In the title complex, [Pt(C11H7BrN)(C5H7O2)], two crystallographically non-equivalent dimers stacked by π–π interactions are arranged antiparallel to each other.
Keywords: crystal structure, platinum(II), cyclometalated complex, acetylacetonato ligand, π–π interactions
Abstract
The title cyclometalated platinum(II) complex with 2-(4-bromophenyl)pyridinato and acetylacetonato ligands, [Pt(C11H7BrN)(C5H7O2)], consists of two crystallographically non-equivalent dimers, each stacked by π–π interactions with distances of ≃ 3.4 Å. In both dimers, the platinum(II) complexes are arranged antiparallel to each other. Each complex exhibits a slightly distorted square-planar coordination environment around the central Pt(II) atom. The dihedral angles between two chelate rings including the PtII atom in these complexes are 0.08 (12) and 1.54 (9)°.
Chemical context
Square-planar cyclometalated platinum(II) complexes with luminescent properties have recently attracted attention because of their potential applications (Chi & Chou, 2010 ▸; Ma et al., 2013 ▸), such as DNA probing, as chemical sensors or as organic light-emitting diodes (OLEDs). In particular, platinum(II) complexes including β-diketonate anions (e.g. acetylacetonate) as an ancillary ligand have been widely studied because of their excellent stabilities and high quantum yields. Although these complexes afford luminescence in the solid state, their crystal structures have not been sufficiently explored. We report herein the crystal structure of the cyclometalated platinum(II) complex with 2-(4-bromophenyl)pyridinato (Brppy, C11H7BrN) and acetylacetonato (acac, C5H7O2) ligands, [Pt(Brppy)(acac)].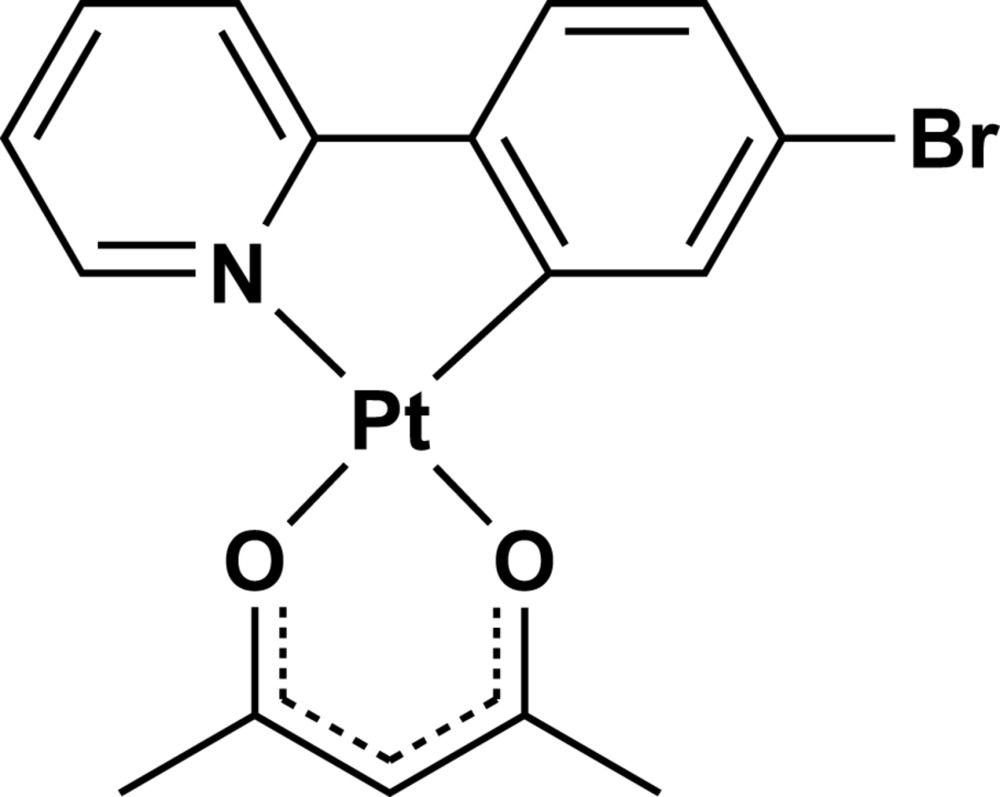
Structural commentary
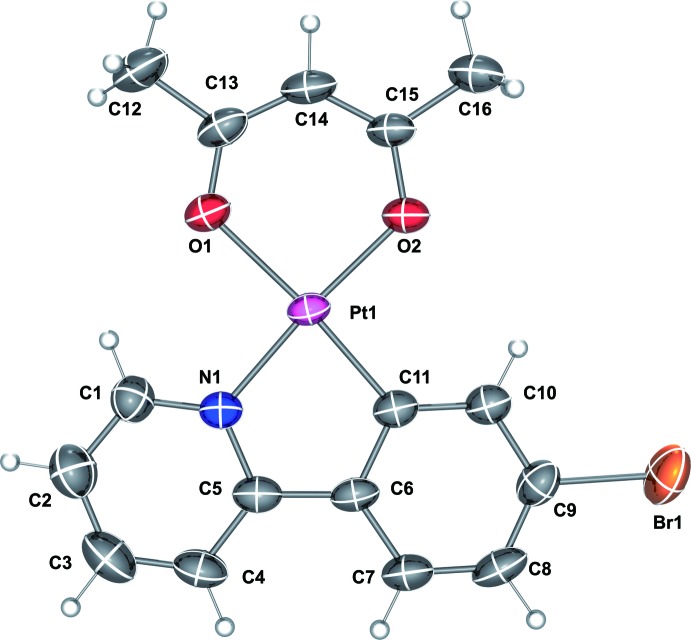
The asymmetric unit of the title compound contains two complex molecules with very similar configurations (r.m.s. deviation of fit of two molecules = 0.07 Å). The structure of one of the complex molecules of the title compound is shown in Fig. 1 ▸. In both complexes, the PtII atom is coordinated by C and N atoms of the bidentate Brppy ligand and two O atoms of the acac ligand. The coordination environments around the central PtII atoms (Pt1 and Pt2) are slightly distorted from an ideal square-planar configuration, with angles around Pt1 in the range 81.89 (18)–93.04 (17)° and around Pt2 in the range 81.73 (18)–93.57 (16)°. The Pt—C bond lengths [Pt1—C11 = 1.970 (5) and Pt2—C27 = 1.969 (5) Å] are slightly shorter than the Pt—N bond lengths [Pt1—N1 = 1.995 (4) and Pt2—N2 = 1.999 (4) Å] due to the stronger electron-donating ability of a C atom compared to that of an N atom. Pt—O bond lengths are compiled in Table 1 ▸. The phenyl and pyridyl rings are approximately coplanar [the dihedral angle between the N1,C1–C5 and C6–C11 rings is 1.31 (17)° while that between the N2,C17–C21 and C22–C27 rings is 3.12 (13)°]. In addition, the dihedral angles between two planes composed of the two chelate rings in the cyclometalated complex are 0.08 (12)° (involving Pt1) and 1.54 (9)° (involving Pt2).
