Abstract
Background
Biofouling, the colonization of artificial and natural surfaces by unwanted microorganisms, has an important economic impact on a wide range of industries. Low cost antifouling strategies are typically based on biocides which exhibit a negative environmental impact, affecting surrounding organisms related and not related to biofouling. Considering that the critical processes resulting in biofouling occur in the nanoscale/microscale dimensions, in this work we present a bionanotechnological approach to reduce adhesion of biofilm-producing bacteria Halomonas spp. CAM2 by introducing single layer graphene coatings. The use of this nanomaterial has been poorly explored for antifouling application.
Results
Our study revealed that graphene coatings modify material surface energy and electrostatic interaction between material and bacteria. Such nanoscale surface modification determine an important reduction over resulting bacterial adhesion and reduces the expression levels of genes related to adhesion when bacteria are in contact with graphene-coated material.
Conclusions
Our results demonstrate that graphene coatings reduce considerably adhesion and expression levels of adhesion genes of biofilm-producing bacteria Halomonas spp. CAM2. Hydrophobic-hydrophilic interaction and repulsive electrostatic force dominate the interactions between Halomonas spp. CAM2 and material surface in saline media, impacting the final adhesion process. In addition no bactericide effect of graphene coatings was observed. The effect over biofilm formation is localized right at coated surface, in contrast to other antifouling techniques currently used, such as biocides.
Keywords: Graphene, Halomonas, Biofilms, Bacterial adhesion, Antifouling
Background
Marine fouling is the accumulation of micro and macroorganisms on underwater surfaces, which provide a favorable mechanism to survive in the environment. The economic impact of fouling on shipping vessels, oceanographic sensors, power plants and aquaculture systems, among others, has been estimated to be in the range of 50 billion euros per year [1–3]. In particular, shipping, fishing and aquaculture industries exhibit extreme fouling cases. On vessels, fouling adds weight to boats and increases hull roughness and hydrodynamic drag, raising fuel consumption by almost 40 %, with the corresponding increase in emissions of greenhouse gases and other pollutants [4]. In aquaculture, settlement of fouling organisms in culture cages causes suffocation of the cultivated species, delaying the time when the cultivated species reaches commercial size [5]. Another singular example is found in boilers cooled with ocean water where the fouling phenomenon causes strong inefficiency in operational parameters and increase in fuel consumption [6].
Current techniques to prevent (antifouling) and fight fouling (fouling-release) include physical (e.g. heat treatments, pulse-power technology, radioactive coatings, flushing, scrubbing and biological control) and chemical methods (e.g. injectable biocides, chlorine, marine bioactive compounds and other form of bactericide coatings such as copper and copper alloys). Although the choice of the right strategy will depend on the cost and application possibilities, antifouling coatings are probably the most cost-effective method for boats and other surfaces. They are typically based on the controlled release of organic solvents into the environment to kill the colonizing microorganism. However this approach offers a non-localized solution, affecting surrounding organisms related and not related to fouling. As a consequence, the use of certain biocides has been restricted by some countries and in Europe a large amount of data has been gathered as part of the Biocidal Products Directive (BPD, 98/8/EC) [7].
The critical processes at the biointerface resulting in biofouling occur in the nanoscale/microscale dimensions: it follows therefore that surface properties which could control biofouling need to be on the same length scales [8]. Search for non-biocidal technology to control the economic and environmental problem caused by biofouling has focused, as a result, on modifying physico-chemical and mechanical properties of surfaces (such as surface free energy, wettability, elasticity and surface topography) at the nanoscale to reduce bacterial attachment [9–14]. As far as coatings for marine antifouling applications are concerned, surfaces with low surface energy or with an optimized surface topography (with patterns in the order of micrometer) have shown promising results [13, 15] opening a new avenue for the development of antifouling coatings.
Graphene is a one-atom thick carbon sheet that has emerged as a new carbon compound with multiple applications in a wide range of industrial processes and products. While graphene is a promising candidate in electronic applications, its use for biological applications, such as antifouling, has been poorly explored. There are many reports of bacterial interaction with graphene oxide (GO), formed by micro- or nano-sized flakes of functionalized graphene in powder, solution or coating [16–18], which induces inactivation of bacterial cells upon direct contact by physical and oxidative damage to cell as its antibacterial mechanism [13, 19–22]. Flake size turns out to be a relevant aspect for the reported antibacterial activity of GO, whether this be in suspension or coating [18, 20]. However in the case of single layer graphene sheets grown on Cu, which as GO are one-atom thick, they have surface areas in the centimeter square range. Hence the mechanism of the bacterial interaction must be different in both cases.
In this report we present a physico-chemical and biological approach to reduce fouling formation in its initial growth stage as biofilm, by introducing graphene coatings that reduce bacterial adhesion to coated surfaces. Nanoscale behavior is discussed in the particular case of biofilm-producing Halomonas spp. CAM2, which is used as a model marine bacterium.
Results and discussion
Preparation and characterization of nanostructured modified samples
Micro and nanoscale characterization of as-grown graphene on Cu and graphene transferred onto SiO2 samples was carried out to evaluate their composition, microstructure, topography with atomic resolution, graphitic quality and contamination.
Scanning electron micrographs of single layer graphene (SLG) grown on Cu showed some contrast at micrometer scale that could be identified as graphene domains (Fig. 1b). Atomic-resolved images of SLG grown on Cu were obtained by scanning tunneling microscopy (STM) in ultra-high vacuum conditions (Fig. 1c, e). STM topographies exhibit the distinctive honeycomb structure with an interatomic distance of 1.4 A, consistent with literature values [23, 24]. Transfer of graphene to SiO2 process is described in materials and methods section.
Fig. 1.
