Abstract
Influenza A virus (IAV) lacks the enzyme for adding 5′ caps to its RNAs and snatches the 5′ ends of host capped RNAs to prime transcription. Neither the preference of the host RNA sequences snatched nor the effect of cap-snatching on host processes is completely defined. Previous studies of influenza cap-snatching used poly(A)-selected RNAs from infected cells or relied on annotated host genes to define the snatched host RNAs, and thus lack details on many noncoding host RNAs including snRNAs, snoRNAs, and promoter-associated capped small (cs)RNAs, which are made by “paused” Pol II during transcription initiation. In this study, we used a nonbiased technique, CapSeq, to identify host and viral-capped RNAs including nonpolyadenylated RNAs in the same samples, and investigated the substrate–product correlation between the host RNAs and the viral RNAs. We demonstrated that noncoding host RNAs, particularly U1 and U2, are the preferred cap-snatching source over mRNAs or pre-mRNAs. We also found that csRNAs are highly snatched by IAV. Because the functions of csRNAs remain mostly unknown, especially in somatic cells, our finding reveals that csRNAs at least play roles in the process of IAV infection. Our findings support a model where nascent RNAs including csRNAs are the preferred targets for cap-snatching by IAV and raise questions about how IAV might use snatching preferences to modulate host-mRNA splicing and transcription.
Keywords: cap-snatching, influenza, noncoding RNA
INTRODUCTION
Influenza A virus (IAV) causes a highly contagious acute respiratory illness responsible for significant morbidity and mortality in humans. The IAV RNA-dependent RNA polymerase (RdRP) comprises three subunits: polymerase base protein 1 (PB1), polymerase base protein 2 (PB2), and polymerase acidic protein (PA). RdRP primes viral mRNA synthesis with 5′ ends (10–15 nt) cleaved and stolen from host Pol II transcripts, a process called “cap-snatching” (Plotch et al. 1979). During this process, PB2 recognizes and binds to the cap structure on host pre-mRNA (Guilligay et al. 2008; Sugiyama et al. 2009). PA carries the endonucleolytic activity, which cleaves the capped RNA fragments used for priming IAV transcription (Fodor et al. 2002; Dias et al. 2009). Viral mRNAs are thus genetic hybrids that include host-derived 5′ ends (Caton and Robertson 1980).
Previous studies have focused on identifying the preferred sequences of host pre-mRNAs snatched by the IAV RdRP (Beaton and Krug 1981; Shaw and Lamb 1984; Rao et al. 2003). Critical nucleotides reside at the cleavage site, as cap fragments terminating with the sequence CA were shown to prime transcription initiation (Rao et al. 2003). Nonetheless, aspects such as the source of the caps are still poorly understood. A recent deep-sequencing analysis identified some host mRNAs as the preferred cap-snatching substrates (Sikora et al. 2014). This study, however, focused entirely on caps snatched from host mRNAs and was based on gene annotations that contain incomplete or inaccurate information on the 5′ ends of RNAs as compared with the coding sequences. Moreover, a myriad of other host RNAs are transcribed by Pol II and bear 5′ caps, including noncoding RNAs such as small nuclear RNAs (e.g., U1 and U2 snRNAs) and the recently identified class of promoter-associated capped small RNAs (csRNAs) (Seila et al. 2008; Affymetrix/Cold Spring Harbor Laboratory ENCODE Transcriptome Project 2009; Nechaev et al. 2010; Gu et al. 2012). Thus, a more comprehensive approach is required.
Our previous work used high-throughput sequencing methods and new bioinformatics and statistical tools to study the evolution of IAV (Foll et al. 2014; Renzette et al. 2014; Zeldovich et al. 2015). Here, we apply novel strategies to demonstrate how IAV snatches caps from host RNAs, both coding and noncoding, to prime the transcription of viral RNAs. To investigate the relationship between the host transcripts and the viral RNA caps in the same samples, we used CapSeq, an enzymatic method for identifying the 5′ ends of capped RNAs in a transcriptome-wide manner (Gu et al. 2012). We find that noncoding RNAs, especially U1 and U2 snRNAs, constitute the main source of snatched caps, whereas protein-coding transcripts contribute fewer than half of the snatched caps. In addition, we find that promoter-associated csRNAs are snatched by IAV. Coupled with increased recognition for the roles of noncoding RNAs in cellular regulation (Clark et al. 2013), our findings that noncoding and csRNAs are preferential targets of IAV cap-snatching suggest their possible impact on the host–virus interaction.
RESULTS
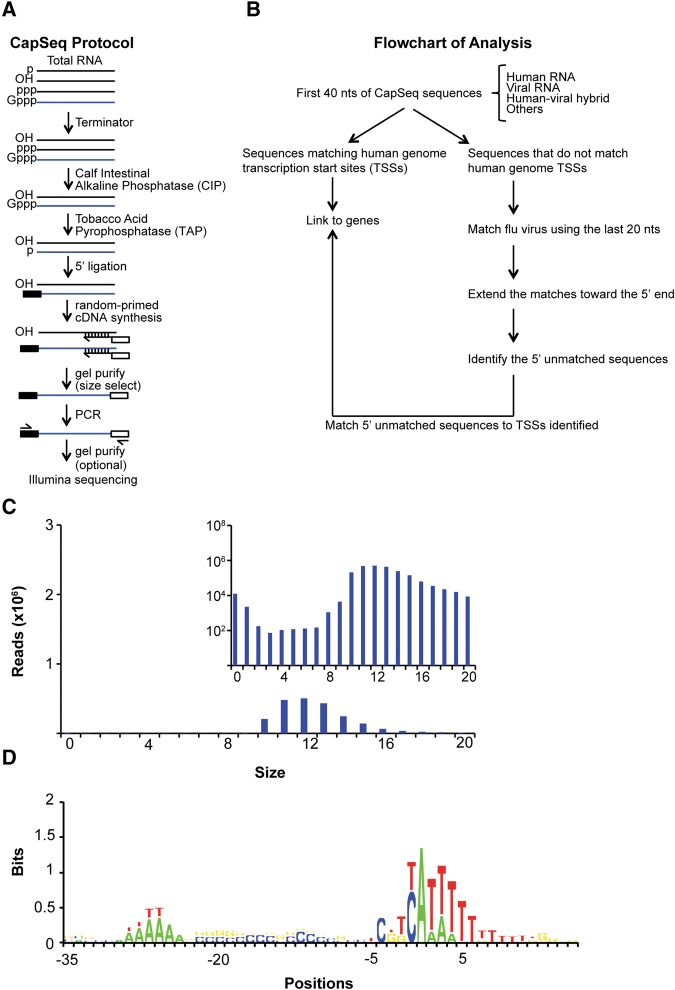
We developed the CapSeq method to profile the 5′ ends of all capped RNAs including poly(A) RNAs, nonpoly(A) RNAs, and csRNAs in eukaryotes. CapSeq uses a series of enzymatic digestion steps to efficiently remove rRNAs and enrich for capped RNAs that are then decapped, ligated to 5′ adaptors, random-primed with octamer primers for first-strand cDNA synthesis, size-selected, and deep sequenced (Fig. 1A; see Gu et al. 2012). We applied the CapSeq method to profile IAV-infected human A549 cells at 24 h (Fig. 1B), which allows for simultaneous sequencing of both host and viral-capped RNAs and subsequent association of the snatched sequences with corresponding host RNAs expressed in the same cell. 5′ caps snatched by IAV were extracted from reads that mapped to the positive strand (mRNA) of IAV; the size distribution revealed a peak at 12 nt (Fig. 1C). As reported previously (Sikora et al. 2014; Koppstein et al. 2015), we found the modal size of snatched caps differs between some viral mRNAs (e.g., PB1 and PB2 peak at 11 nt and NS1 peaks at 12 nt). We used transcription start site (TSS) motif analysis (Fig. 1D) and read distribution to independently confirm that our CapSeq results are highly enriched for TSS-associated reads (>70,000-fold).