Figure 1.
Molecular structure of one of the two independent PtII complexes of the title compound, with displacement ellipsoids drawn at the 50% probability level.
Table 1. Selected bond lengths ().
| O1Pt1 | 2.077(3) | O3Pt2 | 2.081(3) |
| O2Pt1 | 2.007(3) | O4Pt2 | 2.005(3) |
Supramolecular features
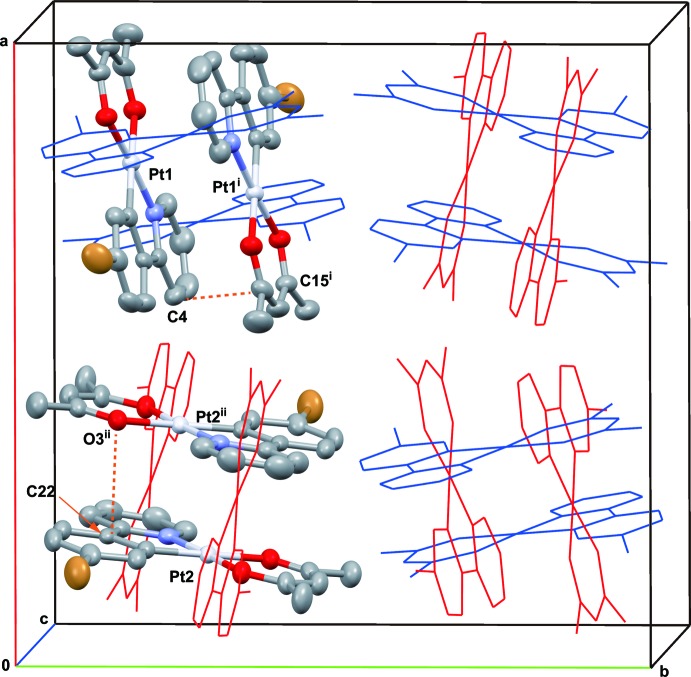
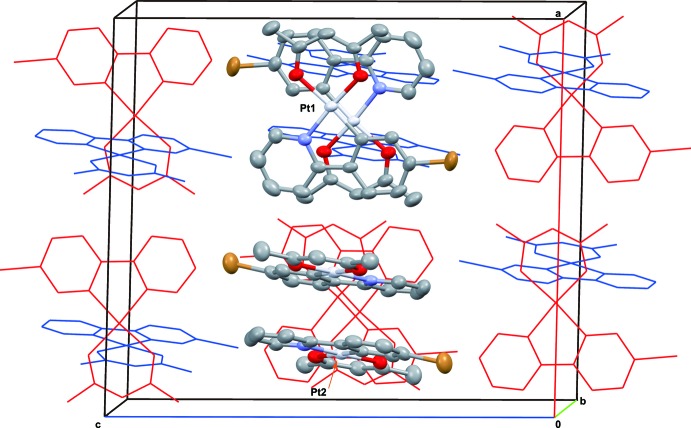
As shown in Figs. 2 ▸ and 3 ▸, in the unit cell two non-equivalent dimers are formed by π–π interactions between individual complexes. Each non-equivalent dimer is in a head-to-tail form. In each unit cell both types of head-to-tail dimers stacked with an intermolecular π–π interaction are perpendicular to each other. The π-plane of one PtII complex (Pt1) is directed to the b axis, on the other hand, that of the other complex (Pt2) is directed to the a axis. The shortest intermolecular contacts are C4⋯C15i = 3.406 (7) and C22⋯O3ii = 3.402 (6) Å [symmetry codes: (i) –x +  , –y +
, –y +  , −z + 1; (ii) –x +
, −z + 1; (ii) –x +  , –y +
, –y +  , –z + 1]. Weak C—H⋯O and C—H⋯Br interactions might also help to consolidate the crystal packing (Table 2 ▸). There is almost no interaction between the two PtII atoms in each dimers because the z-axes of Pt1 and Pt2 are not coaxial. In fact, the Pt—Pt contacts [Pt1⋯Pt1i = 3.688 (1) and Pt2⋯Pt2ii = 3.723 (1) Å] are longer than the van der Waals diameter of the Pt atom (3.5 Å; Bondi, 1964 ▸)
, –z + 1]. Weak C—H⋯O and C—H⋯Br interactions might also help to consolidate the crystal packing (Table 2 ▸). There is almost no interaction between the two PtII atoms in each dimers because the z-axes of Pt1 and Pt2 are not coaxial. In fact, the Pt—Pt contacts [Pt1⋯Pt1i = 3.688 (1) and Pt2⋯Pt2ii = 3.723 (1) Å] are longer than the van der Waals diameter of the Pt atom (3.5 Å; Bondi, 1964 ▸)
Figure 2.
Crystal packing of the title complex, viewed perpendicular to the ab plane. Dashed lines represent the shortest intermolecular contacts. Red wires represent the Pt1 molecule, and blue wires the Pt2 molecule. H atoms are omitted for clarity. [Symmetry codes: (i) –x +  , –y +
, –y +  , –z + 1; (ii) –x +
, –z + 1; (ii) –x +  , –y +
, –y +  , –z + 1.]
, –z + 1.]
Figure 3.
Crystal packing of the title complex, viewed perpendicular to the ac plane. Red wires represent the Pt1 molecule, and blue wires the Pt2 molecule. H atoms are omitted for clarity.
Table 2. Hydrogen-bond geometry (, ).
| DHA | DH | HA | D A | DHA |
|---|---|---|---|---|
| C1H1O1 | 0.95 | 2.40 | 2.999(7) | 121 |
| C4H4O4i | 0.95 | 2.58 | 3.281(6) | 131 |
| C17H17O3 | 0.95 | 2.45 | 3.034(6) | 120 |
| C17H17Br1ii | 0.95 | 2.87 | 3.693(6) | 145 |
Symmetry codes: (i) 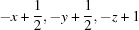 ; (ii)
; (ii) 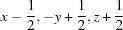 .
.
Synthesis and crystallization
The title complex was synthesized according to a traditional two-step preparation method via the dichlorido-bridged dimer complex [Pt(C11H7BrN)(μ-Cl)]2 (Cockburn et al., 1973 ▸; Liu et al., 2009 ▸), though one-pot synthesis has been reported recently (Hudson et al., 2012 ▸).