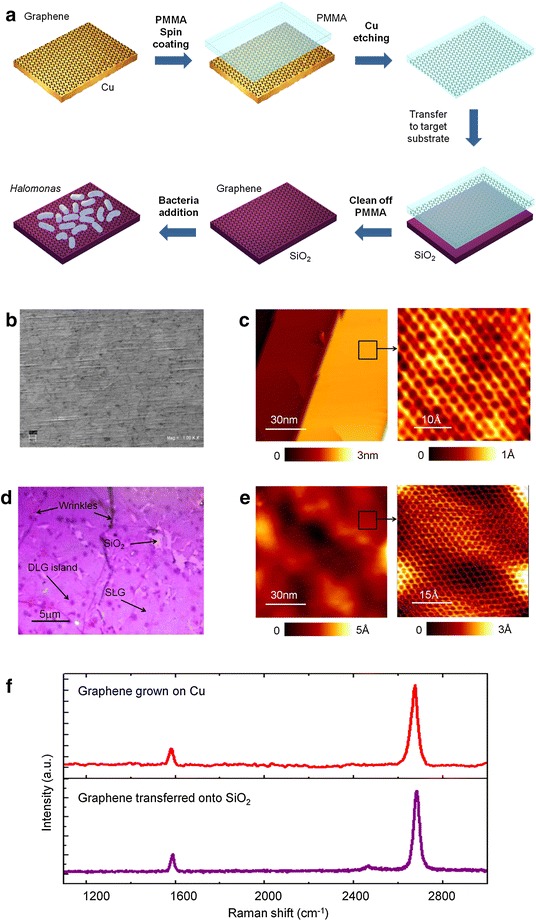
Preparation and characterization of graphene-coated samples. a Illustrative diagram showing PMMA-assisted transfer method used to obtain graphene-coated SiO2 substrates for present study, b SEM image of single layer graphene (SLG) grown on Cu sample, c large-scale STM topographic image (100 × 100 nm2) of SLG grown on Cu. The filtered atomically resolved image (3.5 × 3.5 nm2) shows the hexagonal lattice of SLG, d optical microscopy image of SLG transferred onto SiO2, e STM image of SLG transferred onto SiO2 displays honeycomb lattice and f representative Raman spectra Raman spectra of SLG grown on Cu (up) and SLG transferred onto SiO2 (down). Background caused by the luminescence of the copper was subtracted in the case of SLG grown and transferred onto Cu. Tunnel current and bias voltages for STM images were between 0.1 and 0.6 nA and 0.1–1 V respectively
The morphological characterization of graphene grown on Cu and resulting graphene-coated SiO2 samples prior to bacteria contact was screened by SEM, optical microscopy and STM. Wrinkles in CVD graphene grown on Cu are formed by differential thermal expansion, as the metal contracts more than the graphene during post-growth cooling [25]. Such wrinkles are still present after transfer to SiO2 substrate, as can be clearly seen in optical microscopy (Fig. 1d).
In addition, different contrast in certain areas is observed which can be identified as bilayer islands on top of a monolayer background. The surface of SLG transferred onto SiO2 substrates showed micrometric damages in the graphitic membrane due to the transfer procedure, which leaves some SiO2 areas exposed. Clear visualization of the intrinsic hexagonal structure (honeycomb) of graphene transferred onto SiO2 was possible by STM (Fig. 1c, e). Few signs of surface contamination were found by this atomic-resolved technique.
To verify the graphitic quality of graphene coatings we performed microRaman spectroscopy measurements. Multiple areas of each sample were analyzed and representative spectra are shown in Fig. 1f. SLG grown on Cu and SLG transferred onto SiO2 typically display sharp G (1584 cm-1) and 2D (2680–2693 cm-1) bands, with a small G/2D ratio (0.25 and 0.29 respectively). These results are consistent with single layer graphene, according to values reported in literature [26–28].
Graphene coating effects on bacterial adhesion
We have used SEM and fluorescence microscopy in order to characterize bacterial adhesion to graphene-coated and uncoated SiO2. Morphology of Halomonasspp. CAM2 incubated for 72 h on SiO2 and graphene-coated SiO2 samples are shown in Fig. 2a, b, respectively. Intact and smooth cell surfaces were observed for both substrates, in agreement with previous results, confirming the absence of bactericide effects of graphitic coatings [29]. In addition, SEM micrographs show a notorious difference in the bacterial attachment to both surfaces, which is reduced in the case of graphene-coated material (Fig. 2b).
Fig. 2.
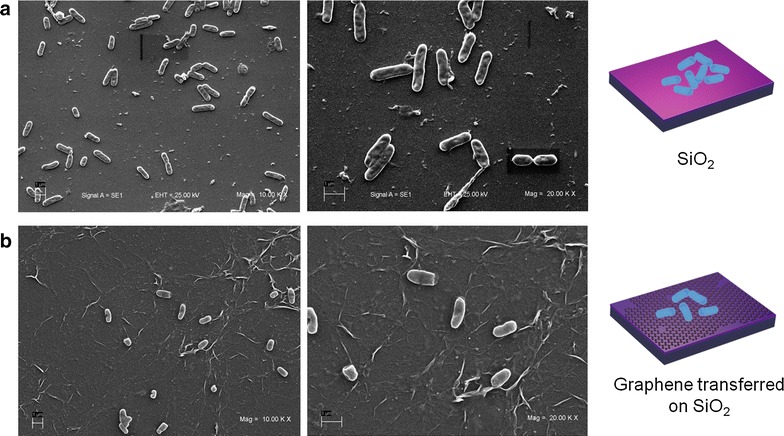
Bacterial attachment to graphene-coated samples. a SEM images of Halomonas spp. CAM2 after 72 h incubation on SiO2 sample and b on graphene-coated SiO2 samples
Representative epifluorescence microscopy images of a partially and completely graphene-coated SiO2 samples are shown in Fig. 3a, b respectively. The SiO2 substrate was partially coated in order to visualize bacterial attachment in response to coated and uncoated surfaces over the same sample. Bacterial bodies were green stained as indicative of live bacteria on sample surface. Interestingly, the highest concentration of live bacteria was found across the uncoated SiO2 surface (upper area in Fig. 3a). In contrast, only few live bacteria were observed across the graphene-coated SiO2 area (lower area). For graphene-coated samples (Fig. 3b) the presence of few cells (bright spots in magnified area) can be attributed to the intrinsic micrometer damage of the graphene membrane caused by transfer process (Fig. 1d), which leads to few SiO2 exposed areas with increased bacterial attachment. Epifluorescene and SEM results suggest graphene coatings suppress dramatically bacterial attachment which is determinant to biofilm and fouling formation. Such behavior is not related to bactericidal activity, according to same results.