FIGURE 1.
CapSeq method and verification of size and motif analysis of snatched caps identified by CapSeq. (A) Schematic of CapSeq method. (B) Flowchart of applying CapSeq to examine IAV cap-snatching. (TSSs) transcription start sites, (nt) nucleotide. (C) Size distribution of the snatched sequences extracted from reads mapped to IAV. The 5′ extra sequences snatched by IAV were extracted from reads mapped to the positive strand (mRNA) of influenza virus. The inset figure uses log scale on the Y-axis to demonstrate the trend of the extra sequences derived from the positive virus strand. (D) Motif analysis of human Pol II TSSs using CapSeq reads. Genomic positions flanking the 5′ end (+1) of each matched CapSeq read are aligned with negative numbers indicating upstream positions. The log2 ratio of the experimental frequency over the expected frequency for each nucleotide was obtained as the bits on the Y-axis. Regardless of the abundance of the CapSeq reads, each mapped genomic locus was only counted once in this analysis.
Identification of host capped RNAs snatched by IAV
To identify the host RNAs snatched by IAV, we mapped snatched sequences ≥10 nt (99% of all snatched sequences) to the host TSSs defined using CapSeq data from the same samples. Of the potential 480,000 TSSs obtained in the IAV-infected sample, the matching rate for a random sequence of 12 nt (the peak size of the snatched sequences) is expected to be very low (480,000/412, i.e., 2.9%). However, by comparing snatched reads to the 5′ ends of host RNAs cloned from the same infected cells, we were able to assign ∼51% of the snatched sequences to host transcripts, and 81% of these were each uniquely assigned to only one host RNA.
The remaining 49% of caps snatched from “unidentified” host capped RNAs are mostly caused by the stringency of our analysis and lack of saturation of CapSeq coverage of host TSSs. We can account for 85% of the snatched caps by including more host TSSs identified in two noninfected samples in addition to the TSSs identified in the infected sample, considering mutations or indels of snatched caps, and including short caps; the remaining 15% of reads could be caused by underestimation of mutation rate, incomplete coverage of the host TSSs by our sequencing, or other factors including csRNAs as described below (see Supplemental file 1 for a detailed breakdown of the possible contribution of each source). Nonetheless, accounting for these sources for snatched 5′ caps does not change the major conclusion from our study (see below).
Cap-snatching prefers donor RNAs starting with a 5′ A
To investigate if cap-snatching prefers certain donor RNA sequence elements, we aligned the CapSeq reads matching the TSSs of human RNAs. As shown in Figure 2A, these reads prefer A (49.7%) at the first position. A similar alignment was performed using the extracted nonvirus 5′ caps of the hybrid sequences. As shown in Figure 2B, we observed a stronger preference for A (∼69.6%) at the first nucleotide of the snatched sequences than at the first nucleotide of host RNA transcripts. This observation suggests that RNAs starting with a 5′ A may be preferred substrates for cap-snatching by IAV. As discussed later, snRNAs, which start with 5′A, contribute a significant fraction of caps snatched by IAV. This at least partially explains why snatched sequences are enriched for 5′ A relative to all host TSSs.
FIGURE 2.
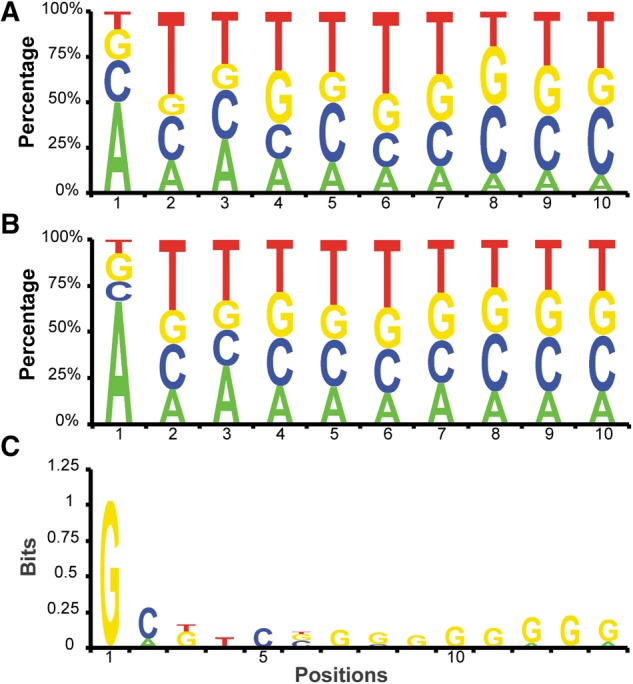
Sequence analysis of snatched caps. (A,B) Human CapSeq reads and those reads snatched by IAV are aligned using the 5′ end as +1 position. The nucleotide frequency at each position was obtained using the CapSeq read number. (C) Analysis of the human sequences immediately downstream from the snatched sequences. The sequences snatched by IAV were mapped to the Pol II transcription start sites (TSSs) in the human genome, as defined by CapSeq here. The human sequences downstream from the snatched sequences were then extracted and aligned as indicated. The log2 ratio of the experimental frequency over the expected frequency was obtained as the bits on the Y-axis. Regardless of the abundance of the snatched sequences or the corresponding human sequences, each mapped locus was only counted once in this analysis.
