Abstract
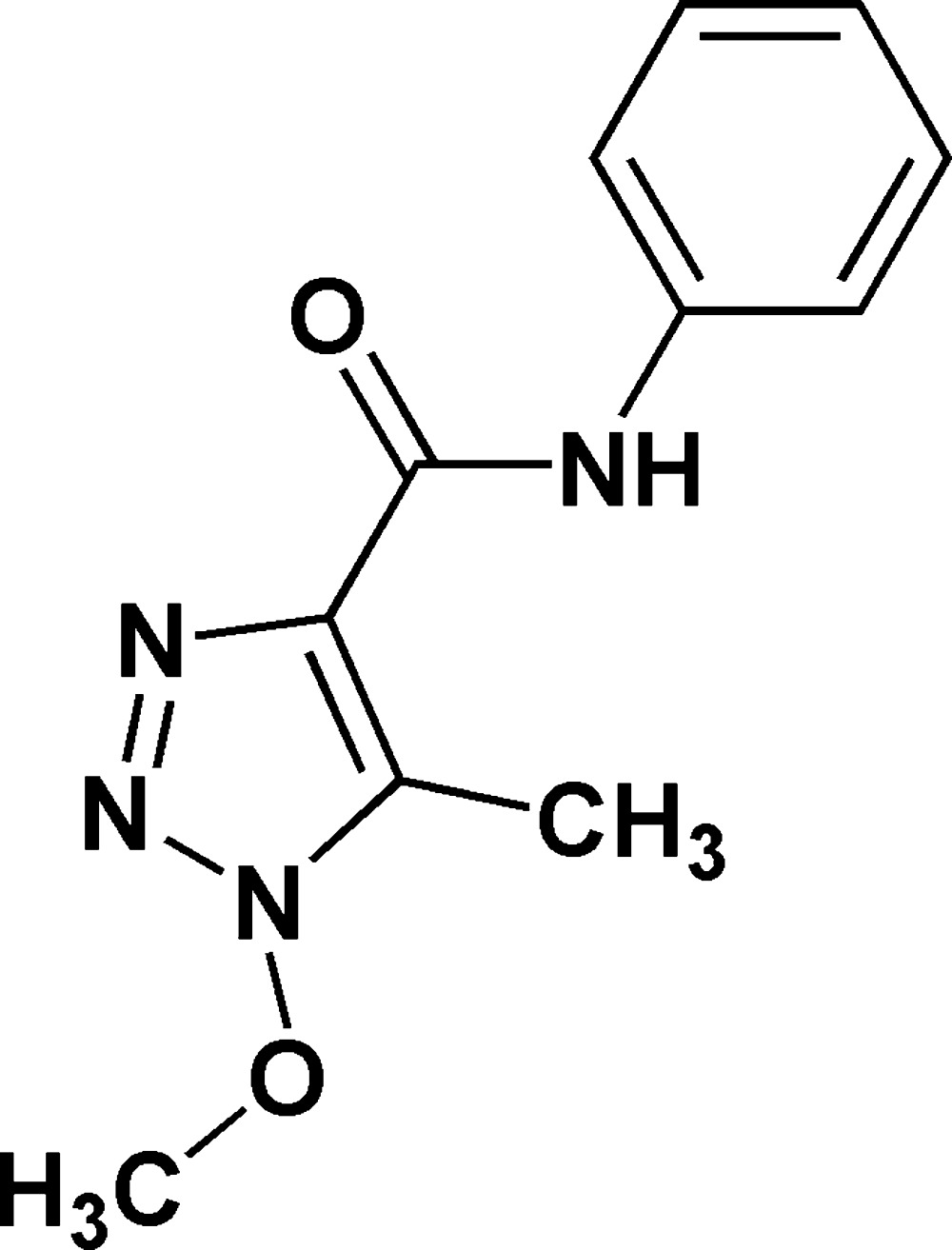
The title compound, C 11H12N4O2,was prepared via the transformation of sodium 4-acetyl-1-phenyl-1H-[1.2.3]triazolate under the action of methoxyamine hydrochloride. The dihedral angle between the triazole and phenyl rings is 25.12 (16)° and the C atom of the methoxy group deviates from the triazole plane by 0.894 (4)Å. The conformation of the CONHR-group is consolodated by an intramolecular N—H⋯N hydrogen bond to an N-atom of the triazole ring, which closes an S(5) ring. In the crystal, weak N—H⋯N hydrogen bonds link the molecules into C(6) [010] chains.