Fig. 3.
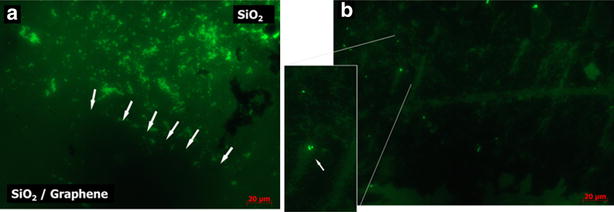
Bacterial distribution in graphene-coated surfaces epifluorescence microscopy image of Halomonas spp. CAM2 incubated on partially graphene-coated SiO2 surfaces. a Partially graphene-coated SiO2 surface and b completely graphene-coated SiO2 surface. White arrows in (a) are indicating boundary between uncoated and graphene-coated areas on SiO2 surface
Graphene coating effects on adhesin gene expression
Relative expression of adhesin gene in Halomonas CAM2 incubated on SiO2 and graphene-coated SiO2 samples was evaluated. Quantitative PCR in real time was performed to quantify the expression levels of four selected genes in Halomonas CAM2 (Fig. 4). Such genes, F7SSV5, F7SSV5, G4f3Q7 and AlgC, are reported to be related to adhesion in other bacterial species (See methods section for detailed information). F7SSV2 and F7SSV5 genes have been reported to codify adhesin transmembrane proteins in Halomonas sp. TD01 [30]. G4f3Q7 codify a polysaccharide intercellular adhesin (PIA) that participates in the biofilm formation in Halomonas sp. HAL1 [31]. AlgC has been previously shown to encode phosphomannomutase, which activity produces a precursor for alginate polymerization and biosynthesis of lipopolysaccaride (LPS), both required for biofilm production in Pseudomonas aeruginosa [32].
Fig. 4.
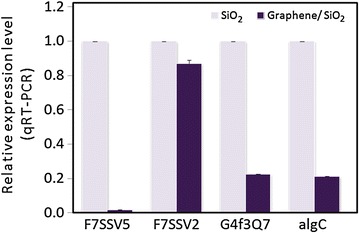
Relative expression levels of 4 adhesin genes. q-PCR results for F7SSV5, F7SSV5, G4f3Q7 and AlgC adhesin genes of Halomonas spp. CAM2 incubated on SiO2 and graphene-coated SiO2 samples
PCR results in Fig. 4 show the expression levels of F7SSV5, G4f3Q7 and AlgC in Halomonas spp. CAM2. They were significantly lower when bacteria were incubated on graphene-coated SiO2 samples compared to the corresponding control (uncoated SiO2 sample). A mild expression reduction was observed for gene F7SSV2. Such expression difference can be understood in terms of protein location at the cell membrane. Although F7SSV2 and F7SSV5 belong to the same protein family “pfam-A adhesion” and containing similar conserved domain (YadA head; ESPR for Extended Signal Peptide Region), they have different functions, according location of amino acids in their sequence, they have different functions, depending on location of amino acids in their sequence (Fig. 5). F7SSV2 is an integral part of inner cell membrane and is not participating in bacterial adhesion behavior at the same molecular level that F7SSV5 does, which is strongly affected by being an outer membrane protein gene.
Fig. 5.
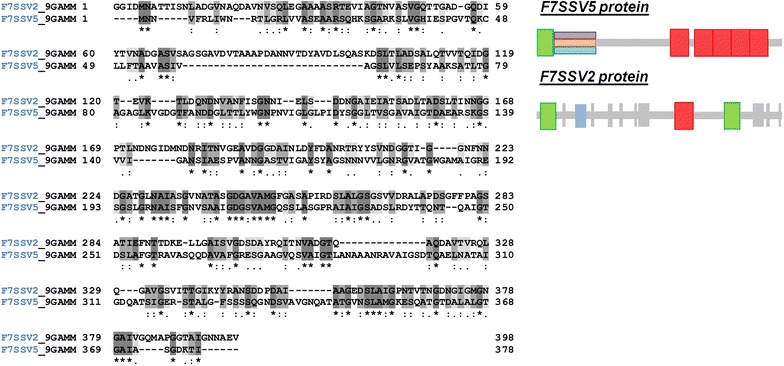
Blast analysis of adhesin genes F7SSV2 and F7SSV5 in Halomonas. Arrangement of the Pfam domains in F7SSV5 and F7SSV2 protein. Domains described are based on searching the Pfam-A family against UniProtKB using hmm search. Letters highlighted in gray are indicating amino acid with similarity properties. (Software used UniprotKB Align, http://www.expasy.com)
Graphene coating effects on surface energy, wettability and electrostatic interaction
Upon approach to a surface, microorganisms will be attracted or repelled, depending on the different non-specific interaction forces [33]. The first relevant interaction in this system is the one related to long-range electrostatic forces between graphene-coated SiO2 surface and cells that might be affecting the initial (and reversible) bacterial adhesion process. In graphene-coated SiO2, SiO2 substrates have a significant surface state density just below the conduction band edge that donates electrons to graphene to balance the chemical potential at the interface. This leads to n-type (or electron-doped) graphene coating [34]. In addition, it has been suggested that bacteria, when introduced into aqueous suspensions, are always negatively charged [35]. To determine if electrostatic long-range interactions between graphene-coated SiO2 and bacteria contribute to an initial repulsion between bacteria and graphene-coated substrate we performed theoretical calculations to obtain electrostatic force F(r) between bacteria and material surface (SiO2 and graphene-coated SiO2) as a function of their separation distance using the expression [36]:
| 1 |
where F is electrostatic force (in N); r is distance between bacteria and surface (in m); is the radius of bacteria (or SiO2 piece or graphene-coated SiO2) (in m), ɛ is the dielectric constant of water [37] (78.43 at 298 K); ɛ0 is the permittivity of free space (8.854 × 10−12C/Jm); kB is Boltzmann’s constant (1.381 × 10−23 J/K), is temperature (293 K), z is the valence of electrolyte ions (1 for NaCl) and e is the charge of an electron (1.602 × 1019 C). The inverse Debye length κ describes the thickness of the electrostatic double layer of counter-ions that surrounds charged parts of the system (bacteria or SiO2) in solution. For monovalent electrolytes (e.g. NaCl), κ−1 is given by 0.304/(c)1/2 (in 1/nm) where c is the concentration of the electrolyte (in mol/L) and contains information of ionic strength of solution [38]. In our case we evaluate 2 and 0.5 % NaCl concentration of suspension media. Surface potential ϕ is described by zeψ/kBT, where ψ is the surface potential of the bacteria, SiO2 piece or graphene-coated SiO2 piece (in V). We considered surface potentials values of SiO2 10 piece, graphene-coated SiO2 10 μm piece and Pseudomonas are −35 mV [39] −77 mV [40] and −9 mV [41] respectively. The theoretical force-distance relationship is shown in Fig. 6g. According to this result the electrostatic force in this system is expected to be repulsive and short range (<5 nm for 0.5 % NaCl and <3 nm for 2 % NaCl). Electrostatic repulsion between bacteria and SiO2 increases when SiO2 is coated with graphene, for both NaCl concentrations, although the effect is higher for lower solution ionic strength.
