Aire−/− mice. Data in E are pooled from three experiments, totaling 7 WT and 11 ACNS1−/− mice. A and D were analyzed by Mann-Whitney rank-sum testing and C was analyzed by χ2 test: ns, not significant; *, P < 0.05; and ****, P < 0.0001.
In conclusion, we have identified a DNA element essential for immune tolerance, the first distal CRE known to regulate Aire. This element, ACNS1, is highly conserved across placental mammals. Consistent with its role in regulating Aire, it is flanked by chromatin enriched in H3K27ac in mTECs, and deletion of endogenous ACNS1 led to abrogation of Aire expression, decreased TSA transcription, and spontaneous autoimmune disease.
In addition, ACNS1 is NF-κB responsive, suggesting that there may be a direct link between NF-κB and Aire expression through this regulatory element, building on previous research on the role of noncanonical NF-κB in Aire-expressing mTECs. NF-κB is active in a variety of cell types, nearly all of which do not express Aire. Consequently, additional factors must be necessary for the induction of Aire expression. The identity of these additional factors and whether they bind at ACNS1 or elsewhere remain important and open questions. Furthermore, additional research is needed to identify other distal Aire CREs. We argue that the variegated GFP expression in mTECs of A4dig mice suggests that such additional elements do exist. Furthermore, the lack of GFP expression in eTACs of A4dig mice may reflect differences in the mechanism of cis-regulation of Aire in mTECs and eTACs.
Although on a population level, Aire induces the expression of thousands of genes, in a given cell only a limited set of these genes is induced, such that any given TSA is only expressed in a small percentage of mTECs (Pinto et al., 2013). It follows that the regulatory region of the A4dig transgene might be able to support sufficient expression of a given antigen to permit effective negative selection and thereby serve as the basis for targeted induction of antigen-specific tolerance in vivo.
Finally, our results here also imply that some patients may develop autoimmunity through a genetic defect whereby ACNS1 is deleted. Indeed, some patients with the clinical features of APS1 have been described without identifiable coding mutations (Owen and Cheetham, 2009) and further investigation of this particular noncoding element is warranted.
MATERIALS AND METHODS
Mice
C57BL/6.Adig mice were generated as previously described (Gardner et al., 2008). FVB.A4dig mice were generated by modification of the JDLNL targeting construct used to generated Adig mice (Gardner et al., 2008). MluI and PvuI sites were inserted immediately upstream of the 5′ Aire homology region in JDLNL using standard cloning methods. Then, a short homology region corresponding to mm9:chr10:77510085-77510611 (− strand) was cloned using PCR and inserted at the MluI site using InFusion (Takara Bio Inc.), adding a SalI site at the 3′ end. The resulting plasmid was linearized with SalI and underwent recombineering-based gap repair in which the plasmid was electroporated into heat-induced SW105 bacteria already transformed with the Adig BAC, followed by kanamycin selection, and then plasmid isolation and sequencing. The resulting A4dig plasmid was isolated using Plasmid Maxi kit (QIAGEN), digested with PvuI to separate the transgene (Fig. 3 A) from the plasmid backbone, purified by gel electrophoresis and QIAquick Gel Extraction kit (QIAGEN), followed by QIAquick PCR Purification kit (QIAGEN), and injected into fertilized FVB/N oocytes by the UCSF Laboratory Animal Resource Center. C57BL/6.ACNS1−/− mice were created by CRISPR-Cas9-mediated deletion in fertilized oocytes. More specifically, the online tool designed by the laboratory of F. Zhang (Massachusetts Institute of Technology, Cambridge, MA) was used to identify candidate guide RNA sequences with a low predicted incidence of off-target mutations. These candidates were cloned by a previously described method (Ran et al., 2013) into pX330-U6-Chimeric_BB-CBh-hSpCas9, which was a gift from F. Zhang (plasmid #42230; Addgene). These candidate guide RNAs were then tested at the UCSF ES Cell Targeting Core by transfecting into ES cells and assessing cutting activity using Cel1-based surveyor assays. Guide RNAs and Cas9 mRNA were in vitro transcribed, isolated, and injected into cytoplasm of fertilized C57BL/6 oocytes at either 50 ng/µl Cas9 and 20 ng/µl each guide RNA or 25 ng/µl Cas9 and 10 ng/µl guide RNAs—both resulted in pups with the desired deletion. All animals were housed and bred in specific pathogen–free conditions at UCSF. All experiments were approved by the Institutional Animal Care and Use Committee of UCSF.