Cap-snatching preferentially cleaves or selects donor RNAs at a 3′ G position
To investigate whether specific sequence motifs on host donor RNAs could affect cap cleavage or selection, we first extracted the nonvirus 5′ cap sequences from the hybrid reads partially matching IAV and mapped these sequences to the host TSSs defined using our human CapSeq reads. Then we analyzed these genomic TSS loci for nucleotide preference 3′ of the snatched caps. As shown in Figure 2C, we observed a strong G preference (bit ∼1, ∼61.2% as compared with a background level of 26.3%) at the genomic position immediately following the snatched caps. In the bit analysis, we counted the nucleotide frequency at genomic loci rather than CapSeq reads snatched by IAV to avoid potential bias introduced by individual donor sequences that make up a large fraction of snatched caps. Alternatively, using the read number of snatched caps by IAV, we found that 73.3% of the reads were derived from donor RNAs containing a G immediately after the snatched cap sequences. We speculate that our algorithm that breaks a hybrid RNA into the virus-matching and human-matching parts caused the G bias. As analyzed above, ∼98% hybrid reads match the virus mRNAs starting at +1, which is G. This G could be generated in two ways: (i) as the first nucleotide added by the viral RdRP or (ii) as the last nucleotide of the snatched human caps, which base pairs with the 3′ C of viral RNAs and primes the synthesis of viral mRNAs from the +2 position. Either way generates functional viral RNAs. Based on our data and previous studies (Plotch et al. 1981; Sikora et al. 2014; Koppstein et al. 2015), we speculate that IAV may preferentially cleave host RNAs after a G or selectively use the 3′ G-containing snatched caps to prime the viral mRNA synthesis.
Cap-snatching prefers noncoding RNAs, especially snRNAs
Our analysis examined all host RNA sequences for their use as caps for IAV transcription. Among the 51% snatched sequences with donor RNAs identified in the 24-h infected sample, 54% are derived from noncoding RNA loci, and only 46% are derived from protein-coding loci, even if we give protein-coding loci a priority over noncoding loci when sequences match both. We observed similar rates in the 12-h infected samples: 56% from noncoding RNAs and 44% from protein-coding RNAs. The rate difference between coding and noncoding RNAs is significant (one-tailed paired t-test, P-value <0.01), despite the limited sampling size. This finding was unaffected by expanding our data set to allow mismatches and/or to include more TSSs defined in the noninfected samples.
snRNAs such as U1 and U2 are highly represented in our viral CapSeq data (Table 1). In fact, of the most abundant host RNAs, snRNAs and snoRNAs have the top snatching rates (snatched reads/[snatched reads + host reads], Table 1), and the snatched U1 and U2 snRNA reads constitute 14.5% and 6.8% of all U1 and U2 reads. Moreover, U1 and U2 snRNAs are the most abundant cap sources of viral RNAs, comprising ∼3.3% and ∼3.5% of all snatched sequences from host RNAs and ∼50-fold higher than the most abundant cap sequences snatched from mRNAs/pre-mRNAs (Fig. 3). A snoRNA, SNORD3, represents the third- and fourth-highest snatched reads. U11 snRNA is snatched at a much lower rate, ∼0.4% of all U11 reads, and makes up only ∼0.07% of all the snatched sequences. As expected, RNA7SL1/2, a highly expressed small noncoding RNA transcribed by Pol III and therefore lacking a cap, was rarely snatched (Fig. 3).
TABLE 1.
Analysis of the snatching rate of top 20 abundant host RNAs in the 24-h infected sample
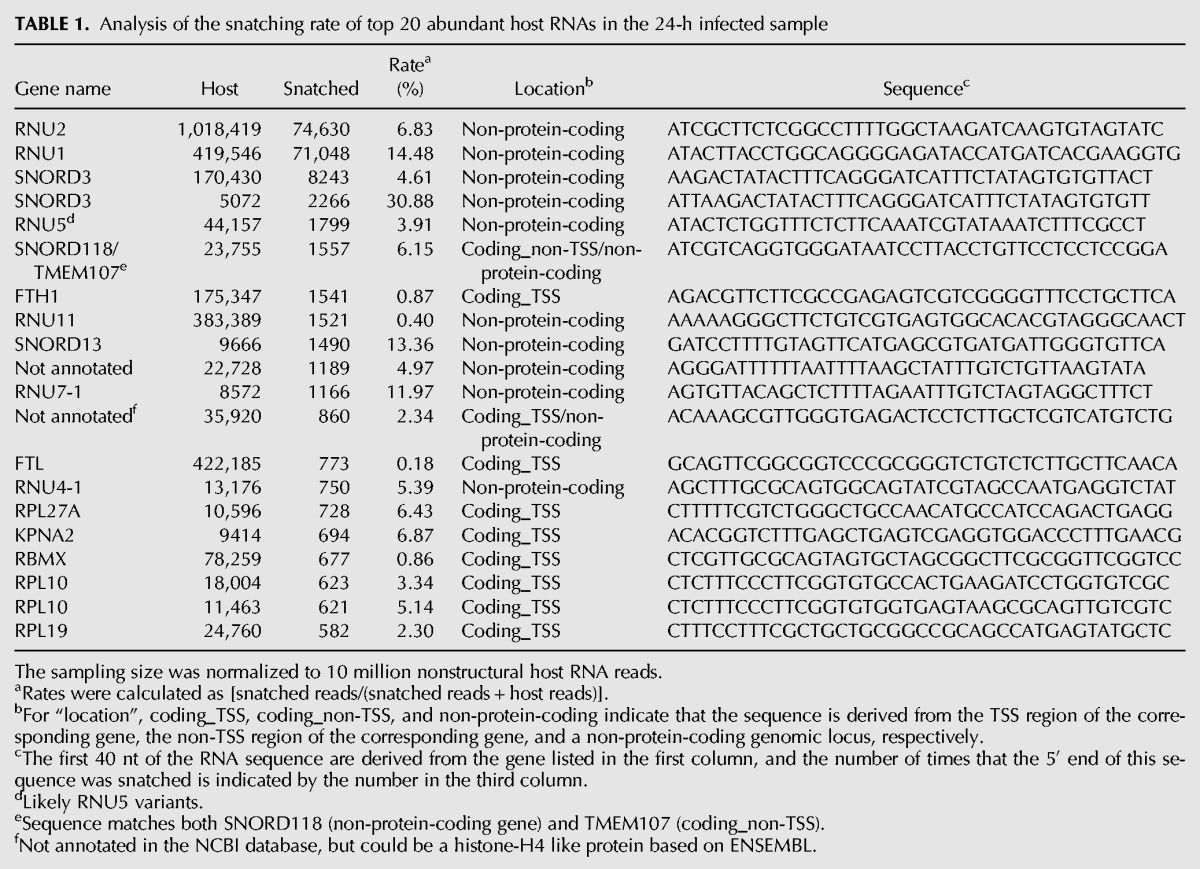
FIGURE 3.