Keywords: crystal structure; 1,2,3-triazole; rearrangements; hydrogen bonding
Related literature
For biological activities of 1.2.3-triazoles, see: Sathish Kumar & Kavitha (2013 ▸); Khazhieva et al. (2015a
▸). For the synthesis, see: Khazhieva et al. (2015b
▸).
Experimental
Crystal data
C11H12N4O2
M r = 232.25
Monoclinic,
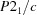
a = 11.4637 (8) Å
b = 6.4345 (13) Å
c = 15.822 (3) Å
β = 100.367 (12)°
V = 1148.0 (3) Å3
Z = 4
Mo Kα radiation
μ = 0.10 mm−1
T = 295 K
0.21 × 0.16 × 0.09 mm
Data collection
Agilent Xcalibur S CCD diffractometer
7259 measured reflections
2302 independent reflections
1077 reflections with I > 2σ(I)
R int = 0.040
Refinement
R[F 2 > 2σ(F 2)] = 0.055
wR(F 2) = 0.147
S = 1.00
2302 reflections
160 parameters
H atoms treated by a mixture of independent and constrained refinement
Δρmax = 0.43 e Å−3
Δρmin = −0.22 e Å−3
Data collection: CrysAlis PRO (Agilent, 2006 ▸); cell refinement: CrysAlis PRO; data reduction: CrysAlis PRO; program(s) used to solve structure: SHELXS97 (Sheldrick, 2008 ▸); program(s) used to refine structure: SHELXS97 (Sheldrick, 2008 ▸); molecular graphics: publCIF (Westrip, 2010 ▸); software used to prepare material for publication: publCIF (Westrip, 2010 ▸).
Supplementary Material
Crystal structure: contains datablock(s) I, exp_221. DOI: 10.1107/S2056989015017776/hb7511sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S2056989015017776/hb7511Isup2.hkl
Supporting information file. DOI: 10.1107/S2056989015017776/hb7511Isup3.cml
. DOI: 10.1107/S2056989015017776/hb7511fig1.tif
The molecular structure of (I), with 50% probability displacement ellipsoids for non-H atoms.
CCDC reference: 1426448
Additional supporting information: crystallographic information; 3D view; checkCIF report
Table 1. Hydrogen-bond geometry (, ).
| DHA | DH | HA | D A | DHA |
|---|---|---|---|---|
| N1H1N2 | 0.86(2) | 2.33(3) | 2.780(4) | 113(2) |
| N1H1N3i | 0.86(2) | 2.41(2) | 3.184(3) | 150(2) |
Symmetry code: (i) 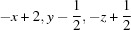 .
.
Acknowledgments
We thank the Russian Foundation for Basic Research (grant 13–03-00137), State task Ministry of Education and Science of the Russian Federation No. 4.560.2014-K and the Project Enhance Competitiveness of the Ural Federal University (Project 5–100-2020)
supplementary crystallographic information
S1. Synthesis and crystallization
The titled compound was prepared as previously reported (Khazhieva et al., 2015b). Crystals were obtained by slow evaporation of a solution in ethanol.
Figures
Fig. 1.