Fig. 6.
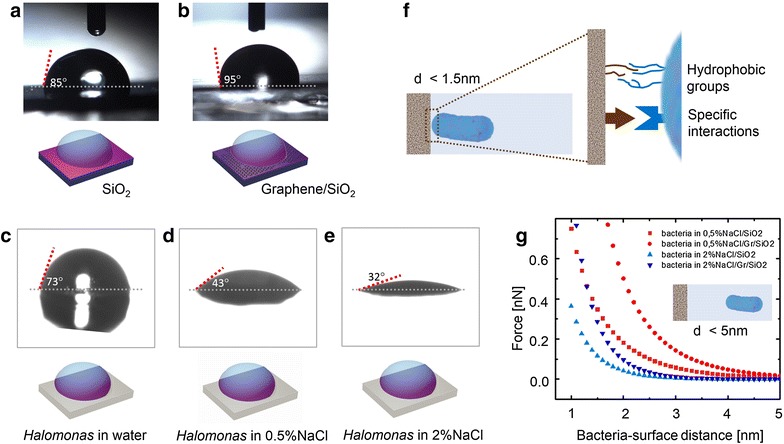
Hydrophobic-hydrophilic nature and electrostatic force between graphene-coated samples and bacteria. Images of contact angle measurements using milliQ water in contact with SiO2 (a), graphene-coated SiO2 (b), lawn of Halomonas spp. CAM2 previously suspended in water (0 % salinity) (c), lawn of Halomonas spp. CAM2 previously suspended in 0.5 % NaCl saline solution (d) and lawn of Halomonas spp. CAM2 previously suspended in 2 % NaCl saline solution (e). Diagram (f) shows a schematic representation of short-distance interaction (<1.5 nm), originated by physico-chemical forces (like the ones related to surface energy other specific interactions) that are relevant for bacterial adhesion. g Theoretical calculations of electrostatic force as function of distance between bacteria and surface (SiO2 and graphene-coated SiO2) confirm repulsive interaction that increases when material is coated with graphene
After overcoming (if) this initial electrostatic repulsion, an even shorter-range hydrophobic interaction (generally when bacteria-surface distance is smaller than 1.5 nm [28]) have a strong impact on bacterial surface adhesion, a characteristic that is mainly determined by physico-chemical surface properties [42].
In order to determine the influence of possible hydrophobic characteristics of graphene coatings over bacterial adhesion, we performed contact angle measurements on SiO2 (Fig. 6a), graphene-coated SiO2 (Fig. 6b) and lawns of Halomonas spp. CAM2 previously suspended in water (0 % NaCl) and saline buffer (0.5 % NaCl and 2 % NaCl) (Fig. 6c, d, e respectively).
Contact angles are related to the surface free energies [43]. Hydrophobic coatings are often used to minimize adhesion since they create a larger contact angle between the bacteria’s glue and the surface. This results in less wettability (lower surface energy) and less fouling since the adhesive is not being able to spread across the surface [44]. According to our measurements a transition from hydrophilic surface (contact angle of ~85° ± 0.7) for SiO2 substrate to hydrophobic surface (contact angle of ~95° ± 0.3) for graphene-coated SiO2 is observed.
In addition, cells that possess a hydrophilic character attach preferentially to hydrophilic surfaces (large surface energy), whereas hydrophobic cells prefer hydrophobic surfaces [3, 45]. The hydrophobic-hydrophilic nature of Halomonas spp. CAM2 surface changes as the bacterium grows in different media. Reported response of Halomonaselongata to saline NaCl media, like the one used in the current experiments, displays an enhanced hydrophilicity [46], which makes the cell more attractive to water molecules in the environment and prevents desiccation. Our contact angle results confirmed such hydrophilic nature of Halomonas spp. CAM2 and a clear trend to increased cell hydrophilicity as a function of salinity of suspension media; 73° for 0 %, 43° for 0.5 % and 32° for 2 % NaCl (Fig. 6c, d, e respectively). In addition these results suggest that the modification from hydrophilic to hydrophobic nature of graphene-coated SiO2 determines the suppression of bacterial attachment for hydrophilic Halomonas spp. CAM2. These results show that both interactions, electrostatic (<5 nm range) and hydrophobic-hydrophilic (<1.5 nm range), are presumably affecting the bacterial attachment process, causing a notorious decrease in adhesin gene expression of Halomonas spp. CAM2, with the corresponding reduction of bacterial adhesion to graphene-coated surfaces.