ChIP-seq
ChIP was performed as previously described (Seumois et al., 2014). In brief, cells were fixed with 1% formaldehyde, washed, snap frozen, and stored at −80°C. Frozen samples were lysed and chromatin was sheared by sonication, using a Bioruptor (Diagenode) to yield 100–500 bp fragments. Chromatin was diluted, incubated overnight with protein A–coated magnetic beads (Invitrogen) precoated with anti-H3K27ac antibody (Abcam; Ab4729). Beads underwent a series of washes. Chromatin was then eluted, treated with RNase and proteinase K, and purified by affinity column (Zymo Research). Whole-genome amplification of DNA was performed using WGA-SEQX (Sigma-Aldrich). Sequencing library was prepared and the samples were sequenced using Illumina Hi-Seq. Sequencing data were processed, mapped, and analyzed as previously described (Seumois et al., 2014). Data have been deposited in the Gene Expression Omnibus database under accession no. GSE74257.
Quantitative PCR
RNA was isolated from sorted mTECs using RNeasy Plus Micro kit (QIAGEN) and reverse transcribed using oligo-dT primers and the SuperScript III kit (Life Technologies). TaqMan gene expression assays (Applied Biosystems) were used for all targets. Quantitative PCR was performed using 7500 Fast Real-Time PCR System (Applied Biosystems). Transcript abundance was normalized to Actb and analyzed using the ΔΔCt method.
Electrophoretic mobility shift assay
293T cells were cultured in high-glucose DMEM, 10% heat-inactivated FCS, and penicillin/streptomycin, at 37°C, 10% CO2. 293T cells were transfected with p52-FLAG using Trans-IT 293 (Mirus). After 24 h, cells were collected and nuclear extracts were prepared using NE-PER kit (Thermo Fisher Scientific). Nuclear extract protein concentration was determined using BCA Protein Assay kit (Thermo Fisher Scientific). p52-FLAG pcDNA3 was a gift from S. Smale (University of California Los Angeles, Los Angeles, CA; Addgene plasmid #20019). EMSAs were performed using LightShift kit (Thermo Fisher Scientific). Binding reactions contained 5 µg of nuclear extract, 10 nM biotinylated probe, 2 µM unlabeled probe, and 1 µg anti-FLAG (M2 clone; Sigma-Aldrich). Binding reactions were electrophoresed on 6% DNA Retardation Gels (Invitrogen) and transferred to Biodyne B nylon membrane (Thermo Fisher Scientific). DNA was cross-linked using UV light and detected using the Chemiluminescent Nucleic Acid Detection Module (Thermo Fisher Scientific). Probes were ordered from IDT. Probe sequences were as follows: WT-CNS1-F: 5′-(biotin)-AATTTTGGACTTTCCATCACACGTGGGGGTTTCCGTGAC-3′, WT-CNS1-R: 5′-GTCACGGAAACCCCCACGTGTGATGGAAAGTCCAAAATT-3′, mut1-CNS1-F: 5′-AATTTCTCACTTTCCATCACACGTGGGGGTTTCCGTGAC-3′, mut1-CNS1-R: 5′-GTCACGGAAACCCCCACGTGTGATGGAAAGTGAGAAATT-3′, mut2-CNS1-F: 5′-AATTTTGGACTTTCCATCACACGTCTCGGTTTCCGTGAC-3′, mut2-CNS1-R: 5′-GTCACGGAAACCGAGACGTGTGATGGAAAGTCCAAAATT-3′, mut1+2-CNS1-F: 5′-AATTTCTCACTTTCCATCACACGTCTCGGTTTCCGTGAC-3′, and mut1+2-CNS1-R: 5′-GTCACGGAAACCGAGACGTGTGATGGAAAGTGAGAAATT-3′.