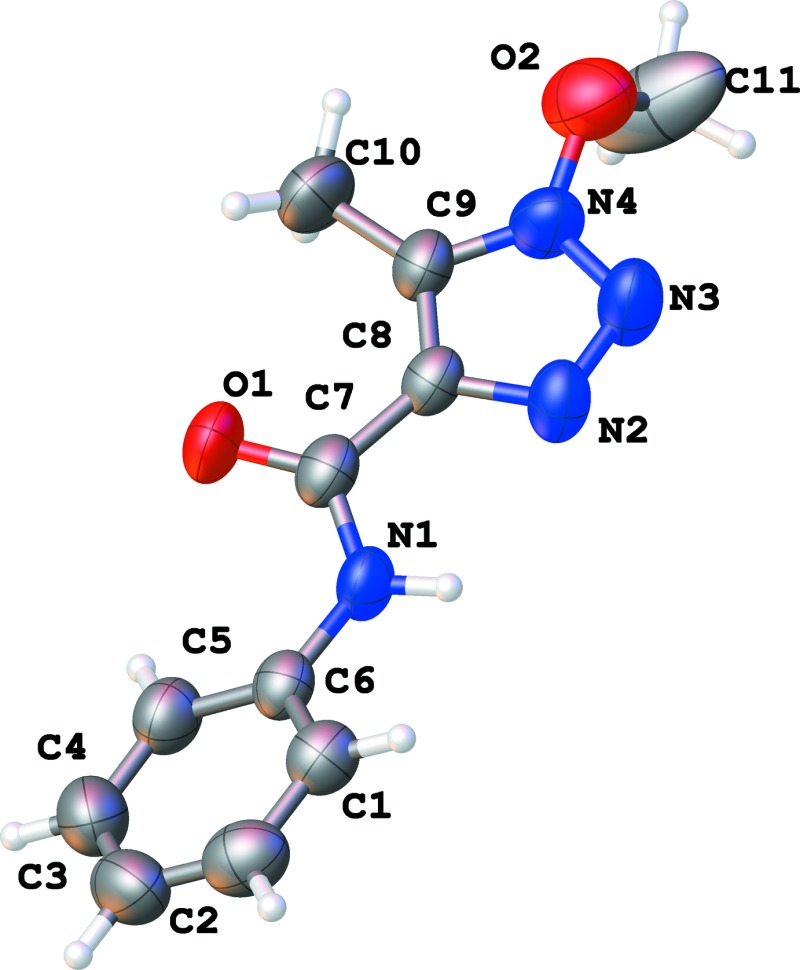
The molecular structure of (I), with 50% probability displacement ellipsoids for non-H atoms.
Crystal data
| C11H12N4O2 | Dx = 1.344 Mg m−3 |
| Mr = 232.25 | Melting point: 310 K |
| Monoclinic, P21/c | Mo Kα radiation, λ = 0.71073 Å |
| a = 11.4637 (8) Å | Cell parameters from 1077 reflections |
| b = 6.4345 (13) Å | θ = 2.9–26.4° |
| c = 15.822 (3) Å | µ = 0.10 mm−1 |
| β = 100.367 (12)° | T = 295 K |
| V = 1148.0 (3) Å3 | Prism, colorless |
| Z = 4 | 0.21 × 0.16 × 0.09 mm |
| F(000) = 488 |
Data collection
| Agilent Xcalibur S CCD diffractometer | 1077 reflections with I > 2σ(I) |
| Radiation source: fine-focus sealed tube | Rint = 0.040 |
| Graphite monochromator | θmax = 26.4°, θmin = 2.9° |
| ω scans | h = −7→14 |
| 7259 measured reflections | k = −5→8 |
| 2302 independent reflections | l = −19→19 |
Refinement
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.055 | Hydrogen site location: inferred from neighbouring sites |
| wR(F2) = 0.147 | H atoms treated by a mixture of independent and constrained refinement |
| S = 1.00 | w = 1/[σ2(Fo2) + (0.0682P)2] where P = (Fo2 + 2Fc2)/3 |
| 2302 reflections | (Δ/σ)max < 0.001 |
| 160 parameters | Δρmax = 0.43 e Å−3 |
| 0 restraints | Δρmin = −0.22 e Å−3 |
Special details
| Geometry. All e.s.d.'s (except the e.s.d. in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell e.s.d.'s are taken into account individually in the estimation of e.s.d.'s in distances, angles and torsion angles; correlations between e.s.d.'s in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell e.s.d.'s is used for estimating e.s.d.'s involving l.s. planes. |
| Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > σ(F2) is used only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R-factors based on ALL data will be even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| O1 | 0.61315 (16) | 0.0563 (3) | 0.15704 (14) | 0.0777 (7) | |
| C8 | 0.7887 (2) | 0.1712 (4) | 0.24262 (18) | 0.0475 (7) | |
| C6 | 0.7399 (2) | −0.2566 (4) | 0.08075 (18) | 0.0496 (7) | |