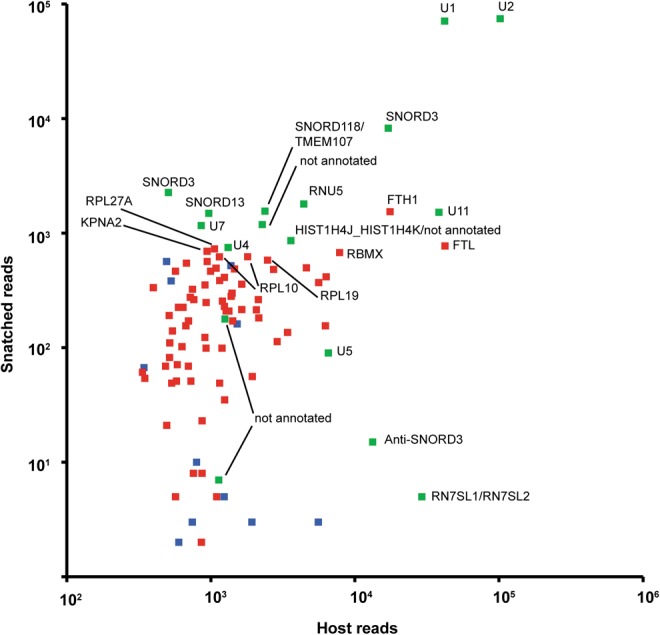
The correlation of the level of snatched sequences with that of the donor RNAs at 24 h. Y-axis, the read number of snatched sequences; X-axis, the read number of the human donor RNAs. Green indicates snatched sequences matching non-protein-coding regions, blue indicates sequences from protein-coding regions but not within known TSS regions, and red indicates snatched sequences in the −100 to +100 regions of the TSSs of protein-coding genes.
U1 snRNA snatching is confirmed by quantitative PCR and in situ hybridization
Because we were surprised by the abundance of noncoding RNAs snatched by IAV, to exclude sequencing artifacts, we used quantitative reverse transcription–polymerase chain reaction (RT-PCR) and in situ hybridization techniques to look for additional evidence. For qRT-PCR, a forward primer specific for either a U1 or U2 snRNA-influenza segment 1 hybrid combined with a reverse primer specific for influenza segment 1 allowed for detection of abundant hybrid RNA species in A549 cells 24 h following IAV infection (Fig. 4A). We also used RNA fluorescence in situ hybridization (FISH) not only to confirm the presence of hybrid human–viral RNA species but also to localize the hybrid sequences in the cell. Representative images from IAV-infected A549 cells at 24 h are shown in Figure 4B. U1-influenza segment 1 hybrids are detected throughout the cytoplasm (white, top row), and are similarly distributed in the cell as segment 1 alone (green). To ensure the specificity of the probes for the U1-influenza segment 1 hybrid, a nontargeting probe with the first half directed against sequence present in low abundance in infected cells (white, bottom row) was used. The nontargeting probe had a slight increase in signal above background, but quantification of the nontargeting probe revealed significantly lower signal compared with the U1-influenza segment 1 probe (P = 0.0001) (Fig. 4C).
FIGURE 4.
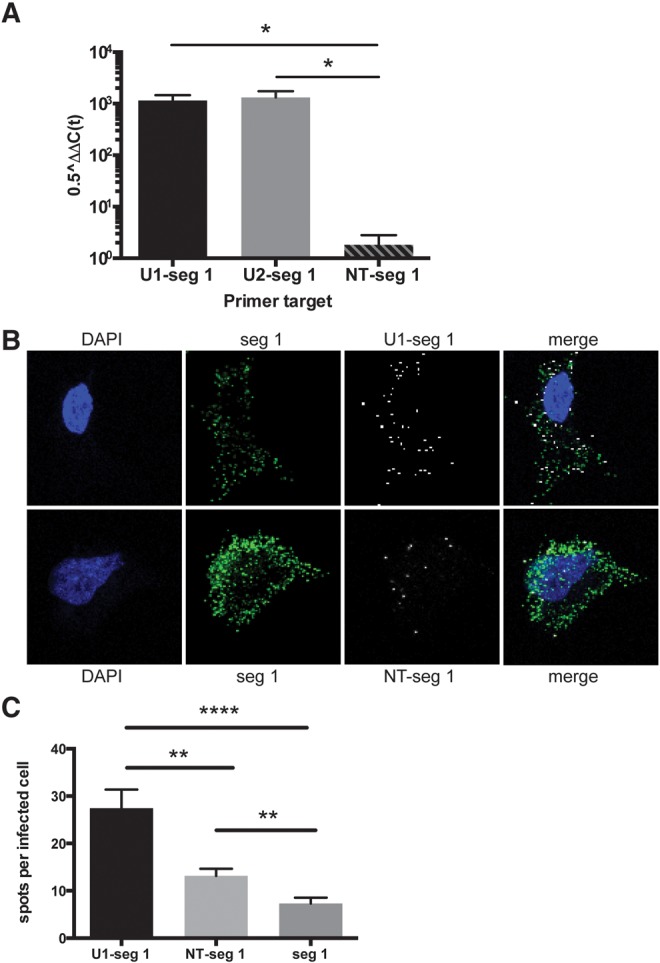
Verification of commonly snatched human RNA sequences by qRT-PCR and RNA fluorescence in situ hybridization (FISH). (A) qRT-PCR analysis was performed on samples from influenza virus–infected cells at 24 h. Host–virus hybrid products are shown as 0.5ΔΔCt normalized to host HPRT. Hybrids of U1-influenza segment 1 and U2-segment 1 are abundant compared with a nontargeting (NT) sequence hybrid, NT-segment 1. Error bars represent the SEM for three independent infection experiments. (*) P < 0.05; two-tailed t-test. (B) FISH was performed on virus-infected cells at 24 h. Blue is DAPI, green is a probe exclusively against segment 1, and white is a probe against the U1-segment 1 hybrid (top row) or an NT-segment 1 hybrid probe (bottom row). (C) Quantification of the number of hybrid probe spots per infected cell as identified by the segment 1 probe staining (n = 47, U1-seg 1; n = 55, NT–seg 1; n = 54, seg 1). Error bars represent the SEM. (**) P = 0.001; (****) P < 0.0001; two-tailed t-test.