In vitro reporter assays
TK-β-gal was a gift from B.L. Black (University of California San Francisco, San Francisco, CA). ACNS1 sequence (mm9, chr10:77509283-77509327) or ACNS1 with mutated kB sites (same mutations as in EMSA probes) was cloned into Kpn-linearized plasmid using standard cloning methods. p52-FLAG pcDNA3 was a gift from S. Smale (plasmid #20019; Addgene). RelB-FLAG pcDNA was also a gift from S. Smale (plasmid #20017; Addgene). pRL-CMV was purchased from Promega. 293T cells were cultured in high-glucose DMEM, 10% heat-inactivated FCS, and penicillin/streptomycin, at 37°C, 10% CO2. 293T cells in 24-well-plates were transfected with 12.5 ng pRL-CMV and 250 ng of β-gal, p52, and RelB plasmids using Trans-IT 293 (Mirus). After 48 h, cells were briefly washed and lysed per Renilla Luciferase Assay System (Promega). Samples were then aliquoted and analyzed for Renilla luciferase using Renilla Luciferase Assay System and for β-gal activity using Luminescent β-galactosidase Detection kit II (Takara Bio Inc.) and luminescence measured using Victor2 1420 Multilabel Counter (PerkinElmer). In each experiment, all conditions were performed in triplicate.
Flow cytometry
mTECs and eTACs were isolated as previously described (Gardner et al., 2008). In brief, thymi or peripheral lymph nodes and spleens were isolated, minced, and digested with DNase I and Liberase TM (Roche) before centrifugation on a gradient of Percoll PLUS (GE Healthcare). The enriched stromal cells were stained with the antibodies against the surface markers indicated in the figure legends (BioLegend). For intracellular staining, cells were fixed using Foxp3 Staining Buffer kit (eBioscience) and stained with anti-Aire (clone 5H12; and eBioscience). Data were collected using an LSRII flow cytometer (BD) and analyzed using FACSDiva (BD) and FlowJo (Tree Star). Cell sorting was performed using a FACS Aria III (BD). mTECs were gated as DAPI−, CD45−, EpCAM+, MHC-II+, Ly51−, cTECs were gated as DAPI−, CD45−, EpCAM+, MHC-II+, Ly51+; and eTACs were gated as DAPI−, CD45int, MHC-IIhi, EpCAM+, and CD86−.
IRBP P2 peptide immunization and tetramer analysis
Mice were immunized with 100 µg P2 peptide (IRBP, aa 271–290), emulsified in complete Freund’s adjuvant, subcutaneously at the chest. Tetramer analysis was performed 9–10 d after immunization on pooled axillary and cervical lymph nodes and spleen using P2-I-Ab tetramer, as described previously (Taniguchi et al., 2012). The tetramer was generated by the National Institutes of Health Tetramer Core Facility. Limit of detection was defined as the mean + 3 SD of the number of P2 tetramer-staining CD8+ T cells observed.