[Pt(C11H7BrN)( μ -Cl)]2: A mixture of 2-(4-bromophenyl)pyridine (0.585 g, 2.5 mmol) and K2PtCl4 (1.00 g, 2.4 mmol) in a 2-ethoxyethanol–water mixture (45 ml/15 ml) was stirred for 6 h at 333 K under an Ar atmosphere. After cooling to room temperature, the yellow–green precipitate was filtered off, washed with dichloromethane, and dried in vacuo. Yield: 0.535 g, (48.2%).
[Pt(C11H7BrN)(C5H7O2)]: A mixture of the dichlorido-bridged dimer complex (0.185 g, 0.20 mmol), acetylacetone (0.020 g, 0.20 mmol) and Na2CO3 (0.211 g, 2.0 mmol) in 2-ethoxyethanol was stirred for 7 h at 323 K under an Ar atmosphere. After cooling to room temperature, the yellow precipitate was filtered off and dried in vacuo. Yield: 0.200 g (47.6%)
Yellow single crystals suitable for X-ray structural analysis were grown by vapor diffusion of hexane into the dichloromethane solution of the title complex.
Analysis found (calculated for C16H14BrNO2Pt): C, 36.15 (36.45); H, 2.25 (2.68); N, 2.59 (2.66). UV–vis [CHCl3, λ max nm−1 (∊ / L mol−1 cm−1)]: 262 (29800), 280 (27500), 317 (sh, 11700), 330 (sh, 9400), 363 (6400), 389 (4200). 1H NMR (CDCl3, 298 K); 8.97 (d, J Pt-H = 40.0 Hz, J = 6.0 Hz, 1H), 7.81 (t, J = 6.0 Hz, 1H), 7.71 (s, J Pt-H = 40.0 Hz, 1H), 7.57 (d, J = 6.0 Hz, 1H), 7.31-7.45 (m, 2H), 7.14 (t, J = 6.0 Hz, 1H), 5.48 (s, 1H), 2.03 (s, 3H), 2.01 (s, 3H).
Refinement
Crystal data, data collection and structure refinement details are summarized in Table 3 ▸. All H atoms were placed in geometrically idealized positions and refined using a riding model, with C—H = 0.95 Å, U iso(H) = 1.2U eq(C) for Csp 2–H, and U iso(H) = 1.5U eq(C) for methyl H atoms.
Table 3. Experimental details.
| Crystal data | |
| Chemical formula | [Pt(C11H7BrN)(C5H7O2)] |
| M r | 527.28 |
| Crystal system, space group | Monoclinic, C2/c |
| Temperature (K) | 200 |
| a, b, c () | 17.557(2), 17.876(2), 19.832(2) |
| () | 91.397(1) |
| V (3) | 6222.4(13) |
| Z | 16 |
| Radiation type | Mo K |
| (mm1) | 11.59 |
| Crystal size (mm) | 0.18 0.06 0.02 |
| Data collection | |
| Diffractometer | Bruker APEXII CCD area detector |
| Absorption correction | Multi-scan (SADABS; Bruker, 2014 ▸) |
| T min, T max | 0.48, 0.80 |
| No. of measured, independent and observed [I > 2(I)] reflections | 35025, 7103, 6001 |
| R int | 0.038 |
| (sin /)max (1) | 0.649 |
| Refinement | |
| R[F 2 > 2(F 2)], wR(F 2), S | 0.027, 0.070, 1.01 |
| No. of reflections | 7103 |
| No. of parameters | 383 |
| H-atom treatment | H-atom parameters constrained |
| max, min (e 3) | 3.66, 1.20 |
Supplementary Material
Crystal structure: contains datablock(s) I. DOI: 10.1107/S2056989015017478/wm5214sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S2056989015017478/wm5214Isup2.hkl
Supporting information file. DOI: 10.1107/S2056989015017478/wm5214Isup3.tif
CCDC reference: 1425736
Additional supporting information: crystallographic information; 3D view; checkCIF report
Acknowledgments
The authors thank Professor Takashi Fujihara (Saitama University) for providing an opportunity for X-ray crystallographic analysis. This work was supported by JSPS KAKENHI Grant-in-Aid for Young Scientists (B) (25810052 to TS), and partially by JSPS KAKENHI Grant No. 2540150.
supplementary crystallographic information
Crystal data
| [Pt(C11H7BrN)(C5H7O2)] | F(000) = 3936 |
| Mr = 527.28 | Dx = 2.251 Mg m−3 |
| Monoclinic, C2/c | Mo Kα radiation, λ = 0.71073 Å |
| a = 17.557 (2) Å | Cell parameters from 9927 reflections |
| b = 17.876 (2) Å | θ = 2.3–27.3° |
| c = 19.832 (2) Å | µ = 11.59 mm−1 |
| β = 91.397 (1)° | T = 200 K |
| V = 6222.4 (13) Å3 | Lath, yellow |
| Z = 16 | 0.18 × 0.06 × 0.02 mm |
Data collection
| Bruker APEXII CCD area detector diffractometer | 7103 independent reflections |
| Radiation source: Bruker TXS fine-focus rotating anode | 6001 reflections with I > 2σ(I) |
| Bruker Helios multilayer confocal mirror monochromator | Rint = 0.038 |