Conclusion
In this paper we present a nano-biotechnological approach to decrease the attachment of marine bacteria Halomonas spp. CAM2 by introducing graphene coatings. According to our theoretical and experimental results graphene coatings modify surface energy and electrostatic interactions with bacterial cells which determines an important reduction of bacterial adhesion, a relevant parameter involved in biofilm formation and consequent biofouling emergence. This nanoscale surface modification affects the expression of genes related to adhesion that are notoriously decreased when bacteria are in contact with graphene-coated SiO2 surfaces instead of uncoated SiO2 surfaces.
No bactericide effects of graphene-coated SiO2 were observed. Such behavior indicates the effect over biofilm formation is localized right at coated surface, in contrast to other antifouling techniques currently used, such as biocides, that exhibit negative effects over all surrounding aquatic species, not necessarily related to biofilm or biofouling formation. We expect this work will contribute to provide new opportunities for designing effective and environmentally friendly antifouling surfaces based on nanoscale modified materials.
Methods
Materials
Silicon dioxide coated wafers and commercial CVD graphene grown on Cu was obtained from Graphene Supermarket Company. Graphene and SiO2 samples used in this study were 1 cm2 in area.
Preparation and characterization of Gr/SiO2
Transfer of graphene films grown on Cu to SiO2 substrates was achieved by the poly(methylmethacrylate) (PMMA) assisted method as shown in Fig. 1a. Thin layer of PMMA on graphene on Cu foil was produced by spin-coating. The polymer provides a supportive framework for graphene before the transfer. The underneath Cu substrate is then etched away by an ammonium persulfate ((NH4)2SO8) solution. After the Cu foil is completely dissolved, the floating membrane can be scooped and placed on SiO2. After drying, the polymeric film is dissolved with acetone.
PMMA and ammonium persulfate ((NH4)2SO8) used for graphene transfer procedure were purchased from Sigma Aldrich.
Scanning Tunneling Microscopy (UHV-VT Omicron) was used to characterize nanoscale morphology. Sample preparation before STM measurements consists of 200 °C annealing in UHV for 30 min. Platinum-iridium tips were used for all STM measurements. The experimental data were analyzed by using WSxM software. Scanning Electron Microscopy (SEM) images were recorded using a Carl Zeiss microscope (EVO MA-10) to characterize microscale morphology and qualitative bacterial adhesion. MicroRaman measurements (Renishaw, 532 nm laser) were used to characterize quality of as-grown graphene and transferred graphene onto SiO2. Contact angle measurements were performed to characterize surface hydrophobicity of coated and uncoated Cu samples. A drop of milliQ water (2μL) was placed on the surface of graphene-coated SiO2 and uncoated samples and images were immediately captured using a high-resolution camera. Bacterial hydrophobicity was measured following standard methods [41] with some modifications. A bacterial strain suspended in 40 mL of water, 0.5 % NaCl and 2 % NaCl were filtered on a micropore cellulose nitrate filter (pore size 0.45 μm, Sartorius Stedim Biotech, Germany) by filtration of the suspension using negative pressure. The filters with a bacteria film were dried at room temperature during 90 min in order to obtain a stable water contact angle measured by sessile drop method using 1 μL of distilled. The contact angle was measured based on image analysis [47] using the image processing software Image J with the plug-in Drop Shape Analysis based on B-spline snakes algorithm developed by [48].
Bacterial strain isolation
A marine strain Halomonas spp. CAM2 was used in the biofilm formation assays. Bacteria was previously isolated from illness larvae of the Chilean scallop Argopecten purpuratus Lamarck, 1891 (bivalvia, pectinidae) and characterized [49]. Stock cultures of the CAM2 strain were maintained at 4 °C on Tryptic soy agar (Difco) supplemented with NaCl (2 %), and subcultured every 2 weeks. For long-term preservation, CAM2 strain cultures were frozen at −80 °C in Tryptic soy broth (Difco) supplemented with 2 % NaCl (w/v) and 20 % glycerol (v/v) [50]. When required, frozen cultures were recovered by streaking onto Tryptic soy agar plates (Difco) supplemented with NaCl (2 %), which were incubated at 20 °C for 24 h.
Bacteria culture and exposure to Gr/SiO2
Surfaces used for biofilms growth were sterilized by rinsing several times with ethanol and sterilized DI water. Bacterial adhesion assays on partially graphene-coated SiO2 were performed in petri dishes. TSA Agar plates were inoculated with freshly growing cells of Halomonas spp. CAM2, so that a lawn of bacteria was grown. After 48 h incubation at 25 °C a half of the surface containing bacteria (in exponential to early stationary phase) was harvested and suspended in 10 ml of sterile saline buffer (0.5 % NaCl). This volume was poured on pieces of 1 cm2 of Gr/SiO2 and SiO2 and incubated for 72 h at 20 °C. An aliquot was removed in order to determine the cell concentration by dilution plating. All experiments were run in triplicate.
Bacterial adhesion study
Analysis of bacterial adhesion was conducted in order to evaluate morphology and viability of microorganisms. For SEM characterization bacteria were fixed on samples with 3 % (v/v) glutaraldehyde and dehydrated by washing with a graded ethanol series (from 10 to 100 %), followed by critical-point drying and gold coating.
Distribution of bacteria on Gr/SiO2 or SiO2 surfaces was determined directly in situ. For epifluorescence analysis Gr/SiO2 or SiO2 pieces were submerged into solution provided by standard LIVE/DEAD BacLight bacterial viability kit (0.01 mM of Syto9 and 0.06 mM of propidium iodide). Samples were kept dark during 15 min and then observed by epifluorescence microscopy Olympus × 71.
Expression of adhesion genes
Halomonas spp. CAM2 has not been previously sequenced. Because this strain was only recently isolated, no genes participating in adhesion for this bacterial specie have been identified and sequenced. Genes F7SSV5, F7SSV2 and G43U7, which have been described to participate in adhesion of Halomonas elongata [30] together with AlgC, involved in the same process for Pseudomonas aureginosa, were used as target. Primers were designed using sequences from GenBank and ApliX software V3.1. Sequence for each gene is described in the Table 1.
Table 1.