Immunofluorescence
For data in Fig. 4 I, thymi were harvested, fixed in 2% PFA, dehydrated in 30% sucrose in PBS, embedded in OCT, and frozen. 25-µm sections were permeabilized in 0.3% Triton X-100, 0.2% BSA, and 0.1% sodium azide in PBS, blocked with BlockAid (Life Technologies), and stained by rabbit polyclonal to cytokeratin 10 (Abcam), goat anti–rabbit conjugated with A546 (Life Technologies), fixed with 0.5% PFA, and stained rabbit monoclonal against cytokeratin 5 directly conjugated to A488 (Abcam). Immunofluorescent slides were imaged using SP5 confocal microscope (Leica). Images are maximum projections of 25-µm z-stacks. For other immunofluorescence data, thymi and lymph nodes were harvested and embedded into OCT media (Tissue-Tek) and frozen. 8-µm sections were fixed in 1:1 acetone/methanol, blocked with 10% normal goat serum, and stained with antibodies against cytokeratin-5 (Abcam), GFP (Invitrogen), or Aire (clone 5H12; eBioscience), followed by goat secondary conjugated with A488 or A594 (Life Technologies). Immunofluorescent slides were imaged using an Axio Imager M2 widefield fluorescence microscope (Carl Zeiss) with CoolSnap HQ2 camera (Photometrics). Images were analyzed using ImageJ (National Institutes of Health).
Histology
Eyes, lacrimal glands, and salivary glands were harvested and fixed overnight with formalin, transferred to 30% ethanol for 30 min, and stored long-term in 70% ethanol. Tissues were embedded in paraffin, sectioned, and stained with hematoxylin and eosin by the UCSF Mouse Pathology Core. Lacrimal and salivary glands were scored by an observer blinded to sample identity using the following criteria: 0, no infiltrate; 0.5, trace infiltrate; 1, minor infiltrate; 2, moderate infiltrate; 3, severe infiltrate; 4, complete tissue destruction. Retinas were scored by an observer blinded to sample identity using the following criteria: 0, no lesion; 1, loss of <50% of photoreceptor layer; 2, loss of >50% of photoreceptor layer; 3, loss of >50% of photoreceptor layer and loss of <50% of the outer nuclear layer; 4, loss of >50% of photoreceptor layer and >50% of outer nuclear layer.
Radioligand binding assay
Murine IRBP cDNA was in vitro transcribed and translated with [35S] labeling, and assays were performed as described (Shum et al., 2009). Autoantibody index was calculated as [cpm sample – cpm negative control]/[cpm positive standard – cpm negative control]. Polyclonal rabbit anti-IRBP (Proteintech) was used as the positive standard. Samples were positive if 3 SD above the mean for the WT mice.
Statistical analysis
Statistical analysis was performed using Prism 6.0 (GraphPad Software). Tetramer analysis and histology scoring were analyzed by Mann-Whitney rank-sum testing, mTEC flow cytometry and in vitro reporter assays were analyzed by Student’s t test, retinopathy incidence was analyzed by χ2 test, and eTAC quantitation was analyzed by Garwood method of Poisson distribution confidence interval calculation; Ns, not significant; *, P < 0.05; **, P < 0.01; ***, P < 0.001; and ****, P < 0.0001.
ACKNOWLEDGMENTS
We thank J. Day of the La Jolla Institute for Allergy and Immunology sequencing facility for assistance with high-throughput sequencing; A.M. Gholani and J. Greenbaum of the La Jolla Institute for Allergy and Immunology bioinformatics core for assistance with ChIP-seq analysis; the National Institutes of Health Tetramer Core Facility for providing tetramer reagent; H. Lu for assistance generating transgenic mice; C. Park and K. Bell for assistance with CRISPR-Cas; J. Bolen of the UCSF Mouse Pathology Core; and G. Haliburton for helpful discussion.
This work was supported by National Institutes of Health grants R01 AI097457 (M.S. Anderson); R01 HL114093 (P. Vijayanand); the UCSF Medical Scientist Training Program, NIDDK Ruth L. Kirschstein Fellowship F30 DK100098, and UCSF Discovery Fellows Program (T.N. LaFlam). Flow cytometry data were generated in the UCSF Parnassus Flow Cytometry Core and microscopy data were generated in the Diabetes and Endocrinology (DERC) Microscopy Core, both of which are supported by the DERC grant, National Institutes of Health P30 DK063720.