| Detector resolution: 8.333 pixels mm-1 | θmax = 27.5°, θmin = 1.6° |
| phi and ω scans | h = −22→22 |
| Absorption correction: multi-scan (SADABS; Bruker, 2014) | k = −23→23 |
| Tmin = 0.48, Tmax = 0.80 | l = −25→25 |
| 35025 measured reflections |
Refinement
| Refinement on F2 | 0 restraints |
| Least-squares matrix: full | Hydrogen site location: inferred from neighbouring sites |
| R[F2 > 2σ(F2)] = 0.027 | H-atom parameters constrained |
| wR(F2) = 0.070 | w = 1/[σ2(Fo2) + (0.0368P)2 + 16.0306P] where P = (Fo2 + 2Fc2)/3 |
| S = 1.01 | (Δ/σ)max = 0.001 |
| 7103 reflections | Δρmax = 3.66 e Å−3 |
| 383 parameters | Δρmin = −1.19 e Å−3 |
Special details
| Geometry. Distance SDEV3.4016 (0.0055) C22 - O3_$6 3.4056 (0.0070) C4 - C15_$5 3.6879 (0.0005) Pt1 - Pt1_$5 3.7230 (0.0005) Pt2 - Pt2_$6$5 1.5 - x, 0.5 - y, 1 - z $6 0.5 - x, 0.5 - y, 1 - zLeast-squares planes (x,y,z in crystal coordinates) and deviations from them (* indicates atom used to define plane)17.0464 (0.0086) x + 3.5524 (0.0343) y - 3.1157 (0.0389) z = 1.9362 (0.0174)* -0.0159 (0.0032) C22 * 0.0106 (0.0035) C23 * 0.0050 (0.0037) C24 * -0.0152 (0.0036) C25 * 0.0096 (0.0034) C26 * 0.0059 (0.0032) C27Rms deviation of fitted atoms = 0.011217.1816 (0.0082) x + 3.3837 (0.0389) y - 2.0681 (0.0436) z = 2.4419 (0.0278)Angle to previous plane (with approximate e.s.d.) = 3.118 (0.129)* -0.0040 (0.0032) N2 * 0.0023 (0.0037) C17 * 0.0000 (0.0041) C18 * -0.0004 (0.0041) C19 * -0.0014 (0.0038) C20 * 0.0036 (0.0033) C21Rms deviation of fitted atoms = 0.00253.5018 (0.0341) x + 17.1159 (0.0108) y - 4.2313 (0.0388) z = 3.1261 (0.0262)Angle to previous plane (with approximate e.s.d.) = 66.846 (0.177)* 0.0047 (0.0032) C6 * -0.0001 (0.0036) C7 * -0.0055 (0.0037) C8 * 0.0064 (0.0036) C9 * -0.0015 (0.0033) C10 * -0.0040 (0.0031) C11Rms deviation of fitted atoms = 0.00433.8621 (0.0359) x + 17.0726 (0.0113) y - 4.0479 (0.0426) z = 3.4669 (0.0355)Angle to previous plane (with approximate e.s.d.) = 1.309 (0.166)* -0.0056 (0.0031) N1 * 0.0048 (0.0038) C1 * -0.0003 (0.0042) C2 * -0.0032 (0.0042) C3 * 0.0024 (0.0037) C4 * 0.0019 (0.0032) C5Rms deviation of fitted atoms = 0.003517.0575 (0.0059) x + 3.8901 (0.0226) y - 2.3217 (0.0295) z = 2.4371 (0.0163)Angle to previous plane (with approximate e.s.d.) = 63.889 (0.149)* 0.0136 (0.0026) O3 * 0.0004 (0.0034) C29 * -0.0121 (0.0038) C30 * -0.0006 (0.0035) C31 * 0.0149 (0.0026) O4 * -0.0162 (0.0018) Pt2Rms deviation of fitted atoms = 0.011717.1494 (0.0057) x + 3.4219 (0.0232) y - 2.3782 (0.0383) z = 2.2709 (0.0209)Angle to previous plane (with approximate e.s.d.) = 1.538 (0.086)* -0.0073 (0.0024) N2 * 0.0020 (0.0029) C21 * 0.0076 (0.0030) C22 * -0.0104 (0.0026) C27 * 0.0081 (0.0018) Pt2Rms deviation of fitted atoms = 0.00763.7521 (0.0218) x + 17.0640 (0.0077) y - 4.2225 (0.0285) z = 3.2745 (0.0235)Angle to previous plane (with approximate e.s.d.) = 65.705 (0.111)* -0.0132 (0.0026) O1 * -0.0080 (0.0035) C13 * 0.0257 (0.0037) C14 * -0.0103 (0.0034) C15 * -0.0121 (0.0026) O2 * 0.0179 (0.0017) Pt1Rms deviation of fitted atoms = 0.01573.7578 (0.0215) x + 17.0576 (0.0090) y - 4.2485 (0.0367) z = 3.2795 (0.0249)Angle to previous plane (with approximate e.s.d.) = 0.080 (0.123)* -0.0089 (0.0023) N1 * 0.0120 (0.0028) C5 * -0.0086 (0.0028) C6 * 0.0027 (0.0024) C11 * 0.0028 (0.0017) Pt1Rms deviation of fitted atoms = 0.0079 |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| Br1 | 0.63794 (4) | 0.11449 (4) | 0.24876 (3) | 0.06869 (19) | |
| Br2 | 0.13287 (5) | 0.08166 (4) | 0.25166 (3) | 0.06997 (19) | |
| C1 | 0.6968 (3) | 0.1984 (3) | 0.6440 (3) | 0.0478 (12) | |
| H1 | 0.7459 | 0.1928 | 0.6646 | 0.057* | |
| C2 | 0.6367 (4) | 0.2211 (3) | 0.6835 (3) | 0.0614 (16) | |
| H2 | 0.6441 | 0.2304 | 0.7304 | 0.074* | |
| C3 | 0.5655 (4) | 0.2296 (3) | 0.6522 (3) | 0.0626 (17) | |
| H3 | 0.5231 | 0.2449 | 0.6777 | 0.075* | |
| C4 | 0.5565 (3) | 0.2159 (3) | 0.5846 (3) | 0.0484 (13) | |