Sequences of oligonucleotide primers used for qPCR
| Gene name | sequence | Amplicon size (bp) |
|---|---|---|
| 16S-F | 5-TCGCGTTAACTTCGCCACAA-3 | 184 |
| 16S-R | 5-AGCGGTGAAATGCGTAGAGA-3 | |
| F7SSV5-F | 5-ATGTCGCCTGATCACCGATA-3 | 197 |
| F7SSV5-R | 5-TGACTCGCTCGCTTTTGGTA-3 | |
| F7SSV2-F | 5-TTTGCTGGCTTGGCTGAGAT-3 | 187 |
| F7SSV2-R | 5-AGTCATTGCTGGCACAAACG-3 | |
| G4F3Q7-F | 5-ACCAGATCGGCAAGCACAAA-3 | 176 |
| G4F3Q7-R | 5-TGGCTGGCGTTTTCATCCAA-3 | |
| algC-F | 5-TGATCTTCGACGTCAAGTGC-3 | 153 |
| algC-R | 5-AAATGTGACCGCTCATCTCG-3 |
Primers were designed for this study using AmplifX V1.3.7 software with genes published on genebank
For RNA isolation, SiO2 surfaces with and without graphene were washed twice with phosphate buffer. Bacteria were scrapped from samples surface with a sterile cotton swab and were stored at −80 °C awaiting RNA isolation [51]. Bacterial RNA was extracted using TRIzol reagent (Ambion by Lifetechnologies).
Samples were defrosted, and genes were extracted using chloroform. RNA was recovered by precipitation with ethanol 70 % and was load on Qiagen RNeasy minElute spin column following manufacturer’s instructions.
RNA quantity was determined by measuring the absorbance at 260 nm using a Nanodrop spectrophotometer ND-1000 (Thermo Fisher Scientific). Contaminated genomic DNA was removed by TURBO DNA-free kit (ambion by lifetechnologies). Further, total RNA samples were analyzed for the presence of DNA contamination by qRT-PCR using 16S rRNA target.
Purified RNA was converted to cDNA using RevertAid™ first strand cDNA synthesis kit (Fermentas) with random hexamer primers according to manufacturer’s instructions.
Real time PCR was performed according to the protocol of the SYBR Green/Rox qPCR Master Mix (Fermentas) with Stratagene Mx3000P real-time PCR system (Stratagene); 16S mRNA levels were used for normalization.
Amplification courses in all genes included the following 3 steps: (1) 1 cycle of an initial denaturation for 10 min at 95 °C, (2) 40 cycles of an initial denaturation for 30 s at 95 °C. Annealing for 1 min at 55 °C and extension for 30 s at 72 °C and, (3) 1 cycle of an initial denaturation for 1 min at 95 °C, annealing for 30 s at 55 °C a final extension for 30 s at 95 °C.
Relative quantification of each target gene (F7SSV5, F7SSV2, G4F3Q7, AlgC) encoding for adhesin in each experimental sample, and control versus reference gene (rDNA16S) was performed in according with Livak Eq. 1: relative expression ratio (R) = 2−[ΔCt sample − ΔCt control] = 2−ΔΔCt.
Authors’ contributions
CP, FD, ED, RR, CR collected research data. CP, FD, RH, CR and PV analyzed the data. CP, CR and PV wrote this article. All authors read and approved the final manuscript.
Acknowledgements
This research was supported by Innova Corfo L1 13IDL1-25496, Chilean Government. PV is grateful for access to Sherwin William facilities, Santiago, Chile.
Competing interests
The authors declare that they have no competing interests.
Contributor Information
Carolina Parra, Email: carolina.parra@usm.cl.
Fernando Dorta, Email: fernando.dorta@usm.cl.
Edra Jimenez, Email: edra.jimenez@usm.cl.
Ricardo Henríquez, Email: ricardo.henriquez@usm.cl.
Cristian Ramírez, Email: cristian.ramirez@usm.cl.
Rodrigo Rojas, Email: rrojas@ucn.cl.
Patricio Villalobos, Email: patricio.villalobos@usm.cl.
References
- 1.Azis PKA, Al-Tisan I, Sasikumar N. Biofouling potential and environmental factors of seawater at the desalination plant intake. Desalination. 2001;135:69–82. doi: 10.1016/S0011-9164(01)00140-0. [DOI] [Google Scholar]
- 2.Fitridge I, Dempster T, Guenther J, de Nys R. The impact and control of biofouling in marine aquaculture: a review. Biofouling. 2012;28:649–669. doi: 10.1080/08927014.2012.700478. [DOI] [PubMed] [Google Scholar]
- 3.Tuson HH, Weibel DB. Bacteria-surface interactions. Soft Matter. 2013;9:4368–4380. doi: 10.1039/c3sm27705d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Townsin RL. The ship hull fouling penalty. Biofouling. 2003;19:9–15. doi: 10.1080/0892701031000088535. [DOI] [PubMed] [Google Scholar]
- 5.Greene JK, Grizzle RE. Successional development of fouling communities on open ocean aquaculture fish cages in the western Gulf of Maine. Aquaculture. 2007;262:289–301. doi: 10.1016/j.aquaculture.2006.11.003. [DOI] [Google Scholar]
- 6.Pronobis M. The influence of biomass co-combustion on boiler fouling and efficiency. Fuel. 2006;85:474–480. doi: 10.1016/j.fuel.2005.08.015. [DOI] [Google Scholar]
- 7.Thomas KV, Brooks S. The environmental fate and effects of antifouling paint biocides. Biofouling. 2010;26:73–88. doi: 10.1080/08927010903216564. [DOI] [PubMed] [Google Scholar]
- 8.Szewczyk P. The role of nanotechnology in improving marine antifouling coatings. Zesz Nauk/Akad Morska Szczecinie. 2010;24:118–123. [Google Scholar]
- 9.Beigbeder PD, Conlan SL, Mutton R, Clare AS, Pettitt ME, Callow ME, et al. Preparation and characterization of silicone-based coatings filled with carbon nanotubes and natural sepiolite and their application as marine fouling-release coating. Biofouling. 2008;24:291. doi: 10.1080/08927010802162885. [DOI] [PubMed] [Google Scholar]