The authors declare no competing financial interests.
Footnotes
Abbreviations used:
- A4dig
- Aire 4.3 kb promoter-driven IGRP-GFP transgene
- Adig
- Aire-driven IGRP-GFP transgene
- Aire
- autoimmune regulator
- ACNS1
- Aire conserved noncoding sequence 1
- BAC
- bacterial artificial chromosome
- β-gal
- β-galactosidase
- ChIP-seq
- chromatin immunoprecipitation and high-throughput sequencing
- CNS
- conserved noncoding sequence
- CRE
- cis-regulatory element
- cTEC
- cortical thymic epithelial cell
- EMSA
- electrophoretic mobility shift assay
- eTAC
- extrathymic Aire-expressing cell
- H3K27ac
- histone 3 with acetylated lysine 27
- IRBP
- interphotoreceptor retinoid-binding protein
- mTEC
- medullary thymic epithelial cell
- TK
- thymidine kinase
- TSA
- tissue-specific antigen
References
- Aaltonen J., Bjorses P., Perheentupa J., Horelli-Kuitunen N., Palotie A., Peltonen L., Lee Y.S., Francis F., Henning S., Thiel C., et al. Finnish-German APECED Consortium . 1997. An autoimmune disease, APECED, caused by mutations in a novel gene featuring two PHD-type zinc-finger domains. Nat. Genet. 17:399–403. 10.1038/ng1297-399 [DOI] [PubMed] [Google Scholar]
- Akiyama T., Shimo Y., Yanai H., Qin J., Ohshima D., Maruyama Y., Asaumi Y., Kitazawa J., Takayanagi H., Penninger J.M., et al. 2008. The tumor necrosis factor family receptors RANK and CD40 cooperatively establish the thymic medullary microenvironment and self-tolerance. Immunity. 29:423–437. 10.1016/j.immuni.2008.06.015 [DOI] [PubMed] [Google Scholar]
- Anderson M.S., Venanzi E.S., Klein L., Chen Z., Berzins S.P., Turley S.J., von Boehmer H., Bronson R., Dierich A., Benoist C., and Mathis D.. 2002. Projection of an immunological self shadow within the thymus by the aire protein. Science. 298:1395–1401. 10.1126/science.1075958 [DOI] [PubMed] [Google Scholar]
- Creyghton M.P., Cheng A.W., Welstead G.G., Kooistra T., Carey B.W., Steine E.J., Hanna J., Lodato M.A., Frampton G.M., Sharp P.A., et al. 2010. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc. Natl. Acad. Sci. USA. 107:21931–21936. 10.1073/pnas.1016071107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derbinski J., Schulte A., Kyewski B., and Klein L.. 2001. Promiscuous gene expression in medullary thymic epithelial cells mirrors the peripheral self. Nat. Immunol. 2:1032–1039. 10.1038/ni723 [DOI] [PubMed] [Google Scholar]
- DeVoss J., Hou Y., Johannes K., Lu W., Liou G.I., Rinn J., Chang H., Caspi R.R., Fong L., Anderson M.S., and Anderson M.S.. 2006. Spontaneous autoimmunity prevented by thymic expression of a single self-antigen. J. Exp. Med. 203:2727–2735. 10.1084/jem.20061864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frazer K.A., Pachter L., Poliakov A., Rubin E.M., and Dubchak I.. 2004. VISTA: computational tools for comparative genomics. Nucleic Acids Res. 32(Web Server):W273-9 10.1093/nar/gkh458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y., Foden J.A., Khayter C., Maeder M.L., Reyon D., Joung J.K., and Sander J.D.. 2013. High-frequency off-target mutagenesis induced by CRISPR-Cas nucleases in human cells. Nat. Biotechnol. 31:822–826. 10.1038/nbt.2623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner J.M., Devoss J.J., Friedman R.S., Wong D.J., Tan Y.X., Zhou X., Johannes K.P., Su M.A., Chang H.Y., Krummel M.F., and Anderson M.S.. 2008. Deletional tolerance mediated by extrathymic Aire-expressing cells. Science. 