| H4 | 0.5080 | 0.2221 | 0.5632 | 0.058* | |
| C5 | 0.6187 (3) | 0.1929 (2) | 0.5470 (3) | 0.0368 (10) | |
| C6 | 0.6190 (3) | 0.1739 (2) | 0.4758 (3) | 0.0355 (10) | |
| C7 | 0.5543 (3) | 0.1762 (3) | 0.4328 (3) | 0.0467 (12) | |
| H7 | 0.5065 | 0.1904 | 0.4502 | 0.056* | |
| C8 | 0.5596 (3) | 0.1583 (3) | 0.3660 (3) | 0.0493 (14) | |
| H8 | 0.5160 | 0.1593 | 0.3367 | 0.059* | |
| C9 | 0.6299 (3) | 0.1388 (3) | 0.3424 (3) | 0.0455 (12) | |
| C10 | 0.6949 (3) | 0.1351 (3) | 0.3833 (2) | 0.0392 (11) | |
| H10 | 0.7421 | 0.1207 | 0.3649 | 0.047* | |
| C11 | 0.6905 (3) | 0.1526 (2) | 0.4509 (2) | 0.0342 (10) | |
| C12 | 0.9693 (4) | 0.1381 (4) | 0.6572 (3) | 0.0652 (17) | |
| H12A | 0.9552 | 0.1822 | 0.6835 | 0.098* | |
| H12B | 1.0234 | 0.1410 | 0.6463 | 0.098* | |
| H12C | 0.9602 | 0.0928 | 0.6837 | 0.098* | |
| C13 | 0.9223 (3) | 0.1355 (3) | 0.5936 (3) | 0.0445 (12) | |
| C14 | 0.9569 (3) | 0.1148 (3) | 0.5327 (3) | 0.0475 (13) | |
| H14 | 1.0104 | 0.1067 | 0.5349 | 0.057* | |
| C15 | 0.9213 (3) | 0.1051 (3) | 0.4705 (3) | 0.0395 (11) | |
| C16 | 0.9671 (3) | 0.0762 (3) | 0.4126 (3) | 0.0514 (14) | |
| H16A | 0.9435 | 0.0305 | 0.3947 | 0.077* | |
| H16B | 1.0192 | 0.0653 | 0.4286 | 0.077* | |
| H16C | 0.9684 | 0.1142 | 0.3770 | 0.077* | |
| C17 | 0.1737 (3) | 0.2277 (3) | 0.6336 (3) | 0.0477 (13) | |
| H17 | 0.1651 | 0.2790 | 0.6435 | 0.057* | |
| C18 | 0.1893 (3) | 0.1793 (4) | 0.6855 (3) | 0.0591 (15) | |
| H18 | 0.1913 | 0.1967 | 0.7308 | 0.071* | |
| C19 | 0.2023 (3) | 0.1049 (4) | 0.6714 (3) | 0.0629 (17) | |
| H19 | 0.2133 | 0.0706 | 0.7069 | 0.075* | |
| C20 | 0.1990 (3) | 0.0805 (3) | 0.6053 (3) | 0.0511 (13) | |
| H20 | 0.2077 | 0.0293 | 0.5951 | 0.061* | |
| C21 | 0.1831 (3) | 0.1312 (3) | 0.5538 (3) | 0.0411 (11) | |
| C22 | 0.1765 (2) | 0.1157 (3) | 0.4814 (3) | 0.0377 (11) | |
| C23 | 0.1874 (3) | 0.0452 (3) | 0.4521 (3) | 0.0452 (12) | |
| H23 | 0.2023 | 0.0040 | 0.4796 | 0.054* | |
| C24 | 0.1767 (3) | 0.0357 (3) | 0.3844 (3) | 0.0482 (13) | |
| H24 | 0.1836 | −0.0121 | 0.3645 | 0.058* | |
| C25 | 0.1556 (3) | 0.0967 (3) | 0.3451 (3) | 0.0444 (12) | |
| C26 | 0.1472 (3) | 0.1677 (3) | 0.3722 (3) | 0.0410 (11) | |
| H26 | 0.1346 | 0.2089 | 0.3438 | 0.049* | |
| C27 | 0.1574 (3) | 0.1782 (3) | 0.4412 (3) | 0.0360 (10) | |
| C28 | 0.1093 (4) | 0.4917 (3) | 0.5815 (3) | 0.0568 (15) | |
| H28A | 0.0791 | 0.4731 | 0.6189 | 0.085* | |
| H28B | 0.0838 | 0.5352 | 0.5611 | 0.085* | |
| H28C | 0.1600 | 0.5062 | 0.5985 | 0.085* | |
| C29 | 0.1167 (3) | 0.4308 (3) | 0.5292 (3) | 0.0435 (12) | |
| C30 | 0.1027 (3) | 0.4486 (3) | 0.4614 (3) | 0.0490 (13) | |
| H30 | 0.0888 | 0.4990 | 0.4520 | 0.059* | |
| C31 | 0.1067 (3) | 0.4011 (3) | 0.4063 (3) | 0.0433 (12) | |
| C32 | 0.0874 (4) | 0.4303 (3) | 0.3369 (3) | 0.0617 (16) | |
| H32A | 0.1263 | 0.4141 | 0.3055 | 0.093* | |
| H32B | 0.0855 | 0.4851 | 0.3379 | 0.093* | |
| H32C | 0.0376 | 0.4108 | 0.3219 | 0.093* | |
| N1 | 0.6882 (2) | 0.1843 (2) | 0.5787 (2) | 0.0349 (8) | |
| N2 | 0.1701 (2) | 0.2051 (2) | 0.5697 (2) | 0.0368 (9) | |
| O1 | 0.85200 (19) | 0.15218 (18) | 0.59970 (18) | 0.0418 (8) | |
| O2 | 0.85083 (18) | 0.11665 (19) | 0.45482 (17) | 0.0402 (8) | |
| O3 | 0.13515 (19) | 0.36673 (18) | 0.55181 (18) | 0.0410 (8) | |
| O4 | 0.1237 (2) | 0.33092 (18) | 0.40711 (17) | 0.0400 (8) | |
| Pt1 | 0.77288 (2) | 0.15171 (2) | 0.52011 (2) | 0.03211 (6) | |
| Pt2 | 0.14668 (2) | 0.27243 (2) | 0.49142 (2) | 0.03308 (6) |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| Br1 | 0.0827 (5) | 0.0827 (5) | 0.0401 (3) | −0.0135 (4) | −0.0115 (3) | 0.0119 (3) |
| Br2 | 0.0957 (5) | 0.0599 (4) | 0.0547 (4) | −0.0007 (3) | 0.0093 (3) | −0.0143 (3) |
| C1 | 0.052 (3) | 0.043 (3) | 0.047 (3) | 0.001 (2) | 0.000 (2) | −0.003 (2) |
| C2 | 0.071 (4) | 0.056 (4) | 0.057 (4) | 0.003 (3) | 0.008 (3) | −0.005 (3) |
| C3 | 0.064 (4) | 0.052 (4) | 0.073 (4) | 0.006 (3) | 0.027 (3) | −0.001 (3) |