CapSeq identifies host mRNAs as source of caps
We also identified sequences snatched from host mRNAs. The cap-snatching frequency of highly expressed host mRNAs is collectively much lower compared with that of U1 and U2 snRNAs. For example, ferritin light chain mRNA (FTL) is expressed at a level comparable to that of U1 snRNA (Fig. 3), but it is snatched at a frequency of only 0.18%, ∼80-fold lower than that of U1 RNA (14.5%). Ferritin heavy chain mRNA (FTH1) is also highly expressed, but is snatched at a frequency of 0.9%, ∼16-fold lower than that of U1 RNA (Table 1).
Although some variability exists between the 12-h (Supplemental file 2; Supplemental Fig. S1) and 24-h (Table 1; Fig. 3) infected samples, significant correlations exist between host RNA levels, snatched cap levels, and snatching rates (Supplemental Fig. S2). In both samples, we observed significantly higher snatching rates for several snRNA/snoRNAs compared with that of mRNAs (one-tailed t-test P < 0.001 and 0.0001 for 12- and 24-h infected samples, respectively).
Promoter-associated csRNAs are snatched by IAV
Promoter-associated sense and anti-sense capped small RNAs (csRNAs) are a noncanonical class of small RNAs that could also serve as a source of caps for IAV. csRNAs are usually generated during transcription initiation of long capped RNAs (referred to as sense strand) by Pol II. csRNAs, identified in flies, worms, mice, and humans, are associated with active promoters and are hypothesized to be important for promoter regulation (Seila et al. 2008; Affymetrix/Cold Spring Harbor Laboratory ENCODE Transcriptome Project 2009; Nechaev et al. 2010; Gu et al. 2012). csRNAs do not encode proteins, likely localize to the nucleus, and are expressed at much lower levels compared with those of corresponding long capped RNAs (Seila et al. 2008; Affymetrix/Cold Spring Harbor Laboratory ENCODE Transcriptome Project 2009; Nechaev et al. 2010; Gu et al. 2012). Sense csRNAs often use the same TSSs as long capped RNAs, making them appear indistinguishable from long capped RNAs when mapped to reference genomes. Anti-sense csRNAs, however, map 100–200 nt upstream of sense csRNAs on the opposite genomic strand (Seila et al. 2008; Affymetrix/Cold Spring Harbor Laboratory ENCODE Transcriptome Project 2009; Nechaev et al. 2010; Gu et al. 2012). Because there are no long capped RNAs annotated in the anti-sense csRNA regions, reads mapped to these regions can be clearly assigned as anti-sense csRNAs rather than a mixture of csRNAs and long capped RNAs.
Our CapSeq method clones all capped RNAs, including csRNAs >100 nt. To clearly identify anti-sense csRNA reads snatched by IAV, we focused on protein-coding gene loci because (i) protein-coding genes are well-expressed and relatively well-annotated; and (ii) the number of protein-coding genes is much greater than the number of snRNA and snoRNA genes. We identified anti-sense reads that map within 300 nt upstream of the TSSs of annotated protein-coding genes. As shown for RPL27A in Figure 5, the anti-sense csRNAs (red arrows) and the more abundant sense capped RNAs including sense csRNAs and mRNAs (blue arrows) were both snatched by IAV at multiple loci (black arrows).
FIGURE 5.
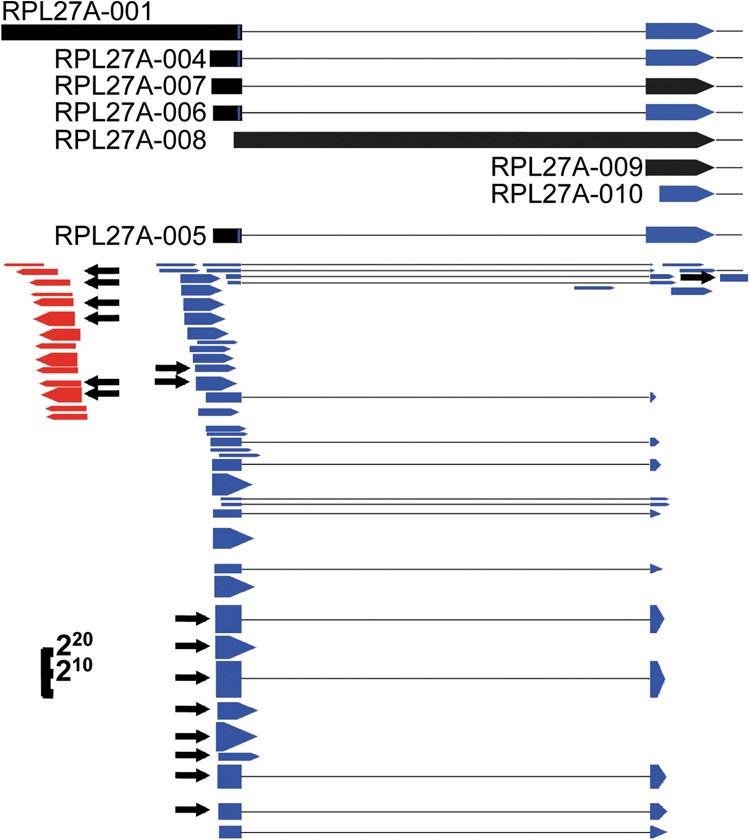
Influenza virus snatches capped small RNAs. At the top are the 5′ end annotations of RPL27A transcripts, as labeled, with blue and black indicating coding regions and 5′ UTR regions, respectively, and black dotted lines indicating introns. Below are the CapSeq RNA reads with blue and red indicating sense reads and anti-sense reads, respectively, and dotted lines indicating introns excluded in the RNA reads. Black arrows indicate reads where the 5′ ends were snatched by influenza virus. The height of the blue and red reads represents the read number using a log scale, as indicated in the lower left corner.
In total, we found that IAV snatched caps from anti-sense csRNAs that map to ∼3000 protein-coding genes and these snatched caps contribute ∼7% of all caps snatched from identified host sources. On average, ∼60% of anti-sense csRNAs were snatched by IAV. In contrast, sense capped RNA reads mapping to protein-coding genes were snatched at a much lower rate, ∼12%, perhaps because host sense reads include mature mRNAs that are unlikely to be cap-snatching substrates because they are in the cytoplasm. Although anti-sense csRNAs were usually expressed at very low levels (Supplemental Fig. S3), we observed a significant correlation between the levels of the abundant anti-sense csRNAs and the caps snatched from them by IAV. The relatively low correlation coefficient (R = 0.29) likely results from the low levels of anti-sense csRNAs and corresponding snatched caps. Sense csRNAs may also be snatched by IAV, although our cloning method cannot distinguish them from mature sense RNAs such as mRNAs/pre-mRNAs. Because of low expression levels and/or size less than 100 nt (Supplemental Fig. S3), many csRNAs may not have been cloned. We suspect that viral mRNA caps without an identified host source could derive from “unidentified” csRNAs, perhaps corresponding to TSSs that do not give rise to long RNAs. Therefore, we may be underestimating the number of csRNAs snatched by the virus.