- 10.Adiga SP, Jin C, Curtiss LA, Monteiro-Riviere NA, Narayan RJ. Nanoporous membranes for medical and biological applications. Nanomed Nanobiotechnol. 2009;1:568–581. doi: 10.1002/wnan.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Callow JA, Callow ME. Trends in the development of environmentally friendly fouling-resistant marine coatings. Nat Commun. 2011;2:1–10. doi: 10.1038/ncomms1251. [DOI] [PubMed] [Google Scholar]
- 12.Sheet I, Holail H, Olama Z, Kabbani A, Hines M. The antibacterial activity of graphite oxide, silver, impregnated graphite oxide with silver and GO-coated sand nanoparticles against waterborne pathogenic E. coli BL21. Int J Curr Microbiol App Sci. 2013;2:1–11. [Google Scholar]
- 13.Banerjee I, Pangule RC, Kane RS. Antifouling coatings: recent developments in the design of surfaces that prevent fouling by proteins, bacteria, and marine organisms. Adv Mater. 2011;23:690–718. doi: 10.1002/adma.201001215. [DOI] [PubMed] [Google Scholar]
- 14.Krishnan S, Weinman CJ, Ober CK. Advances in polymers for anti-biofouling surfaces. J Mater Chem. 2008;18:3405–3413. doi: 10.1039/b801491d. [DOI] [Google Scholar]
- 15.Petronis S, Berntsson K, Gold J, Gatenholm P. Design and microstructuring of PDMS surfaces for improved marine biofouling resistance. J Biomater Sci Polym Ed. 2000;11:1051. doi: 10.1163/156856200743571. [DOI] [PubMed] [Google Scholar]
- 16.Yu L, Zhang Y, Zhang B, Liu J, Zhang H, Song C. Preparation and characterization of HPEI-GO/PES ultrafiltration membrane with antifouling and antibacterial properties. J Membr Sci. 2013;444:452–462. doi: 10.1016/j.memsci.2013.07.042. [DOI] [Google Scholar]
- 17.Yu L, Zhang Y, Zhang B, Liu J. Enhanced antibacterial activity of silver nanoparticles/halloysite nanotubes/graphene nanocomposites with sandwich-like structure. Sci Rep. 2014;4:4551–4555. doi: 10.1038/srep04551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Perrault F, de Faria AF, Nejati S, Elimelech M. Antimicrobial properties of graphene oxide nanosheets: why size matters. ACS Nano. 2015;9:7226–7236. doi: 10.1021/acsnano.5b02067. [DOI] [PubMed] [Google Scholar]
- 19.Perreault F, de Faria FA, Elimelech M. Environmental applications of graphene-based nanomaterials. Chem Soc Rev. 2015;44:5861–5896. doi: 10.1039/C5CS00021A. [DOI] [PubMed] [Google Scholar]
- 20.Perreault F, Tousley ME, Elimelech M. Thin-film composite polyamide membranes functionalized with biocidal graphene oxide nanosheets. Environ Sci Technol Lett. 2014;1:71–76. doi: 10.1021/ez4001356. [DOI] [Google Scholar]
- 21.Romero-Vargas Cartrillón S, Perrault F, de Faria FA, Elimelech M. Interaction of graphene oxide with bacterial cell membranes: insights from force spectroscopy. Environ Sci Technol Lett. 2015;2:112–117. doi: 10.1021/acs.estlett.5b00066. [DOI] [Google Scholar]
- 22.Al-Thani RF, Patan NK, Al-Maadeed MA. Graphene oxide as antimicrobial against two gram-positive and two gram-negative bacteria in addition to one fungus. Online J Biol Sci. 2014;14:230–239. doi: 10.3844/ojbsci.2014.230.239. [DOI] [Google Scholar]
- 23.Park J, Lee J, Liu L, Clark KW, Durand C, Park C, et al. Spatially resolved one-dimensional boundary states in graphene–hexagonal boron nitride planar heterostructures. Nat Commun. 2014;5:5403. doi: 10.1038/ncomms6403. [DOI] [PubMed] [Google Scholar]
- 24.Yu Q, Jauregui LA, Wu W, Colby R, Tian J, Su Z, et al. Control and characterization of individual grains and grain boundaries in graphene grown by chemical vapor deposition. Nat Mater. 2011;10:443–449. doi: 10.1038/nmat3010. [DOI] [PubMed] [Google Scholar]
- 25.Calado VE, Schneider GF, Theulings AMMG, Dekker C, Vandersype LMK. Formation and control of wrinkles in graphene by the wedging transfer method. Appl Phys Lett. 2012;10:103116. doi: 10.1063/1.4751982. [DOI] [Google Scholar]
- 26.Li X, Cai W, An J, Kim S, Nah J, Yang D, et al. Large-area synthesis of high-quality and uniform graphene films on copper foils. Science. 2009;324:1312–1314. doi: 10.1126/science.1171245. [DOI] [PubMed] [Google Scholar]
- 27.Ferrari AC, Meyer JC, Scardaci V, Casiraghi C, Lazzeri M, Mauri F, et al. Raman spectrum of graphene and graphene layers. Phys Rev Lett. 2006;97:187401–187405. doi: 10.1103/PhysRevLett.97.187401. [DOI] [PubMed] [Google Scholar]
- 28.Reina A, Jia X, Ho J, Nezich D, Son H, Bulovic V, et al. Large area, few-layer graphene films on arbitrary substrates by chemical vapor deposition. Nano Lett. 2008;9:30–35. doi: 10.1021/nl801827v. [DOI] [PubMed] [Google Scholar]
- 29.Parra C, Montero-Silva F, Henríquez R, Flores M, Garín C, Ramírez C, et al. Suppressing bacterial interaction with copper surfaces through graphene and hexagonal-boron nitride coatings. ACS Appl Mater Interfaces. 2015;7:6430–6437. doi: 10.1021/acsami.5b01248. [DOI] [PubMed] [Google Scholar]