| C4 | 0.036 (3) | 0.039 (3) | 0.070 (4) | 0.006 (2) | 0.009 (3) | 0.006 (3) |
| C5 | 0.032 (2) | 0.023 (2) | 0.056 (3) | −0.0007 (18) | 0.001 (2) | 0.009 (2) |
| C6 | 0.027 (2) | 0.026 (2) | 0.053 (3) | −0.0041 (18) | −0.004 (2) | 0.009 (2) |
| C7 | 0.031 (3) | 0.049 (3) | 0.060 (3) | 0.000 (2) | −0.004 (2) | 0.008 (3) |
| C8 | 0.038 (3) | 0.045 (3) | 0.063 (4) | −0.007 (2) | −0.019 (3) | 0.017 (3) |
| C9 | 0.050 (3) | 0.046 (3) | 0.040 (3) | −0.008 (2) | −0.007 (2) | 0.009 (2) |
| C10 | 0.037 (3) | 0.035 (2) | 0.046 (3) | −0.004 (2) | −0.001 (2) | 0.009 (2) |
| C11 | 0.033 (2) | 0.024 (2) | 0.045 (3) | −0.0028 (18) | −0.002 (2) | 0.0079 (19) |
| C12 | 0.048 (4) | 0.079 (5) | 0.068 (4) | −0.001 (3) | −0.020 (3) | 0.008 (3) |
| C13 | 0.036 (3) | 0.043 (3) | 0.054 (3) | −0.008 (2) | −0.011 (2) | 0.014 (2) |
| C14 | 0.028 (2) | 0.046 (3) | 0.068 (4) | 0.003 (2) | −0.006 (2) | 0.013 (3) |
| C15 | 0.027 (2) | 0.040 (3) | 0.051 (3) | 0.001 (2) | 0.001 (2) | 0.013 (2) |
| C16 | 0.035 (3) | 0.052 (3) | 0.067 (4) | 0.004 (2) | 0.006 (2) | 0.014 (3) |
| C17 | 0.041 (3) | 0.051 (3) | 0.051 (3) | −0.001 (2) | 0.000 (2) | 0.002 (2) |
| C18 | 0.057 (4) | 0.071 (4) | 0.049 (3) | −0.007 (3) | −0.009 (3) | 0.013 (3) |
| C19 | 0.053 (4) | 0.074 (4) | 0.061 (4) | −0.006 (3) | −0.011 (3) | 0.025 (3) |
| C20 | 0.043 (3) | 0.045 (3) | 0.065 (4) | −0.001 (2) | −0.006 (3) | 0.016 (3) |
| C21 | 0.027 (2) | 0.036 (3) | 0.060 (3) | −0.0026 (19) | 0.000 (2) | 0.010 (2) |
| C22 | 0.022 (2) | 0.028 (2) | 0.064 (3) | −0.0004 (17) | 0.005 (2) | 0.009 (2) |
| C23 | 0.035 (3) | 0.031 (3) | 0.070 (4) | 0.002 (2) | 0.007 (2) | 0.005 (2) |
| C24 | 0.046 (3) | 0.029 (3) | 0.069 (4) | 0.000 (2) | 0.010 (3) | −0.005 (2) |
| C25 | 0.042 (3) | 0.039 (3) | 0.053 (3) | −0.003 (2) | 0.014 (2) | −0.006 (2) |
| C26 | 0.037 (3) | 0.032 (2) | 0.054 (3) | −0.001 (2) | 0.008 (2) | 0.003 (2) |
| C27 | 0.028 (2) | 0.028 (2) | 0.052 (3) | −0.0004 (18) | 0.005 (2) | 0.005 (2) |
| C28 | 0.062 (4) | 0.042 (3) | 0.067 (4) | 0.003 (3) | 0.004 (3) | −0.010 (3) |
| C29 | 0.033 (3) | 0.031 (2) | 0.067 (3) | −0.004 (2) | 0.008 (2) | 0.001 (2) |
| C30 | 0.049 (3) | 0.032 (3) | 0.066 (4) | 0.003 (2) | 0.007 (3) | 0.006 (2) |
| C31 | 0.045 (3) | 0.027 (2) | 0.058 (3) | −0.002 (2) | 0.008 (2) | 0.009 (2) |
| C32 | 0.085 (5) | 0.037 (3) | 0.063 (4) | 0.007 (3) | 0.004 (3) | 0.011 (3) |
| N1 | 0.033 (2) | 0.0284 (19) | 0.043 (2) | 0.0001 (16) | 0.0005 (17) | 0.0037 (16) |
| N2 | 0.0235 (19) | 0.037 (2) | 0.050 (2) | −0.0018 (16) | 0.0014 (16) | 0.0063 (18) |
| O1 | 0.0344 (19) | 0.043 (2) | 0.048 (2) | −0.0013 (14) | −0.0061 (15) | 0.0071 (15) |
| O2 | 0.0295 (17) | 0.0403 (19) | 0.051 (2) | 0.0019 (14) | 0.0007 (14) | 0.0059 (15) |
| O3 | 0.0366 (18) | 0.0320 (17) | 0.055 (2) | −0.0009 (14) | 0.0031 (15) | −0.0027 (15) |
| O4 | 0.045 (2) | 0.0295 (17) | 0.0461 (19) | 0.0016 (14) | 0.0055 (15) | 0.0068 (14) |
| Pt1 | 0.02610 (9) | 0.02845 (10) | 0.04157 (11) | −0.00042 (6) | −0.00348 (7) | 0.00636 (7) |
| Pt2 | 0.02732 (10) | 0.02679 (9) | 0.04527 (11) | −0.00091 (6) | 0.00418 (7) | 0.00340 (7) |
Geometric parameters (Å, º)
| C9—C10 | 1.385 (7) | C16—H16A | 0.9800 |
| C6—C11 | 1.414 (7) | C16—H16B | 0.9800 |
| C10—C11 | 1.380 (7) | C16—H16C | 0.9800 |
| C12—C13 | 1.492 (8) | C17—H17 | 0.9500 |
| C13—C14 | 1.413 (8) | C18—H18 | 0.9500 |
| C14—C15 | 1.382 (7) | C19—H19 | 0.9500 |
| C15—C16 | 1.508 (7) | C2—H2 | 0.9500 |
| C17—C18 | 1.368 (8) | C20—H20 | 0.9500 |
| C18—C19 | 1.378 (9) | C23—H23 | 0.9500 |
| C1—C2 | 1.390 (8) | C24—H24 | 0.9500 |
| C19—C20 | 1.381 (9) | C26—H26 | 0.9500 |
| C20—C21 | 1.389 (7) | C28—H28A | 0.9800 |
| C21—C22 | 1.463 (7) | C28—H28B | 0.9800 |
| C22—C23 | 1.402 (7) | C28—H28C | 0.9800 |
| C23—C24 | 1.362 (8) | C3—H3 | 0.9500 |
| Br2—C25 | 1.905 (5) | C30—H30 | 0.9500 |
| C24—C25 | 1.386 (8) | C32—H32A | 0.9800 |
| C25—C26 | 1.388 (7) | C32—H32B | 0.9800 |
| C22—C27 | 1.407 (6) | C32—H32C | 0.9800 |
| C26—C27 | 1.388 (7) | C4—H4 | 0.9500 |