DISCUSSION
We demonstrate that noncoding RNAs are the preferred source for cap-snatching by IAV. Previous studies of IAV cap-snatching events used protein-coding gene annotations to map the snatched sequences and did not specifically address the issue of noncoding RNAs (Bouloy et al. 1978; Krug et al. 1979; Dhar et al. 1980; Plotch et al. 1981; Shi et al. 1995; Sikora et al. 2014). Because the snatched sequences are only ∼11–12 nt and TSS annotations are neither complete nor accurate, such analyses would miss or improperly categorize at least some target RNAs. Our CapSeq results identified the same sequence length and nucleotide preference at the cleavage site as previous studies (Sikora et al. 2014; Koppstein et al. 2015), thus validating our method. Furthermore, our results were confirmed through an independent experiment (Supplemental file 2; Supplemental Fig. S1) and with independent methods (Fig. 4).
We speculate that nuclear host RNAs are preferentially snatched because the cap-snatching process occurs in the nucleus. However, not all snRNAs in the nucleus are snatched equally. U11 snRNA is highly expressed but snatched at a lower rate compared with U1 and U2 snRNAs. We suspect that other factors, such as a G within the optimal 10–13-nt window of the 5′ cap region, may also affect the efficiency of cleavage and/or priming processes of cap-snatching. However, factors important for cap-snatching from snRNAs may not apply to mRNAs. Deeper sequencing and more in vitro studies will be required to address this issue. Previous studies suggest that influenza virus NS1 inhibits host-mRNA splicing by binding U6 snRNAs or other components of the splicing machinery (Fortes et al. 1995; Wang and Krug 1998). Our findings raise the intriguing possibility that IAV may further suppress host-mRNA splicing by selectively snatching caps from the essential nuclear splicing snRNAs U1 and U2. Further experiments are needed to address whether cap-snatching depletes mature U1 and U2 levels sufficiently to affect splicing.
We also discovered that csRNAs contribute a significant fraction of caps to IAV mRNAs. Although it is difficult to estimate the contribution of sense csRNAs because they often colocalize with long capped RNAs, such as mRNAs or pre-mRNAs, we observed that anti-sense csRNAs from ∼3000 protein-coding genes are snatched and many protein-coding genes provide several anti-sense csRNAs. Moreover, we observed a significant correlation between the levels of csRNAs and the corresponding caps snatched. csRNAs are generated during transcription initiation by “paused” Pol II or by abolished transcription initiation, and are likely localized in the nucleus as noncoding small RNAs. Interestingly, the 5′ ends of csRNAs were snatched at a rate that was several-fold higher than the rate observed for 5′ ends corresponding to mature transcripts. Further experiments are necessary to address whether IAV specifically targets “paused” Pol II or perhaps induces increased Pol II pausing during infection. These findings would also be consistent with the idea that IAV snatches caps cotranscriptionally, perhaps very early in the elongation process.
Recently, Koppstein et al. (2015) published a manuscript proposing that nascent transcripts are the cap-snatching substrates. This nascent transcript model is based on a previous in vitro study suggesting that trimethylated G caps bind to eIF4E less efficiently than m7G caps and thus may not be translated as well as monomethylated mRNAs (Niedzwiecka et al. 2002). However, another study suggests that eIF4E might not be required for influenza mRNA translation (Burgui et al. 2007). The nascent transcript model did not specify the identity of the substrate capped RNAs and was actually very similar to the previous model, which suggests that pre-mRNAs instead of mature mRNAs are snatched by IAV. Moreover, their analysis did not consider csRNAs. In contrast, our analysis provides a more comprehensive profile of caps snatched by IAV with high confidence because we obtained both host and viral RNA profiles in the same samples. Our finding that csRNAs contribute caps at a rate that is disproportionate to their relative abundance strongly supports the notion that nascent transcripts, including csRNAs, are targets for IAV cap-snatching.
Sikora et al. (2014) recently reported that different viral mRNAs may prefer specific host-mRNA species. Although individual viral mRNAs were expressed at different levels in our samples, we did not observe any obvious bias for cap usage (Supplemental file 3; Supplemental Fig. S4). Instead our data indicate that all viral mRNAs prefer the same host RNAs (i.e., snRNAs or snoRNAs). The study by Sikora et al. (2014) used a deep-sequencing approach on cloned 5′ ends of poly(A) containing viral mRNAs and used protein-coding gene annotations from published data without consideration of noncoding RNAs. Although the cap-snatching rate of some individual host RNAs expressed at low levels did change among different IAV segments in our sample, such variations may simply reflect higher standard errors for that data range. Consistent with our findings, Koppstein et al. (2015) also demonstrated that different viral RNAs prefer caps from the same host RNAs.
In conclusion, we demonstrated that noncoding RNAs, especially small noncoding RNAs such as snRNA/snoRNAs and csRNAs, serve as a major cap donor to IAV mRNAs. This also reveals a new function of csRNAs in a pathological condition.
MATERIALS AND METHODS
Viral infection
A549 cells (American Type Culture Collection, ATCC) were seeded at 2.5 × 105 cells per well (0.5 mL per well) in a 24-well plate in F-12K medium supplemented with 10% fetal bovine serum (FBS) and incubated at 37°C overnight. Influenza A/Brisbane/59/2007 (H1N1), originally obtained from the National Institutes of Health (NIH) Biodefense and Emerging Infections Research Resources Repository, National Institute of Allergy and Infectious Diseases (NIAID), NIH (NR-12282; lot 58550257), was propagated in Madin–Darby canine kidney cells (ATCC) for three passages. Virus was added to A549 cells at a multiplicity of infection of 1 in 100 µL of influenza virus growth medium [DMEM with 100 U/mL penicillin, 100 mg/mL streptomycin, 0.2% bovine serum albumin, and 0.2 µg/mL TPCK-treated-trypsin (Sigma-Aldrich)]. After incubation at 37°C for 1 h, cells were washed once with Hank's Balanced Salt Solution and 500 µL of influenza virus growth medium was added to each well. Cells were again incubated at 37°C, and at 24 h following infection, supernatants from four wells were pooled together and cells were harvested in TRIzol reagent. Minimal cytopathic effect was observed in A549 cells postinfection. The experiment was repeated once with cells collected at 12 h postinfection instead of 24 h.