321:843–847. 10.1126/science.1159407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner J.M., Metzger T.C., McMahon E.J., Au-Yeung B.B., Krawisz A.K., Lu W., Price J.D., Johannes K.P., Satpathy A.T., Murphy K.M., et al. 2013. Extrathymic Aire-expressing cells are a distinct bone marrow-derived population that induce functional inactivation of CD4+ T cells. Immunity. 39:560–572. 10.1016/j.immuni.2013.08.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heino M., Peterson P., Sillanp N., Guérin S., Wu L., Anderson G., Scott H.S., Antonarakis S.E., Kudoh J., Shimizu N., et al. 2000. RNA and protein expression of the murine autoimmune regulator gene (Aire) in normal, RelB-deficient and in NOD mouse. Eur. J. Immunol. 30:1884–1893. [DOI] [PubMed] [Google Scholar]
- Hikosaka Y., Nitta T., Ohigashi I., Yano K., Ishimaru N., Hayashi Y., Matsumoto M., Matsuo K., Penninger J.M., Takayanagi H., et al. 2008. The cytokine RANKL produced by positively selected thymocytes fosters medullary thymic epithelial cells that express autoimmune regulator. Immunity. 29:438–450. 10.1016/j.immuni.2008.06.018 [DOI] [PubMed] [Google Scholar]
- Hubert F.-X., Kinkel S.A., Crewther P.E., Cannon P.Z.F., Webster K.E., Link M., Uibo R., O’Bryan M.K., Meager A., Forehan S.P., et al. 2009. Aire-deficient C57BL/6 mice mimicking the common human 13-base pair deletion mutation present with only a mild autoimmune phenotype. J. Immunol. 182:3902–3918. 10.4049/jimmunol.0802124 [DOI] [PubMed] [Google Scholar]
- Khan I.S., Mouchess M.L., Zhu M.-L., Conley B., Fasano K.J., Hou Y., Fong L., Su M.A., and Anderson M.S.. 2014. Enhancement of an anti-tumor immune response by transient blockade of central T cell tolerance. J. Exp. Med. 211:761–768. 10.1084/jem.20131889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loots G.G., and Ovcharenko I.. 2004. rVISTA 2.0: evolutionary analysis of transcription factor binding sites. Nucleic Acids Res. 32:W217-W21 10.1093/nar/gkh383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malchow S., Leventhal D.S., Nishi S., Fischer B.I., Shen L., Paner G.P., Amit A.S., Kang C., Geddes J.E., Allison J.P., et al. 2013. Aire-dependent thymic development of tumor-associated regulatory T cells. Science. 339:1219–1224. 10.1126/science.1233913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagamine K., Peterson P., Scott H.S., Kudoh J., Minoshima S., Heino M., Krohn K.J.E., Lalioti M.D., Mullis P.E., Antonarakis S.E., et al. 1997. Positional cloning of the APECED gene. Nat. Genet. 17:393–398. 10.1038/ng1297-393 [DOI] [PubMed] [Google Scholar]
- Noonan J.P., and McCallion A.S.. 2010. Genomics of long-range regulatory elements. Annu. Rev. Genomics Hum. Genet. 11:1–23. 10.1146/annurev-genom-082509-141651 [DOI] [PubMed] [Google Scholar]
- Owen C.J., and Cheetham T.D.. 2009. Diagnosis and management of polyendocrinopathy syndromes. Endocrinol. Metab. Clin. North Am. 38:419–436: x. 10.1016/j.ecl.2009.01.007 [DOI] [PubMed] [Google Scholar]