| C7 | 0.7204 (2) | 0.0365 (4) | 0.17728 (19) | 0.0533 (7) | |
| N2 | 0.90562 (18) | 0.1400 (4) | 0.27423 (17) | 0.0605 (7) | |
| N4 | 0.8463 (2) | 0.3915 (4) | 0.33811 (19) | 0.0708 (8) | |
| C9 | 0.7489 (2) | 0.3375 (4) | 0.28291 (19) | 0.0553 (8) | |
| N1 | 0.7844 (2) | −0.1083 (4) | 0.14353 (16) | 0.0530 (6) | |
| N3 | 0.9416 (2) | 0.2771 (4) | 0.3343 (2) | 0.0759 (8) | |
| O2 | 0.8515 (2) | 0.5302 (4) | 0.40535 (18) | 0.0956 (8) | |
| C1 | 0.7975 (2) | −0.4450 (5) | 0.0824 (2) | 0.0589 (8) | |
| H1A | 0.8634 | −0.4721 | 0.1246 | 0.071* | |
| C5 | 0.6434 (3) | −0.2172 (5) | 0.0169 (2) | 0.0641 (8) | |
| H5A | 0.6049 | −0.0896 | 0.0148 | 0.077* | |
| C3 | 0.6605 (3) | −0.5520 (6) | −0.0411 (2) | 0.0809 (10) | |
| H3A | 0.6331 | −0.6524 | −0.0821 | 0.097* | |
| C2 | 0.7571 (3) | −0.5924 (5) | 0.0214 (2) | 0.0739 (9) | |
| H2A | 0.7955 | −0.7200 | 0.0225 | 0.089* | |
| C4 | 0.6048 (3) | −0.3651 (6) | −0.0430 (2) | 0.0769 (10) | |
| H4A | 0.5395 | −0.3381 | −0.0857 | 0.092* | |
| C11 | 0.9070 (4) | 0.7045 (6) | 0.3901 (3) | 0.137 (2) | |
| H11A | 0.8970 | 0.8073 | 0.4321 | 0.205* | |
| H11B | 0.8741 | 0.7551 | 0.3337 | 0.205* | |
| H11C | 0.9900 | 0.6765 | 0.3933 | 0.205* | |
| C10 | 0.6343 (3) | 0.4512 (5) | 0.2740 (2) | 0.0818 (10) | |
| H10A | 0.6298 | 0.5214 | 0.3268 | 0.123* | |
| H10B | 0.5699 | 0.3543 | 0.2609 | 0.123* | |
| H10C | 0.6292 | 0.5511 | 0.2284 | 0.123* | |
| H1 | 0.858 (2) | −0.109 (4) | 0.1666 (17) | 0.048 (8)* |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| O1 | 0.0365 (12) | 0.1021 (17) | 0.0930 (17) | 0.0057 (10) | 0.0076 (11) | −0.0186 (13) |
| C8 | 0.0393 (15) | 0.0458 (16) | 0.0589 (18) | 0.0007 (12) | 0.0134 (13) | 0.0056 (14) |
| C6 | 0.0414 (15) | 0.0538 (18) | 0.0549 (19) | −0.0035 (14) | 0.0117 (14) | 0.0045 (16) |
| C7 | 0.0418 (16) | 0.0582 (18) | 0.062 (2) | −0.0016 (14) | 0.0146 (15) | 0.0066 (16) |
| N2 | 0.0444 (14) | 0.0516 (15) | 0.0836 (19) | −0.0032 (11) | 0.0062 (13) | −0.0019 (14) |
| N4 | 0.0607 (17) | 0.0658 (17) | 0.089 (2) | −0.0011 (13) | 0.0206 (15) | −0.0268 (16) |