- 30.Cai L, Tan D, Aibaidula G, Dong XR, Chen JC, Tian WD, Chen GQ. Comparative genomics study of polyhydroxyalkanoates (PHA) and ectoine relevant genes from Halomonas sp. TD01 revealed extensive horizontal gene transfer events and co-evolutionary relationships. Microb Cell Fact. 2011;10:88–103. doi: 10.1186/1475-2859-10-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lin Y, Fan H, Hao X, Johnstone L, Hu Y, Wei G, et al. Draft genome sequence of Halomonas sp. strain HAL1, a moderately halophilic arsenite-oxidizing bacterium isolated from gold-mine soil. J Bacteriol. 2012;194:199–200. doi: 10.1128/JB.06359-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zielinski NA, Chakrabarty AM, Berry A. Characterization and regulation of the Pseudomonas aeruginosa algC gene encoding phosphomannomutase. J Biol Chem. 1991;266:9754–9763. [PubMed] [Google Scholar]
- 33.Gottenbos B, Busscher HJ, Van Der Mei HC, Nieuwenhuis P. Pathogenesis and prevention of biomaterial centered infections. J Mater Sci Mater Med. 2002;13:717–722. doi: 10.1023/A:1016175502756. [DOI] [PubMed] [Google Scholar]
- 34.Romero HE, Shen N, Joshi P, Gutierrez HR, Tadigadapa SA, Sofo JO, Eklund PC. n-type behavior of graphene supported on Si/SiO2 substrates. ACS Nano. 2008;2:2037–2044. doi: 10.1021/nn800354m. [DOI] [PubMed] [Google Scholar]
- 35.Hogt AH, Dankert J, Feijen J. Adhesion of coagulase-negative Staphylococci to methacrylate polymers and copolymers. J Biomed Mater Res. 1986;20:533–545. doi: 10.1002/jbm.820200409. [DOI] [PubMed] [Google Scholar]
- 36.Elimelech M, Gregory J, Jia X, Williams R. Particle deposition and aggregation. Williams RA, Woburn MA, editors. Oxford: Butterworth-Heinemann; 1995.
- 37.Itoh H, Sakuma H. Dielectric constant of water as a function of separation in a slab geometry: a molecular dynamics study. J Chem Phys. 2015;142:184703. doi: 10.1063/1.4919698. [DOI] [PubMed] [Google Scholar]
- 38.Linke D, Goldman A. Bacterial adhesion: chemistry, biology and physics. Berlin: Springer; 2011. [Google Scholar]
- 39.Ducker WA, Senden TJ, Pashley RM. Measurement of forces in liquids using a force microscope. Langmuir. 1992;8:1831–1836. doi: 10.1021/la00043a024. [DOI] [Google Scholar]
- 40.Deshpande Bao, Miao F, Lau CN, LeRoy BJ. Spatially resolved spectroscopy of monolayer graphene on SiO2. Phys Rev B. 2009;79:205411–205416. doi: 10.1103/PhysRevB.79.205411. [DOI] [Google Scholar]
- 41.Bruinsma GM, van der Mei HC, Busscher H. Bacterial adhesion to surface hydrophilic and hydrophobic contact lenses. Biomaterials. 2001;22:3217–3224. doi: 10.1016/S0142-9612(01)00159-4. [DOI] [PubMed] [Google Scholar]
- 42.Busscher HJ, Weerkamp AH. Specific and non-specific interactions in bacterial adhesion to solid substrata. FEMS Microbiol Lett. 1987;46:165–173. doi: 10.1111/j.1574-6968.1987.tb02457.x. [DOI] [Google Scholar]
- 43.Bos R, Van der Mei HC, Busscher HJ. Physico-chemistry of initial microbial adhesive interactio–its mechanisms and methods for study. FEMS Microbiol Rev. 1999;23:179–230. doi: 10.1111/j.1574-6976.1999.tb00396.x. [DOI] [PubMed] [Google Scholar]
- 44.Chapman J, Sullivan T, Regan F. Nanoparticles in anti-microbial materials: use and characterization. Cambridge: The Royal Society of Chemistry; 2012. p. 40–8.
- 45.Montanaro L, Campoccia D, Rizzi S, Donati ME, Breschi L, Prati C, Arciola CR. Evaluation of bacterial adhesion of Streptococcus mutants in dental restorative materials. Biomaterials. 2004;25:4457–4463. doi: 10.1016/j.biomaterials.2003.11.031. [DOI] [PubMed] [Google Scholar]
- 46.Hart DJ, Vreeland RH. Changes in the hydrophobic-hydrophilic cell surface character of Halomonas elongata in response to NaCl. J Bacteriol. 1988;170:132–135. doi: 10.1128/jb.170.1.132-135.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ramírez C, Gallegos I, Ihl M, Bifani V. Study of contact angle, wettability and water vapor permeability in carboxymethylcellulose (CMC) based film with murta leaves (Ugni molinae Turcz) extract. J Food Eng. 2012;109:424–429. doi: 10.1016/j.jfoodeng.2011.11.005. [DOI] [Google Scholar]
- 48.Stalder AF, Kulik G, Sage D, Barbieri L, Hoffmann P. A snake-based approach to accurate determination of both contact points and contact angles. Colloids Surf A. 2006;286:92–103. doi: 10.1016/j.colsurfa.2006.03.008. [DOI] [Google Scholar]
- 49.Rojas R. Pathogenicity of a highly exopolysaccharide-producing Halomonas strain causing epizootics in larval cultures of the chilean scallop Argopecten purpuratus. Microb Ecol. 2009;57:129–139. doi: 10.1007/s00248-008-9401-z. [DOI] [PubMed] [Google Scholar]
- 50.Sambrook J, Russell DW. Molecular cloning: A laboratory manual. New York: Cold Spring Harbor Laboratory Press; 2012.
- 51.Atshan SS, Shamsudin MN, Karunanidhi A, van Belkum A, Lung LT, Sekawi Z, Nathan JJ, Ling KH, Seng JS, Ali AM, Abduljaleel SA, Hamat RA. Quantitative PCR analysis of genes expressed during biofilm development of methicillin resistant Staphylococcus aureus (MRSA) Infect Genet Evol. 2013;18:106–112. doi: 10.1016/j.meegid.2013.05.002. [DOI] [PubMed] [Google Scholar]