CapSeq
Approximately 2 µg of total RNA from influenza virus–infected or uninfected control samples was used to clone the TSSs of human and influenza RNAs by CapSeq, as described previously (Gu et al. 2012).
Bioinformatics analysis
Reads obtained using Illumina HiSeq 2000 were processed and mapped to the human and influenza virus genome and RNA annotations using custom PERL (5.10.1) scripts and Bowtie 0.12.7 (Langmead et al. 2009). The human genome and annotations are Ensembl GRCh37 release 71 and the influenza virus genome and annotations were obtained from the National Center for Biotechnology Information (NCBI) RefSeq database (Pruitt et al. 2014). We also used a custom PERL script to assemble the influenza virus genome with our CapSeq reads. The difference was immediately noticed because the RefSeq sequences miss the 5′ and 3′ ends of noncoding sequences of each viral segment, which usually flanks the coding regions, with sequences as annotated in the RefSeq database. The 5′ ends included in our influenza virus genome start with GC(A/G)AAA, which is the same as previously used (Sikora et al. 2014). The 3′ end of mRNAs is TTTCTACT, which exists in the sequences of other influenza strains in the RefSeq database. Manual BLAST searches were performed to identify the RNA annotations of the top CapSeq read matches using the NCBI BLAST website (Altschul et al. 1990). The Generic Genome Browser Gbrowse 1.70 (Stein et al. 2002) was used to visualize the alignments. Detailed PERL scripts and related database files and analyses used in this study are available upon request. The data set can be accessed at the following link: http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?token=apwnwimmvnupfkr&acc=GSE67493. A detailed description of the bioinformatics analysis is available in Supplemental file 1.
Quantitative real-time PCR
Total RNA from A549 cells infected with IAV for 24 h (two independent experiments + original samples for CapSeq) in TRIzol reagent was recovered following the manufacturer's instructions (Invitrogen). cDNA was generated using the Quantitect reverse transcriptase kit (QIAGEN). Quantitative real-time PCR was performed by SYBR Green (Applied Biosystems) using the DNA Engine Opticon 2 System (Bio-Rad Laboratories) with normalization to the expression of HPRT1. HPRT1 primers were purchased from QIAGEN (forward 5′-TTTGCTTTCCTTGGTCAGGC-3′; reverse 5′-GCTTGCGACCTTGACCATCT-3′). The forward primer sequences, which include influenza segment 1 sequences (underlined), are as follows: hybrid U1-segment 1 (5′-ATACTTACCTGGCAAAAG-3′), hybrid U2-segment 1 (5′-ATCGCTTCTCGGCAAAAG-3′), NT-segment 1 (5′-CAGGTAAGTATGCAAAAG-3′). Sequence of the reverse primer targeting influenza segment 1 is (5′-GTATTTCATTGCCATCATCC-3′).
Fluorescence in situ hybridization
Custom fluorescence in situ hybridization (FISH) probes were designed for use with the Affymetrix View RNA multiplex staining kit. A probe for the positive strand of influenza segment 1 (PB2 gene, nucleotides 333–1342) was made based on the published mRNA sequence of influenza A/Brisbane/59/2007 (GenBank accession number CY163831). The U1-segment 1 probe was designed to detect the first 11 nt of the U1 gene (GenBank accession number NR_104086.2) plus the first 11 nt of influenza segment 1 (PB2) (underlined) (ATACTTACCTGGCAAAAGCAGG). For the nontargeting probe, NT-segment 1, the first 11 nt were chosen from sequences found at low abundance in the deep-sequencing analysis and the remaining nucleotides were against the PB2 gene as above (CAGGTAAGTATGCAAAAGCAGG). A549 cells were seeded in 24-well plates with 12-mm 1.5-thickness coverslips (Fisher) coated with rat-tail collagen (BD Biosciences) at 2 × 105 cells per well. The cells were subsequently cultured and infected with IAV at a multiplicity of infection of 1. After 24 h, cells were fixed in 4% paraformaldehyde and stained according to the Affymetrix protocol using fluorescein isothiocyanate (FITC) conjugation for the segment 1 probe and Cy3 conjugation for U1-segment 1, and NT-segment 1 probes. Fluorescent images were acquired on a Leica SP8 confocal microscope using Leica AS software (version 3.3.0.10134). Images were quantified using FIJI (version 1.49m) using the analyze particles function (Schindelin et al. 2012). Data are represented as the number of spots in the Cy3 channel per infected cell as determined by the presence of segment 1 FITC signal.
DATA DEPOSITION
The data set Influenza A virus preferentially snatches noncoding RNA caps and was generated and deposited in GEO under accession number GSE67493.
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
Supplementary Material
ACKNOWLEDGMENTS
We thank Dr. Melissa Moore and Dr. Darryl Conte for helpful comments and Ashwini Sunkavalli for manuscript preparation.
Author contributions: W.G., G.R.G., D.B.G., C.C.M., J.P.W., and R.W.F. designed research; W.G., G.R.G., W.D., and P.L. performed research; W.G. and R.L. contributed new reagents/analytic tools; W.G., M.I.T., J.P.W., and R.W.F. wrote the paper.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.054221.115.