| C28—C29 | 1.511 (7) | C7—H7 | 0.9500 |
| C2—C3 | 1.392 (9) | C8—H8 | 0.9500 |
| C29—C30 | 1.397 (8) | C1—N1 | 1.325 (6) |
| C30—C31 | 1.387 (8) | C5—N1 | 1.367 (6) |
| C31—C32 | 1.504 (8) | C17—N2 | 1.331 (7) |
| C3—C4 | 1.367 (9) | C21—N2 | 1.378 (6) |
| C4—C5 | 1.400 (7) | C13—O1 | 1.279 (6) |
| C5—C6 | 1.452 (7) | C15—O2 | 1.284 (5) |
| C6—C7 | 1.404 (7) | C29—O3 | 1.270 (6) |
| C7—C8 | 1.369 (8) | C31—O4 | 1.290 (6) |
| Br1—C9 | 1.915 (5) | C11—Pt1 | 1.970 (5) |
| C8—C9 | 1.376 (8) | N1—Pt1 | 1.995 (4) |
| C1—H1 | 0.9500 | O1—Pt1 | 2.077 (3) |
| C10—H10 | 0.9500 | O2—Pt1 | 2.007 (3) |
| C12—H12A | 0.9800 | C27—Pt2 | 1.969 (5) |
| C12—H12B | 0.9800 | N2—Pt2 | 1.999 (4) |
| C12—H12C | 0.9800 | O3—Pt2 | 2.081 (3) |
| C14—H14 | 0.9500 | O4—Pt2 | 2.005 (3) |
| N1—C1—C2 | 122.5 (5) | C21—C20—H20 | 120.1 |
| N1—C1—H1 | 118.8 | N2—C21—C20 | 119.2 (5) |
| C2—C1—H1 | 118.8 | N2—C21—C22 | 113.4 (4) |
| C1—C2—C3 | 117.8 (6) | C20—C21—C22 | 127.4 (5) |
| C1—C2—H2 | 121.1 | C23—C22—C27 | 120.8 (5) |
| C3—C2—H2 | 121.1 | C23—C22—C21 | 124.6 (4) |
| C4—C3—C2 | 119.9 (6) | C27—C22—C21 | 114.6 (4) |
| C4—C3—H3 | 120.0 | C24—C23—C22 | 120.3 (5) |
| C2—C3—H3 | 120.0 | C24—C23—H23 | 119.9 |
| C3—C4—C5 | 120.2 (5) | C22—C23—H23 | 119.9 |
| C3—C4—H4 | 119.9 | C23—C24—C25 | 119.0 (5) |
| C5—C4—H4 | 119.9 | C23—C24—H24 | 120.5 |
| N1—C5—C4 | 119.1 (5) | C25—C24—H24 | 120.5 |
| N1—C5—C6 | 113.4 (4) | C24—C25—C26 | 122.0 (5) |
| C4—C5—C6 | 127.5 (5) | C24—C25—Br2 | 118.9 (4) |
| C7—C6—C11 | 120.6 (5) | C26—C25—Br2 | 119.0 (4) |
| C7—C6—C5 | 124.2 (5) | C27—C26—C25 | 119.5 (5) |
| C11—C6—C5 | 115.2 (4) | C27—C26—H26 | 120.2 |
| C8—C7—C6 | 120.5 (5) | C25—C26—H26 | 120.2 |
| C8—C7—H7 | 119.8 | C26—C27—C22 | 118.3 (5) |
| C6—C7—H7 | 119.8 | C26—C27—Pt2 | 127.0 (4) |
| C7—C8—C9 | 118.2 (5) | C22—C27—Pt2 | 114.7 (4) |
| C7—C8—H8 | 120.9 | C29—C28—H28A | 109.5 |
| C9—C8—H8 | 120.9 | C29—C28—H28B | 109.5 |
| C8—C9—C10 | 123.1 (5) | H28A—C28—H28B | 109.5 |
| C8—C9—Br1 | 118.4 (4) | C29—C28—H28C | 109.5 |
| C10—C9—Br1 | 118.5 (4) | H28A—C28—H28C | 109.5 |
| C11—C10—C9 | 119.6 (5) | H28B—C28—H28C | 109.5 |
| C11—C10—H10 | 120.2 | O3—C29—C30 | 125.6 (5) |
| C9—C10—H10 | 120.2 | O3—C29—C28 | 115.7 (5) |
| C10—C11—C6 | 118.1 (4) | C30—C29—C28 | 118.7 (5) |
| C10—C11—Pt1 | 128.2 (4) | C31—C30—C29 | 127.4 (5) |
| C6—C11—Pt1 | 113.7 (4) | C31—C30—H30 | 116.3 |
| C13—C12—H12A | 109.5 | C29—C30—H30 | 116.3 |
| C13—C12—H12B | 109.5 | O4—C31—C30 | 127.0 (5) |
| H12A—C12—H12B | 109.5 | O4—C31—C32 | 113.3 (5) |
| C13—C12—H12C | 109.5 | C30—C31—C32 | 119.7 (5) |
| H12A—C12—H12C | 109.5 | C31—C32—H32A | 109.5 |
| H12B—C12—H12C | 109.5 | C31—C32—H32B | 109.5 |
| O1—C13—C14 | 125.3 (5) | H32A—C32—H32B | 109.5 |
| O1—C13—C12 | 115.3 (5) | C31—C32—H32C | 109.5 |
| C14—C13—C12 | 119.4 (5) | H32A—C32—H32C | 109.5 |
| C15—C14—C13 | 126.9 (5) | H32B—C32—H32C | 109.5 |
| C15—C14—H14 | 116.5 | C1—N1—C5 | 120.5 (4) |
| C13—C14—H14 | 116.5 | C1—N1—Pt1 | 123.8 (3) |
| O2—C15—C14 | 127.5 (5) | C5—N1—Pt1 | 115.7 (3) |
| O2—C15—C16 | 113.6 (5) | C17—N2—C21 | 120.4 (4) |
| C14—C15—C16 | 119.0 (4) | C17—N2—Pt2 | 124.1 (4) |
| C15—C16—H16A | 109.5 | C21—N2—Pt2 | 115.6 (3) |
| C15—C16—H16B | 109.5 | C13—O1—Pt1 | 123.8 (3) |
| H16A—C16—H16B | 109.5 | C15—O2—Pt1 | 124.2 (3) |
| C15—C16—H16C | 109.5 | C29—O3—Pt2 | 123.8 (4) |
| H16A—C16—H16C | 109.5 | C31—O4—Pt2 | 124.0 (3) |
| H16B—C16—H16C | 109.5 | C11—Pt1—N1 | 81.89 (18) |
| N2—C17—C18 | 121.9 (6) | C11—Pt1—O2 | 93.04 (17) |
| N2—C17—H17 | 119.1 | N1—Pt1—O2 | 174.80 (15) |
| C18—C17—H17 | 119.1 | C11—Pt1—O1 | 174.72 (17) |
| C17—C18—C19 | 119.3 (6) | N1—Pt1—O1 | 92.90 (15) |
| C17—C18—H18 | 120.3 | O2—Pt1—O1 | 92.15 (14) |
| C19—C18—H18 | 120.3 | C27—Pt2—N2 | 81.73 (18) |
| C18—C19—C20 | 119.6 (5) | C27—Pt2—O4 | 92.57 (17) |
| C18—C19—H19 | 120.2 | N2—Pt2—O4 | 174.29 (15) |
| C20—C19—H19 | 120.2 | C27—Pt2—O3 | 175.20 (17) |