- Pinto S., Michel C., Schmidt-Glenewinkel H., Harder N., Rohr K., Wild S., Brors B., and Kyewski B.. 2013. Overlapping gene coexpression patterns in human medullary thymic epithelial cells generate self-antigen diversity. Proc. Natl. Acad. Sci. USA. 110:E3497–E3505. 10.1073/pnas.1308311110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ran F.A., Hsu P.D., Wright J., Agarwala V., Scott D.A., and Zhang F.. 2013. Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 8:2281–2308. 10.1038/nprot.2013.143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronai D., Berru M., and Shulman M.J.. 1999. Variegated expression of the endogenous immunoglobulin heavy-chain gene in the absence of the intronic locus control region. Mol. Cell. Biol. 19:7031–7040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossi S.W., Kim M.-Y., Leibbrandt A., Parnell S.M., Jenkinson W.E., Glanville S.H., McConnell F.M., Scott H.S., Penninger J.M., Jenkinson E.J., et al. 2007. RANK signals from CD4+3− inducer cells regulate development of Aire-expressing epithelial cells in the thymic medulla. J. Exp. Med. 204:1267–1272. 10.1084/jem.20062497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seumois G., Chavez L., Gerasimova A., Lienhard M., Omran N., Kalinke L., Vedanayagam M., Ganesan A.P.V., Chawla A., Djukanović R., et al. 2014. Epigenomic analysis of primary human T cells reveals enhancers associated with TH2 memory cell differentiation and asthma susceptibility. Nat. Immunol. 15:777–788. 10.1038/ni.2937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shum A.K., DeVoss J., Tan C.L., Hou Y., Johannes K., O'Gorman C.S., Jones K.D., Sochett E.B., Fong L., and Anderson M.S. 2009. Identification of an autoantigen demonstrates a link between interstitial lung disease and a defect in central tolerance. Sci Transl Med. 1:9ra20 10.1126/scitranslmed.3000284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su M.A., Giang K., Zumer K., Jiang H., Oven I., Rinn J.L., Devoss J.J., Johannes K.P.A., Lu W., Gardner J., et al. 2008. Mechanisms of an autoimmunity syndrome in mice caused by a dominant mutation in Aire. J. Clin. Invest. 118:1712–1726. 10.1172/JCI34523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taniguchi R.T., DeVoss J.J., Moon J.J., Sidney J., Sette A., Jenkins M.K., and Anderson M.S.. 2012. Detection of an autoreactive T-cell population within the polyclonal repertoire that undergoes distinct autoimmune regulator (Aire)-mediated selection. Proc. Natl. Acad. Sci. USA. 109:7847–7852. 10.1073/pnas.1120607109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visel A., Blow M.J., Li Z., Zhang T., Akiyama J.A., Holt A., Plajzer-Frick I., Shoukry M., Wright C., Chen F., et al. 2009. ChIP-seq accurately predicts tissue-specific activity of enhancers. Nature. 457:854–858. 10.1038/nature07730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Laan M., Bichele R., Kisand K., Scott H.S., and Peterson P.. 2012. Post-Aire maturation of thymic medullary epithelial cells involves selective expression of keratinocyte-specific autoantigens. Front. Immunol. 3:19 10.3389/fimmu.2012.00019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White A.J., Nakamura K., Jenkinson W.E., Saini M., Sinclair C., Seddon B., Narendran P., Pfeffer K., Nitta T., Takahama Y., et al. 2010. Lymphotoxin signals from positively selected thymocytes regulate the terminal differentiation of medullary thymic epithelial cells. J. Immunol. 185:4769–4776. 10.4049/jimmunol.1002151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu M., Chin R.K., Christiansen P.A., Lo J.C., Liu X., Ware C., Siebenlist U., and Fu Y.-X.. 2006. NF-kappaB2 is required for the establishment of central tolerance through an Aire-dependent pathway. J. Clin. Invest. 116:2964–2971. 10.1172/JCI28326 [DOI] [PMC free article] [PubMed] [Google Scholar]