| C9 | 0.0400 (16) | 0.065 (2) | 0.0624 (19) | −0.0028 (14) | 0.0117 (15) | −0.0013 (16) |
| N1 | 0.0354 (13) | 0.0558 (15) | 0.0661 (17) | 0.0045 (11) | 0.0041 (12) | −0.0011 (13) |
| N3 | 0.0465 (15) | 0.0710 (17) | 0.107 (2) | 0.0004 (13) | 0.0059 (14) | −0.0188 (17) |
| O2 | 0.0967 (18) | 0.0961 (18) | 0.102 (2) | −0.0125 (14) | 0.0395 (15) | −0.0152 (16) |
| C1 | 0.0573 (17) | 0.0563 (19) | 0.064 (2) | 0.0021 (15) | 0.0140 (15) | 0.0062 (17) |
| C5 | 0.0512 (18) | 0.073 (2) | 0.067 (2) | 0.0069 (15) | 0.0089 (16) | 0.0035 (19) |
| C3 | 0.077 (2) | 0.093 (3) | 0.075 (3) | −0.018 (2) | 0.018 (2) | −0.024 (2) |
| C2 | 0.081 (2) | 0.061 (2) | 0.085 (3) | −0.0020 (18) | 0.030 (2) | −0.004 (2) |
| C4 | 0.061 (2) | 0.098 (3) | 0.070 (2) | −0.005 (2) | 0.0055 (17) | −0.010 (2) |
| C11 | 0.171 (4) | 0.055 (2) | 0.221 (5) | −0.019 (2) | 0.133 (4) | −0.005 (3) |
| C10 | 0.0566 (19) | 0.098 (2) | 0.092 (3) | 0.0195 (17) | 0.0172 (17) | −0.016 (2) |
Geometric parameters (Å, º)
| O1—C7 | 1.220 (3) | C1—C2 | 1.372 (4) |
| C8—N2 | 1.359 (3) | C1—H1A | 0.9300 |
| C8—C9 | 1.365 (3) | C5—C4 | 1.359 (4) |
| C8—C7 | 1.463 (4) | C5—H5A | 0.9300 |
| C6—C1 | 1.379 (4) | C3—C4 | 1.360 (5) |
| C6—C5 | 1.381 (4) | C3—C2 | 1.370 (5) |
| C6—N1 | 1.405 (3) | C3—H3A | 0.9300 |
| C7—N1 | 1.354 (3) | C2—H2A | 0.9300 |
| N2—N3 | 1.308 (3) | C4—H4A | 0.9300 |
| N4—N3 | 1.327 (3) | C11—H11A | 0.9600 |
| N4—C9 | 1.334 (3) | C11—H11B | 0.9600 |
| N4—O2 | 1.382 (3) | C11—H11C | 0.9600 |
| C9—C10 | 1.488 (4) | C10—H10A | 0.9600 |
| N1—H1 | 0.85 (3) | C10—H10B | 0.9600 |
| O2—C11 | 1.333 (4) | C10—H10C | 0.9600 |
| N2—C8—C9 | 109.5 (2) | C4—C5—C6 | 120.0 (3) |
| N2—C8—C7 | 122.6 (2) | C4—C5—H5A | 120.0 |
| C9—C8—C7 | 127.8 (2) | C6—C5—H5A | 120.0 |
| C1—C6—C5 | 119.6 (3) | C4—C3—C2 | 120.0 (3) |
| C1—C6—N1 | 118.1 (3) | C4—C3—H3A | 120.0 |
| C5—C6—N1 | 122.3 (3) | C2—C3—H3A | 120.0 |
| O1—C7—N1 | 124.1 (3) | C3—C2—C1 | 120.2 (3) |
| O1—C7—C8 | 120.6 (2) | C3—C2—H2A | 119.9 |
| N1—C7—C8 | 115.3 (2) | C1—C2—H2A | 119.9 |
| N3—N2—C8 | 109.2 (2) | C5—C4—C3 | 120.7 (3) |