| C19—C20—C21 | 119.7 (6) | N2—Pt2—O3 | 93.57 (16) |
| C19—C20—H20 | 120.1 | O4—Pt2—O3 | 92.12 (13) |
| N1—C1—C2—C3 | −0.6 (9) | C21—C22—C23—C24 | 177.1 (5) |
| C1—C2—C3—C4 | −0.2 (9) | C22—C23—C24—C25 | 0.6 (8) |
| C2—C3—C4—C5 | 0.4 (9) | C23—C24—C25—C26 | 1.9 (8) |
| C3—C4—C5—N1 | 0.1 (7) | C23—C24—C25—Br2 | −175.1 (4) |
| C3—C4—C5—C6 | 178.0 (5) | C24—C25—C26—C27 | −2.4 (8) |
| N1—C5—C6—C7 | 178.2 (4) | Br2—C25—C26—C27 | 174.6 (4) |
| C4—C5—C6—C7 | 0.2 (8) | C25—C26—C27—C22 | 0.3 (7) |
| N1—C5—C6—C11 | −2.2 (6) | C25—C26—C27—Pt2 | −178.7 (4) |
| C4—C5—C6—C11 | 179.8 (4) | C23—C22—C27—C26 | 2.1 (7) |
| C11—C6—C7—C8 | −0.4 (7) | C21—C22—C27—C26 | −177.6 (4) |
| C5—C6—C7—C8 | 179.2 (5) | C23—C22—C27—Pt2 | −178.8 (3) |
| C6—C7—C8—C9 | −0.6 (8) | C21—C22—C27—Pt2 | 1.5 (5) |
| C7—C8—C9—C10 | 1.2 (8) | O3—C29—C30—C31 | −0.6 (9) |
| C7—C8—C9—Br1 | −179.5 (4) | C28—C29—C30—C31 | 179.8 (5) |
| C8—C9—C10—C11 | −0.9 (8) | C29—C30—C31—O4 | 0.5 (9) |
| Br1—C9—C10—C11 | 179.8 (3) | C29—C30—C31—C32 | 178.3 (5) |
| C9—C10—C11—C6 | −0.1 (7) | C2—C1—N1—C5 | 1.2 (8) |
| C9—C10—C11—Pt1 | 179.7 (4) | C2—C1—N1—Pt1 | 179.9 (4) |
| C7—C6—C11—C10 | 0.7 (7) | C4—C5—N1—C1 | −0.9 (7) |
| C5—C6—C11—C10 | −178.9 (4) | C6—C5—N1—C1 | −179.1 (4) |
| C7—C6—C11—Pt1 | −179.1 (4) | C4—C5—N1—Pt1 | −179.7 (3) |
| C5—C6—C11—Pt1 | 1.3 (5) | C6—C5—N1—Pt1 | 2.1 (5) |
| O1—C13—C14—C15 | −3.6 (9) | C18—C17—N2—C21 | −0.8 (8) |
| C12—C13—C14—C15 | 175.8 (5) | C18—C17—N2—Pt2 | 179.5 (4) |
| C13—C14—C15—O2 | 3.9 (9) | C20—C21—N2—C17 | 0.9 (7) |
| C13—C14—C15—C16 | −174.6 (5) | C22—C21—N2—C17 | 179.7 (4) |
| N2—C17—C18—C19 | 0.4 (9) | C20—C21—N2—Pt2 | −179.4 (4) |
| C17—C18—C19—C20 | −0.1 (9) | C22—C21—N2—Pt2 | −0.6 (5) |
| C18—C19—C20—C21 | 0.3 (8) | C14—C13—O1—Pt1 | 0.3 (7) |
| C19—C20—C21—N2 | −0.6 (8) | C12—C13—O1—Pt1 | −179.0 (4) |
| C19—C20—C21—C22 | −179.2 (5) | C14—C15—O2—Pt1 | −0.7 (7) |
| N2—C21—C22—C23 | 179.7 (4) | C16—C15—O2—Pt1 | 177.9 (3) |
| C20—C21—C22—C23 | −1.7 (8) | C30—C29—O3—Pt2 | −1.2 (7) |
| N2—C21—C22—C27 | −0.6 (6) | C28—C29—O3—Pt2 | 178.4 (3) |
| C20—C21—C22—C27 | 178.0 (5) | C30—C31—O4—Pt2 | 1.5 (7) |
| C27—C22—C23—C24 | −2.6 (7) | C32—C31—O4—Pt2 | −176.5 (4) |
Hydrogen-bond geometry (Å, º)
| D—H···A | D—H | H···A | D···A | D—H···A |
| C1—H1···O1 | 0.95 | 2.40 | 2.999 (7) | 121 |
| C4—H4···O4i | 0.95 | 2.58 | 3.281 (6) | 131 |
| C17—H17···O3 | 0.95 | 2.45 | 3.034 (6) | 120 |
| C17—H17···Br1ii | 0.95 | 2.87 | 3.693 (6) | 145 |
Symmetry codes: (i) −x+1/2, −y+1/2, −z+1; (ii) x−1/2, −y+1/2, z+1/2.
References
- Bondi, A. (1964). J. Phys. Chem. 68, 441–451.
- Bruker (2014). APEX2, SADABS and SAINT. Bruker AXS Inc., Madison, Wisconsin, USA.
- Chi, Y. & Chou, P.-T. (2010). Chem. Soc. Rev. 39, 638–655. [DOI] [PubMed]
- Cockburn, B. N., Howe, V., Keating, T., Johnson, B. F. G. & Lewis, J. (1973). J. Chem. Soc. Dalton Trans. pp. 404–410.
- Farrugia, L. J. (2012). J. Appl. Cryst. 45, 849–854.
- Hudson, Z. M., Blight, B. A. & Wang, S. (2012). Org. Lett. 14, 1700–1703. [DOI] [PubMed]
- Liu, J., Yang, C.-J., Cao, Q.-Y., Xu, M., Wang, J., Peng, H.-N., Tan, W.-F., Lü, X.-X. & Gao, X.-C. (2009). Inorg. Chim. Acta, 362, 575–579.
- Ma, D.-L., He, H.-Z., Leung, K.-H., Chan, D. S.-H. & Leung, C.-H. (2013). Angew. Chem. Int. Ed. 52, 7666–7682. [DOI] [PubMed]
- Macrae, C. F., Bruno, I. J., Chisholm, J. A., Edgington, P. R., McCabe, P., Pidcock, E., Rodriguez-Monge, L., Taylor, R., van de Streek, J. & Wood, P. A. (2008). J. Appl. Cryst. 41, 466–470.
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Sheldrick, G. M. (2015). Acta Cryst. C71, 3–8.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablock(s) I. DOI: 10.1107/S2056989015017478/wm5214sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S2056989015017478/wm5214Isup2.hkl
Supporting information file. DOI: 10.1107/S2056989015017478/wm5214Isup3.tif
CCDC reference: 1425736
Additional supporting information: crystallographic information; 3D view; checkCIF report