REFERENCES
- Affymetrix/Cold Spring Harbor Laboratory ENCODE Transcriptome Project. 2009. Post-transcriptional processing generates a diversity of 5′-modified long and short RNAs. Nature 457: 1028–1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J Mol Biol 215: 403–410. [DOI] [PubMed] [Google Scholar]
- Beaton AR, Krug RM. 1981. Selected host cell capped RNA fragments prime influenza viral RNA transcription in vivo. Nucleic Acids Res 9: 4423–4436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouloy M, Plotch SJ, Krug RM. 1978. Globin mRNAs are primers for the transcription of influenza viral RNA in vitro. Proc Natl Acad Sci 75: 4886–4890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgui I, Yángüez E, Sonenberg N, Nieto A. 2007. Influenza virus mRNA translation revisited: is the eIF4E cap-binding factor required for viral mRNA translation? J Virol 81: 12427–12438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caton AJ, Robertson JS. 1980. Structure of the host-derived sequences present at the 5′ ends of influenza virus mRNA. Nucleic Acids Res 8: 2591–2603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark MB, Choudhary A, Smith MA, Taft RJ, Mattick JS. 2013. The dark matter rises: the expanding world of regulatory RNAs. Essays Biochem 54: 1–16. [DOI] [PubMed] [Google Scholar]
- Dhar R, Chanock RM, Lai CJ. 1980. Nonviral oligonucleotides at the 5′ terminus of cytoplasmic influenza viral mRNA deduced from cloned complete genomic sequences. Cell 21: 495–500. [DOI] [PubMed] [Google Scholar]
- Dias A, Bouvier D, Crépin T, McCarthy AA, Hart DJ, Baudin F, Cusack S, Ruigrok RW. 2009. The cap-snatching endonuclease of influenza virus polymerase resides in the PA subunit. Nature 458: 914–918. [DOI] [PubMed] [Google Scholar]
- Fodor E, Crow M, Mingay LJ, Deng T, Sharps J, Fechter P, Brownlee GG. 2002. A single amino acid mutation in the PA subunit of the influenza virus RNA polymerase inhibits endonucleolytic cleavage of capped RNAs. J Virol 76: 8989–9001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foll M, Poh YP, Renzette N, Ferrer-Admetlla A, Bank C, Shim H, Malaspinas AS, Ewing G, Liu P, Wegmann D, et al. 2014. Influenza virus drug resistance: a time-sampled population genetics perspective. PLoS Genet 10: e1004185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fortes P, Lamond AI, Ortin J. 1995. Influenza virus NS1 protein alters the subnuclear localization of cellular splicing components. J Gen Virol 76(Pt 4): 1001–1007. [DOI] [PubMed] [Google Scholar]
- Gu W, Lee HC, Chaves D, Youngman EM, Pazour GJ, Conte D Jr, Mello CC. 2012. CapSeq and CIP-TAP identify Pol II start sites and reveal capped small RNAs as C. elegans piRNA precursors. Cell 151: 1488–1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guilligay D, Tarendeau F, Resa-Infante P, Coloma R, Crepin T, Sehr P, Lewis J, Ruigrok RW, Ortin J, Hart DJ, et al. 2008. The structural basis for cap binding by influenza virus polymerase subunit PB2. Nat Struct Mol Biol 15: 500–506. [DOI] [PubMed] [Google Scholar]
- Koppstein D, Ashour J, Bartel DP. 2015. Sequencing the cap-snatching repertoire of H1N1 influenza provides insight into the mechanism of viral transcription initiation. Nucleic Acids Res 43: 5052–5064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krug RM, Broni BA, Bouloy M. 1979. Are the 5′ ends of influenza viral mRNAs synthesized in vivo donated by host mRNAs? Cell 18: 329–334. [DOI] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL. 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10: R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nechaev S, Fargo DC, dos Santos G, Liu L, Gao Y, Adelman K. 2010. Global analysis of short RNAs reveals widespread promoter-proximal stalling and arrest of Pol II in Drosophila. Science 327: 335–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niedzwiecka A, Marcotrigiano J, Stepinski J, Jankowska-Anyszka M, Wyslouch-Cieszynska A, Dadlez M, Gingras AC, Mak P, Darzynkiewicz E, Sonenberg N, et al. 2002. Biophysical studies of eIF4E cap-binding protein: recognition of mRNA 5′ cap structure and synthetic fragments of eIF4G and 4E-BP1 proteins. J Mol Biol 319: 615–635. [DOI] [PubMed] [Google Scholar]
- Plotch SJ, Bouloy M, Krug RM. 1979. Transfer of 5′-terminal cap of globin mRNA to influenza viral complementary RNA during transcription in vitro. Proc Natl Acad Sci 76: 1618–1622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plotch SJ, Bouloy M, Ulmanen I, Krug RM. 1981. A unique cap(m7GpppXm)-dependent influenza virion endonuclease cleaves capped RNAs to generate the primers that initiate viral RNA transcription. Cell 23: 847–858. [DOI] [PubMed] [Google Scholar]
- Pruitt KD, Brown GR, Hiatt SM, Thibaud-Nissen F, Astashyn A, Ermolaeva O, Farrell CM, Hart J, Landrum MJ, McGarvey KM, et al. 2014. RefSeq: an update on mammalian reference sequences. Nucleic Acids Res 42: D756–D763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao P, Yuan W, Krug RM. 2003. Crucial role of CA cleavage sites in the cap-snatching mechanism for initiating viral mRNA synthesis. EMBO J 22: 1188–1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renzette N, Caffrey DR, Zeldovich KB, Liu P, Gallagher GR, Aiello D, Porter AJ, Kurt-Jones EA, Bolon DN, Poh YP, et al. 2014. Evolution of the influenza A virus genome during development of oseltamivir resistance in vitro. J Virol 88: 272–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al. 2012. Fiji: an open-source platform for biological-image analysis. Nat Methods 9: 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seila AC, Calabrese JM, Levine SS, Yeo GW, Rahl PB, Flynn RA, Young RA, Sharp PA. 2008. Divergent transcription from active promoters. Science 322: 1849–1851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw MW, Lamb RA. 1984. A specific sub-set of host-cell mRNAs prime influenza virus mRNA synthesis. Virus Res 1: 455–467. [DOI] [PubMed] [Google Scholar]
- Shi L, Summers DF, Peng Q, Galarz JM. 1995. Influenza A virus RNA polymerase subunit PB2 is the endonuclease which cleaves host cell mRNA and functions only as the trimeric enzyme. Virology 208: 38–47. [DOI] [PubMed] [Google Scholar]
- Sikora D, Rocheleau L, Brown EG, Pelchat M. 2014. Deep sequencing reveals the eight facets of the influenza A/HongKong/1/1968 (H3N2) virus cap-snatching process. Sci Rep 4: 6181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein LD, Mungall C, Shu S, Caudy M, Mangone M, Day A, Nickerson E, Stajich JE, Harris TW, Arva A, et al. 2002. The generic genome browser: a building block for a model organism system database. Genome Res 12: 1599–1610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugiyama K, Obayashi E, Kawaguchi A, Suzuki Y, Tame JR, Nagata K, Park SY. 2009. Structural insight into the essential PB1-PB2 subunit contact of the influenza virus RNA polymerase. EMBO J 28: 1803–1811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Krug RM. 1998. U6atac snRNA, the highly divergent counterpart of U6 snRNA, is the specific target that mediates inhibition of AT-AC splicing by the influenza virus NS1 protein. RNA 4: 55–64. [PMC free article] [PubMed] [Google Scholar]
- Zeldovich KB, Liu P, Renzette N, Foll M, Pham ST, Venev SV, Gallagher GR, Bolon DN, Kurt-Jones EA, Jensen JD, et al. 2015. Positive selection drives preferred segment combinations during influenza virus reassortment. Mol Biol Evol 32: 1519–1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.