| N3—N4—C9 | 115.2 (2) | C5—C4—H4A | 119.6 |
| N3—N4—O2 | 118.1 (3) | C3—C4—H4A | 119.6 |
| C9—N4—O2 | 125.9 (2) | O2—C11—H11A | 109.5 |
| N4—C9—C8 | 101.5 (2) | O2—C11—H11B | 109.5 |
| N4—C9—C10 | 123.7 (3) | H11A—C11—H11B | 109.5 |
| C8—C9—C10 | 134.8 (3) | O2—C11—H11C | 109.5 |
| C7—N1—C6 | 126.3 (3) | H11A—C11—H11C | 109.5 |
| C7—N1—H1 | 113.4 (17) | H11B—C11—H11C | 109.5 |
| C6—N1—H1 | 120.2 (17) | C9—C10—H10A | 109.5 |
| N2—N3—N4 | 104.6 (2) | C9—C10—H10B | 109.5 |
| C11—O2—N4 | 111.1 (3) | H10A—C10—H10B | 109.5 |
| C2—C1—C6 | 119.6 (3) | C9—C10—H10C | 109.5 |
| C2—C1—H1A | 120.2 | H10A—C10—H10C | 109.5 |
| C6—C1—H1A | 120.2 | H10B—C10—H10C | 109.5 |
Hydrogen-bond geometry (Å, º)
| D—H···A | D—H | H···A | D···A | D—H···A |
| N1—H1···N2 | 0.86 (2) | 2.33 (3) | 2.780 (4) | 113 (2) |
| N1—H1···N3i | 0.86 (2) | 2.41 (2) | 3.184 (3) | 150 (2) |
Symmetry code: (i) −x+2, y−1/2, −z+1/2.
Footnotes
Supporting information for this paper is available from the IUCr electronic archives (Reference: HB7511).
References
- Agilent (2006). CrysAlis PRO. Agilent Technologies UK Ltd, Yarnton, England.
- Khazhieva, I. S., Glukhareva, T. V., El’tsov, O. S., Morzherin, Yu. Yu., Minin, A. A., Pozdina, V. A. & Ulitko, M. V. (2015b). Khim. Farm. Zh 49, 12–15.
- Khazhieva, I. S., Glukhareva, T. V. & Morzherin, Yu. Yu. (2015a). Chim. Tech. Acta, 2, 52–58.
- Sathish Kumar, S. & Kavitha, H. P. (2013). Mini-Rev. Org. Chem. 10, 40–65.
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Westrip, S. P. (2010). J. Appl. Cryst 43, 920–925.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablock(s) I, exp_221. DOI: 10.1107/S2056989015017776/hb7511sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S2056989015017776/hb7511Isup2.hkl
Supporting information file. DOI: 10.1107/S2056989015017776/hb7511Isup3.cml
. DOI: 10.1107/S2056989015017776/hb7511fig1.tif
The molecular structure of (I), with 50% probability displacement ellipsoids for non-H atoms.
CCDC reference: 1426448
Additional supporting information: crystallographic information; 3D view; checkCIF report


